Abstract
Early embryonic endocardium undergoes endothelial-to-mesenchymal transition to form cardiac cushion mesenchymal cells (MCs). Embryonic endocardium also gives rise to fibroblasts, intramyocardial adipocytes, and coronary mural cells, including smooth muscle cells and pericytes, in development. Whether endocardial cells directly differentiate into fibroblasts, coronary mural cells, and adipocytes or indirectly via an intermediate stage of endocardial-derived cushion MCs remains unknown. In addition to endocardium, epicardium and neural crest also contribute to cardiac cushion MCs. Given the developmental heterogeneity of cushion MCs and the lack of specific markers for endocardial-derived cushion MCs, conventional genetic lineage tracing utilizing Cre recombinase driven by one specific regulatory element is not sufficient to examine the fates of endocardial-derived cushion MCs. Intersectional genetic targeting approaches, which combine regulatory elements from two or more genes, have been employed to increase the specificity of cell targeting. Here, we developed a dual-recombinase intersectional targeting approach using Nfatc1–Dre, Sox9–CreER, and Cre/Dre double-dependent reporter Ai66 to specifically label endocardial-derived cushion MCs. Taking advantage of intersectional lineage tracing, we found that a subset of cardiac cells including fibroblasts, coronary mural cells, and intramyocardial adipocytes in adult hearts were derived from endocardial-derived cushion MCs. Our study suggests that embryonic endocardium contributes to cushion MCs first, and then endocardial-derived cushion MCs migrate into myocardium and differentiate into fibroblasts, coronary mural cells, and adipocytes in development. Understanding developmental origins of cardiac cell lineages will provide us more insights into cardiac development, regeneration, and diseases.
Keywords: heart development, endothelial cell, adipocyte, vascular smooth muscle cells, fibroblast, Endocardium, Mesenchymal cells, pericyte
Introduction
At embryonic day (E)8.5–E9.0,2 a subset of endocardial endothelial cells in atrioventricular canal (AVC) and outflow tract (OFT) undergo endothelial-to-mesenchymal transition (EndoMT), invade cardiac cushion, and transform into mesenchymal cells (MCs). In addition to endocardium, other cell lineages, including epicardium and extracardiac neural crest cells, have also been reported to contribute to the cushion MCs at early and middle embryonic stages, respectively (1–4). Therefore, embryonic cardiac cushion MCs are developmentally heterogeneous, and their fates and function differ profoundly by their originating sources. Understanding the fates of different subpopulations of cushion MCs will provide insights into their roles in cardiac development and regeneration.
In addition to part of cushion MCs, embryonic endocardium has also been reported as precursors of other multiple lineages, including cardiac fibroblasts, coronary vascular mural cells (pericytes and smooth muscle cells (SMCs)), intramyocardial adipocytes, coronary endothelial cells, liver vascular endothelial cells, and blood cells (5). Previous studies identified the endothelial origin of fibroblasts in adult murine hearts using Tie2–Cre and nuclear factor of activated T cells (Nfatc1)–Cre, which label both endocardium and coronary vascular endothelial cells (6, 7). Another study taking advantage of Nfatc1–CreER, CDH5–CreER, and Tie2–Cre revealed endothelial/endocardial contribution to vascular pericytes and SMCs (8). A recent study employing Nfatc1–Dre and Nfatc1–CreER identified endocardial-derived intramyocardial adipocytes in adult hearts (9). Although these studies demonstrated that a population of fibroblasts, coronary mural cells, and adipocytes is derived from an endocardial source, it remains elusive whether endocardium transforms into these cardiac cell types directly or via an intermediate EndoMT stage; that is, via a process named endocardium-cushion MCs-fibroblasts/mural cells/adipocytes. Further lineage-tracing studies are required to specifically label endocardial-derived cushion MCs and to examine their fates in heart development.
Cushion MCs are developmentally heterogeneous, and there are no specific markers for endocardial-, epicardial-, or neural crest-derived cushion MCs to our knowledge. For instance, platelet-derived growth factor receptor β (PDGFRb) and SRY-type box 9 (Sox9) have been reported to be expressed in cushion MCs at embryonic stages (8, 10). However, there is no proof that these two genes are specifically expressed in endocardial-derived cushion MCs. Additionally, PDGFRb and Sox9 are also expressed in embryonic epicardium and epicardial-derived MCs (8, 10, 11). Thus, conventional genetic targeting utilizing Cre recombinase driven by one specific gene promoter cannot specifically label endocardial-derived cushion MCs.
Dual-recombinase intersectional genetic targeting approaches that combine regulatory elements from two or more genes have been utilized to enhance the specificity of cell targeting (12–14). In this study we employed Nfatc1–Dre, Sox9–CreER, and a reporter line Ai66 (Rosa26–rox–stop–rox–loxp–stop–loxp–tdTomato) to produce an intersectional genetic targeting strategy, in which Dre and CreER specifically restricted tdTomato expression to a cell population with overlapping patterns of Nfatc1–Dre and Sox9–CreER labeling, namely endocardial-derived cushion MCs. We utilized this intersectional lineage tracing approach to systematically and comprehensively demonstrate that embryonic endocardium contributes to cardiac fibroblasts, coronary vascular pericytes, SMCs, and intramyocardial adipocytes via an intermediate cushion mesenchymal stage.
Results
Intersectional targeting of endocardial-derived cushion MCs with high specificity
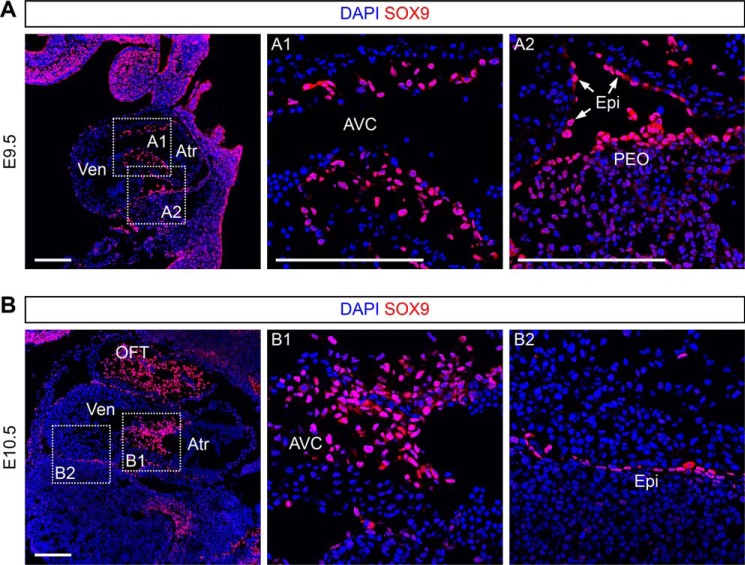
Sox9, which plays critical roles in cardiac valve formation and epicardial-to-mesenchymal transition at embryonic stages, has been reported to be highly expressed in cushion (AVC and OFT) MCs at E9.5–E10.5 and in epicardium at E13.5–E14.5 (11, 15, 16). To further examine the expression map of Sox9 at E9.5 and E10.5 when cardiac cushion MCs have been just formed, we performed immunostaining for SOX9 on embryonic sections and found that SOX9 was detected not only in cushion MCs, but also in epicardium at E9.5 and E10.5 (Fig. 1, A and B). Sox9–CreER mouse line (17) has been used to label embryonic cardiac MCs in our previous studies (10). ESR (estrogen receptor), a surrogate of Sox9, was also detected in cushion MCs and epicardium at E9.5–E11.5 (10). In our previous studies, we crossed Sox9–CreER mouse with a reporter line Rosa26–loxp–stop–loxp–tdTomato (R26–tdTomato) and found that Sox9–CreER;R26–tdTomato labeled cushion MCs, epicardium, and epicardial-derived MCs in embryonic hearts when tamoxifen was administered at E9.5 (10).
Figure 1.
Sox9 is expressed in cushion MCs and epicardium at E9.5 and E10.5. A and B, immunostaining for SOX9 on embryonic sections at E9.5 and E10.5. The boxed regions in the left panel are magnified in the right panels as indicated. Atr, atrium; Ven, ventricle; AVC, atrioventricular canal; PEO, proepicardium; Epi, epicardium; OFT, outflow tract; DAPI, 4′,6′-diamino-2-phenylindole. Scale bars, 200 μm. Each picture is representative of three individual embryonic samples.
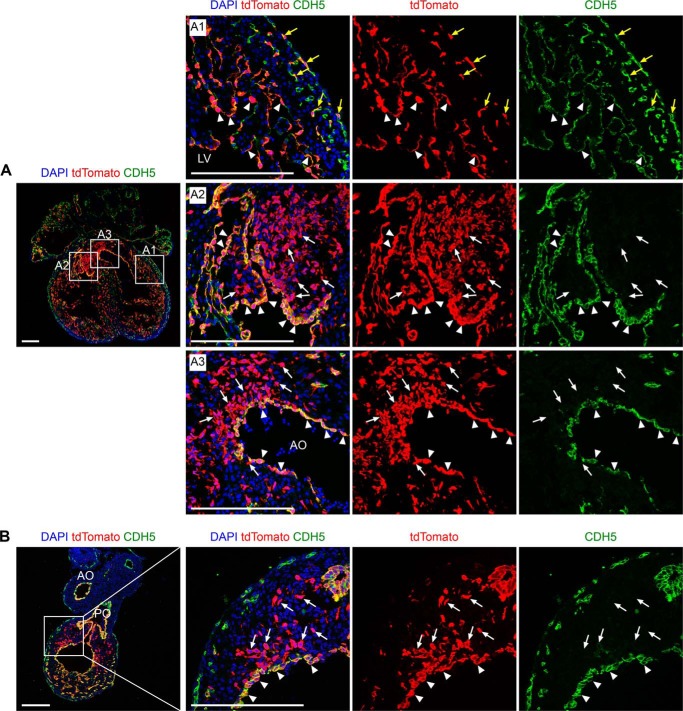
Nfatc1 was reported as a marker for endocardium at early embryonic stages (18–20). The Nfatc1–Dre mouse line has been reported to label embryonic endocardium and its descendants in development (9). Dre, like Cre, is a site-specific recombinase and targets rox sites for recombination (13, 21). We crossed Nfatc1–Dre with a reporter line Rosa26–rox–stop–rox–tdTomato (R26–rsr–tdTomato) and collected the embryos at E13.5. By immunostaining for tdTomato and endothelial marker CDH5 on heart sections, we found that Nfatc1–Dre labeled endocardium, coronary vascular endothelial cells, and endocardial-derived MCs in atrioventricular cushion and OFT cushion (Fig. 2, A and B).
Figure 2.
Nfatc1–Dre labels endocardium, coronary endothelial cells, and endocardial-derived cushion MCs. A and B, immunostaining for tdTomato and CDH5 on heart sections from Nfatc1–Dre;R26–rsr–tdTomato at E13.5. The boxed regions in the left panels are magnified and split channels in the right panels as indicated. The yellow arrows in A1 indicate tdTomato+CDH5+ coronary endothelial cells. The white arrows in A2 indicate tdTomato+CDH5− MCs in atrioventricular cushion. The white arrows in A3 indicate tdTomato+CDH5− MCs in aortic outflow tract (AO). The white arrows in B indicate tdTomato+CDH5− MCs in pulmonary outflow tract (PO). The arrowheads in A and B indicate tdTomato+CDH5+ endocardial cells. DAPI, 4′,6′-diamino-2-phenylindole. Scale bars, 200 μm. Each picture is representative of three individual embryonic samples.
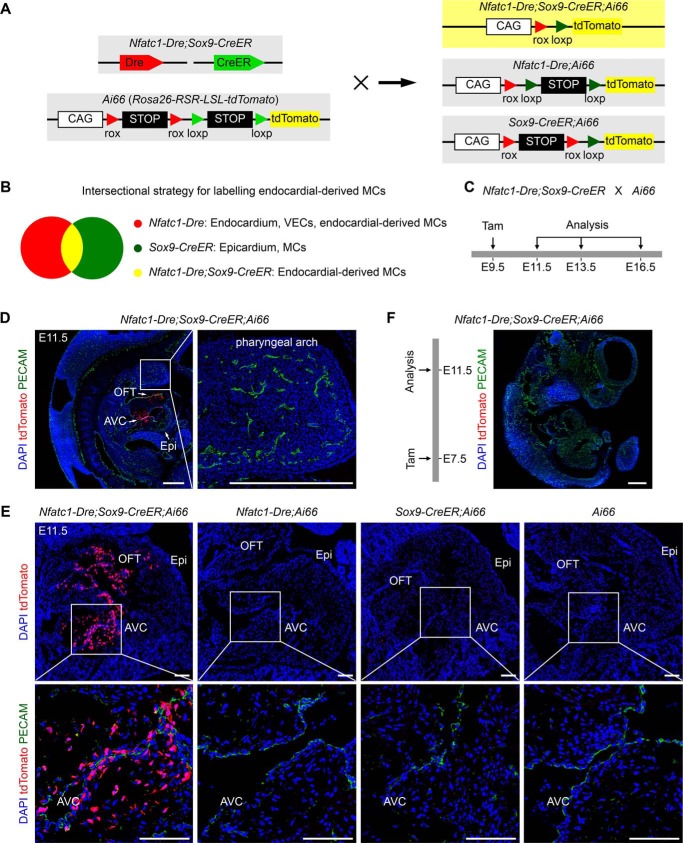
Cushion MCs have heterogenous developmental origins including neural crest, epicardium, and endocardium (22). So far there are no specific markers for endocardial-derived cushion MCs. To specifically label endocardial-derived MCs and directly examine their cellular fates in development, we took advantage of dual-recombinase intersectional genetic targeting approach (13) using Nfatc1–Dre;Sox9–CreER;Rosa26–rox–stop–rox–loxp–stop–loxp–tdTomato (Ai66). In this strategy, two recombination events mediated by Dre and CreER recombinases are required for tdTomato expression (Fig. 3A). The cells labeled by both Sox9–CreER and Nfatc1–Dre should be tdTomato-positive. Sox9–CreER labeled epicardium and cardiac MCs including epicardial-derived MCs and cushion MCs when tamoxifen is administrated at E9.5 (Fig. 3B) (10). Nfatc1–Dre labeled endocardium, coronary vascular endothelial cells, and endocardial-derived MCs (Fig. 3B). Therefore, only the common part, endocardial-derived MCs would be labeled by Nfatc1–Dre;Sox9–CreER;Ai66 (Fig. 3B).
Figure 3.
Dual-recombinase intersectional targeting strategy specifically labels endocardial-derived cushion MCs. A, cartoon figure showing the strategy for dual-recombinase intersectional genetic targeting. Nfatc1–Dre;Sox9–CreER mice were crossed with Cre/Dre double-dependent reporters Ai66. tdTomato will be activated only if the two stop cassettes rox-stop–rox and loxp–stop–loxp are both removed. B, cartoon figure showing the intersectional targeting strategy to specifically label endocardial-derived MCs. At embryonic stages, Nfatc1–Dre labels endocardium, coronary vascular endothelial cells (VECs), and endocardial-derived MCs. Sox9–CreER labels epicardium and MCs, which include both epicardial- and endocardial-derived MCs. Thus, the intersectional strategy should specifically label endocardial-derived MCs. C, schematic diagram showing the strategy for tracing endocardial-derived MCs using the intersectional approach. Tamoxifen (Tam) was administrated at E9.5, and the embryos were harvested for analysis at different embryonic stages. D and E, immunostaining for tdTomato and PECAM on embryonic section from Nfatc1–Dre;Sox9–CreER;Ai66 and littermate controls at E11.5, which were administrated with tamoxifen at E9.5. The boxed regions are magnified in the right panel or in the bottom panels as indicated. Scale bars in D, 400 μm; scale bars in E, 100 μm. F, immunostaining for tdTomato and PECAM on embryonic sections from Nfatc1–Dre;Sox9–CreER;Ai66 at E11.5, which were administrated with tamoxifen at E7.5. Scale bar, 400 μm. OFT, outflow tract; Epi, epicardium; AVC, atrioventricular canal; DAPI, 4′,6′-diamino-2-phenylindole. Each picture is representative of three individual mouse samples.
We crossed Nfatc1–Dre;Sox9–CreER with the reporter line Ai66 and administrated the pregnant mice with tamoxifen at E9.5 when cushion MCs have been just formed. Then we collected the embryos at E11.5 to examine the genetic targeting of Nfatc1–Dre;Sox9–CreER;Ai66 (Fig. 3C). Immunostaining for tdTomato and endothelial marker platelet and endothelial cell adhesion molecule 1 (PECAM) on embryonic sections showed that tdTomato signals were exclusively detected in cardiac OFT and AVC MCs of Nfatc1–Dre;Sox9–CreER;Ai66 embryos (Fig. 3, D and E). No tdTomato+ endothelial or epicardial cells were identified in the hearts of Nfatc1–Dre;Sox9–CreER;Ai66 embryos (Fig. 3, D and E). At this time point, no neural crest-derived tissues, such as the pharyngeal arch, were labeled by tdTomato in Nfatc1–Dre;Sox9–CreER;Ai66 embryos (Fig. 3D), which excludes the possibility that neural crest-derived cushion MCs were labeled by tdTomato. Meanwhile, no tdTomato+ signals were detected in the hearts of littermate controls including Nfatc1–Dre;Ai66, Sox9–CreER;Ai66, and Ai66 (Fig. 3E), suggesting there is no leakiness of Ai66 reporter in littermate controls at E11.5. To further exclude other potential cellular origins for tdTomato-labeled cushion MCs, we administrated the mice with tamoxifen at E7.5 before EndoMT and collected the embryos at E11.5. Few tdTomato+ cells were identified in Nfatc1–Dre;Sox9–CreER;Ai66 embryos (Fig. 3F). Collectively, the above data suggested that the intersectional targeting tool Nfatc1–Dre;Sox9–CreER;Ai66 specifically labeled endocardial-derived cushion MCs when tamoxifen was administrated at E9.5.
Endocardial-derived cushion MCs invade the ventricular myocardium at embryonic stages
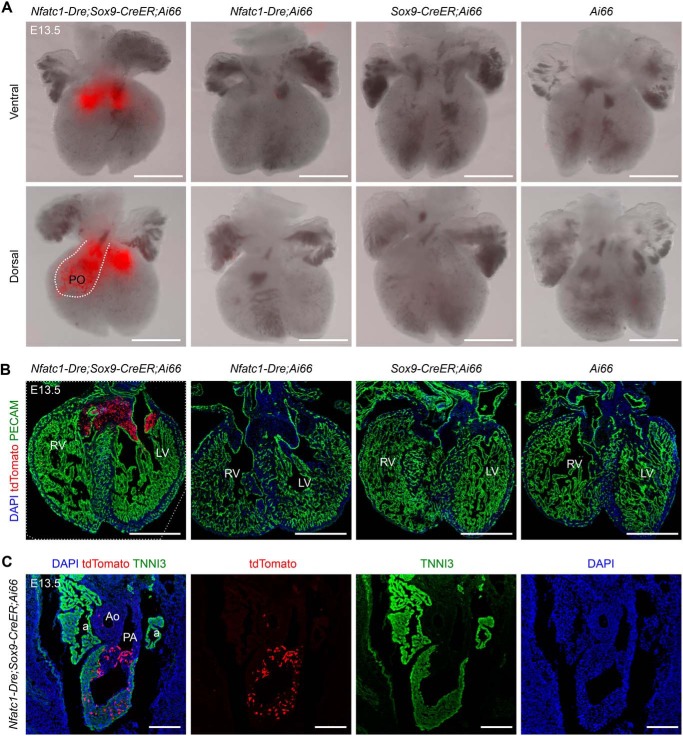
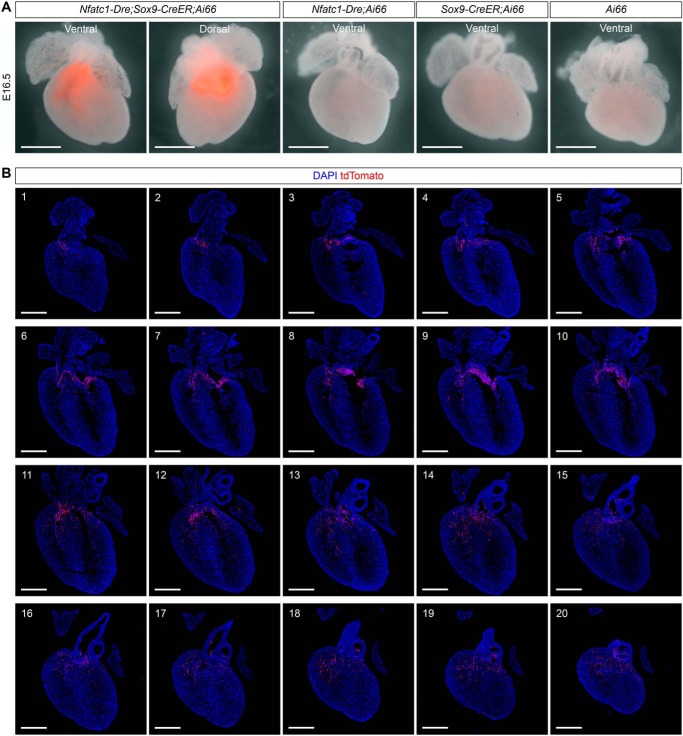
After validating the targeting tool, we followed the fates of tdTomato-labeled MCs to E13.5 (Fig. 3C). Whole-mount fluorescence view showed that tdTomato signals were enriched in the regions of atrioventricular cushion and pulmonary OFT of Nfatc1–Dre;Sox9–CreER;Ai66 embryos (Fig. 4A). No leaky tdTomato signals were observed in the hearts of littermate controls (Fig. 4, A and B). Immunostaining for tdTomato revealed that endocardial-derived MCs in atrioventricular cushion still accumulated together, and few of them migrated into the ventricular free walls or septum (Fig. 4B). In contrast, endocardial-derived MCs in pulmonary OFT had migrated into the myocardium at E13.5 (Fig. 4C). Furthermore, in the hearts of Nfatc1–Dre;Sox9–CreER;Ai66 embryos at E16.5 (Fig. 3C), most of the tdTomato signals in the hearts were still enriched in the regions of atrioventricular cushion and pulmonary OFT (Fig. 5A). Again, no tdTomato signals were observed in the hearts of littermate controls (Fig. 5A). Immunostaining for tdTomato on a serial of heart sections from Nfatc1–Dre;Sox9–CreER;Ai66 embryos showed that more tdTomato-labeled cushion MCs had migrated into the ventricular free walls and septum at E16.5 (Fig. 5B).
Figure 4.
Endocardial-derived MCs in the pulmonary outflow tract invade the myocardium at E13.5. A, whole-mount fluorescence view of the hearts with indicated genotypes, which were administered with tamoxifen at E9.5 and harvested at E13.5. The dotted line indicates pulmonary outflow tract (PO). B, immunostaining for tdTomato and PECAM on embryonic hearts with indicated genotypes at E13.5. C, immunostaining for tdTomato and TNNI3 on embryonic sections from Nfatc1–Dre;Sox9–CreER;Ai66 at E13.5. RV, right ventricle; LV, left ventricle; a, atrium; Ao, aorta; PA, pulmonary artery; DAPI, 4′,6′-diamino-2-phenylindole. Scale bars, 500 μm. Each picture is representative of three individual mouse samples.
Figure 5.
Endocardial-derived MCs have migrated into ventricular free walls and septum at E16. 5. A, whole-mount fluorescence views of the hearts with the indicated genotypes, which were administered with tamoxifen at E9.5 and harvested at E16.5. Scale bars, 2 mm. B, immunostaining for tdTomato on heart sections (sections 1–20, from dorsal to ventral) from Nfatc1–Dre;Sox9–CreER;Ai66 at E16.5. DAPI, 4′,6′-diamino-2-phenylindole. Scale bars, 500 μm. Three individual mouse samples have been examined for each experiment.
Endocardial-derived cushion MCs contribute to fibroblasts, pericytes, SMCs, and intramyocardial adipocytes
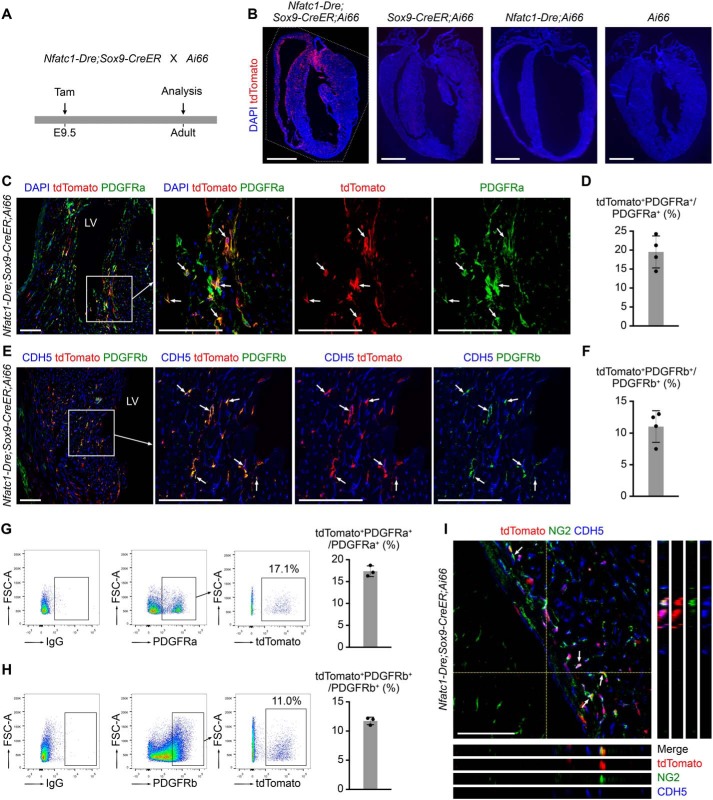
Next, we analyzed the cellular fates of endocardial-derived cushion MCs in adult hearts (Fig. 6A). Immunostaining for tdTomato on heart sections from adult Nfatc1–Dre;Sox9–CreER;Ai66 mice showed that the descendants of endocardial-derived cushion MCs distributed extensively in ventricular free walls and septum (Fig. 6B). No tdTomato+ cells were identified in adult hearts of the controls (Fig. 6B).
Figure 6.
Intersectional lineage tracing reveals the contribution of endocardial-derived MCs to fibroblasts and pericytes in adult hearts. A, the strategy for tracing endocardial-derived cushion MCs in adult hearts. Tamoxifen (Tam) was administrated at E9.5, and the hearts were harvested for analysis at the adult stage. B, immunostaining for tdTomato on adult heart sections with indicated genotypes. Scale bars, 2 mm. C, immunostaining for tdTomato and PDGFRa on heart sections from adult Nfatc1–Dre;Sox9–CreER;Ai66. The arrows indicate tdTomato+PDGFRa+ cells. The boxed regions in the left panel are magnified and split channels in the right panels. Scale bars, 100 μm. D, the cell counting result showing the percentage of tdTomato+ fibroblasts in ventricular free walls and septum (n = 4 mice). E, immunostaining for tdTomato, CDH5, and PDGFRb on heart sections from adult Nfatc1–Dre;Sox9–CreER;Ai66. The arrows indicate tdTomato+PDGFRb+ pericytes. The boxed regions in the left panel are magnified and split channels in the right panels. Scale bars, 100 μm. F, the cell counting result showing the percentage of tdTomato+ pericytes in ventricular free walls and septum (n = 4 mice). G and H, the percentage of tdTomato+ fibroblasts and pericytes in ventricles from adult Nfatc1–Dre;Sox9–CreER;Ai66 were assessed by flow cytometry. IgG were controls for PDGFRa or PDGFRb antibodies (n = 3 mice for each experiment). I, immunostaining for tdTomato, NG2, and CDH5 on heart sections from adult Nfatc1–Dre;Sox9–CreER;Ai66. The arrows indicate tdTomato+NG2+ pericytes. DAPI, 4′,6′-diamino-2-phenylindole; FSC, forward scatter; LV, left ventricle. Scale bars, 100 μm.
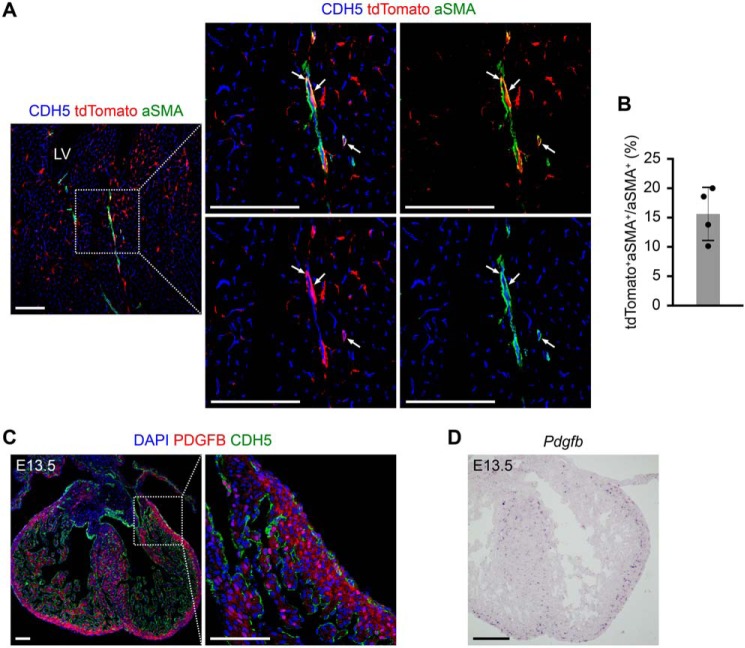
We then examined whether endocardial-derived cushion MCs contributed to fibroblasts, vascular pericytes, SMCs, and adipocytes in adult myocardium. Co-staining for tdTomato and fibroblast marker PDGFRa (6, 11) showed that some PDGFRa+ fibroblasts were labeled by tdTomato (Fig. 6C). The cell counting result showed that 18.55 ± 1.55% of the PDGFRa+ fibroblasts in the ventricular free walls and septum were tdTomato+ (Fig. 6D). To examine the labeling of vascular pericytes, we stained for tdTomato, CDH5, and PDGFRb on cardiac sections and identified tdTomato+PDGFRb+ pericytes in the adult hearts (Fig. 6E). The cell counting result showed that 11.42 ± 1.41% of the PDGFRb+ pericytes were labeled by tdTomato (Fig. 6F). We also collected ventricles from adult Nfatc1–Dre;Sox9–CreER;Ai66 mice, performed flow cytometry to assess the percentage of tdTomato-labeled fibroblasts or pericytes in ventricles, and got similar quantification results compared with those of immunostaining on sections (Fig. 6, G and H). To further confirm the labeling of pericytes, we also stained for another pericyte marker chondroitin sulfate proteoglycan 4 (NG2) with tdTomato and found co-localization of tdTomato and NG2 in adult hearts (Fig. 6I). tdTomato was also detected in perivascular aSMA+ SMCs (Fig. 7A), suggesting the contribution of endocardial-derived cushion MCs to SMCs in adult hearts. The cell counting result showed that 16.85 ± 2.19% of aSMA+ SMCs in adult hearts were labeled by tdTomato (Fig. 7B). Taken together, these results suggest that endocardial-derived cushion MCs contribute to fibroblasts, pericytes, and SMCs in adult hearts.
Figure 7.
Intersectional lineage tracing shows the contribution of endocardial-derived MCs to SMCs in adult hearts. A, immunostaining for tdTomato, CDH5, and aSMA on heart sections from adult Nfatc1–Dre;Sox9–CreER;Ai66 mice, which were treated with tamoxifen at E9.5. The arrows indicate perivascular tdTomato+aSMA+ SMCs. The boxed region in the left panel is magnified in the right panels. LV, left ventricle. Scale bars, 100 μm. B, the cell counting result showing the percentage of tdTomato+ SMCs in ventricular free walls and septum (n = 4 mice). C, immunostaining for PDGFB and CDH5 on heart sections at E13.5. The boxed region in the left panel is magnified in the right panel. Scale bars, 100 μm. D, in situ hybridization for Pdgfb on heart sections at E13.5. Scale bar, 200 μm. DAPI, 4′,6′-diamino-2-phenylindole. Each picture is representative of three individual mouse samples.
The mechanisms regulating the migration of endocardial-derived cushion MCs into the myocardium to form fibroblasts, pericytes, and SMCs are less known. It has been reported that PDGFR signaling is required for the migration of embryonic epicardial cells into the myocardium and tissue-specific loss of PDGFRa and PDGFRb impairs epicardial migration (11). We stained for platelet-derived growth factor B (PDGFB) antibody and found that PDGFB was mostly enriched in the ventricular free walls and septum at E13.5 (Fig. 7C). In situ hybridization for Pdgfb confirmed its expression pattern at E13.5 (Fig. 7D). Given embryonic cushion MCs also express PDGFRa and PDGFRb (8, 10), we suspect that PDGFR signaling may be also necessary for the migration of endocardial-derived cushion MCs into myocardium, which requires further investigation.
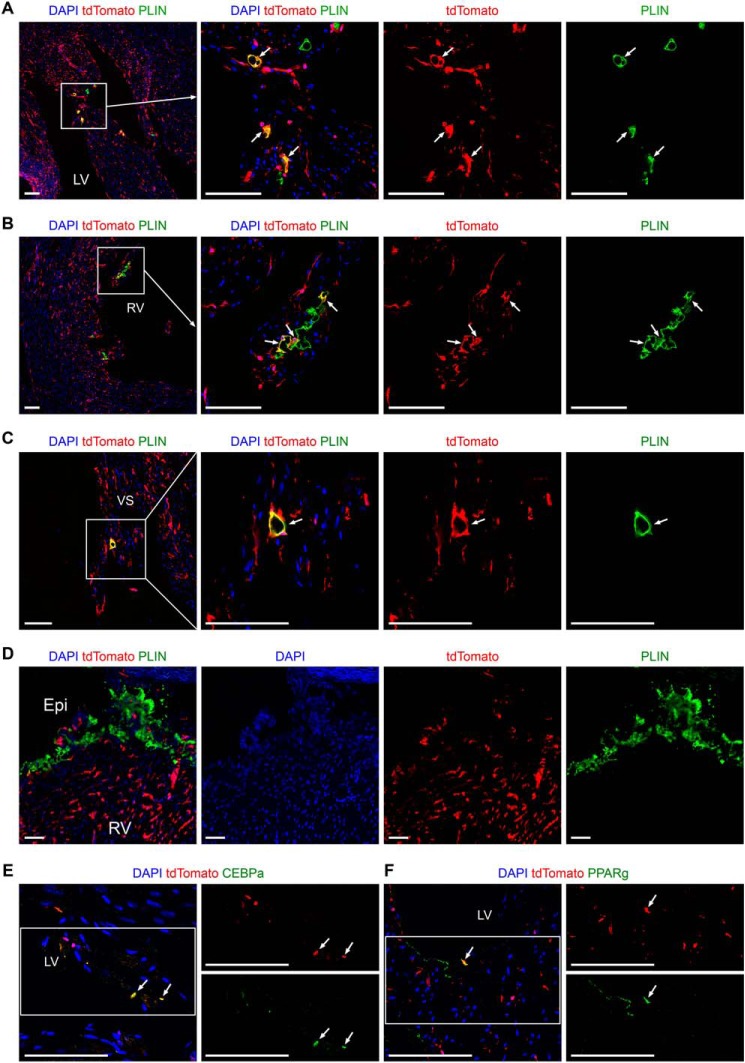
Immunostaining for tdTomato and adipocyte marker perilipin 1 (PLIN) on heart sections revealed that some intramyocardial adipocytes in ventricular free walls and septum were labeled by tdTomato (Fig. 8, A–C), whereas no subepicardial adipocytes that originate from epicardium (23, 24) were tdTomato+ in the adult hearts (Fig. 8D). Quantification results showed that 33.88 ± 3.54% of the intramyocardial adipocytes in ventricular free walls and septum were labeled by tdTomato. The mechanisms regulating the transformation of endocardial-derived cushion MCs to adipocytes are still unknown. We detected the expression of CCAAT enhancer-binding protein α (CEBPα) and peroxisome proliferator activated receptor γ (PPARγ) in some tdTomato+ cells by immunostaining (Fig. 8, E and F), which suggested that these two adipogenic transcription factors may be involved in the cell fate conversion from endocardial-derived cushion MCs to adipocytes. Collectively, these intersectional genetic lineage tracing data suggest that embryonic endocardial-derived MCs first reside in the OFT and AVC cushions and then migrate into ventricular free walls and septum, where they finally differentiate into fibroblasts, pericytes, SMCs, and intramyocardial adipocytes in adult hearts.
Figure 8.
Intersectional lineage tracing shows the contribution of endocardial-derived MCs to intramyocardial adipocytes in adult hearts. A–D, immunostaining for tdTomato and PLIN on heart sections from adult Nfatc1–Dre;Sox9–CreER;Ai66, which were treated with tamoxifen at E9.5. tdTomato+ adipocytes are identified in LV (A), RV (B), and VS (C), but not in subepicardium (D). The boxed regions in the left panels are magnified and split channels in the right panels. The arrows indicate tdTomato+PLIN+ intramyocardial adipocytes. E and F, immunostaining for tdTomato and CEBPα or PPARγ on heart sections from adult Nfatc1–Dre;Sox9–CreER;Ai66. The arrows indicate tdTomato+CEBPα+ or tdTomato+PPARγ+ cells. The boxed regions in the left panel are magnified and split channels in the right panels. LV, left ventricle; RV, right ventricle; VS, ventricular septum; Epi, epicardium. Scale bars, 100 μm. Each picture is representative of three individual mouse samples.
Discussion
Fundamentally, cell types are hardly defined by single genes but rather by intersectional expression of multiple genes. Thus, intersectional genetic targeting approaches combining regulatory elements from two or more genes are more specific comparing with traditional lineage tracing utilizing single recombinase driven by one regulatory element from one gene. In this study, we took advantage of the dual-recombinase intersectional targeting strategy, which specifically labels endocardial-derived cushion MCs, enables us to more unambiguously establish the timing/location of these cells that generates their various descendants in development. The findings that endocardial-derived cushion MCs contribute to fibroblasts, SMCs, pericytes, and intramyocardial adipocytes in adult hearts can provide more insights into valve formation, cardiac development, and even heart diseases involving fibrosis or adipogenesis, such as myocardial infarction and arrhythmogenic right ventricular cardiomyopathy.
The underlying mechanisms that govern the migration of endocardial-derived cushion MCs into myocardium are still unknown. PDGF/PDGFR/Sox9 signaling pathway is required for the migration of epicardial-derived MCs into the ventricular myocardium at embryonic stages (11). Given endocardial-derived cushion MCs and epicardial-derived MCs both express PDGFRa, PDGFRb, and Sox9 (8, 10, 11), we suspect that the expression of PDGF in ventricular myocardium (Fig. 7, C and D) plays essential roles not only in epicardial migration, but also in the recruitment of endocardial-derived cushion MCs into ventricular myocardium. More functional studies, such as genetic inactivation of PDGFRa, PDGFRb, or Sox9 in endocardium or endocardial-derived cushion MCs, are required in future investigations.
The mechanisms regulating cell fate conversions from cushion MCs to vascular pericytes and SMCs have been partially revealed. Previous studies showed that Wnt signaling pathway is necessary for vascular pericyte recruitment from cushion MCs (8). Inactivation of Wnt receptor Frizzled 4 or downstream transcription co-factor β-catenin in PDGFRb+ cushion MCs, or vascular endothelial deletion of Wnt ligand secretion mediator (Wls), all resulted in deficient pericyte recruitment at the late embryonic stage (8). Coronary pericytes are developmental progenitors of coronary SMCs, and the Jagged-1/Notch3 axis is required for transition of pericytes to SMCs (25). Whether endocardial-derived cushion MCs can migrate into myocardium and directly differentiate into SMCs without an intermediate vascular pericyte stage still need further study.
Both endocardial- and epicardial-derived fibroblasts contribute to cardiac fibrosis, and they exhibit similar gene expression profiles and identical proliferation rates after pressure overload-induced injury, suggesting that common signaling mechanisms stimulate their pathological response (6, 7). However, their corresponding phenotypes and cell fate conversions under other pathological conditions, such as injury-induced cardiac calcification (26), are still unknown. Vascular pericytes also give rise to fibroblasts in multiple organs under various pathological conditions (27–30). Whether endocardial-derived pericytes contribute to fibroblasts in homeostasis and injuries has not been examined.
Intramyocardial adipocytes have two developmental origins: embryonic epicardium and embryonic endocardium (9). Here we demonstrate the contribution of endocardial-derived cushion MCs to intramyocardial adipocytes. However, its molecular mechanisms remain unknown. Formation of epicardial-derived cardiac adipocytes requires activation of the PPARγ pathway in the epicardium (24). We also detected the expression of PPARγ and CEBPα in endocardial-derived cells in myocardial layer, which suggests that the differentiation of endocardial-derived cushion MCs to adipocytes may also involve these principal adipogenic pathways. Further functional investigations such as genetic inactivation of PPARγ or CEBPα in endocardium or endocardial-derived cushion MCs will provide insights into this differentiation process.
Experimental procedures
Mouse breeding and genotyping
All animal studies were carried out in accordance with the institutional guidelines of ShanghaiTech University. Sox9–CreER, Nfatc1–Dre, R26–tdTomato, R26–rsr–tdTomato, and Ai66 murine strains were reported previously (13, 17, 31, 32). The R26–rsr–tdTomato mouse line was generated by crossing Ai66 with ACTB–Cre (33), which removed the transcriptional stop cassette flanked by two loxp sites. All mice were maintained on a C57BL6/ICR background. Tamoxifen was administered by oral gavage at the indicated time points (0.1 mg/g of body weight). Cesarean section was performed on pregnant mice that had been administered with tamoxifen to obtain perinatal pups. Genomic DNA was prepared from embryonic yolk sac or mouse tail. Tissues were lysed by incubation with proteinase K overnight at 55 °C, followed by centrifugation at maximum speed for 8 min to obtain supernatant with genomic DNA. DNA was precipitated by adding isopropanol and was washed in 70% ethanol.
Immunofluorescent staining
We performed immunofluorescent staining according to the previous protocols (34). Briefly, embryos or hearts were collected and fixed in 4% paraformaldehyde for 20 min to 1 h based on the size of tissues. After three times washing in PBS, embryonic hearts were observed and photographed using fluorescence microscopy (Nikon SMZ25). Then the tissues were dehydrated in 30% sucrose and embedded in optimum cutting temperature (Sakura). Cryosections, which were collected at 9-μm thickness, were air-dried for 30 min at room temperature and then blocked with blocking buffer (5% donkey serum, 0.1% Triton X-100 in PBS) for 30 min at room temperature. Primary antibodies were incubated over night at 4 °C. The following antibodies were used: tdTomato (Rockland, 600-4010379, 1:1000; Chromotek, 5F8, 1:200), PLIN (Abcam, ab61682, 1:200), PDGFRa (R&D, AF1062s, 1:200 and eBioscience, 14-1401-81, 1:200), PDGFRb (eBioscience, 14-1402-82, 1:200), NG2 (Millipore, AB5302, 1:200), aSMA (Sigma, F3777, 1:400), CDH5 (R&D, AF1002, 1:100), SOX9 (Millipore, AB5535, 1:200), TNNI3 (Abcam, ab56357, 1:200), PDGFB (Thermo Fisher, PA1–27394, 1:200), and PECAM (BD, 553370, 1:500). Signals were developed with Alexa fluorescence antibodies (Invitrogen). Images were acquired by stereomicroscope (Olympus MVX10) or confocal microscope (Nikon A1R).
In situ hybridization
In situ hybridization was performed as previously described (35). Briefly, embryos were harvested in DEPC-treated PBS and fixed overnight in 4% PFA. After washing in PBS, embryos were dehydrated in 30% sucrose, which was dissolved in DEPC-treated PBS. Then the embryos were embedded in OCT and could be stored at −80 °C. Cryosections with 8–10-μm thickness were collected. After air drying for 1 h at room temperature, the slides were incubated with antisense probes (1 μg/ml) overnight at 65 °C. Then the slides were equilibrated in MABT buffer (11.62 g/L maleic acid, 8.15 g/L sodium hydroxide and 8.75 g/L sodium chloride)for 15 min and blocked in blocking buffer (10% sheep serum and 2% blocking reagent in MABT buffer) for 1 h at room temperature. Alkaline phosphatase-coupled antibody to digoxigenin was incubated with sections for 2 h at room temperature at a dilution of 1:2000. After washing in MABT buffer and equilibration in NTMT buffer, sections were developed with 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium in the dark to the desired extent. Slides were mounted in glycerol. Images were acquired using an Olympus microscope (CX23). The primers used to generate Pdgfb probes were 5′-AACTGTATGAAATGCTGAGCGACC-3′ and 5′-AAGTGGAAAGAGGCAGGGACAC-3′.
Flow cytometry
Cardiac tissues were cut into small pieces and then were digested with 100 units/ml collagenase type II and 0.05% trypsin at 37 °C. After digestion, the lysis buffer containing cells were passed through 70-μm cell strainers. Then the cells were centrifuged at 30 × g for 5 min at 4 °C to remove cardiomyocytes pelleted at the bottom of the tube. Noncardiomyocytes in the supernatants were obtained by centrifuging at 500 × g for 10 min at 4 °C. The isolated cells were stained with PDGFRa (Thermo Fisher, 17-1401-81, 1:100) or PDGFRb (eBioscience, 14-1402-82, 1:200) for 1 h at 4 °C. Then the cells were incubated with second antibodies if needed for 30 min at room temperature. Flow cytometry was performed using a BD FACS Aria flow cytometry system (BD Fortessa). Forward and side scatter gating strategy was used in the experiment. 4′,6′-Diamino-2-phenylindole (1:1000) was used for identification of live cells. The data were analyzed using FlowJo software.
Statistical analysis
All data were collected from at least three independent experiments as indicated. For analysis of the percentage of tdTomato-labeled fibroblasts, pericytes, smooth muscle cells, and intramyocardial adipocytes in adult hearts, we collected four heart samples for quantification. Five slides from each heart were stained. Ventricular free walls, ventricular septum, and subepicardium were all included during analysis. Co-localization of tdTomato and cellular markers were determined in 40× field. The data were presented as mean values ± S.D.
Author contributions
X. H., T. F., J. M., and H. Z. data curation; X. H., T. F., Z. J., S. K., W. C., and H. Z. formal analysis; X. H., T. F., Z. J., J. M., S. K., W. C., and H. Z. investigation; X. H. and H. Z. methodology; T. F., S. K., and Z. L. validation; C.-P. L. and B. Z. writing-review and editing; H. Z. supervision; H. Z. writing-original draft; H. Z. project administration.
Acknowledgments
We thank research platforms at School of Life Science and Technology, ShanghaiTech University. We also thank Haojie Chen, Chaohua Zheng, and Hao Feng for animal husbandry.
This work was supported by National Key R&D Program of China Grant 2018YFA0108100; National Science Foundation of China Grants 31871474, 31822034, and 81861128023; Shuguang Program Grant 17SG54, supported by the Shanghai Education Development Foundation and Shanghai Municipal Commission; and the ShanghaiTech University start-up fund. The authors declare that they have no conflicts of interest with the contents of this article.
- En
- embryonic day n
- MC
- mesenchymal cell
- EndoMT
- endothelial-to-mesenchymal transition
- SMC
- smooth muscle cell
- Nfatc1
- nuclear factor of activated T cells
- OFT
- outflow tract
- AVC
- atrioventricular canal
- Sox9
- SRY-type box 9
- aSMA
- α smooth muscle actin
- NG2
- chondroitin sulfate proteoglycan 4
- PECAM
- platelet and endothelial cell adhesion molecule 1
- CEBPα
- CCAAT enhancer binding protein α
- PPARγ
- peroxisome proliferator activated receptor γ
- PDGF
- platelet-derived growth factor
- PLIN
- perilipin 1.
References
- 1. Wessels A., van den Hoff M. J., Adamo R. F., Phelps A. L., Lockhart M. M., Sauls K., Briggs L. E., Norris R. A., van Wijk B., Perez-Pomares J. M., Dettman R. W., and Burch J. B. (2012) Epicardially derived fibroblasts preferentially contribute to the parietal leaflets of the atrioventricular valves in the murine heart. Dev. Biol. 366, 111–124 10.1016/j.ydbio.2012.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. de Lange F. J., Moorman A. F., Anderson R. H., Männer J., Soufan A. T., de Gier-de Vries C., Schneider M. D., Webb S., van den Hoff M. J., and Christoffels V. M. (2004) Lineage and morphogenetic analysis of the cardiac valves. Circ. Res. 95, 645–654 10.1161/01.RES.0000141429.13560.cb [DOI] [PubMed] [Google Scholar]
- 3. Snarr B. S., Kern C. B., and Wessels A. (2008) Origin and fate of cardiac mesenchyme. Dev. Dyn. 237, 2804–2819 10.1002/dvdy.21725 [DOI] [PubMed] [Google Scholar]
- 4. Jiang X., Rowitch D. H., Soriano P., McMahon A. P., and Sucov H. M. (2000) Fate of the mammalian cardiac neural crest. Development 127, 1607–1616 [DOI] [PubMed] [Google Scholar]
- 5. Zhang H., Lui K. O., and Zhou B. (2018) Endocardial cell plasticity in cardiac development, diseases and regeneration. Circ. Res. 122, 774–789 10.1161/CIRCRESAHA.117.312136 [DOI] [PubMed] [Google Scholar]
- 6. Moore-Morris T., Guimarães-Camboa N., Banerjee I., Zambon A. C., Kisseleva T., Velayoudon A., Stallcup W. B., Gu Y., Dalton N. D., Cedenilla M., Gomez-Amaro R., Zhou B., Brenner D. A., Peterson K. L., Chen J., et al. (2014) Resident fibroblast lineages mediate pressure overload-induced cardiac fibrosis. J. Clin. Invest. 124, 2921–2934 10.1172/JCI74783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ali S. R., Ranjbarvaziri S., Talkhabi M., Zhao P., Subat A., Hojjat A., Kamran P., Müller A. M., Volz K. S., Tang Z., Red-Horse K., and Ardehali R. (2014) Developmental heterogeneity of cardiac fibroblasts does not predict pathological proliferation and activation. Circ. Res. 115, 625–635 10.1161/CIRCRESAHA.115.303794 [DOI] [PubMed] [Google Scholar]
- 8. Chen Q., Zhang H., Liu Y., Adams S., Eilken H., Stehling M., Corada M., Dejana E., Zhou B., and Adams R. H. (2016) Endothelial cells are progenitors of cardiac pericytes and vascular smooth muscle cells. Nat. Commun. 7, 12422 10.1038/ncomms12422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang H., Pu W., Liu Q., He L., Huang X., Tian X., Zhang L., Nie Y., Hu S., Lui K. O., and Zhou B. (2016) Endocardium contributes to cardiac fat. Circ. Res. 118, 254–265 10.1161/CIRCRESAHA.115.307202 [DOI] [PubMed] [Google Scholar]
- 10. Zhang H., Huang X., Liu K., Tang J., He L., Pu W., Liu Q., Li Y., Tian X., Wang Y., Zhang L., Yu Y., Wang H., Hu R., Wang F., et al. (2017) Fibroblasts in an endocardial fibroelastosis disease model mainly originate from mesenchymal derivatives of epicardium. Cell Res. 27, 1157–1177 10.1038/cr.2017.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smith C. L., Baek S. T., Sung C. Y., and Tallquist M. D. (2011) Epicardial-derived cell epithelial-to-mesenchymal transition and fate specification require PDGF receptor signaling. Circ. Res. 108, e15–e26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dymecki S. M., Ray R. S., and Kim J. C. (2010) Mapping cell fate and function using recombinase-based intersectional strategies. Methods Enzymol. 477, 183–213 10.1016/S0076-6879(10)77011-7 [DOI] [PubMed] [Google Scholar]
- 13. Madisen L., Garner A. R., Shimaoka D., Chuong A. S., Klapoetke N. C., Li L., van der Bourg A., Niino Y., Egolf L., Monetti C., Gu H., Mills M., Cheng A., Tasic B., Nguyen T. N., et al. (2015) Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–958 10.1016/j.neuron.2015.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He L., Li Y., Li Y., Pu W., Huang X., Tian X., Wang Y., Zhang H., Liu Q., Zhang L., Zhao H., Tang J., Ji H., Cai D., Han Z., et al. (2017) Enhancing the precision of genetic lineage tracing using dual recombinases. Nat. Med. 23, 1488–1498 10.1038/nm.4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Akiyama H., Chaboissier M. C., Behringer R. R., Rowitch D. H., Schedl A., Epstein J. A., and de Crombrugghe B. (2004) Essential role of Sox9 in the pathway that controls formation of cardiac valves and septa. Proc. Natl. Acad. Sci. U.S.A. 101, 6502–6507 10.1073/pnas.0401711101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garside V. C., Cullum R., Alder O., Lu D. Y., Vander Werff R., Bilenky M., Zhao Y., Jones S. J., Marra M. A., Underhill T. M., and Hoodless P. A. (2015) SOX9 modulates the expression of key transcription factors required for heart valve development. Development 142, 4340–4350 10.1242/dev.125252 [DOI] [PubMed] [Google Scholar]
- 17. Xu Z., Wang W., Jiang K., Yu Z., Huang H., Wang F., Zhou B., and Chen T. (2015) Embryonic attenuated Wnt/β-catenin signaling defines niche location and long-term stem cell fate in hair follicle. Elife 4, e10567 10.7554/eLife.10567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ranger A. M., Grusby M. J., Hodge M. R., Gravallese E. M., de la Brousse F. C., Hoey T., Mickanin C., Baldwin H. S., and Glimcher L. H. (1998) The transcription factor NF-ATc is essential for cardiac valve formation. Nature 392, 186–190 10.1038/32426 [DOI] [PubMed] [Google Scholar]
- 19. de la Pompa J. L., Timmerman L. A., Takimoto H., Yoshida H., Elia A. J., Samper E., Potter J., Wakeham A., Marengere L., Langille B. L., Crabtree G. R., and Mak T. W. (1998) Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature 392, 182–186 10.1038/32419 [DOI] [PubMed] [Google Scholar]
- 20. Chang C. P., Neilson J. R., Bayle J. H., Gestwicki J. E., Kuo A., Stankunas K., Graef I. A., and Crabtree G. R. (2004) A field of myocardial-endocardial NFAT signaling underlies heart valve morphogenesis. Cell 118, 649–663 10.1016/j.cell.2004.08.010 [DOI] [PubMed] [Google Scholar]
- 21. Anastassiadis K., Fu J., Patsch C., Hu S., Weidlich S., Duerschke K., Buchholz F., Edenhofer F., and Stewart A. F. (2009) Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice. Dis. Model Mech. 2, 508–515 10.1242/dmm.003087 [DOI] [PubMed] [Google Scholar]
- 22. Liu K., Yu W., Tang M. X., Tang J., Liu X. X., Liu Q. Z., Li Y., He L. J., Zhang L. B., Evans S. M., Tian X. Y., Lui K. O., and Zhou B. (2018) A dual genetic tracing system identifies diverse and dynamic origins of cardiac valve mesenchyme. Development 145 [DOI] [PubMed] [Google Scholar]
- 23. Chau Y. Y., Bandiera R., Serrels A., Martínez-Estrada O. M., Qing W., Lee M., Slight J., Thornburn A., Berry R., McHaffie S., Stimson R. H., Walker B. R., Chapuli R. M., Schedl A., and Hastie N. (2014) Visceral and subcutaneous fat have different origins and evidence supports a mesothelial source. Nat. Cell Biol. 16, 367–375 10.1038/ncb2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yamaguchi Y., Cavallero S., Patterson M., Shen H., Xu J., Kumar S. R., and Sucov H. M. (2015) Adipogenesis and epicardial adipose tissue: a novel fate of the epicardium induced by mesenchymal transformation and PPARγ activation. Proc. Natl. Acad. Sci. U.S.A. 112, 2070–2075 10.1073/pnas.1417232112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Volz K. S., Jacobs A. H., Chen H. I., Poduri A., McKay A. S., Riordan D. P., Kofler N., Kitajewski J., Weissman I., and Red-Horse K. (2015) Pericytes are progenitors for coronary artery smooth muscle. Elife 4, e10036 10.7554/eLife.10036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pillai I. C. L., Li S., Romay M., Lam L., Lu Y., Huang J., Dillard N., Zemanova M., Rubbi L., Wang Y., Lee J., Xia M., Liang O., Xie Y. H., Pellegrini M., et al. (2017) Cardiac fibroblasts adopt osteogenic fates and can be targeted to attenuate pathological heart calcification. Cell Stem Cell 20, 218–232.e5 10.1016/j.stem.2016.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen Y. T., Chang F. C., Wu C. F., Chou Y. H., Hsu H. L., Chiang W. C., Shen J., Chen Y. M., Wu K. D., Tsai T. J., Duffield J. S., and Lin S. L. (2011) Platelet-derived growth factor receptor signaling activates pericyte-myofibroblast transition in obstructive and post-ischemic kidney fibrosis. Kidney Int. 80, 1170–1181 10.1038/ki.2011.208 [DOI] [PubMed] [Google Scholar]
- 28. Humphreys B. D., Lin S. L., Kobayashi A., Hudson T. E., Nowlin B. T., Bonventre J. V., Valerius M. T., McMahon A. P., and Duffield J. S. (2010) Fate tracing reveals the pericyte and not epithelial origin of myofibroblasts in kidney fibrosis. Am. J. Pathol. 176, 85–97 10.2353/ajpath.2010.090517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Henderson N. C., Arnold T. D., Katamura Y., Giacomini M. M., Rodriguez J. D., McCarty J. H., Pellicoro A., Raschperger E., Betsholtz C., Ruminski P. G., Griggs D. W., Prinsen M. J., Maher J. J., Iredale J. P., Lacy-Hulbert A., et al. (2013) Targeting of αv integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat. Med. 19, 1617–1624 10.1038/nm.3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kramann R., Schneider R. K., DiRocco D. P., Machado F., Fleig S., Bondzie P. A., Henderson J. M., Ebert B. L., and Humphreys B. D. (2015) Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell Stem Cell 16, 51–66 10.1016/j.stem.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang H., Pu W., Li G., Huang X., He L., Tian X., Liu Q., Zhang L., Wu S. M., Sucov H. M., and Zhou B. (2016) Endocardium minimally contributes to coronary endothelium in the embryonic ventricular free walls. Circ. Res. 118, 1880–1893 10.1161/CIRCRESAHA.116.308749 [DOI] [PubMed] [Google Scholar]
- 32. Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., Gu H., Ng L. L., Palmiter R. D., Hawrylycz M. J., Jones A. R., Lein E. S., and Zeng H. (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lewandoski M., Meyers E. N., and Martin G. R. (1997) Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb. Symp. Quant. Biol. 62, 159–168 10.1101/SQB.1997.062.01.021 [DOI] [PubMed] [Google Scholar]
- 34. Zhang H., Pu W., Tian X., Huang X., He L. J., Liu Q., Li Y., Zhang L., He L., Liu K., Gillich A., and Zhou B. (2016) Genetic lineage tracing identifies endocardial origin of liver vasculature. Nat. Genet. 48, 537–543 10.1038/ng.3536 [DOI] [PubMed] [Google Scholar]
- 35. Zhang H., von Gise A., Liu Q., Hu T., Tian X., He L., Pu W., Huang X., He L., Cai C. L., Camargo F. D., Pu W. T., and Zhou B. (2014) Yap1 is required for endothelial to mesenchymal transition of the atrioventricular cushion. J. Biol. Chem. 289, 18681–18692 10.1074/jbc.M114.554584 [DOI] [PMC free article] [PubMed] [Google Scholar]