Abstract
Aβ1–42 is involved in Alzheimer's disease (AD) pathogenesis and is prone to glycation, an irreversible process where proteins accumulate advanced glycated end products (AGEs). Nϵ-(Carboxyethyl)lysine (CEL) is a common AGE associated with AD patients and occurs at either Lys-16 or Lys-28 of Aβ1–42. Methyglyoxal is commonly used for the unspecific glycation of Aβ1–42, which results in a complex mixture of AGE-modified peptides and makes interpretation of a causative AGE at a specific amino acid residue difficult. We address this issue by chemically synthesizing defined CEL modifications on Aβ1–42 at Lys-16 (Aβ-CEL16), Lys-28 (Aβ-CEL28), and Lys-16 and -28 (Aβ-CEL16&28). We demonstrated that double-CEL glycations at Lys-16 and Lys-28 of Aβ1–42 had the most profound impact on the ability to form amyloid fibrils. In silico predictions indicated that Aβ-CEL16&28 had a substantial decrease in free energy change, which contributes to fibril destabilization, and a increased aggregation rate. Single-CEL glycations at Lys-28 of Aβ1–42 had the least impact on fibril formation, whereas CEL glycations at Lys-16 of Aβ1–42 delayed fibril formation. We also tested these peptides for neuronal toxicity and mitochondrial function on a retinoic acid-differentiated SH-SY5Y human neuroblastoma cell line (RA-differentiated SH-SY5Y). Only Aβ-CEL16 and Aβ-CEL28 were neurotoxic, possibly through a nonmitochondrial pathway, whereas Aβ-CEL16&28 showed no neurotoxicity. Interestingly, Aβ-CEL16&28 had depolarized the mitochondrial membrane potential, whereas Aβ-CEL16 had increased mitochondrial respiration at complex II. These results may indicate mitophagy or an alternate route of metabolism, respectively. Therefore, our results provides insight into potential therapeutic approaches against neurotoxic CEL-glycated Aβ1–42.
Keywords: Alzheimer disease, amyloid-β (Aβ), glycation, fibril, protein aggregation, mitochondria, neurodegeneration, advanced glycated end product (AGE), N-ϵ-(carboxyethyl)lysine (CEL)
Introduction
Aβ1–42 peptides are implicated in the pathogenesis of Alzheimer's disease (AD).5 Aβ1–42 peptides are released extracellularly after being enzymatically processed from the amyloid precursor protein and are prone to aggregation. Aggregated Aβ1–42 eventually forms amyloid plaques and causes neuronal dysfunction (1). As aging is a major risk factor for AD development, this natural process of aging also ubiquitously produces advanced glycated end products (AGEs), which are a type of post-translational modification (glycation). Several seminal studies provide evidence of increased AGE modifications that are associated with AD brains (2–4). Other disease states, which affect cerebral sugar homeostasis as a consequence of aging, such as Type II diabetes mellitus, may also contribute to the formation of AGEs within the brain (5).
Glycation is an irreversible and nonenzymatically driven process that occurs through a Maillard reaction that conjugates sugars and proteins (6, 7). A particular in vivo sugar derivative, methylglyoxal (MG), is a glycating agent capable of forming different AGEs on Aβ1–42 at lysine residues 16 (Lys-16) and 28 (Lys-28) (8, 9). The increase in in vivo MG production was observed in AD patients (10). Several key studies have showed that a common and major species of AGE, Nϵ-(carboxyethyl)lysine (CEL), had elevated levels in AD brain samples relative to age-matched controls (11). In the cerebrospinal fluid of AD patients, CEL levels had increased and were correlated with a decline in cognitive ability (12). Recently, CEL modifications on AD-associated serum proteins had increased in AD patients relative to age-matched controls (13). Other studies have also demonstrated high levels of CEL in cartilage collagen (14) and eye lenses (15) among aged individuals.
Although CEL is one of the most common forms of MG-derived AGE associated with AD, there are other MG-derived AGEs such as argpyrimidine, MG imidazolone, and MG-lysine dimmer (MOLD) (16). Despite this, the physiological significance of these MG-derived AGEs has not been as extensively explored as CEL. Arginine (Arg-5) on Aβ is also a glycation site, however, there is evidence to suggest a preferential glycation at Lys residues by MG (17). Nϵ-(carboxymethyl)lysine, another non-MG-derived AGE, is also able to attach at Lys residues and was found to increase with CEL modifications (12, 18). Physiologically, Aβ1–42 could be glycated with many forms of AGEs at different amino acid positions. At this stage, however, very little is known about the effect of site-specific CEL modifications and whether there is a major species of AGE that contributes to neuronal toxicity more than another does.
Despite mounting evidence that the increase in CEL modifications on Aβ1–42 were associated with AD, studies have glycated Aβ1–42 with MG to produce unspecific AGEs at different amino acid groups (17, 19). This nonspecific method of glycation makes it difficult to associate a particular AGE modification to toxicity as seen in in vitro studies (20).
Therefore, our current study addresses issues of nonspecific AGE modifications by using organic (21) and peptide chemistry (6) to synthesize unglycated Aβ1–42 and three Aβ1–42 variants with site-specific CEL modifications, namely: a single-CEL modification at Lys-16 (Aβ-CEL16), Lys-28 (Aβ-CEL28), and double-CEL modifications at Lys-16 and Lys-28 (Aβ-CEL16&28). First, we biophysically characterized the CEL modifications relative to unglycated Aβ1–42 by observing peptide morphology with transmission EM (TEM). Following this, we investigated the molecular peptide behavior by calculating the free energy associated with the addition or removal of each peptide from a pentameric stack. We also used the thioflavin T (ThT) assay to measure the kinetics of peptide aggregation, as well as CD to quantify peptide secondary structure.
Next, we assessed the in vitro effect of these three CEL-glycated Aβ1–42 on a retinoic acid (RA)-differentiated human neuroblastoma cell line (RA-differentiated SH-SY5Y), which parallels the behavior and phenotype of mature neurons (22). Our cell culture data investigated overall cell health using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, lactate dehydrogenase (LDH) release (necrosis), and caspase 3/7 activity (apoptosis). We then focused on the effect of the CEL modifications on neuronal mitochondrial function. Mitochondrial dysfunction is an important “cellular switch” that precedes AD pathogenesis (23). It is known that unglycated Aβ affects mitochondrial membrane potential (ΔΨm), mitochondrial swelling, mitochondrial superoxide (O2˙̄) production, and mitochondrial respiration (24). Hence, we chose to investigate the effect of the three CEL-glycated Aβ1–42 on these four mitochondrial parameters.
Results
Double-CEL modifications at Lys-16 and Lys-28 differentially affect Aβ1–42 aggregation over 5 days
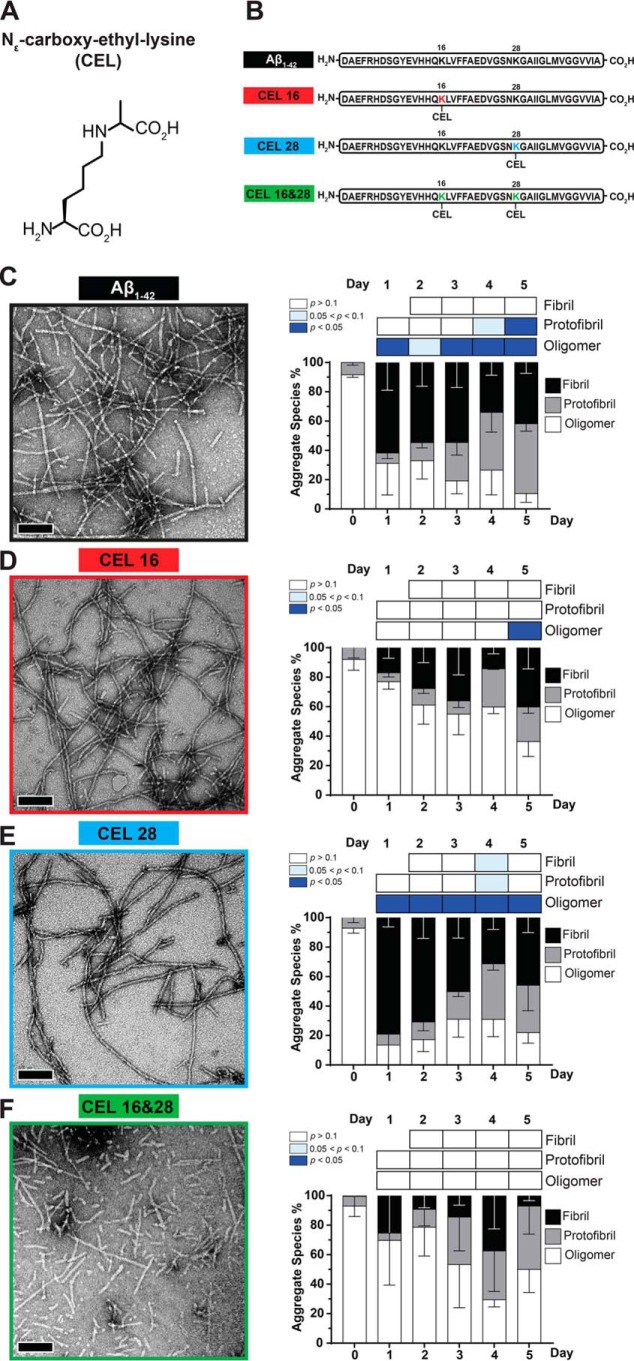
Unglycated Aβ1–42 is known to form fibrils in 10 mm HCl at 37 °C within 24 h (25). We quantified whether site-specific CEL modifications on Aβ1–42 were able to form fibrils to the same extent as unglycated Aβ1–42 under the same experimental conditions. We assessed different aggregate species of the peptides with quantitative TEM across 5 days. The three main aggregate species we considered were fibrils, protofibrils, and oligomers as outlined in Fig. S2.
At day 0, when the lyophilized peptides were freshly resuspended in the fibril-promoting solution, all peptides had >80% oligomers, with protofibrils making up the remaining aggregate species (Fig. 1, C–F). Negligible amounts of fibrils were seen at day 0 across all peptides.
Figure 1.
Double-CEL modifications at Lys-16 and Lys-28 differentially affect Aβ1–42 aggregation over 5 days. A, chemical structure of CEL. B, peptide sequences of Aβ1–42 and the three CEL variants on Aβ1–42. C–F, Aβ-CEL16&28 do not form extensive fibrils and have a high abundance of protofibrils and oligomers after 5 days in a fibril-promoting buffer relative to all CEL-glycated peptides and unglycated Aβ1–42. Representative TEM micrographs at day 5 (left panel) (n = 2–3; scale bar = 100 nm) with the graph of % aggregate species (right panel) showing proportions of fibrils, protofibrils, and oligomers over 5 days. *, p < 0.05. p values for oligomers and protofibrils are relative to day 0 and fibrils are relative to day 1 using Bonferroni's post hoc test in a one-way analysis of variance (ANOVA). See Fig. S1 for synthesis of peptide and Fig. S2 for examples of fibrils, protofibrils, and oligomers.
At day 1, Aβ1–42 and Aβ-CEL28 had a high proportion of fibrils (>60%) followed by a significant decrease (p < 0.05) in oligomers. In contrast, at day 1, Aβ-CEL16 and Aβ-CEL16&28 had a low proportion of fibrils without a significant decrease in oligomers. This high proportion of fibrils and low proportion of oligomers exhibited by Aβ1–42 and Aβ-CEL28 was maintained until day 5 without a significant change in the proportion of protofibrils. However, only Aβ1–42 had a significant increase in protofibrils by day 5 (p < 0.05).
Conversely, Aβ-CEL16 and Aβ-CEL16&28 maintained a low proportion of fibrils and high proportion of oligomers until day 5, with the exception of Aβ-CEL16 displaying a significant decrease (p < 0.05) in oligomers by day 5. The proportion of oligomers for Aβ-CEL16&28 remains unchanged by day 5. The proportion of protofibrils remains unaffected across all days for Aβ-CEL16 and Aβ-CEL16&28. Aβ-CEL16&28 displayed a greater variation in the aggregate species profile across all days compared with all peptides.
There were no observable differences when comparing the TEM micrographs at day 5 between Aβ1–42 and Aβ-CEL28. However, at day 5, Aβ-CEL16 appears to have slightly different aggregate species relative to Aβ1–42, with Aβ-CEL16&28 having the least likeness to Aβ1–42. Thus, a double-CEL modification at both Lys-16 and Lys-28 affects peptide morphology and appears to either prevent the elongation of fibrils or promote the fragmentation of fibrils, whereas a single-CEL modification at Lys-16 slows down the rate of oligomer conversion to either fibrils or protofibrils. Conversely, a single-CEL modification at Lys-28 does not significantly affect peptide morphology relative to the unglycated Aβ1–42.
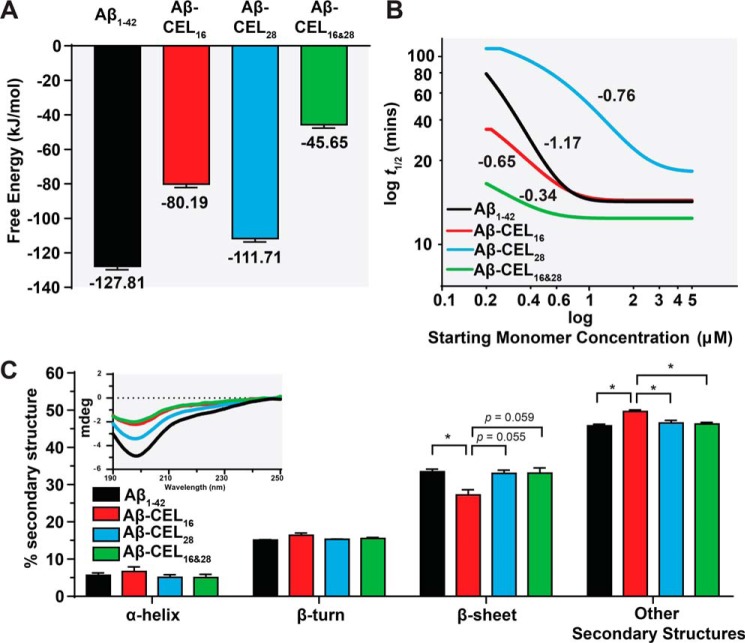
Double-CEL modifications at Lys-16 and Lys-28 decrease free energy release upon elongation
To estimate the propensity of the CEL-glycated Aβ1–42 to aggregate into protofibrillar structures via a simple elongation mechanism, we utilized computational free energy calculations. Umbrella sampling was employed to determine the free energy change, ΔGbind, upon addition of an Aβ1–42 monomer to a 4-monomer protofibril model based on NMR structure PDB code 2BEG (26). The same four single- and double-glycated Aβ variants were tested, with all Aβ1–42 monomers exhibiting the same glycation state for each simulation such that the fibril was homogeneous, analogous to the experimental studies. The results of these simulations are presented in Fig. 2A, with the full potential of mean force curves available in Fig. S3. CEL-glycation impacts ΔGbind in a manner that depends on the location of the CEL. Glycation at Lys-16 results in a ΔGbind that is 37% lower than the unglycated Aβ1–42 ΔGbind, and glycation at Lys-16 and -28 decreases the ΔGbind by 64% of the unglycated Aβ1–42, suggesting that glycation at these positions destabilizes the formation of the fibril or protofibril structure. The proportion of oligomer in the day 5 TEM images (Fig. 1, C–F) mirrors the calculated ΔGbind values, with the two models with less negative ΔGbind values, Aβ-CEL16 and Aβ-CEL16&28, both exhibiting much larger proportions of oligomer relative to Aβ1–42 and Aβ-CEL28.
Figure 2.
Double-CEL modifications at Lys-16 and Lys-28 decrease peptide stability and rate of fibril aggregation without affecting peptide secondary structure. A, Aβ-CEL16&28 has the least negative free energy for binding of a model monomer to a pentameric stack relative to other CEL-glycated peptides and unglycated Aβ1–42. Free energies of binding were computed using umbrella sampling with reported uncertainties determined through bootstrap analysis. B, Aβ-CEL16&28 has the slowest rate of fibril aggregation and Aβ-CEL28 has a higher threshold for maximum rate of fibril aggregation (>3 μm) relative to the CEL-glycated peptides and unglycated Aβ1–42. The rate of fibril aggregation was determined using the aggregation kinetics generated using the ThT assay. The log half-time (t½) is plotted against log starting monomer concentration (micromolars). A plateau at high starting monomer concentrations indicate a saturated (maximum) rate of fibril aggregation. Fitted lines are from 2 to 3 independent experiments and the numeric gradient of each fitted line is displayed. C, Aβ-CEL16&28 has an unchanged secondary structure profile relative to Aβ-CEL28 and Aβ1–42, however, Aβ-CEL16 has a decrease in β-sheet content with an increase in other secondary structures. Peptide secondary structure was estimated using CD (inset) and the spectra deconvoluted using BeStSel. Results are presented as mean ± S.E. (n = 2–3). *, p < 0.05 calculated using Bonferroni's post hoc test in a one-way ANOVA. See Fig. S3 for potential of mean force curves for free energy release. See Fig. S4 for aggregation kinetics of the ThT assay.
Double-CEL modifications at Lys-16 and Lys-28 have a faster rate of fibril aggregation
We also explored the rate of fibril aggregation, another crucial aspect that contributes to peptide morphology and behavior. By utilizing the proposed mathematical models for Aβ1–42 self-assembly in AmyloFit (27), we assessed the kinetics of peptide aggregation using the ThT assay, with representative ThT plots available in Fig. S4. We plotted the half-time (t½; time to reach half the maximum ThT signal) of each peptide against their starting monomeric peptide concentrations (0.2 to 5 μm) (Fig. 2B). The slope of each fitted line indicates the rate of fibril aggregation (28).
Our results show that the rate of fibril aggregation for Aβ1–42 (−1.17) was slower compared with Aβ-CEL16 (−0.65) and Aβ-CEL28 (−0.76), with Aβ-CEL16&28 (−0.34) being the fastest. Furthermore, we note that the rate of aggregation is saturated at >1 μm for Aβ1–42, Aβ-CEL16, and Aβ-CEL28, whereas Aβ-CEL28 is saturated at >3 μm.
Single-CEL modification at Lys-16 has reduced β-sheet content, whereas double-CEL modifications at Lys-16 and Lys-28 remain unchanged
Amyloid fibrils are known to be rich in secondary structures like β-sheets and differences in peptide secondary structure can affect tertiary and quarternary structure. Here, we used CD to collect the far-UV peptide spectra (Fig. 2C, inset). Then, we used a spectral deconvolution software specific to analyzing Aβ1–42 spectra, BeStSel (29), to investigate whether secondary structure changes with CEL modifications on Aβ1–42. Upon spectral deconvolution, our results indicate that the β-sheet secondary structure was the most abundant secondary structure. The “other” secondary structures may include 310-helix, π-helix, β-bridge, bend, loop/irregular, and/or invisible regions of peptide (29). We observed that Aβ-CEL16 had a significant decrease (p < 0.05) of β-sheet content (27.2%) relative to Aβ1–42 (33.5%), and a significant increase (p < 0.05) of other secondary structures (49.7%) relative to all peptides (Fig. 2C). There was no significant difference in α-helix and β-turn secondary structures between CEL-glycated peptides and the unglycated Aβ1–42. Despite Aβ-CEL16&28 having altered aggregate species, free energy of release, and rate of fibril aggregation, the secondary structure of Aβ-CEL16&28 remains unchanged compared with both Aβ1–42 and Aβ-CEL28.
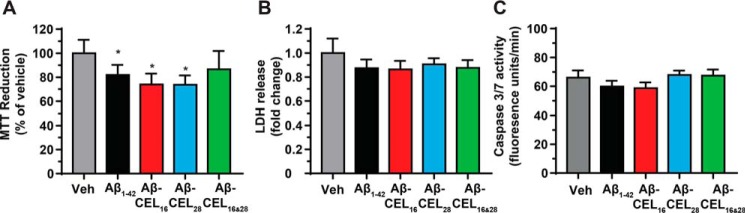
Single-CEL modifications at either Lys-16 or Lys-28 decrease MTT reduction independent of a change in basal levels of necrosis and apoptosis
To investigate the overall status of cell health after 24 h of 10 μm peptide treatment, we measured MTT reduction, extracellular LDH release (necrosis), and extracellular caspase 3/7 activity (apoptosis) in RA-differentiated SH-SY5Y cells. MTT reduction is commonly used to determine both mitochondrial function and cellular redox activity where both parameters contribute to the reduction of the MTT dye (30, 31). The amount of MTT dye reduced is proportional to the colorimetric product. Treatment with Aβ1–42, Aβ-CEL16, and Aβ-CEL28 significantly decreased MTT reduction relative to the vehicle control (Fig. 3A) and this decrease in MTT reduction occurred independent of a change in basal levels of necrosis and apoptosis (Fig. 3, B and C). We did not observe abnormal cell morphologies after peptide treatment. Aβ-CEL16&28 did not affect MTT reduction, LDH release, or caspase 3/7 activity.
Figure 3.
Single-CEL modifications decrease MTT reduction independent of a change in basal levels of necrosis and apoptosis. All experiments performed after 24 h of 10 μm peptide treatment in RA-differentiated SH-SY5Y human neuroblastoma cells. A, Aβ1–42, Aβ-CEL16, and Aβ-CEL28 inhibit MTT reduction, however, Aβ-CEL16&28 does not inhibit MTT reduction. MTT reduction was measured after 24 h of 10 μm peptide treatment. Results are presented as mean ± S.E. (n = 3–4). B and C, no significant difference in LDH release and caspase 3/7 activity was observed for all peptide treatments relative to the vehicle (Veh). Results are presented as mean ± S.E. (n = 4). *, p < 0.05 relative to the vehicle. p value for all experiments were calculated using Bonferroni's post hoc test in a one-way ANOVA from randomized complete block design.
Double-CEL modifications at Lys-16 and Lys-28 depolarize ΔΨm independent of mitochondrial swelling and mitochondrial O2˙̄ production
After investigating the overall cell health, we were interested in whether the CEL-glycated peptides had any specific effects on mitochondrial function. Mitochondrial dysfunction has been implicated as an important molecular initiator that precedes AD pathogenesis (23, 24). We assessed mitochondrial function with a mitochondrial membrane potential (ΔΨm) fluorophore, JC-10, and observed a significant drop in the proportion of cells with polarized ΔΨm upon treatment with Aβ-CEL16&28 relative to Aβ1–42, Aβ-CEL16, and Aβ-CEL28 (Fig. 4A). This drop in ΔΨm was independent of mitochondrial swelling and O2˙̄ production. Mitochondrial swelling and O2˙̄ production are two other parameters implicated in mitochondrial dysfunction as measured by using MitoTracker Green and MitoSOX, respectively (Fig. 4, B and C). MitoTracker Green accumulates proportionally in the matrix of mitochondria and the CEL-glycated peptides did not cause a change in mitochondrial volume (and hence swelling), unlike the unglycated Aβ1–42 (Fig. 4B). Similarly, MitoSOX is specific to O2˙̄ produced in the mitochondria and we did not observe a significant change in mitochondrial O2˙̄ production when treated with the CEL-glycated peptides. However, the unglycated Aβ1–42 caused a significant increase in mitochondrial O2˙̄ production (Fig. 4C).
Figure 4.

Double-CEL modifications at Lys-16 and Lys-28 depolarizes ΔΨm independent of mitochondrial swelling and mitochondrial O2˙̄ production. All experiments performed by flow cytometry after 24 h of 10 μm peptide treatment in RA-differentiated SH-SY5Y human neuroblastoma cells. A, Aβ-CEL16&28 as a decrease in cell population of polarized ΔΨm. ΔΨm was assessed by JC-10. Results are presented as mean ± S.E. (n = 3) with representative flow cytometry plots available in Fig. S5. B, treatment with unglycated Aβ1–42 increased mitochondrial swelling relative to Aβ-CEL16 and the vehicle (Veh). This Aβ1–42 induced mitochondrial swelling is marginally different from Aβ-CEL28 (p = 0.07) and Aβ-CEL16&28 (p = 0.06). Mitochondrial volume was assessed by MitoTracker Green. Results are presented as mean ± S.E. (n = 4) with representative flow cytometry plots in the corresponding histogram (right). A shift of the histogram toward the right indicates an increase in MitoTracker Green within the cell. C, treatment with unglycated Aβ1–42 has increased mitochondrial O2˙̄ production relative to all peptides and the vehicle. Mitochondrial O2˙̄ production was assessed by MitoSOX. Results are presented as mean ± S.E. (n = 3–4) with representative flow cytometry plots shown in Fig. S5. *, p < 0.05; **, p < 0.01. p values were calculated in a one-way ANOVA using a completely randomized block design blocked by different experiments. See Fig. S5 for flow cytometry gating strategy.
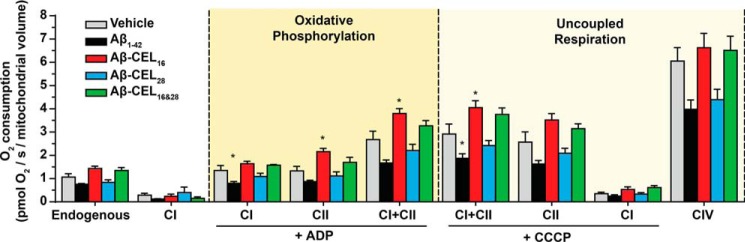
CEL modification at Lys-16 increases mitochondrial respiration at complex II and maximum respiration
Mitochondrial respiration is important for maintaining overall cell health by meeting the bioenergetic demand. Unglycated Aβ1–42 is known to affect mitochondrial respiration (32) and we questioned whether CEL-glycated peptides had an effect to the same extent as unglycated Aβ1–42. We used high-resolution respirometry to measure mitochondrial respiration in permeabilized cells treated with 10 μm peptide for 24 h. We titrated mitochondrial respiration substrates that support the electron transport system (Fig. S6A). Oxygen consumption was also corrected for mitochondrial volume. See Fig. S6B for oxygen consumption relative to cell count.
Our results indicated that unglycated Aβ1–42 lowered mitochondrial respiration across all respiration states, but was only statistically significant at CI under oxidative phosphorylation (OXPHOS) and CI+CII under uncoupled respiration (p < 0.05) (Fig. 5). Aβ-CEL16 increased mitochondrial respiration at CII and CI+CII under OXPHOS and CI+CII under uncoupled respiration. Aβ-CEL28 did not show any significant difference in mitochondrial respiration relative to the vehicle. Aβ-CEL16&28 showed no difference in respiration relative to the vehicle control. Overall, we observed that CEL modifications at Lys-16 of Aβ1–42 caused an increase in respiration at CII, which was in contrast to the decrease in respiration caused by unglycated Aβ1–42.
Figure 5.
CEL modification at Lys-16 increases mitochondrial respiration at complex II and increases maximum respiration. All experiments were performed by oximetry after 24 h of 10 μm peptide treatment in RA-differentiated SH-SY5Y human neuroblastoma cells. Aβ-CEL16 increases mitochondrial respiration at CII and CI+CII under OXPHOS and CI+CII under uncoupled (maximum) respiration. Aβ1–42 decreases mitochondrial respiration at CI under OXPHOS and CI+CII under uncoupled respiration. Shown here is oxygen consumption from intact cells (endogenous) and mitochondria of permeabilized cells at different states of respiration. The two main states of respiration are coupled respiration (OXPHOS) and uncoupled (maximum) respiration. Results are presented as mean ± S.E. (n = 5); CIV (n = 4). *, p < 0.05 relative to vehicle. p value was calculated using Bonferroni's post hoc test in a one-way ANOVA from randomized complete block design. See Fig. S6 for an example of oxygen consumption trace and the mitochondrial substrates added.
Discussion
Post-translational modifications of Aβ1–42, particularly AGEs, remains understudied despite evidence suggesting a connection to, and exacerbation of AD pathogenesis (5, 33). Our study is the first to synthesize CEL-glycated Aβ1–42 at Lys-16 and Lys-28, a major AGE associated with AD, and in doing so, we characterized peptide behavior and investigated the effect of these variants on neuronal function.
From the TEM micrographs, it is evident that Aβ-CEL16&28 had the most profound change in peptide morphology with the presence of many oligomers and protofibrils after 5 days in a fibril promoting solution. TEM micrographs at day 5 show that a single-CEL glycation at Lys-16 slowed fibril formation, whereas a single-CEL glycation at Lys-28 had little effect on the ability to form fibrils. Therefore, double, but not single, CEL glycations of Aβ1–42 affect the ability of fibril formation.
It is important to note that aggregate species exist on a continuum and that size (length) observed through a TEM micrograph may not be a good indicator of aggregate species (34). Accurate quantitation of Aβ1–42 aggregate species have been carried out with conformation-specific antibodies (34). In this case, such conformation-specific antibodies may not recognize the target epitope if a CEL glycation is present, and the nonavailability of antibodies that recognize both CEL and Aβ1–42 makes this antibody approach difficult. Notwithstanding this, size (length) observed through a TEM micrograph is still an appropriate and commonly used technique to distinguish between oligomeric, protofibrillar, and fibrillar Aβ1–42 aggregates (35, 36).
Next, we investigated whether different peptide behaviors may contribute to the inability of Aβ-CEL16&28 to form fibrils as seen with the TEM micrographs. In silico modeling reveals a 2.7-fold decrease in free energy required to dissociate Aβ-CEL16&28 relative to unglycated Aβ1–42, which supports the idea that CEL modifications at Lys-16 and Lys-28 destabilize the fibril structure. We postulate that the net-neutral charge, produced when a negatively charged CEL moiety is conjugated to a positively charged Lys residue, disturbs the adjacent hydrophobic region (Leu-17 to Ala-21) (17, 35), and the ability to form a salt bridge hairpin turn at Asp-23 with Lys-28 (35). Hence, the double-CEL glycation on Lys-16 and Lys-28 may synergistically affect the ability of Aβ-CEL16&28 to form fibrils.
In addition to the decrease in free energy with a double-CEL glycation, we observed the fastest rate of aggregation with Aβ-CEL16&28 relative to the single-CEL glycated Aβ1–42 and unglycated Aβ1–42 peptides. This rate of aggregation is indicative of the time taken to reach maximum fibrillar mass, which suggests that the destabilizing effect of a double-CEL glycated on Aβ1–42 encourages rapid peptide aggregation.
We also undertook CD and spectral deconvolution of each peptide to determine whether there was a change in peptide secondary structure that could explain the different peptide morphologies seen with the TEM micrographs. Interestingly, our CD data indicated that only Aβ-CEL16 had a change in secondary structures, with an increase in other secondary structures and a decrease in β-sheet content. Indeed, the substitution of Lys-16 for Ala-16, a neutral amino acid residue, on Aβ1–42 resulted in a decreased β-sheet content, with an increased α-helix content (9). Although CEL-glycated Lys residues have a net-neutral charge, similar to the substitution of Lys for Ala, the type of increase in secondary structure observed was different than in Ref. 9. This may be due to the different deconvolution software used in Ref. 9, with BeStSel proven to be more accurate when deconvoluting CD spectra of β-sheet–rich structures like Aβ1–42 (29). Despite this change in secondary structure content, Aβ-CEL16 peptides are still able to form fibrils, albeit delayed relative to Aβ1–42 and Aβ-CEL28, and the unchanged secondary structure content of Aβ-CEL16&28 does not reflect on the peptide's inability to form fibrils. Thus, secondary structure is unlikely to affect peptide aggregation and behavior in this instance.
Following the biophysical characterization, we investigated the effects of unglycated and CEL-glycated Aβ1–42 on a neuronal in vitro model, RA-differentiated SH-SY5Y cells. Because the oligomeric species of Aβ1–42 are the most neurotoxic (37), we hypothesized that the propensity for Aβ-CEL16&28 to remain oligomeric would exert more neurotoxicity on RA-differentiated SH-SY5Y cells relative to the unglycated Aβ1–42. Neurotoxicity is, however, a broad term for the outcome of different pathways of neuronal dysfunction, which include mitochondrial and nonmitochondrial pathways. Interestingly, treatment with Aβ-CEL16&28 resulted in a depolarization of ΔΨm independent of a change in neurotoxicity, mitochondrial respiration, O2˙̄ production, and mitochondrial swelling. A depolarization of ΔΨm is implicated in the initiation of the selective degradation of dysfunctional mitochondria, also known as mitophagy (38). Moreover, this ΔΨm depolarization is a well-characterized and key mediator of neuronal mitophagy (39, 40). Recently, mitophagy has also been implicated as an important process that inhibits Aβ-associated pathology (41). Therefore, we suspect that the depolarization of ΔΨm by Aβ-CEL16&28 may be a compensatory mechanism, rather than a mediator of dysfunction, which “tags” aberrant mitochondria for mitophagy to maintain a healthy pool of mitochondria and sustain neuronal function.
Conversely, single-CEL glycated Aβ1–42 are as neurotoxic as unglycated Aβ1–42. The neurotoxic effect of unglycated Aβ1–42 can be attributed to mitochondrial dysfunction. Treatment with unglycated Aβ1–42 caused a 2.2-fold increase in O2˙̄ production and a 1.6-fold increase in mitochondrial volume (swelling). The swelling of mitochondria was also observed when neuronal synaptic vesicles were incubated with Aβ (42). Moreover, our observation that Aβ1–42 causes mitochondrial dysfunction corroborates with findings from Caspersen and colleagues (32). Interestingly, single- and double-CEL glycated Aβ1–42 failed to induce O2˙̄ production relative to the unglycated Aβ1–42. This may be indicative of: 1) the conversion of O2˙̄ to hydrogen peroxide (H2O2) and/or hydroxyl radicals (HO•), which are also thought to be involved in AD-associated oxidative stress (43), or 2) the lack of O2˙̄ production by mitochondria upon treatment with CEL-glycated Aβ1–42. Thus, we postulate that CEL-mediated neurotoxicity may occur independent of O2˙̄ production.
AD-mediated mitochondrial dysfunction is associated with an altered bioenergetic profile (44). We quantified mitochondrial bioenergetics using high-resolution respirometry and noted an increase in respiration primarily at CII of the electron transport system when RA-differentiated SH-SY5Y cells were treated with Aβ-CEL16. Indeed, Bubber and colleagues (45) showed that increased CII activity in AD brains relative to age-matched controls was a potential result of an alternate route of energy metabolism. It is possible that Aβ-CEL16 elicits this energetic response by increasing the number of CII subunits, which favors nonglycolytic energy production (45). Moreover, it may be easier to mediate this switch in energy metabolism by up-regulating CII transcription and translation because CII is comprised of only four nuclear encoded subunits.
Herein, our study has provided insight into the effect of site-specific CEL glycations on Aβ1–42 in terms of peptide behavior and the effect on neuronal mitochondrial function. We have demonstrated that double-CEL glycations at Lys-16 and Lys-28 of Aβ1–42 synergistically affected fibril formation by decreasing the free energy change, which destabilized the fibril structure. This destabilization of Aβ-CEL16&28 also resulted in an increased aggregation rate. Moreover, single-CEL glycations at Lys-28 of Aβ1–42 affected fibril formation and aggregation the least, compared with CEL glycations at Lys-16 of Aβ1–42. Notwithstanding this, only CEL glycations at Lys-16 of Aβ1–42 had an effect on peptide secondary structure, particularly with a decrease in β-sheet content. These results suggest that a single-CEL glycation at Lys-16, but not Lys-28, of Aβ1–42 affected peptide behavior and double-CEL glycations at Lys-16 and Lys-28 had the most profound effect on peptide behavior. Interestingly, only single-CEL glycation at Lys-16 or Lys-28 of Aβ1–42 had a neurotoxic effect similar to unglycated Aβ1–42, whereas double-CEL glycations at Lys-16 and Lys-28 did not have a neurotoxic effect. Neither single- nor double-CEL glycations of Aβ1–42 affected parameters of mitochondrial dysfunction unlike unglycated Aβ1–42. However, double-CEL glycations at Lys-16 and Lys-28 of Aβ1–42 had depolarized ΔΨm, as a potential result of mitophagy, and single-CEL glycations at Lys-16 of Aβ1–42 had an increase in CII respiratory activity, similar to what was observed in AD brains (45). Therefore, our results suggests that the neurotoxicity of single-CEL glycations at Lys-16 or Lys-28 may occur through nonmitochondrial pathways, however, further studies are needed to elucidate the mechanism(s) involved. Last, the neurotoxicity of single-CEL glycated Aβ1–42 highlights the potential for a therapeutic target against these CEL-glycated peptides to potentially slow and prevent the pathogenesis of Aβ1–42.
Experimental procedures
Materials
Thioflavin T (T3516), all-trans-RA (R2625), MTT (M5655), and LDH assay kit (11644793001) were purchased from Sigma. Phenol-free DMEM/F-12 (11039047), TrypLE express enzyme (12604021), ×100 GlutaMAX (3505061), and 10,000 units/ml of penicillin-streptomycin (15140122) was purchased from Invitrogen. Caspase 3 substrate (Ac-Asp-Glu-Val-Asp-AMC; I-1660) was purchased from Bachem (Bubendorf, Switzerland). N,N-Diisopropylethylamine, piperidine, N-methylmorpholine, triisopropylsilane, tetrabutylammonium iodide, and N,N′-diisopropylcarbodiimide were purchased from Sigma. TFA was purchased from Scharlau (Barcelona, Spain) and 1,2-ethanedithiol was supplied by Fluka. Fmoc-Asp(OtBu)-OH, Fmoc-Ala-OH, Fmoc-Glu(OtBu)-OH, Fmoc-Phe-OH, Fmoc-Arg(Pbf)-OH, Fmoc-His(Trt)-OH, Fmoc-Ser(t-Bu)-OH, Fmoc-Gly-OH, Fmoc-Tyr(tBu)-OH, Fmoc-Val-OH, Fmoc-Gln(Trt)-OH, Fmoc-Lys(Boc)-OH, Fmoc-Leu-OH, Fmoc-Asn(Trt)-OH, Fmoc-Ile-OH, and Fmoc-Met-OH were purchased from GL Biochem (Shanghai, China). Fmoc-Ala-HMPP linker was purchased from Polypeptide (Strasbourg, France), O-(7-azabenzotriazol-1-yl)-N,N,N′,N′-tetramethyl-uronium hexafluorophosphate (HATU) was purchased from CS Bio (Shanghai, China), aminomethyl ChemMatrix resin was purchased from PCAS Biomatrix Inc. (Canada), and acetonitrile (MeCN, HPLC grade) was purchased from Merck (Germany).
Peptide synthesis and purifications
Fmoc-CEL(Boc)(OtBu)-CO2H was prepared as previously described (21). All peptides were assembled by automated Fmoc-solid phase peptide synthesis (Fmoc-SPPS) using a TributeTM peptide synthesizer (Protein Technologies Inc.). To aminomethyl ChemMatrix resin (100 mg, 0.062 mmol, 0.62 mmol/g) pre-swollen in CH2Cl2 (5 ml, 10 min) was added a mixture of Fmoc-Ala-HMPP (91 mg, 0.186 mmol) and N,N′-diisopropylcarbodiimide (24 μl, 0.186 mmol) in CH2Cl2:DMF (3:1, v/v, 2.0 ml). The reaction mixture was agitated at room temperature for 1 h, after which the resin was filtered and washed with DMF (3 × 3 ml). Conditions for automated Fmoc-SPPS are as follows.
Fmoc-amino acid couplings
To the peptidyl resin was added a mixture of Fmoc-AA-OH (0.50 mmol), HATU (0.46 mmol), and N-methylmorpholine (1.00 mmol) in DMF (2.0 ml). The reaction mixture was agitated at room temperature for 30 min, after which the resin was filtered and washed with DMF (2 ml, 4 × 30 s).
Coupling of Fmoc-CEL(Boc)(OtBu)-CO2H
To the peptidyl resin was added a mixture of Fmoc-CEL(Boc)(OtBu)-CO2H (119 mg, 0.20 mmol), HATU (72 mg, 0.19 mmol), and N,N-diisopropylethylamine (87 μl, 0.50 mmol) in DMF (2.0 ml). The reaction mixture was agitated at room temperature for 2 h, after which the resin was filtered and washed with DMF (2 ml, 4 × 30 s).
Fmoc-deprotections
To the peptidyl resin was added a solution of 20% piperidine in DMF (3.0 ml), and the reaction mixture was agitated room temperature for 5 min. This procedure was repeated once, after which the resin was filtered and washed with DMF (2 ml, 5 × 30 s). Following the final Fmoc-deprotection, the peptidyl-resin was washed with DMF (3 × 3 ml), CH2Cl2 (3 × 3 ml), and dried under vacuum. To the dry resin-bound peptide was added a mixture of TFA/triisopropylsilane/H2O/EDT (94:1:2.5:2.5; v/v/v/v; 10 ml) and the reaction mixture was agitated at room temperature for 2 h. The TFA mixture was filtered, the resin washed with the cleavage mixture (2 × 2 ml), and the combined filtrates concentrated under a gentle flow of nitrogen. The crude peptides were precipitated with cold Et2O (40 ml), isolated by centrifugation, and this isolation procedure was repeated twice. The crude peptides were air-dried and purified by semi-preparative reverse phase HPLC (RP-HPLC) on a Thermo Scientific Dionex Ultimate 3000 HPLC equipped with a four channel UV Detector at 210, 225, 254, and 280 nm. A semi-preparative column (XTerra MS C18, 125 Å, 250 × 10 mm, 10 μm) was used at either room temperature or 50 °C and at a flow rate of 5 ml min−1. A linear gradient of 8% B to 18% B over 55 min was employed, where solvent A was 0.1% NH4OH in water and B was 0.1% NH4OH in acetonitrile. Any co-eluting Met(O) by-products were consequently reduced by dissolution of the purified peptide (1.0 mg, 2.2 μmol) in neat TFA (1 ml, 1 mg ml−1) and cooled to 0 °C. To this was added a cooled solution of tetrabutylammonium iodide (0.8 mg, 22.2 μmol) dissolved in neat TFA (0.8 ml). The mixture was kept at 0 °C with regular agitation for 5 min, during which a pale brown discoloration was observed. The reaction mixture was then concentrated under a gentle flow of nitrogen, the product was precipitated with cold Et2O, isolated by centrifugation, and re-purified by semi-preparative RP-HPLC as described above.
The purified peptides were characterized by analytical RP-HPLC and low resolution MS. Analytical RP-HPLC was performed on a Thermo Scientific Dionex Ultimate 3000 UHPLC equipped with a four channel UV Detector at 210, 225, 254, and 280 nm. An analytical column (XTerra MS C18, 125 Å, 150 × 4.6 mm, 5 μm) was used at 50 °C and at a flow rate of 1 ml min−1. A linear gradient of 1% B to 41% B over 20 min was employed, where solvent A was 0.1% NH4OH in water and B was 0.1% NH4OH in acetonitrile. Low resolution MS analysis was acquired on an Agilent Technologies 1260 Infinity LC equipped with an Agilent Technologies 6120 Quadrupole mass spectrometer, using 50% B over 3 min, where solvent A was 0.1% formic acid in water and B was 0.1% formic acid in acetonitrile. Synthesis, purification, and characterization afforded Aβ1–42 as white fluffy flakes (66% yield relative to crude reduced peptide, >98% purity), RT: 11.8 min, ESI-MS (deconvoluted mass 4513.42 ± 0.20 Da, calculated mass 4514.08); Aβ1–42-CEL16 as white fluffy flakes (54% yield relative to crude, 94% purity), RT: 10.9 min, ESI-MS (deconvoluted mass 4585.50 ± 0.48 Da, calculated mass 4586.14); Aβ1–42-CEL28 as white fluffy flakes (73% yield relative to crude reduced peptide, >98% purity), RT: 10.7 min, ESI-MS (deconvoluted mass 4585.75 ± 0.75 Da, calculated mass 4586.14); Aβ1–42-CEL16&28 as white fluffy flakes 20% yield relative to crude reduced peptide, >98% purity), RT: 10.3 min, ESI-MS (deconvoluted mass 4657.43 ± 0.29 Da, calculated mass 4658.20). Further details are available in Fig. S1.
Sample pre-treatment and concentration quantitation
Pre-treatment of peptides to remove preformed aggregates were performed as per Ryan and colleagues (46) by treating once with 10% (w/v) NH4OH to a concentration of 0.5 mg/ml in LoBind Protein 1.5-ml microcentrifuge tubes (Eppendorf, Germany). Peptides reconstituted in 10% (w/v) NH4OH were left for 10 min at room temperature (21 °C) followed by bath sonication for 5 min in a Soniclean 160T (Transtek Systems, Australia) at a frequency band of 50–60 Hz. After bath sonication, the peptides in solution were centrifuged at 16,100 × g for 10 min at 21 °C and the supernatant was removed and dispensed into 100-μl aliquots that were lyophilized in a vacuum freeze dryer (Alpha 2–4 LDplus, Martin Christ, Germany) and stored at −30 °C.
Peptide concentration was determined in a Cary 4000 UV spectrophotometer (Varian) at 280 nm using a 10-mm path length quartz cuvette. Briefly, pre-treated NH4OH-lyophilized peptide was reconstituted (155 μl) in 0.22 μm of filtered 0.33 mm NaOH, 1 × DPBS as described by Ryan and colleagues (46). The reconstituted peptide was then centrifuged at 16,100 × g for 10 min at 21 °C and 150 μl of the supernatant was immediately used for spectrophotometric analysis. Concentration of the peptide was determined from the absorbance at 280 nm using the molar extinction coefficient for a single tyrosine residue in Aβ1–42 (1490 m−1 cm−1).
Transmission electron microscopy
To promote fibril formation, NH4OH pre-treated peptides were incubated in 10 mm HCl for up to 5 days at 37 °C (25) in LoBind Protein 1.5-ml microcentrifuge tubes (Eppendorf, Germany) with sampling taken at 0 h and subsequently every 24 h up to 120 h. Samples (2 μl) were adsorbed onto glow-discharged carbon-coated 400-mesh copper grids (Gilder, UK) for 30 s, and then washed in ultrapure water (18.2 mΩ·cm) for 30 s followed by 60 s in 2% (w/v) uranyl acetate. Excess uranyl acetate was wicked from the copper grid using a filter paper and imaged in a CM12 transmission electron miscroscope (Philips, Netherlands) at an operating voltage of 120 kV with a Bioscan 792 camera (Gatan, USA). At least 3 fields of view (×140,000 magnification) were taken for each sample.
Quantitation of electron micrographs
Aggregate ratio of oligomers, protofibrils, and fibrils were manually counted using ImageJ. Classification of oligomers, protofibrils, and fibrils are shown in Fig. S2. The “aggregate species %” is the relative populations of each aggregate species.
Computational modeling
The model for the Aβ fibril chosen for this work was the core of the Aβ1–42 fibril, determined from NMR studies (26). The published PDB structure only contains Aβ17–42 with the N-terminal 16 amino acids missing, due to their structural heterogeneity. For our model, the missing 16 amino acids were added using the tLeaP program from Amber12 (47). The protofibril model consists of 5 Aβ monomers labeled A–E, from the inner to outer surface of the fibril model. All amino acids were assumed to be in their standard protonation states for physiological pH (7.0), resulting in a net charge of −20, which was neutralized by the addition of 20 sodium ions. The protofibril was added to a unit cell with dimensions 12 × 8 × 22 nm, and was fully solvated using TIP3P water (48). Periodic boundary conditions were applied in all directions.
All simulations were conducted using the Gromacs2016.1 software package (49). Simulations were run using the Charmm36m force field (50), with additional terms for CEL generated using CGenFF (51). The CEL parameters were verified by comparison to parameters generated through an alternative method, using a combination of RESP charge fitting using R.E.D Server (52) and ForceGen (53). The lengths of covalent bonds involving hydrogen atoms were restrained using the LINCS algorithm (54), such that a time step of 2 fs could be employed. Short-range nonbonded interactions were calculated using a cut-off of 1.4 nm, with long range electrostatics calculated using the particle mesh Ewald (PME) algorithm (55).
The initial models underwent steepest descent energy minimization for 5000 steps until the change in force was less than 0.02 kJ mol−1 nm−1 followed by a 5000-step conjugate gradient minimization using a 0.01 kJ mol−1 nm−1 tolerance. The models were then heated from 50 to 310 K in the NVT ensemble at a heating rate of 2.1 K ps−1. The Nose-Hoover thermostat (56) was used to maintain the temperature, with the protein and nonprotein atoms coupled to separate temperature baths using a thermostat time constant of 0.1 ps. Following NVT equilibration at 310 K for 250 ps, a short constant pressure equilibration was conducted for 500 ps at 1 atmosphere pressure, using the Parrinello-Rahman barostat with a pressure coupling constant of 2.0 ps.
Following equilibration, restraints were removed from peptide E, with the remaining peptides restrained to their initial positions via harmonic restraints on the heavy atoms with a force constant of 1000 kJ mol−1 to provide an immobile reference for the pulling simulations. Position restraints are frequently employed on the model Aβ structure to mimic the stability of much larger structures, while reducing the computational expense (57, 58). For each glycation state, peptide E was pulled away from the rest of the protofibrillar structure along the z axis over 1 ns at a constant pulling rate of 0.01 nm ps−1 using a spring constant of 1000 kJ mol−1 nm−2. From the generated trajectories, snapshots were taken for the starting configurations of the umbrella sampling windows (59, 60). Snapshots were taken in an asymmetric fashion with the first distribution of windows taken at 0.1-nm increments until the center of mass distance between peptides D and E was 2.0 nm; beyond this distance the remaining snapshots were taken at 0.2-nm intervals. This asymmetric distribution enabled greater resolution at small center of mass distances while reducing the overall number of windows, such that only 45 windows were used. Within each window a 10-ns simulation was performed utilizing the same simulation protocol as above to give a total umbrella sampling simulation time of 450 ns. The weighted histogram analysis method (61) within gromacs2016.1 (62) was utilized to analyze the results. Statistical errors were estimated using bootstrap analysis with the b-hist option.
Thioflavin T assay and half-time (t½) analysis
ThT was prepared fresh before every experiment at a concentration of 1 mm in ultrapure water (18.2 mΩ·cm) and was diluted in ice-cold 10 mm sodium phosphate buffer (pH 7.4) to a final concentration of 10 μm. Peptides were reconstituted in ice-cold 10 μm ThT and immediately diluted (100 μl/well) in a nonbinding solid bottom black 96-well plate (GN655900) on ice. The plate was sealed with a clear plastic cover and placed in an EnVision plate reader (PerkinElmer Life Sciences) at a temperature of 30 °C. Fluorescence intensity was monitored over 300 min (1 min between each read) using an excitation and emission wavelengths of 440 and 485 nm, respectively. Each independent experiment had triplicate wells of each peptide.
Analysis of ThT fluorescence intensity and the time it takes to get to half the maximum fluorescence, half-time (t½), was performed as previously described using the AmyloFit software (27). Double log-log plots of t½ (min) and starting monomeric concentration (μm) was used to assess molecular mechanism of aggregation using the scaling exponent (γ).
The line of best fit used for the double log-log plots is the one-phase decay function in GraphPad Prism (version 7.00) with the model in Equation 1.
| (Eq. 1) |
CD
Frozen lyophilized peptides were left to thaw to room temperature and then diluted into a 1-mm quartz cuvette (Hellman Analytics, Germany) to 10 μm concentration in 10 mm sodium phosphate buffer (pH 7.4). The secondary structure of the peptides was quantified in a Chirascan CD spectrophotometer (Applied Biophysics, UK) at a chamber temperature of 30 °C with a bandwidth of 2.5 nm and a read time of 2 s/1 nm from 260 to 180 nm. Experiments were done on different days. Mdeg values were subtracted from the baseline (10 mm sodium phosphate buffer alone). Spectra deconvolution and estimation of secondary structure were carried out using BeStSel (29).
Cell culture and RA differentiation
Human neuroblastoma SH-SY5Y (ATCC) cells were routinely cultured in complete media (10% heat-inactivated FBS in 1:1 phenol free DMEM/F-12 supplemented with 1% Pen/Strep and 1% Glutamax) in a humidified 37 °C incubator at 5% CO2. The medium was replaced every 2 days. After an overnight period where cells were allowed to attach to cultureware surface, differentiation was initiated by changing the complete media to a reduced serum media (1% heat-inactivated FBS in 1:1 phenol-free DMEM/F-12 supplemented with 1% Pen/Strep and 1% Glutamax). This reduced serum media was supplemented with 10 μm all-trans-RA with a DMSO concentration of 0.5% for up to 4 days with RA medium changes every 2 days.
Peptide treatments on cell culture
Peptides were removed from −30 °C and left to equilibrate to room temperature before being reconstituted in 0.33 mm NaOH, 1× DPBS then centrifuged at 16,100 × g for 10 min at 21 °C. The peptide in the supernatant was carefully removed and diluted to a concentration of 10 μm into pre-warmed 1% FBS (1:1) phenol-free DMEM/F-12 medium supplemented with 10 μm RA. The vehicle was carried through the same process (without peptide) and the 0.33 mm NaOH, 1× DPBS dilution did not exceed 1/8.
MTT assay
Mitochondrial function and cellular redox activity was assessed with the MTT assay. SH-SY5Y cells were seeded into a 96-well plate at a density of 5 × 105 cells/well and left to adhere to the surface overnight followed by the differentiation protocol as described. Treatment with 10 μm peptide commenced on day 4 for 24 h.
After 24 h, 15 μl (5 mg/ml in PBS) of pre-warmed MTT reagent was added to each well and left for 4 h to form formazan crystals. To solubilize the crystals, 200 μl of 40 mm HCl acidified isopropyl alcohol was added to each well and mixed by pipetting. Once solubilized, the calorimetric absorbance was read in an EnVision plate reader (PerkinElmer Life Sciences) at 540 nm with a subtraction of background at 720 nm. All wells were corrected for with a blank containing the same buffer diluent but with no cells. Equation 2 shows the blank and background correction to get a percentage of cell toxicity relative to the vehicle.
| (Eq. 2) |
LDH release and caspase 3/7 activity assay
Cells were seeded in 12-well dishes at a density of 1.0 × 106 and left to adhere to the surface overnight followed by the differentiation protocol as described previously. Treatment with 10 μm peptide commenced on day 4 for 24 h and the cell culture medium was collected and spun at 16,100 × g for 10 min at 4 °C to remove cell debris. The supernatant was used for LDH release and caspase 3/7 activity assays. The cells were used for the mitochondrial respiration assay.
The LDH release assay was performed as per the manufacturer's instructions (Roche, Switzerland). The assay was performed in a clear bottom 96-well plate and the calorimetric absorbance was read in a SpectraMax iD3 plate reader (Molecular Devices) at 540 nm with a subtraction of background at 720 nm. Technical duplicate wells were used for each experiment followed by the blank subtraction (cell-free culture media only). Equation 3 shows blank and background correction to get a normalized LDH release value relative to the vehicle.
| (Eq. 3) |
For the caspase 3/7 activity assay, 25 μl of the supernatant was added to a nonbinding solid bottom black 96-well plate (GN655900, Greiner). Then the fluorogenic caspase substrate (reconstituted in DMSO) was added to a final concentration of 40 μm in buffer (20 mm HEPES, 1 mm EDTA, 5 mm DTT, 0.1% CHAPS, 10% sucrose). The fluorescence was monitored at 37 °C for 3.5 h with 2-min intervals between each read in a SpectraMax iD3 plate reader (Molecular Devices) at excitation and emission of 385 and 460 nm, respectively. A linear line of best fit between fluorescence intensity and time was used to determine the slope (R2 > 0.96 for all slopes) and presented as a function of fluorescence units/min.
Mitochondrial membrane potential (ΔΨm), mitochondrial volume, and mitochondrial superoxide (O2˙̄) production
Cells were seeded in 12-well dishes at a density of 1.0 × 106 and left to adhere overnight before differentiation with 10 μm RA for 4 days. After 4 days, the cells were treated with 10 μm peptide for 24 h the same way as previously mentioned for the MTT assay. After 24 h, the cells were washed twice with 1× PBS and detached from the surface with TrypLE (Invitrogen). TrypLE was neutralized with equal amounts of complete medium containing 10% FBS and the cells were pelleted at 200 × g for 4 min at 21 °C. For ΔΨm, JC-10 was prepared as per manufacturer's instructions (Abcam, UK) and the cells were incubated with JC-10 for 30 min at 37 °C protected from light. For mitochondrial volume and mitochondrial O2˙̄ production, 200 nm MitoTracker Green and 5 μm MitoSOX were used, respectively, and diluted into warm 1% FBS DMEM/F-12. The cells were incubated for 30 min at 37 °C protected from light. After 30 min of dye incubation, the cells were pelleted, washed twice, and suspended with warm 1% FBS DMEM/F-12 before analysis using an Accuri C6 (BD Bioscience). Data were analyzed using FlowJo Version 10.4 (FlowJo LLC). The gating strategy is outlined in Fig. S5. The final gates contained between 77 and 8000 individual events.
Mitochondrial respiration
Cells were seeded in 12-well dishes at a density of 1.0 × 106 and left to adhere to the surface overnight followed by the differentiation protocol as described previously. Treatment with 10 μm peptide commenced on day 4 for 24 h and the cells were dissociated from the surface after 24 h as per flow cytometry methodology. The cell culture medium was saved for the LDH release and Caspase 3/7 activity assays. The cells were counted and immediately placed into the Oxygraph O2k (Oroboros, Austria) containing MiRO5 respiration medium (0.5 mm EGTA, 3 mm MgCl2·6H2O, 60 mm lactobionic acid (pH 7.0 with 5 n KOH), 20 mm taurine, 10 mm KH2PO4, 20 mm HEPES, 110 mm sucrose, 15 mm BSA). All experiments were conducted at 37 °C with a stirrer speed of 750 rpm. A modified multiple substrate uncoupler inhibitor titration (SUIT) protocol was carried out for all experiments (63) as outlined in Fig. S6A.
Statistical analyses
All analyses were done in either GraphPad Prism (version 7.00) or R (version 3.5.1). The types of statistical analyses are listed in the figure legends.
Author contributions
J. N. and H. K. conceptualization; J. N. and H. K. data curation; J. N., H. K., T. C., K. C., A. E. S. B., and J. R. A. formal analysis; J. N., H. K., T. C., and J. R. A. investigation; J. N., H. K., T. C., and J. R. A. methodology; J. N. and H. K. writing-original draft; J. N., H. K., T. C., K. C., A. E. S. B., J. R. A., M. A. B., A. H., and N. P. B. writing-review and editing; J. R. A., M. A. B., A. H., and N. P. B. supervision; J. R. A., M. A. B., A. H., and N. P. B. funding acquisition.
Supplementary Material
Acknowledgments
We thank Dr. Adrian Turner (Imaging Centre, School of Biological Sciences, University of Auckland) for help with the TEM and the flow cytometry facility (Auckland Cytometry, Faculty of Science, University of Auckland), and the statistical consulting facility (University of Auckland).
This work was supported by grants from Brain Research New Zealand Rangahau Roro Aotearoa, the University of Auckland Faculty Research Development Fund, the Lottery Grants Board. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S6.
- AD
- Alzheimer's disease
- AGEs
- advanced glycation end products
- CEL
- Nϵ-(carboxyethyl)lysine
- Aβ1–42
- amyloid β 1–42 fragment
- MG
- methylglyoxal
- ThT
- thioflavin T
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- LDH
- lactate dehydrogenase
- TEM
- transmission electron microscopy
- ΔΨm
- mitochondrial membrane potential
- O2˙̄
- superoxide
- CI
- complex I
- CII
- complex II
- HATU
- O-(7-azabenzotriazol-1-yl)-N,N,N′,N′-tetramethyl-uronium hexafluorophosphate
- SSPS
- solid-phase peptide synthesis
- DMF
- N,N-dimethylformamide
- RP-HPLC
- reverse phase HPLC
- Pen/Strep
- penicillin-streptomycin
- ANOVA
- analysis of variance
- RA
- retinoic acid
- OXPHOS
- oxidative phosphorylation
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- FBS
- fetal bovine serum
- DPBS
- Dulbecco's phosphate-buffered saline.
References
- 1. Goedert M., and Spillantini M. G. (2006) A century of Alzheimer's disease. Science 314, 777–781 10.1126/science.1132814 [DOI] [PubMed] [Google Scholar]
- 2. Sasaki N., Fukatsu R., Tsuzuki K., Hayashi Y., Yoshida T., Fujii N., Koike T., Wakayama I., Yanagihara R., Garruto R., Amano N., and Makita Z. (1998) Advanced glycation end products in Alzheimer's disease and other neurodegenerative diseases. Am. J. Pathol. 153, 1149–1155 10.1016/S0002-9440(10)65659-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Smith M. A., Taneda S., Richey P. L., Miyata S., Yan S. D., Stern D., Sayre L. M., Monnier V. M., and Perry G. (1994) Advanced Maillard reaction end products are associated with Alzheimer disease pathology. Proc. Natl. Acad. Sci. U.S.A. 91, 5710–5714 10.1073/pnas.91.12.5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vitek M. P., Bhattacharya K., Glendening J. M., Stopa E., Vlassara H., Bucala R., Manogue K., and Cerami A. (1994) Advanced glycation end products contribute to amyloidosis in Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 91, 4766–4770 10.1073/pnas.91.11.4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Valente T., Gella A., Fernàndez-Busquets X., Unzeta M., and Durany N. (2010) Immunohistochemical analysis of human brain suggests pathological synergism of Alzheimer's disease and diabetes mellitus. Neurobiol. Dis. 37, 67–76 10.1016/j.nbd.2009.09.008 [DOI] [PubMed] [Google Scholar]
- 6. Kaur H., Kamalov M., and Brimble M. A. (2016) Chemical synthesis of peptides containing site-specific advanced glycation endproducts. Acc. Chem. Res. 49, 2199–2208 10.1021/acs.accounts.6b00366 [DOI] [PubMed] [Google Scholar]
- 7. Kimura T., Takamatsu J., Ikeda K., Kondo A., Miyakawa T., and Horiuchi S. (1996) Accumulation of advanced glycation end products of the Maillard reaction with age in human hippocampal neurons. Neurosci. Lett. 208, 53–56 10.1016/0304-3940(96)12537-4 [DOI] [PubMed] [Google Scholar]
- 8. Degenhardt T. P., Thorpe S. R., and Baynes J. W. (1998) Chemical modification of proteins by methylglyoxal. Cell Mol. Biol. (Noisy-le-grand). 44, 1139–1145 [PubMed] [Google Scholar]
- 9. Sinha S., Lopes D. H. J., and Bitan G. (2012) A key role for lysine residues in amyloid β-protein folding, assembly, and toxicity. ACS Chem. Neurosci. 3, 473–481 10.1021/cn3000247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kuhla B., Lüth H.-J., Haferburg D., Boeck K., Arendt T., and Münch G. (2005) Methylglyoxal, glyoxal, and their detoxification in Alzheimer's disease. Ann. N.Y. Acad. Sci. 1043, 211–216 10.1196/annals.1333.026 [DOI] [PubMed] [Google Scholar]
- 11. Pamplona R., Dalfó E., Ayala V., Bellmunt M. J., Prat J., Ferrer I., and Portero-Otín M. (2005) Proteins in human brain cortex are modified by oxidation, glycoxidation, and lipoxidation: effects of Alzheimer disease and identification of lipoxidation targets. J. Biol. Chem. 280, 21522–21530 10.1074/jbc.M502255200 [DOI] [PubMed] [Google Scholar]
- 12. Ahmed N., Ahmed U., Thornalley P. J., Hager K., Fleischer G., and Münch G. (2005) Protein glycation, oxidation and nitration adduct residues and free adducts of cerebrospinal fluid in Alzheimer's disease and link to cognitive impairment. J. Neurochem. 92, 255–263 10.1111/j.1471-4159.2004.02864.x [DOI] [PubMed] [Google Scholar]
- 13. Lin C.-Y., Sheu J.-J., Tsai I.-S., Wang S.-T., Yang L.-Y., Hsu I.-U., Chang H.-W., Lee H.-M., Kao S.-H., Lee C.-K., Chen C.-H., and Lin Y.-F. (2018) Elevated IgM against Nϵ-(Carboxyethyl)lysine-modified apolipoprotein A1 peptide 141–147 in Taiwanese with Alzheimer's disease. Clin. Biochem. 56, 75–82 10.1016/j.clinbiochem.2018.04.009 [DOI] [PubMed] [Google Scholar]
- 14. Verzijl N., DeGroot J., Oldehinkel E., Bank R. A., Thorpe S. R., Baynes J. W., Bayliss M. T., Bijlsma J. W., Lafeber F. P., and Tekoppele J. M. (2000) Age-related accumulation of Maillard reaction products in human articular cartilage collagen. Biochem. J. 350, 381–387 10.1042/0264-6021:3500381,10.1042/bj3500381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ahmed M. U., Brinkmann Frye E., Degenhardt T. P., Thorpe S. R., and Baynes J. W. (1997) Nϵ-(Carboxyethyl)lysine, a product of the chemical modification of proteins by methylglyoxal, increases with age in human lens proteins. Biochem. J. 324, 565–570 10.1042/bj3240565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Takeuchi M., and Yamagishi S. (2008) Possible involvement of advanced glycation end-products (AGEs) in the pathogenesis of Alzheimer's disease. Curr. Pharm. Des. 14, 973–978 10.2174/138161208784139693 [DOI] [PubMed] [Google Scholar]
- 17. Fica-Contreras S. M., Shuster S. O., Durfee N. D., Bowe G. J. K., Henning N. J., Hill S. A., Vrla G. D., Stillman D. R., Suralik K. M., Sandwick R. K., and Choi S. (2017) Glycation of Lys-16 and Arg-5 in amyloid-β and the presence of Cu2+ play a major role in the oxidative stress mechanism of Alzheimer's disease. J. Biol. Inorg. Chem. 22, 1211–1222 10.1007/s00775-017-1497-5 [DOI] [PubMed] [Google Scholar]
- 18. Pamplona R., Portero-Otín M., Bellmun M. J., Gredilla R., and Barja G. (2002) Aging increases Nϵ-(carboxymethyl)lysine and caloric restriction decreases Nϵ-(carboxyethyl)lysine and Nϵ-(malondialdehyde)lysine in rat heart mitochondrial proteins. Free Radic. Res. 36, 47–54 10.1080/10715760210165 [DOI] [PubMed] [Google Scholar]
- 19. Emendato A., Milordini G., Zacco E., Sicorello A., Dal Piaz F., Guerrini R., Thorogate R., Picone D., and Pastore A. (2018) Glycation affects fibril formation of Aβ peptides. J. Biol. Chem. 293, 13100–13111 10.1074/jbc.RA118.002275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li X.-H., Du L.-L., Cheng X.-S., Jiang X., Zhang Y., Lv B.-L., Liu R., Wang J.-Z., and Zhou X.-W. (2013) Glycation exacerbates the neuronal toxicity of β-amyloid. Cell Death Dis. 4, e673 10.1038/cddis.2013.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gruber P., and Hofmann T. (2005) Chemoselective synthesis of peptides containing major advanced glycation end-products of lysine and arginine. J. Peptide Res. 66, 111–124 10.1111/j.1399-3011.2005.00279.x [DOI] [PubMed] [Google Scholar]
- 22. Kovalevich J., and Langford D. (2013) Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. Methods Mol. Biol. 1078, 9–21 10.1007/978-1-62703-640-5_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin M. T., and Beal M. F. (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443, 787–795 10.1038/nature05292 [DOI] [PubMed] [Google Scholar]
- 24. Reddy P. H., and Beal M. F. (2008) Amyloid β, mitochondrial dysfunction and synaptic damage: implications for cognitive decline in aging and Alzheimer's disease. Trends Mol. Med. 14, 45–53 10.1016/j.molmed.2007.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stine W. B., Jungbauer L., Yu C., and LaDu M. (2011) Preparing synthetic Aβ in different aggregation states. Methods Mol. Biol. 670, 13–32 10.1007/978-1-60761-744-0_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lührs T., Ritter C., Adrian M., Riek-Loher D., Bohrmann B., Döbeli H., Schubert D., and Riek R. (2005) 3D structure of Alzheimer's amyloid-β(1–42) fibrils. Proc. Natl. Acad. Sci. U.S.A. 102, 17342–17347 10.1073/pnas.0506723102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meisl G., Kirkegaard J. B., Arosio P., Michaels T. C., Vendruscolo M., Dobson C. M., Linse S., and Knowles T. P. (2016) Molecular mechanisms of protein aggregation from global fitting of kinetic models. Nat. Protocols. 11, 252–272 10.1038/nprot.2016.010 [DOI] [PubMed] [Google Scholar]
- 28. Meisl G., Rajah L., Cohen S. A. I., Pfammatter M., Šarić A., Hellstrand E., Buell A. K., Aguzzi A., Linse S., Vendruscolo M., Dobson C. M., and Knowles T. P. J. (2017) Scaling behaviour and rate-determining steps in filamentous self-assembly. Chem. Sci. 8, 7087–7097 10.1039/C7SC01965C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Micsonai A., Wien F., Kernya L., Lee Y.-H., Goto Y., Réfrégiers M., and Kardos J. (2015) Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 112, E3095–E3103 10.1073/pnas.1500851112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bernas T., and Dobrucki J. (2002) Mitochondrial and nonmitochondrial reduction of MTT: interaction of MTT with TMRE, JC-1, and NAO mitochondrial fluorescent probes. Cytometry 47, 236–242 10.1002/cyto.10080 [DOI] [PubMed] [Google Scholar]
- 31. Liu Y., Peterson D. A., Kimura H., and Schubert D. (1997) Mechanism of cellular 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction. J. Neurochem. 69, 581–593 [DOI] [PubMed] [Google Scholar]
- 32. Caspersen C., Wang N., Yao J., Sosunov A., Chen X., Lustbader J. W., Xu H. W., Stern D., McKhann G., and Yan S. D. (2005) Mitochondrial Aβ: a potential focal point for neuronal metabolic dysfunction in Alzheimer's disease. FASEB J. 19, 2040–2041 10.1096/fj.05-3735fje [DOI] [PubMed] [Google Scholar]
- 33. Castellani R. J., Harris P. L., Sayre L. M., Fujii J., Taniguchi N., Vitek M. P., Founds H., Atwood C. S., Perry G., and Smith M. A. (2001) Active glycation in neurofibrillary pathology of Alzheimer disease: Nϵ-(carboxymethyl)lysine and hexitol-lysine. Free Radic. Biol. Med. 31, 175–180 10.1016/S0891-5849(01)00570-6 [DOI] [PubMed] [Google Scholar]
- 34. Glabe C. G. (2008) Structural classification of toxic amyloid oligomers. J. Biol. Chem. 283, 29639–29643 10.1074/jbc.R800016200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ahmed M., Davis J., Aucoin D., Sato T., Ahuja S., Aimoto S., Elliott J. I., Van Nostrand W. E., and Smith S. O. (2010) Structural conversion of neurotoxic amyloid-β1–42 oligomers to fibrils. Nat. Struct. Mol. Biol. 17, 561–567 10.1038/nsmb.1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bieschke J., Herbst M., Wiglenda T., Friedrich R. P., Boeddrich A., Schiele F., Kleckers D., Lopez del Amo J. M., Grüning B. A., Wang Q., Schmidt M. R., Lurz R., Anwyl R., Schnoegl S., Fändrich M., et al. (2011) Small-molecule conversion of toxic oligomers to nontoxic β-sheet–rich amyloid fibrils. Nat. Chem. Biol. 8, 93–101 10.1038/nchembio.719 [DOI] [PubMed] [Google Scholar]
- 37. Dahlgren K. N., Manelli A. M., Stine W. B. Jr., Baker L. K., Krafft G. A., and LaDu M. J. (2002) Oligomeric and fibrillar species of amyloid-β peptides differentially affect neuronal viability. J. Biol. Chem. 277, 32046–32053 10.1074/jbc.M201750200 [DOI] [PubMed] [Google Scholar]
- 38. Twig G., Hyde B., and Shirihai O. S. (2008) Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochim. Biophys. Acta 1777, 1092–1097 10.1016/j.bbabio.2008.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kerr J. S., Adriaanse B. A., Greig N. H., Mattson M. P., Cader M. Z., Bohr V. A., and Fang E. F. (2017) Mitophagy and Alzheimer's disease: cellular and molecular mechanisms. Trends Neurosci. 40, 151–166 10.1016/j.tins.2017.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martinez-Vicente M. (2017) Neuronal mitophagy in neurodegenerative diseases. Front. Mol. Neurosci. 10, 64 10.3389/fnmol.2017.00064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fang E. F., Hou Y., Palikaras K., Adriaanse B. A., Kerr J. S., Yang B., Lautrup S., Hasan-Olive M. M., Caponio D., Dan X., Rocktäschel P., Croteau D. L., Akbari M., Greig N. H., Fladby T., et al. (2019) Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer's disease. Nat. Neurosci. 22, 401–412 10.1038/s41593-018-0332-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mungarro-Menchaca X., Ferrera P., Morán J., and Arias C. (2002) β-Amyloid peptide induces ultrastructural changes in synaptosomes and potentiates mitochondrial dysfunction in the presence of ryanodine. J. Neurosci. Res. 68, 89–96 10.1002/jnr.10193 [DOI] [PubMed] [Google Scholar]
- 43. Cheignon C., Tomas M., Bonnefont-Rousselot D., Faller P., Hureau C., and Collin F. (2018) Oxidative stress and the amyloid beta peptide in Alzheimer's disease. Redox Biol. 14, 450–464 10.1016/j.redox.2017.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Grimm A., and Eckert A. (2017) Brain aging and neurodegeneration: from a mitochondrial point of view. J. Neurochem. 143, 418–431 10.1111/jnc.14037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bubber P., Haroutunian V., Fisch G., Blass J. P., and Gibson G. E. (2005) Mitochondrial abnormalities in Alzheimer brain: mechanistic implications. Ann. Neurol. 57, 695–703 10.1002/ana.20474 [DOI] [PubMed] [Google Scholar]
- 46. Ryan T. M., Caine J., Mertens H. D., Kirby N., Nigro J., Breheney K., Waddington L. J., Streltsov V. A., Curtain C., Masters C. L., and Roberts B. R. (2013) Ammonium hydroxide treatment of Aβ produces an aggregate free solution suitable for biophysical and cell culture characterization. Peer J. 1, e73 10.7717/peerj.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Case D. A., Darden T. A., Cheatham T. E. 3rd, Simmerling C. L., Wang J., Duke R. E., and Luo R. (2012) Amber12, University of California, San Francisco, CA [Google Scholar]
- 48. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., and Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926 10.1063/1.445869 [DOI] [Google Scholar]
- 49. Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., and Lindah E. (2015) Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- 50. Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., de Groot B. L., Grubmüller H., and MacKerell A. D. (2017) CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vanommeslaeghe K., Hatcher E., Acharya C., Kundu S., Zhong S., Shim J., Darian E., Guvench O., Lopes P., Vorobyov I., and Mackerell A. D. (2010) CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31, 671–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vanquelef E., Simon S., Marquant G., Garcia E., Klimerak G., Delepine J. C., Cieplak P., and Dupradeau F.-Y. (2011) R.E.D. server: a web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res. 39, W511–W517 10.1093/nar/gkr288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nash A., Collier T., Birch H. L., and de Leeuw N. H. (2017) ForceGen: atomic covalent bond value derivation for Gromacs. J. Mol. Model. 24, 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hess B., Bekker H., Berendsen H. J. C., and Fraaije J. G. E. M. (1997) LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 [DOI] [Google Scholar]
- 55. Darden T., York D., and Pedersen L. (1993) Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089 10.1063/1.464397 [DOI] [Google Scholar]
- 56. Evans D. J., and Holian B. L. (1985) The Nose–Hoover thermostat. J. Chem. Phys. 83, 4069 10.1063/1.449071 [DOI] [Google Scholar]
- 57. Takeda T., and Klimov D. K. (2009) Probing the effect of amino-terminal truncation for Aβ1–40 peptides. J. Phys. Chem. B 113, 6692–6702 10.1021/jp9016773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lemkul J. A., and Bevan D. R. (2010) Assessing the stability of Alzheimer's amyloid protofibrils using molecular dynamics. J. Phys. Chem. B 114, 1652–1660 10.1021/jp9110794 [DOI] [PubMed] [Google Scholar]
- 59. Patey G. N., and Valleau J. P. (1973) The free energy of spheres with dipoles: Monte Carlo with multistage sampling. Chem. Phys. Lett. 21, 297–300 10.1016/0009-2614(73)80139-3 [DOI] [Google Scholar]
- 60. Torrie G. M., and Valleau J. P. (1977) Nonphysical sampling distributions in Monte Carlo free-energy estimation: umbrella sampling. J. Comput. Phys. 23, 187–199 10.1016/0021-9991(77)90121-8 [DOI] [Google Scholar]
- 61. Kumar S., Rosenberg J. M., Bouzida D., Swendsen R. H., and Kollman P. A. (1992) The weighted histogram analysis method for free-energy calculations on biomolecules: I. the method. J. Comput. Chem. 13, 1011–1021 10.1002/jcc.540130812 [DOI] [Google Scholar]
- 62. Hub J. S., de Groot B. L., and van der Spoel D. (2010) g_wham: a free weighted histogram analysis implementation including robust error and autocorrelation estimates. J. Chem. Theory Comput. 6, 3713–3720 10.1021/ct100494z [DOI] [Google Scholar]
- 63. Pesta D., and Gnaiger E. (2012) High-resolution respirometry: OXPHOS protocols for human cells and permeabilized fibers from small biopsies of human muscle. Methods Mol. Biol. 810, 25–58 10.1007/978-1-61779-382-0_3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.