Abstract
Antibacterial strategies sans antibiotic drugs have recently garnered much interest as a mechanism by which to inhibit biofilm formation and growth on surfaces due to the rise of antibiotic-resistant bacteria. Based on the photofluidization of azobenzenes, we demonstrate for the first time the ability achieve up to a 4 log reduction in bacterial biofilms by opto-mechanically activating the disruption and dispersion of biofilms. This unique strategy with which to enable biofilm removal offers a novel paradigm with which to combat antibiotic resistance.
Keywords: azobenzenes, photofluidization, optically responsive materials, antibacterial and antifouling strategies, antibacterial coatings, smart materials, photoresponsive polymers
Graphical Abstract
Biofilm formation is the most potent evolutionary mechanism by which bacteria continue to evade targeted, antibacterial drugs.1 Unlike their planktonic counterparts, bacteria within biofilms are highly organized microbial aggregates embedded within a secreted matrix of extracellular polymeric substances (EPS) that can now confer 10–1000 times more resistance to antibacterial agents.2,3 The evolutionary impetus to adapt to novel biochemical strategies that impair bacterial growth is evidenced by the growing microbial resistance to both traditional and newer antibiotic drugs over the past decade.4,5 Multidrug-resistant bacteria such as Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species, termed the ESKAPE organisms, are the leading cause of nosocomial (hospital-acquired) infections throughout the world and are currently implicated in over 2 million nosocomial infections at costs that exceed $1 billion annually.6–10 Because the incidences of adverse drug reactions to antibiotics account for significant morbidity and mortality,7,11 there is an imperative need to examine nontraditional antibiotic strategies that do not rely solely on a targeted, biochemical mechanism to impair biofilm growth and formation.
Because biofilms can form on biotic and abiotic substrates that vary from tooth surfaces within the oral cavity to indwelling biomedical devices (e.g., hip implants) and plumbing materials, antibacterial strategies that prevent the formation and proliferation of biofilms by controlling the material properties of the bacteria—substrate interface have garnered immense interest.11 The sharkskin-inspired design of micropatterned surfaces and one-way shape memory polymers have been shown to resist bacterial adhesion and disrupt biofilm formation.12,13 An advantage of this approach is that the pattern or stimuli-responsive “smart” shape change is a material surface modification and does not introduce or rely on biochemical additives or antimicrobials to function as an antibacterial surface. While a micropatterned surface has been shown to reduce bacterial colonization, it does not ultimately prevent biofilm growth and formation.14 More recently, Bao et al. formulated a highly hydrophobic surface containing 2,3,5,6-tetrafluoro-p-phenylenedimethanol and demonstrated reduced bacterial adhesion and biofilm formation by Bacillus subtilis and Escherichia coli. The authors were able to demonstrate specific antibacterial activity at one-fifth the fluoride content of more traditional antibacterial surfaces such as polytetrafluoroethylene, making their approach a more environmentally friendly one compared with traditional fluoride-based antibacterial surfaces.2 Efforts toward formulating antibacterial coatings with silver nanoparticles that are both antibacterial and biocompatible have also been impacted by the growing silver particle resistance observed.15–17 Currently, the most effective antimicrobial agents and potent mechanical systems aim to modify the surface interaction between the bacteria and the substrate; however, they do not and cannot physically penetrate the biofilm efficiently enough to disrupt and remove the biofilm. Consequently, effective and stable biofilm control remains elusive.3,18,19
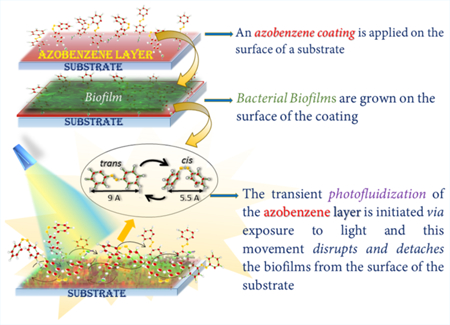
In this communication, the unique property of photo-fluidization demonstrated by azobenzene molecules was implemented to detach biofilms via photoresponsive, dynamic antifouling coatings.20,21 Photofluidization is the photodirected transient flow and softening of the surface of a glassy material (along with a concomitant, transient decrease in the Youngs’ modulus) via the rapid trans–cis–trans isomerization cycles of azobenzene groups that can be achieved when the azobenzene molecules are exposed to multiple wavelengths of light simultaneously (Figure 1a).21–24 Photofluidization presents a unique opportunity to engineer antifouling coatings while otherwise maintaining the high modulus necessary for substrate durability (Figure 1b).
Figure 1.
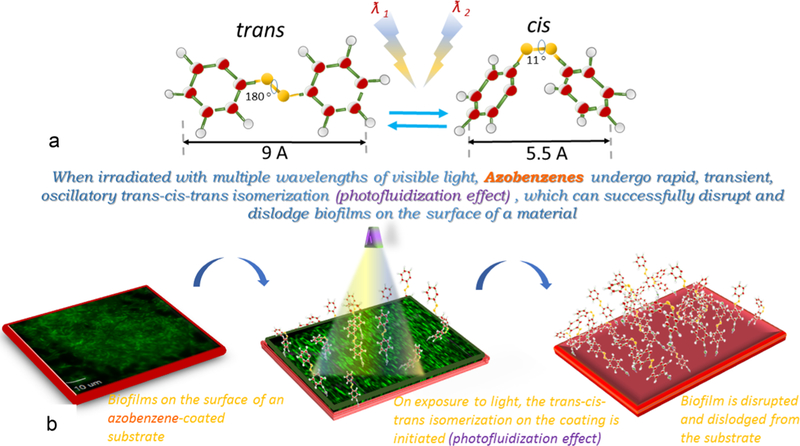
Azobenzene change in conformation from trans to cis upon exposure to UV light (~365 nm) and from cis to trans upon exposure to visible light (~490 nm) or heat. However, when irradiated with intermediate wavelengths (430–480 nm), azobenzenes undergo rapid, transient, oscillatory trans–cis–trans isomerization (photofluidization effect) and can successfully disrupt biofilms on the surface of a material (panel a). The photofluidization effect can be used to disrupt biofilms from the surface of a substrate (panel b).
To test the feasibility of utilizing photofluidization for biofilm removal, P. aeruginosa (PA01) biofilms (green) formed on a covalently tethered, azobenzene-coated substrate (red) were generated (Figure 2a) and imaged on a confocal microscope (3I Marianas, inverted microscope, 1000×). PA01 biofilms with a chromosomal encoded gfp were grown in brain–heart infusion broth at 37 °C on the surface of the polymer substrate, gently washed in 1× phosphate-buffered saline (PBS) to remove planktonic bacteria from the biofilm, and exposed to light from a routinely used clinical dental lamp (3 M Elipar).25,26 A significant portion of the biofilm was ejected during the first and second light exposure as shown in Figure 2b,c. In this instance, the light exposures and gentle washing in PBS resulted in close to 100% removal of the biofilm (Figure 2d).
Figure 2.
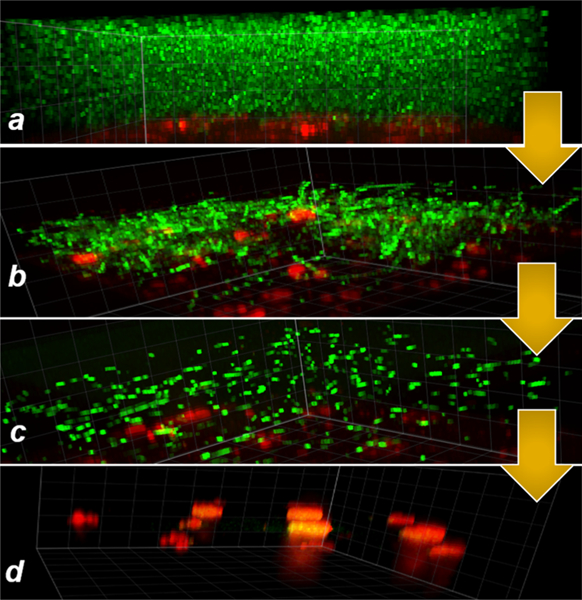
Pseudomonas aeruginosa biofilms (gfp green) grown on an azopolymer coating on the surface of a glassy polymer substrate (red, rhodamine; panel a). On exposure to a clinical dental light at 430–480 nm (3 M Elipar DeepCure-S LED Curing Light) for 45 s at 700 mW/cm2, the photofluidization effect initiated via the rapid trans–cis–trans isomerization of the azobenzenes result in biofilm disruption and ejection (panels b and c). As soon as the light is switched off, the oscillatory dynamics cease, and materials return to its native state. The azocoating after the second exposure shows the further absence of biofilm (panel d).
Because photofluidization is a physical change induced optically on a material surface, we hypothesized that azobenzenes should be able to disrupt and remove diverse biofilms as long as the opto-mechanically induced forces on the azobenzene (AZO) molecule were greater than both the cohesive strength of the biofilm and the adhesive strength between the biofilm and the substrate.27 To examine the versatility of this approach, in addition to P. aeruginosa biofilms, biofilms from bacteria known to create robust biofilms such as the Gram-negative bacteria (uropathogenic Escherichia coli) and two Gram-positive bacteria (Staphylococcus aureus and Streptococcus mutans), both with sucrose-dependent biofilms (SD-SM) and sucrose-independent biofilms (SI-SM), were chosen for this proof-of-concept study.18,25,26,28–30 The growth curves of each bacteria on the polymer substrates were determined to ensure the formation of a robust biofilm prior to light exposure (Figure S1). Static biofilms were grown on the surface of the glassy, polymer substrate without the azobenzene coating (no-AZO control) and with the azobenzene coating (AZO).
The glassy polymer substrate was prepared from a formulation of poly(methyl methacrylate), methyl methacrylate, and triethylene glycol dimethacrylate (Figure S2). To form the covalently tethered azopolymer-coated substrates, the azobenzene molecule, 4-phenyl azophenyl acrylate, was synthesized via a modified procedure previously described in literature. Briefly, a reaction between 4-phenylazophenol and acryloyl chloride was catalyzed by triethylamine in anhydrous dichloromethane to yield an acrylated azobenzene molecule that was then characterized using 1H nuclear magnetic resonance (NMR; Mw = 252 g/mol). The acrylated AZO molecule was then thermally cross-linked as a 20 μm thick covalently cross-linked coating on both surfaces of the 0.90 mm thick substrate (D = 6.5 mm). To form the coating, 1.5 mg of the AZO molecule was dispersed in 40 μL of dimethylformaldehyde (DMF) with 0.2 wt % rhodamine B acrylate and thermally cured (thermal initiator, 1 wt % AIBN).The AZO-coated substrate was further cured thermally in a vacuum to attain ≥99% acrylic double-bond conversion of the substrate and coating surface as observed by Fourier transform infrared spectroscopy (FTIR-Near). The glass transition temperature (Tg) and rubbery modulus of the combined material was measured at 113 ± 1.7 °C and 17.4 ± 3.4 MPa, respectively (via dynamic mechanical analysis), ensuring that the bulk material retained its glassy material properties at room temperature (~21 °C). Cytotoxicity tests on L929 mouse fibroblast cells confirmed that the AZO-coated polymer substrate was not cytotoxic (Figure S4).
The protocol to enable the light-induced fluidization effect and subsequent biofilm removal from the surface of the material was as follows: AZO-coated polymer substrates and the control polymer substrates with biofilms were exposed to a 3 M Elipar DeepCure-S LED curing light (430–480 nm, 700 mW/cm2 exposure for 20 s on each side of the substrate) and gently washed in sterile PBS to remove the unattached bacteria. The total number of light exposures and subsequent washes were limited to 3 per sample for this study. The substrates were imaged before and after the light exposures and the bacteria removed from surface of the substrates via photofluidization were quantified by determining the colony-forming units per milliliter (CFU/mL) in PBS via serial dilutions.
All five biofilms generated on the AZO- coated substrate were imaged on a Zeiss Axioplan II microscope (step size of 0.1 μm and magnification of 630×). Bacterial biofilms were grown on the surface of the AZO-coated substrates and the control substrates and imaged using live dead staining (Invitrogen LIVE/DEAD BacLight Viability kit) before and after three light exposures and subsequent washes. Although biofilm formation and growth was observed on all substrates, it was observed that the biofilms grew and spread more robustly on the uncoated substrates in comparison with the photo-responsive AZO-coated substrates. In addition to P. aeruginosa biofilms (Figure 3A), biofilms of E. coli (Figure. 3B), S. aureus (Figure 3C), and S. mutans sucrose-independent (S. mutans SI) (Figure 3D) and S. mutans sucrose-dependent (S. mutans SD, Figure 3E) were studied. S. mutans, in the presence of sucrose, is known to generate strong, adhesive biofilms, and interestingly, our approach to enable biofilm disruption was unsuccessful for this particular biofilm.29 Currently, our group is working toward quantifying the forces required to enable bacteria-specific biofilm disruption and synthesizing azobenzenes with molecular structures that can apply optimal forces to enable the disruption of strong biofilms.
Figure 3.
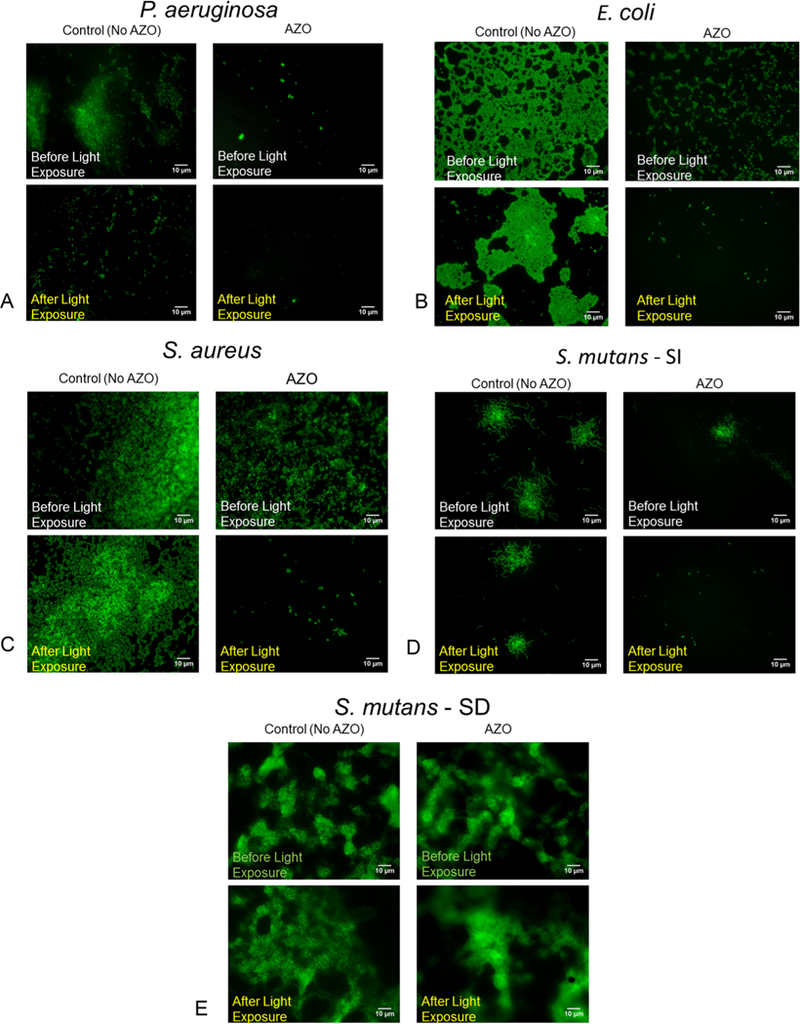
Biofilms on the surface of the AZO substrates and the control substrates (No AZO) were exposed to light from a dental lamp (3 M Elipar DeepCure-S LED Curing Light) to initiate the fluidization effect and subsequently gently washed in PBS to remove the detached bacteria from the biofilm. This process was repeated 3 times for each sample. A live–dead stain before and after the light exposures and subsequent washes captures the ability of the photofluidization effect to remove bacterial biofilms from 4 different biofilms (3A–D). Our approach was not successful in removing biofilms formed via Streptococcus mutans in the presence of sucrose (3E).
The log reduction in bacterial biofilms achieved after three photofluidization events (exposures to light and subsequent washes) was assessed by determining the bacteria removed from the substrates relative to the total bacteria present on the substrates (determined via sonication) (Figure 4). The data indicates that the AZO substrate enabled a greater log reduction (up to 2–4 times greater) of bacterial colonies from the surface of the substrate when the azobenzene photofluidization effect is initiated in the P. aeruginosa, E. coli, S. aureus, and S. mutans SI samples in comparison to the control (no AZO). As initially indicated by the images, the CFU counts show that the S. mutans SD samples were largely unaffected by the photofluidization effect.
Figure 4.
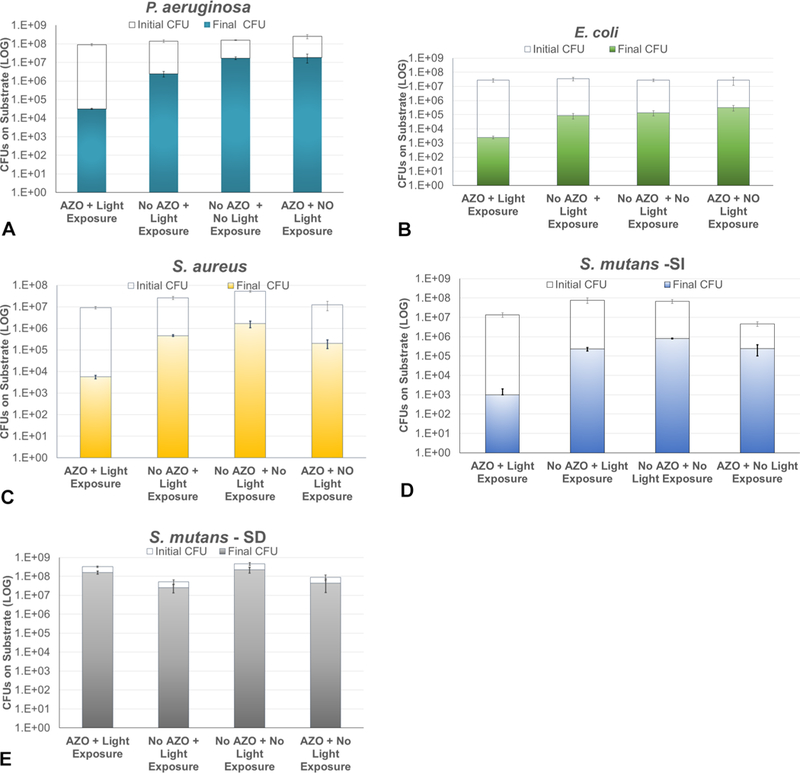
Quantification of the biofilms removed via the photofluidization effect after three light exposures and washes on AZO and no AZO (control subtrates) indicates that a significant loss of biofilm can be achieved in 4 out of 5 biofilms tested: 4A–D (n ≥ 3). The sucrose-dependent S. mutans biofilms (4E), however, did not respond to the photofluidization effect.
There are two key advantages to the antifouling approach outlined in this study. First, the ability to repeatedly elicit an opto-mechanical response from an AZO coating to dislodge biofilms is not pathogen-specific and can be repeatedly initiated to potentially remove multiple pathogens simultaneously. Second, because biofilm disruption and removal is enabled by the rapid, transient, trans–cis–trans isomerization of the azobenzene molecule in response to light, it is highly unlikely that the bacteria can undergo evolutionary adaptations to neutralize and “out-evolve” the photofluidization effect. Clearly, detailed studies are now required to optimize these substrates as ubiquitous antibiofilm surfaces for multispecies biofilms because the biofilms formed via the co-adhesion of DNA, exopolysaccharide and cell debris will have different properties and mechanics than the individual biofilm model used in this study. Although the uniqueness of our approach in delivering forces from the surface of a substrate using a photoisomer, thereby mechanically dislodging the biofilm from the base of the material, is highly effective for most single-species biofilms, it will also require a detailed review of the adhesive and cohesive forces within the biofilm itself and synthesis of novel azobenzene molecules to attain them. We believe that our work thus far indicates that there is immense translational potential in the approach outlined here.
Supplementary Material
ACKNOWLEDGMENTS
D.P.N. acknowledges financial support from NIH-NIDCR (grant no. K25DE027418). The authors thank the Horswill Lab (University of Colorado Department of Immunology and Microbiology) and the Mysorekar Lab (Washington University Department of Obstetrics & Gynecology) for the S. aureus and E. coli bacterial strains, respectively. The authors also thank Gavin Campbell for help with the cytotoxicity studies.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsami.8b21058.
Additional data on the biofilm growth curves (Figure S1 and Table S1), sample fabrication (Figure S2), 1H NMR characterization of the acrylated azobenzenes AAZO molecule (Figure S3), cytotoxicity tests (Figure S4), and the protocol for the CFU quantification and analysis and list of bacterial strains used in the study (Table S2) (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Algburi A; Comito N; Kashtanov D; Dicks LMT; Chikindas ML Control of Biofilm Formation : Antibiotics and Beyond. Appl. Environ. Microbiol. 2017, 83 (3), 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Bao Q; Nishimura N; Kamata H; Furue K; Ono Y; Hosomi M; Terada A Antibacterial and Anti-Biofilm Efficacy of Fluoropolymer Coating by a 2,3,5,6-Tetrafluoro-p-Phenylenedimethanol Structure. Colloids Surf. B 2017, 151, 363–371. [DOI] [PubMed] [Google Scholar]
- (3).Gilbert P; Allison DG; McBain AJ Biofilms in Vitro and in Vivo: Do Singular Mechanisms Imply Cross-Resistance? J. Appl. Microbiol. 2002, 92 (s1), 98S–110S. [PubMed] [Google Scholar]
- (4).Santajit S; Indrawattana N Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. BioMed Res. Int. 2016, 2016, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).O’Neill J Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Rev. Antimicrob. Resist. 2016, May, 1–75. [Google Scholar]
- (6).Rice LB Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis. 2008, 197 (8), 1079–1081. [DOI] [PubMed] [Google Scholar]
- (7).Kanaan H; El-Mestrah M; Sweidan A; As-Sadi F; Bazzal A. Al; Chokr A Screening for Antibacterial and Antibiofilm Activities InAstragalus Angulosus. J. Intercult. Ethnopharmacol 2017, 6 (1), 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Hall-Stoodley L; Costerton JW; Stoodley P Bacterial Biofilms: From the Natural Environment to Infectious Diseases. Nat. Rev. Microbiol. 2004, 2 (2), 95–108. [DOI] [PubMed] [Google Scholar]
- (9).Simoes M; Simoes LC; Vieira MJ A Review of Current and Emergent Biofilm Control Strategies. LWT - Food Sci. Technol 2010, 43 (4), 573–583. [Google Scholar]
- (10).Mah T-FC; Toole GAO Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 2001, 9 (1), 34–39. [DOI] [PubMed] [Google Scholar]
- (11).Mishra B; Lushnikova T; Golla RM; Wang X; Wang G Design and Surface Immobilization of Short Anti-Biofilm Peptides. Acta Biomater. 2017, 49, 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Mann EE; Manna D; Mettetal MR; May RM; Dannemiller EM; Chung KK; Brennan AB; Reddy ST Surface Micropattern Limits Bacterial Contamination. Surface micro-pattern limits bacterial contamination 2014, 3 (1), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Gu H; Lee SW; Buffington SL; Henderson JH; Ren D On-Demand Removal of Bacterial Biofilms via Shape Memory Activation. ACS Appl. Mater. Interfaces 2016, 8 (33), 21140–21144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).May RM; Hoffman MG; Sogo MJ; Parker AE; Brennan AB; Reddy ST Micro-Patterned Surfaces Reduce Bacterial Colonization and Biofilm Formation in Vitro : Potential for Enhancing Endotracheal Tube Designs. Clin. Transl Med. 2014, 3 (1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Makama S; Kloet SK; Piella J; van den Berg H; de Ruijter NCA; Puntes VF; Rietjens IMCM; van den Brink NW Effects of Systematic Variation in Size and Surface Coating of Silver Nanoparticles on Their In Vitro Toxicity to Macrophage RAW 264.7 Cells. Toxicol. Sci. 2018, 162 (1), 79–88. [DOI] [PubMed] [Google Scholar]
- (16).Graves JL; Tajkarimi M; Cunningham Q; Campbell A; Nonga H; Harrison SH; Barrick JE Rapid Evolution of Silver Nanoparticle Resistance in Escherichia Coli. Front. Genet. 2015, 6, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Finley PJ; Norton R; Austin C; Mitchell A; Zank S; Durham P Unprecedented Silver Resistance in Clinically Isolated Enterobacteriaceae: Major Implications for Burn and Wound Management. Antimicrob. Agents Chemother. 2015, 59 (8), 4734–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Wang C; Symington JW; Ma E; Cao B; Mysorekar IU Estrogenic Modulation of Uropathogenic Escherichia Coli Infection Pathogenesis in a Murine Menopause Model. Infect. Immun. 2013, 81 (3), 733–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Sharma G; Rao S; Bansal A; Dang S; Gupta S; Gabrani R Pseudomonas Aeruginosa Biofilm: Potential Therapeutic Targets. Biologicals 2014, 42 (1), 1–7. [DOI] [PubMed] [Google Scholar]
- (20).Fang GJ; MacLennan JE; Yi Y; Glaser MA; Farrow M; Korblova E; Walba DM; Furtak TE; Clark NA Athermal Photofluidization of Glasses. Nat. Commun. 2013, 4, 1–10. [DOI] [PubMed] [Google Scholar]
- (21).Hurduc N; Donose BC; Macovei A; Paius C; Ibanescu C; Scutaru D; Hamel M; Branza-Nichita N; Rocha L Direct Observation of Athermal Photofluidisation in Azo-Polymer Films. Soft Matter 2014, 10 (26), 4640–4647. [DOI] [PubMed] [Google Scholar]
- (22).Yadavalli NS; Loebner S; Papke T; Sava E; Hurduc N; Santer S A Comparative Study of Photoinduced Deformation in Azobenzene Containing Polymer Films. Soft Matter 2016, 12 (9), 2593–2603. [DOI] [PubMed] [Google Scholar]
- (23).Bin J; Oates WS A Unified Material Description for Light Induced Deformation in Azobenzene Polymers. Sci. Rep. 2015, 5 (14654), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Pirani F; Angelini A; Frascella F; Rizzo R; Ricciardi S; Descrovi E; Mahimwalla ZS; Gritsai Y; Goldenberg LM; Kulikovska O Light-Driven Reversible Shaping of Individual Azopolymeric Micro-Pillars. Sci. Rep. 2016, 6 (August), 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Okkotsu Y; Little AS; Schurr MJ The Pseudomonas Aeruginosa AlgZR Two-Component System Coordinates Multiple Phenotypes. Front. Cell. Infect. Microbiol. 2014, 4 (June), 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Pritchett CL; Little AS; Okkotsu Y; Frisk A; Cody WL; Covey CR; Schurr MJ Expression Analysis of the Pseudomonas Aeruginosa AlgZR Two-Component Regulatory System. J. Bacteriol. 2015, 197 (4), 736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Aggarwal S; Hozalski RM Determination of Biofilm Mechanical Properties from Tensile Tests Performed Using a Micro-Cantilever Method. Biofouling 2010, 26 (4), 479–486. [DOI] [PubMed] [Google Scholar]
- (28).Tsutsumi K; Maruyama M; Uchiyama A; Shibasaki K Characterisation of a Sucrose-Independent in Vitro Biofilm Model of Supragingival Plaque. Oral Dis 2018, 24 (3), 465–475. [DOI] [PubMed] [Google Scholar]
- (29).Shemesh M; Tam A; Steinberg D Expression of Biofilm- Associated Genes of Streptococcus Mutans in Response to Glucose and Sucrose. J. Med. Microbiol. 2007, 56 (11), 1528–1535. [DOI] [PubMed] [Google Scholar]
- (30).Herbert S; Ziebandt AK; Ohlsen K; Schafer T; Hecker M; Albrecht D; Novick R; Gotz F Repair of Global Regulators in Staphylococcus Aureus 8325 and Comparative Analysis with Other Clinical Isolates. Infect. Immun. 2010, 78 (6), 2877–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


