Abstract
Natural killer (NK) cells accumulate in adult murine and human uteri during decidualization induced physiologically, pathologically or experimentally. Adoptive transfer studies indicate that uterine NK (uNK) cells arise from circulating progenitors. However, virgin uteri contain few circulating NK1.1+CD49a− conventional NK (cNK) cells while NK1.1+CD49a+ tissue-resident NK (trNK) cells are abundant. Here, we employed a novel, immune competent NK cell-specific reporter mouse to track accumulation of uNK cells during unmanipulated pregnancies. We identified cNK and trNK cells accumulating in both decidua basalis (DB) and myometrium. Only trNK cells showed evidence of proliferation. In parabiosis studies using experimentally-induced deciduomata, the accumulated uNK cells were proliferating trNK cells; migrating NK cells made no contribution. Together, these data suggest proliferating trNK cells are the source of uNK cells during endometrial decidualization.
Keywords: pregnancy, uterine NK cells, tissue-resident NK cells, conventional NK, decidualization
Introduction
Natural killer (NK) cells are components of the innate immune system that provide early protection against infections and developing tumors. Conventional NK (cNK) cells are derived from precursors in bone marrow where their development into mature cNK cells takes several steps and continues in the periphery (1, 2). Circulating cNK cells are widely distributed but many tissues, including liver, skin, kidney, and uterus, possess tissue-resident NK (trNK) cells. Although trNK cells are considered group 1 innate lymphoid cells (ILC1s), in C57BL/6 mice they are all NK1.1+NKp46+CD3–CD19– (3–7), a classic NK cell phenotype. The cNK and trNK cell subsets can be distinguished. First, parabiosis studies established that cNK cells could migrate between parabionts via the circulatory system whereas trNK cells remained in host tissue. Second, they have phenotypic differences, especially DX5 and CD49a that have mutually exclusive expression on cNK and trNK cells, respectively (3, 4, 6). Finally, cNK and trNK cells have differential dependencies on transcription factors, indicating they belong to different lineages. For example, cNK cells require NF-IL3 for development while trNK cells do not. In contrast, development of trNK cells in liver and skin requires T-bet, that has a less profound effect on cNK cell development. Interestingly, NK cells in the virgin uterus are predominantly trNK cells and develop independent of both NF-IL3 and T-bet (4), strongly suggesting that they form a lineage distinct from cNK cells and trNK cells in liver or skin.
Studies of uterine NK (uNK) cells in pregnancy have shown that they constitute the vast majority of maternal leukocytes at sites of implantation in species with hemochorial placentation (8, 9). Early in mouse and human gestations, 70% of the lymphocytes at each implantation site are uNK cells. Their frequency declines after midpregnancy (8, 10). Classically, uNK cells are identified in tissue sections by immunohistochemical (IHC) reactivity with periodic acid Schiff (PAS) and Dolichos biflorus agglutinin (DBA) that has suggested uNK cell heterogeneity (11, 12). The uNK cells are found in anatomically distinct regions of implantation sites, the decidua basalis (DB) induced by implantation and the later developing mesometrial lymphoid aggregate of pregnancy (MLAp), located in the uterine wall (13). The DB and MLAp develop in response to blastocyst implantation via a process termed decidualization that dramatically remodels the endometrium. Decidualization transforms uterine stromal cells into large, endocrinologically active decidual cells (14) and can be artificially induced without a conceptus by wounding of a progesterone-primed uterus. The resulting deciduomata are widely used to study early pregnancy-related events in rodents (15, 16).
All studies to date addressing whether uNK cells in the pregnant uterus develop in situ from precursors in the virgin uterus or home there from the periphery have been addressed using uterine transplant and adoptive cell transfer experiments. Normal uterine horn transplants into alymphoid mice that lacked NK cells suggested that the uterus was populated by progenitors from peripheral sources (17, 18). After adoptive transfer of progenitor cells from SCID mice into alymphoid recipients, donor-derived uNK cells could be detected in the pregnant uterus (11, 19), providing further support for NK progenitor cell homing. In another study, adoptively transferred splenic NK cells did not home to the uterus; instead, there was evidence of expansion of uNK cells resident to the uterus (20). Thus, the contribution of NK cells from the periphery and/or tissue to the expanding uNK cell pool during pregnancy or decidualization remains unresolved.
Here we report a novel NK cell reporter mouse and its use to visualize the emergence of uNK cells during pregnancy. The trNK cell subset proliferated locally in physiological pregnancy and in experimentally-induced deciduomata. Parabiotic studies with deciduomata confirmed that there is minimal contribution from migrating cNKs to the proliferating pool of trNK cells.
Material and Methods
Mice
WT C57BL/6N mice were purchased from Charles River (Wilmington, MA). The Ncr1iCre was previously described (21). The RosamT/mG mouse (22) was purchased from Jackson Laboratory (Bar Harbor, ME). Ncr1iCre mice were crossed to the RosamT/mG and F1 dams were used in all of the experiments. Gestational day (gd) 0.5 was marked on the morning of a copulation plug. The F1 dams were mated with WT C57BL/6N males.
Survival surgery
All mice were housed in a pathogen-free facility and surgical procedures were performed in accordance with the animal protocol approved by the Washington University School of Medicine (WUSM) Animal Studies Committee. To induce artificial decidualization we used the ovariectomized-oil induced decidualization (Ovx-OID) model. Briefly mice were ovariectomized, allowed to recover and received daily s.c. injections of 100ng β-estradiol (E2) (Sigma, St. Louis MO) for 3 consecutive days. After 2 days rest, the mice were given 1mg Progesterone (P4) (Sigma, St. Louis MO) and 10ng E2 for 3 consecutive days. No earlier than 6hs after the last injection, a needle scratch was used to mechanically stimulate the anti-mesometrial side of the uterine horn and 20ul of sesame oil were injected. The next day, each mouse began daily s.c. injections of 1mg of P4, until sacrifice at d5 post Ovx-OID. Parabiosis surgery was performed as previously reported (3, 4). We surgically joined the Ncr1iCre x RosamT/mG reporter to WT C57BL/6N and induced decidualization in one horn of each parabiont. We sacrificed the parabiosed mice d7 post surgery.
Cell isolations
Single cell suspensions were prepared from spleen, uterus, MLAp, DB and labyrinth for flow cytometry. All tissues isolated were mechanically dissociated and put through a 100 or 70um cell strainer. The uterus and MLAp were incubated in 167ug/ml liberase TL (Sigma, St. Louis MO) and 100ug/ml of DNase (Roche Applied Science, Indianapolis) for 1h at 37°C in shaking incubator.
Flow cytometry
All cells were stained with Fixable Viability Dye™ (eBioscience) prior to blocking Fc receptors with 2.4G2 and surface staining with directly conjugated Ab’s. BrdU incorporation was measured as per manufacturer’s instructions (BD Bioscience). The following Ab’s were purchased from eBioscience: CD3 (clone 145–2C11), CD19 (eBio1D3), NK1.1 (PK136), CD49b (DX5), Ly49D (eBio4E5), Ly49H (3D10), Ly49I (YLI-90), CD45 (30-F11), and NKp46 (29A1.4). CD49a (Ha31/8) was purchased from BD Bioscience. The cell surface and intracellular sources of DBA lectin (Vector Labs) and Ki67 were stained using the Foxp3 fix/perm reagents (BD Bioscience). Events were acquired on the Canto (BD Biosciences) using the FACSDiva software (BD Biosciences) and analyzed with FlowJo software (Treestar).
Histology
All tissues were harvested at the indicated gd or days post ovx-OID and fixed in 2% paraformaldehyde-lysine-phosphate fixative (PLP) at 4°C overnight followed by 30% sucrose solution at room temperature for 6hs. Tissues were embedded and frozen at −80 in optimum cutting temperature (O.C.T.) medium (Sakura Finetek, Torrance CA). Frozen samples were cut at 9um, mounted on glass slides and DAPI nuclear stain (Vector Labs, Burlingame, CA) was added prior to acquisition or sections were stained with H&E. A 20× objective was used to generate the images on the NanoZoomer (Hamamatsu Photonics K.K., Japan).
Statistical analysis
The data were analyzed by the unpaired Student t test using GraphPad Prism 6 software. Asterisks indicate statistical significance and p values are denoted as *for p<0.05 and ****for p<0.0001.
Results
Emergence of GFP+ NK cells in the pregnant uterus of reporter mice
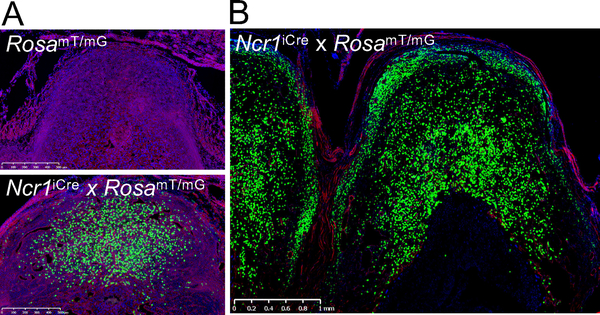
Ncr1 encodes NKp46, a receptor expressed on all NK cells, allowing for improved Cre (iCre) to be restricted to NK cells in Ncr1iCre mice. RosamT/mG mice contain a construct knocked into the Rosa26 locus in which membrane-bound Tomato is constitutively expressed in all tissues. Upon Cre expression, the Tomato cassette and a stop codon are excised, allowing for expression of membrane-bound green fluorescent protein (GFP), i.e., theoretically NKp46+ cells. To establish that we could identify uNK cells in the Ncr1iCre x RosamT/mG reporter mouse, we initially examined implantation sites at gestational day 6.5 (gd6.5) and gd11.5 in comparison to control RosamT/mG implantation sites. At gd6.5, an abundance of GFP+ cells was present at implantation sites when compared to the control (Fig 1A). At gd11.5, the implantation sites of the Ncr1iCre x RosamT/mG mice were rich in GFP+ cells (Fig. 1B). The GFP+ cells segregated to the DB and myometrium, the latter consistent with the MLAp. Thus, anatomically distinct regions in the pregnant uterus harbored GFP+ cells that were either in close association (DB) or separated (myometrium) from the conceptus.
Figure 1: Emergence of GFP+ cells in Ncr1iCre x RosamT/mG dams in pregnancy.
A) Pregnant uterus (gd6.5) of RosamT/mG control mouse and Ncr1iCre x RosamT/mG reporter mouse. A 500 um bar is shown. B) Pregnant Ncr1iCre x RosamT/mG reporter (gd11.5), with accumulation of GFP+ cells in the DB and myometrium. A 1mm bar is shown. Three independent experiments were analyzed at gd6.5 (n=4) and gd11.5 (n=8).
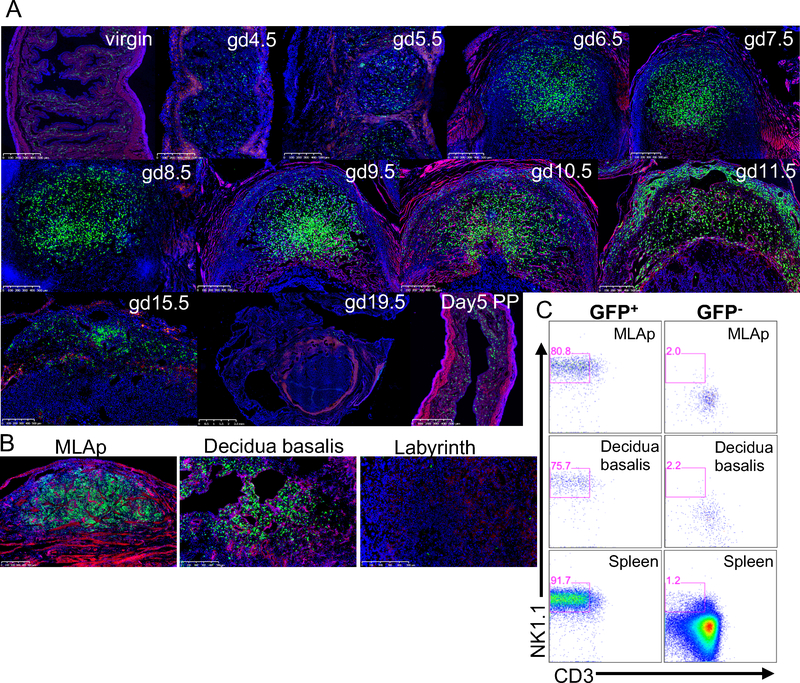
Because our pilot results mimicked previously reported histological studies, we undertook a time course study of GFP+ uterine cells that included virgin, pregnant and postpartum (PP) uteri. GFP+ cells were present in virgin uterus, consistent with our prior studies. During the course of gestation, we found a marked increase in GFP+ cells at gd6.5 in the DB as compared to virgin, gd4.5 and gd5.5 uteri (Fig. 2A). The MLAp appeared by gd9.5 then GFP+ cells decreased in number in both DB and MLAp by gd15.5 and were essentially nonexistent at the implantation site of a dam at gd19.5 (Fig. 2A). The uterus was repopulated with GFP+ cells d5 PP to a pattern resembling the virgin uterus (Fig. 2A). Taken together, these data are consistent with the kinetics of uNK cell accumulation and decline described by IHC in prior studies, although with greater sensitivity, as we found more GFP+ cells and easily detected GFP+ cells within the myometrium.
Figure 2: Time course of GFP+ cells accumulating in Ncr1iCre x RosamT/mG dams during pregnancy.
A) The uteri of pregnant Ncr1iCre x RosamT/mG reporter mice were prepared for histology at the indicated gestational days (gd). B) The MLAp, DB and labyrinth were all physically separated. Each anatomical region was separately processed for histology. A 500um scale is shown on all images. C) At gd11.5 the MLAp and DB were separated and single cell suspensions prepared for flow cytometry, along with spleen from Ncr1iCre x RosamT/mG reporter mouse. The percentages in the gates represent CD45+CD19− that express GFP or are GFP-negative that express NK1.1 and are CD3-negative. Three independent experiments were analyzed at each gd with 6–8 implantation sites each.
To further determine if the GFP+ cells in both the MLAp and DB had phenotypes of NK cells, we studied gd11.5, when the GFP+ cells were most abundant (Fig. 2A). We also isolated and physically separated the MLAp, DB, and placenta labyrinth that were then embedded for analysis. Upon histological evaluation, we verified the presence of GFP+ cells in the anatomically separated MLAp and DB and their absence from the labyrinth of the placenta (Fig. 2B). When we performed flow cytometry on cell suspensions from the isolated MLAp or DB, we determined that GFP+ cells were comprised of cells that phenotypically resembled NK cells (CD45+CD3−CD19− NK1.1+) while the GFP– gate contained insignificant percentages of CD3−CD19−NK1.1+ cells (Fig. 2C), comparable to analysis of GFP+ and GFP– cells in the spleen. Thus, we conclude the GFP+ cells indeed represent uNK cells, residing in both the MLAp and DB in the pregnant uterus.
trNK cells proliferate during pregnancy
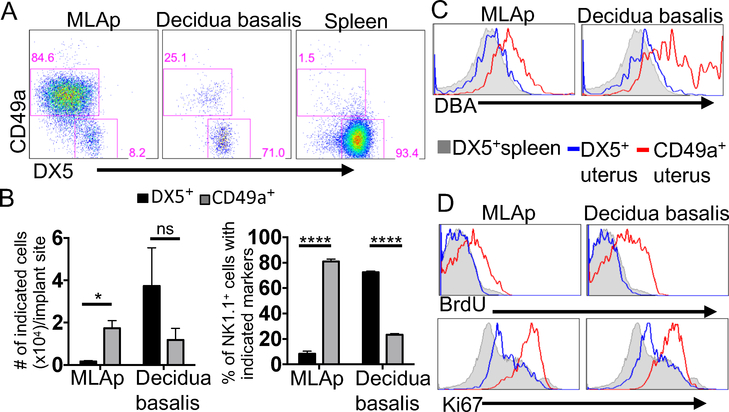
More extensive analysis of gd11.5 implantation site revealed that GFP+ cells with the phenotypic characteristics of cNK cells (CD45+CD3−CD19−NK1.1+DX5+Eomes+CD49a−) and of trNK cells (CD45+CD3−CD19−NK1.1+DX5− Eomes+CD49a+) were present in the MLAp and DB (Fig. 3A, Supplemental Fig.1A, D). A small percentage of CD49a+Eomes− cells, consistent with ILC1s was also present, as previously reported (23). In the MLAp, trNK cells were more prevalent in number and percentage compared to cNK cells (Fig. 3B). In contrast, cNK cells dominated in frequency and cell number in the DB. The trNK cells in the MLAp were uniformly positive at low levels for DBA, known as an uNK cell-specific marker, while trNK cells in the DB showed heterogeneous DBA staining with some cells being more intensely reactive than others (Fig. 3C). Although all GFP+ cells having cNK or trNK cell phenotypes in both MLAp and DB uniformly expressed NKp46 as expected, Ly49 receptor expression varied amongst these uNK subsets (Supplemental Fig.1B). The uNK cells with the cNK phenotype in both the DB and MLAp resembled the cNK cells in the spleen except for DBA reactivity, Ly49H and Ly49I expression. Thus, GFP+ uNK cells at gd11.5 in both the MLAp and DB contain mixtures of cells with cNK and trNK cell phenotypes in different proportions, suggesting each site provides a unique niche for uNK cells.
Figure 3: The DB and MLAp both contain uNK cells that are phenotypically cNK and trNK cells.
The uterus of a pregnant B6 dam was processed at gd11.5. A) The MLAp and DB were physically separated and single cell suspensions were prepared for flow cytometry. The numbers in the gates represent the percentage of CD45+CD3−CD19− NK1.1+ that expresses either CD49a or DX5. B) Cell numbers and percentages were plotted from 3 experiments with 6–8 implantation sites in each experiment. C) The MLAp and DB were prepared for flow cytometry. Histograms were gated on CD45+CD3−CD19NK1.1+ and overlays are of the DX5+ and CD49a+ from the distinct regions and DX5+ spleen. D) The tissues were prepared for flow cytometry and analyzed for BrdU incorporation and Ki67 expression. Representative histograms of 4 independent experiments are shown, (n=4).
The accumulation of uNK cells in pregnant uterus could be due to local proliferation of trNK cells, migration of cNK cells from the periphery, or both. At gd11.5, a high percentage of trNK cells in the MLAp and DB incorporated BrdU while cNK cells in both sites did not incorporate BrdU (Fig 3D). There was a higher expression of Ki67 on trNK cells from both sites than cNK cells in either site in the pregnant uterus, and the spleen (Fig. 3D). Moreover, Ki67 was detected specifically on trNK cells during decidualization at gd6.5 (Supplemental Fig.1F). These results indicate that trNK cells are selectively proliferating early and in both the MLAp and DB during pregnancy.
trNK cell proliferate following artificial decidualization
Our proliferation studies did not exclude the potential contribution of cNK cells homing from the periphery into the pregnant uterus. To evaluate this question, parabiosis experiments were undertaken. However, when we joined mice for parabiosis then mated them soon thereafter, no successful pregnancies ensued, despite resumption of estrus cycles. Similarly, joining mice for parabiosis shortly after impregnation did not result in successful pregnancies anytime before gd6.5 (uNK accumulation, Fig. 2A). Since decidualization can be experimentally induced to mimic physiological decidualization (15, 24) we elected to use this approach. Pilot studies were undertaken to phenotypically assess the cells that were induced by artificial decidualization (Fig. 4A). By histological analysis, the deciduomata in Ncr1iCre x RosamT/mG reporter mice contained GFP+ cells (Fig. 4B). By flow cytometry, deciduomata contained both cNK and trNK cells with ten times more trNK cells than cNK cells (Fig. 4C, 4D). A higher level of Ki67 expression was detected on trNK cells (Fig. 4E) as was seen in pregnancy (Fig. 3D). Additionally, trNK cells in the deciduomata expressed Eomesodermin and DBA (Supplemental Fig.1C and E, respectively) as seen in physiological pregnancy (Supplemental Fig.1D and Fig. 3C). Thus, uNK cells accumulate in the decidualized uterus in the absence of a conceptus, as previously suggested.
Figure 4: The trNK cells accumulate and proliferate in deciduomata.
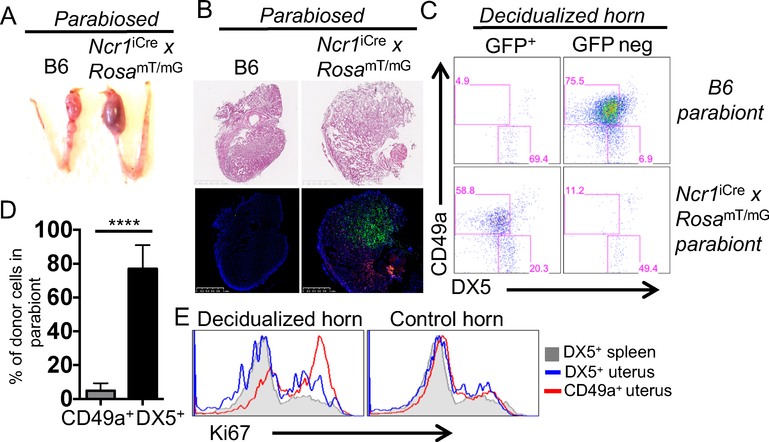
A) A photograph of the decidualized and control horns. B) Ncr1iCre x RosamT/mG NK reporter mice were artificially decidualized and on d5 the decidualized and control horns were processed for histology. A 2.5mm scale is shown. C) The decidualized and control horns were processed for flow cytometry. The numbers in the gates represent the percentage of CD45+CD3−CD19−NK1.1+ that express CD49a or DX5. D) Cell number and percentage plotted of 2 independent experiments, n=10. E) Tissues were processed for flow cytometry and the uNK cells were analyzed for the expression of Ki67.
Minimal contribution of migrating cNK cells to deciduomata in parabionts
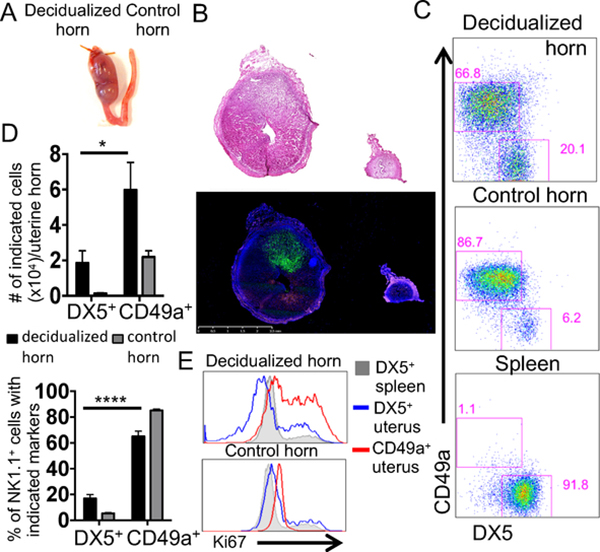
Since both cNK and trNK cells accumulated in deciduomata, the parabiosis approach was deemed to be appropriate to determine if there is a contribution from migrating NK cells from the periphery. We performed parabiosis surgery at the time of artificial decidualization and surgically joined a Ncr1iCre x RosamT/mG reporter mouse to a C57BL/6N animal (Fig. 5A). On d7, the spleen of both parabionts contained GFP+ and GFP– cells with the NK cell phenotype (NK1.1+CD3–CD19–) from the other parabiont and were DX5+ (Supplemental Fig. 2A). This finding is consistent with cNK cells migrating from either parabiont, as previously reported (4). In contrast, when we examined the B6 deciduomata by histology we found no evidence for GFP+ cells (Fig. 5B), indicating that uNK cells were derived only from resident cells and not from migrating cells. These findings were confirmed by flow cytometry (Fig. 5C) that showed no contribution from migrating cells.
Figure 5: CD49a+NK1.1+ uterine trNK cells are resident and proliferate.
Ncr1iCre x RosamT/mG NK reporter and B6 mice were artificially decidualized and parabiosed. A) A photograph of the decidualized and control horns of the parabionts. B) On d7 the decidualized and control horns from each parabiont were processed for histology. A 1mm scale is shown. C) The decidualized and control horns of each parabiont were processed for flow cytometry. The percentages in the gates represent CD45+CD3−CD19−NK1.1+ that express GFP or are GFP negative that express CD49a and DX5. D) Cell percentage of donor cells of 6 decidualized horns from 2 independent parabiosis experiments, (n=6). E) Tissues were processed as in C and the uNK cells were analyzed for the expression of Ki67.
In the Ncr1iCre x RosamT/mG reporter parabiont, uNK cells in the deciduomata were all GFP+ and NK1.1+CD49a+ (Fig. 5C, D), consistent with a trNK cell phenotype, and indicating that CD49a is a reliable marker of trNK cells in deciduomata. The trNK cells were proliferating as indicated by high Ki67 expression (Fig. 5E). Finally, NK1.1+CD49a+ NK cells did not accumulate in the control horn of the reporter mouse, (Supplemental Fig. 2B). Thus, uNK cells in the deciduomata are almost exclusively trNK cells (NK1.1+CD49a+) that proliferate locally.
Discussion
Here we provide clarity to the long debate about the origin of uNK cells during pregnancy. These studies were aided by use of a novel NK reporter mouse, Ncr1iCre x RosamT/mG, to visualize uNK cells during pregnancy. We confirmed GFP+ cells had characteristics of NK cells, including CD3−CD19−NK1.1+NKp46+ phenotype while GFP− cells contained few cells with an NK cell phenotype, also consistent with evaluation of splenic NK cells. The anatomic distribution of uNK cells could be readily ascertained here, revealing their accumulation in the DB and MLAp. In both sites, there was heterogeneity of uNK cells, reflected by subsets having phenotypes consistent with cNK and trNK cells. During pregnancy, we found trNK cells were proliferating. Parabiosis studies were undertaken that revealed the accumulating NK cells consisted of proliferating trNK cells with no contribution from migrating cNK cells, and also established that CD49a is a reliable marker of trNK cells in decidualized tissues. Taken together, these data suggest that trNK cells are the primary source of uNK cells in decidualization and in physiologic pregnancy.
Our findings do not refute several prior studies supporting the recruitment of NK cells to the pregnant uterus (11, 25, 26). Here, we found abundant numbers of uNK cells with cNK cell phenotype in the DB. The cNK cells were not actively proliferating, unlike trNK cells. Since cNK cells were relatively few in the virgin uterus, it is likely that they migrated into the pregnant uterus. However, we could not formally test cell migration since the parabiosis approach was not feasible in pregnant mice. Although we were able to study deciduomata, artificial decidualization tends to recapitulate events in early pregnancy because the deciduomata are resorbed at later times. Nonetheless, cNK cells very likely migrate into the pregnant uterus.
Our data, along with prior studies supporting a role for migrating NK cells, suggest new hypotheses whereby there may be waves of NK cells accumulating at the implantation site during pregnancy. We propose that a first wave of NK cells accrues when the endometrium remodels during the decidualization process. As shown here, uterine trNK cells appear to be the primary responding NK cell subset in this process and migrating cNK cells appear to play a negligible role. We also propose a second wave, consistent with prior studies and data presented here, in which cNK cells are recruited from the periphery during placentation and remodeling of the spiral arteries. Consistent with this hypothesis, mice that lack NF-IL3 are deficient in cNK cells and have abnormal remodeling of the spiral arteries critical for proper placentation (23, 27). However, our prior studies indicated that trNK cells in the uterus are not dependent on NF-IL3. Hence, we suggest that pregnancy orchestrates the movement and retention of NK cell subsets at different times, implying that each subset has distinct functions during pregnancy.
Supplementary Material
Acknowledgements
We thank Sarah England and Yokoyama and England lab members for great discussions, members of Kelle Moley’s lab, Arin Oestreich and Claire Stephens, for providing protocols. We thank Adrian Erlebacher for a helpful discussion regarding experimentally-induced decidualization. We thank Alafi Neuroimaging Laboratory for the use of the Nanozoomer.
Bibliography
- 1.Kim S, Iizuka K, Kang HS, Dokun A, French AR, Greco S, Yokoyama WM 2002. In vivo developmental stages in murine natural killer cell maturation. Nature Immunol 3: 523–528 [DOI] [PubMed] [Google Scholar]
- 2.Chiossone L, Chaix J, Fuseri N, Roth C, Vivier E, Walzer T 2009. Maturation of mouse NK cells is a 4-stage developmental program. Blood 113: 5488–5496 [DOI] [PubMed] [Google Scholar]
- 3.Peng H, Jiang X, Chen Y, Sojka DK, Wei H, Gao X, Sun R, Yokoyama WM, Tian Z 2013. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J Clin Invest 123: 1444–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, Zhong C, Chase JM, Rothman PB, Yu J, Riley JK, Zhu J, Tian Z, Yokoyama WM 2014. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. Elife 3: e01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Victorino F, Sojka DK, Brodsky KS, McNamee EN, Masterson JC, Homann D, Yokoyama WM, Eltzschig HK, Clambey ET 2015. Tissue-Resident NK Cells Mediate Ischemic Kidney Injury and Are Not Depleted by Anti-Asialo-GM1 Antibody. Journal of immunology 195: 4973–4985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diefenbach A, Colonna M, Koyasu S 2014. Development, differentiation, and diversity of innate lymphoid cells. Immunity 41: 354–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, Gilfillan S, Colonna M, Immunological Genome C 2015. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol 16: 306–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bulmer JN, Williams PJ, Lash GE 2010. Immune cells in the placental bed. The International journal of developmental biology 54: 281–294 [DOI] [PubMed] [Google Scholar]
- 9.Moffett A, Colucci F 2014. Uterine NK cells: active regulators at the maternal-fetal interface. The Journal of clinical investigation 124: 1872–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erlebacher A 2013. Immunology of the maternal-fetal interface. Annual review of immunology 31: 387–411 [DOI] [PubMed] [Google Scholar]
- 11.Zhang JH, Yamada AT, Croy BA 2009. DBA-lectin reactivity defines natural killer cells that have homed to mouse decidua. Placenta 30: 968–973 [DOI] [PubMed] [Google Scholar]
- 12.Croy BA, van den Heuvel MJ, Borzychowski AM, Tayade C 2006. Uterine natural killer cells: a specialized differentiation regulated by ovarian hormones. Immunological reviews 214: 161–185 [DOI] [PubMed] [Google Scholar]
- 13.Croy BA, He H, Esadeg S, Wei Q, McCartney D, Zhang J, Borzychowski A, Ashkar AA, Black GP, Evans SS, Chantakru S, van den Heuvel M, Paffaro VA Jr., Yamada AT 2003. Uterine natural killer cells: insights into their cellular and molecular biology from mouse modelling. Reproduction 126: 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lima PD, Zhang J, Dunk C, Lye SJ, Croy BA 2014. Leukocyte driven-decidual angiogenesis in early pregnancy. Cellular & molecular immunology 11: 522–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madjerek ZS 1967. Comparison of the traumatic deciduoma with the normal decidual reaction in mice. Acta physiologica et pharmacologica Neerlandica 14: 520. [PubMed] [Google Scholar]
- 16.McConaha ME, Eckstrum K, An J, Steinle JJ, Bany BM 2011. Microarray assessment of the influence of the conceptus on gene expression in the mouse uterus during decidualization. Reproduction 141: 511–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chantakru S, Miller C, Roach LE, Kuziel WA, Maeda N, Wang WC, Evans SS, Croy BA 2002. Contributions from self-renewal and trafficking to the uterine NK cell population of early pregnancy. Journal of immunology 168: 22–28 [DOI] [PubMed] [Google Scholar]
- 18.Croy BA, Di Santo JP, Greenwood JD, Chantakru S, Ashkar AA 2000. Transplantation into genetically alymphoid mice as an approach to dissect the roles of uterine natural killer cells during pregnancy—a review. Placenta 21 Suppl A: S77–80 [DOI] [PubMed] [Google Scholar]
- 19.Croy BA, Esadeg S, Chantakru S, van den Heuvel M, Paffaro VA, He H, Black GP, Ashkar AA, Kiso Y, Zhang J 2003. Update on pathways regulating the activation of uterine Natural Killer cells, their interactions with decidual spiral arteries and homing of their precursors to the uterus. Journal of reproductive immunology 59: 175–191 [DOI] [PubMed] [Google Scholar]
- 20.Chiossone L, Vacca P, Orecchia P, Croxatto D, Damonte P, Astigiano S, Barbieri O, Bottino C, Moretta L, Mingari MC 2014. In vivo generation of decidual natural killer cells from resident hematopoietic progenitors. Haematologica 99: 448–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narni-Mancinelli E, Chaix J, Fenis A, Kerdiles YM, Yessaad N, Reynders A, Gregoire C, Luche H, Ugolini S, Tomasello E, Walzer T, Vivier E 2011. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proceedings of the National Academy of Sciences of the United States of America 108: 18324–18329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L 2007. A global doublefluorescent Cre reporter mouse. Genesis 45: 593–605 [DOI] [PubMed] [Google Scholar]
- 23.Boulenouar S, Doisne JM, Sferruzzi-Perri A, Gaynor LM, Kieckbusch J, Balmas E, Yung HW, Javadzadeh S, Volmer L, Hawkes DA, Phillips K, Brady HJ, Fowden AL, Burton GJ, Moffett A, Colucci F 2016. The Residual Innate Lymphoid Cells in NFIL3-Deficient Mice Support Suboptimal Maternal Adaptations to Pregnancy. Frontiers in immunology 7: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herington JL, Underwood T, McConaha M, Bany BM 2009. Paracrine signals from the mouse conceptus are not required for the normal progression of decidualization. Endocrinology 150: 4404–4413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chantakru S, Miller C, Roach LE, Kuziel WA, Maeda N, Wang WC, Evans SS, Croy BA 2002. Contributions from self-renewal and trafficking to the uterine NK cell population of early pregnancy. J Immunol 168: 22–28 [DOI] [PubMed] [Google Scholar]
- 26.Croy BA, Xie X 2006. In vivo models for studying homing and function of murine uterine natural killer cells. Methods in molecular medicine 122: 77–92 [DOI] [PubMed] [Google Scholar]
- 27.Redhead ML, Portilho NA, Felker AM, Mohammad S, Mara DL, Croy BA 2016. The Transcription Factor NFIL3 Is Essential for Normal Placental and Embryonic Development but Not for Uterine Natural Killer (UNK) Cell Differentiation in Mice. Biology of reproduction 94: 101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.