Abstract
PURPOSE
Digital pathology (DP), referring to the digitization of tissue slides, is beginning to change the landscape of clinical diagnostic workflows and has engendered active research within the area of computational pathology. One of the challenges in DP is the presence of artefacts and batch effects, unintentionally introduced during both routine slide preparation (eg, staining, tissue folding) and digitization (eg, blurriness, variations in contrast and hue). Manual review of glass and digital slides is laborious, qualitative, and subject to intra- and inter-reader variability. Therefore, there is a critical need for a reproducible automated approach of precisely localizing artefacts to identify slides that need to be reproduced or regions that should be avoided during computational analysis.
METHODS
Here we present HistoQC, a tool for rapidly performing quality control to not only identify and delineate artefacts but also discover cohort-level outliers (eg, slides stained darker or lighter than others in the cohort). This open-source tool employs a combination of image metrics (eg, color histograms, brightness, contrast), features (eg, edge detectors), and supervised classifiers (eg, pen detection) to identify artefact-free regions on digitized slides. These regions and metrics are presented to the user via an interactive graphical user interface, facilitating artefact detection through real-time visualization and filtering. These same metrics afford users the opportunity to explicitly define acceptable tolerances for their workflows.
RESULTS
The output of HistoQC on 450 slides from The Cancer Genome Atlas was reviewed by two pathologists and found to be suitable for computational analysis more than 95% of the time.
CONCLUSION
These results suggest that HistoQC could provide an automated, quantifiable, quality control process for identifying artefacts and measuring slide quality, in turn helping to improve both the repeatability and robustness of DP workflows.
INTRODUCTION
Definitive disease diagnosis routinely takes place via visual inspection of a tissue slide by a pathologist under a microscope. Before this can take place, the tissue slide itself must be created. This process, which involves gross organ dissection, selection and preparation of tissue blocks for slide creation, microtomy (cutting and tissue placement on the slide), staining, and cover slipping, is fraught with multiple preanalytic opportunities for the introduction of artefacts and batch effects.1-3 These artefacts may include improper tissue placement (eg, folding, compressing, tearing, air bubbles), improper reagents (eg, over- or understaining, stain concentration differences, stain batch variation), and poor microtomy (eg, knife chatter, thickness variances). The increasingly popular digitization of these same slides, to take advantage of computational-aided diagnostic approaches,4-6 for example, introduces yet another potential source of artefacts. This digitization process sees the same glass slides routinely used for microscope-based pathology being placed on the equivalent of a digital camera so that digital representations of the slide may be constructed. Scanner manufacturers may employ different approaches for slide digitization, including different hardware (eg, bulbs for lighting, charge-coupled device chips for digitization), algorithms for image manipulation (eg, stitching, compression), and file formats. Therefore, the choice of slide scanner could influence the image appearance, which in turn might have implications for any subsequent image analysis procedure.7 These digital pathology (DP) slides may additionally include digitization artefacts such as blurriness, lighting, and contrast issues. Taken together, there are a number of different combinations of sources of preanalytic variance that may result in substantial differences in appearance and quality of a tissue slide.
These same artefacts and variances could negatively affect downstream clinical and research workflows.8 In the analog workflow in use today, continuous quality control (QC) processes are limited, unlike in laboratory medicine, which relies on continuous statistical process control. Clinically, slides rejected on quality grounds represent a drag on the clinical pathology workflow, because these slides need to be recut or rescanned, in turn causing additional delays and unnecessary costs. From a research standpoint, artefacts represent sources of noise that can adversely affect the development and validation of analytic classifiers for tasks such as disease detection, diagnosis, and prognosis.9,10 This is especially important for increasingly popular deep learning– and machine learning–based approaches,11-16 which rely on well-annotated and relatively artefact-free images to learn underlying disease-specific representations.
Currently, most QC processes for clinical and research applications are performed manually, making the process subjective, laborious, and error prone. For instance, a wide spectrum of artefacts and image qualities (Fig 1) can be seen in many of the 30,000 digitized tissue slides hosted by The Cancer Genome Atlas (TCGA).17 This occurs despite the fact that these slide images undergo manual QC before being introduced into TCGA. In addition, more subtle artefacts such as variations in stain that may not affect a pathologist’s diagnostic interpretation may still have implications for subsequent computational image analysis and machine-learning algorithms. For instance, technical artefacts resulting from the preparation facility (ie, batch effects) may be confounded with the biologic signal under investigation (Data Supplement). Although batch effects remain a well-known issue in the bioinformatics field,18 they have received less attention in the DP domain.
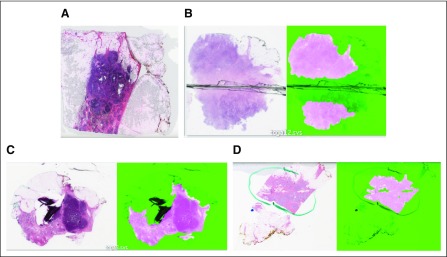
FIG 1.
Original images juxtaposed with corresponding results from HistoQC (fuchsia indicating acceptable tissue): images identified as having (A) a significant air bubble artefact requiring removal from experimental cohort, (B) blurry tissue near a coverslip crack, (C) folded tissue, and (D) pen markings correctly identified as regions to be avoided.
Recently, other groups19-23 have begun to develop DP algorithms for QC tasks, such as blurriness and stain assessment. Unfortunately, there has not been a single, unified user-friendly platform that has included these and other QC approaches for a comprehensive and integrated QC review of DP slide images.
Recognizing the need for a modular, user-friendly QC tool, we present here an open-source QC application, HistoQC, for automated assessment of slide quality alongside a public repository of slides containing artefacts. HistoQC employs a combination of image metrics (eg, color histograms, brightness, contrast), features (eg, edge and smoothness detectors), and supervised classifiers (eg, pen detection) to aid users in identifying slides with gross technical artefacts, artefact-affected regions that may not be suitable for computational analysis (Data Supplemental provides current list of classifiers and metrics), and samples potentially affected by batch effects. The modular nature of HistoQC allows for the facile embedding of additional metrics and artefact-detection algorithms as they become available in the literature.
METHODS
HistoQC functions in the following way. The user supplies a configuration file that defines the parameters of the QC pipeline, such as which modules to execute and in what order. As the python-based pipeline is executed on the slide, relevant output images are created (eg, thumbnail images indicating regions of potential blurriness), with metadata (eg, scanner type, magnification, microns per pixel) and metrics being saved in a tab-separated value file. As designed, image metrics can be computed on the entire slide (ie, including the background) or limited solely to regions containing detected tissue. Although any common data analytic tool may be used to review the tab-separated value output (eg, Matlab [MathWorks, Natick, MA], Excel [Microsoft, Redmond, WA], R [R Foundation, Vienna, Austria]), we have developed an HTML5-based user interface (Fig 2) that seamlessly allows for real-time visualization and filtering of the data. This approach helps identify those specific slides that might require additional scrutiny.
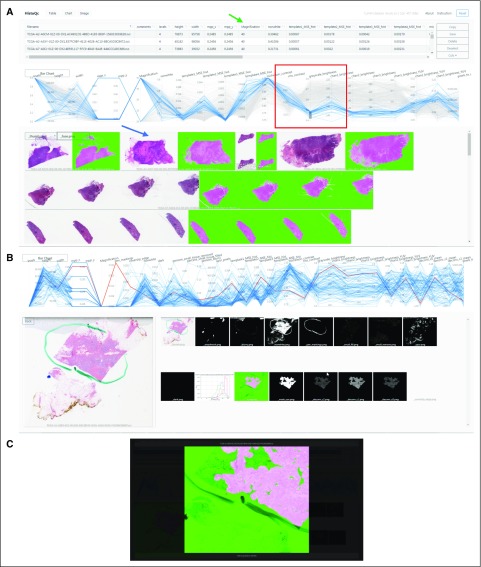
FIG 2.
(A) HistoQC user interface showing table of HistoQC-produced metrics with sortable columns (green arrow), parallel coordinate plot (red box; additional details in Fig 3), and thumbnail images of the cohort alongside HistoQC overlay output indicating artefact-free regions (blue arrow). (B) Selecting a single image highlights the appropriate line in the parallel coordinate graph and shows the series of outputs produced by the modules of the pipeline, allowing for more detailed subsequent review. (C) Double clicking on any image brings up a higher-resolution version with dynamic zoom, allowing for fine-tuned inspection of potential artefacts. Hist, histogram; MPP, microns per pixel; MSE, mean squared error.
Slides requiring additional scrutiny can be discovered in multiple ways: sorting various columns to view outliers with unexpectedly high or low values (Fig 2 green arrow), viewing interactive parallel coordinate plots24 (Fig 2 red box; Fig 3) of metrics that help visualize potential batch effects and outliers, or viewing the original slides juxtaposed with various output masks (Fig 2 blue arrow). For improved user experience and efficiency, clicking a row or image shows the various masks produced by the pipeline (Fig 2 middle), with a subsequent click taking the user to a higher-magnification version of the mask of interest for more detailed review (Fig 2 bottom). After the user has either annotated rows using the comments field or removed rows from the table, the resulting table can be saved and used as a list of samples suitable for downstream experiments. We note that postinstallation, no Internet connection is required, making our approach suitable for nonanonymized clinical data.
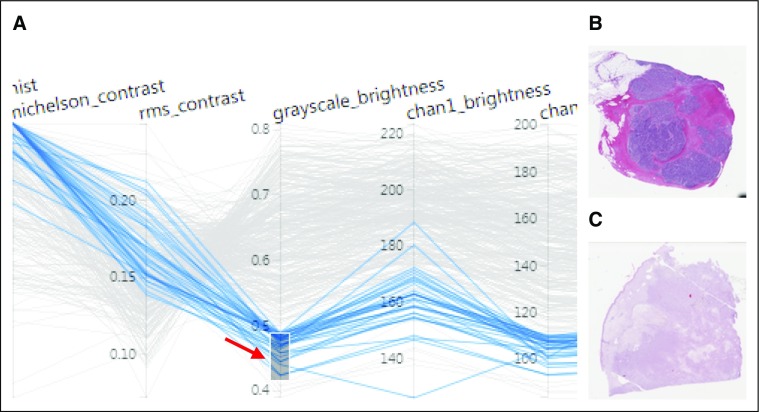
FIG 3.
(A) Higher magnification of the parallel coordinate plot bounded by the red box in Figure 2. The different y-axes correspond to different image metrics determined by HistoQC and may have their own ranges and scales. Each horizontally oriented line, in either gray or blue, represents a whole-slide image (WSI) analyzed by HistoQC. By examining the convergence or divergence of each horizontal WSI line with respect to the rest of the images in the cohort, batch effects and outliers can be more easily visually identified. In the example illustrated, the user has interactively drawn a gray box (red arrow) to select images with a grayscale intensity value between 0.4 and 0.5, resulting in all lines that do not meet this criterion turning gray. This image selection mechanism alters the visibility of slides in both the table (Fig 2 green arrow) and the thumbnails (Fig 2 blue arrow). Furthermore, the user can dynamically drag or extend the gray box upward and downward to update the visible slides in real time. As a result, images with (B) low and (C) high grayscale brightness values are easier to identify and review.
We evaluated the ability of HistoQC to identify regions of artefact-free tissue on a total of 450 randomly selected slides from the TCGA breast cancer cohort at a magnification of 40×. Because of the efficient implementation of HistoQC, the analysis took 130 minutes using a four-hyperthreaded core processor. Representative slides identified by HistoQC as containing artefacts such as air bubbles and slide cracks are illustrated in Figures 1A and 1B. HistoQC successfully identified regions with tissue folding and pen markings, removing them from the outputted masks (Figs 1C and 1D). As derived from HistoQC output, the Data Supplement shows the potential presence of batch effects in microns per pixel and considerable heterogeneity in tissue brightness for the TCGA breast cancer data set, important considerations for downstream experimental design.
RESULTS
To validate the results generated by HistoQC, two pathologists with experience in DP were asked to assign a value of either acceptable or not acceptable to each of the masks produced by HistoQC. Acceptability was defined by at least an 85% area overlap between the pathologists’ visual assessment and the computational assessment by HistoQC of artefact-free tissue. Each pathologist independently reviewed 250 samples. In addition, a total of 50 images from TCGA were evaluated by both pathologists and HistoQC to determine interexpert agreement on HistoQC output. Overall, the agreement between HistoQC and the experts was 94% (235 of 250) for expert 1 and 97% (242 of 250) for expert 2. For the 50 slides evaluated by both experts, interobserver agreement was 96% (48 of 50), comparable to that of HistoQC with the individual readers. The main reasons for the disagreement were faintly stained slides resulting in tissue detection failures and a few regions of predominantly stromal-rich areas being incorrectly identified as blurry (Data Supplement). These failures appeared on the HistoQC user interface as outliers, primarily because of metrics (eg, estimated tissue area) being a number of standard deviations away from those associated with the remainder of the analyzed slides. Slides identified by HistoQC as containing artefacts were uploaded to the Histology Quality Control Repository25 for community review.
The pathologists also provided qualitative feedback regarding patterns of cases that HistoQC seemed to incorrectly identify as being compromised or not. These cases generally fell into three categories: poorly fixed tissue, necrotic tissue, or subtle adipose tissue infiltrate with scant tissue reaction. HistoQC also sometimes struggled to fully identify parenchyma in mucinous tumors. We are working on further improving HistoQC to address these limitations in the next version.
DISCUSSION
To summarize, we presented and have released an open-source QC tool for DP slides called HistoQC. Initial results suggest HistoQC is suitable for delineation of slide level artefacts. Comparison of HistoQC against manual QC by two pathologists on 450 images yielded an average agreement greater than 95%, comparable in range of agreement to that between the two individual human readers. In addition, the image metrics computed by HistoQC could be used by researchers and analytic pipeline developers to precisely define the input image characteristics with which their algorithms have been both trained and validated. Stringent specification of these image characteristic ranges allows for algorithms to be selectively invoked only on the appropriate images, likely improving algorithm confidence.
Taken together, the clinical pathology and DP vendor communities seem to be beginning to appreciate the importance of quantifiable QC processes for engendering DP workflows.26 Before DP systems can be used within a clinical setting, slide scanners themselves must receive regulatory approval. Scanner manufacturers have been attempting to quantitatively assess the reproducibility of scanner-generated images, especially to ensure consistency of image quality over time.27,28 With penetration of these scanners into clinical workflows, these types of validation studies may become enshrined as part of the routine quality assurance and maintenance of the scanners.26 As such, from both regulatory and maintenance standpoints, a single automated QC pipeline like HistoQC can provide quantitative metrics for benchmarking the quality and consistency of scanner-generated DP images.
DP workflows are on the verge of leveraging powerful computer-aided diagnostic (CAD) support algorithms, potentially helping to greatly reduce inter- and intraobserver diagnostic variability. As revealed in a number of recent publications, many CAD and artificial intelligence (AI) algorithms appear to not generalize very well when evaluated on a cohort distinct from the set of images on which they were initially trained.14,29 Consequently, these AI and CAD algorithms must be robustly validated on a large collection of heterogeneous inputs.9,10 Approaches like HistoQC could allow for pre-evaluation of test sets to ensure that the CAD and AI algorithms are evaluated on a sufficiently diverse set of test images.
Although HistoQC is ready for research applications, there remain areas for additional improvement beyond addressing the comments of our pathologists. For example, due to the heterogeneity in compression levels typically present between DP scanners and the evidence that the resulting compression artefacts affect performance of deep-learning and AI algorithms,30 HistoQC could be extended to detect and measure compression effects. Building further on the need to incorporate additional features, we envision HistoQC evolving into a collection of community-driven reference implementations of sophisticated detectors and metrics. For example, Senaras et al22 presented a deep learning–based blur detector, and Avanaki et al19 proposed texture-based image quality metrics. We hope that these types of algorithms will in the future be embedded into HistoQC to enable the comparison of results across different sites and laboratories. The work presented here focused on the evaluation of HistoQC in the context of hematoxylin and eosin bright-field microscopy images. Clearly, there is also a need for the application of QC metrics in other types of multimodal microscopy images, such as immunohistochemical staining and quantitative immunofluorescence.
Last, we hope to aggregate unique artefacts identified by the user community during its use of HistoQC. We have stood up an image quality repository to allow end users to upload slides that contain artefacts.25 This repository will help provide training and validation material needed for the benchmarking of future CAD approaches. The source code of HistoQC (Data Supplement) is freely available for use, modification, and contribution (http://github.com/choosehappy/HistoQC).
Footnotes
Supported by National Cancer Institute, National Institutes of Health, Awards No. 1U24CA199374-01, R01CA202752-01A1, R01CA208236-01A1, R01 CA216579-01A1, and R01 CA220581-01A1; National Center for Research Resources Award No. 1 C06 RR12463-01; Veterans Affairs (VA) Merit Review Award No. IBX004121A from the US Department of VA Biomedical Laboratory Research and Development Service; Nephrology Training Grant No. 5T32DK007470 CWRU; US Department of Defense Lung Cancer Investigator-Initiated Translational Research Award No. W81XWH-18-1-0440; the Ohio Third Frontier Technology Validation Fund, the Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering; and the Clinical and Translational Science Award Program, Case Western Reserve University.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, US Department of Veterans Affairs, US Department of Defense, or US Government.
AUTHOR CONTRIBUTIONS
Conception and design: All authors
Financial support: Anant Madabhushi
Administrative support: Anant Madabhushi
Provision of study material or patients: Michael Feldman, Hanna Gilmore, Anant Madabhushi
Collection and assembly of data: Andrew Janowczyk, Michael Feldman, Hanna Gilmore, Anant Madabhushi
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Andrew R. Janowczyk
Consulting or Advisory Role: Merck
Hannah Gilmore
Travel, Accommodations, Expenses: Sectra
Michael Feldman
Consulting or Advisory Role: Philips Healthcare
Travel, Accommodations, Expenses: Philips Healthcare
Anant Madabhushi
Leadership: Inspirata
Stock and Other Ownership Interests: Inspirata, Elucid Bioimaging
Honoraria: AstraZeneca, Inspirata
Consulting or Advisory Role: Inspirata, AstraZeneca, Merck
Research Funding: Inspirata (Inst), Philips Healthcare (Inst)
Patents, Royalties, Other Intellectual Property: Intellectual property licensed by Inspirata (Inst); intellectual property licensed by Elucid Bioimaging (Inst)
No other potential conflicts of interest were reported.
REFERENCES
- 1. Chatterjee S: Artefacts in histopathology. J Oral Maxillofac Pathol 18:S111-S116, 2014 (suppl 1) [DOI] [PMC free article] [PubMed]
- 2.Rastogi V, Puri N, Arora S, et al. Artefacts: A diagnostic dilemma—A review. J Clin Diagn Res. 2013;7:2408–2413. doi: 10.7860/JCDR/2013/6170.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taqi SA, Sami SA, Sami LB, et al. A review of artifacts in histopathology. J Oral Maxillofac Pathol. 2018;22:279. doi: 10.4103/jomfp.JOMFP_125_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Doyle S, Rodriguez C, Madabhushi A, et al: Detecting prostatic adenocarcinoma from digitized histology using a multi-scale hierarchical classification approach. Conf Proc IEEE Eng Med Biol Soc 1:4759-4762, 2006. [DOI] [PubMed] [Google Scholar]
- 5.Doyle S, Monaco J, Feldman M, et al. An active learning based classification strategy for the minority class problem: Application to histopathology annotation. BMC Bioinformatics. 2011;12:424. doi: 10.1186/1471-2105-12-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee G, Singanamalli A, Wang H, et al. Supervised multi-view canonical correlation analysis (sMVCCA): Integrating histologic and proteomic features for predicting recurrent prostate cancer. IEEE Trans Med Imaging. 2015;34:284–297. doi: 10.1109/TMI.2014.2355175. [DOI] [PubMed] [Google Scholar]
- 7. doi: 10.1016/j.compmedimag.2016.05.003. Janowczyk A, Basavanhally A, Madabhushi A: Stain Normalization using Sparse AutoEncoders (StaNoSA): Application to digital pathology. Comput Med Imaging Graph 57:50-61, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leo P, Elliott R, Shih NNC, et al. Stable and discriminating features are predictive of cancer presence and Gleason grade in radical prostatectomy specimens: A multi-site study. Sci Rep. 2018;8:14918. doi: 10.1038/s41598-018-33026-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madabhushi A, Lee G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med Image Anal. 2016;33:170–175. doi: 10.1016/j.media.2016.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. doi: 10.1146/annurev-bioeng-112415-114722. Bhargava R, Madabhushi A: A review of emerging themes in image informatics and molecular analysis for digital pathology. Annu Rev Biomed Eng 18:387-412, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Janowczyk, Madabhushi A: Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use case. J Pathol Inform 7:29, 2016. [DOI] [PMC free article] [PubMed]
- 12.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–2210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saltz J, Gupta R, Hou L, et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Reports. 2018;23:181–193.e7. doi: 10.1016/j.celrep.2018.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coudray N, Ocampo PS, Sakellaropoulos T, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24:1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bychkov D, Linder N, Turkki R, et al. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci Rep. 2018;8:3395. doi: 10.1038/s41598-018-21758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang H, Cruz-Roa A, Basavanhally A, et al: Cascaded ensemble of convolutional neural networks and handcrafted features for mitosis detection. Presented at the International Society for Optics and Photonics, San Diego, CA, February 15-20, 2014. [Google Scholar]
- 17.Weinstein J. N., Collisson EA, Mills GB, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goh WWB, Wang W, Wong L. Why batch effects matter in omics data, and how to avoid them. Trends Biotechnol. 2017;35:498–507. doi: 10.1016/j.tibtech.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 19. Avanaki AR, Espig KS, Xthona A, et al: Automatic image quality assessment for digital pathology. Presented at the 13th International Workshop on Breast Imaging, Malmö, Sweden, June 19-22, 2016. [Google Scholar]
- 20. Ameisen D, Deroulers C, Perrier V, et al: Towards better digital pathology workflows: Programming libraries for high-speed sharpness assessment of whole slide images. Diagn Pathol 9:S3, 2014 (suppl 1) [DOI] [PMC free article] [PubMed]
- 21. Ameisen D, Deroulers C, Perrier V, et al: Stack or trash? Quality assessment of virtual slides. Diagn Pathol 8:S23, 2013 (suppl 1)
- 22.Senaras C, Niazi MKK, Lozanski G, et al. DeepFocus: Detection of out-of-focus regions in whole slide digital images using deep learning. PLoS One. 2018;13:e0205387. doi: 10.1371/journal.pone.0205387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen S, Kurc TM, Gao Y, et al. A methodology for texture feature-based quality assessment in nucleus segmentation of histopathology image. J Pathol Inform. 2017;8:38. doi: 10.4103/jpi.jpi_43_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edsall RM. The parallel coordinate plot in action: design and use for geographic visualization. Comput Stat Data Anal. 2003;43:605–619. https://www.sciencedirect.com/science/article/pii/S0167947302002955 [Google Scholar]
- 25. Janowczyk A: HistoQCRepo. http://histoqcrepo.com/
- 26. doi: 10.5858/arpa.2018-0378-CP. Bui MM, Riben MW, Allison KH, et al: Quantitative image analysis of human epidermal growth factor receptor 2 immunohistochemistry for breast cancer: Guideline from the College of American Pathologists. Arch Pathol Lab Med [epub ahead of print on January 15, 2019] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shrestha P, Hulsken B. Color accuracy and reproducibility in whole slide imaging scanners. J Med Imaging (Bellingham) 2014;1:027501. doi: 10.1117/1.JMI.1.2.027501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shrestha P, Kneepkens R, Vrijnsen J, et al. A quantitative approach to evaluate image quality of whole slide imaging scanners. J Pathol Inform. 2016;7:56. doi: 10.4103/2153-3539.197205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zech JR, Badgeley MA, Liu M, et al. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLoS Med. 2018;15:e1002683. doi: 10.1371/journal.pmed.1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dodge S, Karam L: Understanding how image quality affects deep neural networks Presented at the Eighth International Conference on Quality of Multimedia Experience, Lisbon, Portugal, June 6-8, 2016. [Google Scholar]