Abstract
Chronic human immunodeficiency virus type 1 (HIV-1) disease is characterized by the retention of provirus within latently infected cells. Anti-HIV-1 CRISPR-Cas9 gene editing is an attractive strategy to excise or inactivate the HIV-1 genome. Recent strategies have focused on designing gRNAs that target the long terminal repeat (LTR) because 5′ and 3′ LTR symmetry can facilitate proviral excision. However, the promiscuity of CRISPR-Cas9 gene editing system necessitates the investigation of potential off-target effects. Here, potential gRNAs designed from HIV-1 phylogenetic subtypes using the CRISPRseek tool were investigated. Across the LTR, it was found that certain regions show higher human homology than others. When using recommended cutoffs, 96.40% of gRNAs were predicted to have no high probability off-target effects. Given this observation, while high-probability off-target effects are a potential danger, they can be avoided with proper gRNA design.
Introduction
Despite recent advances in the treatment of human immunodeficiency virus type 1 (HIV-1) infection and disease, there is no cure. Antiretroviral therapy (ART) is utilized to help mitigate symptoms and impede viral spread within the body.1 Because ART targets multiple aspects of the replication cycle of HIV-1, patient viral loads can be suppressed to undetectable levels in the peripheral blood.2 However, ART cannot remove the integrated HIV-1 genome from latently infected cells. This means that ART cannot cure HIV-1 infection and necessitates a lifelong dependence on this therapy.2
With the discovery of CRISPR*-Cas9, we can now remove the latent HIV-1 proviral genome from infected cells. While there are multiple strategies for targeting HIV-1, the symmetrical nature of the HIV-1 5′ and 3′ long terminal repeat (LTR) sequences makes the LTR an ideal target. Because of this symmetry, a gRNA that targets a sequence in the 5′ LTR will also target the mirrored sequence in the 3′ LTR.3 If both ends were to be simultaneously cleaved by two Cas9 enzymes, the provirus could be excised from the cell and provide a sterilizing cure for HIV infection. Even without excision, the LTR contains many transcription factor binding sites (TFBS)4–11 that could impair HIV transcription if altered, thereby preventing the infected cell from escaping latency.
Multiple experimental systems have been developed using the Streptococcus pyogenes Cas9 (SpCas9) system in an attempt to cure HIV-1. Studies have focused on editing human genes that facilitate HIV-1 entry such as CCR512,13 and CXCR4.14,15 Others targeted gag/pol, two genes essential for viral replication,16 or have targeted the promoter region with the intention of disrupting transcription.17 Alternatively, the CRISPR-Cas9 system has also been utilized to induce HIV-1 reactivation as part of the “shock and kill” cure strategy.18,19 However, because of the symmetric nature of the HIV-1 LTRs, others have focused on using double-stranded breaks on opposite sides of the genome to excise the HIV-1 genome completely.20,21 The field has also begun to consider the effects of interpatient HIV-1 genetic variation in an attempt to develop a broad spectrum targeting anti-HIV-1 gRNA.22,23 To date, there has been little exploration of the potential off-target effects of an HIV-1-targeted Cas9 application. The accidental alteration of host DNA could be detrimental to the patient's survival, even if HIV-1 provirus were excised in the process. This necessitates intense investigation of the potential off-target effects of any CRISPR-Cas9 therapeutic strategy and the selection of an ideal gRNA template that would minimize off-target effects.
Anecdotally, it has been proposed that a likely source of off-target effects from an anti-HIV CRISPR-Cas9 therapy is human endogenous retroviral (HERV) 5′ LTRs or human TFBS that are similar to HIV-1 TFBS. HIV-1 and HERVs share a similar structure and genomic composition because of their retroviral origins. Additionally, HIV-1 Tat has been shown to initiate HERV-K and HERV-W transcription.24,25 This implies that HERV 5′ LTRs might be similar enough to be cleaved by a SpCas9 utilizing a gRNA intended for HIV-1's 5′ LTR. The proviral LTR also recruits transcription factors intended for human binding sites; this implies that a gRNA targeting such a site might hit the human TFBS.
In order to address these concerns, a thorough in silico analysis of potential targets of anti-HIV CRISPR-Cas9 therapy was performed. gRNA templates were designed from every possible 20-mer from a collection of patient HIV-1 5′ LTR sequences. The human genome was then scanned for potential off-target effects. To investigate these off-target effects, two questions were asked. First, are some chromosomes more prone to off-target effects than others? Second, are there regions of the LTR that when they are used as a template to design effective anti-HIV-1 gRNA they result in identifiable off-target effects? The answers to these questions should inform future gRNA design and minimize the possible off-target effects of any CRISPR-Cas9 excision therapeutic strategy.
Materials and Methods
In order to account for the worldwide genetic variability of HIV-1, many different subtype sequences and groups were used as queries from the Los Alamos National Laboratory's (LANL) HIV-1 Sequence Database.26 To account for all gRNA design strategies, 5′ LTR sequences were gathered, and all possible 20-mer segments were constructed. It is known that the efficiency of gRNA cleavage decreases in a non-linear manner as the number of gRNA-to-DNA mismatches increases. This is especially true for mismatches that reside near the PAM.27 To account for this, the CRISPRseek tool (v1.16.0)28 was used to examine these 20-mer segments for human homology, cutting frequency domain (CFD) score, and PAM presence to determine where the human genome is most similar to the HIV-1 5′ LTR.
A clipped and unaligned FASTA file containing the 5′ LTR sequences of all available HIV-1 subtypes/groups and HXB2 was downloaded from the LANL HIV-1 Sequence Database on October 19, 2017. This file was acquired from the “Sequence Search Interface” Web page where the “genomic region” section was changed to “5′ LTR.” Five HIV-1 5′ LTR sequences were randomly selected from each subtype/group that had at least five available sequences along with the HXB2 5′ LTR. We specifically chose to have an equal representation of each HIV-1 subtype in order to weight genetic variation more evenly that occurs across all HIV-1-infected individuals. Additionally, the HIV-1 5′ LTR contains highly conserved regions within HIV-1 subtypes. An ideal gRNA would be designed to target these conserved regions for maximum on-target activity and should appear in all HIV-1 5′ LTRs. This file was then used as an input for a multiple sequence alignment using Clustal Omega (v1.2.4).29 Positional values were then mapped from the aligned HXB2 5′ LTR sequence and were ungapped to restore them to their original form.
CRISPRseek was used to search for off-target effects in the human genome given different gRNA protospacers. To assess the similarity between HIV and the human genome, the randomly selected HIV 5′ LTR sequences were split into all possible 20-mer segments, which were processed using the CRISPRseek offTargetAnalysis function where options were used as described for human usage in scenario 5 in the CRISPRseek vignette.30 These parameters were selected for CRISPRseek to search for off-target effects in the human genome, output their location, and classify the location as an intron, exon, or noncoding DNA. The maximum number of gRNA-to-DNA mismatches was set to four, as any four mismatches would result in a CFD score well below established cutoffs. The outputs for these analyses were stored in OffTargetAnalysis files.
From the CRISPRseek outputs, all off-target effects were grouped together by chromosome location. The total number of base pairs that comprised the off-target effects for each chromosome were summed and divided by the total length of the corresponding chromosome to get the fraction of each chromosome that was similar to the HIV-1 5′ LTR. All outputs from the OffTargetAnalysis files were also intersected with a HERV bed file containing all HERV 5′ LTR sequences in the human genome (hg19)31 using BEDTools (v2.26.0).32 The HERV file was downloaded from UCSC genome browser33 and parsed from the repeatMasker track. After intersection, the same process was applied to the HERV 5′ LTRs to generate the fraction of off-target effects that were comprised of HERV LTRs. Calculating these fractions was completed using in house Python scripts utilizing the BioPython (v1.68),34 pandas (v0.20.3),35 regex (v2017.12.12),36 and NumPy (v1.13.3)37 external libraries. These results were saved and plotted in R (v3.4.3)38 using the readr (v1.1.1),39 ggplot2 (v2.2.1),40 reshape2 (v1.4.2),41 and extrafont (v0.17)42 libraries to generate Figure 1. Libraries used to run CRISPRseek include BSgenome.Hsapiens.UCSC.hg19 (v1.4.0),43 TxDb.Hsapiens.UCSC.hg19.knownGene (v3.3.2),44 and org.Hs.eg.db (v3.4.1).45
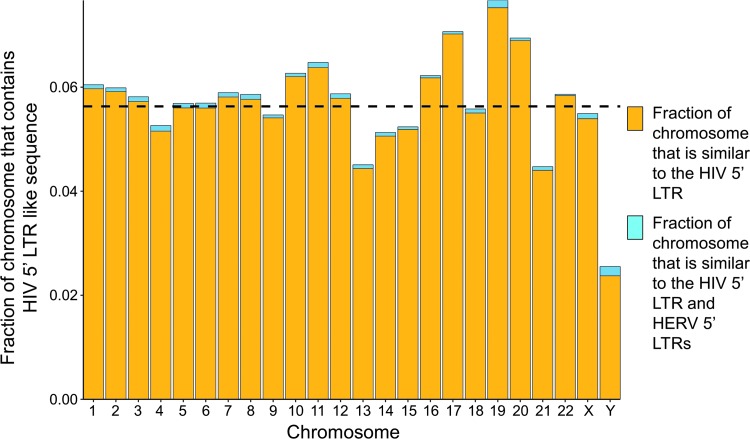
FIG. 1.
Human chromosomes show a low similarity to the human immunodeficiency virus type 1 (HIV-1) 5′ long terminal repeat (LTR) sequence and human endogenous retroviral 5′ LTRs comprise a small fraction of off-target effects. The fraction of each chromosome that shares a high sequence similarity to the HIV-1 5′ LTR is shown. The dashed line indicates the average proportion of sequences similar to the HIV 5′ LTR within each chromosome is 0.0563 (5.63%).
HIV-1 position-specific analysis was performed using the same CRISPRseek output files. Before all CRISPRseek output files were combined, the mapped HXB2 position and HIV subtype/group label were appended to the OffTargetAnalysis files. After concatenation, the file was then modified to aggregate the total number of hits that shared the same name, subtype, and HXB2 position. These were again aggregated for the mean number of hits shared by each subtype and position and compiled into a heat map. To determine the fraction of PAM sequences at each HXB2 position, the number of hits in the concatenated OffTargetAnalysis file was aggregated by unique gRNA sequences and HXB2 positions. A 20-mer was then assigned a PAM status if a “CCN” pattern was found in the first 10 base pairs of the gRNA or a “NGG” pattern in the last 10 base pairs of the gRNA. After assignment, the mean PAM composition was aggregated by HXB2 position. The R libraries used to run the CRISPRseek script were as described in the above paragraph. Additional external R libraries used to generate the heat map were readr, ggplot2, zoo (v1.8-0),46 gridExtra (v2.2.1),47 extrafont, and cowplot (v0.7.0)48 libraries.
These off-target hits were subjected to filters that maintained off-target effects if the hit had a CFD score ≥0.70, a gRNA efficacy ≥0.50, and the gRNA target resided in a region where all corresponding 20-mers had a PAM sequence. The off-target effects were totaled, and the proportion of off-target effects found in exons, introns, and non-coding DNA were calculated. To calculate the proportion of off-target effects residing in promoters, all human promoters were downloaded from the Eukaryotic Promoter Database49 and were defined as up to −2,000 bps from and +50 bps beyond the transcription start site. These were intersected with the off-target file to find the number of off-target effects that resided in promoters. These proportions were then organized into a pie chart. Afterward, the gRNAs that generated these off-target effects were counted and binned based on whether the gRNAs generated one, two, or more than three off-target effects. These numbers were aggregated and subtracted from the total number of unique gRNAs that were subjected to the CRISPRseek pipeline to find the number of gRNAs with no off-target effects. The proportion of gRNAs with off-target effects was calculated and organized in a pie chart.
Results
A total of 732 5′ LTR sequences were downloaded from LANL. However, only subtypes A, B, C, D, F, G, and group O had at least five 5′ LTR sequences in LANL, so these were the only subtypes/groups considered for further analysis. Five 5′ LTRs were randomly selected from these subtypes/groups, yielding a total of 35 5′ LTR sequences used for analysis (Table 1). From these sequences, a total of 21,525 gRNA templates were generated, where 11,959 of those templates were unique.
Table 1.
Summary of 5′ Long Terminal Repeat Sequences Downloaded from Los Alamos National Laboratory by Group and Subtype
| Subtype/group | Number of 5′ LTR sequences downloaded | LANL sample IDs (subtype.country.year.name-accession) |
|---|---|---|
| M | ||
| A | 30 | A1.RW.1993.93RW037A.AB287379 A1.UG.1992.92UG037_A40.AB253429 A1.UG.-.UG275.AB485632 A1.RW.1994.94RW13.AF196742 A1.RW.1992.92RW008.AB253422 |
| B | 245 | B.ES.1989.S61_K1p21.HM469982 B.US.2004.ES10-53.EF363127 B.JP.-.DR1712.AB604947 B.ES.1989.E1_5p11.GQ386793 B.JP.2008.NMC104_clone_01.AB731663 |
| C | 99 | C.KE.1997.97KE46.AF196733 C.ZM.1989.ZAM18.AB485645 C.ZA.1989.pZAC_R3714.JN188292 C.ZA.2007.704010042_CH042.mo6.KC156124 C.MW.2007.703010167_CH167.w8.KC156213 |
| D | 20 | D.UG.1991.UG270.AB485651 D.UG.2007.pSC191727.JX236679 D.CD.2002.LA18ZiAn.KU168272 D.UG.2005.p190049.JX236668 D.CD.1985.Z2Z6_Z2_CDC_Z34.M22639 |
| F | 9 | F1.RO.1996.BCI_R07.AB485659 F1.RO.1996.BCI_R07.AB485658 F1.RO.2003.LA20DuCl.KU168274 F1.RO.1997.97RO203.AF196763 F1.BR.1990.BZ163.AB485657 |
| G | 9 | G.CD.2003.LA23LiEd.KU168277 G.AO.1997.97AN20.AF196748 G.CM.2003.CM44-10.KU168302 G.GH.2003.03GH175G.AB287004 G.-.1993.93CB76.AF196747 |
| O | 27 | O.CM.-.pCMO2_5.AY623602 O.US.-.I_2478B.AB485669 O.FR.2006.LA55RBF206.KU168298 O.FR.1999.LA54BCF120.KU168297 O.FR.1998.LA30RBF125.KU168282 |
Searching for sequences that share common sequence composition within the HIV-1 5′ LTR revealed that only 5.63% of the genome is similar to the HIV-1 LTR (Fig. 1). The distribution of HIV-1-like sequence across each chromosome appears to be stable throughout all chromosomes, with a standard deviation of 1.00%. Intersecting these off-target locations with known HERV LTR locations reveals that HERV 5′ LTRs only account for a small portion of generated off-target effects in this initial analysis. On average, HERV 5′ LTR sequences comprise 0.08% of potential off-target effects from CRISPR-Cas9 gRNAs. Of the off-target effects arising from HERV 5′ LTR sequences, only 8.55% of those effects come from HERV-K 5′ LTR sequences, which corresponds to 0.0068% of total off-target effects.
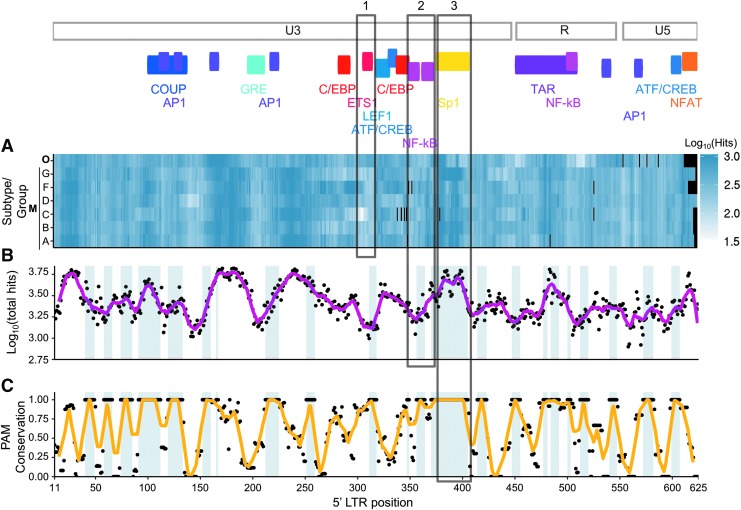
Examining the locations on the HIV-1 LTR of gRNAs for potential off-target effects revealed both subtype-specific as well as region-specific patterns (Fig. 2A and B). For example, gRNAs designed against subtypes B and C have low likelihoods of off-target effects when targeting near ETS1, while gRNAs designed against subtypes D or F would likely have off-target effects (Fig. 2A, box 1). Another type of pattern would be positions that show low homology across all subtypes and groups. For example, the first two AP-1 TFBS, NF-κB, ATF/CREB, GRE, and C/EBP show the lowest amount of human homology across all subtypes and groups (Fig. 2A, less color, and Fig. 2B, low troughs, box 2). The last type of pattern would be regions that show higher proportions of off-target effects across all maintained subtypes and groups (peaks in Fig. 2B) with high conservation of PAM sequences (peaks in Fig. 2C), as observed in the Sp1 and TAR regions (Fig. 2C, box 3). Since PAM sequences are required to achieve cleavage, positions where all sequences at each position containing a PAM sequence (Fig. 2C) are highlighted in the graph.
FIG. 2.
Different HIV subtypes show a distinct yet slightly overlapping pattern of similarity to the human genome. The top of the graph shows a map of the 5′ LTR with selected transcription factor binding site locations. (A) Each heat map tile corresponds to the log10 transform of the average number of homologous human regions of the gRNA. Darker shades of blue indicate a greater number of off-target hits at a particular location. Black tiles indicate that no sequence information is available due to a gap in the reference sequences. (B) Each dot represents the log10 transform of the total number of hits across all subtypes used in (A), with the purple line indicating a 10-sized rolling average window. Blue shading in the background represents areas where all 20-mers for that position contain a PAM sequence. (C) Each dot represents the proportion of 20-mers that contain a PAM sequence. The orange line represents a 10-sized rolling average window. Blue shading in the background represents areas where all 20-mers for that position contain a PAM sequence. Boxes 1, 2, and 3 highlight specific examples presented in the Results section.
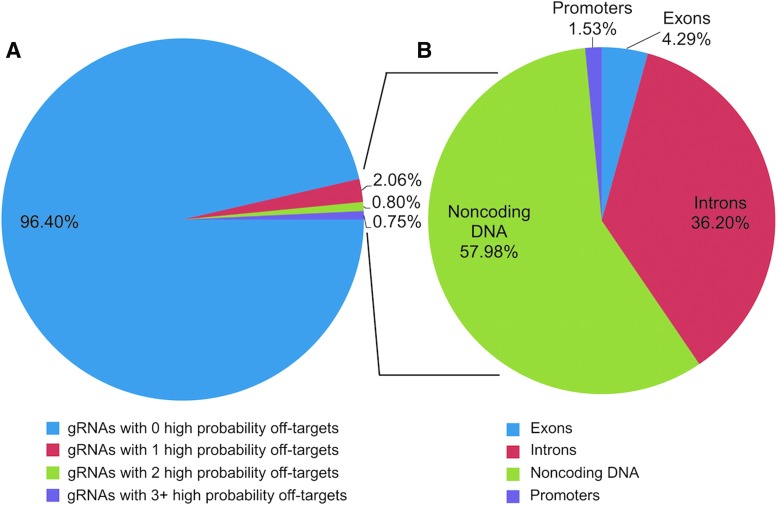
Filtering these CRISPRseek potential off-target hits to maintain only those with a high probability (gRNA efficacy ≥0.50, CFD score ≥0.70, PAM conservation = 1.00) revealed that only 326 of the predicted off-target effects were maintained. These off-target effects originated from 149 unique gRNAs. However, given that 4,114 of the unique gRNAs were considered viable for a CRISPR-Cas9 treatment, this means that 96.40% of the viable gRNAs are predicted to generate no off-target effects (Fig. 3A). Of the 326 off-target effects, only 14 (4.29%) of them were exonic (Fig. 3B). Eight of the off-target effects resided in targeted promoter regions, 189 (57.98%) resided in other non-coding DNA, and 118 (36.20%) off-target hits targeted introns. Due to the chosen promoter size, three of the eight off-target effects that hit promoters also resided in introns (two off-target) or exons (one off-target). A complete list of exonic off-target effects is provided in Table 2. A complete list of off-target effects in promoters is provided in Table 3. A list of all off-target effects can be found in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/crispr).
FIG. 3.
The majority (96.40%) of tested gRNA templates do not cause any off-target effects, and very few exonic off-target effects exist. (A) The total number of gRNAs with predicted off-target cleavage was calculated to be 149 of the 4,114 viable gRNAs that were created, which corresponds to 3.60% of the tested gRNAs. Eighty-five of the gRNAs only have one off-target effect (2.06% of all gRNAs), 33 (0.80%) of the gRNAs have two off-target effects, and 31 of the gRNAs have three or more off-target effects (range 3–24; median = 4). (B) These 149 gRNAs generate 326 off-target effects. Fourteen off-target effects targeted exons (4.29% of all off-targets), 118 (36.20%) targeted introns, and 189 (57.98%) targeted noncoding DNA. Eight of them generated off-target effects in promoter regions, but three of the eight hit within exons or introns. Within the pie chart, those were counted in either exonic or intronic off-target effects, so 5 (1.53%) hit within non-coding DNA.
Table 2.
High Probability Exonic Off-Target Effect Locations from Designed gRNA Templates
| Sample | Position | gRNA template | Gene | UCSC coordinates | CFD score | Mismatches | gRNA efficacy |
|---|---|---|---|---|---|---|---|
| A1- AB287379 | 105 | ACCAGGGCCAGGAATCAGAT | VANGL2 | chr1:160370369-160370391 | 0.758 | 4 | 0.624 |
| G-KU168277 | 375 | GGACTTTCCGGGGAGGCGTG | GLIPR2 | chr9:36144888-36144910 | 0.737 | 4 | 0.520 |
| D- KU168272 | 374 | GGACTTTCCGGGGAGGCGTG | GLIPR2 | chr9:36144888-36144910 | 0.737 | 4 | 0.520 |
| A1-AF196742 | 600 | AGAGATACCTCAGACCAGTG | SVOP | chr12:109366191-109366213 | 0.738 | 4 | 0.644 |
| A1-AF196742 | 103 | ACACCAGGACCAGGGCCCAG | NTSR1 | chr20:61393217-61393239 | 0.867 | 3 | 0.617 |
| C-KC156124 | 112 | CCAGGGGTCAGATTTCCGCT | FAM90A1 | chr12:8374111-8374133 | 0.721 | 3 | 0.914 |
| C-KC156124 | 112 | CCAGGGGTCAGATTTCCGCT | FAM90A25P | chr8:12272282-12272304 | 0.721 | 3 | 0.914 |
| C-KC156124 | 112 | CCAGGGGTCAGATTTCCGCT | FAM90A7P | chr8:7413915-7413937 | 0.721 | 3 | 0.914 |
| C-KC156124 | 112 | CCAGGGGTCAGATTTCCGCT | FAM90A7P | chr8:7421565-7421587 | 0.721 | 3 | 0.914 |
| D-JX236668 | 373 | GGGACTTTCCAGGGAGGAGT | CCDC136 | chr7:128455924-128455946 | 0.867 | 2 | 0.505 |
| O-KU168297 | 157 | GTGCCAGTAACAAAAGAGGA | WDR78 | chr1:67299340-67299362 | 0.714 | 2 | 0.628 |
| B-AB731663 | 544 | TTGCCTTGAGTGCTTTAAAG | CEP41 | chr7:130035972-130035994 | 0.700 | 4 | 0.584 |
| D-M22639 | 374 | GGACTTTCCGGGGAGGCGTG | GLIPR2 | chr9:36144888-36144910 | 0.737 | 4 | 0.520 |
| F1-AB485657 | 382 | CCAGAGGGCGGGCCAGAGGG | LOXL4 | chr10:100022646-100022668 | 0.875 | 4 | 0.551 |
Table 3.
High Probability Off-Target Effects in Promoter Regions from Designed gRNA Templates
| Sample | Position | gRNA template | Promoter | In exon | In intron | Gene | UCSC coordinates | CFD score | Mismatches | gRNA efficacy |
|---|---|---|---|---|---|---|---|---|---|---|
| C-KC156124 | 112 | CCAGGGGTCAGATTTCCGCT | DEFB103B_1 | TRUE | FAM90A7P | chr8:7429210-7429232 | 0.721 | 3 | 0.914 | |
| B-AB731663 | 312 | GCTGTTTCCGGAGTACTACA | ZNF83_1 | chr19:52690031-52690053 | 0.739 | 4 | 0.798 | |||
| O-AB485669 | 383 | AGCGTGGGAGGGACAAGGGG | SLC2A11_3 | chr22:23876264-23876286 | 0.813 | 4 | 0.614 | |||
| D-JX236668 | 373 | GGGACTTTCCAGGGAGGAGT | HILPDA_1 | TRUE | CCDC136 | chr7:128455924-128455946 | 0.867 | 2 | 0.505 | |
| F1-AF196763 | 395 | CAGAGGGCGGGACAAGGGAG | KIAA1967_2 KIAA1967_1 | TRUE | PEBP4 | chr8:22604104-22604126 | 0.913 | 3 | 0.533 | |
| O-KU168297 | 383 | AGCGTGGGAGGGACAAGGGG | SLC2A11_3 | chr22:23876264-23876286 | 0.813 | 4 | 0.614 | |||
| F1-AB485657 | 382 | CCAGAGGGCGGGCCAGAGGG | FBXO2_1 FBXO6_3 FBXO2_2 FBXO44_1 FBXO6_2 FBXO44_2 | chr1:11653942-11653964 | 0.734 | 4 | 0.816 | |||
| B-AB731663 | 607 | CCTCAGACCATTTAAGTCAG | CHP2_1 CHP2_2 | chr16:23754470-23754492 | 0.750 | 3 | 0.566 |
Discussion
Overall, these studies predict that there are few gRNAs that likely exhibit off-target effects from targeting the HIV-1 5′ LTR with gRNAs for CRISPR-Cas9-based gene editing therapy. From this analysis, most off-target sequences have a small, predicted cleavage score when searched across the human genome, implying that the probability of Cas9 cleaving those sites of the human genome is very low. Predicted cleavage scores are generated from overall sequence-to-gRNA homology and mismatch position alone, but there are other factors that contribute to gRNA cleavage ability such as low or high GC content and position-specific base composition.50 When these factors are taken into account, it seems that these gRNAs are predicted to be inefficiently utilized by SpCas9, which results in inefficient cleavage, even when a high sequence-to-gRNA similarity has been identified.
The dissimilarity between HERV 5′ LTRs and HIV 5′ LTRs might be because of accumulated mutations within HERV sequences. Most HERV sequences have lost their ability to guide transcription through random mutations.51 Many years of evolution would make a spectrum of active promoter competent HIV 5′ LTRs substantially different from HERV 5′ LTRs. The cross-activation of HERV-K and HERV-W by HIV-1 Tat is likely explained by sequence homology shorter than the 20-mer homology required by Cas9 systems. While TAR is 58 nucleotides, the bulge in the stem loop structure where Tat binds is <20 nucleotides. Some HERV sequences, such as HERV-K, have been shown to be expressed during pregnancy52 and have been proposed to play a role in the etiology of multiple sclerosis53 and rheumatoid arthritis.54 However, only a very small portion of the off-target activity observed in this study comes from HERV-K.
Predicted exonic off-target effects originate in genes with a myriad of functions. Exonic off-target cleavage for the family with sequence similarity 90, members A7 and A25 (FAM90A7P and FAM90A7P, respectively), are pseudogenes and are likely not to be deleterious if altered. However, Lysyl oxidase-like 4 (LOXL4) and coiled-coil domain-containing protein 136 (CCDC136) are potential tumor suppressor genes,55,56 and glioma pathogenesis-related protein 2 (GLIPR2) has been identified as a potential oncogene.57 gRNAs that are predicted to cleave these genes should not be used. Most promoters that are predicted to be cleaved by anti-HIV-1 gRNAs are from the F-box (FBX) family, which has been shown to participate in ubiquitination.58,59 If left unaccounted for, this could have many potential downstream consequences. However, because these off-target effects are only likely in a small fraction of gRNAs targeting HIV, careful design of anti-HIV-1 gRNAs can easily avoid these issues.
It is important to remember that as quasi-species develop, different regions of the HIV-1 5′ LTR evolve at different rates, which makes some regions more difficult to target than others.60 If a gRNA targets a rapidly evolving genetic region, there is a risk that Cas9 might not cleave every on-target site due to sequence-to-gRNA mismatches. This implies that even if two Cas9 enzymes were to reach their 5′ and 3′ LTR targets simultaneously, one failed cleavage event would prevent excision, and the cleaved site could become an escape mutant, preventing that Cas9 system from targeting the region again with that gRNA.
A limitation of this study is the use of a human reference genome, as it does not reflect the genomic composition of any individual patient. Small differences between individuals can lead to unforeseen off-target effects. Understanding how to account for patient specific single-nucleotide polymorphisms is an open question in the field of in silico off-target searching. Furthermore, it is important to note that while CRISPR-Cas9 scoring matrixes have been shown to correlate with in vitro cleavage events, they do not perfectly predict cleavage events. One proposed reason for this discrepancy is that these matrixes do not account for the chromatin architecture surrounding the cleavage site, which has been shown to hinder CRISPR-Cas9 cleavage efficiency.61,62 The discrepancies between in silico prediction and in vitro behavior demonstrates that in silico predictions must be verified in vitro with unbiased off-target detection methods such as GUIDE-seq63 or CIRCLE-seq,64 which may have to be performed on an patient basis to rule out individual-specific off-target effects.
Conclusions
The results from this research bode well for the potential of a CRISPR-Cas9-mediated HIV-1 therapeutic strategy. With few predicted worrisome gRNA templates, researchers are free to target most sections of the LTR that they propose would be advantageous for CRISPR-Cas9 gene therapy. However, it is important to remember that these off-target predictions were performed in silico and with a reference genome. In the search for a gRNA template that causes minimal off-target effects, it will be critical that the gRNA be tested against patient samples to confirm these in silico predictions.
Supplementary Material
Acknowledgments
These studies were funded in part by the Public Health Service, National Institutes of Health, through grants from the National Institute of Mental Health (NIMH) R01 MH110360 (Contact Principal Investigator Brian Wigdahl), the NIMH Comprehensive NeuroAIDS Center (CNAC) P30 MH092177 (Principal Investigator Kamel Khalili; Brian Wigdahl, Principal Investigator of the Drexel subcontract involving the Clinical and Translational Research Support Core; Will Dampier, Principal Investigator of a developmental grant titled “Functional Evaluation of HIV excision therapy in CNS derived cell lines”), and under the Ruth L. Kirschstein National Research Service Award T32 MH079785 (Brian Wigdahl, Principal Investigator of the Drexel University College of Medicine component and Dr. Olimpia Meucci as Co-Director). The contents of the paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Author Disclosure Statement
The authors declare they have no competing financial interests.
Clustered Regularly Interspaced Short Palindromic Repeats.
References
- 1.Centers for Disease Control and Prevention. Living with HIV. Available online at https://www.cdc.gov/hiv/basics/livingwithhiv/index.html (accessed August4, 2018)
- 2.HIV.gov. HIV treatment overview Available online at https://www.hiv.gov/hiv-basics/staying-in-hiv-care/hiv-treatment/hiv-treatment-overview (accessed August4, 2018)
- 3.Delelis O, Carayon K, Saïb A, et al. Integrase and integration: biochemical activities of HIV-1 integrase. Retrovirology 2008;5:11–4.. DOI: 10.1186/1742-4690-5-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loregian A, Bortolozzo K, Boso S, et al. Interaction of Sp1 transcription factor with HIV-1 Tat protein: looking for cellular partners. FEBS Lett 2003;543:61–65. DOI: 10.1016/S0014-5793(03)00399-5 [DOI] [PubMed] [Google Scholar]
- 5.Kretzschmar M, Meisterernst M, Scheidereit C, et al. Transcriptional regulation of the HIV-1 promoter by NF-kappa B in vitro. Genes Dev 1992;6:761–774. DOI: 10.1101/gad.6.5.761 [DOI] [PubMed] [Google Scholar]
- 6.Raha T, Cheng SWG, Green MR. HIV-1 Tat stimulates transcription complex assembly through recruitment of TBP in the absence of TAFs. PLoS Biol 2005;3:e4–4.. DOI: 10.1371/journal.pbio.0030044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, Nonnemacher MR, Alexaki A, et al. Functional studies of CCAAT/enhancer binding protein site located downstream of the transcriptional start site. Clin Med Insights Pathol 2017;10:11795557176945–5.. DOI: 10.1177/1179555717694556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahiya S, Liu Y, Nonnemacher MR, et al. CCAAT enhancer binding protein and nuclear factor of activated T cells regulate HIV-1 LTR via a novel conserved downstream site in cells of the monocyte-macrophage lineage. PLoS One 2014;9:e8811–6.. DOI: 10.1371/journal.pone.0088116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burdo TH, Nonnemacher M, Irish BP, et al. High-affinity interaction between HIV-1 Vpr and specific sequences that span the C/EBP and adjacent NF-κB sites within the HIV-1 LTR correlate with HIV-1-associated dementia. DNA Cell Biol 2004;23:261–269. DOI: 10.1089/104454904773819842 [DOI] [PubMed] [Google Scholar]
- 10.Nonnemacher MR, Irish BP, Liu Y, et al. Specific sequence configurations of HIV-1 LTR G/C box array result in altered recruitment of Sp isoforms and correlate with disease progression. J Neuroimmunol 2004;157:39–47. DOI: 10.1016/j.jneuroim.2004.08.021 [DOI] [PubMed] [Google Scholar]
- 11.Shah S, Alexaki A, Pirrone V, et al. Functional properties of the HIV-1 long terminal repeat containing single-nucleotide polymorphisms in Sp site III and CCAAT/enhancer binding protein site I. Virol J 2014;11:9–2.. DOI: 10.1186/1743-422X-11-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W, Ye C, Liu J, et al. CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS One 2014;9:e11598–7.. DOI: 10.1371/journal.pone.0115987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu L, Yang H, Gao Y, et al. CRISPR/Cas9-mediated CCR5 ablation in human hematopoietic stem/progenitor cells confers HIV-1 resistance in vivo. Mol Ther 2017;25:1782–1789. DOI: 10.1016/j.ymthe.2017.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu S, Yao Y, Xiao H, et al. Simultaneous knockout of CXCR4 and CCR5 genes in CD4+ T cells via CRISPR/Cas9 confers resistance to both X4- and R5-tropic human immunodeficiency virus type 1 infection. Hum Gene Ther 2018;29:51–67. DOI: 10.1089/hum.2017.032 [DOI] [PubMed] [Google Scholar]
- 15.Hou P, Chen S, Wang S, et al. Genome editing of CXCR4 by CRISPR/cas9 confers cells resistant to HIV-1 infection. Sci Rep 2015;5:1557–7.. DOI: 10.1038/srep15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ueda S, Ebina H, Kanemura Y, et al. Anti-HIV-1 potency of the CRISPR/Cas9 system insufficient to fully inhibit viral replication. Microbiol Immunol 2016;60:483–496. DOI: 10.1111/1348-0421.12395 [DOI] [PubMed] [Google Scholar]
- 17.Wang Z, Pan Q, Gendron P, et al. CRISPR/Cas9-derived mutations both inhibit HIV-1 replication and accelerate viral escape. Cell Rep 2016;15:481–489. DOI: 10.1016/j.celrep.2016.03.042 [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Yin C, Zhang T, et al. CRISPR/gRNA-directed synergistic activation mediator (SAM) induces specific, persistent and robust reactivation of the HIV-1 latent reservoirs. Sci Rep 2015;5:1–14. DOI: 10.1038/srep16277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saayman SM, Lazar DC, Scott TA, et al. Potent and targeted activation of latent HIV-1 using the CRISPR/dCas9 activator complex. Mol Ther 2016;24:488–498. DOI: 10.1038/mt.2015.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaminski R, Chen Y, Fischer T, et al. Elimination of HIV-1 genomes from human T-lymphoid cells by CRISPR/Cas9 gene editing. Sci Rep 2016;6:2255–5.. DOI: 10.1038/srep22555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaminski R, Bella R, Yin C, et al. Excision of HIV-1 DNA by gene editing: a proof-of-concept in vivo study. Gene Ther 2016;23:690–695. DOI: 10.1038/gt.2016.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dampier W, Sullivan NT, Chung C-H, et al. Designing broad-spectrum anti-HIV-1 gRNAs to target patient-derived variants. Sci Rep 2017;7:1441–3.. DOI: 10.1038/s41598-017-12612-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dampier W, Sullivan NT, Mell J, et al. Broad spectrum and personalized gRNAs for CRISPR/Cas9 HIV-1 therapeutics. AIDS Res Hum Retroviruses 2018;AID.2017.0274. DOI: 10.1089/AID.2017.0274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uleri E, Mei A, Mameli G, et al. HIV Tat acts on endogenous retroviruses of the W family and this occurs via Toll-like receptor 4. AIDS 2014;28:2659–2670. DOI: 10.1097/QAD.0000000000000477 [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez-Hernandez MJ, Cavalcoli JD, Sartor MA, et al. Regulation of the human endogenous retrovirus K (HML-2) transcriptome by the HIV-1 Tat protein. J Virol 2014;88:8924–8935. DOI: 10.1128/JVI.00556-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leitner T, Hahn B, Mullins J, et al. (eds) HIV Sequence Compendium. Los Alamos, NM: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, 2017. [Google Scholar]
- 27.Hsu PD, Scott DA, Weinstein JA, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 2013;31:827–832. DOI: 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu LJ, Holmes BR, Aronin N, et al. CRISPRseek: a bioconductor package to identify target-specific guide RNAs for CRISPR-Cas9 genome-editing systems. PLoS One 2014;9:e10842–4.. DOI: 10.1371/journal.pone.0108424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011;7:53–9.. DOI: 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu LJ, Brodsky M. CRISPRseek user's guide. 2017. Available online at https://www.bioconductor.org/packages/devel/bioc/vignettes/CRISPRseek/inst/doc/CRISPRseek.pdf (accessed August4, 2018)
- 31.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature 2001;409:860–921. DOI: 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- 32.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010;26:841–2. DOI: 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Afgan E, Baker D, van den Beek M, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 2016;44:W3–W10. DOI: 10.1093/nar/gkw343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cock PJA, Antao T, Chang JT, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009;25:1422–1423. DOI: 10.1093/bioinformatics/btp163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKinney W. Data structures for statistical computing in Python. Proceedings of the 9th Python in Science Conference 2010:51–56. Available online at http://conference.scipy.org/proceedings/scipy2010/pdfs/mckinney.pdf (accessed August4, 2018) [Google Scholar]
- 36.Barnett M. regex Available online at https://bitbucket.org/mrabarnett/mrab-regex (accessed August4, 2018)
- 37.van der Walt S, Colbert SC, Varoquaux G. The NumPy array: a structure for efficient numerical computation. Comput Sci Eng 2011;13:22–30. DOI: 10.1109/MCSE.2011.37 [DOI] [Google Scholar]
- 38.Team RC. R: a language and environment for statistical computing. Available online at https://www.r-project.org/ (accessed August4, 2018)
- 39.Wickham H, Hester J, Francois R. readr: read rectangular text data. Available online at https://cran.r-project.org/package=readr (accessed August4, 2018)
- 40.Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag, 2009. [Google Scholar]
- 41.Taylor D. Geometrical effects in fatigue: a unifying theoretical model. Int J Fatigue 1999;21:413–420. DOI: 10.1016/S0142-1123(99)00007-9 [DOI] [Google Scholar]
- 42.Chang W. extrafont: Tools for using fonts Available online at https://cran.r-project.org/package=extrafont (accessed August4, 2018)
- 43.The Bioconductor Dev Team. BSgenome.Hsapiens.UCSC.hg19: Full genome sequences for Homo sapiens (UCSC version hg19). DOI: 10.18129/B9.bioc.BSgenome.Hsapiens.UCSC.hg19 (accessed August4, 2018) [DOI]
- 44.Carlson M. and Bioconductor Package Maintainer. TxDb.Hsapiens.UCSC.hg19.knownGene: Annotation package for TxDb object(s). DOI: 10.18129/B9.bioc.TxDb.Hsapiens.UCSC.hg19.knownGene [DOI]
- 45.Carlson M. org.Hs.eg.db: Genome wide annotation for Human. DOI: 10.18129/B9.bioc.org.Hs.eg.db [DOI]
- 46.Zeileis A, Grothendieck G. zoo: S3 infrastructure for regular and irregular time series. J Stat Softw 2005;14:1–30. DOI: 10.18637/jss.v014.i06 [DOI] [Google Scholar]
- 47.Auguie B. gridExtra: Miscellaneous Functions for “Grid” Graphics Available online at https://cran.r-project.org/package=gridExtra (accessed August4, 2018)
- 48.Wilke CO. cowplot: streamlined plot theme and plot annotations for “ggplot2.” Available online at https://cran.r-project.org/package=cowplot (accessed August4, 2018)
- 49.Périer RC, Praz V, Junier T, et al. The eukaryotic promoter database (EPD). Nucleic Acids Res 2000;28:302–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doench JG, Hartenian E, Graham DB, et al. Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat Biotechnol 2014;32:1262–1267. DOI: 10.1038/nbt.3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cohen CJ, Lock WM, Mager DL. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene 2009;448:105–114. DOI: 10.1016/j.gene.2009.06.020 [DOI] [PubMed] [Google Scholar]
- 52.Kämmerer U, Germeyer A, Stengel S, et al. Human endogenous retrovirus K (HERV-K) is expressed in villous and extravillous cytotrophoblast cells of the human placenta. J Reprod Immunol 2011;91:1–8. DOI: 10.1016/j.jri.2011.06.102 [DOI] [PubMed] [Google Scholar]
- 53.Tselis A. Evidence for viral etiology of multiple sclerosis. Semin Neurol 2011;31:307–316. DOI: 10.1055/s-0031-1287656 [DOI] [PubMed] [Google Scholar]
- 54.Freimanis G, Hooley P, Ejtehadi HD, et al. A role for human endogenous retrovirus-K (HML-2) in rheumatoid arthritis: investigating mechanisms of pathogenesis. Clin Exp Immunol 2010;160:340–347. DOI: 10.1111/j.1365-2249.2010.04110.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Asuncion L, Fogelgren B, Fong KSK, et al. A novel human lysyl oxidase-like gene (LOXL4) on chromosome 10q24 has an altered scavenger receptor cysteine rich domain. Matrix Biol 2001;20:487–491. DOI: 10.1016/S0945-053X(01)00161-5 [DOI] [PubMed] [Google Scholar]
- 56.Jiang N, Zhan F, Tan G, et al. A cDNA located on chromosome 7q32 shows loss of expression in epithelial cell line of nasopharyngeal carcinoma. Chin Med J (Engl) 2000;113:650–653 [PubMed] [Google Scholar]
- 57.Huang S, Zhang L, Niu Q, et al. Hypoxia promotes epithelial–mesenchymal transition of hepatocellular carcinoma cells via inducing GLIPR-2 expression. PLoS One 2013;8:e7749–7.. DOI: 10.1371/journal.pone.0077497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kipreos ET, Pagano M. The F-box protein family. Genome Biol 2000;1:reviews300–2..1. DOI: 10.1186/gb-2000-1-5-reviews3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Craig KL, Tyers M. The F-box: a new motif for ubiquitin dependent proteolysis in cell cycle regulation and signal transduction. Prog Biophys Mol Biol 1999;72:299–328. DOI: 10.1016/S0079-6107(99)00010-3 [DOI] [PubMed] [Google Scholar]
- 60.Dampier W, Nonnemacher MR, Mell J, et al. HIV-1 genetic variation resulting in the development of new quasispecies continues to be encountered in the peripheral blood of well-suppressed patients. PLoS One 2016;11:e015538–2.. DOI: 10.1371/journal.pone.0155382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Daer RM, Cutts JP, Brafman DA, et al. The impact of chromatin dynamics on Cas9-mediated genome editing in human cells. ACS Synth Biol 2017;6:428–438. DOI: 10.1021/acssynbio.5b00299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uusi-Mäkelä MIE, Barker HR, Bäuerlein CA, et al. Chromatin accessibility is associated with CRISPR-Cas9 efficiency in the zebrafish (Danio rerio). PLoS One 2018;13:e019623–8.. DOI: 10.1371/journal.pone.0196238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsai SQ, Zheng Z, Nguyen NT, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 2015;33:187–197. DOI: 10.1038/nbt.3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsai SQ, Nguyen NT, Malagon-Lopez J, et al. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR–Cas9 nuclease off-targets. Nat Methods 2017;14:607–614. DOI: 10.1038/nmeth.4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.