Abstract
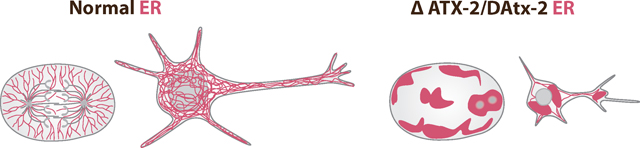
Ataxin-2, a conserved RNA-binding protein, is implicated in the late-onset neurodegenerative disease Spinocerebellar ataxia type-2 (SCA2). SCA2 is characterized by shrunken dendritic arbors and torpedo-like axons within the Purkinje neurons of the cerebellum. Torpedo-like axons have been described to contain displaced Endoplasmic Reticulum (ER) in the periphery of the cell; however, the role of Ataxin-2 in mediating ER function in SCA2 is unclear. We utilized the C. elegans and Drosophila homologs of Ataxin-2 (ATX-2 and DAtx2, respectively) to determine the role of Ataxin-2 in ER function and dynamics in embryos and neurons. Loss of ATX-2 and DAtx2 resulted in collapse of the ER in dividing embryonic cells and germline, and ultrastructure analysis revealed unique spherical stacks of ER in mature oocytes and fragmented and truncated ER tubules in the embryo. ATX-2 and DAtx2 reside in puncta adjacent to the ER in both C. elegans and Drosophila embryos. Lastly, depletion of DAtx2 in cultured Drosophila neurons recapitulated the shrunken dendritic arbor phenotype of SCA2. ER morphology and dynamics were severely disrupted in these neurons. Taken together, we provide evidence that Ataxin-2 plays an evolutionary conserved role in ER dynamics and morphology in C. elegans and Drosophila embryos during development and in fly neurons, suggesting a possible SCA2 disease mechanism.
Keywords: Traffic, Intracellular Transport, Ataxin-2, ATX-2, DAtx-2, mitosis, cytokinesis, neurons, Endoplasmic Reticulum (ER), RNA, RBPs
Graphical Abstract
Introduction
The endoplasmic reticulum (ER) is a continuous network of membrane-bound cisternae and tubules that extends from the nuclear envelope into the cytoplasm. The ER is a dynamic organelle and is the site for regulating various cellular processes, including intracellular calcium levels, protein and lipid synthesis, protein sorting, and cell division (reviewed in references1–3). During the cell cycle, the ER oscillates between a dispersed, tubule state in interphase and a structured, reticulate, sheet state during mitosis 4–6. Additionally, the ER contacts the plasma membrane and other organelles to facilitate its cellular function. Disrupted ER structure and ER stress have been implicated in several types of diseases including cancer and neurological disorders7–10. For example, neoplastic progression and ER stress appear to correlate with reduced protein synthesis in prostate tumors10. In several neurodegenerative diseases, including Parkinson’s disease, ER stress has been suggested to lead to loss of dopaminergic neurons11. In lysosomal storage disorders, such as Niemann-Pick type C disease and Gaucher disease, two thirds of the patients have neurological symptoms and defects in ER and lysosomal function have been proposed to play a direct role in defects in autophagy that are observed12. ER stress markers also appear to increase dramatically over the course of the disease progression in Type 1 Diabetes13. Although many reports have suggested defects in ER function and stress in these diseases, very few groups have characterized the role of the direct disease gene targets with regard to ER dynamics.
While the mechanisms underlying the cell and tissue abnormalities observed in neurogenerative disease are complex, recent work suggests that ER disruption could lead to neuron loss14. To look at this more closely, we sought to determine if Ataxin-2, associated with Spinocerebellar ataxia type-2 (SCA2)15, might function to regulate the ER. Ataxin-2 was initially discovered to be associated with the ER, and the Drosophila homolog, DAtx2, binds to polyribosomes16,17. While polyglutamine expansion models of Ataxin-2 have demonstrated a disruption in the association of Ataxin-2 with the ER, it is unclear how Ataxin-2 modulates ER morphology in the context of cell division and neurobiology.
Here, we characterized the temporal and spatial dynamics of the ER in embryos, germlines, and in cultured neurons in C. elegans and Drosophila. Our data demonstrate that there is a unique disruption of ER morphology during the cell cycle and in the germline. Additionally, we have recapitulated the shrunken dendritic arbor phenotype of SCA2 patients in cultured Drosophila neurons and have shown that the ER dynamics and morphology are disrupted. Taken together, we suggest that proper regulation of ER morphology by the RNA Binding Protein, Ataxin-2 may be critical for neuronal function and maintenance. Lastly, we have uncovered a link between cell division and neurobiology and provide a link between defects in cell division and neurodegenerative disease. Our findings suggest that modulating ER function may provide a novel approach to improving neurological function in neurodegenerative disease patients.
Results and Discussion
Depletion of ATX-2 disrupts ER morphology and dynamics in C. elegans.
In human tissue culture cells, Ataxin-2 localizes to discrete puncta throughout the cytoplasm and shows a preferential association with RER/membrane-bound ribosomes17, but the role of Ataxin-2 in controlling the morphology and dynamics of the ER during cell division are unknown. To determine the role of ATX-2 regulation ER morphology and dynamics, we used a strain expressing the ER marker SP12-GFP, an ER localized signal peptidase4, and imaged the first cell division. In control embryos, the ER oscillates between a dispersed, tubule state in interphase (Fig. 1A;−2:40min) and a structured, reticulate, sheet state during mitosis (Fig. 1A;6:40min; Movie 1)4,18,19. In atx-2 fRNAi-treated embryos, the dispersed ER tubules were absent (Fig. 1A;−4:20min) and sheets persisted as large aggregates of ER throughout the cell cycle, resulting in collapsed ER morphology. In particular, the ER aggregated in large dynamic clusters around the spindle and at the cortex, particularly in the anterior cortex (Fig. 1A; Movie 3). Additionally, this unique ER collapse phenotype was also observed in the C. elegans germline (Fig. 1B; Weak penetrance 16/40 germlines; intermediate penetrance 20/40 germlines, and strong penetrance 4/40 germlines imaged). This ER phenotype was unique to ATX-2 depletion, as car-1, another RBP with described roles in cell division and ER morphology20, only showed a loss of anaphase spindle-associated ER vesicles and aggregates along the anterior cortex of the embryo (Fig. 1A; Movie 2), similar to previous data20. Furthermore, this distinct ER collapse phenotype in atx-2 fRNAi depleted-embryos may be specific to RNA-binding proteins as depletion of ZEN-4, a kinesin required for cell division, did not result in ER collapse (Fig. 1A; Movie 4). To confirm that this ER collapse phenotype was specific to ATX-2 or RBPs, we tested another RBP, PAB-1/PolyA Binding Protein (a known ATX-2 binding partner21). We found that pab-1 fRNAi-treated embryos also exhibited ER morphology defects similar but less severe than the phenotype observed in atx-2 fRNAi depleted-embryos, where numerous ER tubules were still present (Fig. 1A; Movie 5), yet distinct from defects observed in car-1 embryos. Furthermore, in pab-1 fRNAi-treated embryos the ER localized along the furrow and in the cytoplasm in long sheets emanating from the cortex (Fig. 1A; 15:20min), a phenotype we did not observe for any of the other RBPs tested.
Figure 1: ATX-2 depletion leads to a dramatic collapse of the ER in C. elegans embryos.
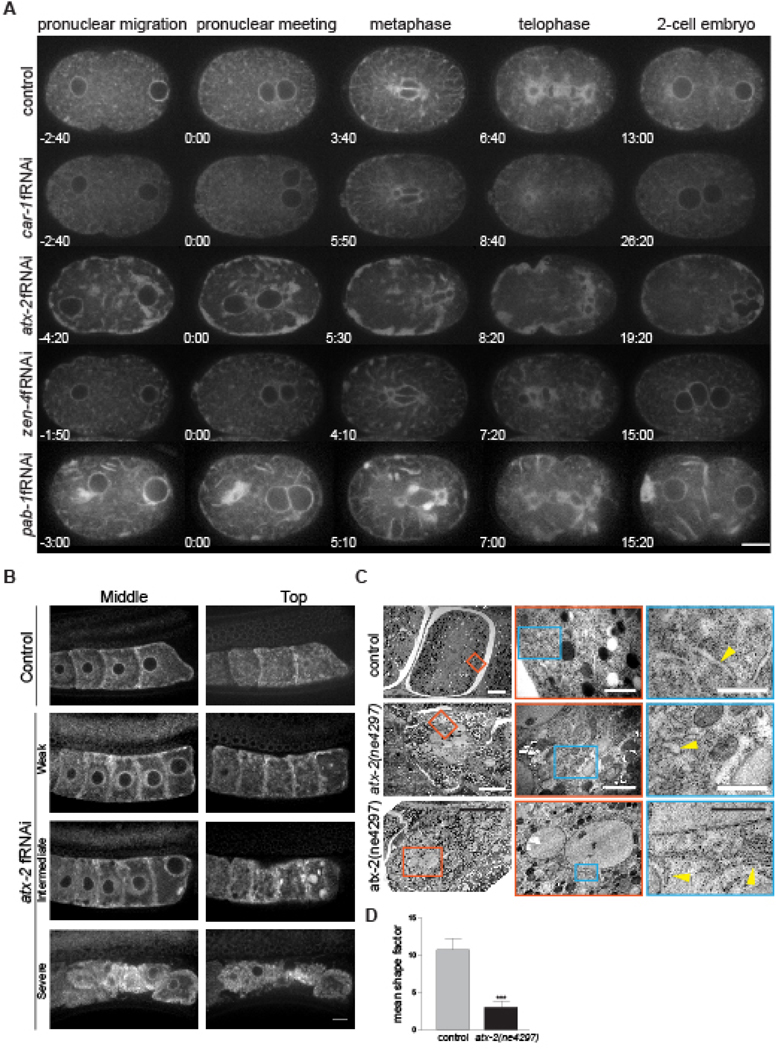
(A) ER dynamics in C. elegans in control, car-1 fRNAi-, atx-2 fRNAi-, zen-4 fRNAi-, and pab-1 fRNAi-treated embryos expressing the ER marker SP12-GFP. In control embryos, the ER oscillates between a tubule state in interphase (−2:40) and sheet state in mitosis (6:40). car-1 fRNAi-treated embryos show a mild phenotype with ER aggregates near the anterior cortex (8:40). In atx-2 fRNAi-treated embryos the ER lacks tubules (−4:20) and the sheets are aggregated near the cortex and collapsed throughout mitosis (8:20). zen-4 fRNAi-treated embryos exhibit a phenotype similar to control embryos with very mild ER aggregation in the anterior. The ER in pab-1 fRNAi-treated embryos is collapsed in the embryo anterior (0:00), and the ER localizes in bands along the furrow and in the cell anterior (15:20). par-5 fRNAi-treated embryos exhibit an intermediate phenotype between zen-4 fRNAi-treated and atx-2 fRNAi-treated embryos, with ER tubules maintained in the cytoplasm (0:00). ER aggregates along anterior cortex (2:40), and ER accumulation at the spindle midzone (6:00). Times, in min:sec, are given relative to pronuclear meeting. Scale bar, 10 μm. (B) Germline of worms expressing SP12:GFP treated with control or atx-2 fRNAi. Left and right represent plane near the center and top of oocyte, respectively. In wild-type, the ER is evenly distributed throughout the cytoplasm as an interconnected network while increased penetrance of atx-2 fRNAi is associated with abnormal clustering and cortical distribution of ER. Weak penetrance represents 16/40 germlines, intermediate penetrance represents 20/40 germlines, and strong penetrance represents 4/40 germlines imaged. Scale bar, 10μm. (C) Transmission electron microscopy images of control (him-5(e1490)) and two atx-2(ne4297) C. elegans embryos. The first column is a view of one-cell stage in the worm uterus, scale bar 5μm. Column two is a magnified view of the red box from column one, scale bar 1μm. Column three is a magnified view of the blue box from column two, scale bar 0.5μm. In control embryos, the ER is organized into long tubules and cisternae (yellow arrowhead). In ATX-2 depleted embryos the ER appears as shortened, collapsed tubules (yellow arrowhead) (middle panels) and/or with evidence of stacked lamellae (bottom panels) similar to much larger ER stacks found in primary oocytes as in Figure S1. (D) Quantification of ER tubule shape factor in control and ATX-2 depleted embryos. Shape factor is determined by dividing the tubule length by tubule width. Results are the mean ± standard error mean (SEM). Asterisks indicate statistical significance for compared data (***p-value<0.001).
To further assess the ER morphology in ATX-2 depleted embryos and germlines, we utilized high resolution, transmission electron microscopy (TEM). Here, whole worm sections of control (him-5(e1490) hermaphrodites) and atx-2(ne4297) worms at the non-permissive temperature (24°C) were imaged, and the germline and the embryos within the uterus were analyzed. In control one-cell embryos, the rough ER is organized into long cisternae that extend throughout the cytoplasm (yellow arrowhead). These cisternae have an average shape factor of 10.74 ± 1.51, measured by dividing the cisternae length by the cisternae width (Fig. 1C,D). In two ATX-2 depleted embryos, the ER appeared as either spherical or tubular bodies (yellow arrowhead, middle panel), rather than as cisternae, and have an average shape factor of 3.06 ± 0.75 (Fig. 1C,D). This fragmented ER is most clearly seen near the nucleus and the cell cortex (periphery) and is indicative of collapsed ER. In the late germline, ultrastructure analysis revealed unique spherical stacks of ER in mature oocytes (Fig. S1, C-D). Additionally, defects in lipid droplets and mitochondria were observed in the atx-2(ne4297) oocytes (Fig. S1). In control oocytes, lipid droplets are dispersed throughout the oocyte and mitochondria appear as round or tubular shapes throughout the oocyte. In atx-2(ne4297) oocytes, the lipid droplets are smaller and more abundant, and the mitochondria appear larger and more elongated (Fig. S1). Given this unique ER morphology observed with ATX-2 depletion in both SP12-GFP embryos and the germline, we suggest that ATX-2 plays a necessary role in the maintenance of ER morphology and dynamics during development. Of the RBPs tested and found in the literature, depletion of ATX-2 resulted in the most severe ER collapse phenotype to date, suggesting that ATX-2 plays a unique role in maintaining proper ER morphology and dynamics.
The role for Ataxin-2/DAtx2 in mediating ER morphology and dynamics is conserved in Drosophila embryos
DAtx2 was identified as the Drosophila homolog of human Ataxin-2 and C. elegans ATX-222. Here, we sought to examine if DAtx2 functioned similarly to C. elegans ATX-2 in maintaining the morphology of the ER. We injected double stranded RNA (dsRNA) corresponding to DAtx2 into the early cortical syncytial cycles of Drosophila embryos co-expressing the ER marker, Pdi-GFP and the DNA marker, H2Av-RFP5. These embryos were imaged using time-lapse confocal microscopy during mitosis of nuclear cycle 12 (Fig. 2). During mitosis, the ER displayed dramatic reorganization and remodeling events. In metaphase the ER localized to the perispindle region and accumulated at the poles 5,23, similar to C. elegans embryos (Fig. 1)4,18,19. Knockdown of DAtx2 displayed several defects in ER organization, including ectopic formation of ER aggregates (Fig. 2, bottom-white arrowheads; Movie 6) and a disruption of ER alignment along the perispindle region in metaphase. Furthermore, depletion of DAtx2 induced defects in nuclear spacing and collision between neighboring nuclei, resulting in a nuclear fallout phenotype (Fig. 2, white arrow; Movie 7). These disruptions in nuclear dynamics in the early cortical syncytial divisions have been demonstrated as indicators of defects in the cytokinetic machinery 24,25. These results suggest that, similar to C. elegans ATX-2, DAtx2 is necessary for proper ER organization and dynamics, and cytokinesis completion (Fig. 2, yellow arrowhead)26, and demonstrates that the Ataxin-2 role is conserved.
Figure 2: DAtx2 depletion leads to a dramatic collapse of the ER in Drosophila embryos.
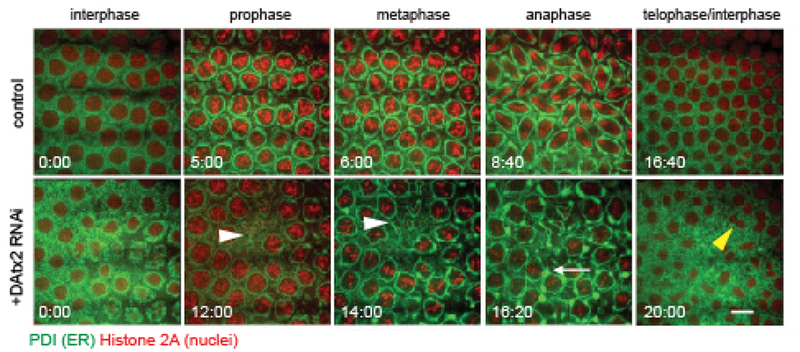
ER dynamics in Drosophila in control injected and DAtx2 RNAi injected cycle 12 embryos co-expressing the ER marker PDI-GFP (green) and the histone marker H2-RFP (red). Embryos injected with DAtx2 dsRNA displayed defects in ER reorganization and nuclear dynamics during mitosis. These defects include formation of ectopic ER aggregates (white arrows) and nuclear fallout and collisions (white arrowheads). Scale bar, 5 μm.
ATX-2 and DAtx2 localizes adjacent to ER structures in early C. elegans and Drosophila embryos.
It has been shown that a fraction of Ataxin-2 associates with the rough ER in HeLa cells and rat primary neurons17. Given that ATX-2 depletion resulted in collapsed ER morphology in C. elegans and Drosophila, we wanted to determine if ATX-2 and DAtx2 shared a similar localization pattern. To do this in C. elegans, we immuno-stained embryos expressing the ER marker SP12-GFP with anti-ATX-2 (magenta) and anti-GFP (cyan) antibodies (Fig. S2, A-C). ATX-2 localized adjacent to the ER puncta during mitosis and adjacent to cortical ER structures (Fig. S2A-C; inset 1) in the embryo. Co-localization between ATX-2 and the ER in the embryo is observed by overlapping (white) fluorescence in the merge images (Fig. S2A-C) which occurred at a few cortical and cytoplasmic regions. Given that there are only some regions where ATX-2 and the ER co-localize during mitosis suggests a possible additional function for ATX-2 in an adjacent compartment. In Drosophila, we labeled stage 10 embryos with anti-DAtx2 antibodies and anti-Rtnl1 (ER) and discovered very similar localization pattern, where DAtx2 is found in a distinct yet adjacent localization to the ER (Fig. S2, D)(N=7 at cycle 12). These results might not be incompatible with previous observed data by immunofluorescence and co-pelleting techniques in mammalian systems. First, the ER network covers a large fraction of the cytoplasm, especially in smaller tissue culture cells making it difficult to resolve by light microscopy whether colocalization is due to a specific interaction or just stochastic colocalization. Second, co-pelleting experiments do not answer if enrichment of a protein in a specific fraction is due to a specific association or not. Our data show that there is not a clear co-localization between Atx2 and the ER in both C. elegans and Drosophila embryos. One possibility given recent literature26, and our results favor that ATX-2/DAtx2 could be associated with the newly discovered TIGER domain (TIS granule and the ER), which is enriched in membrane protein encoding mRNAs, for which RBPs might be also found26(unpublished work, Skop Lab). Given that the loss of ATX-2/DAtx2 in both species leads to distinct ER defects, we hypothesize that this RBP may play a unique and conserved role in the binding of a subset of ER-specific membrane protein transcripts critical for proper ER morphology and dynamics in a compartment distinct from the ER itself.
ATX-2 affects Golgi puncta size in C, elegans
Ataxin-2 has been shown to localize to the Golgi in tissue culture cells, and mutant forms of Ataxin-2 containing a polyglutamine expansion have resulted in disrupted Golgi morphology27,28, but this function remains unclear. To determine if ATX-2 is important for Golgi morphology during C. elegans embryonic development, we utilized GFP-UGTP-1, a nucleotide sugar transporter and Golgi marker29. In control embryos, GFP-UGTP-1 labels dynamic cytoplasmic puncta throughout mitosis (Fig. S3A). To quantify GFP-UGTP-1 puncta size, a region of interest (ROI) was measured in the anterior (blue box) and posterior (orange box) of furrow initiation staged embryos (control: n=8; atx-2 fRNAi: n=10; Fig. S3B). In control embryos, the GFP-UGTP-1 cytoplasmic puncta were on average 0.2 μm2 in size in the anterior and 0.1 μm2 in posterior. In atx-2 fRNAi-treated embryos, GFP-UGTP-1 puncta were larger in size (green arrowhead; Fig. S3A). The GFP-UGTP-1 puncta in the anterior of the embryo averaged 0.6 μm2 in size, and the puncta in the posterior of the embryo averaged 0.5 μm2 in size (Fig. S3B). Taken together, these data suggest that ATX-2 may regulate Golgi stack size or the clustering of Golgi stacks; however, a stronger ER morphology phenotype is observed.
Depletion of DAtx2 affects ER distribution in neurons
Defects in ATX-2 function have been linked to the neurodegenerative disease SCA215,30,31. Given the role of ATX-2/DAtx2 in early worm and fly embryos, we wanted to investigate whether DAtx2 plays a role in ER morphology and dynamics in Drosophila neurons. To deplete DAtx2 specifically in neurons, we drove a UASp-DAtx2 shRNA by a pan-neuronal driver elav>Gal432. By driving DAtx2 dsRNA by elav>Gal4, we depleted DAtx2 post-mitotically in all neurons starting at embryonic stage 11, without affecting its levels in other tissues. Depletion of DAtx2 in neurons did not induce obvious defects in first or second instar larvae, but elav>DAtx2-RNAi third instar larvae appeared sluggish. Furthermore, depletion of DAtx2 in neurons was lethal at pupae stage. 99% of the control pupae eclosed to adult stage (n=495), but none of DAtx2-RNAi pupae did (n=369). For detailed analysis of neurons after depletion of DAtx2, we prepared primary cultures obtained from dissociated brains of third instar larvae. In control cultures, primary neurons developed long axons (average length 55.5 ± 22 μm after 48 hrs in culture), but axons in elav>DAtx2-RNAi neurons were significantly shorter (33.8 ± 22 μm; Fig. 3A,C). Interestingly, dendrite length was also shorter (18.2 ± 8 μm and 11.7 ± 6 μm respectively for control and elav>DAtx2 RNAi dendrites; Fig. 3A). In addition to shorter neurites, axons of elav>DAtx2-RNAi neurons displayed a larger number of bulges (2.28 bulges/axon) in comparison to control (0.46 bulges/axon) (Fig. 3C,D). Such bulges (axonal “jams”) are often seen in neurons that are defective in membrane organelle transport33,34.
Figure 3: DAtx2 depletion impairs ER distribution in Drosophila cultured neurons.
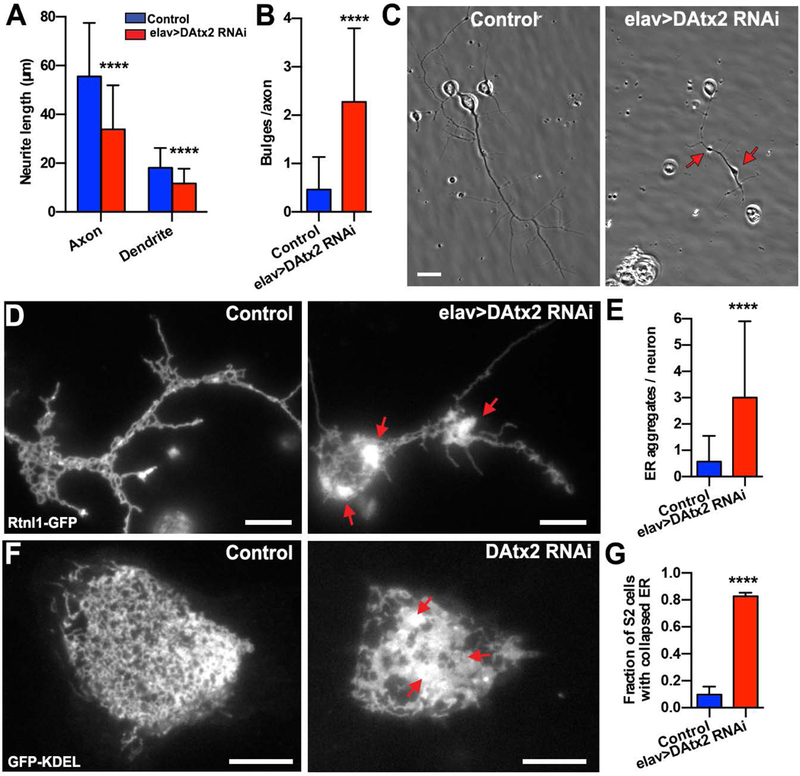
(A) Quantification of axon and dendrite length of 2-days-old neuronal primary cultures (n= 90 and 62 neurons for control and elav>DAtx2-RNAi, respectively) (B) Quantification of number of bulges identified in the axons of the neurons by phase-contrast microscopy (n=80 and 69 neurons for control and elav>DAtx2-RNAi, respectively) (C) Representative examples of 2 days-old-cultured neurons by phase contrast microscopy. Scale bars, 5 μm. (D) Representative near-TIRF images of control (left panel) and elav>DAtx2-RNAi (right panel) neurons expressing the ER marker Rtnl-1-GFP. Note that ER network collapses in elav>DAtx2-RNAi (red arrows). Scale bar 5 μm. (E) Quantification of ER aggregates per neurons. (F) Representative confocal images of control (left panel) and DAtx2-RNAi (right panel) S2 cells expressing GFP-KDEL. Scale bars, 10 μm. (G) Fraction of S2 cells with collapsed ER (n=39 cells for condition). Red arrows indicate ER collapse. Mann-Whitney test ****p<0.0001.
To characterize ER distribution in Drosophila cultured neurons, we used a fly expressing an ER marker (Rtnl-1-GFP) and combined it with elav>Gal4 (Rtnl-1-GFP; elav>Gal4). By crossing this fly stock with UASp-DAtx2 shRNA, we can fluorescently visualize the ER network in DAtx2 RNAi neurons. First, we examined ER distribution using near total internal reflection fluorescence (near-TIRF) microscopy. In control cultured neurons, the ER was organized into long tubules and cisternae (Fig. 3D, left panel). However, we found that the ER in DAtx2-RNAi neurons often formed small aggregates or larger clusters (Fig. 3D, right panel and 3E). The same ER collapse was observed in Ataxin-2 depleted S2 cells expressing the ER marker, GFP-KDEL, indicating that the role of Ataxin-2 in regulating ER distribution is not limited to neurons (Fig. 3F,G). Interestingly, the hallmark of pathology in SCA2 patients is the formation of torpedo-like axons in the Purkinje cells35. This set of neurons has been characterized by disoriented neurofilaments, aggregation of calreticulin and mitochondrial and ER displacement to the cell periphery36. Our data support that misfunction of polyQ Ataxin-2 may induce ER collapse both in neurons and glia in the brains of SCA2 patients.
Depletion of DAtx2 affects ER dynamics in neurons
The collapse of ER found in DAtx2-RNAi neurons indicates that DAtx2 might play an important role in neuronal ER dynamics. First, we decided to visualize ER dynamics using combination of TIRF with structured illumination microscopy5. TIRF-SIM allows us to resolve ER tubular structures in a 100nm focal plane with higher resolution. Time-lapse imaging of control primary neurons expressing Rtnl-1-GFP revealed that ER forms a very dynamic network as previously reported37–39. In agreement with these findings, we observed tubular structures travelling long distances and filling ER-empty areas either in the axons or in the soma of cultured Drosophila neurons (Fig. 4A and Movie 8). Knocking down DAtx2 dramatically impaired not only the distribution but also the dynamics of the ER network. The ER membranes visualized by TIRF-SIM in the knocked-down neurons never displayed large-scale unidirectional movements that were typically observed in the control neurons (Fig. 4B and Movie 8). As TIRF-SIM does not allow for visualization of the whole ER network deeper in the axoplasm, we decided to supplement these observations with fluorescence recovery after photobleaching (FRAP) experiments. We used a scanning confocal microscopy to bleach a small (about 6 μm in length) segment of the axon and watched the entry of non-photobleached Rtnl-1-GFP-labeled ER cisternae into the photobleached zone. To quantify the transport of ER, we measured the GFP signal recovery as a function of time. DIC images revealed that the photobleaching procedure did not damage the neurites (Fig. S4A). In the control, we observed a robust filling of GFP signal in the photobleached zone and completed fluorescence recovery in a few minutes (Fig. 4C and Movie 9). Quantification of the fluorescent recovery revealed that after 60 seconds in the control axons 63% of the signal was recovered (Fig. 4C,E,F) (see material and methods for FRAP quantification details). FRAP values were independent of the distance of the photobleached area to the cell body (data not shown). In Atx2-depleted neurons, FRAP values in the axons was dramatically impaired (only 23% of the signal recovered after 60 seconds) indicating that ER transport is inhibited (Fig. 4D-F and Movie 10). In principle, signal recovery can be caused by either diffusion of non-photobleached ER marker in the ER membrane plane, or active transport of non-photobleached ER network. Fluorescence intensity profiles of the photobleached area at different time points favor the hypothesis that the recovery is caused primarily by the active transport of ER (Fig. S4 and the legend). This interpretation is also consistent with qualitative analysis of ER movement using TIRF-SIM. Taken together these data demonstrate that Ataxin-2 plays an important role in proper ER distribution and dynamics in cell division and interphase, and that may have important implications in both neurodevelopment and neurodegeneration.
Figure 4: DAtx2 controls ER dynamics in Drosophila cultured neurons.
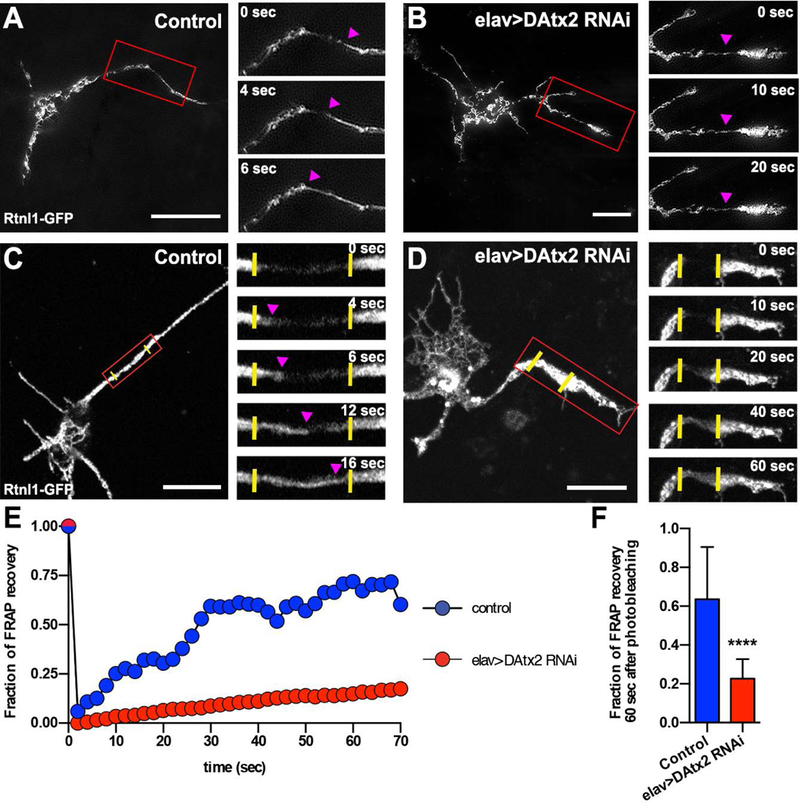
Representative TIRF-SIM images of control (A) and elav>DAtx2-RNAi (B) 2 days-old cultured neurons expressing the ER marker Rtnl-1-GFP. Insets show magnified views (red boxes) of ER at different time points. Magenta arrow heads show a moving ER cisternae. Scale bars, 10 μm. Representative confocal images of control (C) and elav>DAtx2-RNAi (D) 2 days-old cultured neurons expressing Rtnl-1-GFP before photobleaching. Insets show magnified views (red boxes) of Rtnl-1-GFP signal recovery at different time points. Yellow vertical lines represent the original photobleached boundaries and magenta arrow heads indicate position of signal recovery. Note that the time of signal recovery in elav>DAtx2-RNAi axon is slower that in the control. Scale bars, 10 μm. (E) FRAP kinetics of examples of neurons shown in C and D. (F) Quantification of the % signal recovery 60 seconds after photobleaching (n=14 neurons for each group). Mann-Whitney test ****p<0.0001.
How does Ataxin-2 regulate ER morphology and dynamics?
One uniquely interesting hint is the presence of an ER exit signal within the well-described Lsm domain of Ataxin-240. These ER exit signals target proteins to ER exit sites (ERES), which are structures where COPII-coated vesicles are budding and exiting the transitional ER, areas of ER devoid of ribosomes41–43. Although much is known about how these exit sites form and associate with other proteins, little is known about the association and function of RBPs at the ERES. Some RBPs (e.g. p180 in yeast) have been shown to facilitate direct, ribosome-independent mRNA binding in the ER membrane44,45, yet this function has not previously been described for Ataxin-2. In human cells, Ataxin-2 localizes near the plasma membrane27, suggesting an association with cortical ER, which is enriched in RNAs and RBPs45–47. The role of Ataxin-2 at the ER remains unclear, but there exists an exciting possibility that Ataxin-2 might function in the targeting of mRNAs in a spatial and temporal manner via an association with the ER, possibly through the newly discovered TIS granule that sits adjacent to the ER (TIGER domain). This is a biophysically and biochemically distinct environment enriched with membrane protein-encoding transcripts48. Notably, we have also shown that ATX-2/DAtx2 is closely associated but primarily non-overlapping with ER membranes during mitosis, and depletion of ATX-2 leads to a severe collapse of the ER specific to this RNA binding protein. Alternatively the ER collapse phenotype could occur through the modulation of PAR-5/14–3-3σ levels26, or directly via an association with polyribosomes at the ER itself16,44,45. Lastly, ATX-2 could provide a direct structural function to the ER as an ER-associated RNA binding protein. Interestingly, ER-shaping proteins are critical for maintaining the network organization of ER and mitochondria within motor neurons49.
Other ER resident proteins moonlighting as RBPs, such as the OST complex/OSTD-1 and the reticulon complex, have been shown to be necessary for cell division and ER morphology50–52. These data and the similarity among cell division phenotypes observed here suggest the possibility that the ER and RNA binding proteins may act in concert during mitosis. ATX-2/Ataxin-2/DAtx2 could function to mediate and maintain the structure and function of the cortical ER, which is rich in RNA binding proteins45–47. Cortical ER also contacts the plasma membrane at Membrane Contact Sites (MCSs), which are unique sites shown to regulate cellular calcium levels53,54. Depletion of ATX-2/DAtx2 could lead to altered calcium homeostasis within the cell, contributing to delays in the cell cycle, such as the delay during the anaphase-telophase transition we previously observed26. In SCA2 mice models, altered calcium levels have been observed and drug treatments regulating calcium-activated potassium channels have shown promise in the treatment of SCA2 and other ataxias55.
Overall, we demonstrated that the RNA binding protein ATX-2 plays a unique and necessary role in regulating proper ER morphology and dynamics. We hypothesize that Ataxin-2 may function in maintaining proper ER structure by facilitating the translation of mRNAs that encode ER resident proteins or as an integral structural component of the ER itself, facilitating both cytoplasmic and cortical ER function. This connection between SCA2 disease morphology and Ataxin-2 function has not been described and may provide some insight into the mechanism of the disease.
Materials and Methods
Worm and Fly Strains
Worm strains used in this study, include: N2 (wild type;56), DR466 (him-5(e1490);57), WH327 (GFP-SP12;4), WM210 (atx-2(ne4297);26), WH351 (GFP-UGTP-1;29). All strains were maintained at 24°C, except for atx-2(ne4297) which was maintained at 15°C and shifted to 24°C to deplete ATX-2. Some strains above were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). The fly strains used in this study, include: PDI-GFP;His2Av-RFP5, elav-Gal4 (3rd chromosome insertion, Bloomington stock #8760);58, DAtx2-RNAi TRiP line (Valium 20, 2nd chromosome insertion, Bloomington stock # 44012), and elav-Gal4 (generated using standard balancing procedures). Rtnl-1-GFP/ H2 transgenic line was created 5 using the original Rtnl1-GFP line generated in the protein trap screen for endogenous GFP expression of proteins59. Rtnl1-GFP[G00199] stock was obtained from the Flytrap database59. Fly stocks and crosses were cultured on standard cornmeal food at room temperature.
RNA interference in embryos and S2 cells
To knock down ATX-2, CAR-1, PAB-1, and ZEN-4 in vivo in C. elegans, the pL4440 feeding vectors specific to these genes were obtained from the Ahringer feeding RNAi library and sequence verified as previously described60–64. RNAi experiments were performed at 24°C for 16–22 hours. Drosophila embryos were prepared for microinjection and staged according to previously published protocols65,66. DAtx2 dsRNA was injected into Drosophila embryos at 500 ng/μl following previously published protocols 66,67. To knockdown DAtx2 in S2 cells, 5×105 cells cultured in 24 well plate with serum-free InsectXpress medium (Lonza) were treated with 25 μg of DAtx2 dsRNA in day 1 and 3. Cells were examined in the day 5. Primers used to create T7 templates from fly genomic DNA were as follows:
Forward:
TAATACGACTCACTATAGGGGCGCAAATAAGCTAAATGGCGATTCGAAC
Reverse:
TAATACGACTCACTATAGGGAAACTGTCGCAGTTCCTGCATTTGGTTAT
Preparation of Drosophila primary neurons
Primary cultures of dissociated neurons were obtained from brains of 3rd instar larva using a protocol previously described68. Neurons were plated onto Concanavalin A-coated coverslips in Schneider’s medium supplemented with 20% fetal bovine serum, 5 μg/ml insulin, 100 μg/ml penicillin, 100 μg/ml streptomycin, and 10 μg/ml tetracycline. The cultures were incubated at 25°C for two days prior to visualize them in the microscope.
Live-imaging
Worm embryo dissection and slide mounting were performed as previously described in26. Time-lapse images were taken every 10 seconds using a 200M inverted Axioscope microscope (Carl Zeiss) equipped with a spinning disk confocal scan head (QLC100; VisiTech, Sunderland, United Kingdom) and an Orca ER camera (Hamamatsu). The Orca ER camera was operated using MetaMorph software (version 7.7.11.0 Molecular Devices, LLC, Sunnyvale, CA, USA). Image rotating and cropping was performed using Fiji (ImageJ) software 69. Fly embryo imaging was performed on a Zeiss SD observer spinning disc confocal microscope. Images were taken at 20 second intervals from interphase cycle 12 in mitosis. All primary neuron images were acquired using a Nikon (Tokyo, Japan) Eclipse Ti inverted microscope at 25°C equipped with perfect focus controlled by Nikon NIS-Elements software. To visualize neuronal morphology, phase-contrast images were taken with a 100×1.4- Ph3 Plan APO objective and a digital CMOS, ORCA-Flash4.0 V2 C11440–22CU (Hamamatsu Photonics, Japan) and samples were illuminated using a CoolLED PrecisExcite (Hampshire, UK). ER distribution in S2 cells was imaged we used a Nikon Eclipse U2000 inverted stand with a Yokogawa CSU10 spinning disc confocal head and a 100 X 1.45 NA lens. Images were acquired using Evolve EMCCD (Photometrics) driven by Nikon Elements software. TIRF-SIM images were collected on a Nikon Ti-E inverted microscope (Nikon Instruments, Melville, NY) with a SIM illuminator and SIM enclosure equipped with an Apo TIRF 100×/numerical aperture (NA) 1.49 oil objective and electron-multiplying charge-coupled device (EMCCD) camera (iXon DU897; Andor, UK). Time-lapse images were recorded every 2 seconds. For FRAP experiments, a Yokogawa CSU-X1 spinning disk confocal head, and a 60×/ 1.4 objective was used. Images were acquired with an iXon EMCCD. Photobleaching of a small region of the axon was performed using a FRAPPA Photobleaching Module with a 405nm laser (Andor, UK). Time-lapse images were taken every 1 or 2 seconds.
C. elegans germline ER Imaging
Immobilization of worms for in utero imaging was performed using agarose pads with polystyrene beads as previously described70. RNAi Experiments were performed at 24°C for 16–22 hours. Images were acquired using a spinning disk confocal system with a Nikon Eclipse inverted microscope with a 40 X 1.30 oil objective, a CSU-22 spinning disc system, and a Photometrics EM-CCD camera from Visitech International operated by MetaMorph software (Molecular Devices). Image analysis was performed on Fiji (National Institutes of Health).
C. elegans Immunostaining
Immunostaining was performed using a methanol-4%formaldehyde fix as previously described71, and the following primary antibodies were used: anti-ATX-2 mouse monoclonal (1:100;Ciosk, 200421) and anti-GFP rabbit polyclonal (1:100; ab6556; Abcam) diluted in 1X PBS and 1% BSA (PBSB). Following an overnight incubation at 4°C, unbound primary antibodies were washed with PBST (1x PBS, 0.5% Tween). The secondary antibodies used were Alexa Fluor 568 anti-mouse (A-11004; Molecular Probes) and Alexa Fluor 488 anti-rabbit (A-11034; Molecular Probes) diluted 1:200 in PBSB. Secondary antibodies incubated at room temperature for 2 hours, and unbound secondary antibodies were removed with PBST washes. Embryos were mounted with Vectashield containing DAPI (H-1200; Vector Laboratories). Confocal imaging was performed on a departmental Zeiss 510 Confocal LSM operated with ZEN software (Carl Zeiss, Germany).
C. elegans Transmission Electron Microscopy (TEM)
atx-2(ne4297) animals were grown at 15°C then shifted to 24°C 24 hours before fixation to obtain the mutant phenotype. Young adult atx-2(ne4297) animals were subjected to high-pressure freeze fixation using 2% OsO4, 0.1% UAc, 2% H2O in acetone as the primary fixative according to protocol HP-1231. him-5(e1490) hermaphrodite animals were used as a control and were fixed using the same primary fixative according to protocol HP-1210. For each strain, thin sections were collected onto formvar-coated copper slot grids, and then post-stained with uranyl acetate and lead citrate. Sections were examined using a Philips CM10 electron microscope, and images were captured using an Olympus Morada digital camera.
TEM ER Quantification
ER segments were identified from images of embryos at 34000X and measured using FIJI. For both atx-2(ne4297) mutant animals and him-5(e1490) control animals, ER measurements were taken from 3 animals (two embryos from the first two animals and one embryo from the third animal). Shape factors were calculated by dividing the length of an ER segment by its width. An analysis of variance was used to compare the two groups. Means and standard errors were calculated for each group. Analyses were performed using SAS version 9.3.
Drosophila Embryo Immunostaining
Drosophila embryos were collected for 3 hours. Embryos were fixed using a hand devitellinization technique as previously described72. Embryos were incubated in primary antibody, rabbit anti-Atx-2 [1:200]16, and secondary antibody, Alexa Fluor 647 donkey anti-rabbit (1:200, Invitrogen, #A31573). Embryos were imaged using Zeiss LSM 710 confocal microscopy. Zeiss LSM 710 upright scanning confocal microscope built on Axio Imager.Z2 with a 63x/1.4 oil objective. Images were acquired and analyzed using Zen Black 2012 Software (Carl Ziess Microscopy, LLC.)
Quantification of neurite length of Drosophila primary cultures
To quantify the neurite length of primary cultures, each neurite was manually measured using FIJI software.
Quantification of Golgi puncta in C. elegans embryos
To quantify Golgi puncta size, images from control (n=8) and atx-2 fRNAi-treated (n=10) embryos staged at furrow initiation were used. ROI measuring 55×55a.u. (example ROI shown in Figure S3A,B, blue and orange boxes) were selected from the anterior and posterior of each embryo. Puncta within the ROI were thresh-holded using FIJI software and analyzed to determine puncta area.
Quantification of FRAP images
To quantify ER dynamics in axons of primary neurons, the mean intensity value for two different regions of identical areas were taken for each frame: photobleached (Fp) and not photobleached (Fo). An empty region of the frame was used to measure the background (Fb). Pre-photobleaching value was normalized to 1 for each sample. The fraction of fluorescent recovery for each frame was calculated as followed (Fp-Fb)/(Fo-Fb) and plotted as a function of time. Recovery values at 60 seconds were used to compare the effect of different conditions on ER dynamics.
Supplementary Material
Synopsis:
The RNA binding protein, Ataxin-2 has a conserved role controlling ER morphology and dynamics during mitosis, in the germline, and in neurons. ATX-2/DAtx2 localizes in puncta adjacent to the ER in embryos. Electron microscopy revealed that the Rough ER is forced into unique spherical stacks in the oocyte and in embryos leads to shortened and collapsed ER tubules. Disruption of ER dynamics by knocking-down protein levels of DAtx2 in neurons recapitulates phenotypes found in advanced SCA2 patients. We discuss potential mechanisms explaining how Ataxin-2 can regulate ER dynamics and its implications in SCA2 disease.
Acknowledgements
We thank Randall Dahn for advice and for critically reading the manuscript, Jill Willdonger for initially performing DAtx2 knockdown experiments in sensory neurons, and David Kirchenbüechler (Northwestern University Center for Advanced Microscopy),for help with ER FRAP quantification. Drosophila Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. Some C. elegans strains above were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Funding
A.R.S. was supported by the National Science Foundation under award MCB 1158003. C. elegans strains were obtained from the C. elegans Genetics Stock Center which is supported by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). J.N.B. was supported by NIH R01 GM114471. V.I.G was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award R01 GM52111. The imaging work that was performed at the Northwestern University Center for Advanced Microscopy generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center. B.R. was supported by the National Science Foundation CAREER Award 1553695. Imaging work involving the early Drosophila embryo was performed at the San Francisco State University, Cell and Molecular Imaging Center (CMIC) with the generous assistance of Annette Chan. D.H.H. was supported by NIH OD 010943.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Schwarz DS, Blower MD. The endoplasmic reticulum: structure, function and response to cellular signaling. Cell Mol Life Sci. 2016;73(1):79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stevenson J, Huang EY, Olzmann JA. Endoplasmic Reticulum-Associated Degradation and Lipid Homeostasis. Annu Rev Nutr. 2016;36:511–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westrate LM, Lee JE, Prinz WA, Voeltz GK. Form follows function: the importance of endoplasmic reticulum shape. Annu Rev Biochem. 2015;84:791–811. [DOI] [PubMed] [Google Scholar]
- 4.Poteryaev D, Squirrell JM, Campbell JM, White JG, Spang A. Involvement of the actin cytoskeleton and homotypic membrane fusion in ER dynamics in Caenorhabditis elegans. Molecular biology of the cell. 2005;16(5):2139–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergman ZJ, McLaurin JD, Eritano AS, Johnson BM, Sims AQ, Riggs B. Spatial reorganization of the endoplasmic reticulum during mitosis relies on mitotic kinase cyclin A in the early Drosophila embryo. PLoS One. 2015;10(2):e0117859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu L, Ladinsky MS, Kirchhausen T. Cisternal organization of the endoplasmic reticulum during mitosis. Molecular biology of the cell. 2009;20(15):3471–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ozcan L, Tabas I. Role of endoplasmic reticulum stress in metabolic disease and other disorders. Annu Rev Med. 2012;63:317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roussel BD, Kruppa AJ, Miranda E, Crowther DC, Lomas DA, Marciniak SJ. Endoplasmic reticulum dysfunction in neurological disease. Lancet Neurol. 2013;12(1):105–118. [DOI] [PubMed] [Google Scholar]
- 9.Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7(12):1013–1030. [DOI] [PubMed] [Google Scholar]
- 10.Logothetis C, Aparicio A, Thompson TC. ER stress in prostate cancer: A therapeutically exploitable vulnerability? Sci Transl Med. 2018;10(439). [DOI] [PubMed] [Google Scholar]
- 11.Zeng XS, Geng WS, Jia JJ, Chen L, Zhang PP. Cellular and Molecular Basis of Neurodegeneration in Parkinson Disease. Front Aging Neurosci. 2018;10:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lieberman A, Deep A, Shi J, et al. Downward finger displacement distinguishes Parkinson disease dementia from Alzheimer disease. Int J Neurosci. 2018;128(2):151–154. [DOI] [PubMed] [Google Scholar]
- 13.Bhatta M, Chatpar K, Hu Z, Wang JJ, Zhang SX. Reduction of Endoplasmic Reticulum Stress Improves Angiogenic Progenitor Cell function in a Mouse Model of Type 1 Diabetes. Cell Death Dis. 2018;9(5):467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamanaka T, Nukina N. ER Dynamics and Derangement in Neurological Diseases. Front Neurosci. 2018;12:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pulst SM, Nechiporuk A, Nechiporuk T, et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat Genet. 1996;14(3):269–276. [DOI] [PubMed] [Google Scholar]
- 16.Satterfield TF, Pallanck LJ. Ataxin-2 and its Drosophila homolog, ATX2, physically assemble with polyribosomes. Human molecular genetics. 2006;15(16):2523–2532. [DOI] [PubMed] [Google Scholar]
- 17.van de Loo S, Eich F, Nonis D, Auburger G, Nowock J. Ataxin-2 associates with rough endoplasmic reticulum. Experimental neurology. 2009;215(1):110–118. [DOI] [PubMed] [Google Scholar]
- 18.Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA. Mechanisms determining the morphology of the peripheral ER. Cell. 2010;143(5):774–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Audhya A, Hyndman F, McLeod IX, et al. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J Cell Biol. 2005;171(2):267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Squirrell JM, Eggers ZT, Luedke N, et al. CAR-1, a protein that localizes with the mRNA decapping component DCAP-1, is required for cytokinesis and ER organization in Caenorhabditis elegans embryos. Molecular biology of the cell. 2006;17(1):336–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ciosk R, DePalma M, Priess JR. ATX-2, the C. elegans ortholog of ataxin 2, functions in translational regulation in the germline. Development. 2004;131(19):4831–4841. [DOI] [PubMed] [Google Scholar]
- 22.Satterfield TF, Jackson SM, Pallanck LJ. A Drosophila homolog of the polyglutamine disease gene SCA2 is a dosage-sensitive regulator of actin filament formation. Genetics. 2002;162(4):1687–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bobinnec Y, Marcaillou C, Morin X, Debec A. Dynamics of the endoplasmic reticulum during early development of Drosophila melanogaster. Cell Motil Cytoskeleton. 2003;54(3):217–225. [DOI] [PubMed] [Google Scholar]
- 24.Royou A, Field C, Sisson JC, Sullivan W, Karess R. Reassessing the role and dynamics of nonmuscle myosin II during furrow formation in early Drosophila embryos. Molecular biology of the cell. 2004;15(2):838–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sullivan W, Theurkauf WE. The cytoskeleton and morphogenesis of the early Drosophila embryo. Curr Opin Cell Biol. 1995;7(1):18–22. [DOI] [PubMed] [Google Scholar]
- 26.Gnazzo MM, Uhlemann EE, Villarreal A, Shirayama M, Dominguez EG, Skop AR. The RNA-binding protein ATX-2 regulates cytokinesis through PAR-5 and ZEN-4. Molecular biology of the cell. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huynh DP, Yang HT, Vakharia H, Nguyen D, Pulst SM. Expansion of the polyQ repeat in ataxin-2 alters its Golgi localization, disrupts the Golgi complex and causes cell death. Human molecular genetics. 2003;12(13):1485–1496. [DOI] [PubMed] [Google Scholar]
- 28.Ng H, Pulst SM, Huynh DP. Ataxin-2 mediated cell death is dependent on domains downstream of the polyQ repeat. Experimental neurology. 2007;208(2):207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bembenek JN, Richie CT, Squirrell JM, et al. Cortical granule exocytosis in C. elegans is regulated by cell cycle components including separase. Development. 2007;134(21):3837–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Damme P, Veldink JH, van Blitterswijk M, et al. Expanded ATXN2 CAG repeat size in ALS identifies genetic overlap between ALS and SCA2. Neurology. 2011;76(24):2066–2072. [DOI] [PubMed] [Google Scholar]
- 31.Elden AC, Kim HJ, Hart MP, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466(7310):1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campos AR, Rosen DR, Robinow SN, White K. Molecular analysis of the locus elav in Drosophila melanogaster: a gene whose embryonic expression is neural specific. EMBO J. 1987;6(2):425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Correia SC, Perry G, Moreira PI. Mitochondrial traffic jams in Alzheimer’s disease - pinpointing the roadblocks. Biochimica et biophysica acta. 2016;1862(10):1909–1917. [DOI] [PubMed] [Google Scholar]
- 34.Hurd DD, Saxton WM. Kinesin mutations cause motor neuron disease phenotypes by disrupting fast axonal transport in Drosophila. Genetics. 1996;144(3):1075–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neurodegeneration Dickson D. : the molecular pathology of dementia and movement disorders. 2nd ed. Chichester, West Sussex: Wiley-Blackwell; 2011. [Google Scholar]
- 36.Mann DM, Stamp JE, Yates PO, Bannister CM. The fine structure of the axonal torpedo in Purkinje cells of the human cerebellum. Neurological research. 1980;1(4):369–378. [DOI] [PubMed] [Google Scholar]
- 37.Lee C, Chen LB. Dynamic behavior of endoplasmic reticulum in living cells. Cell. 1988;54(1):37–46. [DOI] [PubMed] [Google Scholar]
- 38.Nixon-Abell J, Obara CJ, Weigel AV, et al. Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science. 2016;354(6311). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo Y, Li D, Zhang S, et al. Visualizing Intracellular Organelle and Cytoskeletal Interactions at Nanoscale Resolution on Millisecond Timescales. Cell. 2018;175(5):1430–1442 e1417. [DOI] [PubMed] [Google Scholar]
- 40.Shibata H, Huynh DP, Pulst SM. A novel protein with RNA-binding motifs interacts with ataxin-2. Human molecular genetics. 2000;9(9):1303–1313. [DOI] [PubMed] [Google Scholar]
- 41.Orci L, Ravazzola M, Meda P, et al. Mammalian Sec23p homologue is restricted to the endoplasmic reticulum transitional cytoplasm. Proc Natl Acad Sci U S A. 1991;88(19):8611–8615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stephens DJ. De novo formation, fusion and fission of mammalian COPII-coated endoplasmic reticulum exit sites. EMBO Rep. 2003;4(2):210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Budnik A, Stephens DJ. ER exit sites--localization and control of COPII vesicle formation. FEBS Lett. 2009;583(23):3796–3803. [DOI] [PubMed] [Google Scholar]
- 44.Reid DW, Nicchitta CV. Diversity and selectivity in mRNA translation on the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2015;16(4):221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hermesh O, Jansen RP. Take the (RN)A-train: localization of mRNA to the endoplasmic reticulum. Biochim Biophys Acta. 2013;1833(11):2519–2525. [DOI] [PubMed] [Google Scholar]
- 46.Genz C, Fundakowski J, Hermesh O, Schmid M, Jansen RP. Association of the yeast RNA-binding protein She2p with the tubular endoplasmic reticulum depends on membrane curvature. J Biol Chem. 2013;288(45):32384–32393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doroshenk KA, Tian L, Crofts AJ, Kumamaru T, Okita TW. Characterization of RNA binding protein RBP-P reveals a possible role in rice glutelin gene expression and RNA localization. Plant Mol Biol. 2014;85(4–5):381–394. [DOI] [PubMed] [Google Scholar]
- 48.Ma W, Mayr C. A Membraneless Organelle Associated with the Endoplasmic Reticulum Enables 3’UTR-Mediated Protein-Protein Interactions. Cell. 2018;175(6):1492–1506 e1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fowler PC, O’Sullivan NC. ER-shaping proteins are required for ER and mitochondrial network organization in motor neurons. Human molecular genetics. 2016. [DOI] [PubMed] [Google Scholar]
- 50.Zhang D, Vjestica A, Oliferenko S. The cortical ER network limits the permissive zone for actomyosin ring assembly. Curr Biol. 2010;20(11):1029–1034. [DOI] [PubMed] [Google Scholar]
- 51.Bonner MK, Han BH, Skop A. Profiling of the mammalian mitotic spindle proteome reveals an ER protein, OSTD-1, as being necessary for cell division and ER morphology. PloS one. 2013;8(10):e77051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jagannathan S, Hsu JC, Reid DW, et al. Multifunctional roles for the protein translocation machinery in RNA anchoring to the endoplasmic reticulum. J Biol Chem. 2014;289(37):25907–25924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elbaz Y, Schuldiner M. Staying in touch: the molecular era of organelle contact sites. Trends in biochemical sciences. 2011;36(11):616–623. [DOI] [PubMed] [Google Scholar]
- 54.Hofer AM. Ca signaling at membrane contact sites. Cell Calcium. 2015. [DOI] [PubMed] [Google Scholar]
- 55.Kasumu AW, Liang X, Egorova P, Vorontsova D, Bezprozvanny I. Chronic suppression of inositol 1,4,5-triphosphate receptor-mediated calcium signaling in cerebellar purkinje cells alleviates pathological phenotype in spinocerebellar ataxia 2 mice. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32(37):12786–12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brenner S The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hodgkin J, Horvitz HR, Brenner S. Nondisjunction Mutants of the Nematode CAENORHABDITIS ELEGANS. Genetics. 1979;91(1):67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 1994;8(15):1787–1802. [DOI] [PubMed] [Google Scholar]
- 59.Morin X, Daneman R, Zavortink M, Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(26):15050–15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2(1):RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30(4):313–321. [DOI] [PubMed] [Google Scholar]
- 62.Ahringer J Reverse genetics. WormBook , ed. 2006;The C. elegans Research Community, WormBook, doi/ 10.1895/wormbook.1.47.1, http://www.wormbook.org. [DOI] [Google Scholar]
- 63.Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395(6705):854. [DOI] [PubMed] [Google Scholar]
- 64.Kamath RS, Fraser AG, Dong Y, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421(6920):231–237. [DOI] [PubMed] [Google Scholar]
- 65.Riggs B, Fasulo B, Royou A, et al. The concentration of Nuf, a Rab11 effector, at the microtubule-organizing center is cell cycle regulated, dynein-dependent, and coincides with furrow formation. Molecular biology of the cell. 2007;18(9):3313–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tram U, Riggs B, Koyama C, Debec A, Sullivan W. Methods for the study of centrosomes in Drosophila during embryogenesis. Methods Cell Biol. 2001;67:113–123. [DOI] [PubMed] [Google Scholar]
- 67.Royou A, McCusker D, Kellogg DR, Sullivan W. Grapes(Chk1) prevents nuclear CDK1 activation by delaying cyclin B nuclear accumulation. J Cell Biol. 2008;183(1):63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu W, Lakonishok M, Gelfand VI. Kinesin-1-powered microtubule sliding initiates axonal regeneration in Drosophila cultured neurons. Molecular biology of the cell. 2015;26(7):1296–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nature methods. 2012;9(7):676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim E, Sun L, Gabel CV, Fang-Yen C. Long-term imaging of Caenorhabditis elegans using nanoparticle-mediated immobilization. PloS one. 2013;8(1):e53419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Skop AR, White JG. The dynactin complex is required for cleavage plane specification in early Caenorhabditis elegans embryos. Current biology : CB. 1998;8(20):1110–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rothwell WF, Sullivan W. Drosophila embryo collection. CSH Protoc. 2007;2007:pdb prot4825. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


