Abstract
High-elevation organisms experience shared environmental challenges that include low oxygen availability, cold temperatures, and intense ultraviolet radiation. Consequently, repeated evolution of the same genetic mechanisms may occur across high-elevation taxa. To test this prediction, we investigated the extent to which the same biochemical pathways, genes, or sites were subject to parallel molecular evolution for 12 Andean hummingbird species (family: Trochilidae) representing several independent transitions to high elevation across the phylogeny. Across high-elevation species, we discovered parallel evolution for several pathways and genes with evidence of positive selection. In particular, positively selected genes were frequently part of cellular respiration, metabolism, or cell death pathways. To further examine the role of elevation in our analyses, we compared results for low- and high-elevation species and tested different thresholds for defining elevation categories. In analyses with different elevation thresholds, positively selected genes reflected similar functions and pathways, even though there were almost no specific genes in common. For example, EPAS1 (HIF2α), which has been implicated in high-elevation adaptation in other vertebrates, shows a signature of positive selection when high-elevation is defined broadly (>1,500 m), but not when defined narrowly (>2,500 m). Although a few biochemical pathways and genes change predictably as part of hummingbird adaptation to high-elevation conditions, independent lineages have rarely adapted via the same substitutions.
Keywords: Andes, convergent evolution, hypoxia, respiratory electron transport, transcriptome, Trochilidae
Introduction
A fundamental question in evolutionary biology concerns the range of genetic mechanisms available for species to respond to the same environmental demands. Repeated adaptive evolution may be predictable across divergent taxa because the genetic mechanisms are tightly constrained and restrict adaptive mutations to specific genes or amino acid sites. However, phenotypic convergence may also arise from less predictable processes. For example, mutations at different genes may confer similar functional benefits, particularly when genes have functional similarities, or contribute to the same biochemical pathways (Rosenblum et al. 2014; Bailey et al. 2015). To test the extent and specificity of parallel evolution in genetic mechanisms, we can ask a nested triad of questions for any set of species that have independently evolved under shared environmental conditions. Did positive selection affect 1) the same biochemical pathways, 2) the same genes, or 3) the same amino acid sites?
More predictable genetic mechanisms for adaptation would result in positive answers for each of these three questions (“yes–yes–yes”), reflecting parallelism at all three hierarchical levels (Rosenblum et al. 2014). Intermediate scenarios could involve predictability at certain levels in the hierarchy of biological organization, but not others (e.g., “yes–yes–no” or “yes–no–no”). Convergent phenotypes could arise through shared molecular changes at any of these three levels; for example, changes involving mutation at the same codon in the same gene (Arendt and Reznick 2008; Projecto-Garcia et al. 2013), independent mutations to the same gene (Rosenblum et al. 2010; Linnen et al. 2013; Natarajan et al. 2016), or changes to different genes in the same pathway (Arendt and Reznick 2008). For example, although the same amino acid substitutions in the melanocortin-1 receptor gene affect pigmentation in beach mice and mammoths (Hoekstra et al. 2006; Römpler et al. 2006), independent substitutions in this same gene result in pale skin color for two lizard species (Rosenblum et al. 2010), and changes to different genes in the same pathway can influence melanization in similar ways (Arendt and Reznick 2008). If genetic mechanisms are unpredictable and taxon specific, it could result in “no–no–no,” despite convergent evolution of phenotypes. A range of genetic mechanisms can potentially converge on similar phenotypes, illustrating functional redundancy and flexibility in evolutionary processes. At each hierarchical level, the frequency of parallelism can provide more detailed insights into the degree of predictability. Here, we ask this triad of questions to investigate the predictability of the genetic mechanisms underlying high-altitude adaptation across Andean hummingbird lineages.
Compared with the lowlands, high-elevation environments have less available oxygen (hypoxia), lower ambient temperatures, and greater exposure to ultraviolet (UV) radiation. In these conditions, phenotypes that may result in higher fitness, such as increased hemoglobin-oxygen-binding affinity or lower hemoglobin concentrations to reduce blood viscosity and hypertension, are likely to be favored by natural selection and found in independent taxa that experience the same challenges (Monge and León-Velarde 1991; Zhuang et al. 1993; Beall et al. 1998; Hainsworth and Drinkhill 2007). Conversely, species at lower altitudes may face different challenges that could result in predictable evolution (e.g., heat-dissipation or disease resistance). Whether high or low, describing patterns of molecular evolution associated with elevation has the potential to provide insight on the predictability of evolution and the mechanistic underpinnings of species distributions.
In highland environments, natural selection is expected to act on pathways and genes related to oxygen uptake and transport, and to metabolism and energy production (e.g., in pathways responsible for oxygen sensing and cellular respiration). The literature on the genetics of high-altitude adaptation has uncovered many candidates for adaptive evolution, including repeated identification of specific mutations, genes, and pathways. There are a handful of cases in which the same genes are repeatedly identified as evolving under positive selection across different taxa or populations, and in the latter case this is particularly well-studied across human populations: endothelial PAS domain protein 1 (EPAS1), egl-9 family hypoxia-inducible factor 1 (EGLN1), and peroxisome proliferator activated receptor alpha (PPARA) (Yi et al. 2010; Simonson et al. 2012). Indeed, recent research points to evidence for convergent molecular evolution at EPAS1 and EGLN1 in high-altitude Andean ducks that mirrors their evolution in human populations (Graham and McCracken 2019). However, the explosion of new genomic data sets and research on highland taxa has expanded the list to include many more positively selected genes (PSGs) implicated in high-altitude adaptation (Huerta-Sánchez et al. 2013; Qu et al. 2013, 2015; Foll et al. 2014; Graham et al. 2018). The candidate genes that have been identified by testing for positive selection include genes involved in the hypoxia response (Yi et al. 2010; Simonson et al. 2012; Huerta-Sánchez et al. 2013; Qu et al. 2013; Graham et al. 2018), skeletal development (Qu et al. 2013), energy metabolism (Qu et al. 2013, 2015), response to UV radiation (Zhang et al. 2016), and hemoglobin-oxygen-binding affinity (Storz et al. 2009; McCracken et al. 2010; Muñoz-Fuentes et al. 2013). These studies suggest that oxygen sensing and transport, hypoxia response, and energy metabolism are predictable functional candidates for natural selection, even though the degree of predictability at various hierarchical levels—pathways, genes, and amino acid sites—remains poorly understood. An ancillary challenge when testing predictability is defining “high elevation” and considering the possibility that shifts in natural selection may be sensitive to specific thresholds along altitudinal gradients. To date, the latter issue has been largely ignored by comparative genomic studies (but see Sun et al. 2018).
Hummingbirds (family: Trochilidae) are a useful study system for examining the genetic basis of high-altitude adaptation because eight of the nine major clades include species that occur >2,000 m elevation. Despite having the highest basal metabolic rates of any endothermic vertebrate (Lasiewski 1963), hummingbirds are diverse and abundant at and above tree-line in the Andes (∼3,000 m), with at least a few species regularly occurring above 4,500 m (Parker et al. 1996; Benham et al. 2011). Previous work on morphological, physiological, and biomechanical differences in highland versus lowland hummingbirds provides a foundation for understanding their phenotypic responses to reduced partial pressure of oxygen (Altshuler and Dudley 2006; Stiles 2008). In hummingbirds, research on the genetic mechanisms underlying high-altitude adaptation has focused on the oxygen-binding affinity of hemoglobin. Specifically, two amino acid changes in the beta-A subunit were found to both affect hemoglobin-oxygen-binding affinity and to have undergone predictable amino acid changes in conjunction with evolutionary shifts in elevation (Projecto-Garcia et al. 2013). In contrast, adult-expressed hemoglobin isoforms in other bird clades that have colonized the high Andes have also adapted, but with less predictability as to the specific codons involved (Natarajan et al. 2016).
Here, we identified pathways, PSGs, and positively selected sites involved in high-altitude adaptation by sequencing transcriptomes for 12 Andean hummingbird species. We examined amino acid substitutions at expressed loci and mapped these changes to the phylogeny to test for positive selection. If common regions of the transcriptome showed evidence of selection in sets of species with similar altitudinal ranges, we inferred that those regions were predictable targets of selection associated with shared environmental challenges. Although high-elevation hummingbirds exhibit predictable hemoglobin evolution to the level of the nucleotide (Projecto-Garcia et al. 2013), little is known about the rest of the genome. We asked whether high-elevation species evolve under positive natural selection at the same pathways, genes, and/or sites; whether the same is true of lowland species; and how different definitions of “high elevation” influence the sets of PSGs.
Materials and Methods
Study System
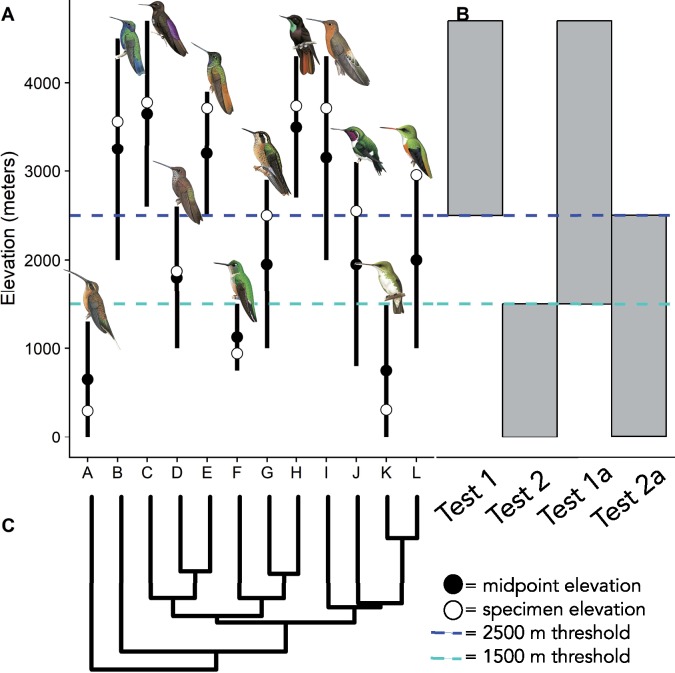
We studied 12 hummingbird species including low-, mid-, and high-elevation species from multiple clades, forming phylogenetic replicates with which to test for convergence across the hummingbird family (fig. 1 and supplementary table S1, Supplementary Material online). We grouped the species according to their altitudinal ranges for analysis (fig. 1A) (Parker et al. 1996; Schulenberg et al. 2007; Benham et al. 2011). The high-elevation species are typical of the temperate zone habitats of the high Andes, where all are most abundant above 3,000 m in elevation and have ranges extending above 4,000 m. The midelevation species are typical of the subtropical zone, with centers of abundance around 2,000 m, and ranges extending from the upper tropical zone to the lower temperate zone. The low-elevation species occur in the tropical zone, almost exclusively below 1,500 m.
Fig. 1.
—(A) Elevation ranges (black lines), midpoint elevations, and specimen sampling elevations for the 12 study species (A, Phaethornis malaris; B, Colibri coruscans; C, Aglaeactis castelnaudii; D, Coeligena coeligena; E, Coeligena violifer; F, Phlogophilus harterti; G, Adelomyia melanogenys; H, Metallura phoebe; I, Patagona gigas peruviana; J, Chaetocercus mulsant; K, Amazilia amazilia; and L, Amazilia viridicauda). We used specimen data, expert knowledge, and Schulenberg et al. (2007) to identify the core elevational ranges of each sampled taxon in our study. Hummingbird illustrations are from del Hoyo et al. (2018). (B) Gray bars mark species groups for the four elevation scenarios to test for high-altitude adaptation in species with midpoint elevations occurring >2,500 m (Test 1), <1,500 m (Test 2), >1,500 m (Test 1a), and <2,500 m (Test 2a). (C) Phylogenetic relationships between study species pruned from McGuire et al. (2014).
RNA Extraction and Sequencing
Hummingbird specimens were collected from sites in Peru under an approved Animal Use Protocol from the University of New Mexico Institutional Animal Care and Use Committee (IACUC Protocol number 08UNM033-TR-100117; Animal Welfare Assurance number A4023-01) and under permits from the management authority of Peru (76-2006-INRENA-IFFS-DCB, 087-2007-INRENA-IFFS-DCB, 135-2009-AG-DGFFS-DGEFFS, 0377-2010-AG-DGFFS-DGEFFS, 0199-2012-AG-DGFFS-DGEFFS, and 006-2013-MINAGRI-DGFFS/DGEFFS). Complete specimen data are available via the ARCTOS online database (arctosdb.org; supplementary table S1, Supplementary Material online). Tissue samples were flash frozen in liquid nitrogen in the field and later preserved in RNAlater in the lab to preserve RNA and increase concentration yield. We extracted RNA from liver and muscle tissue from one specimen per species following the Qiagen RNeasy kit protocol (Qiagen, Valencia, CA). We assessed RNA quality (RIN scores >8) and quantity (10 ng to 1 µg total RNA) using Bioanalyzer traces. We used a NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, Ipswich, MA) to synthesize cDNA from mRNA and prepare libraries, including steps for end repair, adaptor ligation, and PCR enrichment with unique indexing primers for each species. We checked library quality and quantity with Bioanalyzer traces and Qubit DNA concentration readings. Finally, the 12 libraries were pooled and sequenced on one lane of the Illumina HiSeq 2000 with 100-bp paired-end reads.
De Novo Transcriptome Assembly
We created our own pipeline for cleaning, merging, and assembling the demultiplexed transcriptome reads. First, we removed all reads that failed the Illumina CASAVA quality filter (all reads with “Y” in the header). Then, we removed Illumina adapter sequences, low quality reads, and duplicate reads with TRIMMOMATIC v0.32 (Bolger et al. 2014). We merged overlapping paired-end reads to make longer single reads using FLASH (Magoč and Salzberg 2011). Prior to assembly, we compared read quality before and after these filter steps using FastQC v0.11.2 (Andrews 2010). The reads were de novo assembled with TRINITY v2.1.1 (Grabherr et al. 2011).
Exon Annotation and Alignment
We annotated the transcript assemblies with the zebra finch genome (Ensembl release 92) using reciprocal blast searches (BlastX and TBlastN v2.2.31; e-value cut-off = 1e-10; Altschul et al. 1997). For this approach, we defined the coding and untranslated regions of the assemblies with EXONERATE v2.2.0 (Slater and Birney 2005) and generated nonredundant sequence databases with CD-HIT v4.6.4 (Li and Godzik 2006) following Singhal (2013). To increase the number of previously identified candidate genes included in our data set, we conducted additional reciprocal BlastN searches to extract matching sequences from the hummingbird transcriptome assemblies.
Next, we extracted the exons that we could annotate from the assemblies to create sequence alignments. We used BlastP to ensure the reading frame was correct and aligned exons using MAFFT v7.245 (Katoh and Standley 2013). For analysis, we used exon alignments with sequences from all 12 species. We checked all sequence alignments for gaps, premature stop codons, and untrimmed ends in GENEIOUS v7.1.5 (Kearse et al. 2012). Alignments with combinations of isoform (splice variants) or paralog (gene duplicates) sequences were removed from downstream analysis.
Testing for Shared PSGs and Sites
To test for genes and amino acid sites that evolved under positive selection, we used a branch-site model (Models 1A and A) in codeml from PAML v4.9a (Yang 2007). The branch-site model tests for natural selection along branches in the phylogeny and at sites in the peptide sequence alignments using the dN/dS ratio (ω), where a branch or site has evolved under neutral evolution if ω = 1, purifying selection if ω < 1, and positive selection if ω > 1. We used a pruned phylogeny from McGuire et al. (2014) with the 12 study species. The null model (Model 1A) constrains ω to remain below one and assumes that background and foreground branches share the same ω ratio, whereas the test model (Model A) allows ω to exceed one and foreground branch ω can differ from background branch ω values. Following each codeml analysis, we calculated likelihood ratio test (LRT) values from the null and test model likelihoods to identify genes and amino acid sites that evolved under positive selection ().
To identify shared PSGs and amino acid sites across highland or lowland species, we designed four branching scenarios that set multiple highland or lowland species as the foreground branches (fig. 1B). Because the midelevation species occupy elevations at which reductions in arterial oxygen saturation begin to have physiological effects, we grouped them with either highland or lowland species. Thus, the branching scenarios reflect two elevational thresholds set at 1,500 and 2,500 m, allowing us to investigate how natural selection on elevation-related traits varies with elevation. To test for PSGs and sites shared by high-elevation species, foreground branches were defined as species with midpoint elevations above 2,500 m (Test 1, corresponding to 75% of the partial pressure of oxygen when compared with sea level) or midpoint elevations above 1,500 m (Test 1a, corresponding to 84% of the partial pressure of oxygen when compared with sea level). As negative controls to Tests 1 and 1a, we tested for PSGs and sites shared by low-elevation species, for which the elevation ranges remain similar to the putative ancestral state for hummingbirds, and there is no a priori expectation of having evolved high-altitude adaptations. For the negative control tests, we defined foreground branches as species with midpoint elevations below 1,500 m (Test 2), or midpoint elevations below 2,500 m (Test 2a). Genes were retained for analysis if P values were significant at the P < 0.05 level.
To test whether different PSGs occur in the same biochemical pathways across highland or lowland species, we individually tagged one species as the foreground with the remaining 11 as the background branches. In this case, we had to account for multiple hypothesis testing because we tested each branch of the phylogeny (Anisimova and Yang 2007). Therefore, we calculated adjusted P values from the LRT values to reduce the false discovery rate (Benjamini and Hochberg 1995). Genes were retained in the analysis if adjusted P values were still significant at the P < 0.05 level. We then compared the PSGs within each elevation category to determine whether any of the genes act in similar pathways. In addition, we checked for PSGs that were potentially biased if, for example, one lineage in the foreground branches had an especially high ω compared with the other foreground branches by comparing the results from the single-branch foreground codeml analyses to the multibranch foreground analyses. If the same PSGs were found in both foreground branch scenarios, we removed the gene from further analysis to avoid including potential false positives.
To further validate whether PSGs were associated with high elevation, as opposed to other environmental factors, and to eliminate anomalous genes, we ran branch-site models (Model 1A vs. Model A) with eight scrambled foreground branch groups for PSGs identified in all PAML analyses (n = 45) (supplementary fig. S1, Supplementary Material online). These branching scenarios have no known relationship with elevation ranges. The foreground and background branch groups each included six species to approximately match the number of foreground species for Test 1 (n = 5) and Test 1a (n = 9) analyses. We used the Environment for Tree Exploration toolkit to conduct these permutations (Huerta-Cepas et al. 2016). The Environment for Tree Exploration toolkit calculates P values for the LRT between branch-site models. We then assessed whether positive selection was detected for codon sites at PSGs even when the foreground branches were scrambled to remove any association with high elevation. If a given gene had completely nonsignificant results in these permutations, this was interpreted to support the rejection of the null hypothesis of no foreground positive selection. Conversely, one or more significant results implied the rejection of the null hypothesis could arise through factors other than elevation.
Identifying Gene Function
To investigate the functions of PSGs, we searched through several gene and pathway databases. We examined biological process, molecular function, and cellular component gene ontology (GO) terms associated with PSGs using the Panther database (Mi et al. 2017). We searched for GO terms associated with our set of PSGs based on functional information known from well-annotated genomes, such as the chicken (Gallus gallus) or human (Homo sapiens) genomes. For PSGs that only had zebra finch Ensembl IDs and no gene annotation, GO terms were unavailable. For additional gene function descriptions, we gathered information about the biochemical pathways in which gene products are involved in by searching through the Genecards (www.genecards.org; Last accessed on April 22, 2019), Uniprot (UniProt Consortium 2016), and KEGG Pathway (Kanehisa and Goto 2000) databases.
The pipeline for lab work, assembly of transcriptomes and tests for positive selection is summarized in supplementary figure S2, Supplementary Material online, and all scripts for bioinformatics and data analyses are available at https://github.com/marisalim/Transcriptome_pipeline; Last accessed on May 19, 2019. For data analysis, we used computing resources from the National Science Foundation Extreme Science and Engineering Discovery Environment (Blacklight, Greenfield, and Bridges) hosted by the Pittsburgh Supercomputing Center through allocation BIO150018 (Towns et al. 2014) and from Stony Brook University (SeaWulf) hosted by the Institute for Advanced Computational Science.
Results
We successfully extracted RNA and sequenced cDNA for all 12 study species. Across all species, we assembled an average of ∼36 million reads that translated to ∼3,200 transcripts, with N50 of ∼1,000 bp, average transcript length of ∼671 bp, and ∼49% GC content (supplementary table S2, Supplementary Material online). Of the 12,746 annotated genes from the assembled sequence reads, 941 nuclear genes and 6 mitochondrial genes were sequenced for all 12 species. Our sequence alignments included 25 genes that have been identified as candidate genes for altitude adaptation in other vertebrate taxa (supplementary table S3, Supplementary Material online).
In the single-branch foreground analyses, five genes were identified as PSGs based on significant LRTs (ANXA6, EIF4A2, PSAP, GPI, and TCEA3) with a foreground ω = 1 instead of ω > 1 (supplementary table S4, Supplementary Material online). Although we removed these PSGs from downstream analysis, they could be targets for future investigation as their functions overlap with other PSGs retained in the study (supplementary table S4, Supplementary Material online). An additional eight genes from the single-branch analyses had exaggerated foreground ω values, suggesting division by 0 errors (ω = 999), and were also filtered from the data set (GPD1, TMEM38A, and TPM1 in Adelomyiamelanogenys; NDUFB10 in Amaziliaamazilia; RRP8 in Coeligenacoeligena; THRSP in Amaziliaviridicauda; UBXN4 in Metalluraphoebe; and YRDC in Phlogophilusharterti). Two PSGs (GPI and TIMM21) were filtered from further analysis because they were identified in both the single-branch and multibranch foreground codeml analyses, and their inclusion in the latter might bias results from changes in a single branch.
Based on results from the PAML permutation tests, ∼92% of the combinations showed no signature of positive selection with the arbitrary foreground branch groups (supplementary fig. S3, Supplementary Material online). However, ∼8% of the combinations did indicate positive selection for one to seven of the permutations. A functional relationship to elevation adaptation may exist for this subset of PSGs (supplementary table S5, Supplementary Material online), but we focused further interpretation on the PSGs that showed no sign of positive selection for any of the arbitrary foreground branch permutations.
When GO information was available, the gene function descriptions were the same from both chicken and human genomes, suggesting the functions are sufficiently conserved across avian and mammalian species to be useful for inferring gene function in hummingbirds. We used GO terms from the human genome for four genes (CLPB, JPH1, and MLF1), as these were unavailable from the chicken genome. Complete biological process ontology terms for annotated PSGs identified in our study are in supplementary table S6, Supplementary Material online.
Are There Pathways, PSGs, or Sites Shared across High- or Low-Elevation Species?
In the multibranch analysis with high-elevation species >2,500 m in the foreground branches, we identified six shared PSGs and five shared positively selected sites (Test 1: table 1). One of the PSGs is involved in mitochondrial translation and organelle biogenesis and maintenance (MRPS26). The other PSGs are involved in respiratory electron transport and oxidative phosphorylation (UQCRQ; fig. 2), metabolism of proteins (TIMM17A), negative regulation of angiotensin II resulting in increased blood pressure (AGTRAP), and formation of fibrin clots in the blood stream (C1QBP). AGTRAP was previously reported to have undergone positive selection in Andean human populations (Bigham et al. 2010), and in a Tibetan lizard species (Phrynocephalus erythrurus) (Yang et al. 2015). The other PSGs have not been reported previously in the high-altitude genetics literature. However, a few genome-wide association studies of human populations identified C1QBP as an immunity gene candidate for positive selection (Barreiro and Quintana-Murci 2010).
Table 1.
PSGs Shared by Species >2,500 m (Test 1) or >1,500 m (Test 1a)a
| Test | Ensembl ID | Gene Symbol | Foreground ω | Positively Selected Sites for Foreground Lineages | Biochemical Pathway or Gene Function Information |
|---|---|---|---|---|---|
| Test 1 | 00194 | UQCRQ | 11.64 | His30* | Respiratory electron transport, oxidative phosphorylation |
| 01468 | TIMM17A | 22.28 | Metabolism of proteins; mitochondrial protein import | ||
| 01776 | CCNI | 27.48 | Ala324* | Regulation of the cell cycle | |
| 02414 | AGTRAP | 40.04 | Negative regulation of angiotensin II (causes vasoconstriction and increase in blood pressure) signaling | ||
| 04374 | C1QBP | 8.17 | Asp66*; Phe104** | Formation of fibrin clot (clotting cascade) | |
| 11312 | MRPS26 | 7.72 | Ala60* | Mitochondrial translation; organelle biogenesis and maintenance | |
| Test 1a | 01031 | EEF2 | 8.57 | Immune function; viral mRNA translation | |
| 04086 | EPAS1 | 59.96 | Cellular response to hypoxia | ||
| 04383 | CCT3 | 16.57 | Assists with protein folding | ||
| 07661 | NDUFS8 | 11.24 | Ser69** | Respiratory electron transport, oxidative phosphorylation | |
| 09466 | PDHB | 13.76 | Val214* | Pyruvate metabolism, Krebs cycle | |
| 09734 | 37.25 | ||||
| 10265 | EEF1B2 | 12.2 | Viral mRNA translation | ||
| 13074 | CDKN1B | 21.14 | Cell signaling; cell cycle | ||
| 13412 | AAMDC | 3.71 | Little information available; Adipogenesis Associated Mth938 Domain Containing is a Protein Coding gene | ||
| 14844 | HADHB | 13.77 | Glycerophospholipid biosynthesis; metabolism; fatty acid metabolism | ||
| 17602 | MGST3 | 22.43 | Ser141** | Glutathione metabolism |
Ensembl IDs begin with prefix ENSTGUP000000. The associated maximum likelihood estimates for foreground dN/dS (ω) ratios are shown for two site classes. For these site classes, the foreground branches are both evolving under positive selection (ω > 1), but the background branches are either evolving neutrally (ω = 1) or under purifying selection (ω < 1). Positively selected sites inferred based on Bayes Empirical Bayes at P > 95% (*) or P > 99% (**) are shown with the reference amino acid (first sequence in alignment) and site position. Pathway or gene function information for annotated PSGs was compiled from the Panther and Genecards databases.
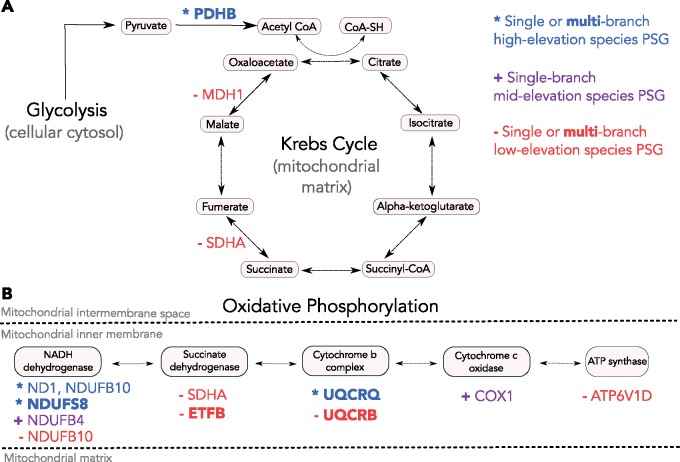
Fig. 2.
—Diagram of the subset of PSGs that are involved with cellular respiration in (A) the Krebs cycle or (B) the oxidative phosphorylation pathway. We used information from the Genecards and KEGG databases to identify the location where PSG gene products act within the pathways. The font color and symbols (*, +, and −) for PSGs indicate the elevation category of the species for which the gene was identified. PSG results from the multibranch foreground PAML analyses are indicated in bold text.
In the negative control to Test 1, the multibranch analysis with low-elevation species <1,500 m in the foreground branches resulted in 11 shared PSGs (Test 2: supplementary table S7, Supplementary Material online). In general, there were limited and nonspecific functional overlaps based on GO information for PSGs from Tests 1 and 2 (supplementary table S8, Supplementary Material online). Three PSGs related to the respiratory electron transport pathway were also found for Test 1 (UQCRQ) and Test 2 (ETFB and UQCRB) (fig. 2). The remaining PSGs for low-elevation species are involved in pathways related to striated muscle contraction (TNNT3), diverse cellular processes (SPG7), iron-sulfur cluster binding (ISCA1), and autophagy initiation (DRAM2). ISCA1 was, however, previously identified in a high-elevation study of human populations from the Ethiopian highlands (Huerta-Sánchez et al. 2013).
How Are the Sets of PSGs Affected by Changing the Definitions of “High Elevation” and “Low Elevation”?
We identified 11 shared PSGs in the multibranch analysis for species with midpoint elevations >1,500 m as foreground branches (Test 1a; table 1). Surprisingly, by shifting the threshold for high-elevation from 2,500 to 1,500 m, we recovered a completely different set of PSGs, with no overlapping genes. One notable result of shifting the midpoint elevation threshold down to 1,500 m was the identification of EPAS1, a component of the hypoxia-inducible factor pathway that has been identified previously in several studies (Wang et al. 2014; Zhang et al. 2014; Graham and McCracken 2019), including those of high-elevation human populations (Beall et al. 2010; Yi et al. 2010). In addition, more of the PSGs shared by species above the 1,500-m threshold were related to metabolic and immune functions, compared with those above the 2,500-m threshold (table 1). Although the sets of PSGs differed between Tests 1 and 1a, there were a few common pathways, such as respiratory electron transport (fig. 2), cell cycle regulation, and mitochondrial translation (table 1), and a few common GO terms including response to hypoxia, transcription, and visual perception (supplementary table S8, Supplementary Material online).
In the negative control to Test 1a, we identified 15 PSGs shared by species with midpoint elevations <2,500 m (Test 2a; supplementary table S7, Supplementary Material online). The shift to a higher threshold for low-elevation species resulted in very few functional overlaps and just one common PSG. In general, genes identified for species <1,500 m have diverse cellular functions, whereas several of the genes for species <2,500 m are involved in metabolic processes (supplementary table S7, Supplementary Material online). There were only broad functional similarities as both analyses identified genes involved in respiratory electron transport (fig. 2) and metabolism (supplementary table S8, Supplementary Material online). The only PSG identified in both analyses was ENSTGUP00000017537, which is uncharacterized, but may be involved in cell redox homeostasis (Uniprot ID H1A3S4). The shared site analysis identified some of the same sites for ENSTGUP00000017537, but none were significant at P < 0.05.
Do Different PSGs Occur in the Same Biochemical Pathways across Foreground Species for Each Elevation Category?
For all but three PSGs from the single-branch foreground analyses (CLPB, JPH1, and NOB1; table 2), we identified different PSGs that share similar function or occur in the same pathway for low-, mid-, and high-elevation species. Considering all elevation categories combined, there were seven unique PSGs related to energy metabolism pathways (fig. 2 and table 2). Two highland species had PSGs from the oxidative phosphorylation pathway (NDUFB10 in Coeligenaviolifer; ND1 in Patagonagigas peruviana). One midelevation species had two PSGs in the oxidative phosphorylation pathway (NDUFB4 and COX1 in Chaetocercusmulsant). Two low-elevation species had PSGs involved in the Krebs cycle (MDH1 in Amaziliaamazilia; SDHA in P. harterti), and two low-elevation species had PSGs involved in oxidative phosphorylation (ATP6V1D in Phaethornismalaris; NDUFB10 and SDHA in P. harterti).
Table 2.
PSGs from the Single-Branch Foreground Analysesa
| Foreground Branch | Ensembl ID | Gene Symbol | Adjusted P Value (P < 0.05) | Foreground ω | Biochemical Pathway or Gene Function Information |
|---|---|---|---|---|---|
| Aglaeactis castelnaudii | 11922 | JPH1 | 0 | 7.53 | Part of structural foundation and cross-talk across cell membrane in skeletal muscle |
| Amazilia amazilia | 02317 | MDH1 | 0.017 | 2.37 | Glucose metabolism, Krebs cycle |
| Amazilia viridicauda | 01501 | DNAJA1 | 0.012 | 281.75 | Heat shock protein cochaperone; regulation of apoptosis in response to cellular stress |
| Amazilia viridicauda | 08371 | NOB1 | 0.003 | 139.67 | rRNA processing in the nucleus and cytosol |
| Chaetocercus mulsant | 00167 | CACTIN | 0.017 | 34.35 | Regulation of innate immune response |
| Chaetocercus mulsant | 18304 | COX1 | 0 | 1.00021 | Respiratory electron transport, oxidative phosphorylation |
| Chaetocercus mulsant | 18150 | FEM1A | 0 | 619.67 | Substrate recognition; anti-inflammatory signaling |
| Chaetocercus mulsant | 13770 | NDUFB4 | 0.038 | 108.64 | Respiratory electron transport, oxidative phosphorylation |
| Coeligena violifer | 05988 | ATG9A | 0.007 | 17.95 | Related to autophagy |
| Coeligena violifer | 06887 | NDUFB10 | 0.008 | 5.91 | Respiratory electron transport, oxidative phosphorylation |
| Colibri coruscans | 05988 | ATG9A | 0.008 | 22.73 | Related to autophagy |
| Colibri coruscans | 13977 | CLPB | 0 | 8.61 | Longevity regulating pathway; diverse cellular activities |
| Patagona gigas peruviana | 18302 | ND1 | 0 | 15.53 | Respiratory electron transport, oxidative phosphorylation |
| Phaethornis malaris | 11919 | ATP6V1D | 0.018 | 1.34 | Respiratory electron transport, oxidative phosphorylation; insulin receptor recycling; innate immune system |
| Phaethornis malaris | 11555 | MLF1 | 0.016 | 13.71 | Transcriptional misregulation in cancer |
| Phlogophilus harterti | 08535 | MFGE8 | 0.036 | 1.13 | Metabolism of proteins; promotes phagocytosis of apoptotic cells; immune function |
| Phlogophilus harterti | 06887 | NDUFB10 | 0.003 | 780.9 | Respiratory electron transport, oxidative phosphorylation |
| Phlogophilus harterti | 08539 | SDHA | 0.04 | 1.09 | Respiratory electron transport, oxidative phosphorylation; Krebs cycle |
Ensembl IDs begin with prefix ENSTGUP000000. The associated foreground branch omega (ω) values are shown for two site classes. For these site classes, the foreground branches are both evolving under positive selection (ω > 1), but the background branches are either evolving neutrally (ω = 1) or under purifying selection (ω < 1). Genes with foreground ω values just above one should be interpreted with caution, as they may represent positive selection or relaxed purifying selection. Pathway or gene function information was compiled from the Panther and Genecards databases.
In all elevation categories, there were PSGs associated with apoptotic programmed cell death (table 2). Two highland species shared the same PSG, but no positively selected sites, related to autophagy (ATG9A in C. violifer and Colibricoruscans), an alternative to cell death that occurs within cells to deliver damaged organelles to the lysosome. One midelevation species had a PSG involved in apoptosis (DNAJA1 in A. viridicauda). In addition, positive selection on immune response genes in a midelevation species was related to inflammatory response and regulation of signaling pathways that mark viruses or infected cells prior to phagocytosis (FEM1A and CACTIN in Ch. mulsant). Two low-elevation species had PSGs involved in cell cycle arrest (MLF1 in Phaethornismalaris) and phagocytosis (MFGE8 in P. harterti), a process through which viruses or infected cells are engulfed and destroyed.
In these single-branch foreground analyses, we identified one previously identified candidate gene (DNAJA1, A. viridicauda) and 18 new species-specific PSGs for Andean hummingbirds (table 2). DNAJA1 was previously identified as a PSG in great tits of the eastern Himalayas (Parus major) (Qu et al. 2015). A test for putative selective sweeps in Tibetan highland chicken populations (Gallus gallus) identified JPH2 (Zhang et al. 2016), a gene involved in cardiac muscle development and paralog of the Aglaeactiscastelnaudii PSG, JPH1. In the context of adaptation to cold temperatures, COX1 was identified as a candidate gene in polar bears (Ursus maritimus) (Welch et al. 2014). Furthermore, key physiological adaptations for bar-headed geese (Anser indicus), which fly over the Himalayas, involve changes to cytochrome c oxidase enzymatic activity and the genes that encode it, including mutations in COX3 that could alter its interaction with COX1 (Scott et al. 2011).
Summary: Parallels across Pathways, PSGs, and Sites
The above results are summarized with respect to the shared pathway, gene, and/or site framework in table 3 for species with midpoint elevations >2,500 m and table 4 for species with midpoint elevations >1,500 m. Although we identified genes that occur in several pathways for both “high”-elevation species definitions, two functional categories were repeatedly identified. In both the narrow (>2,500 m) and broad (>1,500 m) definitions, the most commonly shared pathways or gene functions were related to cellular respiration, metabolism, or mitochondrial biogenesis and translation. Across high-elevation species (>2,500 m), there were two PSGs (MRPS26 and UQCRQ) with shared positively selected sites that are all involved in cellular respiration or mitochondrial processes across high-elevation species. One protein metabolism-related PSG, TIMM17A, was shared across all high-elevation species, but there were no shared sites. We also identified two different PSGs, both involved in oxidative phosphorylation for two highland species (NDUFB10, C. violifer; ND1, Patagonagigas peruviana) (fig. 2). Across mid- and high-elevation species (>1,500 m), we found three PSGs with shared positively selected sites (NDUFS8, PDHB, and MGST3), all involved in cellular respiration or mitochondrial processes. One PSG involved in cellular respiration, HADHB, was shared across all mid- and high-elevation species but with no shared sites. For the midelevation species, Ch. mulsant, we found two different PSGs both involved in oxidative phosphorylation (NDUFB4 and COX1) (fig. 2).
Table 3.
Summary of All Codeml Results for Shared Pathways, PSGs, and/or Positively Selected Sites for High-Elevation Species >2,500 m (Colibri coruscans, Aglaeactis castelnaudii, Coeligena violifer, Metallura phoebe, and Patagona gigas peruviana)a
| Species | Shared Pathway/Gene Function under Selection | Shared PSG | Unique PSG | Shared Positively Selected Site |
|---|---|---|---|---|
| C. violifer | Related to autophagy | ATG9A | — | |
| Col. coruscans | — | |||
| C. violifer | Respiratory electron transport, ATP synthesis, metabolism | NDUFB10 | — | |
| P. gigas peruviana | ND1 | — | ||
| High-elevation species >2,500 m | Regulation of the cell cycle | CCNI | Asp80 | |
| Formation of fibrin clot (clotting cascade) | C1QBP | Ala60 | ||
| Mitochondrial translation; organelle biogenesis and maintenance | MRPS26 | Val275 | ||
| Respiratory electron transport, oxidative phosphorylation | UQCRQ | Gly3, Ser4 | ||
| Metabolism of proteins; mitochondrial protein import | TIMM17A | Lys1, Asp2, Lys3 | ||
| Negative regulation of angiotensin II (causes vasoconstriction and increase in blood pressure) signaling | AGTRAP | — |
From left to right, the table shows the group of species that share pathways, PSGs, and positively selected sites (yes–yes–yes), share pathways and PSGs but not sites (yes–yes–no), and that have different PSGs involved in similar pathways/functions (yes–no–no). For shared sites, we show the reference amino acid (first sequence in alignment) and site position, otherwise a dash is shown if no positively selected sites were identified or shared.
Table 4.
Summary of All Codeml Results for Shared Pathways, PSGs, and/or Positively Selected Sites for High-Elevation Species >1,500 m (Coeligena coeligena, Adelomyia melanogenys, Chaetocercus mulsant, Amazilia viridicauda, Colibri coruscans, Aglaeactis castelnaudii, Coeligena violifer, Metallura phoebe, and Patagona gigas peruviana)a
| Species | Shared Pathway/Gene Function under Selection | Shared PSG | Unique PSG | Shared Positively Selected Site |
|---|---|---|---|---|
| A. viridicauda | Promotes apoptosis in response to cellular stress mediated by exposure to anisomycin or UV | DNAJA1 | — | |
| Ch. mulsant | Involved in regulation of innate immune response; regulation of lipopolysaccharide-mediated signaling pathway (can induce apoptosis) | CACTIN | — | |
| Negative regulation of inflammatory response | FEM1A | — | ||
| Respiratory electron transport, oxidative phosphorylation | COX1 | — | ||
| High-elevation species >1,500 m | Glutathione metabolism | MGST3 | Ser141 | |
| Respiratory electron transport, oxidative phosphorylation | NDUFS8 | Ser69 | ||
| Pyruvate metabolism, citric acid cycle | PDHB | Val214 | ||
| Glycerophospholipid biosynthesis; metabolism; fatty acid metabolism | HADHB | — | ||
| (Not well characterized) | 15014 | Ser20 | ||
| (Not well characterized) | AAMDC | — | ||
| Assists with protein folding | CCT3 | — | ||
| Cell signaling; cell cycle | CDKN1B | — | ||
| Viral mRNA translation | EEF1B2 | — | ||
| Immune function; viral mRNA translation | EEF2 | — | ||
| Cellular response to hypoxia | EPAS1 | — | ||
| (Not well characterized) | 09734 | — |
From left to right, the table shows the group of species that share pathways, PSGs, and positively selected sites (yes–yes–yes), share pathways and PSGs but not sites (yes–yes–no), and that have different PSGs involved in similar pathways/functions (yes–no–no). For shared sites, we show the reference amino acid (first sequence in alignment) and site position, otherwise a dash is shown if no positively selected sites were identified or shared. Unannotated PSGs are labeled with their Ensembl ID (prefix ENSTGUP000000).
Pathways and gene functions related to cell death and immune function were also identified in both definitions of “high” elevation. Two high-elevation species (C. violifer and Col. coruscans) shared the same autophagy-related gene, ATG9A, although no sites were positively selected in either species. For the midelevation species, Ch. mulsant, we found two different PSGs related to cellular immune responses prior to phagocytosis (FEM1A and CACTIN).
Lastly, there were some species-specific PSGs that were only identified in one species and whose functions did not overlap with other PSGs in the data set (CLPB in Col. coruscans; JPH1 in Aglaeactiscastelnaudii; NOB1 in A. viridicauda).
Discussion
If different species have adapted to similar environmental conditions, we can evaluate the extent to which molecular adaptation is limited to specific pathways, genes, or sites, and is therefore constrained and deterministic (Nielsen 2005; Stapley et al. 2010). Using comparative transcriptomics, we studied Andean hummingbird species that independently evolved to live at high altitudes to test for molecular parallelism at three hierarchical levels. With this framework, we found greater predictability for genetic adaptation to high elevation at particular biochemical pathways and biological functions than in specific genes or mutations, regardless of how “high elevation” was defined. Thus, our results suggest that selection on different genes can result in functional convergence, such that the same phenotypic adaptations are attained through different genetic mechanisms. This finding is concordant with recent studies that show repeated selection across different taxa on the same pathways (Arendt and Reznick 2008; Manceau et al. 2010; Sun et al. 2018), or candidate genes (Natarajan et al. 2016), is more common than evolution of the same mutations in a given gene.
Our test for genes under selection in sets of lowland species served as a negative control for our test of selection related to high elevation. In the comparison of PSGs for highland versus lowland hummingbirds, there were only overlaps in general functional categories such as protein binding. The main shared pathway was the respiratory electron transport chain, which is unsurprising given that this energy production pathway has many biological roles and hummingbirds have extremely high metabolism regardless of elevation (Lasiewski 1963). Hence, cellular respiration may represent a pathway for which positive natural selection operates across the elevational gradient. Many PSGs for high-elevation species occur in oxidative phosphorylation and mitochondrial process pathways. Variation in oxygen availability should impact processes such as oxidative phosphorylation, which requires modulation of oxygen supply to sustain cellular energy production while avoiding cell damage. Indeed, other studies have shown that natural selection acts on the mitochondrial genes of organisms living in high-elevation hypoxic environments (Ehinger et al. 2002; Cheviron et al. 2008; Cheviron and Brumfield 2009; Scott et al. 2011; Sun et al. 2018).
In addition, several genes identified for mid- and high-elevation species were related to cell death and immune function. These PSGs could be related to damage resulting from increased UV exposure in highland environments (Alkorta-Aranburu et al. 2012; Qu et al. 2013; Zhang et al. 2016), or to shifts in life-history strategies associated with elevation (Boyce et al. 2015). The physiological response to hypoxia includes tissue inflammation, so selection on genes like FEM1A, which negatively regulate the inflammatory response, and CACTIN, which regulates signaling for immune response, may also be pertinent for highland species (Bartels et al. 2013). Furthermore, the same gene related to autophagy (ATG9A) was independently identified for two high-elevation species, C. violifer and Col. coruscans. ATG9A encodes a protein required for autophagy to occur under low-glucose and low-oxygen situations (Weerasekara et al. 2014).
In some cases, we identified the same genes as other studies of highland taxa (AGTRAP, DNAJA1, and EPAS1), bolstering support for the contribution of these particular genes to high-altitude adaptation (Bigham et al. 2010; Yi et al. 2010; Wang et al. 2014; Zhang et al. 2014; Qu et al. 2015; Yang et al. 2015; Graham and McCracken 2019). Otherwise, most of the PSGs we identified are novel candidates for high-elevation adaptation with similar functions and roles in pathways as candidate genes found by other high-elevation studies. Our results therefore reinforce the observation that there is likely flexibility in the evolutionary machinery for adaptation across divergent taxa. Although testing the functional consequences of these gene variants is outside the scope of this study, repeatedly identified PSGs, or PSGs with functional convergence provide a starting point for integrating physiological and genetic research of pathways involved in blood flow, energy production, and oxygen sensing.
Research concerning the genetic underpinnings of high-elevation adaptation rarely addresses the potential variation in targets of selection at different points along elevational gradients (but see Sun et al. 2018). Although some gene functions were the same, our set of PSGs changed strikingly depending on the elevational threshold applied. By grouping midelevation species with either high- or low-elevation groups, we shifted the threshold at which these categories turn over, from ∼1,500 to ∼2,500-m elevation. This allowed us to coarsely approximate the elevation at which positive natural selection begins to act on particular genes. For example, a previously well-studied candidate gene, EPAS1, was only identified in the analysis that included both mid- and high-elevation species in the foreground branches. This suggests that positive selection on EPAS1 occurs even at relatively modest elevations above 1,500 m. In addition, this result also supports the role for regulatory elements in high-altitude adaptation, as EPAS1 is a transcription factor. Measuring variation in the signature of natural selection along elevational gradients presents an exciting challenge for future research, which may entail stratified sampling schemes along elevation gradients to examine variation in physiological responses, gene expression, protein formation, or allele frequencies.
Our negative controls and permutations confirm that a subset of PSGs related to metabolism—and specifically, oxidative phosphorylation—hypoxia response, and blood pressure were not false positives at either elevational threshold. Thus, based on the physiological demands of high-elevation environments, the biology of Andean hummingbirds, and the findings documented in previous high-elevation studies, we argue that the regions of the transcriptome discussed in this paper are involved in dealing with low oxygen and increased metabolic requirements. The phenotypic effects of this new set of PSGs and sites need to be assessed using experiments to test for physical changes in protein function due to, for example, particular gene mutations (Projecto-Garcia et al. 2013; Hauser et al. 2017). These experiments could also shed light on the potential influence of multinucleotide mutations on detection of PSGs (Venkat et al. 2018). Future studies should be designed to provide additional resolution on key elevational thresholds for natural selection. Because the short- and long-term physiology and genetics of adaptation to high-altitude may differ, comparisons of population- and species-level tests could yield additional insights on the processes involved. A broader taxonomic survey of high-altitude species could also increase the statistical power to detect genetic adaptation of convergent biochemical pathways.
By analyzing patterns of molecular evolution at a large set of expressed genes, our results complement previous research on hummingbirds focused on the ecological, biomechanical, physiological, and functional genetic aspects of high-altitude adaptation. Instead of analyzing single genes in isolation, we gained unique insights from analyzing PSGs according to their functional context in biochemical pathways and within a hierarchical framework (following Cheviron and Brumfield 2012; Rosenblum et al. 2014). Our study design provides a template for future research to test the extent to which genetic mechanisms for adaptation are predictable across species that independently colonized regions with similar environmental challenges. It seems that within certain biochemical pathways, there are numerous possible ways to optimize function after an elevational shift, leading to a diversity of adaptive paths. Alternatively, the diversity of paths could result from lineage-specific constraints arising from different genetic backgrounds and pervasive epistasis. Building on the growing number of genetic examples of convergent evolution, the positively selected pathways, genes, and sites that we identified contribute to the ecological annotation of Andean hummingbird transcriptomes and will inform future research on physiology, ecology, and evolution in organisms that evolved into and out of mountains.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Ke Bi, Sonal Singhal, Lydia Smith, and Mark Phuong for helpful discussions and laboratory support. This work was supported by an American Ornithologists Union Research Award, Wilson Ornithology Society Award, National Science Foundation (NSF) Graduate Research Fellowship, NSF DEB-1442142, NSF DEB-1146491, funds from the Swiss Federal Institute for Forest, Snow and Landscape Research (WSL), and two startup funds from Stony Brook University. Sampling was assisted by Emil Bautista, Andrew B. Johnson, Thomas Valqui, and many students from CORBIDI and the Museum of Southwestern Biology at UNM. This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation Grants (Grant Nos. S10RR029668 and S10RR027303). For data analysis, we used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by NSF (Grant No. ACI-1548562). The authors would also like to thank Stony Brook Research Computing and Cyberinfrastructure, and the Institute for Advanced Computational Science at Stony Brook University for access to the SeaWulf computing system, which was made possible by funding from NSF (Grant No. 1531492).
Data deposition: The raw read data have been deposited in the NCBI Sequence Read Archive under BioProject: PRJNA543673, BioSample: SAMN11774663-SAMN11774674, SRA Study: SRP198856. All scripts used for analyses are available on Dryad: doi:10.5061/dryad.v961mb4.
Literature Cited
- Alkorta-Aranburu G, et al. 2012. The genetic architecture of adaptations to high altitude in Ethiopia. PLoS Genet. 8(12):e1003110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler DL, Dudley R.. 2006. The physiology and biomechanics of avian flight at high altitude. Integr Comp Biol. 46(1):62–71. [DOI] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Cambridge, UK: Babraham Institute. Available from: www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed March 25, 2015.
- Anisimova M, Yang Z.. 2007. Multiple hypothesis testing to detect lineages under positive selection that affects only a few sites. Mol Biol Evol. 24(5):1219–1228. [DOI] [PubMed] [Google Scholar]
- Arendt J, Reznick D.. 2008. Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends Ecol Evol. 23(1):26–32. [DOI] [PubMed] [Google Scholar]
- Bailey SF, Rodrigue N, Kassen R.. 2015. The effect of selection environment on the probability of parallel evolution. Mol Biol Evol. 32(6):1436–1448. [DOI] [PubMed] [Google Scholar]
- Barreiro LB, Quintana-Murci L.. 2010. From evolutionary genetics to human host defence genes. Nat Rev Genet. 11(1):17–30. [DOI] [PubMed] [Google Scholar]
- Bartels K, Grenz A, Eltzschig HK.. 2013. Hypoxia and inflammation are two sides of the same coin. Proc Natl Acad Sci U S A. 110(46):18351–18352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall CM, et al. 1998. Hemoglobin concentration of high-altitude Tibetans and Bolivian Aymara. Am J Phys Anthropol. 106(3):385–400. [DOI] [PubMed] [Google Scholar]
- Beall CM, et al. 2010. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci U S A. 107(25):11459–11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benham PM, et al. 2011. Satellite imagery reveals new critical habitat for endangered bird species in the high Andes of Peru. Endang Species Res. 13(2):145–157. [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300. [Google Scholar]
- Bigham A, et al. 2010. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 6(9):e1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce AJ, Freeman BG, Mitchell AE, Martin TE.. 2015. Clutch size declines with elevation in tropical birds. Auk Ornithol Adv. 132(2):424–432. [Google Scholar]
- Cheviron ZA, Brumfield RT.. 2009. Migration-selection balance and local adaptation of mitochondrial haplotypes in rufous-collared sparrows (Zonotrichia capensis) along an elevational gradient. Evolution 63(6):1593–1605. [DOI] [PubMed] [Google Scholar]
- Cheviron ZA, Brumfield RT.. 2012. Genomic insights into adaptation to high-altitude environments. Heredity 108(4):354–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheviron ZA, Whitehead A, Brumfield RT.. 2008. Transcriptomic variation and plasticity in rufous-collared sparrows (Zonotrichia capensis) along an altitudinal gradient. Mol Ecol. 17(20):4556–4569. [DOI] [PubMed] [Google Scholar]
- del Hoyo J, Elliott A, Sargatal J, Christie DA, de Juana E, editors. 2018. Handbook of the birds of the world alive. Barcelona (Spain: ): Lynx Edicions; Available from: http://www.hbw.com/ (accessed June 28, 2018). [Google Scholar]
- Ehinger M, Fontanillas P, Petit E, Perrin N.. 2002. Mitochondrial DNA variation along an altitudinal gradient in the greater white-toothed shrew, Crocidura russula. Mol Ecol. 11(5):939–945. [DOI] [PubMed] [Google Scholar]
- Foll M, Gaggiotti OE, Daub JT, Vatsiou A, Excoffier L.. 2014. Widespread signals of convergent adaptation to high altitude in Asia and America. Am J Hum Genet. 95(4):394–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham AM, McCracken KG.. 2019. Convergent evolution on the hypoxia-inducible factor (HIF) pathway genes EGLN1 and EPAS1 in high-altitude ducks. Heredity 122(6):819–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham AM, et al. 2018. Migration-selection balance drives genetic differentiation in genes associated with high-altitude function in the Speckled Teal (Anas flavirostris) in the Andes. Genome Biol Evol. 10(1):14–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainsworth R, Drinkhill MJ.. 2007. Cardiovascular adjustments for life at high altitude. Respir Physiol Neurobiol. 158(2-3):204–211. [DOI] [PubMed] [Google Scholar]
- Hauser FE, et al. 2017. Accelerated evolution and functional divergence of the dim light visual pigment accompanies cichlid colonization of Central America. Mol Biol Evol. 34(10):2650–2664. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP.. 2006. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science 313(5783):101–104. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, Serra F, Bork P.. 2016. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol. 33(6):1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Sánchez E, et al. 2013. Genetic signatures reveal high-altitude adaptation in a set of Ethiopian populations. Mol Biol Evol. 30(8):1877–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S.. 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasiewski RC. 1963. Oxygen consumption of torpid, resting, active, and flying hummingbirds. Physiol Zool. 36(2):122–140. [Google Scholar]
- Li W, Godzik A.. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22(13):1658–1659. [DOI] [PubMed] [Google Scholar]
- Linnen CR, et al. 2013. Adaptive evolution of multiple traits through multiple mutations at a single gene. Science 339(6125):1312–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč T, Salzberg SL.. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manceau M, Domingues VS, Linnen CR, Rosenblum EB, Hoekstra HE.. 2010. Convergence in pigmentation at multiple levels: mutations, genes and function. Philos Trans R Soc B 365(1552):2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken KG, Barger CP, Sorenson MD.. 2010. Phylogenetic and structural analysis of the HbA (alphaA/betaA) and HbD (alphaD/betaA) hemoglobin genes in two high-altitude waterfowl from the Himalayas and the Andes: bar-headed (Anser indicus) and Andean goose (Chloephaga melanoptera). Mol Phylogenet Evol. 56(2):649–658. [DOI] [PubMed] [Google Scholar]
- McGuire JA, et al. 2014. Molecular phylogenetics and the diversification of hummingbirds. Curr Biol. 24(8):910–916. [DOI] [PubMed] [Google Scholar]
- Mi H, et al. 2017. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45:183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monge C, León-Velarde F.. 1991. Physiological adaptation to high altitude: oxygen transport in mammals and birds. Physiol Rev. 71(4):1135–1172. [DOI] [PubMed] [Google Scholar]
- Muñoz-Fuentes V, Cortázar-Chinarro M, Lozano-Jaramillo M, McCracken KG.. 2013. Stepwise colonization of the Andes by ruddy ducks and the evolution of novel β-globin variants. Mol Ecol. 22(5):1231–1249. [DOI] [PubMed] [Google Scholar]
- Natarajan C, et al. 2016. Predictable convergence in hemoglobin function has unpredictable molecular underpinnings. Science 354(6310):336–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. 2005. Molecular signatures of natural selection. Annu Rev Genet. 39(1):197–218. [DOI] [PubMed] [Google Scholar]
- Parker T, Stotz D, Fitzpatrick J.. 1996. Database A: zoogeography and ecological attributes of bird species breeding in the Neotropics In: Neotropical birds: ecology and conservation. Chicago (IL: ): University of Chicago Press; p. 131–291. [Google Scholar]
- Projecto-Garcia J, et al. 2013. Repeated elevational transitions in hemoglobin function during the evolution of Andean hummingbirds. Proc Natl Acad Sci U S A. 110(51):20669–20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, et al. 2013. Ground tit genome reveals avian adaptation to living at high altitudes in the Tibetan plateau. Nat Commun. 4:1–9. [DOI] [PubMed] [Google Scholar]
- Qu Y, et al. 2015. Genetic responses to seasonal variation in altitudinal stress: whole-genome resequencing of great tit in eastern Himalayas. Sci Rep. 5:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römpler H, et al. 2006. Nuclear gene indicates coat-color polymorphism in mammoths. Science 313(5783):62. [DOI] [PubMed] [Google Scholar]
- Rosenblum EB, Parent CE, Brandt EE.. 2014. The molecular basis of phenotypic convergence. Annu Rev Ecol Evol Syst. 45(1):203–226. [Google Scholar]
- Rosenblum EB, Römpler H, Schöneberg T, Hoekstra HE.. 2010. Molecular and functional basis of phenotypic convergence in white lizards at White Sands. Proc Natl Acad Sci U S A. 107(5):2113–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulenberg TS, Stotz DF, Lane DF, O’Neill JP, Parker TA.. 2007. Birds of Peru. Princeton (NJ: ): Princeton University Press. [Google Scholar]
- Scott GR, et al. 2011. Molecular evolution of cytochrome C oxidase underlies high-altitude adaptation in the bar-headed goose. Mol Biol Evol. 28(1):351–363. [DOI] [PubMed] [Google Scholar]
- Simonson TS, McClain DA, Jorde LB, Prchal JT.. 2012. Genetic determinants of Tibetan high-altitude adaptation. Hum Genet. 131(4):527–533. [DOI] [PubMed] [Google Scholar]
- Singhal S. 2013. De novo transcriptomic analyses for non-model organisms: an evaluation of methods across a multi-species data set. Mol Ecol Resour. 13(3):403–416. [DOI] [PubMed] [Google Scholar]
- Slater G, Birney E.. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6(1):31.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapley J, et al. 2010. Adaptation genomics: the next generation. Trends Ecol Evol. 25(12):705–712. [DOI] [PubMed] [Google Scholar]
- Stiles GF. 2008. Ecomorphology and phylogeny of hummingbirds: divergence and convergence in adaptations to high elevations. Ornithol Neotrop. 19:511–519. [Google Scholar]
- Storz JF, et al. 2009. Evolutionary and functional insights into the mechanism underlying high-altitude adaptation of deer mouse hemoglobin. Proc Natl Acad Sci U S A. 106(34):14450–14455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y-B, et al. 2018. Species groups distributed across elevational gradients reveal convergent and continuous genetic adaptation to high elevations. Proc Natl Acad Sci U S A. 115(45):E10634–E10641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towns J, et al. 2014. XSEDE: accelerating scientific discovery. Comput Sci Eng. 16(5):62–74. [Google Scholar]
- UniProt Consortium. 2016. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45:158–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkat A, Hahn MW, Thornton JW.. 2018. Multinucleotide mutations cause false inferences of lineage-specific positive selection. Nat Ecol Evol. 2:1280–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G-D, et al. 2014. Genetic convergence in the adaptation of dogs and humans to the high altitude environment of the Tibetan plateau. Genome Biol Evol. 6(8):2122–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerasekara VK, et al. 2014. Metabolic-stress-induced rearrangement of the 14-3-3ζ interactome promotes autophagy via a ULK1- and AMPK-regulated 14-3-3ζ interaction with phosphorylated Atg9. Mol Cell Biol. 34(24):4379–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch AJ, et al. 2014. Polar bears exhibit genome-wide signatures of bioenergetic adaptation to life in the arctic environment. Genome Biol Evol. 6:433–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, et al. 2015. Comparative transcriptomic analysis revealed adaptation mechanism of Phrynocephalus erythrurus, the highest altitude lizard living in the Qinghai-Tibet Plateau. BMC Evol Biol. 15:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yi X, et al. 2010. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329:75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, et al. 2016. Genome resequencing identifies unique adaptations of Tibetan chickens to hypoxia and high-dose ultraviolet radiation in high-altitude environments. Genome Biol Evol. 8:evw032.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, et al. 2014. Hypoxia adaptations in the Grey Wolf (Canis lupus chanco) from Qinghai-Tibet Plateau. PLoS Genet. 10(7):e1004466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang J, et al. 1993. Hypoxic ventilatory responsiveness in Tibetan compared with Han residents of 3,658 m. J Appl Physiol. 74(1):303–311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.