Abstract
N6-methyladenine (6mA or m6dA) is a DNA modification that has long been known to play an important role in a variety of biological functions in prokaryotes. This modification has only recently been described in eukaryotes, where it seems to have evolved species-specific functions ranging from nucleosome positioning to transposon repression. In Drosophila, 6mA has been shown to be important for enforcing the tissue specificity of neuronal genes in the brain and suppressing transposable element expression in the ovaries. In this study, we have analyzed the raw signal data from nanopore sequencing to identify 6mA positions in the D. melanogaster genome at single-base resolution. We find that this modification is enriched upstream from transcription start sites, within the introns and 3′ UTRs of genes, as well as in simple repeats. These 6mA positions are enriched for sequence motifs that are recognized by known transcriptional activators involved in development, such as Bicoid and Caudal, and the genes that carry this modification are enriched for functions involved in development, regulation of transcription, and neuronal activity. These genes show high expression specificity in a variety of tissues besides the brain, suggesting that this modification may play a more general role in enforcing the specificity of gene expression across many tissues, throughout development, and between the sexes.
Keywords: Drosophila methylation epigenetics 6mA nanopore
The DNA modification N6-methyladenine (6mA) is common among prokaryotes and is known to play a role in the restriction/modification systems involved in defense against bacteriophage infection (Luria and Human 1952), as well as in the regulation of gene expression (Oshima et al. 2002; Reisenauer and Shapiro 2002). This modification has only recently been described in eukaryotes, where it has been implicated in a variety of functions. In the ciliate Tetrahymena thermophila and the green alga Chlamydomonas, 6mA is associated with active transcription and may play a role in nucleosome positioning (Fu et al. 2015; Wang et al. 2017; Luo et al. 2018). In humans, the modification is also associated with active transcription and is enriched in exons (Xiao et al. 2018). In C. elegans, 6mA is broadly distributed across the genome and its exact function remains unclear (Greer et al. 2015), whereas in Drosophila and mice, there is evidence that the 6mA modification is involved in suppression of transposable elements and transcriptional repression in neurons (Zhang et al. 2015; Yao et al. 2017; Yao et al. 2018). 6mA is most prevalent in the early Drosophila embryo, where it may play a role in development, but this modification has also been identified in ovary and brain tissues (Zhang et al. 2015; Yao et al. 2018). In Drosophila ovaries, there is evidence that 6mA is involved in transposon suppression, while in the brain, 6mA acts in concert with polycomb group proteins to repress gene expression (Zhang et al. 2015; Yao et al. 2018). A 6mA methyltransferase has not been identified in Drosophila, however the ten-eleven-translocation (TET) protein, DMAD, acts as a demethylase that facilitates the removal of 6mA (Zhang et al. 2015). Recent work has shown that DMAD null mutants accumulate 6mA methylation in the brain and have defects in brain development, suggesting that removal of the 6mA modification by DMAD is required for gene activation and proper brain development (Yao et al. 2018). Based on these results, Yao et al. (2018) describe a model for 6mA function where this modification enforces transcriptional silencing of a set of neuronal genes outside of the brain. Within neurons, DMAD acts in combination with the transcriptional activator Wds to remove 6mA from these genes, leading to their neuron-specific expression.
Based on these results, we hypothesized that the 6mA modification might play a more general role in enforcing the tissue-specific expression of a much larger set of genes across multiple tissues in the adult fly. To test this hypothesis, we analyzed the raw signal data generated by nanopore sequencing of Drosophila melanogaster adult females, to identify a set of high-confidence genomic positions where the 6mA modification is present in the majority of cells in the adult fly. We find that these positions are enriched at simple repeats and within the introns, 3′ UTRs, and upstream regions of genes. The genes that carry these modifications are highly enriched for developmental, regulatory, and neuronal functions, they are expressed in a variety of tissues, and their expression is significantly more tissue specific than unmethylated genes.
Materials & Methods
Nanopore Sequencing
DNA was extracted from 30 adult females using the Qiagen DNeasy Blood & Tissue Kit and prepped for sequencing using the Oxford Nanopore Technologies (ONT) SQK-LSK108 library preparation kit. The PCR-free libraries were constructed using the ONT 1D Genomic DNA by Ligation protocol and the PCR-based library was constructed using the ONT 1D Low Input Genomic DNA with PCR protocol. Libraries were sequenced on the MinION Mk1B device using version r9.4 flow cells and basecalled using the ONT Albacore software package (version 2.1.10).
Genome Assembly
We used Canu (Koren et al. 2017) to assemble the DGRP732 nanopore reads using an estimated genome size of 140 Mb along with the options: overlapper = mhap utgReAlign = true. We generated Hi-C data from ∼200 mg of 8-16 hr embryos using a in situ DNase Hi-C protocol (Ramani et al. 2016), aligned the data to our Canu assembly using Juicer (Durand et al. 2016), and scaffolded the Canu contigs using the 3D-DNA pipeline (Dudchenko et al. 2017). We used 3D-DNA to create a single “megascaffold”, which we then manually separated into chromosome arms based on comparisons to the D. melanogaster reference assembly. We polished our scaffolds using nanopore reads with Racon (Vaser et al. 2017) and then identified uninformative (i.e., reads that did not contain a ligation junction) Illumina reads from our Hi-C data, which we used as single-end reads to polish the assembly with Pilon (Walker et al. 2014). We used Mercator (Dewey 2007) to create a one-to-one orthology map between assemblies.
Wolbachia
We used BLAST (Altschul et al. 1990) to search the wMel Wolbachia genome assembly against our DGRP732 assembly to determine if any assembled contigs were from Wolbachia. We also searched for Wolbachia-derived nanopore sequences by aligning the raw reads to the wMel genome assembly using graphmap (Sović et al. 2016). We observed high rates of cross-mapping of Drosophila reads to two different ∼100kb segments of the wMel assembly when mapping all reads to the wMel assembly alone. We therefore created a concatenated genome assembly composed of both the DGRP732 and wMel assemblies. We used bedtools to mask the two ∼100kb segments of the wMel genome and samtools (Li et al. 2009) to exclude alignments with mapping quality < 20. We then used bedtools to calculate the average read coverage for the remainder of the Wolbachia genome.
6mA Identification
We used the genome assembly described above and aligned the raw signal data to the assembly using the re-squiggle algorithm in Tombo (version 1.4)(Stoiber et al. 2017). We then calculated sequencing coverage for each genomic position, for each dataset, and removed positions whose coverage fell outside of +/− 50% of the genomic mean. We used the Tombo 6mA model (command: tombo detect_modifications alternative_model–alternate-bases 6mA) to identify A/T positions whose signal level matched a 6mA model better than the canonical base. We did this for each of the two PCR-free sequencing replicates and retained positions where at least 70% of reads were inferred to carry the 6mA modification in replicate 1 and whose percentage estimate was replicated in the second sequencing sample (within +/− 10%). We also ran Tombo with a control dataset using the sample_compare algorithm to identify all positions whose signal deviated from the expected level (determined by the control library)(command: tombo detect_modifications sample_compare) and retained positions with at least 70% modified reads. We used bedtools (Quinlan and Hall 2010) to identify the positions that were retained in both the 6mA model and sample_compare approaches, which became our set of 10,467 high confidence 6mA positions. We followed an analogous approach to also identify 1,648,942 A/T positions where we were confident that they did NOT contain the 6mA modification, which we used as control positions for our genome features permutation test.
Genome Features Enrichment
We transferred the coordinates of our high-confidence positions from the DGRP732 assembly to the D. melanogaster reference genome assembly FlyBase version 6 (Hoskins et al. 2015; Thurmond et al. 2019) by using Mercator (Dewey 2007) to create a whole-genome alignment between the two assemblies. We used the longest isoform per gene from the FlyBase r6.22 genome feature annotations along with bedtools to count the number of positions that overlapped each of the features shown in Figure 2A. TE insertion and simple repeat locations will differ between our strain and the iso1 reference assembly. To determine whether these differences affect our enrichment analysis, we identified TE insertions and simple repeats in both the iso1 and DGRP732 assemblies using RepeatMasker (www.repeatmasker.org) and tantan (Frith 2011) and determined the number of 6mA positions that overlap these features in each assembly. We found the same enrichment patterns in both cases. To determine the statistical significance of enriched or depleted features, we randomly sampled a set of 10,467 positions from our control positions (i.e., those that do not contain the 6mA modification) without replacement and counted the number of features for each annotation category that they overlapped. We repeated this resampling procedure 10,000 times and calculated the p-value using the number of times the counts from the random set were greater than or equal to the counts from our high-confidence positions.
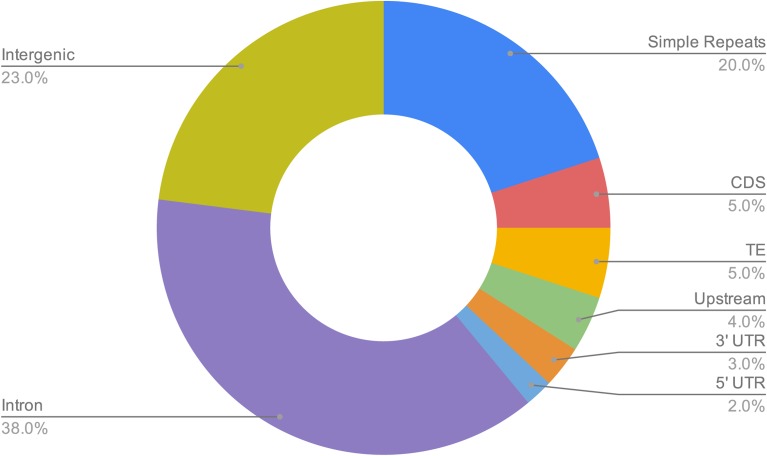
Figure 2.
Genomic features overlapping 6mA positions. The doughnut chart shows the proportion of 6mA positions within each genome feature category. From a permutation test (see Methods), we found that the 6mA positions overlap introns, 3′ UTRs, upstream from transcription start sites, and simple repeats significantly more than expected by chance, whereas they are significantly depleted from transposable elements as well as coding and intergenic sequences (3′ UTR: P = 0.0079, P < 0.0001 in all other cases).
Motif Enrichment
We used bedtools to extract +/− 25 bp of sequence surrounding each high confidence position and the findMotifs.pl script from Homer (Heinz et al. 2010) to identify enriched motifs in these sequences. For the background set of sequences, we used +/− 25 bp surrounding the set of control positions which we determined did not contain the 6mA modification. Similarity to known sequence motifs was reported by findMotifs.pl. To verify the Homer motifs, we used the differential enrichment function from MEME (Bailey et al. 2009) on the same set of sequences that we used for Homer (Heinz et al. 2010). For the MEME search, we used an E-value threshold of 0.1 and min/max motif size of 8 and 12, respectively. We used STAMP (Mahony and Benos 2007) to compare the enriched motifs identified by Homer to those identified by MEME.
6mA-containing Genes
We used bedtools along with the FlyBase gene annotations to identify every gene that overlapped one or more 6mA positions. We identified enriched GO terms for this gene set using GOrilla (Eden et al. 2009) with all FlyBase genes as the background set. We obtained expression enrichment scores for these genes from FlyAtlas (Chintapalli et al. 2007).
Data availability
All sequencing data and the DGRP732 genome assembly are available via the NCBI Sequence Read Archive (SRA) and Whole Genome Shotgun (WGS) databases under Bioproject PRJNA515844. Supplemental material available at FigShare: https://doi.org/10.25387/g3.7841036.
Results
Genome Assembly
We performed nanopore sequencing of DNA extracted from 30 adult females of strain DGRP-732 from the Drosophila Genetic Reference Panel (DGRP)(Mackay et al. 2012)(Table S1). We assembled the nanopore data using Canu (Koren et al. 2017), scaffolded the Canu contigs with Hi-C data using the 3D-DNA pipeline (Dudchenko et al. 2017), and polished the scaffolds using Racon (Vaser et al. 2017) and Pilon (Walker et al. 2014). We obtained contig/scaffold N50 values of 5.4 Mb and 25.7 Mb, respectively and identified one-to-one alignments between our assembly and the D. melanogaster iso1 reference genome (Hoskins et al. 2015) that encompassed 98% of the iso1 assembly. The assembly has been deposited in the NCBI whole genome shotgun (WGS) database (BioProject PRJNA515844) and will be described in more detail in another manuscript (Ellison & Cao, in prep).
Identification and genomic locations of 6mA modifications
We performed nanopore sequencing in two replicates (one flowcell per replicate) and used the 6mA model in the Tombo software package (Stoiber et al. 2017) to estimate the percent of sequences carrying the 6mA modification for each genomic position (percent reads modified, PRM). We found only a moderately strong correlation in PRM values between replicates (Spearman rho = 0.56), which suggests that there is a fair amount of noise in the raw nanopore signal data. For this reason, we used a series of stringent criteria for identifying putative 6mA positions. We started by only considering positions with a PRM value >= 70% in replicate 1 whose PRM value was conserved in our second sequencing experiment (+/− 10%), which resulted in 613,921 genomic positions (Figure 1A).
Figure 1.
Identification of 6mA positions in the Drosophila melanogaster genome. Panel A: Flowchart showing the filtering steps involved in identifying the high-confidence 6mA positions. Panel B: Boxplot showing the difference in signal level between the PCR-free and control sequencing libraries for the high-confidence 6mA positions and unmethylated positions. As expected, the adenine positions that were identified as methylated show a significantly larger difference in signal level compared to the unmethylated positions (Wilcoxon Test P < 2.2e-16).
These two sequencing libraries were generated using a PCR-free protocol in order to preserve the 6mA modifications. We also generated a third library as a control using a PCR-based protocol in order to produce sequences from DNA molecules lacking the 6mA modification. We sequenced this library using a single flowcell and then used the sample compare model in Tombo to identify all genomic positions whose PCR-free current levels deviated from those in the control library. We intersected these positions with those identified using the 6mA model, to create a final set of 10,467 high-confidence 6mA positions (Figure 1A, File S1). We also used an analogous approach to identify a set of positions where we had high-confidence that they did not carry the 6mA modification (6mA-free positions). As expected, we find a much larger difference in current level between the PCR and PCR-free libraries for the 6mA positions compared to the 6mA-free positions (Wilcoxon-test P < 2.2e-16, Figure 1B). To determine whether 6mA positions are conserved between Drosophila strains, we performed nanopore sequencing of a PCR-free library from strain DGRP379 using a single flowcell and identified all genomic positions with a PRM value >= 70%. We found that, for ∼16% of our high-confidence positions from DGRP732, the exact same position is identified as putatively methylated in DGRP379, which is significantly more than expected by chance (hypergeometric P < 2.2e-16). Overall, ∼90% of the high-confidence positions from DGRP732 are within 100bp of a putatively methylated position in DGRP379, suggesting that, between strains, the methylation status of a given genomic region is more conserved than that of specific basepairs.
Both DGRP379 and DGRP732 were previously reported to lack the endosymbiotic bacterium Wolbachia that is common among Drosophila (Richardson et al. 2012). To verify that our stocks were also uninfected, we first confirmed via BLAST that the DGRP732 assembly lacked contigs showing homology to Wolbachia. We then appended the wMel Wolbachia genome assembly (Wu et al. 2004) to the DGRP732 assembly, aligned the raw nanopore reads from each strain to the concatenated assembly, and calculated the average coverage for the Wolbachia genome using only reads with mapping quality >= 20 (see Methods). For DGRP379, we found only three reads that mapped to Wolbachia (average coverage = 0.004x) and for DGRP739, which has more sequencing data, we found 44 reads (average coverage = 0.19x). Based on these results, we conclude that Wolbachia is either absent from these strains, as previously reported, or at very low abundance.
We investigated the genomic enrichment of our DGRP732 high-confidence 6mA-methylated sites by calculating the number of positions that overlap the following features: CDS, introns, 5′ UTR, 3′ UTR, 1kb upstream, intergenic sequence, simple repeats, and transposable elements. Using a permutation test (see Methods), we found that our 6mA positions are significantly enriched at introns, 3′ UTRs, upstream from transcription start sites, and simple repeats, whereas they are significantly depleted from coding sequence, intergenic sequence, and transposable elements (TEs)(3′ UTR: P = 0.0079, P < 0.0001 in all other cases)(Figure 2). A similar enrichment pattern was previously reported for gain-of-6mA positions in the brain of a DMAD mutant (Yao et al. 2018), with the exception of TEs, which we find as being depleted of 6mA whereas they are weakly enriched within TEs in the brain (Yao et al. 2018). In another study, 6mA positions were highly enriched within TEs in the ovaries (Zhang et al. 2015). We also found that, among chromosomes, the 6mA positions showed less than a twofold difference in their density, varying from ∼5 positions per 100kb (chromosomes 4 & 3R) to ∼10 positions per 100kb (chromosome 2L), on average (Figure S1A). Within chromosomes, the 6mA positions were fairly evenly distributed across the euchromatic chromosome arms and depleted from the pericentric heterochromatin (Figure S1B).
Previous work identified gain-of-6mA genomic regions in the brain that showed an accumulation of 6mA methylation in a DMAD null mutant. We found 3% (318 positions) of our high-confidence positions overlap these gain-of-6mA locations, which is significantly more than expected by chance (hypergeometric test P = 3.2e-13).
Our final set of 6mA positions show an enrichment pattern similar to what was previously observed in the brain (Yao et al. 2018). They also show a signficant overlap with the gain-of-6mA positions identified in a DMAD mutant (Yao et al. 2018) and their locations are conserved between DGRP strains. These results suggest that, despite the noise in the raw nanopore signal data, our conservative approach of using replicates and a control dataset has allowed us to accurately identify 6mA positions at single base resolution.
6mA sequence motifs
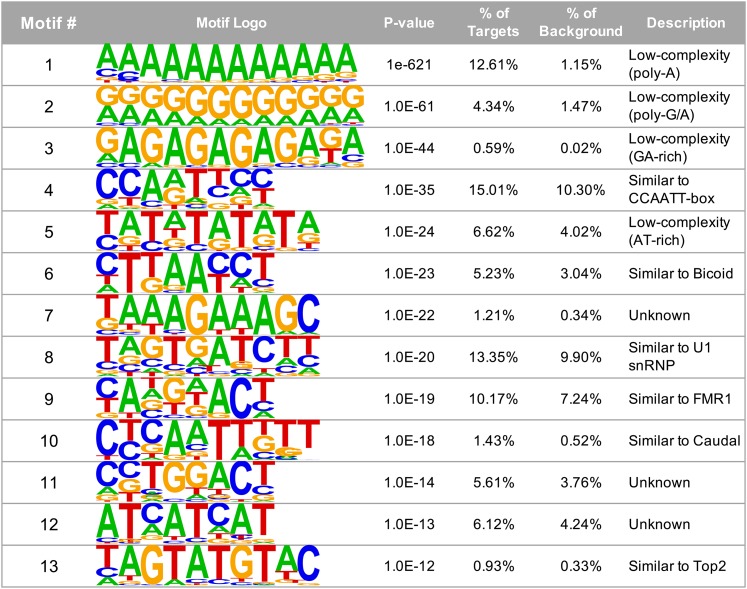
We extracted a total of 50 basepairs surrounding each 6mA position and used Homer (Heinz et al. 2010) to identify enriched motifs present in the sequences. We identified a total of 13 significantly enriched motifs (Figure 3). Four of these motifs consist of low-complexity sequences, including a poly-A motif, two GA-rich motifs, and a TA-repeat (Figure 3: motifs 1, 2, 3, and 5). Interestingly, the 6mA-associated sequence motifs previously identified in C. elegans (AGAAGAAGAAGA)(Greer et al. 2015) and the Drosophila brain (AGAAGGAG)(Yao et al. 2018) are also GA-rich elements. Two of the motifs that we identify (motifs 6 and 10) are very similar to the motifs that are recognized by Bicoid (CTAATCT) and Caudal (AAATTTTT), which are both homeobox transcription factors involved in anterior/posterior patterning (Mlodzik and Gehring 1987; Driever and Nusslein-Volhard 1989). We also find a motif (motif 8) that resembles the sequence recognized by the small ribonucleoprotein particle U1 (subunit 70K)(TCTTGATC)(Mancebo et al. 1990), which is part of the spliceosome, and another motif (motif 4) that contains the sequence CCAAT, which is commonly found in eukaryotic promoters and is recognized by CBF domain transcription factors (Bucher 1990). In Drosophila, the three CBF transcription factors (Nf-YA, Nf-YB, and Nf-YC) have been shown to play a role in eye and thorax development (Yoshioka et al. 2008; Ly et al. 2013). Another of our enriched motifs (motif 9) is similar to that recognized by Fmr1 (ANGGACA), which is involved in RNA trafficking and translation, and whose loss causes Fragile X syndrome (Ishizuka et al. 2002), while another motif (motif 13) is similar to that recognized by Topoisomerase 2 (Top2; TACATATGTATGTA), which is well-known for its role in DNA replication, but has also been found to play a role in transcription and insulator function (Lupo et al. 2001; Ramos et al. 2011). To verify these motifs, we also ran MEME (Bailey et al. 2009) using the same input sequences. We compared the Homer vs. MEME enriched motifs using STAMP (Mahony and Benos 2007) and found that 12 of the 13 Homer motifs show significant similarity to one or more motifs identified by MEME (Table S2). The only motif that was not identified by MEME is Homer motif #8 (Figure 3). Together, these motifs suggest that the 6mA modification plays a role in regulating development and transcription and raise the possibility that DMAD may interact with other transcriptional activators besides Wds.
Figure 3.
Sequence motifs associated with 6mA positions. We extracted 50 bp of sequence surrounding each 6mA position and used Homer (Heinz et al. 2010) to search for enriched motifs within these sequences. Four of the motifs identified by Homer are low-complexity sequences, including two GA-rich sequences (motifs 1, 2, 3, and 5). Three motifs are similar to those recognized by developmental transcription factors including the CBF family, Bicoid, and Caudal (motifs 4, 6, and 10), and three others are similar to motifs recognized by genes involved in RNA processing: U1 snRNP, Fmr1, Top2 (motifs 8, 9, and 13).
6mA is enriched at developmental genes
Our set of 6mA positions overlap a total of 2,624 genes (File S1). We performed a Gene Ontology (GO) enrichment analysis on this set of genes and found a total of 549 significantly enriched terms with FDR q-value <= 0.02 across the three GO categories (COMPONENT: 57, FUNCTION: 71, PROCESS: 421)(Table S3). The enriched terms are heavily biased toward categories involved in development, regulation of gene expression, and neuron-specific functions (Table 1), in agreement with the previously identified roles for this modification in Drosophila (Zhang et al. 2015; Yao et al. 2018).
Table 1. GO term enrichment for genes containing the 6mA modification.
| GO term ID* | Description | FDR q-value |
|---|---|---|
| PROCESS | ||
| GO:0032502 | developmental process | 5.49E-36 |
| GO:0007411 | axon guidance | 1.25E-22 |
| GO:0050793 | regulation of developmental process | 2.35E-20 |
| GO:0009886 | post-embryonic animal morphogenesis | 3.63E-19 |
| GO:0010646 | regulation of cell communication | 5.03E-16 |
| GO:0023051 | regulation of signaling | 4.82E-16 |
| GO:0022414 | reproductive process | 1.39E-07 |
| GO:0010468 | regulation of gene expression | 2.17E-06 |
| FUNCTION | ||
| GO:0005261 | cation channel activity | 3.07E-06 |
| GO:0003700 | DNA-binding transcription factor activity | 5.50E-06 |
| GO:0004672 | protein kinase activity | 1.63E-04 |
| GO:0003779 | actin binding | 1.54E-04 |
| GO:0140110 | transcription regulator activity | 2.40E-04 |
| GO:0038023 | signaling receptor activity | 8.22E-04 |
| COMPONENT | ||
| GO:0005886 | plasma membrane | 9.23E-21 |
| GO:0030054 | cell junction | 7.72E-12 |
| GO:0097458 | neuron part | 5.91E-09 |
| GO:1902495 | transmembrane transporter complex | 1.40E-08 |
| GO:0045202 | synapse | 1.40E-06 |
Selected GO terms are shown here. See Table S3 for full list of all 549 enriched terms.
6mA-containing genes show high tissue-specificity in a variety of tissues
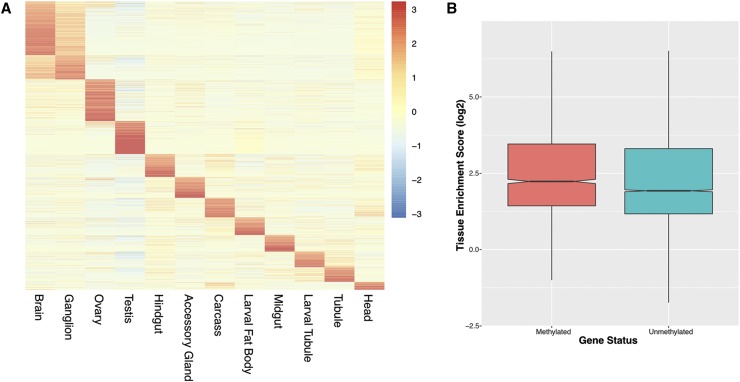
We used the FlyAtlas resource (Chintapalli et al. 2007) to examine the expression patterns of the 2,624 genes that overlap one or more 6mA positions. The FlyAtlas project used microarrays to measure gene expression in a variety of D. melanogaster tissues as well as in the whole fly. Part of the project involved the calculation of enrichment scores that measure the tissue specificity of each gene by comparing its expression level in a given tissue to its expression level in the whole fly (Chintapalli et al. 2007). Enrichment scores for 2,051 of the 2,624 genes were available from FlyAtlas and visualization of these scores shows that most genes are strongly expressed in a single tissue, with the exception of the brain and ganglia whose profiles overlap because these tissues are both part of the central nervous system (Figure 4A). Overall, we find that the 6mA-containing genes show significantly higher tissue specificity compared to the rest of the genes in the D. melanogaster genome (Wilcoxon test P < 2.2e-16)(Figure 4B). While many 6mA-containing genes are highly expressed in either the brain/ganglion or ovaries, both of which are tissues where 6mA-mediated gene regulation has previously been identified, we also find many genes whose expression is highly enriched in other tissues from both adult flies and larvae, including the testes, which suggests that 6mA may play a more general role in regulating the specificity of gene expression across many tissues and developmental stages, as well as between sexes.
Figure 4.
6mA positions are found in genes with high tissue specificity. The FlyAtlas project calculated enrichment scores (tissue vs. whole fly gene expression) for twelve different D. melanogaster tissues (Chintapalli et al. 2007). 2,051 of the 2,624 genes that overlap 6mA positions had enrichment scores, which are shown in the heatmap in Panel A, where each row represents a 6mA-containing gene and each column corresponds to one of the 12 tissues. Individual cells are colored according to enrichment score. Genes with the 6mA modification show significantly higher tissue enrichment scores compared to the D. melanogaster genes that do not contain any 6mA positions (Panel B; Wilcoxon Test P < 2.2e-16).
Discussion
We have used nanopore sequencing to identify 6mA methylation at single base-pair resolution across the Drosophila genome. These positions tend to be located upstream from transcription start sites and within introns, 3′ UTRs, and simple repeats and they are associated with genes whose functions are related to transcriptional regulation, development, and neuronal activity. These findings corroborate those from previous studies where ChIP-seq was used to identify 6mA in Drosophila brains and ovaries (Zhang et al. 2015; Yao et al. 2018), and provide additional support for the importance of this modification in the repression of developmental and neuronal genes. One difference between this study and previous studies of 6mA in Drosophila is that our 6mA positions were significantly depleted from transposable elements whereas 6mA positions in ovaries were enriched within TEs (Zhang et al. 2015), as were gain-of-6mA positions in the brain that became methylated in the DMAD mutant (Yao et al. 2018). This difference could be due to the fact that TE repression is reduced in somatic cells compared to the germline. For example, the gain-of-6mA at TEs in the brain of DMAD mutants suggests that, in wild-type flies, DMAD plays a role in actively removing 6mA from TEs in the brain (Yao et al. 2018). Other studies have shown that, in the embryo, TE insertions are strongly enriched for repressive histone modifications, but this enrichment weakens throughout development into adulthood (Lee 2015; Lee and Karpen 2017). Because our data are from whole, adult flies, we may not be able to identify the 6mA positions whose presence within TEs is restricted to the germline.
Our results also expand the role of 6mA in several important ways. A previous study in the Drosophila brain identified a single GA-rich sequence motif associated with 6mA and showed that the 6mA demethylase DMAD interacts with the regulatory protein Wds to remove 6mA and activate gene expression (Yao et al. 2018). While we find a similar GA-rich motif, we also find a variety of additional enriched motifs, several of which are similar to motifs recognized by regulatory proteins involved in development (Bicoid, Caudal, and CBF transcription factors) and RNA processing (U1 snRP, Fmr1, and Top2). These results suggest that 6mA may be found in different sequence contexts in different tissues and raise the possibility that DMAD may work in concert with other regulatory proteins besides Wds.
Our results also suggest that, rather than only regulating the expression of neuronal genes, the 6mA modification may play a more general role in enforcing the tissue specificity of gene expression across many different tissues, and throughout development. Given that our sequencing data were from DNA extracted from whole flies, the 6mA positions that we identify are those that carry the 6mA modification in the majority of cells in the organism. The fact that our 6mA-containing genes show high expression specificity is consistent with a model where 6mA is involved in the repression of these genes in the majority of cells. Such genes would only be expressed in a minority of cells (e.g., in specific tissues, sexes, and/or developmental stages) when DMAD, in concert with one or more activation proteins, removes the modification and activates the gene. Future work involving nanopore sequencing of different Drosophila tissues would provide insight into this model.
ACKNOWLEDGMENTS
The authors thank two anonymous reviewers for their insightful comments and acknowledge the Office of Advanced Research Computing (OARC) at Rutgers, The State University of New Jersey for providing access to the Amarel cluster and associated research computing resources that have contributed to the results reported here.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25387/g3.7841036.
Communicating editor: M. Arbeitman
Literature Cited
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., et al. , 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37: W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher P., 1990. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 212: 563–578. 10.1016/0022-2836(90)90223-9 [DOI] [PubMed] [Google Scholar]
- Chintapalli V. R., Wang J., Dow J. A., 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39: 715–720. 10.1038/ng2049 [DOI] [PubMed] [Google Scholar]
- Dewey C. N., 2007. Aligning multiple whole genomes with Mercator and MAVID. Methods Mol. Biol. 395: 221–236. 10.1007/978-1-59745-514-5_14 [DOI] [PubMed] [Google Scholar]
- Driever W., Nusslein-Volhard C., 1989. The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature 337: 138–143. 10.1038/337138a0 [DOI] [PubMed] [Google Scholar]
- Dudchenko O., Batra S. S., Omer A. D., Nyquist S. K., Hoeger M., et al. , 2017. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356: 92–95. 10.1126/science.aal3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand N. C., Shamim M. S., Machol I., Rao S. S., Huntley M. H., et al. , 2016. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst. 3: 95–98. 10.1016/j.cels.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z., 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith M. C., 2011. A new repeat-masking method enables specific detection of homologous sequences. Nucleic Acids Res. 39: e23 10.1093/nar/gkq1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Luo G. Z., Chen K., Deng X., Yu M., et al. , 2015. N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell 161: 879–892. 10.1016/j.cell.2015.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer E. L., Blanco M. A., Gu L., Sendinc E., Liu J., et al. , 2015. DNA Methylation on N6-Adenine in C. elegans. Cell 161: 868–878. 10.1016/j.cell.2015.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., et al. , 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins R. A., Carlson J. W., Wan K. H., Park S., Mendez I., et al. , 2015. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 25: 445–458. 10.1101/gr.185579.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka A., Siomi M. C., Siomi H., 2002. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 16: 2497–2508. 10.1101/gad.1022002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Walenz B. P., Berlin K., Miller J. R., Bergman N. H., et al. , 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27: 722–736. 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. C., 2015. The Role of piRNA-Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in Drosophila melanogaster. PLoS Genet. 11: e1005269 10.1371/journal.pgen.1005269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. C. G., Karpen G. H., 2017. Pervasive epigenetic effects of Drosophila euchromatic transposable elements impact their evolution. eLife 6: e25762 10.7554/eLife.25762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G. Z., Hao Z., Luo L., Shen M., Sparvoli D., et al. , 2018. N(6)-methyldeoxyadenosine directs nucleosome positioning in Tetrahymena DNA. Genome Biol. 19: 200 10.1186/s13059-018-1573-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo R., Breiling A., Bianchi M. E., Orlando V., 2001. Drosophila chromosome condensation proteins Topoisomerase II and Barren colocalize with Polycomb and maintain Fab-7 PRE silencing. Mol. Cell 7: 127–136. 10.1016/S1097-2765(01)00161-7 [DOI] [PubMed] [Google Scholar]
- Luria S. E., Human M. L., 1952. A nonhereditary, host-induced variation of bacterial viruses. J. Bacteriol. 64: 557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly L. L., Suyari O., Yoshioka Y., Tue N. T., Yoshida H., et al. , 2013. dNF-YB plays dual roles in cell death and cell differentiation during Drosophila eye development. Gene 520: 106–118. 10.1016/j.gene.2013.02.036 [DOI] [PubMed] [Google Scholar]
- Mackay T. F., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster Genetic Reference Panel. Nature 482: 173–178. 10.1038/nature10811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony S., Benos P. V., 2007. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 35: W253–W258. 10.1093/nar/gkm272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancebo R., Lo P. C., Mount S. M., 1990. Structure and expression of the Drosophila melanogaster gene for the U1 small nuclear ribonucleoprotein particle 70K protein. Mol. Cell. Biol. 10: 2492–2502. 10.1128/MCB.10.6.2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodzik M., Gehring W. J., 1987. Expression of the caudal gene in the germ line of Drosophila: formation of an RNA and protein gradient during early embryogenesis. Cell 48: 465–478. 10.1016/0092-8674(87)90197-8 [DOI] [PubMed] [Google Scholar]
- Oshima T., Wada C., Kawagoe Y., Ara T., Maeda M., et al. , 2002. Genome-wide analysis of deoxyadenosine methyltransferase-mediated control of gene expression in Escherichia coli. Mol. Microbiol. 45: 673–695. 10.1046/j.1365-2958.2002.03037.x [DOI] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani V., Cusanovich D. A., Hause R. J., Ma W., Qiu R., et al. , 2016. Mapping 3D genome architecture through in situ DNase Hi-C. Nat. Protoc. 11: 2104–2121. 10.1038/nprot.2016.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos E., Torre E. A., Bushey A. M., Gurudatta B. V., Corces V. G., 2011. DNA topoisomerase II modulates insulator function in Drosophila. PLoS One 6: e16562 10.1371/journal.pone.0016562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisenauer A., Shapiro L., 2002. DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. EMBO J. 21: 4969–4977. 10.1093/emboj/cdf490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson M. F., Weinert L. A., Welch J. J., Linheiro R. S., Magwire M. M., et al. , 2012. Population genomics of the Wolbachia endosymbiont in Drosophila melanogaster. PLoS Genet. 8: e1003129 10.1371/journal.pgen.1003129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sović I., Sikic M., Wilm A., Fenlon S. N., Chen S., et al. , 2016. Fast and sensitive mapping of nanopore sequencing reads with GraphMap. Nat. Commun. 7: 11307 10.1038/ncomms11307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoiber M. H., Quick J., Egan R., Lee J. E., Celniker S. E., et al. , 2017. De novo Identification of DNA Modifications Enabled by Genome-Guided Nanopore Signal Processing. bioRxiv. [Google Scholar]
- Thurmond J., Goodman J. L., Strelets V. B., Attrill H., Gramates L. S., et al. , 2019. FlyBase 2.0: the next generation. Nucleic Acids Res. 47: D759–D765. 10.1093/nar/gky1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaser R., Sovic I., Nagarajan N., Sikic M., 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27: 737–746. 10.1101/gr.214270.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B. J., Abeel T., Shea T., Priest M., Abouelliel A., et al. , 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9: e112963 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chen X., Sheng Y., Liu Y., Gao S., 2017. N6-adenine DNA methylation is associated with the linker DNA of H2A.Z-containing well-positioned nucleosomes in Pol II-transcribed genes in Tetrahymena. Nucleic Acids Res. 45: 11594–11606. 10.1093/nar/gkx883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Sun L. V., Vamathevan J., Riegler M., Deboy R., et al. , 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2: E69 10.1371/journal.pbio.0020069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C. L., Zhu S., He M., Q. Zhang Chen, et al. , 2018. N(6)-Methyladenine DNA Modification in the Human Genome. Mol. Cell 71: 306–318 e307. 10.1016/j.molcel.2018.06.015 [DOI] [PubMed] [Google Scholar]
- Yao B., Cheng Y., Wang Z., Li Y., Chen L., et al. , 2017. DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat. Commun. 8: 1122 10.1038/s41467-017-01195-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B., Li Y., Wang Z., Chen L., Poidevin M., et al. , 2018. Active N(6)-Methyladenine Demethylation by DMAD Regulates Gene Expression by Coordinating with Polycomb Protein in Neurons. Mol. Cell 71: 848–857 e846. 10.1016/j.molcel.2018.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka Y., Suyari O., Yamaguchi M., 2008. Transcription factor NF-Y is involved in regulation of the JNK pathway during Drosophila thorax development. Genes Cells 13: 117–130. 10.1111/j.1365-2443.2007.01155.x [DOI] [PubMed] [Google Scholar]
- Zhang G., Huang H., Liu D., Cheng Y., Liu X., et al. , 2015. N6-methyladenine DNA modification in Drosophila. Cell 161: 893–906. 10.1016/j.cell.2015.04.018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequencing data and the DGRP732 genome assembly are available via the NCBI Sequence Read Archive (SRA) and Whole Genome Shotgun (WGS) databases under Bioproject PRJNA515844. Supplemental material available at FigShare: https://doi.org/10.25387/g3.7841036.