Abstract
The temporal coding of action potential activity is fundamental to nervous system function. Here we consider how gene expression in neurons is regulated by specific patterns of action potential firing, with an emphasis on new information on epigenetic regulation of gene expression. Patterned action potential activity activates intracellular signaling networks selectively in accordance with the kinetics of activation and inactivation of second messengers, phosphorylation and dephosphorylation of protein kinases, and cytoplasmic and nuclear calcium dynamics, which differentially activate specific transcription factors. Increasing evidence also implicates activity-dependent regulation of epigenetic mechanisms to alter chromatin architecture. Changes in three-dimensional chromatin structure, including chromatin compaction, looping, double-stranded DNA breaks, histone and DNA modification, are altered by action potential activity to selectively inhibit or promote transcription of specific genes. These mechanisms of activity-dependent regulation of gene expression are important in neural development, plasticity, and in neurological and psychological disorders.
Keywords: chromatin remodeling, activity-dependent plasticity, oscillation, temporal coding, transcranial magnetic stimulation, behavior-dependent gene expression
Environmental information is encoded by neurons through the firing of action potentials with specific spiking patterns (temporal coding) (Kayser and others 2009); therefore, gene expression in the nervous system must be regulated by the temporal features of action potential firing to produce adaptive responses. In addition to temporal coding, neural activity across populations of neurons can summate to create local field potentials that fluctuate in intensity at specific frequencies, transiently coupling activity in networks of neurons to coordinate information processing by the degree of coherence and synchrony of brainwaves and neuronal oscillations (Buzsáki and Draghun 2004; Bonnefond and others 2017). Neural oscillation patterns have correlates with specific behaviors and are associated with aspects of cognitive function such as memory, attention, and skill learning (Corlier and others 2016; Di Nota and others 2017; Friese and others 2013), suggesting the possibility of changes in gene expression associated with specific frequencies of neuronal oscillation are essential components of frequency-specific behaviors.
Neuronal activity-dependent changes in gene expression are classically attributed to intracellular calcium kinetics activating calcium-dependent protein kinase cascades, ultimately recruiting activated transcription factors in the nucleus. However, this is only one aspect of how context-specific action potential patterns can regulate expression of an appropriate gene network. Along with calcium-dependent and calcium-independent cytosolic signaling from neuronal firing, intranuclear events that may be regulated by the temporal features of neuronal firing have been much less studied, but increasing evidence suggests the importance of activity-dependent modification of chromatin structure in regulating gene expression.
Expression of genes requires a network of interactions between DNA, which is wrapped into a dynamic three-dimensional chromatin structure, along with heterogenous transcription machinery composed of protein factors, and non-coding RNA. Furthermore, a diverse array of epigenetic modifications come together to create biochemical marks on proteins or DNA nucleotide bases to allow for the specialized gene expression in individual cells adapted to unique and stimulating environments. In this review, we summarize the current understanding of neural information transduction into the nucleus with temporal specificity, with an emphasis on the connection between unique neuronal activity patterns and interactions within the epigenome to produce and maintain a stimulus-specific transcriptome.
Evidence of Action Potential Pattern-Specific Gene Expression
Rhythmic Magnetic and Optogenetic Stimulation
Transcranial magnetic stimulation (TMS) provides compelling evidence that gene expression is regulated by specific patterns of neuronal firing and neural oscillations in vivo. For example, repetitive TMS using an intermittent pattern of theta-burst frequency for 2 weeks following a stroke injury in rat upregulates 52 genes involved in angiogenesis, inflammation, neuroprotection and neuronal plasticity, while repetitive TMS at a constant 1 Hz or 5 Hz frequency had no effect on gene expression (Ljubisavljevic and others 2015). In another study, the immediate early genes (IEGs) c-fos and zif268, both of which are implicated in synaptic plasticity, were found to be differentially expressed in response to distinct patterns of TMS (Aydin-Abidin and others 2008).
An in vitro study used 5 different patterns of repetitive magnetic stimulation in a study of gene expression and intracellular calcium transients. Coils positioned outside culture dishes were used to drive magnetic fields through the preparation to excite neuronal firing in primary cell cultures isolated from mouse cerebral cortex (Grehl and others 2015). All patterns of stimulation elevated intracellular calcium to a similar extent, but the pattern of gene expression was highly dependent on the stimulation pattern. Thus, regulation of gene expression by neuronal firing is not simply explained by the amplitude of intracellular calcium concentration generated by different stimulus patterns. These different patterns of stimulation had functional consequences, as shown by phenotypic effects on neuronal morphology and survival, which were consistent with the expression changes in genes implicated in neuron morphology and survival (Grehl and others 2015).
Other studies using electrical stimulation of awake adult rats (Ryan and others 2012) or rat hippocampal slices (Bukalo and others 2016; Lee and others 2005), in patterns that induce long-term potentiation (LTP) or longterm depression (LTD), show that the temporal pattern of gene expression in hippocampal neurons is altered differently by the different stimulus patterns (Bukalo and others 2016; Lee and others 2005; Ryan and others 2012). LTP-associated gene expression profiles also differ when stimulation is applied via synaptic input or action potential firing (Dudek and Fields 2002). Taken together, these findings illustrate the importance of both the temporal pattern and spatial component of neuronal activity in the subsequent activity-induced gene expression.
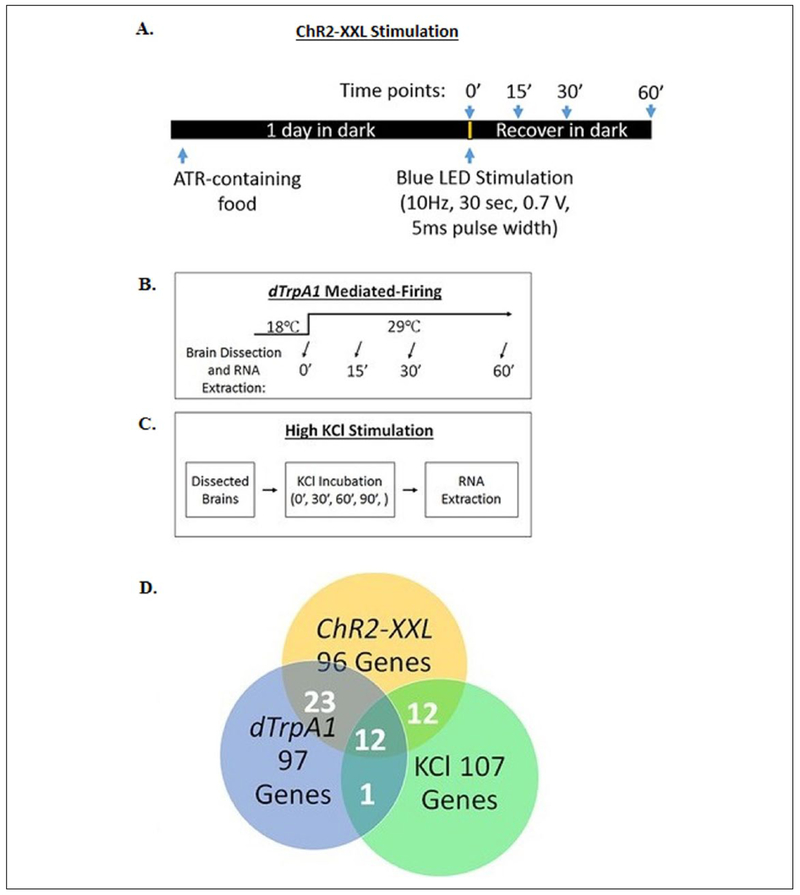
More recently, studies using optogenetic stimulation of Drosophila to activate different patterns of firing in the fly’s nervous system, have been used to investigate this question. RNA-sequence data from Drosophila neurons activated by two stimulation paradigms indicate that each stimulation paradigm produces unique expression of activity-regulated genes (Chen and others 2016) (Fig. 1). To explain mechanisms driving activity-dependent expression of genes, Chen and others determined the kinetics of activity-regulated gene enrichment at different time-points. The majority of the transcripts sampled did not reach maximal expression until 60 minutes after robust, non-patterned LED-light induced depolarization. Analysis of chromatin compaction, by using transposase-accessible sequencing (ATAC-seq) indicates that the transcription start sites of activity-related genes were more open prior to stimulation (Chen and others 2016). Interestingly, this group also reports that different neuron populations each generate different sets of activity-regulated transcripts after undergoing the same stimulus paradigm.
Figure 1.
Activity-dependent transcripts in Drosophila vary based on neuronal stimulation type. (A) Schematic for optogenetic stimulation of channelrhodopsin-2-XXL (ChR2) expressing neurons under Gal4 control with blue LED light. (B) Schematic for heat induced activation with dTrpA1 heat-sensitive cation channel. (C) Schematic for robust depolarization with KCl treatment. (D) Overlapping expression of activity induced genes based on stimulation paradigm. Data generated from high throughput deep sequencing of mRNA libraries isolated from stimulated Drosophila. Figure adapted from Chen and others (2016).
However, using these various approaches of stimulating neural networks in vivo and in vitro, it is difficult to control the precise pattern of action potential firing, because the neurons are interconnected by excitatory and inhibitory synapses, and they typically exhibit spontaneous firing in complex patterns and bursts, making these methods an insufficient test of the hypothesis that gene expression is regulated by the temporal pattern of action potential firing. With these experimental approaches, the frequency-dependent effects of stimulation on gene expression are most likely related to changes in overall network excitability that is influenced by the different patterns of applied stimulation. In general, low-frequency TMS (1 Hz) decreases cortical network excitability and higher frequency stimulation (5 Hz and above, and theta-burst stimulation) increase network activity (Aydin-Abidin and others 2008).
Patterned Electrical Stimulation of Axons
To test directly the hypothesis that gene expression is regulated by the temporal pattering of action potential firing, cell cultures of mouse dorsal root ganglion (DRG) neurons have been stimulated by electrodes in combination with calcium imaging and analysis of gene expression. DRG neurons do not have dendrites nor do they form synapses on other DRG neurons. These neurons are not spontaneously active in cell culture and in response to brief pulses of electrical stimulation, they fire a single action potential, rather than a train of action potentials (Fields and others 1992). With the ability to precisely control the pattern of action potential firing, DRG neuron cultures are therefore an ideal method to test whether temporal patterns of action potentials affect gene expression. Using four stimulation paradigms, Fields and others (1997) demonstrate that specific action potential patterns cause differential gene expression in neurons. The experimenters determined that expression of c-fos correlates inversely with the length of interval between consecutive stimuli presented at different frequencies and does not correlate with the net concentration of cytosolic calcium (Fields and others 1997). To explain this phenomenon, the authors analyzed the calcium-dependent intracellular signaling cascades and activation of the transcription factor cyclic-AMP response element binding protein (CREB), in response to the four firing patterns that differentially regulate expression of c-fos. The results indicate that changes in gene expression induced by the temporal features of action potential firing are in part a consequence of differences in the kinetics of activation and inactivation of calcium-dependent protein kinases and transcription factors controlling gene transcription in response to membrane depolarization. This and additional mechanisms for regulating gene expression by temporal coding of action potential firing will be considered in the next section.
Encoding Transients of Neuronal Activity in the Cytosol and Nucleus
Intracellular Signaling from Membrane Depolarization to Transcription Factors
Stimulus-specific changes in gene expression require the transduction of synaptic activity patterns into the nucleus with accurate temporal integrity. Calcium signaling activated by membrane depolarization is highly implicated in expression of genes, in part through activation of protein kinase C and ERK/MAPK signaling pathways to modulate many downstream transcription factors (Cohen and Greenberg 2008; Flavell and Greenberg 2008; Adams and others 2000). Transcriptome analysis following pharmacological depolarization of neurons has been valuable in identifying and characterizing many “activity-dependent genes” (Coba and others 2008; Hunsberger and others 2005; Pham and others 1999). In various cell types, IEGs, such as c-fos and c-jun, increase expression rapidly following neuronal stimulation, without the need for de novo protein synthesis. This rapid transcription occurs through activation of specific transcription factors (Bahrami and Drabløs 2016), the most widely studied being phosphorylation of the calcium-responsive transcription factor CREB (Pham and others 2000). Synaptic activity mediated through the excitatory neuronal receptor NMDA (N-methyl-d-aspartate) can activate the ERK signaling pathway and result in downstream activation of calcium-calmodulin kinase IV (CaMKIV) (Bito and others 1996). CaMKIV enters the nucleus to phosphorylate CREB allowing the CREB-binding protein (CBP) to form a complex and mediate transcription (Bito and others 1996; Impey and others 2002). This mechanism was thought to dominate activity dependent gene expression as it provided a clear connection between calcium kinetics and nuclear transcription factor activation (Impey and others 2002). However, while phosphorylation of CREB and the CBP complex mediate plasticity-related gene expression (Barco and others 2002; Impey and others 2002), electrical stimulation of DRG neurons by different frequencies and patterns of action potentials readily induces prolonged phosphorylation of CREB, but with kinetics that would be unable to maintain temporal-specific integrity of many stimulation patterns (Fields and others 1997).
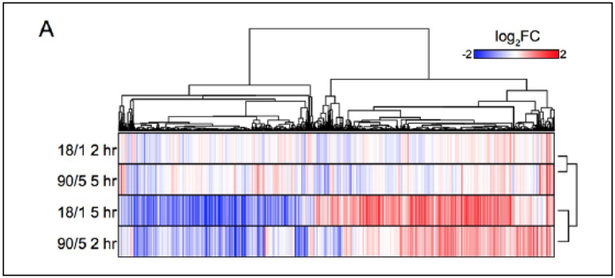
Stimulating action potentials in mouse DRG axons at 10 Hz in 1.8-second bursts, (18 action potentials, with a 1-minute interburst interval, referred to as 18/1) or for 9 seconds (90 action potentials, with a 5 minute interburst interval, referred to as 90/5), both deliver the same total number of action potentials at the same 10 Hz frequency during the experimental time course, but expression of many genes was found to be differentially regulated by these two action potential firing patterns. Gene expression analysis by genome-wide microarray performed after 2 and 5 hours of stimulation at these two patterns detected 2901 mRNA transcripts that were differentially expressed between the two stimulus patterns. Interestingly, genes that were upregulated by one stimulus pattern were typically downregulated by the second pattern (Fig. 2). Classically, gene expression that is regulated by neuronal firing has been considered a special property of IEGs, but this study shows that thousands of genes coding a wide range of proteins are regulated by the temporal features of action potential firing. Given this evidence that thousands of neuronal genes are not only activity-regulated but firing pattern-regulated suggests that many other signaling pathways, in conjunction with calcium signaling resulting in phosphorylation of CREB, must be involved. In support of this, after categorizing the differentially regulated genes based on their known functions, the authors conclude that 18 canonical signaling pathways were activated depending on the stimulus pattern. Similar to intracellular signaling pathways in the cytoplasm, intranuclear events are also sensitive to the pattern of action potential firing.
Figure 2.
Specific patterns of action potentials induce differential gene expression in mammalian dorsal root ganglion neurons (DRGs). Heat map represents RNA-seq data from DRGs after 10 Hz stimulation for 1.8 seconds with a 1-minute interval (18/1) or for 9 seconds with a 5-minute (90/5) interval for 2 or 5 hours (Lee and others 2017). Blue indicates downregulation of mRNA and red indicates upregulation of mRNA. Reprinted from Lee and others (2017).
Nuclear Calcium
In addition to cytosolic signaling, neuronal firing patterns are encoded into corresponding nuclear calcium transients (Bengtson and others 2010; Hardingham and others 2001). Repetitive high frequency or theta burst stimulation paradigms used to experimentally induce LTP, produce waves of nuclear calcium (Bengtson and others 2010). Nuclear calcium dynamics correlate to the temporal pattern of environmental stimulus (Bengtson and others 2010; Hardingham and others 2001). Therefore, changes in nuclear calcium concentration might contribute to the processes by which environmental stimuli manifest in alternative gene transcription (Bading and others 2000). For example, nuclear calcium has been shown to bind nucleosomes and stabilize DNA-histone interactions (Yang and Hayes 2011). (Activity-dependent chromatin remodeling will be considered in greater depth separately in this review.) In particular, calcium ions modify nucleosome compaction, promoting chromosome condensation (Phengchat and others 2016). Nuclear calcium dynamics can act independently of cytosolic signaling to produce expression of CREB-dependent genes (Hardingham and others 2001). It is important to note, however, that the role of nuclear calcium transients as a medium for encoded synaptic information is debated, as some findings indicate that nuclear calcium dynamics do not appear to be tightly influenced by robust cytosolic calcium changes (Al-Mohanna and others 1994; Leite and others 2003), unlike the studies identified above. Taken together, the literature suggests that environmental stimuli may be converted into nuclear calcium dynamics, which have the ability to modulate and induce gene expression.
Transcription Factor Binding Sites
The study mentioned previously, Lee and others (2017), determined that activity-dependent expression of genes in DRG neurons is regulated on a genome-wide scale according to temporal features of action potential firing. By hypothesis, genes may be differentially sensitive to distinct action potential firing patterns by virtue of having multiple transcription factor binding sites in the regulatory elements throughout enhancer and promotor regions. If transcription factor activation occurs in response to only a subset of action-potential patterns, then pattern-dependent gene expression may arise when a transcription factor is preferentially recruited based on the stimulus provided, and subsequently binds to its motif inducing expression of only a certain set of downstream genes. To test this hypothesis, the authors analyzed the regulatory regions of the genes that were differentially affected by the two stimulus patterns applied. Using distant regulatory elements of co-regulated genes (DIRE), the authors found enrichment of certain transcription factor binding sites in the two sets of genes that were differentially regulated by the two patterns of stimulation. For example, transcription factors associated with activation of ERK/MAPK pathways were enriched in genes responding to the 18/1 stimulus pattern while canonical calcium signaling transcription factors are overrepresented in genes responding to the 90/5 stimulation (Lee and others 2017).
The authors conclude that differential transcription in response to distinct neuronal firing patterns is the result of cytosolic signaling pathway activation and enrichment of transcription factor bindings sites in different genes. Beyond these important considerations, more questions remain. If the expression of a gene or a gene network is altered based on the pattern of neuronal activity, and the expression of gene is dependent on the local and distal epigenetic architecture, what is the role of dynamic epigenetic modifications in neural pattern specific gene expression?
Neuronal Activity and Epigenetic Interactions
Experience-Dependent Epigenetic Modification
The new field of “neuroepigenetics” characterizes how the epigenetic landscape allows neurons to respond to unique environmental stimuli (reviewed further by Cholewa-Waclaw and others 2016; Grigorenko and others 2016; Riccio 2010). The most well-characterized epigenetic marks include histone methylation (Allfrey and others 1964), acetylation (Allfrey and others 1964), phosphorylation (Gutierrez and Hnilica 1967; Stevely and Stocken 1966), sumoylation (Kang and others 2001), and DNA methylation (Gold and others 1963). Each of these marks are a molecular addition to the protein or DNA to alter its biophysical interactions, such as introducing steric hindrance or by altering the strength of ionic interactions between charged histone tails and the negatively charged DNA sugar phosphate-backbone (recently reviewed by Yao and others 2016). While many features of the epigenome have been studied, continued research into known modifications as well as identification of novel and biologically elusive modifications is an active, ongoing field of research. In Table 1, we briefly summarize the most characterized histone tail and DNA modifications in neuroscience literature (Cao and Yan 2012; Cubenas-Potts and Matunis 2013; Huang and Dixit 2016; McConnell and Wadzinski 2009; Rossetto and others 2012; Roth and Sweatt 2009).
Table 1.
Biochemical Regulation of Chromatin Architecture with Histone and DNA Modifications.
| Modification Class |
Biochemical Action | Effect
on Chromatin Architecture |
Effect on Transcription | Inactivation | Pharmacology | |
|---|---|---|---|---|---|---|
| Histone | Acetylation | Transfer of acetyl group to NH3 groups of lysine residues by histone acetyltransferase, e.g., cyclic-AMP response element binding protein (CREB) | Most commonly, decreases affinity of histones with DNA and relaxes chromatin structure | Usually associated with transcriptional activation by increasing recruitment of transcription factors | Histone deacetylation via histone deacetylases (HDACs) | HDAC inhibitors Trichostatin A, sodium butyrate, valproic acid, suberoylanilide hydroxamic acid |
| Phosphorylation | Phosphorylation of serine, threonine, and tyrosine residues by protein kinases | Chromatin relaxation or compaction | Usually associated with transcriptional activation by proteins containing phosphorbinding modules, e.g., 14-3-3 and BRCT (BRCA1 C-terminus) domains, regulates histone acetylation and methylation | Phosphatases | To promote phosphatase activity: protein
serine/threonine phosphatase (PSTPs), protein tyrosine phosphatase
(PTPs) or dual specific phosphatase (DSPs). To inhibit: phosphatase inhibitor cocktails |
|
| Methylation | Methylation of lysine, arginine residues of histones (H3 and H4) by histone methyltransferase | Generally associated with relaxing compaction | Promote or inhibit gene transcription depending on methylation site and number of methyl groups attached | Generally irreversible, but histone demethylases are known | Histone methyltransferase inhibitors and demethylases | |
| Ubiquitination | Addition of ubiquitin to lysine residues by ubiquitin ligases | Chromatin condensation by histone degradation | Gene silencing or transcriptional activation | Deubiquitinating enzymes (DUBs) | Ubiquitin E1 enzyme inhibitors: PYR-41 and PYD-4409 | |
| SUMOylation | Small ubiquitin-related modifiers (SUMOs) | Multifaceted regulator of DNA methylation, histones, and transcription regulators | Generally associated with gene inactivation, but also gene activation by enhancing chromatin accessibility | Desumoylating enzymes, metalloproteases | SUMOylation inhibitor: 2-D08 | |
| DNA | Cytosine methylation | Covalent addition of CH3 group to cytosine adjacent to guanine nucleotides (CpG islands) by DNA methyltransferases (DNMTs) | Recruits methyl-DNA binding proteins; e.g., HDACs to compact chromatin | Usually associated with suppression of gene transcription | Generally irreversible, demethylation enzymes not well established | DNMT inhibitors include azacytidine (Vidaza), decitabine (Dacogen), RG108, zebularine |
Epigenetic regulation can occur in response to environmental factors such as stress (Fuchikami and others 2010) and visual experience (Ruan and others 2016). Learning and memory are associated with histone modifications (Bredy and others 2007; Gupta and others 2010; Stefanko and others 2009; Blank and others 2014) and altered chromatin structure due to DNA double stranded breaks at the promoters of immediate early genes (Madabhushi and others 2015; Watson and Tsai 2017). Trans-generational effects of environmental enrichment on memory are associated with DNA methylation and histone modification (Arai and Feig 2011). Long-term memory is regulated by histone acetylation, and disruption of histone acetyltransferase (HAT) activity impairs long-term memory (Halder and others 2016; Pandey and others 2015; Rossetto and others 2012; Roth and Sweatt 2009). Augmenting acetylation by histone deacetylase (HDAC) inhibitors can enhance memory formation (Bieszczad and others 2015; Roth and Sweatt 2009). Conversely, eliminating the metabolic enzyme that synthesizes acetyl-CoA (acetyl-CoA synthetase 2), thus reducing acetyl group availability, reduces transcription of canonical memory-related neuronal genes and impairs long-term spatial memory (Mews and others 2017). Fear conditioning is hallmarked by rapid methylation of memory-repressive genes and demethylation of memory-associated genes (Roth and Sweatt 2009).
A wide range of cognitive disorders are associated with epigenetic regulation, including alcohol and drug addiction (Basavarajappa and Subbanna 2016; Kim and others 2017), psychotic disorders including schizophrenia (Costa and others 2003; Ruzicka 2015), and Alzheimer’s disease (Cuadrado-Tejedor and others 2015). Epigenetic remodeling is similarly reported with developmental and physiological changes such as pain (Géranton and Tochiki 2015), nervous system development (Yoo and Crabtree 2009), aging (Pina and others 1988; Sen 2015; Singh and Thakur 2017), and synaptic plasticity (Maze and others 2015; Zhu and others 2016).
While features of the epigenetic architecture are widely associated with both neurological behaviors and pathology, the molecular interplay between neuronal activity and the epigenome in neuronal populations remains largely unexplored. As a note, the limitation of the following studies, similar to activity-dependent findings discussed above, is that many have yet to examine the temporal kinetics of action potentials that allow neuronal responses to be biologically relevant to the experiences and behaviors encoded by said action potentials. A summary of notable publications providing evidence of epigenetic interactions regulating stimulation pattern specific neuronal activity-dependent gene expression are featured in Table 2 and synthesized in Figure 3.
Table 2.
Evidence for Neuronal Activity Pattern–Specific Changes to Gene Expression or Epigenetic Modifications.
| Publication | Cell Type | Stimulation Type | Analogous Behavior or Disease |
Molecular or Epigenetic Feature |
Primary Technique | Important Findings |
|---|---|---|---|---|---|---|
| Su and others (2017) | Adult mouse dentate granule neurons | Whole animal in vivo electroconvulsive stimulation | Drug-resistant depression treatment | Genome-wide chromatin compaction | ATAC-seq | Neuronal activity induced 11,438 gained-open and 1739 gained-closed sites by 1 hour after stimulation. Notably, gained-open and gained-closed sites are enriched at active enhancers. C-Fos binding involved in initial chromatin opening at opened sites, but not maintenance of open site over time |
| Guo and others (2011) | Adult mouse dentate granule neurons | Whole animal in vivo electroconvulsive stimulation | Drug-resistant depression treatment | Genome-wide DNA methylation | Methyl-Sensitive Cut Counting (MSCC) to measure single nucleotide CpG methylation levels | Neuronal activity induced de novo methylation at 1,892 CpG sites and demethylation at 1158 CpG sites |
| Maag and others (2017) | Neurons isolated from dentate gyrus | LTP induced with in vivo high-frequency stimulation (HFS) to dentate gyrus for 30 minutes, 2 hours, or 5 hours | Hippocampal long-term potentiation | DNA methylation in LTP | Methylated DNA immunoprecipitation (MeDIP)-array | DNA methylation changes occurred in 48 regions after 30 minutes, in 699 regions after 2 hours and in 448 regions after 5 hours. DNA methylation changes due to HFS-induced LTP were uniquely associated with 353 genes that were not associated differential methylation following other stimulation types including contextual fear therapy with and without shock, or electroconvulsion therapy |
| Lee and others (2017) | Dorsal root ganglion (DRG) | Two patterns (18/1 or 90/5) and two duration lengths (2 hours or 5 hours) for in vitro stimulation: 10 Hz for 1.8 seconds with 1-minute intervals (termed 18/1) or 10 Hz for 9 seconds with 5-minute intervals (termed 90/5) | Patterned neuronal information | Differential expression of gene networks and transcription factor binding | Genome wide microarray in combination with distant regulatory elements of co-regulated genes (DIRE) and gene set enrichment analysis (GSEA) | Networks of genes, beyond the classically defined immediate response genes, are regulated by both kinetics and duration of stimulus pattern. 2501 genes up-regulated by electrical stimulation and 3424 were down-regulated by electrical stimulation. Pattern specific changes attributed to alternative enrichment of transcription factor binding in each stimulation condition. |
| Chen and others (2016) | Drosophila neurons | Optogenetic stimulation for 30 seconds at 10 Hz in vivo; in vivo heat activated dTrpA1 cation channel opening; ex vivo KCl (90mM) stimulation | Patterned neuronal information | Stimulus-dependent activity-induced gene expression | mRNA high-throughput sequencing, ATAC-seq | Expression of activity-related genes in Drosophila alters based on stimulation paradigm. Within a single stimulation paradigm, activity-regulated genes varied between differentiated neuron types |
| Bukalo and others (2016) | CA1 neurons from hippocampal slices | Theta-burst stimulation delivered antidromically producing action-potential induced long-term depression (AP-LTD) | Memory consolidation during Slow-wave sleep | Differential expression of exons within the same gene after AP-LTD stimulation | RT-PCR | Antidromic AP-LTD induction significantly decreased mRNA transcripts containing BDNF exon I or exon II after 15 minutes, while mRNA transcripts containing exons IV or IX were unaffected at the same time-point |
| Zhao and others (2017) | Dorsal root ganglion (DRG) | Spinal nerve ligation (SNL) and constriction injury of sciatic nerve (CCI) | Neuropathic pain | DNA methylation | Bisulfite pyro-sequencing assay for DNA methylation | DNA methyltransferase (DNMT) 3a, but not DNMT3b, mRNA and protein increased in DRG neurons following peripheral nerve injury. SNL-induced increase in DNMT3a produces increased methylation in the promoter region of Kcna2 gene, decreasing Kcna2 expression |
| Liang and others (2016) | Dorsal root ganglion (DRG) | Spinal nerve ligation (SNL) and constriction injury of sciatic nerve (CCI) | Neuropathic pain | Histone methylation | Chromatin immunoprecipitation, RT-PCR | mRNA for G9a, the enzyme responsible for the methylation of histone H3 at lysine 9, increases with SNL and CCI, but not sham surgery |
| Fields and others (1997) | Dorsal root ganglion (DRG) | Four stimulation paradigms each delivering 540 impulses at 10 Hz for 30 minutes. Each paradigm stimulated for 1.8, 3.6, 5.4, or 9.0 seconds with 1-, 2-, 3-, or 5-minute intervals, respectively. | Neuronal bursting activity in utero | Temporal specificity of cytoplasmic and nuclear signaling | Western blot | Phosphorylation of CREB occurs with neuronal activity but does not maintain temporal specifically Therefore, other factors must account for pattern-specific changes in gene expression |
| Worley and others (1993) | Hippocampal granule neurons | In vivo hippocampal stimulation in 10 train and 50 train paradigms. “10 train”: 10 repetitions at 400 Hz each lasting 25 ms (grouped into 10 pulses, and stimulated for 400 pulses total); “50 train:” 50 repetitions at 400 Hz each lasting 20 ms (grouped into 8 pulses and stimulated for 400 pulses total) | Hippocampal long-term enhancement | Relationship between temporally unique stimulation patterns and “activity dependent” transcription factor mRNA abundance | In situ hybridization | 10-train stimulation induced significant increase in zif268 and junB but not c-fos or c-jun mRNA levels, by 30 minutes post-stimulation. 50-train stimulation alternatively induced significant increase in zif268, junB, c-fos, and c-jun at the same time-point |
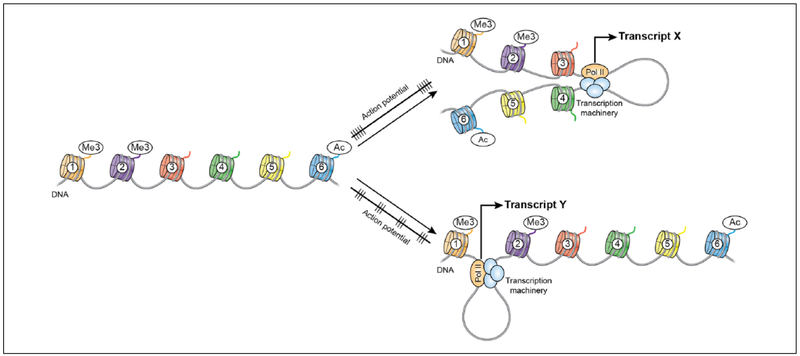
Figure 3.
Dynamic structures of the neuronal epigenome allow for encoded neural activity to produce targeted and controlled pattern-dependent gene expression in response to temporally specific neuronal activation. The unknown molecular mechanisms leading to pattern-specific gene expression present exciting questions for molecular, cellular and cognitive neuroscience and biology fields. Schematic illustrates DNA wrapped around histones (colored cylinders) each marked with representative histone protein modifications (labeled circles attached to histone tail). Straight line with perpendicular dashes beside arrows indicate action potential patterns.
Chromatin Structure and Dynamics
Three-dimensional chromatin structure is defined on a small scale by spacing of nucleosomes, which wrap ~200 DNA nucleotides, and on a larger scale by topographically associated domains (TADs) which span hundreds to thousands of kilobases (Dixon and others 2012). TADs can be defined at their boundaries by the insulating DNA binding protein CCCTC-binding factor (CTCF) (Narendra and others 2015). Hi-C, a chromatin conformation capture method used to illustrate DNA-DNA interactions, reveals DNA-DNA interactions are most enriched between sequences within the same TAD (Rao and others 2014). The boundaries of TADs as defined by binding of CTCF at the CTCF binding motif largely influence interactions such as those occurring between enhancers and promoters (Dixon and others 2016). The interactions between DNA, RNA, and protein used to first identify, then transcribe the appropriate sequence necessary for cell function requires epigenetic marks on histones and DNA to shape the physical architecture and accessibility of regions within the genome. The theory of a universal histone code, such that each mark is associated with a consistent biological function like “activating” or “inhibiting,” has been frequently debated (Rando 2012). Despite this, each mark is highly informational to the appropriate architecture of chromatin allowing sequences of nucleotides, whether they may act as enhancer or promoter elements, or are transcribed into non-coding RNA or protein coding regions, to become accessible or hidden depending on tissue and developmental stage context. One important context is the ability to respond and “remember” incoming environmental stimuli (Ravi and Kannan 2013).
Neuronal Activity and Chromatin Remodeling
Analysis of genome-wide chromatin compaction generated from ATAC-seq data using dentate granule neurons before and after acute electroconvulsive stimulation reveals ~50,000 new open chromatin regions primarily occurring in introns and intergenic regions (Su and others 2017). To associate the alternative chromatin accessibility regions with TADs previously characterized from cortical neurons, the ATAC-seq data was compared to previously compiled CTCF chromatin immunoprecipitation (ChIP) and histone modification ChIP data to reveal colocalization of newly opened sites with the known activating marks: methylated lysine 4 in histone 3 (referred to as H3K4me1) and acetylated lysine 27 of histone 3 (H3K27Ac) (Su and others 2017). RNA-seq of stimulated neurons reveals overlap of upregulated mRNA expression and regions with gained chromatin opening (Su and others 2017). ChromHMM analysis, used to characterize chromatin states, demonstrated active enhancer regions experience the most robust effects of chromatin remodeling following neural stimulation (Su and others 2017). It is important to note that this article does not explore how neuronal activity translates from cytosolic calcium signaling kinetics, into a change in nuclear protein interactions. While this study demonstrates crucial advancements in our understanding of neuronal activity and constant chromatin remodeling, the experimenters do not account for temporal specificity of neural activity that is essential to encode environment and behavior-specific information. It is therefore essential to first update the CTCF ChIP databases to include stimulated neurons, because it has not yet been shown that stimulation alters CTCF-DNA binding, and secondly apply physiologically relevant patterned stimuli to understand how these genome-wide features maintain temporal integrity.
At specific genomic regions, neuronal activity can induce relocation of gene loci and specific enhancer-promoter looping contacts for transcriptional regulation (Madabhushi and others 2015; Watson and Tsai 2017). Neuronal activity-dependent DNA double-stranded break formation in the promoter of immediate early genes can overcome repressive topological constraints to allow for rapid activity-induced transcription (Madabhushi and others 2015; Watson and Tsai 2017).
Neuronal Activity and Histone Modifications
Genome wide changes in histone modifications have also been associated with neuronal activity. Robust depolarization with KCl resulted in increased H3K27ac and H3K4m3 marks and decreased H3K9m3 and H3K27m3 marks at the tyrosine hydroxylase promoter of neural precursor cell prior to KCl-induced differentiation into dopaminergic neurons (He and others 2011). Studies thus suggest neuronal activity can drive differentiation of cells through regulation of histone modifications. Interestingly, in vitro experiments in rat liver cells demonstrate that histone H1 responds to elevated calcium with a conformational change (Tarkka and others 1997). Taken together with the nuclear calcium signaling dynamics that may regulate transcription (Bengtson and others 2010), binding between temporally regulated nuclear calcium concentration in response to action potential activity and H1 histone binding present an intriguing and plausible mechanism for activity-induced, potentially pattern-specific, changes in chromatin structure.
Emerging evidence demonstrates enzymatic activity of histone modifying proteins are regulated by RNAs (Bose and Berger 2017). As whole genome sequencing improves, the existence and function of non-coding RNAs (ncRNAs) is becoming a major consideration. ncRNA transcribed from enhancer regions (eRNAs) is found to act as a scaffold between enhancers and promoters, and assists in chromatin looping and transcription factor binding ultimately affecting the abundance of transcripts (Bose and Berger 2017; Rajarajan and others 2016). eRNAs have widespread influence on the genome evidenced by RNA-seq data from E14.5 whole mouse tissue analysis that reveals about 70% of enhancers identified in DNA isolated from brain tissue transcribe eRNAs (Cheng and others 2015).
Important new experiments pairing photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) to analyze RNA-protein interactions with in vitro enzyme activity assays reveal ncRNAs, including eRNAs, bind CREB-binding protein (CBP) and enhance its histone acetyltransferase (HAT) activity in the CBP active site (Bose and others 2017). HAT activity and H3K27ac marks are often associated with increased transcription (Bose and Berger 2017). Ultimately, the involvement of eRNAs in epigenetic modification regulation presents an intriguing layer of regional or target gene specificity in response to neuronal activity. With many recent advancements to understand eRNA function, temporal or localized characteristics of eRNA transcription and binding is hot topic in molecular biology and may have important implications for intricacies within pattern specific activity-dependent neuronal gene expression.
Neuronal Activity and DNA Modifications
Neuronal activity applied in vitro produces de novo methylation and rapid demethylation in a reported 1.4% of CpG dinucleotides in neurons isolated from dentate gyrus neurons (Guo and others 2011). Alterations in methylation due to activity lasted weeks after stimulation (Guo and others 2011). Interestingly, de novo DNA methylation can negatively regulate CTCF binding on DNA (Bell and Felsenfeld 2000). In addition to effects on CTCF binding, gene-ontology analysis by Guo and others (2011) suggest activity-modified CpGs were most enriched at genes and motifs involved with splicing variants (Guo and others 2011). Altered DNA methylation genome-wide may be explained by the activity of DNA methyltransferases (DNMT) DNMT1 and DNMT3a (Day and others 2013; Sharma and others 2008). DNMT3a ChIP assay results indicate neuronal activity induced with the sodium channel agonist alters DNMT3a-DNA binding as well as subsequent IEG expression (Day and others 2013). Depolarization of cultured cortical neurons with KCl and sodium channel agonist veratridine results in decreased mRNA of DNMT1 and DNMT3a (Sharma and others 2008). Thus, differences in DNA methylation due to neuronal activity may occur via an indirect process. Changes in methylation have also been reported in vivo. Induction of LTP with high-frequency stimulation in rats produce differential methylation of LTP-associated genes as measured with a methylated DNA immunoprecipitation assay (Maag and others 2017). Levels of methylation were correlated with RNA-seq data at multiple timepoints after stimulation and indicated both up and down regulation of LTP-associated genes where alternative methylation occurred (Maag and others 2017). Importantly, the experiments performed by Maag and others present compelling evidence connecting a behaviorally relevant stimulus with pattern-dependent epigenetic alterations. While these authors focus on a predetermined set of genes, eventually performing a genome-wide analysis of sequences with altered epigenetic interactions will allow for a broader understanding of the pattern-specific epigenetic changes associated with many behaviors.
Future Questions and Conclusion
The majority of research in this field has demonstrated that generalized neuronal depolarization affects both gene expression and the epigenetic landscape (reviewed by Cortés-Mendoza and others 2013; Flavell and Greenberg 2008; West and Greenberg 2011). Yet, environmental information is encoded in patterns of action potentials and further, recent findings indicate that alternative gene expression occurs on a neuronal firing pattern–dependent manner (Lee and others 2017). To understand how specific populations of neurons maintain context dependent role, it is essential to look to the interactions between the epigenetic architecture and the intracellular propagation of neuronal activity into the nucleus. Specifically, addressing novel and essential questions surrounding how the temporal integrity of distinct frequencies of stimulation is maintained in the interplay between activity and epigenetic modifications to produce pattern specific alternative gene expression will prove invaluable to the field. Which epigenetic modifications are altered with stimulus pattern integrity? Do these malleable protein, DNA, and RNA constructs work alone or in combination? How do protein activation cascades or calcium transients propagate from the cytoplasm to the nucleus with temporal specificity? How are protein and nucleotide structures modified by cytoplasmic and nuclear signaling cascades? Are there certain genes or genomic regions that are more highly modified by activity? Are these regions conserved across differentiated neuron populations? How do separate differentiated cells respond differently to the same stimulus? What aspects of the genomic architecture maintain a differentiated specialized neuron state versus what aspects are highly malleable based on cellular activity? Furthermore, intricacies in the type of stimulus and the networks of genes expressed suggests that the currently characterization of “activity-dependent genes” due to robust depolarization might miss genomic regions that are opened and expressed during more specific or intermediate types of neuronal stimulation.
From the neuroscience perspective, activity dependent gene expression is an important and fascinating phenomenon underlying the neuronal plasticity critical for learning and memory. From the molecular biology perspective, epigenetic remodeling in response to constant and persistent temporally unique stimuli presents an ideal system to discover stimuli-induced modulation of interactions between three-dimensional chromatin structures, subsequent DNA-DNA interactions, as well as DNA-protein interactions. Expanding the knowledge of epigenetics mediating neuronal cell responses to activity presents exciting and major interdisciplinary questions in cognitive neuroscience, cellular neuroscience and molecular biology.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: JB and RDF are supported by the Intramural Program of the Eunice Kennedy Shriver National Institute of Child Health & Human Development (NICHD) of the National Institutes of Health (NIH) ZIA HD000713-23.
Footnotes
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- Adams JP, Roberson ED, English JD, Selcher JC, Sweatt JD. 2000. MAPK regulation of gene expression in the central nervous system. Acta Neurobiol Exp (Wars) 60(3):377–94. [DOI] [PubMed] [Google Scholar]
- Al-Mohanna FA, Caddy KW, Bolsover SR. 1994. The nucleus is insulated from large cytosolic calcium ion changes. Nature 367(6465):745–50. [DOI] [PubMed] [Google Scholar]
- Allfrey VG, Faulkner R, Mirsky AE. 1964. Acetylation and methylation of histones and their possible role in regulation of RNA synthesis. Proc Natl Acad Sci U S A 51(5):786–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai JA, Feig LA. 2011. Long-lasting and transgenerational effects of an environmental enrichment on memory formation. Brain Res Bull 85(1–2):30–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin-Abidin S, Trippe J, Funke K, Eysel UT, Benali A. 2008. High- and low-frequency repetitive transcranial magnetic stimulation differentially activates cFos and zif268 protein expression in the rat brain. Exp Brain Res 188(2):249–61. [DOI] [PubMed] [Google Scholar]
- Barco A, Alarcon JM, Kandel ER. 2002. Expression of constitutively active CREB protein facilitates the late phase of long-term potentiation by enhancing synaptic capture. Cell 108(5):689–703. [DOI] [PubMed] [Google Scholar]
- Bading H 2000. Transcription-dependent neuronal plasticity. Eur J Biochem 267(17):5280–3. [DOI] [PubMed] [Google Scholar]
- Bahrami S, Drabløs F. 2016. Gene regulation in the immediate-early response process. Adv Biol Regul 62:37–49. [DOI] [PubMed] [Google Scholar]
- Basavarajappa BS, Subbanna S. 2016. Epigenetic mechanisms in developmental alcohol-induced neurobehavioral deficits. Brain Sci 6(2):E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 405(6785):482–5. [DOI] [PubMed] [Google Scholar]
- Bengtson CP, Freitag HE, Weislogel JM, Bading H. 2010. Nuclear calcium sensors reveal that repetition of trains of synaptic stimuli boosts nuclear calcium signaling in CA1 pyramidal neurons. Biophys J 99(12):4066–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieszczad KM, Bechay K, Rusche JR, Jacques V, Kudugunti S, Miao W, and others. 2015. Histone deacetylase inhibition via RGFP966 releases the brakes on sensory cortical plasticity and the specificity of memory formation. J Neurosci 35(38):13124–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bito H, Deisseroth K, Tsien RW. 1996. CREB phosphorylation and dephosphorylation: a Ca2+- and stimulus duration dependent switch for hippocampal gene expression. Cell. 87(7):1203–14. [DOI] [PubMed] [Google Scholar]
- Blank M, Dornelles AS, Werenicz A, Velho LA, Pinto DF, Fedi AC, and others. 2014. Basolateral amygdala activity is required for enhancement of memory consolidation produced by histone deacetylase inhibition in the hippocampus. Neurobiol Learn Mem 111:1–8. [DOI] [PubMed] [Google Scholar]
- Bonnefond M, Kastner S, Jensen O. 2017. Communication between brain areas based on nested oscillations. eNeuro 4(2). March 10 [Epub ahead of print]. doi: 10.1523/ENEURO.0153-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose DA, Berger SL. 2017. eRNA binding produces tailored CBP activity profiles to regulate gene expression. RNA Biol 14(12):1655–9. doi: 10.1080/15476286.2017.1353862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose DA, Donahue G, Reinberg D, Shiekhattar R, Bonasio R, Berger SL. 2017. RNA binding to CBP stimulates histone acetylation and transcription. Cell 168(1–2):135–49. doi: 10.1016/j.cell.2016.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredy TW, Wu H, Crego C, Zellhoefer J, Sun YE, Barad M. 2007. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn Mem 14(4):268–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukalo O, Lee PR, Fields RD. 2016. BDNF mRNA abundance regulated by antidromic action potentials and AP-LTD in hippocampus. Neurosci Lett 635:97–102. doi: 10.1016/j.neulet.2016.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsáki G, Draguhn A. 2004. Neuronal oscillations in cortical networks. Science 304(5679):1926–9. [DOI] [PubMed] [Google Scholar]
- Cao J, Yan Q. 2012. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front Oncol 2:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Rahman R, Guo F, Rosbash M. 2016. Genome-wide identification of neuronal activity-regulated genes in Drosophila. Elife. 5:e19942. doi: 10.7554/eLife.19942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JH, Pan DZ, Tsai ZT, Tsai HK. 2015. Genome-wide analysis of enhancer RNA in gene regulation across 12 mouse tissues. Sci Rep 5:12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholewa-Waclaw J, Bird A, von Schimmelmann M, Schaefer A, Yu H, Song H, and others. 2016. The role of epigenetic mechanisms in the regulation of gene expression in the nervous system. J Neurosci 36(45):11427–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coba MP, Valor LM, Kopanitsa MV, Afinowi NO, Grant SG. 2008. Kinase networks integrate profiles of N-methyl-d-aspartate receptor-mediated gene expression in hippocampus. J Biol Chem 283(49):34101–7. doi: 10.1074/jbc.M804951200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Greenberg ME. 2008. Communication between the synapse and the nucleus in neuronal development, plasticity, and disease. Annu Rev Cell Dev Biol. 24:183–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corlier J, Valderrama M, Navarrete M, Lehongre K, Hasboun D, Adam C, and others. 2016. Voluntary control of intracortical oscillations for reconfiguration of network activity. Sci Rep 6:36255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés-Mendoza J, Díaz de León-Guerrero S, Pedraza-Alva G, Pérez-Martínez L. 2013. Shaping synaptic plasticity: the role of activity-mediated epigenetic regulation on gene transcription. Int J Dev Neurosci 31(6):359–69. [DOI] [PubMed] [Google Scholar]
- Costa E, Grayson DR, Mitchell CP, Tremolizzo L, Veldic M, Guidotti A. 2003. GABAergic cortical neuron chromatin as a putative target to treat schizophrenia vulnerability. Crit Rev Neurobiol. 15:121–42. [DOI] [PubMed] [Google Scholar]
- Cuadrado-Tejedor M, Garcia-Barroso C, Sanzhez-Arias J, Mederos S, Rabal O, Ugarte A, and others. 2015. Concomitant histone deacetylase and phosphodiesterase 5 inhibition synergistically prevents the disruption in synaptic plasticity and it reverses cognitive impairment in a mouse model of Alzheimer’s disease. Clin Epigenetics 7:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubenas-Potts C, Matunis MJ. 2013. SUMO: A multifaceted modifier of chromatin structure and function. Dev Cell 24:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Childs D, Guzman-Karlsson MC, Kibe M, Moulden J, Sweatt JD, and others. 2013. DNA methylation regulates associative reward learning. Nat Neurosci 16(10):1445–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nota PM, Chartrand JM, Levkov GR, Montefusco-Siegmund R, DeSouza JF. 2017. Experience-dependent modulation of alpha and beta during action observation and motor imagery. BMC Neurosci 18(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Gorkin DU, Ren B. 2016. Chromatin domains: the unit of chromosome organization. Mol Cell 62(5):668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, and others. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485(7398):376–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek SM, Fields RD. 2002. Somatic action potentials are sufficient for late-phase LTP-related cell signaling. Proc Natl Acad Sci U S A. 99(6):3962–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields RD, Eshete F, Stevens B, Itoh K. 1997. Action potential-dependent regulation of gene expression: temporal specificity in Ca2+, cAMP-responsive element binding proteins, and mitogen-activated protein kinase signaling. J Neurosci 17(19):7252–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields RD, Yu C, Neale EA, Nelson PG. 1992. Recording chambers in cell culture In: Kettenmann H, Grantyn R, editors. Electrophysiological methods for in vitro studies in vertebrate neurobiology. New York, NY: Wiley-Liss; p.67–76. [Google Scholar]
- Friese U, Köster M, Hassler U, Martens U, Trujillo-Barreto N, Gruber T. 2013. Successful memory encoding is associated with increased cross-frequency coupling between frontal theta and posterior gamma oscillations in human scalp-recorded EEG. Neuroimage 66:642–7. doi: 10.1016/j.neu-roimage.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. 2008. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci 31:563–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchikami M, Yamamoto S, Morinobu S, Takei S, Yamawaki S. 2010. Epigenetic regulation of BDNF gene in response to stress. Psychiatry Investig 7(4):251–6. doi: 10.4306/pi.2010.7.4.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Géranton SM, Tochiki KK. 2015. Regulation of gene expression in pain states by epigenetic mechanisms. Prog Mol Biol Transl Sci 131:147–83. doi: 10.1016/bs.pmbts.2014.11.012. [DOI] [PubMed] [Google Scholar]
- Gold M, Hurwitz J, Anders M. 1963. The enzymatic methylation of RNA and DNA, II. On the species specificity of methylation enzymes. Proc Natl Acad Sci U S A 50(1):164–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grehl S, Viola HM, Fuller-Carter PI, Carter KW, Dunlop SA, Hool LC, and others. 2015. Cellular and molecular changes to cortical neurons following low intensity repetitive magnetic stimulation at different frequencies. Brain Stimul 8(1):114–23. doi: 10.1016/j.brs.2014.09.012. [DOI] [PubMed] [Google Scholar]
- Grigorenko EL, Kornilov SA, Naumova OY. 2016. Epigenetic regulation of cognition: a circumscribed review of the field. Dev Psychopathol. 28(4 Pt 2):1285–304. doi: 10.1017/S0954579416000857. [DOI] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang M-H, Bonaguidi MA, and others. 2011. Neuronal activity modifies DNA methylation landscape in the adult brain. Nat Neurosci 14(10):1345–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, and others. 2010. Histone methylation regulates memory formation. J Neurosci 30(10):3589–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez RM, Hnilica LS. 1967. Tissue specificity of histone phosphorylation. Science 157(3794):1324–5. [DOI] [PubMed] [Google Scholar]
- Halder R, Hennion M, Vidal RO, Shomroni O, Rahman RU, Rajput A, and others. 2016. DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nat Neurosci 19(1):102–10. doi: 10.1038/nn.4194. [DOI] [PubMed] [Google Scholar]
- Hardingham GE, Arnold FJ, Bading H. 2001. Nuclear calcium signaling controls CREB-mediated gene expression triggered by synaptic activity. Nat Neurosci 4(3):261–7. [DOI] [PubMed] [Google Scholar]
- He XB, Yi SH, Rhee YH, Kim H, Han YM, Lee SH, and others. 2011. Prolonged membrane depolarization enhances midbrain neuron differentiation via epigenetic histone modifications. Stem Cells 29(11):1861–73. [DOI] [PubMed] [Google Scholar]
- Huang X, Dixit VM. 2016. Drugging the undruggables: exploring the ubiquitin system for drug development. Cell Res 26(4):484–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunsberger JG, Bennett AH, Selvanayagam E, Duman RS, Newton SS. 2005. Gene profiling the response to kainic acid induced seizures. Brain Res Mol Brain Res 141(1):95–112. doi: 10.1016/j.molbrainres.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Impey S, Fong AL, Wang Y, Cardinaux JR, Fass DM, Obrietan K, and others. 2002. Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron 34(2):235–44. [DOI] [PubMed] [Google Scholar]
- Kang ES, Park CW, Chung JH. 2001. Dnmt3b, de novo DNA methyltransferase, interacts with SUMO-1 and Ubc9 through its N-terminal region and is subject to modification by SUMO-1. Biochem Biophys Res Commun 289(4):862–8. [DOI] [PubMed] [Google Scholar]
- Kayser C, Montemurro MA, Logothetis NK, Panzeri S. 2009. Spike-phase coding boosts and stabilizes information carried by spatial and temporal spike patterns. Neuron 61(4):597–608. [DOI] [PubMed] [Google Scholar]
- Kim HD, Call T, Magazu S, Ferguson D. 2017. Drug addiction and histone code alterations. Adv Exp Med Biol 978:127–43. [DOI] [PubMed] [Google Scholar]
- Leite MF, Thrower EC, Echevarria W, Koulen P, Hirata K, Bennett AM, and others. 2003. Nuclear and cytosolic calcium are regulated independently. Proc Natl Acad Sci U S A 100(5):2975–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PR, Cohen JE, Becker KG, Fields RD. 2005. Gene expression in the conversion of early-phase to late-phase longterm potentiation. Ann N Y Acad Sci 1048:259–71. [DOI] [PubMed] [Google Scholar]
- Lee PR, Cohen JE, Iacobas DA, Iacobas S, Fields RD. 2017. Gene networks activated by specific patterns of action potentials in dorsal root ganglia neurons. Sci Rep 7:43765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Gu X, Zhao J-Y, Wu S, Miao X, Xiao J, and others. 2016. G9a participates in nerve injury-induced Kcna2 downregulation in primary sensory neurons. Sci Rep 6:37704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljubisavljevic MR, Javid A, Oommen J, Parekh K, Nagelkerke N, Sheshab S, and others. 2015. The effects of different repetitive transcranial magnetic stimulation (rTMS) protocols on cortical gene expression in a rat model of cerebral ischemic-reperfusion injury. PLoS One 10(10):e139892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag JL, Kaczorowski DC, Panja D, Peters TJ, Bramham CR, Wibrand K, and others. 2017. Widespread promoter methylation of synaptic plasticity genes in long-term potentiation in the adult brain in vivo. BMC Genomics 18(1):250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madabhushi R, Gao F, Pfenning AR, Pan L, Yamakawa S, Seo J, and others. 2015. Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell 161(7):1592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Wenderski W, Noh KM, Bagot RC, Tzavaras N, Purushothaman I, and others. 2015. Critical role of histone turnover in neuronal transcription and plasticity. Neuron 87(1):77–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JL, Wadzinski BE. 2009. Targeting protein serine/threonine phosphatases for drug development. Mol Pharmacol 75(6):1249–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mews P, Donahue G, Drake AM, Luczak V, Abel T, Berger SL. 2017. Acetyl-CoA synthetasae regulates histone acetylation and hippocampal memory. Nature 546(7658):381–6. doi: 10.1038/nature22405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra V, Rocha PP, An D, Raviram R, Skok JA, Mazzoni EO, and others. 2015. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347(6225):1017–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey K, Sharma KP, Sharma SK. 2015. Histone deacetylase inhibition facilitates massed pattern-induced synaptic plasticity and memory. Learn Mem 22(10):514–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham TA, Impey S, Storm DR, Stryker MP. 1999. CRE-mediated gene transcription in neocortical neuronal plasticity during the developmental critical period. Neuron 22(1):63–72. [DOI] [PubMed] [Google Scholar]
- Phengchat R, Takata H, Morii K, Inada N, Murakoshi H, Uchiyama S, and others. 2016. Calcium ions function as a booster of chromosome condensation. Sci Rep 6:38281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina B, Martinez P, Suau P. 1988. Differential acetylation of core histones in rat cerebral cortex neurons during development and aging. Eur J Biochem 174(2):311–5. [DOI] [PubMed] [Google Scholar]
- Rajarajan P, Gil SE, Brennand KJ, Akbarian S. 2016. Spatial genome organization and cognition. Nat Rev Neurosci 17(11):681–91. doi: 10.1038/nrm.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ. 2012. Combinatorial complexity in chromatin structure and function: revisiting the histone code. Curr Opin Genet Dev 22(2):148–55. doi: 10.1016/j.gde.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, and others. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159(7):1665–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi B, Kannan M. 2013. Epigenetics in the nervous system: an overview of its essential role. Indian J Hum Genet 19(4):384–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio A 2010. Dynamic epigenetic regulation in neurons: enzymes, stimuli and signaling pathways. Nat Neurosci 13(11):1330–7. [DOI] [PubMed] [Google Scholar]
- Roth TL, Sweatt JD. 2009. Regulation of chromatin structure in memory formation. Curr Opin Neurobiol 19(3):336–42. doi: 10.1016/j.conb.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetto D, Avvakumov N, Cote J. 2012. Histone phosphorylation and chromatin modification involved in diverse nuclear events. Epigenetics 7(10):1098–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan H, Gao J, Qi X, Tao Y, Guo X, Guo Z, and others. 2016. Visual experience dependent regulation of neuronal structure and function by histone deacetylase 1 in developing Xenopus tectum in vivo. Dev Neurobiol 77(8):947–62. doi: 10.1002/dneu.22480. [DOI] [PubMed] [Google Scholar]
- Ruzicka WB. 2015. Epigenetic mechanisms in the pathophysiology of psychotic disorders. Harv Rev Psychiatry 23(3):212–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MM, Ryan B, Kyrke-Smith M, Logan B, Tate WP, Abraham WC, and others. 2012. Temporal profiling of gene networks associated with the late phase of long-term potentiation in vivo. PLoS One, 7(7):e40538. doi: 10.1371/journal.pone.0040538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen H 2015. Epigenetic regulation of memory by acetylation and methylation of chromatin: implications in neurological disorders, aging, and addiction. Neuromolec Medicine 17(2):97–110. [DOI] [PubMed] [Google Scholar]
- Sharma RP, Tun N, Grayson DR. 2008. Depolarization induces downregulation of DNMT1 and DNMT3 in primary cortical cultures. Epigenetics 3(2):74–80. doi: 10.4161/epi.3.2.6103. [DOI] [PubMed] [Google Scholar]
- Singh P, Thakur MK. 2017. Histone deacetylase 2 inhibition attenuates downregulation of hippocampal plasticity gene expression during aging. Mol Neurobiol. March 31 [Epub ahead of print]. doi: 10.1007/s12035-017-0490-x. [DOI] [PubMed] [Google Scholar]
- Stefanko DP, Barrett RM, Ly AR, Reolon GK, Wood MA. 2009. Modulation of long-term memory for object recognition via HDAC inhibition. Proc Natl Acad Sci U S A 106(23):9447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevely WS, Stocken LA. 1966. Phosphorylation of rat-thymus histone. Biochem J 100(2):20C–21C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Shin J, Zhong C, Wang S, Roychowdhury P, Lim J, and others. 2017. Neuronal activity modifies the chromatin accessibility landscape in the adult brain. Nat Neurosci 20(3):476–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarkka T, Oikarinen J, Grundström T. 1997. Nucleotide and calcium-induced conformational changes in histone H1. FEBS Lett 406(1–2):56–60. [DOI] [PubMed] [Google Scholar]
- Watson LA, Tsai HH. 2017. In the loop: how chromatin topology links genome structure to function in mechanisms underlying learning and memory. Curr Opin Neurobiol 43:48–55. doi: 10.1016/j.conb.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Greenberg ME. 2011. Neuronal activity–regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol 3(6):a005744. doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley PF, Bhat RV, Baraban JM, Erickson CA, McNaughton BL, Barnes CA. 1993. Thresholds for synaptic activation of transcription factors in hippocampus: correlation with long-term enhancement. J Neurosci 13(11):4776–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Hayes JJ. 2011. The divalent cations Ca2+ and Mg2+ play specific roles in stabilizing histone-DNA interactions within nucleosomes that are partially redundant with the core histone tail domains. Biochemistry 50(46):9973–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B, Christian KM, He C, Jin P, Ming GL, Song H. 2016. Epigenetic mechanisms in neurogenesis. Nat Rev Neurosci 17(9):537–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Crabtree GR. 2009. ATP-dependent chromatin remodeling in neural development. Curr Opin Neurobiol 19(2):120–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J-Y, Liang L, Gu X, Li Z, Wu S, Sun L, and others. 2017. DNA methyltransferase DNMT3a contributes to neuropathic pain by repressing Kcna2 in primary afferent neurons. Nat Commun 2017;8:14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Liang C, Li D, Tian M, Liu S, Gao G, and others. 2016. Histone methyltransferase Ash1L mediates activity-dependent repression of neurexin-1α. Sci Rep 6:26597. [DOI] [PMC free article] [PubMed] [Google Scholar]