Abstract
A major challenge associated with biochemical and cellular analysis of pseudokinases is a lack of target-validated small-molecule compounds with which to probe function. Tribbles 2 (TRIB2) is a cancer-associated pseudokinase with a diverse interactome, including the canonical AKT signaling module. There is substantial evidence that human TRIB2 promotes survival and drug resistance in solid tumors and blood cancers and therefore is of interest as a therapeutic target. The unusual TRIB2 pseudokinase domain contains a unique cysteine-rich C-helix and interacts with a conserved peptide motif in its own carboxyl-terminal tail, which also supports its interaction with E3 ubiquitin ligases. We found that TRIB2 is a target of previously described small-molecule protein kinase inhibitors, which were originally designed to inhibit the canonical kinase domains of epidermal growth factor receptor tyrosine kinase family members. Using a thermal shift assay, we discovered TRIB2-binding compounds within the Published Kinase Inhibitor Set (PKIS) and used a drug repurposing approach to classify compounds that either stabilized or destabilized TRIB2 in vitro. TRIB2 destabilizing agents, including the covalent drug afatinib, led to rapid TRIB2 degradation in human AML cancer cells, eliciting tractable effects on signaling and survival. Our data reveal new drug leads for the development of TRIB2-degrading compounds, which will also be invaluable for unraveling the cellular mechanisms of TRIB2-based signaling. Our study highlights that small molecule–induced protein down-regulation through drug “off-targets” might be relevant for other inhibitors that serendipitously target pseudokinases.
INTRODUCTION
The human protein kinome encodes ~60 protein pseudokinases, which lack at least one conventional catalytic residue but often control rate-limiting signaling outputs within cellular networks (1). Like canonical kinases, pseudokinases drive conformation-dependent signaling associated with both physiology and disease (2, 3). The human “pseudokinome” includes cancer-associated signaling proteins such as human epidermal growth factor receptor 3 (HER3), Janus kinase 2 (JAK2; JH2 domain), and Tribbles 2 (TRIB2), which have received much less attention than their conventional, catalytically active counterparts even though pseudokinase domains represent rational targets for drug discovery (4). Discovering or repurposing biologically and/or clinically active compounds that target atypical conformations of canonical kinases or pseudokinases is an area of active research (2, 3, 5–9). Moreover, the burgeoning pseudokinase field is strongly placed to benefit from the decades of research undertaken on canonical protein kinases, which has seen the approval of more than 40 kinase inhibitors for human cancer and inflammatory diseases (10, 11). This includes understanding how adenosine triphosphate (ATP)–competitive, allosteric, or covalent inhibitors might influence pseudokinase-based signaling mechanisms that are relevant to health and disease (3, 12).
The three human TRIB pseudokinases (TRIB1, TRIB2, and TRIB3) and the related pseudokinase STK40 (serine/threonine kinase 40, also known as SgK495) are homologs of the Drosophila melanogaster pseudokinase termed Tribbles, which controls ovarian border cell and neuronal stem cell physiology in flies (13, 14). These proteins contain a catalytically impaired pseudokinase domain. Adaptations in the pseudokinase fold, including a highly unusual αC-helix, are thought to support a competitive regulatory interaction in cis with a unique C-terminal tail DQLVP motif (15–17). Through a still obscure mechanism, TRIB and STK40 function as adaptor proteins that recruit ubiquitin E3 ligases, such as constitutive photomorphogenesis protein 1 homolog, through interaction with the conserved C-tail peptide (15, 17), which is also required for signaling and cellular transformation (18). Mechanistically, the signaling outputs of Tribbles proteins are controlled through the ubiquitylation and subsequent proteasomal destruction of Tribbles “pseudosubstrates”; in vertebrates, these include the transcription factor CCAAT/enhancer binding protein α (C/EBPα), the cell cycle–associated phosphatase CDC25C, and the enzyme acetyl coenzyme A carboxylase (19–21).
A long-standing goal in cancer research is drug-induced degradation of oncogenic proteins. Progress toward this objective has been transformed by the synthesis of proteolysis-targeting chimeras (PROTACs), which induce proteasome-dependent degradation of their targets. Multifunctional small-molecule PROTACS often have protein-binding regions derived from kinase inhibitors (22, 23), and multiple classes of non-PROTAC kinase inhibitors also induce kinase target degradation, although typically at higher (micromolar) concentrations than those required for enzymatic inhibition (7). Recent reports also disclose classes of covalent compounds that bind and disable cysteine (Cys)–containing small guanine nucleotide–binding proteins such as mutant human RAS, permitting covalent inactivation of this previously “undruggable” oncoprotein (24,25). Cys residues are widespread and highly conserved in kinases (26), and the conservation of Cys residues both inside and outside the catalytic domain provides kinome-wide opportunities for exploitation using chemical biology (27). In this context, covalent targeting methodologies involving compound-accessible Cys residues in kinases (8, 28–31) and pseudokinases (3) have attracted substantial attention for small-molecule design, due to the potential for gains in target specificity and durability of responses, combined with tractability in experimental systems.
Tribbles pseudokinases are implicated in many physiological signaling pathways, often in the context of protein stability, but also through regulation of key modules, such as the canonical AKT pathway (32). TRIB2 is also implicated in the etiology of human cancers, including leukemia, melanoma, and lung and liver cancers (33). In particular, TRIB2 is a potential drug target in subsets of acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL), which are in urgent need of targeted therapeutics to help treat untargeted or drug-resistant patient populations (34). TRIB2 protein abundance has also been linked to drug resistance mechanisms, where an ability to modulate the prosurvival AKT signaling module underlies a central regulatory role in cell proliferation, differentiation, metabolism, and apoptosis (32, 35–39).
Here, we found that the low-affinity ATP-binding site in TRIB2 (40) was druggable with small molecules previously described as ATP-competitive pan–epidermal growth factor receptor (EGFR) family kinase inhibitors. Biochemical analysis confirmed the existence of distinct compound-induced TRIB2 conformations, and a compound screen identified known EGFR family inhibitors that stabilize or destabilize TRIB2 in vitro. TRIB2-binding compounds included the clinically used breast cancer therapeutic lapatinib (marketed as Tykerb) (41) and the U.S. Food and Drug Administration–approved irreversible electrophilic covalent inhibitors afatinib (Giotrif) (42, 43) and neratinib (Nerlynx) (44, 45). In the case of the latter two destabilizing agents, binding led to uncoupling of the pseudokinase domain from its own C-terminal tail. Consistently, afatinib exposure led to rapid TRIB2 degradation in cells, driven by an interaction with the Cys-rich pseudokinase domain, which interfered with AKT signaling and decreased cell survival in a TRIB2-expressing leukemia model. The availability of target-validated compounds that act as rapid TRIB2 pseudokinase down-regulators through a direct effect on the pseudokinase represents a new way to evaluate TRIB2 physiology and cell signaling. It might also have a broader impact on the rapidly developing pseudoenzyme field (46), where the concept of pseudokinase destabilization or elimination by targeted kinase “inhibitors” (7) has a number of potentially useful applications.
RESULTS
Analysis of human TRIB2 using a thermal stability assay
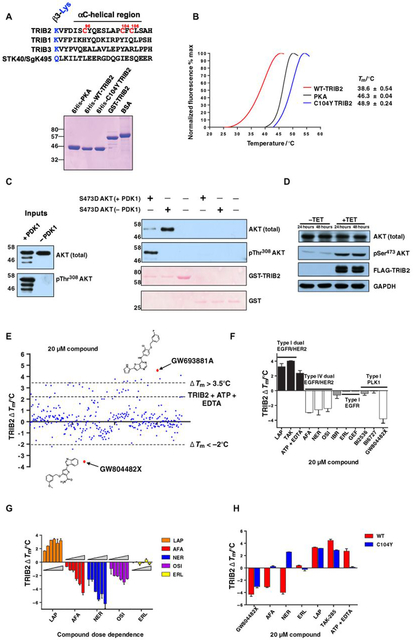
Human TRIB2 differs from TRIB1, TRIB3, and STK40 in the pseudokinase domain because of a Cys-rich region at the end of the β3-Lys–containing motif leading into the truncated αC-helix in the N-lobe (Fig. 1A, top). We developed a differential scanning fluorimetry (DSF) assay (47–49) to examine thermal stability of full-length (1 to 343) His-tagged TRIB2 proteins and compared it to either full-length adenosine 3′,5′-monophosphate (cAMP)–dependent protein kinase (PKAc) catalytic subunit, which is a model for canonical kinases, or full-length C104Y TRIB2, in which Cys104 was replaced with the Tyr residue conserved in human TRIB1 and TRIB3 (Fig. 1A). Proteins were purified to homogeneity (Fig. 1A, bottom), and thermal stability based on unfolding profiles was determined for each protein (reported as a Tm value, Fig. 1B). As previously demonstrated (40), TRIB2 (Tm = ~39°C) was much less thermostable than the canonical protein kinase (PKA, Tm = 46.3°C). Remarkably, the C104Y single substitution induced stabilization of TRIB2, with the Tm value increasing to ~49°C, comparable to that of human TRIB1 (15), suggesting an important structural role for this unique Cys residue in TRIB2 (un) folding dynamics. To confirm that recombinant TRIB2 binds to a known physiological target, we demonstrated that glutathione S-transferase (GST)–tagged TRIB2 interacted preferentially with catalytically inactive (non–Thr308-phosphorylated) AKT1 in vitro (Fig. 1C). Consistent with a functional regulatory interaction between TRIB2 and AKT in cells (50), transient overexpression of tetracycline (TET)-inducible FLAG-tagged TRIB2 in HeLa cells led to a large increase in endogenous AKT phosphorylation at the hydrophobic motif (Ser473, Fig. 1D), an established marker for AKT catalytic activity and generation of a downstream cellular antiapoptotic signal (51).
Fig. 1. Full-length TRIB2 is a target for protein kinase inhibitors in vitro.
(A) Top: Sequence alignment of human TRIB2, TRIB1, TRIB3, and STK40/SgK495, highlighting Cys-rich residues (numbered in red) in the TRIB2 pseudokinase domain. Bottom: Blot of the recombinant proteins used for in vitro analysis. The indicated purified proteins (5 μg each) were resolved by SDS–polyacrylamide gel electrophoresis (SDS-PAGE). WT, wild-type; BSA, bovine serum albumin. (B) Thermal denaturation profiles of recombinant proteins. A representative unfolding profile is shown. Tm values (±SD) were obtained from three separate fluorescence profiling experiments, each point assayed in duplicate. (C) The ability of GST-TRIB2 to interact with active [3-phosphoinositide-dependent protein kinase 1 (PDK1)–phosphorylated] or inactive (non–PDK1-phosphorylated) S473D AKT1 was assessed by glutathione-Sepharose pulldown followed by immunoblotting. Left: “Master-mix”input. (D) Transient transfection of TET-inducible FLAG-TRIB2 leads to increased AKT phosphorylation on Ser473. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (E) TRIB2 DSF screen using PKIS. His-TRIB2 (5 μM) was used for all DSF analysis. ΔTm values were calculated for each compound (N = 2). Scattergraph of data highlights a wide variety of compounds that either stabilize or destabilize TRIB2 in vitro. Cutoff values of >+3.5°C and <−2°C were used to designate hits. (F) Comparative DSF analysis of clinical and preclinical kinase inhibitors as potential TRIB2-binding compounds. LAP, lapatinib; TAK, TAK-285; AFA, afatinib; NER, neratinib; OSI, osimertinib; IBR, ibrutinib; ERL, erlotinib; GEF, gefitinib. (G) Dose-dependent analysis of thermal shifts induced by clinical TRIB2-binding compounds. Compounds were tested at 5, 10, 20, 40, 80, and 160 μM. (H) Profiling of TRIB2 and C104Y with selected inhibitors by DSF.
A DSF screen for TRIB2-binding compounds using a kinase inhibitor library
The ability of full-length recombinant human TRIB2 to bind to ATP in the presence of EDTA (40, 48) confirms that a vestigial nucleotide-binding site is present within the pseudokinase domain. Moreover, our previous work established that an analog-sensitive (F190G) TRIB2 variant could be stabilized by bulky pyrimidine analogs in vitro (40). To discover drug-like compounds for WT (full-length) TRIB2, we screened the Published Kinase Inhibitor Set (PKIS), a collection of high-quality class-annotated kinase inhibitors (52). We enforced cutoff values of ~ΔTm = <−2°C and >+3.5°C (therefore eliminating ~97% of the library) to define “hit” compounds that had the ability to destabilize or stabilize TRIB2 in a thermal stability assay at a 1:4 TRIB2/compound molar ratio (Fig. 1E and table S1). The top “stabilizing” compound identified was GW693881A, a dual EGFR and HER2 thienopyrimidine inhibitor with a ΔTm of +4.7°C. The top “destabilizing” compound was GW804482X, a thiophene polo-like kinase (PLK) inhibitor that induced a ΔTm of −3.4°C (Fig. 1E, red symbols). Most of the top stabilizing and destabilizing compounds belonged to well-known ATP-competitive pyrimidine or quinazoline EGFR family chemotypes (fig. S1) (53–55), suggesting broad structural cross-reactivity between TRIB2 and a compound-binding EGFR and HER2 conformation. To build upon these findings, we screened a larger panel of known dual EGFR and HER2 inhibitors (fig. S2) and established that the clinical type I EGFR family inhibitors TAK-285 and lapatinib also stabilized TRIB2 in vitro (Fig. 1F). The ATP-competitive covalent EGFR family inhibitors afatinib, neratinib, and osimertinib (but not the unrelated covalent Bruton’s tyrosine kinase inhibitor ibrutinib or the type I EGFR-specific inhibitors erlotinib and gefitinib) destabilized TRIB2, similar to the PLK inhibitor GW804482X (Fig. 1F) and the dual EGFR family inhibitor GW569530A (fig. S1). As expected (15), purified TRIB1 (fig. S3A) was more thermostable than TRIB2 in the absence of kinase inhibitors (fig. S3B). However, it was not destabilized by afatinib, neratinib, or osimertinib, although like TRIB2, destabilization was evident in the presence of GW804482X (fig. S3C), in agreement with independent findings (56). Compound effects were caused through pseudokinase targeting in the thermal shift assay, because no shift was elicited when the canonical kinase PKAc was compared in a side-by-side counterscreen, with dasatinib as a positive control (fig. S3D). The preclinical PLK inhibitors BI2536 and BI6727 (volasertib) had no discernible effects on TRIB2 stability in this assay, in contrast to the chemically distinct thiophene PLK inhibitor GW804482X (Fig. 1F).
Afatinib, neratinib, and osimertinib are covalent (type IV) inhibitors of EGFR family tyrosine kinases, interacting irreversibly with a conserved Cys residue in the canonical ATP-binding site (57). The stabilization of TRIB2 by lapatinib, and the destabilization of TRIB2 by all three covalent inhibitors, occurred in a dose-dependent manner (Fig. 1G and fig. S4). A C104Y TRIB2 mutant was no longer destabilized by either afatinib or neratinib but remained “sensitive” to lapatinib TAK-285 and GW804482X on the basis of thermal protection (Fig. 1H and fig. S5A). None of these latter compounds contain the electrophilic “warhead” required for covalent interactions (fig. S2). Elution profiles from Superdex 200 were identical for WT and C104Y TRIB2 (fig. S5B), confirming that both proteins were monomeric species in solution, with an estimated relative molecular mass (Mr) of 45.3 kDa. C104Y TRIB2 was also insensitive to thermal shift in the presence of ATP and EDTA (Fig. 1H and fig. S5C). These data suggest that the amino acid identity at TRIB2 position 104 is likely important for both ATP binding and interaction with covalent kinase inhibitors that induce TRIB2 destabilization in vitro.
Mechanistic analysis of TRIB2 structural stability and TRIB2 compound binding
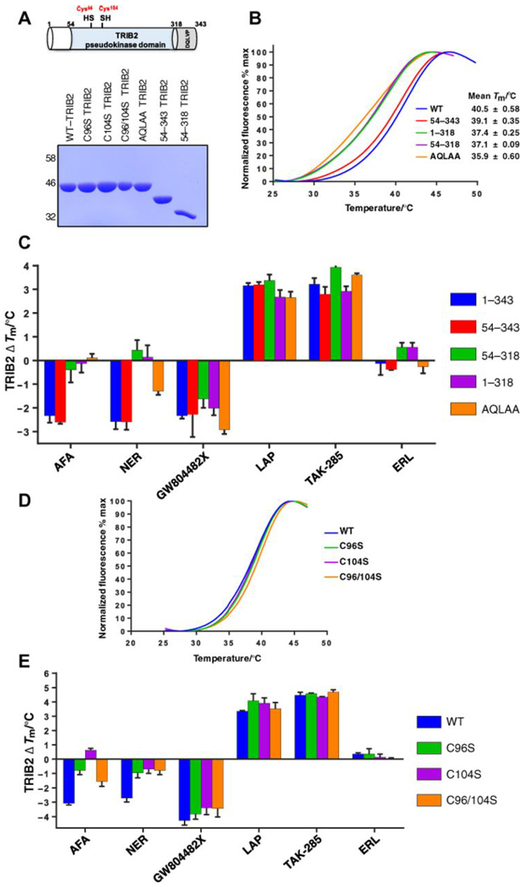
Structural analysis of TRIB1 by x-ray crystallography and small-angle x-ray scattering led to the proposal of an in cis self-assembly model, whereby the unique C-terminal tail region, which contains the conserved “DQLVP” motif, binds directly to the pseudokinase domain adjacent to the short αC-helix of TRIB1 (15,16, 56). To investigate whether this mechanism is also relevant in TRIB2, we generated a series of truncated proteins. These lacked either the N-terminal extension, which was predicted to be disordered by both I-TASSER (Iterative Threading ASSEmbly Refinement) (58) and VSL2 (59), the C-terminal tail, or both N- and C-terminal regions. We also generated a triple-point mutant in which the DQLVP tail motif, which is required for TRIB1 and TRIB2 cellular transformation in vivo (18), was mutated to a nonfunctional “AQLAA” sequence (Fig. 2A). Both full-length (1 to 343) TRIB2 and TRIB2 lacking the N-terminal domain (TRIB2 54 to 343) exhibited similar Tm values of 39° to 40°C (Fig. 2B). In contrast, deletion of the C-tail (TRIB2 1 to 318) changed TRIB2 stability, with Tm values falling to ~37°C, diagnostic of a destabilized TRIB2 conformation (Fig. 2B). Mutation of DQLVP to AQLAA further destabilized TRIB2, leading to a Tm value of ~36°C (Fig. 2B). Using this panel of recombinant TRIB2 proteins, we measured the relative effects of kinase inhibitors on TRIB2 stability. Consistent with a lack of effect on compound interactions, deletion of the TRIB2 N-terminal region had no effect on ΔTm values induced by any compound. However, removal of the C-tail region (54 to 318 and 1 to 318 mutants) abolished afatinib- and neratinib-induced TRIB2 destabilization but had a negligible effect on GW804482X binding (Fig. 2C). Consistently, destabilization by afatinib was also completely abolished in the AQLAA triple mutant, whereas neratinib effects were reduced by >50%. Notably, neither the destabilizing effect of GW804482X nor the stabilizing effects of lapatinib or TAK-285 differed between any of the TRIB2 proteins evaluated. These results suggest a very similar destabilizing mechanism induced by covalent EGFR family compounds via displacement of the TRIB2 C-tail, which is a unique feature of Tribbles pseudokinases (13, 15).
Fig. 2. TRIB2 thermal stability is modulated through Cys binding to covalent inhibitors.
(A) Top: Schematic cartoon of TRIB2 with domain boundaries numbered and cysteine residues highlighted (red). Bottom: SDS-PAGE of 5-μg recombinant TRIB2 proteins. (B) Thermal denaturation profiles of 5 μM WT-TRIB2 (amino acids 1 to 343), three truncated variants, and an AQLAA triple-point mutant. Representative curves for each protein and average Tm values (±SD) are shown, calculated from N = 3 experiments. (C) Thermal shift analysis of TRIB2 deletion and AQLAA proteins measured in the presence of a panel of compounds (20 μM). The change in Tm value (ΔTm) is reported from N = 3 experiments, each performed in triplicate. (D) Thermal denaturation profiles for purified TRIB2 and C96S, C104S, and C96/104S proteins. (E) Thermal shift analysis of TRIB2 Cys-mutated proteins measured in the presence of a panel of compounds (20 μM). The change in Tm value (ΔTm) is reported from N = 3 experiments.
To evaluate whether afatinib targeted unique Cys residues in the TRIB2 pseudokinase domain (Fig. 1A), we performed mass spectrometry (MS) analysis to evaluate protein modification. As detailed in fig. S6A, incubation of TRIB2 with a fivefold molar excess of afatinib led to covalent interaction with Cys96 in TRIB2. A doubly charged chymotryptic product ion representing the TRIB2-derived DISC96Y: afatinib peptide adduct at mass/charge ratio (m/z) 543.2 (fig. S6A) and the isotopic ratios of the 35Cl- or 37Cl-containing peptide ions unequivocally confirmed Cys96 as a site of TRIB2 binding (fig. S6B). Having confirmed an intact mass for recombinant full-length TRIB2 of 43,587.09 Da, very similar to the predicted mass of 43,587.22 Da (fig. S6C), we were also able to ascertain that preincubation with afatinib generated covalent adducts containing predominantly either one or two molecules of afatinib. There was also some evidence for tri- and tetra-modified TRIB2 adducts (fig. S6D). We next examined afatinib interaction with TRIB2 by MicroScale Thermophoresis (MST), a biophysical technique for quantification of reversible bio-molecular interactions (60). This revealed an interaction between fluorescent nitrilotriacetic acid-coupled His-TRIB2 and two compounds, which could be fitted to reversible binding with dissociation constant (Kd) values of ~16 μM for afatinib and ~20 μM for TAK-285 (fig. S7, A and B). A sub-micromolar interaction between myeloid cell leukemia–1 (MCL-1) and A1210477 served as a positive control (61). In agreement with MS data (fig. S6), we therefore propose initial (reversible) binding of ATP-competitive afatinib (62, 63), before subsequent formation of a covalent adduct (or adducts) with the TRIB2 pseudokinase domain. We also evaluated the potential interaction of afatinib and neratinib using TRIB2 modeled on the C/EBP-bound “SLE-in” conformation from TRIB1 (13, 56) and compared it to the known afatinib target EGFR (in a “DFG-in” conformation) using AutoDock Vina (64). Docking of covalent inhibitors revealed a putative binding pocket formed by residues from the vestigial TRIB2 C-helix, including Cys96, and the β3 strand (fig. S8A). Afatinib and neratinib dock in a structurally similar pose in which the enamine β carbon orients toward the sulfhydryl group of Cys96. This docking pose, however, is distinct from the afatinib-bound crystal structure of EGFR, wherein Cys797 is located in the D-helix. We performed molecular dynamics simulations (fig. S8B) on the docked complexes to assess the feasibility of the binding poses. Both afatinib and neratinib remained stably bound to TRIB2 for nearly 17 ns, suggesting that the binding poses are favorable and feasible (65). The enamine β carbon remained within 3 to 5 A of the Cys96 sulfur atom for the majority of the simulation. Given the rapid nature of the thiol-ene reaction coupled with the proximity of reactants, we believe that this time frame offers sufficient time for the formation of a covalent bond (66), which we confirmed by MS analysis.
To validate the biochemical importance of Cys residues for afatinib binding, we next examined interaction with TRIB2 in which two Cys amino acids were mutated to non–thiol-containing Ser residues. Individual or combined mutation of Cys96 and Cys104 to a Ser residue had no effect on the thermal stability (Tm) of the purified TRIB2 proteins (Fig. 2D), in contrast to the highly stabilizing effect of a Tyr at position 104 (Fig. 1B and fig. S5C). However, individual or joint mutation of Cys96 and Cys104 to Ser severely blunted the destabilizing effect of afatinib and neratinib on TRIB2, in contrast to the noncovalent TRIB2 compound GW804482X or the EGFR family stabilizing compounds lapatinib and TAK-285 (Fig. 2E). Together, our findings confirm that these covalent compounds elicit effects on TRIB2 through Cys96 and/or Cys104.
Evaluation of TRIB2 interactors in human cells
To evaluate TRIB2 targeting by compounds in living cells, we generated a polyclonal TRIB2 antibody and an isogenic stable HeLa cell line expressing TET-inducible FLAG-tagged human TRIB2. Polyclonal TRIB2 antibodies were equally efficient at recognizing recombinant His-FLAG-TRIB2 and cellular FLAG-TRIB2 as a commercial monoclonal FLAG antibody (fig. S9A). We also constructed a stable HeLa cell line expressing low levels of inducible FLAG-TRIB2 (fig. S9B). We estimated that ~1 ng of TRIB2 was expressed in 40 μg of whole-cell lysate in the presence of TET, by comparing known amounts of recombinant His-FLAG-TRIB2 protein (fig. S9B).
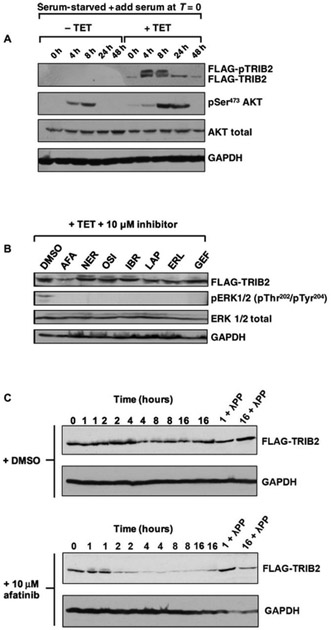
To quantify effects of TRIB2 expression on canonical signaling in model cells, we evaluated AKT phosphorylation. AKT became phosphorylated in stable HeLa cells after serum stimulation in the absence of TET (Fig. 3A). TRIB2 expression led to the appearance of a TRIB2 doublet (phosphorylation confirmed in the upper band by λ-phosphatase treatment, fig. S10) and a marked increase in the extent and duration of AKT phosphorylation at Ser473. Using this HeLa cell model, we next evaluated the effects of small-molecule inhibitors on TRIB2 signaling, by comparing a panel of in vitro TRIB2 destabilizing compounds discovered by DSF (afatinib, neratinib, and osimertinib) with a series of control compounds. Brief (4 hours) exposure of TRIB2-expressing cells to afatinib led to a specific decrease in TRIB2 protein expression. All of the compounds evaluated were specific EGFR or EGFR family signaling pathway inhibitors, and consistently they all blocked extracellular signal–regulated kinase (ERK) phosphorylation at these concentrations (Fig. 3B); any unique effects of drugs among this panel are therefore likely to be “off-target” to EGFR and HER2. We next incubated cells for increasing lengths of time with afatinib. A rapid, time-dependent, elimination of TRIB2 protein was evident when cells were exposed to this drug, in contrast to dimethyl sulfoxide (DMSO) controls in which TRIB2 protein remained relatively stable during the experiment (Fig. 3C), as expected (19). To evaluate intracellular interaction between TRIB2 and kinase inhibitors, we exposed HeLa cells expressing TET-inducible FLAG-TRIB2 to each compound and quantified TRIB2 thermal stability in the cell extracts using a cellular thermal stability assay (CETSA) (fig. S11) (67). Consistently, FLAG-TRIB2 was destabilized more rapidly than DMSO and erlotinib-treated controls in the presence of afatinib, becoming undetectable in extracts heated to 45°C (fig. S11). This was distinct from lapatinib, which partially stabilized TRIB2 in cell extracts, consistent with our in vitro analysis (Fig. 1F).
Fig. 3. Afatinib promotes rapid degradation of FLAG-TRIB2 in an inducible HeLa model.
(A) Uninduced (−TET) or TET-induced (+TET) HeLa cells containing a stably integrated FLAG-TRIB2 transgene were serum-starved for 16 hours before the addition of serum and lysed at the indicated times. Whole-cell extracts were blotted with FLAG antibody to detect FLAG-TRIB2 and pSer473 AKT. Total AKT and total GAPDH served as loading controls. (B) A selection of clinically approved kinase inhibitors, including dual EGFR/HER2- and EGFR-specific compounds, were added to TET-induced cells at a final concentration of 10 μM. Stable cells were induced to express FLAG-tagged TRIB2 with TET for 16 hours before inhibitor treatment for 4 hours. Whole-cell extracts were immunoblotted with FLAG, phosphorylated ERK (pERK 1/2), total ERK, or GAPDH antibodies. (C) Stable HeLa cells were incubated with TET for 16 hours and then incubated with DMSO (top panels) or 10 μM afatinib (bottom panels) before lysis at the indicated time. One-hour and 16-hour samples were pretreated with λ protein phosphatase (λPP) before SDS-PAGE. Whole-cell extracts were immunoblotted with FLAG or GAPDH antibodies. Blots are representative of three independent experiments.
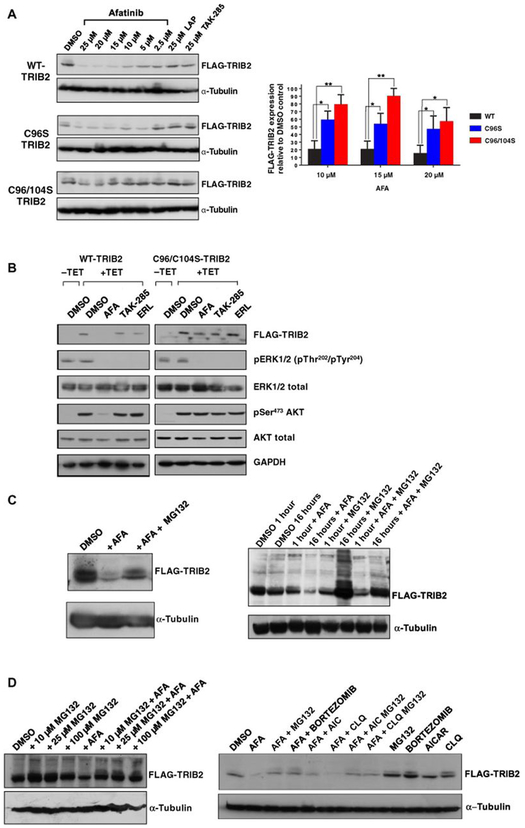
Afatinib interacts with TRIB2 through a biochemical Cys-based mechanism, so we generated isogenic TET-inducible C96S or C96/104S TRIB2-mutant stable cell lines (fig. S12) and evaluated the effects of afatinib on exogenous TRIB2 stability. Afatinib (but not lapatinib or TAK-285) induced dose-dependent loss of TRIB2 in WT-TRIB2 cells, which was partially prevented for C96S and completely abolished in C96/104S TRIB2-expressing cells (quantified in Fig. 4A, right panel), demonstrating unequivocally that afatinib binds to TRIB2 in cells. Furthermore, afatinib (but not TAK-285 or erlotinib)–treated WT-TRB2 cells exhibited a marked decrease in AKT Ser473 phosphorylation, and this effect was abrogated in C96/104S TRIB2-expressing cells (Fig. 4B). Afatinib, TAK-285, and erlotinib all blocked ERK phosphorylation in WT and C96/104S-TRIB2 isogenic cell lines, consistent with on-target inhibition of their shared EGFR target (Fig. 4B).
Fig. 4. “On-target” degradation of TRIB2 by afatinib: C96/104S TRIB2 double-point mutant is resistant to degradation.
(A) The indicated concentration of afatinib, lapatinib, or TAK-285 was incubated for 4 hours with isogenic stable HeLa cells expressing FLAG-tagged WT-TRIB2, C96S, or C96/104S TRIB2 (induced by TET exposure for 16 hours). After lysis, whole-cell extracts were immunoblotted with the indicated antibodies. Right: FLAG-TRIB2 abundance was quantified after exposure to 10, 15, and 20 μM afatinib relative to DMSO controls using ImageJ densitometry software. Data are means ± SD from N = 3 independent biological replicates. (B) WT and C96/104S stable HeLa cell lines were subjected to serum block-and-release protocol in the presence (+TET) or absence (−TET) of TET. Subsequently, the indicated compounds (10 μM) were added for 4 hours before cell lysis and immunoblotting with the indicated antibodies. (C) FLAG-tagged TRIB2-expressing HeLa cells were incubated with 0.1% (v/v) DMSO or afatinib (10 μM), in the presence or absence of MG132 (10 μM for 4 hours, left) or at the indicated time points (right) before lysis and processing for immunoblotting. (D) Left: FLAG-TRIB2–expressing stable cells were incubated with the indicated concentration of MG132 in the presence or absence of 10 μM afatinib for 4 hours before cell lysis and immunoblotting. Right: FLAG-TRIB2–expressing stable cells were incubated for 1 hour with MG132 (10 μM), bortezomib (BOR; 10 μM), AICAR (AIC; 1 mM), or chloroquine (CLQ; 50 μM) before the addition of afatinib (10 μM) for an additional 4 hours followed by lysis and immunoblotting with the indicated antibodies. Blots are representative of three independent experiments. *P ≤ 0.05 , **P ≤ 0.01.
To investigate the mechanism of TRIB2 destabilization by afatinib, we added the drug to HeLa cells expressing WT-TRIB2 in the presence of the proteasome inhibitor MG132, which partially rescued TRIB2 degradation after both rapid and prolonged exposure to afatinib (Fig. 4C). This finding is consistent with previously reported proteasome-dependent TRIB2 turnover (68). We further evaluated this mechanism using a range of MG132 concentrations and the clinical proteasome inhibitor bortezomib (Fig. 4D) (21); under both conditions, afatinib-mediated destabilization of TRIB2 was decreased. This effect was in contrast to afatinib-induced TRIB2 destabilization, which was still observed in the presence of nonspecific inhibitors of autophagy [5-aminoimidazole-4-carboxamide ribonucleotide (AICAR)] and lysosomal degradation (chloroquine) (Fig. 4D). Our observations are in line with published findings, which suggest that TRIB2 fate is dependent on turnover by the ubiquitin proteasome system in human cancer cells (19, 69). However, the (noncovalent) TRIB1- and TRIB2-destabilizing compound GW804482X did not induce TRIB2 degradation in cells after a 4-hour incubation period, in contrast to proteasome-dependent degradation induced by the same concentration of afatinib (fig. S13).
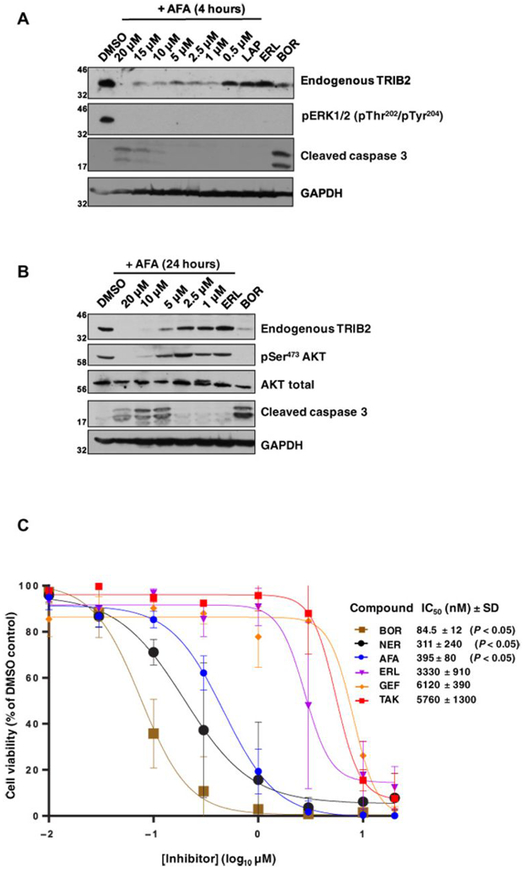
High expression of TRIB2 drives AML in vivo by inhibiting myeloid differentiation and promoting proliferation (18, 35). However, endogenous TRIB2 expression has been analyzed previously only in a few cell types, in large part because of a lack of reliable TRIB2 reagents. Using our TRIB2 antibody, we evaluated endogenous expression of TRIB2 in the clinically relevant U937 AML cell model (Fig. 5A) and established that acute (4 hours) exposure to afatinib, but not lapatinib or erlotinib, decreased TRIB2 protein abundance in a dose-dependent manner (Fig. 5A). Consistent with their ability to inhibit EGFR signaling, all three compounds completely blocked ERK phosphorylation. We next established dose-dependent effects on both TRIB2 expression and AKT Ser473 phosphorylation in afatinib-treated U937 cells (Fig. 5B). Notably, these effects were tightly correlated with apoptotic induction of caspase 3 cleavage, but only at afatinib concentrations that also induced TRIB2 degradation and concomitant loss of AKT Ser473 phosphorylation (Fig. 5B). To determine the impact of afatinib treatment on U937 cell viability, we quantified cellular cytotoxicity after 72 hours of exposure (Fig. 5C). Afatinib (and neratinib) reduced cell viability with sub-micromolar half-maximal inhibitory concentration (IC50) values, whereas EGFR inhibitors erlotinib and gefitinib and the dual EGFR family inhibitor TAK-285 were comparably 10- to 20-fold less effective (Fig. 5C). Because these compounds do not induce cellular TRIB2 destabilization, AKT activation, or caspase 3 activation, our data suggest that the TRIB2-destabilizing compounds afatinib and neratinib have an enhanced ability to kill AML-derived cells (50).
Fig. 5. Afatinib rapidly destabilizes endogenous TRIB2 and specifically induces caspase 3 cleavage and U937 cytotoxicity.
(A) Endogenous TRIB2 is destabilized in human U937 cells in a dose-dependent manner after a 4-hour exposure to afatinib. Cells were incubated with either 0.1% (v/v) DMSO or the indicated concentrations of afatinib, lapatinib (10 μM), erlotinib (10 μM), or the proteasome inhibitor bortezomib (10 μM) for 4 hours before lysis and immunoblotting of endogenous TRIB2, pERK, or cleaved caspase 3. GAPDH served as a loading control. (B) Endogenous TRIB2 is destabilized after exposure to afatinib for 24 hours, concomitant with reduced AKT phosphorylation at Ser473. After cell collection and lysis, whole-cell extracts were immunoblotted with the indicated antibodies. AKT and GAPDH served as loading controls; erlotinib and bortezomib were used at 10 μM. (C) The cytotoxicity of a panel of TRIB2-destabilizing small molecules (neratinib) was compared to EGFR inhibitors or the TRIB2 stabilizer TAK-285. MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assays were performed after 72 hours of compound exposure, with bortezomib used as a positive control. IC50 values (nM, mean ± SD) were derived from N = 3 independent experiments, each performed in triplicate. Statistical analysis confirmed a significant difference in cytotoxicity between afatinib and erlotinib (Student’s t test; P = 0.0104), between bortezomib and erlotinib (P = 0.0072), and between neratinib and erlotinib (P = 0.0104). Blots are representative of three independent experiments.
DISCUSSION
A drug-targetable regulatory mechanism for human TRIB2
Cell signaling can be controlled by nonenzymatic components, exemplified by mechanistically related families of pseudoenzymes such as pseudokinases (46). Here, we sought to target the cancer-associated pseudokinase TRIB2 with a chemical small molecule. To discover TRIB2-binding compounds, we exploited an unbiased TRIB2 thermal shift assay (40, 48) and identified multiple chemical classes of EGFR family member (typically EGFR and HER2, but never monovalent EGFR) kinase inhibitors. Several compounds induced stereotypical (positive) thermal shifts in TRIB2, consistent with binding in the atypical nucleotide-binding site (48). A second compound group led to marked TRIB2 (but not TRIB1) destabilization, and this could be explained through a Cys-directed covalent effect, in which the TRIB2 pseudokinase domain became uncoupled from its own C-tail by the compound. A similar mechanism has recently been proposed to explain thermally distinct conformers in TRIB1, which shares ~70% identity with TRIB2 in the pseudokinase domain and C-tail (16). Deletion of the TRIB1 C-tail also leads to destabilization of the TRIB1 pseudokinase domain (15), similar to the effects observed for TRIB2 described here. These data allow us to propose that TRIB2 pseudokinase-domain docking to the C-terminal tail is a dynamic interaction, which can be targeted with covalent ATP-competitive covalent inhibitors that induce a structural conformation associated with decreased stability in vitro.
How do covalent EGFR family inhibitors target TRIB2?
We focused our studies on electrophilic type IV kinase (covalent) inhibitors, because they are amenable to comparative chemical and mutational analyses in their targets (31, 70). We confirmed that at least two highly conserved TRIB2 Cys residues interact with afatinib (71) in vitro. These Cys residues are unique within the distorted TRIB2 αC-helix (Fig. 1A). In most canonical kinases, including EGFR family members, helix positioning is critical for catalysis and switching between inactive and active enzyme conformations, both of which are targets for small molecules (72). Dynamic helix positioning is also important for small-molecule interactions, including “inactive” conformations in kinases in which the αC-helix adapts upon binding, as described for HER2 (73). The conformation of the flexible C-helix relative to the ATP binding P-loop can also create an allosteric binding pocket in canonical kinases such as ERK1/2, which can be targeted with selective non-ATP-competitive inhibitors such as SCH772984 (74). In TRIB2, the unique disposition of Cys residues (Fig. 1A) makes it vulnerable to destabilization by the covalent inhibitors afatinib, neratinib, and osimertinib in vitro. These compounds also undergo a Michael addition to a distinct, but conserved, Cys residue in the ATP-binding site of EGFR-family tyrosine kinases (Cys797 in EGFR, Cys805 in HER2, and Cys803 in HER4) (61). Afatinib also demonstrates low, but detectable, affinity for the pseudokinase domain of HER3 (75, 76), which has a nonmodifiable Ser residue (Ser794) at the equivalent position as Cys797 of EGFR, and assumes a pseudo “active” catalytic conformation, despite having exceptionally low kinase activity (76, 77). At nanomolar concentrations, afatinib is relatively specific for EGFR and HER2 in cells, although at micromolar concentrations, other canonical kinases also appear to be targeted (9). Moreover, osimertinib has cross-reactivity with kinases (9) and nonkinases such as lysosomal cathepsins (78). In the absence of a high-resolution TRIB2 pseudokinase structure, we can only model the conformation(s) relevant for small-molecule interaction, although the N-lobe has shared (SLE motif) and distinct (Cys96 and Cys104) features compared to TRIB1, TRIB3, and STK40 (15), and TRIB1 adopts at least two conformations, one of which (SLE-in) appears vulnerable to small molecules (56). This is confirmed by biochemical differences in the ability of TRIB2 to bind weakly to ATP, which likely predisposes it to C-helix targeting by the kinase inhibitors identified here (fig. S8). None of the EGFR family members have a Cys residue in the C-helical region conserved in TRIB2, and the overall identity between HER2 and TRIB2 in the (pseudo)kinase domain is very low indeed (~22%). We believe that our discovery of TRIB2 binding to covalent compounds like afatinib owes as much to the availability of reactive Cys residues adjacent to the unique allosteric pocket in the pseudokinase domain as to their weak affinity for the vestigial ATP-binding site. Our work also builds on previous studies in which pseudokinase domains were targeted with compounds (3), such as TX1-85-1 (8), which covalently binds Cys721 in the roof of the ATP site (inducing HER3 degradation in cells) and noncovalent JAK2 JH2 (pseudokinase) domain ligands (79, 80).
In the course of compound screening, we discovered that unrelated classes of ATP-competitive kinase inhibitors, including thienopyrimidines (55) and thiazolylquinazolines (53), also bound to TRIB2 in vitro. In contrast to destabilization (a feature of covalent TRIB2-binding compounds), these compounds stabilize TRIB2, similar to lapatinib and the pyrrolo(3,2-d)pyrimidine EGFR-family inhibitor TAK-285, which are known to bind to HER2 in an active-like conformation (81). Of relevance, lapatinib and TAK-285 did not induce TRIB2 degradation in cells, although CETSA suggested an interaction with lapatinib (fig. S11). Previous studies found that lapatinib can also deprive HER2 of an interaction with the Hsp90-Cdc37 system, leading to time-dependent HER2 degradation at micromolar concentrations (82). Our data prove that the cellular mechanism by which TRIB2 stability is regulated by compounds is proteasome-based, and we speculate that an afatinib-induced conformational change might induce TRIB2 ubiquitination, or negatively regulate interaction with an unknown stabilizing factor (or factors), similar to effects of Hsp90 inhibitors on Cdc37 kinome clients (83–85). Consistently, TRIB2 abundance is regulated by ubiquitination (19, 68, 69), and future work will attempt to correlate TRIB2 small-molecule interactions with dynamic changes in ubiqutination status.
The future: Targeting TRIB2 with kinase inhibitors in cancer cells and beyond
The finding that TRIB2 stabilizing and destabilizing small-molecule compounds can be discovered by DSF profiling is an important advance for the analysis of compounds that target catalytically deficient pseudokinases and confirms that TRIB2 represents a bona fide pseudokinase drug target (5). We found that conserved Cys residues in TRIB2 make it vulnerable to families of small-molecule kinase inhibitors originally developed as dual nanomolar covalent EGFR and HER2 inhibitors. These residues also represent an additional form of selectivity filter, permitting targeting of TRIB2 by these compounds at micromolar concentrations in cells. On the basis of this mechanism, a lack of equivalent Cys residues in other pseudokinases, including TRIB1, TRIB3, and STK40, likely precludes in teraction with covalent compounds such as afatinib through this mechanism.
Using a chemical genetic approach, we provide evidence that afatinib induces on-target effects through TRIB2 stability and AKT signaling in a variety of human cancer cells. Experimental on-target effects of covalent TRIB2-destabilizing agents were confirmed using chemical genetics and a mutant TRIB2 allele, similar to “drug resistance” approaches developed for compound target validation in the kinase inhibitor field (86–89). Drug effects were prevented by mutation of two TRIB2 Cys C-helix residues, confirming a meaningful cellular interaction. Afatinib binding was also validated by DSF, MS, and MST and by using in-cell thermal stability assay (CETSA) approaches with exogenous TRIB2. Finally, we established that afatinib (and neratinib) exhibit sub-micromolar toxicity in the human AML model cell line U937, where they are 10- to 20-fold more effective at cell killing compared to equipotent EGFR-family or EGFR-specific inhibitors (72), which do not degrade TRIB2 but still block ERK activation. U937 cells are known to be hypersensitive to TRIB2 small interfering RNA knockdown and to require TRIB2 for cell survival (68).
Our work demonstrates that covalent inhibitors such as afatinib have TRIB2-degrading activity in human cells at micromolar concentrations, in similar ranges to those reported for other destabilizing kinase inhibitors, such as the related TRIB2-destabilizing quinazoline neratinib (90). Although we cannot rule out simultaneous dual effects of afatinib on TRIB2 and ERK/AKT pathways contributing to cellular phenotypes, we provide direct evidence that TRIB2 binding is required for TRIB2 destabilization and AKT regulation. Should an appropriate concentration of drug permit a direct TRIB2 interaction, an off-target TRIB2-dependent phenotype might be relevant to compound efficacy (or side effects) in patient groups exposed to high concentrations of these covalent compounds. Many severe side effects of drugs are only detected after long-term clinical use, potentially leading to their withdrawal (91), and covalent drugs have the potential to accumulate to relatively high concentrations in cells. Side effects of lapatinib (92) and afatinib (93) are already well established in patients. On the basis of our mechanistic studies, it will be interesting to develop enzyme-linked immunosorbent assay–based procedures to quantify effects of these drugs on TRIB2 protein stability in clinical samples obtained from patients, as part of broader proteomics approaches to establish all the intracellular targets of such compounds. Our study also provides impetus for generating improved (ideally TRIB2-specific) covalent compounds that induce TRIB2 degradation at lower (nanomolar) concentrations, ideally by synthesizing compounds in which the effects of eliminating or preserving tyrosine kinase inhibition can be compared side by side. In the latter case, simultaneous elimination of TRIB2 and inhibition of the ERK signaling pathway could be a polypharmacological asset, especially if TRIB2-dependent drug resistance in tumor cells (39, 50) can be modulated by tyrosine kinase inhibition. Together, our data establish a new paradigm for the pharmacological evaluation of agents that interfere with TRIB2-based signaling and raise the intriguing possibility that clinical inhibitors might be used as TRIB2-degrading agents in research, and possibly clinical, contexts. This information can also be exploited in the future for targeting a variety of TRIB2-overexpressing solid (94, 95) and hematological (21, 68) tumors.
MATERIALS AND METHODS
Chemicals, reagents, antibodies, and TRIB2 small-molecule screen
TET, doxycycline, MG132, AICAR, and chloroquine were purchased from Sigma-Aldrich. Afatinib, neratinib, osimertinib, ibrutinib, erlotinib, lapatinib, TAK-285, BI2536, BI6727, gefitinib, and bortezomib were purchased from LC Laboratories or Selleck. Total AKT, pSer473 AKT, pThr308 AKT, total ERK1/2, dual pThr202/pTyr204 ERK1/2, cleaved caspase 3, and α-tubulin antibodies were purchased from New England BioLabs and used as previously described (51, 96). 6His–horseradish peroxidase (HRP) and α-FLAG antibodies were purchased from Sigma-Aldrich GAPDH antibody was purchased from Proteintech, and a polyclonal rabbit α-TRIB2 antibody was raised toward a unique N-terminal human TRIB2 sequence and affinity-purified before evaluation with recombinant TRIB2 and a variety of human cellular extracts.
The PKIS chemical library (designated as SB, GSK, or GW compounds) comprises 367 ATP-competitive kinase inhibitors, covering ~30 chemotypes (~70% with molecular mass <500 Da and clogP values <5) that were originally designated as ATP-competitive inhibitors of 24 distinct protein kinase targets, including multiple EGFR and HER2 tyrosine kinase classes (52). Compounds were stored frozen as 10 mM stocks in DMSO. For initial screening, compounds were preincubated with TRIB2 for 10 min and then used for DSF, which was initiated by the addition of fluorescent SYPRO Orange. For dose-dependent thermal shift assays, a compound range was prepared by serial dilution in DMSO and added directly into the assay to the appropriate final concentration, as previously described (97). All control experiments contained 2% (v/v) DMSO, which had essentially no effect on TRIB2 stability.
Cloning, site-directed mutagenesis, and recombinant protein production
pET30 6His-TRIB2 (and various deletion or amino acid substitution constructs, including an N-terminal FLAG-tagged TRIB2, termed His-FLAG-TRIB2) and pET30a 6His-PKAc, which encodes catalytically active cAMP-dependent protein kinase domain, have been described previously (40, 49). Full-length TRIB2 was also cloned into pOPINJ to generate a His-GST-TRIB2 encoding construct for GST pulldown assays. MCL-1 and A-1210477 were prepared as previously described (61). TRIB1 (84 to 372) was a gift from P. Mace and was purified as described previously for TRIB2 (40). Briefly, protein expression was induced in BL21(DE3) pLysS bacteria with 0.4 mM isopropyl-β-d-thiogalactopyranoside, and after overnight culture at 18°C, proteins were purified to near homogeneity using an initial affinity step (immobilized metal affinity chromatography or glutathione-Sepharose chromatography) followed by size exclusion chromatography (16/600 Superdex 200) in appropriate buffers. S473D 6His-AKT1 (DU1850, amino acids 118 to 470), either active (PDK1-phosphorylated to a specific activity of 489 U/mg) or inactive, were purchased from the Division of Signal Transduction Therapy (University of Dundee) and stored at −80°C before analysis. Site-directed mutagenesis was performed as previously described (98), using KOD Hot Start DNA Polymerase (Millipore) and appropriate mutagenic primer pairs (sourced from IDT). All plasmids were Sanger-sequenced across the entire coding regions to confirm expected codon usage.
Differential scanning fluorimetry
Thermal shift assays were performed using an Applied Biosystems StepOnePlus Real-Time PCR instrument using a standard DSF procedure previously developed and validated for the analysis of kinases (47, 49) and pseudokinases (48). All proteins were diluted in 20 mM tris-HCl (pH 7.4), 100 mM NaCl, and 1 mM DTT to a concentration of 5 μM and then incubated with the indicated concentration of compound in a total reaction volume of 25 μl, with a final concentration of 2% (v/v) DMSO. SYPRO Orange (Invitrogen) was used as a fluorescence probe. The temperature was raised in regular 0.3°C intervals from 25° to 95°C. Compound binding experiments were assessed in duplicate and then reported relative to DMSO controls.
MS analysis of TRIB2 afatinib binding
To evaluate TRIB2 binding in vitro, afatinib was incubated for 15 min with purified 6His-TRIB2 at a 1:10 molar ratio, then denatured with 0.05% (w/v) RapiGest SF (Waters Corporation), and digested with chymotrypsin [1:20 protease/protein (w/w) ratio] for 16 hours at 25°C. RapiGest hydrolysis was induced by the addition of triflouroacetic acid (TFA) to 1% (v/v), incubated at 37°C for 1 hour. Insoluble product was removed by centrifugation (13,000g, 20 min). Reversed-phase high-performance liquid chromatography separation was performed using an UltiMate 3000 nano system (Dionex) coupled in-line with a Thermo Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific). Digested peptides (500 fmol) were loaded onto the trapping column (PepMap100 C18, 300 μm × 5 mm), using partial loop injection, for 7 min at a flow rate of 9 μl/min with 2% (v/v) MeCN and 0.1% (v/v) TFA and then resolved on an analytical column (Easy-Spray C18, 75 μm × 500 mm, 2-μm bead diameter column) using a gradient of 96.2% A [0.1% (v/v) formic acid (FA)]/3.8% B [80% (v/v) MeCN, 0.1% (v/v) FA] to 50% B over 35 min at a flow rate of 300 nl/min. MS1 spectra were acquired over m/z 400 to 1500 in the orbitrap (60 K resolution at m/z 200). Data-dependent MS2 analysis was performed using a top-speed approach (cycle time of 3 s), using higher-energy collisional dissociation and electron-transfer and higher-energy collision dissociation for fragmentation, with product ions being detected in the ion trap (rapid mode). Data were processed using Thermo Proteome Discoverer (v. 1.4), and spectra were searched in MASCOT against the Escherichia coli International Protein Index database with the added sequence of full-length 6His-TRIB2 (1 to 343). Parameters were set as follows: MS1 tolerance of 10 ppm, MS/MS mass tolerance of 0.6 Da, with oxidation of methionine and afatinib binding at cysteine as variable modifications. MS2 spectra were interrogated manually.
To evaluate the interaction between afatinib and intact TRIB2 protein, TRIB2 was incubated with afatinib as above and then desalted using a C4 desalting trap (Waters MassPREP Micro desalting column, 2.1 mm × 5 mm, 20 μm particle size, 1000 Å pore size). TRIB2 was eluted with 50% (v/v) MeCN and 0.1% (v/v) FA. Intact TRIB2 mass analysis was performed using a Waters nanoACQUITY UPLC (ultra performance liquid chromatography) system coupled to a Waters SYNAPT G2. Samples were eluted from a C4 trap column at a flow rate of 10 μl/min using three repeated 0 to 100% acetonitrile gradients. Data were collected between m/z 400 and 3500 and processed using MaxEnt1 (maximum entropy software, Waters Corporation).
TRIB2 modeling and compound docking
The structure model of human TRIB2 pseudokinase domain (13) (UniProt ID: Q92519) (residues 58 to 308) was built with MODELLER (99) using a crystal structure of TRIB1 pseudokinase in the open SLE-in conformation (56) as a template. The homology model was subjected to energy minimization using the Rosetta Relax protocol (100). The chemical structures of afatinib (CID 10184653) and neratinib (CID 9915743) were retrieved from PubChem. Afatinib and neratinib were docked to our TRIB2 model using AutoDock Vina (62) with an exhaustiveness of 100, rigid body, and a search space centered on Cys96. Top poses were identified where the enamide group localizes near the Cys96 sulfhydryl moiety. For comparison, we used a crystal structure of EGFR in complex with afatinib (Protein Data Bank: 4g5j). Missing disordered loop regions in the structure were modeled using RosettaRemodel (101) with default settings. The covalently bound afatinib was removed from the structure and allowed to redock with AutoDock Vina using the same parameters from TRIB2, with the exception of search space being centered on EGFR Cys797. Top poses were identified where the enamide group localized near the Cys797 sulfhydryl moiety. For molecular dynamics simulations, TRIB2 and EGFR binding modes were parameterized with GROMACS 5.1.3. Afatinib and neratinib were parameterized with GAFF (General AMBER Force Field) (102) using ACPYPE (AnteChamber PYthon Parser interfacE) (103). Protein was fixed with the AMBER99SB-ILDN force field and solvated with the TIP3P water model in a dodecahedron box at least 1 nm larger than the protein in all directions. Sodium and chloride ions were added to neutralize the system. Neighbor lists for nonbonded interactions were maintained by the Verlet cutoff scheme, and long-range electrostatics were calculated by the particle mesh Ewald method. Energy minimization was performed with steepest descent followed by conjugate gradient until the total energy was under 100 kJ/mol per nanometer. The system was heated from 0 to 310 K and then pressurized to 1 bar. Position restraint was applied to nonhydrogen atoms during equilibration. The production run was performed in 2-fs time steps. Data visualization was performed in VMD and PyMOL.
MicroScale Thermophoresis
A Monolith NT.115 instrument (NanoTemper Technologies GmbH) was used for MST analysis. His-TRIB2 was initially labeled with a NanoTemper labeling kit; the fluorescent red dye NT-647 was coupled via NHS chemistry to the N-terminal His-tag, placing the fluorophore away from the pseudokinase domain. For MST, the reaction was performed in 20 mM bicine (pH 9.0), 100 mM NaCl, 5% glycerol, 0.05% Tween 20, and 2% (v/v) DMSO. Fluorescent TRIB2 (~5 μM) was kept constant in the assay, whereas final afatinib and TAK-285 concentrations were titrated over a 3-nM and 50- to 100-μM range. Near-saturation binding was achieved, allowing for an affinity to be estimated for the reversible interaction between afatinib, TAK-285, and fluorescent TRIB2. NT-647–linked MCL-1 and A-1210477 were used as a positive control.
Cell lines and reagents
Flp-In T-REx parental HeLa cells (Invitrogen) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 4 mM l-glutamine, 10% (v/v) fetal bovine serum (FBS), penicillin, and streptomycin (Gibco) as previously described (86). To engineer TET-controlled expression of FLAG-tagged full-length TRIB2 in human Flp-In T-REx cell lines, the host plasmid pcDNA5/FRT/TO, encoding full-length TRIB2 sequences [or appropriate amino acid substitution(s)], with a single N-terminal FLAG tag (1 μg of DNA per well in a six-well plate of cells) was cotransfected with 9 μg of pOG44 Flp-recombinase expression vector using Lipofect-amine. Cells that had successfully integrated the FLAG-tagged TRIB2 sequence were stably selected with hygromycin B (200 μg/ml), according to the manufacturer’s instructions. TET was added to medium at a final concentration of 1 μg/ml to induce FLAG-TRIB2 expression. For transient transfection, 50% confluent HeLa cells were transfected with 40 μg of DNA per 10-cm dish for 48 hours before lysis.
For serum starvation, stable HeLa cells were grown until ~60% confluent in complete medium (+FBS), washed with phosphate-buffered saline (PBS), and replaced with serum-free DMEM for 16 hours. Cells were then incubated with DMEM containing 10% (v/v) FBS ± TET (1 μg/ml) for 16 hours, followed by the addition of an appropriate inhibitor for 4 hours. All whole-cell lysates were generated with modified radioimmunoprecipitation assay (RIPA) buffer (see below). Nonadherent AML-derived human U937 cells (which express high levels of endogenous TRIB2 protein) were supplied by K. Keeshan (University of Glasgow) and were cultured as previously described (21).
MTT cytotoxicity assay
U937 cells were seeded in a 96-well plate at a concentration of 0.2 × 106 cells/ml, 18 hours before compound addition, which was performed in triplicate, with all experiments including a final concentration of 0.1% DMSO (v/v). To quantify U937 cell viability, metabolic activity was assessed 72 hours after compound exposure using an MTT assay (Abcam), as described previously (104). Briefly, thiazolyl blue tetrazolium bromide was dissolved in PBS and added to cells at a final concentration of 0.25 mg/ml and incubated at 37°C for 3 hours. The reaction was stopped by the addition of 50 μl of acidified 10% SDS, followed by reading of absorbance at 570 nm. Viability was defined relative to DMSO-containing controls incubated for the same period of time.
Immunoblotting and CETSA
HeLa and U937 whole-cell lysates were generated using a modified RIPA buffer [50 mM tris-HCl (pH 7.4), 1% (v/v) NP-40, 1% (v/v) SDS, 100 mM NaCl, and 100 nM okadaic acid] supplemented with protease and phosphatase inhibitors (Roche) and briefly sonicated. For Western blotting, samples were boiled for 5 min in sample buffer [50 mM tris (pH 6.8), 1% SDS, 10% glycerol, 0.01% bromophenol blue, and 10 mM DTT]. Subsequently, between 40 and 120 μg of total protein was resolved by SDS-PAGE followed by transfer onto a nitrocellulose membrane (Bio-Rad). After blocking in tris-buffered saline + 0.1% (v/v) Tween 20 (TBS-T) + 5% milk (pH 7.4), primary and secondary antibodies were incubated under the same condition, and proteins were detected using HRP-conjugated antibodies and enhanced chemiluminesence reagent. Immunoblots were quantified using ImageJ software, as previously described (96). For λ-phosphatase treatment, 40 μg of FLAG-TRIB2–expressing stable cell extracts in lysis buffer without SDS and phosphatase inhibitors was incubated with 10 ng of purified λ-phosphatase for 30 min at 37°C before processing for Western blotting.
For in-cell CETSA, we used a previously published procedure (105). Briefly, stable HeLa cells were incubated with TET for 16 hours to induce expression of FLAG-TRIB2 and, at ~90% confluency, were incubated with 0.1% (v/v) DMSO or 100 μM of the indicated compound for 1 hour. Intact cells were isolated by trypsinization (1 min), resuspended in PBS, and then aliquoted into individual polymerase chain reaction (PCR) tubes before heating at the indicated temperature in a PCR thermal cycler for 3 min. Cells were then placed on ice for 2 min and lysed by sonication, before centrifugation at 16,000g for 20 min at 4°C. The soluble lysate was analyzed for the presence of FLAG-TRIB2 and α-tubulin by immunoblotting.
Statistical analysis
All experimental procedures were repeated in at least three separate experiments with matched positive and negative controls (unless stated otherwise). Results are expressed as means ± SD for all in vitro experiments, and data are expressed as means ± SD. When applied, statistical significance of differences (*P ≤ 0.05) was assessed using a Student’s t test for normally distributed data. All statistical tests were performed using Prism 7 (GraphPad Software).
Supplementary Material
Fig. S1. Discovery of multiple PKIS compounds as TRIB2-binding compounds.
Fig. S2. Chemical structure of preclinical and clinical compounds evaluated in this study.
Fig. S3. Validation of DSF assay using TRIB1 and a PKAc counterscreen.
Fig. S4. Thermal melting profiles of TRIB2 using DSF.
Fig. S5.Analysis of C104Y mutation in TRIB2.
Fig. S6. MS-based analysis of the covalent TRIB2:afatinib complex.
Fig. S7. MST assay.
Fig. S8. Molecular docking analysis.
Fig. S9. A new TRIB2 antibody for quantitative analysis of TRIB2 expression levels and stability.
Fig. S10. Analysis of TRIB2 dephosphorylation in cell extracts.
Fig. S11. TRIB2 binding to afatinib induces destabilization relative to DMSO in a whole-cell thermal shift assay (CETSA).
Fig. S12. Comparative protein expression analysis of stable HeLa FLAG-TRIB2 cell lines.
Fig. S13. Lack of effect of the noncovalent TRIB1- and TRIB2-destabilizing compound GW804482X on TRIB2 stability in TRIB2-expressing HeLa cells.
Table S1. PKIS compound screening data for full-length TRIB2.
Acknowledgments:
We acknowledge P. Mace (University of Otago, New Zealand) for the gift of TRIB1 (84 to 372) plasmid. We also thank S. Evans for excellent technical support, including media preparation.
Funding: This work was funded by two UK Biotechnology and Biological Sciences Research Council Doctoral Training Partnership studentships (to D.M.F. and S.F.), a Tools and Resources Development Fund award (BB/N021703/1, to P.A.E.), Royal Society Research Grants (to P.A.E. and C.E.E.), and North West Cancer Research grants (CR1088 and CR1097, to P.A.E.). Funding for N.K. from the NIH (5RO1GM114409) is also acknowledged. The Structural Genomics Consortium (SGC) is a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, the Eshelman Institute for Innovation, Genome Canada, the Innovative Medicines Initiative (European Union/European Federation of Pharmaceutical Industries and Associations) [ULTRA-DD grant no. 115766], Janssen, Merck KGaA (Darmstadt, Germany), MSD, Novartis Pharma AG, the Ontario Ministry of Economic Development and Innovation, Pfizer, the Sao Paulo Research Foundation (FAPESP), Takeda, and The Wellcome Trust [106169/ZZ14/Z].
Footnotes
Competing interests: There are no perceived conflicts of interest from any authors. The SGC receives direct funds from various pharmaceutical companies (see above), although it remains entirely independent.
Data and materials availability: Original TRIB2 PKIS screening data are presented in table S1. Further compound information can be obtained by contacting D.H.D. or W.J.Z. All data needed to evaluate the conclusions made are available in the main section or the supplementary section of the paper.
SUPPLEMENTARY MATERIALS
www.sciencesignaling.org/cgi/content/full/11/549/eaat7951/DC1
REFERENCES AND NOTES
- 1.Eyers PA, Murphy JM, Dawn of the dead: Protein pseudokinases signal new adventures in cell biology. Biochem. Soc. Trans 41,969–74 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Kung JE, Jura N, Structural basis for the non-catalytic functions of protein kinases. Structure 24, 7–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrne DP, Foulkes DM, Eyers PA, Pseudokinases: Update on their functions and evaluation as new drug targets. Future Med. Chem 9, 245–265 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Reiterer V, Eyers PA, Farhan H, Day of the dead: Pseudokinases and pseudophosphatases in physiology and disease. Trends Cell Biol. 24, 489–505 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Foulkes DM, Byrne DP, Bailey FP, Eyers PA, Tribbles pseudokinases: Novel targets for chemical biology and drug discovery? Biochem. Soc. Trans 43, 1095–103 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Hari SB, Merritt EA, Maly DJ, Conformation-selective ATP-competitive inhibitors control regulatory interactions and noncatalytic functions of mitogen-activated protein kinases. Chem. Biol 21 , 628–635 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones LH, Small-molecule kinase downregulators. Cell Chem. Biol 25, 30–35 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Xie T, Min Lim S, Westover KD, Dodge ME, Ercan D, Ficarro SB, Udayakumar D, Gurbani D, Seop Tae H, Riddle SM, Sim T, Marto JA, Jänne PA, Crews CM, Gray NS, Pharmacological targeting of the pseudokinase Her3. Nat. Chem. Biol 10, 1006–1012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klaeger S, Heinzlmeir S, Wilhelm M, Polzer H, Vick B, Koenig P-A, Reinecke M, Ruprecht B, Petzoldt S, Meng C, Zecha J, Reiter K, Qiao H, Helm D, Koch H, Schoof M, Canevari G, Casale E, Re Depaolini S, Feuchtinger A, Wu Z, Schmidt T, Rueckert L, Becker W, Huenges J, Garz A-K, Gohlke B-O, Paul Zolg D, Kayser G, Vooder T, Preissner R, Hahne H, Tõnisson N, Kramer K, Götze K, Bassermann F, Schlegl J, Ehrlich H-C, Aiche S, Walch A, Greif PA, Schneider S, Rudolf Felder E, Ruland J, Médard G, Jeremias I, Spiekermann K, Kuster B, The target landscape of clinical kinase drugs. Science 358, eaan4368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Yang PL, Gray NS, Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer 9, 28–39 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferguson FM, Gray NS, Kinase inhibitors: The road ahead. Nat. Rev. Drug Discov 17, 353–377 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Bailey FP, Byrne DP, McSkimming D, Kannan N, Eyers PA, Going for broke: Targeting the human cancer pseudokinome. Biochem. J 465, 195–211 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Eyers PA, Keeshan K, Kannan N, Tribbles in the 21st century: The evolving roles of tribbles pseudokinases in biology and disease. Trends Cell Biol. 27, 284–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otsuki L, Brand AH, Cell cycle heterogeneity directs the timing of neural stem cell activation from quiescence. Science 360, 99–102 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy JM, Nakatani Y, Jamieson SA, Dai W, Lucet IS, Mace PD, Molecular mechanism of CCAAT-enhancer binding protein recruitment by the TRIB1 pseudokinase. Structure 23, 2111–2121 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Eyers PA, TRIBBLES: A twist in the pseudokinase tail. Structure 23, 1974–1976 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Durzynska I, Xu X, Adelmant G, Ficarro SB, Marto JA, Sliz P, Uljon S, Blacklow SC, STK40 is a pseudokinase that binds the E3 ubiquitin ligase COP1. Structure 25, 287–294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keeshan K, Bailis W, Dedhia PH, Vega ME, Shestova O, Xu L, Toscano K, Uljon SN, Blacklow SC, Pear WS, Transformation by Tribbles homolog 2 (Trib2) requires both the Trib2 kinase domain and COP1 binding. Blood 116, 4948–4957 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang KL, Paredes R, Carmody R, Eyers PA, Meyer S, McCarthy TV, Keeshan K, Human TRIB2 oscillates during the cell cycle and promotes ubiquitination and degradation of CDC25C. Int. J. Mol. Sci 17, E1378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salomè M, Campos J, Keeshan K, TRIB2 and the ubiquitin proteasome system in cancer. Biochem. Soc. Trans 43, 1089–1094 (2015). [DOI] [PubMed] [Google Scholar]
- 21.O’Connor C, Lohan F, Campos J, Ohlsson E, Salomè M, Forde C, Artschwager R, Liskamp RM, Cahill MR, Kiely PA, Porse B, Keeshan K, The presence of C/EBPα and its degradation are both required for TRIB2-mediated leukaemia. Oncogene 35, 5272–5281 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Cromm PM, Crews CM, Targeted protein degradation: From chemical biology to drug discovery. Cell Chem. Biol 24, 1181–1190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai AC, Crews CM, Induced protein degradation: An emerging drug discovery paradigm. Nat. Rev. Drug Discov 16, 101–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM, K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim SM, Westover KD, Ficarro SB, Harrison RA, Geun Choi H, Pacold ME, Carrasco M, Hunter J, Doo Kim N, Xie T, Sim T, Jänne PA, Meyerson M, Marto JA, Engen JR, Gray NS, Therapeutic targeting of oncogenic K-Ras by a covalent catalytic site inhibitor. Angew. Chem. Int. Ed. Engl 53, 199–204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corcoran A, Cotter TG, Redox regulation of protein kinases. FEBS J. 280, 1944–1965 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Chaikuad A, Koch P, Laufer SA, Knapp S, The cysteinome of protein kinases as a target in drug development. Angew. Chem 57, 4372–4385 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Liu Q, Sabnis Y, Zhao Z, Zhang T, Buhrlage SJ, Jones LH, Gray NS, Developing irreversible inhibitors of the protein kinase cysteinome. Chem. Biol 20, 146–159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Z, Liu Q, Bliven S, Xie L, Bourne PE, Determining cysteines available for covalent inhibition across the human kinome. J. Med. Chem 60, 2879–2889 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatcher JM, Wu G, Zeng C, Zhu J, Meng F, Patel S, Wang W, Ficarro SB, Leggett AL, Powell BE, Marto JA, Zhang K, Chi Ki Ngo J, Fu X-D, Zhang T, Gray NS, SRPKIN-1: A covalent SRPK1/2 Inhibitor that potently converts VEGF from pro-angiogenic to anti-angiogenic ISOFORM. Cell Chem. Biol 25, 460–470 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang T, Kwiatkowski N, Olson CM, Dixon-Clarke SE, Abraham BJ, Greifenberg AK, Ficarro SB, Elkins JM, Liang Y, Hannett NM, Manz T, Hao M, Bartkowiak B, Greenleaf AL, Marto JA, Geyer M, Bullock AN, Young RA, Gray NS, Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors. Nat. Chem. Biol 12, 876–884 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du K, Herzig S, Kulkarni RN, Montminy M, TRB3: A tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science 300, 1574–1577 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Zanella F, Renner O, García B, Callejas S, Dopazo A, Peregrina S, Carnero A, Link W, Human TRIB2 is a repressor of FOXO that contributes to the malignant phenotype of melanoma cells. Oncogene 29, 2973–2982 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Liang KL, Rishi L, Keeshan K, Tribbles in acute leukemia. Blood 121, 4265–4270 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Keeshan K, Rishi L, Keeshan K, Tribbles homolog 2 inactivates C/EBPalpha and causes acute myelogenous leukemia. Cancer Cell 10, 401–411 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salazar M, Lorente M, García-Taboada E, Pérez Gómez E, Dávila D, Zúñiga-García P, Flores JM, Rodríguez A, Hegedus Z, Mosén-Ansorena D, Aransay AM, Hernández-Tiedra S, López-Valero I, Quintanilla M, Sánchez C, Iovanna JL, Dusetti N, Guzmán M, Francis SE, Carracedo A, Kiss-Toth E, Velasco G, TRIB3 suppresses tumorigenesis by controlling mTORC2/AKT/FOXO signaling. Mol. Cell. Oncol 2, e980134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salazar M, Lorente M, García-Taboada E, Pérez Gómez E, Dávila D, Zúñiga-García P, María Flores J, Rodríguez A, Hegedus Z, Mosén-Ansorena D, Aransay AM, Hernández-Tiedra S, López-Valero I, Quintanilla M, Sánchez C, Iovanna JL, Dusetti N, Guzmán M, Francis SE, Carracedo A, Kiss-Toth E, Velasco G, Loss of Tribbles pseudokinase-3 promotes Akt-driven tumorigenesis via FOXO inactivation. Cell Death Differ. 22, 131–144 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naiki T, Saijou E, Miyaoka Y, Sekine K, Miyajima A, TRB2, a mouse Tribbles ortholog, suppresses adipocyte differentiation by inhibiting AKT and C/EBPβ. J. Biol. Chem 282, 24075–24082 (2007). [DOI] [PubMed] [Google Scholar]
- 39.O’Connor C, Yalla K, Salomé M, Ananyambica Moka H, Gómez Castañeda E, Eyers PA, Keeshan K, Trib2 expression in granulocyte-monocyte progenitors drives a highly drug resistant acute myeloid leukaemia linked to elevated Bcl2. Oncotarget 9, 14977–14992 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey FP, Byrne DP, Oruganty K, Eyers CE, Novotny CJ, Shokat KM, Kannan N, Eyers PA, The Tribbles 2 (TRB2) pseudokinase binds to ATP and autophosphorylates in a metal-independent manner. Biochem. J 467, 47–62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Azambuja E, de Azambuja E, Holmes AP, Piccart-Gebhart M, Holmes E, Di Cosimo S, Swaby RF, Untch M, Jackisch C, Lang I, Smith I, Boyle F, Xu B, Barrios BH, Perez EA, Azim HA Jr., Azim KSB, Kuemmel S, Huang CS, Vuylsteke P, Hsieh RK, Gorbunova V, Eniu A, Dreosti L, Tavartkiladze N, Gelber RD, Eidtmann H, Baselga J, Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): Survival outcomes of a randomised, open-label, multicentre, phase 3 trial and their association with pathological complete response. Lancet Oncol. 15, 1137–1146 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Singh M, Jadhav HR, Targeting non-small cell lung cancer with small-molecule EGFR tyrosine kinase inhibitors. Drug Discov. Today 23, 745–753 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Kosaka T, Tanizaki J, Paranal RM, Endoh H, Lydon C, Capelletti M, Repellin CE, Choi J, Ogino A, Calles A, Ercan D, Redig AJ, Bahcall M, Oxnard GR, Eck MJ, Jänne PA, Response heterogeneity of EGFR and HER2 Exon 20 insertions to covalent EGFR and HER2 inhibitors. Cancer Res. 77, 2712–2721 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sequist LV, Besse B, Lynch TJ, Miller VA, Wong KK, Gitlitz B, Eaton K, Zacharchuk C, Freyman A, Powell C, Ananthakrishnan R, Quinn S, Soria J-C, Neratinib, an irreversible pan-ErbB receptor tyrosine kinase inhibitor: Results of a phase II trial in patients with advanced non-small-cell lung cancer. J. Clin. Oncol 28, 3076–3083 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Bose P, Ozer H, Neratinib: An oral, irreversible dual EGFR/HER2 inhibitor for breast and non-small cell lung cancer. Expert Opin. Investig. Drugs 18, 1735–1751 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Murphy JM, Mace PD, Eyers PA, Live and let die: Insights into pseudoenzyme mechanisms from structure. Curr. Opin. Struct. Biol 47, 195–104 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Rudolf AF, Skovgaard T, Knapp S, Juhl Jensen L, Berthelsen J, A comparison of protein kinases inhibitor screening methods using both enzymatic activity and binding affinity determination. PLOS ONE 9, e98800 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murphy JM, Zhang Q, Young SN, Reese ML, Bailey FP, Eyers PA, Ungureanu D, Hammaren H, Silvennoinen O, Varghese LN, Chen K, Tripaydonis A, Jura N, Fukuda K, Qin J, Nimchuk Z, Beth Mudgett M, Elowe S, Gee CL, Liu L, Daly RJ, Manning G, Babon JJ, Lucet IS, A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem. J 457, 323–334 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Byrne DP, Vonderach M, Ferries S, Brownridge PJ, Eyers CE, Eyers PA, cAMP-dependent protein kinase (PKA) complexes probed by complementary differential scanning fluorimetry and ion mobility-mass spectrometry. Biochem. J 473, 3159–3175 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hill R, Madureira PA, Ferreira B, Baptista I, Machado S, Colaço L, dos Santos M, Liu N, Dopazo A, Ugurel S, Adrienn A, Kiss-Toth E, Isbilen M, Gure AO, Link W, TRIB2 confers resistance to anti-cancer therapy by activating the serine/threonine protein kinase AKT. Nat. Commun 8, 14687 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Bago R, Sommer E, Castel P, Crafter C, Bailey FP, Shpiro N, Baselga J, Cross D, Eyers PA, Alessi DR, The hVps34-SGK3 pathway alleviates sustained PI3K/Akt inhibition by stimulating mTORC1 and tumour growth. EMBO J. 35, 1902–1922 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elkins JM, Fedele V, Szklarz M, Abdul Azeez KR, Salah E, Mikolajczyk J, Romanov S, Sepetov N, Huang X-P, Roth BL, Al Haj Zen A, Fourches D, Muratov E, Tropsha A, Morris J, Teicher BA, Kunkel M, Polley E, Lackey KE, Atkinson FL, Overington JP, Bamborough P, Müller S, Price DJ, Willson TM, Drewry DH, Knapp S, Zuercher WJ, Comprehensive characterization of the Published Kinase Inhibitor Set. Nat. Biotechnol 34, 95–103 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Waterson AG, Stevens KL, Reno MJ, Zhang YM, Boros EE, Bouvier F, Rastagar A, Uehling BE, Dickerson SH, Reep B, McDonald OB, Wood ER, Rusnak DW, Alligood KJ, Rudolph SK, Alkynyl pyrimidines as dual EGFR/ErbB2 kinase inhibitors. Bioorg. Med. Chem. Lett 16, 2419–2422 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Gaul MD, Guo Y, Affleck K, Cockerill GS, Gilmer TM, Griffin RJ, Guntrip S, Keith BR, Knight WB, Mullin RJ, Murray DM, Rusnak DW, Smith K, Tadepalli S, Wood ER, Lackey K, Discovery and biological evaluation of potent dual ErbB-2/EGFR tyrosine kinase inhibitors: 6-thiazolylquinazolines. Bioorg. Med. Chem. Lett 13, 637–40 (2003). [DOI] [PubMed] [Google Scholar]
- 55.Rheault TR, Caferro TR, Dickerson SH, Donaldson KH, Gaul MD, Goetz AS, Mullin RJ, McDonald OB, Petrov KG, Rusnak DW, Shewchuk LM, Spehar GM, Truesdale AT, Vanderwall DE, Wood ER, Uehling DE, Thienopyrimidine-based dual EGFR/ErbB-2 inhibitors. Bioorg. Med. Chem. Lett 19, 817–820 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Jamieson SA, Ruan Z, Burgess AE, Curry JR, McMillan HD, Brewster JL, Dunbier AK, Axtman AD, Kannan N, Mace PD, Substrate binding allosterically relieves autoinhibition of the pseudokinase TRIB1. Sci. Signal 11, eaau0597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Solca F, Dahl G, Zoephel A, Bader G, Sanderson M, Klein C, Kraemer O, Himmelsbach F, Haaksma E, Adolf GR, Target binding properties and cellular activity of afatinib (BIBW 2992), an irreversible ErbB family blocker. J. Pharmacol. Exp. Ther 343, 342–350 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Roy A, Kucukural A, Zhang Y, I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vucetic S, Brown CJ, Keith Dunker A, Obradovic Z, Flavors of protein disorder. Proteins 52, 573–584 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S, Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol 9, 342–353 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Milani M, Byrne DP, Greaves G, Butterworth M, Cohen GM, Eyers PA, Varadarajan S, DRP-1 is required for BH3 mimetic-mediated mitochondrial fragmentation and apoptosis. Cell. Death. Dis 8, e2552 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller RM, Paavilainen VO, Krishnan S, Serafimova IM, Taunton J, Electrophilic fragment-based design of reversible covalent kinase inhibitors. J. Am. Chem. Soc 135, 5298–5301 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwartz PA, Kuzmic P, Solowiej J, Bergqvist S, Bolanos B, Almaden C, Nagata A, Ryan K, Feng J, Dalvie D, Kath JC, Xu M, Wani R, William Murray B, Covalent EGFR inhibitor analysis reveals importance of reversible interactions to potency and mechanisms of drug resistance. Proc. Natl. Acad. Sci. U.S.A 111, 173–178 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trott O, Olson AJ, AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem 31, 455–461 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bianco G, Forli S, Goodsell DS, Olson AJ, Covalent docking using autodock: Two-point attractor and flexible side chain methods. Protein Sci. 25, 295–301 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Northrop BH, Coffey RN, Thiol–ene click chemistry: Computational and kinetic analysis of the influence of alkene functionality. J. Am. Chem. Soc 134, 13804–13817 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Martinez Molina D, Jafari R, Ignatushchenko M, Seki T, Andreas Larsson E, Dan C, Sreekumar L, Cao Y, Nordlund P, Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341,84–87 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Rishi L, Hannon M, Salomè M, Hasemann M, Frank A-K, Campos J, Timoney J, O’Connor C, Cahill MR, Porse B, Keeshan K, Regulation of Trib2 by an E2F1-C/EBPα feedback loop in AML cell proliferation. Blood 123, 2389–2400 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Qiao Y, Zhang Y, Wang J, Ubiquitin E3 ligase SCF(β-TRCP) regulates TRIB2 stability in liver cancer cells. Biochem. Biophys. Res. Commun 441,555–559 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Gao Y, Zhang T, Terai H, Ficarro SB, Kwiatkowski N, Hao MF, Sharma B, Christensen CL, Chipumuro E, Wong KK, Marto JA, Hammerman PS, Gray NS, George RE, Overcoming resistance to the THZ series of covalent transcriptional CDK inhibitors. Cell Chem. Biol 25, 135–142.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Modjtahedi H, Chul Cho B, Michel MC, Solca F, A comprehensive review of the preclinical efficacy profile of the ErbB family blocker afatinib in cancer. Naunyn Schmiedebergs Arch. Pharmacol 387, 505–521 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wodicka LM, Ciceri P, Davis MI, Hunt JP, Floyd M, Salerno S, Hua XH, Ford JM, Armstrong RC, Zarrinkar PP, Treiber DK, Activation state-dependent binding of small molecule kinase inhibitors: Structural insights from biochemistry. Chem. Biol 17, 1241–1249 (2010). [DOI] [PubMed] [Google Scholar]
- 73.Aertgeerts K, Skene R, Yano J, Sang B-C, Zou H, Snell G, Jennings A, Iwamoto K, Habuka N, Hirokawa A, Ishikawa T, Tanaka T, Miki H, Ohta Y, Sogabe S, Structural analysis of the mechanism of inhibition and allosteric activation of the kinase domain of HER2 protein. J. Biol. Chem 286, 18756–18765 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chaikuad A, Tacconi E, Zimmer J, Liang Y, Gray NS, Tarsounas M, Knapp S, A unique inhibitor binding site in ERK1/2 is associated with slow binding kinetics. Nat. Chem. Biol 10, 853–860 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber CK, Zarrinkar PP, Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol 29, 1046–1051 (2011). [DOI] [PubMed] [Google Scholar]
- 76.Jura N, Shan Y, Cao X, Shaw DE, Kuriyan J, Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc. Natl. Acad. Sci. U.S.A 106, 21608–21613 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shi F, Telesco SE, Liu Y, Radhakrishnan R, Lemmon MA, ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc. Natl. Acad. Sci. U.S.A 107, 7692–7697 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Niessen S, Dix MM, Barbas S, Potter ZE, Lu S, Brodsky O, Planken S, Behenna D, Almaden C, Gajiwala KS, Ryan K, Ferre R, Lazear MR, Hayward MM, Kath JC, Cravatt BF, Proteome-wide map of targets of T790M-EGFR-directed covalent inhibitors. Cell Chem. Biol 24, 1388–1400.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Newton AS, Deiana L, Puleo DE, Cisneros JA, Cutrona KJ, Schlessinger J, Jorgensen WL, JAK2 JH2 fluorescence polarization assay and crystal structures for complexes with three small molecules. ACS Med. Chem. Lett 8, 614–617 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Puleo DE, Kucera K, Hammarén HM, Ungureanu D, Newton AS, Silvennoinen O, Jorgensen WL, Schlessinger J, Identification and characterization of JAK2 pseudokinase domain small molecule binders. ACS Med. Chem. Lett 8, 618–621 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Linhardt RJ, Rice KG, Merchant ZM, Kim YS, Lohse DL, Structure and activity of a unique heparin-derived hexasaccharide. J. Biol. Chem 261, 14448–14454 (1986). [PubMed] [Google Scholar]
- 82.Polier S, Samant RS, Clarke PA, Workman P, Prodromou C, Pearl LH, ATP-competitive inhibitors block protein kinase recruitment to the Hsp90-Cdc37 system. Nat. Chem. Biol 9, 307–312 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schneider C, Sepp-Lorenzino L, Nimmesgern E, Ouerfelli O, Danishefsky S, Rosen N, Ulrich Hartl F , Pharmacologic shifting of a balance between protein refolding and degradation mediated by Hsp90. Proc. Natl. Acad. Sci. U.S.A 93, 14536–14541 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Taipale M, Krykbaeva I, Whitesell L, Santagata S, Zhang J, Liu Q, Gray NS, Lindquist S, Chaperones as thermodynamic sensors of drug-target interactions reveal kinase inhibitor specificities in living cells. Nat. Biotechnol 31,630–637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Taipale M, Krykbaeva I, Koeva M, Kayatekin C, Westover KD, Karras GI, Lindquist S, Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell 150, 987–1001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Scutt PJ, Chu MLH, Sloane DA, Cherry M, Bignell CR, Williams DH, Eyers PA, Discovery and exploitation of inhibitor-resistant aurora and polo kinase mutants for the analysis of mitotic networks. J. Biol. Chem 284, 15880–15893 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sloane DA, Trikic MZ, Chu MLH, Lamers MBAC, Mason CS, Mueller I, Savory WJ, Williams DH, Eyers PA, Drug-resistant aurora A mutants for cellular target validation of the small molecule kinase inhibitors MLN8054 and MLN8237. ACS Chem. Biol 5, 563–576 (2010). [DOI] [PubMed] [Google Scholar]
- 88.Eyers PA, van den IJssel P, Quinlan RA, Goedert M, Cohen P, Use of a drug-resistant mutant of stress-activated protein kinase 2a/p38 to validate the in vivo specificity of SB 203580. FEBS Lett. 451,191–196 (1999). [DOI] [PubMed] [Google Scholar]
- 89.Zhang T, Inesta-Vaquera F, Niepel M, Zhang J, Ficarro SB, Machleidt T, Xie T, Marto JA, Kim ND, Sim T, Laughlin JD, Park H, LoGrasso PV, Patricelli M, Nomanbhoy TK, Sorger PK, Alessi DR, Gray NS, Discovery of potent and selective covalent inhibitors of JNK. Chem. Biol 19, 140–154 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang Y, Zhang J, Liu C, Du S, Feng L, Luan X, Zhang Y, Shi Y, Wang T, Wu Y, Cheng W, Meng S, Li M, Liu H, Neratinib induces ErbB2 ubiquitylation and endocytic degradation via HSP90 dissociation in breast cancer cells. Cancer Lett. 382, 176–185 (2016). [DOI] [PubMed] [Google Scholar]
- 91.Uetrecht J, Naisbitt DJ, Idiosyncratic adverse drug reactions: Current concepts. Pharmacol. Rev 65, 779–808 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moy B, Goss PE, Lapatinib-associated toxicity and practical management recommendations. Oncologist 12, 756–765 (2007). [DOI] [PubMed] [Google Scholar]
- 93.Yap TA, Vidal L, Adam J, Stephens P, Spicer J, Shaw H, Ang J, Temple G, Bell S, Shahidi M, Uttenreuther-Fischer M, Stopfer P, Futreal A, Calvert H, de Bono JS, Plummer R, Phase I trial of the irreversible EGFR and HER2 kinase inhibitor BIBW 2992 in patients with advanced solid tumors. J. Clin. Oncol 28, 3965–3972 (2010). [DOI] [PubMed] [Google Scholar]
- 94.Hill R, Kiran R, Kalathur R, Colaço L, Brandão R, Ugurel S, Futschik M, Link W, TRIB2 as a biomarker for diagnosis and progression of melanoma. Carcinogenesis 36, 469–477 (2015). [DOI] [PubMed] [Google Scholar]
- 95.Grandinetti KB, Stevens TA, Ha S, Salamone RJ, Walker JR, Zhang J, Agarwalla S, Tenen DG, Peters EC, Reddy VA, Overexpression of TRIB2 in human lung cancers contributes to tumorigenesis through downregulation of C/EBPα. Oncogene 30, 3328–3335 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tyler RK, Chu MLH, Johnson H, McKenzie EA, Gaskell SJ, Eyers PA, Phosphoregulation of human Mps1 kinase. Biochem. J 417, 173–181 (2009). [DOI] [PubMed] [Google Scholar]
- 97.Byrne DP, Li Y, Ngamlert P, Ramakrishnan K, Eyers CE, Wells C, Drewry DH, Zuercher WJ, Berry NG, Fernig DG, Eyers PA, New tools for evaluating protein tyrosine sulphation: Tyrosyl Protein Sulphotransferases (TPSTs) are novel targets for RAF protein kinase inhibitors. Biochem. J 475, 2435–2455 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McSkimming DI, Dastgheib S, Baffi TR, Byrne DP, Ferries S, Thomas Scott S, Newton AC, Eyers CE, Kochut KJ, Eyers PA, Kannan N, KinView: A visual comparative sequence analysis tool for integrated kinome research. Mol. Biosyst 12, 3651–3665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Webb B, Sali A, Protein structure modeling with MODELLER. Methods Mol. Biol 1654, 39–54 (2017). [DOI] [PubMed] [Google Scholar]
- 100.Nivon LG, Moretti R, Baker D, A Pareto-optimal refinement method for protein design scaffolds. PLOS ONE 8, e59004 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huang PS, Andrew Ban Y-E, Richter F, Andre I, Vernon R, Schief WR, Baker D, RosettaRemodel: A generalized framework for flexible backbone protein design. PLOS ONE 6, e24109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA, Development and testing of a general amber force field. J. Comput. Chem 25, 1157–1174 (2004). [DOI] [PubMed] [Google Scholar]
- 103.Sousa da Silva AW, Vranken WF, ACPYPE—AnteChamber PYthon Parser interfacE.. BMC. Res. Notes 5, 367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bailey FP, Clarke K, Kalirai H, Kenyani J, Shahidipour H, Falciani F, Coulson JM, Sacco JJ, Coupland SE, Eyers PA, Kinome-wide transcriptional profiling of uveal melanoma reveals new vulnerabilities to targeted therapeutics. Pigment Cell Melanoma Res. 31,253–266 (2018). [DOI] [PubMed] [Google Scholar]
- 105.Savitski MM, Reinhard FBM, Franken H, Werner T, Fälth Savitski M, Eberhard D, Martinez Molina D, Jafari R, Bakszt Dovega R, Klaeger S, Kuster B, Nordlund P, Bantscheff M, Drewes G, Tracking cancer drugs in living cells by thermal profiling of the proteome. Science 346, 1255784 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Discovery of multiple PKIS compounds as TRIB2-binding compounds.
Fig. S2. Chemical structure of preclinical and clinical compounds evaluated in this study.
Fig. S3. Validation of DSF assay using TRIB1 and a PKAc counterscreen.
Fig. S4. Thermal melting profiles of TRIB2 using DSF.
Fig. S5.Analysis of C104Y mutation in TRIB2.
Fig. S6. MS-based analysis of the covalent TRIB2:afatinib complex.
Fig. S7. MST assay.
Fig. S8. Molecular docking analysis.
Fig. S9. A new TRIB2 antibody for quantitative analysis of TRIB2 expression levels and stability.
Fig. S10. Analysis of TRIB2 dephosphorylation in cell extracts.
Fig. S11. TRIB2 binding to afatinib induces destabilization relative to DMSO in a whole-cell thermal shift assay (CETSA).
Fig. S12. Comparative protein expression analysis of stable HeLa FLAG-TRIB2 cell lines.
Fig. S13. Lack of effect of the noncovalent TRIB1- and TRIB2-destabilizing compound GW804482X on TRIB2 stability in TRIB2-expressing HeLa cells.
Table S1. PKIS compound screening data for full-length TRIB2.