Abstract
Background
Salmonella enterica subsp. enterica contains more than 2,600 serovars of which four are of major medical relevance for humans. While the typhoidal serovars (Typhi and Paratyphi A) are human-restricted and cause enteric fever, non-typhoidal Salmonella serovars (Typhimurium and Enteritidis) have a broad host range and predominantly cause gastroenteritis.
Methodology/Principle findings
We compared the core proteomes of Salmonella Typhi, Paratyphi A, Typhimurium and Enteritidis using contemporary proteomics. For each serovar, five clinical isolates (covering different geographical origins) and one reference strain were grown in vitro to the exponential phase. Levels of orthologous proteins quantified in all four serovars and within the typhoidal and non-typhoidal groups were compared and subjected to gene ontology term enrichment and inferred regulatory interactions. Differential expression of the core proteomes of the typhoidal serovars appears mainly related to cell surface components and, for the non-typhoidal serovars, to pathogenicity.
Conclusions/Significance
Our comparative proteome analysis indicated differences in the expression of surface proteins between Salmonella Typhi and Paratyphi A, and in pathogenesis-related proteins between Salmonella Typhimurium and Enteritidis. Our findings may guide future development of novel diagnostics and vaccines, as well as understanding of disease progression.
Author summary
With an estimated 20 million typhoid cases and an even higher number of non-typhoid cases the health burden caused by salmonellosis is huge. Salmonellosis is caused by the bacterial species Salmonella enterica and over 2500 different serovars exist, of which four are of major medical relevance for humans: Typhi and Paratyphi A cause typhoid fever while Typhimurium and Enteritidis are the dominant cause of non-typhoidal Salmonella infections. The proteome is the entire set of proteins that is expressed by a genome and the core proteome are all orthologous proteins detected in a given sample set. In this study we have investigated differential expression of the core proteomes of the Salmonella serovars Typhi, Paratyphi A, Typhimurium and Enteritidis, as well as the regulating molecules. Our comparative proteome analysis indicated differences in the expression of surface proteins between the serovars Typhi and Paratyphi A, and in pathogenesis-related proteins between Typhimurium and Enteritidis. Our findings in proteome-wide expression may guide the development of novel diagnostics and vaccines for Salmonella, as well as understanding of disease.
Introduction
The gram-negative bacterial genus Salmonella is divided in two species, Salmonella enterica and Salmonella bongori. Only the Salmonella enterica subspecies enterica is of clinical relevance for humans and is further classified into more than 2,600 serovars. The human restricted serovar Typhi (STY) and the closely related serovar Paratyphi A (SPTA) cause enteric fever [1], while the generalist serovars Typhimurium (STM) and Enteritidis (SENT) are the most important causes of non-typhoidal salmonellosis [2]. Enteric fever is a systemic disease that affects more than 27 million people worldwide and leads to more than 200,000 deaths annually [3,4]. While STY and SPTA both cause a systemic disease, SPTA causes a milder disease with a shorter incubation time [5]. In the last 20 years, the number of infections with SPTA has significantly increased in Asia [6]. The global burden of non-typhoidal Salmonella, a common cause of food poisoning that is usually characterized by localized gastroenteritis, is even higher with an estimated 93.8 million cases and 155,000 deaths each year [2]. Moreover, invasive non-typhoidal Salmonella has emerged as an important cause of bloodstream infection in Sub-Saharan Africa in both adults and children, and the incidence of invasive non-typhoidal Salmonella is estimated at 3.4 million cases with more than 600,000 deaths each year [7].
Comparative genomics of Salmonella enterica has revealed specific genetic fingerprints associated with invasive disease and host adaptation [8,9]. A comparative analysis of 8 typhoidal and 27 non-typhoidal Salmonella genomes demonstrated presence of typhoid-specific protein families which include virulence factors such as Vi polysaccharide pilus related proteins [10]. In addition, an in silico comparative analysis of Salmonella genomes identified 469 genes involved in the central anaerobic metabolism which was intact in gastrointestinal pathogens (SENT and STM among others) but decaying in extra-intestinal pathogens, such as STY and SPTA. This metabolic advantage might have a role in competing with other bacteria in the inflamed gut, thereby enhancing transmission of the gastrointestinal pathogens [11]. However, not all phenotypic differences in typhoidal and non-typhoidal Salmonella can be explained by presence or absence of functional genes. Investigating differential expression of the core proteomes (defined as all orthologous proteins quantified in a given sample set) between Salmonella serovars [12], and the regulating molecules involved, can reveal additional insights in the adaptations to different host environments and pathogenesis, as well as reveal the expression of potential vaccine and diagnostic targets.
In the last decade, mass spectrometry (MS) based proteomics has advanced rapidly and provides a comprehensive view on the proteins that are expressed by an organism. In clinical microbiology laboratories, MALDI-TOF MS is routinely used for bacterial genus and species identification [13]. In research, proteomics was used to characterize the proteomes of Salmonella Typhimurium and Enteritidis under specific in vitro culture conditions mimicking the phagosome [14,15], to identify proteins that were expressed by Salmonella Typhimurium isolated from infected macrophages [16], and to study antimicrobial resistance and virulence in Salmonella Typhimurium [17–19]. Next to proteome analysis within single serovars, comparative proteome studies have been conducted to assess the proteome variability between different Salmonella serovars. However, these studies used laboratory reference strains which may not represent the currently circulating clinical strains [20–22].
Here, we conducted a comparative analysis of the core proteomes of the clinically most relevant Salmonella enterica serovars: Typhi, Paratyphi A, Typhimurium and Enteritidis, using 20 Salmonella strains isolated from patients covering various geographical origins, as well as one reference strain per serovar. Our findings show that differential expression of the core proteome of the typhoidal serovars is mainly related to cell surface components and, for the non-typhoidal serovars, to pathogenicity.
Methods
Bacterial strains and growth conditions
Five clinical isolates per Salmonella serovar Typhi, Paratyphi A, Typhimurium and Enteritidis were selected from the strain collection at the clinical laboratory of the travel clinic of the Institute of Tropical Medicine, Antwerp, Belgium for shotgun proteome analysis. One ATCC reference strain for each Salmonella serovar was added to the sample set and for the Salmonella Typhi reference strain, a clinical strain was certified (Table 1). Given that the burden of typhoid fever and invasive non-typhoidal salmonellosis is highest in Asia and Africa respectively, we have selected representative strains from different countries covering both continents. All in vitro incubation was done at 37°C. Minimum and maximum temperatures were recorded and ranged between 35°C and 37°C. As all clinical strains have been isolated from patients, the strains were revived from Microbank cryogenic vials (Pro-Lab Diagnostics) on blood agar (BD Columbia Agar, 5% sheep blood) and grown overnight at 37°C. Single colonies were sub-cultured on MacConkey agar (BD MacConkey II Agar) and grown overnight at 37°C. Colonies were further solubilized into 3 ml of synthetic growth medium and supplemented with 1% glucose (Teknova HI-DEF Azure Media) until the OD was 0.06, and 250 μl of this suspension was inoculated into 5 ml of synthetic medium supplemented with 1% glucose and grown at 37°C with shaking at 220 rpm until mid-log phase (OD 0.5-OD 0.6). The Teknova HI-DEF Azure synthetic medium (S1 File) is based on the medium described by Neidhardt et al. [23].
Table 1. Geographical origin and year of isolation of the Salmonella enterica Typhi, Paratyphi A, Typhimurium and Enteritidis strains.
| ID strain | Salmonella enterica serovar | Geographic origin | Year of isolation |
|---|---|---|---|
| Clinical isolates | |||
| 9092306 | Typhi | Bangladesh | 2009 |
| 9121199 | Typhi | Burkina Faso | 2009 |
| 2427† | Typhi | Cambodia | 2010 |
| 3182/3† | Typhi | DRC* | 2010 |
| 12091815 | Typhi | Thailand | 2012 |
| 8041131 | Paratyphi A | India | 2008 |
| 8121108 | Paratyphi A | Senegal | 2008 |
| 1964† | Paratyphi A | Cambodia | 2010 |
| 12082646 | Paratyphi A | India | 2012 |
| 12122069 | Paratyphi A | Myanmar | 2012 |
| 3011187 | Typhimurium | Ethiopia | 2003 |
| 2371 | Typhimurium | Cambodia | 2010 |
| 11082746 | Typhimurium | Malawi | 2011 |
| HRG039VD28 | Typhimurium | The Gambia | 2013 |
| 11185/3† | Typhimurium | DRC* | 2014 |
| 9001877 | Enteritidis | Cambodia | 2009 |
| 3252/3† | Enteritidis | DRC* | 2010 |
| 10080748 | Enteritidis | Nigeria | 2010 |
| 12050236 | Enteritidis | Senegal | 2012 |
| 12080487 | Enteritidis | Indonesia | 2012 |
| Reference isolates | |||
| ITM00032304‡ | Typhi | Senegal | 2000 |
| ATCC9150 | Paratyphi A | Malaysia | 1993 |
| ATCC14028 | Typhimurium | unknown | 1960# |
| ATCC13076 | Enteritidis | unknown | unknown |
* Democratic Republic of the Congo
# ATCC 14028 is a descendant of CDC 60–6516, which is a strain isolated in 1960 from pools of hearts and livers of 4-week-old chickens.
†Obtained from microbiological surveillance studies in the respective countries. The other strains were obtained from patients at the travel clinic of ITM.
‡Clinical strain certified by the Belgian National Reference Centre for Salmonella and Shigella (ISP-WIV, currently Sciensano, Brussels).
Protein extraction and in-solution digestion
Upon harvesting the bacteria, duplicate samples of 1 ml were taken from each culture and centrifuged at 5000 x g for 10 min at 4°C and the cell pellets were washed twice with phosphate buffered saline (PBS). Duplicate samples are thus further considered as technical replicates. Proteins were extracted from the bacterial pellets with the Qproteome Bacterial Protein Prep Kit (Qiagen) following the manufacturer’s instructions. Briefly, after snap-freezing on dry ice, bacterial cell pellets were thawed on ice for 15 minutes. Cell pellets were re-suspended 750 μl of lysis buffer supplemented with lysozyme and Benzonase Nuclease, all included in the extraction kit. EDTA-free protease inhibitor (Roche) was added to a final concentration of 2%. After incubation on ice for 30 minutes, lysates were centrifuged at 14,000 for 30 minutes to pellet the cellular debris, and the supernatant was collected. The protein concentration was determined with the BCA Protein Assay Kit (Pierce) (S1 Table). Proteins were reduced with 15 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP-HCl) and alkylated with 30 mM iodoacetamide (IAM) for 15 min in the dark while shaking at 37°C. The buffer was exchanged to digestion buffer (50 mM ammonium bicarbonate, pH 7.9) using G-25 illustra NAP-5 gel filtration columns (GE Healthcare). The eluates were then heated at 99°C for 5 min, put immediately on ice and, after cooling, sequencing grade modified trypsin (Promega) was added to a 1:100 trypsin to protein ratio upon which digestion proceeded at 37°C for 16 h. The trypsin activity was stopped by adding 60 μl of 10% trifluoroacetic acid (TFA) (0.6% final concentration).
LC-MS/MS analysis
The peptide mixtures were subjected to LC−MS/MS analysis using an Ultimate 3000 RSLC nano LC (Thermo Scientific, Bremen, Germany) in-line connected to a Q Exactive mass spectrometer (Thermo Fisher Scientific). The sample mixture was first loaded on a trapping column (made in-house, 100 μm internal diameter (I.D.), 20 mm long, filled with 5 μm C18 Reprosil-HD beads, Dr. Maisch, Ammerbuch-Entringen, Germany). After flushing from the trapping column, the peptides were loaded on an analytical column (75 μm I.D., 400 mm long and filled with 3 μm C18 Reprosil-HD beads (Dr. Maisch)) packed in the needle PicoFrit SELF/P PicoTip emitter (PF360-75-15-N-5 (NewObjective, Woburn, USA)). Peptides were loaded with loading solvent (0.1% TFA in water) and separated with a linear gradient from 98% solvent A’ (0.1% formic acid in water) to 40% solvent B′ (0.1% formic acid in water/acetonitrile, 20/80 (v/v)) in 130 min at a flow rate of 300 nL/min. This was followed by a 15 min wash reaching 99% solvent B’. The mass spectrometer was operated in data-dependent, positive ionization mode, automatically switching between MS and MS/MS acquisition for the 10 most abundant peaks in a given MS spectrum. The source voltage was 3.4 kV and the capillary temperature was at 275°C. One MS1 scan (m/z 400−2000, AGC target 3 × 106 ions, maximum ion injection time 80 ms) acquired at a resolution of 70,000 (at 200 m/z) was followed by up to 10 tandem MS scans (resolution 17,500 at 200 m/z) of the most intense ions fulfilling the defined selection criteria (AGC target 5 × 104 ions, maximum ion injection time 60 ms, isolation window 2 Da, fixed first mass 140 m/z, spectrum data type: centroid, underfill ratio 2%, intensity threshold 1.7xE4, exclusion of unassigned 1, 5–8, >8 charged precursors, peptide match preferred, exclude isotopes: on, dynamic exclusion time 20 s). The HCD collision energy was set to 25% normalized collision energy and the polydimethylcyclosiloxane background ion at 445.120025 Da was used for internal calibration (lock mass). The mass spectrometry proteomics data have been deposited to the PRIDE Archive (http://www.ebi.ac.uk/pride/archive/) via the PRIDE partner repository with the data set identifier PXD011154 (username: reviewer00797@ebi.ac.uk; password: hN5SqXtY).
MS data processing
Raw MS files were analyzed by MaxQuant [24] version 1.5.0.25 and MS/MS spectra were searched against the translated protein sequences of the annotated genomes of Salmonella Typhi CT18 (NCBI accession number AL513382.1) [25], Paratyphi A ATCC 9150 (CP000026.1) [26], Typhimurium 14028S (CP001363.1) [27], and Enteritidis PT4/P125109 (AM933172.1) [28]. The following parameters were applied for the database search: enzyme specificity was set to trypsin/P allowing for a maximum of two missed cleavages; carbamidomethylation of cysteine was set as a fixed modification; methionine oxidation, N-terminal formylation on the protein level and conversion of N-terminal glutamine to pyroglutamate were set as variable modifications. The first search for precursor ions was performed with a mass tolerance of 20 ppm for calibration, while 6 ppm was applied for the main search. For protein identification, at least two unique peptides were required per protein group and the minimum peptide length was set to 7. The false discovery rate for peptide and protein identification was set to 1%. The minimum score threshold for both modified and unmodified peptides was set to 30. MS runs were analyzed with the “match between runs” option between samples of a given serovar. For matching, a retention time window of 42 s was selected. Protein quantification was based on the MaxQuant label-free (MaxLFQ) algorithm. For all other parameters, default settings were applied as advised by the developers.
Comparative analysis of core proteomes
The MaxQuant output file “proteinGroups.txt” was loaded into Perseus 1.5.0.8. The protein entries were filtered to remove potential contaminants, reverse hits and proteins only identified by site. Then, the LFQ intensities were log2 transformed and data were filtered for proteins containing a minimum number of valid values in 9 out of 12 samples. The log2 transformed data were then normalized by subtracting the median per sample within the dataset. To compare the different Salmonella serovars we used orthology mapping. Orthologous genes within the four serovars were retrieved from the Orthologous Matrix (OMA) database [29] with NCBI Taxonomy IDs 220341 (STY), 295319 (SPTA), 550537 (SENT) and 588858 (STM). Statistical significant differences in LFQ intensities were assessed using a two-sided t-test with Bonferroni adjusted P values using R. Proteins were considered differentially expressed if they showed a minimal 2-fold change in their overall levels with an adjusted P-value lower than 0.05. Principal component analysis (PCA) was done in Perseus 1.5.0.8 using default settings as advised by the developers.
Functional enrichment analysis
Differentially expressed proteins were subjected to gene ontology (GO) term enrichment to investigate biological processes, molecular function and cellular compartment using the Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics resources 6.7 [30]. Briefly, we have uploaded the differentially expressed core proteins as an input list and performed GO term enrichment analysis against a background list with default settings (count threshold is 2 and EASE threshold is 0.1).
Regulatory network analysis
To infer regulatory interactions that can explain differential expression profiles we used the PheNetic web server (http://bioinformatics.intec.ugent.be/phenetic/#/index) with default settings (Cost is 0.1, Pathlength is 4 and k-best paths is 20) and upstream run mode [31]. Input data consisted of the available interaction network for Salmonella Typhimurium LT2 (http://bioinformatics.intec.ugent.be/phenetic/index.html#/network), the list of detected proteins that are shared by two groups, and the list of differentially expressed proteins with P<0.05.
Ethics statement
The clinical Salmonella isolates were obtained through the project “Surveillance of antimicrobial resistance among consecutive blood culture isolates in tropical settings”, within the Third Framework Agreement between the Belgian Directorate of Development Cooperation (DGD) and the Institute of Tropical Medicine (ITM), Antwerp, Belgium. The partner institutes involved in this surveillance project that provided strains were: Sihanouk Hospital Centre of Hope, Phnom Penh, Cambodia and Institut National de Recherche Biomédicale, Kinshasa, Democratic Republic of the Congo. Ethical approval for the Microbiological Surveillance was granted by the Institutional Review Board at the ITM in Antwerp, by the Ethics Committees of the Antwerp University (Belgium). Ethical approval for the Microbiological Surveillance Study was granted by the Institutional Review Board of ITM, the Ethics Committee of Antwerp University and the competent ethical committees from the DR Congo and Cambodia respectively. The Salmonella Typhimurium isolate from The Gambia was received from the Medical Research Council (MRC) Keneba, MRC The Gambia. Ethical approval was granted by the Gambia Government/MRC Joint Ethics Committee. The remaining strains were obtained from patients presenting at the travel clinic at ITM, ethical approval was granted by the Institutional Review Board of ITM. All isolates and subsequent biological samples were anonymized.
Results
Salmonella proteins identified by LC-MS/MS
The reference genomes of STY, SPTA, SENT and STM used in our analysis contain 4,600, 4,095, 4,318 and 5,372 protein-encoding genes, respectively. In total, 3596 orthologous genes in the four serovars were retrieved from the OMA database and 1,414, 1,558, 1,222 and 1,099 proteins were detected by LC-MS/MS analysis in the STY, SPTA, SENT and STM strains, respectively. Protein detection in technical replicates showed Pearson correlation coefficients higher than 0.92 for all samples, except for the STM strain from Ethiopia with a Pearson correlation of 0.86 (S2 Table). Intra-serovar PCA of the LFQ intensities of expressed proteins show little variation in expression levels between strains within the same serovar (S2 File). However, in order to conduct reliable intra-serovar comparisons, more strains should have been included per serovar.
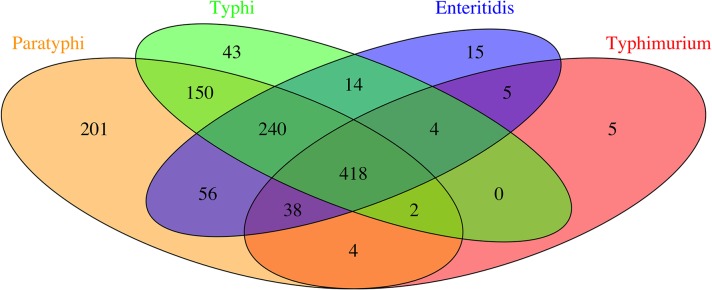
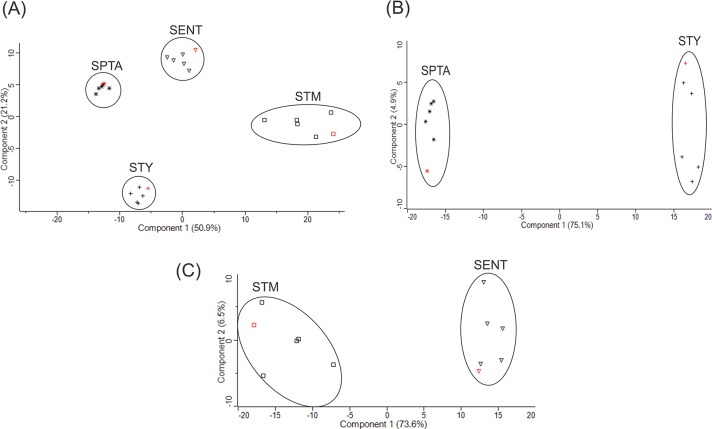
In total, 418 orthologous proteins were detected in all serovars (Fig 1) and expression levels in the typhoidal (STY and SPTA) and non-typhoidal (STM and SENT) Salmonella serovars were compared by PCA of the LFQ intensities (Fig 2A). The first two components capture ~72% of the variability in the dataset and show that the typhoidal serovars do not separate from the non-typhoidal serovars based on the observed variability in LFQ intensities. When we compared the typhoidal with the non-typhoidal Salmonella strains, a total of 128 proteins showed a minimal 2-fold change in their overall levels with an adjusted P-value lower than 0.05 (S3 Table). GO term enrichment of these 128 proteins showed that all GO terms with a P value lower than 0.05 are related to translation and structural components of the ribosomes (Table 2).
Fig 1. Venn diagram of the orthologous proteins detected by LC-MS/MS in 6 Salmonella Typhi, 6 Salmonella Paratyphi A, 6 Salmonella Enteritidis and 6 Salmonella Typhimurium strains.
Fig 2. Principal component analysis (PCA) separate serovars based on LFQ intensities.
The PCA plots show that the first and second principle components capture ~72% of the variability among the Salmonella serovars Typhi (STY), Paratyphi A (STPA), Typhimurium (STM) and Enteritidis (SENT) (A), 80% of the variability between the serovars STY and SPTA (B), and ~80% of the variability between the serovars STM and SENT (C). Reference strains for each serovar are presented in red.
Table 2. Gene ontology functional enrichment analysis of differentially expressed core proteins between typhoidal and non-typhoidal, Typhi and Paratyphi A, Enteritidis and Typhimurium Salmonella.
| Category | Term | Count | % | P-value |
|---|---|---|---|---|
| Typhoidal versus non-typhoidal | ||||
| GOTERM_MF_FAT | GO:0005198~structural molecule activity | 38 | 29.46 | 2.56E-10 |
| GOTERM_MF_FAT | GO:0003735~structural constituent of ribosome | 37 | 28.68 | 3.13E-10 |
| GOTERM_BP_FAT | GO:0006412~translation | 42 | 32.56 | 5.99E-07 |
| GOTERM_CC_FAT | GO:0005840~ribosome | 37 | 28.68 | 9.52E-06 |
| GOTERM_CC_FAT | GO:0030529~ribonucleoprotein complex | 37 | 28.68 | 2.17E-05 |
| GOTERM_MF_FAT | GO:0003723~RNA binding | 29 | 22.48 | 2.39E-05 |
| GOTERM_CC_FAT | GO:0043232~intracellular non-membrane-bounded organelle | 38 | 29.46 | 9.00E-05 |
| GOTERM_CC_FAT | GO:0043228~non-membrane-bounded organelle | 38 | 29.46 | 9.00E-05 |
| GOTERM_MF_FAT | GO:0019843~rRNA binding | 21 | 16.28 | 2.82E-04 |
| GOTERM_CC_FAT | GO:0033279~ribosomal subunit | 12 | 9.3 | 0.024948 |
| GOTERM_MF_FAT | GO:0000049~tRNA binding | 7 | 5.42 | 0.048571 |
| STY versus SPTA | ||||
| GOTERM_BP_FAT | GO:0016051~carbohydrate biosynthetic process | 10 | 4.4 | 5.55E-04 |
| GOTERM_BP_FAT | GO:0008610~lipid biosynthetic process | 10 | 4.4 | 0.001 |
| GOTERM_BP_FAT | GO:0034637~cellular carbohydrate biosynthetic process | 8 | 3.52 | 0.006 |
| GOTERM_BP_FAT | GO:0000271~polysaccharide biosynthetic process | 7 | 3.08 | 0.009 |
| GOTERM_BP_FAT | GO:0009103~lipopolysaccharide biosynthetic process | 6 | 2.64 | 0.014 |
| GOTERM_BP_FAT | GO:0008653~lipopolysaccharide metabolic process | 6 | 2.64 | 0.014 |
| GOTERM_BP_FAT | GO:0005976~polysaccharide metabolic process | 7 | 3.08 | 0.016 |
| GOTERM_BP_FAT | GO:0044264~cellular polysaccharide metabolic process | 6 | 2.64 | 0.026 |
| GOTERM_BP_FAT | GO:0033692~cellular polysaccharide biosynthetic process | 6 | 2.64 | 0.026 |
| GOTERM_CC_FAT | GO:0030312~external encapsulating structure | 7 | 3.08 | 0.033 |
| STM versus SENT | ||||
| GOTERM_CC_FAT | GO:0019861~flagellum | 8 | 4.14 | 0.037 |
| GOTERM_CC_FAT | GO:0042995~cell projection | 8 | 4.14 | 0.037 |
| GOTERM_BP_FAT | GO:0009405~pathogenesis | 9 | 4.66 | 0.044 |
Differentially expressed proteins in Salmonella Typhi (STY) and Paratyphi A (SPTA) are associated with the cell surface
A set of 810 core proteins were detected in Typhi and Paratyphi A and their LFQ intensities were used as input for PCA (Fig 2B). The first two components allow a clear separation of the STY from the SPTA strains, covering 80% of the total variation in expression levels. In addition, the PCA shows that clinical isolates do not separate from the reference strains in both serovars. A total of 230 proteins with a minimal 2-fold change in their overall levels and an adjusted P-value lower than 0.05 were considered significantly differentially expressed between STY and SPTA strains (S4 Table). GO functional enrichment analysis of these proteins indicated an enrichment of biological pathways that are related to carbohydrate and polysaccharide biosynthesis and metabolism, as well as the external encapsulating structure (Table 2). We have plotted our differential expression data set on the wide interaction network for Salmonella Typhimurium LT2. Using the upstream run mode, PheNetic searches for regulatory mechanisms that can explain our observed data set. The inferred sub-network (Fig 3) shows that many differentially expressed proteins are connected to each other by outer membrane, stress and carbohydrate metabolism regulatory proteins such as CpxR, YjeB and CRP, which are not necessarily differentially expressed themselves, but might have a post-translational serovar-specific effect. Moreover, the small regulatory RNAs OmrA and OmrB connect differentially expressed proteins involved in carbohydrate metabolism.
Fig 3. Phenetic sub-network inference analysis of differential protein expression in STY versus SPTA.
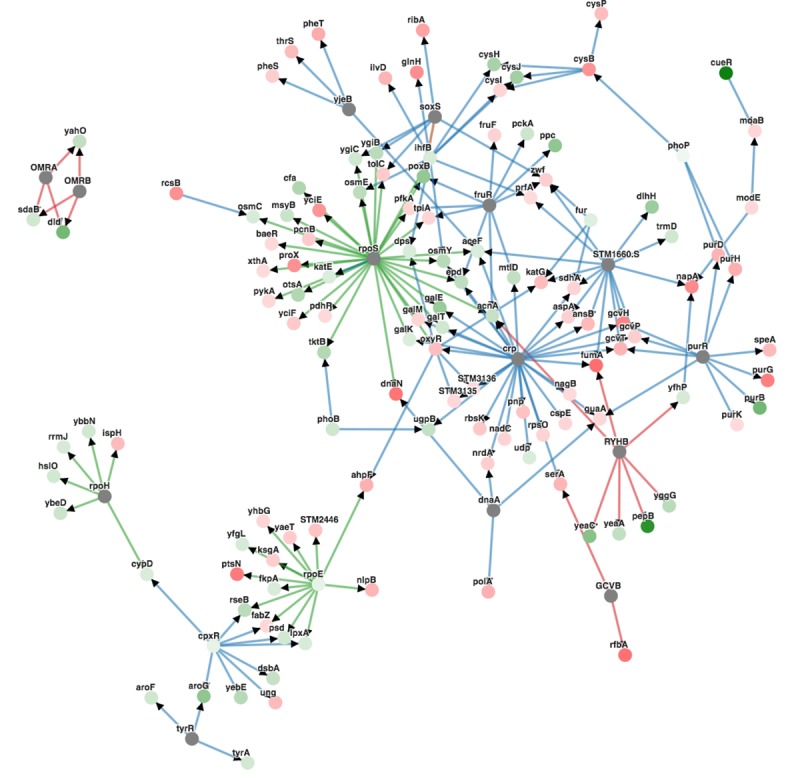
122 out of 230 differentially expressed proteins are shown in this sub-network. Red nodes represent proteins with higher expression in SPTA versus STY. Green nodes represent proteins with higher expression in STY versus SPTA. The more intense the color, the higher the level of differential expression. Gray nodes have no differential expression. The color of the edge indicates the interaction type with blue referring to metabolic, green to protein-protein and red to protein-DNA interactions.
Differentially expressed proteins in Salmonella Typhimurium (STM) and Enteritidis (SENT) are associated with pathogenicity
A set of 465 core proteins were detected in all strains of STM and SENT. PCA of the LFQ intensities of these proteins showed a clear separation of the STM isolates from the SENT isolates based on the observed protein expression levels where the first two components cover ~80% of the total variation in expression levels (Fig 2C). The PCA also shows that the reference strains and the clinical isolates do not separate in STM and SENT. A total of 192 proteins with a minimal 2-fold change in their overall levels and an adjusted P-value lower than 0.05 were considered significantly differentially expressed between STM and SENT strains (S5 Table). GO enrichment analysis of these proteins showed that all GO terms with P<0.05 are related to pathogenesis (Table 2). The inferred subnetwork (Fig 4) revealed that the flagellar biosynthesis sigma factor FliA and the flagellar transcriptional regulators FlhD and FlhC (STM1924.S) connect the upregulated flagellar synthesis and motility proteins in STM. HilA, the main regulator of Salmonella Pathogenicity Island 1 (SPI-1), is possibly involved in the upregulation of the type 3 secretion system (T3SS) structural protein Prgl and effector protein SipA in STM.
Fig 4. Phenetic sub-network inference analysis of differential protein expression in STM versus SENT.
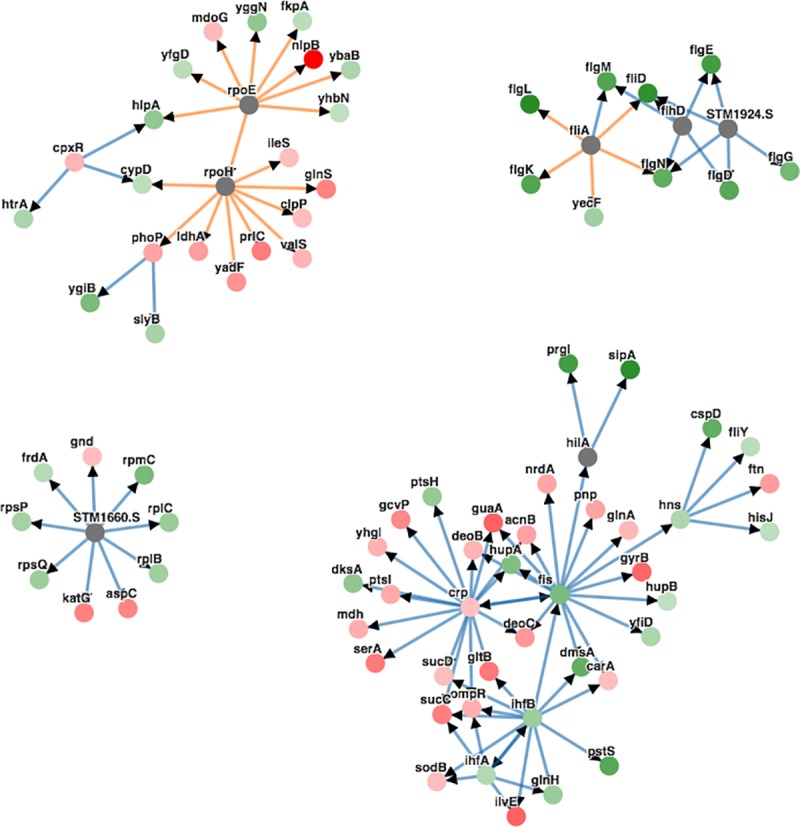
78 out of 192 differentially expressed proteins are shown in the sub-network. Red nodes represent proteins with higher expression in SENT versus STM. Green nodes represent proteins with higher expression in STM versus SENT. The more intense the color, the higher the level of differential expression. Gray nodes have no differential expression. The color of the edge indicates the interaction type with blue referring to metabolic and orange to protein-DNA interactions.
Discussion
The genomes of typhoidal and non-typhoidal Salmonella have a high level of similarity with more than 98% of sequence identity [32]. However, these two groups cause different diseases, host-pathogen interactions and immune responses. Here, we conducted the first comprehensive analysis of the proteomes of the Salmonella serovars Typhi, Paratyphi A, Typhimurium and Enteritidis using five clinical isolates that cover different geographical regions and one reference strain per Salmonella serovar. We have compared the expression levels of proteins from the core proteome under in vitro conditions and identified regulators that may help to explain the differences between different Salmonella serovars.
The classification of the four serovars into typhoidal and non-typhoidal groups is largely based on clinical presentation, with systemic and gastrointestinal disease, respectively. However, PCA of the LFQ intensities of the 418 detected proteins shared by all four serovars did not separate the typhoidal from the non-typhoidal serovars. Out of these 418 detected core proteins, 128 were significantly differentially expressed between typhoidal and the non-typhoidal serovars. However, GO analysis showed enrichment for proteins involved in translation and ribosomal activity, and thus largely represent the house keeping machinery of the bacterial cells. PCA showed that the LFQ intensities of the reference and clinical isolates within the STY, SPTA, STM and SENT serovars do not cluster separately, and the reference strains can thus be considered as representative for the serovar.
Further analysis showed that 230 proteins were differentially expressed between STY and SPTA. GO analysis revealed that proteins involved in carbohydrate and lipopolysaccharide metabolism, and proteins involved in external encapsulating structures were most enriched. The regulators in the sub-network analysis connecting the differentially expressed proteins are implicated in the cell envelope stress response and in polysaccharide metabolism. For example, OmrA/B connect Dld and SdaB, two proteins that are involved in transport of sugars and carbohydrate biosynthesis in E.coli, respectively. It is plausible that a serovar-specific effect acts at the sRNA-level, which is not detected in our proteomic analysis. CpxR that is known to have a role in the response to alterations in the cell envelope in Salmonella [33], explains the expression of Psd and LpxA required for phospholipid and glycolipid metabolism, respectively [34,35]. RpoS, RpoE and RpoH are involved in the stress response to different environmental conditions and contribute to Salmonella virulence [36–38]. CRP regulates the transcription of different operons involved in the transport of sugars and in catabolic functions [39], and FruR is required for carbohydrate metabolism [40]. The observation that cell surface proteins are significantly differently expressed between STY and SPTA is relevant for the diagnosis of Salmonella as well as for vaccination purposes. While the reference diagnostic method for typhoid fever is microbiological culture (blood, bone marrow or stool) and subsequent serotyping, rapid diagnostic tests (RDTs) have been developed and are commercially available for STY antigen and antibody detection [41]. However, diagnostic accuracy of the current RDTs is low, ranging from 31–97% [42] and more performant RDTs are urgently needed, including RDTs for SPTA. It has recently been shown that Salmonella antigen-based RDTs can be successfully applied to blood culture broths for Salmonella identification [43]. Three currently available typhoid vaccines are recommended by the WHO: an oral vaccine based on a live attenuated mutant strain of STY Ty21a (Ty21a), the injectable Vi capsular polysaccharide (ViCPS) vaccine and the typhoid conjugate vaccine (TCV) (http://www.who.int/immunization/policy/position_papers/typhoid/en/). However, these Typhi vaccines do not provide protection against paratyphoid fever caused by SPTA [44], and hence, a vaccine that protects against typhoid and paratyphoid fever would be of high value. When selecting antigens for developing new diagnostics or vaccines for both STY and SPTA, one should take into account that although encoded in both serovars, membrane proteins can be differentially expressed between both serovars and this should be tested in vitro and in vivo.
Upon comparing the proteomes of STM and SENT, 465 core proteins were detected, of which 192 were differentially expressed between the two serovars. GO enrichment analysis revealed that flagellar proteins and proteins involved in pathogenesis were most differentially expressed between both serovars. Among the higher expressed proteins in STM over SENT, six proteins are directly related to Salmonella pathogenicity island 1-encoded Type III secretion system (InvJ, SipA, SipD, SipC, PrgI, SipB). The T3SS-1 is an important virulence machinery that controls penetration of the gut epithelium during the infection by injecting effector proteins directly into the cytoplasm of epithelial cells through a needle-like appendages [45]. The regulator proteins InvJ and PrgI are known to be involved in needle and inner rod assembly [46], while SipA induces actin cytoskeletal rearrangements [47] and the translocases SipB and SipC form a translocation pore into the host cell membrane which is connected to the needle complex [48]. The sub-network also shows that HilA is possibly involved in the observed activation of the invasion proteins (SipA and PrgI) in STM. In addition, in the inferred sub-network the regulators FlhC (STM1924.S), FlhD and FliA were identified as regulators that connect 8 differentially expressed flagellar proteins (FlgL, FliD, FlgE, FlgM, FlgK, FlgD, FlgN, FlgG), showing higher expression profiles in Typhimurium strains. Besides their role in motility, flagellins were shown to stimulate both the innate and adaptive immune system and to cause inflammation upon STM infection [49]. Moreover, loss of flagellin expression in Salmonella has been linked to increased virulence in mice [50].
Some limitations in our study should be considered. The Salmonella strains were grown in standard in vitro conditions which may not be representative for protein expression in the infected host [51]. The addition of glucose to the medium may have induced catabolite repression. However, the addition of glucose as carbon source in needed to permit the growth of bacteria. Moreover, growth temperatures ranged between 35°C and 37°C and may have impacted expression levels. For instance, pathogenicity related gene expression is known to be temperature-sensitive [52]. In addition, the protein extraction procedure might have minorly affected the observed protein profiles although all steps have been performed on ice or 4°C. However, all strains have been grown using the same in vitro culture conditions and underwent the same extraction procedure and any possible effects are thus very likely averaged out in the comparative analysis. In addition, our mass spectrometry set-up is not as sensitive as the newest instruments currently available, and we captured around 20 to 40% of the proteomes. Poorly expressed proteins in the standard in vitro culture conditions used may thus have been missed, such as virulence related proteins [53]. Finally, the aim of our study was to conduct a comparative analysis of orthologous proteins shared between the four Salmonella serovars, and as such, we do not present information on serovar-specific (non-orthologous) proteins.
In conclusion, to the best of our knowledge this is the first study that compared the core proteomes of a large panel of clinical Salmonella isolates, covering the four clinically most relevant Salmonella enterica serovars: Typhi, Paratyphi A, Typhimurium and Enteritidis. Our comparative proteome analysis indicated differences in the expression of surface proteins between STY and SPTA, and in pathogenesis-related proteins between STM and SENT. Our insights may guide future developed of novel diagnostics and vaccines, and understanding of disease progression.
Supporting information
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank Tessa De Block for the technical assistance in the study. The partner institutes involved in this surveillance project that provided strains were: Sihanouk Hospital Centre of Hope, Phnom Penh, Cambodia and Institut National de Recherche Biomédicale, Kinshasa, Democratic Republic of the Congo. The stool isolate from The Gambia was received from the Medical Research Council (MRC) Keneba, MRC The Gambia, Keneba, The Gambia. The remaining strains were obtained from the travel clinic at ITM.
Data Availability
The mass spectrometry proteomics data have been deposited to the PRIDE Archive (http://www.ebi.ac.uk/pride/archive/) via the PRIDE partner repository with the data set identifier PXD011154
Funding Statement
This work was supported by the Flemish Ministry of Sciences (EWI, SOFI project IDIS) (SD) and the InBev-Baillet Latour (IBL) (SD) Fund. The clinical isolates were obtained through the project “Surveillance of antimicrobial resistance among consecutive blood culture isolates in tropical settings”, that was funded by the Belgian Directorate of Development. Cooperation (DGD) (JJ) through the Third Framework Agreement between the Belgian DGD and the Institute of Tropical Medicine (ITM), Belgium. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Crump JA, Mintz ED. Global trends in typhoid and paratyphoid fever. Clin Infect Dis. 2010. January 15;50(2):241–6. 10.1086/649541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, et al. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010. March 15;50(6):882–9. 10.1086/650733 [DOI] [PubMed] [Google Scholar]
- 3.Buckle GC, Walker CLF, Black RE. Typhoid fever and paratyphoid fever: Systematic review to estimate global morbidity and mortality for 2010. J Glob Health. 2012;2: 010401 10.7189/jogh.02.010401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull World Health Organ. 2004. May;82(5):346–53. [PMC free article] [PubMed] [Google Scholar]
- 5.Bhan MK, Bahl R, Bhatnagar S. Typhoid and paratyphoid fever. Lancet. 2005. September 27;366(9487):749–62. 10.1016/S0140-6736(05)67181-4 [DOI] [PubMed] [Google Scholar]
- 6.Ochiai RL, Wang X, von Seidlein L, Yang J, Bhutta ZA, Bhattacharya SK, et al. Salmonella Paratyphi A Rates, Asia. Emerg Infect Dis. 2005. November;11(11):1764–6. 10.3201/eid1111.050168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ao TT, Feasey NA, Gordon MA, Keddy KH, Angulo FJ, Crump JA. Global burden of invasive nontyphoidal Salmonella disease, 2010(1). Emerging Infect Dis. 2015. June;21(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feasey NA, Hadfield J, Keddy KH, Dallman TJ, Jacobs J, Deng X, et al. Distinct Salmonella Enteritidis lineages associated with enterocolitis in high-income settings and invasive disease in low-income settings. Nat Genet. 2016;48(10):1211–7. 10.1038/ng.3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okoro CK, Kingsley RA, Connor TR, Harris SR, Parry CM, Al-Mashhadani MN, et al. Intra-continental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat Genet. 2012. November;44(11):1215–21. 10.1038/ng.2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou Q-H, Li R-Q, Liu G-R, Liu S-L. Comparative genomic analysis between typhoidal and non-typhoidal Salmonella serovars reveals typhoid-specific protein families. Infect Genet Evol. 2014. August;26:295–302. 10.1016/j.meegid.2014.06.008 [DOI] [PubMed] [Google Scholar]
- 11.Nuccio S-P, Bäumler AJ. Comparative Analysis of Salmonella Genomes Identifies a Metabolic Network for Escalating Growth in the Inflamed Gut. mBio. 2014. May 1;5(2):e00929–14. 10.1128/mBio.00929-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L, Tan J, O’Brien EJ, Monk JM, Kim D, Li HJ, et al. Systems biology definition of the core proteome of metabolism and expression is consistent with high-throughput data. Proc Natl Acad Sci USA. 2015. August 25;112(34):10810–5. 10.1073/pnas.1501384112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Idelevich EA, Schüle I, Grünastel B, Wüllenweber J, Peters G, Becker K. Rapid identification of microorganisms from positive blood cultures by MALDI-TOF mass spectrometry subsequent to very short-term incubation on solid medium. Clin Microbiol Infect. 2014. October;20(10):1001–6. 10.1111/1469-0691.12640 [DOI] [PubMed] [Google Scholar]
- 14.Brown RN, Sanford JA, Park JH, Deatherage BL, Champion BL, Smith RD, et al. A Comprehensive Subcellular Proteomic Survey of Salmonella Grown under Phagosome-Mimicking versus Standard Laboratory Conditions. Int J Proteomics. 2012;2012:123076 10.1155/2012/123076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim K, Yang E, Vu G-P, Gong H, Su J, Liu F, et al. Mass spectrometry-based quantitative proteomic analysis of Salmonella enterica serovar Enteritidis protein expression upon exposure to hydrogen peroxide. BMC Microbiology. 2010. June 8;10:166 10.1186/1471-2180-10-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi L, Adkins JN, Coleman JR, Schepmoes AA, Dohnkova A, Mottaz HM, et al. Proteomic analysis of Salmonella enterica serovar typhimurium isolated from RAW 264.7 macrophages: identification of a novel protein that contributes to the replication of serovar typhimurium inside macrophages. J Biol Chem. 2006. September 29;281(39):29131–40. 10.1074/jbc.M604640200 [DOI] [PubMed] [Google Scholar]
- 17.Coldham NG, Randall LP, Piddock LJV, Woodward MJ. Effect of fluoroquinolone exposure on the proteome of Salmonella enterica serovar Typhimurium. J Antimicrob Chemother. 2006. December;58(6):1145–53. 10.1093/jac/dkl413 [DOI] [PubMed] [Google Scholar]
- 18.Niemann GS, Brown RN, Gustin JK, Stufkens A, Shaikh-Kidwai AS, Li J, et al. Discovery of novel secreted virulence factors from Salmonella enterica serovar Typhimurium by proteomic analysis of culture supernatants. Infect Immun. 2011. January;79(1):33–43. 10.1128/IAI.00771-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Webber MA, Coldham NG, Woodward MJ, Piddock LJV. Proteomic analysis of triclosan resistance in Salmonella enterica serovar Typhimurium. J Antimicrob Chemother. 2008. July;62(1):92–7. 10.1093/jac/dkn138 [DOI] [PubMed] [Google Scholar]
- 20.Charles RC, Harris JB, Chase MR, Lebrun LM, Sheikh A, LaRocque RC, et al. Comparative proteomic analysis of the PhoP regulon in Salmonella enterica serovar Typhi versus Typhimurium. PLoS ONE. 2009. September 10;4(9):e6994 10.1371/journal.pone.0006994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Y, Chien K-Y, Chen H-L, Chiu C-H. Pseudogene recoding revealed from proteomic analysis of Salmonella serovars. J Proteome Res. 2012. March 2;11(3):1715–9. 10.1021/pr200904c [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Huang K-Y, Huo Y. Proteomic comparison between Salmonella Typhimurium and Salmonella Typhi. J Microbiol. 2014. January;52(1):71–6. 10.1007/s12275-014-3204-3 [DOI] [PubMed] [Google Scholar]
- 23.Neidhardt FC, Bloch PL, Smith DF. Culture Medium for Enterobacteria. J Bacteriol. 1974. September;119(3):736–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotech. 2008. December;26(12):1367–72. [DOI] [PubMed] [Google Scholar]
- 25.Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001. October 25;413(6858):848–52. 10.1038/35101607 [DOI] [PubMed] [Google Scholar]
- 26.McClelland M, Sanderson KE, Clifton SW, Latreille P, Porwollik S, Sabo A, et al. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat Genet. 2004. December;36(12):1268–74. 10.1038/ng1470 [DOI] [PubMed] [Google Scholar]
- 27.Jarvik T, Smillie C, Groisman EA, Ochman H. Short-Term Signatures of Evolutionary Change in the Salmonella enterica Serovar Typhimurium 14028 Genome. J Bacteriol. 2010. January;192(2):560–7. 10.1128/JB.01233-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomson NR, Clayton DJ, Windhorst D, Vernikos G, Davidson S, Churcher C, et al. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res. 2008. October;18(10):1624–37. 10.1101/gr.077404.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altenhoff AM, Škunca N, Glover N, Train C-M, Sueki A, Piližota I, et al. The OMA orthology database in 2015: function predictions, better plant support, synteny view and other improvements. Nucleic Acids Res. 2015. January;43(Database issue):D240–249. 10.1093/nar/gku1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 31.De Maeyer D, Weytjens B, Renkens J, De Raedt L, Marchal K. PheNetic: network-based interpretation of molecular profiling data. Nucl Acids Res. 2015. April 15;gkv347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001. October 25;413(6858):852–6. 10.1038/35101614 [DOI] [PubMed] [Google Scholar]
- 33.Nandre RM, Mahajan P. Molecular Significance of lon and cpxR Genes in the Pathogenicity of Salmonella. Open Journal of Animal Sciences. 2015. September 23;05(04):429. [Google Scholar]
- 34.Dowhan W. A retrospective: Use of Escherichia coli as a vehicle to study phospholipid synthesis and function. Biochim Biophys Acta. 2013. March;1831(3):471–94. 10.1016/j.bbalip.2012.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou P, Zhao J. Structure, Inhibition, and Regulation of Essential Lipid A Enzymes. Biochim Biophys Acta. 2017. November;1862(11):1424–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bang I-S, Frye JG, McClelland M, Velayudhan J, Fang FC. Alternative sigma factor interactions in Salmonella: sigma and sigma promote antioxidant defences by enhancing sigma levels. Mol Microbiol. 2005. May;56(3):811–23. 10.1111/j.1365-2958.2005.04580.x [DOI] [PubMed] [Google Scholar]
- 37.Cho Y, Park YM, Barate AK, Park S-Y, Park HJ, Lee MR, et al. The role of rpoS, hmp, and ssrAB in Salmonella enterica Gallinarum and evaluation of a triple-deletion mutant as a live vaccine candidate in Lohmann layer chickens. Journal of Veterinary Science. 2015. June;16(2):187 10.4142/jvs.2015.16.2.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kazmierczak MJ, Wiedmann M, Boor KJ. Alternative Sigma Factors and Their Roles in Bacterial Virulence. Microbiol Mol Biol Rev. 2005. December;69(4):527–43. 10.1128/MMBR.69.4.527-543.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Botsford JL, Harman JG. Cyclic AMP in prokaryotes. Microbiol Rev. 1992. March;56(1):100–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vartak NB, Reizer J, Reizer A, Gripp JT, Groisman EA, Wu LF, et al. Sequence and evolution of the FruR protein of Salmonella typhimurium: a pleiotropic transcriptional regulatory protein possessing both activator and repressor functions which is homologous to the periplasmic ribose-binding protein. Res Microbiol. 1991. December;142(9):951–63. [DOI] [PubMed] [Google Scholar]
- 41.Rahman M, Siddique AK, Tam FC-H, Sharmin S, Rashid H, Iqbal A, et al. Rapid detection of early typhoid fever in endemic community children by the TUBEX O9-antibody test. Diagnostic Microbiology and Infectious Disease. 2007. July 1;58(3):275–81. 10.1016/j.diagmicrobio.2007.01.010 [DOI] [PubMed] [Google Scholar]
- 42.Thriemer K, Ley B, Menten J, Jacobs J, van den Ende J. A systematic review and meta-analysis of the performance of two point of care typhoid fever tests, Tubex TF and Typhidot, in endemic countries. PLoS ONE. 2013;8(12):e81263 10.1371/journal.pone.0081263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuijpers LMF, Chung P, Peeters M, Phoba M-F, Kham C, Barbé B, et al. Diagnostic accuracy of antigen-based immunochromatographic rapid diagnostic tests for the detection of Salmonella in blood culture broth. PLoS One 13(3): e0194024 10.1371/journal.pone.0194024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahon BE, Newton AE, Mintz ED. Effectiveness of typhoid vaccination in US travelers. Vaccine. 2014. June 17;32(29):3577–9. 10.1016/j.vaccine.2014.04.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galán JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006. November 30;444(7119):567–73. 10.1038/nature05272 [DOI] [PubMed] [Google Scholar]
- 46.Monjarás Feria JV, Lefebre MD, Stierhof Y-D, Galán JE, Wagner S. Role of autocleavage in the function of a type III secretion specificity switch protein in Salmonella enterica serovar Typhimurium. MBio. 2015. October 13;6(5):e01459–01415. 10.1128/mBio.01459-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patel JC, Galán JE. Manipulation of the host actin cytoskeleton by Salmonella—all in the name of entry. Curr Opin Microbiol. 2005. February;8(1):10–5. 10.1016/j.mib.2004.09.001 [DOI] [PubMed] [Google Scholar]
- 48.Kim JS, Eom JS, Jang JI, Kim HG, Seo DW, Bang I-S, et al. Role of Salmonella Pathogenicity Island 1 protein IacP in Salmonella enterica serovar typhimurium pathogenesis. Infect Immun. 2011. April;79(4):1440–50. 10.1128/IAI.01231-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olsen JE, Hoegh-Andersen KH, Casadesús J, Rosenkranzt J, Chadfield MS, Thomsen LE. The role of flagella and chemotaxis genes in host pathogen interaction of the host adapted Salmonella enterica serovar Dublin compared to the broad host range serovar S. Typhimurium. BMC Microbiol. 2013. March 25;13:67 10.1186/1471-2180-13-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ikeda JS, Schmitt CK, Darnell SC, Watson PR, Bispham J, Wallis TS, et al. Flagellar Phase Variation of Salmonella enterica Serovar Typhimurium Contributes to Virulence in the Murine Typhoid Infection Model but Does Not Influence Salmonella-Induced Enteropathogenesis. Infect Immun. 2001. May;69(5):3021–30. 10.1128/IAI.69.5.3021-3030.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y, Zhang Q, Hu M, Yu K, Fu J, Zhou F, et al. Proteomic Analyses of Intracellular Salmonella enterica Serovar Typhimurium Reveal Extensive Bacterial Adaptations to Infected Host Epithelial Cells. Infect Immun. 2015. July;83(7):2897–906. 10.1128/IAI.02882-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lam O, Wheeler J, Tang CM. Thermal control of virulence factors in bacteria: A hot topic. Virulence. 2014. December 10;5(8):852–62. 10.4161/21505594.2014.970949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Espadas G, Borràs E, Chiva C, Sabidó E. Evaluation of different peptide fragmentation types and mass analyzers in data-dependent methods using an Orbitrap Fusion Lumos Tribrid mass spectrometer. Proteomics. 2017. May;17(9). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the PRIDE Archive (http://www.ebi.ac.uk/pride/archive/) via the PRIDE partner repository with the data set identifier PXD011154