Organismal physiology emerges from metabolic pathways and structures that can vary across development and among individuals. Matoo, Julick, and Montooth found significant variation, both genetic and ontogenetic, in mitochondrial physiology in wild-type and mitochondrial-nuclear...
Keywords: development, mtDNA, metabolism, oxidative phosphorylation, reactive oxygen species
Abstract
Organismal physiology emerges from metabolic pathways and subcellular structures like the mitochondria that can vary across development and among individuals. Here, we tested whether genetic variation at one level of physiology can be buffered at higher levels of biological organization during development by the inherent capacity for homeostasis in physiological systems. We found that the fundamental scaling relationship between mass and metabolic rate, as well as the oxidative capacity per mitochondria, changed significantly across development in the fruit fly Drosophila. However, mitochondrial respiration rate was maintained at similar levels across development. Furthermore, larvae clustered into two types—those that switched to aerobic, mitochondrial ATP production before the second instar, and those that relied on anaerobic, glycolytic production of ATP through the second instar. Despite genetic variation for the timing of this metabolic shift, metabolic rate in second-instar larvae was more robust to genetic variation than was the metabolic rate of other instars. We found that larvae with a mitochondrial-nuclear incompatibility that disrupts mitochondrial function had increased aerobic capacity and relied more on anaerobic ATP production throughout development relative to larvae from wild-type strains. By taking advantage of both ways of making ATP, larvae with this mitochondrial–nuclear incompatibility maintained mitochondrial respiratory capacity, but also had higher levels of whole-body reactive oxygen species and decreased mitochondrial membrane potential, potentially as a physiological defense mechanism. Thus, genetic defects in core physiology can be buffered at the organismal level via physiological plasticity, and natural populations may harbor genetic variation for distinct metabolic strategies in development that generate similar organismal outcomes.
METABOLISM is the sum total of biochemical processes that organisms use to sustain life and fuel reproduction, and an individual’s metabolic rate is often interpreted as an integrated measure of its “pace of life” (Glazier 2005, 2014, 2015). Early surveys of natural molecular variation revealed a surprising amount of variation at loci encoding these biochemical processes (Harris 1966; Hubby and Lewontin 1966; Lewontin and Hubby 1966)—a pattern that has been historically used to advocate both for the predominance of classical mutation, drift, and purifying selection forces (Kimura 1983), and for the maintenance of variation through selection (Gillespie 1999; reviewed by Charlesworth and Charlesworth 2016). Subsequent surveys revealed substantial quantitative genetic variation in metabolic enzyme activities within species, arising from molecular variation at enzyme-encoding loci, as well as trans-acting and epistatic variation throughout the genome (Laurie-Ahlberg et al. 1980, 1982; Clark et al. 1995a,b; Clark and Wang 1997; Mitchell-Olds and Pedersen 1998; Montooth et al. 2003). In a few cases, this biochemical variation has been linked to variation at higher levels of physiological performance (Laurie-Ahlberg et al. 1982; Watt et al. 1983; Montooth et al. 2003; Crawford and Oleksiak 2007), and, in some cases, may be adaptive (Watt 1977; Tishkoff et al. 2001; Verrelli and Eanes 2001; Watt et al. 2003). However, we lack a mechanistic understanding of how genetic variation in the pathways of metabolism is transformed up the hierarchical levels of biological organization to result in variation in organismal performance traits that determine fitness. This is important for consideration of metabolism as an adaptive phenotype and for predicting how selection on metabolic performance will shape variation in genomes.
A challenge to connecting genetic variation in biochemical processes to metabolic performance, is that metabolism is an emergent property of interacting biochemical, structural, regulatory, and physiological systems, often arranged in hierarchical functional modules (Jeong et al. 2000; Strogatz 2001; Ravasz et al. 2002; Barabási and Oltvai 2004). In addition, metabolic enzymes and metabolites have potential “moonlighting” functions in the signaling that underlies metabolic homeostasis (Marden 2013; Boukouris et al. 2016). The capacity for homeostasis in physiological systems also suggests that genetic variation in biochemical processes may not necessarily result in organismal fitness variation. The regulatory processes that maintain energy homeostasis may provide stability in metabolic trajectories in an analogous way to the canalized developmental trajectories envisioned by Waddington (1942, 1957) and Meiklejohn and Hartl (2002). Furthermore, multiple biochemical pathways may be available to achieve similar energetic outputs. Finally, the hierarchical biological processes that contribute to metabolism are influenced by both extrinsic (e.g., temperature, resource availability, habitat) and intrinsic (e.g., genotype, life stage, sex, reproductive status) factors (reviewed by Glazier 2005), such that genetic variation in biochemical processes may affect organismal performance and fitness in only a subset of conditions. Such conditionally neutral variation is expected to experience less effective selection (Van Dyken and Wade 2010).
Development is a potentially important context for the expression of genetic variation in metabolism. During development, organisms partition resources between the competing demands of growth, development, maintenance, and storage for future reproduction. Energy homeostasis during development is largely achieved by feedback controls where energy-demand processes increase the concentration of ADP, which is then available for energy-supply processes to generate ATP. The mitochondria play a central role in the energy supply-demand balance. Not only the abundance and activity of mitochondria, but also the surface area, membrane composition, and network structure of mitochondria have been reported to affect metabolism (Porter and Brand 1993; Porter et al. 1996; Miettinen and Björklund 2017). Oxidative phosphorylation (OXPHOS), which drives aerobic ATP production, requires proteins from both the mitochondrial and nuclear genomes, creating the potential for intergenomic epistasis to underlie variation in metabolic phenotypes. Our understanding of the underlying genetic architecture of metabolism is incomplete, but studies indicate that both nuclear DNA (nDNA) (Montooth et al. 2003; Nespolo et al. 2007; Tieleman et al. 2009), mitochondrial DNA (mtDNA) (Martin 1995; Ballard and Rand 2005; Arnqvist et al. 2010; Kurbalija Novičić et al. 2015), and interactions between genomes and environment affect metabolism (Hoekstra et al. 2013, 2018).
Energy balance is particularly challenging for holometabolous species with rapid and massive larval growth that requires simultaneous accumulation of the resources needed to fuel metamorphosis. Drosophila melanogaster is an especially powerful system to study developmental metabolism, given the genetic resources and an ∼200-fold increase in body mass across three larval instars (Church and Robertson 1966). There is evidence of significant genetic variation for metabolic rate within Drosophila (Montooth et al. 2003; Hoekstra et al. 2013), and individuals with mitochondrial–nuclear genotypes that disrupt mitochondrial function have increased larval metabolic rate and delayed development (Hoekstra et al. 2013; Meiklejohn et al. 2013). Transcriptomic and metabolic profiling in D. melanogaster reveal the dynamic nature of energy metabolism that draws on both aerobic and anaerobic energy production, as well as the presence of proliferative metabolic programs during larval development (Graveley et al. 2011; Tennessen et al. 2011). Despite this wealth of data, we lack a detailed understanding of the links between genome variation, mitochondrial function, and organismal metabolic rate during development in Drosophila.
In the present study, we tested whether metabolic strategies in D. melanogaster varied across larval instars, using both wild-type larvae and larvae with a mitochondrial-nuclear genotype that compromises mitochondrial function. We observed significant variation in the ontogeny of metabolism at the level of mitochondrial aerobic capacity and ATP production, but also observed that this variation was buffered at higher levels of metabolic performance via physiological plasticity. Thus, there may be multiple genotypic and physiological paths to equivalent organismal outcomes within populations.
Methods
Drosophila stocks and maintenance
We used four Drosophila mitochondrial-nuclear (hereafter referred to as mito-nuclear) genotypes generated by Montooth et al. (2010). Individuals with the (mtDNA);nuclear genotype (simw501);OreR have a genetic incompatibility that decreases OXPHOS activity putatively via compromised mitochondrial protein translation, resulting in delayed development, decreased immune function, and reduced female fecundity (Hoekstra et al. 2013, 2018; Meiklejohn et al. 2013; Holmbeck et al. 2015; Zhang et al. 2017; Buchanan et al. 2018). The mito-nuclear incompatibility is between naturally occurring single nucleotide polymorphisms (SNPs) in the mt-tRNATyr gene and the nuclear-encoded mt-tyrosyl-tRNA synthetase gene Aatm that aminoacylates the mt-tRNATyr (Meiklejohn et al. 2013). Individuals with the mito-nuclear genotypes—(ore);OreR, (simw501);Aut, and (ore);Aut—serve as genetic controls that enable us to test for the effects of mitochondrial and nuclear genotypes, and their interaction on developmental physiology. Additionally, we measured traits in two inbred isofemale fly strains sampled in Vermont (VT4 and VT10) as representatives of natural populations that were not manipulated to generate specific mito-nuclear genotypes (for details see Cooper et al. 2014).
Flies from all strains were raised on standard cornmeal-molasses-yeast Drosophila medium and acclimated to 25° with a 12:12 hr dark:light cycle for at least three generations prior to all experiments. To collect first-, second- and third-instar larvae, adults were allowed to lay eggs for 3–4 hr on standard media, and larvae from these cohorts were staged based on developmental time and distinguishing morphological features.
Larval metabolic rate
Routine metabolic rate was measured as the rate of CO2 produced by groups of 20 larvae of the same instar and strain using a flow-through respirometry system (Sable Systems International, Henderson, NV) (Hoekstra et al. 2013). Groups of larvae were collected onto the cap of 1.7 ml tube containing 0.5 ml of fly medium and placed inside one of four respirometry chambers that were housed in a temperature-controlled cabinet (Tritech Research, Los Angeles, CA) maintained at 25°. Between 8 and 13 biological replicates for each strain and instar were randomized across chambers and respirometry runs, during which each group of larvae was sampled for CO2 production for two 10-min periods. CO2 that accumulated in the chambers as a result of larval metabolism was detected using an infrared CO2 analyzer (Li-Cor 7000 CO2/H2O Analyzer; LI-COR, Lincoln, NE). was calculated from the mean fractional increase in CO2 at a constant air-flow rate of 100 ml/min over a 10-min time interval for each replicate after baseline-drift correction. The wet weight of the group of larvae was recorded using a Cubis microbalance (Sartorius AG, Göttingen, Germany) at the beginning of each respirometry run.
Larval water and lipid content
We measured larval whole-body water content using a protocol modified from Grefen et al. (2006). Six groups of 10 larvae each were collected for each strain at each instar, rinsed with larval wash buffer (0.7% NaCl and 0.1% Triton X-100), and blotted dry. The wet weight of the group of larvae was recorded (±0.01 mg) using a Cubis microbalance (Sartorius AG, Göttingen, Germany). Larvae were dried at 60° overnight and reweighed. The mass of whole-body water was calculated by subtracting dry mass from wet mass.
Larval whole-body lipid content was measured using a protocol modified from Bligh and Dyer (1959). Six groups of 10 larvae each were collected for each strain at each instar, rinsed with larval wash buffer, blotted dry, and weighed (±0.01 mg). Larvae were homogenized in a chloroform/methanol mixture (2:1 v/v) using tissue:solvent proportion of 1:20 w/v. Samples were sonicated for 1 min using a Sonic Dismembrators (ThermoFisher Scientific), vortexed for 1 hr at room temperature, and then centrifuged for 5 min at 13,000 × g. The supernatant was transferred in a new tube, mixed with ultrapure water (0.25 vol of the supernatant), vortexed for 2 min, and centrifuged for 5 min at 13,000 × g. The lower phase (chloroform) was transferred into a preweighed microcentrifuge tube and chloroform was allowed to evaporate to determine the dry mass of extracted lipids using a microbalance.
Isolation of mitochondria
Mitochondria were isolated from larvae following a protocol modified from Aw et al. (2016); 100–200 larvae for each biological replicate were collected and rinsed with larval wash buffer (0.7% NaCl and 0.1% Triton X-100). Larvae were gently homogenized in 300–500 µl of chilled isolation buffer (154 mM KCl, 1 mM EDTA, pH 7.4) in a glass-teflon Thomas® homogenizer on ice. The homogenate was filtered through a nylon cloth into a clean, chilled, microcentrifuge tube. The homogenate was then centrifuged at 1500 × g for 8 min at 4°. The resulting mitochondrial pellet was suspended in 40–50 µl of ice-cold mitochondrial assay solution (MAS: 15 mM KCl, 10 mM KH2PO4, 2 mM MgCl2, 3 mM HEPES, 1 mM EGTA, FA-free BSA 0.2%, pH 7.2). Unless otherwise stated, all chemicals were purchased from Sigma Aldrich (St Louis, MO) or Fisher Scientific (Pittsburgh, PA) and were of reagent grade or higher.
Mitochondrial respiration
Oxygen consumption of freshly isolated mitochondria was measured using the Oxygraph Plus System (Hansatech Instruments, Norfolk, UK) in 3 ml water-jacketed glass chambers equipped with a magnetic stirrer and Clark-type oxygen electrodes. Temperature of the respiration chambers was kept constant at 25° using a Fisher Isotemp 4100 R20 refrigerated water circulator (Fisher Scientific, Hampton, NH). A two-point calibration of electrodes using air-saturated distilled water and sodium sulfite was done for establishing 100% and zero oxygen levels in the chamber, respectively. The assay was completed within 2 hr of mitochondrial isolation, and six or seven biological replicates were measured for each larval stage of each strain. Mitochondrial suspension (50 µl; ∼1.5 mg protein) was added to 950 µl of MAS in the respiration chamber. Pyruvate (5 mM) and malate (2.5 mM) were used as respiratory substrates at saturating amounts. Maximum respiration (State 3) was achieved by adding 400 µM of ADP, and State 4ol respiration was calculated as described by Chance and Williams (1955) by adding 2.5 µg ml−1 oligomycin. Oligomycin is an ATPase inhibitor and State 4ol gives an estimate of oxygen consumption linked to mitochondrial proton leak, rather than to ATP production, at high membrane potential (Brand et al. 1994). Uncoupled respiration (State 3u), indicative of maximum respiration or electron transport system (ETS) capacity, is achieved by adding 0.5 µM of carbonyl cyanide m-chlorophenyl hydrazone (CCCP). CCCP is a protonophore that increases proton permeability in mitochondria and effectively disconnects ETS from ATPase. Data were acquired and respiration rates were corrected for electrode drift using the OxyTrace+ software. The respiratory control ratio (RCR+) was calculated as the ratio of State 3 over State 4ol (Estabrook 1967). Respiration rates were normalized by unit mitochondrial protein added. Protein concentrations were determined using Bio-Rad Protein Assay Dye Reagent Concentrate (5000006; Bio-Rad) and bovine serum albumin (BSA) as a standard.
Mitochondrial membrane potential (ΔΨm)
Mitochondrial membrane potential was measured using the JC-1 indicator dye (Fisher Scientific) following a protocol modified from Villa-Cuesta et al. (2014); 100 mg of larvae were weighed and used to isolate mitochondria as described above. Approximately 1.4 mg of mitochondrial protein was added, and the final volume was increased to 300 µl using MAS. Next, 3 µl of a 1 µg/µl solution of JC-1 dissolved in dimethyl sulfoxide (DMSO) was added to the suspension. Mitochondrial samples were incubated for 30 min at 37° protected from light. At the end of incubation, samples were centrifuged for 3 min at 6000 × g and suspended in 600 µl of fresh MAS. Mitochondrial membrane potential was expressed as the ratio of fluorescence for aggregate:monomeric forms of JC-1 at red (excitation 485 nm, emission 600 nm) and green (excitation 485 nm, emission 530 nm) wavelengths respectively; 50 μM of CCCP was added to collapse membrane potential as a negative control.
Citrate synthase activity
Citrate synthase activity was measured following the protocol from Meiklejohn et al. (2013); 100–200 larvae were homogenized in 1 ml chilled isolation buffer (225 mM mannitol, 75 mM sucrose, 10 mM MOPS, 1 mM EGTA, 0.5% fatty acid-free BSA, pH 7.2) using a glass-teflon Thomas® homogenizer. The homogenate was centrifuged at 300 × g for 5 min at 4°. The supernatant was transferred into a clean tube and centrifuged again at 6000 × g for 10 min at 4°. The resulting mitochondrial pellet was resuspended in 50 µl of respiration buffer (225 mM mannitol, 75 mM sucrose, 10 mM KCl, 10 mM Tris-HCl, and 5 mM KH2PO4, pH 7.2). All samples were stored at −80° till further analysis.
Maximum citrate synthase activity (Vmax) of the mitochondrial extracts was measured spectrophotometrically at 25° using a Synergy 2 plate reader (BioTek); 6 µg of mitochondrial protein was added to the assay mixture containing 100 mM Tris-HCl (pH 8.0), 2.5 mM EDTA, 100 µM Acetyl Co-A, and 100 µM of DTNB [5,5′-dithiobis (2-nitrobenzoic acid)]. The reaction was monitored for 2 min as a background reading. The reaction was then started by adding 500 µM oxaloacetate to the assay to generate CoA-SH. CoA-SH was detected by its reaction with DTNB to form a yellow product (mercaptide ion) that was measured using absorbance at 412 nm. Enzyme activity was normalized by protein concentration of the sample added. Six biological samples per strain and instar were measured, each with two technical replicates.
Lactate quantification
Whole-body lactate concentrations were measured by an NAD+/NADH-linked fluorescent assay following the protocol of Callier et al. (2015); 100–200 larvae were homogenized in 100–500 µl of 17.5% perchloric acid and centrifuged at 14,000 × g for 2 min at 4°. Following precipitation of proteins, the clear supernatant was transferred into a clean tube and neutralized with a buffer containing 2 M KOH and 0.3 M MOPS, and again centrifuged at 14,000 × g for 2 min at 4°. Neutralized sample (20–50 µl) was added to the assay buffer (pH 9.5) containing a final concentration of 1000 mM hydrazine, 100 mM Tris-base, 1.4 mM EDTA, and 2.5 mM NAD+ in a 96-well plate. The assay was performed in fluorescence mode (Ex/Em = 360/460 nm) using a Synergy 1H Hybrid Reader (BioTek). After incubating the plate for 5 min at room temperature, a background reading was taken. Lactate dehydrogenase (17.5 U/well; L3916; Sigma) diluted with Tris buffer was then added to each sample, and the reaction mixture was allowed to incubate at 37° for 30 min protected from light. A second reading was taken to measure NADH levels, after correcting for background fluorescence. Six biological samples per strain and instar were measured, each with two technical replicates. Sodium lactate was used as a standard for the assay. Lactate concentrations in the samples were normalized by the wet weight of the larvae.
Hydrogen peroxide quantification
Larvae (100–200) were weighed, rinsed with larval wash buffer (0.7% NaCl and 0.1% Triton X-100), and homogenized in 500 µl of prechilled assay buffer (pH 7.5) containing 20 mM HEPES, 100 mM KCl, 5% glycerol, 10 mM EDTA, 0.1% Triton X-100, 1 mM PMSF (P7626; Sigma), and 1:10 (v/v) protease inhibitor cocktail (P2714; Sigma) using a glass-teflon Thomas homogenizer. The homogenate was centrifuged at 200 × g for 5 min at 4°, and the supernatant was stored at −80°. Hydrogen peroxide (H2O2) concentration was determined with a fluorometric H2O2 Assay Kit (MAK 165; Sigma) following the manufacturer’s protocol in a 96-well plate using the Synergy H1 Hybrid Reader. Six biological samples per strain and instar were measured, each with two technical replicates. H2O2 concentrations in the samples were expressed as nM/µg of protein. The mitochondria serve as both source and sink of reactive oxygen species (ROS) in the organism (Munro and Treberg 2017). Thus, we used this whole-body measure of H2O2 as an estimate of the organismal consequences of mitochondrial function.
Statistical analyses
All statistical analyses used the statistical package R version 2.15.1 (R Development Core Team 2011). We implemented standard major-axis regression in the R-package SMATR (Warton et al. 2006; Hoekstra et al. 2013) to estimate the relationship between log-transformed mass and , and to test for larval-instar and genetic effects on the slope of this relationship. When there was statistical evidence for a common slope among genotypes or strains, we fit the common slope to test for effects of genotype or strain on the y-intercept (i.e., genetic effects on the mass-specific metabolic rate). We removed a single observation where a first-instar replicate had a value less than zero. We also used SMATR to analyze the scaling of total lipid plus water content with dry body mass. ANOVA was used to test for the fixed effects of mtDNA, nuclear genome, larval instar, and all interactions on lactate accumulation, H2O2 concentration, mitochondrial physiology (State3, State4ol, uncoupled respiration, RCR+, and ΔΨm) and citrate synthase activity using averages across technical replicates. Post hoc comparisons among instars within a genotype or strain, and among genotypes or strains within instar were evaluated using Tukey HSD tests that were corrected for multiple testing.
Data availability
Stocks and strains are available upon request. Supplemental files including all phenotype data are available at FigShare. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.7946795.
Results
Metabolic rate scaling with mass varies across larval instars and strains
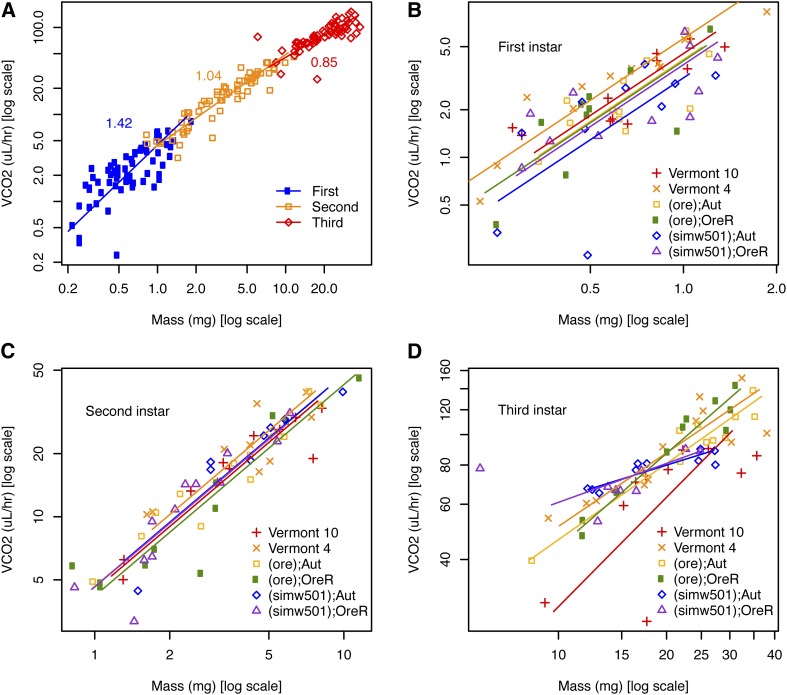
Metabolic rate scales with mass according to the power function R = aMb, where a is the constant scaling coefficient, M is mass, and b is the scaling exponent (Kleiber 1932). The scaling exponent b, estimated by the slope of the relationship between log-transformed metabolic rate and mass, differed significantly across larval instars (Figure 1A) (LR = 18.1, df = 2, P = 0.0001). Metabolic scaling with body mass was hypermetric in first-instar larvae [b (CI) = 1.42 (1.21, 1.67)], isometric in second-instar larvae [b = 1.04 (0.95, 1.15)], and hypometric in third-instar larvae [b = 0.85 (0.71, 1.01)]. Within first- and second-instar larvae, there was no evidence that metabolic scaling with mass differed significantly among strains, nor were there significant effects of strain on the elevation of the fitted relationship (i.e., on the mass-specific metabolic rate) (Figure 1, B and C and Table 1). However, there was more variance among strains in mass-specific metabolic rate in first-instar larvae relative to second-instar larvae (Figure 1, B and C and Table 1). Metabolic scaling with mass in third-instar larvae differed significantly among strains, as evidenced by significantly different slopes (Figure 1D and Table 1). The variation in metabolic scaling with mass did not result from natural strains differing from mito-nuclear genotypes, but rather from variation in the scaling exponent within both groups. The pattern was significant regardless of the inclusion of several data points that, while not statistical outliers, did appear as outliers in the relationship between metabolic rate and mass (Figure 1D and Supplemental Material, Table S1).
Figure 1.
Metabolic scaling with mass varied across larval development and among strains. (A) The mass-scaling exponent for routine metabolic rate differed significantly among larvae from different instars (LR = 18.1, df = 2, P = 0.0001), with the relationship between metabolic rate and mass becoming more shallow across development. (B and C) There was more genetic variation for metabolic rate in first-instar larvae, relative to second-instar larvae. (D) Mass-scaling exponents differed significantly among strains in the third instar of development (Table 1 and Table S1).
Table 1. Ontogenetic and genetic effects on the scaling of routine metabolic rate (RMR) as a function of mass.
| Phenotype | Strain | Slope (95% CI)a | Y-interceptb |
|---|---|---|---|
| First-instar RMR | H0: equal slopes (LR = 4.61, df = 5, P = 0.46) | H0: no elevation difference (Wald = 9.28, df = 5, P = 0.10) | |
| Common slope | 1.29 (1.10, 1.54) | ||
| VT10 | 0.66 (0.54, 0.77) | ||
| VT4 | 0.75 (0.64, 0.86) | ||
| (ore);Aut | 0.62 (0.51, 0.73) | ||
| (ore);OreR | 0.61 (0.43, 0.79) | ||
| (simw501);Aut | 0.50 (0.26, 0.75) | ||
| (simw501);OreR | 0.59 (0.42, 0.75) | ||
| Second-instar RMR | H0: equal slopes (LR = 3.99, df = 5, P = 0.55) | H0: no elevation difference (Wald = 2.58, df = 5, P = 0.76) | |
| Common slope | 1.01 (0.90, 1.13) | ||
| VT10 | 0.65 (0.55, 0.75) | ||
| VT4 | 0.71 (0.6, 0.81) | ||
| (ore);Aut | 0.67 (0.58, 0.75) | ||
| (ore);OreR | 0.62 (0.51, 0.73) | ||
| (simw501);Aut | 0.67 (0.55, 0.79) | ||
| (simw501);OreR | 0.67 (0.57, 0.76) | ||
| Third-instar RMR | H0: equal slopes (LR = 20.1, df = 5, P = 0.001) | ||
| VT10 | 1.16 (0.65, 2.06) | ||
| VT4 | 0.78 (0.54, 1.10) | ||
| (ore);Aut | 0.81 (0.65, 1.00) | ||
| (ore);OreR | 1.00 (0.81, 1.24) | ||
| (simw501);Aut | 0.36 (0.25, 0.52) | ||
| (simw501);OreR | 0.41 (0.19, 0.89) |
Either strain-specific slopes or a common slope with confidence interval, when justified by the test for equal slopes among strains.
In no case was there evidence that there was a shift in mass along the x-axis among strains (P > 0.09).
To get an estimate of the extent to which changes in body composition were underlying ontogenetic differences in the mass scaling of metabolic rate, we tested whether the relationship between lipid plus water content and dry mass differed among instars. Lipid and water are metabolically less active components of body mass, and we predicted that the decreased scaling of metabolic rate with mass in third-instar larvae could be the result of a greater amount of this metabolically less-active mass. We found a significant effect of instar on the y-intercept of the relationship between lipid plus water content and dry mass (Figure S1 and Table S2). Third-instar larvae did have greater lipid plus water content as a function of dry mass relative to second-instar larvae. However, first-instar larvae had values of this parameter similar to third-instar larvae.
Mitochondrial respiration is similar across larval instars and strains
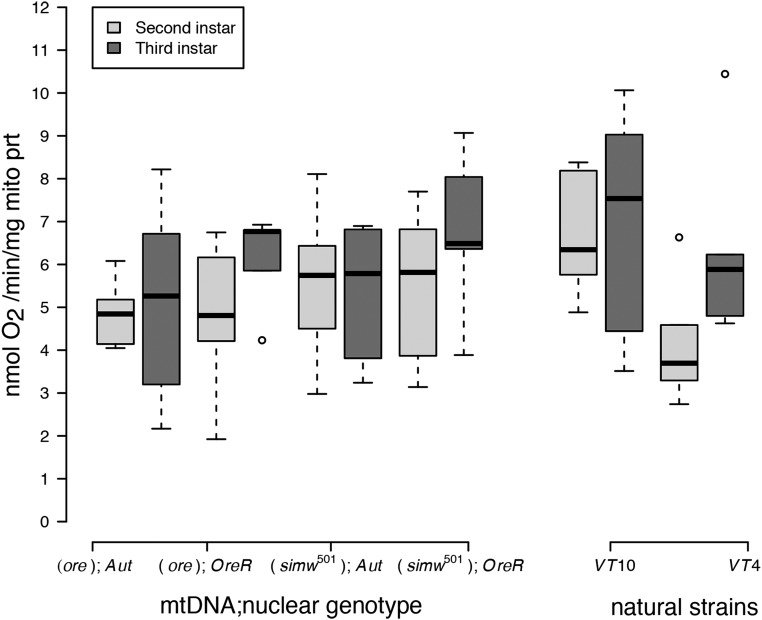
Despite these ontogenetic and genetic differences in the scaling of organismal metabolic rate with mass, second- and third-instar larvae had similar rates of mitochondrial oxygen consumption linked to ATP production (i.e., State 3 respiration) per unit of mitochondrial protein in both mito-nuclear genotypes (instar, P = 0.13) and natural strains (instar, P = 0.12) (Figure 2) (Table S3). Measures of State 3 respiration from first-instar larvae mitochondria were either below our detection limits or of low-quality, even when including similar amounts of larval mass in the preparation. This indicates that there is an increase in mitochondrial quantity or functional capacity between the first- and second-larval instars. Furthermore, larval State 3 respiration did not differ significantly among mito-nuclear genotypes or natural strains, nor were there any significant interactions between instar and genetic factors (Table S2). Maximum respiratory capacity of mitochondria (or CCCP- induced uncoupled respiration, State 3u) was also maintained across larval instars in all mito-nuclear genotypes (instar, P = 0.18) (Figure S1A and Table S2). However, the natural strain VT10 had a significantly elevated maximal respiratory capacity in the second instar that resulted in a significant instar-by-genotype interaction (P = 0.001) (Figure S2A and Table S3).
Figure 2.
Oxygen-coupled ATP production, measured by the State 3 mitochondrial oxygen consumption per unit of mitochondrial protein, was maintained at statistically similar levels across strains and instars (Table S3).
Healthy mitochondria have high rates of oxygen consumption and ATP production when ADP is abundant (i.e., State 3 respiration), but low rates of oxygen consumption in the absence of ATP synthesis (i.e., State 4ol respiration). The ratio of these two measures is called the respiratory control ratio (RCR+). While the RCR+ was generally maintained at a ratio of 2–3 across strains and instars, mitochondria of larvae from two strains, (ore);OreR and VT10, had elevated RCR+ in the third instar that contributed to a significant instar-by-genotype interaction in both mito-nuclear genotypes (instar × nuclear, P = 0.004) and natural strains (instar × strain, P = 0.0001) (Figure S2B and Table S3). This was due to decreased State 4ol respiration in second-instar mitochondria from these strains (Figure S2C and Table S3).
Certain genotypes and strains use anaerobic ATP production further into development
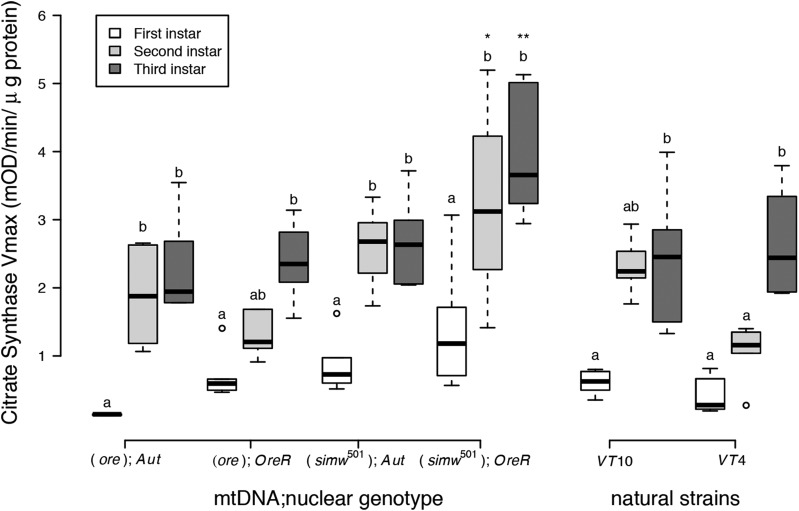
We measured the activity of citrate synthase, a nuclear-encoded enzyme located in the mitochondrial matrix. As the first step in the tricarboxylic acid (TCA) cycle, the activity of this enzyme is often used as an indicator of oxidative capacity. Citrate synthase activity per unit of mitochondrial protein increased across development in larvae of all mito-nuclear genotypes and strains (Figure 3) (mito-nuclear genotypes: instar, P < 0.0001; natural strains: instar, P < 0.0001). There were also genotype-specific effects on citrate synthase activity. Larvae with the incompatible (simw501);OreR genotype had elevated citrate synthase activity relative to other genotypes across all three instars (Figure 3), resulting in significant mito-nuclear variance for this measure of oxidative capacity (mito × nuclear, P = 0.022) (Table S4). Interactions between instar and strain significantly affected citrate synthase activity in the natural strains, as well (instar × strain, P = 0.010). Larvae of particular genotypes and strains could be categorized as those for which citrate synthase reaches its maximal level by the second instar (e.g., VT10 and (ore); Aut) and those for which second-instar mitochondria have citrate synthase activity levels intermediate to first- and third-instar mitochondria [e.g., VT4 and (ore);OreR] (Figure 3).
Figure 3.
Oxidative capacity, measured by citrate synthase activity (Vmax) per unit of mitochondrial protein, increased significantly across instars and was largest in larvae with the incompatible (simw501);OreR genotype. While larvae of all mito-nuclear genotypes increased oxidative capacity throughout development, there was significant variation among genotypes. (simw501);OreR larvae had significantly higher oxidative capacity than larvae with the nuclear genetic control (ore);OreR in the second (*PTukey’s = 0.015) and third instars (**PTukey’s = 0.008). The simw501 mtDNA had no effect in the Aut background (P Tukey’s > 0.833 in both instars), resulting in a significant mtDNA × nuclear interaction (P = 0.022). Natural strains from Vermont also varied significantly in the extent to which oxidative capacity reached its maximal level in the second vs. third instar of development (Table S4). Different letters within mito-nuclear genotypes and strains denote significantly different means at P Tukey’s < 0.006, and asterisks designate significant differences between mito-nuclear genotypes and strains of the same larval instar.
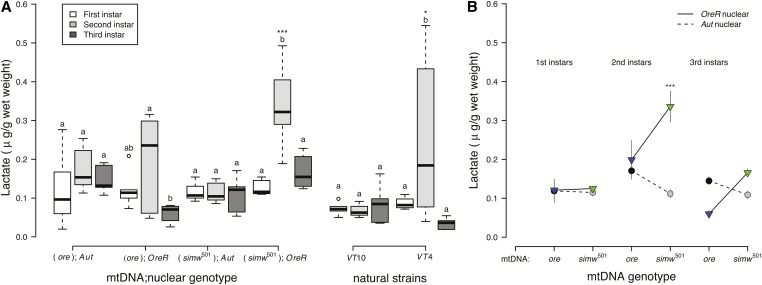
In addition to aerobic, oxidative ATP production, D. melanogaster larvae use anaerobic, glycolytic ATP production that results in the production of lactate. There was significant genetic variation in the extent to which larvae accumulated lactate during development. Second-instar larvae of some strains accumulated lactate, while larvae of other strains did not accumulate any lactate across development (Figure 4A). This variation was observed within both the mito-nuclear genotypes (instar × mtDNA × nuclear, P = 0.033) and the natural strains (instar × strain, P = 0.009). Larvae with the incompatible (simw501);OreR genotype accumulated the highest amounts of lactate in the second instar, relative to other genotypes, resulting in a strong mito-nuclear interaction (Figure 4B and Table S5). Larvae from the natural strain VT4 also accumulated high levels of lactate in the second instar (Figure 4A). Furthermore, larvae from strains that had intermediate levels of citrate synthase activity during the second instar [e.g., VT4 and (ore);OreR] also tended to have increased lactate accumulation during the second instar.
Figure 4.
(A) Lactate levels per gram weight of larvae varied significantly among strains in second-instar larvae and were highest in larvae with the incompatible (simw501);OreR genotype. Genetic variation for second-instar larval lactate levels was also observed among natural strains from Vermont (instar × strain, P = 0.009) (Table S5), with VT4 larvae having significantly more lactate than VT10 larvae (*PTukey’s = 0.014). (B) There was a significant instar × mtDNA × nuclear interaction effect for lactate levels (P = 0.033) (Table S5). (simw501);OreR larvae had significantly higher lactate levels than did larvae from all other mito-nuclear genotypes and strains in the second instar (***P Tukey’s < 0.0003). Different letters within mito-nuclear genotypes and strains denote significantly different means at P Tukey’s < 0.036, and asterisks designate significant differences between mito-nuclear genotypes and strains of the same larval instar.
Larvae with a mito-nuclear incompatibility accumulated more ROS
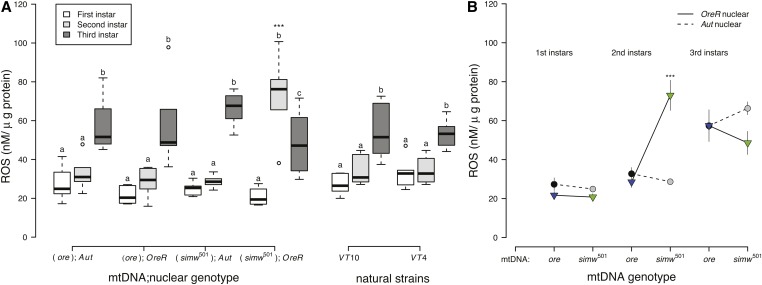
Larvae from all strains had significantly increased levels of H2O2 by the third instar, relative to earlier instars (P < 0.0001) (Figure 5A and Table S6). However, larvae with the incompatible (simw501);OreR genotype had significantly elevated levels of H2O2 in the second instar, both relative to other strains and to first- and third-instar larvae of the same genotype. This resulted in a significant effect of the instar × mtDNA × nuclear interaction on levels of H2O2 (P < 0.0001) (Figure 5B and Table S6).
Figure 5.
(A) ROS levels, measured as the concentration of H2O2 per gram wet weight of larvae, increased significantly across instars, and were highest in second-instar larvae with the incompatible (simw501);OreR genotype. (B) There was a strong effect of instar on ROS levels (instar, P = 2.347e−12), but this pattern varied among mito-nuclear genotypes (instar × mtDNA × nuclear, P = 5.166e−05) (Table S5). Second-instar (simw501);OreR larvae had significantly higher ROS levels relative to larvae from all other mito-nuclear genotypes (***PTukey’s < 0.0001), while larvae from all other mito-nuclear genotypes had similar patterns of increasing ROS throughout development. The interaction between instar and strain did not affect ROS levels among larvae from natural strains (Table S5), which had a similar pattern to larvae from the control mito-nuclear genotypes. Different letters within mito-nuclear genotypes and strains denote significantly different means at P Tukey’s < 0.041, and asterisks designate significant differences between mito-nuclear genotypes and strains of the same larval instar.
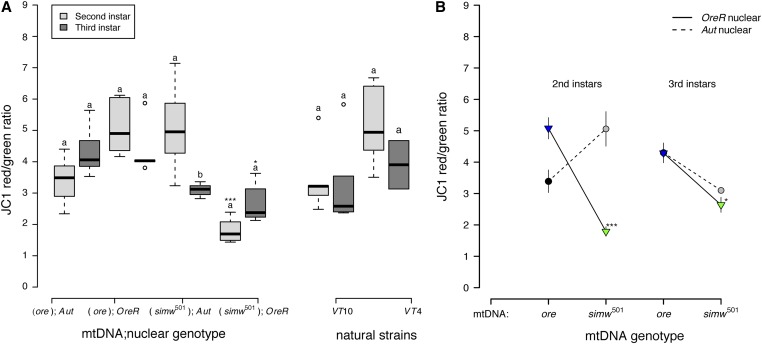
We tested whether mitochondrial membrane potential (ΔΨm) was disrupted in (simw501);OreR larvae. ΔΨm provides the driving force that is utilized by complex V of OXPHOS to make ATP, and is used as an indicator of mitochondrial viability and cellular health. Larvae from all strains, except (simw501);OreR, maintained high levels of mitochondrial membrane potential in the second and third instar (Figure 6A and Table S6). Larvae with the incompatible (simw501);OreR genotype had significantly lower ΔΨm relative to control genotypes in both the second and third instars. The effect of the mito-nuclear interaction on ΔΨm was particularly pronounced in second-instar larvae (instar × mtDNA × nuclear P < 0.0001) (Figure 6B and Table S7).
Figure 6.
Larvae with the incompatible (simw501);OreR genotype had significantly decreased mitochondrial quality, as measured by the mitochondrial membrane potential (ΔΨm). (A) Both mito-nuclear control genotypes and natural strains from Vermont generally maintained high membrane potential in second- and third-instar larvae. However, (simw501);OreR larvae had significantly lower mitochondrial membrane potential than did larvae with the nuclear genetic control (ore);OreR in the second (***PTukey’s < 0.0001) and third instars (*PTukey’s = 0.016). (B) This effect of the simw501 mtDNA was not evident in the Aut background, where it increased membrane potential in second-instar larvae and had no effect in third-instar larvae (PTukey’s = 0.167). This resulted in a significant instar × mtDNA × nuclear interaction effect (P = 1.580e−05) (Table S6). Values >2 typically indicate healthy mitochondria. Different letters within mito-nuclear genotypes and strains denote significantly different means at P Tukey’s < 0.002, and asterisks designate significant differences between mito-nuclear genotypes and strains of the same larval instar.
In summary, the maintenance of mitochondrial respiration in larvae with the incompatible (simw501);OreR genotype across second and third instars was coincident with significant increases in oxidative capacity of mitochondria, increased lactate and ROS production during the second instar, and decreased mitochondrial membrane potential, relative to larvae with control genotypes.
Discussion
Ontogenetic shifts in the relationship between metabolic rate and mass
Metabolic rates scale allometrically with mass, but the parameters that define this relationship vary among taxa, genotypes, life stages and environments (Glazier 2005; Greenlee et al. 2014). We found that the relationship between mass and metabolic rate differed significantly among larval instars of D. melanogaster. Metabolic scaling in developing animals has been described as an “impasse of principles,” wherein the basic tenant of metabolic allometry—that the physiological principles of organisms are relatively conserved—is at odds with the basic tenant of development that the physiological state of organisms is dynamic across ontogeny (Burggren 2005). Insect development involves complex changes in cellular energy demand and body composition that likely affect how metabolic rate scales with mass. Thus, models and principles of interspecific allometric scaling may not be applicable to ontogenetic scaling.
We observed a shift from hypermetric scaling in first-instar larvae (b > 1), to isometric scaling in second-instar larvae (b = 1), followed by hypometric scaling in third-instar larvae (b < 1). This shift in metabolic scaling toward lower mass-specific metabolic rates in larger instars, was in spite of our observation that larger instars had seemingly greater oxidative capacity, as indicated by increased levels of citrate synthase activity per unit of mitochondria. Nevertheless, mitochondrial oxygen consumption linked to ATP production was maintained at similar levels across second- and third-instar larvae. These patterns suggest that although there may be increased oxidative capacity of mitochondria as development progresses, mitochondrial respiration and organismal respiration are not simple reflections of oxidative capacity, but rather are emergent properties of organellar, cellular, and organismal processes.
The ontogenetic change in metabolic scaling that we observed may reflect a change in energy demand across development as larval growth transitions from cell proliferation to cell growth. Hypermetric metabolic-scaling exponents (b > 1), where metabolic rates of larger individuals are greater per unit mass, could result from the increased energetic costs associated with the rapid cell proliferation and increase in cell number early in Drosophila development (O’Farrell 2004; Vollmer et al. 2017). Later in development, larval accumulation of mass occurs primarily via increases in cell volume (O’Farrell 2004), reducing the surface area to volume ratio of cells and potentially limiting metabolism. These observations support studies, collectively grouped under Resource Demand (RD) models, that suggest that metabolic scaling is driven by an intrinsic metabolic demand from the cellular level to tissue growth potential (Von Bertalanffy and Pirozynski 1953; Shin and Yasuo 1984, 1993; Ricklefs 2003; Glazier 2005). In this way, an organism’s metabolic rate across development is a reflection of the potential of tissues for proliferation and growth (Ricklefs 2003). Our observations also contribute to a small, but growing, number of insect studies that support a conceptual framework where completion of growth in holometabolous insects is correlated with decreased mass-specific metabolic rates (Glazier 2005). In both the tobacco hornworm Manduca sexta, and the silkworm Bombyx mori, metabolic-scaling exponents also decrease across ontogeny (Blossman-Myer and Burggren 2010; Callier and Nijhout 2011, 2012; Sears et al. 2012).
Metabolic scaling with mass may also be influenced by the biochemical composition of the body. System Composition (SC) models hypothesize that ontogenetic changes in metabolic scaling reflect shifts in body composition and the relative proportions of metabolically active vs. inert or “sluggish” tissues (Glazier 2005; Isler and VanSchaik 2006; Greenlee et al. 2014). Lipid composition and storage change across development in Drosophila, with a net increase of metabolically inert storage lipids like triacylglycerides across development (Carvalho et al. 2012). While third-instar larvae in our study did have increased lipid plus water content per unit dry mass relative to second instars, first-instar larvae had values of this parameter similar to third-instar larvae. Thus, accumulating more water and lipid per unit dry mass cannot fully explain the observed pattern of increasingly hypometric metabolic scaling with mass across development. Another possibility is that changes in relative tissue sizes contribute to ontogenetic change in how metabolic rate scales with mass. In Manduca, the contribution of metabolically active gut tissue to the body decreases across development, which may contribute to an increasingly hypometric metabolic scaling with mass across development (Callier and Nijhout 2012).
Genetic variation in body composition across development could also underlie the genetic variation in metabolic scaling that we observed in third-instar larvae. If larvae with different genotypes differ in the degree to which they accumulate mass in the third instar via different types of energy storage, this could generate genetic variation for how metabolic rate scales with mass. Midway through the third instar, D. melanogaster membrane-lipid accumulation is paused, while levels of storage lipids like triacylglycerides increase (Carvalho et al. 2012). This suggests a transition in the third instar from metabolism supporting membrane synthesis and cell proliferation to metabolism supporting mass accumulation via lipid storage. If larvae with different genotypes vary in the timing or extent of this switch, this could contribute to the greater genetic variation for metabolic scaling that we observed in this developmental stage.
Physiological compensation in larvae with a mito-nuclear incompatibility comes at a cost
Mitochondrial respiration coupled to ATP production was maintained in larvae with the mito-nuclear incompatible genotype at in vitro levels similar to control genotypes, despite compromised OXPHOS in flies with this genotype (Meiklejohn et al. 2013). The maintenance of mitochondrial respiration in these larvae was accompanied by increases in mitochondrial oxidative capacity, measured by citrate synthase activity, and glycolytic ATP production, measured by lactate accumulation, relative to larvae with control genotypes. These increases may reflect physiological compensation to maintain ATP levels in larvae whose mitochondria consume similar levels of oxygen but are less efficiently generating ATP. This compensation does not appear to occur in adult males of this genotype that have decreased mitochondrial respiration and decreased citrate synthase activity (Pichaud et al. 2019). We suggest that by using the functional complementation of both glycolytic and mitochondrial ATP production (i.e., both substrate-level and oxidative phosphorylation), larvae of this mito-nuclear incompatible genotype are able to synthesize the ATP needed to support development.
Physiological compensation can sometimes have counterintuitive costs paid over the lifespan. While larvae with the (simw501);OreR genotype appear to use physiological compensation to support larval development, these larvae have significantly delayed development and compromised pupation height, immune function, and female fecundity (Meiklejohn et al. 2013; Zhang et al. 2017; Buchanan et al. 2018). Additionally, while in vitro mitochondrial respiration in larvae of this genotype was maintained similar to other strains, whole-organism metabolic rate in larvae with this genotype was elevated (Hoekstra et al. 2013), potentially via compensatory upregulation of citrate synthase and the TCA cycle to supply ATP. Thus, even when drawing on both glycolytic and oxidative ATP production, individuals with this mito-nuclear incompatibility may produce energy supplies very close to energetic demand. Previous results from our laboratory support this model; conditions that normally accelerate larval development, significantly magnified the developmental delay of (simw501);OreR larvae, suggesting that larvae with this genotype have limited capacity to compensate the defect in OXPHOS (Hoekstra et al. 2013, 2018). Larvae with the incompatible (simw501);OreR genotype may use a majority of their aerobic scope to complete normal development, limiting the resources available to allocate to other aspects of fitness. Once the demands of growth are removed, adults with this mito-nuclear incompatibility appear to regain some aerobic scope, as larvae that survived to pupation also completed metamorphosis and had normal adult size and metabolic rates (Hoekstra et al. 2013, 2018). However, the costs paid out during development appear to have significant impacts on adult fecundity. Both female and male fecundity were severely compromised in adults with this genotype that were developed at warmer temperatures that increase biological rates and energy demand (Hoekstra et al. 2013; Zhang et al. 2017).
At the cellular level, physiological compensation in (simw501);OreR larvae may be a source of oxidative stress, indicated by higher levels of H2O2 in larvae of this genotype, relative to other genotypes. H2O2 is a byproduct of the mitochondrial electron transport chain (ETC) that supports OXPHOS in healthy cells, and we observed increases in H2O2 as oxidative capacity increased across development in all larvae of all strains. However, compromised electron flow through the ETC can increase H2O2 levels and generate oxidative stress (Somero et al. 2017), and isolated mitochondria from adults with the mito-nuclear incompatible genotype did have higher rates of H2O2 production (Pichaud et al. 2019). There are two ways that this may be occurring. First, upregulation of the TCA cycle to supply more NADH for ATP production via the ETC may increase production of superoxide anion at Complex I. Second, there may be stoichiometric imbalance in the ETC due to presumably normal levels of cytoplasmically translated Complex II but compromised levels of the mitochondrially translated downstream OXPHOS complexes in larvae with this genotype. This could result in backflow of electrons that can produce superoxide ions when the ratio of reduced:unreduced coenzyme Q becomes elevated. The idea that individuals with the incompatible (simw501);OreR genotype are experiencing oxidative stress suggests an alternative interpretation of the elevated citrate synthase activity that we observed in larvae with this genotype. Levels of citrate synthase were increased in the blue mussel Mytilus trossulus in response to heat stress, a change that was coupled with increases in isocitrate dehydrogenase (IDH), which generates NADPH to support H2O2-scavenging reactions in the mitochondria (Tomanek and Zuzow 2010). This highlights the importance of considering that TCA cycle enzymes provide important functions beyond their role in OXPHOS, as they provide substrates for biosynthesis, support antioxidant reactions, and act as signaling molecules (Marden 2013; Boukouris et al. 2016; Somero et al. 2017).
Finally, we observed that mitochondria from larvae with the incompatible (simw501);OreR genotype could support mitochondrial oxygen consumption linked to ATP production at wild-type levels despite the fact that their membrane potential was significantly reduced. There is precedence for this observation. Mitochondrial diseases with OXPHOS defects are correlated with a suite of metabolic phenotypes that include upregulated glycolysis, lactate accumulation, elevated ROS, and decreased mitochondrial membrane potential, but stable ATP levels (Szczepanowska et al. 2012; Frazier et al. 2019). ROS act as essential secondary messengers in cellular homeostasis, but above a certain threshold level can be dangerous and lead to apoptosis (Giorgio et al. 2007; Bigarella et al. 2014). A potential defense mechanism is to decrease the mitochondrial membrane potential (e.g., by uncoupling) to reduce further ROS production and protect the cell from oxidative damage (Dlasková et al. 2006). Our data cannot distinguish whether upregulation of citrate synthase and decreased membrane potential in the mitochondria are the cause or the consequence of oxidative stress in larvae with the mito-nuclear incompatibility. However, new models from ecophysiology (Tomanek and Zuzow 2010), developmental physiological genetic (Tennessen et al. 2011, 2014; Li et al. 2017, 2019), and disease (Ward and Thompson 2012) systems provide promising paths for future dissection of the mechanisms by which mitochondrial-nuclear genetic variation scales up to organismal fitness variation.
Genetic variation in cellular metabolism supports similar organismal outcomes
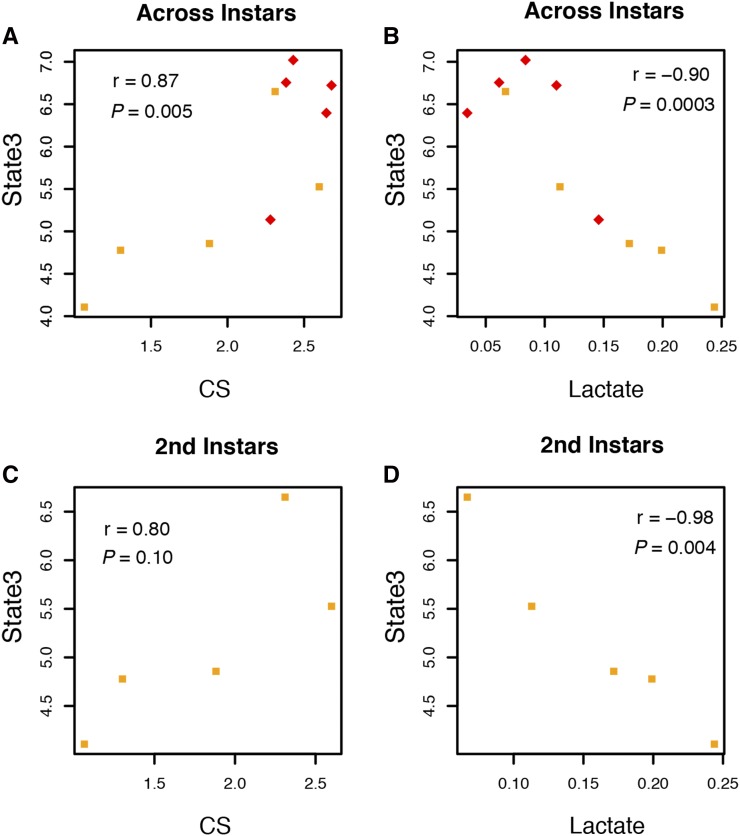
Aerobic organisms can generate ATP via mitochondrial OXPHOS, but also anaerobically via glycolytic pathways that are supported by fermentation-generated NAD+ (e.g., by lactate production). While larvae with (simw501);OreR genotype showed a striking pattern of both increased oxidative capacity and increased reliance on lactate metabolism, we wanted to determine the extent to which components of aerobic and anaerobic metabolism were correlated among the wild-type control and natural strains that we measured. Across instars there was a significant positive correlation between mitochondrial State 3 respiration and citrate synthase activity among wild-type strains (r = 0.87, P = 0.005), consistent with multiple aspects of oxidative capacity increasing in concert across development (Figure 7A). Within the second instar there was a similar magnitude, but nonsignificant correlation between State 3 respiration and citrate synthase activity among strains (r = 0.8, P = 0.107) (Figure 7C) that was not evident in the third instar (r = 0.49 P = 0.406). Thus, both aspects of oxidative metabolism increase together across development, but there is also a signature of genetic covariance for multiple aspects of oxidative metabolism within the second instar.
Figure 7.
Components of metabolism were strongly correlated among wild-type strains, particularly in second-instar larvae. Each point represents the mean phenotype for the five wild-type strains (three mito-nuclear control genotypes and two natural strains) in second-instar (squares) and third-instar (diamonds) larvae. The Pearson’s statistic r and associated P value are provided for each correlation. State 3, mitochondrial O2 consumption linked to ATP production; CS, citrate synthase activity. (A and B) Across instars, (C and D) 2nd instars.
Across instars, there was a significant negative correlation between citrate synthase activity and lactate levels (r = −0.87, P = 0.001) and between State 3 respiration and lactate levels among strains (r = −0.90, P = 0.0003) (Figure 7B), consistent with high reliance on anaerobic glycolytic ATP production when oxidative capacity is low. These correlations were most evident within the second instar (CS-lactate: r = −0.91, P = 0.033, State3-lactate: r = −0.98, P = 0.004) (Figure 7D), and in the same direction, although not significant, within the third instar (CS-lactate: r = −0.45, P = 0.452, State3-lactate: r = −0.61, P = 0.278). Including traits from larvae with the (simw501); OreR genotype weakened these correlations (State3-CS: r = 0.43, P = 0.163; CS-lactate: r = −0.07, P = 0.829). Thus, the mito-nuclear incompatibility breaks the genetic-physiological negative correlation between oxidative and glycolytic ATP production by significantly increasing both citrate synthase activity and lactate levels, relative to wild-type strains.
Finally, wild-type strains differed significantly in the amount of variance specifically for lactate accumulation in second instar larvae (Levene’s test, P = 0.002). Thus, the second instar appears to be a time in development when both strains and individuals within strains differ in their reliance on glycolytic ATP production as they switch over to oxidative metabolism. This pattern is consistent with genetic variation for a metabolic switch from glycolytic to mitochondrial production of ATP regulated by the Drosophila estrogen-related receptor dERR (Tennessen and Thummel 2011; Tennessen et al. 2011). Yet, despite this genetic variation for how second-instar larvae generate ATP, the organismal metabolic rate of second-instar larvae appeared more robust to genetic variation than were the metabolic rates of other instars. We also observed that, despite this developmental switch from glycolytic to mitochondrial ATP production, in vitro mitochondrial respiration rates per unit mitochondrial protein remained constant across second- and third-instar larvae. Again, this highlights that organellar and organismal metabolic rates are not simple reflections of the underlying metabolic pathways being used. In this way, higher levels of biological organization may buffer and potentially shelter genetic variation in metabolism from selection.
dERR is responsible for a vital transcriptional switch of carbohydrate metabolism in second-instar larvae (Tennessen et al. 2011) that coincides with increases in lactate dehydrogenase (dLDH) and lactate accumulation (Li et al. 2017). dLDH activity recycles NAD+ which allows for continued glycolytic ATP production and supports the TCA cycle in generating cellular building blocks to support normal cell proliferation and larval growth during development (Tennessen and Thummel 2011). Furthermore, dLDH expression and lactate production results in the accumulation of the metabolic signaling molecule L-2-hydroxyglutarate (L-2HG). L-2HG affects genome-wide DNA methylation and coordinates glycolytic flux through epigenetic modification, heterochromatin formation, and changes in gene expression (Li et al. 2017). We found that lactate accumulation in second-instar larvae was strongly affected by genotype, suggesting differential timing of this switch among both natural strains and mito-nuclear genotypes. Investigating potential epigenetic, bioenergetic, and life-history consequences of this genetic variation may reveal whether different metabolic strategies at the subcellular level fund similar or distinct fitness outcomes at the organismal level. This is critical for understanding whether populations harbor genetic variation in biochemical pathways that ultimately has similar fitness outcomes, or whether we should expect to see the signatures of selection acting on enzymes that control shifts in metabolic flux (e.g., Flowers et al. 2007; Pekny et al. 2018).
In conclusion, the dramatic and rapid growth of Drosophila during ontogeny requires a precise and genetically determined metabolic program that enhances biosynthesis and proliferation coupled with a tight temporal coordination. Here, we have shown how genetic variation influence patterns of metabolism in larvae of both natural strains and mito-nuclear genotypes of Drosophila during developmental. Our study supports that genetic defects in core physiology can be buffered at the organismal level via physiological plasticity (Li et al. 2019), and that natural populations likely harbor genetic variation for distinct metabolic strategies in development that may generate similar organismal outcomes.
Acknowledgments
We would like to thank Madeleine Koenig for her assistance with sample preparation. M. Koenig was supported by the Undergraduate Creative Activities and Research Experience (UCARE) program at University of Nebraska-Lincoln (UNL). The research was supported by National Science Foundation-Integrative Organismal Systems (NSF-IOS) CAREER Award 1149178, NSF Experimental Program to Stimulate Competitive Research (EPSCoR) Track II Award 1736249, and funds from UNL to O.B.M., C.R.J., and K.L.M.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.7946795.
Communicating editor: M. Wolfner
Literature Cited
- Arnqvist G., Dowling D., Paul E., Laurene G., Tom T., et al. , 2010. Genetic architecture of metabolic rate: environment specific epistasis between mitochondrial and nuclear genes in an insect. Evolution. 64: 3354–3363. 10.1111/j.1558-5646.2010.01135.x [DOI] [PubMed] [Google Scholar]
- Aw W., Bajracharya R., Towarnicki S., Ballard J., 2016. Assessing bioenergetic functions from isolated mitochondria in. J. Biol. Methods 3: e42 10.14440/jbm.2016.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard W., Rand D., 2005. The population biology of mitochondrial DNA and its phylogenetic implications. Annu. Rev. Ecol. Evol. Syst. 36: 621–642. 10.1146/annurev.ecolsys.36.091704.175513 [DOI] [Google Scholar]
- Barabási A., Oltvai Z., 2004. Network biology: understanding the cell’s functional organization. Nat. Rev. Genet. 5: 101–113. 10.1038/nrg1272 [DOI] [PubMed] [Google Scholar]
- Bigarella C., Liang R., Ghaffari S., 2014. Stem cells and the impact of ROS signaling. Development 141: 4206–4218. 10.1242/dev.107086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh E. G., Dyer W. J., 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. 10.1139/y59-099 [DOI] [PubMed] [Google Scholar]
- Blossman-Myer B., Burggren W., 2010. Metabolic allometry during development and metamorphosis of the silkworm Bombyx mori: analyses, patterns, and mechanisms. Physiol. Biochem. Zool. 83: 215–231. 10.1086/648393 [DOI] [PubMed] [Google Scholar]
- Boukouris A., Zervopoulos S., Michelakis E., 2016. Metabolic enzymes moonlighting in the nucleus: metabolic regulation of gene transcription. Trends Biochem. Sci. 41: 712–730. 10.1016/j.tibs.2016.05.013 [DOI] [PubMed] [Google Scholar]
- Brand M. D., Chien L. F., Ainscow E. K., Rolfe D. F., Porter R. K., 1994. The causes and functions of mitochondrial proton leak. Biochim. Biophys. Acta 1187: 132–139. 10.1016/0005-2728(94)90099-X [DOI] [PubMed] [Google Scholar]
- Buchanan J., Meiklejohn C., Montooth K., 2018. Mitochondrial dysfunction and infection generate immunity-fecundity tradeoffs in Drosophila. Integr. Comp. Biol. 58: 591–603. 10.1093/icb/icy078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burggren W., 2005. Developing animals flout prominent assumptions of ecological physiology. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 141: 430–439. 10.1016/j.cbpb.2005.03.010 [DOI] [PubMed] [Google Scholar]
- Callier V., Nijhout F., 2011. Control of body size by oxygen supply reveals size-dependent and size-independent mechanisms of molting and metamorphosis. Proc. Natl. Acad. Sci. USA 108: 14664–14669. 10.1073/pnas.1106556108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callier V., Nijhout H. F., 2012. Supply-side constraints are insufficient to explain the ontogenetic scaling of metabolic rate in the tobacco hornworm, Manduca sexta. PLoS One 7: e45455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callier V., Hand S., Campbell J., Biddulph T., Harrison J., 2015. Developmental changes in hypoxic exposure and responses to anoxia in Drosophila melanogaster. J. Exp. Biol. 218: 2927–2934. 10.1242/jeb.125849 [DOI] [PubMed] [Google Scholar]
- Carvalho M., Sampaio J., Palm W., Brankatschk M., Eaton S., et al. , 2012. Effects of diet and development on the Drosophila lipidome. Mol. Syst. Biol. 8: 600 10.1038/msb.2012.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chance B., Williams G., 1955. Respiratory enzymes in oxidative phosphorylation: I. kinetics of oxygen utilization. J. Biol. Chem. 217: 383–394. [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 2016. Population genetics from 1966 to 2016. Heredity 118: 2–9. 10.1038/hdy.2016.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church R., Robertson F., 1966. A biochemical study of the growth of Drosophila melanogaster. J. Exp. Zool. 162: 337–351. 10.1002/jez.1401620309 [DOI] [Google Scholar]
- Clark A., Wang L., 1997. Epistasis in measured genotypes: Drosophila p-element insertions. Genetics 147: 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A., Wang L., Hulleberg T., 1995a P-element-induced variation in metabolic regulation in Drosophila. Genetics 139: 337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A., Wang L., Hulleberg T., 1995b Spontaneous mutation rate of modifiers of metabolism in Drosophila. Genetics 139: 767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper B. S., Hammad L. A., Montooth K. L., 2014. Thermal adaptation of cellular membranes in natural populations of Drosophila melanogaster. Funct. Ecol. 28: 886–894. 10.1111/1365-2435.12264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford D., Oleksiak M., 2007. The biological importance of measuring individual variation. J. Exp. Biol. 210: 1613–1621. 10.1242/jeb.005454 [DOI] [PubMed] [Google Scholar]
- Dlasková A., Špaček T., Škobisová E., Šantorová J., Ježek P., 2006. Certain aspects of uncoupling due to mitochondrial uncoupling proteins in vitro and in vivo. Biochim. Biophys. Acta 1757: 467–473. 10.1016/j.bbabio.2006.05.005 [DOI] [PubMed] [Google Scholar]
- Estabrook R., 1967. Mitochondrial respiratory control and the polarographic measurement of ADP : O ratios. Methods Enzymol. 10: 41–47. 10.1016/0076-6879(67)10010-4 [DOI] [Google Scholar]
- Flowers J., Sezgin E., Kumagai S., Duvernell D., Matzkin L., et al. , 2007. Adaptive evolution of metabolic pathways in Drosophila. Mol. Biol. Evol. 24: 1347–1354. 10.1093/molbev/msm057 [DOI] [PubMed] [Google Scholar]
- Frazier A., Thorburn D. R., Compton A. G., 2019. Mitochondrial energy generation disorders: genes, mechanisms, and clues to pathology. J. Biol. Chem. 294: 5386–5395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gefen E., Marlon A. J., Gibbs A. E., 2006. Selection for desiccation resistance in adult Drosophila melanogaster affects larval development and metabolite accumulation. J. Exp. Biol. 209: 3293–3300. 10.1242/jeb.02397 [DOI] [PubMed] [Google Scholar]
- Gillespie J., 1999. The role of population size in molecular evolution. Theor. Popul. Biol. 55: 145–156. 10.1006/tpbi.1998.1391 [DOI] [PubMed] [Google Scholar]
- Giorgio M., Trinei M., Migliaccio E., Pelicci P., 2007. Hydrogen peroxide: a metabolic by-product or a common mediator of ageing signals? Nat. Rev. Mol. Cell Biol. 8: 722–728. 10.1038/nrm2240 [DOI] [PubMed] [Google Scholar]
- Glazier D., 2005. Beyond the ‘3/4-power law’: variation in the intra- and interspecific scaling of metabolic rate in animals. Biol. Rev. Camb. Philos. Soc. 80: 611–662. 10.1017/S1464793105006834 [DOI] [PubMed] [Google Scholar]
- Glazier D., 2014. Metabolic scaling in complex living systems. Systems 2: 451–540. 10.3390/systems2040451 [DOI] [Google Scholar]
- Glazier D. S., 2015. Is metabolic rate a universal ‘pacemaker’ for biological processes? Biol. Rev. Camb. Philos. Soc. 90: 377–407. 10.1111/brv.12115 [DOI] [PubMed] [Google Scholar]
- Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M., et al. , 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479. 10.1038/nature09715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenlee K., Montooth K., Helm B., 2014. Predicting performance and plasticity in the development of respiratory structures and metabolic systems. Integr. Comp. Biol. 54: 307–322. 10.1093/icb/icu018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris H., 1966. Genetics of man enzyme polymorphisms in man. Proc. R. Soc. Lond. B. Biol. Sci. 164: 298–310. [DOI] [PubMed] [Google Scholar]
- Hoekstra L., Siddiq M., Montooth K., 2013. Pleiotropic effects of a mitochondrial–nuclear incompatibility depend upon the accelerating effect of temperature in Drosophila. Genetics 195: 1129–1139. 10.1534/genetics.113.154914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra L. A., Julick C. R., Katelyn K. M., Montooth K. L., 2018. Energy demand and the context-dependent effects of genetic interactions underlying metabolism. Evol. Lett. 2: 102–113. 10.1002/evl3.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmbeck M., Donner J., Villa-Cuesta E., Rand D., 2015. A Drosophila model for mito-nuclear diseases generated by an incompatible interaction between tRNA and tRNA synthetase. Dis. Model. Mech. 8: 843–854. 10.1242/dmm.019323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubby J., Lewontin R., 1966. A molecular approach to the study of genic heterozygosity in natural populations. I. the number of alleles at different loci in Drosophila pseudoobscura. Genetics 54: 577–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isler K., VanSchaik C., 2006. Metabolic costs of brain size evolution. Biol. Lett. 2: 557–560. 10.1098/rsbl.2006.0538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H., Tombor B., Albert R., Oltvai Z., Barabási A., 2000. The large-scale organization of metabolic networks. Nature 407: 651–654. 10.1038/35036627 [DOI] [PubMed] [Google Scholar]
- Kimura M., 1983. The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge: 10.1017/CBO9780511623486 [DOI] [Google Scholar]
- Kleiber M., 1932. Body size and metabolism. Hilgardia 6: 315–353. 10.3733/hilg.v06n11p315 [DOI] [Google Scholar]
- Kurbalija Novičić Z., Immonen E., Jelić M., AnÐelković M., Stamenković-Radak M., et al. , 2015. Within-population genetic effects of mtDNA on metabolic rate in Drosophila subobscura. J. Evol. Biol. 28: 338–346. 10.1111/jeb.12565 [DOI] [PubMed] [Google Scholar]
- Laurie-Ahlberg C., Maroni G., Bewley G., Lucchesi J., Weir B., 1980. Quantitative genetic variation of enzyme activities in natural populations of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 77: 1073–1077. 10.1073/pnas.77.2.1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie-Ahlberg C., Wilton A., Curtsinger J., Emigh T., 1982. Naturally occurring enzyme activity variation in Drosophila melanogaster. I. sources of variation for 23 enzymes. Genetics 102: 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin R., Hubby J., 1966. A molecular approach to the study of genic heterozygosity in natural populations. II. amount of variation and degree of heterozygosity in natural populations of Drosophila pseudoobscura. Genetics 54: 595–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Chawla G., Hurlburt A., Sterrett M., Zaslaver O., et al. , 2017. Drosophila larvae synthesize the putative oncometabolite L-2-hydroxyglutarate during normal developmental growth. Proc. Natl. Acad. Sci. USA 114: 1353–1358. 10.1073/pnas.1614102114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Buddika K., Sterrett M. C., Julick C. R., Pletcher R. C., et al. , 2019. Lactate and glycerol-3-phosphate metabolism cooperatively regulate growth and redox balance during Drosophila melanogaster larval development. bioRxiv: 517532. 10.1101/517532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marden J., 2013. Nature’s inordinate fondness for metabolic enzymes: why metabolic enzyme loci are so frequently targets of selection. Mol. Ecol. 22: 5743–5764. 10.1111/mec.12534 [DOI] [PubMed] [Google Scholar]
- Martin A., 1995. Metabolic rate and directional nucleotide substitution in animal mitochondrial DNA. Mol. Biol. Evol. 12: 1124–1131. [DOI] [PubMed] [Google Scholar]
- Meiklejohn C., Hartl D., 2002. A single mode of canalization. Trends Ecol. Evol. 17: 468–473. 10.1016/S0169-5347(02)02596-X [DOI] [Google Scholar]
- Meiklejohn C., Holmbeck M., Siddiq M., Abt D., Rand D., et al. , 2013. An incompatibility between a mitochondrial tRNA and its nuclear-encoded tRNA synthetase compromises development and fitness in Drosophila. PLoS Genet. 9: e1003238 10.1371/journal.pgen.1003238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen T., Björklund M., 2017. Mitochondrial function and cell size: an allometric relationship. Trends Cell Biol. 27: 393–402. 10.1016/j.tcb.2017.02.006 [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T., Pedersen D., 1998. The molecular basis of quantitative genetic variation in central and secondary metabolism in Arabidopsis. Genetics 149: 739–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montooth K. L., Marden J. H., Clark A. G., 2003. Mapping determinants of variation in energy metabolism, respiration and flight in Drosophila. Genetics 165: 623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montooth K. L., Meiklejohn C. D., Abt D. N., Rand D. M., 2010. Mitochondrial-nuclear epistasis affects fitness within species but does not contribute to fixed incompatibilities between species of Drosophila. Evolution 64: 3364–3379. 10.1111/j.1558-5646.2010.01077.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro D., Treberg J. R., 2017. A radical shift in perspective: mitochondria as regulators of reactive oxygen species. J. Exp. Biol. 220: 1170–1180. 10.1242/jeb.132142 [DOI] [PubMed] [Google Scholar]
- Nespolo R., Castañeda L., Roff D., 2007. Quantitative genetic variation of metabolism in the nymphs of the sand cricket, Gryllus firmus, inferred from an analysis of inbred-lines. Biol. Res. 40: 5–12. 10.4067/S0716-97602007000100001 [DOI] [PubMed] [Google Scholar]
- O’Farrell P., 2004. How metazoans reach their full size: the natural history of bigness, pp. 1–22 in Cell Growth: Control of Cell Size, edited by Hall M., Raff M., Thomas G. Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- Pekny J. E., Smith P. B., Marden J. H., 2018. Enzyme polymorphism, oxygen and injury: a lipidomic analysis of flight-induced oxidative damage in a succinate dehydrogenase d (Sdhd)-polymorphic insect. J. Exp. Biol. 221: pii: jeb171009 10.1242/jeb.171009 [DOI] [PubMed] [Google Scholar]
- Pichaud N., Bérubé R., Côté G., Belzile C., Dufresne F., et al. , 2019. Age dependent dysfunction of mitochondrial and ROS metabolism induced by mitonuclear mismatch. Front. Genet. 10: 130 10.3389/fgene.2019.00130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter R., Brand M., 1993. Body mass dependence of H+ leak in mitochondria and its relevance to metabolic rate. Nature 362: 628–630. 10.1038/362628a0 [DOI] [PubMed] [Google Scholar]
- Porter R., Hulbert A., Brand M., 1996. Allometry of mitochondrial proton leak: influence of membrane surface area and fatty acid composition. Am. J. Physiol. Integr. Comp. Physiol. 271: R1550–R1560. 10.1152/ajpregu.1996.271.6.R1550 [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2011. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Ravasz E., Somera A., Mongru D., Oltvai Z., Barabási A., 2002. Hierarchical organization of modularity in metabolic networks. Science 297: 1551–1555. 10.1126/science.1073374 [DOI] [PubMed] [Google Scholar]
- Ricklefs R., 2003. Is rate of ontogenetic growth constrained by resource supply or tissue growth potential? A comment on West et al.’s model. Funct. Ecol. 17: 384–393. 10.1046/j.1365-2435.2003.00745.x [DOI] [Google Scholar]
- Sears K., Kerkhoff A., Messerman A., Itagaki H., 2012. Ontogenetic scaling of metabolism, growth, and assimilation: testing metabolic scaling theory with Manduca sexta larvae. Physiol. Biochem. Zool. 85: 159–173. 10.1086/664619 [DOI] [PubMed] [Google Scholar]
- Shin O., Yasuo I., 1984. Allometric relationship between tissue respiration and body mass in the carp. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 77: 415–418. 10.1016/0300-9629(84)90205-6 [DOI] [Google Scholar]
- Shin O., Yasuo I., 1993. Allometric relationship between tissue respiration and body mass in a marine teleost, porgy Pagrus major. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 105: 129–133. [Google Scholar]
- Somero G., Lockwood B., Tomanek L., 2017. Biochemical Adaptation: Response to Environmental Challenges, from Life’s Origins to the Anthropocene. Sinauer Associates, Sunderland, MA [Google Scholar]
- Strogatz S. H., 2001. Exploring complex networks. Nature 410: 268–276. 10.1038/35065725 [DOI] [PubMed] [Google Scholar]
- Szczepanowska J., Malinska D., Wieckowski M., Duszynski J., 2012. Effect of mtDNA point mutations on cellular bioenergetics. Biochim. Biophys. Acta 1817: 1740–1746. 10.1016/j.bbabio.2012.02.028 [DOI] [PubMed] [Google Scholar]
- Tennessen J., Baker K., Lam G., Evans J., Thummel C., 2011. The Drosophila estrogen-related receptor directs a metabolic switch that supports developmental growth. Cell Metab. 13: 139–148. 10.1016/j.cmet.2011.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen J. M., Thummel C. S., 2011. Coordinating growth and review maturation — insights from Drosophila. Curr. Biol. 21: R750–R757. 10.1016/j.cub.2011.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen J. M., Bertagnolli N. M., Evans J., Sieber M. H., Cox J., et al. , 2014. Coordinated metabolic transitions during Drosophila embryogenesis and the onset of aerobic glycolysis. G3 (Bethesda) 4: 839–850. 10.1534/g3.114.010652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieleman I., Versteegh M., Fries A., Helm B., Dingemanse N., et al. , 2009. Genetic modulation of energy metabolism in birds through mitochondrial function. Proc. R. Soc. B Biol. Sci. 276: 1685–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff S. A., Varkonyi R., Cahinhinan N., Abbes S., Argyropoulos G., et al. , 2001. Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science 293: 455–462. 10.1126/science.1061573 [DOI] [PubMed] [Google Scholar]
- Tomanek L., Zuzow M., 2010. The proteomic response of the mussel congeners Mytilus galloprovincialis and M. trossulus to acute heat stress: implications for thermal tolerance limits and metabolic costs of thermal stress. J. Exp. Biol. 213: 3559–3574. 10.1242/jeb.041228 [DOI] [PubMed] [Google Scholar]
- Van Dyken J. D., Wade M. J., 2010. The genetic signature of conditional expression. Genetics 184: 557–570. 10.1534/genetics.109.110163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrelli B., Eanes W., 2001. The functional impact of Pgm amino acid polymorphism on glycogen content in Drosophila melanogaster. Genetics 159: 201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa-Cuesta E., Holmbeck M., Rand D., 2014. Rapamycin increases mitochondrial efficiency by mtDNA-dependent reprogramming of mitochondrial metabolism in Drosophila. J. Cell Sci. 127: 2282–2290. 10.1242/jcs.142026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer J., Casares F., Iber D., 2017. Growth and size control during development. Open Biol. 7: pii: 170190. 10.1098/rsob.170190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Bertalanffy L., Pirozynski W., 1953. Tissue respiration, growth, and basal metabolism. Biol. Bull. 105: 240–256. 10.2307/1538640 [DOI] [Google Scholar]
- Waddington C., 1942. Canalization of development and the inheritance of acquired characters. Nature 150: 563–565. 10.1038/150563a0 [DOI] [PubMed] [Google Scholar]
- Waddington C., 1957. The Strategy of the Genes: A Discussion of Some Aspects of Theoretical Biology. George Allen & Unwin, Ltd., London. [Google Scholar]
- Ward P. S., Thompson C. B., 2012. Signaling in control of cell growth and metabolism. Cold Spring Harb. Perspect. Biol. 4: a006783 10.1101/cshperspect.a006783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warton D., Wright I., Falster D., Westoby M., 2006. Bivariate line-fitting methods for allometry. Biol. Rev. Camb. Philos. Soc. 81: 259–291. 10.1017/S1464793106007007 [DOI] [PubMed] [Google Scholar]
- Watt W., 1977. Adaptation at specific loci. I. natural selection on phosphogluco isomerase of Colias butterflies: biochemical and population aspects. Genetics 87: 177–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt W., Cassin R., Swan M., 1983. Adaptation at specific loci. III. field behavior and survivorship differences among Colias PGI genotypes are predictable from in vitro biochemistry. Genetics 103: 725–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt W., Wheat C., Meyer E., Martin J., 2003. Adaptation at specific loci. VII. natural selection, dispersal and the diversity of molecular–functional variation patterns among butterfly species complexes (Colias: Lepidoptera, Pieridae). Mol. Ecol. 12: 1265–1275. 10.1046/j.1365-294X.2003.01804.x [DOI] [PubMed] [Google Scholar]
- Zhang C., Montooth K., Calvi B., 2017. Incompatibility between mitochondrial and nuclear genomes during oogenesis results in ovarian failure and embryonic lethality. Development 144: 2490–2503. 10.1242/dev.151951 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Stocks and strains are available upon request. Supplemental files including all phenotype data are available at FigShare. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.7946795.