Precise activity of a gene requires its promoter to be matched with an appropriate enhancer. Insulators are DNA elements which can limit inappropriate enhancer-promoter interactions. More recently, their ability to stimulate gene activity has been also recognized...
Keywords: Drosophila, insulators, transvection, E(spl), locus, homolog pairing
Abstract
Transvection is the phenomenon where a transcriptional enhancer activates a promoter located on the homologous chromosome. It has been amply documented in Drosophila where homologs are closely paired in most, if not all, somatic nuclei, but it has been known to rarely occur in mammals as well. We have taken advantage of site-directed transgenesis to insert reporter constructs into the same genetic locus in Drosophila and have evaluated their ability to engage in transvection by testing many heterozygous combinations. We find that transvection requires the presence of an insulator element on both homologs. Homotypic trans-interactions between four different insulators can support transvection: the gypsy insulator (GI), Wari, Fab-8 and 1A2; GI and Fab-8 are more effective than Wari or 1A2. We show that, in the presence of insulators, transvection displays the characteristics that have been previously described: it requires homolog pairing, but can happen at any of several loci in the genome; a solitary enhancer confronted with an enhancerless reporter is sufficient to drive transcription; it is weaker than the action of the same enhancer-promoter pair in cis, and it is further suppressed by cis-promoter competition. Though necessary, the presence of homotypic insulators is not sufficient for transvection; their position, number and orientation matters. A single GI adjacent to both enhancer and promoter is the optimal configuration. The identity of enhancers and promoters in the vicinity of a trans-interacting insulator pair is also important, indicative of complex insulator-enhancer-promoter interactions.
TRANSGENIC reporter genes are powerful tools for studying gene regulation. However, a transgene is often susceptible to interactions with surrounding chromatin, leading to significant variations in expression patterns and levels, depending on the site of insertion in the genome (Levis et al. 1985). In order to factor-out these “position effects,” multiple lines of transgenic animals must be studied. Recently, this problem has been tackled by two strategies now widely used in Drosophila. One relies on the ability to guide integration of a transgene to a specific location in the genome via the use of the ΦC31-mediated integration system (Thorpe and Smith 1998; Groth et al. 2004; Markstein et al. 2008; Pfeiffer et al. 2008; Kvon 2015). By targeting all reporters to the same “landing site,” they are directly comparable, and only a single transgenic line per construct needs to be analyzed. The second way to minimize genomic position effects utilizes insulator sequences in transgenesis vectors. Insulator DNA elements were first identified by their potential to block gene function when interposed between enhancer and promoter (Kellum and Schedl 1992; Cai and Levine 1995; Kuhn et al. 2003), and to insulate transgenes from effects of surrounding chromatin (Kellum and Schedl 1991; Roseman et al. 1993, 1995). These faculties of insulators seem to derive from their strong propensity for interaction with each other (Kuhn et al. 2003; Chetverina et al. 2008; Kyrchanova et al. 2008b, 2011). Such interactions are proposed to form chromatin loops bridging even distant loci at the base of the loop and isolating interactions inside the loop from interactions outside the loop (Blanton et al. 2003; Byrd and Corces 2003; Doyle et al. 2014; Cubeñas-Potts and Corces 2015).
We started using both of these approaches to characterize enhancer modules of two neighboring Drosophila genes E(spl)m7 and E(spl)m8. To that end we generated a series of reporter constructs flanked by two copies of the insulator sequence from the gypsy transposon (gypsy insulator, GI) and integrated each construct into the same attP locus. When we tested two different reporters in a heterozygous configuration, we noted that they markedly affected each other’s expression in trans. The ability of enhancers to activate promoters in the homologous locus is called transvection and was first reported in Drosophila, as a phenomenon of pairing-dependent intragenic (unexpected) complementation in loci like Ultrabithorax (Ubx) (Lewis 1954; Martínez-Laborda et al. 1992), decapentaplegic (dpp) (Gelbart 1982), yellow (Geyer et al. 1990) and white (Babu and Bhat 1980). Subsequent studies employing randomly integrated P-element transgenes showed that the Drosophila genome is generally permissive to enhancer action in trans (Chen et al. 2002; Kravchenko et al. 2005) offering a first glimpse of the molecular basis of this phenomenon and reconfirming the need for somatic homolog pairing (or synapsis). In dipterans, like Drosophila, homolog synapsis is not limited to germline meiotic cells, but is very common in somatic tissues. This is in contrast to mammals, where somatic homolog synapsis seems to happen only in special occasions, and, accordingly, only sporadic cases of transvection have been reported (McKee 2004; Heride et al. 2010; Apte and Meller 2012; Stratigi et al. 2015; Joyce et al. 2016). The development of site-specific integration methods in Drosophila (such as the ΦC31-based recombination method) has recently rekindled the interest in transvection (Lee and Wu 2006; Bateman et al. 2012a; Mellert and Truman 2012; Fujioka et al. 2016). However, the mechanism of transvection is still poorly understood.
Using our series of reporter transgenes, we decided to search for sequence determinants of transvection. We found that this interaction is dependent on the presence of homotypic insulator DNA elements on both homologs. Our transgenesis vectors contained two such insulators: the Gypsy Insulator (GI) commonly utilized to protect transgenes from genomic position effects, and the Wari Insulator (WI) carried in the 3′ part of the mini-white marker gene. Two other insulators were also found to support transvection, while insulator removal from either of the trans-interacting transgenes abrogated transvection at five discrete genomic loci. While necessary, the presence of insulators was not sufficient to produce a robust transvection outcome. Parameters like the number, position and orientation of GIs relative to the trans-interacting enhancers and promoters proved to be of paramount importance. The implications of these results on the design of transgenes and on the broader role of insulators in transcription will be discussed.
Materials and Methods
Plasmid constructs
See attached Supplemental Material.
Fly maintenance and stocks
Flies were maintained under standard conditions at 25°. Stocks containing attP docking sites used for the integration of attB plasmids: attP40 (RRID:BDSC_25709), attP2 (RRID:BDSC_25710) (Groth et al. 2004; Markstein et al. 2008), VK2 (RRID:BDSC_9723), VK13 (RRID:BDSC_24864), VK37 (RRID:BDSC_24872), VK40 (RRID:BDSC_35568) (Venken et al. 2006); each carrying, and, if not, crossed to, a chromosome expressing ΦC31 integrase under the control of nanos derived from the attP40 stock (RRID:BDSC_25709) (Bischof et al. 2007). The su(Hw) mutant effects were assayed in the animals transheterozygous for su(Hw)e04061 null allele (RRID:BDSC_18224) (Thibault et al. 2004) and su(Hw)2 strong hypomorphic allele resulting in the 10 times decreased su(Hw) expression (RRID:BDSC_983) (Parkhurst et al. 1988; Harrison et al. 1993; Georgiev et al. 1997).
Integration of attB plasmids into attP fly lines
All plasmids in this study were integrated into a unique attP landing site, as specified in the text and figure legends for each transgene. Microinjection was performed as previously described (Ringrose 2009). A solution of 500 ng/µl plasmid DNA was microinjected into nanos- ΦC31; attP fly embryos. Flies that grew to adulthood were crossed with y w flies. Depending on the injected DNA construct, the mini-white or 3xP3-dsRed marker was used for subsequent screening, and for tracking the transgene.
Immunostaining and microscopy
Fixation and immunohistochemistry of embryos and larval tissues was performed according to standard protocols. CNSs and imaginal disks were dissected from late third-instar larvae, fixed with 4% paraformaldehyde, and labeled with rabbit polyclonal anti-GFP (Minotech Biotechnology) and mouse anti-β-galactosidase (Cat# Z3781, RRID:AB_430877; Promega) primary antibodies. Goat anti-rabbit IgG secondary antibody, Alexa488-conjugated (Cat# A-11034, RRID:AB_2576217; Thermo Fisher Scientific) was used for GFP detection. Goat anti-mouse, Alexa633-conjugated (Cat# A-21052, RRID:AB_2535719; Thermo Fisher Scientific) or donkey anti-mouse, Alexa647-conjugated (Cat# A-31571, RRID:AB_162542; Thermo Fisher Scientific) secondary antibodies were used for β-galactosidase detection. Samples were imaged on a Leica SP8 confocal platform using a 20× oil immersion objective with fixed zoom levels for each tissue type (CNS, wing, and eye disks). The images were pseudocolored in green (GFP), red (LacZ), and blue (DsRed). All samples within each figure were fixed and immunostained at the same time. Scanning of all figure samples was performed using identical microscope and software settings, and, when possible, completed within one imaging session to enable semi-quantitative comparison. Where scanning of all figure samples within one session was not possible, replicates of the samples from two chosen genotypes were rescanned together with the remaining samples in the next scanning session ensuring that the replicated samples are comparable. For each genotype, at least 10 wing disks, five CNSs, and three eye-antennal disks were scanned. Images were manipulated using ImageJ (pseudocoloring, rotation, and maximum intensity projection z-stacks) and arranged into data sets using Adobe Photoshop CC 2017 and Microsoft PowerPoint. Note that we used two different z-projections for some wing disk images. For the top part, containing the wing pouch, a full z-projection of the sample was done, while the bottom part, containing the notum and hinge, encompassed only sections containing the adult muscle precursors (AMPs). This was done to avoid confusing AMP expression with expression in the overlying tegula, a sensory organ primordium. Whereas AMPs and tegula can be easily distinguished in the 3D confocal stacks, they merge to one cluster upon z-projection. Since enhancer e7 activity is specific for the AMPs and not the tegula, we excluded the tegula sections from the z-projections shown.
Luciferase assays
Luciferase activity was measured using the Promega Luciferase Assay System Kit (Cat# E153A). CNSs and imaginal disks from 10 late-third-instar larvae were collected in 200 μl of 1× lysis reagent CCLR for each sample. Samples were collected over a series of days and stored at −80° until five independent samples were collected for each genotype. Samples were defrosted, put on ice, and homogenized using Kontes pestles. Homogenized samples were incubated at room temperature for 10 min and then centrifuged for 5 min to pellet tissue remains. The obtained homogenates were subsequently measured for luciferase activity and total protein content for normalization, then 20 µl of each homogenized sample was mixed with 50 μl of Promega Luciferase Assay Reagent and promptly measured on single tube luminometer (TD-20/20; Turner Designs). Total protein was measured using the Pierce BCA Protein Assay Kit (Cat# 23225); 10 µl of each homogenized sample was mixed with 200 µl BCA Working Reagent on clear-bottomed 96-well plates (Costar) and incubated at 37° for 1 hr. The plates were allowed to equilibrate to room temperature for 10 min before measuring absorbance on an Awareness Technology ChroMate Microplate Reader at 562 nm. Three replica plates were averaged for each sample. A standard curve was produced with BSA dilutions in Promega 1× lysis reagent CCLR.
Data availability
All plasmids and fly strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental Material (Figures S1–S11 and Supplemental Materials and Methods) were deposited to GSA Figshare https://gsajournals.figshare.com/s/d548db1426e0efad9e9f. Supplemental material available at https://doi.org/10.25386/genetics.7925462.
Results
Enhancer element analysis of E(spl)m7 and E(spl)m8 reveals a transvection phenomenon
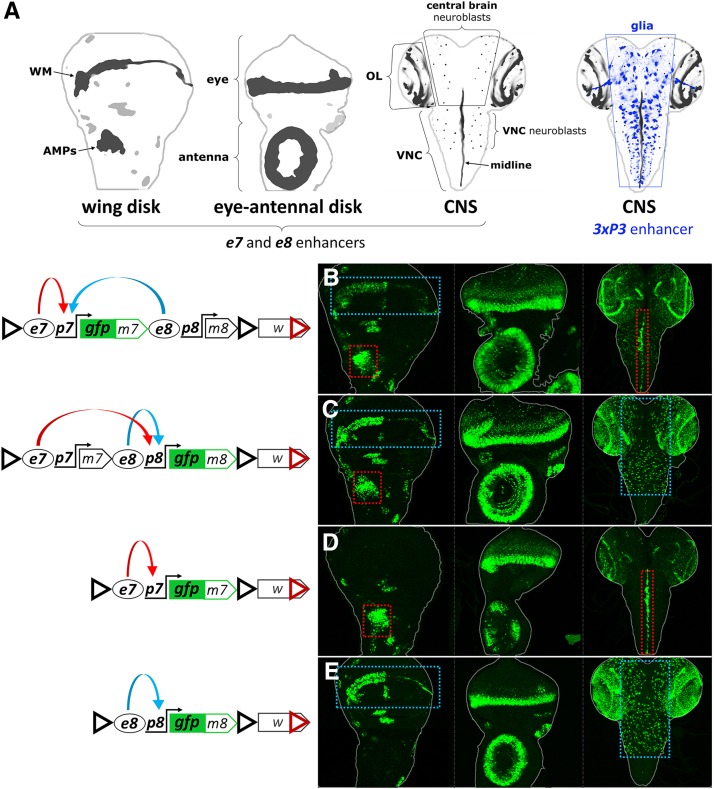
This work began with the aim to dissect the transcriptional regulation of two adjacent genes in the E(spl) complex, E(spl)m7 and E(spl)m8, both encoding transcription factors involved in many aspects of the response to Notch signaling. These are small, intronless genes and are thought to be regulated by their proximal flanking sequences (Delidakis et al. 2014). We cloned the 7 kb genomic fragment encompassing these two genes and tagged them with EGFP independently in two otherwise identical genomic constructs: GFPm7-m8 [EGFP fused to the open reading frame (ORF) of E(spl)m7, Figure 1B] and m7-GFPm8 [EGFP fused to the E(spl)m8 ORF, Figure 1C] (see Supplemental Materials for further features of these transgenes). Consistent with the known in situ hybridization expression patterns (de Celis et al. 1996), GFP-E(spl)m7 and GFP-E(spl)m8 displayed the same pattern in wing imaginal disks from third instar larvae: (1) in the region of wing margin (WM), and (2) in the adult muscle precursors (AMPs, or adepithelial cells) of the thorax, among other cells (Figure 1, B and C). Likewise, both constructs expressed GFP similarly in eye-antennal imaginal disks, whereas their central nervous system (CNS) patterns were different, especially apparent in the ventral nerve cord (VNC) where GFPm7 was expressed strongly in the midline, while GFPm8 was expressed mainly in the neuroblasts.
Figure 1.
e7p7 and e8p8 interact in cis. (A) Schematics of a wing disk, an eye-antennal disk, and two central nervous systems (CNSs), with the areas of m7 and m8 expression marked in shades of black. WM, wing margin; AMPs, adult muscle precursors; OL, optic lobe; VNC, ventral nerve cord. In the second CNS, a set of glial cells is indicated in shades of blue, corresponding to the expression pattern of the artificial 3xP3 enhancer (see later in Figure 4). (B and D) GFPm7 and (C and E) GFPm8 expression in wing disk, eye-antennal disk, and CNS (in left, middle, and right columns, respectively) from the EGFP-tagged constructs shown in the diagrams on the left panel; red dotted rectangles highlight e7-specific expression in the AMPs and in the midline of the VNC; blue dotted rectangles indicate e8-specific expression in the WM and in neuroblasts of the central brain and VNC. Also note that both enhancers drive expression in some common areas, e.g., the eye morphogenetic furrow. In the constructs’ schematics enhancers are shown as ovals (e7 and e8), promoters as bent arrows (p7 and p8), and insulators as triangles: black triangle: gypsy insulator (GI); red triangle: Wari insulator (WI), included in the 3′ of the mini-white marker gene. Blue and red curved arrows in the diagrams depict, respectively, e7 and e8 activities, which are shared between p7 (B) and p8 (C) in the wing disk.
We went on to characterize the patterns produced from individual enhancers located immediately upstream of E(spl)m7 and E(spl)m8 by generating shorter genomic constructs, GFPm7 and GFPm8 (Figure 1, D and E, respectively). GFPm7 contains the 2.1 kb sequence upstream of E(spl)m7 containing its putative enhancer, e7, and promoter, p7. GFPm8 contains the 1.3 kb 5′ flanking E(spl)m8 sequence, containing its putative enhancer, e8, and promoter, p8. GFPm7 and GFPm8 recapitulated the expression patterns seen for these genes in the longer m7-m8 transgenes, with two notable exceptions in the wing disk: GFPm7 lacked the wing margin (WM) (Figure 1D) and GFPm8 lacked the muscle precursors (AMPs) (Figure 1E). Additional, less conspicuous, differences in the bristle proneural clusters and the antennal primordium were noted, but we did not consider these any further. We conclude that these E(spl) genes contain two upstream enhancers, e7 and e8, which drive distinct expression patterns in the CNS and in the wing disk, and similar patterns in the eye disk. In the context of the genomic fragment encompassing both genes, e7 and e8 are shared between promoters of the two genes, p7 and p8, in the WM and AMPs (both genes expressed), but they act exclusively on their downstream gene in the VNC midline (only m7) and the neuroblasts (only m8).
We also created an e7p7-lacZ construct to permit simultaneous detection of e7p7-driven expression of β-galactosidase with e8p8-driven GFP from GFPm8 genomic constructs. Both constructs were inserted into the same chromosomal locus via the ΦC31 integration method (Markstein et al. 2008; Pfeiffer et al. 2010). e7p7-lacZ faithfully recapitulated the expression pattern of GFPm7. When we made heterozygous animals containing both transgenes (e7p7-lacZ and e8p8-GFPm8), we noticed that e7p7-lacZ displayed novel expression in the wing margin (characteristic of e8), while e8p8-GFP was expressed in the AMPs (characteristic of e7) (Figure 2C, cf. Figure 2, A and B). This effect was observed when both transgenes were inserted into the same attP landing site, either attP40 (chromosome 2) or attP2 (chromosome 3) (Figure 2, C and D). No such intertransgene interaction was observed when one transgene was inserted in attP40 and the other in attP2 (Figure 2, E and F), suggesting that homolog pairing is required for this interaction; in other words, we are observing a transvection phenomenon.
Figure 2.
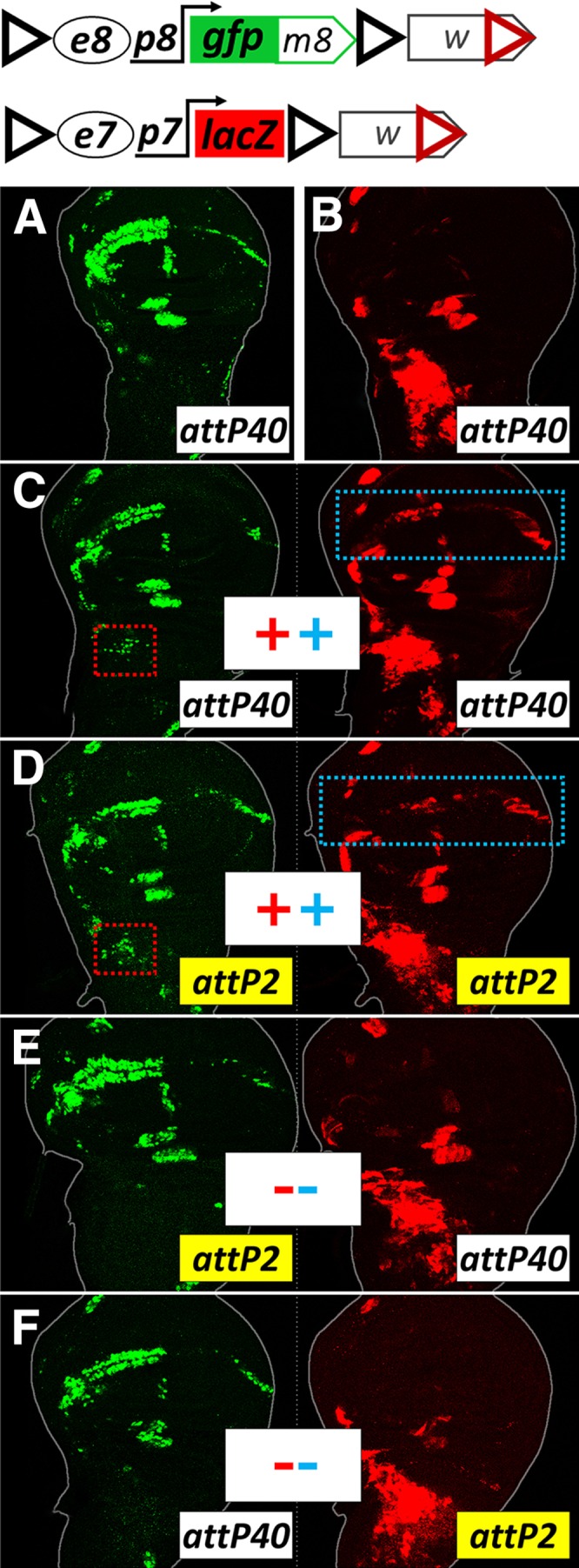
e7p7 and e8p8 interact in trans. (A and B) cis-expression patterns in wing disks isolated from hemizygous animals carrying e8p8-GFPm8 (A) or e7p7-lacZ (B) transgenes in the attP40 locus. (C–F) e8p8-GFPm8 and e7p7-lacZ crossed in the same animal. When present as heterozygotes in the same locus [(C) in attP40, (D) in attP2], both transgenes expand their pattern to locations dictated by their homologous transgene (marked with dotted rectangles). Occurrence or not of transvection is marked by a + or – symbol, respectively, in the middle of the panel. Red symbols refer to e7→p8 transvection, also marked by red dotted rectangles. Blue symbols refer to e8→p7 transvection, also marked by blue dotted rectangles. No transvection is observed when the e8p8-GFPm8 and e7p7-lacZ transgenes are placed in the same animal, but in nonhomologous loci (E and F).
Transvection is mediated by homotypic interactions between GIs or Wari insulators
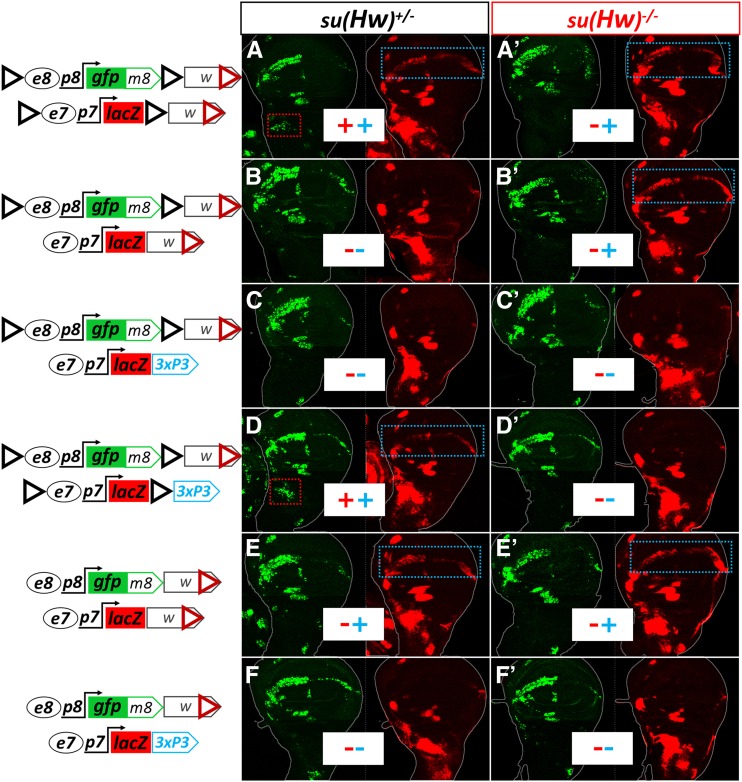
Having encountered robust bidirectional gene activation in trans (e8p8 activated by e7 in the AMPs and e7p7 activated by e8 at the WM), we decided to dissect the sequences that mediate this phenomenon. First, we asked whether the observed transvection is a general phenomenon requiring only an enhancer paired to a promoter, or whether it depends on additional transgene sequences. The constructs in Figure 2 were based on the pPelican vector (Barolo et al. 2000), which contains a pUC8 backbone, the mini-white gene, two gypsy insulators (GI) flanking a multiple cloning site (MCS), as well as a ΦC31 attB site inserted by us. In order to systematically address the role of vector sequences we recloned the e7p7-lacZ reporter construct into a minimal pBluescript backbone to which we added a ΦC31 attB site to enable fly transgenesis and either the mini-white gene or a 3xP3-dsRed marker which expresses DsRed in the adult eye in response to an artificial Pax6 (Toy/Eyeless) responsive enhancer (Berghammer et al. 1999; Horn et al. 2000). All constructs for this analysis were introduced into the attP40 locus.
Both pBluescript-based e7p7-lacZ constructs (mini-white and 3xP3-dsRed) expressed LacZ in the same pattern as the original pPelican-based reporter. However, when they were tested in trans to the original e8p8-GFPm8, no transvection was observed, namely we detected neither LacZ in the WM nor GFP in the AMPs (Figure 3, B and C, cf. A). We concluded that transvection at attP40 does not happen whenever an enhancer-promoter pair is placed in trans, but needs additional sequences from the vectors. Upon flanking the e7p7-lacZ-3xP3-dsRed reporter with two GIs in the same orientation as in pPelican, bidirectional transvection (e8→p7, blue dotted rectangle and e7→p8, red dotted rectangle in Figure 3D) was restored implicating the paired GIs as mediators of the effect. Because the GI is known to be bound by the Suppressor of Hairy wing Su(Hw) zinc-finger protein, which mediates its insulator activity (Parkhurst et al. 1988; Spana et al. 1988; Spana and Corces 1990; Holdridge and Dorsett 1991; Geyer and Corces 1992; Gerasimova et al. 1995; Ramos et al. 2006), we tested our transgene pairs in a su(Hw) mutant background. Indeed, e7→p8 (AMP) transvection was lost in this background, but, surprisingly, e8→p7 (WM) transvection was now apparent in all transgene combinations that had a mini-white marker in both homologs (Figure 3, A’, B’, and E’), even the one that had not displayed this effect in wild type (wt) genetic background (Figure 3, B vs. B’). We then modified the pPelican-based e8p8-GFPm8 construct by removing its GIs. This construct was capable of supporting e8→p7 transvection with the mini-white but not the 3xP3-dsRed version of the pBluescript-e7p7-lacZ reporter in both wt and su(Hw) genetic backgrounds (Figure 3, E–F and E′–F′). The simplest conclusion from these results is that e8→p7 transvection in the WM occurs when mini-white is present on both homologs, but this effect is annulled when GIs are placed nearby. Only upon removal of GIs from both homologs, or their inactivation by su(Hw) loss, is this effect observed. On the other hand, the presence of GIs in both homologs can sustain transvection in both directions, not only e8→p7 but also e7→p8.
Figure 3.
GIs and mini-white mediate transvection. Wing imaginal disks from animals heterozygous for variations of the e8p8-GFP-m8 and e7p7-lacZ transgenes. The two GIs in forward orientation, flanking the two reporters, are indicated by black triangles; marker genes are located downstream of the second (3′) GI: either the mini-white gene (“w”) or the 3xP3-dsRed cassette (‘3xP3′). An image of the same disk from each genotype is split into two channels: p8-driven GFP-m8 in green and p7-driven LacZ in red. Dotted rectangles (red and blue) indicate cells (AMPs and WM, respectively) exhibiting transvection – also indicated by + or – in the same color (red or blue). Each combination of transgenes (in rows) is tested in the presence of Su(Hw) [su(Hw)+/−, first column, (A–F)] and absence of Su(Hw) [su(Hw)−/−, second column, (A’–F’)]. All transgenes are inserted in the attP40 locus.
We mapped the transvection-mediating element of the white locus within its 3′ part: out of three subfragments derived from mini-white in Figure S1, A–C, only its 3′−most 0.9 kb recapitulated white-white-mediated transvection (Figure S1C). It was previously reported that this sequence contains the 3′-UTR as well as an insulator element, dubbed Wari (hereafter referred to as WI) (Chetverina et al. 2008). Therefore, either of two different insulators, GI and WI, can promote transvection when placed in a paired configuration (in both homologs) near an enhancer-promoter pair. WI has been shown to have su(Hw)-independent insulator activity, but also to interact with GI in cis (Chetverina et al. 2008). When we confronted a WI-containing e7p7-lacZ with a WI-containing (mini-white) e8p8-GFPm8 flanked by GIs (Figure S1D), transvection was abolished, consistent with what we had observed earlier with the entire mini-white (Figure 3B). We hypothesized that this inhibition could result from presumptive insulation imposed by the GI located between e8 and WI in e8p8-GFPm8, thereby restricting access of WI to e8. This was not the case, as the inhibition of WI-mediated transvection was sustained even when we deleted the 3′ GI, leaving only the 5′ GI intact (Figure S1E). Thus, a heterotypic GI-WI interaction in cis can disable the homotypic WI-WI-mediated transvection but not the GI-GI-mediated transvection. However, we should emphasize that this inhibition was context-dependent and quite an opposite action of GI, namely enhancement of WI-WI-mediated transvection, was also possible in a different context (described later in this manuscript – see Figure 9).
Figure 9.
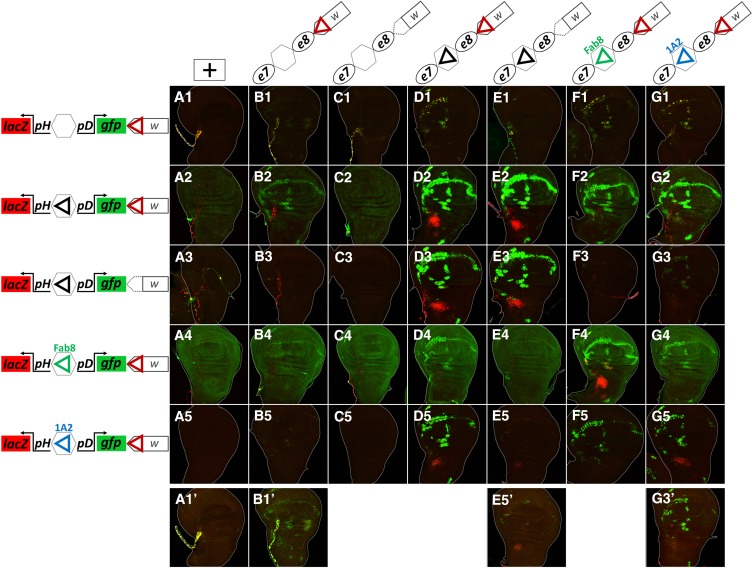
1A2 and Fab-8 insulators also mediate transvection. All panels show merged GFP (green) and LacZ (red) channels of confocal z-projections from third instar wing disks. (A1–A5) Wing disks from animals hemizygous for responder transgenes; insulators are shown as triangles. (B1–G5) Wing disks from heterozygotes for sender transgenes (as indicated in each column) with responder transgenes (as indicated in each row). All transgenes are inserted in attP40. (A1–G5) were imaged at the same intensity settings. (A1’, B1’, E5′ and G3′) are higher-sensitivity images of the respective panels, to reveal very low levels of transvection. Note that the responder construct in row 3, as well as the sender constructs in columns (C and E), are deleted for WI (red triangle).
GI-mediated transvection is promiscuous, whereas WI-mediated transvection is highly selective
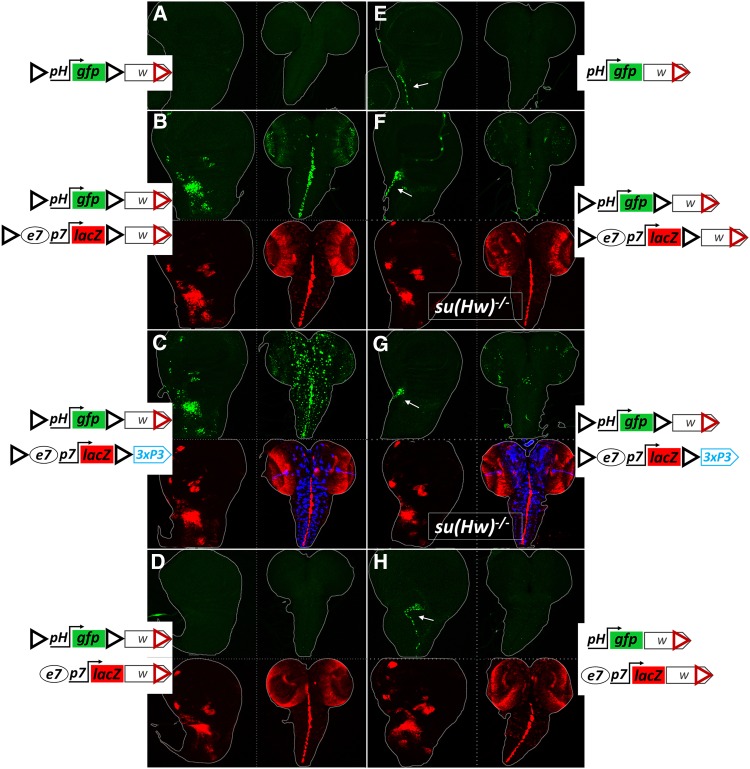
Because e7p7 and e8p8 interact in cis in their native context (Figure 1, B and C), it raises the possibility that the transvection we observed is tied to the specificity of these two regulatory modules for each other. Thus, we sought to determine whether GIs and WIs could mediate transvection of e7p7 and e8p8 to an unrelated, heterologous promoter. To address this question, we generated an enhancerless construct containing a basal promoter commonly used in assaying enhancer activity, derived from the Drosophila hsp70 gene (hereafter referred to as pH). As expected, a transgene carrying this minimal pH promoter fused to GFP, flanked by GIs and marked with mini-white displayed, on its own, no expression in wing disks or CNS (Figure 4A). When crossed to an e7p7-lacZ construct, congruent in two flanking GIs and mini-white, GFP was detected with the e7-specific pattern [Figure 4B cf. GFP (trans) to LacZ (cis)]. This activation of pH in trans relies on homotypic interaction between GIs, as disparity between transgenesis markers (but retention of congruent GIs) did not affect transvection of e7 (Figure 4C), while removal of GIs from one of the transgenes (with congruent mini-white markers) abolished the effect (Figure 4D). Unlike the e8p8→e7p7 transvection, removing GIs entirely, but keeping congruent mini-white markers did not support transvection (Figure 4H). Therefore, paired WIs cannot support transvection between e7 and pH.
Figure 4.
GIs, but not WIs, mediate transvection of e7 and 3xP3 enhancers to a heterologous, enhancerless hsp70 promoter (pH). Representative late larval wing disks and CNSs from the indicated genotypes are shown. (A–H) Projections of trans (GFP, green) and cis (LacZ red and DsRed blue, when present), expression from the same wing disk or CNS of each sample. ph-gfp produces no (cis) expression as a hemizygote (A and E), other than tracheal branch in the uninsulated version [white arrow in (E)], probably originating from enhancer trapping. The same is observed in the insulated version upon GI inactivation in the su(Hw)−/− background (F and G). Only combinations of transgenes with GIs in both homologs support transvection [GFP in AMPs and CNS in (B and C)], while congruency in white in the absence of GIs in one or both homologs does not (D and H). Depletion of Su(Hw) protein abolishes transvection [(F and G), which bear the same transgenes as (B and C), respectively]. Note a dotted pattern in the CNS in (C), manifested by the 3xP3 activity in cis (DsRed, in blue) and in trans (GFP, in green), which comes from a glial cell population (see blue pattern in Figure 1A). The artificial 3xP3 enhancer does not drive expression in wing disks.
Interestingly, in this set of experiments, GIs mediated trans-activation of the pH promoter not only by the e7 enhancer located in between the two GIs, but also by an enhancer exterior to the two GIs: the 3xP3, which displays strong expression in a subset of glia in the CNS (Figure 4C). Depletion of Su(Hw) suppressed transvection of both enhancers, e7 (Figure 4, F and G) and 3xP3 (CNS in Figure 4G), and allowed pH to trap a tracheal enhancer (in cis) in the vicinity of attP40 (white arrows in Figure 4, F and G). The fact that two unrelated enhancers, e7 and 3xP3, can transvect to a heterologous promoter, pH, suggested that GI-mediated transvection is unselective for enhancer-promoter pairs. This prompted us to use the GIs-containing transgene system to screen for putative enhancer elements across the 50 kb long E(spl) locus. A collection of 18 fragments of this locus inserted in a transgene between two GIs activated specific patterns of expression in trans from the GI-flanked pH-gfp transgene (Figure S2) – a full 15 out of these 18 fragments displayed robust enhancer activity in third-instar larval disk/CNS tissue. Additionally, using this system, we were able to recapitulate in trans the cis-pattern of another enhancer unrelated to the E(spl) locus, the vestigial quadrant enhancer (last column, Figure S2).
Unlike GI, WI-mediated transvection was specific for the e8p8/e7p7 combination and mediated trans-activation of p7 by e8 unidirectionally. To test the possibility that WI-mediated transvection is specific for the E(spl)m7 promoter (p7), we tested various minimal GFP reporters driven by different basal promoters, pH, p7, and E(spl)m8 promoter (p8). We confronted these enhancerless nonexpressing reporters with an e8p8-lacZ transgene. We made sure that all combinations were congruent for both GIs and mini-white, which enabled us to simultaneously test GI-mediated transvection in a wt background and WI-mediated transvection in the absence of Su(Hw) (Figure S3). All three basal reporters responded to e8 enhancer in a su(Hw)+/− background (GI-dependent transvection); the p7 was by far the strongest responder, with pH following and p8 showing a very weak activation (Figure S3, G–I). However, in the su(Hw)−/− background (WI-dependent transvection), the e8 enhancer did not transvect to any of the three promoters (Figure S3, M–O, compare to Figure S3, G–I). Therefore, the unidirectional e8p8→e7p7 transvection supported by WIs was not due to a selectivity of WI for p7. When the three basal promoters were fused to the e7 enhancer (Figure S3, D–F), these reporters started expressing (as hemizygotes) in the AMPs and some proneural clusters, but not in the WM, consistent with the activity of the e7 enhancer (Figure S3, D–F). Once confronted in trans with e8p8-lacZ, these e7-bearing reporters were able to robustly express GFP in the WM (Figure S3, J–L) and this expression was retained in the su(Hw) mutant genetic background (Figure S3, P–R). Thus, the transvection mediated by the interaction between WIs was not promoter context specific, but rather enhancer context specific, with the responding gene requiring the presence of the e7 enhancer in cis in order to sustain WI-mediated transvection. In conclusion, whereas trans-paired GIs mediated transvection between any enhancer-promoter pair tested, trans-paired WIs were more selective and mediated only e8→e7 transvection, an effect that was dominantly suppressed by the presence of GI elements. This e8→e7 transvection may reflect some intrinsic affinity of these two enhancers for each other, but it still requires the presence of GIs or WIs in order to materialize.
Transvection is weaker than cis enhancer-promoter activity and is suppressed by promoter cis-preference
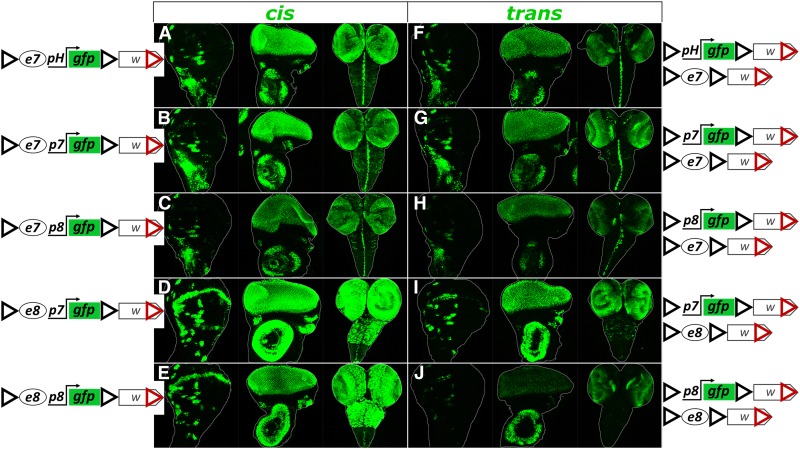
To gauge the relative strength of transvection compared to cis enhancer-promoter (e-p) interaction we generated e-p pairs driving GFP expression in cis and compared them to the same e-p pairs driven in trans. All constructs designed for this purpose were based on the backbone of pPelican/pStinger vectors (Barolo et al. 2000), which contain two GIs to enable GI-mediated transvection, and subsequently inserted into the attP40 locus. These flanking GIs also provided efficient insulation: all enhancerless promoter-reporter constructs had undetectable levels of expression in all three larval tissues tested (wing disk, eye disk, and CNS; data not shown). In all cases we observed lower GFP levels from transvection than from the cis-combination (Figure 5). We tested three promoters, p7, p8, and pH. Regardless of the enhancer assayed (e7 or e8) the strongest expression levels, both in cis and in trans, were produced by p7, whereas p8 was the weakest out of the three promoters. This suggests that a promoter’s strength for driving transcription is its intrinsic property and does not depend on the enhancer activating it, at least for the two enhancers tested.
Figure 5.
Transcriptional activity of different enhancer-promoter (e-p) pairs interacting in cis or in trans in the presence of GIs. Each genotype is examined for GFP expression in the third instar wing disk, eye disk and CNS. (A–E) Samples from hemizygotes of a transgene containing a given e-p pair linked in cis show higher levels of GFP expression than samples from heterozygotes containing same e and p, in a trans configuration (F–J). All transgenes are inserted in attP40.
We also studied the embryonic cis vs. trans expression of e7 using the pH promoter constructs. In cis, e7 displayed very dynamic expression, starting at the mesectoderm in stage 7 (3 hr after egg laying, AEL), then in a neuroectodermal cluster pattern up to stage 10 (5 hr AEL) and later in the VNC midline and the epidermis in a complex segmentally repeated pattern. Interestingly, the earlier patterns could not be transvected or were transvected only in sporadic cells. From stage 11 (6 hr AEL) onward, this sporadic transvection gave way to a more complete one, where the e7 trans pattern recapitulated the cis pattern (Figure S4). This correlates nicely with what is known about somatic homolog pairing: whereas in early embryonic stages paternal and maternal homologs start out unpaired, they gradually increase their pairing and reach maximum levels by about stage 11 (Hiraoka et al. 1993; Fung et al. 1998; Gemkow et al. 1998). Therefore, this observation, corroborates the need for homolog pairing in order for transvection to take place.
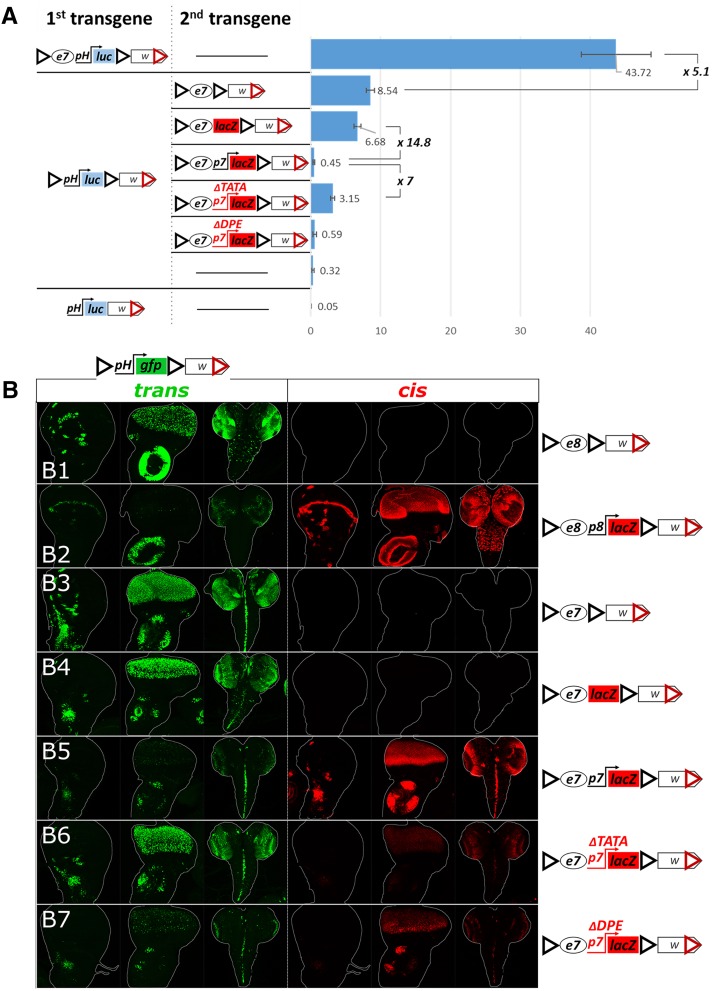
To obtain a more quantitative measure of the cis vs. trans activity of an enhancer, we used a luciferase (luc) reporter (instead of GFP) and measured its activity in extracts of larval disk-brain complexes. The GI-flanked e7-pH-luc reporter showed five times higher activity in cis than the e7 driving an enhancerless pH-luc reporter in trans (both GI-flanked, Figure 6A). This trans activity was still much higher (∼26×) than the basal levels of the pH-luc reporter. Interestingly, in this assay, pH-luc basal levels were low, yet detectable, even though the GFP counterpart had undetectable levels of GFP in the same tissues – this probably reflects the higher sensitivity of the luc vs. the GFP reporter (Arnone et al. 2004). Upon removal of the GIs, luc reporter activity dropped to almost undetectable levels, which is consistent with previous observations that GIs can stimulate basal transcription from some promoters (Wei and Brennan 2001; Golovnin et al. 2005; Markstein et al. 2008; Soshnev et al. 2008).
Figure 6.
Regulation of GI-mediated transvection by cis-preference. (A) The chart shows levels of basal and e7-induced (in cis and in trans) pH-driven luciferase activity. Levels of luciferase activity were measured from third instar larval disk-brain complexes. Luciferase values normalized to total protein are shown as arbitrary units (a.u.). The mean and SD of five replicates is shown. The activity of luciferase transgenes (first column in the construct panel) is assayed on their own (as hemizygotes; horizontal line in the second column) or in combination with a second transgene in trans. (B1–B7) Transgenes containing e8 or e7, with or without a cis-linked promoter (depicted on the right), were placed in trans to pH-gfp. For each genotype (each row) the third-instar wing disk, eye disk and CNS were examined for (1) GFP expression (green), reflecting trans activity of the enhancer-containing transgenes on pH-gfp and (2) β-galactosidase expression (red), reflecting cis activity of the enhancer-linked promoter, when lacZ is present. All transgenes contain GIs and mini-white and are inserted in attP40.
In the above experiments, we noticed that confronting an enhancerless reporter in trans to a solitary enhancer gave more robust expression compared to all our previous experiments, where the transvecting enhancer was linked in cis to a promoter (Figure 6, cf. B1 and B2 for e8, and B3 and B4 with B5 for e7). This came as no surprise, since numerous earlier studies on transvection have indicated that an enhancer’s action in trans is suppressed by the presence of a promoter in cis (Geyer et al. 1990; Martínez-Laborda et al. 1992; Hendrickson and Sakonju 1995; Casares et al. 1997; Sipos et al. 1998; Morris et al. 1999a,b, 2004; Bateman et al. 2012a; Kravchuk et al. 2017). Different promoters of varying core element composition have been reported to display cis-preference (i.e., to attenuate transvection). As a general rule, mutations compromising transcriptional strength of a cis-promoter usually release the enhancer toward trans action (Morris et al. 1999b, 2004; Lee and Wu 2006). We made two mutations on the p7 promoter in an attempt to compromise its strength without completely inactivating it. p7 is a multi-element promoter, containing a TATA box, an initiator (Inr) and a downstream promoter element (DPE) (Klämbt et al. 1989; Kutach and Kadonaga 2000). We introduced two deletions into the e7p7-lacZ construct aiming to disrupt each of these activities; one, e7p7-ΔTATA-lacZ, removed the TATA box [deletion of −41 to −22 bp relative to the transcription start site (TSS)] and another, e7p7-ΔDPE-lacZ, removed the Inr and DPE elements (deletion of −16 to +67 bp relative to TSS). Both of these promoter mutations retained weak yet detectable transcriptional activity (Figure 6, B5–B7, cis column). Even though the reduction in cis promoter activity was comparable between ΔTATA and ΔDPE, the two had dramatically different effects on transvection of the linked e7 enhancer (Figure 6, B5–B7, trans column). p7-ΔTATA partially relieved e7 from cis-preference inhibition, leading to much higher trans-activation of pH-gfp than that observed with p7-ΔDPE. These effects of the mutant cis-p7 promoters on e7 transvection were independent of the identity of the trans promoter, as p7 and p8 –based enhancerless reporters responded with a similar trend (Figure S5). When the various e7p7-lacZ versions were confronted with the pH-luc reporter, we confirmed that the DPE deletion was comparable to the unmutated promoter in strongly suppressing transvection (11–15× weaker than a promoterless e7-lacZ), whereas the TATA deletion released e7 from cis-preference giving 6–7× stronger trans reporter expression (compared to the unmutated e7p7 or the e7p7ΔDPE) (Figure 6A). Interestingly, in this assay the intact e7p7 (and e7p7-ΔDPE) produced very low trans-activation of the pH-luc reporter, only 1.4–2× higher than its basal levels attained in the absence of a transvecting enhancer. We speculate that this reflects the ability of the transvecting e7 enhancer to activate pH-luc in a number of cells (as visualized by the pH-GFP reporter), but, at the same time, to repress the basal pH-luc activity in the remaining cells. Unfortunately, we can only measure the resultant luciferase activity in the whole brain-disk extract with no cell-to-cell resolution, so we cannot test this scenario. Regardless, these luciferase constructs enabled us to obtain a quantitative measure of transvection strength, which ranged from 5× to almost 100× lower than the cis output of the same promoter-enhancer pair, depending on the presence of a cis-linked promoter, and, in particular, the integrity of its TATA box.
Relative position, number, and orientation of GIs determine transvection effects
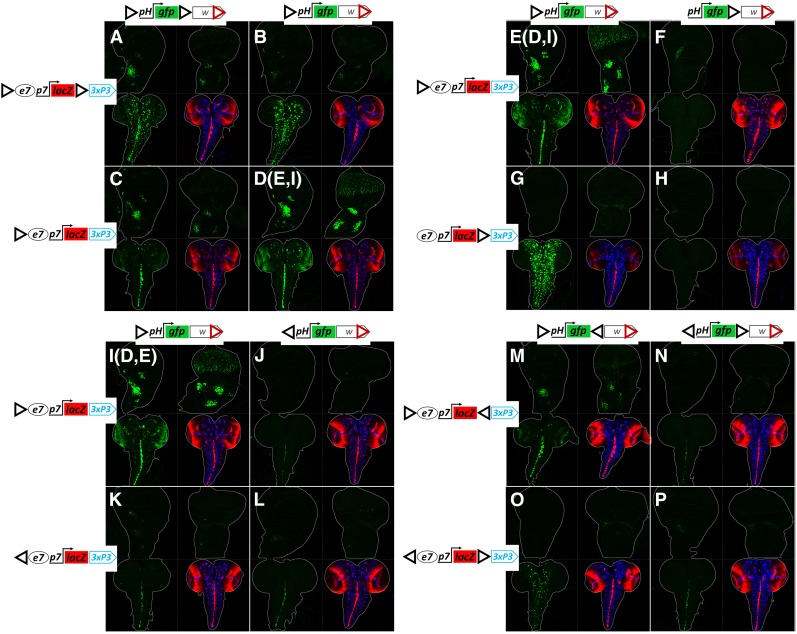
In all the previous experiments, all our GI-based transgenes contained two GIs each in forward orientation (GIsFOR) as in the pPelican and pStinger series vectors: the “5′ GIFOR” at the 5′ end of each construct (following only the ΦC31 attB integration site), and the “3′ GIFOR,” 3′ to the lacZ or gfp reporters (preceding the 3xP3 or mini-white marker genes, Figure S6). GI is 367 bp long and its “forward” orientation is the same as the one found in the original gypsy transposon, where GI is located shortly downstream of the 5′ LTR (Spana et al. 1988, Figure S7). The two transgenes (e7p7-lacZ and pH-gfp) in this starting configuration of GIs (hereafter referred to as the “dual-GIsFOR” configuration), when presented to each other in trans, result in expression of GFP in two distinct patterns: e7-specific in all tested tissues (wing disk, eye disk, and CNS) and 3xP3-specific in the CNS (Figure 4 and Figure S6). Thus, the pH located in between the two GIsFOR receives input from two enhancers in trans: e7 – located downstream of the 5′ GIFOR, and 3xP3 located downstream of the 3′ GIFOR. We have introduced a series of modifications in the configuration of the GIs within these two constructs in order to understand how their relative number, position, and orientation influence transvection. All resultant constructs for this analysis were introduced into the attP40 locus.
Deletion of the 3′ GIFOR in the transvection-receiving construct, pH-gfp, while preserving the dual-GIsFOR configuration in the “sending” e7p7-lacZ construct, caused a reduction in transvection of e7, with concomitant increase of 3xP3 transvection (compare Figure 7, A to B). When the 3′ GIFOR was deleted in the “sending” construct, and the “receiving” construct was kept in its initial dual GIs configuration, the transvection of e7 seemed unaffected, while transvection of 3xP3 was nearly lost (compare Figure 7, C to A). Finally, deletion of the 3′ GIsFOR in both constructs led to augmented GFP expression with an e7 pattern, and an almost undetectable 3xP3 pattern (cf. Figure 7, D and A). These data demonstrate that a robust trans-activation of the pH promoter by the e7 enhancer is mediated via an interaction between the 5′ GIsFOR of the two transgenes. This interaction seems to be weakened by the presence of a 3′ GIFOR in either or both of the interacting transgenes. However, the 3′ GIFOR in the sending construct is required for effective transvection of the 3xP3, suggesting that the 3xP3 enhancer, like e7, needs an adjacent GI in order to robustly act in trans.
Figure 7.
The relative position, number, and orientation of GIs determine transvection effects. (A–P) Confocal z-projection of GFP expression in third instar larval wing disk (top left panel for each genotype), eye disk (top right), and CNS (bottom left). Bottom right shows the merged e7p7-driven LacZ (red) and 3xP3-pH-driven, DsRed expression (blue) in the same CNS as the bottom left panel; for patterns of e7p7-lacZ expression in disks, see previous figures (Figure 2, Figure 3, Figure 4, and Figure 6); 3xP3-pH-dsRed shows no expression in third instar imaginal disks. Each genotype contains a pH-gfp transgene with mini-white in trans to an e7p7-lacZ transgene with 3xP3-dsRed and various arrangements of GIs (black triangles), as indicated. Note that (D, E, and I) represent different samples obtained from the same genotype. All transgenes are inserted in attP40.
Consistently, when the dual-enhancer construct (e7p7-lacZ-3xP3-dsRed) contained a single GIFOR, only its downstream enhancer was transvected to a GIFOR–preceded pH-gfp: e7 (with weak sporadic activity of 3xP3) (Figure 7, D, E, and I) or 3xP3 (Figure 7G). Moreover, the presence of the GIFOR upstream of the pH promoter was essential for its activation by a trans-enhancer, as its deletion abolished transvection altogether, even when another GIFOR was present at the 3′ position (Figure 7, F and H). It therefore seems that the trans-activity of both enhancers (e7 and 3xP3) obtained from the dual-GIsFOR e7p7-lacZ transgene (Figure 7, A and B) resulted from the interaction between the 5′ GIFOR preceding pH with the two GIsFOR of e7p7-lacZ in trans, each upstream of each enhancer.
We next addressed the significance of the orientation of GIs in mediating transvection. Surprisingly, the presence of a reversed GI in the 5′ position (5′ GIREV) in either of the two transgenes strongly reduced transvection effects, even when both transgenes contained 5′ GIREV (Figure 7, J–L compared to I). Therefore, trans interaction between enhancers and promoters is favored when both are located on the 3′ side of GI. Weak transvection, on the other hand, can be sustained regardless of 5′ GI orientation. In fact, even incongruent combinations of 5′ GIs (one FOR and the other REV, Figure 7, J and K) displayed weak transvection, suggesting that it is not so much the congruence of the trans-insulator pair, but rather the absolute orientation of both GIs that is needed for robust transvection.
The fact that the preferred position of the trans-interacting enhancer-promoter is on the 3′ side of the two GIs made us consider the possibility that placing the 5′ and 3′ GIs in a convergent orientation (i.e., 5′ GIFOR/3′ GIREV) in both constructs might strengthen trans-interaction. However, this was not the case, as such transgenes produced equal levels of transvection to those with 5′ and 3′ GIs in the forward orientation (Figure 7M compared to A) and less than the combination where 3′ GIs are absent altogether (Figure 7D). These results suggest that the interaction between 5′ GIFOR-preceded enhancer and trans-promoter is weakened by the presence of a second (3′) GI in cis, irrespective of its orientation. Moreover, transgenes with a divergent configuration of GIs (5′ GIREV/3′ GIFOR) did not improve the weak transvection observed between transgenes with a single 5′ GIREV (compare Figure 7, P to L), nor were incongruent 5′ GI configurations improved by a 3′ GI (Figure 7, N and O) (note, however, that 3xP3 was efficiently transvected to pH in Figure 7O as both 3xP3 and pH are preceded by GIFOR).
Is the ability of a single 5′ GIFOR to support transvection a peculiarity of the attp40 locus or can it happen in more genomic loci? We tested constructs with a single 5′ GIFOR in four more attP loci and we got robust transvection in all cases (Figure S8). Importantly, removing the 5′ GIsFOR from the transgenes abolished transvection in all loci, reconfirming the need for paired homotypic insulators in both homologs. As a corollary, we conclude that, at least in the five genomic loci we tested, nearby endogenous genomic insulators were not capable of mediating transvection from the GI-less constructs, suggesting that “insulator trapping” is probably not a common phenomenon in the Drosophila genome. On the contrary, enhancer trapping is very common; we were able to detect some non-e7 dependent patterns of expression of our transgenes in four out of the five loci tested (all except VK40; see Figure S8). Why nearby insulators are unable to support transvection in the GI-less constructs is not clear. Although the chromatin occupancy of many insulator-binding proteins has been described, only a fraction of these bound sites act as insulators in functional assays (Soshnev et al. 2008; Schwartz et al. 2012; Van Bortle et al. 2014). With this caveat in mind, the putative insulator landscape of each of the landing sites used is shown in Figure S9: the closest putative insulator could map anywhere from 1 (VK40) to 25 kb (VK37) away from our GI-less transgenic reporters and yet no transvection is observed.
In summary, transvection needs homotypic insulators in both homologs, but having homotypic insulators is not sufficient. The outcome is also influenced by the position, orientation, and number of these insulators. In the context of the e7p7-lacZ-3xP3 → pH-gfp transvection, both GIs have to be 5′ of the pH promoter and directly adjacent to the transvecting enhancer, e7 and/or 3xP3. The FOR orientation is greatly favored for both homologs; the REV orientation produces a much weaker effect. Finally, adding another GI in one or both transgenes weakens the 5′GIFOR mediated transvection. It should be kept in mind, however, that the transcriptional outcome of a trans-interacting insulator pair, is also greatly dependent on the enhancers and promoters located in the vicinity of these GIs: when the same e7p7-lacZ series of transgenes was tested in trans to an e8p8-m8GFP series (instead of the pH-GFP), the e7p7→e8p8 transvection largely obeyed the above rules, but the e8p8→e7p7 transvection was detectable even with a single 3′ GI, regardless of orientation (Figure S10). Still, the single 5′ GIFOR configuration gave the strongest trans-effect even with this transgene combination. The removal of GI from either homolog completely abolished the effect, as expected.
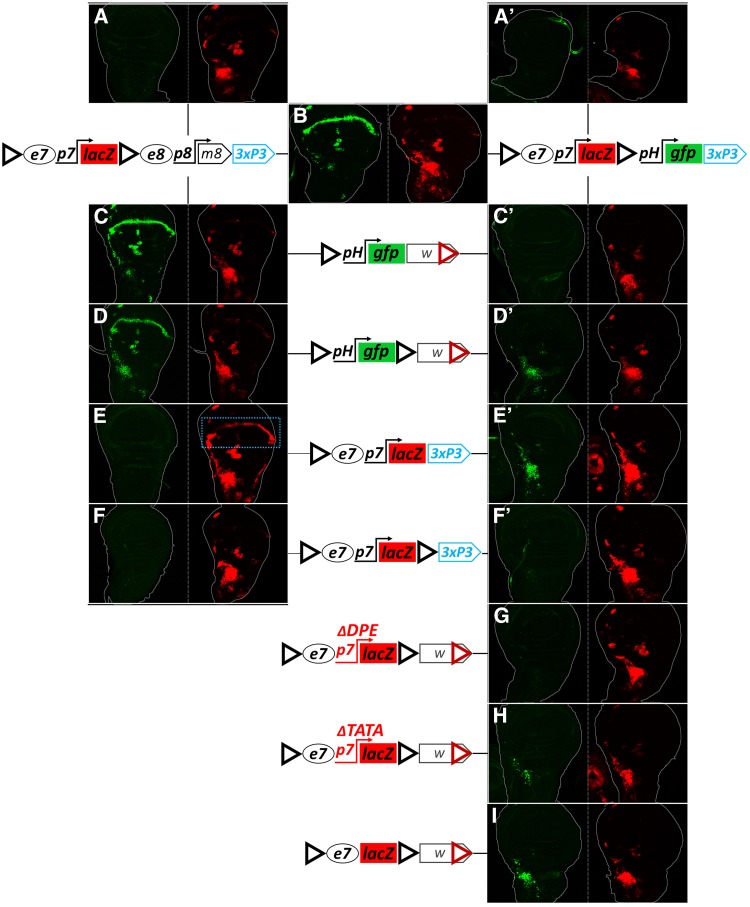
Why was the addition of a 3′ GI detrimental to e7p7→ pH transvection (Figure 7, A–D and M–P)? Could it potentially engage in homotypic interactions with either the cis or the trans 5′ GI that might compete with the ability of these 5′ GIs to support transvection? To gain insight into the activity of this 3′ GIFOR, we appended a “tester” module to our GIFOR-e7p7-lacZ-GIFOR-3xP3-dsRed transgene. Two different modules were cloned immediately 3′ of the 3′ GIFOR: (1) a “receiving” pH-gfp module or (2) a “sending,” e8p8-m8 module (e8p8 driving an untagged E(spl)m8 CDS). Figure 8 presents the results obtained in wing disks, which are consistent with those obtained in the CNS and eye-antennal disk (data not shown). The tester modules did not influence e7p7-lacZ expression: both transgenes, on their own, expressed LacZ in the AMPs, as expected (Figure 8, A and A’); an e8-specific wing margin LacZ pattern was not detected, consistent with insulation of e7p7-lacZ from e8 (Figure 8A); similarly, the transgene containing the insulated enhancerless pH-gfp module showed no GFP expression (Figure 8A’), suggesting that the 3′ GI in this construct insulates e7p7 from pH, instead of enabling their interaction. It therefore seems that the two GIsFOR in these tester constructs do not productively interact in cis (see also Cai and Shen 2001; Kyrchanova et al. 2008a). When tested against each other, we observed a robust activity of the pH-gfp module in the WM, indicative of a trans interaction between the two 3′ GIsFOR resulting in e8p8-m8→pH-gfp transvection (Figure 8B). Therefore, 3′ GIs prefer to homotypically interact in trans. The presence of only few GFP positive AMPs, and no apparent WM LacZ expression in this combination (Figure 8B), demonstrate that the “diagonal” interactions between GIsFOR (i.e., 5′ GIsFOR −3′ GIFOR) are less favored than the “vertical” ones (5′-5′ and 3′-3′).
Figure 8.
In dual GI transgenes, both 5′ and 3′ GIs participate in trans-interactions. Two tester transgenes, GIFOR-e7p7-lacZ-GIFOR-e8p8-m8 (left column, (A–F)), and GIFOR-e7p7-lacZ-GIFOR-pH-gfp (right column, A’–F’, G–I) were tested as hemizygotes (A and A’), combined inter se (B) or combined with various other GI-containing transgenes in attP40 (C–I). Third instar wing imaginal disks are shown for each genotype in two channels (GFP in green, LacZ in red). Any GFP expression is caused by transvection, since the hemizygotes of all GFP constructs used express no GFP, other than a piece of trachea in (A’), probably due to enhancer trapping. LacZ expression in the AMPs may come from cis or trans, but WM LacZ expression, marked by a blue dotted rectangle in (E), is triggered by the e8 enhancer in trans.
Placing the 5′ GIFOR pH-gfp transgene in trans to the testers gave strong transvection of e8 (via the 3′ GIFOR; Figure 8C) but only weak or no transvection of e7 (via the 5′ GIFOR; Figure 8, C and C’). Interestingly, e7→pH transvection with both testers was augmented when a 3′ GIFOR was added to the responding pH-GFP transgene, as evidenced by broadly expressed GFP in the AMPs (Figure 8, D and D’). Therefore, when confronted with dual GIsFOR, a single GIFOR preferentially interacts with the trans 3′ GIFOR, but this shifts to the 5′ GIFOR when a second GIFOR is added, resulting in a dual/dual configuration. This conclusion was supported by confronting the tester transgenes with a single vs. dual GIFOR configured e7p7-lacZ reporter. Only the single 5′ GIFOR was able to interact in trans with the two tester modules, as evidenced by e8p8-m8→e7p7-lacZ and e7p7-lacZ→pH-gfp transvection (Figure 8, E and E’), whereas both of these transvection effects were lost in the dual/dual configuration (Figure 8, F and F’). Surprisingly, e7p7-lacZ→pH-gfp transvection was regained when the p7 promoter was compromised or (better) deleted in the dually GIFOR flanked e7p7-lacZ transgene (Figure 8, G–I). We therefore conclude that GIs sample different homotypic interactions in trans, both vertical and diagonal. The latter are disfavored, but can still occur and their ability to support transvection is influenced by the enhancer-promoter interactions in their vicinity, in agreement with recent live imaging data that show that insulator–insulator interactions (both cis and trans) are stabilized when accompanied by productive enhancer-promoter interactions (Chen et al. 2018; Lim et al. 2018). For example, the e7p7-pH interaction is not sufficient to sustain diagonal transvection (Figure 8F’), unless relieved from cis promoter preference (Figure 8, H and I). As another example, a similar diagonal GI–GI interaction can sustain transvection of e8 (from the e8p8-m8 tester) to pH-gfp (Figure 8D), but not to e7p7-lacZ (Figure 8F). Such alternative trans-interactions in dual-GI combinations probably compete with the more favorable 5′GI–5′GI interactions and could underlie the suppression of transvection produced by the addition of 3′ GIs in Figure 7, A–D.
Finally, we note that the 3xP3→pH transvection in Figure 7A is also mediated by a 5′ – 3′ (diagonal, less favored) GIFOR interaction. We tested the same configuration of enhancers and insulators and changed only the responding promoter on the dual GIsFOR p-gfp construct. Only pH was able to strongly respond to 3xP3, whereas two other promoters, p7 and p8, showed no or very weak response (Figure S11). Therefore, when alternative GI–GI interactions are possible, they can be biased positively or negatively by the affinity that their nearby enhancer/promoter elements have for each other.
Other insulators also mediate transvection
To determine whether other Drosophila insulators also mediate transvection, we generated two series of constructs based on the pLacZattB vector (Bischof et al. 2007): “sender” constructs containing an insulator cloned in between the e7 and e8 enhancers, and “responder” constructs containing an insulator between divergently oriented pH-lacZ and pD-gfp, a reporter driven by the Drosophila Synthetic Core Promoter (DSCP, abbreviated as pD) (Pfeiffer et al. 2008). Besides the 367 bp GI, we tested two new insulators: (1) the 540 bp Fab-8 insulator isolated from the Abdominal-B region of the bithorax complex (Figure S7); Fab-8 insulator activity depends on dCTCF, the ortholog of the vertebrate CTCF protein (Barges et al. 2000; Moon et al. 2005; Kyrchanova et al. 2008a, 2016), and (2) the 454 bp 1A2 insulator located downstream of the yellow gene (Figure S7), containing two Su(Hw) binding sites (Golovnin et al. 2003; Parnell et al. 2003). Fab-8 and 1A2 exemplify two major classes of endogenous insulators [centered around binding of dCTCF and Su(Hw), respectively], which are abundantly represented in the Drosophila genome (Parnell et al. 2006; Adryan et al. 2007; Nègre et al. 2010; Schwartz et al. 2012; Baxley et al. 2017). In addition, the resultant constructs contain WI carried in the mini-white marker gene. All transgenes were inserted into the attP40 locus, and we present the results from wing disks that are consistent with the results obtained from the CNS and eye-antennal disks (data not shown).
The responder transgene without an insulator between pH and pD promoters (“blank” responder), on its own, showed trachea-specific activity of both promoters (both LacZ and GFP, Figure 9A1), similar to the expression of the uninsulated pH-gfp reporter at the attP40 locus (Figure 4E). Inserting GI between the two promoters insulated pD-gfp from the tracheal enhancer, and resulted in the trapping of (an)other enhancer(s) at the pD promoter, ubiquitously active in all cells of the disk’s epithelium (weak ubiquitous GFP expression in Figure 9A2). This latter, ubiquitous, activity was abolished by a deletion of the WI from the mini-white (Figure 9A3), indicating cooperation between GI and WI in mediating the cis activity of this enhancer onto pD. The Fab-8 responder, similarly to the GI responder (Figure 9A2), also trapped the epithelial enhancer via the pD promoter (Figure 9A4), while the 1A2 responder did not show any activity from any of the promoters (Figure 9A5).
Heterozygotes between the “blank” sender and “blank” responder transgenes produced extremely faint but visible expression of GFP in the WM, indicative of a WI-mediated trans interaction between e8 and pD (Figure 9, B1 and B1’). This interaction was augmented in the GI responder (WM in Figure 9B2). This enhancement of WI–WI mediated transvection by GI, was confirmed by deleting one WI, which abolished GFP expression in the WM (Figure 9B3). This is in contrast to our previous result where GI–WI interaction in cis had an inhibitory effect on the WI-mediated transvection (Figure 3), suggesting that the transcriptional outcome of GI–WI cis interaction is context-dependent. Fab-8 also showed a detectable, albeit weaker, enhancement of e8→pD transvection at the WM, whereas 1A2 had no detectable effect (Figure 9, B4 and B5). Consistent with the conclusion that this transvection was mediated by homotypic WI/WI, when we deleted WI from the sender transgene, no transvection was observed at the WM in combination with any of the responders (Figure 9, C1–C5). WI-mediated transvection was also weakly enhanced by adding a heterologous insulator in the sender homolog and testing against the “blank” responder (Figure 9, D1, F1, G1). Again, only e8 was transvected, although this time both pH and pD responded, consistent with the fact that no insulator lies between the two. In this case 1A2 was able to enhance transvection comparably to GI and Fab-8 (Figure 9G1, compare to D1 and F1). Removal of WI from the GI sender abolished transvection (Figure 9E1).
Unlike the weak WI-mediated transvection effects discussed so far with the “blank” senders (Figure 9, column B) or responders (Figure 9, row 1), we got very strong transvection of both e7 and e8, when we combined homotypic insulators in sender and responder, i.e., GI/GI, Fab-8/Fab-8 or 1A2/1A2 (Figure 9, D2, D3, E2, E3, F4, and G5). In all cases, e7 transactivated the pH-lacZ reporter (in the AMPs) and e8 transactivated pD-gfp (in the WM), consistent with orientation-dependent function of all three insulators. GI produced the strongest effect and 1A2 the weakest. When the WI was deleted from either the sender or the responder GI construct, no difference in transvection efficiency was seen (Figure 9, D2, E2, D3, and E3), thus confirming, in a different context, our earlier conclusion that WIs do not affect GI-mediated transvection.
Moderately strong WM GFP expression (e8→pD transvection) was also seen in apparently heterotypic insulator combinations, specifically Fab-8 or 1A2 senders with a GI responder (Figure 9, F2 and G2), and the reciprocal, i.e., a GI sender with Fab-8 or 1A2 responders (Figure 9, D4 and D5). Upon deleting the WI from either the GI sender or responder, however, all of these effects were abolished (Figure 9, F3, G3, E4, and E5), consistent with being mediated via WI/WI and enhanced by the presence of the heterologous insulators, similar to the effects noted earlier with “blank” sender/responder constructs. On the other hand, the AMP lacZ and WM GFP expression seen in both 1A2/GI combinations (Figure 9, G2 and D5) was maintained, albeit much more weakly, upon WI deletion (Figure 9, G3, G3′, E5, and E5′), suggesting that this results from a trans heterotypic interaction between GI and 1A2, which is not surprising, since both are Su(Hw)-binding insulators.
In conclusion, we provide evidence that insulator landscape can affect enhancer-promoter communication both in cis (enhancer trapping) and in trans. All homotypic insulator combinations tested support transvection. The presence of additional heterotypic insulators in one or both homologs can augment (or, in other contexts, suppress) this effect. We finally provide evidence that heterotypic insulators can weakly promote transvection if they belong to the same class.
Discussion
Transvection is the ability of an enhancer to activate transcription from an unlinked promoter located at the same locus of the homologous chromosome. Using a collection of enhancers and promoters driving GFP and LacZ reporters and targeted to specific genomic loci, we were able to study transvection and characterize parameters influencing its outcome (summarized in Figure 10). The salient features of this phenomenon borne out by our results are the following: (1) We confirmed that homolog pairing is a prerequisite for transvection, as already known from classical studies. In Drosophila, homolog pairing in nonmeiotic cells is widespread, but seems to evolve gradually during the first half of embryogenesis (Hiraoka et al. 1993; Fung et al. 1998; Gemkow et al. 1998)—we showed that transvection unfolds in a similarly gradual manner, being stochastic and erratic during the early stages of embryogenesis, while recapitulating precisely the cis-activity of the enhancer at later embryonic and larval stages, once homolog pairing has been completed. (2) Insulators are needed for transvection. At least four different insulators, GI, WI, Fab-8, and 1A2, are capable of mediating transvection. We focused on GI and WI, which are commonly found in transgenesis vectors. (3) GI-mediated transvection is strong, resulting in ∼20% of the expression level the same enhancer-promoter combination would give in a cis configuration, and can work with all enhancer-promoter combinations tested. In contrast, WI-mediated transvection is weak, and is detectable only if the responding promoter is accompanied by another enhancer (Figure S3) or another insulator (Figure 9B2). (4) While necessary, the presence of GIs in both homologs is not sufficient for transvection. (4a) The most important parameters in determining the transvection outcome are the number and position of GIs in both homologs—the strongest effects are seen when a single GI is present in each homolog, one immediately upstream of the responding promoter and another adjacent to the interacting enhancer. Additional GIs at other positions of homologously inserted transgenes have a detrimental effect, probably by competing against the 5′GI/5′GI enhancer-promoter enabling interaction. (4b) The presence of a promoter in cis to the enhancer reduces its effectiveness in transvection. This cis-preference phenomenon, which has been described before (Geyer et al. 1990; Martínez-Laborda et al. 1992; Hendrickson and Sakonju 1995; Casares et al. 1997; Sipos et al. 1998; Morris et al. 1999a,b, 2004; Bateman et al. 2012a; Kravchuk et al. 2017), depends on the activity of the cis-linked promoter: we have shown that the TATA element seems to play a more important role than the DPE in inhibiting a cis-linked enhancer from acting in trans. (4c) The GI is highly asymmetric with 12 Su(Hw) binding sites all in the same orientation: the GI orientation with respect to the enhancer-promoter interacting pair is important, although not crucial for the transvection outcome. The 3′ side of the GI is the optimal for promoting transvection, but the 5′ side is also functional albeit less effectively. (5) Nonhomotypic insulators generally do not promote transvection, with the exception of GI/1A2, both of which bind Su(Hw) and showed a weak interaction. (6) Nonhomotypic insulators do, however, cis-influence the transvection produced by homotypic insulators in a context-dependent manner. For example GIs can enable or inhibit WI/WI-mediated transvection. However, WI seems unable to affect GI/GI-mediated transvection.
Figure 10.
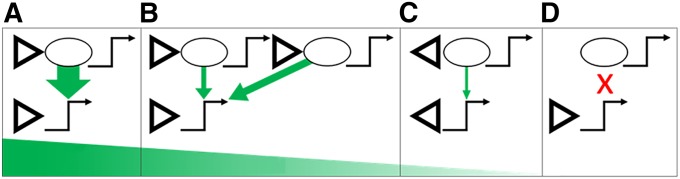
Parameters of transvection. A schematic summary of the parameters determining the ability of enhancers (ovals) to communicate with promoters (bent arrows) in trans. The top and bottom schematics in each panel depict elements present in the two paired homologs. Triangles represent gypsy insulators. A graded series of examples is shown with transvection ranging from high (A), through (B) medium, (C) low, to undetectable (D). For more details see Discussion.
Based on these observations, one should be careful when planning to use heterozygous transgene combinations in the same landing site. If one wishes to minimize transvection, one should preferably not include GI or any other insulator in the transgenes, at the expense of losing shielding from position effects. If shielding is desired, GIs can be used in only one of the transgenes. If shielding of both transgenes is desired, GIs can be placed in different orientations, and as far as possible from the transgenes’ enhancers and promoters. If, on the other hand, one wishes to promote transgene transvection, one should place GIs in the forward orientation directly upstream of the transgenes’ cis-regulatory elements.
A role of insulators in transvection has been described, but the mechanism been at best unclear (Fukaya and Levine 2017). Whereas some studies propose that insulators are needed for transvection (Lim et al. 2018), others propose that they only have an accessory role (Kravchenko et al. 2005; Schoborg et al. 2013), or that they affect transvection by promoting homolog pairing (Fujioka et al. 2016). And, finally, other studies ignore them altogether (Bateman et al. 2012a; Mellert and Truman 2012; Blick et al. 2016). Instead, the classical proposed role of insulators is to insulate, i.e., to inhibit enhancer-promoter communication in cis, although sometimes they can enable such enhancer-promoter communication, e.g., via their so-called “bypass” activity (Cai and Shen 2001; Muravyova et al. 2001; Kyrchanova et al. 2008b; Fujioka et al. 2013). These two apparently contradictory activities have been reconciled by the “looping” model (reviewed in Chetverina et al. 2017; Schwartz and Cavalli 2017), which is based on evidence that insulators mediate homotypic, or sometimes heterotypic, interactions (Kyrchanova et al. 2008b; Li et al. 2011; Vogelmann et al. 2014; Bonchuk et al. 2015; Fujioka et al. 2016). Via these interactions, insulators can form chromatin loops, and these loops can either bring enhancers and promoters in proximity (e.g., when both are near the loop’s anchor points) or avert their proximity (e.g., when one is within one loop and the other is outside that loop). These activities occur in cis and shape chromosomal architecture in parallel to affecting transcriptional regulation. The same insulator-insulator interactions can occur in trans (Kravchenko et al. 2005; Fujioka et al. 2016; Lim et al. 2018) and this could mediate interactions of enhancers on one homolog with promoters on the other (transvection).
One model proposes that insulators promote transvection by mediating homolog pairing in somatic cells. This hypothesis is supported by the fact that different classes of insulators are distributed widely in the Drosophila genome (Bartkuhn et al. 2009; Bushey et al. 2009; Nègre et al. 2010; Schwartz et al. 2012), and their congruent matching could underlie paternal–maternal homolog alignment from end to end. Alternatively, insulators may not mediate homolog pairing per se; rather, prior homolog pairing is a prerequisite for allowing insulators to mediate transvection. We favor the latter model: although we did not directly assay pairing, we have encountered numerous instances where addition of extra copies of the GI has a detrimental effect on transvection (Figure 7 and Figure 8). This result would be hard to reconcile with a model where insulators promote pairing, as we would expect pairing, and thus transvection, to locally increase as more insulators are added. Consistent with the view that homolog pairing precedes transvection is the fact that screens designed to identify somatic homolog pairing factors did not reveal any of the numerous insulator binding proteins (Bateman et al. 2012b; Joyce et al. 2012). Moreover, a recent study imaged two homologously inserted transgenes in live embryos and found the same frequency of colocalization (pairing) whether an insulator was included or not (Lim et al. 2018) – yet transvection between these genes required an insulator on both homologs, in agreement with our results. Another recent study mapped DNA elements mediating pairing and transvection from the ss locus: the two activities were found to map on two different fragments (Viets et al. 2018).
If insulators do not mediate homolog pairing, their role could be to enable the productive interaction of enhancers with certain promoters. Several of our observations support such a more active role: (1) In order to promote trans-interaction between e8 and either pH or p7, the WI requires the presence of another enhancer (e7) nearby. (2) Transvection supported by single 5′ GIs is orientation-dependent: the FOR orientation is much more effective than the REV orientation (Figure 7); if insulator–insulator interactions were the only parameter influencing transvection, having congruently disposed insulators in both homologs would most likely produce an identical result, whether the configuration were FOR/FOR or REV/REV. (3) When two GIs are present in each homolog, they exhibit a strong bias for “vertical” trans GI/GI interactions (Figure 8). The fact that this bias can be alleviated by promoter mutations is consistent with more direct insulator-promoter communication. (4) A forward GI exhibits a strong promoter preference: it transvects the 3xP3 enhancer only to pH and not to p7 or p8 in a certain transgene combination (Figure S11).
Recent data agree with such a more direct role of insulators in enhancer-promoter communication. A genome-wide chromatin occupancy analysis for >15 insulator binding proteins showed that a large proportion of their binding sites is near a promoter or an enhancer (Cubeñas-Potts et al. 2016). Direct contacts between insulators and nearby enhancers and promoters has been detected in transgenes via 3C (Kyrchanova et al. 2013). Live imaging of two loci separated (in cis) by >100 kb has shown that homotypic insulators promote proximity between these loci, but they do it much more effectively in the presence of a promoter in the one locus that gets activated by enhancers on the other (Chen et al. 2018). Finally, studies replacing specific insulator elements in the Bithorax Complex with other insulators, strongly support the ability of the resident insulators, like Fab-7 and Fab-8, to interact with neighboring enhancers (the iab-6 and iab-7 elements) to bring them in the proximity of the AbdB promoter (Kyrchanova et al. 2016; Postika et al. 2018). How insulators select which enhancers to pair with which promoters is an important question that still remains to be elucidated.
Why has the necessity for insulators been overlooked in some of the studies on transvection? Most probably because the fly genome and common transgenesis cloning vectors are rich in insulators. For example, in one study (Mellert and Truman 2012), all transgenes used contained mini-white and its associated WI, which we have shown is capable of selectively mediating enhancer action in trans; consistently, transvection was observed only with a subset of enhancers in that study. Two other studies (Bateman et al. 2012a; Blick et al. 2016) used recombinase-mediated cassette exchange, which allows for transgene integration without vector sequences, making these instances of transvection harder to reconcile with the need for an insulator. One possible hypothesis would be that the inserted transgenes trapped nearby insulators. Our GI-less transgenes were never able to trap nearby insulators, but we used five landing sites (attP40, VK2, VK13, VK37, VK40) distinct from those used in the above two studies (JB53F and JB37B), so our results cannot be compared. Given the strong dependence of transvection on insulator position, orientation and the nature of the interacting enhancers and promoters, it is likely that all of these factors will also influence insulator trapping. The large diversity of insulators in Drosophila (currently >15 binding factors identified; Maksimenko et al. 2015; Chetverina et al. 2017) is suggestive of a potentially high degree of selectivity in their interactions both with each other and with enhancer and promoter elements in their vicinity. Further work is needed to characterize these interactions and their importance in transcriptional regulation.
The association between insulators and transcriptional cis-regulatory elements (enhancers and promoters) is not a peculiarity of Drosophila; it has been reported also for vertebrates (Guo et al. 2015; reviewed in Hnisz et al. 2016). On the other hand, Drosophila (dipterans in general) seem to be unique in establishing somatic homolog pairing early in development and maintaining it throughout life (Abed et al. 2018; Erceg et al. 2018). Transvection could be an epiphenomenon of these two biological processes: insulator interactions with enhancers/promoters and homolog pairing. This would explain why it is more often encountered in Drosophila, but is only sporadic in mammals (Apte and Meller 2012; Stratigi et al. 2015). Does transvection also serve a role in regulating transcriptional output and accordingly could it be positively selected in dipteran evolution? Some studies have suggested that it increases transcription from the two alleles, or that it coordinates their transcriptional on/off decisions (Goldsborough and Kornberg 1996; Johnston et al. 2014). How widespread this effect is across the genome and whether it contributes to organism adaptation to fluctuations in environmental conditions or response to stressful stimuli is not known. At the least, transvection would ensure robustness of gene expression levels in the face of genetic variation, specifically heterozygosity for mutations in promoters or enhancers.
Acknowledgments
We thank Michalis Averof for pMinos{3xP3-dsRed}, Maria Monastirioti for pGL3-hsp70-luc, Chrysoula Pitsouli, and Norbert Perrimon for providing attP2 and attP40 flies; Marina Gkantia, Giorgos Samantsidis, Alexis Molfetas, Babis Galouzis, and Vasilis Ntasis for their help in generating plasmid constructs and isolating transgenic flies. Many thanks to Charalampos Spilianakis and Eva Zacharioudaki for critical reading of the manuscript. This work was supported by the European Union (EU) Marie Curie Early stage research training (EST) program No 7295/FAMED, ARISTEIA grants (No 4436 and 1967) from the General Secretariat for Research and Technology of Greece (cofunded by the EU European Social Fund), the Fondation Santé and intramural Institute of Molecular Biology and Biotechnology (IMBB) support.
Footnotes
Supplemental material available at https://doi.org/10.25386/genetics.7925462.
Communicating editor: P. Geyer
Literature Cited
- Abed J. A., Erceg J., Goloborodko A., Nguyen S. C., McCole R. B., et al. , 2018. Highly structured homolog pairing reflects functional organization of the Drosophila genome. bioRxiv. 10.1101/443887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adryan B., Woerfel G., Birch-Machin I., Gao S., Quick M., et al. , 2007. Genomic mapping of Suppressor of Hairy-wing binding sites in Drosophila. Genome Biol. 8: R167 10.1186/gb-2007-8-8-r167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apte M. S., Meller V. H., 2012. Homologue pairing in flies and mammals: gene regulation when two are involved. Genet. Res. Int. 2012: 430587 10.1155/2012/430587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnone M. I., Dmochowski I. J., Gache C., 2004. Using reporter genes to study cis-regulatory elements. Methods Cell Biol. 74: 621–652 [DOI] [PubMed] [Google Scholar]
- Babu P., Bhat S., 1980. Effect of zeste on white complementation. Basic Life Sci. 16: 35–40. [DOI] [PubMed] [Google Scholar]
- Barges S., Mihaly J., Galloni M., Hagstrom K., Müller M., et al. , 2000. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127: 779–790. [DOI] [PubMed] [Google Scholar]
- Barolo S., Carver L. A., Posakony J. W., 2000. GFP and B-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques 29: 726–732. 10.2144/00294bm10 [DOI] [PubMed] [Google Scholar]
- Bartkuhn M., Straub T., Herold M., Herrmann M., Rathke C., et al. , 2009. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 28: 877–888. 10.1038/emboj.2009.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman J. R., Johnson J. E., Locke M. N., 2012a. Comparing enhancer action in cis and in trans. Genetics 191: 1143–1155. 10.1534/genetics.112.140954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman J. R., Larschan E., D’Souza R., Marshall L. S., Dempsey K. E., et al. , 2012b. A genome-wide screen identifies genes that affect somatic homolog pairing in Drosophila. G3 (Bethesda) 2: 731–740. 10.1534/g3.112.002840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxley R. M., Bullard J. D., Klein M. W., Fell A. G., Morales-Rosado J. A., et al. , 2017. Deciphering the DNA code for the function of the Drosophila polydactyl zinc finger protein Suppressor of Hairy-wing. Nucleic Acids Res. 45: 4463–4478. 10.1093/nar/gkx040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghammer A. J., Klingler M., Wimmer E. A., 1999. A universal marker for transgenic insects. Nature 402: 370–371. 10.1038/46463 [DOI] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104: 3312–3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton J., Gaszner M., Schedl P., 2003. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 17: 664–675. 10.1101/gad.1052003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blick A. J., Mayer-Hirshfeld I., Malibiran B. R., Cooper M. A., Martino P. A., et al. , 2016. The capacity to act in trans varies among drosophila enhancers. Genetics 203: 203–218. 10.1534/genetics.115.185645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonchuk A., Maksimenko O., Kyrchanova O., Ivlieva T., Mogila V., et al. , 2015. Functional role of dimerization and CP190 interacting domains of CTCF protein in Drosophila melanogaster. BMC Biol. 13: 63 10.1186/s12915-015-0168-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey A. M., Ramos E., Corces V. G., 2009. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 23: 1338–1350. 10.1101/gad.1798209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd K., Corces V. G., 2003. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J. Cell Biol. 162: 565–574. 10.1083/jcb.200305013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Levine M., 1995. Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo. Nature 376: 533–536. 10.1038/376533a0 [DOI] [PubMed] [Google Scholar]
- Cai H. N., Shen P., 2001. Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science 291: 493–495. 10.1126/science.291.5503.493 [DOI] [PubMed] [Google Scholar]
- Casares F., Bender W., Merriam J., Sánchez-Herrero E., 1997. Interactions of Drosophila Ultrabithorax regulatory regions with native and foreign promoters. Genetics 145: 123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Levo M., Barinov L., Fujioka M., Jaynes J. B., et al. , 2018. Dynamic interplay between enhancer–promoter topology and gene activity. Nat. Genet. 50: 1296–1303. 10.1038/s41588-018-0175-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. L., Huisinga K. L., Viering M. M., Ou S. A., Wu C. T., et al. , 2002. Enhancer action in trans is permitted throughout the Drosophila genome. Proc. Natl. Acad. Sci. USA 99: 3723–3728. 10.1073/pnas.062447999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetverina D., Savitskaya E., Maksimenko O., Melnikova L., Zaytseva O., et al. , 2008. Red flag on the white reporter: a versatile insulator abuts the white gene in Drosophila and is omnipresent in mini-white constructs. Nucleic Acids Res. 36: 929–937. 10.1093/nar/gkm992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetverina D., Fujioka M., Erokhin M., Georgiev P., Jaynes J. B., et al. , 2017. Boundaries of loop domains (insulators): determinants of chromosome form and function in multicellular eukaryotes. BioEssays 39: 1600233 10.1002/bies.201600233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubeñas-Potts C., Corces V. G., 2015. Architectural proteins, transcription, and the three-dimensional organization of the genome. FEBS Lett. 589: 2923–2930. 10.1016/j.febslet.2015.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubeñas-Potts C., Rowley M. J., Lyu X., Li G., Lei E. P., et al. , 2016. Different enhancer classes in Drosophila bind distinct architectural proteins and mediate unique chromatin interactions and 3D architecture. Nucleic Acids Res. 45: 1714–1730. 10.1093/nar/gkw1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis J. F., de Celis J., Ligoxygakis P., Preiss A., Delidakis C., et al. , 1996. Functional relationships between Notch, Su(H) and the bHLH genes of the E(spl) complex: the E(spl) genes mediate only a subset of Notch activities during imaginal development. Development 122: 2719–2728. [DOI] [PubMed] [Google Scholar]
- Delidakis C., Monastirioti M., Magadi S. S., 2014. E(spl): genetic, developmental, and evolutionary aspects of a group of invertebrate Hes proteins with close ties to Notch signaling. Curr. Top. Dev. Biol. 110: 217–262. 10.1016/B978-0-12-405943-6.00006-3 [DOI] [PubMed] [Google Scholar]
- Doyle B., Fudenberg G., Imakaev M., Mirny L. A., 2014. Chromatin loops as allosteric modulators of enhancer-promoter interactions. PLoS Comput. Biol. 10: e1003867 10.1371/journal.pcbi.1003867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erceg J., Abed J. A., Goloborodko A., Lajoie B. R., Fudenberg G., et al. , 2018. The genome-wide, multi-layered architecture of chromosome pairing in early Drosophila embryos. bioRxiv. 10.1101/443028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M., Sun G., Jaynes J. B., 2013. The Drosophila eve insulator homie promotes eve expression and protects the adjacent gene from repression by polycomb spreading. PLoS Genet. 9: e1003883 10.1371/journal.pgen.1003883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M., Mistry H., Schedl P., Jaynes J. B., 2016. Determinants of chromosome architecture: insulator pairing in cis and in trans. PLoS Genet. 12: e1005889 10.1371/journal.pgen.1005889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya T., Levine M., 2017. Transvection. Curr. Biol. 27: R1047–R1049. 10.1016/j.cub.2017.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung J. C., Marshall W. F., Dernburg A., Agard D. A., Sedat J. W., 1998. Homologous chromosome pairing in Drosophila melanogaster proceeds through multiple independent initiations. J. Cell Biol. 141: 5–20. 10.1083/jcb.141.1.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart W. M., 1982. Synapsis-dependent allelic complementation at the decapentaplegic gene complex in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 79: 2636–2640. 10.1073/pnas.79.8.2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemkow M. J., Verveer P. J., Arndt-Jovin D. J., 1998. Homologous association of the Bithorax-Complex during embryogenesis: consequences for transvection in Drosophila melanogaster. Development 125: 4541–4552. [DOI] [PubMed] [Google Scholar]
- Georgiev P., Tikhomirova T., Yelagin V., Belenkaya T., Gracheva E., et al. , 1997. Insertions of hybrid P elements in the yellow gene of Drosophila cause a large variety of mutant phenotypes. Genetics 146: 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova T. I., Gdula D. A., Gerasimov D. V., Simonova O., Corces V. G., 1995. A drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell 82: 587–597. 10.1016/0092-8674(95)90031-4 [DOI] [PubMed] [Google Scholar]
- Geyer P. K., Corces V. G., 1992. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 6: 1865–1873. 10.1101/gad.6.10.1865 [DOI] [PubMed] [Google Scholar]
- Geyer P. K., Green M. M., Corces V. G., 1990. Tissue-specific transcriptional enhancers may act in trans on the gene located in the homologous chromosome: the molecular basis of transvection in Drosophila. EMBO J. 9: 2247–2256. 10.1002/j.1460-2075.1990.tb07395.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsborough A. S., Kornberg T. B., 1996. Reduction of transcription by homologue asynapsis in Drosophila imaginal discs. Nature 381: 807–810. 10.1038/381807a0 [DOI] [PubMed] [Google Scholar]
- Golovnin A., Biryukova I., Romanova O., Silicheva M., Parshikov A., et al. , 2003. An endogenous Su(Hw) insulator separates the yellow gene from the Achaete-scute gene complex in Drosophila. Development 130: 3249–3258 [corrigenda: Development 135: 787 (2008)] 10.1242/dev.00543 [DOI] [PubMed] [Google Scholar]
- Golovnin A., Melnick E., Mazur A., Georgiev P., 2005. Drosophila Su(Hw) insulator can stimulate transcription of a weakened yellow promoter over a distance. Genetics 170: 1133–1142. 10.1534/genetics.104.034587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A. C., Fish M., Nusse R., Calos M. P., 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166: 1775–1782. 10.1534/genetics.166.4.1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Xu Q., Canzio D., Shou J., Li J., et al. , 2015. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162: 900–910. 10.1016/j.cell.2015.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison D. A., Gdula D. A., Coyne R. S., Corces V. G., 1993. A leucine zipper domain of the suppressor of Hairy-wing protein mediates its repressive effect on enhancer function. Genes Dev. 7: 1966–1978. 10.1101/gad.7.10.1966 [DOI] [PubMed] [Google Scholar]
- Hendrickson J. E., Sakonju S., 1995. Cis and trans interactions between the iab regulatory regions and abdominal-A and abdominal-B in Drosophila melanogaster. Genetics 139: 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heride C., Ricoul M., Kieu K., von Hase J., Guillemot V., et al. , 2010. Distance between homologous chromosomes results from chromosome positioning constraints. J. Cell Sci. 123: 4063–4075. 10.1242/jcs.066498 [DOI] [PubMed] [Google Scholar]
- Hiraoka Y., Dernburg A. F., Parmelee S. J., Rykowski M. C., Agard D. A., et al. , 1993. The onset of homologous chromosome pairing during Drosophila melanogaster embryogenesis. J. Cell Biol. 120: 591–600. 10.1083/jcb.120.3.591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Day D. S., Young R. A., 2016. Insulated neighborhoods: structural and functional units of mammalian gene control. Cell 167: 1188–1200. 10.1016/j.cell.2016.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdridge C., Dorsett D., 1991. Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol. Cell. Biol. 11: 1894–1900. 10.1128/MCB.11.4.1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn C., Jaunich B., Wimmer E. A., 2000. Highly sensitive, fluorescent transformation marker for Drosophila transgenesis. Dev. Genes Evol. 210: 623–629. 10.1007/s004270000111 [DOI] [PubMed] [Google Scholar]
- Johnston R. J., Desplan C., Johnston R. J., Jr, Desplan C., Johnston R. J., et al. , 2014. Interchromosomal communication coordinates intrinsically stochastic expression between alleles. Science 343: 661–665. 10.1126/science.1243039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce E. F., Williams B. R., Xie T., Wu C. T., 2012. Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS Genet. 8: e1002667 10.1371/journal.pgen.1002667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce E. F., Erceg J., Wu C. T., 2016. Pairing and anti-pairing: a balancing act in the diploid genome. Curr. Opin. Genet. Dev. 37: 119–128. 10.1016/j.gde.2016.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R., Schedl P., 1991. A position-effect assay for boundaries of higher order chromosomal domains. Cell 64: 941–950. 10.1016/0092-8674(91)90318-S [DOI] [PubMed] [Google Scholar]
- Kellum R., Schedl P., 1992. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol. 12: 2424–2431. 10.1128/MCB.12.5.2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klämbt C., Knust E., Tietze K., Campos-Ortega J. A., 1989. Closely related transcripts encoded by the neurogenic gene complex enhancer of split of Drosophila melanogaster. EMBO J. 8: 203–210. 10.1002/j.1460-2075.1989.tb03365.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravchenko E., Savitskaya E., Kravchuk O., Parshikov A., Georgiev P., et al. , 2005. Pairing between gypsy insulators facilitates the enhancer action in trans throughout the Drosophila genome. Mol. Cell. Biol. 25: 9283–9291. 10.1128/MCB.25.21.9283-9291.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravchuk O., Kim M., Klepikov P., Parshikov A., Georgiev P., et al. , 2017. Transvection in Drosophila: trans-interaction between yellow enhancers and promoter is strongly suppressed by a cis-promoter only in certain genomic regions. Chromosoma 126: 431–441. 10.1007/s00412-016-0605-6 [DOI] [PubMed] [Google Scholar]
- Kuhn E. J., Viering M. M., Rhodes K. M., Geyer P. K., 2003. A test of insulator interactions in Drosophila. EMBO J. 22: 2463–2471. 10.1093/emboj/cdg241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutach A. K., Kadonaga J. T., 2000. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell. Biol. 20: 4754–4764. 10.1128/MCB.20.13.4754-4764.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon E. Z., 2015. Using transgenic reporter assays to functionally characterize enhancers in animals. Genomics 106: 185–192. 10.1016/j.ygeno.2015.06.007 [DOI] [PubMed] [Google Scholar]
- Kyrchanova O., Toshchakov S., Podstreshnaya Y., Parshikov A., Georgiev P., 2008a. Functional interaction between the Fab-7 and Fab-8 boundaries and the upstream promoter region in the Drosophila Abd-B gene. Mol. Cell. Biol. 28: 4188–4195. 10.1128/MCB.00229-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O., Chetverina D., Maksimenko O., Kullyev A., Georgiev P., 2008b. Orientation-dependent interaction between Drosophila insulators is a property of this class of regulatory elements. Nucleic Acids Res. 36: 7019–7028. 10.1093/nar/gkn781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O., Ivlieva T., Toshchakov S., Parshikov A., Maksimenko O., et al. , 2011. Selective interactions of boundaries with upstream region of Abd-B promoter in Drosophila bithorax complex and role of dCTCF in this process. Nucleic Acids Res. 39: 3042–3052. 10.1093/nar/gkq1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O., Maksimenko O., Stakhov V., Ivlieva T., Parshikov A., et al. , 2013. Effective blocking of the white enhancer requires cooperation between two main mechanisms suggested for the insulator function. PLoS Genet. 9: e1003606 10.1371/journal.pgen.1003606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O., Mogila V., Wolle D., Deshpande G., Parshikov A., et al. , 2016. Functional dissection of the blocking and bypass activities of the fab-8 boundary in the Drosophila bithorax complex. PLoS Genet. 12: e1006188 10.1371/journal.pgen.1006188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. M., Wu C. T., 2006. Enhancer-promoter communication at the yellow gene of Drosophila melanogaster: diverse promoters participate in and regulate trans interactions. Genetics 174: 1867–1880. 10.1534/genetics.106.064121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Hazelrigg T., Rubin G. M., 1985. Effects of genomic position on the expression of transduced copies of the white gene of Drosophila. Science 229: 558–561. 10.1126/science.2992080 [DOI] [PubMed] [Google Scholar]
- Lewis E. B., 1954. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am. Nat. 88: 225–239. 10.1086/281833 [DOI] [Google Scholar]
- Li H. B., Muller M., Bahechar I. A., Kyrchanova O., Ohno K., et al. , 2011. Insulators, not Polycomb response elements, are required for long-range interactions between Polycomb targets in Drosophila melanogaster. Mol. Cell. Biol. 31: 616–625. 10.1128/MCB.00849-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B., Heist T., Levine M., Fukaya T., 2018. Visualization of transvection in living Drosophila embryos. Mol. Cell 70: 287–296.e6. 10.1016/j.molcel.2018.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimenko O., Bartkuhn M., Stakhov V., Herold M., Zolotarev N., et al. , 2015. Two new insulator proteins, Pita and ZIPIC, target CP190 to chromatin. Genome Res. 25: 89–99. 10.1101/gr.174169.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M., Pitsouli C., Villalta C., Celniker S. E., Perrimon N., 2008. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40: 476–483. 10.1038/ng.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Laborda A., González-Reyes A., Morata G., 1992. Trans regulation in the Ultrabithorax gene of Drosophila: alterations in the promoter enhance transvection. EMBO J. 11: 3645–3652. 10.1002/j.1460-2075.1992.tb05449.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee B. D., 2004. Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim. Biophys. Acta 1677: 165–180. 10.1016/j.bbaexp.2003.11.017 [DOI] [PubMed] [Google Scholar]
- Mellert D. J., Truman J. W., 2012. Transvection is common throughout the Drosophila genome. Genetics 191: 1129–1141. 10.1534/genetics.112.140475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon H., Filippova G., Loukinov D., Pugacheva E., Chen Q., et al. , 2005. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 6: 165–170. 10.1038/sj.embor.7400334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. R., Chen J., Filandrinos S. T., Dunn R. C., Fisk R., et al. , 1999a. An analysis of transvection at the yellow locus of Drosophila melanogaster. Genetics 151: 633–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. R., Geyer P. K., Wu C. T., 1999b. Core promoter elements can regulate transcription on a separate chromosome in trans. Genes Dev. 13: 253–258. 10.1101/gad.13.3.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. R., Petrov D. A., Lee A. M., Wu C. T., 2004. Enhancer choice in cis and in trans in Drosophila melanogaster: role of the promoter. Genetics 167: 1739–1747. 10.1534/genetics.104.026955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muravyova E., Golovnin A., Gracheva E., Parshikov A., Belenkaya T., et al. , 2001. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science 291: 495–498. 10.1126/science.291.5503.495 [DOI] [PubMed] [Google Scholar]
- Nègre N., Brown C. D., Shah P. K., Kheradpour P., Morrison C. A., et al. , 2010. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 6: e1000814 10.1371/journal.pgen.1000814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst S. M., Harrison D. A., Remington M. P., Spana C., Kelley R. L., et al. , 1988. The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Genes Dev. 2: 1205–1215. 10.1101/gad.2.10.1205 [DOI] [PubMed] [Google Scholar]
- Parnell T. J., Viering M. M., Skjesol A., Helou C., Kuhn E. J., et al. , 2003. An endogenous suppressor of hairy-wing insulator separates regulatory domains in Drosophila. Proc. Natl. Acad. Sci. USA 100: 13436–13441. 10.1073/pnas.2333111100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell T. J., Kuhn E. J., Gilmore B. L., Helou C., Wold M. S., et al. , 2006. Identification of genomic sites that bind the Drosophila suppressor of Hairy-wing insulator protein. Mol. Cell. Biol. 26: 5983–5993. 10.1128/MCB.00698-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer B. D., Jenett A., Hammonds A. S., Ngo T.-T. B., Misra S., et al. , 2008. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA 105: 9715–9720. 10.1073/pnas.0803697105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer B. D., Ngo T.-T. B., Hibbard K. L., Murphy C., Jenett A., et al. , 2010. Refinement of tools for targeted gene expression in Drosophila. Genetics 186: 735–755. 10.1534/genetics.110.119917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postika N., Metzler M., Affolter M., Müller M., Schedl P., et al. , 2018. Boundaries mediate long-distance interactions between enhancers and promoters in the Drosophila Bithorax complex. PLoS Genet. 14: e1007702 10.1371/journal.pgen.1007702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos E., Ghosh D., Baxter E., Corces V. G., 2006. Genomic organization of gypsy chromatin insulators in Drosophila melanogaster. Genetics 172: 2337–2349. 10.1534/genetics.105.054742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L., 2009. Transgenesis in Drosophila melanogaster. Methods Mol. Biol. 561: 3–19. 10.1007/978-1-60327-019-9_1 [DOI] [PubMed] [Google Scholar]
- Roseman R. R., Pirrotta V., Geyer P. K., 1993. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 12: 435–442. 10.1002/j.1460-2075.1993.tb05675.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman R. R., Johnson E. A., Rodesch C. K., Bjerke M., Nagoshi R. N., et al. , 1995. A P element containing suppressor of hairy-wing binding regions has novel properties for mutagenesis in Drosophila melanogaster. Genetics 141: 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoborg T., Kuruganti S., Rickels R., Labrador M., 2013. The Drosophila gypsy insulator supports transvection in the presence of the vestigial enhancer. PLoS One 8: e81331 10.1371/journal.pone.0081331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y. B., Cavalli G., 2017. Three-dimensional genome organization and function in Drosophila. Genetics 205: 5–24. 10.1534/genetics.115.185132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y. B., Linder-Basso D., Kharchenko P. V., Tolstorukov M. Y., Kim M., et al. , 2012. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 22: 2188–2198. 10.1101/gr.138156.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipos L., Mihály J., Karch F., Schedl P., Gausz J., et al. , 1998. Transvection in the Drosophila Abd-B domain: extensive upstream sequences are involved in anchoring distant cis-regulatory regions to the promoter. Genetics 149: 1031–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnev A. A., Li X., Wehling M. D., Geyer P. K., 2008. Context differences reveal insulator and activator functions of a Su(Hw) binding region. PLoS Genet. 4: e1000159 10.1371/journal.pgen.1000159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana C., Corces V. G., 1990. DNA bending is a determinant of binding specificity for a Drosophila zinc finger protein. Genes Dev. 4: 1505–1515. 10.1101/gad.4.9.1505 [DOI] [PubMed] [Google Scholar]
- Spana C., Harrison D. A., Corces V. G., 1988. The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 2: 1414–1423. 10.1101/gad.2.11.1414 [DOI] [PubMed] [Google Scholar]
- Stratigi K., Kapsetaki M., Aivaliotis M., Town T., Flavell R. A., et al. , 2015. Spatial proximity of homologous alleles and long noncoding RNAs regulate a switch in allelic gene expression. Proc. Natl. Acad. Sci. USA 112: E1577–E1586. 10.1073/pnas.1502182112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault S. T., Singer M. A., Miyazaki W. Y., Milash B., Dompe N. A., et al. , 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36: 283–287. 10.1038/ng1314 [DOI] [PubMed] [Google Scholar]
- Thorpe H. M., Smith M. C., 1998. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc. Natl. Acad. Sci. USA 95: 5505–5510. 10.1073/pnas.95.10.5505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bortle K., Nichols M. H., Li L., Ong C. T., Takenaka N., et al. , 2014. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 15: R82 10.1186/gb-2014-15-5-r82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J. T., He Y., Hoskins R. A., Bellen H. J., 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314: 1747–1751. 10.1126/science.1134426 [DOI] [PubMed] [Google Scholar]
- Viets K., Sauria M., Chernoff C., Anderson C., Tran S., et al. , 2018. TADs pair homologous chromosomes to promote interchromosomal gene regulation. bioRxiv. 10.1101/445627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelmann J., Le Gall A., Dejardin S., Allemand F., Gamot A., et al. , 2014. Chromatin insulator factors involved in long-range DNA interactions and their role in the folding of the Drosophila genome. PLoS Genet. 10: e1004544 10.1371/journal.pgen.1004544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Brennan M. D., 2001. The gypsy insulator can act as a promoter-specific transcriptional stimulator. Mol. Cell. Biol. 21: 7714–7720. 10.1128/MCB.21.22.7714-7720.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All plasmids and fly strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental Material (Figures S1–S11 and Supplemental Materials and Methods) were deposited to GSA Figshare https://gsajournals.figshare.com/s/d548db1426e0efad9e9f. Supplemental material available at https://doi.org/10.25386/genetics.7925462.