Abstract
Remodeling of the extracellular matrix supports tissue and organ development, by regulating cellular morphology and tissue integrity. However, proper extracellular matrix remodeling requires spatiotemporal regulation of extracellular metalloproteinase activity. Members of the ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) family, including MIG-17 and GON-1, are evolutionarily conserved, secreted, zinc-requiring metalloproteinases. Although these proteases are required for extracellular matrix remodeling during gonadogenesis in Caenorhabditis elegans, their in vivo regulatory mechanisms remain to be delineated. Therefore, we focused on the C. elegans tissue inhibitors of metalloproteinases (TIMPs), TIMP-1 and CRI-2. Analysis of the transcription and translation products for GFP/Venus fusions, with TIMP-1 or CRI-2, indicated that these inhibitors were secreted and localized to the basement membrane of gonads and the plasma membrane of germ cells. A timp-1 deletion mutant exhibited gonadal growth defects and sterility, and the phenotypes of this mutant were fully rescued by a TIMP-1::Venus construct, but not by a TIMP-1(C21S)::Venus mutant construct, in which the inhibitor coding sequence had been mutated. Moreover, genetic data suggested that TIMP-1 negatively regulates proteolysis of the α1 chain of type IV collagen. We also found that the loss-of-function observed for the mutants timp-1 and cri-2 involves a partial suppression of gonadal defects found for the mutants mig-17/ADAMTS and gon-1/ADAMTS, and that this suppression was canceled upon overexpression of gon-1 or mig-17, respectively. Based on these results, we propose that both TIMP-1 and CRI-2 act as inhibitors of MIG-17 and GON-1 ADAMTSs to regulate gonad development in a noncell-autonomous manner.
Keywords: a disintegrin and metalloproteinase with thrombospondin motifs, tissue inhibitors of metalloproteinases, extracellular matrix, type IV collagen, gonad
THE extracellular matrix (ECM) supports cellular structure and function, and tissue integrity, by modulating the tissue-specific extracellular microenvironment during organ development and remodeling. Aberrant ECM remodeling leads to various pathological states including tissue degeneration, cancer progression, and tumor growth (Fata et al. 2000; Vu and Werb 2000; Page-McCaw et al. 2007; Kessenbrock et al. 2010; Jackson et al. 2017). During tissue remodeling, ECM proteins are processed by extracellular metalloproteinases including matrix metalloproteases, a disintegrin and metalloproteinase, and a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) (Vu and Werb 2000; Porter et al. 2005; Page-McCaw et al. 2007; Apte 2009). The metalloproteinases, including ADAMTS family members, are secreted, require zinc for activity, and play a central role in remodeling of the ECM during development and under normal physiological conditions in mammals. Humans have 19 ADAMTS-encoding genes (Kuno et al. 1997; Porter et al. 2005; Dubail and Apte 2015). Mutations in human ADAMTSs cause tissue malformations and diseases related to connective tissue remodeling, such as Weill–Marchesani and Ehlers–Danlos syndromes (Colige et al. 1999; Dagoneau et al. 2004; Morales et al. 2009). Gene inactivation experiments have indicated that ADAMTS family members are required for development of the ovaries, palate, and limbs in mice (Shindo et al. 2000; Shozu et al. 2005; Brown et al. 2006; McCulloch et al. 2009; Enomoto et al. 2010). These proteases are also required for cell migration, gonadal morphogenesis, proper gonad function, regulation of pharynx length, and synapse formation in Caenorhabditis elegans and detachment of cells from the apical ECM of salivary glands in Drosophila melanogaster (Blelloch et al. 1999; Blelloch and Kimble 1999; Nishiwaki et al. 2000; Ismat et al. 2013; Kurshan et al. 2014; Qin et al. 2014; Shibata et al. 2016). Despite the apparent importance of ADAMTSs in the regulation of ECM remodeling, how their activities are regulated in vivo is not well understood.
The gonads of C. elegans have a very simple architecture and develop in a stereotypical pattern (Kimble and Hirsh 1979). During gonad development, sheet-like somatic cells undergo dynamic changes that involve, for example, migration and elongation. At the tip of the two gonadal arms, distal tip cells lead the directional elongation of each arm, with the arms turning twice in a 90°, stage-specific manner, thereby resulting in U-shaped arms by the young-adult stage (Hedgecock et al. 1987; Su et al. 2000). Within the gonadal somatic sheets, germ cells proliferate and differentiate to produce gametes. At the gonadal surface, remodeling of the basement membrane—a specialized ECM network—supports the gonadal morphogenesis process. Two ADAMTSs are involved in gonadal morphogenesis: GON-1, which is essential for gonadal growth, and MIG-17, which is required for directional elongation of the gonadal arms (Blelloch and Kimble 1999; Nishiwaki et al. 2000). GON-1 acts antagonistically with basement membrane fibulin-1 to regulate gonadal growth (Hesselson et al. 2004). MIG-17 recruits, removes, and/or activates the basement membrane components fibulin-1, type IV collagen, and nidogen-1 and, by doing so, regulates the directional elongation of the gonad arms (Kubota et al. 2004, 2008). MIG-17 localization on the basement membrane depends on the basement membrane protein MIG-6/papilin (Kawano et al. 2009). Despite the importance of these proteases in gonadal morphogenesis, how their activities are regulated is unknown.
The tissue inhibitors of metalloproteinases (TIMPs) are a conserved family of proteins that regulate matrix metalloprotease activities (Brew et al. 2000; Fata et al. 2000; Jackson et al. 2017). Mammalian TIMPs have been shown to negatively regulate the activity of matrix metalloproteases both in vitro and in an organ culture system, and they also negatively regulate the activity of ADAMTSs in vitro (Hashimoto et al. 2001; Kashiwagi et al. 2001; Wang et al. 2006). These observations led us to hypothesize that TIMPs may act as regulators of ADAMTSs as part of ECM remodeling during C. elegans gonadal development. For the present study, we therefore characterized the in vivo expression patterns and functional roles of two C. elegans TIMPs, namely TIMP-1 and CRI-2, by genetic manipulation. We found that these TIMPs are secreted from nongonadal cells, and localize to the basement membrane of gonads and the plasma membrane of germ cells during gonad development. We also found that TIMP-1 is required for gonadal morphogenesis, that eliminating TIMP-1 and CRI-2 activities via mutagenesis or RNA interference (RNAi)-mediated knockdown could partially suppress the gonadal morphogenesis defects in mig-17/ADAMTS and gon-1/ADAMTS mutants that had lost metalloproteinase activity, and that these effects were canceled upon overexpression of gon-1 or mig-17, respectively. We propose that these two TIMPs negatively regulate MIG-17 and GON-1 activities during C. elegans gonadogenesis.
Materials and Methods
C. elegans strains
C. elegans strains were derived from the wild-type (WT) Bristol strain N2 (Brenner 1974). Worms were incubated at 20° except for those that were fed the feeding RNAi bacteria, which were maintained at 24.5°. To isolate L3-stage larvae, newly hatched L1 worms were incubated at 20° for 30 hr and then observed under a differential interference contrast (DIC) microscope using a Zeiss ([Carl Zeiss], Thornwood, NY) Axioplan2 microscope with an Orca-R2 12 bit/16 bit cooled charge-coupled device (CCD) camera (Hamamatsu Photonics).
The following alleles were used for mutant construction: mig-17(k174), mig-17(k113) (Nishiwaki 1999), gon-1(q518), gon-1(e1254) (Blelloch et al. 1999), unc-42(e270), unc-119(ed3), unc-119(e2498), cri-2(gk314) (obtained from the C. elegans Gene Knockout Consortium), timp-1(tk71) (this work), nT1[qIs51], tkTi1[emb-9p::emb-9::mCherry + Cbr-unc-119(+)] (Shibata et al. 2016), oxIs322[myo-2p::mCherry::H2B + myo-3p::mCherry::H2B + Cbr-unc-119(+)] (Frøkjær-Jensen et al. 2014), Ex[gon-1p::gon-1 + sur-5;;GFP + punc-119(+)], Ex[mig-17p::mig-17::Venus + punc-119(+)], bkcEx1[timp-1p::timp-1::Venus + punc-119(+)] (this work), and bkcEx2[cri-2p::cri-2::Venus + punc-119(+)] (this work). k174 and q518 are the null alleles of mig-17 and gon-1, respectively.
Transgenic worms were prepared by microinjection of the targeted gene (Mello et al. 1991). Single-copy transgenic-insertion worms were generated using the miniMos method (Frøkjær-Jensen et al. 2014) for genomic Venus fusion expression and tissue-specific rescue experiments with unc-119(ed3) as the host strain. For the microinjections, the following mixtures were used: 10 μg/ml each of GFP::HIS-58/Venus-tagged miniMos-target transgene (timp-1p::GFP::his-58 + Cbr-unc-119(+) plasmid pYK60, timp-1p::timp-1::Venus + Cbr-unc-119(+) plasmid pYK37, cri-2p::cri-2::Venus + Cbr-unc-119(+) plasmid pYK82, dpy-7p::timp-1::Venus + Cbr-unc-119(+) plasmid pYK87, myo-3p::timp-1::Venus + Cbr-unc-119(+) plasmid pYK56, and timp-1p::timp-1(C21S)::Venus + Cbr-unc-119(+) plasmid pYK74); transposase pCFJ601 (50 μg/ml); injection markers Prab-3::mCherry::unc-54 3′-UTR plasmid pGH8 (10 μg/ml), Pmyo-2::mCherry::unc-54 3′-UTR plasmid pCFJ90 (2.5 μg/ml), Pmyo-3::mCherry::unc-54 3′-UTR plasmid pCFJ104 (5 μg/ml), and pBluescript II KS(−) (30 μg/ml); and negative selection marker Phsp-16.41::peel-1::tbb-2 3′-UTR plasmid pMA122 (10 μg/ml). The insertion sites were confirmed by sequencing and are listed in Supplemental Material, Table S1. To create the extrachromosomal array for timp-1::Venus and cri-2::Venus, all of the following were injected: 5 μg/ml of a Venus-tagged transgene (pSK1-timp-1p::timp-1::Venus::timp-1 3′-UTR plasmid pYK71 or pSK1-cri-2p::cri-2::Venus::cri-2 3′-UTR plasmid pYK76), pBluescript II KS(−) (95 μg/ml), and punc-119(+) (50 μg/ml).
The timp-1(tk71) allele was isolated using a PCR-based deletion screen of our original trimethylpsoralen (TMP)–UV-mutagenized library (Kubota et al. 2004). To obtain timp-1(tk71)/+mig-17(k174)V hermaphrodites, timp-1(tk71)mig-17(k174)/hT2[gIs48]V males were mated with unc-42(e270)mig-17(k174)V hermaphrodites and GFP-negative progeny with coordinated movement were scored. To obtain gon-1(e1254)IV;timp-1(tk71)/+V hermaphrodites, gon-1(e1254)/hT2[gIs48]IV males were mated with unc-119(ed3)III;gon-1(e1254)/hT2[gIs48]IV;timp-1(tk71)/hT2[gIs48]V hermaphrodites and GFP-negative progeny with coordinated movement were scored. To obtain gon-1(q518)IV;timp-1(tk71)/+V hermaphrodites, gon-1(q518)/hT2[gIs48]IV males were mated with unc-119(ed3)III;gon-1(q518)/hT2[gIs48]IV;timp-1(tk71)/hT2[gIs48]V hermaphrodites and GFP-negative progeny with coordinated movement were scored.
Amino acid sequence alignment
Amino acid sequence alignment was performed using the Clustal multiple sequence alignment algorithm in Muscle (3.8) (European Molecular Biology Laboratory-European Bioinformatics Institute). The GenBank accession numbers for the C. elegans precursor TIMP-1 and CRI-2 sequences are NP_505115.1 and Q21265.1, respectively. The GenBank accession numbers for the Homo sapiens TIMP-2 and TIMP-3 precursor sequences are AAB19474.1 and AAB34532.1, respectively.
Plasmid construction
The plasmids constructed for this study are listed in Table S2. A miniMos backbone vector, denoted pYK14, was made by inserting a 376-bp fragment containing a multicloning site into a StuI site in pCFJ909. To construct the targeting vectors, the following fragments were amplified, and individually subcloned into pYK14 at its NotI and AscI sites. To construct transgenes that expressed Venus fusion proteins and the GFP::HIS-58 reporter from putative endogenous 5′ cis-regulatory regions of timp-1, the genomic fragments, timp-1::Venus derived from the timp-1 regulatory region (3200 bp), the timp-1 regulatory region (3200 bp) and coding region, and the timp-1 3′-UTR (1056 bp), were each PCR amplified and then fused with Venus/GFP::HIS-58. To isolate the timp-1p::timp-1(C21S)::Venus plasmid, codon 21 (TGT) was changed to TCT via PCR-mediated substitution. cri-2::Venus: the cri-2 regulatory region (4854 bp) and coding region, and the cri-2 3′-UTR region (1233 bp). For overexpression experiments, timp-1p::timp-1::Venus and cri-2p::cri-2::Venus were subcloned into pSK1, a modified vector from a pBluescript II KS(−) backbone.
Expression of TIMP-1::Venus was controlled by the regulatory region of dpy-7 (448 bp) in epidermal cells and by the regulatory region of myo-3 (2500 bp) in body wall muscle cells.
Strain construction for rescue experiments
Strains that expressed timp-1 under putative endogenous 5′ cis-regulatory regions of timp-1 and tissue-specific promoters, and strains that expressed TIMP-1(C21S), were constructed using miniMos methods (Frøkjær-Jensen et al. 2014). The resultant integrated alleles were: bkcSi8[dpy-7p::timp-1::Venus], bkcSi3[myo-3p::timp-1::Venus], bkcSi1[timp-1p::timp-1::Venus], bkcSi2[timp-1p::timp-1::Venus], bkcSi7[cri-2p::cri-2::Venus], bkcSi5[timp-1p::timp-1(C21S)::Venus], and bkcSi6[timp-1p::timp-1(C21S)::Venus]. The alleles were transferred into unc-119(ed3);timp-1(tk71))/nT1[qIs51] animals upon mating (Table S1). Young adult worms (just after final molting) that were homozygous for tk71 were used to score gonad morphology.
Staining with 4’,6-diamidino-2-phenylindole
Young adult animals were washed with 0.1% (v/v) Tween 20 in 0.01 M PBS and fixed with ice-cold methanol for 10 min. Then, their germ cell nuclei were visualized by staining with 0.2 μg/ml 4’,6-diamidino-2-phenylindole (Dojindo) at room temperature for 15 min. After washing with the Tween/PBS solution three times to remove excess stain, the animals were mounted on glass slides with VECTASHIELD (Vector Laboratories, Burlingame, CA).
Feeding RNAi
The worms were fed on freshly prepared RNAi-feeding plates, as described previously (Kamath et al. 2001). Full-length timp-1 and cri-2 cDNAs were isolated from a C. elegans cDNA library, and inserted into the feeding RNAi vector, L4440. An L4440 vector lacking an insert was used as control(RNAi). After confirming that each inserted sequence was correct, the feeding vectors were individually transformed into Escherichia coli HT115(DE3) samples, which were then seeded separately onto plates of nematode growth medium agar containing Luria-Bertani medium and 50 μg/ml ampicillin, and incubated for 12 hr at 24.5°. Then, each culture was seeded onto a 60-mm feeding agar plate containing 50 μg/ml ampicillin, 1 mM isopropyl β-d-1-thiogalactopyranoside, and 12.5 µg/ml tetracycline and incubated at 25° for 8 hr. L4-stage worms were transferred onto a feeding plate and cultured at 24.5°. Phenotypes for the F1 worms were determined at the young-adult stage.
Microscopy
Fluorescence and Nomarski microscopy procedures were performed using a Zeiss Axioplan2 microscope with an Orca-R2 12-bit/16-bit cooled CCD camera (Hamamatsu Photonics), and a Plan-Apochromat X63 water NA1.2 objective lens or a Plan-Apochromat X40 NA0.95 objective lens. The microscope system was controlled by MetaMorph software (Molecular Devices). Images were processed with Image J (National Institutes of Health) or Adobe Photoshop CS6.
Analysis of brood size
Brood size was defined as the number of larvae hatched from the eggs of each of 20 adult WT, cri-2(gk314), or timp-1(tk71) hermaphrodites.
Statistical analyses
P-values for Fisher’s exact test were used to assess the significance of observed differences in gonadal defects. For the analysis of gonadal defects of the timp-1(tk71), timp-1(tk71);control(RNAi), gon-1(e1254), gon-1(e1254);control(RNAi), gon-1(q518), gon-1(q518);control(RNAi), gon-1(q518);Ex[mig-17p::mig-17::Venus], and gon-1(q518);control(RNAi);Ex[mig-17p::mig-17::Venus] animals, the percentage of the gonadal growth defect (the sum of the class 1 and class 2 defects) was compared. For the analysis of the gonadal defects of the mig-17(k113);control(RNAi), mig-17(k174), mig-17(k174);control(RNAi), mig-17(k174);Ex[gon-1p::gon-1], and mig-17(k174);control(RNAi)Ex[gon-1p::gon-1] animals, the percentage of the directional gonadal elongation defect (class 4 defect) was compared. The P-value for the Student’s t-test of the timp-1(tk71) and cri-2(gk314) mutants was calculated for comparison with WT animals.
Data availability
Strains and reagents cited in Tables S1 and S2 are available upon request. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.7981535.
Results
The C. elegans TIMP-encoding genes timp-1 and cri-2
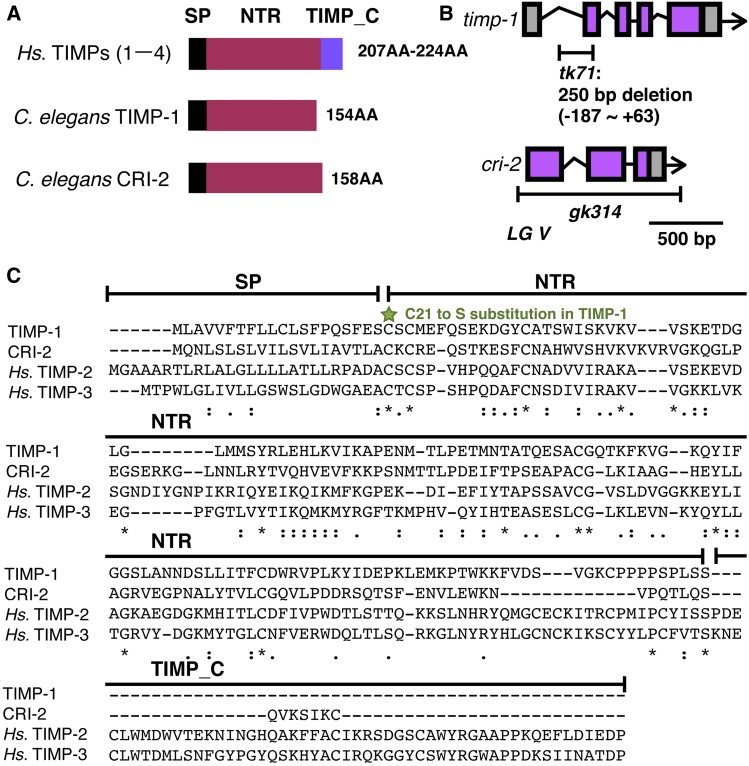
To further understand the regulation of ECM remodeling during gonad development, we focused on the TIMPs, TIMP-1 and CRI-2, as they inhibit metalloproteinase activity. The H. sapiens and Drosophila TIMP sequences contain an N-terminal signal peptide, a netrin-like domain, and a TIMP_C-terminal domain (Figure 1A) (Brew et al. 2000; Kucera et al. 2011). The C. elegans genome has two genes encoding a TIMP, namely timp-1/K07C11.3 and cri-2/K07C11.5, which are located very close to each other on linkage group V (Figure 1B). Both genes lack the TIMP_C domain, as reported for other nematode species (Kucera et al. 2011).
Figure 1.
C. elegans TIMP genes (A) Domain structures of H. sapiens TIMPs, and C. elegans TIMP-1 and CRI-2. (B) Genomic structures and deletion sites in timp-1 and cri-2. In tk71, nucleotides between positions −187 and +63 in the TIMP-1 gene are deleted. The gk314 mutation is a complete deletion of the cri-2 coding region. (C) Multiple sequence alignment of the TIMP-1, CRI-2, and Hs. TIMP-2 and -3 precursors. The green star indicates the C21S mutation site in TIMP-1. Asterisks indicate positions at which a residue is fully conserved. Colons indicate positions at which residues have very similar physical properties (score > 0.5 in the Gonnet PAM 250 matrix). Periods indicate positions at which residues have weakly similar physical and chemical properties (score ≤ 0.5 in the Gonnet PAM 250 matrix). Hs., H. sapiens; NTR, netrin-like domain; PAM, point accepted mutation; SP, signal peptide; TIMPs, tissue inhibitors of metalloproteinases; TIMP_C, TIMP C-terminal domain.
To analyze the function of TIMP-1, we isolated the mutant timp-1(tk71) using the TMP–UV method (Kubota et al. 2004). The resulting mutated worm genome contains a deleted initiation codon for timp-1. Because timp-1(tk71) lacks an initiation codon, we assumed that it would act as a null mutation for timp-1. To assess the function of CRI-2, we obtained the mutant cri-2(gk314) from the C. elegans Gene Knockout Consortium. Its genome has a completely deleted coding region in cri-2 (Figure 1B). The netrin-like domain that has essential roles in metalloproteinase inhibition is conserved in TIMP-1, CRI-2, H. sapiens TIMP-2, and TIMP-3 (Figure 1C; (Brew et al. 2000).
TIMP-1 and CRI-2 are required for gonad development
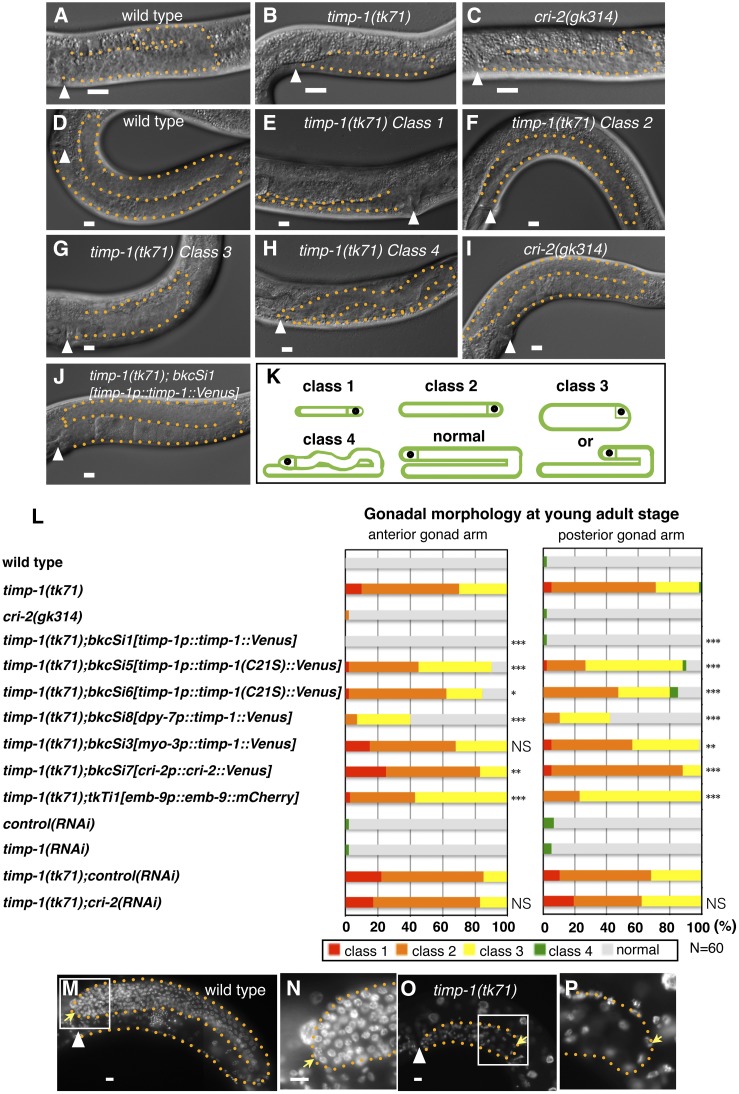
To establish whether TIMPs are required for gonad development, we first examined the morphologies of timp-1(tk71) and cri-2(gk314) gonads via DIC microscopy at 30 hr after hatching (L3-stage larvae), and at the young-adult stage (just after final molting). Although the arms of the timp-1(tk71) gonads were shorter than those of WT worms, gonad primordia were seen in WT, timp-1(tk71), and cri-2(gk314) larvae (Figure 2, A–C). At the young-adult stage, almost all WT gonads had formed U-shaped arms that appeared normal (Figure 2D), whereas the gonads in timp-1(tk71) had four different morphological defects (classes 1–4; Figure 2, E–H, K, and L). The class 1–3 defects were originally reported for gon-1/ADAMTS mutants (Blelloch et al. 1999; Blelloch and Kimble 1999). Class 1 is the most severe defect, for which very short and slender arms were observed on the ventral muscle (Figure 2, E and K). Class 2 is a moderate defect, for which short gonad arms of normal width were observed on the ventral muscle (Figure 2, F and K). Class 3 is a mild defect, for which slightly shortened gonad arms or short gonad arms with approximately double the normal width were observed. For the class 3 defects, the gonad arms reached the dorsal muscle (Figure 2, G and K). A class 4 defect involves a meandering gonad arm of normal elongation that follows an abnormal direction after the first turn. Class 4 defects were found in mig-17/ADAMTS mutants (Figure 2, H and K) (Nishiwaki et al. 2000). Penetrance of the class 1, 2, and 3 defects in the young-adult timp-1(tk71) mutants was ∼10, 60, and 30%, respectively, for the anterior gonad arms (Figure 2L), and 5, 65, and 30% for the posterior arms. A small number of the young adults had gonads with class 4 defects (Figure 2L). Staining of the germ cells with 4’,6-diamidino-2-phenylindole revealed that their number was substantially decreased in timp-1(tk71) compared with WT worms (Figure 2, M–P), and all worms appeared to be sterile (Table 1). The timp-1(RNAi) worms did not have gonadal growth defects, suggesting that a small amount of timp-1 was sufficient for proper gonadal morphogenesis (Figure 2L). In contrast, cri-2(gk314) worms had no gonadal morphological defects (Figure 2I) and cri-2(RNAi) did not enhance the gonadal phenotype of timp-1(tk71) (Figure 2L), indicating that CRI-2 is not essential for gonad morphogenesis. Moreover, overexpression of CRI-2 from the transgene bkcSi7[cri-2p::cri-2::Venus] did not rescue the gonadal phenotype of timp-1(tk71) (Figure 2L). Thus, of the two TIMPs, TIMP-1 was more important for proper gonadal morphogenesis. Although, the cri-2(gk314) mutant did not show morphogenesis defects, lethality, or sterility, its brood size was decreased by ∼40%, indicating that both timp-1 and cri-2 are required for germline development (Table 1).
Figure 2.
TIMP-1, but not CRI-2, is required for gonadal morphogenesis. (A–J) Gonad morphology of wild-type (A and D), timp-1(tk71) (B and E–H), cri-2(gk314) (C and I), and timp-1(tk71);bkcSi1[timp-1p::timp-1::Venus] (J) L3-stage larvae 30 hr after hatching (A–C), in young adults (D–J), and in the hermaphrodite posterior (A–D and F–J) and anterior (E) gonads. (K) The four classes of gonadal morphogenesis defects that were observed at the young-adult stage. Class 1, a very short and slender gonad arm on the ventral muscle; class 2, a short gonad arm of normal width on the ventral muscle; class 3, a short-arm gonad with an approximately twofold width or a slightly shortened gonad arm (in both types of class 3 defect, the gonad arm reaches the dorsal muscle). Class 4, a meandering gonad arm. (L) Percentages of abnormal gonad morphologies found for young-adult wild-type and mutant worms. P-values are indicated for Fisher’s exact test in comparison with timp-1(tk71) or timp-1(tk71):control(RNAi). * P < 0.05, ** P < 0.01, and *** P < 0.005. NS, not significant; (M–P) Staining (4’,6-diamidino-2-phenylindole) of gonadal nuclei in young-adult wild-type (M and N) and timp-1(tk71) (O and P) hermaphrodites. (N and P) show enlargements of the enclosed areas shown in (M and O), respectively. Posterior gonads are shown. The orange dotted lines mark the gonad boundaries. Arrowheads indicate the vulvae. Yellow arrows indicate distal tip cells. In all panels, the anterior region of the gonad is to the left and the dorsal region is at the top of the image. Bars, 10 μm. The animals were cultured at 20° except for those used in the RNAi experiments (24.5°). RNAi, RNA interference.
Table 1. Brood sizes of the wild-type animals, and cri-2 and timp-1 mutants.
| Genotype | Wild-type | cri-2(gk314) | timp-1(tk71)# |
|---|---|---|---|
| Brood size | 288 ± 22a | 177 ± 55*** | 0 ± 0*** |
N = 20; ± SD. *** P < 0.005. #, homozygous timp-1 mutant hermaphrodites from heterozygous hermaphrodites were scored.
Results for Student’s t-test vs. wild-type.
TIMP-1 and CRI-2 localize to the gonadal, and the pharyngeal, basement and plasma membranes
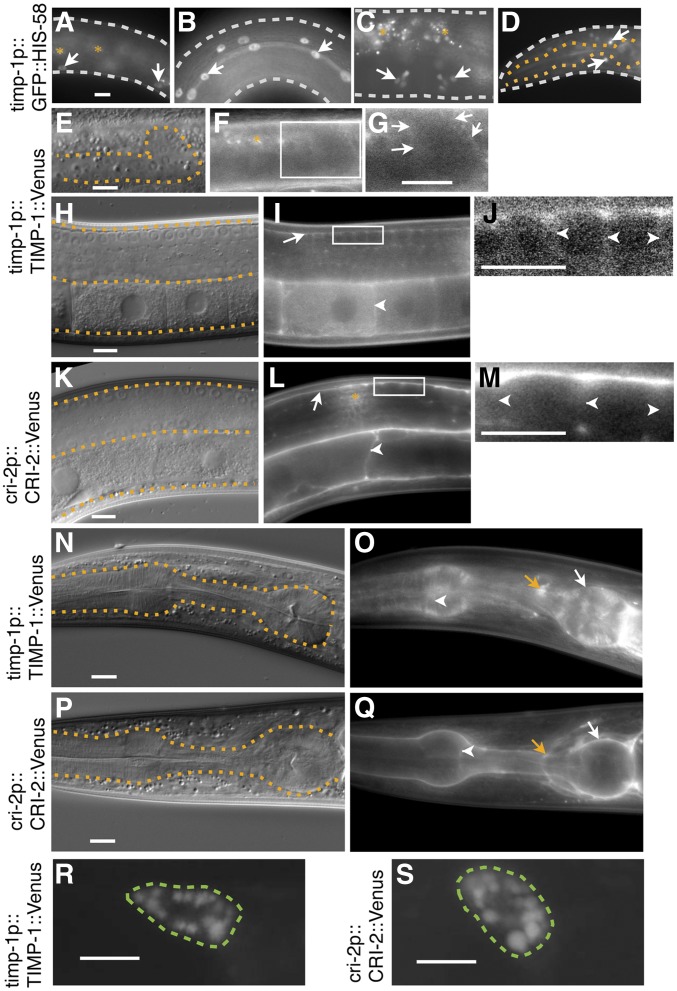
The expression patterns of TIMP-1 and CRI-2 were characterized using transgenic lines that expressed Venus fusion proteins under the control of their putative endogenous 5′ cis-regulatory regions. The integrated transgene bkcSi1[timp-1p::timp-1::Venus] completely rescued the gonadal defects and sterility of timp-1(tk71) (Figure 2, J and L), indicating that the TIMP-1::Venus fusion protein was functional and that its expression pattern would be expected to be the same as that of endogenous TIMP-1. TIMP-1::Venus was detected on the gonadal basement membrane of the L3-stage larvae (Figure 3, E–G), the basement membrane of somatic gonads, and the plasma membrane of germ cells, including oocytes, of young adults (Figure 3, H–J). When CRI-2::Venus was expressed from the transgene bkcSi7[cri-2p::cri-2::Venus], it also had a similar localization pattern (Figure 3, K–M). In the head region, TIMP-1::Venus and CRI-2::Venus localized to the basement and plasma membranes of the pharyngeal cells, and at the nerve ring (Figure 3, N–Q). We then asked whether TIMP-1 and CRI-2 are secreted by examining the endocytic uptake of TIMP-1::Venus and CRI-2::Venus by coelomocytes, which are scavenger cells in the pseudocoelom. Accumulation of TIMP-1::Venus and CRI-2::Venus was observed in coelomocytes of these transgenic strains (Figure 3, R and S), suggesting that TIMP-1 and CRI-2 were secreted.
Figure 3.
Expression and localization patterns of TIMP-1 and CRI-2. (A–D) Expression of the transcriptional reporter GFP::HIS-58 under the control of the putative endogenous timp-1 5′ cis-regulatory region. The bkcSi4[timp-1p::GFP::his-58] allele was used. In young adults, signals were detected in epidermal cells (arrows, A), seam cells (arrows, B), the lateral region of the vulva (arrows, C), and a subpopulation of neuronal cells in the nerve ring (arrows, D). DIC (E, H, K, N, and P) and fluorescence (F, G, I, J, L, M, O, and Q) images of L3-stage larvae (E–G) and young adults (H–Q). Images of the posterior gonads (E–M) and pharynges (N–Q) of worms with timp-1(tk71);bkcSi1[timp-1p::timp-1::Venus] (E–J, N, and O) and bkcSi7[cri-2p::cri-2::Venus] (K–M, P, and G). (G, J, and M) display, at a higher magnification, the squares enclosed by the white boundaries in (F, I, and L). The contrast was enhanced to visualize the basement and plasma membrane localization of the signals. (E–Q) White arrows, white arrowheads, and orange arrows indicate the basement membrane, plasma membrane, and nerve ring, respectively. Fluorescence images of the coelomocyte of (R and S) young adult worms with bkcEx1[timp-1p::timp-1::Venus] (R) and bkcEx2[cri-2p::cri-2::Venus] (S). The white, orange, and green dotted lines outline the worm, the gonad or pharynx, or coelomocyte, respectively. The orange asterisks indicate autofluorescence signals (A, C, and F) or signal cross talk from the coelomocyte (L), a scavenger cell that takes up secreted molecules such as CRI-2::Venus. In all panels, the anterior region of the gonad is to the left and its dorsal region is at the top of the image. Three independent timp-1p::GFP::his-58 transgenic lines exhibited similar expression patterns. Bars, 10 μm.
timp-1 is transcribed in epidermal cells, seam cells, a subpopulation of vulval cells, and head neurons
With the expression of GFP::his-58 under the control of the putative endogenous timp-1 5′ cis-regulatory region, we found green fluorescence in different types of cells, i.e., epidermal cells, seam cells (lateral epidermal cells), lateral region vulval cells, and a subpopulation of head neurons (Figure 3, A–D), which suggested that timp-1 had been transcribed in these cells; conversely, green fluorescence was not detected in muscle cells. We confirmed that the timp-1 promoter was inactive in muscle cells by showing that GFP::HIS-58 under the control of the timp-1 promoter was not coexpressed with the mCherry muscle marker oxIs322[myo-3p::mCherry::H2B] (data not shown). Although TIMP-1::Venus was detected on the gonadal basement and plasma membrane of the germ cells (see above), GFP::HIS-58 under the control of the timp-1 promoter was not detected in gonadal somatic cells or in germ cells.
Epidermal expression of TIMP-1::Venus rescues the gonadal defects found in the timp-1(tk71) mutant
As noted above, although TIMP-1::Venus was expressed when under the control of the putative endogenous timp-1 5′ cis-regulatory region and was detected on the gonadal basement membrane (Figure 3I), expression of the transcription reporter GFP::HIS-58 (also under the control of the timp-1 promoter) was not detected in gonadal cells, raising the possibility that TIMP-1 was synthesized in and secreted from nongonadal tissues, and then localized to the gonadal basement membrane. Therefore, we assessed whether TIMP-1 that had been expressed in nongonadal tissues was responsible for gonad development. The single-copy integrated transgene that expressed TIMP-1::Venus under the control of the epidermal-specific promoter dpy-7p bkcSi8[dpy-7p::timp-1::Venus]) significantly rescued defects in the gonad and the sterile state of timp-1(tk71) (Figure 2L). Conversely, the single-copy integrated transgene of TIMP-1::Venus that was expressed under the control of the muscle-specific promoter myo-3p (bkcSi3[myo-3p::timp-1::Venus]) did not rescue the gonadal growth defect phenotype of the anterior gonad, and it only partially rescued the gonadal growth defect phenotype of the posterior gonad (Figure 2L). Thus, TIMP-1 expressed in epidermal cells is secreted and localizes to the gonadal basement and plasma membrane, where it is responsible for gonadal morphogenesis and germ cell development.
Inhibition of metalloproteinase activity is required for gonadal morphogenesis
We next examined the relationship between the ability of TIMP-1 to inhibit MIG-17 and GON-1, and, consequently, gonad development. Previous studies revealed that the first cysteine residue in TIMPs is essential for their protease inhibitor activity (Caterina et al. 1997; Kucera et al. 2011). Therefore, we constructed two integrated transgenic lines (bkcSi5[timp-1p::timp-1(C21S)::Venus] and bkcSi6[timp-1p::timp-1(C21S)::Venus]) to express a mutant TIMP-1::Venus in which residue 12, a cysteine in the TIMP-1 precursor and the first residue in mature TIMP-1, was replaced with a serine (Figure 1C). In both lines, TIMP-1(C21S)::Venus localized to the basement and plasma membrane of the gonad and the pharynx, respectively (Figure S1); however, notably, the mutated protein rescued the WT gonadal phenotype only partially in timp-1(tk71) (Figure 2L), which suggested that the inhibitor activity of TIMP-1 is essential for gonadal morphogenesis and germ cell proliferation.
Accumulation of the type IV collagen α1 chain construct EMB-9::mCherry in the gonadal basement membrane is defective in the timp-1(tk71) mutant
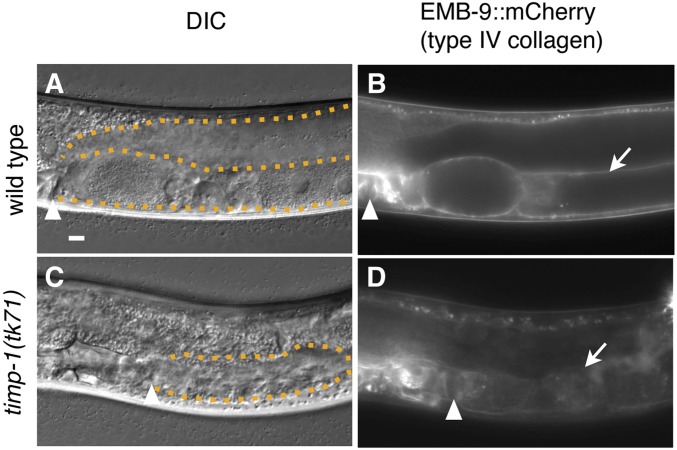
We previously demonstrated that emb-9(null)/+ partially suppresses the directional gonad elongation defect of the mig-17(k174) null mutant (Kubota et al. 2008) and proposed a model in which GON-1 negatively regulates the accumulation of type IV collagen (Kubota et al. 2012). Consequently, if TIMPs negatively regulate the activities of MIG-17 and GON-1, then accumulation of type IV collagen should be decreased in the timp mutants. Therefore, we examined the relationship between TIMP activity and the amount of gonadal type IV collagen. C. elegans type IV collagen is composed of two α1 chains, denoted EMB-9, and one α2 chain, denoted LET-2 (Guo et al. 1991; Sibley et al. 1993). Consistent with previous studies, the type IV collagen α1 chain construct EMB-9::mCherry, when expressed from the integrated transgene tkTi1[emb-9::mCherry], localized to the gonadal basement membrane in WT C. elegans (Figure 4, A and B) (Ihara et al. 2011; Shibata et al. 2016). Conversely, localization of EMB-9::mCherry to the gonadal basement membrane was disrupted in timp-1(tk71) (Figure 4, C and D). This mislocalization of EMB-9::mCherry could be rescued by tjTi54[timp-1p::timp-1::Venus] (Figure S2). Thus, TIMP-1 appeared to be required for proper regulation of EMB-9 accumulation on the gonadal basement membrane.
Figure 4.
EMB-9::mCherry localization on the gonadal basement membrane in the timp-1(tk71) mutant. DIC (A and C) and fluorescence (B and D) images of gonads in WT (A and B) and timp-1(tk71) (C and D) young adults with tkTi1[emb-9/type IV collagen α1 chain::mCherry]. The arrowheads in (A–D) indicate vulvae. The arrows in (B and D) indicate the gonadal basement membrane. The orange dotted lines outline the gonad in (A and C). The photographic exposure time was the same in (B and D). In all panels, the anterior region of the gonad is to the left and its dorsal region is at the top of the image. Bar, 10 μm.
Furthermore, overexpression of EMB-9::mCherry from the transgene tkTi1[emb-9::mCherry] partially rescued the gonadal defect of timp-1(tk71) (Figure 2L). These results suggested that TIMP is involved in the inhibition of EMB-9 proteolysis, which appears essential for the regulation of gonadal morphogenesis.
Gonadal morphogenesis defects in mig-17/ADAMTS and gon-1/ADAMTS mutants are partially suppressed by depletion of TIMP-1 and CRI-2
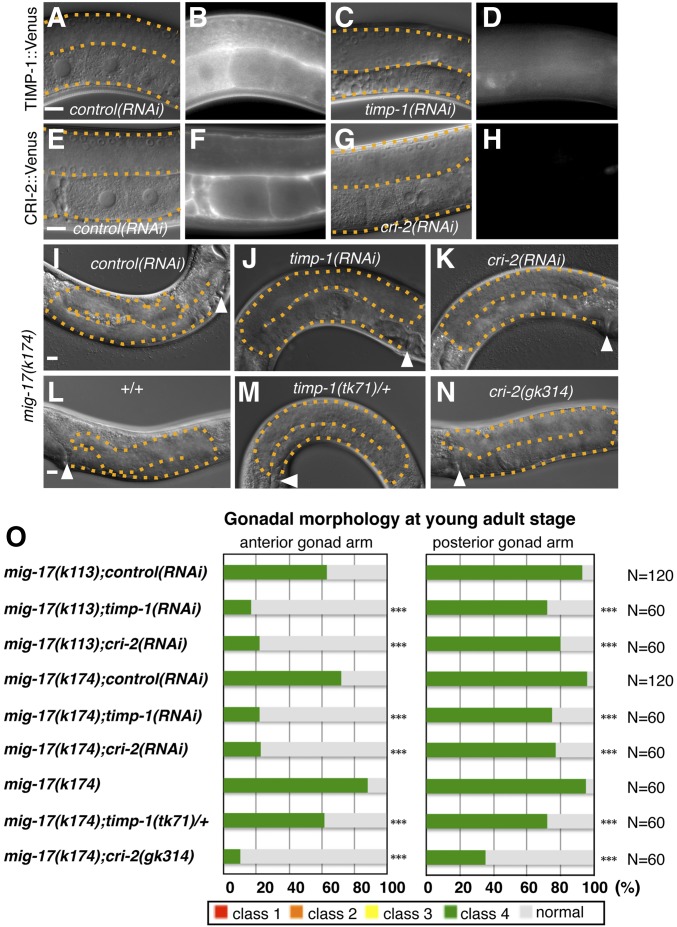
Because a mammalian TIMP inhibits ADAMTS activity in vitro (Hashimoto et al. 2001; Kashiwagi et al. 2001; Wang et al. 2006), we next characterized the relationship between the ADAMTSs MIG-17 and GON-1, and the protease inhibitors TIMP-1 and CRI-2, in C. elegans. We used RNAi to knock down TIMP expression; alternatively, we eliminated TIMP expression by deleting the corresponding TIMP gene in the mig-17 and gon-1 mutant backgrounds. The effectiveness of the RNAi-mediated knockdown was confirmed by the observation that accumulation of TIMP-1::Venus and CRI-2::Venus was reduced at the gonadal basement membrane in the young-adult worms (Figure 5).
Figure 5.
timp-1 and cri-2 partially suppresses the gonadal morphogenesis defects in mig-17/ADAMTS mutants. (A–N) DIC (A, C, E, G, and I–N) and fluorescence (B, D, F, and H) images of gonads in young adults. Localization of posterior gonads in TIMP-1::Venus (B and D), CRI-2::Venus (F and H), control(RNAi);timp-1(tk71);bkcSi1[timp-1p::timp-1::Venus] (A and B), timp-1(RNAi);timp-1(tk71);bkcSi1[timp-1p::timp-1::Venus] (C and D), control(RNAi);bkcSi7[cri-2p::cri-2::Venus] (E and F), and cri-2(RNAi);bkcSi7[cri-2p::cri-2::Venus] worms (G and H). DIC images of anterior gonads of mig-17(k174) subjected to control RNAi (I), mig-17(k174);timp-1(RNAi) (J), and mig-17(k174);cri-2(RNAi) (K). DIC images of posterior gonads of mig-17(k174) (L), mig-17(k174);timp-1(tk71)/+ (M), and mig-17(k174);cri-2(gk314) (N). The arrowheads in (I–N) indicate vulvae. The orange dotted lines outline the gonad in (A, C, E, G, and I–N). The photographic exposure time was the same for (B and D) and for (F and H). In all panels, the anterior region of the gonad is to the left and its dorsal region is at the top of the image. (O) Percentages of abnormal gonad morphology in the young adults. The four classes of gonadal defects were scored as in Figure 2K. P-values are indicated for Fisher’s exact test in comparison with mig-17(k113);control(RNAi), mig-17(k174);control(RNAi), or mig-17(k174). *** P < 0.005. Bars, 10 μm. The animals were cultured at 20° except for those used in the RNAi experiments (24.5°). RNAi, RNA interference.
We then assessed the effects of TIMP reduction in the mig-17 mutants (Figure 5). In the gonads of the worms subjected to RNAi-mediated knockdown, penetrance of the class 4 phenotype (meandering gonadal arms) in the weak-allele mig-17(k113) and null-allele mig-17(k174) mutants decreased (Figure 5, I–K and O). In addition, the class 4 defect in mig-17(k174) was partially suppressed by timp-1(tk71)/+ (Figure 5, L, M, and O). The cri-2(gk314) allele also significantly suppressed the class 4 defect in the mig-17(k174) mutant (Figure 5, L, N, and O).
Next, we assessed the effects of TIMP knockdown in the gon-1 mutants (Figure 6). When the weak-allele gon-1(e1254) and the null-allele gon-1(q518) mutants were subjected to RNAi-mediated knockdown of timp-1 and cri-2, the effects on gonadal growth and elongation defects were ameliorated (Figure 6, A–C and G). In particular, ∼20% of the anterior and posterior gonad arms in the gon-1(e1254);cri-2(RNAi) animals were fully elongated (Figure 6G). In addition, the timp-1(tk71)/+ and cri-2(gk314) alleles also partially suppressed the gonadal growth defects of gon-1(e1254) and gon-1(q518) (Figure 6, D–G). The cri-2(gk314) mutation substantially suppressed the gonadal defects of gon-1(e1254) (Figure 6G). These results suggested that TIMP-1 and CRI-2 regulate gonad development by acting antagonistically with the activities of MIG-17 and GON-1 (Figure 8).
Figure 6.
timp-1 and cri-2 partially suppress the gonadal morphogenesis defects in the gon-1/ADAMTS mutants. (A–F) DIC images of posterior gonads of gon-1(e1254);control(RNAi) (A), gon-1(e1254);timp-1(RNAi) (B), gon-1(e1254);cri-2(RNAi) (C), gon-1(q518) (D), gon-1(q518);timp-1(tk71)/+ (E), and gon-1(q518);cri-2(gk314) (F). (G) Percentages of abnormal gonad morphology in the young adults. The arrowheads in the panels indicate vulvae. The orange dotted lines outline the gonads in the panels. The four classes of gonadal defects were scored as in Figure 2K. P-values are indicated for Fisher’s exact test in comparison with gon-1 (e1254);control(RNAi), gon-1 (e1254), gon-1(q518);control(RNAi), or gon-1(q518). * P < 0.05, *** P < 0.005. In all panels, the anterior region of the gonad is to the left and its dorsal region is at the top of the image. Bar, 10 μm. The animals were cultured at 20° except for those used in the RNAi experiments (24.5°). RNAi, RNA interference.
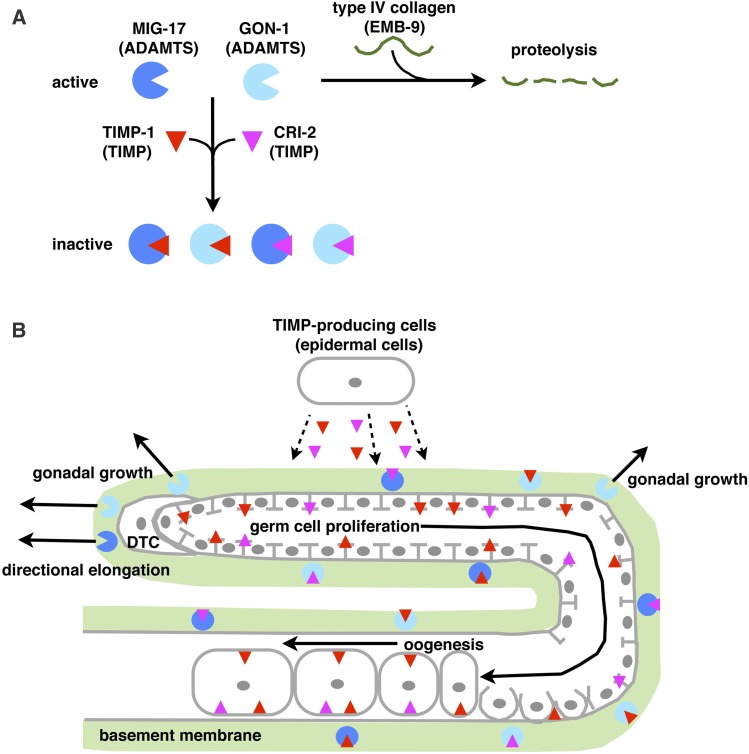
Figure 8.
Interaction of TIMPs and ADAMTSs during C. elegans gonad development. (A) On the gonadal basement membrane, the ADAMTSs MIG-17 and GON-1 degrade the type IV collagen α1 chain (EMB-9) of the basement membrane. The TIMPs TIMP-1 and CRI-2 inhibit the proteolytic activities of MIG-17 and GON-1. (B) TIMPs are produced by and secreted from epidermal cells, and then localize to the gonadal basement membrane and the plasma membranes of germ cells. Within the gonadal basement membrane, TIMP-1 and CRI-2 regulate gonadal morphogenesis by locally inhibiting the proteolytic activity of MIG-17 and GON-1. Around the DTCs, MIG-17 activity regulates directional gonad arm elongation (Ihara and Nishiwaki 2007). Because GON-1 is produced from DTCs and muscle cells, the proteolytic activity of GON-1 around DTCs is greater than at the lateral region of the gonad arms (Blelloch and Kimble 1999). At the plasma membrane of the germ cells and oocytes, TIMP-1 and CRI-2 may promote germ cell proliferation and oogenesis in a metalloproteinase-independent manner. ADAMTS, a disintegrin and metalloproteinase with thrombospondin motifs; DTCs, distal tip cells; TIMPs, tissue inhibitors of metalloproteinases.
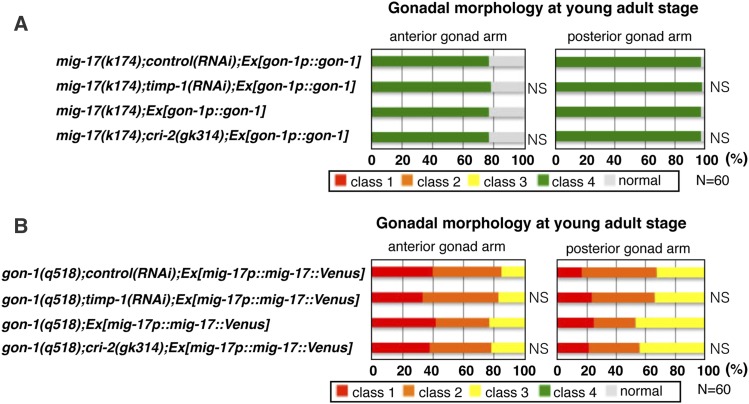
The suppression of gonadal defects found in the mig-17 and gon-1 null mutants upon depletion of the TIMP genes is canceled by overexpression of gon-1 or mig-17, respectively
The fact that depletion of timp-1 and cri-2 rescued the directional gonadal elongation defect of the mig-17 null mutant suggested the possibility that both TIMP-1 and CRI-2 may have an activity that negatively regulates another ADAMTS proteinase, GON-1. We analyzed the effect of gon-1 overexpression in the mig-17 null mutant depleted of timp-1 and cri-2, and found that the class 4 phenotype was reversed (Figure 7A). Similarly, because depletion of timp-1 and cri-2 rescued the gonadal growth defect of the gon-1 null mutant, both TIMP-1 and CRI-2 may negatively regulate MIG-17. We therefore examined whether overexpression of the functional mig-17::Venus could reverse the suppression effect of the gon-1 null mutant depleted of timp-1 and cri-2; the gonadal growth defect (sum of the class 1 and class 2 defects) phenotype was indeed reversed (Figure 7B). Taken together, these results support the idea that TIMP-1 and CRI-2 act as negative regulators of the GON-1 and MIG-17 ADAMTS proteinases.
Figure 7.
Suppression effects of timp-1 and cri-2 on mig-17 and gon-1 null mutants are abrogated by overexpression of gon-1 or mig-17::Venus, respectively (A and B). Percentages of abnormal gonad morphology in young adults. The four classes of gonadal defects were scored as in Figure 2K. P-values are indicated for Fisher’s exact test in comparison with mig-17(k174);control(RNAi);Ex[gon-1p::gon-1], mig-17(k174) Ex[gon-1p::gon-1], gon-1(q518);control(RNAi);Ex[mig-17p::mig-17::Venus], or gon-1(q518);Ex[mig-17p::mig-17::Venus]. The animals were cultured at 20° except for those used in the RNAi experiments (24.5°). NS, not significant; RNAi, RNA interference.
Discussion
C. elegans TIMP-1 and CRI-2 are secreted, and localize to the basement membrane of gonads and the plasma membrane of germ cells
The product of the TIMP-1::Venus construct localizes to the basement membrane of gonads (Graham et al. 1997; Fitzgerald and Schwarzbauer 1998; Kang and Kramer 2000; Ackley et al. 2001; Huang et al. 2003; Hesselson et al. 2004; Kubota et al. 2004; Muriel et al. 2005; Kawano et al. 2009). In addition, it localizes to the plasma membrane of germ cells and oocytes of young-adult worms. The CRI-2::Venus construct yielded a localization pattern similar to that of the TIMP-1::Venus construct. The localization patterns of these C. elegans TIMPs are similar to that of mammalian TIMP-3, which interacts with the ECM, and cell surface signaling molecules such as VEGF and focal adhesion kinase (Yu et al. 2000; Qi et al. 2003; Gill et al. 2006; Kassiri et al. 2009). Possible roles for the TIMPs at the plasma membrane of germ cells may be to indirectly regulate ECM remodeling, or to directly promote signaling necessary for germ cell development and oogenesis. Although epidermal-derived TIMP-1::Venus significantly rescued the gonadal growth defect of the timp-1(tk71) mutant, muscle-derived TIMP-1::Venus only partially rescued the defect. It is possible that an unknown, epidermally expressed cofactor may be necessary for TIMP-1 activation. Alternatively, TIMP-1 may be mostly inactive when it is produced in muscle cells; for example, it may exist in a misfolded state.
Both TIMPs and ADAMTSs regulate gonadal development in a noncell-autonomous manner
How does epidermally expressed TIMP-1 regulate gonadal development? Similar to previous observations that muscle-derived MIG-17::Venus accumulates in coelomocytes (Kubota et al. 2006), the accumulation of TIMP-1::Venus and CRI-2::Venus in coelomocytes was observed in these transgenic strains. These results suggest that both of these TIMPs regulate gonadal morphogenesis and germline development in a noncell-autonomous manner. This noncell-autonomous regulation of gonadal development has also been observed both in mig-17/ADAMTS and gon-1/ADAMTS mutants (Blelloch and Kimble 1999; Nishiwaki et al. 2000). In addition to these observations, it was previously proposed that MIG-17/ADAMTS and GON-1/ADAMTS regulate type IV collagen level on the gonadal basement membrane during gonadal development (Kubota et al. 2008, 2012), which is consistent with our finding that timp-1 is required for type IV collagen accumulation on the gonadal basement membrane. Thus, competition between the ADAMTS-mediated promotion of gonadal basement membrane remodeling and its inhibition by TIMPs maintains appropriate levels of type IV collagen to achieve proper gonadal morphogenesis. When we focused on the roles of TIMPs in germ cell development, although the timp-1(tk71) mutant was completely sterile, the cri-2(gk314) mutant was fertile and viable. These results suggest that specific or divergent functions of TIMP-1 and CRI-2 regulate germ cell development. Taken together, our results demonstrate that the noncell-autonomous actions of the TIMPs and ADAMTSs are essential for gonadal development.
C. elegans TIMP-1 regulates gonad development in a metalloproteinase inhibitor activity-dependent manner
The product of the TIMP-1(C21S)::Venus construct, which was expected to lack protease inhibitor activity, localized to the gonadal basement membrane where an ADAMTS is found (Nishiwaki et al. 2000). However, the fact that the TIMP-1(C21S)::Venus construct did not rescue TIMP activity in the timp-1 mutant suggests that TIMP-1 regulates gonad development by inhibiting metalloproteinase activity. One possible explanation for the partial loss of inhibitor activity by TIMP-1(C21S) is that the mutated protein may have partially retained activity against metalloproteinases.
C. elegans TIMPs negatively regulate multiple ADAMTSs to control gonad development
Although it has been shown that ADAMTSs regulate ECM remodeling (Russell et al. 2003; Brown et al. 2006; McCulloch et al. 2009; Enomoto et al. 2010; Dubail and Apte 2015; Kim and Nishiwaki 2015), the regulatory mechanisms of these proteinases in vivo have not been delineated. Our results suggest that TIMP-1 and CRI-2 inhibit MIG-17 and GON-1 activities so as to regulate proteolytic remodeling of the gonadal basement membrane during C. elegans gonadogenesis. This conclusion is consistent with our previous observation that a reduction in the amount of the type IV collagen α1 chain partially suppresses the directional elongation of the gonad arm phenotype in the mig-17 mutant (Kubota et al. 2008) and fits our model that partial reduction of type IV collagen accumulation by GON-1 is important in promoting gonadal elongation (Kubota et al. 2012). The α1 chain of type IV collagen might be a substrate for MIG-17 and GON-1. These observations are consistent with previous findings that excessive proteolysis of type IV collagen occurred in the Drosophila timp mutant (Pearson et al. 2016). Taken together, we propose that partial proteolytic remodeling of type IV collagen by ADAMTSs is required for directional elongation of the gonad arm and gonadal growth, and that this proteolytic activity is negatively regulated by TIMP-1 and CRI-2 (Figure 8).
Reduction of TIMP-1 or CRI-2 partially suppressed the defects of directional elongation of the gonad arm and gonadal growth in the null mutants, mig-17 and gon-1. This suppression of gonadal defects in mig-17 and gon-1 null mutants upon depletion of TIMPs was completely abrogated by overexpression of gon-1 or mig-17, respectively. These results suggest that TIMP-1 and CRI-2 inhibit both MIG-17 and GON-1. Alternatively, these TIMPs may negatively regulate the protease activities of matrix proteinases other than MIG-17 and GON-1. However, proteases negatively regulated by TIMPs have not been identified. In humans, the 19 ADAMTSs and four TIMPs exhibit tissue-specific expression patterns, and regulate tissue remodeling during various developmental processes (Porter et al. 2005; Arpino et al. 2015; Dubail and Apte 2015). Among the four mammalian TIMPs, only TIMP-3 significantly inhibits ADAMTS-2, ADAMTS-4, and ADAMTS-5 in an in vitro proteinase activity assay (Hashimoto et al. 2001; Kashiwagi et al. 2001; Wang et al. 2006), and therefore the extent of phenotype suppression in ADAMTS mutants of C. elegans by depletion of the TIMPs may reflect their inhibition activity against the ADAMTSs. Specific preferences for individual ADAMTSs by TIMPs should, therefore, be characterized in future studies.
Negative regulation of the numerous ADAMTSs by TIMPs is evolutionarily conserved. For example, the human-derived ADAMTS transgenes ADAMTS-4 and ADAMTS-9 could partially rescue the gonadal elongation-defective phenotype of the C. elegans gon-1 mutant (Hesselson et al. 2004). C. elegans may serve as a model organism for ADAMTS-related human diseases. Our data demonstrate that reduction of C. elegans TIMPs partially suppressed gonadal morphogenesis defects in ADAMTS-defective mutants. Therefore, we suggest that TIMPs may potentially be therapeutic targets for ADAMTS-related human congenital disorders and other pathologies.
Acknowledgments
Some of the worm strains used in this study were provided by the Caenorhabditis Genetics Center, which is funded by the USA National Institutes of Health Office of Research Infrastructure Programs (P40 OD-010440) and by the C. elegans Gene Knockout Consortium. This work was supported by the JSPS KAKENHI (grant number 22111005 to K.N).
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.7981535.
Communicating editor: H. Bülow
Literature Cited
- Ackley B. D., Crew J. R., Elamaa H., Pihlajaniemi T., Kuo C. J., et al. , 2001. The NC1/endostatin domain of Caenorhabditis elegans type XVIII collagen affects cell migration and axon guidance. J. Cell Biol. 152: 1219–1232. 10.1083/jcb.152.6.1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apte S. S., 2009. A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (ADAMTS) superfamily: functions and mechanisms. J. Biol. Chem. 284: 31493–31497. 10.1074/jbc.R109.052340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpino V., Brock M., Gill S. E., 2015. The role of TIMPs in regulation of extracellular matrix proteolysis. Matrix Biol. 44–46: 247–254. 10.1016/j.matbio.2015.03.005 [DOI] [PubMed] [Google Scholar]
- Blelloch R., Kimble J., 1999. Control of organ shape by a secreted metalloprotease in the nematode Caenorhabditis elegans. Nature 399: 586–590. 10.1038/21196 [DOI] [PubMed] [Google Scholar]
- Blelloch R., Anna-Arriola S. S., Gao D., Li Y., Hodgkin J., et al. , 1999. The gon-1 gene is required for gonadal morphogenesis in Caenorhabditis elegans. Dev. Biol. 216: 382–393. 10.1006/dbio.1999.9491 [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brew K., Dinakarpandian D., Nagase H., 2000. Tissue inhibitors of metalloproteinases: evolution, structure and function. Biochim. Biophys. Acta 1477: 267–283. 10.1016/S0167-4838(99)00279-4 [DOI] [PubMed] [Google Scholar]
- Brown H. M., Dunning K. R., Robker R. L., Pritchard M., Russell D. L., 2006. Requirement for ADAMTS-1 in extracellular matrix remodeling during ovarian folliculogenesis and lymphangiogenesis. Dev. Biol. 300: 699–709. 10.1016/j.ydbio.2006.10.012 [DOI] [PubMed] [Google Scholar]
- Caterina N. C., Windsor L. J., Yermovsky A. E., Bodden M. K., Taylor K. B., et al. , 1997. Replacement of conserved cysteines in human tissue inhibitor of metalloproteinases-1. J. Biol. Chem. 272: 32141–32149. 10.1074/jbc.272.51.32141 [DOI] [PubMed] [Google Scholar]
- Colige A., Sieron A. L., Li S. W., Schwarze U., Petty E., et al. , 1999. Human Ehlers-Danlos syndrome type VII C and bovine dermatosparaxis are caused by mutations in the procollagen I N-proteinase gene. Am. J. Hum. Genet. 65: 308–317. 10.1086/302504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagoneau N., Benoist-Lasselin C., Huber C., Faivre L., Megarbane A., et al. , 2004. ADAMTS10 mutations in autosomal recessive Weill-Marchesani syndrome. Am. J. Hum. Genet. 75: 801–806. 10.1086/425231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubail J., Apte S. S., 2015. Insights on ADAMTS proteases and ADAMTS-like proteins from mammalian genetics. Matrix Biol. 44–46: 24–37. 10.1016/j.matbio.2015.03.001 [DOI] [PubMed] [Google Scholar]
- Enomoto H., Nelson C. M., Somerville R. P., Mielke K., Dixon L. J., et al. , 2010. Cooperation of two ADAMTS metalloproteases in closure of the mouse palate identifies a requirement for versican proteolysis in regulating palatal mesenchyme proliferation. Development 137: 4029–4038. 10.1242/dev.050591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fata J. E., Ho A. T., Leco K. J., Moorehead R. A., Khokha R., 2000. Cellular turnover and extracellular matrix remodeling in female reproductive tissues: functions of metalloproteinases and their inhibitors. Cell. Mol. Life Sci. 57: 77–95. 10.1007/s000180050500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald M. C., Schwarzbauer J. E., 1998. Importance of the basement membrane protein SPARC for viability and fertility in Caenorhabditis elegans. Curr. Biol. 8: 1285–1288. 10.1016/S0960-9822(07)00540-4 [DOI] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Sarov M., Taylor J., Flibotte S., et al. , 2014. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat. Methods 11: 529–534. 10.1038/nmeth.2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S. E., Pape M. C., Leco K. J., 2006. Tissue inhibitor of metalloproteinases 3 regulates extracellular matrix–cell signaling during bronchiole branching morphogenesis. Dev. Biol. 298: 540–554. 10.1016/j.ydbio.2006.07.004 [DOI] [PubMed] [Google Scholar]
- Graham P. L., Johnson J. J., Wang S., Sibley M. H., Gupta M. C., et al. , 1997. Type IV collagen is detectable in most, but not all, basement membranes of Caenorhabditis elegans and assembles on tissues that do not express it. J. Cell Biol. 137: 1171–1183. 10.1083/jcb.137.5.1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X. D., Johnson J. J., Kramer J. M., 1991. Embryonic lethality caused by mutations in basement membrane collagen of C. elegans. Nature 349: 707–709. 10.1038/349707a0 [DOI] [PubMed] [Google Scholar]
- Hashimoto G., Aoki T., Nakamura H., Tanzawa K., Okada Y., 2001. Inhibition of ADAMTS4 (aggrecanase-1) by tissue inhibitors of metalloproteinases (TIMP-1, 2, 3 and 4). FEBS Lett. 494: 192–195. 10.1016/S0014-5793(01)02323-7 [DOI] [PubMed] [Google Scholar]
- Hedgecock E. M., Culotti J. G., Hall D. H., Stern B. D., 1987. Genetics of cell and axon migrations in Caenorhabditis elegans. Development 100: 365–382. [DOI] [PubMed] [Google Scholar]
- Hesselson D., Newman C., Kim K. W., Kimble J., 2004. GON-1 and fibulin have antagonistic roles in control of organ shape. Curr. Biol. 14: 2005–2010. 10.1016/j.cub.2004.11.006 [DOI] [PubMed] [Google Scholar]
- Huang C. C., Hall D. H., Hedgecock E. M., Kao G., Karantza V., et al. , 2003. Laminin alpha subunits and their role in C. elegans development. Development 130: 3343–3358. 10.1242/dev.00481 [DOI] [PubMed] [Google Scholar]
- Ihara S., Nishiwaki K., 2007. Prodomain-dependent tissue targeting of an ADAMTS protease controls cell migration in Caenorhabditis elegans. EMBO J. 26: 2607–2620. 10.1038/sj.emboj.7601718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara S., Hagedorn E. J., Morrissey M. A., Chi Q., Motegi F., et al. , 2011. Basement membrane sliding and targeted adhesion remodels tissue boundaries during uterine-vulval attachment in Caenorhabditis elegans. Nat. Cell Biol. 13: 641–651. 10.1038/ncb2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismat A., Cheshire A. M., Andrew D. J., 2013. The secreted AdamTS-A metalloprotease is required for collective cell migration. Development 140: 1981–1993. 10.1242/dev.087908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson H. W., Defamie V., Waterhouse P., Khokha R., 2017. TIMPs: versatile extracellular regulators in cancer. Nat. Rev. Cancer 17: 38–53. 10.1038/nrc.2016.115 [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Martinez-Campos M., Zipperlen P., Fraser A. G., Ahringer J., 2001. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2: RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S. H., Kramer J. M., 2000. Nidogen is nonessential and not required for normal type IV collagen localization in Caenorhabditis elegans. Mol. Biol. Cell 11: 3911–3923. 10.1091/mbc.11.11.3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi M., Tortorella M., Nagase H., Brew K., 2001. TIMP-3 is a potent inhibitor of aggrecanase 1 (ADAM-TS4) and aggrecanase 2 (ADAM-TS5). J. Biol. Chem. 276: 12501–12504. 10.1074/jbc.C000848200 [DOI] [PubMed] [Google Scholar]
- Kassiri Z., Defamie V., Hariri M., Oudit G. Y., Anthwal S., et al. , 2009. Simultaneous transforming growth factor beta-tumor necrosis factor activation and cross-talk cause aberrant remodeling response and myocardial fibrosis in Timp3-deficient heart. J. Biol. Chem. 284: 29893–29904. 10.1074/jbc.M109.028449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano T., Zheng H., Merz D. C., Kohara Y., Tamai K. K., et al. , 2009. C. elegans mig-6 encodes papilin isoforms that affect distinct aspects of DTC migration, and interacts genetically with mig-17 and collagen IV. Development 136: 1433–1442. 10.1242/dev.028472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessenbrock K., Plaks V., Werb Z., 2010. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 141: 52–67. 10.1016/j.cell.2010.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. S., Nishiwaki K., 2015. Control of the basement membrane and cell migration by ADAMTS proteinases: lessons from C. elegans genetics. Matrix Biol. 44–46: 64–69. 10.1016/j.matbio.2015.01.001 [DOI] [PubMed] [Google Scholar]
- Kimble J., Hirsh D., 1979. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 70: 396–417. 10.1016/0012-1606(79)90035-6 [DOI] [PubMed] [Google Scholar]
- Kubota Y., Kuroki R., Nishiwaki K., 2004. A fibulin-1 homolog interacts with an ADAM protease that controls cell migration in C. elegans. Curr. Biol. 14: 2011–2018. 10.1016/j.cub.2004.10.047 [DOI] [PubMed] [Google Scholar]
- Kubota Y., Sano M., Goda S., Suzuki N., Nishiwaki K., 2006. The conserved oligomeric Golgi complex acts in organ morphogenesis via glycosylation of an ADAM protease in C. elegans. Development 133: 263–273. 10.1242/dev.02195 [DOI] [PubMed] [Google Scholar]
- Kubota Y., Ohkura K., Tamai K. K., Nagata K., Nishiwaki K., 2008. MIG-17/ADAMTS controls cell migration by recruiting nidogen to the basement membrane in C. elegans. Proc. Natl. Acad. Sci. USA 105: 20804–20809. 10.1073/pnas.0804055106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y., Nagata K., Sugimoto A., Nishiwaki K., 2012. Tissue architecture in the Caenorhabditis elegans gonad depends on interactions among fibulin-1, type IV collagen and the ADAMTS extracellular protease. Genetics 190: 1379–1388. 10.1534/genetics.111.133173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucera K., Harrison L. M., Cappello M., Modis Y., 2011. Ancylostoma ceylanicum excretory-secretory protein 2 adopts a netrin-like fold and defines a novel family of nematode proteins. J. Mol. Biol. 408: 9–17. 10.1016/j.jmb.2011.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno K., Kanada N., Nakashima E., Fujiki F., Ichimura F., et al. , 1997. Molecular cloning of a gene encoding a new type of metalloproteinase-disintegrin family protein with thrombospondin motifs as an inflammation associated gene. J. Biol. Chem. 272: 556–562. 10.1074/jbc.272.1.556 [DOI] [PubMed] [Google Scholar]
- Kurshan P. T., Phan A. Q., Wang G. J., Crane M. M., Lu H., et al. , 2014. Regulation of synaptic extracellular matrix composition is critical for proper synapse morphology. J. Neurosci. 34: 12678–12689. 10.1523/JNEUROSCI.1183-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch D. R., Nelson C. M., Dixon L. J., Silver D. L., Wylie J. D., et al. , 2009. ADAMTS metalloproteases generate active versican fragments that regulate interdigital web regression. Dev. Cell 17: 687–698. 10.1016/j.devcel.2009.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. 10.1002/j.1460-2075.1991.tb04966.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales J., Al-Sharif L., Khalil D. S., Shinwari J. M., Bavi P., et al. , 2009. Homozygous mutations in ADAMTS10 and ADAMTS17 cause lenticular myopia, ectopia lentis, glaucoma, spherophakia, and short stature. Am. J. Hum. Genet. 85: 558–568. 10.1016/j.ajhg.2009.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriel J. M., Dong C., Hutter H., Vogel B. E., 2005. Fibulin-1C and Fibulin-1D splice variants have distinct functions and assemble in a hemicentin-dependent manner. Development 132: 4223–4234. 10.1242/dev.02007 [DOI] [PubMed] [Google Scholar]
- Nishiwaki K., 1999. Mutations affecting symmetrical migration of distal tip cells in Caenorhabditis elegans. Genetics 152: 985–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki K., Hisamoto N., Matsumoto K., 2000. A metalloprotease disintegrin that controls cell migration in Caenorhabditis elegans. Science 288: 2205–2208. 10.1126/science.288.5474.2205 [DOI] [PubMed] [Google Scholar]
- Page-McCaw A., Ewald A. J., Werb Z., 2007. Matrix metalloproteinases and the regulation of tissue remodelling. Nat. Rev. Mol. Cell Biol. 8: 221–233. 10.1038/nrm2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J. R., Zurita F., Tomas-Gallardo L., Diaz-Torres A., Diaz de la Loza Mdel C., et al. , 2016. ECM-regulator timp is required for stem cell Niche Organization and Cyst production in the Drosophila ovary. PLoS Genet. 12: e1005763 10.1371/journal.pgen.1005763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter S., Clark I. M., Kevorkian L., Edwards D. R., 2005. The ADAMTS metalloproteinases. Biochem. J. 386: 15–27. 10.1042/BJ20040424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J. H., Ebrahem Q., Moore N., Murphy G., Claesson-Welsh L., et al. , 2003. A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat. Med. 9: 407–415. 10.1038/nm846 [DOI] [PubMed] [Google Scholar]
- Qin J., Liang J., Ding M., 2014. Perlecan antagonizes collagen IV and ADAMTS9/GON-1 in restricting the growth of presynaptic boutons. J. Neurosci. 34: 10311–10324. 10.1523/JNEUROSCI.5128-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D. L., Doyle K. M., Ochsner S. A., Sandy J. D., Richards J. S., 2003. Processing and localization of ADAMTS-1 and proteolytic cleavage of versican during cumulus matrix expansion and ovulation. J. Biol. Chem. 278: 42330–42339. 10.1074/jbc.M300519200 [DOI] [PubMed] [Google Scholar]
- Shibata Y., Kawakado Y., Hori N., Tanaka K., Inoue R., et al. , 2016. Organ length control by an ADAMTS extracellular protease in Caenorhabditis elegans. G3 (Bethesda) 6: 1449–1457. 10.1534/g3.116.028019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo T., Kurihara H., Kuno K., Yokoyama H., Wada T., et al. , 2000. ADAMTS-1: a metalloproteinase-disintegrin essential for normal growth, fertility, and organ morphology and function. J. Clin. Invest. 105: 1345–1352. 10.1172/JCI8635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shozu M., Minami N., Yokoyama H., Inoue M., Kurihara H., et al. , 2005. ADAMTS-1 is involved in normal follicular development, ovulatory process and organization of the medullary vascular network in the ovary. J. Mol. Endocrinol. 35: 343–355. 10.1677/jme.1.01735 [DOI] [PubMed] [Google Scholar]
- Sibley M. H., Johnson J. J., Mello C. C., Kramer J. M., 1993. Genetic identification, sequence, and alternative splicing of the Caenorhabditis elegans alpha 2(IV) collagen gene. J. Cell Biol. 123: 255–264. 10.1083/jcb.123.1.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M., Merz D. C., Killeen M. T., Zhou Y., Zheng H., et al. , 2000. Regulation of the UNC-5 netrin receptor initiates the first reorientation of migrating distal tip cells in Caenorhabditis elegans. Development 127: 585–594. [DOI] [PubMed] [Google Scholar]
- Vu T. H., Werb Z., 2000. Matrix metalloproteinases: effectors of development and normal physiology. Genes Dev. 14: 2123–2133. 10.1101/gad.815400 [DOI] [PubMed] [Google Scholar]
- Wang W. M., Ge G., Lim N. H., Nagase H., Greenspan D. S., 2006. TIMP-3 inhibits the procollagen N-proteinase ADAMTS-2. Biochem. J. 398: 515–519. 10.1042/BJ20060630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W. H., Yu S., Meng Q., Brew K., Woessner J. F., Jr, 2000. TIMP-3 binds to sulfated glycosaminoglycans of the extracellular matrix. J. Biol. Chem. 275: 31226–31232. 10.1074/jbc.M000907200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and reagents cited in Tables S1 and S2 are available upon request. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.7981535.