Abstract
It becomes increasingly important in using genome-wide association studies (GWAS) to select important genetic information associated with qualitative or quantitative traits. Currently, the discovery of biological association among SNPs motivates various strategies to construct SNP-sets along the genome and to incorporate such set information into selection procedure for a higher selection power, while facilitating more biologically meaningful results. The aim of this paper is to propose a novel Bayesian framework for hierarchical variable selection at both SNP-set (group) level and SNP (within group) level. We overcome a key limitation of existing posterior updating scheme in most Bayesian variable selection methods by proposing a novel sampling scheme to explicitly accommodate the ultrahigh-dimensionality of genetic data. Specifically, by constructing an auxiliary variable selection model under SNP-set level, the new procedure utilizes the posterior samples of the auxiliary model to subsequently guide the posterior inference for the targeted hierarchical selection model. We apply the proposed method to a variety of simulation studies and show that our method is computationally efficient and achieves substantially better performance than competing approaches in both SNP-set and SNP selection. Applying the method to the Alzheimers Disease Neuroimaging Initiative (ADNI) data, we identify biologically meaningful genetic factors under several neuroimaging volumetric phenotypes. Our method is general and readily to be applied to a wide range of biomedical studies.
Keywords: imaging genetics, genome-wide association studies, SNP-set, Bayesian variable selection, Markov chain Monte Carlo
IN modern genetics, genome-wide association studies (GWAS) has become a popular tool to study complex human diseases (Walsh et al. 2014; Wang et al. 2014; Hibar et al. 2015). The goal of GWAS is to identify single nucleotide polymorphisms (SNPs) associated with complex traits. Over the last few years, improvement in genotyping technology has enriched the measurements of SNPs to >1 million (Altshuler et al. 2008). Due to the ultrahigh-dimensionality of SNPs, most GWAS approaches analyze one SNP at a time to test marginal association of the SNP with phenotype. However, in many scenarios, large differences can exist between marginal effects of SNPs and their joint effects (He and Lin 2011). Thus, it is imperative to carry out whole-genome GWAS that considers all SNPs together.
There are at least two challenges associated with whole-genome GWAS. The first one is the “large p small n” problem. To address this issue, various regularization or screening methods (Tibshirani 1996; Fan and Li 2001; Efron et al. 2004; Zou and Hastie 2005; Zou 2006; Fan and Lv 2008) were proposed and recently extended to the context of GWAS (Hoggart et al. 2008; Wu et al. 2009; Cho et al. 2010; He and Lin 2011; Sampson et al. 2013; Jiang et al. 2016; Bao and Wang 2017; Huang et al. 2017). As an alternative, Bayesian methods also play a prominent role in solving variable selection problem. O’Hara and Sillanpää (2009) provided an overview of several commonly used Bayesian variable selection methods and posterior simulation algorithms, such as the Gibbs variable selection (GVS) (Dellaportas et al. 2002) and the stochastic search variable selection (SSVS) (George and McCulloch 1993). Compared with regularization methods, Bayesian models have the natural advantage to quantify uncertainty and combine prior information. Their recent application on GWAS also showed a higher detection power by simultaneously fitting multiple marker effects and implicitly correcting biological structures (Sahana et al. 2010; Dashab et al. 2012; Kärkkäinen and Sillanpää 2012). One major limitation to apply Bayesian variable selection models on GWAS is the intensive computation. Therefore, existing approaches made attempts to explore different inference mechanisms to reduce the computational cost or improve the mixing of Markov chain. For instance, besides traditional Markov chain Monte Carlo (MCMC) algorithm (Guan et al. 2011), variational Bayes (Carbonetto et al. 2012), evolutionary stochastic search (Bottolo et al. 2013) and other variations of stochastic searching algorithms (Briollais et al. 2016; Yang et al. 2017) were developed under Bayesian sparse models for multi-SNP analysis. Alternatively, Bayesian lasso (Li et al. 2010; Jiang et al. 2016) and Bayesian mixed model (Zhou et al. 2013; Zhou 2014) were also considered to improve the scalability in the presence of ultrahigh-dimensional SNP data.
Another challenge is caused by biological architecture. Multiple causal SNPs may be located in a single region, each with a small effect. In order to increase power to map them, it is desirable to consider them simultaneously and perform SNP-set analysis. A variety of methods were proposed for SNP-set analysis, including, but not limited to, Kwee et al. (2008), Wang and Abbott (2008), Wu et al. (2010) and a more recent Bayesian latent sparse model by Lu et al. (2015). Tzeng et al. (2011) provided an overview of different marker-set approaches for gene-trait association and appealing features of set-based methods compared with those using individual SNPs. For the marker-set methods, grouping strategies to define SNP-sets play a vital role in practice. As suggested by Wu et al. (2011), the ones incorporating biological information, for example grouping SNPs by genes, pathways or haplotype/linkage disequilibrium (LD) blocks, tend to gain more power. Based on the defined SNP-sets, analysis could be carried out through weighted sum of genotypes (Wang and Elston 2007; Price et al. 2010), U-statistics (Tzeng et al. 2003; Wei et al. 2008), or variance-component methods (Tzeng and Zhang 2007; Wu et al. 2010). Meanwhile, the use of SNP-sets can also reduce the number of predictors as well as alleviate the collinearity issue since the correlations are much smaller among SNP-sets than SNPs.
The appealing features of SNP-set analysis motivate us to incorporate set-wise information into the selection procedure. In this paper, we develop a Bayesian hierarchical variable selection model to carry out whole-genome association analysis and achieve SNP/SNP-set trait association mapping. Our variable selection approach is inherently hierarchical, and involves selection at both SNP-set level and individual SNP level. Although there is a broad literature on Bayesian variable selection under high or ultrahigh-dimensional feature space (Bottolo et al. 2010; Johnson and Rossell 2012; Johnson 2013), few efficient hierarchical variable selection methods have been developed. Stingo et al. (2011) considered gene expression data, and adopted Bayesian spike-and-slab priors to simultaneously select genes and pathways. Rockova et al. (2014) proposed a two-step procedure to carry out hierarchical variable selection under Bayesian group Lasso. Combining spike-and-slab priors with shrinkage priors, Duan and Thomas (2013), Zhang et al. (2014a,b), Liquet et al. (2017) proposed similar modeling frameworks, and achieved group level selection and within-group shrinkage. Though the existing approaches are promising, they suffered with two limitations. First, the methods employ traditional MCMC algorithm, which are computational intensive and difficult to scale up. Second, the group level selection does not utilize any structural information, which could lead to poor performance. Recently, Tang et al. (2017) proposed a Bayesian hierarchical generalized linear models incorporating group information, and developed an EM algorithm for parameter estimation. However, the method does not impose group-level sparsity and may be less powerful under sparse signal like GWAS application.
In this paper, we develop a Bayesian hierarchical selection model, named Sparse Group Hierarchical Sampling (SGHS), with an efficient posterior inference algorithm. The key idea of SGHS is to reallocate posterior computation spending on the potential signal and noise parts via a “smart” proposal distribution. The sampling scheme for the proposal distribution is specified by an auxiliary model constructed under set-wise variable selection using factor regression. Such a modeling scheme allows the MCMC algorithm to explore the entire sample space more efficiently and dramatically mitigate the computational burden of updating large-scale unknown parameters. Simultaneously, grouping and structural information are integrated within the posterior inference, leading to the improvement on selection accuracy and results interpretability.
The remainder of this article is organized as follows. In Model specification, we present our basic model for variable selection, prior specifications, and a standard posterior computation algorithm. In Sparse group hierarchical sampling, we propose our SGHS for hierarchical variable selection. We conduct simulation studies in Simulation studies to assess the performance of our proposed approach, and in The Alzheimer’s Disease neuroimaging initiative, we apply the method to the ADNI data to identify disease related genetic information. Finally, we conclude with a Discussion.
Materials and Methods
Model specification
We introduce the variable selection model in a Bayesian framework. Our description is based on GWAS, but the proposed method is general and readily extended to other applications. Assume there are n subjects in the data. For subject , is a trait of interest, such as the brain volume of a region of interest (ROI), and is a -vector of clinical variables, including an intercept term. Suppose that the whole genome contains J SNPs, based on which, K SNP-sets are defined with SNPs included in SNP-set k. We assume the SNP-sets are mutually exclusive, then the total number of SNPs . We let denote the genotype of SNP j within SNP-set k representing by the minor alleles, and we also conduct a subsequent normalization for each SNP.
We consider a standard regression model for hierarchical variable selection given by
| (1) |
with the residual error . Here is the set-wise selection indicator describing the selection status for SNP-set k, and is the individual one for SNP j within SNP-set k. Moreover, and are regression coefficients with . Our goal is to identify risk SNP-sets along with specific SNPs by using the selection indicators . Particularly, is included in model (1) if both and are nonzero. We can further write model (1) in a more compact form
| (2) |
where and . Here “” is the individual-to-set mapping matrix with “” representing the Hadamard product (Styan 1973). As is the general case in practice, we assume that “signals” are sparse with the cardinality of the active set . It is worth noting that, in practice, SNP-sets can be overlapped based on certain biologically grouping strategy, e.g., genes, pathways. In such cases, we can randomly assign a SNP to one of its overlapped SNP-sets.
We introduce priors for all the parameters in model (1). Specifically, we assign conjugate Gaussian priors to the regression coefficients as
| (3) |
and conjugate hyper-priors for the variance parameters and . An equivalent model formulation is to define a coefficient , which results in the well-known point mass mixture prior with a point mass at zero. Compared with mixture prior, the above specification enables a more efficient updated scheme for coefficients, which we will explain later. In terms of the selection indicators and , we assume separate independent Bernoulli priors
| (4) |
where and with controlling the proportion of SNP-sets in the model, and determining the proportion of significant SNPs. We finally assign a prior for the variance of residual error .
Parameters included in the posterior inference are , , , , , and η with the joint conditional posterior distribution
| (5) |
Standard MCMC algorithm can be implemented to update each parameter from its full conditional distribution. The details of updating scheme for all the parameters are provided in Appendix A1. As noted, to update , we resort to a block updating scheme by dividing the whole long vector into two blocks, and corresponding to the selected () and unselected () predictors. Thus, the conditional distributions of and are given by
| (6) |
where and with corresponding to the active set. Such vector-wise updating scheme can dramatically reduce computational cost and lead to better mixing than the single-site Gibbs sampling.
Sparse group hierarchical sampling
Standard MCMC algorithm becomes inefficient in the presence of high-dimensional data, and even computationally infeasible under GWAS with tens of thousands of predictors. Therefore, in this section, we propose the SGHS scheme to overcome the computational complexity in the posterior computation. The key idea of SGHS is to construct an auxiliary model based on set-wise variable selection, which subsequently realizes a reweight of posterior computational effort on potential signals and noises in the target model to efficiently search among model spaces.
Specifically, we first introduce an auxiliary set-wise indicator and define each of their elements
| (7) |
Through this definition, the auxiliary set-wise indicator is completely determined by the SNP-wise selection indicator, by the selection consistency between two levels, and the structure of stays the same as that of the set-wise selection indicator . Accordingly, the posterior distribution for the parameters becomes
| (8) |
where only if (7), and zero otherwise.
In the right-hand side of (8), the first term consistent with Equation 5 is the main probability part, while the second term captures the connection of the selection information between SNP-set level and SNP level, inducing the incorporation of group membership. A comparison between (5) and (8) shows that the posterior probabilities and sampling schemes for parameters , , η, and hyper-parameters , keep consistent with those in the standard MCMC algorithm. As for the selection indicators and and auxiliary indicator , we take a novel approach to jointly update via a Metropolis-Hastings (M-H) step by constructing a proposal distribution
| (9) |
where the subscripts “c” and “*” denote the current value and the proposed value, and “” represents all the other parameters. Here specifies the sampling scheme for the auxiliary indicator , and we design it to depend only on the data as shown in Equation 9. Such specification removes the interference of other parameters, which allows function to be achieved by a separate model with variable selection only at SNP-set level. We refer to this model as an auxiliary model (distinguished from the target model) with the goal to help the posterior simulation of the target model. Given the sampled value , the sampling scheme for is specified by function which induces the incorporation of information from SNP-set level selection. Under such a proposal distribution, function is a receiver with the set-wise selection information “copied” from the auxiliary model, and function is a transmitter to pass such information from to the target model. In the following sections, we will discuss the choice of and that could lead to a more efficient posterior sample procedure.
Receiver function :
As discussed before, the sampling procedure of receiver function is induced by an auxiliary SNP-set level selection model. In other words, realization of can be directly simulated from the posterior distribution of the auxiliary model. As a natural choice of an auxiliary model, following model (1), we have
| (10) |
where . We use a superscript “a” in (10) to distinguish the corresponding parameters in the auxiliary model from the target model.
Model (10) turns out to be less attractive in general cases where not all the SNPs within a SNP-set are predictive for phenotype of interest. Since the assumption of nonzero coefficients in the risk SNP-sets is conflicted with the within-group level sparsity, it is difficult for model (10) to approximate the truth. To address this issue, we resort to an alternative set-wise selection model based on an empirical factor (principal component) regression under each SNP-set. Specifically, we adopt a reduced rank singular-value decomposition (SVD) on each , namely , for . Here, with is an factor matrix subject to , where is a diagonal matrix formed by positive singular values of , and is the loadings matrix subject to . By replacing each with the reduced SVD representation, the new auxiliary model becomes
| (11) |
where is the coefficient for factor l within SNP-set k. In GWAS data where there are strong correlations among predictors, to avoid overfitting, we could also allow certain truncation by specifying a cutoff on the number of factors included with . Comparing with , under model (11), we realize a dimension reduction of the predictors from J to L with , where . For selected SNP sets, the overall nonzero assumption of vector also fits many scenarios in real applications. Thus, we use model (11) as the auxiliary model in our applications. Again, the choice of an auxiliary model is not uniquely determined since it serves only as a platform to influence posterior sampling of the main model, but model (11), for example, has been shown to perform well in a wide range of scenarios.
Following the prior specification in (3) for the target model, we assign similar conjugate priors for the coefficients
| (12) |
and for the variance parameters , and . We assign independent Bernoulli priors for the auxiliary indicators
| (13) |
where and with controls the proportion of selection. We further introduce a hyper-prior for each proportion parameter as follows:
| (14) |
with the shape parameters. The hyper-prior (14) distinguishes the posterior probabilities between risk and nonrisk SNP-sets via a mixture of a zero point mass and a spread Beta distribution. By combining priors (13) and (14), we can integrate out which results in independent Bernoulli priors for indicator with proportion parameter . In this case, we use (13) and (14) to ensure sparsity in the group level auxiliary model, which subsequently brings impact to the target model.
Transmitter function :
To efficiently sample from the large feature space, we propose as follows:
| (15) |
with
| (16) |
where ϕ, and are tuning parameters to make sure detailed balance in the M-H step is satisfied, as well as to allow enough information to be borrowed from proposals. Specifically, ϕ determines the amount of difference between SNP-set selection and the proposal, and if a SNP-set is currently unselected, controls the sparsity for the selection of its SNPs; otherwise, influences the agreement of the SNP-level selection between proposed and current status. In practice, we specify , and to allow an efficient transmission of the selection information.
To implement this M-H step, we first draw with simulated by the posterior distribution of in the auxiliary model. Then, we draw . Finally, we calculate
| (17) |
and set when with . To further improve the mixing of Markov chains, in addition to the M-H step, we also conduct a further moving step for under from its full conditional (32). Since the true signal is sparse, such a moving step does not require a heavy computation. Under the specification of this proposal distribution, in our SGHS scheme, each SNP has a positive probability to be selected or unselected. However, instead of updating all the SNPs at each iteration of the posterior simulation, a large amount of nonrisk variables (noises) have been directly “labeled” iteration by iteration (without updating), which allows us to spend most of the computation updating potential signal part. As a result, the computational efficiency is dramatically improved compared with the existing MCMC algorithm. A detailed MCMC algorithm for the SGHS scheme is provided in Appendix A2.
Data availability
We used ADNI1 genetics and MRI image data that are available through application (http://adni.loni.usc.edu/about/adni1/). All the ADNI data are shared without embargo through the LONI Image and Data Archive (IDA).
Results
Simulation studies
We conduct simulation studies to evaluate the finite-sample performance of SGHS. Our goal is to select genetic markers that are highly associated with outcome of interest. We focus on the comparison between the proposed SGHS and existing methods include lasso (Lasso) (Tibshirani 1996), smoothly slipped absolute deviation (SCAD) (Fan and Li 2001), sparse-group Lasso (SGL) (Friedman et al. 2010), functional genome-wide association analysis (FGWAS) (Huang et al. 2017), Bayesian variable selection with posterior inference via model averaging and subset selection (piMASS) (Guan et al. 2011), and genome-wide efficient mixed model association (GEMMA) based on Bayesian sparse linear mixed model (Zhou 2014). To better assess each method under GWAS, all the simulation scenarios are designed to mimic real genetic data.
We generate subjects with the genetic information simulated from the Hapmap projects 2009-02 phaseIII data (International HapMap 3 Consortium 2010). Specifically, for each subject, we randomly combine two haplotypes from the CEPH population to form its genotypes. We consider both a low dimensional scenario by randomly selecting 5000 SNPs, and a high-dimensional one with 100,000 SNPs. Under each scenario, we determine SNP-sets (LD blocks) by starting from an initial SNP m with a putative block of SNPs and considering sub-blocks with until >50% of elements in the corresponding matrix of value surpass the threshold κ. To further assess the impact of LD structure, we set different thresholds with and 0.1 to construct the grouping information, based on which, we consider the following two cases of signal patterns to evaluate the robustness of variable selection.
Case 1: We randomly select 10 risk SNP-sets. Within each of them, we randomly set 10 SNPs as risk ones.
Case 2: We randomly select 10 risk SNP-sets. Within SNP-set k with , we randomly set k SNPs as risk ones; and within SNP-sets with , all the SNPs are risk ones. We denote as the number of risk SNPs in the SNP-set k.
Based on the two signal patterns, we consider a variety of settings for the value of nonzero regression coefficients in model (1), i.e., the regression coefficients of risk SNPs denoted by , as shown in Table 1. Specifically, in Case 1, where sparse signals exist, we consider six different settings. In Settings 1 and 2, as the starting point, we assign a unified genetic effect under different magnitudes. In Settings 3 and 4, the associations between the risk SNPs and the phenotype become more general but the majority of them are in the same sign. Finally, in Settings 5 and 6, we allow both positive and negative genetic effects within each SNP set to purposely create barriers for the internal SNP-set level selection. In Case 2, we further dilute the signals in the later half of risk SNP-sets while keep sparsity in the former half in Settings I and II, and our design guarantees the average strength of signals in each risk SNP-set is comparable. For each setting, we generate 100 Monte Carlo (MC) datasets to assess feature selection performance among all the methods.
Table 1. Simulation design: different settings of nonzero coefficients under two cases.
| Setting | |
|---|---|
| Case 1 | 1. |
| 2. | |
| 3. | |
| 4. | |
| 5. | |
| 6. | |
| Case 2 | I. |
| II. |
To implement SGHS, we conduct posterior inference with random initials for 10,000 iterations with 5000 burn-in for both the auxiliary and target models. The average computational time per dataset is 3.4 min for low dimensional scenarios and 26.5 min for high dimensional ones (Matlab implementation, 3.4 GHz CPU, 8 GB Memory, Windows System) to finish the whole posterior inference and the convergence is checked by GR method (Gelman and Rubin 1992) as well as trace plots. To allow for a noninformative hyper-prior, we assign a relative large value for and as 10. We also set to accommodate sparse signals in the auxiliary model, and less informative and in the target model. Finally, we truncate the number of factors in each subset by looking for the minimum number of singular vectors that can explain ∼ of the total variance. For the competing methods, we use R packages glmnet, ncvreg, and SGL to implement Lasso, SCAD, and SGL, and public released pipelines for FGWAS, GEMMA, and piMASS. For the Bayesian algorithms GEMMA and piMASS, we set all the tuning parameters as recommended by the manuals, and the average computational time is 10.9/48.6 min for GEMMA and 2.1/16.6 min for piMASS under low/high dimensional scenarios. Finally, the feature selection performance is assessed by sensitivity (Sens); specificity (Spec); Youden’s J statistic (J Stat), which equals sensitivity specificity; and area under the receiver operating characteristic curve (AUC) for all the methods.
The simulation results are summarized in Table 2 and Table 3 under low and high dimensions. To determine selection status for the Bayesian methods, i.e., GEMMA, piMASS, and SGHS, we use 0.5 as cutoff on the marginal posterior probability of selection indicator (Barbieri and Berger 2004). We first compare our method to the competing approaches. For all the settings under different LD thresholds κ and dimensions, our proposed method outperforms all competing methods in selection accuracy with a higher sensitivity, J Stat and AUC. Specifically, Settings 1 and 2 are constructed with a unified genetic effect among risk SNPs, and SGHS achieves the highest AUC and a much higher J Stat than its competitors under different dimensions and κ. In Settings 3 and 4, with a more general genetic effect, SGHS maintains its satisfactory performance in feature selection. In Settings 5 and 6, where positively and negatively associated SNPs within a causal SNP-set are almost equally distributed, while we notice decreases on AUC and J Stat for almost all the methods, SGHS still achieves a superior selection performance compared to other methods along with a satisfactory AUC. This reflects, to some extent, the robustness of SGHS to the extreme settings. Finally, the diluted signal patterns in Settings I and II further deteriorates the performance for LASSO, SCAD, SGL, piMASS, and GEMMA, but brings little impact on FGWAS and SGHS, with the latter remaining the best performer. Overall, Table 2 and Table 3 show a general pattern that SGHS works considerably better on detecting true risk features, as indicated by much higher sensitivities. Even though false positives have also been brought in as a consequence, we still obtain a remarkable feature selection accuracy.
Table 2. Simulation results: feature selection performance with .
| Setting | Method | Sens | Spec | J Stat | AUC | Sens | Spec | J Stat | AUC | Sens | Spec | J Stat | AUC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Lasso | 0.500 | 0.922 | 0.422 | 0.796 | 0.501 | 0.932 | 0.433 | 0.810 | 0.515 | 0.939 | 0.454 | 0.803 |
| SCAD | 0.168 | 0.963 | 0.131 | 0.698 | 0.161 | 0.968 | 0.129 | 0.717 | 0.167 | 0.970 | 0.137 | 0.695 | |
| SGL | 0.684 | 0.904 | 0.588 | 0.815 | 0.688 | 0.926 | 0.614 | 0.817 | 0.648 | 0.936 | 0.584 | 0.798 | |
| FGWAS | 0.444 | 0.957 | 0.401 | 0.748 | 0.541 | 0.950 | 0.491 | 0.773 | 0.599 | 0.951 | 0.550 | 0.801 | |
| piMASS | 0.128 | 0.998 | 0.126 | 0.816 | 0.146 | 0.998 | 0.144 | 0.816 | 0.112 | 0.998 | 0.110 | 0.839 | |
| GEMMA | 0.178 | 0.993 | 0.171 | 0.721 | 0.176 | 0.995 | 0.171 | 0.717 | 0.144 | 0.996 | 0.140 | 0.727 | |
| SGHS | 0.806 | 0.919 | 0.725 | 0.957 | 0.809 | 0.945 | 0.754 | 0.966 | 0.803 | 0.951 | 0.754 | 0.974 | |
| 2 | Lasso | 0.544 | 0.912 | 0.456 | 0.811 | 0.588 | 0.928 | 0.516 | 0.829 | 0.573 | 0.933 | 0.506 | 0.816 |
| SCAD | 0.171 | 0.966 | 0.137 | 0.713 | 0.160 | 0.970 | 0.130 | 0.717 | 0.182 | 0.970 | 0.152 | 0.703 | |
| SGL | 0.688 | 0.910 | 0.598 | 0.814 | 0.688 | 0.927 | 0.515 | 0.815 | 0.644 | 0.935 | 0.579 | 0.802 | |
| FGWAS | 0.443 | 0.941 | 0.384 | 0.751 | 0.553 | 0.949 | 0.502 | 0.779 | 0.606 | 0.951 | 0.557 | 0.797 | |
| piMASS | 0.196 | 0.997 | 0.193 | 0.813 | 0.196 | 0.998 | 0.194 | 0.982 | 0.160 | 0.998 | 0.158 | 0.840 | |
| GEMMA | 0.298 | 0.978 | 0.276 | 0.701 | 0.284 | 0.979 | 0.263 | 0.689 | 0.296 | 0.978 | 0.274 | 0.722 | |
| SGHS | 0.850 | 0.915 | 0.765 | 0.947 | 0.853 | 0.941 | 0.794 | 0.974 | 0.861 | 0.949 | 0.810 | 0.977 | |
| 3 | Lasso | 0.522 | 0.918 | 0.440 | 0.800 | 0.555 | 0.933 | 0.488 | 0.813 | 0.597 | 0.934 | 0.531 | 0.815 |
| SCAD | 0.175 | 0.966 | 0.141 | 0.710 | 0.164 | 0.970 | 0.134 | 0.698 | 0.175 | 0.971 | 0.146 | 0.697 | |
| SGL | 0.653 | 0.915 | 0.568 | 0.795 | 0.661 | 0.929 | 0.590 | 0.806 | 0.628 | 0.938 | 0.566 | 0.789 | |
| FGWAS | 0.468 | 0.946 | 0.414 | 0.697 | 0.566 | 0.949 | 0.515 | 0.696 | 0.579 | 0.952 | 0.531 | 0.797 | |
| piMASS | 0.216 | 0.998 | 0.214 | 0.842 | 0.182 | 0.998 | 0.180 | 0.839 | 0.148 | 0.998 | 0.146 | 0.841 | |
| GEMMA | 0.262 | 0.977 | 0.239 | 0.671 | 0.274 | 0.979 | 0.253 | 0.677 | 0.240 | 0.981 | 0.221 | 0.656 | |
| SGHS | 0.821 | 0.913 | 0.734 | 0.952 | 0.811 | 0.945 | 0.756 | 0.962 | 0.803 | 0.949 | 0.752 | 0.968 | |
| 4 | Lasso | 0.603 | 0.924 | 0.527 | 0.822 | 0.601 | 0.942 | 0.543 | 0.840 | 0.593 | 0.947 | 0.540 | 0.827 |
| SCAD | 0.170 | 0.967 | 0.137 | 0.714 | 0.161 | 0.970 | 0.131 | 0.713 | 0.165 | 0.971 | 0.136 | 0.690 | |
| SGL | 0.688 | 0.911 | 0.599 | 0.815 | 0.677 | 0.927 | 0.604 | 0.810 | 0.652 | 0.937 | 0.589 | 0.805 | |
| FGWAS | 0.430 | 0.953 | 0.383 | 0.755 | 0.550 | 0.950 | 0.500 | 0.727 | 0.599 | 0.951 | 0.500 | 0.805 | |
| piMASS | 0.326 | 0.993 | 0.319 | 0.818 | 0.274 | 0.994 | 0.268 | 0.815 | 0.284 | 0.995 | 0.279 | 0.821 | |
| GEMMA | 0.402 | 0.953 | 0.355 | 0.698 | 0.350 | 0.962 | 0.312 | 0.682 | 0.442 | 0.960 | 0.402 | 0.716 | |
| SGHS | 0.827 | 0.919 | 0.746 | 0.964 | 0.847 | 0.937 | 0.784 | 0.974 | 0.828 | 0.947 | 0.775 | 0.975 | |
| 5 | Lasso | 0.465 | 0.916 | 0.381 | 0.489 | 0.423 | 0.926 | 0.349 | 0.475 | 0.424 | 0.925 | 0.349 | 0.503 |
| SCAD | 0.179 | 0.966 | 0.145 | 0.504 | 0.185 | 0.969 | 0.154 | 0.482 | 0.170 | 0.969 | 0.149 | 0.499 | |
| SGL | 0.523 | 0.909 | 0.432 | 0.512 | 0.448 | 0.935 | 0.383 | 0.498 | 0.495 | 0.938 | 0.433 | 0.502 | |
| FGWAS | 0.431 | 0.958 | 0.509 | 0.526 | 0.443 | 0.964 | 0.407 | 0.622 | 0.490 | 0.963 | 0.453 | 0.719 | |
| piMASS | 0.182 | 0.997 | 0.179 | 0.765 | 0.162 | 0.998 | 0.160 | 0.767 | 0.154 | 0.998 | 0.152 | 0.709 | |
| GEMMA | 0.178 | 0.992 | 0.170 | 0.652 | 0.224 | 0.995 | 0.219 | 0.684 | 0.172 | 0.996 | 0.168 | 0.662 | |
| SGHS | 0.795 | 0.923 | 0.718 | 0.952 | 0.784 | 0.951 | 0.735 | 0.961 | 0.782 | 0.957 | 0.739 | 0.957 | |
| 6 | Lasso | 0.390 | 0.939 | 0.329 | 0.450 | 0.362 | 0.949 | 0.311 | 0.494 | 0.376 | 0.948 | 0.324 | 0.505 |
| SCAD | 0.186 | 0.959 | 0.145 | 0.490 | 0.162 | 0.965 | 0.127 | 0.487 | 0.168 | 0.965 | 0.133 | 0.499 | |
| SGL | 0.554 | 0.894 | 0.448 | 0.502 | 0.542 | 0.918 | 0.460 | 0.504 | 0.539 | 0.926 | 0.465 | 0.507 | |
| FGWAS | 0.433 | 0.956 | 0.509 | 0.528 | 0.436 | 0.963 | 0.399 | 0.575 | 0.446 | 0.966 | 0.412 | 0.621 | |
| piMASS | 0.100 | 0.999 | 0.099 | 0.897 | 0.104 | 0.999 | 0.103 | 0.804 | 0.080 | 0.999 | 0.079 | 0.805 | |
| GEMMA | 0.124 | 0.997 | 0.123 | 0.728 | 0.120 | 0.998 | 0.118 | 0.750 | 0.092 | 0.998 | 0.090 | 0.770 | |
| SGHS | 0.660 | 0.960 | 0.620 | 0.927 | 0.646 | 0.963 | 0.609 | 0.942 | 0.714 | 0.956 | 0.660 | 0.965 | |
| I | Lasso | 0.428 | 0.902 | 0.330 | 0.733 | 0.445 | 0.927 | 0.472 | 0.749 | 0.456 | 0.930 | 0.386 | 0.743 |
| SCAD | 0.098 | 0.913 | 0.011 | 0.660 | 0.093 | 0.936 | 0.029 | 0.640 | 0.091 | 0.942 | 0.033 | 0.637 | |
| SGL | 0.419 | 0.920 | 0.339 | 0.658 | 0.435 | 0.935 | 0.370 | 0.686 | 0.497 | 0.936 | 0.433 | 0.697 | |
| FGWAS | 0.387 | 0.994 | 0.381 | 0.812 | 0.602 | 0.993 | 0.595 | 0.827 | 0.640 | 0.992 | 0.632 | 0.836 | |
| piMASS | 0.116 | 0.999 | 0.115 | 0.831 | 0.128 | 0.999 | 0.127 | 0.835 | 0.141 | 0.999 | 0.140 | 0.847 | |
| GEMMA | 0.218 | 0.989 | 0.207 | 0.659 | 0.240 | 0.990 | 0.230 | 0.664 | 0.257 | 0.987 | 0.244 | 0.666 | |
| SGHS | 0.782 | 0.965 | 0.747 | 0.977 | 0.772 | 0.972 | 0.744 | 0.982 | 0.787 | 0.978 | 0.765 | 0.981 | |
| II | Lasso | 0.443 | 0.904 | 0.347 | 0.744 | 0.469 | 0.931 | 0.400 | 0.765 | 0.465 | 0.932 | 0.397 | 0.755 |
| SCAD | 0.099 | 0.913 | 0.011 | 0.659 | 0.108 | 0.937 | 0.045 | 0.656 | 0.091 | 0.942 | 0.033 | 0.640 | |
| SGL | 0.431 | 0.919 | 0.350 | 0.670 | 0.448 | 0.935 | 0.383 | 0.676 | 0.479 | 0.937 | 0.416 | 0.700 | |
| FGWAS | 0.388 | 0.993 | 0.381 | 0.808 | 0.602 | 0.993 | 0.595 | 0.822 | 0.571 | 0.993 | 0.564 | 0.756 | |
| piMASS | 0.147 | 0.999 | 0.146 | 0.825 | 0.168 | 0.998 | 0.166 | 0.840 | 0.165 | 0.999 | 0.164 | 0.831 | |
| GEMMA | 0.214 | 0.986 | 0.200 | 0.628 | 0.264 | 0.986 | 0.250 | 0.656 | 0.273 | 0.989 | 0.262 | 0.656 | |
| SGHS | 0.878 | 0.951 | 0.829 | 0.952 | 0.846 | 0.935 | 0.781 | 0.972 | 0.836 | 0.960 | 0.796 | 0.981 | |
Sens, the average sensitivity; Spec, the average specificity; J Stat, the average Youden’s J statistic; and AUC, the average area under the receiver operating characteristic curve.
Table 3. Simulation results: feature selection performance with .
| Setting | Method | Sens | Spec | J Stat | AUC | Sens | Spec | J Stat | AUC | Sens | Spec | J Stat | AUC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Lasso | 0.469 | 0.997 | 0.466 | 0.816 | 0.45 | 0.997 | 0.447 | 0.793 | 0.466 | 0.997 | 0.463 | 0.797 |
| SCAD | 0.155 | 0.997 | 0.152 | 0.738 | 0.161 | 0.998 | 0.159 | 0.712 | 0.147 | 0.998 | 0.145 | 0.709 | |
| SGL | 0.738 | 0.993 | 0.731 | 0.853 | 0.674 | 0.995 | 0.669 | 0.821 | 0.670 | 0.997 | 0.667 | 0.832 | |
| FGWAS | 0.458 | 0.998 | 0.456 | 0.524 | 0.516 | 0.998 | 0.514 | 0.718 | 0.549 | 0.998 | 0.547 | 0.801 | |
| piMASS | 0.084 | 0.999 | 0.083 | 0.824 | 0.108 | 0.999 | 0.107 | 0.825 | 0.108 | 0.999 | 0.107 | 0.825 | |
| GEMMA | 0.072 | 0.999 | 0.071 | 0.552 | 0.094 | 0.999 | 0.093 | 0.564 | 0.098 | 0.999 | 0.097 | 0.579 | |
| SGHS | 0.858 | 0.995 | 0.853 | 0.993 | 0.886 | 0.996 | 0.882 | 0.998 | 0.904 | 0.996 | 0.900 | 0.999 | |
| 2 | Lasso | 0.546 | 0.996 | 0.542 | 0.835 | 0.552 | 0.996 | 0.548 | 0.827 | 0.553 | 0.996 | 0.549 | 0.829 |
| SCAD | 0.175 | 0.998 | 0.173 | 0.738 | 0.148 | 0.999 | 0.147 | 0.712 | 0.154 | 0.999 | 0.153 | 0.706 | |
| SGL | 0.732 | 0.996 | 0.728 | 0.857 | 0.676 | 0.997 | 0.673 | 0.821 | 0.684 | 0.997 | 0.681 | 0.834 | |
| FGWAS | 0.485 | 0.998 | 0.483 | 0.649 | 0.512 | 0.998 | 0.510 | 0.786 | 0.564 | 0.998 | 0.562 | 0.799 | |
| piMASS | 0.138 | 0.999 | 0.137 | 0.833 | 0.168 | 0.999 | 0.167 | 0.843 | 0.168 | 0.999 | 0.167 | 0.843 | |
| GEMMA | 0.128 | 0.999 | 0.127 | 0.586 | 0.152 | 0.999 | 0.151 | 0.615 | 0.182 | 0.999 | 0.181 | 0.633 | |
| SGHS | 0.945 | 0.983 | 0.928 | 0.996 | 0.941 | 0.985 | 0.926 | 0.997 | 0.971 | 0.977 | 0.948 | 0.999 | |
| 3 | Lasso | 0.528 | 0.996 | 0.524 | 0.811 | 0.508 | 0.997 | 0.505 | 0.807 | 0.510 | 0.996 | 0.506 | 0.799 |
| SCAD | 0.175 | 0.999 | 0.174 | 0.745 | 0.153 | 0.999 | 0.152 | 0.699 | 0.151 | 0.999 | 0.150 | 0.689 | |
| SGL | 0.696 | 0.996 | 0.692 | 0.839 | 0.658 | 0.997 | 0.655 | 0.798 | 0.674 | 0.997 | 0.671 | 0.829 | |
| FGWAS | 0.446 | 0.998 | 0.444 | 0.592 | 0.520 | 0.998 | 0.518 | 0.657 | 0.532 | 0.998 | 0.530 | 0.796 | |
| piMASS | 0.148 | 0.999 | 0.147 | 0.852 | 0.170 | 0.999 | 0.169 | 0.847 | 0.170 | 0.999 | 0.169 | 0.847 | |
| GEMMA | 0.106 | 0.999 | 0.105 | 0.569 | 0.180 | 0.999 | 0.179 | 0.612 | 0.146 | 0.999 | 0.145 | 0.595 | |
| SGHS | 0.924 | 0.982 | 0.906 | 0.996 | 0.929 | 0.979 | 0.908 | 0.991 | 0.969 | 0.971 | 0.940 | 0.995 | |
| 4 | Lasso | 0.606 | 0.995 | 0.601 | 0.862 | 0.596 | 0.996 | 0.592 | 0.845 | 0.596 | 0.996 | 0.592 | 0.839 |
| SCAD | 0.165 | 0.998 | 0.163 | 0.729 | 0.157 | 0.999 | 0.156 | 0.715 | 0.149 | 0.999 | 0.148 | 0.708 | |
| SGL | 0.728 | 0.996 | 0.724 | 0.853 | 0.664 | 0.997 | 0.661 | 0.813 | 0.686 | 0.997 | 0.683 | 0.836 | |
| FGWAS | 0.468 | 0.998 | 0.466 | 0.580 | 0.510 | 0.998 | 0.508 | 0.787 | 0.564 | 0.998 | 0.562 | 0.809 | |
| piMASS | 0.186 | 0.999 | 0.185 | 0.844 | 0.242 | 0.999 | 0.241 | 0.849 | 0.242 | 0.999 | 0.241 | 0.849 | |
| GEMMA | 0.126 | 0.999 | 0.125 | 0.588 | 0.196 | 0.999 | 0.195 | 0.642 | 0.196 | 0.999 | 0.195 | 0.626 | |
| SGHS | 0.955 | 0.950 | 0.905 | 0.988 | 0.967 | 0.949 | 0.916 | 0.990 | 0.978 | 0.954 | 0.932 | 0.996 | |
| 5 | Lasso | 0.353 | 0.997 | 0.350 | 0.505 | 0.337 | 0.997 | 0.334 | 0.501 | 0.354 | 0.997 | 0.351 | 0.494 |
| SCAD | 0.182 | 0.996 | 0.178 | 0.497 | 0.165 | 0.997 | 0.162 | 0.503 | 0.167 | 0.998 | 0.165 | 0.484 | |
| SGL | 0.578 | 0.991 | 0.569 | 0.488 | 0.490 | 0.987 | 0.477 | 0.446 | 0.464 | 0.986 | 0.450 | 0.551 | |
| FGWAS | 0.372 | 0.998 | 0.370 | 0.307 | 0.442 | 0.998 | 0.440 | 0.477 | 0.465 | 0.999 | 0.464 | 0.409 | |
| piMASS | 0.146 | 0.999 | 0.145 | 0.772 | 0.136 | 0.999 | 0.135 | 0.791 | 0.136 | 0.999 | 0.135 | 0.791 | |
| GEMMA | 0.104 | 0.999 | 0.103 | 0.562 | 0.120 | 0.999 | 0.119 | 0.574 | 0.116 | 0.999 | 0.115 | 0.574 | |
| SGHS | 0.875 | 0.994 | 0.869 | 0.998 | 0.866 | 0.997 | 0.863 | 0.994 | 0.908 | 0.995 | 0.903 | 0.998 | |
| 6 | Lasso | 0.345 | 0.997 | 0.342 | 0.484 | 0.320 | 0.998 | 0.318 | 0.513 | 0.318 | 0.998 | 0.316 | 0.506 |
| SCAD | 0.157 | 0.997 | 0.154 | 0.494 | 0.139 | 0.997 | 0.136 | 0.501 | 0.150 | 0.997 | 0.147 | 0.506 | |
| SGL | 0.578 | 0.987 | 0.565 | 0.486 | 0.566 | 0.993 | 0.559 | 0.542 | 0.504 | 0.992 | 0.496 | 0.493 | |
| FGWAS | 0.388 | 0.998 | 0.386 | 0.397 | 0.405 | 0.998 | 0.403 | 0.552 | 0.456 | 0.998 | 0.454 | 0.472 | |
| piMASS | 0.064 | 0.999 | 0.063 | 0.807 | 0.108 | 0.999 | 0.107 | 0.820 | 0.108 | 0.999 | 0.107 | 0.820 | |
| GEMMA | 0.068 | 0.999 | 0.067 | 0.543 | 0.098 | 0.999 | 0.097 | 0.571 | 0.090 | 0.999 | 0.089 | 0.554 | |
| SGHS | 0.737 | 0.997 | 0.734 | 0.998 | 0.757 | 0.997 | 0.754 | 0.983 | 0.800 | 0.997 | 0.797 | 0.987 | |
| I | Lasso | 0.436 | 0.994 | 0.430 | 0.732 | 0.444 | 0.996 | 0.440 | 0.783 | 0.500 | 0.996 | 0.496 | 0.802 |
| SCAD | 0.087 | 0.996 | 0.083 | 0.638 | 0.079 | 0.997 | 0.076 | 0.685 | 0.086 | 0.997 | 0.083 | 0.700 | |
| SGL | 0.414 | 0.996 | 0.410 | 0.693 | 0.414 | 0.997 | 0.411 | 0.692 | 0.426 | 0.997 | 0.423 | 0.674 | |
| FGWAS | 0.500 | 0.999 | 0.499 | 0.772 | 0.511 | 0.999 | 0.510 | 0.683 | 0.606 | 0.999 | 0.605 | 0.809 | |
| piMASS | 0.073 | 0.999 | 0.072 | 0.863 | 0.089 | 0.999 | 0.088 | 0.863 | 0.089 | 0.999 | 0.088 | 0.863 | |
| GEMMA | 0.087 | 0.999 | 0.086 | 0.562 | 0.080 | 0.999 | 0.079 | 0.556 | 0.131 | 0.999 | 0.130 | 0.588 | |
| SGHS | 0.943 | 0.951 | 0.894 | 0.990 | 0.934 | 0.955 | 0.889 | 0.984 | 0.954 | 0.958 | 0.912 | 0.991 | |
| II | Lasso | 0.472 | 0.994 | 0.466 | 0.751 | 0.475 | 0.996 | 0.471 | 0.784 | 0.510 | 0.997 | 0.507 | 0.818 |
| SCAD | 0.084 | 0.996 | 0.080 | 0.654 | 0.070 | 0.997 | 0.067 | 0.671 | 0.076 | 0.997 | 0.073 | 0.692 | |
| SGL | 0.416 | 0.996 | 0.412 | 0.690 | 0.428 | 0.997 | 0.425 | 0.688 | 0.488 | 0.997 | 0.485 | 0.691 | |
| FGWAS | 0.465 | 0.999 | 0.464 | 0.566 | 0.532 | 0.999 | 0.531 | 0.785 | 0.609 | 0.999 | 0.608 | 0.718 | |
| piMASS | 0.080 | 0.999 | 0.079 | 0.857 | 0.066 | 0.999 | 0.065 | 0.863 | 0.066 | 0.999 | 0.065 | 0.863 | |
| GEMMA | 0.073 | 0.999 | 0.072 | 0.554 | 0.082 | 0.999 | 0.081 | 0.565 | 0.113 | 0.999 | 0.112 | 0.578 | |
| SGHS | 0.937 | 0.934 | 0.871 | 0.985 | 0.953 | 0.944 | 0.897 | 0.990 | 0.960 | 0.953 | 0.913 | 0.992 | |
Sens, the average sensitivity; Spec, the average specificity; J Stat, the average Youden’s J statistic; and AUC, the average area under the receiver operating characteristic curve.
When comparing among the competing methods, we find the results are somewhat mixed across different settings. Specifically, SGL is the one we originally expected to outperform the rest of the competing methods as it also works under a hierarchical selection framework. Although SGL obtains higher sensitivities than the other competing methods in most of the scenarios, a lower specificity due to large number of false positives deteriorates the overall performance, especially in extreme situations. The two Bayesian methods piMASS and GEMMA achieve similar feature selection performance. Different from SGL, they lack power to detect true signals with very low sensitivities but high specificities. The performances of LASSO, SCAD, and FGWAS fluctuate in between, and none of them generally outperform the others.
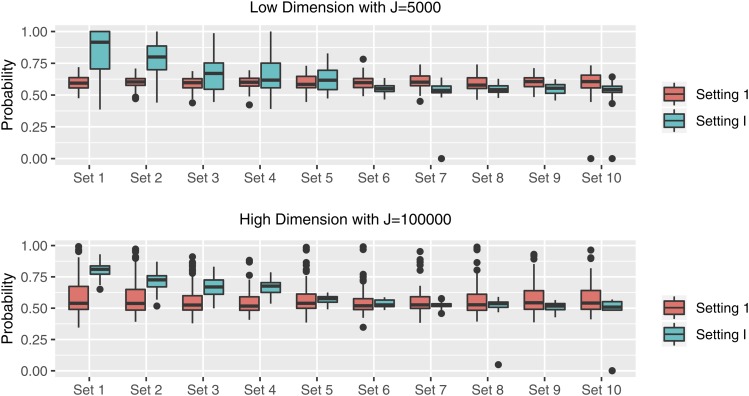
In terms of comparison among different settings, dimensions, and LD structures, as illustrated previously, methods generally work better to detect unified or clustered signal patterns but may encounter difficulty when both positive and negative effects exist. We further calculate the average marginal posterior probabilities for the risk SNPs in causal SNP-sets obtained from SGHS under both cases with . Figure 1 shows the boxplots representing these posterior probabilities for Setting 1 of Case 1 and Setting I of Case 2 under different dimensions. As shown in Figure 1, under Case 1, the possibilities to identify risk SNPs in each SNP-set are comparable. Under Case 2, where relatively sparse signals exist in the first five SNP-sets, we notice a higher probability in identifying signals within these SNP-sets. However, when it comes to SNP-sets 6–10 with diluted signals, the majority of genetic effect is lost during the selection. Finally, SGHS achieves an equally good, or even better, performance under high-dimensional scenarios compared with lower dimensions, and the impact of κ is not strong in our simulation settings.
Figure 1.
Simulation results: the average marginal posterior probabilities of risk SNPs over each risk SNP-set under Setting 1 and Setting I with different dimensions.
The Alzheimer’s Disease neuroimaging initiative
There has been substantial interest in investigating neurodegenerative diseases such as Alzheimer’s disease (AD) based on neuroimaging and genetic markers. Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies, and nonprofit organizations, as a $60 million, 5-year public–private partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California, San Francisco. ADNI is the result of efforts of many coinvestigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the US and Canada. The initial goal of ADNI was to recruit 800 subjects but ADNI has been followed by ADNI-GO and ADNI-2. To date, these three protocols have recruited over 1500 adults, aged 55–90 years, to participate in the research, consisting of cognitively normal older individuals, people with early or late MCI, and people with early AD. The follow-up duration of each group is specified in the protocols of ADNI-1, ADNI-2, and ADNI-GO. Subjects originally recruited by ADNI-1 and ADNI-GO had the option to be followed in ADNI-2. For up-to-date information, see www.adni-info.org.
We conduct GWAS analysis on imaging phenotypes related to AD, and our goal is to identify genetic markers that are associated with imaging traits, and to further assess their predictive power. The advantage of using imaging phenotypes in GWAS is that imaging measurements tend to benefit the identification of pathogenic genes due to their close relationship with the biological etiology of multiple neurodegenerative and neuropsychiatric diseases, e.g., AD (Cannon and Keller 2006; Turner et al. 2006; Peper et al. 2007; Paus 2010; Scharinger et al. 2010; Chiang et al. 2011a,b). For imaging traits, the raw MRI data were collected through 1.5 Tesla MRI scanners with protocols individualized for each scanner, including standard T1-weighted images obtained using volumetric three-dimensional (3D) sagittal MPRAGE or equivalent protocols with varying resolutions. The T1-weighted MRI images were preprocessed by standard steps including anterior commissure and posterior commissure correction, skull-stripping, cerebellum removing, intensity inhomogeneity correction, segmentation, and registration (Shen and Davatzikos 2004). Subsequently, 93 ROIs were labeled automatically by labeling the template and transferring the labels following the deformable registration of subject images (Wang et al. 2011). After calculating the volume of each ROI for each subject, we consider nine of them as phenotypes: six subcortical regions, including left and right hippocampal volumes, left and right lateral ventricular volumes, and left and right amygdala volumes; and three global volumetric measures, including whole gray matter volume, whole white matter volume, and whole brain volume.
A total of 818 subjects were genotyped using the Human 610-Quad BeadChip (Illumina, San Diego, CA). For data quality control, we focused on the 760 Caucasian subjects and removed ones identified as (i) sex check failure, (ii) >10% missing SNP, and (iii) outliers in the phenotypes/genotypes stratification, resulting in 745 subjects. The original SNPs data were generated from the Human Genome reference sequence build hg18, which were lifted over to hg19 in the current analysis. As a typical step in GWAS, we removed SNPs with (i) >5% missing values, (ii) minor allele frequency smaller than 5%, and (iii) Hardy-Weinberg equilibrium P-value . We also calculated the LD blocks to form the SNP-sets, and removed SNP-sets with single SNP. Eventually, 421,823 SNPs are left in our analysis, grouped into 16,084 SNP-sets with the number of SNPs varying from 2 to 100. We also include gender, age, and the first five principle component calculated by EIGENSOFT (Price et al. 2006) into the analysis. We adopt the SGHS approach to investigate the joint association of SNPs with each of the nine MRI phenotypes in light of the autosomal LD blocks information. All the posterior simulation and hyper-parameters settings follow a similar line as the simulation studies.
We first list all the selected SNP-sets associated with the nine imaging phenotypes along with the numbers of total SNPs and selected SNPs belonging to each set in Table 4. Based on the number of selected SNPs (columns 6 and 11), we observe the phenomenon that none of SNPs is selected within certain risk SNP-sets (the 0s in columns 6 and 11). This demonstrates the fact that some genetic information is diluted as it is widely studied in GWAS.
Table 4. ADNI data analysis results: list of selected SNP-sets associated with the phenotypes with the total number of SNPs and number of selected SNPs.
| ROIs | Chr | Begin BP | End BP | Total #a | Selected #b | Chr | Begin BP | End BP | Total # | Selected # |
|---|---|---|---|---|---|---|---|---|---|---|
| LA | 3 | 41021263 | 41272081 | 31 | 0 | 9 | 121993508 | 122159267 | 37 | 8 |
| 4 | 20956632 | 20827274 | 44 | 0 | 11 | 50057854 | 55275456 | 99 | 0 | |
| 8 | 55302231 | 55441025 | 32 | 0 | 12 | 21970019 | 22242951 | 86 | 21 | |
| 8 | 84373550 | 84826056 | 59 | 0 | 20 | 12888105 | 12982672 | 29 | 0 | |
| RA | 2 | 81058605 | 81607450 | 51 | 0 | 8 | 53851670 | 54119394 | 50 | 2 |
| 6 | 89741617 | 89985379 | 72 | 4 | 9 | 20625875 | 21101230 | 99 | 0 | |
| 7 | 104491613 | 105158228 | 57 | 5 | 14 | 73223110 | 73425315 | 47 | 0 | |
| 7 | 150167583 | 150491084 | 85 | 11 | 15 | 58109235 | 349838 | 97 | 6 | |
| 8 | 13013625 | 13253219 | 99 | 7 | 17 | 18879649 | 19697976 | 75 | 3 | |
| 8 | 32221412 | 32465554 | 66 | 0 | ||||||
| LH | 2 | 38140126 | 38328300 | 44 | 0 | 6 | 118538069 | 119102035 | 99 | 13 |
| 4 | 89677537 | 90116432 | 85 | 0 | 10 | 84680499 | 85193946 | 90 | 0 | |
| 4 | 89677537 | 90116432 | 85 | 0 | 11 | 85908537 | 86220724 | 77 | 0 | |
| 5 | 106230898 | 106426493 | 25 | 0 | 16 | 12625171 | 12776281 | 99 | 0 | |
| 6 | 38712688 | 38712688 | 57 | 0 | ||||||
| RH | 2 | 333497170 | 33623720 | 32 | 0 | 3 | 177565144 | 177970694 | 38 | 0 |
| 2 | 212224689 | 212385723 | 48 | 0 | 4 | 186063341 | 186375488 | 42 | 0 | |
| 3 | 85591467 | 86298087 | 99 | 24 | 8 | 5781984 | 5968366 | 72 | 0 | |
| 3 | 146758405 | 147093600 | 41 | 4 | 8 | 86886950 | 87362706 | 73 | 0 | |
| LL | 5 | 178303311 | 178436190 | 31 | 0 | 11 | 52097415 | 52595115 | 79 | 24 |
| 6 | 146559200 | 146971848 | 51 | 0 | 12 | 9042343 | 9362931 | 66 | 0 | |
| 7 | 15157688 | 15413574 | 50 | 11 | 15 | 46876803 | 47310628 | 74 | 0 | |
| RL | 1 | 173854659 | 175094025 | 99 | 0 | 11 | 46198841 | 47293457 | 80 | 0 |
| 5 | 151739218 | 152417867 | 93 | 0 | 16 | 10019899 | 10292060 | 99 | 42 | |
| 6 | 32304085 | 32395036 | 99 | 35 | ||||||
| GM | 2 | 105454590 | 105798292 | 55 | 7 | 6 | 81868051 | 82061044 | 47 | 21 |
| 6 | 169471078 | 169589925 | 40 | 0 | ||||||
| WM | 2 | 46537604 | 46763587 | 70 | 0 | 8 | 98930457 | 99109800 | 48 | 19 |
| 6 | 31434111 | 31518354 | 99 | 0 | 12 | 106625131 | 106950695 | 51 | 0 | |
| 8 | 4766370 | 4893353 | 52 | 0 | 20 | 35487159 | 35925296 | 33 | 0 | |
| WB | 6 | 167287772 | 167537594 | 50 | 17 | 9 | 4767677 | 4904969 | 39 | 0 |
LA/RA, left/right amygdala volumes; LH/RH, left/right hippocampal volumes; LL/RL, left/right lateral ventricle volumes; GM, gray matter volume; WM, white matter volume; WB, whole brain volume.
Total number of SNPs in the SNP-set.
Number of selected SNPs in the SNP-set.
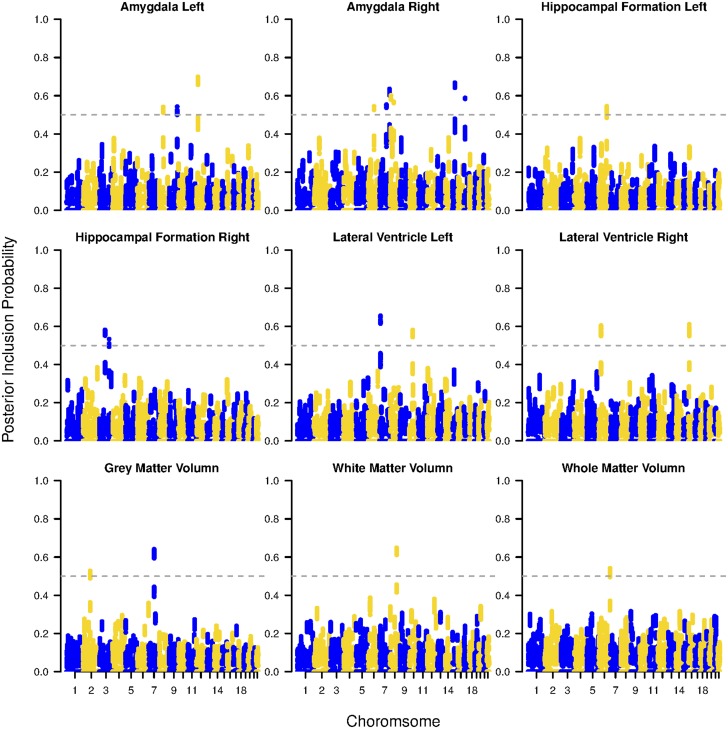
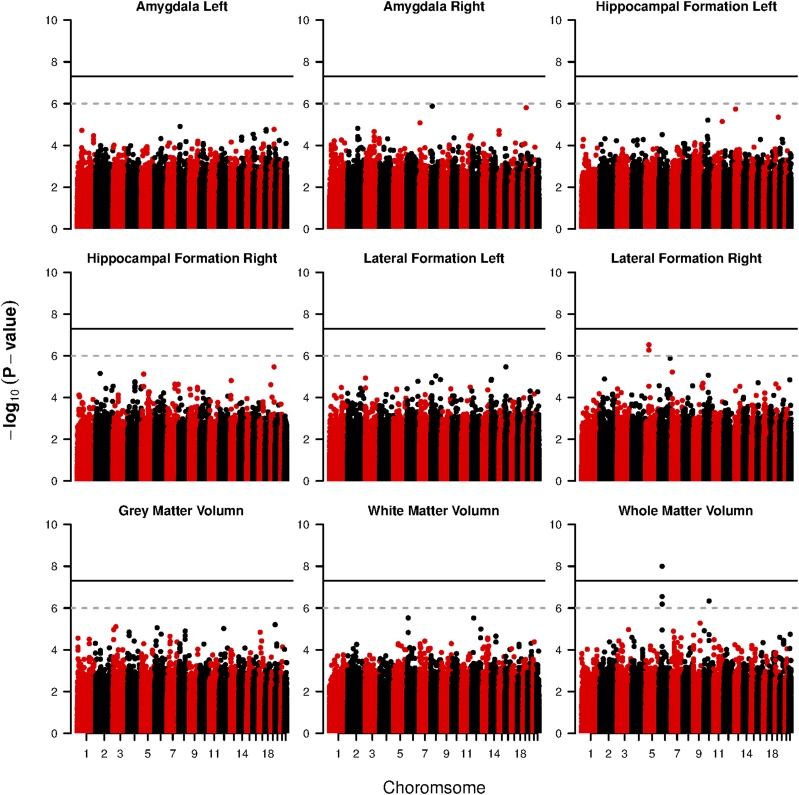
We further consider the final SNP-level selection. The Manhattan plots in Figure 2 provide the SNP-wise inclusion probability with respective to each imaging phenotype. Under a 0.5 cutoff, for each phenotype, we map the selected SNPs to their associated genes, and summarize these risk genes along with the number of total/selected SNPs in Table 5. A further comparison between the number of selected SNPs and the total number of SNPs belonging to the risk gene (columns 3 and 4) demonstrates that our method is capable of identifying both sparse and diluted genetic information. Among the selected genes, a number of them have been reported previously in the literature. Such genes include ASAH2B (Avramopoulos et al. 2007), SGMS1 (Hsiao et al. 2013), GRIN2A (Leuba et al. 2014), Tmem176b (Melchior et al. 2010). In addition, several other genes have been shown to be related to the brain dysfunction or implicitly associated with Alzheimer’s disease. For instance, BRINP1 has been shown to highly express in various brain regions, and a lack of BRINP1 may lead to human psychiatric disorders (Kobayashi et al. 2014). CCR6 has been implicated as an important biomarker associated with the inflammatory process of AD-like diseases (Subramanian et al. 2010). RNASET2 deficiency interferes with brain development and myelination (Henneke et al. 2009). Genes like CADM2, DLC1, and ABCC9 are related to Autism spectrum disorder (ASD) or Parkinson’s disease (Casey et al. 2012; Jones et al. 2013; Lin et al. 2014), which may also serve as potential biomarkers for AD. Based on the selected genes, we also conduct a gene annotation analysis based on the enrichment for Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto 2000). The associated pathways are calcium signaling pathway, which is a key component to regulate the neuronal excitability and processes related to the development neural diseases such as AD (Berridge 2013), and neuroactive ligand-receptor interaction, which is a well-known biomarker for cognitive ability (Antonell et al. 2013; Kong et al. 2015). As a comparison, we also perform GWAS based on single SNP analysis via PLINK (Purcell et al. 2007) by performing quantitative trait association, and provide the Manhattan plots for value under each imaging phenotype in Figure 3. As a result, there are considerably less risk SNPs [associated with the human collagen alpha 1 (XIII) chain gene COL13A1] identified under both the well accepted threshold and the threshold suggested by Li et al. (2012) compared with the result obtained by SGHS.
Figure 2.
ADNI data analysis results: Manhattan plots for inclusion probabilities of all SNPs in all autosomes based on SGHS.
Table 5. ADNI data analysis results: list of selected genes associated with the phenotypes with the total number of SNPs and the number of selected SNPs.
| ROIs | Gene | Total Number of SNPs | Number of Selected SNPs | Chromosome |
|---|---|---|---|---|
| Amygdala left | BRINP1 | 45 | 6 | 9 |
| ABCC9 | 68 | 16 | 12 | |
| CMAS | 6 | 1 | 12 | |
| Amygdala right | ALDH1A2 | 91 | 4 | 15 |
| DLC1 | 139 | 7 | 8 | |
| GIMAP4 | 5 | 2 | 7 | |
| GIMAP7 | 4 | 3 | 7 | |
| KMT2E | 5 | 2 | 7 | |
| LHFPL3 | 131 | 1 | 7 | |
| TMEM176B | 5 | 1 | 7 | |
| GABRR1 | 25 | 1 | 6 | |
| Hippocampal formation left | CEP85L | 33 | 4 | 6 |
| SLC35F1 | 103 | 1 | 6 | |
| PLN | 1 | 1 | 6 | |
| Hippocampal formation right | CADM2 | 116 | 12 | 3 |
| Lateral ventricle left | A1CF | 14 | 1 | 10 |
| ASAH2B | 2 | 1 | 10 | |
| SGMS1 | 67 | 17 | 10 | |
| AGMO | 65 | 6 | 7 | |
| Lateral ventricle right | BTNL2 | 18 | 12 | 6 |
| C6orf10 | 109 | 14 | 6 | |
| GRIN2A | 123 | 40 | 16 | |
| Gray matter volume | CACNA2D1 | 122 | 21 | 7 |
| MRPS9 | 8 | 4 | 2 | |
| White matter volume | ERICH5 | 5 | 1 | 8 |
| MATN2 | 44 | 16 | 8 | |
| Whole matter volume | CCR6 | 9 | 2 | 6 |
| FGFR1OP | 8 | 2 | 6 | |
| RNASET2 | 3 | 3 | 6 |
Figure 3.
ADNI data analysis results: Manhattan plots of P-values of SNPs in all autosomes based on single SNP analysis.
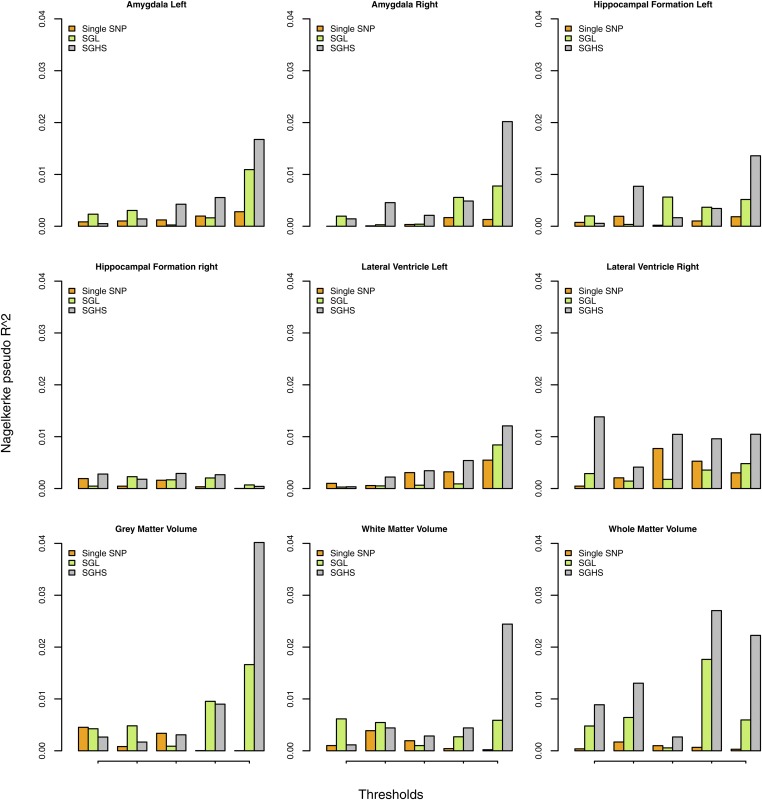
Finally, we assess the capacity of the selected genetic markers to predict imaging phenotypes using polygenic score (The International Schizophrenia Consortium 2009). Besides single SNP analysis and SGHS, we also apply SGL as another competing method. We use twofold cross validation by randomly splitting the dataset into equally sized ones, and perform the analyses on each as the training one. Under the corresponding testing sets, the Nagelkerke pseudo is calculated first for single SNP analysis based on the selected SNPs under different thresholds of P-values, and the grid of selected SNP numbers is further used as the thresholds for SGL and SGHS to obtain their scores. The final is averaged over two testing sets and we repeat the procedure five times to remove splitting bias. We present the results for all autosomes in Figure 4, which clearly shows a dramatic improvement of prediction by using SGHS compared with single SNP analysis and SGL in almost all the autosomes and thresholds. The predictive power for the selected risk profiles varies across different imaging phenotypes, and we also see a general pattern of an increase number of selected SNPs leading to a higher polygenic score.
Figure 4.
ADNI data analysis results: Polygenic score under different thresholds in all autosomes based on single SNP analysis, SGL and SGHS.
Discussion
In this paper, we develop a unified Bayesian framework to realize hierarchical variable selection, while inducing grouping effect among predictors. Motivated by GWAS, our proposed method incorporates SNP-set information into the variable selection procedure, and facilitates selection at both SNP-set level and SNP level. Furthermore, by introducing a novel sampling scheme based on an auxiliary model for group-level selection, our approach is computationally efficient under high-dimensional feature space. We show in the simulation studies that the proposed method achieves considerably better performance than a number of competing methods under a wide range of settings. By applying the proposed method to the ADNI data set, we identify important genetic information that is highly associated with the volumes of ROIs in the brain.
While our method is applied to an imaging-genetics study with a quantitative trait as phenotype, it is directly applicable to a dichotomous variable (e.g., case or control). As discussed in Albert and Chib (1993), one could use a probit regression model for the binary outcome, which leads to few modifications on the current posterior sampling scheme. In addition, we can also consider an incorporation of more biological information in the selection procedure. For instance, it is interesting to conduct Bayesian variable selection by incorporating the information on pathways and gene networks in microarray data (Li et al. 2010; Stingo et al. 2011) or functional connectivity for neuroimaging studies (Huang et al. 2013; Goldsmith et al. 2014). Similarly, we may introduce Ising or binary Markov random field (MRF) priors to the two levels of selection indicators in order to incorporate hierarchical biological information.
After a realization of whole genome-wide association analysis, one extension of our work is to move forward to the whole-brain and whole GWAS. In this case, we need to use a multiple multivariate regression model to further capture the association among phenotypes (Zhu et al. 2014). Besides the potential low detection power, the prohibitive computational cost will be the biggest issue of such analysis. A different direction is to extend the current model to the longitudinal data, which will increase the power to detect genetic association with neuroimaging phenotypes (Xu et al. 2014). In this case, we need to modify our method to model the temporal association between responses and predictors while accounting for complex temporal correction structure.
Appendix A
Standard MCMC Algorithm
Below is the standard MCMC algorithm for posterior computation of model (2).Sampling scheme for α. Draw
| (18) |
where and .Sampling scheme for η. Draw
| (19) |
Sampling scheme for . Draw
| (20) |
Sampling scheme for . Draw
| (21) |
Sampling scheme for . The full conditional of is
| (22) |
Draw (the coefficients corresponding to the selected predictors) and (the coefficients corresponding to the unselected predictors) separately from
| (23) |
where , and includes the columns of corresponding to the important voxels defined by and .Sampling scheme for . For , the full conditional of is given by
| (24) |
with .Sampling scheme for . For and , the full conditional of is given by
with .
Appendix B
SGHS Algorithm
Parameters in the auxiliary model
Sampling scheme for . Draw
| (26) |
where and .Sampling scheme for . Draw
| (27) |
Sampling scheme for . Draw
| (28) |
Sampling scheme for . Draw
| (29) |
Sampling scheme for The full conditional of is
| (30) |
Draw (the coefficients corresponding to the selected predictors) and (the coefficients corresponding to the unselected predictors) separately from
| (31) |
where and and includes the columns of corresponding to the selected entries defined by .Sampling scheme for For , the full conditional of is given by
| (32) |
with .
Parameters in the main model
The updating scheme for , , , and η follows the Standard MCMC Algorithm in Appendix A1.Sampling scheme for and M-H Step For ,• Draw ;• Draw ;• Draw . Set when with R defined by Equation 17.Moving Step For , with , , draw with the full conditional distribution defined by (32).
Footnotes
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/themes/freshnews-dev-v2/documents/policy/ADNI_Acknowledgement_List%205-29-18.pdf
Communicating editor: S. Mikko
Literature Cited
- Albert J., Chib S., 1993. Bayesian analysis of binary and polychotomous response data. J. Am. Stat. Assoc. 88: 669–679. 10.1080/01621459.1993.10476321 [DOI] [Google Scholar]
- Altshuler D., Daly M. J., Lander E. S., 2008. Genetic mapping in human disease. Science 322: 881–888. 10.1126/science.1156409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonell A., Lladó A., Altirriba J., Botta-Orfila T., Balasa M., et al. , 2013. A preliminary study of the whole-genome expression profile of sporadic and monogenic early-onset Alzheimer’s disease. Neurobiol. Aging 34: 1772–1778. 10.1016/j.neurobiolaging.2012.12.026 [DOI] [PubMed] [Google Scholar]
- Avramopoulos D., Wang R., Valle D., Fallin M. D., Bassett S. S., 2007. A novel gene derived from a segmental duplication shows perturbed expression in Alzheimer’s disease. Neurogenetics 8: 111–120. [DOI] [PubMed] [Google Scholar]
- Bao M., Wang K., 2017. Genome-wide association studies using a penalized moving-window regression. Bioinformatics 33: 3887–3894. 10.1093/bioinformatics/btx522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri M. M., Berger J. O., 2004. Optimal predictive model selection. Ann. Stat. 32: 870–897. 10.1214/009053604000000238 [DOI] [Google Scholar]
- Berridge M. J., 2013. Dysregulation of neural calcium signaling in Alzheimer disease, bipolar disorder and schizophrenia. Prion 7: 2–13. 10.4161/pri.21767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottolo L., Richardson S., 2010. Evolutionary stochastic search for Bayesian model exploration. Bayesian Anal. 5: 583–618. 10.1214/10-BA523 [DOI] [Google Scholar]
- Bottolo L., Chadeau-Hyam M., Hastie D. I., Zeller T., Liquet B., et al. , 2013. GUESS-ing polygenic associations with multiple phenotypes using a GPU-based evolutionary stochastic search algorithm. PLoS Genet. 9: e1003657 10.1371/journal.pgen.1003657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briollais L., Dobra A., Liu J., Friedlander M., Ozcelik H., et al. , 2016. A Bayesian graphical model for genome-wide association studies (GWAS). Ann. Appl. Stat. 10: 786–811. 10.1214/16-AOAS909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon T. D., Keller M. C., 2006. Endophenotypes in the genetic analyses of mental disorders. Annu. Rev. Clin. Psychol. 2: 267–290. 10.1146/annurev.clinpsy.2.022305.095232 [DOI] [PubMed] [Google Scholar]
- Carbonetto P., Stephens M., 2012. Scalable variational inference for Bayesian variable selection in regression, and its accuracy in genetic association studies. Bayesian Anal. 7: 73–108. 10.1214/12-BA703 [DOI] [Google Scholar]
- Casey J. P., Magalhaes T., Conroy J. M., Regan R., Shah N., et al. , 2012. A novel approach of homozygous haplotype sharing identifies candidate genes in autism spectrum disorder. Hum. Genet. 131: 565–579. 10.1007/s00439-011-1094-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang M.-C., Barysheva M., Toga A. W., Medland S. E., Hansell N. K., et al. , 2011a BDNF gene effects on brain circuitry replicated in 455 twins. Neuroimage 55: 448–454. 10.1016/j.neuroimage.2010.12.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang M.-C., McMahon K. L., de Zubicaray G. I., Martin N. G., Hickie I., et al. , 2011b Genetics of white matter development: a DTI study of 705 twins and their siblings aged 12 to 29. Neuroimage 54: 2308–2317. 10.1016/j.neuroimage.2010.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S., Kim K., Kim Y. J., Lee J.-K., Cho Y. S., et al. , 2010. Joint identification of multiple genetic variants via elastic-net variable selection in a genome-wide association analysis. Ann. Hum. Genet. 74: 416–428. 10.1111/j.1469-1809.2010.00597.x [DOI] [PubMed] [Google Scholar]
- Dashab G. R., Kadri N. K., Shariati M. M., Sahana G., 2012. Comparison of linear mixed model analysis and genealogy-based haplotype clustering with a Bayesian approach for association mapping in a pedigreed population. BMC Proc. 6: S4 10.1186/1753-6561-6-S2-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaportas P., Forster J. J., Ntzoufras I., 2002. On Bayesian model and variable selection using MCMC. Stat. Comput. 12: 27–36. 10.1023/A:1013164120801 [DOI] [Google Scholar]
- Duan L., Thomas D. C., 2013. 2013 A Bayesian hierarchical model for relating multiple SNPs within multiple genes to disease risk. Int. J. Genomics 406217 10.1155/2013/406217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B., Hastie T., Johnstone I., Tibshirani R., 2004. Least angle regression. Ann. Stat. 32: 407–499. 10.1214/009053604000000067 [DOI] [Google Scholar]
- Fan J., Li R., 2001. Variable selection via nonconcave penalized likelihood and its oracle properties. J. Am. Stat. Assoc. 96: 1348–1360. 10.1198/016214501753382273 [DOI] [Google Scholar]
- Fan J., Lv J., 2008. Sure independence screening for ultrahigh dimensional feature space. J. R. Stat. Soc. Series B Stat. Methodol. 70: 849–911. 10.1111/j.1467-9868.2008.00674.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J., Hastie T., Tibshirani R., 2010. A note on the group lasso and a sparse group lasso. arXiv: 1001.0736v1.
- Gelman A., Rubin D. B., 1992. Inference from iterative simulation using multiple sequences. Stat. Sci. 7: 457–472. 10.1214/ss/1177011136 [DOI] [Google Scholar]
- George E., McCulloch R., 1993. Variable selection via Gibbs sampling. J. Am. Stat. Assoc. 88: 881–889. 10.1080/01621459.1993.10476353 [DOI] [Google Scholar]
- Goldsmith J., Huang L., Crainiceanu C. M., 2014. Smooth scalar-on-image regression via spatial Bayesian variable selection. J. Comput. Graph. Stat. 23: 46–64. 10.1080/10618600.2012.743437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Stephens M., 2011. Bayesian variable selection regression for genome-wide association studies and other large-scale problems. Ann. Appl. Stat. 5: 1780–1815. 10.1214/11-AOAS455 [DOI] [Google Scholar]
- He Q., Lin D.-Y., 2011. A variable selection method for genome-wide association studies. Bioinformatics 27: 1–8. 10.1093/bioinformatics/btq600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneke M., Diekmann S., Ohlenbusch A., Kaiser J., Engelbrecht V., et al. , 2009. RNASET2-deficient cystic leukoencephalopathy resembles congenital cytomegalovirus brain infection. Nat. Genet. 41: 773–775. 10.1038/ng.398 [DOI] [PubMed] [Google Scholar]
- Hibar D. P., Stein J. L., Renteria M. E., Arias-Vasquez A., Desrivières S., et al. , 2015. Common genetic variants influence human subcortical brain structures. Nature 520: 224–229. 10.1038/nature14101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggart C. J., Whittaker J. C., De Iorio M., Balding D. J., 2008. Simultaneous analysis of all SNPs in genome-wide and re-sequencing association studies. PLoS Genet. 4: e1000130 10.1371/journal.pgen.1000130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao J.-H. T., Fu Y., Hill A. F., Halliday G. M., Kim W. S., 2013. Elevation in sphingomyelin synthase activity is associated with increases in amyloid-beta peptide generation. PLoS One 8: e74016 10.1371/journal.pone.0074016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Thompson P., Wang Y., Yu Y., Zhang J., et al. , 2017. FGWAS: functional genome wide association analysis. Neuroimage 159: 107–121. 10.1016/j.neuroimage.2017.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Goldsmith J., Reiss P. T., Reich D. S., Crainiceanu C. M., 2013. Bayesian scalar-on-image regression with application to association between intracranial DTI and cognitive outcomes. Neuroimage 83: 210–223. 10.1016/j.neuroimage.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap 3 Consortium; D. M. Altshuler, R. A. Gibbs, L. Peltonen, D. M. Altshuler, R. A. Gibbs et al, 2010 Integrating common and rare genetic variation in diverse human populations. Nature 467: 52–58. 10.1038/nature09298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., He Y., Zhang H., 2016. Variable selection with prior information for generalized linear models via the prior lasso method. J. Am. Stat. Assoc. 111: 355–376. 10.1080/01621459.2015.1008363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson V. E., 2013. On numerical aspects of Bayesian model selection in high and ultrahigh-dimensional settings. Bayesian Anal. 7: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson V. E., Rossell D., 2012. Bayesian model selection in high-dimensional settings. J. Am. Stat. Assoc. 107: 649–660. 10.1080/01621459.2012.682536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. R., Huang A. L., Ptáček L. J., Fu Y.-H., 2013. Genetic basis of human circadian rhythm disorders. Exp. Neurol. 243: 28–33. 10.1016/j.expneurol.2012.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28: 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärkkäinen H. P., Sillanpää M. J., 2012. Robustness of Bayesian multilocus association models to cryptic relatedness. Ann. Hum. Genet. 76: 510–523. 10.1111/j.1469-1809.2012.00729.x [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Nakatani T., Koda T., Matsumoto K., Ozaki R., et al. , 2014. Absence of BRINP1 in mice causes increase of hippocampal neurogenesis and behavioral alterations relevant to human psychiatric disorders. Mol. Brain 7: 12 10.1186/1756-6606-7-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y., Liang X., Liu L., Zhang D., Wan C., et al. , 2015. High throughput sequencing identifies MicroRNAs mediating α-synuclein toxicity by targeting neuroactive-ligand receptor interaction pathway in early stage of drosophila Parkinson’s disease model. PLoS One 10: e0137432 10.1371/journal.pone.0137432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwee L. C., Liu D., Lin X., Ghosh D., Epstein M. P., 2008. A powerful and flexible multilocus association test for quantitative traits. Am. J. Hum. Genet. 82: 386–397. 10.1016/j.ajhg.2007.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuba G., Vernay A., Kraftsik R., Tardif E., Michel Riederer B., et al. , 2014. Pathological reorganization of NMDA receptors subunits and postsynaptic protein PSD-95 distribution in Alzheimer’s disease. Curr. Alzheimer Res. 11: 86–96. 10.2174/15672050113106660170 [DOI] [PubMed] [Google Scholar]
- Li J., Das K., Fu G., Li R., Wu R., 2010. The Bayesian lasso for genome-wide association studies. Bioinformatics 27: 516–523. 10.1093/bioinformatics/btq688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Yeung J. M., Cherny S. S., Sham P. C., 2012. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet. 131: 747–756. 10.1007/s00439-011-1118-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B., Wang Y., Wang Z., Tan H., Kong X., et al. , 2014. Uncovering the rare variants of DLC1 isoform 1 and their functional effects in a Chinese sporadic congenital heart disease cohort. PLoS One 9: e90215 10.1371/journal.pone.0090215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liquet B., Mengersen K., Pettitt A., Sutton M., 2017. Bayesian variable selection regression of multivariate responses for group data. Bayesian Anal. 12: 1039–1067. 10.1214/17-BA1081 [DOI] [Google Scholar]
- Lu Z.-H., Zhu H., Knickmeyer R. C., Sullivan P. F., Williams S. N., et al. , 2015. Multiple SNP set analysis for genome-wide association studies through Bayesian latent variable selection. Genet. Epidemiol. 39: 664–677. 10.1002/gepi.21932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior B., Garcia A. E., Hsiung B.-K., Lo K. M., Doose J. M., et al. , 2010. Dual induction of TREM2 and tolerance-related transcript, Tmem176b, in amyloid transgenic mice: implications for vaccine-based therapies for Alzheimer’s disease. ASN Neuro 2: AN20100010 10.1042/AN20100010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara R. B., Sillanpää M. J., 2009. A review of Bayesian variable selection methods: what, how and which. Bayesian Anal. 4: 85–117. 10.1214/09-BA403 [DOI] [Google Scholar]
- Paus T., 2010. Population neuroscience: why and how. Hum. Brain Mapp. 31: 891–903. 10.1002/hbm.21069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peper J. S., Brouwer R. M., Boomsma D. I., Kahn R. S., Pol H., et al. , 2007. Genetic influences on human brain structure: a review of brain imaging studies in twins. Hum. Brain Mapp. 28: 464–473. 10.1002/hbm.20398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L., Patterson N. J., Plenge R. M., Weinblatt M. E., Shadick N. A., et al. , 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38: 904–909. 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- Price A. L., Kryukov G. V., de Bakker P. I., Purcell S. M., Staples J., et al. , 2010. Pooled association tests for rare variants in exon-resequencing studies. Am. J. Hum. Genet. 86: 832–838. 10.1016/j.ajhg.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., et al. , 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockova V., Lesaffre E., 2014. Incorporating grouping information in Bayesian variable selection with applications in genomics. Bayesian Anal. 9: 221–258. 10.1214/13-BA846 [DOI] [Google Scholar]
- Sahana G., Guldbrandtsen B., Janss L., Lund M. S., 2010. Comparison of association mapping methods in a complex pedigreed population. Genet. Epidemiol. 34: 455–462. 10.1002/gepi.20499 [DOI] [PubMed] [Google Scholar]
- Sampson J. N., Chatterjee N., Carroll R. J., Müller S., 2013. Controlling the local false discovery rate in the adaptive Lasso. Biostatistics 14: 653–666. 10.1093/biostatistics/kxt008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharinger C., Rabl U., Sitte H. H., Pezawas L., 2010. Imaging genetics of mood disorders. Neuroimage 53: 810–821. 10.1016/j.neuroimage.2010.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D., Davatzikos C., 2004. Measuring temporal morphological changes robustly in brain MR images via 4-dimensional template warping. Neuroimage 21: 1508–1517. 10.1016/j.neuroimage.2003.12.015 [DOI] [PubMed] [Google Scholar]
- Stingo F., Chen Y., Tadesse M., Vannucci M., 2011. Incorporating biological information into linear models: a Bayesian approach to the selection of pathways and genes. Ann. Appl. Stat. 5: 1978–2002. 10.1214/11-AOAS463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styan G. P., 1973. Hadamard products and multivariate statistical analysis. Linear Algebra Appl. 6: 217–240. 10.1016/0024-3795(73)90023-2 [DOI] [Google Scholar]
- Subramanian S., Ayala P., Wadsworth T. L., Harris C. J., Vandenbark A. A., et al. , 2010. CCR6: a biomarker for Alzheimer’s-like disease in a triple transgenic mouse model. J. Alzheimers Dis. 22: 619–629. 10.3233/JAD-2010-100852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Shen Y., Li Y., Zhang X., Wen J., et al. , 2017. Group spike-and-slab lasso generalized linear models for disease prediction and associated genes detection by incorporating pathway information. Bioinformatics 34: 901–910. 10.1093/bioinformatics/btx684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Schizophrenia Consortium , 2009. Common polygenic variation contributes to risk of schizophrenia that overlaps with bipolar disorder. Nature 460: 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R., 1996. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. B 58: 267–288. [Google Scholar]
- Turner J. A., Smyth P., Macciardi F., Fallon J. H., Kennedy J. L., et al. , 2006. Imaging phenotypes and genotypes in schizophrenia. Neuroinformatics 4: 21–49. 10.1385/NI:4:1:21 [DOI] [PubMed] [Google Scholar]
- Tzeng J.-Y., Zhang D., 2007. Haplotype-based association analysis via variance-components score test. Am. J. Hum. Genet. 81: 927–938. 10.1086/521558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng J.-Y., Devlin B., Wasserman L., Roeder K., 2003. On the identification of disease mutations by the analysis of haplotype similarity and goodness of fit. Am. J. Hum. Genet. 72: 891–902. 10.1086/373881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng J.-Y., Zhang D., Pongpanich M., Smith C., McCarthy M. I., et al. , 2011. Studying gene and gene-environment effects of uncommon and common variants on continuous traits: a marker-set approach using gene-trait similarity regression. Am. J. Hum. Genet. 89: 277–288. 10.1016/j.ajhg.2011.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh K. M., Codd V., Smirnov I. V., Rice T., Decker P. A., et al. , 2014. Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat. Genet. 46: 731–735. 10.1038/ng.3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Abbott D., 2008. A principal components regression approach to multilocus genetic association studies. Genet. Epidemiol. 32: 108–118. 10.1002/gepi.20266 [DOI] [PubMed] [Google Scholar]
- Wang T., Elston R. C., 2007. Improved power by use of a weighted score test for linkage disequilibrium mapping. Am. J. Hum. Genet. 80: 353–360. 10.1086/511312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Nie J., Yap P. T., Shi F., Guo L., et al. , 2011. Robust deformable-surface-based skull-stripping for large-scale studies, (Med. Image Comput. Comput. Assist. Interv.), Vol. 14, pp. 635–642. [DOI] [PubMed] [Google Scholar]
- Wang, Y., J. D. McKay, T. Rafnar, Z. Wang, M. N. Timofeeva et al., 2014 Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat. Genet. 46: 736–741 [corrigenda: Nat. Genet. 49: 651 (2017)]. 10.1038/ng.3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z., Li M., Rebbeck T., Li H., 2008. U-Statistics-based tests for multiple genes in genetic association studies. Ann. Hum. Genet. 72: 821–833. 10.1111/j.1469-1809.2008.00473.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M. C., Kraft P., Epstein M. P., Taylor D. M., Chanock S. J., et al. , 2010. Powerful SNP-set analysis for case-control genome-wide association studies. Am. J. Hum. Genet. 86: 929–942. 10.1016/j.ajhg.2010.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M. C., Lee S., Cai T., Li Y., Boehnke M., et al. , 2011. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 89: 82–93. 10.1016/j.ajhg.2011.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. T., Chen Y. F., Hastie T., Sobel E., Lange K., 2009. Genome-wide association analysis by lasso penalized logistic regression. Bioinformatics 25: 714–721. 10.1093/bioinformatics/btp041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z., X. Shen, and W. Pan; Alzheimer’s Disease Neuroimaging Initiative, 2014 Longitudinal analysis is more powerful than cross-sectional analysis in detecting genetic association with neuroimaging phenotypes. PLoS One 9: e102312. 10.1371/journal.pone.0102312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Fritsche L. G., Zhou X., Abecasis G., International Age-Related Macular Degeneration Genomics Consortium , 2017. A scalable Bayesian method for integrating functional information in genome-wide association studies. Am. J. Hum. Genet. 101: 404–416. 10.1016/j.ajhg.2017.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Baladandayuthapani V., Mallick B. K., Manyam G. C., Thompson P. A., et al. , 2014a Bayesian hierarchical structured variable selection methods with application to molecular inversion probe studies in breast cancer. J. R. Stat. Soc. Ser. C Appl. Stat. 63: 595–620. 10.1111/rssc.12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Morris J. S., Zhang J., Orlowski R. Z., Baladandayuthapani V., 2014b Bayesian joint selection of genes and pathways: applications in multiple myeloma genomics. Cancer Inform. 13: 113–123. 10.4137/CIN.S13787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., 2014. GEMMA User Manual, University of Chicago, Chicago. [Google Scholar]
- Zhou X., Carbonetto P., Stephens M., 2013. Polygenic modeling with Bayesian sparse linear mixed models. PLoS Genet. 9: e1003264 10.1371/journal.pgen.1003264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H., Z. S. Khondker, Z. Lu, and J. G. Ibrahim; Alzheimer’s Disease Neuroimaging Initiative, 2014 Bayesian generalized low rank regression models for neuroimaging phenotypes and genetic markers. J. Am. Stat. Assoc. 109: 1084–1098. 10.1080/01621459.2014.881742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H., 2006. The adaptive lasso and its oracle properties. J. Am. Stat. Assoc. 101: 1418–1429. 10.1198/016214506000000735 [DOI] [Google Scholar]
- Zou H., Hastie T., 2005. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Series B Stat. Methodol. 67: 301–320. 10.1111/j.1467-9868.2005.00503.x [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We used ADNI1 genetics and MRI image data that are available through application (http://adni.loni.usc.edu/about/adni1/). All the ADNI data are shared without embargo through the LONI Image and Data Archive (IDA).