Faithful segregation of mammalian X and Y chromosomes in male meiosis depends on pairing and recombination in a short interval of residual sequence homology known as the pseudoautosomal region (PAR)...
Keywords: meiosis, recombination, pseudoautosomal region, sex chromosome evolution
Abstract
Faithful segregation of homologous chromosomes at meiosis requires pairing and recombination. In taxa with dimorphic sex chromosomes, pairing between them in the heterogametic sex is limited to a narrow interval of residual sequence homology known as the pseudoautosomal region (PAR). Failure to form the obligate crossover in the PAR is associated with male infertility in house mice (Mus musculus) and humans. Yet despite this apparent functional constraint, the boundary and organization of the PAR is highly variable in mammals, and even between subspecies of mice. Here, we estimate the genetic map in a previously documented expansion of the PAR in the M. musculus castaneus subspecies and show that the local recombination rate is 100-fold higher than the autosomal background. We identify an independent shift in the PAR boundary in the M. musculus musculus subspecies and show that it involves a complex rearrangement, but still recombines in heterozygous males. Finally, we demonstrate pervasive copy-number variation at the PAR boundary in wild populations of M. m. domesticus, M. m. musculus, and M. m. castaneus. Our results suggest that the intensity of recombination activity in the PAR, coupled with relatively weak constraints on its sequence, permit the generation and maintenance of unusual levels of polymorphism in the population of unknown functional significance.
PAIRING, synapsis, and crossing-over between homologous chromosomes is a requirement for orderly progression through meiosis that is broadly conserved in eukaryotes (Mather 1938; Mirzaghaderi and Hörandl 2016). In most organisms, pairing is facilitated by sequence homology. The sex chromosomes of mammals—which are divergent in sequence and structure over most of their length (Graves 2006)—thus pose a unique challenge for males, the heterogametic sex. Homology between the X and Y chromosomes is limited to remnant “pseudoautosomal regions” (PARs) that are a vestige of the sex chromosomes’ origins as an ancestral autosome pair (Ohno 1966). Synapsis and crossing-over in this relatively small territory appears to be necessary for faithful segregation of the sex chromosomes in both humans and mice (Rouyer et al. 1986; Hassold et al. 1991; Matsuda et al. 1991; Burgoyne et al. 1992), although not in some other mammals (Ashley and Moses 1980; Wolf et al. 1988; Carnero et al. 1991). We limit our discussion here to the placental mammals; the sex chromosomes of marsupials and monotremes are distinct, and have more complex meiotic behavior (Sharman et al. 1950; Sharp 1982; Graves and Watson 1991; Page et al. 2005).
Despite the putative functional importance of the PAR, its boundary and structure appear to evolve rapidly in eutherian mammals (Raudsepp and Chowdhary 2015). PARs have been defined at the sequence level for only a handful of organisms, due in large part to the challenges of assembling repeat- and GC-rich sequences. The human X chromosome has two PARs: PAR1 on Xp and PAR2 on Xq (Figure 1A). Only PAR1 represents the remnant of the ancestral autosome, while PAR2 arose via several rearrangements within the primate lineage (Charchar et al. 2003); PAR1 recombines avidly in male meiosis (Hinch et al. 2014), while PAR2 recombines infrequently and has sequence properties similar to the rest of the X chromosome (Cotter et al. 2016). At least one well-described structural polymorphism of PAR1 segregates in human populations (Mensah et al. 2014). The allele is a translocation of X-linked sequence onto the Y chromosome, and arose via nonhomologous recombination between repetitive elements present on the X and the male-specific portion of the Y chromosome. The duplicated segment recombines in male meiosis at rates similar to single-copy PAR1 (Poriswanish et al. 2018). The PARs of simian primates, such as the rhesus macaque, have sequence homology and conserved synteny with human PAR1 on Xp (Ellis et al. 1990), and are in the order of 3 Mb in physical size, while those of ruminants extend further into the ancestral X chromosome and are closer to 10 Mb in size (Das et al. 2009).
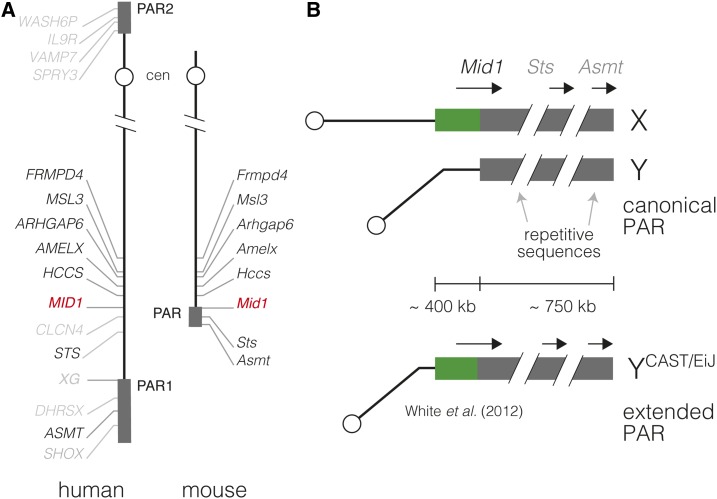
Figure 1.
Schematic organization of the pseudoautosomal region (PAR) in human and the house mouse. (A) Synteny map of the human and mouse X chromosomes. Orthologous genes are shown in black, nonorthologous genes in gray, and the gene marking the end of the human–mouse synteny block (MID1) in red. PARs shown as gray boxes. Not all human genes are shown. (B) The mouse PAR. The canonical PAR defined in the C57BL/6J reference strain (top) encompasses at least two genetically mapped protein-coding genes (Sts and Asmt) and the 3′ fragment of Mid1. Its physical size is ∼750 kb, most of which consists of unassembled repetitive sequence. The extended PAR of CAST/EiJ (bottom) involves the duplication of ∼400 kb of ancestrally X-unique sequence onto the Y, including the 5′ fragment of Mid1 (White et al. 2012a).
However, the mouse PAR is dissimilar to those of humans or other mammals. Its basic organization is shown in Figure 1B. Its boundary falls in the third intron of the mouse ortholog of human Opitz syndrome (Online Mendelian Inheritance in Man #300000) gene MID1 (Palmer et al. 1997). Restriction mapping has shown that the mouse PAR is ∼750 kb in size in the C57BL/6J inbred strain, most of which consists of arrays of GC-rich satellite and other repetitive sequences (Perry et al. 2001), and is exquisitely prone to internal rearrangement in male meiosis (Kipling et al. 1996a,b). Two genes, Sts (Kipling et al. 1996a) and Asmt (Kasahara et al. 2010), have been genetically mapped distal to Mid1. Two further genes (Ngln4x and Sfrs17a) have been localized to the PAR in comparative studies (Bellott et al. 2014) but lie in an assembly gap, the sequence of which has not yet been finished (https://www.ncbi.nlm.nih.gov/grc/mouse/issues/MG-3246). The representation of the PAR in the current version of the reference genome (mm10/GRCm38) comprises bases (of which lie in two gaps whose sizes represent a rough estimate by the Genome Reference Consortium), and includes the sequences of Mid1 and Asmt.
The PAR boundary is variable even within the Mus genus. It falls between the eighth and ninth exons of Mid1 in the Mus spretus-derived strain SPRET/Ei (Perry et al. 2001). White et al. (2012a) showed that in the Mus musculus castaneus-derived strain CAST/EiJ, the PAR boundary is shifted ∼400-kb proximally so that the entire sequence of Mid1 is present on both sex chromosomes. This implies translocation of previously X-unique sequence into the Y chromosome at the PAR boundary. Recombinants in this “extended PAR” were recovered from F2 crosses with the M. m. domesticus-derived WSB/EiJ strain, and heterozygosity in the PAR was associated with spermatogenic failure (White et al. 2012b). Hereafter, we refer to the PAR defined in C57BL/6J as the “canonical PAR” and the CAST/EiJ allele as an extended PAR (Figure 1B). Unlike the canonical PAR, the extended portion of the PAR in CAST/EiJ consists mostly of unique sequence, with GC content and repeat density similar to the genomic background.
Gain or loss of pseudoautosomal identity is predicted to have several important consequences for molecular evolution. First, since pseudoautosomal genes are diploid in both sexes, they are subject to a different dosage-compensation regime than the rest of the X chromosome and are more likely to escape X-inactivation (Graves et al. 1998). Second, to the extent that recombination is mutagenic (Hellmann et al. 2003), the intensity of pseudoautosomal recombination in male meiosis means that pseudoautosomal sequences are expected to diverge more quickly across species than strictly X- or Y-linked sequences. Diversity within populations should also be higher than at sex-linked sites, due to both higher mutational input, and mitigation of hitchhiking and background-selection effects by the high local recombination rate in males. Finally, the spectrum of mutations in pseudoautosomal sequences should be biased by the locally increased effects of GC-biased gene conversion (Lamb 1984; Eyre-Walker 1993). These expectations have generally been borne out in empirical studies of the human (Schiebel et al. 2000; Filatov and Gerrard 2003; Cotter et al. 2016) and mouse (Perry and Ashworth 1999; Huang et al. 2005; White et al. 2012a) PARs.
In this work, we use crosses between inbred strains from the three major subspecies of house mice to create a genetic map for the both the extended PAR of CAST/EiJ and the proximal portion of the canonical PAR. We use whole-genome sequencing to confirm the presence of a 14-kb extension of the PAR in the M. m. musculus-derived PWK/PhJ strain and show that, although it involves a complex structural rearrangement, it too recombines in heterozygous males. Finally, we uncover a remarkable degree of polymorphism at, and likely across, the PAR boundary in natural populations of all three subspecies. These findings add to a growing body of evidence that the degree of homology required for male fertility is relatively weak (Dumont 2017; Acquaviva et al. 2019). Relaxed constraints on PAR structure, combined with the intensity of double-strand break activity in male meiosis, permit the generation and maintenance of unusual levels of diversity in this peculiar region of the sex chromosomes.
Materials and Methods
Mouse breeding
The crosses described herein were bred as part of ongoing studies of aging, recombination, and infertility in hybrid males. Standard mouse strains (CAST/EiJ, PWK/PhJ, WSB/EiJ, and FVB/NJ) were purchased from the Jackson Laboratory (Bar Harbor, ME) between 2008 and 2011. Reciprocal F1 hybrid males were generated in all pairwise combinations between CAST/EiJ (abbreviated with strain code F), PWK/PhJ (G), and WSB/EiJ (H) in 2008–2009. F1 males were bred with multiple FVB/NJ females to generate N2 progeny, which were killed at birth by cervical dislocation. N2 progeny were named as follows: sire identifier (PAE_xxx), two-character encoding of sire genotype (e.g., HF for WSB/EiJ CAST/EiJ), one-character encoding of approximate sire age at conception (E = elderly, Y = young), and two- or three-character pup identifier. All breeding was completed by 2011. The study was approved by the Institutional Animal Care and Use Committee of the University of North Carolina at Chapel Hill, and all animal husbandry was conducted in the university’s Association for Assessment and Accreditation of Laboratory Animal Care-accredited facility (#A3410-01; Animal Welfare Assurance) in accordance with institutional and federal regulations.
DNA extraction, genotyping, and quality control
Genomic DNA was extracted from tail clips of adult representatives of parental inbred strains, or whole tails or heads of N2 progeny using either a standard phenol–chloroform method (Green and Sambrook 2012), or DNeasy Blood & Tissue Kits (catalog no. 69506; QIAGEN, Valencia, CA). Approximately 1.5 μg of DNA per sample was shipped to Neogen (Lincoln, NE) for hybridization to one of two Mouse Universal Genotyping Arrays, MegaMUGA (77K probes) or MiniMUGA (11K probes). Both arrays are custom-designed on the Illumina Infinium HD platform (Illumina, San Diego, CA) (Steemers et al. 2006), with content optimized for information content in classical inbred strains and the eight founder strains of the Collaborative Cross (CC) (A/J, C57BL/6J, 129S1/SvImJ, NOD/ShiLtJ, NZO/HlLtJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ), as described in Morgan et al. (2016). Genotypes were called by the vendor from hybridization intensity signals using the semisupervised clustering algorithm implemented in the Illumina BeadStudio software.
Genotype data were processed in R v3.3.2 (R Foundation for Statistical Computing, http://www.r-project.org/) using the argyle package v0.2.2 (Morgan 2016). Samples with <10% missing genotype calls were retained for analysis; there were no genotyping failures. Samples were genetically sexed by comparing average hybridization intensities for X- and Y-linked markers. We identified a single XO individual among the PWK/PhJ CAST/EiJ N2s, and none in the WSB/EiJ CAST/EiJ or WSB/EiJ PWK/PhJ N2s. To confirm correct assignment of N2s to F1 sires, we examined N2 genotypes at sites on the X chromosome that were informative between the parent strains. We identified 24 progeny representing two consecutive litters from the same sire (PAE_018) that were initially labeled as WSB/EiJ CAST/EiJ. Males were homozygous for WSB/EiJ genotypes on the X, and females heterozygous at sites informative between CAST/EiJ and FVB/NJ. This pattern was consistent with a (CAST/EiJ × WSB/EiJ)F1 sire, and we updated our pedigrees accordingly.
A complete list of N2 progeny, their parents, genetically confirmed sexes, and the platform on which they were genotyped is provided in Supplemental Material, File S1.
Refinement of genotypes on the X chromosome
Because the aim of this study was to identify crossovers in the PAR, we focused particular attention on the quality of genotype calls at markers in this region. The extended PAR of CAST/EiJ begins (in the mm10 assembly) at chromosome X:169,542,082 and the canonical PAR at chromosomeX:169969759. We augmented MegaMUGA genotypes and hybridization intensities ( N2s parental strains) with 111 representatives of the eight CC founder strains, and the 26 viable reciprocal F1 hybrids between them (these control genotypes are available for public download from http://csbio.unc.edu/CCstatus/index.py). Of 19 markers whose probe sequences align to the PAR in mm10, only one (JAX00723372) yields automated calls in the heterozygous and both homozygous states among the 111 control samples. We manually inspected hybridization intensities at each of the remaining 18 markers to find any with cluster patterns from which reliable genotypes might be recovered. Three clusters—putatively corresponding to the two homozygous and one heterozygous states—were observed at a single marker, UNC31596457. No heterozygotes are called at this marker by the automated algorithm in BeadStudio; however, it is clear that CAST/EiJ has a private allele at this marker and that one of the clusters corresponds to CAST/(non-CAST) heterozygotes in the control samples (Figure S1). Therefore, we recalled genotypes at this marker by fitting a three-component bivariate Gaussian mixture model to hybridization intensities of the control samples and N2s using the R package mclust v5.2.1 (Fraley and Raftery 2002). Samples with cluster assignment probability were marked as missing. The result is shown in Figure S2.
Following this genotype refinement for MegaMUGA, informative markers in or beyond the extended PAR of CAST/EiJ were identified separately on the two array platforms by comparing genotypes between single representatives of the relevant inbred strains. This left seven and six markers on MegaMUGA for the PWK/PhJ CAST/EiJ and WSB/EiJ CAST/EiJ crosses, respectively, and one and two markers on MiniMUGA for the same crosses. A list of informative markers for each array-cross combination is provided in File S2. Refined genotype calls for all N2 progeny and controls in the study are provided in File S3.
Crossovers in the PAR were identified as illustrated in Figure 2. Genetic lengths of intermarker intervals were calculated assuming an identity map function (i.e., no crossover interference) and SE obtained from equation 2.12 in Xu (2013).
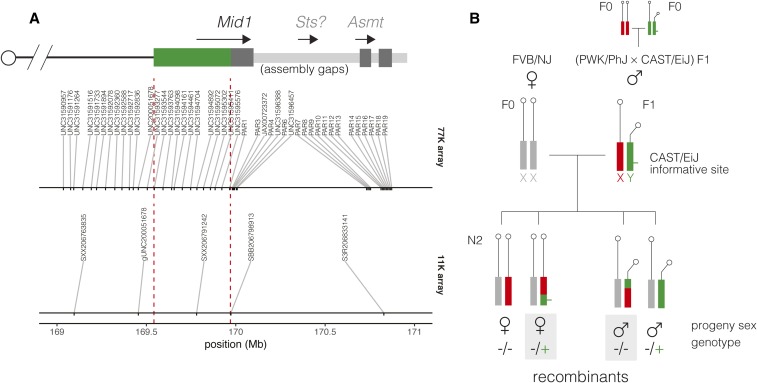
Figure 2.
Experimental design for detecting recombination in the extended pseudoautosomal region (PAR) of CAST/EiJ. (A) Marker map near the pseudoautosomal boundary. A schematic representation of the CAST/EiJ extended PAR (green) and canonical PAR (dark gray), with its internal assembly gaps (light gray) and protein-coding genes, is shown in the top panel. Nominal physical positions (in the mm10 assembly) of SNP markers from the 77K and 11K arrays are shown in the bottom panel. CAST/EiJ and canonical PAR boundaries are indicated by red dashed lines. (B) Multiple (PWK/PhJ CAST/EiJ)F1 males were bred with FVB/NJ females, and N2 progeny genotyped with commercial SNP arrays. In the absence of recombination, female progeny inherit an maternal FVB/NJ allele and a paternal PWK/PhJ allele at X chromosome markers; nonrecombinant male progeny inherit a maternal FVB/NJ allele and a paternal CAST/EiJ allele. Other combinations require recombination between the X and Y chromosomes.
Whole-genome sequencing
New whole-genome sequence data were generated for 87 N2 progeny of the WSB/EiJ PWK/PhJ cross at the University of North Carolina High-Throughput Sequencing Facility. Genomic DNA was fragmented by ultrasonication on a Covaris instrument (Covaris, Woburn, MA) and enriched for fragments ∼300 bp in size on a PippinPrep system (Sage Science, Beverly, MA). Paired-end libraries were prepared using the Illumina TruSeq PE v1 chemistry. Samples were multiplexed in two pools (with 45 and 42 samples, respectively) and sequenced 2 × 100 bp on an Illumina HiSeq 2500 instrument to a depth of ∼. Demultiplexing and postprocessing were performed with Illumina CASAVA v1.8.2. Data has been submitted to the European Nucleotide Archive (accession PRJEB32247).
Reads for inbred strains PWK/PhJ, FVB/NJ, and WSB/EiJ were obtained from the public FTP site of the Sanger Mouse Genomes Project, February 2015 release (ftp://ftp-mouse.sanger.ac.uk/REL-1502-BAM) (Keane et al. 2011). Reads for CC strains CC001/Unc, CC022/GeniUnc, CC032/GeniUnc, CC044/GeniUNC, and CC045/GeniUnc were generated by our research group and have been previously published (European Nucleotide Archive accession PRJEB14673) (Srivastava et al. 2017). Reads for wild M. musculus and M. spretus have been previously published (Harr et al. 2016), and were obtained from the European Nucleotide Archive (accessions PRJEB9450, PRJEB11742, PRJEB14167, and PRJEB2176). Reads for M. spicilegus have been previously published (Neme and Tautz 2016) and were obtained from the European Nucleotide Archive (accession PRJEB11513). Reads for M. caroli have been previously published (Thybert et al. 2018) and were obtained from the European Nucleotide Archive (accession PRJEB14895). A complete list of samples used in whole-genome sequencing analysis is provided in File S4.
The standard mouse reference assembly is a haploid representation of the genome, except in the PAR, whose sequence appears on both sex chromosomes. This induces ambiguous sequence alignments and effectively halves the observed read depth over each copy of the PAR. To avoid these artifacts, we masked the sequence of the Y PAR (chromosome Y:90745845–91644698) in the mm10/GRCm38 assembly with Ns. Reads were aligned to this masked reference with bwa mem 0.7.15-r1140 (Li 2013) with flags -t 16 -YK100000000 and default settings otherwise. Optical duplicates were marked with samblaster v0.1.24 (Faust and Hall 2014).
Ascertainment of copy-number variants and their boundaries
Copy-number variants (CNVs) in the distal X chromosome were identified by examining read depths in windows of varying sizes, as described in the main text. Paired-end reads are expected to align with a characteristic orientation: one read on the forward strand and its mate on the reverse (configuration “FR”). Clusters of pairs with other orientations (RR, FF, or RF) may be a signal of underlying structural variation. Summaries of total and FR read depths were calculated with pysamstats v1.1.0 (https://github.com/alimanfoo/pysamstats), and normalized against read depth in single-copy autosomal sequence. CNV boundaries were refined by manual inspection of read alignments in the Integrative Genomics Viewer (IGV) v2.4.16 (Robinson et al. 2011). Clusters of three or more reads with soft-clipped alignments (that is, alignments of less than the full read length, indicated by the presence of S in the Concise Idiosyncratic Gapped Alignment Report (CIGAR) string) all terminating at exactly the same base pair, and coinciding with an abrupt change in both total read depth and the proportion of FR reads, were deemed candidate CNV breakpoints. Such reads, as well as their mates (whether the mate was mapped or not), were extracted and converted to fastq format with samtools v1.9 (Li et al. 2009). For the breakpoints of the PWK/PhJ Y CNV, reads crossing each of three candidate breakpoints were assembled de novo with SPAdes v3.10.1 (Bankevich et al. 2012) with flags --12 (for paired-end reads) and --only-assembler (skipping error correction, which requires higher coverage). The longest contig assembled for each breakpoint was aligned back to the mm10 reference with blat (Kent 2002) at the University of California Santa Cruz (UCSC) Genome Browser (https://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10). Contigs were screened for possible ORFs with the ExPASy Translate tool (https://web.expasy.org/translate/). Their sequences are provided in File S5.
To identify potential sources of breakpoint sequence that did not align in mm10, we performed blat searches against recent de novo assemblies of M. spretus [GenBank accession GCA_001624865.1 (Lilue et al. 2018)], M. caroli [GCA_900094665.2 (Thybert et al. 2018)], and M. pahari [GCA_900095145.2 (Thybert et al. 2018)]. This contig has gapped alignments to these outgroup genomes that resemble alignments of a spliced transcript (Figure S3 and File S6), but the aligned segments do not correspond to annotated exons. The longest ORF it encodes is 66-aa long, and Basic Local Alignment Search Tool (BLAST) searches of six-frame translations (blastx) against the National Center for Biotechnology Information (NCBI) nonredundant protein database yield no hits with e-values (search performed January 31, 2019). We conclude that despite having the appearance of a retrotransposition event, the sequence is unlikely to represent an ancestral coding sequence.
Single-nucleotide variant calling and filtering
For analyses of recombination within the PWK/PhJ extended PAR, we used bcftools v1.9 to ascertain biallelic single-nucleotide variants (SNVs) in the distal X chromosome (chromosome X:169400000–169969759) with call rates in 169 samples including N2 progeny, inbred parental strains, wild mice, and representative CC strains with a PWK/PhJ Y chromosome. We retained 84 sites homozygous in the PWK/PhJ female and heterozygous in the CC044/Unc strain (carrying a PWK/PhJ Y chromosome), and segregating at absolute allele count in the N2s. A single N2 individual (PAE_033_HG_E_463) had an outlying number of heterozygous calls and appeared to be cross-contaminated with another individual; it was excluded from this and all other analyses. Genotype calls at these sites are provided in VCF format in File S7.
Principal components analysis (PCA) on genotypes in the distal X chromosome was performed with akt v3beb346 (Arthur et al. 2017), treating all samples as nominally diploid (i.e., substituting homozygous for hemizygous genotypes.) Because we did not have access to whole-genome sequence data for an inbred PWK/PhJ male, we created a pseudodiploid individual homozygous for PWK/PhJ Y-linked alleles to perform ancestry assignment for the PWK/PhJ Y PAR.
Reanalysis of chromatin immunoprecipitation sequencing data
Raw reads from published chromatin immunoprecipitation sequencing (ChIPseq) against histone 3 lysine 4 trimethylation (H3K4me3) in 12 days postpartum testes from reciprocal F1 hybrid males between C57BL/6J and CAST/EiJ (Baker et al. 2015) were downloaded from the NCBI Sequence Read Archive (accession PRJNA259788), and aligned to the mm10 reference with bwa mem as described above. Postprocessed read alignments from ChIPseq against DMC1 from the same genotypes (but different laboratories), representing single-stranded DNA fragments at double-strand break (DSB)-free ends (Smagulova et al. 2016), were downloaded from the NCBI Gene Expression Omnibus (accession GSE75419).
Tag reads were assigned to the CAST/EiJ or C57BL/6J haplotype according to the base called at known informative sites between these strains (using biallelic SNVs from the Mouse Genomes Project VCF, May 2015 release; ftp://ftp-mouse.sanger.ac.uk/REL-1505-SNPs_Indels/). Given n informative sites spanned by a read, the log-likelihood of the read originating from each parental haplotype was calculated as the binomial probability of observing k bases consistent with that haplotype and bases inconsistent. A read was deemed assignable if log-likelihood of maternal vs. paternal assignments differed by units. Tag density was calculated with the bamCoverage tool from the bamTools suite (Barnett et al. 2011) using 100-bp bins and a 500-bp smoothing window. No ChIP background subtraction or peak-calling was performed.
We note that, since reads were aligned to the standard mm10 reference, reads arising from the CAST/EiJ haplotype are somewhat less likely to be aligned, and so the count of CAST/EiJ reads will be an underestimate. This “reference bias” causes our analyses to be conservative.
Analysis of gene expression
Assay of gene expression by paired-end, unstranded RNA sequencing (RNAseq) in adult brains from 90 progeny of a complete diallel cross (e.g., all possible reciprocal crosses) between CAST/EiJ, PWK/PhJ, and WSB/EiJ has been described previously (Crowley et al. 2015). We obtained the raw reads for these 90 samples (File S8) and quantified expression using kallisto v0.44.0 (Bray et al. 2016) with default options. Similar to the approach described for whole-genome sequencing, we removed eight redundant transcripts annotated in the Y-encoded copy of the PAR (retaining X-encoded copies) and performed quantification against the remaining transcripts in the Ensembl version 96 annotation. Transcript-level estimates were aggregated to gene level with the R package tximport (Soneson et al. 2015) and analyses of differential expression were performed with limma-voom (Ritchie et al. 2015), using the method of Benjamini and Hochberg to adjust P-values for control of the false discovery rate.
To investigate allele-specific expression and splicing patterns, we also aligned reads to our masked version of the reference genome and masked transcript annotation with STAR v2.7.0e (Dobin et al. 2013). Read pairs were aligned to parental haplotypes as described above for the ChIPseq data. Coverage along annotated exons of Mid1 was calculated with bamCoverage using 5-bp bins and a 25-bp smoothing window. Reads were pooled across biological replicates of the same genotype and sex.
Mid1 transcript isoforms were assembled from read alignments in the parental strains (CAST/EiJ, PWK/PhJ, and WSB/EiJ), pooling reads from both male and female samples, using StringTie v1.3.5 (Pertea et al. 2015) with options -f 0.1 -a 4 -c 0.1 -g 500 and the Ensembl version 96 annotation as a guide. Assembled transcripts are provided in File S9.
Data availability
Whole-genome sequence data from N2 progeny have been submitted to the European Nucleotide Archive (accession PRJEB32247). The inferred genetic map for the extended PAR is in File S10; informative markers near the PAR boundary in File S2; assembled breakpoint sequences of the PWK/PhJ PAR variant in File S5; a list of N2 progeny used for recombination analysis in File S1; array genotype and intensity data, with marker locations, in File S3; and genotype calls in the PWK/PhJ PAR variant from the whole-genome sequence in File S7. Supporting code is available at https://github.com/andrewparkermorgan/mouse_pseudoautosomal_region. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.7798889.
Results
Recombination in the extended PAR of CAST/EiJ
To estimate a recombination map in the CAST/EiJ extended PAR, we used genotype data from two crosses segregating for the CAST/EiJ Y chromosome: 219 N2 progeny of (PWK/PhJ CAST/EiJ)F1 males and 48 N2 progeny of (WSB/EiJ CAST/EiJ)F1 males. The experimental design is shown in Figure 2. For notational convenience, we will refer to these crosses by the sire genotype throughout. Progeny were genotyped on one of two SNP array platforms (one with ∼ markers, hereafter “77K,” and one with ∼ markers, hereafter “11K”) whose contents are optimized for populations derived from eight inbred strains, including those used here (Morgan et al. 2016). The nominal position of informative markers in the CAST/EiJ extended PAR in the mm10 reference genome in is shown in Figure 2A. To capture recombination in the canonical PAR, we identified two additional informative markers beyond the PAR boundary (see Materials and Methods).
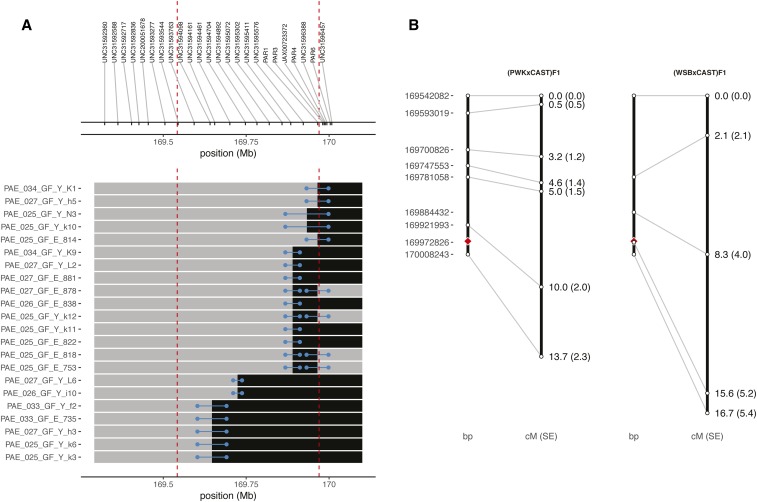
Recombinants in the PAR were then identified in curated genotypes using the rules illustrated in Figure 2B. Sex (i.e., presence or absence of the male-specific region of the Y chromosome) anchors the proximal end of the genetic map of the PAR by definition. Aggregating across both array platforms, 26 of 219 progeny of (PWK/PhJ CAST/EiJ)F1 males and 8 of 48 of progeny of (WSB/EiJ CAST/EiJ)F1 males have evidence of recombination. Of these, at least four (all from the PWK/PhJ CAST/EiJ cross) are double recombinants (Table 1). Example recombinant chromosomes from the PWK/PhJ CAST/EiJ cross are shown in Figure 3A and the estimated genetic maps for both crosses in Figure 3B (and in tabular form in File S10). The genetic lengths of the portions of the PAR spanned by our array markers are cM (point estimate SE) and cM in the PWK/PhJ CAST/EiJ and WSB/EiJ CAST/EiJ crosses, respectively (most of this map– and cM, respectively–lies in the “extended” portion of the PAR due to limited marker coverage in the canonical PAR). Given the region’s physical size of 466 kb (spanning chromosome X:169,542,082–169,969,759 in the mm10 assembly), this translates to recombination rates of and cM/Mb, respectively, assuming negligible crossover interference (Soriano et al. 1987). In contrast, male-specific autosomal rates in these crosses have been estimated at ∼0.4 cM/Mb (Dumont and Payseur 2011a,b). This 100-fold elevation of the recombination rate in the CAST/EiJ extended PAR over the autosomal background is consistent with obligatory crossing-over in the PAR at every meiosis (Burgoyne 1982), despite its relatively small physical size (Perry et al. 2001). We find no evidence for transmission bias against recombinants (pooled odds ratio from both crosses by logistic regression, C.I. , by likelihood-ratio test), and a rate of sex-chromosome nondisjunction (XOs: , ) similar to prior estimates for these F1s (Dumont 2017).
Table 1. Summary of crossovers detected in the PAR in N2 progeny.
| Recombinants | ||||||
|---|---|---|---|---|---|---|
| Cross | Platform | Sex | n | NR | Single | Double |
| PWK/PhJ CAST/EiJ | 77K | XX | 90 | 78 | 8 | 4 |
| XY | 90 | 80 | 10 | 0 | ||
| 11K | XX | 18 | 17 | 1 | 0 | |
| XY | 20 | 17 | 3 | 0 | ||
| XO | 1 | 1 | 0 | 0 | ||
| WSB/EiJ CAST/EiJ | 77K | XX | 2 | 2 | 0 | 0 |
| XY | 4 | 3 | 1 | 0 | ||
| 11K | XX | 23 | 20 | 3 | 0 | |
| XY | 19 | 15 | 4 | 0 | ||
NR, non-recombinant.
Figure 3.
Genetic map of the CAST/EiJ extended pseudoautosomal region (PAR). (A) Recombinant chromosomes in the extended PAR in the PWK/PhJ CAST/EiJ cross. The expected nonrecombinant haplotype for sex is shown in gray and the recombinant haplotype in black. Blue bars show the interval between flanking markers (with points nudged to avoid overplotting). CAST/EiJ and canonical PAR boundaries are indicated by red dashed lines. Physical positions of markers from the 77K array are shown in the top panel. (B) Physical (left) and genetic (right) positions are shown for informative markers in the PWK/PhJ CAST/EiJ and WSB/EiJ CAST/EiJ crosses, pooling across both array platforms. Red diamond indicates boundary of canonical PAR.
Two biochemical surrogates of meiotic recombination—H3K4me3, which marks sites for DSB formation, and DMC1 protein, which binds free ends of DSBs—both accumulate on the CAST/EiJ Y haplotype in spermatocytes. We reanalyzed ChIPseq performed against H3K4me3 and DMC1 in testes of reciprocal F1 hybrids between C57BL/6J and CAST/EiJ (Baker et al. 2015; Smagulova et al. 2016). Reads were assigned to parental haplotypes using known SNVs that are informative between the parental strains. In the extended PAR, H3K4me3 and DMC1 reads are attributed almost exclusively to the X chromosome in CAST/EiJ C57BL/6J males, but to both haplotypes in C57BL/6J CAST/EiJ males (Figure S4). This suggests that recombination is initiated from both haplotypes in the extended PAR when the CAST/EiJ Y chromosome is present, and proceeds through the normal homologous recombination pathway.
Extension of the PAR in PWK/PhJ
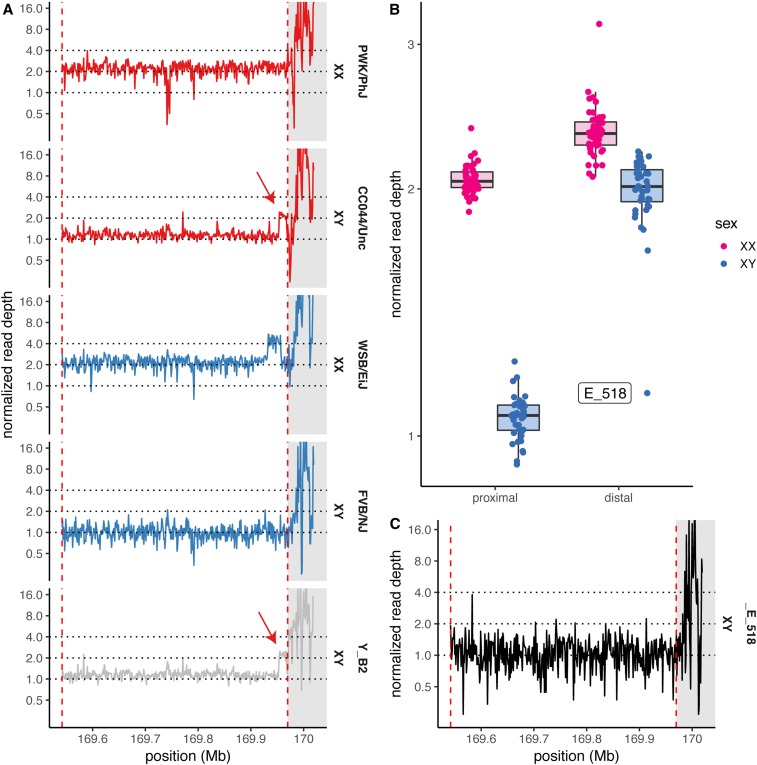
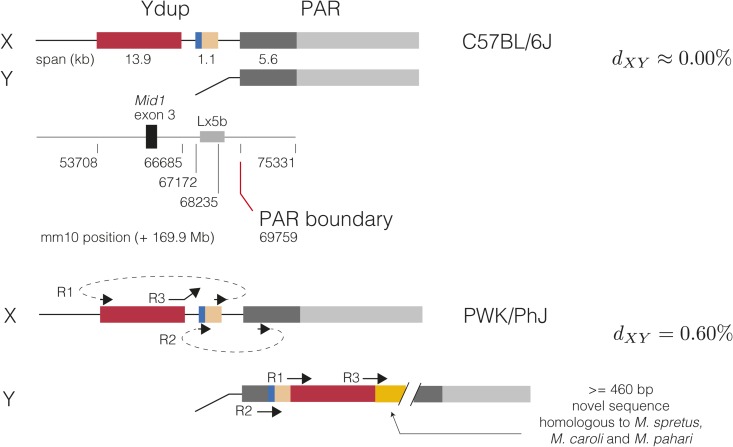
We next attempted to estimate the rate of recombination in the canonical PAR. For this, we turned to low-coverage whole-genome sequence data from 86 progeny of (WSB/EiJ PWK/PhJ)F1 sires and FVB/NJ dams (hereafter referred to as the N2 generation). It was immediately apparent that the complex repetitive sequence in the assembled portion of the PAR, to say nothing of the unassembled portion, would preclude accurate genotyping of SNVs from short reads. However, we noticed a CNV at the PAR boundary segregating among the N2s that is not present in published whole-genome sequence data from the founders of the cross (FVB/NJ, male; WSB/EiJ, female; PWK/PhJ, female) (Keane et al. 2011), but that is present in a CC strain carrying a PWK/PhJ Y chromosome [CC044/Unc (Srivastava et al. 2017)] (Figure 4A). The copy gain segregates almost perfectly with male sex; 39 of 40 males have the expected copy number 1 over the adjacent segment of the X chromosome but two copies of the CNV segment, while 46 of 46 females have two copies across the entire region (Figure 4B). This pattern is consistent with duplication of X-linked sequence onto the Y chromosome in PWK/PhJ. The segment is thus a candidate for an extension of the PAR. This agrees with the observation of White et al. (2012a) that primers in the second intron of Mid1 amplify from both X and Y chromosomes in PWD/PhJ, a strain closely related to PWK/PhJ (Yang et al. 2011).
Figure 4.
A translocation of X-linked sequence onto the Y in PWK/PhJ. (A) Copy number in the distal X chromosome, estimated from whole-genome sequencing read depths in 1-kb bins, for the parental inbred strains (with sex-chromosome karyotypes as shown) of the WSB/EiJ CAST/EiJ cross and a representative N2 male (PAE_041_HG_Y_B2). Duplicated segment indicated by red arrow. CAST/EiJ and canonical pseudoautosomal region (PAR) boundaries are indicated by red dashed lines. Canonical PAR shaded in gray. (B) Copy number over the proximal X chromosome (“proximal”) and chromosome X:169.96 Mb [“distal,” at red arrow in (A)] among N2 males (blue) and females (pink). (C) Copy number profile of N2 male PAE_033_HG_E_518, demonstrating de novo loss of the duplication.
Closer examination of the duplication shows that it lies near but not immediately adjacent to the PAR boundary in the mm10 reference assembly, and that it is not contiguous (Figure S5). The boundaries of the duplication correspond to clusters of read alignments with aberrant mate-pair orientation. We manually inspected read alignments near the putative duplication boundaries in the IGV and obtained base pair resolution of the boundaries by identifying multiple of reads with partial alignments that terminated at the same position. The coordinates of the duplicated segments in mm10 are chromosome X:169,953,707–169,966,684 and chromosome X:169,967,172–169,968,235; the nominal size of the duplicated region is 14,040 bp. Reads from fragments overlapping the duplication boundary were pooled and de novo-assembled with SPAdes. This yielded three contigs (File S5) whose alignments back to the reference (with blat), shown in Figure S5, define the breakpoints of the duplication. The distal boundary coincides with a cluster of ancient long interspersed L1 elements, but other boundaries do not appear to correspond to either specific repetitive elements or exons of the Mid1 gene.
We used the copy number of subsegments of the CNV and the constraints imposed by the breakpoint alignments to reconstruct the organization of the PAR boundary on the PWK/PhJ Y chromosome (Figure 5). In this proposed configuration, the previously X-linked sequence lies internal to the PWK/PhJ Y PAR; this is supported by direct evidence from single reads whose alignments cross the breakpoint. The duplication also contains 460 bp of novel sequence (in breakpoint contig region_3) that is absent from the mm10 assembly, including unplaced contigs, but is found in Mid1 introns in de novo assemblies for M. spretus, M. caroli, and M. pahari. Recovery of this sequence from multiple outgroup species suggests that it was present in the common ancestor of the genus Mus (5 MYA; Thybert et al. 2018) and has since been lost in some lineages of M. musculus. Note that the duplication may be much larger than its nominal size in the mm10 reference, as it may contain other novel sequences that lie too far from the breakpoints to be captured by our assembly strategy.
Figure 5.
Proposed organization of the PWK/PhJ Y-linked duplication. X-linked segments involved in the duplication are shown in their position in C57BL/6J (mm10 assembly) in the top panel. Breakpoint coordinates and selected annotations are shown in the middle panel. The bottom panel shows alignments of breakpoint-spanning contigs to the PWK/PhJ X chromosome (which is near-identical to C57BL/6J), along with one possible reconstruction of the Y chromosome consistent with copy-number and alignment constraints. , absolute sequence divergence between X- and Y-linked copies of the pseudoautosomal region (PAR).
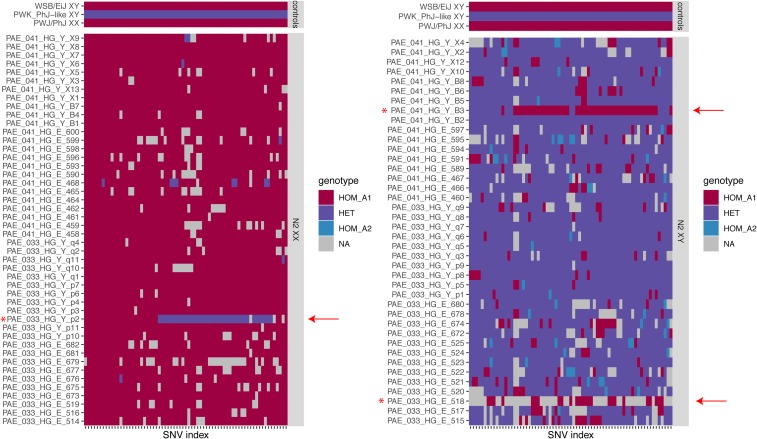
Having established that the duplication is genetically and physically linked to the PAR on the PWK/PhJ Y chromosome, we next tested whether it is truly pseudoautosomal in the sense that it recombines in male meiosis. Biallelic SNVs in the duplicated region were ascertained in the N2 progeny, the parental strains of the cross, and a collection of wild mice with publicly available whole-genome sequence data. Only sites segregating at moderate frequency (minor allele count ) in the N2s and heterozygous in a CC male carrying the PWK/PhJ Y chromosome were retained. This conservative ascertainment strategy yielded 84 high-confidence sites informative for recombination in (WSB/EiJ PWK/PhJ)F1 males. To our surprise, the PWK/PhJ X and Y chromosomes carry different alleles at all 84 sites; we revisit this finding below. Manual review of genotype calls in the N2s revealed two likely recombinants (Figure 6): one male (PAE_041_HG_Y_B3) with a run of homozygous genotypes and one female (PAE_033_HG_Y_p2) with a run of heterozygous genotypes. The male with copy number 1 (PAE_033_HG_E_518) has reverted to homozygosity across the entire region. We conclude that this individual represents a de novo loss of the PWK/PhJ Y-linked copy of the duplicated sequence. Further support for the de novo loss is provided by the absence of aberrantly oriented read alignments at the boundaries of the allele in PAE_033_HG_E_518, but not the male recombinant (Figure S6). The recombination fraction in the duplicated region in this cross, excluding the individual with the de novo loss, is () in 28 kb, for an estimated recombination rate of 84 cM/Mb. The duplication is thus a bona fide extension of the PAR.
Figure 6.
Genotypes of N2 females (left) and males (right) at informative sites between the PWK/PhJ Y chromosome and the PWK/PhJ X or WSB/EiJ X haplotypes under the PWK/PhJ Y-linked duplication. The same set of parental inbred strain controls is shown in both top panels. Genotype calls are color-coded as homozygous for the mm10 reference allele (HOM_A1), heterozygous (HET), or homozygous for the alternate allele (HOM_A2). Missing calls (NA) are shown in gray. Recombinant individuals shown with red arrowheads. SNV, single-nucleotide variant.
Origin of the extended PAR in PWK/PhJ
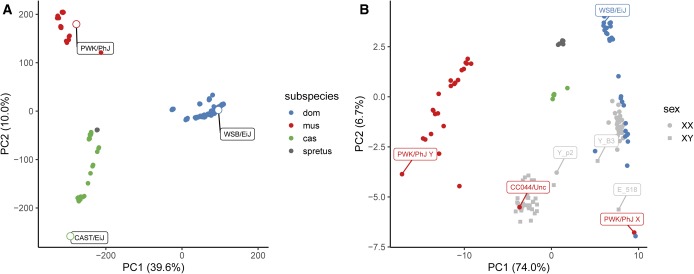
Free recombination is difficult to reconcile with the divergence between the X- and Y-linked copies in PWK/PhJ. The two haplotypes differ at 84 sites in 14 kb (), within the range expected for autosomal sequences from different mouse subspecies (Geraldes et al. 2008). PWK/PhJ is a wild-derived strain whose founders were trapped in the Czech Republic (Gregorová and Forejt 2000), has M. m. musculus ancestry [Yang et al. (2011) and Figure 7A], and a M. m. musculus haplotype in the male-specific portion of the Y chromosome (Morgan and Pardo-Manuel de Villena 2017). However, both PWK/PhJ and the related strain PWD/PhJ are known to carry a M. m. domesticus haplotype in the distal portion of the X chromosome (Yang et al. 2011). PCA on SNV genotypes, both at autosomal sites and within the Y-linked duplication, clearly separates wild and wild-derived M. m. domesticus, M. m. castaneus, and M. m. musculus individuals by subspecies (Figure 7, A and B). A pseudodiploid PWK/PhJ Y PAR haplotype, constructed from the consensus PWK/PhJ paternal allele in N2 progeny, clusters with wild M. m. musculus, while the PWK/PhJ X chromosome clusters with wild M. m. domesticus (Figure 7B). N2 female progeny, expected to be homozygous for two M. m. domesticus haplotypes (WSB/EiJ and FVB/NJ), cluster with M. m. domesticus. N2 male progeny and the CC044/Unc inbred male, expected to be heterozygous between an M. m. musculus- and an M. m. domesticus-like haplotype, fall in between the two subspecies-specific clusters. Recombinant N2 progeny lie between the heterozygote and homozygote clusters. The sequence in the PWK/PhJ extended PAR appears to be of M. m. musculus origin.
Figure 7.
Determining the ancestry of the PWK/PhJ Y-linked duplication. (A) Principal components analysis (PCA) on genotype matrix at autosomal SNPs from the 77K array platform, in a collection of 346 wild mice of M. m. domesticus (dom) M. m. musculus (mus), M. m. castaneus (cas), and M. spretus ancestry, plus the three wild-derived inbred strains used in this study (open points with labels). Data from Didion et al. (2016). (B) PCA on 84 sites informative between the PWK/PhJ X and Y chromosomes in wild mice (colored by subspecies) and (WSB/EiJ PWK/PhJ)N2 progeny.
When did the duplication occur? If it occurred during the establishment of the PWK/PhJ line, it must have been early in the inbreeding process, before the distal X chromosome was fixed for an M. m. domesticus haplotype. We examined read alignments from wild-caught M. m. musculus and identified a male from the Czech Republic (CR16) with clusters of aberrantly oriented read pairs coinciding with the boundaries of the PWK/PhJ duplication (Figure S7). Reads with partial alignments from this male supported precisely the same breakpoints as in PWK/PhJ. Although we cannot exclude the possibility of recurrent mutation, the most parsimonious explanation is that the duplication in PWK/PhJ is segregating at moderate frequencies in wild populations of M. m. musculus.
Gene expression across the pseudoautosomal boundary
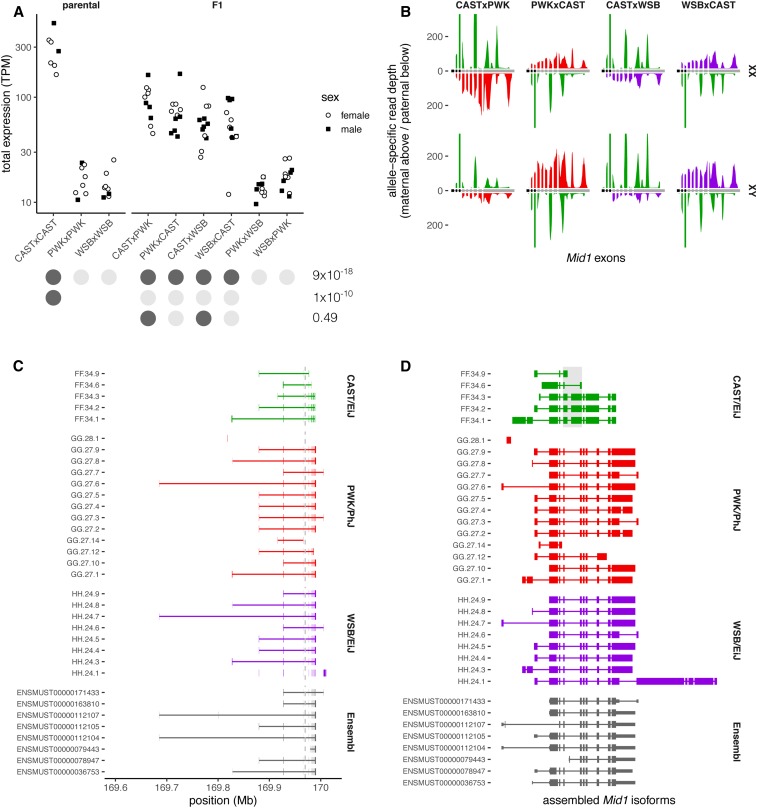
The gene that spans the pseudoautosomal boundary, Mid1, is broadly expressed in cell lineages of all three germ layers during embryonic development (Dal Zotto et al. 1998), and mouse knockouts have abnormal migration and branching of axons (Lu et al. 2013). Therefore, we investigated the effects of shifts in the PAR boundary on transcriptional regulation of Mid1. Our group has previously assayed gene expression by RNAseq in adult brains in male and female progeny of a full diallel cross between CAST/EiJ, PWK/PhJ, and WSB/EiJ (Crowley et al. 2015). We requantified expression in a total of 90 individuals from nine genotypes with kallisto (Bray et al. 2016), using the Ensembl version 96 annotation after removal of redundant gene models present on both the X- and Y-encoded copies of the PAR. To characterize patterns of differential splicing, we also aligned reads to the mm10 reference genome after masking the Y-encoded copy of the PAR to avoid mapping artifacts (since the PAR is the only locus that deviates from the conventional haploid representation in the reference genome). Expression of Mid1 is 11.2-fold higher when at least one CAST/EiJ haplotype is present ( C.I. , adjusted ), and there is no difference between males and females (1.2-fold higher expression in females, C.I. , adjusted ) (Figure 8A). In reciprocal F1 hybrids between CAST/EiJ and either PWK/PhJ or WSB/EiJ, there is no apparent effect of maternal vs. paternal inheritance of the CAST/EiJ haplotype on Mid1 expression (1.3-fold higher when maternally inherited, C.I. , adjusted ).
Figure 8.
Expression of pseudoautosomal gene Mid1 in F1 hybrids. (A) Total expression in adult whole brains, quantified as transcripts per million (TPM), in all nine possible reciprocal crosses between CAST/EiJ, PWK/PhJ, and WSB/EiJ. Group assignments for several tests of differential expression are shown as color-coded dots beneath the x-axis, with corresponding adjusted P-values at right. (B) Aggregate allele-specific read depth over Ensembl-annotated exons of Mid1 (transcript ENSMUST00000112107) in F1 females (top row) and males (bottom row), with a CAST/EiJ X or Y chromosome. Maternal reads are shown above the horizontal axis and paternal reads below, shaded according to strain origin (green, CAST/EiJ; red, PWK/PhJ; and purple, WSB/EiJ). Vertical axis truncated at ± 200. (C) Transcript isoforms assembled from RNA sequencing read alignments in the parental strains compared to Mid1 transcript models from Ensembl, plotted in genomic coordinates. Gray dashed line shows canonical pseudoautosomal region boundary. (D) Same transcripts as (C), but drawn with introns compressed, exons to scale, and vertically aligned to facilitate comparison of splice sites. Coding region of Ensembl transcripts shown as tall boxes. CAST/EiJ splicing differences in exons corresponding to canonical exons 4 and 5 emphasized by light gray-shaded box.
Next, we used known sequence variants to assign reads to parental haplotypes, and examined coverage on the maternal and paternal alleles in males and females from reciprocal F1 hybrids with a CAST/EiJ parent (Figure 8B). Reads from biological replicates were pooled for improved visualization. In females, Mid1 is transcribed from both X chromosomes. In males, transcription of the exons in the canonical PAR (exons 4–10, gray boxes) occurs on both X and Y. This 3′ fragment of the gene does not encode an ORF, so the roles of the Y-linked transcripts are unclear. Transcription of the full-length transcript, including the 5′ fragment proximal to the PAR boundary (exons 1–3, black boxes), occurs only from the X, unless a CAST/EiJ Y chromosome is present. Depth of coverage is qualitatively more heterogeneous across exons on the CAST/EiJ haplotype relative to the other two parental haplotypes.
Finally, we learned transcript models from read alignments in the parental strains and compared them to existing annotations in Ensembl (Figure 8, C and D). Transcripts containing the full-length coding sequence of the MID1 protein were recovered from PWK/PhJ and WSB/EiJ, but not from CAST/EiJ, despite having nearly reads aligned in the Mid1 locus in that strain. Although we observed hundreds of reads originating from exons 6–10 in CAST/EiJ (Figure 8B), none had spliced alignments across the junction between exons 5 and 6, nor between 7 and 8. Several CAST/EiJ isoforms have a 3′ extension of exon 4 and a 5′ extension of exon 5 (gray highlighted box in Figure 8D); the coding potential of these isoforms is uncertain. The difficulty of assembling full-length transcripts in CAST/EiJ may be due to mapping artifacts if the organization of this portion of the PAR is divergent from the reference genome. Moreover, it is difficult to assess the accuracy of our transcript assembly in the presence of abundant copy-number variation of segments that include exons 4–10 (next section).
Extreme structural diversity at the pseudoautosomal boundary
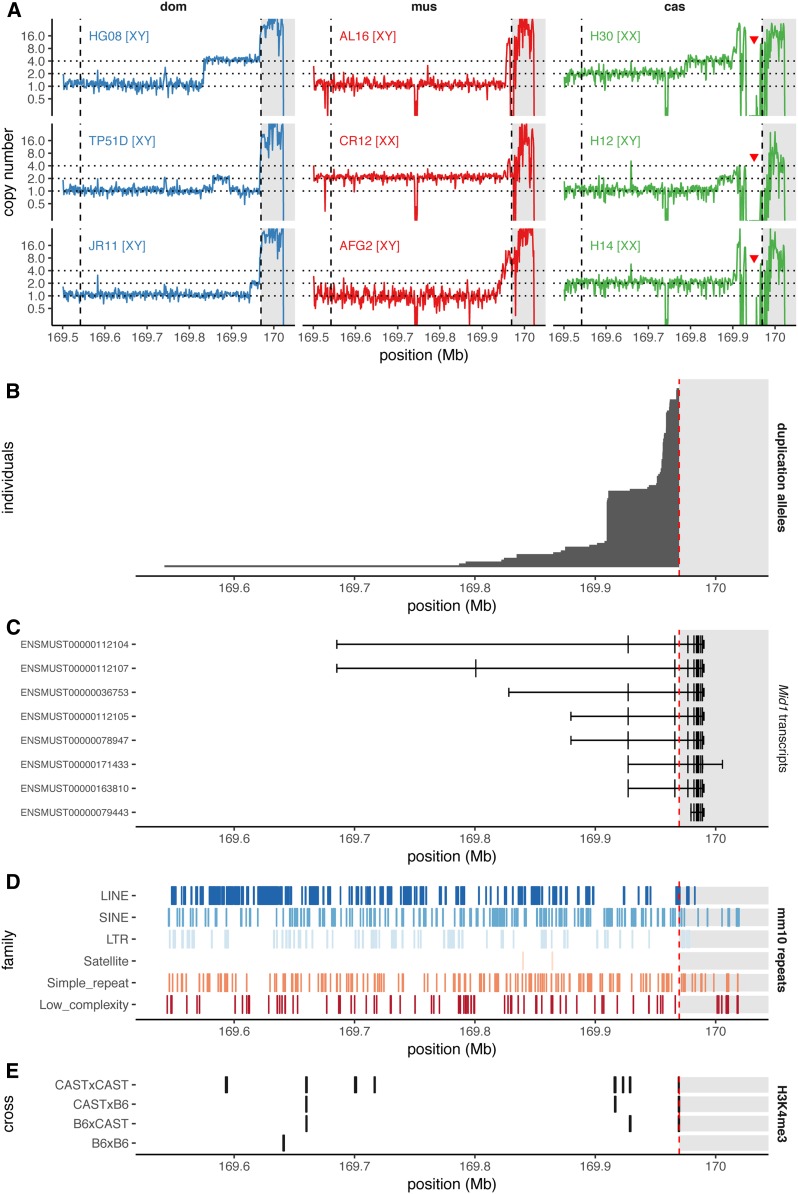
Inspection of sequencing data from 67 wild mice revealed abundant copy-number variation in the 1 Mb adjacent to the PAR boundary as well as within the canonical PAR itself (Figure 9A and Figure S8). Nominal copy number varies from zero to > 20 for some segments, and individual mice may have four or more distinct copy-number states across the region. As for the PWK/PhJ allele, we sought to identify breakpoints of copy gains and losses by manual review of read alignments in the IGV for 38 mice with evidence of CNVs in the distal X chromosome (excluding the canonical PAR). Clusters of three or more reads with partial alignments terminating at the same base pair in the mm10 reference were deemed sufficient evidence for a breakpoint. This analysis yielded 32 distinct breakpoints within 200 kb of the PAR boundary, none of which are shared across subspecies. Their spatial distribution appears nonrandom: their density seems to decrease moving outward from the PAR boundary (Figure 9B and File S11). However, tests for enrichment of breakpoints in proximity to exons and repetitive elements in the reference genome, and known recombination hotspots, all returned null results (cf. Figure 9, C–E). The density of repetitive elements in the 500 kb of sequence proximal to the pseudoautosomal boundary is not different from the subtelomeric regions of the autosomes (mean , SD ). The previously described ∼466-kb expansion of the PAR in the CAST/EiJ strain (White et al. 2012a), of predominantly M. m. castaneus ancestry and derived from founders trapped in Thailand, is not found in wild M. m. castaneus from northern India. The allele may be confined to southeast Asia. Several large deletions adjacent to and possibly involving the canonical PAR boundary that were also identified in CAST/EiJ appear fixed in the Indian population (red arrowheads in Figure 9A).
Figure 9.
Abundant copy-number variation at the pseudoautosomal boundary in wild house mice. (A) Copy number in the distal X chromosome, estimated from whole-genome sequencing read depth in 1-kb bins, for representative individuals of M. m. domesticus (dom, blue), M. m. musculus (mus, red), and M. m. castaneus (cas, green). Inferred sex-chromosome karyotype is shown for each individual. Dashed lines show the boundaries of the CAST/EiJ extended and canonical pseudoautosomal regions (PARs), respectively. Canonical PAR is shaded gray. Deletions adjacent to the PAR boundary in M. m. castaneus are indicated with red arrowheads. (B) Distinct copy-number alleles ascertained from breakpoint-spanning reads in 67 wild mice. Each allele is represented as a rectangle extending from the proximal breakpoint up to the pseudoautosomal boundary, and is shown in proportion to the number of times it is observed in our sample. (C) Transcripts of the Mid1 gene from Ensembl release 96. (D) Repeat elements annotated in the mm10 reference by the University of California Santa Cruz Genome Browser. (E) Recombination hotspots ascertained in C57BL/6J (B6), CAST/EiJ (CAST), and F1 males (Baker et al. 2015).
To gain some insight into the evolutionary trajectory of the PAR in the genus Mus, we estimated copy number in the distal X chromosome and the canonical PAR in eight M. spretus, one M. spicilegus, and one M. caroli. Both males and females have the expected copy number for the X chromosome in the vicinity of the PAR boundary (Figure S9). Within the most proximal region of the canonical PAR, from the PAR boundary to just past the eighth exon of Mid1 (chromosome X: 169986134), males have one copy and females two copies, suggesting that the region is not pseudoautosomal in these species [similarly, no evidence of heterozygosity in the proximal exons of Mid1 was found in whole-exome data from wild-caught male representatives of more distant outgroups M. cookii and Nannomys minutoides (Sarver et al. 2017); data not shown]. This agrees with previous findings that the PCR marker DXYCbl1, whose sequence aligns at chromosome X: 169974684–169974964, is X-unique in offspring of (C57BL/6JxSPRET/Ei)F1 males (Breen et al. 1994; Perry et al. 2001). Given the conservation of copy number on either side of the PAR boundary in three outgroup species, we conclude that the duplications observed in each M. musculus subspecies are derived alleles. Without pedigree data, we cannot be certain of X-, Y-, or pseudoautosomal linkage for these duplications, but similarity to the pattern we observe for the PWK/PhJ allele, which itself segregates in the wild, strongly suggests that at least some involve the pseudoautosomal boundary.
Discussion
Here, we combine crosses involving intersubspecific hybrid males with whole-genome sequencing data from wild mice to clarify and extend previous observations on the lability of the mouse PAR. We show that the previously described extension of the PAR in M. m. castaneus-derived CAST/EiJ—that is, translocation of X-unique sequence onto the Y chromosome—undergoes crossing-over in heterozygous males at a nearly 100-fold-higher rate than autosomal sequences. Our estimate of the recombination rate in the extended PAR ( cM/Mb) agrees with a recent estimate from whole-genome sequencing of single recombinant sperm (24 cM/Mb) from a C57BL/6J CAST/EiJ male (Hinch et al. 2019). The nominal marker order in the reference genome is supported by haplotypes reconstructed for recombinant progeny. Nonetheless, we observe several double recombinants within <500 kb, consistent with prior observations that double recombinants are common in the canonical PAR (Soriano et al. 1987; Pardo-Manuel de Villena and Sapienza 1996).
We further confirm that a shorter extension of the PAR has occurred in PWK/PhJ (White et al. 2012a). Unlike the CAST/EiJ allele, the X- and Y-linked copies in PWK/PhJ are divergent in sequence and structure. This suggests that they do not recombine freely in the inbred strain, although we find that they do recombine in intersubspecific hybrid males. The X-linked copy is of M. m. domesticus ancestry, evidence of either introgression in the wild or contamination in a laboratory colony, while the Y-linked copy comes from M. m. musculus and segregates in the wild in the Czech Republic.
In fact, the PWK/PhJ Y-linked allele is just one of numerous structural variants at the PAR boundary that segregate in natural populations of all three subspecies. All of these are derived alleles: both the PAR boundary and a proximal segment of the canonical PAR are single-copy in M. spretus, M. spicilegus, and M. caroli, and therefore most likely were single-copy in the common Mus ancestor. A summary of the major evolutionary events at the pseudoautosomal boundary in Mus is provided in Figure 10. Using breakpoint-spanning reads to uniquely identify structural alleles, we find ≥32 alleles in a sample of 67 mice. None of them are shared across subspecies. The unusual level of standing variation suggests that the mutation rate near the PAR boundary is quite high. Indeed, we recover a de novo deletion on the PWK/PhJ Y haplotype in a sample of only 86 mice, for an estimated mutation rate of per generation ( Poisson C.I. ). To our surprise, none of these rearrangements seem to prevent robust expression of an essential gene (Mid1) that flanks the pseudoautosomal boundary [Dal Zotto et al. (1998) and Figure 8].
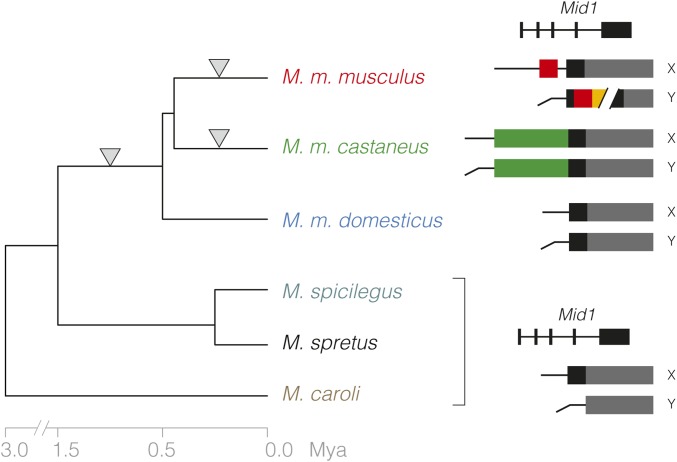
Figure 10.
Changes in the pseudoautosomal boundary mapped onto the Mus phylogeny. Ancestral pseudoautosomal region (PAR), including unassembled repetitive regions, represented in gray. X-unique sequence translocated to the Y, expanding the PAR, shown as colored blocks. There are now at least three independent alterations of the boundary and/or organization of the PAR (inverted triangles): one in the ancestor of M. musculus (after the divergence from the M. spretus–M. spicilegus clade), expanding the PAR to include exons 4–8 of Mid1; the previously documented expansion in CAST/EiJ; and the rearrangement we describe in PWK/PhJ. Approximate divergence times are shown below the tree, and schematics of the organization of each allele at right.
Taken together, our results indicate that the peculiar properties of the PAR in male meiosis can “leak” across the PAR boundary. This is consistent with previous evidence that the regime governing DSB formation in the PAR and near the PAR boundary is distinct from that on autosomes. DSBs are formed later in meiosis in the PAR than on autosomes (Kauppi et al. 2011), at far higher density (Lange et al. 2016), by a distinct isoform of the SPO11 protein (α rather than β) (Keeney et al. 1997; Bellani et al. 2010; Kauppi et al. 2011; Smagulova et al. 2013; Faisal and Kauppi 2016), independently of PRDM9 (at least in mice) (Brick et al. 2012), and along a proportionately longer chromosomal axis (Acquaviva et al. 2019; Papanikos et al. 2019). It has recently been shown that DSBs in the canonical PAR are formed preferentially at minisatellite arrays, and that it is the chromatin configuration of the repeat-rich sequence, rather than the sequence per se, that is essential for X–Y recombination (Acquaviva et al. 2019). It seems plausible that any nearby sequence drawn into the protein–DNA complex at the chromosomal axis could be susceptible to unequal exchange. Once the PAR is extended by transposition of sequence from the X onto the Y chromosome, the transposed region is subject to PAR-like levels of recombination, despite not sharing the sequence features of the canonical PAR. It follows that these sequence features (such as elevated GC content) are indeed a consequence rather than a cause of the PAR’s high recombination rate in males.
Our findings also support the notion that while the requirement for X–Y pairing in the first meiotic prophase is strictly enforced, the degree of structural homology in the PAR necessary to achieve pairing is relatively weak in mice. It has been known for several decades that failure to pair the sex chromosomes is associated with meiotic arrest in interspecific (Matsuda et al. 1992; Hale et al. 1993) and intersubspecific mouse hybrids (Forejt 1996). The unpaired sex chromosomes do not undergo meiotic sex-chromosome inactivation (Royo et al. 2010), and misexpression of specific X- and Y-linked genes is toxic to germ cells (Good et al. 2010; Vernet et al. 2011; Campbell et al. 2013). However, absence of X–Y pairing in sterile hybrids occurs in the context of widespread asynapsis of both autosomes and sex chromosomes (Bhattacharyya et al. 2013). More direct evidence of the requirement for recombination in the PAR comes from Spo11 (Keeney et al. 1997; Bellani et al. 2010; Kauppi et al. 2011; Smagulova et al. 2013; Faisal and Kauppi 2016) and Ankrd31 (Boekhout et al. 2019; Papanikos et al. 2019) mutants, in which males form crossovers in the autosomes but not the PAR and are sterile. The best evidence for a specific role of PAR structural divergence in infertility comes from an experiment by White et al. (2012b): F2 males heterozygous for a WSB/EiJ and a CAST/EiJ haplotype in the PAR have a variety of spermatogenic defects, independent of genotypes at other loci.
However, Dumont (2017) has shown that, although the number of X–Y crossovers (assayed by immunostaining against the MLH1 protein) in intersubspecific F1 hybrid males is reduced relative to the parental strains, an absence of stable X–Y pairing is pervasive in both inbreds and F1s. Failure to pair the sex chromosomes is associated with meiotic arrest and subsequent apoptosis of ≤ of spermatocytes in some intersubspecific hybrids (Matsuda et al. 1992), but also ≤ 25% of spermatocytes in some inbred strains (Imai et al. 1981; Krzanowska 1989). Even this level of X–Y dissociation is compatible with fertility, at least under laboratory conditions. Two Y-chromosome variants with large extensions of the PAR—the Yaa allele, a 4-Mb proximal extension of the PAR on the SB/Le background (Hudgins et al. 1985; Subramanian et al. 2006), and the Y* allele, a PAR-to-PAR fusion of a Y and an X chromosome (Eicher et al. 1991)—are readily propagated in laboratory colonies. Given these observations and the number of segregating PAR-boundary alleles we observe in natural populations, it is reasonable to conclude that the organization of the region is not under strong purifying selection in house mice. However, we stress that our study provides limited direct information on the fitness consequences of X–Y exchanges in the PAR in intersubspecific hybrids. These events are compatible with viability through at least the neonatal period (when progeny were killed for genotyping), but details of their effects on male and female fertility are unknown.
Our study has several further limitations. First, all of our analyses, whether of array genotypes or whole-genome sequence data, are predicated on the representation of the PAR in the mm10 reference genome. The assembly of the PAR has proven difficult because of its repetitive content, and is thus fragmented and incomplete. The PAR of C57BL/6J is ∼750-kb long by restriction mapping (Perry et al. 2001) but has only 161 kb of nongap bases in mm10. We are blind to any sequence that does not have at least partial representation in the reference assembly. It is extremely difficult, and in many cases impossible, to accurately reconstruct complex structural variants from short-read data alone. The noisy alignment patterns in the canonical PAR indicate that its organization is much more complex than the reference assembly would suggest, but we can say little more than that. Detailed characterization of repetitive loci in mammalian genomes (e.g., Soh et al. 2014; Cantsilieris et al. 2018; Lilue et al. 2018) with orthogonal technologies, including long-read single-molecule sequencing and high-throughput physical mapping, has shown that the view from short reads may be a faint shadow of the truth. Despite its technical limitations, our study provides a useful foundation for future efforts to characterize the sequence, structure, and function of this enigmatic region of the genome.
Acknowledgments
The authors thank Grace Clark, Justin Gooch, the late Mark Calaway, and Darla Miller for assistance with DNA preparation and submission of samples for genotyping; Randy Nonneman for assistance with mouse husbandry; and Chris Baker, Kevin Brick, and Fatima Smagulova for their generous technical assistance in reanalyzing their published ChIPseq data. The authors benefited from fruitful correspondence with Beth Dumont while preparing the manuscript. This work was supported in part by National Institutes of Health grants F30 MH-103925 (A.P.M.), R01 HD-065024 (F.P.-M.d.V.), R21 MH-096261 (F.P.-M.d.V.), K01 MH-094406 (J.J.C.), P50 MH-090338 (F.P.-M.d.V.), P50 HG-006582 (F.P.-M.d.V.), and U42 OD-010924 (Terry Magnuson).
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.7798889.
Communicating editor: J. Schimenti
Literature Cited
- Acquaviva L., Boekhout M., Karasu M. E., Brick K., Pratto F., et al. , 2019. Ensuring meiotic DNA break formation in the mouse pseudoautosomal region. bioRxiv. Available at https//.org/10.1101/536136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur R., Schulz-Trieglaff O., Cox A. J., O’Connell J., 2017. AKT: ancestry and kinship toolkit. Bioinformatics 33: 142–144. 10.1093/bioinformatics/btw576 [DOI] [PubMed] [Google Scholar]
- Ashley T., Moses M. J., 1980. End association and segregation of the achiasmatic X and Y chromosomes of the sand rat, Psammomys obesus. Chromosoma 78: 203–210. 10.1007/BF00328392 [DOI] [PubMed] [Google Scholar]
- Baker C. L., Kajita S., Walker M., Saxl R. L., Raghupathy N., et al. , 2015. PRDM9 drives evolutionary erosion of hotspots in Mus musculus through Haplotype-Specific initiation of meiotic recombination. PLoS Genet. 11: e1004916 10.1371/journal.pgen.1004916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., et al. , 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19: 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett D. W., Garrison E. K., Quinlan A. R., Strömberg M. P., Marth G. T., 2011. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 27: 1691–1692. 10.1093/bioinformatics/btr174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellani M. A., Boateng K. A., McLeod D., Camerini-Otero R. D., 2010. The expression profile of the major mouse SPO11 isoforms indicates that SPO11beta introduces double strand breaks and suggests that SPO11alpha has an additional role in prophase in both spermatocytes and oocytes. Mol. Cell. Biol. 30: 4391–4403. 10.1128/MCB.00002-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellott D. W., Hughes J. F., Skaletsky H., Brown L. G., Pyntikova T., et al. , 2014. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508: 494–499 (erratum: Nature 514: 126) 10.1038/nature13206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya T., Gregorova S., Mihola O., Anger M., Sebestova J., et al. , 2013. Mechanistic basis of infertility of mouse intersubspecific hybrids. Proc. Natl. Acad. Sci. USA 110: E468–E477. 10.1073/pnas.1219126110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekhout M., Karasu M. E., Wang J., Acquaviva L., Pratto F., et al. , 2019. REC114 partner ANKRD31 controls number, timing, and location of meiotic DNA breaks. Mol. Cell. S1097–2765: 30227-8. 10.1016/j.molcel.2019.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N. L., Pimentel H., Melsted P., Pachter L., 2016. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34: 525–527 (erratum: Nat. Biotechnol. 34: 888) 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Breen M., Deakln L., Macdonald B., Miller S., Sibson R., et al. , 1994. Towards high resolution maps of the mouse and human genomes--a facility for ordering markers to 0.1 cM resolution. European Backcross Collaborative Group. Hum. Mol. Genet. 3: 621–627. 10.1093/hmg/3.4.621 [DOI] [PubMed] [Google Scholar]
- Brick K., Smagulova F., Khil P., Camerini-Otero R. D., Petukhova G. V., 2012. Genetic recombination is directed away from functional genomic elements in mice. Nature 485: 642–645. 10.1038/nature11089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne P. S., 1982. Genetic homology and crossing over in the X and Y chromosomes of mammals. Hum. Genet. 61: 85–90. 10.1007/BF00274192 [DOI] [PubMed] [Google Scholar]
- Burgoyne P. S., Mahadevaiah S. K., Sutcliffe M. J., Palmer S. J., 1992. Fertility in mice requires X-Y pairing and a y-chromosomal “spermiogenesis” gene mapping to the long arm. Cell 71: 391–398. 10.1016/0092-8674(92)90509-B [DOI] [PubMed] [Google Scholar]
- Campbell P., Good J. M., Nachman M. W., 2013. Meiotic sex chromosome inactivation is disrupted in sterile hybrid male house mice. Genetics 193: 819–828. 10.1534/genetics.112.148635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantsilieris S., Nelson B. J., Huddleston J., Baker C., Harshman L., et al. , 2018. Recurrent structural variation, clustered sites of selection, and disease risk for the complement factor H (CFH) gene family. Proc. Natl. Acad. Sci. USA 115: E4433–E4442. 10.1073/pnas.1717600115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnero A., Jiménez R., Burgos M., Sánchez A., Díaz de la Guardia R., 1991. Achiasmatic sex chromosomes in Pitymys duodecimcostatus: mechanisms of association and segregation. Cytogenet. Cell Genet. 56: 78–81. 10.1159/000133054 [DOI] [PubMed] [Google Scholar]
- Charchar F. J., Svartman M., El-Mogharbel N., Ventura M., Kirby P., et al. , 2003. Complex events in the evolution of the human pseudoautosomal region 2 (PAR2). Genome Res. 13: 281–286. 10.1101/gr.390503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter D. J., Brotman S. M., Wilson Sayres M. A., 2016. Genetic diversity on the human X chromosome does not support a strict pseudoautosomal boundary. Genetics 203: 485–492. 10.1534/genetics.114.172692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley J. J., Zhabotynsky V., Sun W., Huang S., Pakatci I. K., et al. , 2015. Analyses of allele-specific gene expression in highly divergent mouse crosses identifies pervasive allelic imbalance. Nat. Genet. 47: 353–360 (erratum: Nat. Genet. 47: 690) 10.1038/ng.3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Zotto L., Quaderi N. A., Elliott R., Lingerfelter P. A., Carrel L., et al. , 1998. The mouse mid1 gene: implications for the pathogenesis of Opitz syndrome and the evolution of the mammalian pseudoautosomal region. Hum. Mol. Genet. 7: 489–499. 10.1093/hmg/7.3.489 [DOI] [PubMed] [Google Scholar]
- Das P. J., Chowdhary B. P., Raudsepp T., 2009. Characterization of the bovine pseudoautosomal region and comparison with sheep, goat, and other mammalian pseudoautosomal regions. Cytogenet. Genome Res. 126: 139–147. 10.1159/000245913 [DOI] [PubMed] [Google Scholar]
- Didion J. P., Morgan A. P., Yadgary L., Bell T. A., McMullan R. C., et al. , 2016. R2d2 drives selfish sweeps in the house mouse. Mol. Biol. Evol. 33: 1381–1395. 10.1093/molbev/msw036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., et al. , 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont B. L., 2017. Meiotic consequences of genetic divergence across the murine pseudoautosomal region. Genetics 205: 1089–1100. 10.1534/genetics.116.189092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont B. L., Payseur B. A., 2011a. Evolution of the genomic recombination rate in murid rodents. Genetics 187: 643–657. 10.1534/genetics.110.123851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont B. L., Payseur B. A., 2011b. Genetic analysis of Genome-Scale recombination rate evolution in house mice. PLoS Genet. 7: e1002116 10.1371/journal.pgen.1002116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicher E. M., Hale D. W., Hunt P. A., Lee B. K., Tucker P. K., et al. , 1991. The mouse y* chromosome involves a complex rearrangement, including interstitial positioning of the pseudoautosomal region. Cytogenet. Cell Genet. 57: 221–230. 10.1159/000133152 [DOI] [PubMed] [Google Scholar]
- Ellis N., Yen P., Neiswanger K., Shapiro L. J., Goodfellow P. N., 1990. Evolution of the pseudoautosomal boundary in old world monkeys and great apes. Cell 63: 977–986. 10.1016/0092-8674(90)90501-5 [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., 1993. Recombination and mammalian genome evolution. Proc. Biol. Sci. 252: 237–243. 10.1098/rspb.1993.0071 [DOI] [PubMed] [Google Scholar]
- Faisal I., Kauppi L., 2016. Sex chromosome recombination failure, apoptosis, and fertility in male mice. Chromosoma 125: 227–235. 10.1007/s00412-015-0542-9 [DOI] [PubMed] [Google Scholar]
- Faust G. G., Hall I. M., 2014. SAMBLASTER fast duplicate marking and structural variant read extraction. Bioinformatics 30: 2503–2505. 10.1093/bioinformatics/btu314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov D. A., Gerrard D. T., 2003. High mutation rates in human and ape pseudoautosomal genes. Gene 317: 67–77. 10.1016/S0378-1119(03)00697-8 [DOI] [PubMed] [Google Scholar]
- Forejt J., 1996. Hybrid sterility in the mouse. Trends Genet. 12: 412–417. 10.1016/0168-9525(96)10040-8 [DOI] [PubMed] [Google Scholar]
- Fraley C., Raftery A. E., 2002. Model-Based clustering, discriminant analysis, and density estimation. J. Am. Stat. Assoc. 97: 611–631. 10.1198/016214502760047131 [DOI] [Google Scholar]
- Geraldes A., Basset P., Gibson B., Smith K. L., Harr B., et al. , 2008. Inferring the history of speciation in house mice from autosomal, x–linked, y-linked and mitochondrial genes. Mol. Ecol. 17: 5349–5363. 10.1111/j.1365-294X.2008.04005.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good J. M., Giger T., Dean M. D., Nachman M. W., 2010. Widespread Over-Expression of the X chromosome in sterile F1 hybrid mice. PLoS Genet. 6: e1001148 10.1371/journal.pgen.1001148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves J. A., 2006. Sex chromosome specialization and degeneration in mammals. Cell 124: 901–914. 10.1016/j.cell.2006.02.024 [DOI] [PubMed] [Google Scholar]
- Graves J. A., Watson J. M., 1991. Mammalian sex chromosomes: evolution of organization and function. Chromosoma 101: 63–68. 10.1007/BF00357055 [DOI] [PubMed] [Google Scholar]
- Graves J. A., Disteche C. M., Toder R., 1998. Gene dosage in the evolution and function of mammalian sex chromosomes. Cytogenet. Cell Genet. 80: 94–103. 10.1159/000014963 [DOI] [PubMed] [Google Scholar]
- Green M. R., Sambrook J., 2012. Molecular Cloning: A Laboratory Manual (Fourth Edition): Three-volume set, Ed. 4 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Gregorová S., Forejt J., 2000. PWD/Ph and PWK/Ph inbred mouse strains of Mus m. musculus subspecies--a valuable resource of phenotypic variations and genomic polymorphisms. Folia Biol. (Praha) 46: 31–41. [PubMed] [Google Scholar]
- Hale D. W., Washburn L. L., Eicher E. M., 1993. Meiotic abnormalities in hybrid mice of the C57BL/6J x Mus spretus cross suggest a cytogenetic basis for Haldane’s rule of hybrid sterility. Cytogenet. Cell Genet. 63: 221–234. 10.1159/000133539 [DOI] [PubMed] [Google Scholar]
- Harr B., Karakoc E., Neme R., Teschke M., Pfeifle C., et al. , 2016. Genomic resources for wild populations of the house mouse, Mus musculus and its close relative Mus spretus. Sci. Data 3: 160075 10.1038/sdata.2016.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T. J., Sherman S. L., Pettay D., Page D. C., Jacobs P. A., 1991. XY chromosome nondisjunction in man is associated with diminished recombination in the pseudoautosomal region. Am. J. Hum. Genet. 49: 253–260. [PMC free article] [PubMed] [Google Scholar]
- Hellmann I., Ebersberger I., Ptak S. E., Pääbo S., Przeworski M., 2003. A neutral explanation for the correlation of diversity with recombination rates in humans. Am. J. Hum. Genet. 72: 1527–1535. 10.1086/375657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinch A. G., Altemose N., Noor N., Donnelly P., Myers S. R., 2014. Recombination in the human pseudoautosomal region PAR1. PLoS Genet. 10: e1004503 10.1371/journal.pgen.1004503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinch A. G., Zhang G., Becker P. W., Moralli D., Hinch R., et al. , 2019. Factors influencing meiotic recombination revealed by whole-genome sequencing of single sperm. Science 363: eaau8861 10.1126/science.aau8861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.-W., Friedman R., Yu N., Yu A., Li W.-H., 2005. How strong is the mutagenicity of recombination in mammals? Mol. Biol. Evol. 22: 426–431. 10.1093/molbev/msi025 [DOI] [PubMed] [Google Scholar]
- Hudgins C. C., Steinberg R. T., Klinman D. M., Reeves M. J., Steinberg A. D., 1985. Studies of consomic mice bearing the Y chromosome of the BXSB mouse. J. Immunol. 134: 3849–3854. [PubMed] [Google Scholar]
- Imai H. T., Matsuda Y., Shiroishi T., Moriwaki K., 1981. High frequency fo X-Y chromosome dissociation in primary spermatocytes of F1 hybrids between Japanese wild mice (Mus musculus molossinus) and inbred laboratory mice. Cytogenet. Cell Genet. 29: 166–175. 10.1159/000131565 [DOI] [PubMed] [Google Scholar]
- Kasahara T., Abe K., Mekada K., Yoshiki A., Kato T., 2010. Genetic variation of melatonin productivity in laboratory mice under domestication. Proc. Natl. Acad. Sci. USA 107: 6412–6417. 10.1073/pnas.0914399107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L., Barchi M., Baudat F., Romanienko P. J., Keeney S., et al. , 2011. Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science 331: 916–920. 10.1126/science.1195774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane T. M., Goodstadt L., Danecek P., White M. A., Wong K., et al. , 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294. 10.1038/nature10413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S., Giroux C. N., Kleckner N., 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. 10.1016/S0092-8674(00)81876-0 [DOI] [PubMed] [Google Scholar]
- Kent W. J., 2002. BLAT–the BLAST-like alignment tool. Genome Res. 12: 656–664. 10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipling D., Salido E. C., Shapiro L. J., Cooke H. J., 1996a. High frequency de novo alterations in the long–range genomic structure of the mouse pseudoautosomal region. Nat. Genet. 13: 78–82. 10.1038/ng0596-78 [DOI] [PubMed] [Google Scholar]
- Kipling D., Wilson H. E., Thomson E. J., Lee M., Perry J., et al. , 1996b. Structural variation of the pseudoautosomal region between and within inbred mouse strains. Proc. Natl. Acad. Sci. USA 93: 171–175. 10.1073/pnas.93.1.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzanowska H., 1989. X-Y chromosome dissociation in mouse strains differing in efficiency of spermatogenesis: elevated frequency of univalents in pubertal males. Gamete Res. 23: 357–365. 10.1002/mrd.1120230312 [DOI] [PubMed] [Google Scholar]
- Lamb B. C., 1984. The properties of meiotic gene conversion important in its effects on evolution. Heredity 53: 113–138. 10.1038/hdy.1984.68 [DOI] [PubMed] [Google Scholar]
- Lange J., Yamada S., Tischfield S. E., Pan J., Kim S., et al. , 2016. The landscape of mouse meiotic Double-Strand break formation, processing, and repair. Cell 167: 695–708.e16. 10.1016/j.cell.2016.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., 2013 Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: 1303.3997 [q-bio].
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilue J., Doran A. G., Fiddes I. T., Abrudan M., Armstrong J., et al. , 2018. Sixteen diverse laboratory mouse reference genomes define strain-specific haplotypes and novel functional loci. Nat. Genet. 50: 1574–1583. 10.1038/s41588-018-0223-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T., Chen R., Cox T. C., Moldrich R. X., Kurniawan N., et al. , 2013. X-linked microtubule-associated protein, mid1, regulates axon development. Proc. Natl. Acad. Sci. USA 110: 19131–19136. 10.1073/pnas.1303687110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather K., 1938. CROSSING-OVER. Biol. Rev. Camb. Philos. Soc. 13: 252–292. 10.1111/j.1469-185X.1938.tb00516.x [DOI] [Google Scholar]
- Matsuda Y., Hirobe T., Chapman V. M., 1991. Genetic basis of X-Y chromosome dissociation and male sterility in interspecific hybrids. Proc. Natl. Acad. Sci. USA 88: 4850–4854. 10.1073/pnas.88.11.4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y., Moens P. B., Chapman V. M., 1992. Deficiency of X and Y chromosomal pairing at meiotic prophase in spermatocytes of sterile interspecific hybrids between laboratory mice (Mus domesticus) and Mus spretus. Chromosoma 101: 483–492. 10.1007/BF00352471 [DOI] [PubMed] [Google Scholar]
- Mensah M. A., Hestand M. S., Larmuseau M. H. D., Isrie M., Vanderheyden N., et al. , 2014. Pseudoautosomal region 1 length polymorphism in the human population. PLoS Genet. 10: e1004578 (erratum: PLoS Genet. 10: e1004889) 10.1371/journal.pgen.1004578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaghaderi G., Hörandl E., 2016. The evolution of meiotic sex and its alternatives. Proc. Biol. Sci. 283: 20161221 10.1098/rspb.2016.1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. P., 2016. argyle: an R package for analysis of Illumina genotyping arrays. G3 (Bethesda) 6: 281–286. 10.1534/g3.115.023739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. P., Pardo-Manuel de Villena F., 2017. Sequence and structural diversity of mouse Y chromosomes. Mol. Biol. Evol. 34: 3186–3204. 10.1093/molbev/msx250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. P., Fu C.-P., Kao C.-Y., Welsh C. E., Didion J. P., et al. , 2016. The mouse universal genotyping array: from substrains to subspecies. G3 (Bethesda) 6: 263–279. 10.1534/g3.115.022087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neme R., Tautz D., 2016. Fast turnover of genome transcription across evolutionary time exposes entire non-coding DNA to de novo gene emergence. Elife 5: e09977 10.7554/eLife.09977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S., 1966. Sex Chromosomes and Sex-Linked Genes, Springer-Verlag, Berlin: 10.1007/978-3-662-35113-0 [DOI] [Google Scholar]
- Page J., Berríos S., Parra M. T., Viera A., Suja J. A., et al. , 2005. The program of sex chromosome pairing in meiosis is highly conserved across marsupial species: implications for sex chromosome evolution. Genetics 170: 793–799. 10.1534/genetics.104.039073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S., Perry J., Kipling D., Ashworth A., 1997. A gene spans the pseudoautosomal boundary in mice. Proc. Natl. Acad. Sci. USA 94: 12030–12035. 10.1073/pnas.94.22.12030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanikos F., Clément J. A. J., Testa E., Ravindranathan R., Grey C., et al. , 2019. Mouse ANKRD31 regulates spatiotemporal patterning of meiotic recombination initiation and ensures recombination between X and Y sex chromosomes. Mol. Cell. S1097–2765: 30226–6. 10.1016/j.molcel.2019.03.022 [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F., Sapienza C., 1996. Genetic mapping of DXYMov15-associated sequences in the pseudoautosomal region of the C57BL/6J strain. Mamm. Genome 7: 237–239. 10.1007/s003359900067 [DOI] [PubMed] [Google Scholar]
- Perry J., Ashworth A., 1999. Evolutionary rate of a gene affected by chromosomal position. Curr. Biol. 9: 987–989. 10.1016/S0960-9822(99)80430-8 [DOI] [PubMed] [Google Scholar]
- Perry J., Palmer S., Gabriel A., Ashworth A., 2001. A short pseudoautosomal region in laboratory mice. Genome Res. 11: 1826–1832. 10.1101/gr.203001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Pertea G. M., Antonescu C. M., Chang T.-C., Mendell J. T., et al. , 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33: 290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poriswanish N., Neumann R., Wetton J. H., Wagstaff J., Larmuseau M. H. D., et al. , 2018. Recombination hotspots in an extended human pseudoautosomal domain predicted from double-strand break maps and characterized by sperm-based crossover analysis. PLoS Genet. 14: e1007680 10.1371/journal.pgen.1007680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudsepp T., Chowdhary B. P., 2015. The eutherian pseudoautosomal region. CGR 147: 81–94. [DOI] [PubMed] [Google Scholar]
- Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., et al. , 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43: e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., et al. , 2011. Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouyer F., Simmler M. C., Johnsson C., Vergnaud G., Cooke H. J., et al. , 1986. A gradient of sex linkage in the pseudoautosomal region of the human sex chromosomes. Nature 319: 291–295. 10.1038/319291a0 [DOI] [PubMed] [Google Scholar]
- Royo H., Polikiewicz G., Mahadevaiah S. K., Prosser H., Mitchell M., et al. , 2010. Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr. Biol. 20: 2117–2123. 10.1016/j.cub.2010.11.010 [DOI] [PubMed] [Google Scholar]
- Sarver B. A. J., Keeble S., Cosart T., Tucker P. K., Dean M. D., et al. , 2017. Phylogenomic insights into mouse evolution using a pseudoreference approach. Genome Biol. Evol. 9: 726–739. 10.1093/gbe/evx034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebel K., Meder J., Rump A., Rosenthal A., Winkelmann M., et al. , 2000. Elevated DNA sequence diversity in the genomic region of the phosphatase PPP2R3L gene in the human pseudoautosomal region. Cytogenet. Cell Genet. 91: 224–230. 10.1159/000056849 [DOI] [PubMed] [Google Scholar]
- Sharman G. B., McIntosh A. J., and H. N. Barber, 1950. Multiple sex chromosomes in the marsupials. Nature 166: 996 10.1038/166996a0 [DOI] [PubMed] [Google Scholar]
- Sharp P., 1982. Sex chromosome pairing during male meiosis in marsupials. Chromosoma 86: 27–47. 10.1007/BF00330728 [DOI] [PubMed] [Google Scholar]
- Smagulova F., Brick K., Pu Y., Sengupta U., Camerini-Otero R. D., et al. , 2013. Suppression of genetic recombination in the pseudoautosomal region and at subtelomeres in mice with a hypomorphic spo11 allele. BMC Genomics 14: 493 10.1186/1471-2164-14-493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F., Brick K., Pu Y., Camerini-Otero R. D., Petukhova G. V., 2016. The evolutionary turnover of recombination hot spots contributes to speciation in mice. Genes Dev. 30: 266–280. 10.1101/gad.270009.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh Y. Q. S., Alföldi J., Pyntikova T., Brown L. G., Graves T., et al. , 2014. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell 159: 800–813. 10.1016/j.cell.2014.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C., Love M. I., Robinson M. D., 2015. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000 Res. 4: 1521 10.12688/f1000research.7563.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P., Keitges E. A., Schorderet D. F., Harbers K., Gartler S. M., et al. , 1987. High rate of recombination and double crossovers in the mouse pseudoautosomal region during male meiosis. Proc. Natl. Acad. Sci. USA 84: 7218–7220. 10.1073/pnas.84.20.7218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A., Morgan A. P., Najarian M. L., Sarsani V. K., Sigmon J. S., et al. , 2017. Genomes of the mouse collaborative cross. Genetics 206: 537–556. 10.1534/genetics.116.198838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steemers F. J., Chang W., Lee G., Barker D. L., Shen R., et al. , 2006. Whole-genome genotyping with the single-base extension assay. Nat. Methods 3: 31–33. 10.1038/nmeth842 [DOI] [PubMed] [Google Scholar]
- Subramanian S., Tus K., Li Q.-Z., Wang A., Tian X.-H., et al. , 2006. A tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc. Natl. Acad. Sci. USA 103: 9970–9975. 10.1073/pnas.0603912103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thybert D., Roller M., Navarro F. C. P., Fiddes I., Streeter I., et al. , 2018. Repeat associated mechanisms of genome evolution and function revealed by the Mus caroli and Mus pahari genomes. Genome Res. 28: 448–459. 10.1101/gr.234096.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet N., Mahadevaiah S. K., Ojarikre O. A., Longepied G., Prosser H. M., et al. , 2011. The y-encoded gene zfy2 acts to remove cells with unpaired chromosomes at the first meiotic metaphase in male mice. Curr. Biol. 21: 787–793. 10.1016/j.cub.2011.03.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. A., Ikeda A., Payseur B. A., 2012a. A pronounced evolutionary shift of the pseudoautosomal region boundary in house mice. Mamm. Genome 23: 454–466. 10.1007/s00335-012-9403-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. A., Stubbings M., Dumont B. L., Payseur B. A., 2012b. Genetics and evolution of hybrid male sterility in house mice. Genetics 191: 917–934. 10.1534/genetics.112.140251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K. W., Baumgart K., Winking H., 1988. Meiotic association and segregation of the achiasmatic giant sex chromosomes in the male field vole (Microtus agrestis). Chromosoma 97: 124–133. 10.1007/BF00327369 [DOI] [Google Scholar]
- Xu S., 2013. Principles of Statistical Genomics, Springer, New York: 10.1007/978-0-387-70807-2 [DOI] [Google Scholar]
- Yang H., Wang J. R., Didion J. P., Buus R. J., Bell T. A., et al. , 2011. Subspecific origin and haplotype diversity in the laboratory mouse. Nat. Genet. 43: 648–655. 10.1038/ng.847 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Whole-genome sequence data from N2 progeny have been submitted to the European Nucleotide Archive (accession PRJEB32247). The inferred genetic map for the extended PAR is in File S10; informative markers near the PAR boundary in File S2; assembled breakpoint sequences of the PWK/PhJ PAR variant in File S5; a list of N2 progeny used for recombination analysis in File S1; array genotype and intensity data, with marker locations, in File S3; and genotype calls in the PWK/PhJ PAR variant from the whole-genome sequence in File S7. Supporting code is available at https://github.com/andrewparkermorgan/mouse_pseudoautosomal_region. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.7798889.