Abstract.
Soil-transmitted helminth (STH) infections and malaria are parasitic diseases with enormous global health burdens. Research has demonstrated a relationship between each of these parasites and the gut microbiome, suggesting that the gut microbiota may be implicated in governing host susceptibility to diverse pathogens, and perhaps even coinfection by different pathogens, through similar microbiome-influenced pathways. Here, we have derived a first microbiome community profile associated with STH infections in Odisha, India, and tested the hypothesis that the gut microbiome can modulate host susceptibility to multiple parasite infections through the same pathways. This study revealed several bacterial taxa negatively associated with specific STH infections, including Lactobacillus and Lachnospiracaea. Our results also suggest that relative abundance of Lactobacillus is driven by the STH infection status more so than by the Plasmodium infection status. This study contributes to efforts to understand the effects of the microbiome on host susceptibility to parasitic infections in endemic communities.
The gut microbiome consists of trillions of bacteria, which exert powerful effects on host nutrition, development, and immunity.1,2 Dysbiotic microbiomes, often with reduced diversity, have been implicated in numerous chronic inflammatory conditions, including obesity, allergies, irritable bowel syndrome, and type I diabetes.3,4 Gut microbiomes are also known to modulate host susceptibility to infectious pathogens, particularly intestinal parasites, via effects on both local and systemic immune responses.5–7 Recent human field studies, for example, have reported increased diversity among microbiomes of helminth-infected subjects, suggesting a protective role against inflammatory disease in the host.7–10 Helminths that reside in the gut can directly affect the immune system and are thought to benefit indirectly from mutualistic relationships with enteric microbes that can influence immune responses at the intestinal mucosa.10,11 Although less intuitive, there is evidence that gut microbiota can also associate with Plasmodium parasites, perhaps through interactions mediated by the immune system.12 A mouse study reported a negative association of Bifidobacterium and Lactobacillus with severe malaria, and a human field study reported a negative association of Bifidobacterium, Streptococcus, and Enterobacteriaceae with Plasmodium falciparum infection.13,14 Another study has indicated that gut helminths may also be associated with enteric Lactobacilli species,6 suggesting the intriguing possibility that the gut microbiota may be implicated in governing host susceptibility to diverse pathogens, and perhaps even coinfection by different pathogens, through similar microbiome-influenced pathways.
A pilot study was conducted in Odisha, India, where both soil-transmitted helminth (STH) infections and malaria are endemic.15,16 The study was approved by the Institutional Review Boards at the University of Notre Dame and the Institute of Life Sciences, Bhubaneswar. Informed consent was obtained from all adult subjects and from a guardian of all minor subjects. The study was conducted in two phases; the first aimed to provide an initial profile of the association between microbiota and STH infections in this region. The second phase specifically examined the association between Lactobacillus and parasite coinfection. For the STH phase, stool samples were taken from subjects aged 2–5 years recruited from urban slums in Bhubaneswar, Odisha (n = 16). Stool collection kits were distributed to parents of potential subjects and collected 24 hours later. Common STHs Ascaris lumbricoides, Necator americanus, and Trichuris trichiura were detected through microscopy, flotation, and polymerase chain reaction (PCR) (for N. americanus and T. trichiura only). DNA was extracted from stool samples, and bacterial 16S rRNA was sequenced using the Illumina platform (Illumina Inc., San Diego, CA). Sequences were dereplicated, denoised, and clustered de novo into operational taxonomic units (OTUs) using UPARSE.17 Data were analyzed using R version 3.4.3 (R Core Team, Vienna, Austria) and the Linear discriminant analysis Effect Size online interface.18
For the Plasmodium phase, blood and stool samples were taken from subjects of all ages recruited from rural villages near Rourkela, Odisha (n = 68). Stool collection kits were distributed and collected the same day. DNA extracted from blood and stool samples was used for PCR detection of P. falciparum, Plasmodium vivax, N. americanus, and T. trichiura. Fecal DNA from each sample in the Plasmodium phase was used as the template in one qPCR using Universal Bacteria primers and one quantitative PCR (qPCR) using Lactobacillus-specific primers. Cycle threshold (CT) values were recorded as measures of total bacteria and Lactobacillus abundance. The cycle threshold of Universal Bacteria was subtracted from the CT of Lactobacillus for each sample, yielding a value representing the inverse relative abundance of Lactobacillus.
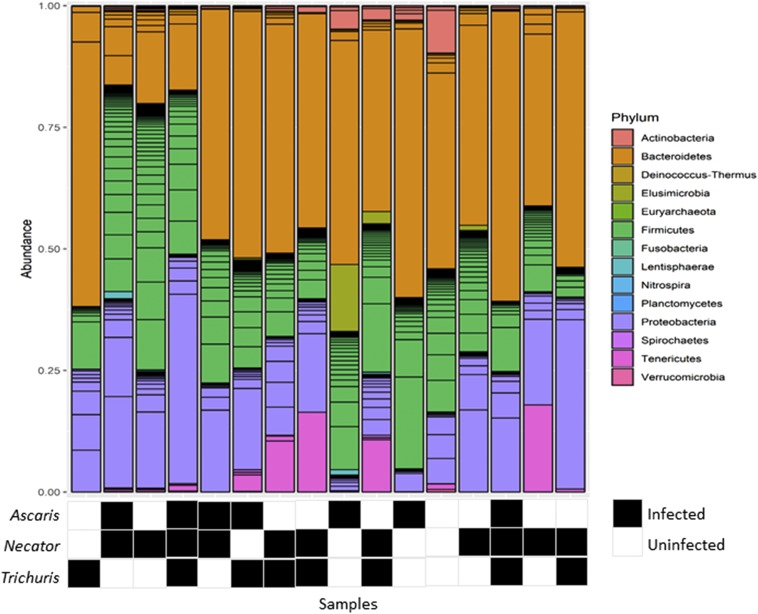
One or more species of STH was detected in 87.5% of samples in the STH phase of the study. Each of the common STHs, A. lumbricoides, N. americanus, and T. trichiura, was present in this subset of samples. Necator americanus was the most prevalent STH (63%) followed by A. lumbricoides and T. trichiura (both 44%). Bacteria from 14 phyla were detected. The relative abundance of bacteria in each phylum varied among samples; however, Bacteroides, Firmicutes, and Proteobacteria consistently made up the largest portions of the microbiomes (Figure 1). The most abundant genus was Prevotella with a mean relative abundance of 40.3%.
Figure 1.
Relative abundance of phyla in the human gut microbiome. Lines within phyla show the contribution of individual operational taxonomic units. The relative abundance of phyla varied among samples.
Shannon diversity did not vary in Ascaris- (t = 0.21, P = 0.84, 2-tailed t-test), Necator- (t = 0.31, P = 0.76, 2-tailed t-test), and Trichuris-infected (t = 0.137, P = 0.19, 2-tailed t-test) individuals versus uninfected controls. Average Bray–Curtis beta diversity (between individual diversity) was greater among Necator-infected (M = 0.54) than Necator-uninfected (M = 0.45) individuals (t = 2.21, P = 0.03, 2-tailed t-test). Approximately 13% of variation in Bray–Curtis dissimilarity could be explained by the Necator infection status (R2 = 0.13). However, average Bray–Curtis beta diversity did not vary significantly in STH-, Ascaris-, and Trichuris-infected individuals versus controls.
Linear discriminant analysis Effect Size analysis (α = 0.05) identified several taxa as bacterial biomarkers of STH infection status. The mean relative abundance of Lachnospiracaea_OTU_2 was reduced among Ascaris-infected (n = 7) versus Ascaris-uninfected samples (n = 9). The mean relative abundance of Lactobacillus_OTU_2, Lachcnospiracaea_OTU_3, and Lachnospiracaea_OTU_4 was reduced among Necator-infected (n = 10) versus Necator-uninfected samples (n = 6). The mean relative abundance of Lachnospiracaea_OTU_1, Dorea, Bifidobacterium, and Olsenella was reduced among Trichuris-infected (n = 7) versus Trichuris-uninfected samples (n = 9).
Of the 68 subjects sampled in the Plasmodium phase of the study, 43% were female (Tables 1 and 2). According to the WHO BMI and BMI-for-age classification guidelines, 23% of subjects were underweight and 13% of subjects were overweight or obese. The STHs N. americanus and T. trichiura were detected in 8.5% and 3.4% of samples, respectively. Using rapid diagnostic tests, Plasmodia were detected in 46% of blood samples. Using the PCR diagnostic method, P. vivax was detected in 18% of blood DNA samples and P. falciparum was detected in 15% of blood DNA samples. Both P. vivax and P. falciparum were more prevalent among younger subjects than older subjects.
Table 1.
Characteristics of Plasmodium study subjects by age group
| Characteristic | Age group | All (n = 68) | |||||
|---|---|---|---|---|---|---|---|
| 0–17 (n = 9) | 18–25 (n = 15) | 26–35 (n = 15) | 36–45 (n = 15) | 46–55 (n = 9) | 56–60 (n = 5) | ||
| Female | 2 (22%) | 7 (47%) | 6 (40%) | 6 (40%) | 6 (67%) | 2 (40%) | 29 (43%) |
| RDT positive for Plasmodium | 6 (67%) | 8 (53%) | 9 (60%) | 6 (40%) | 0 | 2 (40%) | 31 (46%) |
| PCR positive for Plasmodium | |||||||
| Plasmodium vivax | 4 (44%) | 4 (27%) | 3 (20%) | 1 (7.7%) | 0 | 0 | 12 (18%) |
| Plasmodium falciparum | 4 (44%) | 3 (20%) | 3 (20%) | 0 | 0 | 0 | 10 (15%) |
| PCR positive for soil-transmitted helminths (number positive/number tested) | |||||||
| Necator americanus | 0/7 | 0/13 | 1/14 (7.1%) | 0/13 | 0/7 | 1/5 (20%) | 2/59 (3.4%) |
| Trichuris trichiura | 0/7 | 3/13 (23%) | 1/14 (7.1%) | 0/13 | 0/7 | 1/5 (20%) | 5/59 (8.5%) |
| Nutritional status by BMI (number/number tested) | |||||||
| Underweight | 3/8 (38%) | 2/12 (17%) | 4/12 (33%) | 2/14 (14%) | 1/9 (11%) | 2/5 (40%) | 14/60 (23%) |
| Overweight | 1/8 (13%) | 0/12 | 3/12 (25%) | 1/14 (7.1%) | 1/9 (11%) | 1/5 (20%) | 7/60 (13%) |
Table 2.
Multivariate logistic regression analysis of risk factors associated with Plasmodium infection in rural villages in Odisha, India
| Variable | COR | 95% CI | P-value |
|---|---|---|---|
| Education | |||
| Less than primary school | Ref | – | – |
| Completed primary school | 0.21 | 0.01–1.73 | 0.194 |
| Number of children | 0.51 | 0.18–0.98 | 0.098 |
| Lactobacillus measure CT (Lacto)-CT (Universal) | 0.89 | 0.71–1.09 | 0.279 |
F(45,48) = −8.15, P = 0.043 < 0.05, Nagelkerke R2 = 0.29, AIC = 36.29. COR = crude odds ratio.
Stepwise logistic regression analysis indicated that education, number of children, and Lactobacillus were the best predictors of the Plasmodium infection status in our sample (akaike information criterion [AIC] = 36.29). All three variables were associated with reduced odds of infection; however, none were significant predictors at α = 0.05. The true predictors of Plasmodium infection were likely too numerous and complex for our limited survey to detect.
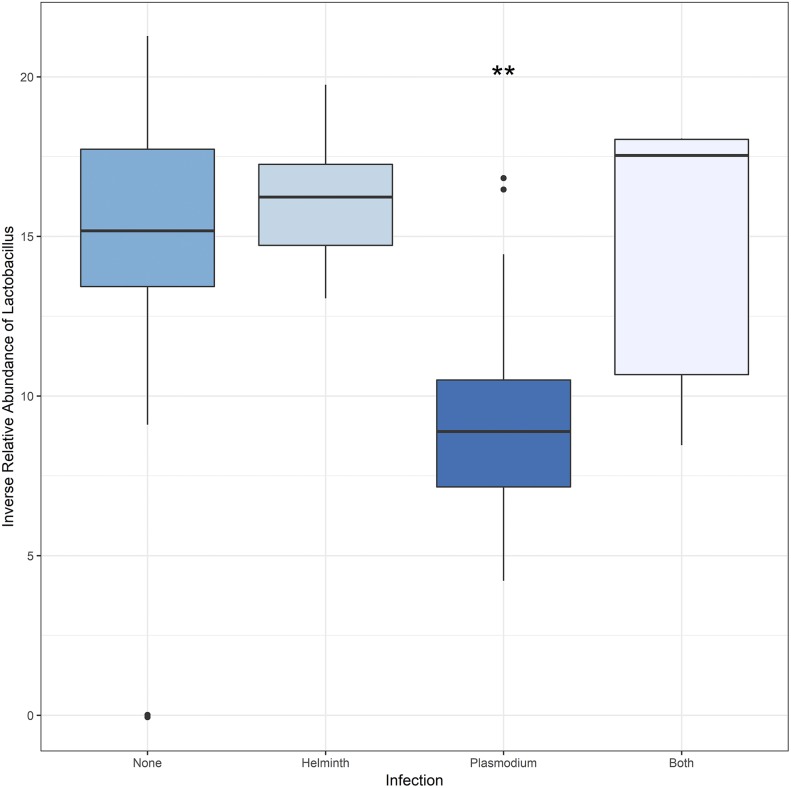
Analysis of the uninfected, helminth-only, Plasmodium-only, and coinfection status revealed that the median relative abundance of Lactobacillus was highest in microbiome samples from Plasmodium-only subjects (Figure 2). An analysis of variance (ANOVA) test confirmed that the mean relative abundance of Lactobacillus differed significantly in at least one group (F(3,96) = 18.96, P < 0.001). A Tukey test revealed significant differences in the relative abundance of Lactobacillus between the Plasmodium-only and each other group (Plasmodium-none p-adj < 0.001, Plasmodium-helminth p-adj < 0.001, Plasmodium-both p-adj = 0.02). The test did not detect a significant difference between the helminth-only group and those with coinfection or no infection (Helminth-both p-adj = 0.85; Helminth-none p-adj = 0.83).
Figure 2.
Relative abundance of Lactobacillus in gut microbiome samples by the Plasmodium and soil-transmitted helminth infection status. Distributions of CT (Lactobacillus)—CT (Universal Bacteria) values are shown for each infection status: uninfected, helminth-only, Plasmodium-only, helminth/Plasmodium coinfection. The relative abundance of Lactobacillus was significantly greater for Plasmodium infection–only samples than any other group (Plasmodium-None: P < 0.001; Plasmodium-helminth: P < 0.001; Plasmodium-both: P < 0.02). None, helminth, and both groups displayed statistically similar relative abundances of Lactobacillus. This figure appears in color at www.ajtmh.org.
The gut microbiomes described in this study of Bhubaneswar children match the profile of gut microbiomes previously reported in children living in rural India and Burkina Faso. All three studies found that gut microbiomes were dominated by the phyla Proteobacteria, Firmicutes, Bacteroidetes, and the genus Prevotella.19,20 Prevotella have been functionally linked to diets high in plant fiber, as they secrete enzymes that help metabolize cellulose into short-chain fatty acids known to protect against gut inflammation.20
This study also identified numerous microbiome characteristics associated with STH infections. Beta diversity was generally higher in STH-infected subjects, although a significance difference was observed only for Necator infections, in agreement with the findings of Jenkins, Lee, Kay, and Rosa.7–10 The direction of this relationship and the mechanism of action are not yet understood; however, the association between STH infection and increased microbiome beta diversity has been observed repeatedly. Perhaps helminth invasion of the intestine disturbs its ecological balance, causing once-similar microbiomes to diverge as they adjust.21 Additionally, we also found greater relative abundance of bacteria belonging to Lactobacillus, Lachnospiracea, and Olsenella taxa in uninfected samples than in infected samples. Our findings regarding Lachnospiracea are consistent with those reported by Rosa et al.,10 but our findings regarding Olsenella show disagreement. This disagreement may be the result of geographic variability in the microbiome behavior or a type II error due to the small sample size of this study. The STH phase was further limited by the low resolution of 16S sequence data, which cannot identify species or genes.
As sample collection was conducted in the dry season associated with low malaria transmission, the proportion of subjects infected with Plasmodium was predictably low. Because of reports that P. falciparum was the more prevalent Plasmodium species in Odisha, we were surprised to observe higher rates of P. vivax in this sample.16 There were also high rates of coinfection. Each subject infected with P. falciparum also carried P. vivax. Analysis of coinfections revealed that Lactobacillus abundance was similar in uninfected, helminth-only, and Plasmodium/helminth coinfected microbiomes. With greater sample size, we might have distinguished the uninfected controls from the other groups. Our sample size was sufficient to determine that Lactobacillus was significantly more abundant in Plasmodium-only microbiomes. This suggests that Lactobacillus abundance observed in coinfected microbiomes is driven by helminths more so than Plasmodium, possibly due to the direct contact between helminths and microbiota in the GI tract. However, the association found with the two social factors related to educational status and number of children in a family investigated in this study point to the existence of more complicated pathways which may govern this relationship (Table 2). The results of this study are based on observational data and must not be used to draw conclusions about cause and effect. Further, because of convenient sampling, our small sample may have included only a narrow swath of the Rourkela and Bhubaneswar populations.
This study contributes to efforts to understand effects of the microbiome on host susceptibility to parasitic infections in endemic communities. We have derived a first microbiome community profile associated with STH infections in Odisha and tested the hypothesis that the gut microbiome can modulate host susceptibility to multiple parasite infections through the same pathways. We observed increased Lactobacillus among Plasmodium-only subjects but reduced Lactobacillus among STH-only subjects and those carrying coinfections (Figure 2). This indicates that relative abundance of Lactobacillus may influence susceptibility to either type of parasite, but in opposite directions. It may also govern coinfection with STH infections and malaria in this setting. A larger study is required to confirm these first findings and to identify the mechanisms behind the observed associations between the gut microbiome and host infection.
REFERENCES
- 1.Brown RL, Clarke TB, 2016. The regulation of host defences to infection by the microbiota. Immunology 150: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanton LV, et al. 2016. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis CD, 2016. The gut microbiome and its role in obesity. Nutr Today 51: 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honda K, Littman DR, 2012. The microbiome in infectious disease and inflammation. Annu Rev Immunol 30: 759–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Partida-Rodríguez O, Serrano-Vázquez A, Nieves-Ramírez ME, Moran P, Rojas L, Portillo T, González E, Hernández E, Finlay BB, Ximenez C, 2017. Human intestinal microbiota: interaction between parasites and the host immune response. Arch Med Res 48: 690–700. [DOI] [PubMed] [Google Scholar]
- 6.Brosschot TP, Reynolds LA, 2018. The impact of a helminth-modified microbiome on host immunity. Mucosal Immunol 11: 1039–1046. [DOI] [PubMed] [Google Scholar]
- 7.Jenkins TP, Rathnayaka Y, Perera PK, Peachey LE, Nolan MJ, Krause L, Rajakaruna RS, Cantacessi C, 2017. Infections by human gastrointestinal helminths are associated with changes in faecal microbiota diversity and composition. PLoS One 12: e0184719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kay GL, Millard A, Sergeant MJ, Midzi N, Gwisai R, Mduluza T, Ivens A, Nausch N, Mutapi F, Pallen M, 2015. Differences in the faecal microbiome in Schistosoma haematobium infected children vs. uninfected children. PLos Negl Trop Dis 9: e0003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SC, et al. 2014. Helminth colonization is associated with increased diversity of the gut microbiota PLoS Negl Trop Dis 8: e2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosa BA, et al. 2018. Differential human gut microbiome assemblages during soil-transmitted helminth infections in Indonesia and Liberia. Microbiome 6: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maizels RM, McSorley HJ, 2016. Regulation of the host immune system by helminth parasites. J Allergy Clin Immunol 138: 666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yilmaz B, et al. 2014. Gut microbiota elicits a protective immune response against malaria transmission. Cell 159: 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villarino NF, LeCleir GR, Denny JE, Dearth SP, Harding CL, Sloan SS, Gribble JL, Campagna SR, Wilhelm SW, Schmidt NW, 2016. Composition of the gut microbiota modulates the severity of malaria. Proc Natl Acad Sci USA 113: 2235–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yooseph S, et al. 2015. Stool microbiota composition is associated with the prospective risk of Plasmodium falciparum infection. BMC Genomics 16: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salam N, Azam S, 2017. Prevalence and distribution of soil-transmitted helminth infections in India. BMC Public Healt 17: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anvikar AR, Shah N, Dhariwal AC, Sonal GS, Pradhan MM, Ghosh SK, Valecha N, 2016. Epidemiology of Plasmodium vivax malaria in India. Am J Trop Med Hyg 95: 108–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edgar RC, 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10: 996. [DOI] [PubMed] [Google Scholar]
- 18.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C, 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh TS, Sen Gupta S, Bhattacharya T, Yadav D, Barik A, Chowdhury A, Das B, Mande SS, Nair GB, 2014. Gut microbiomes of Indian children of varying nutritional status. PLoS One 9: e95547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P, 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA 107: 14691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banks SC, Cary GJ, Smith AL, Davies ID, Driscoll DA, Gill AM, Lindenmayer DB, Peakall R, 2013. How does ecological disturbance influence genetic diversity? Trends Ecol Evol 28: 670–679. [DOI] [PubMed] [Google Scholar]