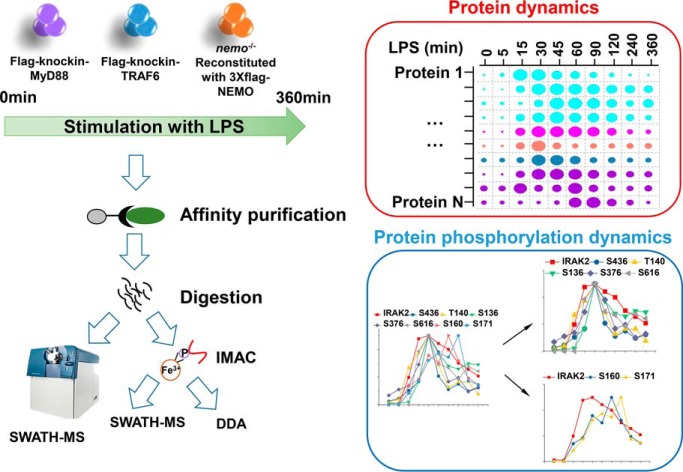
The LPS-induced dynamic interactions and phosphorylation in MYD88, TRAF6, and NEMO complexes have been systematically mapped by SWATH-based quantitative proteomics. The results reported reveal highly dynamic complex assembly and complex regulation network in LPS signaling pathway.
Keywords: SWATH-MS, Immunoaffinity, Immunology*, Quantification, Phosphoproteome
Graphical Abstract
Highlights
Intensive investigation of dynamic interactions in MyD88, TRAF6, and NEMO complexes.
Mechanistic insights into IRAKs proteins' assembly.
A signal amplification mechanism for MyD99-denpendent TLR signaling disclosed by stoichiometry of complexes.
Quantitative measurement of multiple phosphorylation sites on the key components in TLR signaling pathway.
Abstract
Lipopolysaccharide (LPS)-induced macrophage activation is a prototype of innate immune response. Although key effector proteins in LPS signaling pathway have been revealed, the map of dynamic protein interactions and phosphorylation as well as the stoichiometry of protein complexes are lacking. Here we present a dynamic map of protein interactions and phosphorylation in MyD88, TRAF6 and NEMO complexes obtained by SWATH-MS. The comprehensive MS measurement leads to quantification of up to about 3,000 proteins across about 21–40 IP samples. We detected and quantified almost all known interactors of MyD88, TRAF6 and NEMO. By analyzing these quantitative data, we uncovered differential recruitment of IRAK family proteins to LPS-induced signaling complexes and determined the stoichiometry of the Myddosome complex. In addition, quantitative phosphoproteomics analysis identified a number of unreported high-confidence phosphosites on the key proteins in LPS signaling pathway. Collectively, data of dynamic protein interactions and phosphorylation presented by this study could be a resource for further study of the LPS signaling pathway.
Lipopolysaccharides (LPS)1 are major components of the outer membrane of Gram-negative bacteria. Ligation of Toll-like receptor 4 (TLR4) with LPS ultimately leads to activation of multiple signaling pathways, such as MAPK, NF-κB, and IRFs pathways (1). The activated signaling pathways regulate expression of a variety of cytokines, chemokines, and type I IFNs, which are essential in inflammation and host defense against microbial infection. MyD88 and TRIF are proximal signaling effectors of TLR4 that elicit MyD88-dependent and MyD88-independent (TRIF-dependent) pathways (2, 3). The activation of the MyD88-dependent pathway leads to the expression of pro-inflammatory cytokines, whereas the MyD88-independent pathway mediates the induction of Type I interferons and interferon-inducible genes.
On LPS stimulation, TIRAP is first recruited to TLR4 and MyD88 is then recruited to TIRAP, highlighting the importance of TIRAP in MyD88 recruitment to TLR4 (4, 5). The IRAK family proteins, including IRAK1–4, are then recruited to MyD88. Downstream of IRAK1 and IRAK4, TRAF6 is critical to the MyD88-dependent pathway (6). TRAF6 forms a complex with UBC13 and UEV1A, which activates TAK1 (7). Subsequently TAK1 activates downstream NF-κB and MAPK pathways.
The MyD88-indepent signaling pathway is mediated by TRIF, an important TIR-containing adaptor protein. TRIF recruits TRAF3 which then associates with TANK (TRAF family member-associated NF-κB activator), TBK1 and IKKi to mediate IRF3 dimerization (8–10). IRF3, cooperating with NF-κB, activates the transcription of target genes, such as Type I interferons and interferon-inducible genes (11).
Although TLR4 signaling pathway has been intensively studied in the past few years, several important questions still remain unsolved. Although LPS-induced NF-κB activation is abolished in TAK1-deficient MEF cells, it is not affected in TAK1-deficient macrophages (12, 13). These reports indicate that other unknown players downstream of TRAF6, but not TAK1, are essential for LPS-induced NF-κB activation in macrophages. Another intriguing question is how TIRAP-MyD88-IRAK complex (named Myddosome) is assembled and what the stoichiometry of this complex is.
SWATH-MS (sequential window acquisition of all theoretical fragment ion spectra mass spectrometry) is a data-independent acquisition MS technique, which combines the advantages of shotgun MS and SRM (Selected Reaction Monitoring) (14, 15). SWATH-MS enables consistent and accurate protein quantification across multiple samples. Besides, SWATH-MS can offer relative protein quantification within one sample, allowing for calculation of protein complex stoichiometry (16).
In this study, we used immunoprecipitation to purify time-resolved protein complexes from macrophage cell line RAW 264.7 on a serial of LPS treatments, and quantified proteins and phosphorylation events in these complexes using SWATH-MS. Almost all known players involved in LPS pathway have been identified and quantified in one experiment. Dynamic key protein profiles and their phosphorylation events in MyD88, TRAF6, and NEMO complexes under LPS treatment are shown, revealing the complex regulation network in LPS signaling pathway. In addition, we showed the stoichiometry of Myddosome, which can offer an insight into how TIRAP and MyD88 cooperate to ensure signal transduction and how IRAK complex are assembled.
MATERIALS AND METHODS
Experimental Design and Statistical Rationale
3xFlag-KI TRAF6 RAW 264.7 cell line and 3xFlag-KI MyD88 RAW 264.7 cell line were treated with LPS for ten time points. 3xFlag-NEMO-reconstituted RAW 264.7 cell line was treated with LPS for seven time points. RAW 264.7 cell line expressing 3xFlag-vector was treated with LPS for ten time points. Protein complexes were purified with M2 beads. Four biological replicates were performed for 3xFlag-KI TRAF6 cells, and three biological replicates were performed for 3xFlag-KI MyD88 cells and 3xFlag-NEMO cells. Two biological replicates were performed for 3xFlag-vector cells. One biological replicate is the repeat of the whole experiment from cell culture to MS analysis under the same conditions. No technical replicates (multiple injections for one peptide sample) was performed in the study. In the differential expression analysis, proteins with log2(fold change) >1 and -log10(p value) >1.5 were considered as significantly changed.
Generation of 3xflag-knockin TRAF6 and 3xflag-knockin Myd88 RAW 264.7 Cell Lines
The 3xFlag tag knock-in cell line was generated by using the CRISPR-Cas9 and rAAV-assisted repair template delivery. The rAAV targeting vector was constructed by insertion of left homologous arm (∼ 1 kb genomic DNA sequences upstream of the start codon of the target gene) and right homologous arm (∼1 kb genomic DNA sequences downstream of the star codon of the target gene) into the rAAV-Neo-Lox P-3xFLAG Knockin vector. Targeting rAAV viruses were packaged in 293T cells. Guide RNAs were designed to cut proximal to the start codon in the 5′ UTR or intron of target genes. The target sequences used were AACTCCACAGGCGAGCGTAC for Myd88 and TAAATAACATTGAAACATTA for TRAF6. The plasmids harboring the gene gRNA sequences and Cas9 gene were transfected into the RAW 264.7 cells. Those cells were infected with the targeting rAAV virus 24h post transfection and then selected for neomycin-resistant clones. Those clones were then screened for homologous recombination by genomic PCR and the positive clones were infected with adenovirus expressing Cre-recombinase to excise the neomycin gene cassette. The final successful 3xFlag knock-in clones were confirmed by genomic PCR and Western blotting.
Generation of 3xFlag-NEMO-reconstituted and 3xFlag-vector-expressing RAW 264.7 Cell Lines
NEMO knockout cell line was generated using CRISPR-Cas9. The target sequence used was tgagaccctccagcgctgcc. The plasmids harboring the gene gRNA sequences and Cas9 gene were transfected into the RAW 264.7 cells. The cell clones were screened by genomic PCR and positive clones were confirmed by DNA sequencing and Western blotting. NEMO KO RAW 264.7 was infected with lentivirus contained 3xFlag-NEMO. The positive single clones were confirmed by Western blotting.
The wildtype RAW 264.7 cell line was infected using 3xFlag-vector-expressing lentivirus. After 24 h, the cells were employed in negative-control experiment.
Immunoprecipitation, Digestion, and IMAC
Cells were seeded at 1 × 107 cells per 15 cm dish in DMEM supplemented with 10% FBS. After cells reached 80% confluency, the cells were stimulated with 100 ng/ml LPS for various time points. For 3xFlag-knockin TRAF6 RAW 264.7 cells, we treated cells for ten-time points (0, 5, 15, 30, 45, 60, 90, 120, 240, 360 min) in four biological replicates, which resulted in 40 IP samples. 3xFlag-knockin Myd88 cells were stimulated with LPS for ten-time points in three biological replicates. For Nemo−/− RAW 264.7 cells reconstituted with 3xFlag-NEMO, LPS treatment time points were 0, 15, 30, 45, 60, 90, 120 min in biological triplicates, which resulted in 21 IP samples. For each IP sample, ten 15 cm dishes of cells were collected. Cell pellets were collected in ice-cold PBS and lysed with HBS lysis buffer (12.5 mm HEPES, pH7.5, 150 mm NaCl, 1% Nonidet P-40, Protease inhibitor mixture and phosphatase inhibitor mixture) on ice for 30 min. The cell lysates were then centrifuged at 20,000 × g for 30 min at 4 °C. The supernatants were collected for immunoprecipitated overnight with anti-Flag M2 antibody-conjugated agarose at 4 °C. Beads containing protein complexes were washed four times with HBS lysis buffer. Bound Flag-immune complexes were eluted twice with 0.15 mg/ml of 3xFlag peptide with N-terminal biotin tag in HBS lysis buffer and then precipitated with 20% trichloroacetic acid (TCA). The protein pellets were washed three times with 1-ml cold acetone and dried in speedvac.
TCA-precipitated proteins were re-suspended in 50 μL8 M urea in 50 mm NH4HCO3, and 10 mm Tris(2-carboxyethyl) phosphine hydrochloride (TCEP) and 40 mm chloroacetamide (CAA) were added into reactions for 30 min at 37 °C for cysteine reduction and alkylation. Next, 8 m urea were diluted to 1.6 m urea with 50 mm NH4HCO3 and trypsin was added at the protein/trypin ratio of 50:1. Digestion was performed overnight at 37 °C. The biotin-3xFlag peptide was removed by the avidin beads. Peptides were acidified to a final concentration of 1% formic acid (FA), followed by desaltion using C18 StageTips. After desaltion, peptides were eluted with 70% acetonitrile/1% formic acid and dried.
For phosphopeptide enrichment with IMAC, peptides were dissolved in 50 μl 60%ACN/1%AA and incubated with 5 μl bead volume of IMAC beads. The peptides with IMAC beads was shaken for 30 min at room temperature. Nonphopshopeptides were washed with 25% ACN/0.1 M NaCl/0.1%AA for three times followed by one-time water wash. Phosphopeptides were then eluted with 6% NH3·H2O and dried in speedvac.
Mass Spectrometry
Peptides were analyzed on a TripleTOF 5600 instrument (Sciex, Concord, Canada) in SWATH (Sequential window acquisition of all theoretical mass spectra) mode. The peptides were separated by NanoLC Ultra 2D Plus (Eksigent) HPLC system with a flow of 300 nl/min. An in-house pulled emitter-integrated column (inner diameter 75 μm) packed with about 35 cm Magic C18 AQ 3-μm 200- Å resin was used to separate the peptides along a linear 120 or 240 min gradient from 2% to 35% Buffer B (0.1% (V/V) formic acid, 5% DMSO in acetonitrile) in Buffer A (0.1% (V/V) formic acid, 5% DMSO in H2O) (17). A 250-ms survey scan (TOF-MS), which was collected in 350–1500 m/z, was performed followed by 80, 50-ms MS2 experiments or 100, 33-ms MS2 experiments that were collected in 100–1800 m/z to cover 400–1200 m/z, and the cycle time is about 3.6 s.
The 80 fixed isolation windows are: 400–410, 409–420, 419–430, 429–440, 439–450, 449–460, 459–470, 469–480, 479–490, 489–500, 499–510, 509–520, 519–530, 529–540, 539–550, 549–560, 559–570, 569–580, 579–590, 589–600, 599–610, 609–620, 619–630, 629–640, 639–650, 649–660, 659–670, 669–680, 679–690, 689–700,699–710,709–720, 719–730, 729–740, 739–750, 749–760, 759–770, 769–780, 779–790, 789–800, 799–810, 809–820, 819–830, 829–840, 839–850, 849–860, 859–870, 869–880, 879–890, 889–900, 899–910, 909–920, 919–930, 929–940, 939–950, 949–960, 959–970, 969–980, 979–990, 989–1000, 999–1010, 1009–1020, 1019–1030, 1029–1040, 1039–1050, 1049–1060, 1059–1070, 1069–1080, 1079–1090, 1089–1100, 1099–1110, 1109–1120, 1119–1130, 1129–1140, 1139–1150, 1149–1160, 1159–1170, 1169–1180, 1179–1190, 1189–1200.
The 100 variable isolation windows are: 399.5–409.9, 408.9–418.9, 417.9–427.4, 426.4–436, 435–443.6, 442.6–450.8, 449.8–458, 457–464.8, 463.8–471.1, 470.1–476.9, 475.9–482.8, 481.8–488.6, 487.6–494, 493–499, 498–504.4, 503.4–509.3, 508.3–514.3, 513.3–519.2, 518.2–524.2, 523.2–529.1, 528.1–534.1, 533.1–539, 538–543.5, 542.5–548.5, 547.5–553, 552–558, 557–562.5, 561.5–567, 566–571.5, 570.5–576, 575–580.5, 579.5–585, 584–589.5, 588.5–594, 593–598, 597–602.5, 601.5–607, 606–611.1, 610.1–615.6, 614.6–620.1, 619.1–624.6, 623.6–628.6, 627.6–633.1, 632.1–637.6, 636.6–642.1, 641.1–646.6, 645.6–651.1, 650.1–655.6, 654.6–660.1, 659.1–665.1, 664.1–669.6, 668.6–674.5, 673.5–679, 678–684, 683–688.5, 687.5–693.4, 692.4–698.4, 697.4–703.3, 702.3–708.7, 707.7–713.7, 712.7–719.1, 718.1–724.5, 723.5–729.9, 728.9–735.3, 734.3–740.7, 739.7–746.5, 745.5–751.9, 750.9–757.8, 756.8–763.6, 762.6–769.5, 768.5–775.3, 774.3–781.2, 780.2–787, 786–793.3, 792.3–800.1, 799.1–806.4, 805.4–813.1, 812.1–820.3, 819.3–827.5, 826.5–835.2, 834.2–843.3, 842.3–851.4, 850.4–859.9, 858.9–868.9, 867.9–878.4, 877.4–888.3, 887.3–899.1, 898.1–910.3, 909.3–922.9, 921.9–936, 935–949.5, 948.5–963.4, 962.4–978.7, 977.7–994.9, 993.9–1015.6, 1014.6–1042.2, 1041.2–1070.1, 1069.1–1100.7, 1099.7–1140.7, 1139.7–1196.5.
Ions were fragmented for MS2 experiment in the collision cell using a collision energy according to the equation of a doubly charged peptide, ramped ±15 V from the calculated collision energy.
For phosphopeptides, half of IMAC samples were analyzed using shotgun MS and half of them for SWATH-MS analysis. For both DDA and SWATH-MS, the HPLC gradient was 30 min. In shotgun MS, MS1 scan was 250 ms and range was 350–1250 m/z, and MS2 scan was 50 ms and MS2 range was 100–1800 m/z. In one cycle, a MS1 scan was followed by 20 MS2 scans, resulting in 2.5s cycle time.
Generation of Internal Spectral Library from SWATH-MS Data Using Group-DIA
SWATH-MS wiff files were converted to profile mzXML files using proteoWizard MSConvert V.3.0.447 (18), and the profile mzXML files were split into a number of MS2 mzXML files and 1 MS1 mzXML file according to the SWATH window using the in-house script. For MyD88 data set, thirty 200-min runs were collectively analyzed. For TRAF6 data set, forty 120-min gradient runs were analyzed together. For NEMO data set, twenty-one 240-min runs were analyzed. Group-DIA software was composed of four modules: alignment, analysis, identification, and validation. For generation of internal library, only “alignment” and “analysis” modules were performed. Retention time in multiple runs were first aligned using MS1 intensity. MS1 the MS2 features were first extracted in single run and then concatenated across all runs. The similarity in precursors and product ions' XICs were compared, and the pair of precursor and product ions were then extracted. The generated presudo-spectra were stored in mgf and mzML formats. The mgf files were converted to mzXML files, which were analyzed with TPP (Trans-Proteomic Pipeline, Version 4.8) software (19). mzXML files were subjected to database search using Comet (Version 2017.01) (20) and X!tandem (Version 2013.06.15.1, native and k-score) (21) against the full nonredundant, canonical mouse genome as annotated by UniprotKB/Swiss-Prot (downloaded in September, 2014) appendant with common contaminants and reversed sequence decoys (33,864 sequences includes decoys). The search parameters were set as followed, parent monoisotopic tolerance 50 ppm, modification 57.021464@C, potential modification mass 15.994915@M and maximum missed cleavage sites 2. The pep.xml search results were validated and scored using PeptideProphet (22) with parameters -p0.05 -l7 -PPM -OAdPE -dDECOY and combined by iProphet (23) with parameters DECOY = DECOY. Mayu (version 1.07) (24) was used to determine iProphet probability corresponding to 1% peptide FDR. The peptide ions passing the 1% FDR were input into SpectraST (25) for library building with CID-QTOF setting. The retention time of peptides in sptxt file was replaced with iRT time using spectrast2spectrast_irt.py script (downloaded from www.openswath.org), and iRT peptides used for retention time normalization were endogenous peptides. The sptxt file was made consensus nonabundant spectral library with the iRT retention time using spectraST. Combination of different libraries was performed by SpectraST software with options of “-cJU” and “-cAC.”
Analysis of Phosphoproteomics Data
SWATH-MS wiff files were first analyzed by Group-DIA for generation of mgf files as described above. These mgf files were converted to mzXML files, and DDA wiff files were converted to centroid mzXML files using qtofpeakpicker tool in TPP. These mzXML files were searched with Comet and X!Tandem (native and k-score plugin) altogether. The search parameters were set as followed: parent monoisotopic tolerance 50 ppm, modification 57.021464@C, potential modification mass 15.994915@M 79.966331@STY and maximum missed cleavage sites 2. The pep.xml files were scored using PeptideProphet with parameters -p0.05 -l7 -PPM -OAdPE -dDECOY and combined by iProphet with parameters DECOY = DECOY. The ipro.pep.xml files were subsequently analyzed by PTMProphet for phosphosites localization scoring. The phosphopeptides were first filtered at the iProphet values corresponding to 1% peptide FDR which was determined by Mayu (Version 1.07), and then filtered at localization score 0.7. After these filters, the phosphopeptide ions were for library building by SpectraST with CID-QTOF setting. The retention time of peptides in sptxt file were replaced with iRT time using spectrast2spectrast_irt.py script (downloaded from www.openswath.org), and iRT peptides used for retention time normalization were endogenous peptides. The sptxt file was made consensus nonabundant spectral library with the iRT retention time using spectraST.
Reprocessing of Murine Cell Line L929 Total Spectral Library
As described previously, we used Mascot and X!tandem to process the L929 cell line DDA data. However, we found Comet performed better than Mascot. Therefore, we reprocessed 248 DDA runs with Comet and X!tandem. Database searches and spectral library building were performed using the same method as described above. Briefly, the DDA wiff files were first converted to centroid mzXML files using qtofpeakpicker tool in TPP followed by database searches with Comet and X!Tandem (native and k-score plugin). The pepXML files were validated with PeptideProphet followed by iProphet analysis. An iProphet probability cutoff (p = 0.956653) was determined by MAYU (version 1.07), resulting in 110,211 peptides (FDR = 0.22%) which correspond to 8599 proteins (FDR = 1.08%). The pepXML file was filtered to 1% FDR at protein level and converted to sptxt file using SpectraST with CID-QTOF setting. The retention time of peptides in sptxt file were replaced with iRT time using spectrast2spectrast_irt.py script, where ciRT were used for retention time normalization. After removing contaminant and decoy proteins hits, the sptxt file was made consensus nonabundant spectral library with the iRT retention time using spectraST.
OpenSWATH-Pyprophet-Tric Workflow
The consensus sptxt files were converted to tsv using spectrast2tsv.py script which was then converted to TraML file with TargetedFileConverter tool which is integrated into OpenMS software (Version 2.2.0) (26). All detected peptides of the bait proteins by Group-DIA were utilized for iRT peptides for retention time normalization. An extended version of Pyprophet (Pyprophet-cli v0.19 — https://github.com/PyProphet) was employed for FDR estimation (27, 28). One percent protein FDR at global level is applied in all analyses. The filtered results were input into TRIC software (29) for cross-run alignment. The parameters in TRIC were set as followed: -method LocalMST -realign_method lowess_cython -max_rt_diff 60 -mst: useRTCorrection True -mst: Stdev_multiplier 3.0 -target_fdr 0.01 -max_fdr_quality 0.05.
Protein Inference and Quantification
The TRIC results were used for protein inference and quantification. First, proteins with proteotypic peptides were considered as “uniquely identified,” and the proteotypic peptides accounted for about 90% of all identified peptides (supplemental Table S1). Second, the peptides mapped to more than one protein entry were handled as followed.
The peptides shared with the proteins with proteotypic peptides were excluded for protein inference and quantification.
The peptides without evidence of unique protein mapping were considered as “from one protein representing the gene locus and expressed as the alphabetically first entry of the protein database (gene locus identification)” (30).
To generate complete quantitative matrix of IP data set, peptides only identified in all biological replicates of at least one time point were kept for extraction of quantitative information.
Peptide intensities were directly from TRIC output results, where peptide intensities were calculated by summing the top five most intense fragment ion peak areas. In each data set, all identified peptides from the specific protein were ranked by the average intensity in all runs. Subsequently, top 3 most intense peptides of the specific protein were selected and sum of these three peptides' intensities represented the protein intensity in each run. Where <3 peptides were detected, the available peak groups were summed.
We found that many missing values of protein quantitation were detected when the comprehensive library was used. This issue was not caused by stochastic precursor ion selection as in shotgun MS because SWATH-MS recorded all precursor ions, nor by false positive identification as 1% protein FDR at global level was applied. As shown in Fig. 3C, the peptide peak group of TIRAP was unambiguously detected at the 30 min run in TRAF6 data set, but not at the 0 min run. This result demonstrates that the peptide was present in the 30 min run but not in the 0 min run. Therefore, the peptide intensity in 0 min run was calculated as zero, and “zero” values were obviously inconvenient for downstream bioinformatics processing. Instead of missing value imputation, we used “background intensity strategy” to address this issue. The background intensity strategy was performed as followed.
Fig. 3.
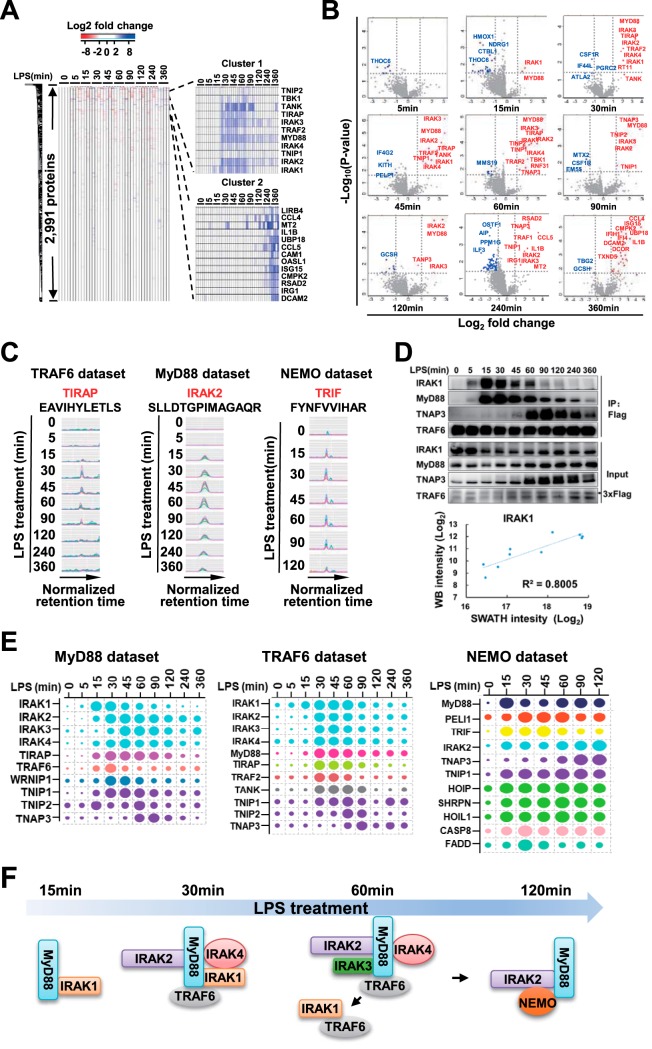
Identification of high-confidence interactors of TRAF6, MyD88 and NEMO. A, Heatmap of protein-abundance change after LPS treatment relative to untreated sample is shown on the left. Hierarchical clusters are shown on the right. B, Differential expression analysis in TRAF6 data set. Proteins with Log2(fold change) >1 and -Log10(p value) >1.5 were considered significantly changed. Upregulated proteins were labeled in red, and downregulated proteins were labeled in blue. C, MS2 XICs of representative peptides of a representative high-confidence interactor of TRAF6, MyD88, and NEMO. Traces in different colors mean different product ions of given peptides. D, Comparison of band intensities in Western blotting and protein intensities in TRAF6 IP SWATH-MS. The panel above shows IRAK1, MyD88, TNAP3, and TRAF6 detected by Western blotting in TRAF6 IP samples, and the panel below shows the comparison of IRAK1 band intensities in Western blotting and IRAK1 protein intensities in SWATH-MS. The coefficient of correlation is shown. E, Relative levels of interactors without induced protein expression by LPS in MyD88, TRAF6, and NEMO data set. The size of each dot is proportional to the relative abundance of the indicated protein. In each data set, proteins labeled in the same color mean one subcomplex they belong to. F, Assembling and de-assembling of IRAK family proteins into LPS signaling complexes, a model deduced from MyD88, TRAF6, and NEMO data sets.
If one peptide was detected in run A but not in run B, the RT (retention time) of the peptide in run A was used for location of the peptide in run B. The peptide RT in run A was transformed in iRT value, which was also considered as iRT value in run B. iRT in run B was then transformed into the actual RT in run B. Considering this RT maybe not precise, RT window of the peptide in B run was taken. The RT window of the peptide in B run was calculated by extending 10 min at the center of RT value.
In the RT window of the peptide in B run, the product ion mz intensities were extracted and summed at each cycle because no peaks were detected across the window. The summed m/z intensity values were ranked, and the median value was taken. Because there was no peak in the retention time range, the summed intensity of base line was significantly lower than other peptide peak intensities. Thus, we compared intensities of 100 peaks with the summed intensities of their base line in a 20 min retention time window. We found the ratios of “peak intensity” to “the summed intensity of base line” was about 5.5–6.0, and we took the average number “5.7.” The median value was multiplied with 5.7 and considered as background intensity.
Targeted Analysis of Phopshoproteomics SWATH-MS Data Using Peakview Software
The spectray library was generated from DDA and pseudo-spectra files, and converted to Peakview-compatible file. The parameters of Peakveiw software were set as followed. “Number of transitions per peptide” was “6.” “False Discovery Rate” was “1%.” “XIC Extraction window” was “10 min,” and “XIC width (Da)” was “0.05.”
The Heatmap Generation and Differential Expression Analysis
The protein intensities were input into Perseus software (31). The Hierarchical clustering and differential expression analysis were conducted with default settings.
Manual Inspection of Peptide XICs
The XICs of peptides of significantly changed proteins were manually inspected, and several rules were used to remove falsely identified peptides by the software. First, we removed the peptides whose XICs had poor quality. Second, we removed the proteins whose peptide XICs showed different quantitative trends in biological replicates over LPS treatment. Third, the peptides with no obvious peak groups were removed (supplementary File S1).
Western Blotting
Proteins of the cell lysates or immunoprecipitants were separated by SDS-PAGE and then transferred onto polyvinylidene difluoride membranes. The membranes were blocked with 5% bovine serum albumin at room temperature for 1 h and then incubated with primary antibodies overnight at 4 °C, followed by incubation with secondary antibodies for 1 h at room temperature. The luminescent signals of immunoblotting were analyzed using an ImageQuant LAS 4000 Scanner (GE Healthcare). Antibodies for Phospho-p65(3033S), Phospho-p38(9216S), Phospho-JNK(9251S), Phospho-ERK(9101), IκBα(9242S), MyD88(4283S),TNAP3(5630S) were purchased from Cell Signaling Technology; Antibodies for NEMO(18474–1-AP) and IRAK1(10478–2-AP) were purchased from Proteintech Group, Inc. Antibody for TRAF6(sc-7221) was purchased from Santa Cruz Biotechnology, Inc.
Luciferase Reporter Assay
HEK293T cells were transiently transfected with the NF-κB firefly luciferase reporter plasmid, pRL-TK-Renilla-luciferase plasmid and the indicated expression constructs. After 24 h, the luciferase activity in the total cell lysate was measured with the Dual-luciferase reporter assay system (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity.
RNA Extraction and Quantitative RT-PCR
Total RNA was isolated from cells using RNAiso Plus (Takara) according to the manufacturer's instructions. cDNA was prepared with M-MLV reverse transcriptase and oligo-dT primers. Quantitative RT-PCR was performed in a CFX96 RealTime System (Bio-Rad) by using SYBR Green reagent along with gene-specific primers. All the results were analyzed by relative quantification by normalizing to the internal control GAPDH RNA level. Primer sequences are available on request.
RESULTS
An AP-SWATH-MS Workflow Using Combination of the Prebuilt Spectral Library and the Internal Library Generated Directly from SWATH-MS Data
Affinity purification (AP)-SWATH workflow has been utilized to investigate dynamic protein interactions in signaling pathways (32–34). A spectral library prebuilt from existing shotgun MS data was commonly employed for interpretation of SWATH-MS data in those studies. Meanwhile, an internal library directly generated from SWATH-MS data can also be used. The former often identifies more peptides but the latter can reveal novel peptides not detected by prebuilt spectral library (35). This is most likely because of that prebuilt library could be more complete but the internal library contains more specific peptide information derived directly from given SWATH-MS analysis. Therefore, more complete interpretation of SWATH-MS data can be achieved by combining the comprehensive prebuilt library and the internal library built from SWATH-MS data.
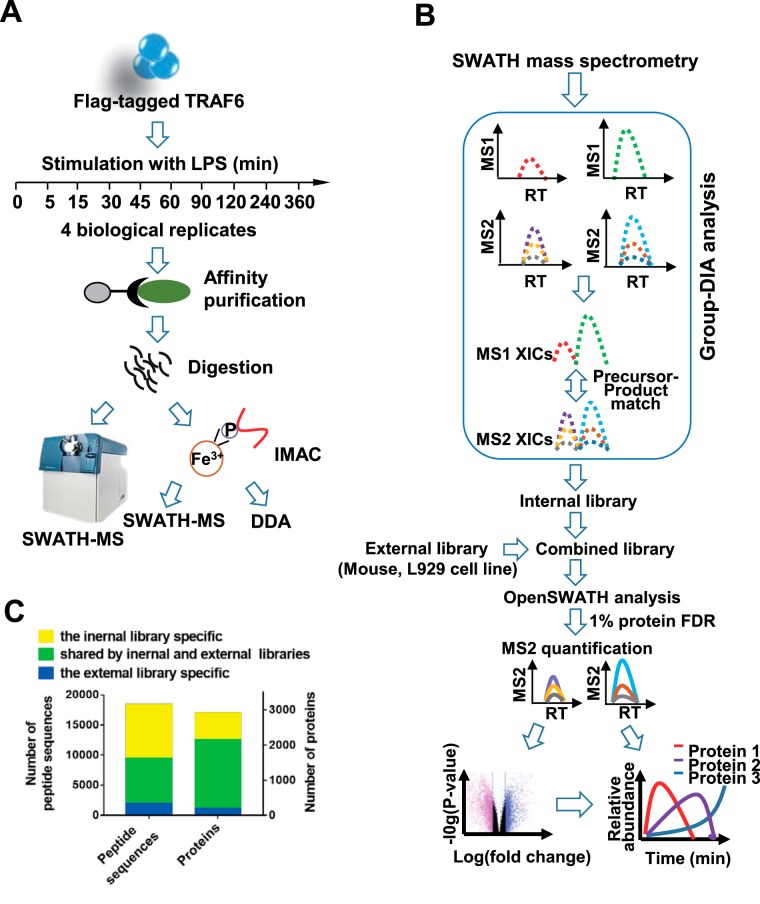
As TRAF6 is an effector essential for LPS signal transduction, we used measurement of dynamic interactions between TRAF6 and other proteins in LPS-stimulated macrophages as an example to describe the AP-SWATH workflow. To validate the crucial role of TRAF6 in LPS signaling pathway, we first knocked out Traf6 in macrophage cell line RAW 264.7. As anticipated, deletion of TRAF6 impaired LPS responses as indicated by poor activation of MAP kinases (p38, JNK and ERK) and NF-κB (supplemental Fig. S1A). To purify endogenous TRAF6 complex, we generated Flag-knock-in TRAF6 RAW 264.7 cell line. Because of the similar LPS responsiveness between wildtype (WT) RAW264.7 and Flag-knock-in TRAF6 cells (supplemental Fig. S1B and S1F), we used SWATH-MS to quantify proteins in anti-Flag immunoprecipitation samples from the Flag-KI TRAF6 cell line. The cells were treated with LPS for 10 different time periods in biological quadruplicates and TRAF6 complexes were then purified by anti-Flag immunoprecipitation and subjected to SWATH-MS analysis (Fig. 1A).
Fig. 1.
The AP-SWATH workflow. A, The TRAF6 complexes were immunoprecipitated from RAW 264.7 cells treated with (100 ng/ml) LPS for indicated time-points, followed by digestion and IMAC enrichment. Tryptic peptides were analyzed by SWATH-MS, and phosphopeptides were analyzed by SWATH-MS and shotgun MS based DDA (data-dependent acquisition). B, Data processing procedures for SWATH-MS data. Group-DIA (data-independent acquisition) was used to construct pseudo-spectra from SWATH-MS data, and an internal library was made by these pseudo-spectra. The internal library and a premade external library of murine cell line were used either independently or in combination (combined library) in OpenSWATH analysis of SWATH-MS data. The final results were filtered at 1% global protein level. C, Comparison of the numbers of peptides and proteins detected in TRAF6 complexes (TRAF6 data set) by using internal, external or combined library.
We have previously built a spectral library (prebuilt library) containing 109,323 peptide sequences which were assigned to 8599 mouse proteins by extensive fractionation at protein and peptide level of murine cell line L929 (36). Untargeted analysis software, Group-DIA (36), was used to generate internal library directly from TRAF6 SWATH-MS data (Fig. 1B). The internal library or the prebuilt library was input into OpenSWATH (13) for detection of peptides. At 1% protein global FDR level, 16,418 peptide sequences were detected with prebuilt library, whereas 9423 peptides were identified with internal library. Among the identified peptides, 6720 peptides were detected by both libraries (Fig. 1C and supplemental Table S1). Indeed, prebuilt library-based strategy detected more peptides than internal library-based approach, and the internal library-based strategy did detect peptides that were not available in the prebuilt library. Importantly, TRAF1, TIRAP and TANK, which are known to play a role in LPS signaling pathway, were identified by the internal library, but not by prebuilt library (supplemental Table S1). To comprehensively analyze TRAF6 SWATH-MS data, we combined these two libraries and detected about double the amount of peptides and 50% more proteins compared with those detected by the internal library alone at 1% protein FDR (Fig. 1C and (supplemental Table S1). With the combined library, 16,519 unique proteotypic peptide sequences assigned to 2921 proteins were identified. To evaluate whether we achieved an in-depth exploration of TRAF6 SWATH-MS data, we searched literatures and found 18 of the identified proteins were previously reported to be involved in LPS-induced cell activation (1). Some of them have not been reported to be associated with TRAF6. Of these 18 proteins, 15 proteins were identified with more than one peptide (supplemental Table S1). Thus, using combination of comprehensive prebuilt library and internal library in AP-SWATH workflow provides in-depth exploration into protein interactions.
In addition to analyzing the TRAF6 complex, we also analyzed MyD88 and NEMO complexes using AP-SWATH workflow as described in Fig. 1. Flag-knock-in MyD88 and 3xFlag-NEMO-reconstituted Nemo−/− RAW 264.7 cell lines behaved similarly to WT RAW 264.7 cells in response to LPS stimulation (supplemental Fig. S1C, S1D, S1E, and S1F) and were used in our AP-SWATH analysis. In total, 2641, 2991, and 2657 proteins were quantified across 30, 40, and 21 IP samples in MyD88, TRAF6, and NEMO data sets at 1% global protein FDR, respectively (supplemental Table S2). About 85% of the proteins identified by the combined library can be found in microarray data from RAW 264.7 cells (37) (supplemental Table S2), suggesting the spectral library prebuilt from L929 cell line can be employed for targeted analysis of SWATH-MS data from RAW 264.7 cell lines.
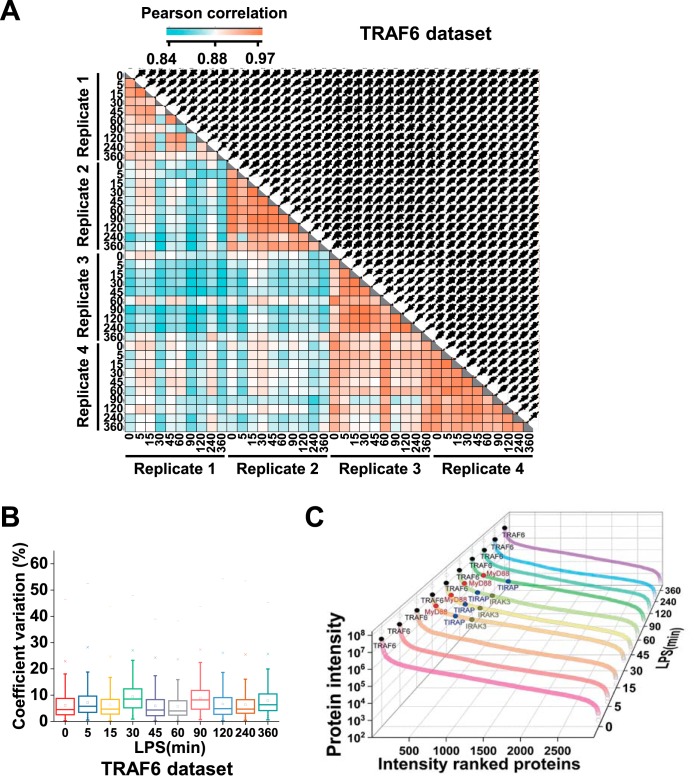
To evaluate the quality of our SWATH-MS analysis, we quantitatively compared protein abundance across different samples in each data set. Pearson's correlation coefficients were about 0.84–0.97 between any two different samples for TRAF6 data set, 0.80–0.96 for MyD88 data set and about 0.78–0.96 for NEMO data set (Fig. 2A, supplemental Fig. S2A and S3A). To further determine quantitative reproducibility in biological replicates, we computed the coefficient of variation (CV) at each time point. For all LPS treatment time points in three data sets, median CVs of log2-transformed protein abundance were below 10% (Fig. 2B, supplemental Fig. S2B and S3B). In TRAF6 data set, the minimal CV was 5.16 ± 3.66% (expressed as median ± standard deviation) at the first time point, and the maximal CV was 7.768 ± 4.082% at the fourth time point. Taken together, this demonstrates excellent reproducibility of the entire experiment.
Fig. 2.
Quality of the quantitative data of TRAF6 data set. A, Correlation analysis of protein intensities between any two samples. The matrix of correlation plots is shown, and the colors represent the indicated correlation coefficients. B, Quantitative reproducibility of protein intensities in biological quadruplicates. The coefficient of variation (CV), which is calculated by dividing the standard deviation of protein profiles by the mean, is reported as percentages. C, Dynamic range of all identified proteins in TRAF6 data set. The log10 abundances of proteins calculated using “TOP 3” approach in ten time-points are shown. Some interactors as well as the bait protein are highlighted.
To evaluate the dynamic range of the MyD88, TRAF6, and NEMO interactome, we elected to estimate the range of their abundances via the “TOP3 peptides” approach. Top3 label-free quantification method shows high sensitivity and accuracy, which also is the preferred method in the absence of reference protein measurements (38, 39). We plotted the estimated log10 protein abundances for the interacting proteins ordered from high to low abundance. The interacting protein abundances in TRAF6 data set were spread over about 4.5–5 orders of magnitude in each time point (Fig. 2C, supplemental Fig. S2C and S3C). The wide dynamic range of protein abundances showed the effective sensitivity of our AP-SWATH workflow.
Identification of High-confidence Interactors of MyD88, TRAF6 and NEMO
To obtain an overview of interacting proteins, we performed hierarchical clustering of MyD88, TRAF6 and NEMO interactions across different timepoints of LPS treatment (Fig. 3A, supplemental Fig. S4A and S5A). Majority of detected proteins in TRAF6 immunoprecipitates did not change in abundance across 10 time points of LPS treatment, implying that these proteins either consistently bound to TRAF6 or were nonspecifically pulled down (Fig. 3A). Some proteins in TRAF6 data set showed dynamic changes and were clustered tightly into two clusters (Fig. 3A). Cluster 1 included the proteins recruited to TRAF6 complex at 30–60 min after LPS treatment. TNIP1, TNIP2, TIRAP, IRAK1, IRAK2, IRAK3, IRAK4, TRAF2, and MyD88 were in this cluster. Cluster 2 contained the proteins whose intensities peaked at 120–360 min after LPS treatment. Those were MRP, OASL1, RSAD2, TNAP3, CMPK2, IRG1, TRAF1, IL1B, and CCL5. Similarly, clusters were identified from MyD88 and NEMO data sets (supplemental Fig. S4A and S5A). To further analyze the interactors of the bait proteins, we conducted a differential expression analysis individually at each time point (Fig. 3B, supplemental Fig. S4B and S5B). Based on fold of changes (log2(fold change) >1) and p value (-log10(p value) >1.5) at all time points, we deduced 91, 102 and 54 proteins from 2641, 2991, and 2657 quantified proteins in MyD88, TRAF6 and NEMO data sets respectively as the dynamic interactors of baits (supplemental Table S2). To further confirm these proteins were really increased or decreased in the given IPs at different time points of LPS treatment, we manually checked the XICs of all peptides of the proteins (Fig. 3C and supplementary File S1), and confirmed 24, 25, and 12 protein abundances increased in MyD88, TRAF6 and NEMO IP complexes respectively (supplemental Table S2). Western blotting results were also highly consistent with quantitative SWATH-MS data (Fig. 3D, supplemental Fig. S4C and S5C). Of these proteins, some were exclusively detected in the bait protein complexes at LPS treatment for a long time, such as at 240–360 min. To determine whether the detection of these proteins were associated with induction of their expression, we measured the mRNA expression levels of some of the proteins and found they were markedly upregulated by LPS treatment (supplemental Fig. S6A), implying that detection of these proteins in the bait protein complexes at 240–360 min of LPS treatment could be resulted from the increase of these protein levels rather than specific recruitment to bait proteins. To obtain a protein list of background binders of M2 beads, we generated a RAW 264.7 cell line expressing 3xFlag-vector and conducted immunoprecipitations using M2 beads in this cell line. The data of ten timepoints of LPS treatment in biological duplicates were collected. LPS-dependent background binders of M2 beads were determined by differential expression analysis (supplemental Fig. S6B) and were removed from the interactors of the bait protein IPs. Ultimately, we obtained 20, 24 and 12 high-confidence MyD88, TRAF6 and NEMO interactors, respectively (supplemental Table S2). As expected, majority of these proteins were well-established to be involved in LPS signaling pathway, but we identified an unknown protein, WRNIP1, which was recruited to MyD88 in an LPS-dependent way. WRNIP1 was recently reported to be involved in RIG-I-mediated antiviral signaling pathway (40), but the role of WRNIP1 in LPS signaling pathway remains undefined.
Dynamics of the Recruitment of Different IRAKs in the Initial LPS Signaling Cascade
Dynamic profiles of high-confidence interactors were shown in the Fig. 3E. In MyD88 and TRAF6 complexes, the highest abundance protein was IRAK2. The relative ratio of IRAK4:IRAK2 in MyD88 complex were consistent with that in TRAF6 complex (supplemental Table S3). However, the abundance of IRAK3 was 100 time lower than that of IRAK2 in MyD88 complex at 45–60 min, whereas its abundance was about one tenth of IRAK2 abundance at 30–60 min in TRAF6 complex. The difference of relative IRAK3 abundance suggested that it might have had performed distinct functions in these two complexes. In addition, IRAK family proteins exhibited distinct recruitment patterns in different complexes. In MyD88 complex, IRAK1 and IRAK3 abundance reached the maximum value at 15–30 min and 45–60 min respectively, whereas IRAK2 and IRAK4 abundances peaked at 30–45 min almost simultaneously. This suggested that IRAK1 was first recruited to MyD88, and IRAK2 and IRAK4 were subsequently recruited to MyD88 followed by IRAK3 recruitment. Previous work showed that IRAK4 was required for LPS-induced MAPK and NF-κB activation and cytokine production (41), whereas IRAK1 and IRAK2 functioned redundantly because only double deficiency of IRAK1 and IRAK2 blocked LPS signaling (42). Our dynamic analysis revealed that the timing of IRAK1 and IRAK2 to execute their function could be different. The interactions of TRAF6 with IRAK1, IRAK2, IRAK3, and IRAK4 occurred at the same time point of LPS stimulation. IRAK1 was dissociated from MyD88 and increased in abundance in TRAF6 complex at 45–60 min, suggesting that IRAK1 was recruited to MyD88 at LPS 15–30 min and shifted to TRAF6 at 45–60 min. It was believed that IRAK1-TRAF6 interaction enabled the release of TRAF6 from the receptor and initialized the downstream signaling (43–45). Intriguingly, IRAK2 was increasing from 0 min to 120 min in NEMO complex. At the meantime it was dissociated from MyD88 and TRAF6 from 60 min. This result implied that IRAK2 was recruited to MyD88 at 30–45 min, dissociated from it at 60–90 min and then translocated to NEMO at 90–120 min (Fig. 3F). IRAK2-NEMO association has not been documented, and biological implication of the interaction requires more research.
The Stoichiometry of MyD88 and TRAF6 Complexes Revealed a Signal Amplification Mechanism for MyD88-dependent TLR4 Signaling
Besides providing relative quantification of proteins across different samples, label-free SWATH-MS intensity can be used to estimate subunit stoichiometry in one sample (16). It should be noted that “Top3 peptide approach” label-free absolute quantification was employed in this study, which was not as accurate as stable-isotope labeled peptide methods.
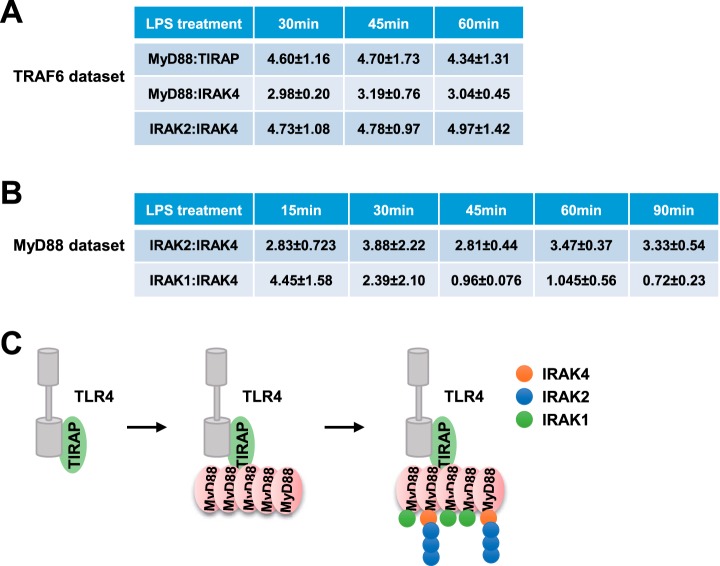
We first determined the stoichiometry of TIRAP and MyD88 in TRAF6 data set. MyD88 and TIRAP both harbor TIR (Toll-Interleukin-I receptor) domains, which can associate directly with TIR domains of TLR (Toll-Like Receptors). Indeed, MyD88 was reported to directly interact with the TIR domains of TLR3, 7 and 9, but the binding of MyD88 to TLR4 requires TIRAP (46, 47). TRAF6 was recruited to MyD88 complex at 30 min of LPS treatment and dissociated at 60 min (Fig. 3E), therefore we analyzed the TIRAP:MyD88 stoichiometry in TRAF6 data set during 30–60 min after LPS treatment (supplemental Table S3). As shown in Fig. 4A, the stoichiometry of TIRAP: MyD88 was about 1:5. This data suggested that TIRAP binds to TLR4, thereby allowing multiple MyD88 recruitment to TIRAP, which subsequently leads to signal cascade amplification.
Fig. 4.
The stoichiometry of MyD88 and TRAF6 complexes. A, The ratios of MyD88:TIRAP, MyD88:IRAK4, and IRAK2:IRAK4 in TRAF6 data set. The values are represented with the median value ± standard errors in biological quadruplicates. B, The ratio of IRAK2:IRAK4 and IRAK1:IRAK4 in MyD88 data set. The values are represented with the median value ± standard errors in biological triplicates. C, The proposed model for Myddosome assembly.
Next, we attempted to investigate the stoichiometry of IRAK family proteins. MyD88:IRAK4 in TRAF6 data set was about 3:1. The stoichiometry of IRAK1-IRAK4 and IRAK2-IRAK4 complexes was calculated in MyD88 data set where IRAK3 abundance was about 100-time lower than that of the other three proteins (supplemental Table S3). As shown in Fig. 4B, the ratio of IRAK1:IRAK4 changed significantly during 15–90 min after LPS stimulation. In contrast, the stoichiometry of IRAK2-IRAK4 complex showed more stability and remained about “3:1.” In TRAF6 complex, the IRAK2-IRAK4 stoichiometry was about “5:1,” slightly larger than “3:1” in MyD88 complex (Fig. 4A). Considering that MyD88 acts upstream of IRAK proteins and TRAF6 functions downstream, IRAK protein complex might have disassembled in TRAF6 complex. Therefore, the 3:1 stoichiometry of IRAK2-IRAK4 should be more reasonable. A previous study described a “1:1” of IRAK2:IRAK4 based on the structure of death domains of IRAK2 and IRAK4 (48), but our data demonstrated that it is “3:1” in MyD88 complex. These observations suggested IRAK2 was tightly associated with IRAK4 during recruitment to and dissociation from MyD88 protein.
Based on these results, we proposed a signal amplification model in MyD88-mediated LPS pathway (Fig. 4C). Although “Top3” quantification method has been reported to be about 2-fold error (38), some conclusions are not affected by the error. Specifically, ratio of “MyD88:TIRAP” was more than “1,” suggesting the signal was amplified in TIRAP-MyD88 complex. Additionally, ratio of “IRAK2-IRAK4” in TRAF6 and MyD88 complex should be greater than “1,” which also indicates potential signal amplification in this complex.
LPS-induced Dynamic Phosphorylation of High-confidence Interactors of TRAF6, NEMO and MyD88
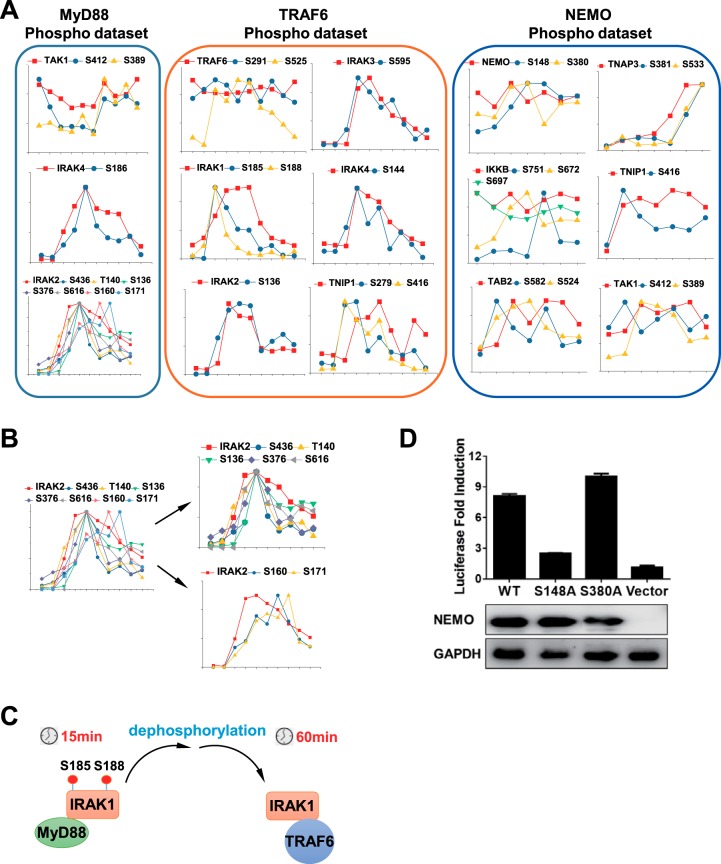
In addition to identification of key components in MyD88, TRAF6 and NEMO complexes, we also identified and quantified the phosphorylation events in these complexes. Phosphopeptides in IP samples were enriched using IMAC, followed by shotgun MS and SWATH-MS analysis separately. The phosphopeptides identified by these two methods were combined. Filtered at 1% peptide level and localization score of 0.7, 1893, 1269, and 1122 distinct phosphopeptides were identified in MyD88, TRAF6 and NEMO complexes, respectively (supplemental Table S4). Among these phosphopeptides, 1006, 1022, and 935 phosphopeptides can be quantified across the timepoints of LPS treatment in MyD88, TRAF6 and NEMO complexes, respectively (supplemental Table S4). We then focused on quantified phosphopeptides of aforementioned high-confidence interactors. To remove weak intensity phosphopeptides, we manually checked and extracted their XICs and excluded ambiguously localized phosphopeptides (supplemental Table S4). We identified 103 phosphosites in these high-confidence interactors and 41 of them have not been reported (supplemental Table S4). We first compared the phosphopetide profiles with their corresponding nonphophopeptide profiles. To obtain the stoichiometry of the phosphorylation sites, temporal phosphorylation levels of each site were subsequently compared with their protein levels (Fig. 5A). S389 and S412 on TAK1, S136, T140, S160, S171, S376, S436, and S616 on IRAK2, and S186 on IRAK4 were quantified across ten LPS timepoints in MyD88 data set. S412 on TAK1 and S186 on IRAK4 showed a similar pattern with their proteins, suggesting the changed phosphorylation probably were attributed to varied protein levels. S389 on TAK1 exhibited distinct profile compared with the protein profile, suggesting S389 was phosphorylated in an LPS-dependent way. Of seven quantified phosphosites on IRAK2, 5 phosphosites showed a similar pattern with IRAK2 protein. S160 and S171 phosphorylation in IRAK2 apparently showed different profiles from that of their protein, suggesting the phosphorylation on these sites occurred after IRAK2 recruitment to MyD88 (Fig. 5B). In TRAF6 complex, phosphorylation of S525 on TRAF6 was LPS-induced, whereas S291 phosphorylation was LPS-independent. Notably, S185 and S188 phosphorylation on IRAK1 in TRAF6 complex peaked at 15 min and decreased sharply, whereas IRAK1 protein abundance in TRAF 6 complex increased from 15 min and stayed at the plateau from 30–60 min. This discrepancy between IRAK1 protein and S185 and S188 phosphorylation of IRAK1 suggested the two Serine amino acids on IRAK1 in TRAF6 complex were not phosphorylated. Given that IRAK1 was first recruited to MyD88 and then shifted to TRAF6, S185 and S188 phosphorylation on IRAK1 in MyD88 complex were probably dephosphorylated followed by translocation to TRAF6 (Fig. 5C). In NEMO complex, phosphorylation of S148, but not S380, was regulated in an LPS-dependent manner. Similarly, S672 and S751 on IKKB, S389 on TAK1, and S381 and S533 on TNAP3 (also named as A20) were phosphorylated in an LPS-dependent manner. S381 on A20 was reported to be phosphorylated by IKKβ, which enhanced the function of A20 to downregulate pro-inflammatory cytokines (49).
Fig. 5.
Identification of phosphosites on high-confidence interactors in MyD88, TRAF6 and NEMO phospho-data set. A, Dynamic profiles of proteins and their phosphosites in MyD88, TRAF6, and NEMO phospho-data set. B, Dynamic profiles of seven phosphosites on IRAK2 in MyD88 phospho-data set. C, Model of dephosphorylation of S185 and S188 on IRAK1 in TRAF6 complex derived from A. D, The ability of NEMO and its mutants in inducing NF-κB activation. The NF-κB firefly luciferase reporter plasmid and renilla luciferase transfection control plasmid were co-transfected into 293T cells along with the plasmid encoding wildtype NEMO, S148A NEMO, or S380A NEMO. Luciferase activities were measured 24 h post transfection. Values represent the fold of luciferase activity induction relative to cells transfected with empty vector.
Because TRAF6, IRAK1, IKKΒ, NEMO, and TAK1 were involved in NF-κB activation, we sought to examine whether the identified phosphosites on them affected the ability to induce NF-κB activation. To this end, we transfected wildtype and phosphorylation site mutant constructs into 293T cells and determined their ability to induce NF-κB luciferase reporter activity. Disruption of most phosphorylation sites did not affect these protein function to induce NF-κB activity (supplemental Fig. S7). However, we did find that substitution of S148, but not 380, to alanine impaired NEMO overexpression induced NF-κB reporter gene expression (Fig. 5D). This result showed that S148 phosphorylation on NEMO most likely played a role in NF-κB activation.
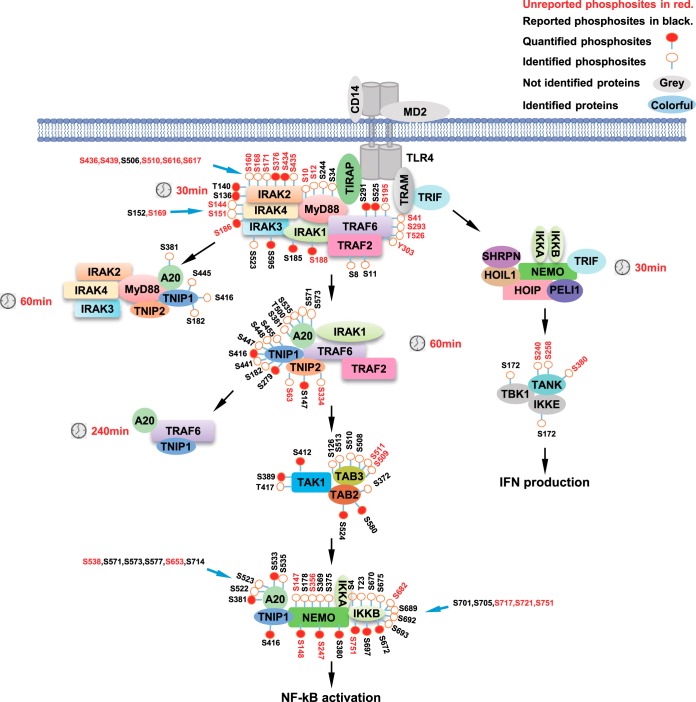
Overview of LPS-induced Signaling Events of Protein Interactions and Phosphorylation
To have an overview of LPS-induced signaling events, we overlaid the data of temporal interaction of signaling molecules and the phosphoproteomics data onto a literature-derived LPS signaling pathway and summarized them in Fig. 6. Almost all known key players in LPS signaling pathway are in the identified proteins using our AP-SWATH workflow. We also identified several high-confidence LPS-induced phosphorylation sites, in which about half were not reported before (phosphosites in red in Fig. 6). This study may serve as a resource for in-depth understanding of LPS signaling pathway.
Fig. 6.
Summary of all identified phosphosites in LPS signaling pathway.
DISCUSSION
In this study, we quantitatively measured time-resolved protein interactions and phosphorylation in LPS signaling pathway using SWATH-MS. This approach is critically dependent on the peptide assay library that is used to identify and quantify them. The peptide libraries are usually constructed by DDA data sets. In this study, we used a comprehensive spectral library containing 8599 murine proteins to analyze the IP SWATH-MS data. Even with this library, not all peptides in SWATH-MS data from IP samples were identified. Targeted analysis using an internal library directly from SWATH-MS led to the identification of extra key proteins that are involved in LPS signaling pathway. This suggested that in-depth exploration of IP SWATH-MS necessitates a combination of the external library and the internal library. Whether this combination is required for the full interpretation of SWATH-MS data from complex samples such as cell lysates or tissues needs further research.
We quantified about 2500–3000 proteins in TRAF6, MyD88, and NEMO IP data sets across about 21–40 IP samples. Of these proteins, the majority were probably background binders of M2 beads as their abundances did not change during LPS treatment. To identify LPS-dependent background binders, we performed immunoprecipitations in 3xFlag-vector-expressing cells at a serial of time points of LPS stimulation. We detected 943 proteins in IP samples using 3xFlag-vector-expressing cells (supplemental Table S2). These proteins contained “normal background binders” and “LPS-dependent background binders,” which were both excluded from interactors of the bait proteins. Importantly, the number of identified proteins in negative-control experiments were apparently less than those identified in MyD88, TRAF6 and NEMO data sets. This suggested that pull-downs in negative-control cells represent only part of the background proteins in the IP using the bait protein.
We selected the proteins whose abundance in the IPs significantly changed on LPS treatment. Selection based on this criterion has successfully retrieved high-confidence interactors of the bait proteins, and most of the interactors have been proved in other studies. Nevertheless, there are other reasons except for LPS-induced recruitment that can lead to our detection of changes in protein abundances in the complexes. One is that the amounts of several proteins were increased after LPS stimulation, which could then increase the background binding of these proteins. Another reason is that proteins would nonspecifically bind to M2 beads if they formed aggregates after LPS stimulation. Although the formation of large protein aggregates was not reported in LPS-treated RAW 264.7 cells, we had employed high speed centrifugation to remove the insoluble large aggregates.
The criterion will miss the interactors that are always binding to bait proteins. Indeed, IKKA and IKKB that are well-established binders of NEMO are not selected in this study. However, the percent of these proteins of all interactors (2/34 = 5.88%) is relatively low. In addition, we showed that automated analysis by OpenSWATH requires manual inspection. Peak groups may be wrongly assigned or the XICs of correctly assigned peptides may be of poor quality, which are the reasons for us to remove the peptide results by OpenSWATH analysis. Further, we removed the proteins whose peptide XICs showed different quantitative trends in biological replicates over LPS treatment (supplementary File S1).
Based on the temporal interactor profiles, we demonstrated differential recruiting pattern of IRAK family proteins in MyD88, TRAF6 and NEMO complexes, and proposed a model of assembling and de-assembling of IRAKs in the signaling complexes of LPS (Fig. 3F and 4C). Our data suggest IRAK1 recruitment and phosphorylation was not necessarily related at least in RAW 264.7 cells (Fig. 5C). It is unexpected that IRAK2 associated with NEMO at 15min-120min after LPS treatment. Because it is well known that IRAK2 and IRAK4 formed a tight complex in Myddosome, the IRAK2 in NEMO complex may perform unknown functions.
Stoichiometry of a given complex was often deduced from the structure data. However, the stoichiometry in biological reactions could be different or dynamically changeable. SWATH-MS enabled the calculation of relative protein quantitation in one sample, and thus allowed us to determine the ratio of MyD88, TIRAP, IRAK2, and IRAK4 in Myddosome.
We also identified many high-confidence phosphosites in the interactors in TRAF6, MyD88 and NEMO complexes. Many of them were not reported before. Especially, 12 phosphosites on IRAK2 were identified, only one of which is included in the Phosphosite.org database. We also showed that one of the identified phosphosites on NEMO affected the ability of NEMO to induce NF-κB activation.
Finally, we believe the data of our AP-SWATH-MS analysis will offer an important resource for further LPS pathway research.
DATA AVAILABILITY
All SWATH MS raw data, all spectral libraries and XICs figures have been made available through the PeptideAtlas database (www.peptideatlas.org/PASS/PASS01283).
Supplementary Material
Acknowledgments
We thank Dr. Zhuobin Xu and Dr. Yuwei Yu for help in using high performance computer.
Footnotes
* This work was supported by the National Natural Science Foundation of China (81788101), National Basic Research Program of China (973 Program 2015CB553800), the National Natural Science Foundation of China (31420103910 and 81630042), the 111 Project (B12001), the National Science Foundation of China for Fostering Talents in Basic Research (J1310027) and the Young Scientists Fund of the National Natural Science Foundation of China (31601115).
 This article contains supplemental Figures, Tables and Files.
This article contains supplemental Figures, Tables and Files.
1 The abbreviations used are:
- LPS
- lipopolysaccharides
- TLR4
- Toll-like receptor 4.
REFERENCES
- 1. Lu Y. C., Yeh W. C., and Ohashi P. S. (2008) LPS/TLR4 signal transduction pathway. Cytokine 42, 145–151 [DOI] [PubMed] [Google Scholar]
- 2. Lord K. A., Hoffman-Liebermann B., and Liebermann D. A. (1990) Nucleotide sequence and expression of a cDNA encoding MyD88, a novel myeloid differentiation primary response gene induced by IL6. Oncogene 5, 1095–1097 [PubMed] [Google Scholar]
- 3. Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., and Akira S. (2003) Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301, 640–643 [DOI] [PubMed] [Google Scholar]
- 4. Horng T., Barton G. M., and Medzhitov R. (2001) TIRAP: an adapter molecule in the Toll signaling pathway. Nat. Immunol. 2, 835–841 [DOI] [PubMed] [Google Scholar]
- 5. Fitzgerald K. A., Palsson-McDermott E. M., Bowie A. G., Jefferies C. A., Mansell A. S., Brady G., Brint E., Dunne A., Gray P., Harte M. T., McMurray D., Smith D. E., Sims J. E., Bird T. A., and O'Neill L. A. (2001) Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature 413, 78–83 [DOI] [PubMed] [Google Scholar]
- 6. Lomaga M. A., Yeh W. C., Sarosi I., Duncan G. S., Furlonger C., Ho A., Morony S., Capparelli C., Van G., Kaufman S., van der Heiden A., Itie A., Wakeham A., Khoo W., Sasaki T., Cao Z., Penninger J. M., Paige C. J., Lacey D. L., Dunstan C. R., Boyle W. J., Goeddel D. V., and Mak T. W. (1999) TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 13, 1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C., and Chen Z. J. (2000) Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103, 351–361 [DOI] [PubMed] [Google Scholar]
- 8. Hacker H., Redecke V., Blagoev B., Kratchmarova I., Hsu L. C., Wang G. G., Kamps M. P., Raz E., Wagner H., Hacker G., Mann M., and Karin M. (2006) Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature 439, 204–207 [DOI] [PubMed] [Google Scholar]
- 9. Oganesyan G., Saha S. K., Guo B., He J. Q., Shahangian A., Zarnegar B., Perry A., and Cheng G. (2006) Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature 439, 208–211 [DOI] [PubMed] [Google Scholar]
- 10. Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., and Maniatis T. (2003) IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4, 491–496 [DOI] [PubMed] [Google Scholar]
- 11. Moynagh P. N. (2005) TLR signalling and activation of IRFs: revisiting old friends from the NF-kappaB pathway. Trends Immunol. 26, 469–476 [DOI] [PubMed] [Google Scholar]
- 12. Sato S., Sanjo H., Takeda K., Ninomiya-Tsuji J., Yamamoto M., Kawai T., Matsumoto K., Takeuchi O., and Akira S. (2005) Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat. Immunol. 6, 1087–1095 [DOI] [PubMed] [Google Scholar]
- 13. Ajibade A. A., Wang Q., Cui J., Zou J., Xia X., Wang M., Tong Y., Hui W., Liu D., Su B., Wang H. Y., and Wang R. F. (2012) TAK1 negatively regulates NF-kappaB and p38 MAP kinase activation in Gr-1+CD11b+ neutrophils. Immunity 36, 43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gillet L. C., Navarro P., Tate S., Rost H., Selevsek N., Reiter L., Bonner R., and Aebersold R. (2012) Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell Proteomics 11, O111 016717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ludwig C., Gillet L., Rosenberger G., Amon S., Collins B. C., and Aebersold R. (2018) Data-independent acquisition-based SWATH-MS for quantitative proteomics: a tutorial. Mol. Syst. Biol. 14, e8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heusel M., Bludau I., Rosenberger G., Hafen R., Frank M., Banaei-Esfahani A., van Drogen A., Collins B. C., Gstaiger M., and Aebersold R. (2019) Complex-centric proteome profiling by SEC-SWATH-MS. Mol. Syst. Biol. 15, e8438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hahne H., Pachl F., Ruprecht B., Maier S. K., Klaeger S., Helm D., Medard G., Wilm M., Lemeer S., and Kuster B. (2013) DMSO enhances electrospray response, boosting sensitivity of proteomic experiments. Nat. Methods 10, 989–991 [DOI] [PubMed] [Google Scholar]
- 18. Chambers M. C., Maclean B., Burke R., Amodei D., Ruderman D. L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., Hoff K., Kessner D., Tasman N., Shulman N., Frewen B., Baker T. A., Brusniak M. Y., Paulse C., Creasy D., Flashner L., Kani K., Moulding C., Seymour S. L., Nuwaysir L. M., Lefebvre B., Kuhlmann F., Roark J., Rainer P., Detlev S., Hemenway T., Huhmer A., Langridge J., Connolly B., Chadick T., Holly K., Eckels J., Deutsch E. W., Moritz R. L., Katz J. E., Agus D. B., MacCoss M., Tabb D. L., and Mallick P. (2012) A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deutsch E. W., Mendoza L., Shteynberg D., Farrah T., Lam H., Tasman N., Sun Z., Nilsson E., Pratt B., Prazen B., Eng J. K., Martin D. B., Nesvizhskii A. I., and Aebersold R. (2010) A guided tour of the Trans-Proteomic Pipeline. Proteomics 10, 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eng J. K., Jahan T. A., and Hoopmann M. R. (2013) Comet: an open-source MS/MS sequence database search tool. Proteomics 13, 22–24 [DOI] [PubMed] [Google Scholar]
- 21. Craig R., and Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 22. Keller A., Nesvizhskii A. I., Kolker E., and Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 23. Shteynberg D., Deutsch E. W., Lam H., Eng J. K., Sun Z., Tasman N., Mendoza L., Moritz R. L., Aebersold R., and Nesvizhskii A. I. (2011) iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol. Cell Proteomics 10, M111 007690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reiter L., Claassen M., Schrimpf S. P., Jovanovic M., Schmidt A., Buhmann J. M., Hengartner M. O., and Aebersold R. (2009) Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol. Cell Proteomics 8, 2405–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lam H., Deutsch E. W., Eddes J. S., Eng J. K., Stein S. E., and Aebersold R. (2008) Building consensus spectral libraries for peptide identification in proteomics. Nat. Methods 5, 873–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rost H. L., Sachsenberg T., Aiche S., Bielow C., Weisser H., Aicheler F., Andreotti S., Ehrlich H. C., Gutenbrunner P., Kenar E., Liang X., Nahnsen S., Nilse L., Pfeuffer J., Rosenberger G., Rurik M., Schmitt U., Veit J., Walzer M., Wojnar D., Wolski W. E., Schilling O., Choudhary J. S., Malmstrom L., Aebersold R., Reinert K., and Kohlbacher O. (2016) OpenMS: a flexible open-source software platform for mass spectrometry data analysis. Nat. Methods 13, 741–748 [DOI] [PubMed] [Google Scholar]
- 27. Reiter L., Rinner O., Picotti P., Huttenhain R., Beck M., Brusniak M. Y., Hengartner M. O., and Aebersold R. (2011) mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nat. Methods 8, 430–435 [DOI] [PubMed] [Google Scholar]
- 28. Rosenberger G., Bludau I., Schmitt U., Heusel M., Hunter C. L., Liu Y., MacCoss M. J., MacLean B. X., Nesvizhskii A. I., Pedrioli P. G. A., Reiter L., Rost H. L., Tate S., Ting Y. S., Collins B. C., and Aebersold R. (2017) Statistical control of peptide and protein error rates in large-scale targeted data-independent acquisition analyses. Nat. Methods 14, 921–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rost H. L., Liu Y., D'Agostino G., Zanella M., Navarro P., Rosenberger G., Collins B. C., Gillet L., Testa G., Malmstrom L., and Aebersold R. (2016) TRIC: an automated alignment strategy for reproducible protein quantification in targeted proteomics. Nat. Methods 13, 777–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Claassen M., Reiter L., Hengartner M. O., Buhmann J. M., and Aebersold R. (2012) Generic comparison of protein inference engines. Mol. Cell Proteomics 11, O110 007088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., and Cox J. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 [DOI] [PubMed] [Google Scholar]
- 32. Caron E., Roncagalli R., Hase T., Wolski W. E., Choi M., Menoita M. G., Durand S., Garcia-Blesa A., Fierro-Monti I., Sajic T., Heusel M., Weiss T., Malissen M., Schlapbach R., Collins B. C., Ghosh S., Kitano H., Aebersold R., Malissen B., and Gstaiger M. (2017) Precise temporal profiling of signaling complexes in primary cells using SWATH mass spectrometry. Cell reports 18, 3219–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Collins B. C., Gillet L. C., Rosenberger G., Rost H. L., Vichalkovski A., Gstaiger M., and Aebersold R. (2013) Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14–3-3 system. Nat. Methods 10, 1246–1253 [DOI] [PubMed] [Google Scholar]
- 34. Lambert J. P., Ivosev G., Couzens A. L., Larsen B., Taipale M., Lin Z. Y., Zhong Q., Lindquist S., Vidal M., Aebersold R., Pawson T., Bonner R., Tate S., and Gingras A. C. (2013) Mapping differential interactomes by affinity purification coupled with data-independent mass spectrometry acquisition. Nat. Methods 10, 1239–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Navarro P., Kuharev J., Gillet L. C., Bernhardt O. M., MacLean B., Rost H. L., Tate S. A., Tsou C. C., Reiter L., Distler U., Rosenberger G., Perez-Riverol Y., Nesvizhskii A. I., Aebersold R., and Tenzer S. (2016) A multicenter study benchmarks software tools for label-free proteome quantification. Nat. Biotechnol. 34, 1130–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li Y., Zhong C. Q., Xu X., Cai S., Wu X., Zhang Y., Chen J., Shi J., Lin S., and Han J. (2015) Group-DIA: analyzing multiple data-independent acquisition mass spectrometry data files. Nat. Methods 12, 1105–1106 [DOI] [PubMed] [Google Scholar]
- 37. Murray R. Z., Wylie F. G., Khromykh T., Hume D. A., and Stow J. L. (2005) Syntaxin 6 and Vti1b form a novel SNARE complex, which is up-regulated in activated macrophages to facilitate exocytosis of tumor necrosis Factor-alpha. J. Biol. Chem. 280, 10478–10483 [DOI] [PubMed] [Google Scholar]
- 38. Ahrne E., Molzahn L., Glatter T., and Schmidt A. (2013) Critical assessment of proteome-wide label-free absolute abundance estimation strategies. Proteomics 13, 2567–2578 [DOI] [PubMed] [Google Scholar]
- 39. Al Shweiki M. R., Monchgesang S., Majovsky P., Thieme D., Trutschel D., and Hoehenwarter W. (2017) Assessment of Label-Free Quantification in Discovery Proteomics and Impact of Technological Factors and Natural Variability of Protein Abundance. J. Proteome Res. 16, 1410–1424 [DOI] [PubMed] [Google Scholar]
- 40. Tan P., He L., Cui J., Qian C., Cao X., Lin M., Zhu Q., Li Y., Xing C., Yu X., Wang H. Y., and Wang R. F. (2017) Assembly of the WHIP-TRIM14-PPP6C Mitochondrial Complex Promotes RIG-I-Mediated Antiviral Signaling. Mol. Cell 68, 293–307.e5 [DOI] [PubMed] [Google Scholar]
- 41. Suzuki N., Suzuki S., Duncan G. S., Millar D. G., Wada T., Mirtsos C., Takada H., Wakeham A., Itie A., Li S., Penninger J. M., Wesche H., Ohashi P. S., Mak T. W., and Yeh W. C. (2002) Severe impairment of interleukin-1 and Toll-like receptor signalling in mice lacking IRAK-4. Nature 416, 750–756 [DOI] [PubMed] [Google Scholar]
- 42. Kawagoe T., Sato S., Matsushita K., Kato H., Matsui K., Kumagai Y., Saitoh T., Kawai T., Takeuchi O., and Akira S. (2008) Sequential control of Toll-like receptor-dependent responses by IRAK1 and IRAK2. Nat. Immunol. 9, 684–691 [DOI] [PubMed] [Google Scholar]
- 43. Wesche H., Henzel W. J., Shillinglaw W., Li S., and Cao Z. (1997) MyD88: an adapter that recruits IRAK to the IL-1 receptor complex. Immunity 7, 837–847 [DOI] [PubMed] [Google Scholar]
- 44. Cao Z., Xiong J., Takeuchi M., Kurama T., and Goeddel D. V. (1996) TRAF6 is a signal transducer for interleukin-1. Nature 383, 443–446 [DOI] [PubMed] [Google Scholar]
- 45. Qian Y., Commane M., Ninomiya-Tsuji J., Matsumoto K., and Li X. (2001) IRAK-mediated translocation of TRAF6 and TAB2 in the interleukin-1-induced activation of NFkappa B. J. Biol. Chem. 276, 41661–41667 [DOI] [PubMed] [Google Scholar]
- 46. Horng T., Barton G. M., Flavell R. A., and Medzhitov R. (2002) The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors. Nature 420, 329–333 [DOI] [PubMed] [Google Scholar]
- 47. Yamamoto M., Sato S., Hemmi H., Sanjo H., Uematsu S., Kaisho T., Hoshino K., Takeuchi O., Kobayashi M., Fujita T., Takeda K., and Akira S. (2002) Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature 420, 324–329 [DOI] [PubMed] [Google Scholar]
- 48. Lin S. C., Lo Y. C., and Wu H. (2010) Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature 465, 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hutti J. E., Turk B. E., Asara J. M., Ma A., Cantley L. C., and Abbott D. W. (2007) IkappaB kinase beta phosphorylates the K63 deubiquitinase A20 to cause feedback inhibition of the NF-kappaB pathway. Mol. Cell Biol. 27, 7451–7461 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All SWATH MS raw data, all spectral libraries and XICs figures have been made available through the PeptideAtlas database (www.peptideatlas.org/PASS/PASS01283).