The proteome and phosphoproteome have been determined for BRAF inhibitor resistant and sensitive cells by applying quantitative MS-based proteomics. We identified the intermediate filament protein nestin as one of the highest downregulated proteins in melanoma cells and tumors. The results reported reveal a link between loss of nestin and an invasive phenotype and provide a quantitative view of PI3K/Akt and integrin pathways involved in resistant and nestin knockout cells.
Keywords: Melanoma, Drug resistance, Mass Spectrometry, SILAC, Phosphoproteome, Cytoskeletal proteins, Nestin, Quantitative proteomics, Signaling pathways, Skin cancer
Graphical Abstract
Highlights
Quantitative phosphoproteome of BRAF drug-resistance in melanoma cells.
Cytoskeletal proteins are downregulated in resistant vs. sensitive cells.
Nestin is associated with an invasive phenotype and resistance to MEK and BRAF inhibitors.
Nestin depletion affects PI3K/AKT and integrin signaling similar to resistant cells.
Abstract
Targeted inhibition of mutated kinases using selective MAP kinase inhibitors in malignant melanoma often results in temporary improvement of clinical symptoms followed by rapid development of resistance. To gain insights in molecular processes that govern resistance, we performed SILAC-based quantitative proteomics profiling of vemurafenib-resistant and -sensitive melanoma cells. Among downregulated proteins in vemurafenib-resistant cell lines we detected multiple proteins involved in cytoskeletal organization and signaling, including the intermediate filament nestin, which was one of the most downregulated proteins. Previous studies showed that nestin is expressed in various types of solid tumors and its abundance correlates with malignant phenotype of transformed cells. However, the role of nestin in cancer cells regarding acquired resistance is still poorly understood. We performed CRISPR/Cas9 knockout of the nestin gene (NES) in vemurafenib-sensitive cells and showed that loss of nestin leads to increased cellular proliferation and colony formation upon treatment with BRAFV600E and MEK inhibitors. Moreover, nestin depletion led to increased invasiveness and metalloproteinase activity like the phenotype of melanoma cells with acquired resistance to the BRAF inhibitor. Finally, phosphoproteome analysis revealed that nestin depletion influenced signaling through integrin and PI3K/AKT/mTOR pathways and led to increased focal adhesion kinase abundance and phosphorylation. Taken together, our results reveal that nestin is associated with acquired vemurafenib resistance in melanoma cells.
Malignant melanoma is the 19th most common cancer worldwide, accounting for ∼232,000 new cases in 2012 (1). Although melanoma accounts for less than one percent of skin cancer cases, it is responsible for 79% of skin cancer-related deaths (2). Therapies for advanced melanoma have changed greatly in recent years with the US Food and Drug Administration approval of several immunotherapy and targeted drugs, including cobimetinib, an inhibitor of MEK kinases (3), and vemurafenib, a BRAF inhibitor, for patients carrying the BRAFV600E mutation (4). These drugs have a response rate of ∼50% as monotherapies and result in an average survival benefit of four to six months (5–7). However, almost all patients ultimately develop resistance to drug treatment (5, 8). Therefore, an understanding of the acquired resistance is essential for the development of effective therapies for malignant melanoma. Multiple cellular pathways have been postulated to explain drug resistance, ranging from signal transduction networks to transcriptional pathways (9). The majority of kinase inhibitor resistance development is caused by molecular or genetic alterations that lead to MAPK pathway reactivation. A variety of genetic causes, including NRAS (10, 11), KRAS (12) and MEK mutations (13), alternative splicing or amplification of BRAF and loss of NF1 (9, 11, 12), have been identified in tumors with acquired resistance. In addition, activation of the PI3K/AKT pathway can be responsible for BRAF inhibitor resistance—because of downregulation of PTEN through loss or mutational inactivation, or somatic mutations in AKT1/3 and PIK3CA (11, 12).
Nestin, a member of the type VI intermediate filament protein family, was originally described as a stem cell/progenitor cell marker, especially during migration and proliferation phases in early embryonic development (14). Expression of nestin is also associated with the regulation of cell death in neural progenitor cells, podocytes of kidneys and neuromuscular junction development in a CDK5-dependent manner (15). In adult tissue, it plays an important role in regeneration processes where it is localized to tissue/organ-specific sites (16). Previous studies have reported that nestin is expressed in various human malignancies, including pancreatic cancer (17, 18), prostate cancer (19), breast cancer (20), glioblastomas (21), gastrointestinal stromal tumors (22), trichoblastoma (23), angiosarcoma (22) and malignant melanoma (24, 25). In some tumor types, nestin expression correlates with aggressive growth, metastasis, migration and poor prognosis (18); however, the roles of nestin in cancer cells have not been characterized at a molecular level. In advanced stages of melanoma, nestin- and CD133-positive melanoma cells were detected in the peripheral blood of patients, at the invading front and at sites of melanoma metastases (26–28). These studies indicate that nestin could be significantly involved in the invasion and distant metastasis of melanomas. In a large-scale proteomic approach, nestin was found to be a useful diagnostic and prognostic biomarker that can potentially distinguish melanoma subtypes and can help to predict melanoma aggressiveness in these different subgroups (29). Interestingly, depletion of nestin in melanoma was shown to increase expression of matrix metalloproteinases (MMP)1 and enhance melanoma invasion (30). Recent evidence indicates that nestin downregulation in prostate cancer cell lines triggers an expression pattern of phosphorylated focal adhesion kinase (FAK). Phosphorylated FAK (pFAK) localizes at the cell membrane and promotes integrin clustering. This results in pFAK- and integrin-dependent matrix degradation and an invasive phenotype (31). In the context of targeted BRAFV600E and MEK inhibitor therapy in melanoma, a loss of nestin expression in tumor cells was identified immediately after treatment therapy (32). All these findings suggest that nestin is associated with tumorigenesis, however, little is known about the role of nestin in melanoma and the relationship of nestin and acquired resistance.
In this study, we use quantitative proteomics to identify proteome and phosphoproteome alterations in A375 melanoma cells resistant to BRAFV600E inhibitor vemurafenib. Our analysis identified nestin as one of the most downregulated proteins in resistant cells. Extensive biological follow-up revealed its connection with invasiveness and cell survival of resistant melanoma cells. Finally, phosphoproteome analysis revealed that nestin depletion influenced signaling through integrin and PI3K/AKT/mTOR pathways.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
The (phospho)proteomics data is derived from three sets of samples prepared and analyzed by LC-MS/MS. A total of 143 runs analyses were performed with 230 min gradient for proteome, 42 min gradient for fractionated proteome and 90 min gradient for phosphoproteome measurements on a Q Exactive HF mass spectrometer. In part 1, SILAC labeled A375 S and A375 R cells (“light,” “medium,” and vice versa) were used in two different screens (123 samples); screen 1, proteome and phosphoproteome measurements (33 samples, three biological replicates (11 per replicate), ten rounds of phosphopeptide enrichment for each replicate), whereas in screen 2, the proteome was fractionated into 30 fractions (90 samples, three biological replicates (30 per replicate)). In part 2, SILAC labeled Nes-KO, A375 S and A375 R cells were used (“light,” “medium,” “heavy”) (22 samples, two biological replicates (eleven per replicate), ten rounds of phosphopeptide enrichment per replicate). Raw data was processed by MaxQuant software as described below. Statistical analysis was performed with Perseus (t test, FDR < 0.01, s0 = 1) (version 1.5.0.31), STRING: functional protein association networks analysis (STRING Consortium 2018) and GraphPad Prism (version 7.04). For detailed description of statistical analysis of each experiment see MS data analysis and statistical analysis in the section methods.
All biological assays were performed in three biological and technical replicates, so that appropriate statistical analysis could be performed. Statistical analysis was performed with two-tailed unpaired t test in GraphPad Prism. p values < 0.05 were considered statistically significant, with * for p < 0.05, ** for p < 0.01, *** for p < 0.001 and **** for p < 0.0001. In each experiment, separate controls were included. Images of experiments and Western blotting were quantified using ImageJ software.
Chemicals
Stock solutions of the BRAFV600E inhibitor PLX4720 (vemurafenib analog, Selleckchem, Houston, TX) and the MEK inhibitor cobimetinib (Hycultec, Beutelsbach, Germany) were prepared in dimethyl sulfoxide (DMSO).
Cell Culture and SILAC Labeling
The use of human tissue from an internal biobank was approved by the local ethical committee (781/2018BO2) and experiments were performed in accordance with the declaration of Helsinki Principles. The human metastatic BRAFV600E-mutated melanoma cell lines A375, Mel1617, 451lu, SKMel28 and SKMel19 were used in this study (33, 34). The generation of the cell lines with acquired resistance to vemurafenib analogue PLX4720 (for simplicity referred to as “vemurafenib” in the Results section) was conducted as described previously (33) (List of used cell lines in supplementary Information S1). Cells were either grown in RPMI 1640 (Sigma-Aldrich, Darmstadt, Germany) containing penicillin/streptomycin (100 U/ml, PAN) and FBS (10%, PAN) or RPMI 1640 SILAC (Sigma-Aldrich) lacking arginine, lysine and leucine. Leucine (12.5 mg/ml, Sigma-Aldrich), penicillin/streptomycin (100 U/ml, PAN, Aidenbach, Germany), dialyzed FBS (10%, PAN) and stable isotope-encoded arginine and lysine were added to the SILAC medium. The “light” SILAC media was further supplemented with l-[12C6,14N2] lysine (Lys0) and l-[12C6,14N4] arginine (Arg0) (Cambridge Isotope Laboratories, Tewksbury, MA), whereas l-[2H4] lysine (Lys4) and l-[13C6] arginine (Arg6) were added to the “medium” SILAC media and l-[13C6,15N2] lysine (Lys8) and l-[13C6,15N4] arginine (Arg10) to “heavy” SILAC media. Cells were grown in a humidified atmosphere, 5% CO2 at 37 °C in either RPMI 1640 or “light,” “medium,” or “heavy” RPMI 1640 SILAC media.
Protein Extraction of Cultured Cells
Cell lysis was performed in lysis buffer (6 m urea, 2 m thiourea, 10 mm Tris pH 8.0) supplemented with protease inhibitor (complete Mini EDTA-free tablets, Roche, Basel, Switzerland), phosphatase inhibitor buffers (5 mm glycerol-2-phosphate, 5 mm sodium fluoride, and 1 mm sodium orthovanadate) and 1% N-Octylglucoside (NOG) on ice for 10 min. DNA and RNA in the lysate was removed using benzonase (Merck, Darmstadt, Germany) for 10 min on room temperature (RT) followed by centrifugation at 2800 × g (10 °C, 20 min). Remaining NOG detergent was removed by acetone precipitation. Briefly, four volumes of acetone (−20 °C), one volume of methanol were added, and the proteins were precipitated overnight at −20 °C. After centrifugation (2800 × g, 4 °C, 20 min), the detergent-containing supernatant was removed, and the protein pellet was washed with 80% acetone (−20 °C). Protein pellets were then resolved in lysis buffer without NOG and protein concentration was measured using Bradford assay.
Sample Preparation for MS Analysis
Extracted proteins from each cell line were mixed 1:1 (“light” to “medium”) or 1:1:1 (“light” to “medium” to “heavy”) and reduced with 10 mm DTT for 1 h, alkylated with 55 mm iodoacetamide for an additional hour and digested with Lys-C (Lysyl Endopeptidase, Wako Chemicals, Neuss, Germany) for 3 h at RT. After adding of four volumes of 50 mm of ammonium bicarbonate, trypsin (Promega Corporation, Madison, Wisconsin) was added and tryptic digestion was carried out overnight. To stop the digestion, 1% TFA was added, peptides were purified on Sep-Pak C18 Cartridge (Waters, Milford, MA) and either eluted in 80% acetonitrile (ACN) for high pH reverse phase chromatography or desalted on C18 StageTips (as described previously (36)).
High pH Reverse Phase Chromatography
1–2 mg of peptides were fractionated on an off-line Ultimate 3000 high-pressure liquid chromatography (HPLC) system (Dionex, Thermo Fischer Scientific, Waltham, MA) equipped with xBridge BEH130 C18 130A, 3.5 μm, 4.6 × 250 mm column (Waters), as described previously (37). The system was operated under high pH conditions using buffer A (5 mm NH4OH) and buffer B (5 mm NH4OH in 90% ACN) at pH 10. Peptides were eluted using an 80 min gradient at a flow rate of 1 ml/min. The gradient consisted of 5% to 25% B over 45 min, followed by 40% B during 10 min and finally 70% B for 5 min. The gradient was held at 70% B for 5 min, reduced to 5% B within 5 min and the column equilibrated for 10 min. One-minute fractions were collected from 0 to 60 min. The 60 fractions were concatenated into 30 pools and dried by vacuum centrifugation. Peptide pools were reconstituted in 1 ml of 80% ACN, 10 μg of the pool were concentrated and desalted on StageTips prior LC-MS/MS measurements for proteome analysis.
Phosphopeptide Enrichment
Phosphopeptides were enriched using TiO2 beads. TiO2 beads (Titansphere, 10 μm, GL Sciences, Tokyo, Japan) were resuspended in DHB solution (80% ACN, 1% TFA, 3% 2,5-dihydroxybenzoic acid (DHB)) and incubated for 20 min. Purified peptides were added to the TiO2 beads in DHB solution and incubated for 10 min for each enrichment round. This step was repeated nine to ten times. Phosphopeptide-bound TiO2 beads were sequentially washed with 30% ACN in 1% TFA, 50% ACN in 1% TFA followed by 80% ACN in 1% TFA. Peptides were eluted with 5% NH4OH in 60% ACN into 20% TFA followed by 80% ACN in 1% FA. The eluate was reduced by vacuum centrifugation, pH was adjusted to < 2.7 with TFA and peptides were desalted on C18 StageTips prior LC-MS/MS measurements.
Formalin Fixed Paraffin Embedded Tissue Preparation for MS Analysis
FFPE tissue of pre-and posttreated patients with the kinase inhibitor vemurafenib (2 serial sections, 5 μm thick; List of FFPE specimens used in this study in supplementary Information S3) were first deparaffinized by two washes in xylene (5 min, 50 °C) followed by three serial washes in ethanol (100%, 95% to 70%) for 10 min each. Ethanol was removed completely, and sections air-dried. Lysis was carried out in 4% SDS, 50 mm DTT, 100 mm HEPES pH 7.5 supplemented with protease inhibitor at 95 °C for 60 min and by sonication for 15 min. 10 μg of proteins were purified by acetone precipitation and protein pellet was resolved in lysis buffer (6 m urea, 2 m thiourea, 10 mm Tris pH 8.0). Proteins in the cleared lysate (13,000 × g, 10 min) were reduced with 10 mm DTT for 60 min, alkylated with 55 mm iodoacetamide for an additional 60 min and LysC digestion was carried with 1 μg of LysC for 3 h at RT. After adding four volumes of 50 mm ammonium bicarbonate, 1 μg of trypsin was added for tryptic digestion overnight. Digestion was stopped by adding 1% TFA and peptides were loaded onto C18 StageTips for desalting and subsequent dimethylation labeling (35). Briefly, peptides were labeled with either 4% CH2O, 0.6 m NaBH3CN (“light”) or 4% 13CD2O, 0.6 m NaBD3CN (“heavy”) in phosphate buffer (50 mm NaH2PO4, 50 mm Na2HPO4). After washing with solvent A, peptides were eluted with solvent B and analyzed by LC-MS/MS.
Liquid Chromatography-MS Analysis
All samples were analyzed on an Easy-nLC 1200 UHPLC (Thermo Fischer Scientific) coupled to an Q Exactive HF mass spectrometer (Thermo Fischer Scientific) equipped with a nanoelectrospray source. Peptides were separated on a 20 cm analytical column (75 μm ID PicoTip fused silica emitter (New Objective, Woburn, MA)) in-house packed with ReproSil-Pur C18-AQ 1.9 μm resin (Dr Maisch GmbH (Ltd.), Ammerbuch, Germany). Peptides were eluted using a 90 min gradient for phosphoproteome, 230 min gradient for proteome analysis and 42 min gradients for fractionated peptide pools. Gradient was generated by solvent A (0.1% FA) and solvent B (80% ACN in 0.1% acetic acid) at 40 °C and 200 nl/min flow rate. The mass spectrometer was operated in data dependent mode. Full MS scans were acquired with a resolution of 120,000 and within a mass range of m/z 300 to 1650. For higher-energy collisional dissociation (HCD), the 12 most intensive peptides were selected, and MS/MS spectra were recorded with a resolution of 60,000. For 45 min gradients, fast scanning top20 method was used with a resolution of 15,000 for HCD scans and maximum fill time of 30 ms. For the analysis of TiO2 enriched phosphopeptides, full MS were acquired in the range of 300–1500 m/z at a resolution of 120,000. Seven most abundant precursor ions from a survey scan were selected for HCD fragmentation (isolation width of 1.20 m/z; 27% normalized collision energy and activation time of 0.1 ms were allowed) and MS/MS spectra were acquired at a resolution of 60,000 on the Orbitrap analyzer.
MS Data Analysis and Statistical Analysis
The raw data files were processed with the MaxQuant software suite (version 1.5.2.8) (38). The Andromeda search engine (39) searched MS/MS data against Uniprot Homo sapiens (release 2015_10_23; 91,646 entries) database containing commonly observed contaminants. Carbamidomethylation of cysteine (C) was set as fixed modification and oxidation of methionine, phosphorylation at serine (S), threonine (T) or tyrosine (Y) were defined as variable modifications. Trypsin/P was selected as a protease. For quantification, the amino acids (Lys4)/(Arg6) and (Lys8)/(Arg10) were defined as “medium” and “heavy” labels for the comparison of cell lines. Dimethylation on peptide N termini and lysine residues was defined as “light” (+28.03 Da) and “heavy” (+36.08 Da) labels for the comparison of pre-and posttreated tumors (FFPE specimens). No more than two missed cleavages were allowed. The MS tolerance was set at 4.5 ppm and MS/MS tolerance at 20 ppm for the analysis using HCD fragmentation method. The false discovery rate (FDR) for peptides and proteins was set to 1%. For all other parameters, the default settings were used. Only protein groups with at least two peptides were included in the final data sets and all contaminants were removed. Protein groups were kept for further statistical analysis only if quantified in 3 out of 3 replicates (for the first experiment) and 2 out of 2 replicates (for the second experiment). To find significant differences between sensitive and resistant A375 cells, log2-transformed SILAC ratios were subjected to t test in Perseus, with a permutation-based FDR threshold of 0.01 and s0 value of 1. The SILAC ratio of identified phosphorylation sites were normalized to the ratios of corresponding protein groups. All term enrichment analyses were performed using Perseus, by mapping reference lists or subsets of proteins with annotation terms. The resources used for annotation of proteins were Gene Ontology (GO), Biological Processes (GOBP), GO Cellular Compartment (GOCC), GO Molecular Functions (GOMF) and Kyoto Encyclopedia of Genes and Genomes (KEGG). In addition, significantly changing proteins determined by t test analysis (FDR ≤ 0.1, s0 value of 1) were mapped to pathways and network associations using STRING (40) against the whole genome as the statistical background. The top 20 significant pathways were selected. The median of the t test difference of assigned significantly changing proteins was calculated for each pathway. A list of all peptide sequences is provided in the supplementary Peptide Table and a list of all protein identifications and phosphorylation site identifications are provided in supplementary Table S1.
Analysis of Microarray Data from Pre- and Posttreated Patient Tumors
To confirm the relevance of our findings regarding nestin protein abundance profile in A375, we reanalyzed NES gene expression profile in human patients with melanoma pre- and posttreatment. The publically available microarray data with accession number GSE50509 and GSE61992 were retrieved from the NCBI GEO database. These data were acquired on Illumina Sentrix HumanHT12 v.4.0 Expression BeadChip as described in the original publications (41, 42). We reanalyzed the microarray datasets within the R environment (43). Each probe was annotated using the illuminaHumanv4.db package (44). Quality control analysis (probe intensity density plot) was performed on all microarrays to check for normalization bias between samples or batches. The normalized data were filtered based on probe quality using illuminaHumanv4.db package. Differentially expressed genes were identified from the filtered normalized data using the LIMMA package (45). Briefly, this was performed by blocking samples on a per-patient basis, fitting a linear model and generating Empirical Bayes statistics to compare pre- and posttreatment samples. The Benjamini-Hochberg multiple testing correction method was applied to all differentially expressed genes (supplementary Table S2).
Immunohistochemistry
Immunohistochemistry staining of clinical FFPE specimens (List of FFPE specimens used in this study in supplementary Information S3) was performed with a nestin specific polyclonal rabbit antibody (N5413, Sigma-Aldrich) diluted 1:50 in PBS containing 0.3% Triton-X100 and 1% BSA. Briefly, 5 μm FFPE tissue sections were de-paraffinized and antigen retrieval was performed in citrate buffer pH 6 in a pressure cooker for 2 min under pressure before a slow cooling down of the samples. Afterward tissue sections were stained according to the manufacturer's protocol (Thermo Scientific, UltraVision LP Detection System: AP Polymer) using FastRed (Thermo Scientific, Liquid Fast-Red Substrate System) as substrate.
Generation of Genome Edited Melanoma Cell Lines
NES gene knockout was carried out by CRISPR/Cas9-mediated genome editing according to the published protocol (46). The SpCas9 plasmid PX459 (Plasmid 62988) was obtained from a nonprofit plasmid share repository (Addgene, Watertown, MA). Suitable CRISPR target sites within NES Exon 1 positive strand were identified using the “CRISPR Design Tool” (http://crispr.mit.edu/). The respective oligonucleotide inserts (Biomers, Ulm, Germany) (5′-cctcgacggcgcgccggttg-3′ (forward), 5′-caaccggcgcgccgtcgagg-3′ (reverse)) were designed with overhangs compatible for ligation into PX459 linearized by digestion with BbsI (New England BioLabs, Frankfurt, Germany). Oligonucleotides were phosphorylated with polynucleotide kinase T4 PNK (New England BioLabs), annealed and inserted into the plasmid using T4 DNA ligase (New England BioLabs) and transformed into chemocompetent DH5α E. coli cells (New England Biolabs). Oligonucleotide inserts (ITD) (5′-gtattactgatattggtggg-3′ (forward), 5′-cccaccaatatcagtaatac-3′ (reverse)) were designed as CRISPR/Cas9 nontargeting (NonTar) control sgRNA and cloned into the SpCas9 plasmid PX459. Melanoma cells were seeded with low density (100,000 cells per ml), grown for 24 h in RPMI- 1640 medium without FBS. Transfection of the SpCas9/sgRNA plasmid or SpCas9/NonTar plasmid was carried out with Lipofectamine 2000 (Thermo Fischer Scientific) according to the manufacturer's instructions. On the next day, cells were selected using puromycin (2 μg/ml, Invivogen, Toulouse, France) and incubated for 2 days. Once individual colonies formed, single colonies were picked, cultured in separate wells and expanded in 6-well plates until cell number was enough for further analysis. Genomic DNA was isolated using GeneElute Mammalian Genomic DNA MiniPrep Kit (Sigma-Aldrich) and PCR amplification was performed using primers (5′-agatgtggggagctcaatcgg-3′ (forward) and 5′-tccaacctctgttccaacgc-3′ (reverse)) and sequenced by Sanger sequencing. Off-target effects of the guide sequence were predicted using Cas-OFFinder online tool (47). A mismatch of three bases were allowed. (List of predicted off-target sites in supplementary Information S4).
siRNA Knockdown
Nestin knockdown was carried out by transfecting A375 S cells with a pool of four FlexiTube siRNA oligos (100 nm, Qiagen, Hilden, Germany) against human nestin. NonSilencing siRNA (NonSil, Dharmacon) was used as control. siRNA transfections were performed using Lipofectamine RNAiMAX (Invitrogen) according to manufacturer's protocol. The medium was changed the following day. All assays were performed at 48–72 h posttransfection.
Western Blotting
Cells were harvested in lysis buffer and proteins were precipitated overnight with acetone/methanol (−20 °C). Cell lysates from SkMel28 S/R, SkMel19 S/R, 451lu S/R and Mel1617 S/R were used for immunoblotting analysis. Protein extracts were separated on 4–12% NuPAGE Bis-Tris gels (Novex, Life Technologies, Carlsbad, CA), transferred to PVDF membranes (0.2 μm, Sigma-Aldrich). The blot membranes were blocked in 1% Tween-20 and probed with primary antibody followed by horseradish peroxidase-conjugated secondary antibodies. Primary antibodies used were anti-Nestin (N5413, Sigma-Aldrich; sc-23927, Santa-Cruz Technologies), anti-EGFR (#2232, Cell Signaling Technologies, Danvers, MA), anti-Integrin β1 (E11) (sc-374430, Santa-Cruz Technolgies), anti-Integrin β4 (B4) (sc377523, Santa-Cruz Technologies), anti-GAPDH (MA5–15738, Thermo Fischer Scientific), anti-AKT (SAB4500797, Sigma Aldrich), anti-AKT (phospho-S124) (SAB4301497, Sigma Aldrich), anti-ERK1/2 (SAB1305560, Sigma Aldrich), anti-ERK1/2 (phospho-T202/Y204) (SAB1306604, Sigma Aldrich), anti-FAK (D1) (sc-271126, Santa-Cruz Technologies) and anti-Histone H3 antibody (D1H2) (#4499, Cell Signaling Technologies). Secondary antibodies used were anti-rabbit IgG, HRP-conjugated (#7074, Cell Signaling Technologies) and anti-mouse IgG, HRP-conjugated (#7075, Cell Signaling Technologies). ECL was detected by exposure with the Fusion SL instrument (Vilber Lourmat, Eberhardzell, Germany). For quantification, ImageJ software was used.
Clonogenic Assay
Cells were seeded in low density with 200 cells/cavity in 12-well plates and treated with BRAFV600E and MEK inhibitors (1 μm PLX4720, 0.1 μm cobimetinib). After 7–10 days, cells were fixed with 4% formaldehyde and stained with 0.05% Coomassie Brilliant Blue solution (Bio-Rad Laboratories, Hercules, CA) containing 80% methanol and 10% acetic acid. Stained colonies were washed twice with DPBS and counted under the microscope.
Proliferation/Cell Viability Assay
A MTS assay was used to analyze the proliferation and survival of melanoma cells. Cells were seeded (2 × 103) into 96-well plates. On the next day, media was changed, and cells were incubated for 96 h with increasing concentrations of respective inhibitors (0.1 μm to 20 μm of PLX4720 or cobimetinib) or with DMSO. After washing with DPBS, cell viability was assessed by CellTiter 96® Aqueous One Solution Cell Proliferation Assay (Promega Corporation), according to the manufacturer's instructions. The IC50 values were determined from the dose-response curves using log inhibitor versus response (three-parameter) test in GraphPad Prism.
Gelatin Zymography
For gelatin zymography, melanoma cells were cultivated to 70–80% confluence, washed twice with DPBS and grown for 24 h in FBS-free RPMI 1640 medium in presence or absence of inhibitors (1 μm PLX4720, 0.1 μm cobimetinib). Conditioned media was collected, concentrated using SpeedVac and nonreducing sample buffer was added (4% SDS, 20% glycerol, 0.01% bromphenol blue, 125 mm Tris-HCL pH 6.8). Samples were separated on Novex 10% Zymogram Plus (Gelatin) gels (Thermo Fischer Scientific). Gels were washed twice with washing buffer (2.5% Triton X-100, 50 mm Tris-HCl, 5 mm CaCl2, 1 μm ZnCl2), incubated for 24 h with incubation buffer (1% Triton X-100, 50 mm Tris-HCl, 5 mm CaCl2, 1 μm ZnCl2) at 37 °C, stained with staining solution (40% methanol, 10% acetic acid, 0.5% Coomassie Blue) for 1 h at RT and incubated with destaining solution (40% methanol, 10% acetic acid) until white bands appeared. For quantification of bands, ImageJ Software was used.
3D Melanoma Spheroid Culture
Cells were seeded on 1.5% agar noble (VWR) coated 96-well plates and incubated for 3 days to form spheroids. Spheroids were embedded into collagen I rat tail protein (1 mg/ml, Thermo Fischer) diluted in RPMI medium and cultured for four to ten more days. A total of 15 to 25 spheroids from each experiment were analyzed per condition. For quantification of outgrowth of spheroids, images were taken on day 0 and day 10. The outgrowth length for 15 spheroids was quantified using ImageJ software.
RESULTS
We used high-resolution mass spectrometry to characterize the proteome and phosphoproteome changes associated with resistance to BRAF inhibitor therapy in vitro (supplemental Fig. S1A). To study acquired drug resistance, the BRAFV600E mutant melanoma cell lines sensitive (A375 S) and resistant to vemurafenib (A375 R) were used (33). For proteomic and phosphoproteomic analysis, A375 S and A375 R cells were subjected to stable isotope labeling by amino acids in cell culture (SILAC) using Lys4/Arg6 supplemented in growth medium. The resulting peptide mixture was measured using on-line liquid chromatography coupled to high-resolution mass spectrometry (LC-MS/MS). In addition, phosphopeptide enrichment was performed and analyzed by LC-MS/MS (supplemental Fig. S1A and supplementary Table S1 and supplementary Peptide Table).
Cytoskeletal Proteins Are Downregulated in Vemurafenib-resistant Melanoma Cells
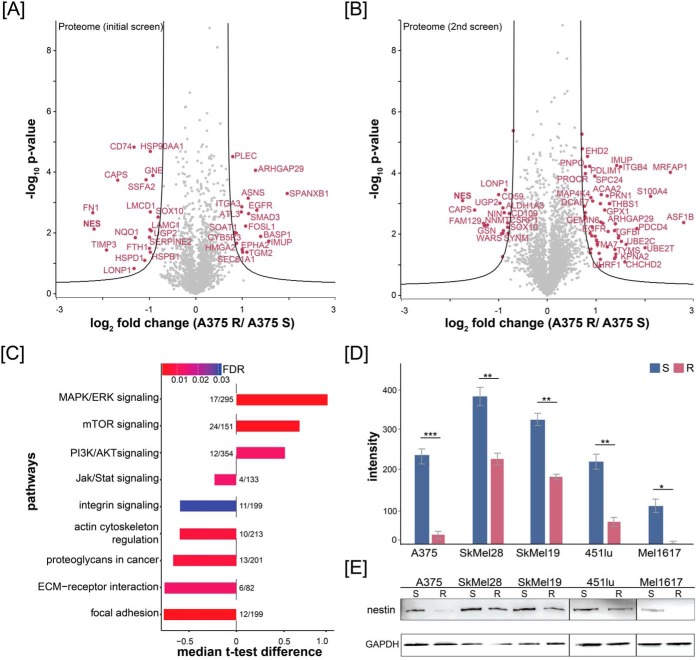
We performed two proteomic screens (each in three biological replicates) that resulted in detection of 129,485 unique peptide sequences corresponding to 9453 distinct protein groups at an estimated FDR of 0.27% at the peptide and 1.28% at the protein level (Fig. 1A, 1B; supplementary Table S1). Protein differential abundance analysis indicated widespread regulation of protein abundance that occurs during the development of BRAF resistance in vitro. The significantly downregulated proteins were involved in focal adhesion, integrin signaling, and actin cytoskeleton regulation (Fig. 1C), upregulated proteins in the activation of PI3K/AKT, mTOR, and MAPK/ERK signaling pathway (Fig. 1C). In the phosphoproteome measurement, we identified 13,354 phosphorylation sites, of which 10,444 were localized to a specific Ser/Thr/Tyr residue (supplemental Fig. S1B and supplementary Table S1) and 9106 phosphorylation sites were quantified reproducibly in all three replicates. We mapped our dataset to the known resistance mechanisms MAPK/ERK and PI3K/AKT pathway and observed a good coverage of regulated key proteins (supplemental Fig. S1C). For example, the RTKs EGFR, PDGFR, and IGF1R were identified upregulated in A375 R versus A375 S cells, as well as NRAS, BRAF, ERK1, MITF, AKT2, and mTOR (supplemental Fig. S1C). In addition, we identified phosphorylation sites on AKT (S124) and ERK1/2 (T202/Y204) significantly upregulated in A375 R cells (supplemental Fig. S1C).
Fig. 1.
Quantitative proteomics of melanoma cells identifies downregulation of nestin in drug-resistant cells. Volcano plot of vemurafenib-resistant and -sensitive A375 proteomes for (A) the initial proteomic screen and (B) the second proteomic screen. Fold change of SILAC ratios between A375 R and A375 S (log2) are plotted against p value (−log10) (n = 3). Black lines indicate the significance threshold (FDR < 0.01; s0 = 1). Significantly up- and downregulated proteins are highlighted in magenta. C, Over-representation of selected KEGG signaling pathways of A375 R compared with A375 S cells using String database analysis. The t test difference of SILAC ratios between A375 R and A375 S (log2) were plotted for each pathway (t test, FDR < 0.1; s0 = 1). Numbers represent identified significantly changing proteins mapped to the pathway by the total protein count involved in that pathway. Color of the bars represents the FDR. D, and E, Western blot analysis of different drug-resistant and drug-sensitive melanoma cell lines (A375, SkMel28, SkMel19, 451lu and Mel1617) against nestin and quantification of signal intensities using ImageJ software.
Among pathways activated in the A375 R cells, multiple proteins involved in cytoskeletal organization were significantly over-represented (Fig. 1A and 1B). Reorganization of the cytoskeleton is often associated with migratory and invasive phenotype of tumor cells and contribute to cancer's aggressiveness. In this dataset, a remarkable number of differentially regulated proteins were involved in cytoskeleton and adhesion pathways (Fig. 1A, 1B, and 1C), and several key proteins, such as nestin, vimentin and gelsolin, showed significant changes in abundance. The type VI intermediate filament protein nestin showed a median log2 ratio of −2.71 in resistant cells in all replicates and was one of the highest changing proteins in the whole dataset. To validate these findings, we assessed expression of nestin by Western blotting in several widely used melanoma cell lines, such as SkMel28 and Mel1617 (Fig. 1D). Quantification of the Western blotting by densitometry revealed significant changes of nestin abundance between drug-sensitive and drug-resistant cells (Fig. 1E).
To further address the clinical relevance of the downregulation of nestin in patients, we used two public transcriptomic datasets available from NCBI GEO repository. We selected all microarray data from patients with matching pre- and posttreatment tumors (vemurafenib, dabrafenib and dabrafenib/trametinib); i.e. 21 and 9 patients from microarray datasets GSE50509 and GSE61992, respectively (41, 42). The bioinformatics reanalysis of these microarray data revealed 23, 0 and 17 significantly changing probe sets in vemurafenib, dabrafenib and dabrafenib/trametinib treated tumors versus pretreatment tumors (FDR ≤ 0.1, supplementary Table S2). The mRNA expression of nestin (probeset ID = ILMN_1738147) was not significantly changing across posttreatment tumors versus pretreatment tumors. However, we did observe a tendency toward nestin downregulation between paired tumors (supplemental Fig. S1D and S1E). We could confirm these findings in FFPE specimens of two patients pre- and posttherapy with BRAF inhibitor using immunohistochemistry against nestin protein (supplemental Fig. S1F). The abundance of nestin in pretreated tumors differed between patients; however, there was a clear tendency toward nestin downregulation between post- and pretreated tumors within each individual patient. Proteomic analysis of FFPE specimens of one patient pre- and post-BRAF inhibitor therapy identified nestin as one of the most downregulated proteins in the post-BRAF inhibitor treated tissue (supplemental Fig. S1G) with a log2 ratio of −2.5. In total, we identified 14 peptides of nestin in both tissue specimens with a sequence coverage of 36.5%. Taken together, these data show that acquired resistance is associated with alterations in cytoskeletal and adhesion molecules and leads to the downregulation of the intermediate protein nestin in melanoma cell lines and metastases of human patients.
Nestin Expression Correlates with Invasive Properties of A375 Cells
To evaluate the functional role of nestin in resistance toward BRAF inhibition, we induced a NES gene knockout in vemurafenib sensitive A375 S cells using the CRISPR/Cas9 system (supplemental Fig. S2A). Single clones (A375 Nes-Ko #1–5) were selected for further analysis based on their effective NES knockout (supplemental Fig. S2B–S2E). As a control we used a nontargeting (NonTar) control guide sequence and transfected A375 S cells with this construct. A375 NonTar cells showed no InDels in the NES gene and no change in nestin protein abundance (supplemental Fig. S2F and S2G).
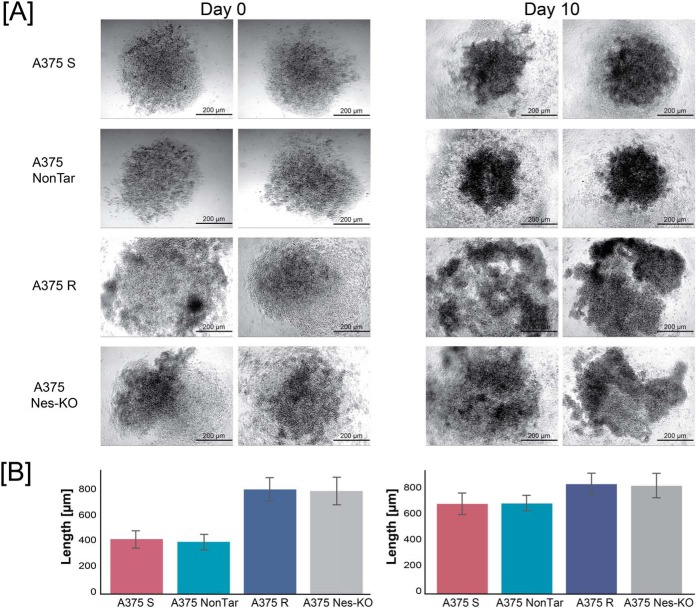
To address the relationship between nestin abundance and the invasive properties of melanoma cells, we performed three-dimensional spherical outgrowth assay. Melanoma spheroids formed from A375 S and A375 NonTar cells increased in size over 10 days (Fig. 2A and 2B). However, vemurafenib-resistant and A375 Nes-KO spheroids were not compact, showed less cellular adhesion and exhibited cells that progressively infiltrated into the surrounding collagen gel. In contrast, A375 S and A375 NonTar spheroids showed restricted invasive movement of a few cells away from the spheroid edge. Loss of nestin expression reduced the spheroid-forming ability of A375 Nes-KO cells like A375 R cells. Collectively, these data show that expression of nestin correlates with an invasive phenotype of resistant melanoma cells.
Fig. 2.
Nestin expression correlates with invasive properties of melanoma cell lines. A, Anchorage-independent growth assays of A375 S, A375 NonTar, A375 R and A375 Nes-KO clone #1 cells. Cells were seeded on agar, incubated for 3 days and spheroids were embedded into a collagen I matrix and further incubated for 10 days. Images are representative of three independent experiments of day 0 and day 10 (15–20 spheroids per cell line and day). All images were acquired with a light microscope at 5-fold magnification. Scale bar: 200 μm. B, Quantification of spheroids outgrowth. Analysis of outgrowth length was performed by measuring the total length from the center for 15 spheroids each on microscopic images using ImageJ software. Results are displayed as mean values ± S.E.
Depletion of Nestin Affects Cell Proliferation and Colony Formation On Treatment with BRAFV600E and MEK Inhibitors
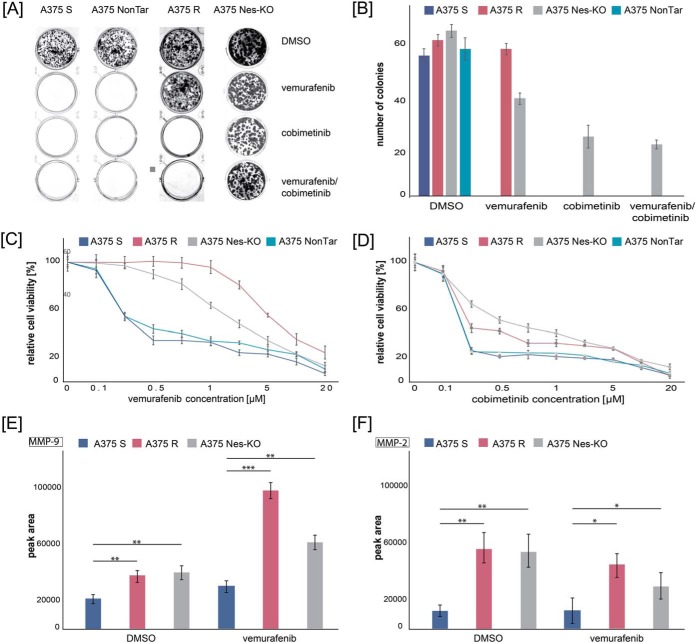
To investigate the effects on cell proliferation in drug-sensitive, drug-resistant and genome edited A375 Nes-KO cell lines, we treated cells with the MAPK signaling pathway inhibitors vemurafenib (BRAFV600E inhibitor) and cobimetinib (MEK inhibitor) and performed in vitro colony formation and cell proliferation (MTS) assays. First, we examined the colony formation ability of A375 Nes-KO compared with A375 S and A375 R controls. A375 NonTar cells were used as CRISPR/Cas9 control cell line. In absence of inhibitors, cell lines showed similar colony formation ability in terms of colony number and size (Fig. 3A). A375 S and A375 NonTar single cells were not able to grow into a colony after treatment with the BRAFV600E inhibitor vemurafenib. In contrast, A375 R showed the same colony formation ability with vemurafenib. For the genome edited A375 Nes-KO cell lines, we observed a 2-fold decrease in colony number (Fig. 3A and 3B). However, the size of the BRAFi treated nestin knockout cell line colonies were smaller (data not shown). When cells were treated with the MEK inhibitor cobimetinib or in combination with BRAFi, the A375 S and the A375 R cell lines showed no ability to grow into colonies (Fig. 3A and 3B). Interestingly, after both treatments, the A375 Nes-KO cell lines colonies were observed in similar numbers and sizes (Fig. 3A and 3B). Next, we examined whether different concentrations of BRAFV600E and MEK inhibitors impair cell survival and proliferation of melanoma cells. We calculated the IC50 value for each cell line and treatment using a three-parameter dose-response test. When treated with vemurafenib, the IC50 of A375 R, A375 S, A375 NonTar, and A375 Nes-KO was 2.46 μm, 0.268 μm, 0.259 μm and 2.091 μm, respectively (Fig. 3C). This confirmed that A375 Nes-KO cells tolerate vemurafenib treatment better than the A375 S cells. In addition, the observed effect was constant over different concentrations of BRAF and MEK inhibitors. No difference was observed between A375 S and A375 NonTar cells. For the cobimetinib treatment, the IC50 of A375 Nes-KO cells (IC50 0.31 μm) was 7.7-times higher than for A375 R cells (IC50 0.04 μm) (Fig. 3D). A375 S and A375 NonTar cells were not able to grow under these conditions.
Fig. 3.
Depletion of nestin affects cell proliferation and colony formation on treatment with BRAFV600E and MEK inhibitors. A, Clonogenic assay of A375 S, A375 NonTar, A375 R and NES knockout (A375 Nes-KO) cells after 10 days treatment with signaling pathway inhibitors. Cells were treated with vemurafenib (1 μm) and cobimetinib (0.1 μm), either alone or in combinations. Controls were treated with DMSO. Cultures were fixed with formaldehyde and stained with crystal blue. Images are representative of three biological and three technical replicates. B, Quantification of single colonies per condition. Results represent the mean of three biological experiments and three technical replicates (9 wells per cell line). Error bar represents standard deviations C–D, A375 S, A375 NonTar, A375 R, and A375 Nes-KO were cultured for 24 h, and then treated with vemurafenib (C) or cobimetinib (D) at the indicated concentrations (0, 0.1, 0.25, 0.5, 1, 2.5, 5, 10 and 20 μm) or DMSO as control. Cell viability was determined by MTS assay 96 h later. Results expressed as percentage of control represent the mean of three biological experiments and six technical replicates (n = 24). Error bar represents standard deviations of replicates. E–F, Activity of MMP9 (E) and MMP2 (F) using gelatin zymography. Supernatants of A375 S, A375 R and A375 Nes-KO cell lines were treated with DMSO or vemurafenib for 24 h, analyzed by zymography gelatin plus gels and stained with Coomassie. ImageJ software was used for quantification of signals. Results represent the mean of three biological experiments ± S.E.
To confirm this finding, we used a siRNA loss-of-function model system and investigated the phenotype of a conditional nestin knockdown (A375 Nes-Kd) with regards to vemurafenib sensitivity. Downregulation of nestin expression was effectively achieved in A375 S cells as opposed to cells harboring nonsilencing siRNA (NonSil) (supplemental Fig. S3A). In agreement to NES gene knockout (Nes-KO), nestin RNA downregulation resulted in increased cell proliferation on BRAFV600E kinase inhibition compared with A375 S cells as seen in the MTS assay (supplemental Fig. S3B). The IC50 of vemurafenib in A375 Nes-Kd cells was similar (1.454 μm) to the NES knockout cells; pointing to the fact that nestin affects cell survival on stimulation with different BRAFV600E and MEK inhibitors.
Depletion of Nestin Induces Matrix Metalloproteinase Activity
Downregulation of nestin was previously connected with increased activity of matrix metalloproteinases (MMP) MMP-9 and MMP-2, which are involved in migratory potential and invasiveness of cancer cells (30). We hypothesized that A375 R with a significantly downregulated expression of nestin will induce melanoma matrix metalloproteinases and that this effect will be mimicked by our functional knockout cell line A375 Nes-KO. To maintain enzymatic activity of MMP-9 and MMP-2, we treated A375 S, A375 R and A375 Nes-KO cells with vemurafenib or DMSO as a control for 24 h and analyzed the supernatants using gelatin zymography (supplemental Fig. S3C). All cell lines secreted MMP-2 and MMP-9; however, A375 R and A375 Nes-KO cells showed a significantly stronger signal for both metalloproteinases compared with the supernatant of A375 S cells (supplemental Fig. S3C). This pattern was like that observed in cells cultured with the BRAFV600E inhibitor. A 2-fold difference in MMP-9 activity and 3-fold difference in MMP-2 activity was observed between untreated supernatants of drug-sensitive and drug-resistant lines (Fig. 3E and 3F). Interestingly, quantification of signals resulted in a similar peak area for bands of MMP-9 and MMP-2 in A375 Nes-KO and A375 R supernatants (Fig. 3E and 3F), highlighting the similarity of their phenotypes. Treatment of cells with the BRAFV600E inhibitor led to significant increased activation of MMP-9 in A375 R and A375 Nes-KO supernatants and decreased activation of MMP-2 in genome edited A375 Nes-KO lines (Fig. 3E and 3F). These results suggest a relationship between expression of nestin and activity of certain MMPs known to enhance tumor invasion.
Loss of Nestin Expression Is Associated with PI3K/AKT and Integrin Signaling
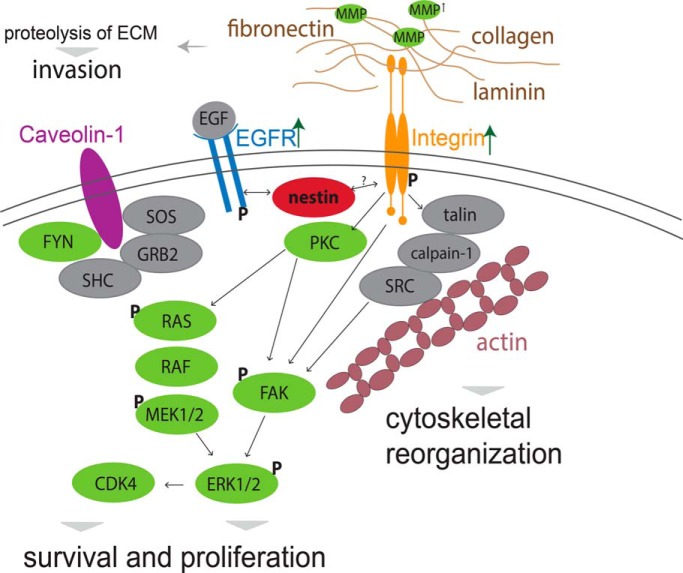
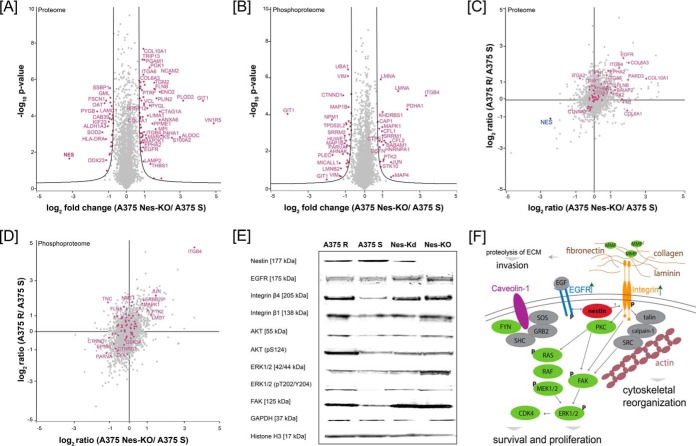
To understand how the loss of nestin protein abundance may alter cellular protein homeostasis, we performed a quantitative (phospho)proteome analysis of A375 Nes-KO versus A375 S versus A375 R cells using a SILAC approach and LC-MS/MS. In two biological replicates, we identified 5965 protein groups and 7524 phosphorylation sites, of which 91 showed significant changes in abundance at a FDR < 0.01 (s0 = 1) in the A375 Nes-KO compared with A375 S (Fig. 4A and 4B, supplementary Table S1 and supplementary Peptide Table). The comparison of A375 R to A375 Nes-KO cells showed significant differences between these two cell lines (supplemental Fig. S4A). Biological replicates showed a good correlation of the proteome and phosphoproteome (supplemental Fig. S4B and S4C). Interestingly, ECM interacting proteins, such as vinculin, fibronectin, integrin β4 and integrin α6, as well as Proteinkinase C, focal adhesion kinase FAK and other downstream signaling proteins were significantly upregulated in the genome-edited cells (Fig. 4A). Enrichment analysis of the significantly regulated proteins for GO cellular component indicated an involvement in adhesion junctions, extracellular region and focal adhesion. (Supplemental Fig. S4D). The KEGG pathways PI3K/AKT signaling, remodeling of focal adhesions, actin cytoskeleton signaling and integrin signaling were significantly over-represented in A375 Nes-KO cells compared with A375 S cells (supplemental Fig. S4D). Furthermore, the phosphoproteome analysis revealed differentially regulated phosphorylation sites on key members of the integrin signaling pathway and downstream proteins. Phosphorylation sites on FAK, ERK1/2 and Integrin β4 were significantly upregulated in the genome edited cell line compared with A375 S cells (Fig. 4B). To identify the (phospho)proteomic overlap between A375 Nes-KO and A375 R cells, we correlated the ratios of A375 Nes-KO versus A375 S against ratios of A375 R versus A375 S (Fig. 4C and 4D). We identified differentially regulated proteins and phosphorylation sites on key members of the integrin signaling pathway in A375 Nes-KO and A375 R cells with similar ratios (Fig. 4C and 4D). For example, abundance of EGFR, Filamin-B and Collagen 6A3 and 10A1, as well as phosphorylation sites on Integrin β4, FAK and ERK1. Focal adhesion kinase FAK plays a central role in cancer cell motility, adhesion, and invasion (48, 49). In melanoma cells, we found Nes-KO to be associated with increased FAK phosphorylation and protein levels (Figs. 4B). Phosphorylation of FAK at S910 was identified with high quality by LC-MS/MS (supplemental Figs. S4E). We confirmed these results by immunoblot using antibodies against key signaling molecules in NES knockout (A375 Nes-KO), nestin knockdown (A375 Nes-Kd), A375 S and A375 R cells (Fig. 4E). Immunoblot revealed a higher expression of EGFR, Integrin β4 and FAK in A375 R, A375 Nes-KO and Nes-Kd cells. Signals for phosphorylation sites of ERK1/2 (T202/Y204) and AKT (S124) were also increased in A375 R, knockout and knockdown cells compared with A375 S. Taken together, these results link nestin with integrin and PI3K/AKT signaling pathways in melanoma; while also revealing new molecular events in context of acquired resistance to vemurafenib (Fig. 4F).
Fig. 4.
Loss of nestin is associated with to PI3K/AKT signaling. Volcano plot of nestin CRISPR/Cas9 knockout (Nes-KO) versus vemurafenib-sensitive (A375 S) cells (A) proteome and (B) phosphoproteome. Fold change of SILAC ratios between A375 Nes-KO and A375 S (log2) are plotted against p value (−log10) (n = 3). Black lines indicate the significance threshold (FDR < 0.01; s0 = 1). Significantly up- and downregulated proteins are highlighted in magenta. C, Proteome correlation of Nes-KO relative to A375 S against A375 R relative to A375 S. Magenta: proteins involved in integrin signaling using GO annotation. D, Correlation of phosphorylation sites of Nes-KO relative to A375 S against A375 R relative to A375 S. Magenta: proteins involved in integrin signaling using GO annotation. E, Western blot analysis of A375 R, A375 S, siRNA knockdown cells Nes-Kd and CRISPR/Cas9 knockout cells Nes-KO against key molecules of the integrin signaling, MAPK/ERK and PI3K/AKT signaling pathways. F, Schematic overview of proposed interaction of nestin with key molecules of integrin, MAPK/ERK signaling pathway. Green: upregulated in A375 Nes-KO versus A375 S cells, red: downregulated in A375 Nes-KO versus A375 S cells, gray: not quantified; P: identified upregulated phosphorylation site in A375 Nes-KO versus A375 S cells; arrows: indicate upregulated receptors in A375 Nes-KO versus A375 S cells.
DISCUSSION
To identify additional resistance mechanisms and reveal new molecular targets to overcome resistance, we investigated two melanoma cell lines (A375 S and A375 R) with differing phenotypes of acquired resistance to the BRAFV600E inhibitor vemurafenib. In this context, we utilized SILAC coupled to mass spectrometry to characterize the global proteomic and phosphoproteomic changes in melanoma cell lines. This study is, to our knowledge, one of the largest global (phospho)proteomic analyses assessing the differentially expressed proteins in drug-sensitive and drug-resistant melanoma cells. We identified several pathways to be over-represented in resistant cells including PI3K/AKT/mTOR signaling, integrin signaling and MAPK/ERK signaling pathways. The MAPK/ERK and PI3K/AKT/mTOR signaling pathways are known resistance mechanisms and constitutively activated in malignant melanoma. Here, several key proteins and phosphorylation sites within these pathways were identified with high confidence like EGFR or phosphorylated ERK1/2. In addition to signaling pathways, the tumor microenvironment and remodeling of the cytoskeletal organization have been reported to play an important role in the development of acquired resistance. For example, Kim and colleagues showed that actin signaling through YAP/TAZ activation confers BRAF inhibitor resistance in melanoma (50). Similarly, we detected several cytoskeletal proteins such as nestin, vimentin and gelsolin to be downregulated in resistant cells. In the present study, we investigated whether the intermediate filament nestin may contribute to resistance in melanoma cells. Expression of nestin in various cancer cell types has been studied, however the mechanistic basis of the function of nestin is still unknown. Nestin was reported to be involved in cancer cell migration, invasion, and metastasis (18, 31, 51). Quendro and colleagues showed in a large-scale proteomic study that nestin and vimentin are both upregulated in melanoma cells and tissue material compared with control melanocytes (29). We could confirm this in A375 melanoma cells and further show that nestin and vimentin are downregulated in resistant cells compared with sensitive cells. Nestin and vimentin are interaction partners with important functions in cell migration, cytoskeletal reorganization and apoptosis (52). Doxie and colleagues showed that nestin expression was completely depleted in nestin-expressing cells in human tumors after BRAF and MEK inhibitor therapy, which highlights the loss of nestin expression in human tumors. In previous reports, NES expression has been reported to be regulated by the transcription factors SOX9 and SOX10 and nestin and SOX9 may be negative prognostic markers in melanoma (53). In agreement with this, both transcription factors were also identified in our data set with the same abundance trend as nestin. Because nestin is a known stemness marker (27) we investigated the presence of other stemness markers in our data set. We could identify the stemness marker ABCG2, which is known to enhance tumorigenic potential of melanoma cells (54). Although ABCG2 was not significantly regulated, we identified the potential cancer stem cell (CSC) marker of ALDH1 to be upregulated in drug-resistant and in nestin CRISPR/Cas9 knockout cells compared with sensitive cells. ALDH1 is associated with multidrug resistance in different types of human melanoma tumors (55) and therefore may influence the stemness of melanoma cells. Taken together, this highlights the good coverage of the dataset and utility as a resource for the melanoma community.
To confirm the clinical significance of our findings, we re-analyzed public microarray datasets for matching pre- and posttreatment tumors. We could only observe a tendency toward NES downregulation, however in none of the treatments did it reach significance. Several hypothetical reasons could explain this. The first reason may be that nestin clinical relevance is patient specific and depends on the patient genetic background or tumor microenvironment. The second contributing factor may be that, contrary to A375 cell line where we have a clear BRAFi sensitive/resistant phenotype, in pre- and posttreatment tumors the response to BRAFi is not well defined (i.e. nonresponder, partial responder, full responder). In addition, we could confirm downregulation of nestin in FFPE specimens using immunohistochemistry; however, expression of nestin differed between tumor specimens. Quantitative proteomics of one pair of pre- and post-treated tumors identified nestin as one of the most downregulated proteins in the dataset with a good sequence coverage. These results highlight the significance of nestin expression in human tumors.
To study the effect of nestin, we used CRISPR/Cas9 approach to generate a NES gene knockout in drug-sensitive melanoma cells. We identified two peptides at the N terminus of the protein by high-resolution mass spectrometry because the knockout occurred at the end of Exon 1 of the genomic sequence. However, the knockout cell line was considered as an effective functional knockout of nestin because both interaction and functional active domains were absent from the resulting protein. To confirm that our results are not because of cell manipulation in terms of Cas9 expression, guide RNA transfection or single cell picking, we generated a CRISPR/Cas9 control cell line (A375 NonTar) using a nontargeting guide sequence. We could not observe differences in the cell proliferation and colony formation between A375 NonTar and drug-sensitive cell lines. This study provides novel data showing that nestin expression significantly correlates with cell survival and colony formation on MAPK signaling pathway inhibitor treatment. Indeed, we are not only describing an increased cell survival and colony formation ability in knockout cells under BRAF inhibitor treatment, but we also show a direct effect of NES expression on the growth of melanoma cells on inhibition with the MEK inhibitor cobimetinib. Several studies have suggested that combined therapy with BRAF and MEK inhibitors are promising trials to delay MAPK-driven acquired resistance and may activate other resistance mechanisms like PI3K/AKT/mTOR signaling pathways (56). Depletion of nestin may activate these resistance mechanisms and increase cell survival on mono- or combined therapy.
Our results indicate a phenotypic difference in invasion and proliferation of drug-sensitive and drug-resistant cells. Interestingly, the genome edited A375 Nes-KO cell lines derived from drug-sensitive cells showed a similarly invasive phenotype to drug-resistant cells. However, expression of nestin has been reported to mediate both, three-dimensional tumorigenesis and cell invasiveness (19, 25). It is also reported that depletion of nestin using shRNA results in an invasive phenotype of melanoma cell lines, which is mediated through upregulation of specific matrix metalloproteinases (MMPs) (30). In this study, we could also show that nestin gene knockout induces activity of MMP2 and MMP9 like resistant phenotype. This falls in line with the hypothesis proposed by Lee and colleagues, who identified nestin depletion to be associated with the activation of MMP-2 (gelatinase A, type IV collagenase) and MMP-9 (gelatinase B, type IV collagenase) (30). The invasion and metastasis of tumor cells have been shown to require proteolytic activity to degrade components of the extracellular matrix (57) and to involve FAK/integrin-mediated cell/matrix adhesion pathways (58). We provide evidence that nestin depletion is associated with signaling through focal adhesion, integrin and PI3K/AKT/mTOR pathways. Interestingly, ECM interacting proteins, like Laminin-B or Filamin-B, the integrins β1 and β4, Proteinkinase C, FAK and other downstream signaling proteins were significantly upregulated in the genome edited cells compared with drug sensitive cells at the proteome level. FAK activated by integrins plays a central role in cell invasion and adhesion by triggering several signaling pathways. Furthermore, the phosphoproteome analysis revealed differentially regulated phosphorylation sites on the key players of the integrin signaling pathway and downstream proteins. In this regard, recent evidence in prostate cancer research indicate that nestin depletion is associated with an expression pattern of phosphorylated FAK (pFAK) at the cell membrane. Phosphorylated FAK promotes integrin clustering, which results in pFAK- and integrin-dependent matrix degradation and an invasive phenotype (31). In melanoma cells, Hyder and colleagues could also observe increased levels of phosphorylated FAK and protein levels in nestin knockdown cells and a localization of phosphorylated FAK at the cell membrane like the studies in prostate cancer (31). In conclusion, we could link nestin protein levels, not only with an invasive phenotype, but also with acquired drug resistance in melanoma.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org/) via the PRIDE partner repository (59) with the data set identifier PXD010683.
Excel files containing the analyzed data are provided in Supplementary Materials.
Supplementary Material
Acknowledgments
We thank Bianca Kuhn for her help in the initial stages of the project.
Footnotes
* This work was supported by a grant from DFG (INST 37/741-1 FUGG) to B.M.
 This article contains supplemental Figures, Tables, and Information. The authors declare no conflicts of interest.
This article contains supplemental Figures, Tables, and Information. The authors declare no conflicts of interest.
1 The abbreviations used are:
- MMP
- matrix metalloproteinase
- BRAFi
- BRAF inhibitor
- CRISPR
- clustered regularly interspaced short palindromic repeats
- DSB
- double strand break
- DTT
- dithiothreitol
- ECM
- extracellular matrix
- FDR
- false discovery rate
- FBS
- fetal bovine serum
- FFPE
- formaldehyde fixed paraffin embedded
- IAA
- iodoacetamide
- LC-MS/MS
- liquid chromatography combined with tandem mass spectrometry
- NOG
- N-ocetylglucosamine
- NonSil
- nonsilencing siRNA
- PAM
- protospacer adjacent motif
- SDS
- sodium deoxycholate
- SILAC
- stable isotope labeling in cell culture
- sgRNA
- single guided RNA
- NonTar
- nontargeting control RNA
- siRNA
- small interfering RNA.
REFERENCES
- 1. Chang A. E., Hynds Karnell L., and Menck H. R. (1998) The National Cancer Data Base Report on cutaneous and noncutaneous melanoma Cancer 83, 1664–1678 [DOI] [PubMed] [Google Scholar]
- 2. American Cancer Society (2017) Cancer Facts & Figures 2017. Atlanta: American Cancer Society [Google Scholar]
- 3. Larkin J., Ascierto P. A., Dreno B., Atkinson V., Liszkay G., Maio M., Mandalà M., Demidov L., Stroyakovskiy D., Thomas L., Cruz-merino, Dutriaux L. D. C., Garbe C., Sovak M. A., Chang I., Choong N., Hack S. P., McArthur G., and Ribas A. (2014) Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J. Med. 371, 1867–1876 [DOI] [PubMed] [Google Scholar]
- 4. Davies H., Bignell G. R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M. J., Bottomley W., Davis N., Dicks E., Ewing R., Floyd Y., Gray K., Hall S., Hawes R., Hughes J., Kosmidou V., Menzies A., Mould C., Parker A., Stevens C., Watt S., Hooper S., Wilson R., Jayatilake H., Gusterson Ba Cooper C., Shipley J., Hargrave D., Pritchard-Jones K., Maitland N., Chenevix-Trench G., Riggins G. J., Bigner D. D., Palmieri G., Cossu A., Flanagan A., Nicholson A., Ho J. W. C., Leung S. Y., Yuen S. T., Weber B. L., Seigler H. F., Darrow T. L., Paterson H., Marais R., Marshall C. J., Wooster R., Stratton M. R., and Futreal P. A. (2002) Mutations of the BRAF gene in human cancer. Nature 417, 949–954 [DOI] [PubMed] [Google Scholar]
- 5. Chapman P. B., Hauschild A., Robert C., Haanen J. B., Ascierto P., Larkin J., Dummer R., Garbe C., Testori A., Maio M., Hogg D., Lorigan P., Lebbe C., Jouary T., Schadendorf D., Ribas A., O'Day S. J., Sosman J. A., Kirkwood J. M., Eggermont A. M. M., Dreno B., Nolop K., Li J., Nelson B., Hou J., Lee R. J., Flaherty K. T., and McArthur G. A. (2011) Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Eng. J. Med. 364, 2507–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sosman J. A., Kim K. B., Schuchter L., Gonzales R., Pavlick A. C., Weber J. S., Joe A. K., and Ribas A. (2012) Survival in BRAF V600–mutant advanced melanoma treated with vemurafenib. N. Eng. J. Med. 366, 707–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Flaherty K. T., Robert C., Hersey P., Nathan P., Garbe C., Milhem M., Demidov L. V., Hassel J. C., Rutkowski P., Mohr P., Dummer R., Trefzer U., Larkin J. M. G., Utikal J., Dreno B., Nyakas M., Middleton M. R., Becker J. C., Casey M., Sherman L. J., Wu F. S., Ouellet D., Martin A.-M., Patel K., and Schadendorf D. (2012) Improved survival with MEK inhibition in BRAF-mutated melanoma. N. Engl. J. Med. 367, 107–114 [DOI] [PubMed] [Google Scholar]
- 8. Roesch A. (2015) Tumor heterogeneity and plasticity as elusive drivers for resistance to MAPK pathway inhibition in melanoma. Oncogene 34, 2951–2957 [DOI] [PubMed] [Google Scholar]
- 9. Kemper K., Krijgsman O., Cornelissen-Steijger P., Shahrabi A., Weeber F., Song J.-Y., Kuilman T., Vis D. J., Wessels L. F., Voest E. E., Schumacher T. N., Blank C. U., Adams D. J., Haanen J. B., and Peeper D. S. (2015) Intra- and inter-tumor heterogeneity in a vemurafenib-resistant melanoma patient and derived xenografts. EMBO Mol. Med. 7, e201404914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nazarian R., Shi H., Wang Q., Kong X., Koya R. C., Lee H., Chen Z., Lee M.-K., Attar N., Sazegar H., Chodon T., Nelson S. F., McArthur G., Sosman J. A., Ribas A., and Lo R. S. (2010) Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 468, 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Allen E. M. V., Wagle N., Sucker A., Treacy D., Goetz E. M., Place C. S., Taylor-weiner A., Kryukov G., Hodis E., Rosenberg M., McKenna A., Cibulskis K., Farlow D., Zimmer L., Hillen U., Gutzmer R., Goldinger M., Ugurel S., Gogas H. J., Egberts F., and Berking C. (2014) The genetic landscape of clinical resistance to RAF inhibition in metastatic melnaoma. Cancer Discov. 4, 94–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shi H., Hugo W., and Kong X. (2014) Acquired resistance and clonal evolution in melanoma during BRAF inhibior therapy. Cancer Discov. 144, 724–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wagle N., Emery C., Berger M. F., Davis M. J., Sawyer A., Pochanard P., Kehoe S. M., Johannessen C. M., MacConaill L. E., Hahn W. C., Meyerson M., and Garraway L. A. (2011) Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J. Clin. Oncol. 29, 3085–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lendahl U., Zimmermann L. B., and McKay R. D. (1990) CNS stem cells express a new class of intermediate filament protein. Cell 60, 585–595 [DOI] [PubMed] [Google Scholar]
- 15. Sahlgren C. M., Pallari H. M., He T., Chou Y. H., Goldman R. D., Eriksson J. E. (2006) A nestin scaffold links Cdk5/p35 signaling to oxidant-induced cell death. EMBO J. 25, 4808–4819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wiese C., Rolletschek A., Kania G., Blyszczuk P., Tarasov K. V., Tarasova Y., Wersto R. P., Boheler K. R., and Wobus A. M. (2004) Nestin expression–a property of multi-lineage progenitor cells? Cell Mol. Life Sci. 61, 2510–2522 [DOI] [PubMed] [Google Scholar]
- 17. Kawamoto M., Ishiwata T., Cho K., Uchida E., Korc M., Naito Z., and Tajiri T. (2009) Nestin expression correlates with nerve and retroperitoneal tissue invasion in pancreatic cancer. Hum. Pathol. 40, 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matsuda Y., Naito Z., Kawahara K., Nakazawa N., Korc M., and Ishiwata T. (2011) Nestin is a novel target for suppressing pancreatic cancer cell migration, invasion and metastasis. Cancer Biol Ther. 11, 512–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kleeberger W., Bova G. S., Nielsen M. E., Herawi M., Chuang A. Y., Epstein J. I., and Berman D. M. (2011) Roles for the stem cell associated intermediate filament Nestin in prostate cancer migration and metastasis. Cancer Res. 67, 9199–9206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li H., Cherukuri P., Li N., Cowling V., Spinella M., Cole M., Godwin A. K., Wells W., and Direnzo J. (2007) Nestin is expressed in the basal/myoepithelial layer of the mammary gland and is a selective marker of basal epithelial breast tumors. Cancer Res. 67, 501–511 [DOI] [PubMed] [Google Scholar]
- 21. Chinnaiyan P., Wang M., Rojiani A. M., Tofilon P. J., Chakravarti A., Ang K. K., Zhang H.-Z., Hammond E., Curran W. Jr, and Mehta M. P.. The prognostic value of nestin expression in newly diagnosed glioblastoma: Report from the Radiation Therapy Oncology Group. Radiat. Oncol. 8, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang X. H., Wu Q. L., Yu X. B., Xu C. X., Ma B. F., Zhang X. M., Li S. N., Lahn B. T., and Xiang A. P. (2001) Nestin expression in different tumours and its relevance to malignant grade. J. Clin. Pathol. 6, 467–474 [DOI] [PubMed] [Google Scholar]
- 23. Misago N., Mori T., and Narisawa Y. (2010) Nestin expression in stromal cells of trichoblastoma and basal cell carcinoma. J. Eur. Acad. Dermatol. Venereol. 24, 1354–1358 [DOI] [PubMed] [Google Scholar]
- 24. Akiyama M., Matsuda Y., Ishiwata T., Naito Z., and Kawana S. (2013) Nestin is highly expressed in advanced-stage melanomas and neurotized nevi. Oncol. Rep. 29, 1595–1599 [DOI] [PubMed] [Google Scholar]
- 25. Akiyama M., Matsuda Y., Ishiwata T., Naito Z., and Kawana S. (2013) Inhibition of the stem cell marker nestin reduces tumor growth and invasion of malignant melanoma. J. Investigative Dermatol. 133, 1384–1387 [DOI] [PubMed] [Google Scholar]
- 26. Brychtova S., Fiuraskova M., Hlobilkova A., Brychta T., and Hirnak J. (2007) Nestin expression in cutaneous melanomas and melanocytic nevi. J. Cutan. Pathol. 34, 370–375 [DOI] [PubMed] [Google Scholar]
- 27. Piras F., Perra M. T., Murtas D., Minerba L., Floris C., Maxia C., Demurtas P., Ugalde J., Ribatti D., and Sirigu P. (2010) The stem cell marker nestin predicts poor prognosis in human melanoma. The stem cell marker nestin predicts poor prognosis in human melanoma. Oncol. Reports 23, 17–24 [PubMed] [Google Scholar]
- 28. Klein W. M., Wu B. P., Zhao S., Wu H., Klein-szanto A., JP, and Tahan S. R. (2007) Increased expression of stem cell markers in malignant melanoma. Mod. Pathol. 133, 102–107 [DOI] [PubMed] [Google Scholar]
- 29. Qendro V., Lundgren D. H., Rezaul K., Mahony F., Ferrell N., Bi A., Latifi A., Chowdhury D., Gygi S., Haas W., Wilson L., Murphy M., and Han D. K. (2014) Large-scale proteomic characterization of melanoma expressed proteins reveals nestin and vimentin as biomarkers that can potentially distinguish melanoma subtypes. J. Proteome Res. 13, 5031–5040 [DOI] [PubMed] [Google Scholar]
- 30. Lee C.-W., Zhan Q., Frank M., Huang J., Larson A., Jie S., and Xiao D. (2015) Nestin depletion induces melanoma matrix metalloproteinases and invasion. Lab. Invest. 94, 1382–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hyder C. L., Lazaro G., Pylva J. W., Qvarnstro S. M., and Eriksson J. E. (2014) Nestin regulates prostate cancer cell invasion by influencing the localisation and functions of FAK and integrins. J. Cell Sci. 127, 2161–2173 [DOI] [PubMed] [Google Scholar]
- 32. Doxie D. B., Greenplate A. R., Gandelman J. S., Diggins K. E., Roe C. E., Dahlman K. B., Sosman J. A., Kelley M. C., and Irish J. M. (2018) BRAF and MEK inhibitor therapy eliminates Nestin-expressing melanoma cells in human tumors. Pigment Cell Melanoma Res. 31, 708–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sinnberg T., Makino E., Krueger M. A., Velic A., Macek B., Rothbauer U., Groll N., Potz O., Czemmel S., Niessner H., Meier F., Ikenberg K., Garbe C., and Schittek B. (2016) A Nexus Consisting of Beta-Catenin and Stat3 Attenuates BRAF Inhibitor Efficacy and Mediates Acquired Resistance to Vemurafenib. EBioMedicine 8, 132–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Herlyn D., Iliopoulos D., Jensen P. J., Parmiter A., Baird J., Hotta H., Adachi K., Ross A. H., Jambrosic J., Koprowski H., and Herlyn M. (1990) In Vitro properties of human melanoma cells metastatic in nude mice. Cancer Res. 50, 2296–2302 [PubMed] [Google Scholar]
- 35. Boersema P. J., Raijmakers R., Lemeer S., Mohammed S., and Heck A. J. (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 [DOI] [PubMed] [Google Scholar]
- 36. Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature Protocols 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 37. Batth T. S., and Olsen J. V. (2016) Offline high pH reversed-phase peptide fractionation for deep phosphoproteome coverage. In: von Stechow L., ed. Phospho-Proteomics: Methods Protocols, pp. 179–192, Springer New York, New York: [DOI] [PubMed] [Google Scholar]
- 38. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 39. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 40. Szklarczyk D., Morris J. H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N. T., Roth A., Bork P., Jensen L. J., and von Mering C. (2017) The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Long G. V., Fung C., Menzies A. M., Pupo G. M., Carlino M. S., Hyman J., Shahheydari H., Tembe V., Thompson J. F., Saw R. P., Howle J., Hayward N. K., Johansson P., Scolyer R. A., Kefford R. F., and Rizos H. (2014) Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nat. Commun. 5, 5694. [DOI] [PubMed] [Google Scholar]
- 42. Rizos H., Menzies A. M., Pupo G. M., Carlino M. S., Fung C., Hyman J., Haydu L. E., Mijatov B., Becker T. M., Boyd S. C., Howle J., Saw R., Thompson J. F., Kefford R. F., Scolyer R. A., and Long G. V. (2014) BRAF inhibitor resistance mechanisms in metastatic melanoma: spectrum and clinical impact. Clin. Cancer. Res. 20, 1965–1977 [DOI] [PubMed] [Google Scholar]
- 43. RCore Team. (2018) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 44. Dunning M., Lynch A., and Eldridge M. (2015) illuminaHumanv4.db: Illumina HumanHT12v4 annotation data (chip illuminaHumanv4). R package version 1.26.0 Ed. [Google Scholar]
- 45. Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., and Smyth G. K. (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ran F. A., Hsu P. D. P., Wright J., Agarwala V., Scott D. A., Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protocols 8, 2281–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bae S., Park J., and Kim J.-S. (2014) Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Turečková J., Vojtěchová M., Krausová M., Šloncová E, Korínek V. (2009) Focal adhesion kinase functions as an Akt downstream target in migration of colorectal cancer cells. Translational Oncol. 2, 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhao X., and Guan J. L. (2011) Focal adhesion kinase and its signaling pathways in cell migration and angiogenesis. Adv. Drug Deliv. Rev. 63, 610–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kim M. H., Kim J., Hong H., Lee S. H., Lee J. K., Jung E., and Kim J. (2016) Actin remodeling confers BRAF inhibitor resistance to melanoma cells through YAP/TAZ activation. EMBO J. 35, 462–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Narita K., Matsuda Y., Seike M., Naito Z., Gemma A., and Ishiwata T. (2014) Nestin regulates proliferation, migration, invasion and stemness of lung adenocarcinoma. Int. J. Oncol. 44, 1118–1130 [DOI] [PubMed] [Google Scholar]
- 52. Chernoivanenko I. S., Minin A. A., and Minin A. A. (2013) [Role of vimentin in cell migration]. Ontogenez 44, 186–202 [DOI] [PubMed] [Google Scholar]
- 53. Bakos R. M., Maier T., Besch R., Mestel D. S., Ruzicka T., Sturm R. A., and Berking C. (2010) Nestin and SOX9 and SOX10 transcription factors are coexpressed in melanoma. Exp. Dermatol. 19, e89–e94 [DOI] [PubMed] [Google Scholar]
- 54. Monzani E., Facchetti F., Galmozzi E., Corsini E., Benetti A., Cavazzin C., Gritti A., Piccinini A., Porro D., Santinami M., Invernici G., Parati E., Alessandri G., and La Porta C. A. M. (2007) Melanoma contains CD133 and ABCG2 positive cells with enhanced tumourigenic potential. Eur. J. Cancer 43, 935–946 [DOI] [PubMed] [Google Scholar]
- 55. Luo Y., Dallaglio K., Chen Y., Robinson W. A., Robinson S. E., McCarter M. D., Wang J., Gonzalez R., Thompson D. C., Norris D. A., Roop D. R., Vasiliou V., and Fujita M. (2012) ALDH1A isozymes are markers of human melanoma stem cells and potential therapeutic targets. Stem Cells 30, 2100–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Griffin M., Scotto D., Josephs D. H., Mele S., Crescioli S., Bax H. J., Pellizzari G., Wynne M. D., Nakamura M., Hoffmann R. M., Ilieva K. M., Cheung A., Spicer J. F., Papa S., Lacy K. E., Karagiannis S. N. (2017) BRAF inhibitors: resistance and the promise of combination treatments for melanoma. Oncotarget 8, 78174–75192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ludwig T. (2005) Local proteolytic activity in tumor cell invasion and metastasis. Bioessays 27, 1181–1191 [DOI] [PubMed] [Google Scholar]
- 58. Moreno-Layseca P., and Streuli C. H. (2014) Signalling pathways linking integrins with cell cycle progression. Matrix Biol. 34, 144–153 [DOI] [PubMed] [Google Scholar]
- 59. Vizcaíno J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q.-W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org/) via the PRIDE partner repository (59) with the data set identifier PXD010683.
Excel files containing the analyzed data are provided in Supplementary Materials.