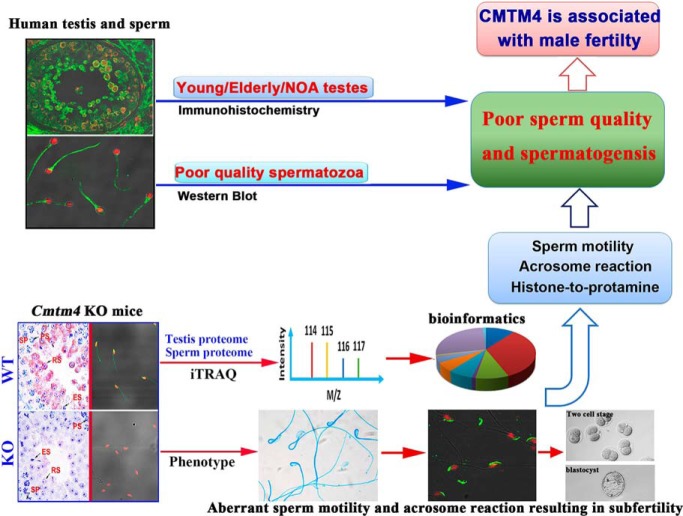
CMTM4 is aberrantly expressed in spermatozoa from infertility patients and testes from elderly adults and NOA patients. Cmtm4 KO mouse was generated to study its involvement in male fertility in vivo. Integrative phenotypic and quantitative proteomics analyses reveal an association of CMTM4 with histone-to-protamine exchange, sperm motility and induction of the acrosome reaction, and demonstrate that CMTM4 is associated with spermatogenesis and sperm quality.
Keywords: Cytokines*, iTRAQ, Knockouts*, Mouse models, Absolute quantification, Cas9, CMTM4, Male fertility, Spermatogenesis, Testis
Graphical Abstract
Highlights
CMTM4 is associated with human spermatogenesis and sperm quality.
Cmtm4 knockout mouse were generated by CRISPR/Cas9 technology for male fertility research.
CMTM4 is required for male fertility but not female fertility.
Phenotype and quantitative proteomics of Cmtm4 KO mice reveal an association of CMTM4 with histone-to-protamine exchange, sperm motility and induction of the acrosome reaction.
Abstract
The chemokine-like factor (CKLF)-like MARVEL transmembrane domain-containing family (CMTM) is a gene family that has been implicated in male reproduction. CMTM4 is an evolutionarily conserved member that is highly expressed in the testis. However, its function in male fertility remains unknown. Here, we demonstrate that CMTM4 is associated with spermatogenesis and sperm quality. Using Western blotting and immunohistochemical analyses, we found CMTM4 expression to be decreased in poor-quality human spermatozoa, old human testes, and testicular biopsies with nonobstructive azoospermia. Using CRISPR-Cas9 technology, we knocked out the Cmtm4 gene in mice. These Cmtm4 knockout (KO) mice showed reduced testicular daily sperm production, lower epididymal sperm motility and increased proportion of abnormally backward-curved sperm heads and bent sperm midpieces. These mice also had an evident sub-fertile phenotype, characterized by low pregnancy rates on prolonged breeding with wild type female mice, reduced in vitro fertilization efficiency and a reduced percentage of acrosome reactions. We then performed quantitative proteomic analysis of the testes, where we identified 139 proteins to be downregulated in Cmtm4-KO mice, 100 (71.9%) of which were related to sperm motility and acrosome reaction. The same proteomic analysis was performed on sperm, where we identified 3588 proteins with 409 being differentially regulated in Cmtm4-KO mice. Our enrichment analysis showed that upregulated proteins were enriched with nucleosomal DNA binding functions and the downregulated proteins were enriched with actin binding functions. These findings elucidate the roles of CMTM4 in male fertility and demonstrates its potential as a promising molecular candidate for sperm quality assessment and the diagnosis or treatment of male infertility.
The human chemokine-like factor (CKLF)1-like MARVEL transmembrane domain-containing (CMTM) family is a gene family encoding proteins that link classical chemokines and the transmembrane 4 superfamily (TM4SF). In humans, the nine members include CKLF and CMTM1–8, which encode proteins that play important roles in the immune system, tumorigenesis, and the male reproductive system (1, 2). Our previous studies have reported that CMTM3, CMTM4, CMTM5, and CMTM7 function as tumor suppressors in the development and progression of carcinomas (3–6). CMTM3 and CMTM7 colocalize with RAB5 in early endosomes and facilitate epidermal growth factor receptor (EGFR) internalization and degradation by enhancing RAB5 activity and early endosome fusion (7, 8). CMTM3 and CMTM4 mediate cell-cell adhesion by involvement in VE-cadherin turnover, and this process is involved in the regulation of angiogenesis (9, 10). CMTM3 and CMTM7 also initiate B-cell linker (BLNK)-mediated signal transduction (11, 12). CMTM6 and CMTM4 have been identified as programmed death-1 (PD-L1) regulators that inhibit immune function (13, 14). These studies showed that the CMTM family has important regulatory effects on the trafficking, degradation, and signal transduction of membrane molecules. Interestingly, CMTM1, CMTM2, and CMTM4 are highly expressed in the human testis, implying biological roles in male reproduction (2). CMTM1 is predominantly expressed in the human testis, with at least 23 alternative splicing isoforms (15). However, Cmtm1 knockout (KO) has no significant influence on male fertility (16). CMTM2 is highly expressed in the testis and is closely correlated with spermatogenic defects (17, 18). Its two homologs in the mouse, Cmtm2a and Cmtm2b, serve as androgen receptor corepressor and enhancer, respectively (19–22). Coexpression of Cmtm2a and Cmtm2b is essential for male fertility in mice (16). These findings indicate that CMTM family members may play important roles in spermatogenesis or testicular development.
CMTM4 is the most conserved member of the CMTM family, and forms a gene cluster with CKLF and CMTM1–3 on chromosome 16q22.1 (2). Our previous studies showed higher expression of CMTM4 in the testis than in other tissues (2, 23), which warrants exploration of its significance in male reproduction. Given its sequence structure and expression characteristics, CMTM4 might also play crucial roles in male fertility as do CMTM2 (16, 17). Previous studies have indicated that CMTM4 acts as a tumor suppressor through its involvement in cell growth and cell cycle regulation (4, 23, 24). However, its roles in male reproduction remain unknown.
In the present study, we first assessed the expression of CMTM4 in the spermatozoa and testes of patients with male infertility to characterize its association with spermatogenesis and sperm quality. Because the amino acid sequences are highly homologous between human and mouse CMTM4, the functions of CMTM4 in male fertility were examined in a KO mouse model, and the underlying mechanism was investigated using isobaric tags for relative and absolute quantification (iTRAQ)-based proteomics. Consistent with the association of CMTM4 expression with sperm quality in patients, Cmtm4 KO mice showed male subfertility with phenotypes of decreased sperm motility and aberrant acrosome reaction. Gene ontology (GO) term analysis revealed that proteins downregulated in KO mice testis and spermatozoa compared with wild-type (WT) were mainly involved in spermatogenesis and sperm functions including motility, the acrosome reaction, and histone-to-protamine exchange. This study also provided in-depth proteomic mapping of the mouse testis and sperm that will facilitate to understand of spermatogenesis and sperm functions. Combining phenotypic characteristics and proteomic analyses of Cmtm4 KO mice, we have shown that CMTM4 plays key roles in regulating sperm function and male fertility by affecting sperm motility and the acrosome reaction.
EXPERIMENTAL PROCEDURES
Ethical Statement
The present study was approved by the Medical Ethical Committee of the YuHuangDing hospital and all participants provided written informed consent. All experiments were performed in accordance with the guidelines provided by YuHuangDing hospital. All animal experiments were carried out in according with the guidelines of the care and use of laboratory animals. All mice were kept under light/dark cycles of 12/12 h with free access to food and water.
Sample Preparation
Samples of human semen were collected from the YuHuangDing hospital, and were classified into four groups without leukocytospermia as follows: the normozoospermia group (24–37 years old, sperm count >39 × 106, progressive motile spermatozoa >40%), asthenozoospermia group (27–36 years old, progressive motile spermatozoa <32%), oligoasthenozoospermia group (27–39 years old, sperm count <39 × 106, progressive motile spermatozoa <32%), and teratozoospermia (27–40 years old, normal morphology <4%). Young testes were obtained from five young fathers (23–27 years old) who died in car accidents, had no history of pathology that could affect reproductive function, and had previously indicated a willingness to donate their bodies to medical research. Donations of their organs for medical research were approved by their immediate family members. Aged testes samples were obtained from five elderly fathers (70–74 years old) who were prostate cancer patients with no anti-androgen treatment before orchiectomy and who had given written informed consent. All procedures were approved by the Ethics Committee of YuHuangDing Hospital. A total of 25 patients with azoospermia (26–37 years old) who underwent surgical testicular sperm extraction were recruited and divided into the obstructive azoospermia (OA) group (n = 5) and the nonobstructive azoospermia (NOA) group (n = 20). In OA, spermatozoa were produced normally inside the testicle, whereas in NOA, spermatogenesis problems were observed with a very low level of sperm production or a total lack of production. Patients with abnormal karyotypes and those who had previously suffered from an injury to the genitals were excluded. Human testicular quality was evaluated by the modified Johnsen score according to our previous studies (25, 26).
Protein Extraction
Human sperm samples exhibiting normozoospermia, asthenozoospermia, oligoasthenozoospermia, and teratozoospermia were collected for protein extraction. As in our previous reports (26), after centrifugation at 800 × g for 20 min at 4 °C in a 50% step Percoll gradient in phosphate buffered saline (PBS) media, the seminal plasma and other contaminating cells in semen were removed. Sperm pellets were dissolved in lysis solution and sonicated for three times at 5 s intervals. Then they were kept at 4 °C for 2 h before the solution was centrifuged at 12,000 × g for 45 min, and the supernatant was collected. After determination of protein concentration, protein samples were stored at −80 °C.
In Vitro Synthesis of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-associated Protein 9 (Cas9) mRNA and sgRNAs
CRISPR-Cas9 constructs were synthesized as described previously (27). Briefly, Cas9 mRNA was synthesized in vitro from linear DNA templates by using the mMESSAGE mMACHINE T7 Ultra kit (Thermo Fisher Scientific Inc, CA) according to the manufacturer's instructions. DNA templates for single guide RNAs (sgRNAs) were also synthesized in vitro by polymerase chain reaction (PCR); RNA was synthesized from these templates with a MEGA shortscript T7 kit (Thermo Fisher Scientific) according to the manufacturer's instructions. The constructs were diluted with RNase-free injection buffer (0.25 mm EDTA, 10 mm Tris at pH 7.4) before microinjection. The following sequence information was used for sgRNA synthesis: sgRNA#1: AGCGCGCCGCGCAGGTAGTC; sgRNA#2: GAGGAGCTGGACGGCTTCGA. CRISPR-Cas9 constructs and sgRNAs were microinjected into fertilized C57BL/6J oocytes. Knockout of Cmtm4 was confirmed by Sanger sequencing.
Genotyping of Cmtm4 Mouse Lines
Genotypes were confirmed by the PCR with the following primers
Cmtm4, forward primer: 5′-GGTCTGGGCCTTTCCTTGC-3′, reverse primer: 5′-GCCCAAGGACCTCGGAGTA-3′; Gapdh was internal control, Gapdh, forward primer: 5′-CACTCATTGCCCCCGTGTTT-3′, reverse primer: 5′-GTCAGGTTTCCCATCCCCACATA-3′. PCR was performed as follows: denaturation at 98 °C for 2 min, 30 cycles of 98 °C for 10 s, 62 °C for 10 s and 72 °C for 50 s; extension at 72 °C for 5 min. Electrophoresis was conducted on 1.2% (w/v) agarose gels and visualized with a Bio-Rad ChemiDOC MP system (Bio-Rad, Hercules, CA 94547).
Assessment of KO Mouse Fertility and Fecundity
To assess fertility and fecundity, one littermate male (6 weeks old) was placed in cages with two mature WT females for two months or more. Two littermate females were caged with a WT fertile male for the same period. The number of female mice achieving pregnancy and the number of offspring were recorded.
Mouse sperm suspensions were prepared by mincing the cauda epididymidis of a mature male mouse (WT or KO) in 1 ml IVF medium for about 1 h. Part of this sperm suspension was divided into 30 μl pellets using a micropipette in a plastic culture dish (35 mm × 11 mm) covered with paraffin oil, and the rest was treated by 10 μm calcium ionophore A23187 (Sigma-Aldrich, St. Louis, MO) for assessing the induction of the acrosome reaction. Mature female mice (WT or KO) were induced to super-ovulate by i.p. injection of 10 i.u. pregnant mare serum gonadotrophin (PMSG) and human chorionic gonadotrophin (hCG) 48 h apart. The oocytes were recovered from the oviducts 14 h after injection of hCG and were introduced into each sperm suspension. The sperm concentration was counted by using improved-Neubauer counting chamber, and at least 200 cells were counted. About 106 spermatozoa/ml was used for insemination. The development stage and morphology of embryos were recorded at 24 h interval after zygote collection.
Evaluation of KO Mouse Sperm Quality
Epididymal spermatozoa was collected from the caudal part of the epididymis. The cauda epididymidis was placed in PBS supplemented with 10% (w/v) bovine serum albumin (BSA) and cut into small pieces followed by incubation on a warming metal plate at 35 °C for 5 min. Sperm motility was measured with Computer-aided Semen Analysis system (CASA) System (Medealab™, Erlangen, Germany). For each sample, 12 fields were examined.
To evaluate the acrosome reaction, spermatozoa were suspended in Biggers-Whitten-Whittingham (BWW) and incubated at 37 °C, 5% CO2 for 3 h to get capacitation. Capacitated sperm was exposed to vehicle alone (DMSO, dimethyl sulfoxide) or calcium ionophore A23187 (10 μm) for 30 min, subjected to Coomassie brilliant blue staining (2% w/v G250) for 3 min, and then washed with PBS three times before mounting on slides for observation. Four hundred spermatozoa were evaluated under a light microscope (×400). Spermatozoa were scored as acrosome-intact when a bright blue staining was observed in the dorsal region of the head or as acrosome-reacted when no labeling was observed.
Daily Sperm Production (DSP)
DSP was estimated according to a previous report (28). Briefly, after the testes were crushed in 600 μl SMT solution containing 0.05% (v/v) Triton X-100, 0.9% NaCl and 0.01% Azide by ultrasonic pulverizer, the solution was homogenized again. Aliquots of 10 μl were placed on a makler counting chamber and sperm heads were counted twice under microscopic visualization to determine the average number of spermatids per sample. Sperm heads in 80 small squares were assessed. These values were divided by testicular weight to give the number of spermatids per gram of testis. Because developing spermatids spend 4.84 days in steps 14–16 of spermatogenesis, the number of spermatids per gram of testis was divided by 4.84 to provide the DSP.
iTRAQ Labeling of Mouse Testicular Samples
The iTRAQ procedures were described in previous publication (29, 30). Briefly, the testicular or spermatozoa protein samples from the wild and Cmtm4 KO mice were separately pooled. The pH was adjusted to 8.5 by adding 25 μl triethylammonium bicarbonate (TEAB) buffer and samples were treated with 20 mm dithiothreitol (DTT) at 56 °C for 60 min and 50 mm iodoacetamide in dark for 30 min. After digestion with 3 μg trypsin (sequencing grade, Promega, Madison, WI) at 37 °C overnight, the peptides were labeled with iTRAQ tags and dried.
2-D Reversed-phase (RP) Separation and Mass Spectrometer Data Acquisition
The first dimension RP separation by microLC was performed by using a Durashell RP column (5 μm, 150 Å, 250 mm ×4.6 mm i.d., Agela, Tianjin, China). Mobile phases A (2% v/v acetonitrile, 20 mm ammonium formate, adjusted to pH 10.0 with NH4OH) and B (98% acetonitrile, 20 mm ammonium formate, adjusted to pH 10.0 using NH4OH) were used to develop a gradient. The peptides were eluted over 45 min by a gradient of 100% buffer A to 100% buffer B at a flow rate of 800 μl/min. In total, 10 fractions were generated for further LC-MS/MS analysis, for which a nanoflow HPLC instrument (EASY-nLC 1000 system, Thermo Fisher Scientific, Waltham, MA) coupled to an on-line Q Exactive mass spectrometer (Thermo Fisher Scientific, Waltham, MA) with a nanoelectrospray ion source (Thermo Fisher Scientific) was used. The chromatography columns were packed in-house with Ultimate XB-C18 3 μm resin (Welch Materials, Shanghai, China). The peptide mixtures were loaded onto the C18 RP column (10-cm length, 75 μm inner diameter) with buffer A (99.5% water and 0.5% formic acid, v/v) and separated with a 75-min linear gradient of 5–100% buffer B (99.5% acetonitrile and 0.5% formic acid, v/v) at a flow rate of 300 nl/min. Including the loading and washing steps, the total time for an LC-MS/MS run was 90 min. The electrospray voltage was 2.0 kV. Peptides were analyzed by data-dependent MS/MS acquisition with a dynamic exclusion duration of 18 s. In MS1, the resolution was 70,000, the automatic gain control (AGC) target was 3e6, and the maximum injection time was 20 ms. In MS2, the resolution was 17,500, the AGC target was 1e6, and the maximum injection time was 60 ms. The scan range was 300–1400 m/z, and the top 75 intensive precursor ions were selected for MS/MS analysis.
Identification and Quantification of Mouse Testicular or Spermatozoa Proteins
The raw data were processed by using the proteomic workflow of Proteome Discoverer 1.3. The fragmentation spectra were searched against the UniProt reviewed mouse database (2017_12, 16950 sequences) by the Mascot search engine (version 2.2.06) with the precursor and fragment mass tolerances set to 15 ppm and 20 milli-mass units (mmu), respectively. The algorithm was set to use trypsin as the enzyme, allowing for two missed cleavage sites. The fixed modification was carbamidomethylation (cysteine), and the variable modifications were oxidation (methionine), acetylation (protein N terminus), and iTRAQ labeling (tyrosine and lysine, N-terminal residues). Peptide ions were filtered from the cut-off scores of Percolator based on p < 0.01. The false discovery rate was set to 1% for peptide identifications. An additional filter was applied with the removal of spectrum matches with scores lower than 10. The iTRAQ quantization values were automatically calculated based on the intensity of the iTRAQ reporter ions in the dissociation scans with higher collision energy using Proteome Discoverer. All protein iTRAQ ratios were exported to an Excel file, the Gaussian distribution of ratios was recalculated manually, and all ratios were transformed to base 10 logarithm values. A confidence interval of 99% (testis proteome, 0.754–1.24; sperm proteome, 0.84–1.19) was used to determine the cut-off values for statistically significant changes.
Bioinformatics
Broad functional classification of molecular function and biological process were performed by using Protein Analysis Through Evolutionary Relationships (PANTHER) tools (http://www.pantherdb.org/). The over-representation analyses of GO (http://www.geneontology.org/) for main biological processes were performed by using the Database for Annotation, Visualization and Integrated Discovery (DAVID) tools (https://david.ncifcrf.gov/). The GO level 2 and 3 categories and a p value cut-off of 0.01 were selected. Knockout and mutant phenotypes related to male reproduction and fertility were obtained by batch query from Mouse Genome Informatics (http://www.informatics.jax.org).
Western Blotting
For protein separation, 50 μg proteins from each sample were loaded on 12% gels for sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS)-PAGE. This was transferred to polyvinylidene difluoride membranes at 100 v for 1 h, blocked with 5% (w/v) skimmed milk for 1 h, and incubated with primary antibodies (diluted 1:800 in blocking solution) (supplemental Table S1) at 4 °C overnight with gentle agitation. The membranes were washed with 0.5% (v/v) Tween-20 in Tris-buffered saline (TBS) three times and incubated with horseradish peroxidase (HRP)-conjugated anti-IgG for 1 h at room temperature (RT). The immune-reactive complexes were detected by an enhanced chemiluminescence (ECL) kit (Amersham Biosciences Life Science, Cleveland, OH). The images were analyzed by commercial image analysis software (Gene Tools, version 4.02; Syngene, Cambridge, UK). The integrated optical density (IOD) of positive immune-staining was calculated, and the IOD ratio of target protein to housekeeping proteins was used to express the results of the Western blot analysis.
Immunohistochemistry (IHC)
Testicular tissue blocks were fixed in Bouin's solution for 12 h, and then embedded in paraffin according to conventional methods. Sections of 4 μm thickness were de-waxed. Antigen retrieval was performed in a microwave oven above 94 °C for 20 min. Endogenous peroxidases of sections were inhibited by incubation with 3% (v/v) H2O2 for 10 min. Then, 3% (v/v) BSA in TBS was used to block nonspecific binding with antibodies at RT for 1 h. Sections were then incubated with corresponding primary antibodies (diluted 1:100 in blocking solution) overnight at 4 °C. After washing three times with TBS, the sections were incubated with horseradish peroxidase-conjugated IgG (Zhong-Shan Biotechnology, Beijing, China) at a final dilution of 1:400 for 1 h at 37 °C. A 3,3′-diaminobenzidine (DAB) kit (Zhong-Shan Biotechnology, Beijing, China) was used to visualize peroxidase activity at the binding sites. Hematoxylin was used to counterstain the sections. After dehydration, the sections were mounted for bright-field microscopy (DM LB2, Leica, Nussloch, Germany). Preimmune IgG was used as a negative control. Image-Pro analysis software was used to analyze the images of immunostained sections.
Immunofluorescence Analysis
The mouse spermatozoa were smeared on slides precoated with 1% (w/v) gelatin. The air-dried slides were fixed with cold methanol for 10 min, blocked for 1 h at room temperature with 3% (w/v) BSA in PBS and incubated at 4 °C overnight with primary antibody against CMTM4, ODF2 (diluted 1:50 in 3% BSA). After three washes with PBS, the anti-rabbit IgG (Alexa Fluor® 488 Conjugate) (1:100 in 3% BSA) was applied and incubated at room temperature for 1 h; rabbit IgG was used as a negative control. Samples were subsequently washed with PBS. Propidiumiodide (0.01 mg/ml, Invitrogen, Carlsbad, CA) counterstaining visualized the nuclei and sections were mounted in 80% (v/v) glycerol. Quantitative assessment of protein expression in spermatozoa was achieved by a Zeiss LSM 510 laser confocal microscope (LSM, Carl Zeiss Microimaging, Thornwood, NY). Slides were systematically examined at 400× magnification according to the World Health Organization manual 2010. Inspection was performed in sequence until a total of 200 spermatozoa had been assessed. By using Zeiss LSM510 Meta software (LSM5 version 3.2, Carl Zeiss Microimaging, Thornwood, NY), the fluorescence intensity value per stained sperm was calculated automatically by subtracting the background fluorescence intensity, which was determined by scanning a sperm-free area.
Experimental Design and Statistical Rationale
The expression of CMTM4 in spermatozoa from normozoospermic, asthenozoospermic, oligoasthenozoospermic, and teratozoospermic men was evaluated by Western blotting. Its cellular localization and expression intensity in testis from young adult, elderly adult, and NOA patients were detected by IHC. Cmtm4 KO mice were constructed to investigate CMTM4 function in male infertility. The testis proteomes, epididymal sperm proteome from Cmtm4 WT and KO mice were compared by iTRAQ, each group was a pool from 10 mice and performed in 2 technical replicates. The MS data were processed by using Proteome Discoverer 1.3 and searched against the UniProt (2017_12, 16950 sequences) with the Mascot search engine (version 2.2.06). The false discovery rate was set to 1% for peptide identifications. Data are reported as means±S.D. Means of two groups were analyzed with Student's t test, and more than two group means were analyzed by one-way analysis of variance (ANOVA). A commercial software package (SPSS 18.0; SPSS, Chicago, IL) was used to perform the correlation analysis. A value of p < 0.05 was statistically significant.
RESULTS
Association of CMTM4 with Human Spermatogenesis and Sperm Quality
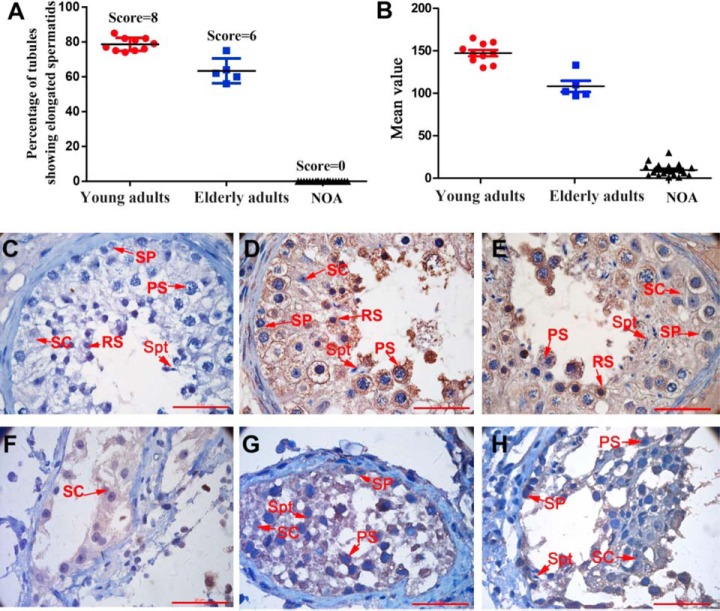
Western blots were performed on protein extracts from the spermatozoa of fertile and infertile men. Spermatozoa from patients diagnosed with asthenozoospermia, oligoasthenozoospermia, or teratozoospermia exhibited obvious poor sperm quality with decreased sperm motility or sperm count (Fig. 1A) (supplemental Table S2). As shown in Fig. 1C–1D, the expression of CMTM4 in spermatozoa from asthenozoospermic, oligoasthenozoospermic, and teratozoospermic men was significantly lower than that in spermatozoa from normazoospermic men. Especially, significantly decreased expression of CMTM4 was observed in spermatozoa from teratozoospermic patients. An analysis of gene expression omnibus (GEO) data from GSE6968, which shows gene expression profiles of normazoospermic and teratozoospermic spermatozoa, demonstrated significant downregulation of CMTM4 in teratozoospermic spermatozoa (Fig. 1B). To further explore the association of CMTM4 with spermatogenesis, we compared the expression levels of CMTM4 in the testes of young adults with those of elderly men, who have poor spermatogenesis associated with poor sperm quality as described in our previous study (26, 31), and with NOA patients, who have impaired spermatogenesis because of Sertoli cell-only pattern (Fig. 2F), maturation arrest (Fig. 2G), and hypospermatogenesis (Fig. 2H) (supplemental Table S2). Testes from 10 young adults (including 5 biopsies from OA patients) (score = 8), 5 elderly adults (score = 6), and 20 NOA patients (score = 0) were evaluated as shown in Fig. 2A. Morphologically, elderly testes showed hyperplasia of the interstitial layer and vacuolization of seminiferous tubules. Compared with young adults, the testes of elderly adults and NOA patients had significantly decreased CMTM4 expression by immunohistochemical analysis (Fig. 2B).
Fig. 1.
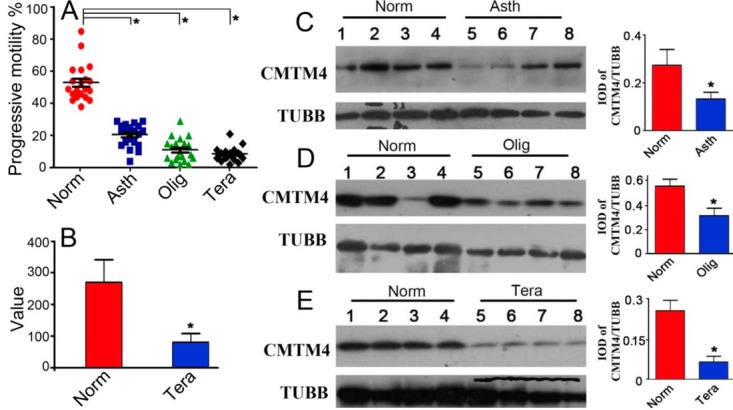
Expression of CMTM4 in human spermatozoa. Human spermatozoa were collected from normozoospermic, asthenozoospermic, oligoasthenozoospermic, teratozoospermic patients and showed significant differences in progressive sperm motility (A); comparison of CMTM4 expression in spermatozoa in normozoospermia and teratozoospermia was analyzed by retrieving GEO data from GSE6968 (B); Western blot analysis of CMTM4 in human spermatozoa in normozoospermia, asthenozoospermia, oligoasthenozoospermia and teratozoospermia. The quantification of CMTM4 expression was calculated by normalizing band intensities to the respective TUBB expressions by Image Pro software (C–E). Asth, asthenozoospermia, Olig, oligoasthenozoospermia, Tera, teratozoospermia; Lines (A) indicate mean values, whiskers indicate standard deviation; Data were compared by one-way analysis of variance (ANOVA); *, p < 0.05.
Fig. 2.
Expression of CMTM4 in human testes. Johnsen score evaluation of testicular sections from young adults, elderly adults and patients with nonobstructive azoospermia (NOA) (A); quantitative evaluation of CMTM4 expression in testes by Image-Pro (B); representative image of testicular cellular localization of negative control (C) and CMTM4 in young adults (D), elderly adults (E) and NOA patients (F–H); SP, spermatogonia; PS, pachytene spermatocyte; RS, round spermatid; SC, Sertoli cell; Spt, spermatids; Lines (A, B) indicate mean values, whiskers indicate standard deviation; Data were compared by one-way analysis of variance (ANOVA); *, p < 0.05; Each bar represents 20 μm.
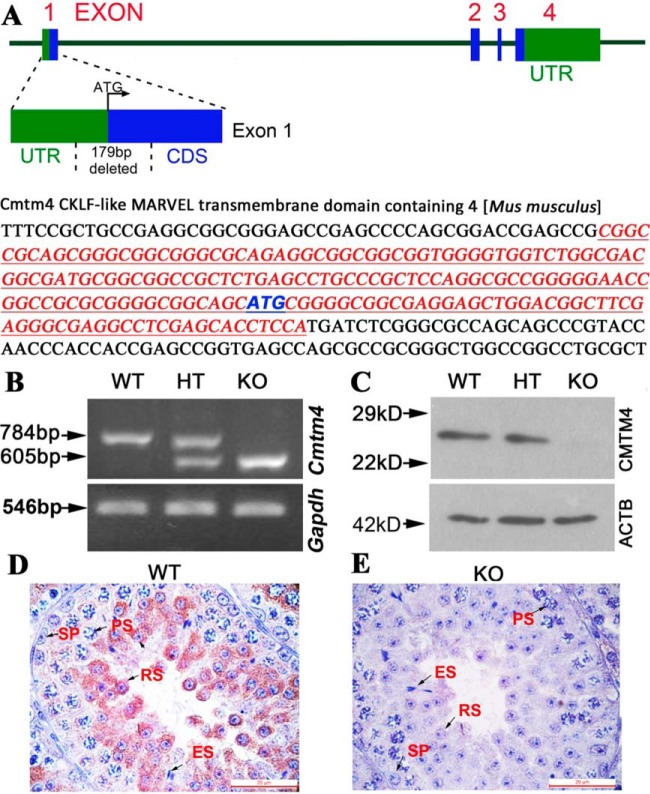
Cmtm4 KO Mice Were Constructed by Cas9 Microinjection
To further investigate the role of CMTM4 in spermatogenesis and male fertility, we constructed Cmtm4 KO mice using CRISPR-Cas9. A pair of independent sgRNAs targeting exon 1 of the Cmtm4 locus was designed to establish the homozygous KO mouse model. Coinjection of the two independent sgRNAs with Cas9 mRNA caused a 179-bp deletion spanning the 5′ untranslated region (UTR) and exon 1 in the open reading frame of the Cmtm4 locus, resulting in a frameshift mutation and KO of Cmtm4 (Fig. 3A). Deletion of the Cmtm4 gene was confirmed by PCR and sequencing analyses (Fig. 3B). Western blot analysis confirmed the absence of CMTM4 protein in KO mice (Fig. 3C). IHC analysis showed that CMTM4 was mainly localized in the cytoplasm of spermatocytes and round spermatids of WT mice, whereas no signal was detected in the testis of KO mice (Fig. 3D, 3E).
Fig. 3.
Construction of Cmtm4 KO mice. Scheme of the genomic target sites in theCmtm4 (A). The untranslated region (UTR) of Cmtm4 is highlighted in green and the coding sequence (CDS) of Cmtm4 is indicated in blue. 179bp sequences in exon 1 were deleted in Cmtm4 KO mice. Polymerase Chain Reaction (PCR) (B), Western blotting (C) and immunohistochemistry analysis (D, E) were performed on mouse testis samples. WT, wild type; HT, heterozygous; KO, knockout; SP, spermatogonia; PS, pachytene spermatocytes; RS, round spermatids; ES, elongated spermatids. Each bar represents 20 μm.
CMTM4 Requirement for Male and Female Fertility
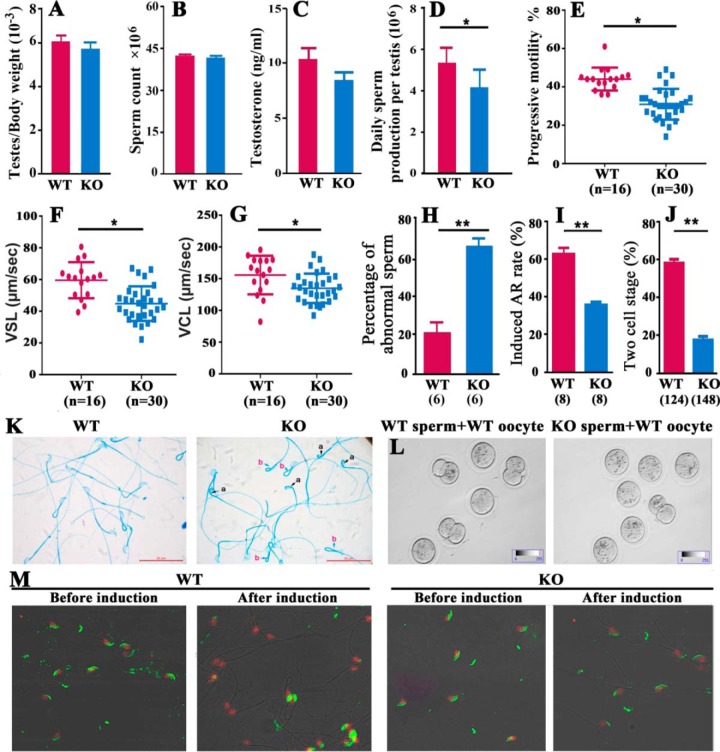
All germ cells (i.e. spermatogonia, primary spermatocytes, round spermatids, elongated spermatids, and Sertoli cells) were examined in the seminiferous tubules of Cmtm4 KO mouse testes, and no obvious morphological abnormalities were found.
No significant differences were observed in the testis/body weight ratio, cauda epididymal sperm content, or serum testosterone concentration between WT and KO mice (Fig. 4A–4C). DSP and progressive motility were significantly reduced in adult KO males compared with adult WT males (Fig. 4D–4G). A higher percentage of abnormal epididymal spermatozoa with backward-curved heads and bent midpieces were observed in Cmtm4 KO mice (Fig. 4H, 4K, supplemental Table S3).
Fig. 4.
Characteristics of fertility of Cmtm4 WT and KO male mice. No variance in testicular weight (A), sperm concentration (B) and serum testosterone level (C) between WT and KO mice was observed. Significantly reduced daily sperm production (D) and sperm motility (E, F, G) and increased abnormal sperm morphology (H, K) were observed in Cmtm4 KO mice compared with WT mice. In vitro fertility analyses showed a decreased percentage of induced acrosome reactions (I, M), the numbers in brackets represent the number of mice examined, and the decreased percentage of oocytes developed to two-cell stages. The numbers in brackets represent the number of WT oocytes used (J, L). Values are shown as mean ± S.D. a, curved back sperm head; b, bent sperm midpiece; WT, wild type; KO, knockout; VSL, straight line velocity; VCL, curvilinear velocity; Lines (E, F, G) indicate mean values, whiskers indicate standard deviation; Data were compared by one-way analysis of variance (ANOVA); *, p < 0.05; **, p < 0.01. Each bar represents 20 μm.
KO males mating with WT females produced fewer pregnancies after 2 months of continuous cohabitation. The Cmtm4+/− littermates were all fertile, and produced the same number of offspring as WT littermates. Cmtm4 KO females gestated normally when mated with WT males (Table I). We then examined the in vitro fertilization (IVF) capacities of spermatozoa obtained from the epididymides of WT and KO male mice. IVF results showed that the spermatozoa of KO mice had low fertilization abilities (Table II, Fig. 4J, 4L), implying impaired sperm function. There was no difference in the percentage of intact acrosomes between spermatozoa from WT and KO mice, whereas a significantly lower percentage of acrosome reactions was induced by the calcium ionophore A23187 in spermatozoa from KO mice (Fig. 4I, 4M).
Table I. Fertility and fecundity of WT and Cmtm4 KO mice.
| Genotype | Male fertility | Litter numbers | Female fertility | Litter numbers |
|---|---|---|---|---|
| +/+ | 15/15 | 8.46 ± 1.85 | 15/15 | 8.33 ± 2.14 |
| +/− | 15/15 | 8.60 ± 1.72 | 15/15 | 7.47 ± 1.30 |
| −/− | 4/15 | 4.00 ± 0.82 | 12/15 | 7.67 ± 2.02 |
Mice of the indicated genotypes were caged with WT mice of the opposite sex. Male or female fertility was indicated as the number of fertile mice/total number of mice, values are shown as mean ± S.D.
Table II. In vitro fertilization assay of WT and Cmtm4 KO mice.
| Group | Total No. of oocytes inseminated | Total No. of oocytes fertilized | Fertilized oocytes developed to |
|
|---|---|---|---|---|
| Two cells (%) | Blastocysts (%) | |||
| WT sperm + WT oocyte | 124 | 73 | 73 (100) | 37 (50.7) |
| WT sperm + KO oocyte | 130 | 82 | 78 (95.1) | 38 (48.7) |
| KO sperm + WT oocyte | 148 | 26 | 26 (100) | 2 (7.7) |
| KO sperm + KO oocyte | 132 | 18 | 18 (100) | 1 (5.6) |
Differentially Expressed Proteins in KO Mouse Testes Identified Through Quantitative Proteomics
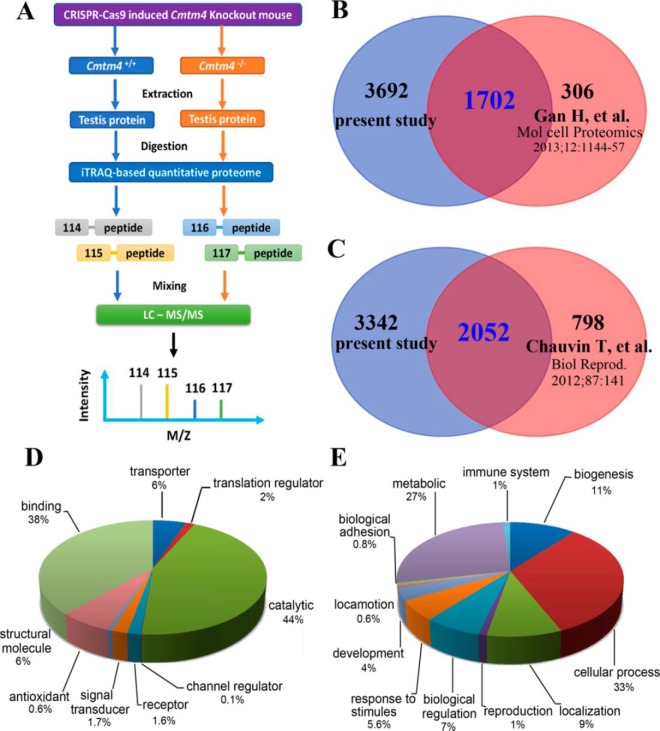
To identify testicular proteins and biological processes that were associated with CMTM4 in male fertility, we performed comprehensive differential proteomics analysis of WT and Cmtm4 KO mouse testes. iTRAQ was applied to compare the proteomes of whole testes from 8-week-old WT and Cmtm4 KO mice, and each group was pooled from 10 mice and performed in duplicate (Fig. 5A). A total of 5394 testicular proteins were identified (supplemental Table S5). There was an overlap of 1702(31.6%) proteins with the reported mouse germ cell proteome (32) (Fig. 5B) and 2052 proteins with the reported mouse sperm cell proteome (33) (Fig. 5C). A broad functional category classification was performed (Fig. 5D, 5E).
Fig. 5.
iTRAQ-based quantitative proteomic analysis of Cmtm4 KO mice testis. Flow chart of iTRAQ analysis of the mouse testicular proteome (A); Venn diagram of the current mouse testis proteome and a previously reported mouse germ cell proteome (B), and the current mouse testicular proteome and a previously reported mouse sperm proteome (C); broad functional analysis of the current mouse testicular proteome in GO terms of molecular functions (D) and biological processes (E).
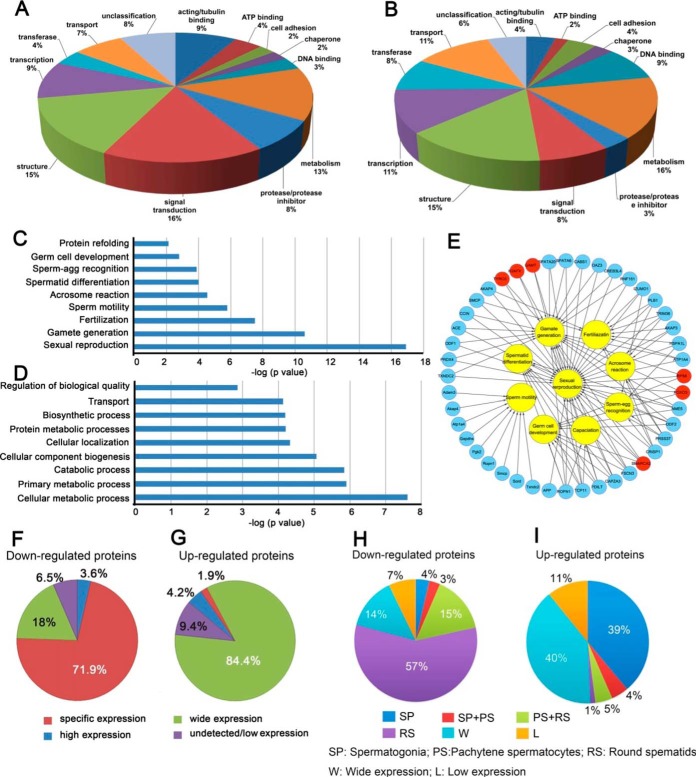
In total, 351 proteins were differentially expressed between WT and Cmtm4 KO testes, including 212 upregulated and 139 downregulated proteins in the KO testes (supplemental Table S6). A broad functional classification showed that majority of proteins related to signal transduction (16%), actin/tubulin binding (9%), and protease/protease inhibitor (8%) functions were downregulated in KO mice, whereas more proteins involved in transport (11%) and DNA binding (9%) functions were upregulated in KO mice (Figs. 6A, 6B). Enrichment analysis indicated that most of the downregulated proteins were involved in spermatogenesis and regulation of sperm quality, such as sperm motility and the acrosome reaction (Fig. 6C), whereas the upregulated proteins were mainly involved in metabolic processes (Fig. 6D). Several well-known proteins involved in the acrosome reaction (IZUMO1, CRISP1, AKAP1, AKAP3, AKAP4, PLB1, and TRIM36) and sperm motility (ODF1, ODF2, ATP1A4, PGK2, SORD, AKAP3, AKAP4, SMCP, and GAPDHS) were downregulated in the KO mice (Fig. 6E), whereas some upregulated proteins belonged to the chromatin components involved in the DNA packaging complex (e.g. HIST1H1B, H2AFX, HIST1H3A, H1FX, and H3F3C). Using retrieved GEO data set GDS3142 that includes mouse transcriptome of different tissues, differentially expressed gene products in the KO testis was grouped into specific, high, wide, and undetectable (low) expression patterns. Notably, 100 (71.9%) of the downregulated proteins were specifically expressed in the testis (Figs. 6F, 6G). Characterization of germ cell-type specific expression of the differentially expressed proteins showed that 57% of downregulated proteins were specifically expressed in round spermatids (Figs. 6H, 6I). The proteomics results indicated that CMTM4 deficiency mainly affected the expression of proteins involved in sperm motility and the acrosome reaction.
Fig. 6.
Bioinformatic analysis of differentially expressed proteins in WT and Cmtm4 KO mice testes. Broad functional classification of downregulated proteins (A) and upregulated proteins (B) in KO mice, and biological processes enriched in downregulated proteins (C) and in upregulated proteins (D) in KO mice; interactional network between differential proteins and major biological processes of sperm function (E); tissue expression patterns of downregulated proteins (F) and upregulated proteins (G); testicular germ cell expression patterns of downregulated proteins (H) and upregulated proteins (I) in the normal mouse testis.
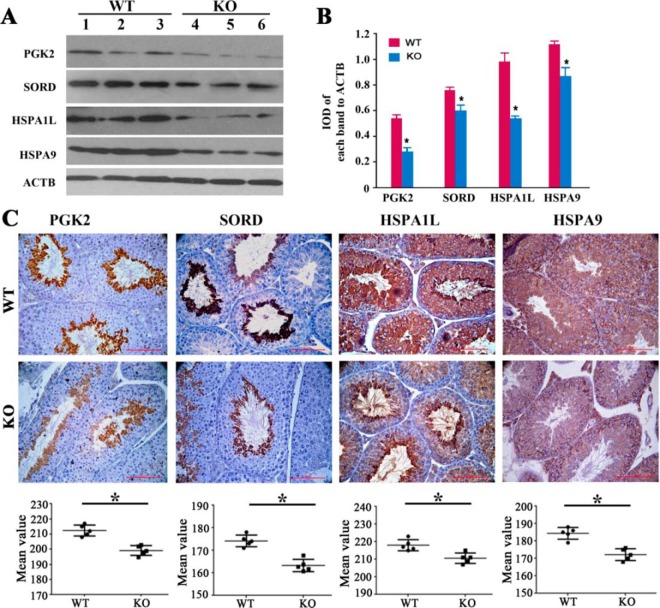
The differential expression of selected proteins (PGK2, SORD, HSPA1L, and HSPA9) was confirmed by Western blotting and IHC analyses. The IHC results indicated that PGK2 and SORD were mainly expressed in round spermatids, whereas HAPA1L and HSPA9 were mainly expressed in spermatocytes (Fig. 7). All these proteins showed downregulated expression in the KO mouse testis. In agreement with the IHC results, lower levels of these proteins were observed in the KO mice in Western blot analysis (Fig. 7).
Fig. 7.
Western blotting (A, B) and immunohistochemical (C) analysis of PGK2, SORD, HSPA1L and HSPA9 in WT and Cmtm4 KO mice. The integrated optical density (IOD) ratio of target protein to ACTB was used to express the results of the Western blot analysis. Lines (C) indicate mean values, whiskers indicate standard deviation; Data were compared by Student's t test; *, p < 0.05; Each bar represents 50 μm.
Differentially Expressed Proteins in KO Mouse Sperm Identified Through Quantitative Proteomics
To identify differentially expressed sperm proteins affected by Cmtm4 KO, we next performed proteomic analysis of Cmtm4 KO sperm, and identified 162 upregulated and 247 downregulated proteins relative to Cmtm4 WT sperm (supplemental Table S7). A total of 3588 sperm proteins were identified (supplemental Table S8), which included 667 (18.6%) and 2103 (58.6%) proteins overlapped with previously reported mouse sperm proteomes (33, 34) (Fig. 8A, 8B). 1410 sperm proteins were newly identified in the present study. Most of these proteins were related to broad functions of metabolism (30%), catalysis (11%), structural (11%) and energetics (11%), which provided complementary information for mouse sperm proteome (Fig. 8C).
Fig. 8.
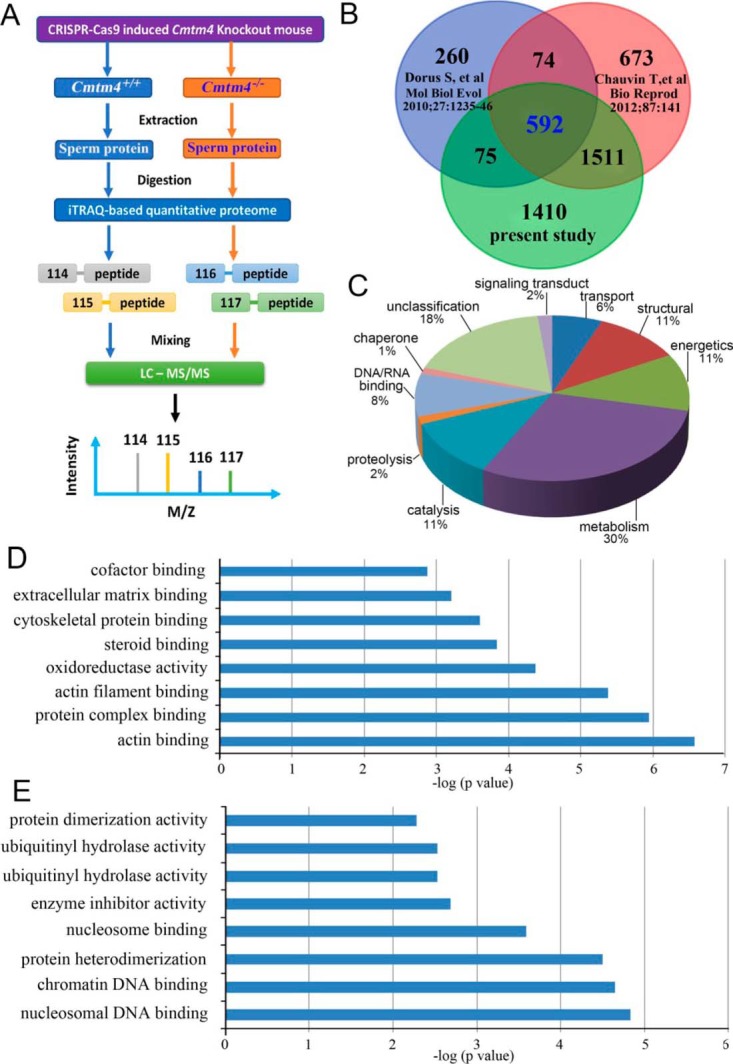
iTRAQ-based quantitative proteomic analysis of Cmtm4 KO mice sperm. Flow chart of iTRAQ analysis of the mouse sperm proteome (A); Venn diagram of the current mouse sperm proteome and previously reported mouse sperm proteomes (B); broad function of current mouse sperm proteomes (C); enriched molecular functional analysis of downregulated (D) and upregulated proteins (E) in KO sperm.
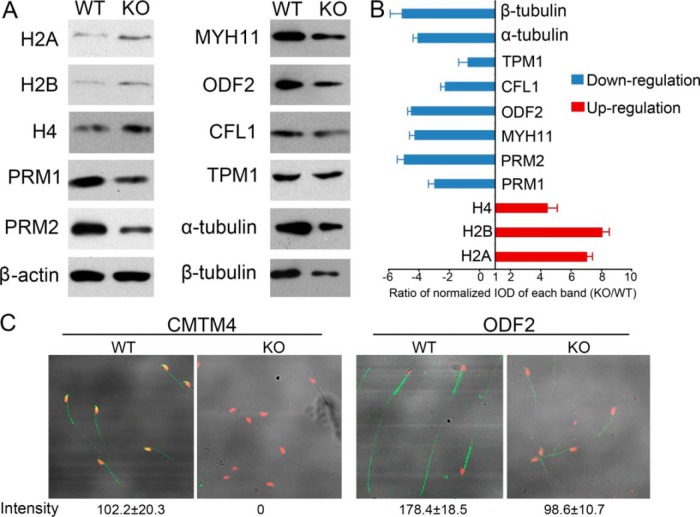
Enrichment analysis indicated that the upregulated proteins were mainly related to functions of DNA binding including histones H2A, H2B, H4 and their variants in KO sperm (Fig. 8D). The abnormal upregulated levels of H2A, H2B, H4 were validated by Western blotting. Given that nucleosomal histones were involved histone-to-protamine exchange, the expression of PRM1, PRM2 were also validated. PRM1 and PRM2 showed downregulated expressions in KO sperm (Fig. 9A, 9B). Downregulated proteins in KO sperm were mainly related to actin binding functions, which are well-known related to sperm motility and acrosome reaction (Fig. 8E). 15 proteins are characterized with male reproductive phenotypes of abnormal sperm function (supplemental Table S9). ODF2, CFL1, MYH11, TPM1, α-tubulin, and β-tubulin were validated to be lower expressed in KO sperm. Immunofluorescence analysis demonstrated that Cmtm4 was localized in sperm acrosome and middle piece, and ODF2 showed low intensity in KO sperm (WT sperm:178 ± 18.5; KO sperm:98.6 ± 10.7, p < 0.05) (Fig. 9C).
Fig. 9.
Western blotting and immunofluorescence staining of selected proteins. Western blot analysis of H2A, H2B, H4, PRM1, PRM2. MYH11, ODF2, CFL1, TPM1, α-tubulin, β-tubulin, and ACTB serving as a control (A); quantification of blotting intensity is shown as ratio of normalized IOD of each band (KO/WT) (B); quantitative immunofluorescence of CMTM4 and ODF2 in WT and KO sperm (C), intensity was calculated by using Zeiss LSM 510 Meta software.
DISCUSSION
Sperm quality is the decisive factor in male fertility (35). Most infertile male patients are characterized by poor sperm quality (36, 37, 38). Because they are likely to affect spermatogenesis or sperm quality, genes and gene products that are highly expressed in the testis are promising targets for the diagnosis or treatment of male infertility (39–42). Our previous studies have identified CMTM4 is highly expressed in the human testes (2, 23); thus, we hypothesized that CMTM4 was correlated with spermatogenesis, and its altered testicular expression would consequently be related to sperm quality. In the present study, we investigated the association of CMTM4 with spermatogenesis and sperm quality and demonstrated a role of CMTM4 in male fertility by phenotypic and quantitative proteomic analyses of the Cmtm4 KO mouse model. This study provides novel information in the functional research of the CMTM family in male reproduction.
In the present study, CMTM4 was observed to be aberrantly expressed in the spermatozoa of infertile individuals, indicating its association with sperm quality. As in our previous reports, the elderly testis showed decreased spermatogenesis, resulting in poor sperm quality (26, 31). Here, IHC analysis showed downregulated expression of CMTM4 in elderly and NOA testes, with especially low expression in NOA testes. The combined results imply that CMTM4 is a potential key protein in spermatogenesis and sperm quality regulation.
There is a high homology of the amino acid sequences (95.7%) between human and mouse CMTM4, and thus the Cmtm4 KO mouse is an ideal model for investigating the functions of CMTM4 in male reproduction in vivo. The Cmtm4 KO mouse revealed subfertility in males but no significant morphological or behavioral abnormalities. No significant difference in serum testosterone level in KO mice compared with WT mice was observed. This indicates that Cmtm4 is not actively involved in androgen regulation, whereas other members of the CMTM family, such as mouse Cmtm2a and Cmtm2b and human CMTM1-v17, CMTM2, and CMTM3, are reported to affect androgen receptor activity (19, 22, 43). However, Cmtm4 KO mice showed an obvious phenotype of reduced progressive sperm motility, abnormal spermatozoa with backward-curved heads or bent midpieces, leading to male subfertility. A previous study reported that CMTM4 knockdown in HeLa cell resulted in cytokinesis defects (44). Thus, Cmtm4 deficiency may affect male meiotic cytokinesis, thereby resulting in lower DSP and abnormal spermatozoa. These distinct phenotypes between WT and KO mice suggest that they may alter sperm functions in KO mouse. Observation of the acrosome reaction indicated that a significantly reduced percentage of acrosome reactions was induced in KO mice sperm by the calcium ionophore A23187. These findings imply that CMTM4 is actively involved in spermatogenesis and sperm quality regulation. Further in vivo fertility assessment showed that male KO mice were subfertile, whereas female KO mice had normal fertility. In vitro fertility experiments also confirmed that spermatozoa from KO mice was inefficient in producing a two-cell stage. Collectively, these results indicate that Cmtm4 is associated with spermatogenesis and sperm quality and is essential for male fertility.
Further, quantitative proteomic analyses were performed to detect the molecules and biological processes in testes affected by Cmtm4 deficiency. Our study identified 5840 mouse testis proteins, which provides an inventory of mouse testicular proteins useful for the understanding of spermatogenesis. Of these, 351 proteins showed differential expression between WT and KO mice. Bioinformatic analysis revealed notable effects of Cmtm4 deficiency on male infertility. Downregulated proteins in KO mice included those that function in spermatogenesis and sperm functions. Of the downregulated proteins, 100 (71.9%) were testis-specifically expressed, reflecting an association with impaired spermatogenesis, and 57% of downregulated proteins were exclusively expressed in round spermatids, indicating the association with sperm quality might involve regulation of post-meiotic biological processes. These downregulated proteins are involved in fertilization, sperm motility, the acrosome reaction, and sperm-oocyte recognition. Several of the downregulated proteins are known to be involved in spermatogenesis; for example, PKG2 and GAPDHS are more abundant in round spermatids, and are testis-specific glycolytic enzymes positively correlated with sperm motility (45–47). Odf1 and Odf2 are major components of the outer dense fibers that modulate sperm motility (48). Spata20, Spata3, Spata6, Spata7, and Spert are well-known spermatogenesis-related proteins (49). Inactivation of Spata6 in mice leads to acephalic spermatozoa and male sterility (50). IZUMO1, CRISP1, AKAP3, AKAP4, PLB1, and TRIM36 are classified in the literatures by bioinformatic analyses as having functional involvement in the acrosome reaction (51–54), and their downregulated expression may contribute to the deficient acrosome reaction of KO mouse spermatozoa. The downregulation of these spermatogenesis-related proteins may contribute to lower sperm quality and reduced fertility. To further understand the effect of CMTM4 knockout on mouse sperm quality, a quantitative proteomics was performed to detect the affected sperm molecules by Cmtm4 deficiency. A total of 3588 proteins were identified and 1997 proteins were included in current testis proteome, which provides complementary information for sperm protein profiles (supplemental Fig. S1). Chromatin DNA binding function that was overrepresented in upregulated proteins of KO mouse sperm are known to participate in chromatin remodeling and meiosis during spermatogenesis (55). Particularly, Western blotting demonstrated the increased levels of H2A, H2B, and H4 in KO sperm, and conversely, reduced levels of PRM1 and PRM2 in KO sperm. The results suggest that impaired histone-to-protamine exchange exists in KO sperm, which may contribute to abnormal sperm morphology and function (56). Most of downregulated proteins in KO sperm perform actin binding functions, which are well-known to play crucial roles in maintenance of sperm morphology and motility (57). Cofilin in sperm is involved in acrosome reaction and its downregulation in sperm is correlated with poor sperm quality (58). ODF2 is related to sperm motility (59). Interestingly, α-tubulin and β-tubulin are downregulated in KO sperm, suggesting the impaired sperm motility and acrosome function (57). Collectively, proteomic alterations in KO sperm indicate its influence on sperm motility and acrosome reaction, which is consistent with its functions of localization in acrosome and midpiece, and implications of proteomic alterations in KO testes.
This study revealed an important role of CMTM4 in male reproduction. CMTM4 expression level was associated with human spermatogenesis and sperm quality. Phenotypic and quantitative proteomic analyses of Cmtm4 KO mice revealed that CMTM4 mainly affects histone-to-protamine exchange during spermatogenesis process, and functions in sperm motility and acrosome reaction. Proteomic analysis also identified a group of essential proteins as being affected by Cmtm4 deficiency. Their interactions and functions in male fertility are warranted to be further study in the future. This study provides an impetus for investigating the roles of other members of the CMTM family in male reproduction, and shows that CMTM4 is a promising molecular candidate for the assessment of sperm quality and the diagnosis or treatment of male infertility.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD011936 and 10.6019/PXD011936, PXD013092 and 10.6019/PXD013092 (https://www.ebi.ac.uk/pride/archive).
Supplementary Material
Acknowledgments
We thank Dr. Ying WanTao (National Center for Protein Sciences, State Key Laboratory of Proteomics, Beijing, China.) for the MS data analysis.
Footnotes
* The current study was supported by the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2018PT31039), National Natural Science Foundation of China (81571490), Shandong Provincial Natural Science Foundation, China (grant no. ZR2014HQ068).
 This article contains supplemental material.
This article contains supplemental material.
The authors declare no conflict of interest.
1 The abbreviations used are:
- CKLF
- chemokine-like factor
- KO
- knockout
- WT
- wild type
- CMTM
- CKLF-like MARVEL transmembrane domain-containing family
- TM4SF
- transmembrane 4 superfamily
- BLNK
- B-cell linker
- ANOVA
- one-way analysis of variance
- iTRAQ
- isobaric tags for relative and absolute quantification
- FDR
- false discovery rate
- GO
- gene ontology
- EGFR
- epidermal growth factor receptor
- PD-L1
- programmed death-1
- OA
- obstructive azoospermia
- NOA
- non-obstructive azoospermia
- CRISPR
- clustered regularly interspaced short palindromic repeats
- CAS9
- CRISPR-associated protein 9
- sgRNAs
- single guide RNAs
- PCR
- polymerase chain reaction
- hCG
- human choionic gonadotrophin
- PMSG
- pregnant mare serum gonadotrophin
- PBS
- phosphate buffered saline
- BSA
- bovine serum albumin
- DMSO
- dimethyl sulfoxide
- IHC
- immunohistochemistry
- CASA
- computer-aided sperm analysis
- DSP
- daily sperm production
- TEAB
- triethylammonium bicarbonate
- DTT
- dithiothreitol
- RP
- reversed-phase
- LC-MS
- liquid chromatograph mass spectrometer
- DAVID
- database for annotation, visualization and integrated discovery
- SDS-PAGE
- sodium dodecyl sulphate polyacrylamide gel electrophoresis
- TBS
- Tris–buffered saline
- HRP
- horseradish peroxidase
- ECL
- enhanced chemiluminescence
- IOD
- integrated optical density
- DAB
- 3,3′-diaminobenzidine
- UTR
- untranslated region
- CDS
- coding sequence
- PCR
- polymerase chain reaction
- IVF
- in vitro fertilization
- VSL
- straight line velocity
- VCL
- curvilinear velocity
- GEO
- gene expression omnibus
- AR
- androgen receptor.
REFERENCES
- 1. Han W., Lou Y., Tang J., Zhang Y., Chen Y., Li Y., Gu W., Huang J., Gui L., Tang Y., Li F., Song Q., Di C., Wang L., Shi Q., Sun R., Xia D., Rui M., Tang J., and Ma D. (2001) Molecular cloning and characterization of chemokine-like factor 1 (CKLF1), a novel human cytokine with unique structure and potential chemotactic activity. Biochem. J. 357, 127–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Han W., Ding P., Xu M., Wang L., Rui M., Shi S., Liu Y., Zheng Y., Chen Y., Yang T., and Ma D. (2003) Identification of eight genes encoding chemokine-like factor superfamily members 1–8 (CKLFSF1–8) by in silico cloning and experimental validation. Genomics 81, 609–617 [DOI] [PubMed] [Google Scholar]
- 3. Wang Y., Li J., Cui Y., Li T., Ng K. M., Geng H., Li H., Shu X. S., Li H., Liu W., Luo B., Zhang Q., Mok T. S., Zheng W., Qiu X., Srivastava G., Yu J., Sung J. J., Chan A. T., Ma D., Tao Q., and Han W. (2009) CMTM3, located at the critical tumor suppressor locus 16q22.1, is silenced by CpG methylation in carcinomas and inhibits tumor cell growth through inducing apoptosis. Cancer Res. 69, 5194–5201 [DOI] [PubMed] [Google Scholar]
- 4. Li T., Cheng Y., Wang P., Wang W., Hu F., Mo X., Lv H., Xu T., and Han W. (2015) CMTM4 is frequently downregulated and functions as a tumour suppressor in clear cell renal cell carcinoma. J. Exp. Clin. Cancer Res. 34, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shao L., Cui Y., Li H., Liu Y., Zhao H., Wang Y., Zhang Y., Ng K. M., Han W., Ma D., and Tao Q. (2007) CMTM5 exhibits tumor suppressor activities and is frequently silenced by methylation in carcinoma cell lines. Clin. Cancer Res. 13, 5756–5762 [DOI] [PubMed] [Google Scholar]
- 6. Li H., Li J., Su Y., Fan Y., Guo X., Li L., Su X., Rong R., Ying J., Mo X., Liu K., Zhang Z., Yang F., Jiang G., Wang J., Zhang Y., Ma D., Tao Q., and Han W. (2014) A novel 3p22.3 gene CMTM7 represses oncogenic EGFR signaling and inhibits cancer cell growth. Oncogene 33, 3109–3118 [DOI] [PubMed] [Google Scholar]
- 7. Yuan W., Liu B., Wang X., Li T., Xue H., Mo X., Yang S., Ding S., and Han W. (2017) CMTM3 decreases EGFR expression and EGF-mediated tumorigenicity by promoting Rab5 activity in gastric cancer. Cancer Lett. 386, 77–86 [DOI] [PubMed] [Google Scholar]
- 8. Liu B., Su Y., Li T., Yuan W., Mo X., Li H., He Q., Ma D., and Han W. (2015) CMTM7 knockdown increases tumorigenicity of human non-small cell lung cancer cells and EGFR-AKT signaling by reducing Rab5 activation. Oncotarget. 6, 41092–41107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chrifi I., Louzao-Martinez L., Brandt M., van Dijk C. G. M., Burgisser P., Zhu C., Kros J. M., Duncker D. J., and Cheng C. (2017) CMTM3 (CKLF-Like Marvel Transmembrane Domain 3) Mediates angiogenesis by regulating cell surface availability of VE-Cadherin in endothelial adherensjunctions. Arterioscler. Thromb. Vasc. Biol. 37, 1098–1114 [DOI] [PubMed] [Google Scholar]
- 10. Chrifi I., Louzao-Martinez L., Brandt M. M., van Dijk C. G. M., Bürgisser P. E., Zhu C., Kros J. M., Verhaar M. C., Duncker D. J., and Cheng C. (2019) CMTM4 regulates angiogenesis by promoting cell surface recycling of VE-cadherin to endothelial adherens junctions. Angiogenesis. 22, 75–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Imamura Y., Katahira T., and Kitamura D. (2004) Identification and characterization of a novel BASH N terminus-associated protein, BNAS2. J. Biol. Chem. 279, 26425–26432 [DOI] [PubMed] [Google Scholar]
- 12. Miyazaki A., Yogosawa S., Murakami A., and Kitamura D. (2012) Identification of CMTM7 as a transmembrane linker of BLNK and the B-cell receptor. PLoS One 7, e31829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mezzadra R., Sun C., Jae L. T., Gomez-Eerland R., de Vries E., Wu W., Logtenberg M. E. W., Slagter M., Rozeman E. A., Hofland I., Broeks A., Horlings H. M., Wessels L. F. A., Blank C. U., Xiao Y., Heck A. J. R., Borst J., Brummelkamp T. R., and Schumacher T. N. M. (2017) Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 549, 106–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burr M. L., Sparbier C. E., Chan Y. C., Williamson J. C., Woods K., Beavis P. A., Lam E. Y. N., Henderson M. A., Bell C. C., Stolzenburg S., Gilan O., Bloor S., Noori T., Morgens D. W., Bassik M. C., Neeson P. J., Behren A., Darcy P. K., Dawson S. J., Voskoboinik I., Trapani J. A., Cebon J., Lehner P. J., and Dawson M. A. (2017) CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 549, 101–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang L., Wu C., Zheng Y. Qiu X., Wang L., Fan H., Han W., Lv B., Wang Y., Zhu X., Xu M., Ding P., Cheng S., Zhang Y., Song Q., and Ma D. (2004) Molecular cloning and characterization of chemokine-like factor super family member 1 (CKLFSF1), a novel human gene with at least 23 alternative splicing isoforms in testis tissue. Int. J. Biochem. Cell Biol. 36, 1492–1501 [DOI] [PubMed] [Google Scholar]
- 16. Fujihara Y., Oji A., Kojima-Kita K., Larasati T., and Ikawa M. (2018) Coexpression of sperm membrane proteins CMTM2A and CMTM2B is essential for ADAM3 localization and male fertility in mice. J. Cell Sci. 131, jcs221481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shi S., Rui M., Han W., Wang Y., Qiu X., Ding P., Zhang P., Zhu X., Zhang Y., Gan Q., and Ma D. (2005) CKLFSF2 is highly expressed in testis and can be secreted into the seminiferous tubules. Int. J. Biochem. Cell Biol. 37, 1633–1640 [DOI] [PubMed] [Google Scholar]
- 18. Liu G., Xin Z. C., Chen L., Tian L., Yuan Y. M., Song W. D., Jiang X. J., and Guo Y. L. (2007) Expression and localization of CKLFSF2 in human spermatogenesis. Asian. J. Androl. 9, 189–198 [DOI] [PubMed] [Google Scholar]
- 19. Jeong B. C., Hong C. Y., Chattopadhyay S., Park J. H., Gong E. Y., Kim H. J., Chun S. Y., and Lee K. (2004) Androgen receptor corepressor-19 kDa (ARR19), a leucine-rich protein that represses the transcriptional activity of androgen receptor through recruitment of histone deacetylase. Mol. Endocrinol. 18, 13–25 [DOI] [PubMed] [Google Scholar]
- 20. Qamar I., Gong E. Y., Kim Y., Song C. H., Lee H. J., Chun S. Y., and Lee K. (2010) Anti-steroidogenic factor ARR19 inhibits testicular steroidogenesis through the suppression of Nur77 transactivation. J. Biol. Chem. 285, 22360–22369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qamar I., Ahmad M. F., and Narayanasamy A. (2015) A time-course study of long term over-expression of ARR19 in mice. Sci. Rep. 5, 13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li T., Han W., Yang T., Ding P., Rui M., Liu D., Wang Y., and Ma D. (2006) Molecular cloning and identification of mouse Cklfsf2a and Cklfsf2b, two homologues of human CKLFSF2. Int. J. Biochem. Cell Biol. 38, 420–429 [DOI] [PubMed] [Google Scholar]
- 23. Plate M., Li T., Wang Y., Mo X., Zhang Y., Ma D., and Han W. (2010) Identification and characterization of CMTM4, a novel gene with inhibitory effects on HeLa cell growth through Inducing G2/M phase accumulation. Mol. Cells. 29, 355–361 [DOI] [PubMed] [Google Scholar]
- 24. Bei C., Zhang Y., Wei R., Zhu X., Wang Z., Zeng W., Chen Q., and Tan S. (2017) Clinical significance of CMTM4 expression in hepatocellular carcinoma. Onco Targets Ther. 10, 5439–5443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bergmann M., and Kliesch S. (1998) Hodenbiopsie In: Krause W, Weidner W (eds) Andrologie, pp 66–71, Enke Verlag, Stuttgart [Google Scholar]
- 26. Liu F. J., Liu X., Han J. L., Wang Y. W., Jin S. H., Liu X. X., Liu J., Wang W. T., and Wang W. J. (2015) Aged men share the sperm protein PATE1 defect with young asthenozoospermia patients. Hum. Reprod. 30, 861–869 [DOI] [PubMed] [Google Scholar]
- 27. Wang Y., Ding Y., and Li J. (2017) CRISPR-Cas9-Mediated Gene Editing in Mouse Spermatogonial Stem Cells. Methods. Mol. Biol. 1622, 293–305 [DOI] [PubMed] [Google Scholar]
- 28. VomSaal F. S., Cooke P. S., Buchanan D. L., Palanza P., Thayer K. A., Nagel S. C., Parmigiani S., and Welshons W. V. (1998) A physiologically based approach to the study of bisphenol A and other estrogenic chemicals on the size of reproductive organs, daily sperm production, and behavior. Toxicol. Ind. Health. 14, 239–260 [DOI] [PubMed] [Google Scholar]
- 29. Sun W., Xing B., Guo L., Liu Z., Mu J., Sun L., Wei H., Zhao X., Qian X., Jiang Y., and He F. (2016) Quantitative Proteomics Analysis of Tissue Interstitial Fluid for Identification of Novel Serum Candidate Diagnostic Marker for Hepatocellular Carcinoma. Sci. Rep. 6, 26499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shen X., Liu X., Zhu P., Zhang Y., Wang J., Wang Y., Wang W., Liu J., Li N., and Liu F. (2017) Proteomic analysis of human follicular fluid associated with successful in vitro fertilization. Reprod. Biol. Endocrinol. 15, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu X. X., Zhang H., Shen X. F., Liu F. J., Liu J., and Wang W. J. (2016) Characteristics of testis-specific phosphoglycerate kinase 2 and its association with human sperm quality. Hum. Reprod. 31, 273–279 [DOI] [PubMed] [Google Scholar]
- 32. Gan H., Cai T., Lin X., Wu Y., Wang X., Yang F., and Han C. (2013) Integrative proteomic and transcriptomic analyses reveal multiple post-transcriptional regulatory mechanisms of mouse spermatogenesis. Mol. Cell. Proteomics 12, 1144–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chauvin T., Xie F., Liu T., Nicora C. D., Yang F., Camp D. G., Smith R. D., and Roberts K. P. (2012) A systematic analysis of a deep mouse epididymal sperm proteome. Biol. Reprod. 87, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dorus S., Wasbrough E. R., Busby J., Wilkin E. C., and Karr T. L. (2010) Sperm proteomics reveals intensified selection on mouse sperm membrane and acrosome genes. Mol. Biol. Evol. 27, 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Netherton J. K., Hetherington L., Ogle R. A., Velkov T., and Baker M. A. (2018) Proteomic analysis of good- and poor-quality human sperm demonstrates that several proteins are routinely aberrantly regulated. Biol. Reprod. 99, 395–408 [DOI] [PubMed] [Google Scholar]
- 36. Gatimel N., Moreau J., Parinaud J., and Léandri R. D. (2017) Sperm morphology: assessment, pathophysiology, clinical relevance, and state of the art in. Andrology 5, 845–862 [DOI] [PubMed] [Google Scholar]
- 37. Virtanen H. E., Jørgensen N., and Toppari J. (2017) Semen quality in the 21st century. Nat. Rev. Urol. 14, 120–130 [DOI] [PubMed] [Google Scholar]
- 38. Ma Q., Li Y., Luo M., Guo H., Lin S., Chen J., Du Y., Jiang Z., and Gui Y. (2017) The expression characteristics of FAM71D and its association with sperm motility. Hum. Reprod. 32, 2178–2187 [DOI] [PubMed] [Google Scholar]
- 39. Liu F., Wang H., and Li J. (2011) An integrated bioinformatics analysis of mouse testis protein profiles with new understanding. BMB. Rep. 44, 347–351 [DOI] [PubMed] [Google Scholar]
- 40. Liu F., Jin S., Li N., Liu X., Wang H., and Li J. (2011) Comparative and functional analysis of testis-specific genes. Biol Pharm Bull. 34, 28–35 [DOI] [PubMed] [Google Scholar]
- 41. Li J., Liu F., Liu X., Liu J., Zhu P., Wan F., Jin S., Wang W., Li N., Liu J., and Wang H. (2011) Mapping of the human testicular proteome and its relationship with that of the epididymis and spermatozoa. Mol. Cell. Proteomics 10, M110.004630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miyamoto T., Minase G., Shin T., Ueda H., Okada H., and Sengoku K. (2017) Human male infertility and its genetic causes. Reprod. Med. Biol. 16, 81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang Y., Li Qiu T. X., Mo X., Zhang Y., Song Q., Ma D., and Han W. (2008) CMTM3 can affect the transcription activity of androgen receptor and inhibit the expression level of PSA in LNCaP cells. Biochem. Biophys. Res. Commun. 371, 54–58 [DOI] [PubMed] [Google Scholar]
- 44. Kittler R., Putz G., Pelletier L., Poser I., Heninger A. K., Drechsel D., Fischer S., Konstantinova I., Habermann B., Grabner H., Yaspo M. L., Himmelbauer H., Korn B., Neugebauer K., Pisabarro M. T., and Buchholz F. (2004) Anendoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature 432, 1036–1040 [DOI] [PubMed] [Google Scholar]
- 45. Huang Z., Danshina P. V., Mohr K., Qu W., Goodson S. G, O'Connell T. M., and O'Brien D. A. (2017) Sperm function, protein phosphorylation, and metabolism differ in mice lacking successive sperm-specific glycolytic enzymes. Biol. Reprod. 97, 586–597 [DOI] [PubMed] [Google Scholar]
- 46. Park H.-J., Lee W.-Y., Park C., Hong K.-H., Kim J.-H., and Song H. (2018) Species-specific expression of phosphoglycerate kinase 2 (PGK2) in the developing porcine testis. Theriogenology 110, 158–167 [DOI] [PubMed] [Google Scholar]
- 47. Fu J., Yao R., Luo Y., Yang D., Cao Y., Qiu Y., Song W., Miao S., Gu Y., and Wang L. (2016) Anti-GAPDHS antibodies: a biomarker of immune infertility. Cell Tissue Res. 364, 199–207 [DOI] [PubMed] [Google Scholar]
- 48. Lehti M. S., and Sironen A. (2017) Formation and function of sperm tail structures in association with sperm motility defects. Biol. Reprod. 97, 522–536 [DOI] [PubMed] [Google Scholar]
- 49. Malcher A., Rozwadowska N., Stokowy T., Kolanowski T., Jedrzejczak P., Zietkowiak W., and Kurpisz M. (2013) Potential biomarkers of nonobstructiveazoospermia identified in microarray gene expression analysis. Fertil. Steril 100, 1686–1694 [DOI] [PubMed] [Google Scholar]
- 50. Yuan S., Stratton C. J., Bao J., Zheng H., Bhetwal B. P., Yanagimachi R., and Yan W. (2015) Spata6 is required for normal assembly of the sperm connecting piece and tight head-tail conjunction. Proc. Natl. Acad. Sci. U.S.A. 112, E430–E439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Inoue N., and Wada I. (2018) Monitoring dimeric status of IZUMO1 during the acrosome reaction in living spermatozoon. Cell. Cycle 17, 1279–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Busso D., Goldweic N. M., Hayashi M., Kasahara M., and Cuasnicú P. S. (2007) Evidence for the involvement of testicular protein CRISP2 in mouse sperm-egg fusion. Biol. Reprod. 76, 701–708 [DOI] [PubMed] [Google Scholar]
- 53. Vizel R., Hillman P., Ickowicz D., and Breitbart H. (2015) AKAP3 degradation in sperm capacitation is regulated by its tyrosine phosphorylation. Biochim. Biophys. Acta 1850, 1912–1920 [DOI] [PubMed] [Google Scholar]
- 54. Kitamura K., Tanaka H., and Nishimune Y. (2003) Haprin, a novel haploid germ cell-specific RING finger protein involved in the acrosome reaction. J. Biol. Chem. 278, 44417–44423 [DOI] [PubMed] [Google Scholar]
- 55. Gou L. T., Kang J. Y., Dai P., Wang X., Li F., Zhao S., Zhang M., Hua M. M., Lu Y., Zhu Y., Li Z., Chen H., Wu L. G., Li D., Fu X. D., Li J., Shi H. J., and Liu M. F. (2017) Ubiquitination-deficient mutations in human PiwiCause male infertility by impairing histone-to-protamine exchange during spermiogenesis. Cell. 169, 1090–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhong H. Z., Lv F. T., Deng X. L., Hu Y., Xie D. N., Lin B., Mo Z. N., and Lin F. Q. (2015) Evaluating γH2AX in spermatozoa from male infertility patients. Fertil. Steril. 104, 574–581 [DOI] [PubMed] [Google Scholar]
- 57. Breitbart H., and Finkelstein M. (2018) Actin cytoskeleton and sperm function. Biochem. Biophys. Res. Commun. 506, 372–377 [DOI] [PubMed] [Google Scholar]
- 58. Chen S. M., Chen X. M., Lu Y. L., Liu B., Jiang M., and Ma Y. X. (2016) Cofilin is correlated with sperm quality and influences sperm fertilizing capacity in humans. Andrology 4, 1064–1072 [DOI] [PubMed] [Google Scholar]
- 59. Zhao W., Li Z., Ping P., Wang G., Yuan X., and Sun F. (2018) Outer dense fibers stabilize the axoneme to maintain sperm motility. J. Cell. Mol. Med. 22, 1755–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD011936 and 10.6019/PXD011936, PXD013092 and 10.6019/PXD013092 (https://www.ebi.ac.uk/pride/archive).