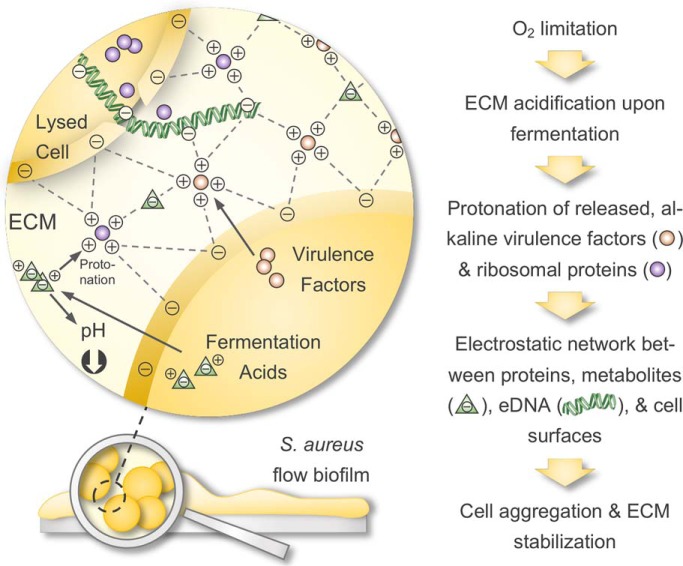
We comprehensively profiled intracellular and ECM proteomes of S. aureus flow biofilms and complemented these data by metabolic footprint analysis and phenotypic assays. We show that moonlighting, secreted virulence factors and ribosomal proteins within the ECM contribute to biofilm stabilization. Mechanistically, we propose that these alkaline proteins get protonated in an acidified ECM (because of the release of acids upon fermentation) mediating electrostatic interactions with anionic cell surface components, eDNA, and metabolites, which leads to cell aggregation and ECM stabilization.
Keywords: Virulence, Metabolomics, Multifunctional Proteins*, Pathogens, Bacteria, biofilm, extracellular matrix, Staphylococcus aureus
Graphical Abstract
Highlights
Establishment of a flow system allowing multi-omics analysis of S. aureus biofilms.
Biofilm proteome profiling (intracellular and ECM) plus metabolic footprint analysis.
Virulence factors and ribosomal proteins stabilize the ECM as moonlighting proteins.
They act as electrostatic bridges between anionic cell surfaces, eDNA and metabolites.
Abstract
Staphylococcus aureus is the causative agent of various biofilm-associated infections in humans causing major healthcare problems worldwide. This type of infection is inherently difficult to treat because of a reduced metabolic activity of biofilm-embedded cells and the protective nature of a surrounding extracellular matrix (ECM). However, little is known about S. aureus biofilm physiology and the proteinaceous composition of the ECM. Thus, we cultivated S. aureus biofilms in a flow system and comprehensively profiled intracellular and extracellular (ECM and flow-through (FT)) biofilm proteomes, as well as the extracellular metabolome compared with planktonic cultures. Our analyses revealed the expression of many pathogenicity factors within S. aureus biofilms as indicated by a high abundance of capsule biosynthesis proteins along with various secreted virulence factors, including hemolysins, leukotoxins, and lipases as a part of the ECM. The activity of ECM virulence factors was confirmed in a hemolysis assay and a Galleria mellonella pathogenicity model. In addition, we uncovered a so far unacknowledged moonlighting function of secreted virulence factors and ribosomal proteins trapped in the ECM: namely their contribution to biofilm integrity. Mechanistically, it was revealed that this stabilizing effect is mediated by the strong positive charge of alkaline virulence factors and ribosomal proteins in an acidic ECM environment, which is caused by the release of fermentation products like formate, lactate, and acetate because of oxygen limitation in biofilms. The strong positive charge of these proteins most likely mediates electrostatic interactions with anionic cell surface components, eDNA, and anionic metabolites. In consequence, this leads to strong cell aggregation and biofilm stabilization. Collectively, our study identified a new molecular mechanism during S. aureus biofilm formation and thus significantly widens the understanding of biofilm-associated S. aureus infections - an essential prerequisite for the development of novel antimicrobial therapies.
Bacteria in nature can predominantly be found as surface-attached, multicellular aggregates called biofilms, which are embedded in a self-produced, extracellular matrix (ECM)1 (1, 2). This matrix mainly contains polymeric substances like polysaccharides, extracellular DNA (eDNA) as well as proteins and varies in its composition depending on environmental conditions and among different bacterial species (3). Compared with planktonically cultivated bacteria, which poorly represent natural conditions, biofilms show an altered growth rate, metabolism, and gene expression profile (2). The reduced metabolic activity of biofilm-embedded cells and the protective nature of the ECM, acting as a barrier against antimicrobial agents and the host immune system, make biofilms inherently difficult to treat thereby causing major healthcare problems (4, 5). Notably, it is estimated that 65 to 80% of all human bacterial infections are biofilm-associated underlining the need for novel therapeutic strategies (6, 7).
The opportunistic pathogen Staphylococcus aureus represents a major cause of nosocomial infections and is well known for its capacity to form biofilms on host tissues and implants. This often leads to chronic infections, e.g. in patients suffering from osteomyelitis, endocarditis, cystic fibrosis or during catheterization (8–12). Generally, biofilm development is characterized by three phases: primary surface attachment, maturation, and biofilm dispersal (13). Moormeier and colleagues suggested two more phases during S. aureus biofilm development, which can be observed between the attachment and maturation phase and are characterized by intense cellular division (termed “multiplication”) followed by a premature detachment of a biofilm subpopulation (termed “exodus”) (14). Numerous studies mainly focused on elucidating individual molecular factors, which are crucial during these biofilm formation steps of S. aureus. Thus, important roles in adhesion to abiotic surfaces and host cells, as well as in biofilm integrity and structuring have been described for proteinaceous and nonproteinaceous factors. This includes microbial surface components recognizing adhesive matrix molecules (MSCRAMMs, e.g. CflAB, SdrCDE), fibronectin-binding proteins FnBPAB, the autolysin AtlA, protein A, biofilm-associated protein Bap, phenol-soluble modulins (PSMs), proteases, nucleases, teichoic acids, the polysaccharide intercellular adhesin (PIA), and eDNA. A complex regulatory network of specific and global regulators tightly controls the expression and synthesis of these molecular factors during the different stages of biofilm growth. Major regulators involved in biofilm formation are: AgrA and RNAIII, Rot, SigB, SarA, SaeRS, MgrA, IcaR, CodY, CcpA, Spx, CidR, Rbf, LytSR, and TcaR (reviewed and summarized by (4, 15–18).
Global approaches can be particularly useful to unravel the intricate interaction within these regulatory networks and to identify proteins with important roles in biofilm formation. However, so far only a few studies applied different omics techniques to analyze S. aureus biofilm physiology (18–27). These studies focused either on (1) static biofilm cultivation models, (2) used obsolete omics technologies, or (3) lacked a multi-omics approach. Most importantly, the composition of the S. aureus ECM, particularly at the protein level, and the role of these proteins during biofilm growth are still largely unexplored.
Consequently, the here presented study aims at a comprehensive characterization of the intra- and extracellular (ECM and flow-through (FT)) proteome, as well as the extracellular metabolome of S. aureus biofilms cultured in a physiologically highly relevant flow system (28) applying state-of-the-art omics technologies. Moreover, we compared these biofilm protein and metabolite profiles to planktonic cells. Our analyses identified a so far nondescribed, molecular mechanism during biofilm formation, which uncovers moonlighting virulence factors and ribosomal proteins in the ECM as key players in mediating biofilm integrity.
EXPERIMENTAL PROCEDURES
Strain and Growth Medium
All experiments were performed using S. aureus HG001, which was described by Herbert et al., 2010 (29). For all cultivations, RPMI 1640 medium (R7509, Sigma-Aldrich, St. Louis) was used. The medium was supplemented with 2 mm glutamine, 140 μm citrate, 7.5 μm FeCl3 and trace metals according to Gertz et al., 1999 (30) and Dörries et al., 2013 (31).
Planktonic and Biofilm Cultivation for Omics Analyses
Planktonic overnight cultures were grown in Erlenmeyer flasks filled with 10 ml medium (liquid to air ratio 1:10) with vigorous agitation at 180 rpm and 37 °C for 18 h.
Planktonic main cultures were grown in Erlenmeyer flasks after inoculation of 100 ml prewarmed medium (liquid to air ratio of 1:5) to an OD500 nm of 0.06 using a fresh overnight culture. The cultures were subsequently incubated at 180 rpm and 37 °C for 12 h.
Biofilm cultivation was performed at 37 °C in a continuous flow reactor system adapted from Dohnt et al., 2011 (32) and Brady et al., 2006 (33), which consists of a medium bottle, a multichannel pump, a drop trap, a silicon tube for biofilm growth (4 mm inner diameter, 1 m length, VWR, Darmstadt, Germany) and a waste bottle (supplemental Fig. S1). Before inoculation, the system was filled with growth medium and equilibrated overnight. To inoculate the system, S. aureus cells were precultivated for 5 h in a planktonic main culture as described above, diluted in fresh, prewarmed medium to an OD500 nm of 0.1 and injected into the silicon tube using a luer-lock syringe. This precultivation step increased attachment reproducibility compared with inoculation using cells of an overnight culture. After inoculation, the system was left without medium flow for 1 h to allow attachment of the cells. Subsequently, biofilm cultivation was started with a continuous medium flow of 25 ml/h (corresponding to 2 m/h and a retention time of 30 min) for 12 h.
Sample Collection and Protein Extraction
Sample collection for intracellular (planktonic and biofilm) and extracellular (planktonic, biofilm ECM and biofilm flow-through (FT)) proteome analysis of the cultures was carried out after 12 h. All sampling and protein extraction steps were performed on ice.
Planktonic cells were harvested and pelleted by centrifugation at 7000 × g and 4 °C for 10 min, and the supernatant was collected. The cell pellet was washed twice (resuspended and vortexed for 2 min (34)) using ice-cold high salt Tris-buffer according to Rice et al., 2007 (35) and Bose et al., 2012 (36): 50 mm Tris-HCl, 10 mm EDTA, 500 mm NaCl, pH 8. The resulting supernatants of the washing steps were pooled with the supernatant of the initial centrifugation step and filter-sterilized (0.45 μm cut off, Sarstedt AG, Nürnberg, Germany). The supernatants and the remaining cell pellets were stored at −70 °C for further analysis.
For biofilm FT samples, the medium eluting from the flow system was collected on ice for 20 min, centrifuged, sterile filtered and stored as described above.
To obtain intracellular and ECM samples of the biofilm, the biofilm tube content was transferred in a centrifugation tube, centrifuged as described above and the supernatant was collected. In a next step, the remaining cell pellet was washed twice with high salt TE-buffer as described for planktonic samples, to effectively separate the ECM from the cells. The cell suspension was centrifuged again and the resulting supernatants containing ECM proteins were pooled with the supernatant of the initial centrifugation step, filter sterilized and stored for further analysis as described above. Applying this method, intracellular protein and metabolite contaminations caused by cell lysis were avoided, which was verified by CFU counting and metabolic footprint analysis comparing samples before and after the separation step (data not shown).
Intracellular protein extracts from the planktonic and biofilm cell pellets were prepared as already described (37, 38) with slight modifications: cells were resuspended in 1 ml TE-buffer followed by mechanical disruption with 500 μl glass beads (0.1 to 0.11 mm, Sartorius Stedim Biotech, Göttingen, Germany) in 3 homogenization cycles at 6.5 m/s for 30 s using a FastPrep-25 homogenizer (MP Biologicals, Santa Ana, California). Cell debris and glass beads were removed by centrifugation at 20,000 × g and 4 °C for 10 min. Insoluble and aggregated proteins were removed by an additional centrifugation step at 20,000 × g and 4 °C for 30 min (37, 38). The resulting intracellular protein extracts were stored at −70 °C for further analysis.
Planktonic extracellular proteins, as well as biofilm ECM and biofilm FT proteins were enriched using StrataClean beads as described by Bonn et al., 2014 (39) with slight modifications. Briefly, 20 μl aliquots of the beads were washed twice with 500 μl TE-buffer for StrataClean beads (TESC, 50 mm Tris-HCl, 10 mm EDTA, pH 8) with intermitted centrifugation at 6000 × g and room temperature (RT) for 2 min. Subsequently, beads were primed by incubation in 200 μl HCl at 100 °C for 6 h and washed twice with 1 ml TESC-buffer. For protein enrichment, beads were incubated with culture supernatants at 4 °C on a rotating tube shaker overnight. Loaded beads were pelleted at 10,000 × g and 4 °C for 15 min, washed twice with 500 μl PBS buffer (137 mm NaCl, 0.2 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, pH 7.4) and dried in a vacuum centrifuge (Eppendorf AG, Hamburg, Germany) for 20 min.
Sample Preparation for MS/MS Analysis
The concentration of intracellular protein extracts was determined according to Bradford et al., 1967 (40) using Roti®-Nanoquant (Roth, Karlsruhe, Germany) following the manufacturer's instructions. Extracellular/ECM/FT protein amounts loaded on the beads were determined by BCA-assay (ThermoFisher Scientific, Waltham, MA) described by Smith et al., 1985 (41) according to manufacturer's instructions using the standard protocol with additional shaking at 1500 rpm to avoid bead sedimentation. Protein amounts of 25 μg per sample were boiled for 10 min at 95 °C and subsequently separated on a 4–12% SDS-polyacrylamide gradient gel (Criterion, BioRad, Munich, Germany). The gel was fixed for 30 min in fixation solution (10% (v/v) acetic acid, 40% (v/v) ethanol), washed twice for 10 min in water and stained with Colloidal Coomassie Brilliant Blue G-250 as previously described (42, 43). After removing excessive Coomassie stain from the gel using water, gel lanes of intracellular and extracellular/ECM/FT samples were fractionated into 10 and 5 gel pieces, respectively, cut into gel blocks of ∼1 mm3 and prepared for MS/MS analysis as follows. Gel blocks were transferred into low binding tubes and washed/destained 5 times with 900 μl gel washing buffer (0.2 m NH4CO3 in 30% (v/v) acetonitrile) at 37 °C in a tube shaker (Eppendorf AG) at 1500 rpm for 15 min. Destained gel blocks were dried in a vacuum centrifuge for 30 min, covered with trypsin solution (2 μg/ml, Promega, Madison) and incubated at 37 °C overnight for protein digest. Subsequently, 100 μl water was added, and peptides were eluted by treatment in an ultrasonic bath for 15 min. The supernatant was transferred into a fresh low binding tube, desiccated in a vacuum centrifuge and peptides were stored at −70 °C. Before MS/MS analysis, peptides were resolubilized adding 10 μl of 0.1% (v/v) acetic acid and transferred into glass vials.
MS/MS Analysis
Tryptic peptides were subjected to liquid chromatography (LC) separation and electrospray ionization-based mass spectrometry (MS) measurement applying adjusted injection volumes for each gel fraction of intracellular samples to reach maximum intensities. Injection volumes for extracellular samples were kept constant. Peptides were loaded on a self-made analytical column (Aeris PEPTIDE 3.6 μm XB - C18 (phenomenex), OD 360 μm, ID 100 μm, length 20 cm) and eluted by a binary nonlinear gradient of 5–75% acetonitrile in 0.1% acetic acid over 80 min with a flow rate of 300 nL/min. LC-MS/MS analyses were performed on an LTQ-Orbitrap-Velos mass spectrometer (ThermoFisher Scientific) coupled to an EASY-nLC 1000. For MS analysis, a full scan in the Orbitrap with a resolution of 30.000 and mass deviation of 0.5 Da was followed by CID MS/MS experiments of the 20 most abundant precursor ions acquired in the linear ion trap with a mass tolerance of 20 ppm.
MS Data Analysis
Database search, as well as label-free quantification (LFQ) and intensity-based absolute quantification (iBAQ), was performed using the MaxQuant software suite (version 1.6.0.16, Max Planck Institute of Biochemistry, Martinsried, Germany) running the built-in Andromeda search engine (Max Planck Institute of Biochemistry) (44–47). The database used was downloaded from UniProt on January 24th, 2018 (Proteome ID UP000008816) and contains 2889 protein sequences of S. aureus NCTC8325 (representing the parental strain S. aureus HG001 was derived from). The entry for protein RsbU was replaced by the corresponding sequence of S. aureus Newman because rsbU has been repaired in strain HG001 by Herbert and colleagues (29). Common laboratory contaminants and reversed sequences were included by MaxQuant. Database search parameters were set as follows: Trypsin/P specific digestion (KR) with two allowed missed cleavages, peptide tolerance 20 ppm, fragment ion tolerance 4.5 Da, methionine oxidation (15.99 Da) as a variable modification, peptide spectral match FDR 1% and protein FDR 1%. No fixed modifications were included. LFQ was performed using the following settings: LFQ minimum ratio count 2 considering unique and razor peptides for quantification. Match between runs was enabled with a match time window of 0.7 min and an alignment time window of 20 min.
Results were filtered for proteins identified with 2 or more unique peptides in at least 2 out of 3 biological replicates. LFQ intensities of intracellular and extracellular/ECM/FT proteome data were used to calculate log2 ratios between biofilm and planktonic samples. No further adjustments for systematic errors were applied. For proteins of intracellular samples, which were identified in 3 out of 3 biological replicates, these log2 ratios were then visualized in Voronoi treemaps (48, 49) using the Paver software (DECODON GmbH, Greifswald, Germany) based on functional assignment of the SEED database (50) or based on regulon maps (51) created by Moche et al., 2014 (18), respectively. For extracellular/ECM/FT data, treemaps were created based on subcellular localization predicted by PSORTb (52) and theoretical pI values extracted from AureoWiki database (53). Predictions by PSORTb were manually cured based on published localization studies, and LocateP (54) and SignalP (55) predictions. riBAQ values were calculated (47) and used to correlate treemap cell sizes with protein abundance.
Metabolic Footprint Analysis
For metabolic footprint analysis using 1H-nuclear magnetic resonance (1H-NMR) spectroscopy, planktonic and biofilm samples were harvested on ice after 12 h of cultivation, sterile filtered (0,45 μm cut off, Sarstedt AG, Nürnberg, Germany) and stored at −20 °C before measurement. For biofilm cultures, ECM samples were collected as described above. Moreover, 1 ml of the FT was collected on ice and prepared as described above. Samples were then thawed at RT, and 400 μl were mixed with 200 μl of sodium hydrogen phosphate buffer (0.2 mol/L, pH 7.0) made up with 50% D2O, including 1 mm 3-trimethylsilyl-[2,2,3,3-D4]-1-propanoic acid for 1H-NMR analysis (31). Samples were analyzed using a Bruker AVANCE-II 600 NMR spectrometer operated by TOPSPIN 3.2 software. Qualitative and quantitative data analyses were carried out using AMIX v3.9.12 software (Bruker Biospin GmbH, Rheinstetten, Germany).
Nitrate concentrations were measured by applying 10 μl cell-free culture supernatant on colorimetric nitrate test stripes (Merck, Darmstadt, Germany). Self-made standards with nitrate concentrations ranging from 0–250 mg/L NO3− served as controls.
Biofilm cultivation for CLSM analysis. To analyze biofilms using confocal laser scanning microscopy (CLSM), flow chambers with three individual growth channels (1 × 4 × 40 mm per channel) were prepared as described by Sternberg and Tolker-Nielsen, 2006 (56). Flow chambers were filled with growth medium overnight to equilibrate the chambers. Each channel of the flow chambers was inoculated with 300 μl of an overnight culture, which was diluted to an OD500 nm of 0.01 using fresh, prewarmed growth medium. For inoculation, a small syringe was used. The flow chambers were left without flow for 1 h to allow bacterial attachment. Subsequent biofilm cultivation was performed with a flow rate of 3 ml/h (0.2 mm/s) for 12 h using a Watson-Marlow 205S peristaltic pump (Watson-Marlow GmbH, Rommerskirchen, Germany). Biofilms were stained with 5 μm Syto9 (prepared in 0.9% NaCl, ThermoFisher Scientific) without flow for 15 min. Subsequently, the biofilms were washed under flow for 15 min.
To elucidate the influence of ECM components on biofilm stability, established biofilms were treated as follows. For enzymatic digestion of ECM proteins, biofilms were incubated with Proteinase K (100 μg/ml in growth medium, ThermoFisher Scientific) without flow at 37 °C for 2 h according to Seidl et al., 2008 (57). Treated biofilms were challenged by elevated shear forces applying an increased flow rate (30 ml/h) for 5 min before CLSM analysis. To analyze biofilm stability under alkaline conditions, established biofilms were subjected to an increased flow of growth medium adjusted to pH 12 as described above. For all treatments, fresh growth medium served as controls. CLSM images were acquired after 0, 2 and 5 min of increased flow using a Zeiss LSM 510 CLSM (Carl Zeiss, Jena, Germany) equipped with a water corrected 63×/NA1.2 objective and filter and detector settings for monitoring Syto9 fluorescence (excitation at 488 nm using an Ar-laser, emission light selected with a 505–550 nm bandpass filter). Image acquisition was performed using the ZEN 2009 software (Carl Zeiss) with z-stack sections of 0.5 μm. Three-dimensional reconstruction of z-stacks was done using the AMIRA software (version 6.0.1, ThermoFisher Scientific).
To visualize eDNA within the ECM, freshly grown biofilms were stained with the eDNA-specific stain Toto-1 (5 μm) and Syto62 (10 μm) as a counterstain (both prepared in 0.9% NaCl, ThermoFisher Scientific). Biofilm staining and washing was carried out in the dark for 15 min as described above. For biofilm and eDNA visualization, image acquisition was performed as described above with filter and detector settings for monitoring of Syto62 fluorescence (excitation at 633 nm using an HeNe-laser, emission light selected with a 650 nm longpass filter) and Toto-1 fluorescence (excitation at 514 nm with an Ar-laser, emission light selected with a 505–550 nm bandpass filter).
Cell Aggregation Test
The cell aggregation test was performed using biofilm-grown cells, which were cultivated, harvested, and pelleted as described for omics analysis. Subsequently, cell pellets were resuspended in the following solutions, which were adjusted to either pH 8 or pH 5.5: (1) planktonic or biofilm culture supernatant containing extracellular/ECM proteins, (2) fresh medium supplemented with bovine serum albumin (BSA, pI = 4.7, Sigma-Aldrich) or bovine cytochrome C (CytC, pI = 10.0, Sigma-Aldrich), (3) BSA or CytC supplemented growth medium plus DNA (S. aureus cDNA). The concentrations of control proteins and DNA used were: proteins = 50 μg/ml, DNA = 25 μg/ml. Cell suspensions of OD500 nm 10 were prepared and incubated at RT for 1 min. Cells were stained with 5 μm Toto-1 and 10 μm Syto62 (ThermoFisher Scientific) for 15 min in the dark. Four microliters of these cell suspensions were applied on a thin layer of 1.5% agarose in 0.9% NaCl, which was mounted on a microscope slide. Fluorescence microscopy images were acquired and processed using a Zeiss Imager M2 (Carl Zeiss) equipped with a 100x/NA 1.3 oil immersion objective and the ZEN 2011 software package (Carl Zeiss).
Test for Osmotic Stress Resistance
Planktonic and biofilm cells were cultivated as described for omics analyses and used to test osmotic stress resistance. To this end, Erlenmeyer flasks filled with 10 ml prewarmed RPMI-Medium supplemented with 3 and 4 m NaCl, respectively, were inoculated to an OD500 nm of 0.05. It was microscopically verified that no cell aggregates were used for inoculation or developed during the experiment ensuring meaningful CFU counting results. Subsequently, cultures were incubated at 37 °C and 180 rpm following counting of surviving CFU every 24 h for 3 days.
pH Determination
Samples were collected as described for omics analysis. pH values of the biofilm FT and planktonic cultures were measured using a pH meter (pHenomenal pH1000L, VWR). To be able to determine the pH directly within the ECM, growth medium within silicon tubes was carefully removed by allowing air to enter the tube at a flow rate of 3 ml/h followed by biofilm sampling and centrifugation as described above. The pH of the resulting supernatant was measured using pH-indicator stripes ranging from pH 4.0–7.0 (Merck).
Galleria mellonella Pathogenicity Model
G. mellonella experiments were carried out according to Hill et al., 2014 with slight modifications (58). Larvae weighing ∼300–400 mg were disinfected in 70% ethanol for 3 s and allowed to dry. Subsequently, 5 μl of cell-free culture supernatant (corresponding to 40 ng protein) were injected in the last left proleg of 15 larvae per sample using a 10 μl Hamilton syringe. Five microliters growth medium supplemented with 8 μg/ml BSA and noninfected larvae served as controls. The larvae were incubated at 37 °C for 68 h with intermediate counting of surviving animals.
Hemolysis Assay
Hemolysis assays were performed as described by Lauderdale et al., 2009 using samples collected as described above for omics analysis (59). Briefly, 0.5 ml rabbit blood (Fiebig Nährstofftechnik, Idstein-Niederauroff, Germany) was pelleted (6000 × g, RT, 5 min) and resuspended in 12.5 ml growth medium. This suspension was mixed 1:1 with biofilm samples followed by an incubation at 37 °C for 10 min. Growth medium and 0.2% SDS prepared in growth medium served as negative and positive controls, respectively. Samples were centrifuged (6000 × g, RT, 5 min), the supernatant was transferred in a 96-well microtiter plate and absorbance was measured at 540 nm in a microtiter plate reader (Synergy MX, BioTek Instruments, Winooski).
Experimental Design and Statistical Rationale
We decided to use an integrated multi-omics approach combining state-of-the-art proteomics and metabolomics techniques and phenotypic verification experiments to shed light on vital physiological processes in the cytoplasm and the ECM of biofilm forming cells (Fig. 1). Therefore, intra- and extracellular proteome analysis of planktonic cultures and intracellular, ECM and FT proteome analysis of biofilm cultures were performed of three biological replicates resulting in a total of 15 individual samples. All samples were prepared in parallel before LC-MS/MS analysis, which was performed in a randomized order. For metabolic footprint analysis, samples of four biological replicates were measured. Phenotypic experiments, including testing of osmotic stress resistance and hemolytic activity, as well as measurement of OD values and pH values were carried out in three biological replicates. In the G. mellonella pathogenicity model, culture supernatants of one biological replicate were used to inject 15 larvae per sample. All microscopy analyses were performed in biological duplicates. Images of 15 randomly selected areas were acquired. Thereof, a representative image of each condition and time point is shown.
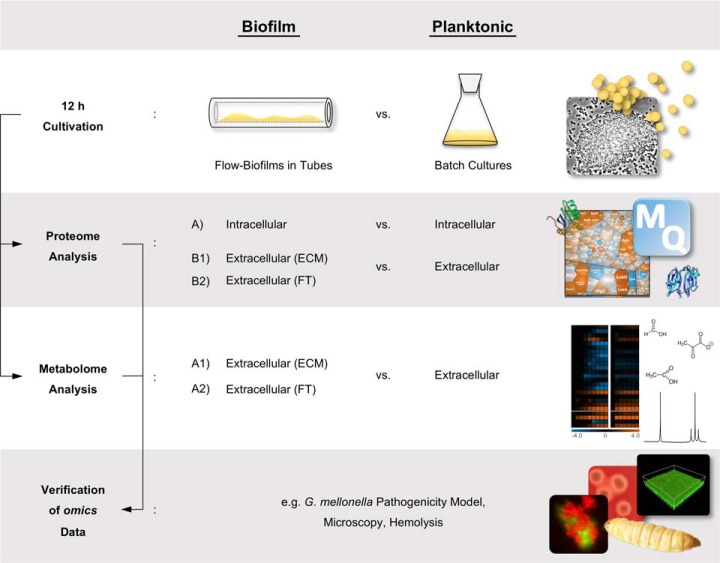
Fig. 1.
Schematic overview of the experimental design. Intracellular, ECM and FT protein profiles of 12 h biofilm cultures grown under flow conditions were compared with intracellular and extracellular protein profiles of 12 h planktonic batch cultures. These data were complemented by metabolic footprint analysis followed by verification of the omics data by different phenotypic analyses e.g. including a G. mellonella pathogenicity model and microscopy. ECM = Extracellular matrix, FT = Flow-through.
Statistical significance was assessed by Student's t test and ANOVA using Excel (version 15.32, Microsoft Corporation, Redmond) for normally distributed proteome data by applying p values of 0.05. GraphPad PRISM (version 8, GraphPad Software, La Jolla) was used for data analysis of metabolome and phenotypic analyses. p values are provided in each figure legend.
RESULTS AND DISCUSSION
Low metabolic activity of biofilm-embedded cells, as well as the ECM acting as a protective barrier against antimicrobial compounds and the host immune system, make biofilm-associated infections extremely difficult to treat. Although S. aureus has been recognized as one of the most frequent causes of biofilm-associated infections, its biofilm physiology and particularly the ECM composition are poorly understood.
Experimental Design
To comprehensively characterize important physiological processes during biofilm formation of S. aureus, we used an integrated multi-omics approach, which combines state-of-the-art proteomics and metabolomics techniques. We compared intracellular and extracellular/ECM/FT protein profiles, as well as ECM and FT metabolite profiles of S. aureus biofilms with intracellular and extracellular protein profiles and extracellular metabolites of their planktonic counterparts. Moreover, we complemented these omics data by phenotypic analyses of biofilm and planktonic cells. A workflow summarizing our approach is shown in Fig. 1. All identified intracellular, extracellular/ECM/FT proteins together with detailed information on peptide and protein identification have been deposited to the ProteomeXchange consortium (http://proteomecentral.proteomexchange.org/cgi/GetDataset) via the PRIDE partner repository with the data set identifier PXD011157 (see data availability section).
To this end, we established a cultivation system enabling us to investigate S. aureus biofilms grown under flow conditions, which reflects the natural environment more accurately compared with static biofilm cultivation models and which allows us to cultivate high amounts of biomass needed for proteome analysis (especially of the ECM).
It is generally believed that the physiology of mature biofilms resembles that of stationary, planktonic batch cultures (60–63), which are therefore an adequate reference for comparative omics-analyses of biofilm and planktonic cells. Hence, as a starting point, we were testing after which time period our biofilm and planktonic cultures represented mature biofilms and stationary cells, respectively. In contrast to other studies that analyzed biofilms over a time period of several days (21, 22, 64), we found that our biofilms matured relatively early, i.e. after 12 h of growth. This might be because we used, in contrast to the above-mentioned studies, a nutrient-rich medium leading to a quick maturation of S. aureus biofilms in our experimental flow setting. Planktonic cultures were harvested 12 h post-inoculation as well, after which they reached the stationary phase. Interestingly, 36 h old flow cultures revealed a dying biofilm characterized by increased cell lysis as indicated by a high abundance of the major autolysin Atl (20, 90), decreased biofilm mass and a substantially higher amount of intracellular proteins within the ECM (data not shown).
Biofilm Cells Show a Similar but Less Pronounced Nutrient Limitation Response Compared with Planktonic Stationary Cells
To establish our biofilm flow system, we optimized cultivation settings (e.g. tube length and flow rate) in preliminary experiments to avoid nutrient depletion and concomitant physiological heterogeneities of the cells along the biofilm tube. According to our metabolome data, this was successfully achieved, because neither glucose nor the amino acids of the growth medium were completely depleted in the biofilm FT (supplemental Fig. S2).
A clear starvation response was observed in planktonic cells, because our metabolome data revealed depletion of glucose and amino acids (supplemental Fig. S2). In agreement with previously published studies (68, 91), this starvation response was indicated by our proteome data showing an increased abundance of proteins belonging to regulons controlled by the carbon catabolite protein A (CcpA), the pleiotropic repressor CodY and CymR, a regulator of sulfur metabolism (65–67) (supplemental Table S1, supplemental Fig. S3). For example, gluconeogenesis (e.g. GpmA, PckA, PycA) and TCA cycle enzymes (e.g. PdhABC, SucABD, SdhA, FumC), as well as amino acid biosynthesis proteins (e.g. LysAC, MetCEFI, LeuABCD, SerA, TrpBCDE), and oligopeptide uptake proteins (e.g. Opp-3ABCDF) were strongly upregulated in stationary planktonic cells compared with biofilm cells. Furthermore, as expected under glucose-limited conditions, glycolysis enzymes were less abundant in planktonic cultures (e.g. Pgi, PfkA, TpiA, GapA, Eno), (supplemental Table S1, Fig. 2). However, differences in abundance of glycolysis/gluconeogenesis and TCA cycle enzymes were rather small. This observation agrees with the hypothesis that deeper layers of the biofilm might also face glucose starvation because of nutrient competition within the biofilm.
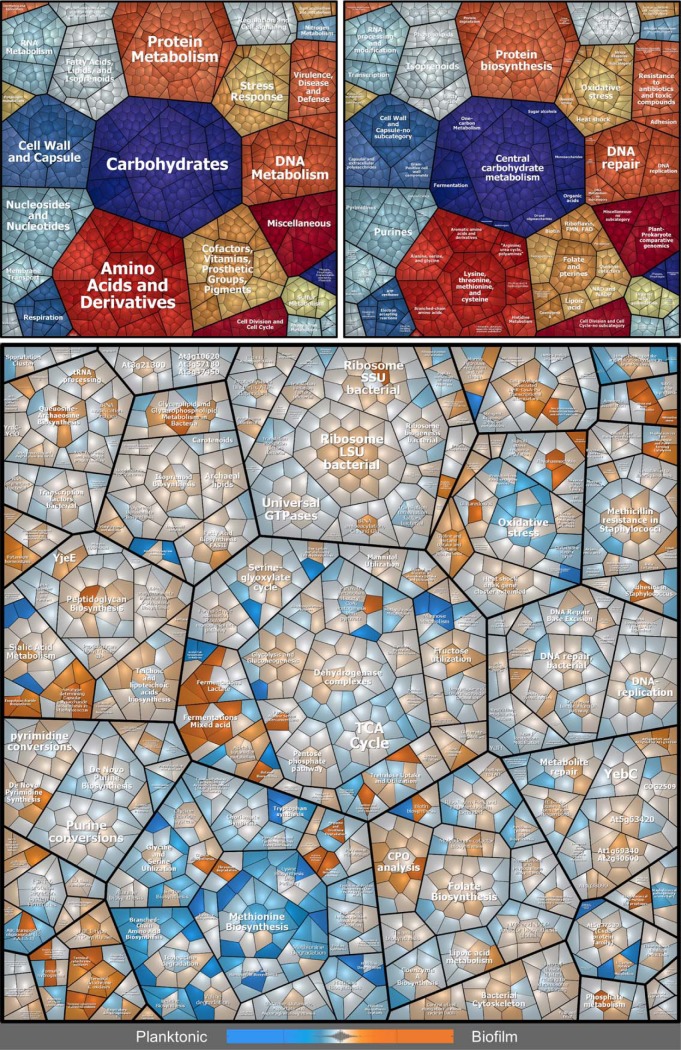
Fig. 2.
Voronoi treemaps visualizing expression profiles of intracellular proteins in biofilms compared with planktonic cultures. Proteins found in MS analyses are displayed as single cells, which are functionally clustered in three hierarchical levels according to the Seed database: first level = upper left panel, second level = upper right panel, third level = bottom panel. Differences in protein abundance are indicated in the bottom panel by a color code based on LFQ intensities: orange = proteins more abundant in biofilms, blue = proteins more abundant in planktonic cultures, light gray: no difference in protein abundance.
Interestingly, iron acquisition proteins of the Fur regulon were found more abundant in planktonic cultures (supplemental Table S1, supplemental Fig. S3), including e.g. iron transporters (SirA, HtsA) and proteins for biosynthesis of the siderophore staphylobactin (SbnEF). Consequently, it can be concluded that iron limitation is more pronounced in planktonic cultures compared with biofilms.
Collectively, these results suggest that the biofilm cells in our setting also experience nutrient limitation albeit to a lesser degree than the stationary planktonic cells. This can intuitively be explained by the nutrient supplying properties of a flow system, which supports the outer cells of the biofilm with enough nutrients during the entire experiment. The deeper layers of the biofilm, however, are likely to suffer from nutrient limitation. Because it was not possible in our experimental setup to analyze outer and inner biofilm cells separately, our proteome data reflect the mean abundance of the detected proteins for a rather heterogeneous population of cells. Nevertheless, dominant physiological effects that are characteristic for the biofilm cells can still be identified. This was particularly obvious for fermentation pathways, which will be explained in the following section.
Oxygen Limitation in Biofilms Leads to ECM Acidification
Biofilms are organized as densely packed communities making it intuitive that nutrient and oxygen availability throughout the biofilm is gradually decreasing and limited in deeper layers because of consumption and diffusion impairment. Especially oxygen limitation has been described for biofilms of various species (18, 21, 24, 68–70). Because of the higher cell densities reached during biofilm growth compared with planktonic growth, we expected to observe oxygen limitation also in our flow biofilms (supplemental Fig. S4).
Indeed, the proteome and metabolome data clearly revealed oxygen limitation in S. aureus biofilms indicated by the strong and significant accumulation of enzymes involved in mixed acid and 2,3-butanediol fermentation (e.g. lactate dehydrogenase Ldh1, alcohol dehydrogenase Adh1, pyruvate formate lyase PflB, acetolactate synthase BudB), and fermentation products like formate, lactate, ethanol, and 2,3-butanediol (supplemental Table S1, Fig. 2, supplemental Fig. S5A). In addition to fermentation, S. aureus can use nitrate respiration for energy production during oxygen-limited conditions. Our proteome data clearly showed an accumulation of enzymes required for nitrate respiration in biofilms (NarJGH, NasDEK) (supplemental Table S1, Fig. 2). Furthermore, nitrate consumption was observable only in the biofilm but not in planktonic cultures (supplemental Fig. S5B). The fermentation and nitrate respiration proteins are part of the Rex and the NreC regulon, respectively. Both regulators were also found upregulated in biofilms underlining the importance of these regulons during biofilm growth (supplemental Fig. S3).
We further observed that the reduced oxygen availability in biofilms impacts proteins of the oxidative stress response, which were found less abundant compared with planktonic cells. This includes superoxide dismutase SodA, catalase KatA, alkyl hydroperoxide reductase AhpC, DNA protection protein Dps, and the glutathione peroxidase BsaA (supplemental Table S1, Fig. 2). Furthermore, proteins of the nitrosative stress response accumulated in biofilm cells (e.g. Hmp, ScdA, SrrAB) probably because of increased reactive nitrogen species production during nitrate respiration (71). As an additional consequence of oxygen limitation and nitrosative stress within biofilms, proteins of the SrrAB regulon involved in electron transport chain maintenance (cytochrome c and quinol oxidase assembly, as well as heme biosynthesis: CydAB, QoxABC, and HemBCDEHLQ) were found more abundant in biofilms (supplemental Table S1, Fig. 2). The importance of SrrAB in static S. aureus biofilms was also shown by Kinkel et al., 2013 (71).
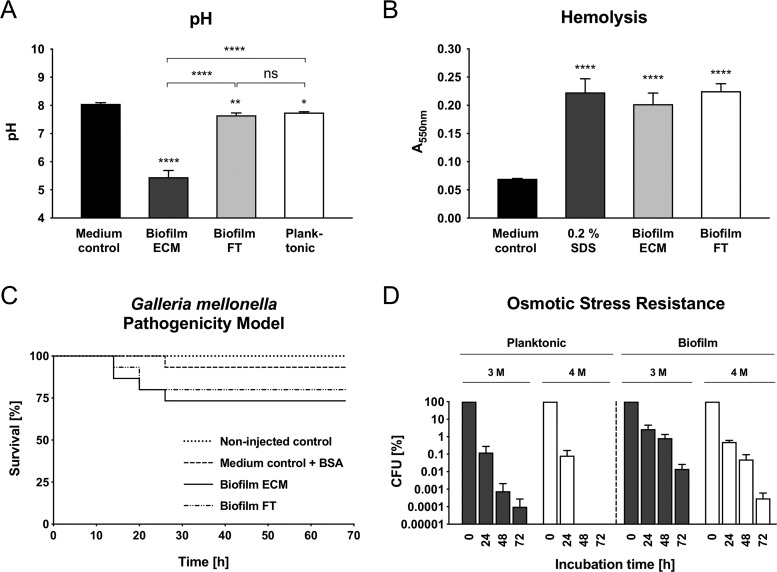
Most importantly, the accumulation of strong acids upon fermentative metabolism in S. aureus biofilms leads to local acidification, which was confirmed by pH measurements revealing a local pH of ∼5.5 in the ECM compared with a pH of 7.6 in the FT, 7.5 in planktonic cultures and 8 in fresh growth medium (Fig. 3A). No acidification of the biofilm FT and planktonic cultures can be explained by dilution effects and the buffering properties of the growth medium. Notably, the observed acidification effect within the ECM is perfectly in line with results of other studies investigating S. aureus biofilms, where decreased pH values were observed (24, 26, 72–74). Interestingly, we did not observe an upregulation of proteins involved in the arginine deiminase and urease pathway, which were reported to counteract local acidification in S. aureus biofilms (supplemental Table S1, Fig. 2) (21, 26, 75, 76). This supports our proposed model of an acidified ECM environment playing a major role in mediating biofilm stability as explained in the following two sections.
Fig. 3.
Phenotypic analyses of S. aureus cells and culture supernatants derived from flow biofilms and planktonic cultures grown for 12 h. A, pH values of a medium control, planktonic, biofilm ECM, and biofilm FT samples are indicated as the mean ± S.D. of quadruplicate experiments. B, Hemolysis assay of a growth medium control, 0.2% SDS as a positive control, cell-free biofilm ECM and FT samples, respectively. Data are displayed as mean values ± S.D. of triplicate experiments. C, G. mellonella larvae were injected with 5 μl of cell-free biofilm ECM or FT samples, respectively. Noninjected larvae and growth medium supplemented with BSA served as controls. The survival of 15 larvae per experiment was monitored for 68 h. D, Freshly cultivated biofilm and planktonic cells were used to inoculate fresh growth medium supplemented with 3 m and 4 m NaCl, respectively. CFU were determined after 0, 24, 48, and 72 h. Data are displayed as mean values ± S.D. of triplicate experiments relative to the CFU (100%) of time point 0 h. * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001. ECM = Extracellular matrix, FT = Flow-through.
S. aureus Biofilms Express High Amounts of Virulence Factors
Next, we were interested in the exact composition of the ECM at the proteome level because it has been frequently reported that proteins are an important component of the ECM of clinical biofilm forming strains (26, 37, 38, 121–125). Moreover, the ECM represents a permeability barrier for many antimicrobial molecules and thus understanding its composition can help to develop novel antimicrobial strategies.
Approximately 30% of the total protein amount we found in the S. aureus ECM represented secreted extracellular proteins. However, the most abundant protein class in the ECM were intracellular proteins (∼60%), which are primarily represented by ribosomal proteins (∼42% thereof, which corresponds to 25% of total ECM protein) (Fig. 4, Fig. 5A and supplemental Fig. S6). This can probably be explained by cell lysis within the biofilm or alternatively, as recently suggested, via nonclassical protein export by a yet unknown pathway (77). Interestingly, it has been proposed that intracellular proteins might contribute to pathogenicity by mediating binding to host matrix proteins and host cells (78). Cell lysis in biofilms is a well-reported phenomenon, which is mediated by the major autolysin Atl and the holin/antiholin system CidABC and LrgAB in S. aureus (35, 79–81). Interestingly, these proteins were not upregulated in our 12 h biofilms (supplemental Table S1). However, Atl was significantly upregulated in the 36-h biofilm, which was accompanied by strong lysis and accumulation of intracellular proteins in the ECM (data not shown).
Fig. 4.
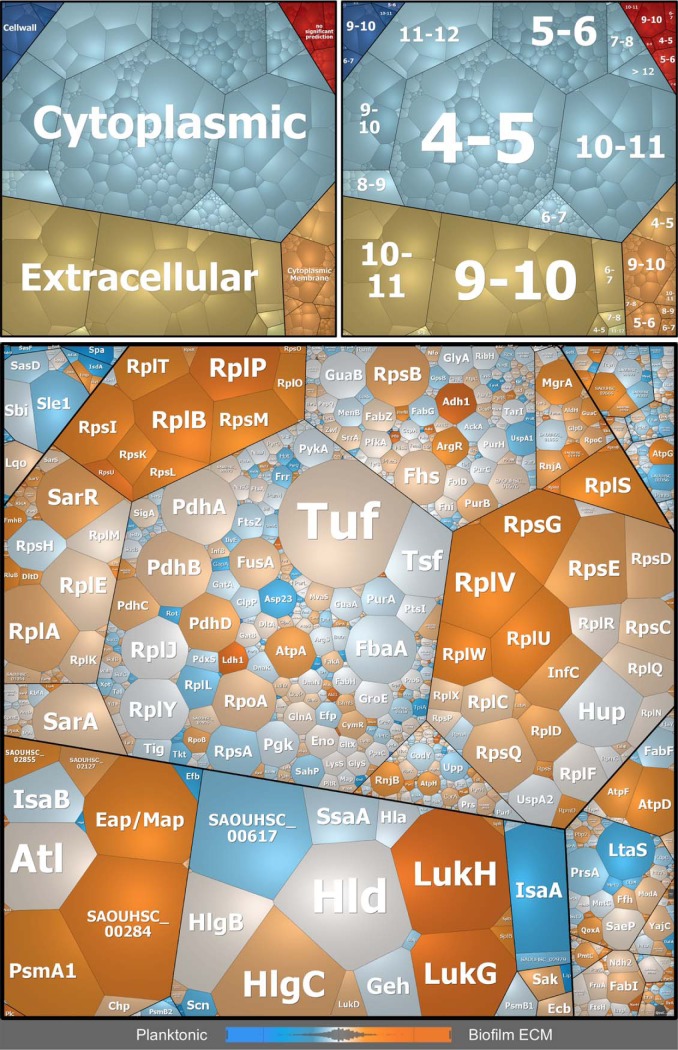
Differences in protein profiles of the biofilm ECM and extracellular planktonic samples visualized by Voronoi treemaps. Proteins found in MS analyses are displayed as single cells, which sizes correlate with protein abundance based on riBAQ values of ECM proteins. Proteins are clustered according to their subcellular localization predicted by PSORTb (upper left panel), and isoelectric point (pI) according to AureoWiki (upper right panel). Bottom panel: Differences in protein abundance between biofilm ECM and extracellular planktonic samples are indicated by a color code, which is based on LFQ intensities: orange = proteins more abundant in ECM, blue = proteins more abundant in planktonic cultures, light gray = no difference in protein abundance. ECM = Extracellular matrix.
Fig. 5.
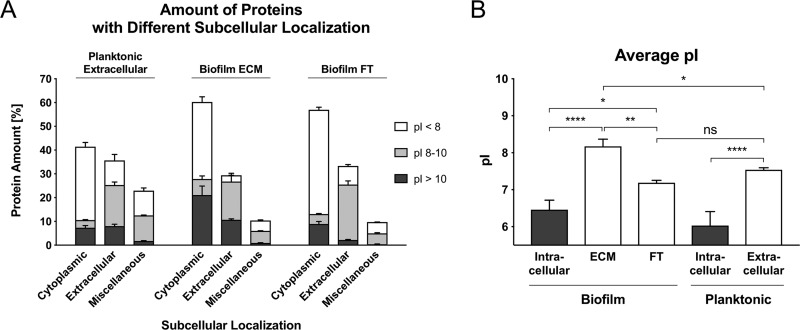
Amount of proteins with different subcellular localizations and differences in the average pI. A, Relative abundance of proteins with different subcellular localizations (predicted by PSORTb and manually cured: Cytoplasmic, Extracellular and Miscellaneous = Cytoplasmic membrane, Cell wall, no significant prediction) and different pI values, which were identified in extracellular planktonic samples, biofilm ECM and biofilm FT samples. The relative protein abundance was calculated based on riBAQ values. B, The average isoelectric point (pI) was calculated and normalized by protein abundance based on riBAQ values. Data are displayed as mean values ± S.D. of triplicate experiments. pI values were extracted from the AureoWiki database. ns = not significant, * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001. ECM = Extracellular matrix, FT = Flow-through.
However, proteins with the highest abundance level in the ECM were extracellular proteins, which are primarily represented by virulence factors including e.g. hemolysins (Hla, HlgBC, Hld), a phenol-soluble modulin (Psmα1), leukotoxins (LukGH), lipases (Geh, Plc), and the extracellular adherence protein (Eap/Map) (supplemental Table S2, Fig. 4, supplemental Fig. S6). Compared with extracellular planktonic samples, most of these virulence factors accumulate at a significantly higher amount in biofilm ECM samples (supplemental Table S2, Fig. 4). All these virulence factors were attributed to fulfill specific functions in S. aureus pathogenesis (82–85). Our findings are consistent with other studies, which observed an accumulation of LukAB and Hla in in vivo S. aureus biofilm models (86–88). In contrast, studies using static biofilm cultivation models often did not identify elevated expression of virulence factors (18, 25, 26, 72), which seems to be an important difference between biofilms cultivated under static and flow conditions, respectively.
Many virulence factors that we found more abundant in biofilms are controlled by the quorum sensing responsive agr locus and the transcription factor SarA (18, 64, 89). However, proteins of the agr system or SarA were not significantly more abundant in biofilms compared with planktonic cells (supplemental Table S1, supplemental Fig. S3). We speculate that protein expression of the agr system reached its maximum in both, biofilm and planktonic cells, so the increased amount of virulence factors in the ECM might be explained by a passive accumulation effect. Alternatively, a local accumulation of the quorum sensing peptide within the ECM compartment could lead to a higher activation of the Agr two component system in biofilm cells as compared with planktonic cells. In addition, other global regulators (SigB, SaeRS, SrrAB, ArlRS, and Rot) known to control virulence gene expression, as well as their corresponding regulons were also more abundant in biofilms identifying these regulators as important players, which might balance virulence gene expression in S. aureus biofilms (supplemental Table S1 and S2, supplemental Fig. S3).
To prove that the identified, secreted virulence factors are functional, we tested cell-free supernatant derived from the ECM and the biofilm FT in hemolysis assays and in a G. mellonella pathogenicity model. Both samples indeed showed hemolytic activity and killed G. mellonella larvae in contrast to medium and BSA controls, respectively, which confirms the pathogenic potential of S. aureus biofilms (Fig. 3B and 3C).
Besides the high abundance of secreted virulence factors in the ECM, our intracellular proteome data also revealed that biofilm cells express higher amounts of capsule biosynthesis proteins CapABCDEFGHMNO compared with planktonic cells (supplemental Table S1, Fig. 2). Interestingly, the same phenomenon was reported by Beenken et al., 2004, who also investigated biofilms grown under constant medium flow (21), but not in other studies comparing planktonic cells and biofilms, which were cultivated as static colony biofilms (18, 25, 26).
Taken together, our findings strongly support the study of Lei et al., 2017 (88), who proposed that S. aureus biofilms exhibit a high virulence potential and apply multiple strategies simultaneously to evade the host immune system. These strategies include protection by the ECM, capsule biosynthesis, and secretion of virulence factors like hemolysins, leukotoxins, lipases, and proteases.
S. aureus Virulence Factors and Ribosomal Proteins Exhibit a Moonlighting Function Contributing to Biofilm Integrity
Interestingly, most of the virulence factors we identified in the ECM are characterized by a high isoelectric point (pI) between 8 and 12 (supplemental Table S2, Fig. 4 and 5A). The same accounts for cytoplasmic proteins found within the ECM. In fact, nearly half of these cytoplasmic proteins have an isoelectric point between 8 and 12 and are primarily represented by ribosomal proteins (Fig. 4 and 5A). Moreover, the average pI of ECM samples is significantly higher compared with planktonic cultures and to the FT (Fig. 4 and 5B, supplemental Fig. S7).
We suspected that these alkaline virulence factors and ribosomal proteins will carry a strong positive charge in an acidic ECM environment created by formate, lactate, acetate, and pyruvate produced during glucose fermentation under oxygen-limited conditions (Fig. 3A, supplemental Fig. S5A).
As a consequence, these cationic proteins in the ECM might interact with anionic cell surfaces (90) and eDNA and thereby act as electrostatic bridges between cells providing physical strength to the biofilm, similarly as was proposed by Foulston et al., 2014 and Dengler et al., 2015 for intracellular proteins (72, 73). To further test this hypothesis, we used freshly cultivated biofilm cells, which were harvested, washed, pelleted and resuspended in either planktonic or biofilm ECM supernatant of pH 8 and pH 5.5, respectively, and investigated cell aggregation using fluorescence microscopy after staining with the eDNA stain Toto-1 and Syto62 as a counterstain. Strikingly, biofilm supernatant indeed induced the formation of cell aggregates under low pH conditions, which could not be observed for planktonic supernatant or a medium control at pH 5.5. At pH 8, no cell aggregates could be observed at all (Fig. 6A).
Fig. 6.
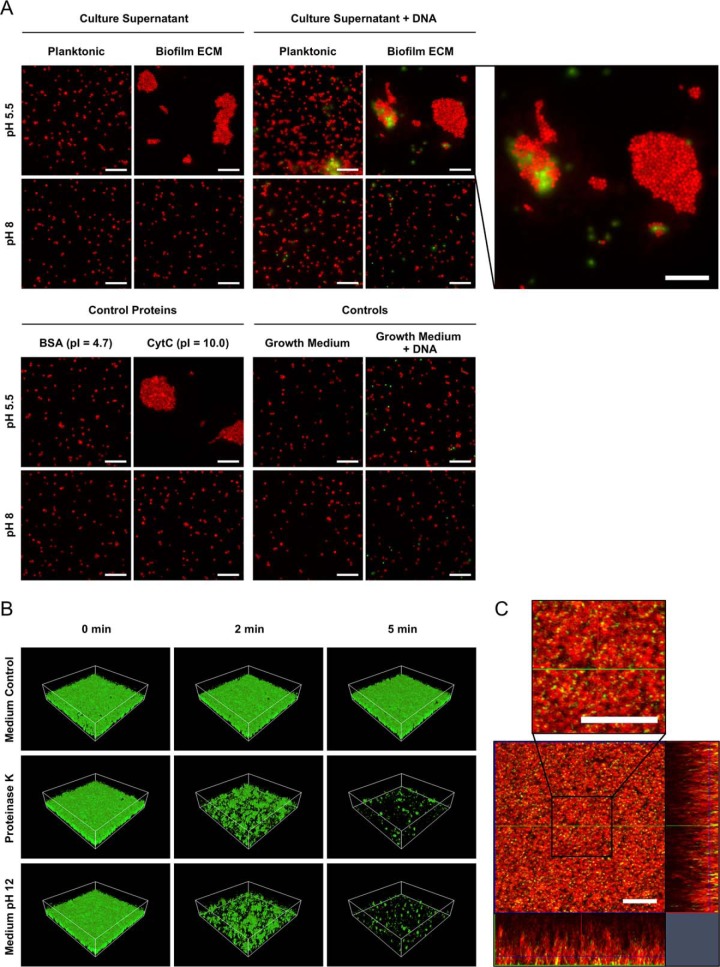
Microscopic analyses of the impact of cationic proteins and eDNA on S. aureus cell aggregation and biofilm stability. A, Freshly cultivated biofilm cells were harvested, washed, pelleted, and resuspended in the following solutions, which were adjusted to either pH 5.5 or pH 8, and investigated by fluorescence microscopy after staining with the eDNA stain Toto-1 and Syto62 as a counterstain: planktonic or biofilm ECM supernatant, fresh growth medium supplemented with BSA or CytC as control proteins, planktonic or biofilm ECM supernatant supplemented with cDNA isolated from S. aureus, medium control, medium control plus cDNA. Representative images of 15 randomly selected areas of duplicate experiments are shown. The scale bar indicates 10 μm. B, Biofilms were cultivated in flow cells for 12 h followed by a treatment with a growth medium control, Proteinase K, and alkalized growth medium of pH 12, respectively. Biofilms were subsequently challenged by elevated shear forces to test biofilm stability and analyzed by confocal laser scanning microscopy (CLSM). For each biofilm, 3 CLSM images of randomly selected areas (spanning 100 μm × 100 μm) were acquired after 0, 2, and 5 min of elevated shear stress in duplicate experiments. A representative image of each treatment and time point is shown. C, Biofilms were cultivated in flow cells for 12 h, stained with the eDNA stain Toto-1 (green) and Syto62 (red) as a counterstain, washed and analyzed by CLSM. A representative image of 5 randomly selected areas of duplicate experiments is shown. The scale bar indicates 10 μm.
To verify that cationic proteins induce cell aggregation, we repeated the experiment using fresh medium supplemented with either cytochrome C (CytC, pI = 10.0) or BSA (pI = 4.7) as a negative control. We observed that only CytC and not BSA induced cell aggregation at pH 5.5 (Fig. 6A). These findings indicate, that cell aggregation in S. aureus biofilms is mediated by cationic proteins and is eDNA independent, because no eDNA was present in this experiment. However, because our CLSM analysis revealed high amounts of eDNA within the ECM (Fig. 6C), we tested if eDNA supplementation enhances the cell aggregation effect and repeated the experiment with planktonic and ECM supernatant in the presence of cDNA isolated from S. aureus to mimic eDNA. Although we observed binding of cDNA to cell aggregates, we did not observe an enhanced cell aggregation effect after addition of cDNA (Fig 6A). A possible explanation for this observation could be that the protein concentration used in our assay was in a range masking an enhancing aggregation eDNA effect besides the fact that ECM protein concentration used in this experiment corresponded to the concentration as determined for our biofilm ECM fraction.
Interestingly, we observed that other anionic metabolites, namely glutamate, aspartate and pyruvate, tend to accumulate within the ECM because they were found in higher concentrations in the ECM compared with the biofilm FT (supplemental Fig. S5A). This accumulation effect was not observed for acetate and lactate, because we measured high concentrations in the ECM, but even higher concentrations in the biofilm FT. This might be because of a saturation effect, because acetate and lactate represent the most abundant negatively charged metabolites within the ECM (supplemental Fig. S5A). We hypothesize that these anionic metabolites located in the ECM act as electrostatic counterparts to cationic proteins and thereby functionally act like eDNA.
To further characterize the role of cationic proteins in biofilm integrity, we treated established flow biofilms with Proteinase K according to Seidl et al., 2008 (57), or alkaline medium adjusted to pH 12 to eliminate positive protein charges, followed by challenging the biofilms with elevated shear forces. The effects on biofilm integrity were visualized using CLSM. Strikingly, Proteinase K-treated and alkalized biofilms clearly showed impaired integrity because it was possible to almost completely eradicate the biofilm within 5 min of elevated shear stress (Fig. 6B). Importantly, alkaline growth medium with pH 12 does not kill S. aureus within 5 min, which was verified by CFU counting (data not shown). Supporting our findings, inhibiting effects of alkaline pH against Staphylococcal biofilm maturation were already reported, without significant inhibition of planktonic growth (91). Of note, DNase treatment of S. aureus biofilms did not impair integrity, which might be because of proteins protecting eDNA within the ECM from digestion (data not shown). In summary, these results strongly support the idea that cationic proteins play a major role in biofilm integrity under the tested conditions.
At present, S. aureus ECM stability is mainly attributed to PIA, an N-acetylglucosamine-based exopolysaccharide found in biofilms of numerous S. aureus strains. PIA is partly de-acetylated, which introduces positive charges at neutral and acidic pH suggesting that PIA mediates cell aggregation via electrostatic interactions with anionic cell surfaces and possibly eDNA (4). A similar concept was also shown in P. aeruginosa biofilms, where Pel, an abundant positively charged exopolysaccharide, interacts with eDNA (92). However, we were not able to identify any of the PIA biosynthesis proteins IcaABCD in our proteome data and the regulator IcaR was slightly less abundant in biofilms compared with planktonic cells, which points to a PIA-independent biofilm (supplemental Table S1). Interestingly, more recent studies reported an increasing number of S. aureus isolates including community- and hospital-acquired MRSA strains, which form PIA-independent, Proteinase K-sensitive biofilms (59, 64, 93–98). Biofilms of these strains were stated to be protein-dependent, which was attributed to adhesive surface proteins (59, 97, 99) and intracellular proteins (72, 73). More precisely, Foulston et al., 2014 showed that cationic, intracellular proteins derived from S. aureus biofilms reversibly bind to cell surfaces upon drop in pH, which contributes to multicellular behavior (72). Moreover, Dengler et al., 2015 speculated that the function of cationic PIA within biofilms could be replaced by cationic proteins, which interact with anionic cell surfaces and eDNA. Thereby, they emphasized the crucial role of eDNA as an electrostatic bridge (73). Both studies suggested local acidification within biofilms following the release of fermentation products, which is perfectly in line with our proteome, metabolome and pH data. In agreement, we also identified a large proportion of cytosolic proteins within the ECM. Importantly, we were able to provide a more detailed view on the proteinaceous composition of the S. aureus ECM, thereby identifying specific proteins dominating the ECM (Fig. 4). Furthermore, we demonstrated that eDNA is a highly abundant component of the ECM (Fig. 6C), which is in line with numerous studies showing an important role of eDNA during both early and later stages of biofilm development (14, 35, 79, 100). However, compared with Dengler et al., 2015 we were not able to observe enhanced cell aggregation after the addition of cDNA to cells mixed with ECM proteins, suggesting that the stability of our biofilm grown under flow-through conditions is predominantly depending on the proteins within the ECM (Fig. 6A). Interestingly, the negative cell surface charge, which seems to be key for cell aggregation, was previously suggested to be caused by teichoic acids in the S. aureus cell wall (4, 101). Supporting this idea, we found a slight but consistent upregulation of many proteins for teichoic acid biosynthesis in biofilms compared with planktonic cells (supplemental Table S1, Fig. 2).
It remains debatable if S. aureus actively increases secretion of alkaline virulence factors as an evolutionary favorable mechanism to stabilize biofilm structures, or if the high abundance of virulence factors within the ECM is caused by a passive accumulation because of the described electrostatic interactions (supplemental Table S1, Fig. 2).
Accumulation of Cationic Proteins and Anionic Metabolites Within the ECM Causes Osmotic Stress in Biofilm-embedded Cells
According to our model of ECM architecture, cells are electrostatically linked in the ECM environment by highly abundant proteins, eDNA and metabolites (Fig. 7). Because each of these molecules is osmotically active and theoretically elevates osmotic pressure, we hypothesized that biofilm-embedded S. aureus cells experience osmotic stress. Supportively, we found several proteins associated with osmotic stress resistance more abundant in biofilm cells. This includes proteins for the uptake and biosynthesis of osmoprotectants like OpuBCD and BetAB (supplemental Table S1, Fig. 2). In addition, we found elevated levels of both cardiolipin synthases Cls1 and Cls2 in biofilms. Cardiolipin was shown to be important for S. aureus during long-term survival under osmotic stress conditions (102). Furthermore, we found the two-component system KdpDE and one protein of a potassium uptake system, KtrA, more abundant in biofilm cells, which mediate osmotic stress resistance (supplemental Table S1) (103, 104). This is in line with results of Price-Whelan et al., 2013, who identified elevated transcript levels of kdpDE and a protective role of the Ktr potassium uptake system under osmotic stress conditions (104). Consequently, biofilm cells might be more osmotolerant than their planktonic counterparts.
Fig. 7.
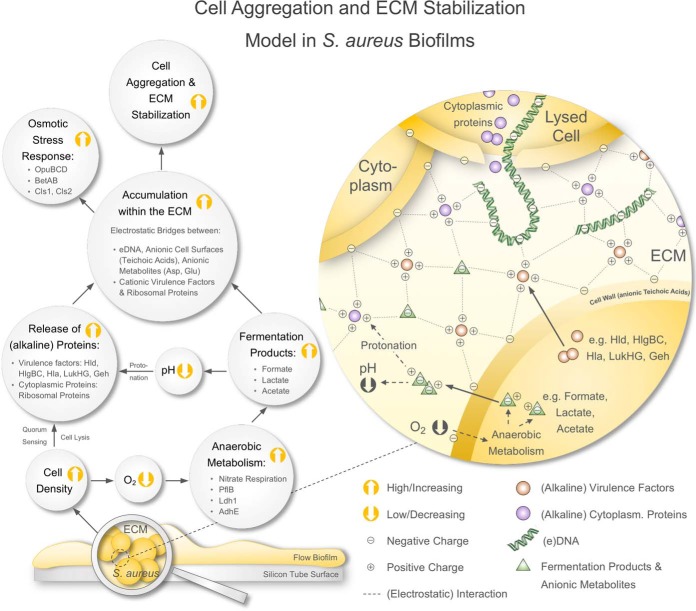
Proposed model of cell aggregation and ECM stabilization mediated by moonlighting virulence factors and ribosomal proteins in S. aureus biofilms. Biofilms cultivated in a continuous flow system on a silicon tube surface grow to high cell densities, which leads to oxygen limitation. Consequently, biofilm-embedded cells are driven toward anaerobic metabolism and secrete high amounts of fermentation products lowering the local pH within the ECM. Furthermore, S. aureus biofilm cells release high amounts of eDNA, virulence factors and ribosomal proteins (besides other cytoplasmic proteins). These proteins get protonated in the acidic ECM environment because of their alkaline character. The accumulation of these cationic proteins, eDNA and anionic fermentation products along with other anionic metabolites (I) creates an electrostatic network involving eDNA and anionic cell surfaces (harboring anionic teichoic acids), which leads to cell aggregation and ECM stabilization and (II) leads to an osmotic stress response in biofilm-embedded cells. ECM = Extracellular matrix.
To test this hypothesis, fresh medium supplemented with elevated NaCl concentrations (3 m and 4 m, respectively) was inoculated with cell suspensions derived from either planktonic or biofilm cultivations and subsequently analyzed for survival using CFU counting. We used cell suspensions instead of intact biofilms in this assay to separate effects of increased cell resistance from potential interference of a protective ECM. Strikingly, biofilm-grown cells show an increased survival rate compared with planktonic cells under both tested NaCl concentrations. Biofilm derived cells even survived in 4 m NaCl medium for 72 h, whereas planktonic cells already died after 48 h (Fig. 3D). Importantly, cells did not aggregate during the experiment (usually caused by the chaotropic properties of high salt concentrations) as we assessed by phase contrast microscopy, which ensured reliable CFU counting results (data not shown) (105).
To our knowledge, high osmotic pressure in S. aureus biofilms has not been reported yet, although it has been described that high osmolarity has a positive effect on biofilm formation, which is mediated by the alternative sigma factor B (106). Furthermore, others have also described an induction of osmotic stress protection systems in S. aureus biofilms. For example, Resch et al., 2015 and Moche et al., 2015 found genes and proteins induced in colony biofilms, which are associated with osmoprotectant uptake, but did not find elevated expression of the kdp system or cls (18, 25). Beenken and colleagues reported elevated transcript levels of kdpDE and kdpABC, as well as cls in flow biofilms, but not of genes associated with osmoprotectant uptake (21). In addition, a transposon mutant library screening by Boles et al., 2010 revealed that a defect in genes involved in osmoregulation results in impaired biofilm formation (98). Contrary to these studies, metabolic profiling of Junka et al., 2013 revealed an accumulation of osmoprotectants in planktonic cells but not in static biofilms (24).
Interestingly, studies on Bacillus subtilis and Vibrio cholera biofilms proposed that ECM components generate osmotic pressure thereby helping biofilm-embedded cells to spread over the growth substratum (107, 108). If a comparable mechanism also exists in S. aureus biofilms and which specific ECM components might contribute to elevated osmotic pressure remains elusive. However, there are reports suggesting a connection of K+/osmolarity sensing by the KdpDE two-component system and regulation of virulence factor expression, which includes positive regulation of capsule biosynthesis genes cap, and negative regulation of invasion factors like lipase Geh, the proteinase Aur and the hemolysins Hla and HlgB (109–111). Notably, in our experiment all of these proteins were found to be more abundant in S. aureus biofilms compared with their planktonic counterparts (supplemental Table S1, S2, Fig. 2 and 4). Because expression of these virulence factors is primarily controlled by other major regulators including e.g. Agr/RNAIII, Rot, CodY, SarA, SaeRS, SrrAB, and ArlRS (64, 89, 112–119), it could be speculated that the KdpDE system has a fine-tuning function for virulence gene expression in S. aureus biofilms.
CONCLUSIONS
Establishing a flow system for highly reproducible cultivation of S. aureus biofilms enabled us to grow biofilms under conditions, which are relevant for different clinical scenarios, i.e. endocarditis or catheter-associated infections, thereby complementing studies employing static biofilm cultivation models. This flow system allowed the cultivation of high amounts of biofilm biomass, which was a prerequisite to apply a multi-omics approach investigating intracellular and ECM proteome profiles in combination with extracellular metabolome profiles.
Using this multi-omics approach, we showed that S. aureus biofilms secrete high amounts of functional virulence factors like hemolysins, leukotoxins, and lipases, which are part of the ECM but can also be found in the biofilm FT. Applying a G. mellonella pathogenicity model and a hemolysis assay, we demonstrated that these virulence factors are active. Furthermore, we show that the S. aureus biofilm ECM consists to a large extent of ribosomal proteins. We demonstrate that secreted virulence factors and ribosomal proteins play a so far unacknowledged role as moonlighting proteins, which contribute to biofilm integrity. This stabilizing effect is mediated by an acidic ECM environment caused by the release of fermentation products like formate, lactate, and acetate. Positive charges on alkaline proteins introduced by the acidic environment promote the interaction of proteins with negatively charged cell surfaces, eDNA and anionic metabolites.
Moreover, we suggest that the proteins and metabolites, which are accumulating within the ECM cause osmotic stress in biofilm-embedded cells. Our proposed model is summarized in Fig. 7. Taken together, our study provides a comprehensive map of the intracellular and ECM proteome of S. aureus flow biofilms.
Data availability
MS raw data, MaxQuant output files and the used database were deposited to the ProteomeXchange consortium (http://proteomecentral.proteomexchange.org/cgi/GetDataset) via the PRIDE partner repository (120) with the data set identifier PXD011157.
Supplementary Material
Acknowledgments
We thank Anica Graf, Susanne Sievers, Daniela Zühlke, and Jörg Bernhardt for fruitful discussions. Moreover, we thank Silvia Dittmann for excellent technical assistance as well as Claudia Hirschfeld and Pierre Mücke for help with MS analysis. Furthermore, we are grateful to Rabea Schlüter and Stefan Bock (Imaging Center of the Faculty of Biology, University of Greifswald) for providing the Amira software and help with 3D biofilm image reconstruction.
Footnotes
* This work was funded by the German Research Foundation (Collaborative Research Center Transregio 34, subprojects A3, A8, and Z4, and the Research Training Group 1870).
 This article contains supplemental Figures and Tables. We have declared no conflict of interest.
This article contains supplemental Figures and Tables. We have declared no conflict of interest.
1 The abbreviations used are:
- ECM
- extracellular matrix
- FT
- flow-through
- eDNA
- extracellular DNA
- CLSM
- confocal laser scanning microscopy
- pI
- isoelectric point
- LFQ
- label-free quantification
- (r)iBAQ
- (relative) intensity-based absolute quantification.
REFERENCES
- 1. Costerton J. W., Geesey G. G., and Cheng K. J. (1978) How bacteria stick. Sci. Am. 238, 2–11 [DOI] [PubMed] [Google Scholar]
- 2. Donlan R. M. (2002) Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8, 881–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Flemming H. C., and Wingender J. (2010) The biofilm matrix. Nat. Rev. Microbiol. 8, 1–11 [DOI] [PubMed] [Google Scholar]
- 4. Otto M. (2008) Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 322, 207–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stewart P. S., and William Costerton J. (2001) Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138 [DOI] [PubMed] [Google Scholar]
- 6. Joo H. S., and Otto M. (2012) Molecular basis of in vivo biofilm formation by bacterial pathogens. Chem. Biol. 19, 1503–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Römling U., and Balsalobre C. (2012) Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Int. Med. 272, 541–561 [DOI] [PubMed] [Google Scholar]
- 8. Lister J. L., and Horswill A. R. (2014) Staphylococcus aureus biofilms: recent developments in biofilm dispersal. Front Cell Infect Microbiol. 4, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chatterjee S., Maiti P., Dey R., Kundu A., and Dey R. (2014) Biofilms on indwelling urologic devices: microbes and antimicrobial management prospect. Ann. Med. Health Sci. Res. 4, 100–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kiedrowski M. R., and Horswill A. R. (2011) New approaches for treating staphylococcal biofilm infections. Ann. N.Y. Acad. Sci. 1241, 104–121 [DOI] [PubMed] [Google Scholar]
- 11. Barrett L., and Atkins B. (2014) The clinical presentation of prosthetic joint infection. J. Antimicrob. Chemother. 69, i25–i27 [DOI] [PubMed] [Google Scholar]
- 12. Parsek M. R., and Singh P. K. (2003) Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57, 677–701 [DOI] [PubMed] [Google Scholar]
- 13. O'Toole G. A., Kaplan H. B., and Kolter R. (2000) Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 1–37 [DOI] [PubMed] [Google Scholar]
- 14. Moormeier D. E., Bose J. L., Horswill A. R., and Bayles K. W. (2014) Temporal and stochastic control of Staphylococcus aureus biofilm development 5, e01341–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Speziale P., Pietrocola G., Foster T. J., and Geoghegan J. A. (2014) Protein-based biofilm matrices in Staphylococci. Front Cell Infect Microbiol. 4, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Payne D. E., and Boles B. R. (2016) Emerging interactions between matrix components during biofilm development. Curr. Genet. 62, 137–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paharik A. E., and Horswill A. R. (2016) The Staphylococcal biofilm: adhesins, regulation, and host response. Microbiol. Spectr. 4, 1–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moche M., Schlüter R., Bernhardt J., Plate K., Riedel K., Hecker M., and Becher D. (2015) Time-resolved analysis of cytosolic and surface-associated proteins of Staphylococcus aureusHG001 under planktonic and biofilm conditions. J. Proteome Res. 14, 3804–3822 [DOI] [PubMed] [Google Scholar]
- 19. Ammons M. C. B., Tripet B. P., Carlson R. P., Kirker K. R., Gross M. A., Stanisich J. J., and Copié V. (2014) Quantitative NMR Metabolite Profiling of Methicillin-Resistant and Methicillin-Susceptible Staphylococcus aureusDiscriminates between Biofilm and Planktonic Phenotypes. J. Proteome Res. 13, 2973–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Atshan S. S., Shamsudin M. N., Sekawi Z., Thian Lung L. T., Barantalab F., Liew Y. K., Alreshidi M. A., Abduljaleel S. A., and Hamat R. A. (2015) Comparative proteomic analysis of extracellular proteins expressed by various clonal types of Staphylococcus aureus and during planktonic growth and biofilm development. Front. Microbiol. 6, 1081–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Beenken K. E., Dunman P. M., McAleese F., Macapagal D., Murphy E., Projan S. J., Blevins J. S., Smeltzer M. S. (2004) Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186, 4665–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bénard L., Litzler P. Y., Cosette P., Lemeland J. F., Jouenne T., and Junter G. A. (2009) Proteomic analysis of Staphylococcus aureus biofilms grown in vitroon mechanical heart valve leaflets. J. Biomed. Mater. Res. 88, 1069–1078 [DOI] [PubMed] [Google Scholar]
- 23. Islam N., Ross J. M., and Marten M. R. (2015) Proteome analyses of Staphylococcus aureus biofilm at elevated levels of NaCl. Clin. Microbiol. 4, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Junka A. F., Deja S., Smutnicka D., Szymczyk P., Ziółkowski G., Bartoszewicz M., and Młynarz P. (2013) Differences in metabolic profiles of planktonic and biofilm cells in Staphylococcus aureus - (1)H Nuclear Magnetic Resonance search for candidate biomarkers. Acta Biochim. Pol. 60, 701–706 [PubMed] [Google Scholar]
- 25. Resch A., Leicht S., Saric M., Pásztor L., Jakob A., Götz F., Nordheim A. (2006) Comparative proteome analysis of S. aureus biofilm and planktonic cells and correlation with transcriptome profiling. Proteomics 6, 1867–1877 [DOI] [PubMed] [Google Scholar]
- 26. Resch A., Rosenstein R., Nerz C., and Götz F. (2005) Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl. Environ. Microbiol. 71, 2663–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tan X., Qin N., Wu C., Sheng J., Yang R., Zheng B., Ma Z., Liu L., Peng X., and Jia A. (2015) Transcriptome analysis of the biofilm formed by methicillin-susceptible Staphylococcus aureus. Sci. Rep. 5, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seper A., Pressler K., Kariisa A., Haid A. G., Roier S., Leitner D. R., Reidl J., Tamayo R., and Schild S. (2014) Identification of genes induced in Vibrio cholerae in a dynamic biofilm system. Int. J. Med. Microbiol. 304, 749–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Herbert S., Ziebandt A. K., Ohlsen K., Schäfer T., Hecker M., Albrecht D., Novick R., and Götz F. (2010) Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infection Immunity 78, 2877–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gertz S., Engelmann S., Schmid R., Ohlsen K., Hacker J., and Hecker M. (1999) Regulation of SigmaB-dependent transcription of sigB and asp23 in two different S. aureus strains. Mol Gen Genet. 61, 558–566 [DOI] [PubMed] [Google Scholar]
- 31. Dörries K., and Lalk M. (2013) Metabolic footprint analysis uncovers strain specific overflow metabolism and D-isoleucine production of Staphylococcus Aureus COL and HG001. PLOS ONE 8, e81500–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dohnt K., Sauer M., Müller M., Atallah K., Weidemann M., Gronemeyer P., Rasch D., Tielen P., and Krull R. (2011) An in vitro urinary tract catheter system to investigate biofilm development in catheter-associated urinary tract infections. J. Microbiol. Methods 87, 302–308 [DOI] [PubMed] [Google Scholar]
- 33. Brady R. A., Leid J. G., Camper A. K., Costerton J. W., and Shirtliff M. E. (2006) Identification of Staphylococcus aureus proteins recognized by the antibody-mediated immune response to a biofilm infection. Infection Immunity 74, 3415–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Toyofuku M., Roschitzki B., Riedel K., and Eberl L. (2012) Identification of proteins associated with the Pseudomonas aeruginosa biofilm extracellular matrix. J. Proteome Res. 11, 4906–4915 [DOI] [PubMed] [Google Scholar]
- 35. Rice K. C., Mann E. E., Endres J. L., Weiss E. C., Cassat J. E., Smeltzer M. S., and Bayles K. W. (2007) The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 104, 8113–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bose J. L., Lehman M. K., Fey P. D., and Bayles K. W. (2012) Contribution of the Staphylococcus aureus Atl AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PLOS ONE 7, e42244–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Becher D., Hempel K., Sievers S., Zühlke D., Pané-Farré J., Otto A., Fuchs S., Albrecht D., Bernhardt J., Engelmann S., Völker U., van Dijl J. M., and Hecker M. (2009) A proteomic view of an important human pathogen - towards the quantification of the entire Staphylococcus aureus proteome. PLOS ONE 4, e8176–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zühlke D., Dörries K., Bernhardt J., Maaβ S., Muntel J., Liebscher V., Pané-Farré J., Riedel K., Lalk M., Völker U., Engelmann S., Becher D., Fuchs S., and Hecker M. (2016) Costs of life - Dynamics of the protein inventory of Staphylococcus aureus during anaerobiosis. Sci. Rep. 6, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bonn F., Bartel J., Büttner K., Hecker M., Otto A., and Becher D. (2014) Picking vanished proteins from the void: how to collect and ship/share extremely dilute proteins in a reproducible and highly efficient manner. Anal. Chem. 86, 7421–7427 [DOI] [PubMed] [Google Scholar]
- 40. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 41. Smith P. K., Krohn R. I., Hermanson G. T., and Mallia A. K. (1985) Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 42. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 43. Neuhoff V., Arold N., Taube D., and Ehrhardt W. (1988) Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9, 255–262 [DOI] [PubMed] [Google Scholar]
- 44. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 45. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 46. Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tyanova S., Temu T., and Cox J. (2016) The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protocols 11, 2301–2319 [DOI] [PubMed] [Google Scholar]
- 48. Bernhardt J., Funke S., Hecker M., and Siebourg J.. Visualizing gene expression data via Voronoi treemaps. 2009 Sixth International Symposium on Voronoi Diagrams, pp. 233–241, Copenhagen [Google Scholar]
- 49. Liebermeister W., Noor E., Flamholz A., Davidi D., Bernhardt J., and Milo R. (2014) Visual account of protein investment in cellular functions. Proc. Natl. Acad. Sci. U.S.A. 111, 8488–8493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Overbeek R. (2005) The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33, 5691–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Otto A., Bernhardt J. O. R., Meyer H., Schaffer M., Herbst F. A., Siebourg J., Mader U., Lalk M., Hecker M., and Becher D.. 2010. Systems-wide temporal proteomic profiling in glucose-starved Bacillus subtilis. Nat. Communications 1, 137–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yu N. Y., Wagner J. R., Laird M. R., Melli G., Rey S., Lo R., Dao P., Sahinalp S. C., Ester M., Foster L. J., Brinkman F. S. L. (2010) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fuchs S., Mehlan H., Bernhardt J., Hennig A., Michalik S., Surmann K., Pané-Farré J., Giese A., Weiss S., Backert L., Herbig A., Nieselt K., Hecker M., Völker U., and Mäder U. (2017) AureoWiki - The repository of the Staphylococcus aureus research and annotation community. Int. J. Med. Microbiol. 308, 558–568 [DOI] [PubMed] [Google Scholar]
- 54. Zhou M., Boekhorst J., Francke C., and Siezen R. J. (2008) LocateP: Genome-scale subcellular-location predictor for bacterial proteins. BMC Bioinformatics 9, 173–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Petersen T. N., Brunak S., Heijne von G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Rev. Microbiol. 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 56. Sternberg C., and Tolker-Nielsen T. (2006) Growing and analyzing biofilms in flow cells. Curr. Protoc. Microbiol. Chapter 1:Unit 1B.2–1B.2.15 [DOI] [PubMed] [Google Scholar]
- 57. Seidl K., Goerke C., Wolz C., Mack D., Berger-Bachi B., and Bischoff M. (2008) Staphylococcus aureus CcpA affects biofilm formation. infection and immunity. Infect. Immun. 76, 2044–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hill L., Veli N., and Coote P.J. (2014) Evaluation of Galleria mellonella larvae for measuring the efficacy and pharmacokinetics of antibiotic therapies against Pseudomonas aeruginosa infection. Int. J. Antimicrobial Agents 43, 254–261 [DOI] [PubMed] [Google Scholar]
- 59. Lauderdale K. J., Boles B. R., Cheung A. L., and Horswill A. R. (2009) Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infection Immunity 77, 1623–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stoodley P., Sauer K., Davies D. G., and Costerton J. W. (2002) Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56, 187–209 [DOI] [PubMed] [Google Scholar]
- 61. Spoering A. L., and Lewis K. (2001) Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183, 6746–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Waite R. D., Papakonstantinopoulou A., Littler E., and Curtis M. A. (2005) Transcriptome analysis of Pseudomonas aeruginosa growth: comparison of gene expression in planktonic cultures and developing and mature biofilms. J. Bacteriol. 187, 6571–6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Folsom J. P., Richards L., Pitts B., Roe F., Ehrlich G. D., Parker A., Mazurie A., and Stewart P. S. (2010) Physiology of Pseudomonas aeruginosa in biofilms as revealed by transcriptome analysis. BMC Microbiol. 10, 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Boles B. R., and Horswill A. R. (2008) agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4, e1000052–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Egeter O., and Bruckner R. (1996) Catabolite repression mediated by the catabolite control protein CcpA in Staphylococcus xylosus. Mol. Microbiol. 21, 739–749 [DOI] [PubMed] [Google Scholar]
- 66. Geiger T., and Wolz C. (2014) Intersection of the stringent response and the CodY regulon in low GC Gram-positive bacteria. Int. J. Med. Microbiol. 304, 150–155 [DOI] [PubMed] [Google Scholar]
- 67. Soutourina O., Poupel O., Coppée J. Y., Danchin A., Msadek T., and Martin-Verstraete I. (2009) CymR, the master regulator of cysteine metabolism in Staphylococcus aureus, controls host sulphur source utilization and plays a role in biofilm formation. Mol. Microbiol. 73, 194–211 [DOI] [PubMed] [Google Scholar]
- 68. Prigent-Combaret C., Vidal O., Dorel C., and Lejeune P. (1999) Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181, 5993–6002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Borriello G., Werner E., Roe F., Kim A. M., Ehrlich G. D., and Stewart P. S. (2004) Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrobial Agents Chemother. 48, 2659–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sønderholm M., Bjarnsholt T., Alhede M., Kolpen M., Jensen P., Kühl M., and Kragh K. (2017) The consequences of being in an infectious biofilm: microenvironmental conditions governing antibiotic tolerance. IJMS 18, 2688–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kinkel T. L., Roux C. M., Dunman P. M., and Fang F. C. (2013) The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia. mBio 4, e00696–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Foulston L., Elsholz A. K. W., DeFrancesco A. S., and Losick R. (2014) The extracellular matrix of Staphylococcus aureus biofilms comprises cytoplasmic proteins that associate with the cell surface in response to decreasing pH. Mbio 5, e01667–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dengler V., Foulston L., DeFrancesco A. S., and Losick R. (2015) An electrostatic net model for the role of extracellular DNA in biofilm formation by Staphylococcus aureus. J. Bacteriol. 197, 3779–3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Xu Y., Maltesen R. G., Larsen L. H., Schønheyder H. C., Le V. Q., Nielsen J. L., Nielsen P. H., Thomsen T. R., and Nielsen K. L. (2016) In vivo gene expression in a Staphylococcus aureus prosthetic joint infection characterized by RNA sequencing and metabolomics: a pilot study. BMC Microbiol. 16, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Li Y. H., Chen Y. Y. M., and Burne R. A. (2000) Regulation of urease gene expression by Streptococcus salivarius growing in biofilms. Environmental Microbiol. 2, 169–177 [DOI] [PubMed] [Google Scholar]
- 76. Lassek C., Burghartz M., Chaves-Moreno D., Otto A., Hentschker C., Fuchs S., Bernhardt J., Jauregui R., Neubauer R., Becher D., Pieper D. H., Jahn M., Jahn D., and Riedel K. (2015) A metaproteomics approach to elucidate host and pathogen protein expression during catheter-associated urinary tract infections (CAUTIs). Mol. Cell. Proteomics 14, 989–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ebner P., Prax M., Nega M., Koch I., Dube L., Yu W., Rinker J., Popella P., Flötenmeyer M., and Götz F. (2015) Excretion of cytoplasmic proteins (ECP) in Staphylococcus aureus. Mol. Microbiol. 97, 775–789 [DOI] [PubMed] [Google Scholar]
- 78. Ebner P., Rinker J., Nguyen M. T., Popella P., Nega M., Luqman A., Schittek B., Di Marco M., Stevanovic S., and Götz F. (2016) Excreted cytoplasmic proteins contribute to pathogenicity in Staphylococcus aureus. Infection Immunity 84, 1672–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Houston P., Rowe S. E., Pozzi C., Waters E. M., and O'Gara J.P. (2011) Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infection Immunity 79, 1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Mashruwala A.A., van de Guchte A., and Boyd J.M. (2017) Impaired respiration elicits SrrAB-dependent programmed cell lysis and biofilm formation in Staphylococcus aureus. eLife 6, e23845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Pásztor L., Ziebandt A. K., Nega M., Schlag M., Haase S., Franz-Wachtel M., Madlung J., Nordheim A., Heinrichs D. E., and Götz F. (2010) Staphylococcal major autolysin (Atl) is involved in excretion of cytoplasmic proteins. J. Biol. Chem. 285, 36794–36803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dinges M. M., Orwin P. M., and Schlievert P. M. (2000) Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 13, 16–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Peschel A., and Otto M. (2013) Phenol-soluble modulins and staphylococcal infection. Nat. Rev. Microbiol. 11, 667–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Alonzo F., and Torres V. J. (2014) The bicomponent pore-forming leucocidins of Staphylococcus aureus. Microbiol. Mol. Biol. Rev. 78, 199–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Nguyen M. T., Luqman A., Bitschar K., Hertlein T., Dick J., Ohlsen K., Bröker B., Schittek B., and Götz F. (2017) Staphylococcal (phospho)lipases promote biofilm formation and host cell invasion. Int. J. Med. Microbio. 308, 1–11 [DOI] [PubMed] [Google Scholar]
- 86. Scherr T. D., Hanke M. L., Huang O., James D. B. A., Horswill A. R., Bayles K. W., Fey P. D., Torres V. J., and Kielian T. (2015) Staphylococcus aureus biofilms induce macrophage dysfunction through leukocidin AB and alpha-toxin. mBio 6, e01021-15-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Reijer den P. M., Haisma E. M., Lemmens-den Toom N. A., Willemse J., Koning R. A., Demmers J. A. A., Dekkers D. H. W., Rijkers E., Ghalbzouri El A., Nibbering P. H., and van Wamel W. (2016) Detection of alpha-toxin and other virulence factors in biofilms of Staphylococcus aureus on polystyrene and a human epidermal model. PLOS ONE 11, e0152544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lei M. G., Gupta R. K., and Lee C. Y. (2017) Proteomics of Staphylococcus aureus biofilm matrix in a rat model of orthopedic implant-associated infection. PLOS ONE 12, e0187981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Bayer M. G., Heinrichs J. H., and Cheung A. L. (1996) The molecular architecture of the sar locus in Staphylococcus aureus. J. Bacteriol. 178, 4563–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Sonohara R., Muramatsu N., Ohshima H., and Kondo T. (1995) Difference in surface-properties between Escherichia-coli and Staphylococcus-aureus as revealed by electrophoretic mobility measurements. Biophys. Chem. 55, 273–277 [DOI] [PubMed] [Google Scholar]
- 91. Nostro A., Cellini L., Di Giulio M., D'Arrigo M., Marino A., Blanco A. R., Favaloro A., Cutroneo G., and Bisignano G. (2012) Effect of alkaline pH on staphylococcal biofilm formation. APMIS 120, 733–742 [DOI] [PubMed] [Google Scholar]
- 92. Jennings L. K., Storek K. M., Ledvina H. E., Coulon C., Marmont L. S., Sadovskaya I., Secor P. R., Tseng B. S., Scian M., Filloux A., Wozniak D. J., Howell P. L., and Parsek M. R. (2015) Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosabiofilm matrix. Proc. Natl. Acad. Sci. U.S.A. 112, 11353–11358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Beenken K. E., Blevins J. S., and Smeltzer M. S. (2003) Mutation of sarA in Staphylococcus aureus limits biofilm formation. infection and immunity. Infect. Immun. 71, 4206–4211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. O'Neill E., Pozzi C., Houston P., Smyth D., Humphreys H., Robinson D. A., and O'Gara J. P.. 2007. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J. Clin. Microbiol. 45, 1379–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lauderdale K. J., Malone C. L., Boles B. R., Morcuende J., and Horswill A. R. (2009) Biofilm dispersal of community-associated methicillin-resistant Staphylococcus aureuson orthopedic implant material. J. Orthop. Res. 81, 55–61 [DOI] [PubMed] [Google Scholar]
- 96. Tsang L. H., Cassat J. E., Shaw L. N., Beenken K. E., and Smeltzer M. S. (2008) Factors Contributing to the Biofilm-Deficient Phenotype of Staphylococcus aureus sarA Mutants. PLOS ONE 3, e3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. O'Neill E., Pozzi C., Houston P., Humphreys H., Robinson D. A., Loughman A., Foster T. J., and O'Gara J. P. (2008) A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 190, 3835–3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Boles B. R., Thoendel M., Roth A. J., Horswill A. R. (2010) Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLOS ONE 5, e10146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Foster T.J., Geoghegan J.A., Ganesh V.K., and Höök M. (2014) Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12, 49–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Mann E. E., Rice K. C., Boles B. R., Endres J. L., Ranjit D., Chandramohan L., Tsang L. H., Smeltzer M. S., Horswill A. R., and Bayles K. W. (2009) Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLOS ONE 4, e5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Peschel A., Otto M., Jack R. W., Kalbacher H., Jung G., and Götz F. (1999) Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274, 8405–8410 [DOI] [PubMed] [Google Scholar]
- 102. Tsai M., Ohniwa R. L., Kato Y., Takeshita S. L., Ohta T., Saito S., Hayashi H., and Morikawa K. (2011) Staphylococcus aureus requires cardiolipin for survival under conditions of high salinity. BMC Microbiol. 11, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Gries C. M., Bose J. L., Nuxoll A. S., Fey P. D., and Bayles K. W. (2013) The Ktr potassium transport system in Staphylococcus aureus and its role in cell physiology, antimicrobial resistance and pathogenesis. Mol. Microbiol. 89, 760–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Price-Whelan A., Poon C. K., Benson M. A., Eidem T. T., Roux C. M., Boyd J. M., Dunman P. M., Torres V. J., and Krulwich T. A. (2013) Transcriptional profiling of Staphylococcus aureus during growth in 2 M NaCl leads to clarification of physiological roles for Kdp and Ktr K+ uptake systems. MBio 4, e00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Jonsson P., and Wadström T. (1984) Cell-surface hydrophobicity of Staphylococcus-aureus measured by the salt aggregation test (Sat). Curr. Microbiol. 10, 203–209 [Google Scholar]
- 106. Rachid S., Ohlsen K., Wallner U., Hacker J., Hecker M., and Ziebuhr W. (2000) Alternative transcription factor sigma(B) is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 182, 6824–6826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Seminara A., Angelini T. E., Wilking J. N., Vlamakis H., Ebrahim S., Kolter R., Weitz D. A., and Brenner M. P.. 2012. Osmotic spreading of Bacillus subtilis biofilms driven by an extracellular matrix. Proc. Natl. Acad. Sci. U.S.A. 109, 1116–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Yan J., Nadell C. D., Stone H. A., Wingreen N. S., and Bassler B. L. (2017) Extracellular-matrix-mediated osmotic pressure drives Vibrio cholerae biofilm expansion and cheater exclusion. Nat. Commun. 8, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Freeman Z. N., Dorus S., and Waterfield N. R. (2013) The KdpD/KdpE two-component system: integrating K+ homeostasis and virulence. PLoS Pathog. 9, e1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Xue T., You Y., Hong D., Sun H., and Sun B. (2011) The Staphylococcus aureus KdpDE two-component system couples extracellular K+ sensing and Agr signaling to infection programming. Infection Immunity 79, 2154–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Zhao L., Xue T., Shang F., Sun H., and Sun B. (2010) Staphylococcus aureus AI-2 quorum sensing associates with the KdpDE two-component system to regulate capsular polysaccharide synthesis and virulence. Infection Immunity 78, 3506–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Coelho L. R., Souza R. R., Ferreira F. A., Guimaraes M. A., Ferreira-Carvalho B. T., and Figueiredo A. M. S. (2008) agr RNAIII divergently regulates glucose-induced biofilm formation in clinical isolates of Staphylococcus aureus. Microbiology 154, 3480–3490 [DOI] [PubMed] [Google Scholar]
- 113. Queck S. Y., Jameson-Lee M., Villaruz A. E., Bach T. H. L., Khan B. A., Sturdevant D. E., Ricklefs S. M., Li M., and Otto M. (2008) RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32, 150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Said-Salim B., Dunman P. M., McAleese F. M., Macapagal D., Murphy E., McNamara P. J., Arvidson S., Foster T. J., Projan S. J., and Kreiswirth B. N. (2003) Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185, 610–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Majerczyk C. D., Sadykov M. R., Luong T. T., Lee C., Somerville G. A., and Sonenshein A. L. (2008) Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 190, 2257–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Mrak L. N., Zielinska A. K., Beenken K. E., Mrak I. N., Atwood D. N., Griffin L. M., Lee C. Y., and Smeltzer M. S. (2012) saeRS and sarA act synergistically to repress protease production and promote biofilm formation in Staphylococcus aureus. PLOS ONE 7, e38453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Rogasch K., Ruhmling V., Pané-Farré J., Hoper D., Weinberg C., Fuchs S., Schmudde M., Broker B.M., Wolz C., Hecker M., and Engelmann S. (2006) Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J. Bacteriol. 188, 7742–7758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Pragman A. A., Yarwood J. M., Tripp T. J., and Schlievert P. M. (2004) Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J. Bacteriol. 186, 2430–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Fournier B., and Hooper D. C. (2000) A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182, 3955–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Vizcaíno A., Côté R.G., Csordas A., Dianes J.A., Fabregat A., Foster J.M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Pérez-Riverol Y., Reisinger F., Ríos D., Wang R., and Hermjakob H. (2012) The proteomics identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MS raw data, MaxQuant output files and the used database were deposited to the ProteomeXchange consortium (http://proteomecentral.proteomexchange.org/cgi/GetDataset) via the PRIDE partner repository (120) with the data set identifier PXD011157.