Summary
The Mixed Lineage Leukemia gene (MLL) is altered in leukemia by chromosomal translocations to produce oncoproteins comprised of the MLL N-terminus fused to the C-terminus of a partner protein. Here, we used domain-focused CRISPR screening to identify ZFP64 as an essential transcription factor in MLL-rearranged leukemia. We show that the critical function of ZFP64 in leukemia is to maintain MLL expression via binding to the MLL promoter, which is the most enriched location of ZFP64 occupancy in the human genome. The specificity of ZFP64 for MLL is accounted for by an exceptional density of ZFP64 motifs embedded within the MLL promoter. These findings demonstrate how a sequence anomaly of an oncogene promoter can impose a transcriptional addiction in cancer.
Introduction
Chromosomal translocations are a common genetic aberration found in human cancer, which drive oncogenesis through the production of a fusion oncoprotein and/or by altering cis regulatory elements (Rabbitts, 1994). Both types of mechanisms act in a dominant manner to promote tumorigenesis, which inevitably render cancer cells addicted to the oncoprotein product of the rearranged locus. This phenomenon, widely known as ‘oncogene addiction’, has motivated efforts to target fusion oncoproteins as a therapeutic intervention and led to the approval of drugs targeting BCR-ABL and PML-RARA in leukemia (Weinstein, 2002). Despite these successes, the majority of recurrent chromosomal translocations found in human cancer are not clinically actionable, highlighting a need to expose vulnerabilities associated with specific gene rearrangements.
Translocations involving the Mixed Lineage Leukemia gene (MLL; also known as MLL1 or KMT2A) are found in ~5% of acute myeloid and lymphoid leukemias and are associated with high-risk patient subgroups (Krivtsov and Armstrong, 2007). MLL translocations result in the expression of a fusion oncoprotein that retains the N-terminus of MLL fused to the C-terminus of the translocation partner. While the wild-type MLL encodes a large chromatin regulator with several domain modules, the MLL fusion protein only contains a subset of DNA binding domains (e.g. a CXXC domain) located at the MLL N-terminus. Since MLL fusion proteins lack the normal effector domains of the wild-type protein, they instead depend on the C-terminal fusion partner to activate transcription. AF4, AF9, ENL, and ELL are the most common fusion partners of MLL in leukemia, with each being a different subunit of the SEC transcription elongation complex (Lin et al., 2010; Yokoyama et al., 2010). Hence, one function of the MLL fusion protein is to mislocalize the SEC to the normal target genes of wild-type MLL, resulting in an aberrant elevation in the rate of transcription elongation (Lin et al., 2010; Yokoyama et al., 2010). MLL fusion proteins also rely on interactions with MENIN and DOT1L to execute their transcriptional functions in leukemia (Bernt et al., 2011; Yokoyama et al., 2005). The key target genes of MLL fusion proteins include HOXA9, MEIS1, and MEF2C, which encode leukemogenic transcription factors (Armstrong et al., 2002; Krivtsov et al., 2006). Both MLLfusion and MLLWT proteins are expressed in MLL-rearranged leukemia, since the MLL rearrangements is heterozygous. However, discrepancies exist in the literature regarding the essentiality of MLLWT for leukemogenesis, which may reflect different genetic targeting strategies used in these studies (Cao et al., 2014; Chen et al., 2017; Mishra et al., 2014; Thiel et al., 2010). However, recent studies have shown that MLLWT is essential to support a leukemogenic transcriptional program in the NPM1-mutant and NUP98-rearranged subtypes of leukemia (Kuhn et al., 2016; Xu et al., 2016). In the current study, we sought to identify genetic vulnerabilities in MLL-rearranged leukemia.
Zinc Finger Protein 64 (ZFP64) is a putative transcriptional regulator containing eleven zinc fingers in a tandem configuration, which is a domain architecture common to many sequence-specific DNA binding proteins (Lambert et al., 2018). To date, only two published studies have investigated the molecular function of ZFP64 (Sakamoto et al., 2008; Wang et al., 2013). In one study, ZFP64 was shown to associate with Notch1 and regulate mesenchymal differentiation in the C2C12 myoblast cell line (Sakamoto et al., 2008). Another study identified an association between ZFP64 and NF-kB in macrophages, which promotes cytokine expression following Toll-like receptor activation (Wang et al., 2013). While both studies implicate ZFP64 as a transcriptional regulator, the sequence-specific DNA-binding and transcriptional effector functions of ZFP64 were left uncharacterized. In addition, we are unaware of any prior study demonstrating a role for ZFP64 in cancer.
Results
CRISPR screen identifies ZFP64 as an essential transcription factor in MLL-rearranged leukemia
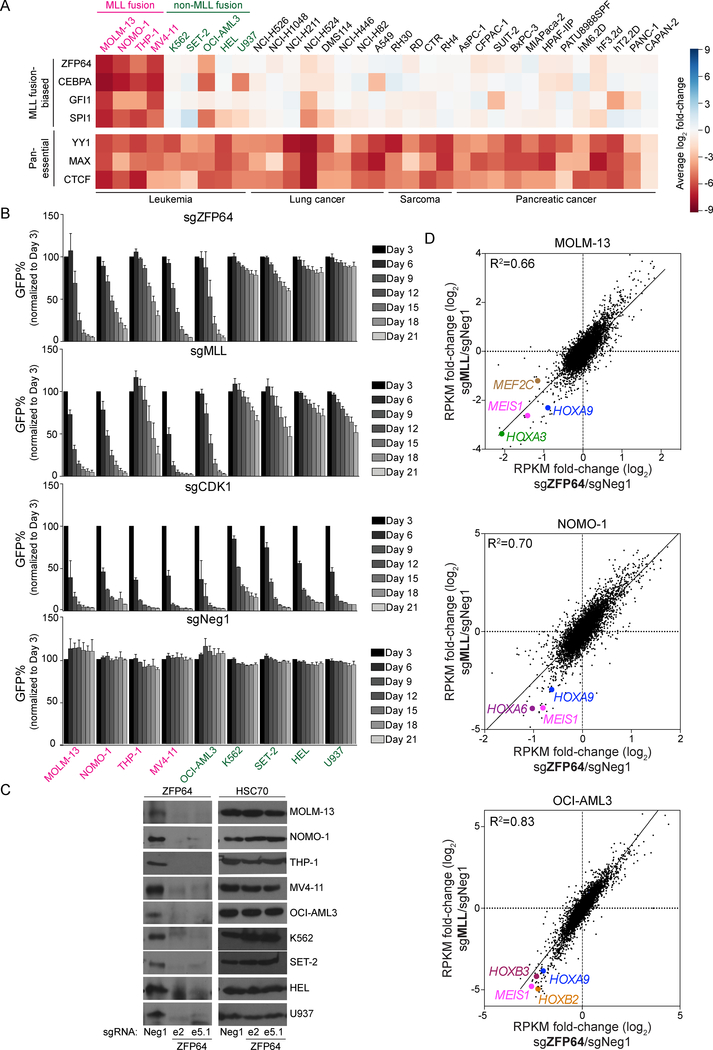
We previously described domain-focused CRISPR-Cas9 screening as a strategy for identifying essential transcriptional regulators in cancer (Shi et al., 2015). Here, we applied this method in an effort to expose transcription factor (TF) vulnerabilities in MLL-rearranged leukemia. For this purpose, we employed a pooled library of 8,658 sgRNAs targeting 1,427 DNA-binding domains of TFs and performed negative-selection ‘dropout’ screening in 33 human cancer cell lines representing leukemia, sarcoma, lung cancer, or pancreatic cancer. Among the nine leukemia cell lines included in the screen were four possessing an MLL translocation (Figure 1A and S1A). The relative impact of each sgRNA on cell growth was assessed via DNA sequencing-based quantification of sgRNA abundance over 14 population doublings. Spike-in positive and negative control sgRNAs included in the library validated the overall accuracy of this screening approach (Table S1). As expected, most of the TF dependencies identified in the screen were present in all 33 cell lines, such as YY1, CTCF, and MAX (Figure 1A and Table S1).
Figure 1. CRISPR dropout screen identifies ZFP64 as essential in MLL-rearranged leukemia.
(A) Heatmap depicts the log2 fold-change of sgRNA abundance (averaging each independent sgRNA targeting a gene) after 14 population doublings. The MLL fusion-biased hits were identified and ranked by subtracting the average of log2 fold-change of the four MLLfusion cell lines from average log2 fold-change of five MLLWT leukemia cell lines. (B) Competition-based proliferation assay of individual sgRNAs performed in the indicated Cas9-expressing cell lines. sgRNA expression is linked to a GFP reporter. Plotted is the GFP% cells (normalized to the day 3 measurement) at the indicated timepoints during culturing. A sgRNA targeting the kinase domain of CDK1 was included as a positive control. sgZFP64 and sgMLL data are the average of two independent sgRNAs. (n=3) (C) Western blotting of ZFP64 or HSC70 (loading control) in whole cell lysates prepared from the indicated Cas9+ leukemia cell lines transduced with the indicated sgRNAs. Lysates were prepared on day 5 post-infection. ‘e’ represents the exon targeted by the sgRNA. (D) RNA-seq scatterplot analysis comparing fold-change of mRNA levels following MLL and ZFP64 knockout in MOLM-13, NOMO-1, and OCI-AML3 cells. Log2 transformed fold-change were calculated based on the effects of two independent sgRNAs targeting ZFP64 or MLL compared to Neg1 negative control sgRNA. RPKM, Reads Per Kilobase of transcript per Million mapped reads. Known MLL target genes are highlighted. All bar graphs represent the mean ± SEM. See also Figure S1.
We ranked each TF based on its relative essentiality in MLLfusion versus MLLWT leukemia lines, which revealed ZFP64, CEBPA, GFI1, and SPI1 as the top candidates (Figure 1A). While CEBPA, GFI1, and SPI1 are known to play a role in MLL fusion leukemia (Maiques-Diaz et al., 2018; Ohlsson et al., 2014; Ye et al., 2015; Zhou et al., 2014), ZFP64 has not been previously studied in this disease. In our pooled screens, sgRNAs targeting ZFP64 led to a potent and selective growth-arrest phenotype in each of the four MLLfusion leukemia lines (MOLM-13, NOMO-1, THP-1, MV4–11), but also in the MLLWT leukemia line OCI-AML3, which harbors an NPM1 mutation. Notably, OCI-AML3 is known to be addicted to MLLWT (Kuhn et al., 2016). Hence, our CRISPR screening results led us to hypothesize that ZFP64 supports the function of both MLLfusion and MLLWT proteins in leukemia.
To validate the results of our pooled screens, we performed competition-based proliferation assays of individual ZFP64 sgRNAs in nine leukemia cell lines. As a control, we included sgRNAs targeting the CXXC domain of MLL, which are expected to inactivate both MLLWT and MLLfusion proteins (Ayton et al., 2004) (Figure S1B). Consistent with the results of our screen, targeting ZFP64 selectively impaired the growth of four MLLfusion leukemia lines and of OCI-AML3 cells in a pattern that correlated with MLL dependence across these cell lines (Figure 1B). In contrast, an sgRNA targeting CDK1 impaired proliferation in all nine leukemia lines with similar efficiency (Figure 1B). We validated that ZFP64 was essential for MOLM-13 growth in vivo (Figure S1C) and verified the on-target effect of ZFP64 sgRNAs by performing Western blotting and rescue experiments with a CRISPR-resistant ZFP64 cDNA (Figure 1C and S1D). ZFP64 expression was detected broadly across cancer cell lines and human tissues, which did not correlate with its requirement for growth (Figure 1C and S1E–G).
To evaluate a possible connection between ZFP64 and MLL as essential transcriptional regulators in leukemia, we performed RNA-seq to compare the global transcriptional changes incurred following ZFP64 or MLL inactivation. In these experiments, RNA was collected in sgRNA-transduced leukemia lines just prior to the onset of the growth arrest phenotype. This analysis revealed a striking global correlation of mRNA changes following ZFP64 and MLL perturbation in four MLLfusion leukemia cell lines and in OCI-AML3 cells (Figure 1D and Figure S1H). This included suppression of HOXA9, MEIS1, or MEF2C following ZFP64 or MLL knockout in each context (Figure 1D–F). We did not observe this same correlation when comparing the knockout of ZFP64 with knockout of SIK3 or RUNX1, which are also essential in MOLM-13 cells (Tarumoto et al., 2018) (Figure S1H). Targeting of ZFP64 or MLL in MOLM-13 cells also resulted in myeloid differentiation (Figure S1I). Together, these results suggest that ZFP64 and MLL are functionally linked with one another.
Epigenomic analysis of ZFP64 reveals outlier levels of enrichment at the MLL promoter
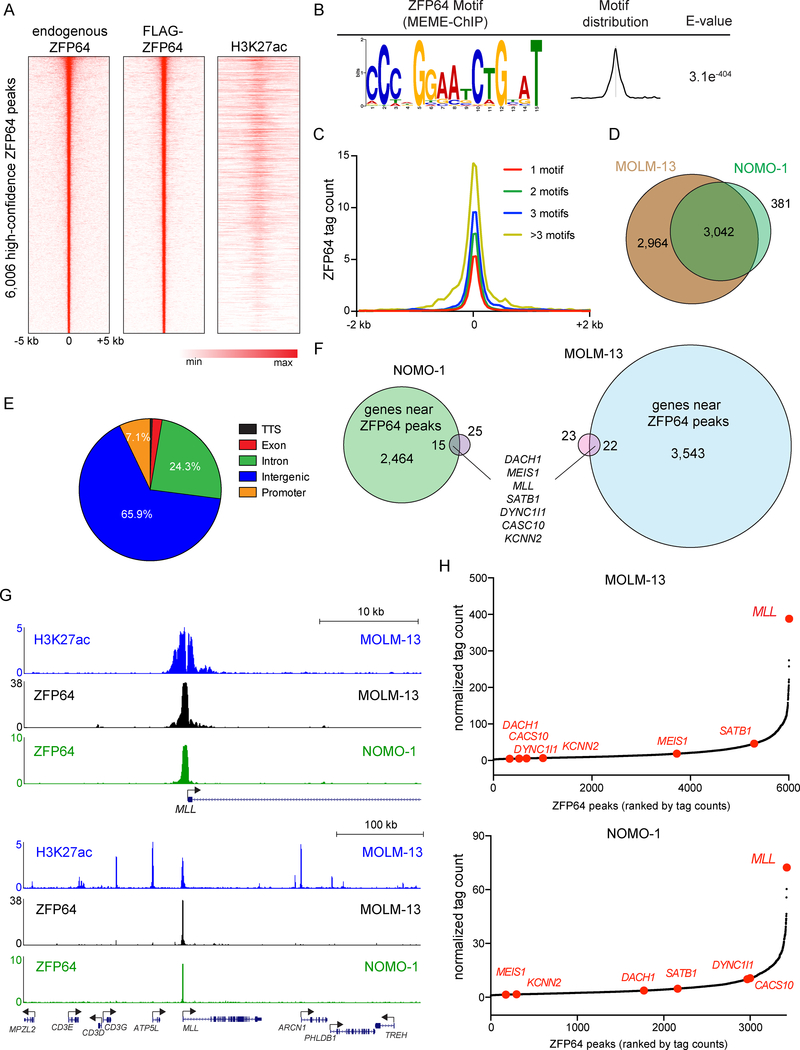
While prior studies have implicated ZFP64 as a transcriptional regulator (Sakamoto et al., 2008; Wang et al., 2013), the sequence-specificity of ZFP64 DNA binding has yet to be characterized. To address this, we performed chromatin immunoprecipitation of endogenous ZFP64 and FLAG-tagged ZFP64 in the MLLfusion leukemia cell line MOLM-13, followed by DNA sequencing (ChIP-seq) to define ZFP64-occupied regions. We observed a high concordance between endogenous ZFP64 and FLAG-ZFP64 occupancy, which allowed us to define 6,006 high-confidence binding sites in this cell type (Figure 2A and S2A).
Figure 2. Epigenomic analysis of ZFP64 in MLL-rearranged leukemia cells.
(A) Density plot showing endogenous ZFP64, FLAG-ZFP64, and H3K27ac enrichment surrounding the summit of 6,006 high confidence ZFP64 peaks in MOLM-13 cells, ranked by ZFP64 peak intensity. (B) ZFP64 ChIP-seq derived de novo motif logo, distribution and E-value of the ZFP64 binding motif derived using MEME-ChIP. Expectancy value (E-value) represents the enrichment of the motif around the center of the peak binding regions and was calculated using the binomial test. (C) Meta-profile comparing ZFP64 occupancy around the summit of peaks with different motif counts. ZFP64 binding intensity is shown as sequencing depth normalized tag count. (D) Overlap of high confidence ZFP64 peak regions between MOLM-13 and NOMO-1 cells. (E) Pie chart showing the distribution of 6,006 high confidence ZPF64 peaks in MOLM-13 cells. TTS, transcription termination site. (F) Venn diagram depicting the genes significantly down-regulated after knockout of ZFP64 in MOLM-13 (blue) and NOMO-1 (green) that are also located near high-confidence ZFP64 occupancy identified using ChIP-seq (pink). Down-regulated genes were defined by log2 fold change < −0.5; p<0.05; q<0.05 in two independent biological replicates of RNA-seq analysis. (G) Gene track of H3K27ac and ZFP64 ChIP-seq occupancy at the MLL locus at two different scales in the indicated leukemia cell lines. The x axis shows genomic position and y axis shows signal of ChIP-seq occupancy in units of reads per million mapped reads. (H) Ranking of ZFP64 binding intensity at high confidence peaks from ChIP-seq in MOLM-13 and NOMO-1 cells. ZFP64 target genes from (F) are indicated in red. See also Figure S2.
Using a de novo motif discovery analysis (Machanick and Bailey, 2011), we derived a 15 nucleotide sequence motif that closely correlated with ZFP64 occupancy (Figure 2B). Using an algorithm that predicts the DNA-binding specificity of zinc fingers based on primary amino acid sequence (Persikov and Singh, 2014), we deduced that the tandem zinc fingers 3 through 7 of ZFP64 were likely to be responsible for direct recognition of this motif (Figure S2B). Importantly, the levels of ZFP64 occupancy detected by ChIP-seq scaled with the density of this motif (Figure 2C). ChIP-seq analysis of ZFP64 in the MLLfusion cell line NOMO-1 revealed a similar pattern of genomic occupancy as observed in MOLM-13 (Figure 2D and Figure S2C–D). Consistent with a role as a transcriptional activator, we found that many ZFP64 binding sites overlapped with active chromatin marks histone H3 lysine 27 acetylation (H3K27ac) and H3K4 methylation (Figure 2A and S2E). Only 7.1% of ZFP64 binding sites were located at promoter regions, with the remainder found at distal locations, which likely represent enhancer elements (Figure 2E). Taken together, these findings demonstrate that ZFP64 is a sequence-specific DNA-binding TF.
Contrary to our expectations, we did not detect ZFP64 occupancy at the canonical MLL binding sites at the HOXA cluster (Guenther et al., 2008; Milne et al., 2005) (Figure S2F). In addition, our immunoprecipitation-mass spectrometry analysis of ZFP64-associated proteins failed to detect an interaction with the MLL protein (data not shown). These findings suggested that ZFP64 was not linked to MLL at the level of a protein-protein interaction. To investigate alternative mechanisms, we compared the ZFP64 ChIP-seq and ZFP64 knockout RNA-seq datasets to identify the core set of ZFP64 target genes in MLLfusion leukemia cells (Figure 2F). Using this approach, we identified seven direct ZFP64 target genes common to both MOLM-13 and NOMO-1 cell lines (Figure 2F).
Remarkably, one of the seven ZFP64 target genes we identified was MLL. We reasoned that maintenance of MLL expression by ZFP64 provided a plausible explanation for the selective essentiality of ZFP64 in MLLfusion leukemia lines and in the MLLWT-addicted OCI-AML3 cell line contexts, which share an addiction to endogenous MLL expression for their growth (Figure 1B). A selective role for ZFP64 in maintaining MLL expression could also explain the similar pattern of transcriptional changes observed after targeting ZFP64 and MLL (Figure 1D–F), since the timepoint of RNA collection would be expected to contain a mixture of primary and secondary effects on transcription.
A link between ZFP64 and regulation of MLL expression became even more compelling upon examination of ZFP64 binding at the MLL locus. A strong peak of ZFP64 occupancy was detected at the MLL promoter and this peak was an outlier as the most highly enriched location of ZFP64 occupancy in the genome of both MOLM-13 and NOMO-1 cells (Figure 2G–H). Notably, the enrichment of ZFP64 detected at the other six target genes identified above was significantly weaker (Figure 2H and S2G). Based on these observations, we investigated whether maintenance of MLL expression underlies the addiction to ZFP64 in leukemia.
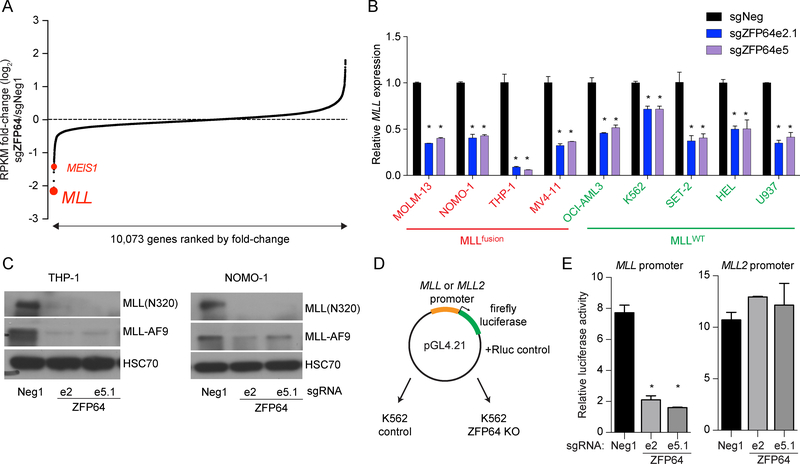
ZFP64 maintains expression of MLLfusion and MLLWT alleles via promoter regulation
Our RNA-seq analysis of five leukemia cell lines revealed that MLL expression was consistently decreased after ZFP64 knockout (Figure 3A and S3A). We confirmed this finding using RT-qPCR, which showed a significant decrease in MLL expression in both MLLfusion and MLLWT leukemia contexts (Figure 3B). RT-qPCR and Western blotting revealed that the protein levels of MLLWT and MLLfusion were reduced following ZFP64 inactivation (Figure 3C and S3B). We next cloned the MLL promoter into a luciferase reporter plasmid, which was transfected into control or ZFP64 knockout K562 cells (Figure 3D). Luciferase activity measurements confirmed that ZFP64 is required to activate the MLL promoter, but is dispensable to activate the promoter of MLL2/KMT2B, the closest homolog of MLL (Figure 3E). An analysis of RNA-seq data obtained from normal and malignant human hematopoietic cells revealed that ZFP64 and MLL are both expressed at comparable levels in leukemia and normal stem and progenitor cells, however both become down-regulated during terminal granulocyte differentiation (Figure S3C) (Bagger et al., 2016). In contrast, MLL2 becomes upregulated during this same transition (Figure S3C). Taken together, these results validate that ZFP64 selectively regulates MLL expression via its promoter element.
Figure 3. ZFP64 maintains MLL expression via promoter activation.
(A) RNA-seq analysis of ZFP64 knockout MOLM-13 cells. Genes are ranked by log2 fold-change in mRNA levels, calculated using two independent sgRNAs targeting ZFP64 compared to Neg1 negative control sgRNA. (B) Reverse transcriptase quantitative PCR (RT-qPCR) analysis of MLL mRNA level in the indicated leukemia cell lines after transduction of sgRNAs targeting ZFP64 compared to a control sgRNA. MLL primers were used to amplify a N-terminal region found on both the fusion and wild-type alleles. Relative mRNA levels for MLL were normalized to GAPDH levels. (n=3) (C) Western blot of MLLWT and MLL-AF9 in THP-1 and NOMO-1 cells transduced with the indicated sgRNAs. Whole cell extracts were prepared on day 5 post-infection with the indicated sgRNA. HSC70 was used as a loading control, ‘e’ represents the exon number. (D) Experimental strategy of luciferase reporter gene assay to measure promoter activity. (E) Results of luciferase reporter assays to measure promoter activity. MLL and MLL2 promoter constructs were transfected into K562 cells containing indicated sgRNAs. Cells were collected at 24 hours post-transfection followed by measurement of luciferase activity. Renilla luciferase activity was used as a normalization for transfection efficiency. All bar graphs represent the mean ± SEM. * denotes unpaired T test, p<0.05. See also Figure S3.
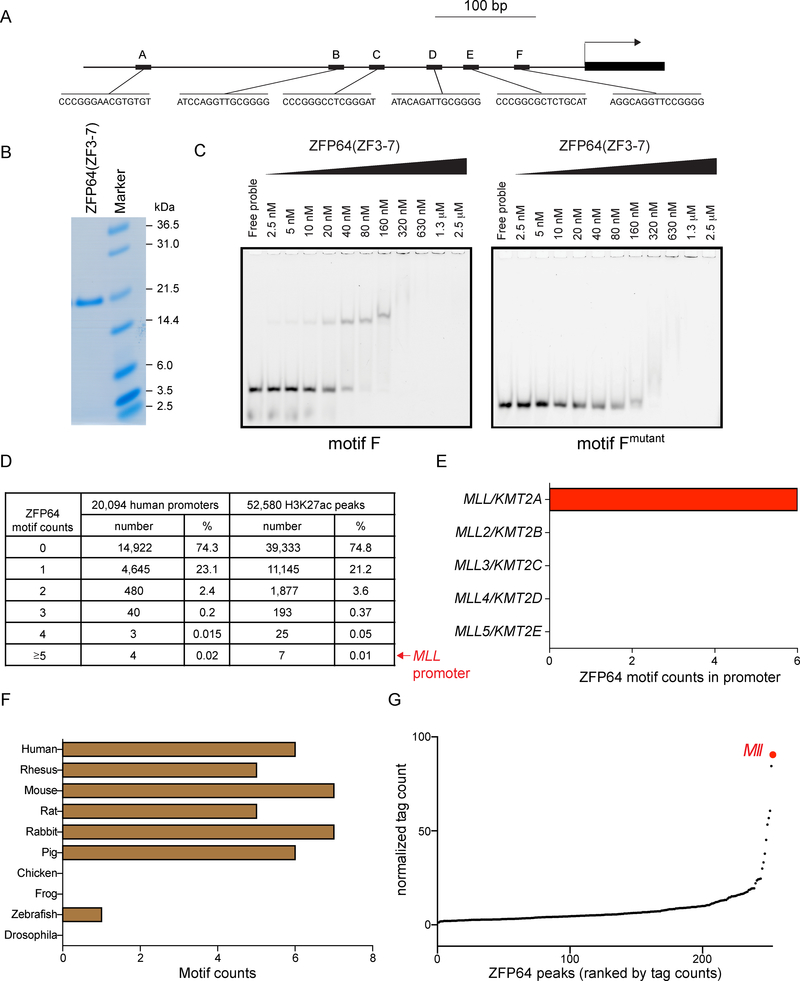
An exceptional density of ZFP64 motifs within the MLL promoter is conserved across mammalian species
We next pursued the mechanism underlying the outlier levels of ZFP64 enrichment observed at the MLL promoter. By analyzing the MLL promoter sequence using the FIMO tool (Grant et al., 2011), we discovered six matches to the ZFP64 binding motif located within a 500 bp interval upstream of the MLL transcription start site (Figure 4A). Using electromobility shift assays, we validated that a zinc finger 3–7 fragment of ZFP64 binds in a sequence-specific manner to all six motifs (Figure 4B–C and S4A). CRISPR tiling experiments suggest that motifs B, C, and F were the most critical for sustaining growth of MOLM-13 and OCI-AML3 cells (Figure S4B). We next analyzed the human genome to determine the frequency of finding a cluster of six ZFP64 motifs within such a short sequence interval. When considering 20,094 unique human promoter regions or 52,580 H3K27ac-enriched regions in MOLM-13 cells (representing active promoter and enhancer regions), the MLL promoter was consistently in the top 0.02% of genomic intervals with regard to its high density of ZFP64 motifs (Figure 4D). Of note, not every promoter with a high density of motifs is occupied by ZFP64, suggesting that other features might preclude ZFP64 occupancy at specific sites (Figure S4C). We did not detect any ZFP64 motifs within the promoters of the MLL homologs MLL2, MLL3, MLL4, or MLL5 (Figure 4E). While ZFP64 and MLL orthologs are present in a wide range of animal species (data not shown), the high ZFP64 motif density at the MLL promoter was only conserved in mammalian species, including mouse (Figure 4F and S4D). A ChIP-seq analysis of ZFP64 in mouse leukemia cells confirmed that the Mll promoter was also the most enriched location in this genome (Figure 4G). Taken together, these finding suggest that regulation of the MLL promoter by ZFP64 is encoded by an evolutionarily conserved homotypic motif cluster at this cis-regulatory element.
Figure 4. An exceptional density of ZFP64 motifs at the MLL promoter.
(A) Schematic of ZFP64 motif matches (p < 10−4 using FIMO) in the MLL promoter (500 bp upstream TSS). (B) Coomassie staining of purified Zinc finger 3 to 7 (ZF3–7) fragment of ZFP64. (C) Electromobility shift assay (EMSA) evaluating the affinity of ZFP64 (ZF3–7) protein with the ZFP64 motif F from the MLL promoter. Titration of ZF3–7 polypeptides were incubated with indicated fluorescein-labelled DNA probes. The DNA-protein complexes were separated on polyacrylamide gels. A mutant of motif F is used to show sequence specificity. Representative gel image is shown. (D) Summary of ZFP64 motif density frequency in the human genome. The ZFP64 motif frequencies were calculated in either all annotated human promoter regions (n=73,182) or in the 1 kb vicinity of H3K27ac enriched regions (n= 52,580) in MOLM-13 cell line. Considering the presence of multiple promoter sequences per gene we picked one representative promoter with the highest count per gene leaving n=20,094 unique gene promoters to calculate the frequencies. (E) Number of matches to the ZFP64 motif in promoter of each MLL family member. (F) ZFP64 motif counts in the MLL ortholog promoter of different species. (G) Ranking of ZFP64 binding intensity at high confidence regions from ChIP-seq in mouse Mll-AF9 knock-in leukemia cells. See also Figure S4.
As a control for the analysis above, we used FIMO to quantify the density of all 519 motifs in the JASPAR mammalian TF database at 20,094 human protein coding gene promoters in search of outlier densities at other human oncogenes (Figure S4E). In accord with prior findings (Gotea et al., 2010), this analysis revealed that outlier homotypic motif densities are quite common among human promoters and can be identified at several human oncogenes (Figure S4E–F). For example, the promoters of CDK4 and MET are in the top 0.1% of human promoters with regard to their density of motifs recognized by the transcription factors MITF and FOS, respectively, which are known to regulate these two oncogene loci (Mathas et al., 2002; Wellbrock et al., 2008) (Figure S4F). This analysis suggests that dense homotypic clusters of TF binding motifs are not entirely unique to MLL, but may have relevance at other loci.
ZFP64 employs an essential C-terminal transactivation domain to maintain MLL expression and leukemia cell growth
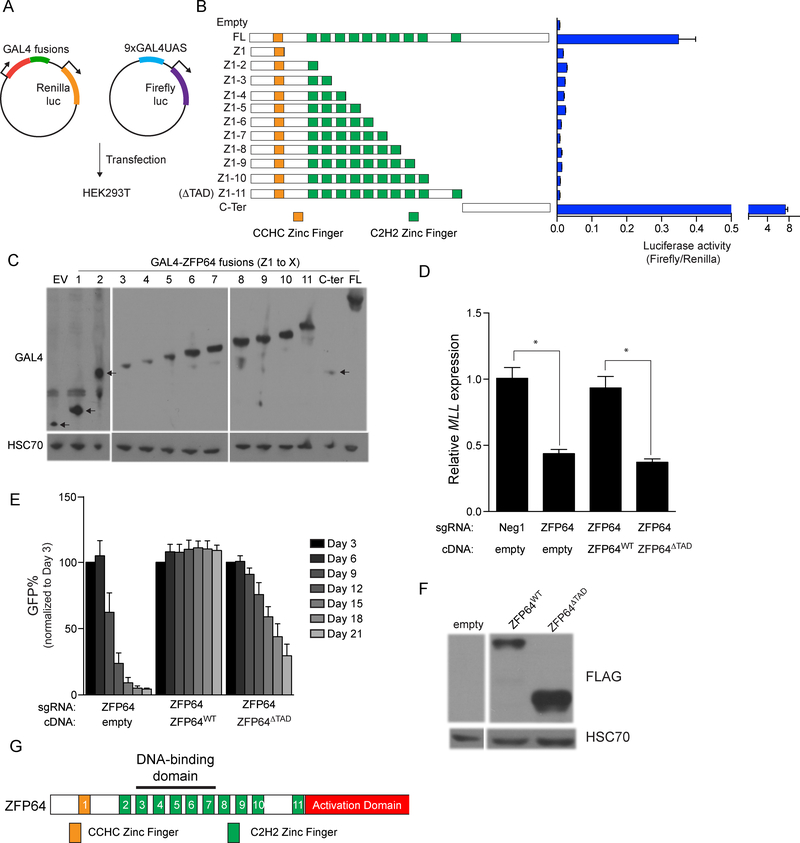
Having elucidated the DNA-binding specificity of ZFP64 and its link to the MLL promoter, we next sought to define the regions of ZFP64 involved in transcriptional activation. To this end, we cloned fragments of ZFP64 in-frame with the GAL4 DNA-binding domain and evaluated the ability of each protein to activate expression of a luciferase reporter harboring GAL4 motifs upstream of a minimal promoter (Figure 5A). Using this approach, we mapped the ZFP64 transactivation domain (TAD) to the C-terminal 229 amino acids, which was necessary and sufficient for full transcriptional activation (Figure 5B–C). Lentiviral transduction of various CRISPR-resistant cDNA constructs into ZFP64 knockout MOLM-13 cells verified that the TAD of ZFP64 is required to maintain MLL expression and to sustain leukemia growth (Figure 5D–G). These results further link activation of MLL expression to the essential function of ZFP64 in leukemia cells.
Figure 5. ZFP64 employs a C-terminal transactivation domain to maintain MLL expression and leukemia growth.
(A) Experimental strategy used to map the trans-activation domain (TAD) of ZFP64 using GAL4 fusion reporter assays. (B) A series of GAL4-ZFP64 fusion proteins tested in the luciferase assay. FL: full length, ZF: Zinc finger. Number Z1-Z11 describes the number of Zinc finger contained in the construct. Transactivation activity of different ZFP64 mutant by luciferase assay is plotted. Renilla luciferase internal control normalized. (n=3) (C) Western blot confirming detectable protein levels of GAL4-ZFP64 fusion proteins after transfected into 293T cells. (D) RT-qPCR analysis of MLL expression in MOLM-13 cells transduced with various combination of sgRNAs or CRISPR-resistant ZFP64 cDNA. Results were normalized to GAPDH. (n=3) (E) Competition-based proliferation assay in MOLM-13 after transduction with various combination of sgRNAs or CRISPR-resistant ZFP64 cDNA. (n=3) (F) Western blot confirming detectable protein levels of CRISPR resistant ZFP64 cDNA (ZFP64WT) and C-terminal deleted ZFP64 mutant (ZFP64ΔTAD) in MOLM-13. (G) Domain structure of ZFP64. All bar graphs represent the mean ± SEM. * denotes unpaired T test, p<0.05.
Activation of MLL expression is the essential function of ZFP64 in leukemia
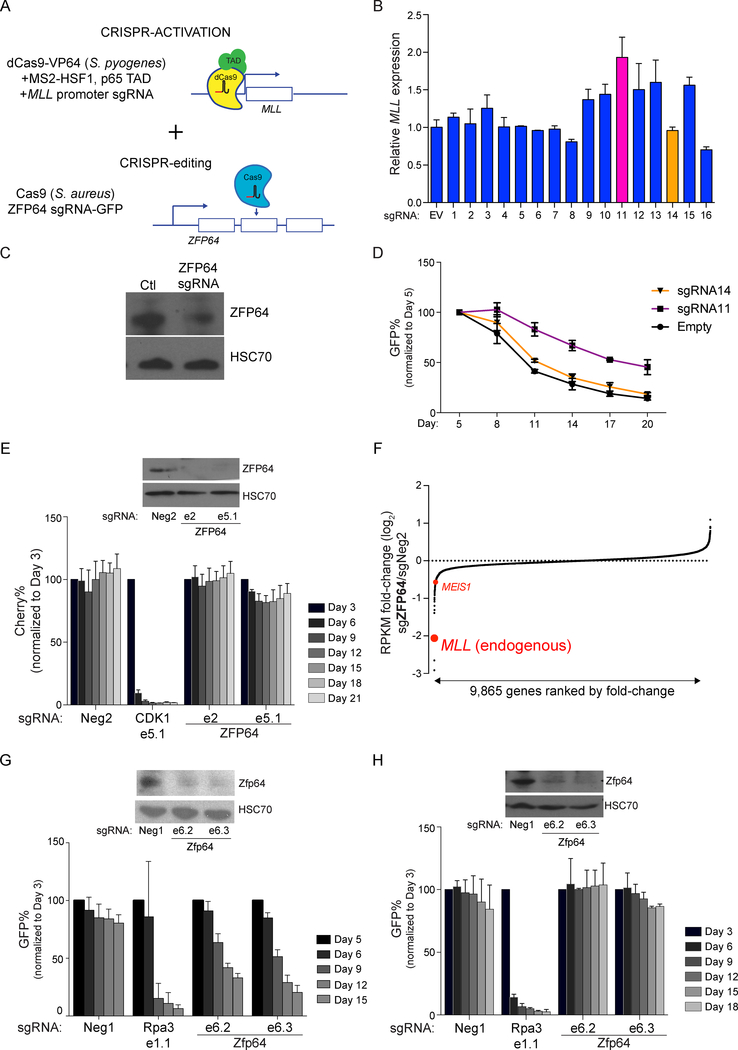
Having established that MLL is a direct target gene of ZFP64, we next investigated if this regulatory function accounts for the ZFP64 addiction in leukemia cells. We tested this hypothesis using a dual CRISPR-activation/mutagenesis approach, in which we tethered transcriptional activation domains to the endogenous MLL promoter. We reasoned that this might bypass the growth-arrest phenotype caused by inactivating ZFP64 (Figure 6A and S5A). We transduced MOLM-13 cells with a catalytically-dead S. pyogenes Cas9 linked to transcriptional activation domains, which allows for sgRNA-directed transcriptional activation of endogenous promoter regions (Konermann et al., 2015) (Figure S5A–B). We then tested 16 independent sgRNAs targeting the MLL promoter, and identified sgRNA#11 as the most effective, causing a two-fold increase in MLL expression (Figure 6B). These cells were then transduced with a lentiviral vector that co-expresses a catalytically-active S. aureus Cas9, a ZFP64-targeting S. aureus sgRNA, and a GFP reporter (Figure 6A, 6C, and S5B). In contrast to the control sgRNA#14 (that fails to activate MLL), expression of sgRNA#11 alleviated the cell growth defect observed following ZFP64 inactivation (Figure 6D). These findings suggest that the essential function of ZFP64 in leukemia is, at least in part, to activate MLL expression.
Figure 6. MLL promoter activation is the critical function of ZFP64 underlying its role as a dependency in MLL-rearranged leukemia.
(A) Experimental strategy for dual CRISPR-activation/mutagenesis. (B) RT-qPCR analysis of MLL after expressing the indicated sgRNAs targeting the MLL promoter in the CRISPR-activation experiment. (C) Western blotting of ZFP64 after transduction of sgRNAs targeting ZFP64 in MOLM-13 using S. aureus CRISPR indel mutagenesis. (D) Competition-based proliferation assay after SaCas9 based inactivation of ZFP64 and Sp_dCas9-based activation of MLL using sgRNA#11 or sgRNA#14. (n=3) (E) Western blotting and competition-based proliferation assay evaluating the effect of ZFP64 sgRNAs in human retroviral MLL-AF9/NrasG12D AML cells. sgRNA expression is linked to mCherry reporter in this experiment, since the cells were already GFP+. (n=3) (F) RNA-seq analysis of human retroviral MLL-AF9/NrasG12D AML cells after transduction with sgRNA targeting ZFP64. To evaluate endogenous MLL, a custom transcript was inserted into the analysis representing the C-terminal portion of the gene (absent from the fusion cDNA). (G) Western blotting and competition-based proliferation assay evaluating the effect of Zfp64 sgRNAs in mouse MLL-AF9 knock-in AML cells. (n=3) (H) Western blotting and competition-based proliferation assay evaluating the effect of Zfp64 sgRNAs in mouse retroviral MLL-AF9/NrasG12D AML cells (n=3). Neg2 is a negative control. All bar graphs represent the mean ± SEM. See also Figure S5.
Since the dual CRISPR strategy above resulted in only ~2-fold MLL activation and only partially bypassed the growth arrest caused by ZFP64 knockout, we devised an alternative approach for evaluating the relevance of the native MLL promoter to the essential function of ZFP64. Retroviral expression of the MLL-AF9 cDNA into normal hematopoietic cells, with or without cooperating oncogenes, is a widely used system for modeling acute myeloid leukemia (Milne, 2017). We reasoned that such models provide an ideal system to test our hypothesis, since expressing the MLLfusion from retroviral LTR promoter would be expected to bypass the endogenous MLL promoter to maintain oncogene expression. We obtained AML lines derived by transducing human cord blood derived CD34+ hematopoietic cells with retroviral vectors that expression MLL-AF9/NrasG12D or MLL-AF9/FLT3ITD cDNAs via a the LTR promoter and performed CRISPR-based targeting of ZFP64 (Wunderlich et al., 2010; Wunderlich et al., 2013). Remarkably, in both cell lines we did not detect any significant defect in cell growth, despite effective knockout of ZFP64 (Figure 6E and S5C–D). We performed ChIP-seq and RNA-seq in the human retroviral MLL-AF9/NrasG12D AML cells and confirmed the exceptional level of ZFP64 occupancy at the endogenous MLL promoter and verified the ZFP64 requirement to express endogenous MLLWT, which is dispensable in MLL fusion leukemia models (Chen et al., 2017) (Figure 6F and S5E).To further corroborate this finding, we evaluated the effect of Zfp64 knockout in a mouse leukemia model in which MLL-AF9/NrasG12D oncogenes are expressed via a retroviral LTR promoter (RN2 cells) and in a knock-in leukemia model in which MLL-AF9 is expressed from the endogenous Mll promoter (Corral et al., 1996). Consistent with the findings in human cells, we found that ZFP64 was essential for cell growth in the MLL-AF9 knock-in mouse leukemia cells, but dispensable in RN2 retroviral leukemia cells (Figure 6G–H and S5F). RNA-seq analysis in RN2 cells showed that the endogenous Mll, but not the MLL-AF9 cDNA, became suppressed following ZFP64 inactivation (Figure S5G). Taken together, these experiments suggest that the essential function of ZFP64 in MLL-rearranged leukemia is to maintain endogenous MLL expression.
Discussion
Cancer cells are addicted to driver oncogenes to maintain a transformed cell state (Weinstein, 2002). Therefore, it is to be expected that cancer cells will also be addicted to transcriptional machineries that sustain oncogene expression. Using a genetic screen, we have identified the zinc finger protein ZFP64 as a critical TF in MLL-rearranged leukemia. Our epigenomic characterization of ZFP64 in leukemia cells leads us to conclude that the vital function of ZFP64 in this context is to maintain the expression of the MLL oncogene. We have shown that six ZFP64 motifs within the promoter region place MLL uniquely under the control of ZFP64, a rare TF motif density among cis-regulatory DNA elements in the human genome. Our study highlights how sequence features of cis-regulatory elements at an oncogene locus can lead to an addiction to the transcriptional regulatory machinery in cancer.
While our genetic-rescue experiments suggest that MLL is the major target of ZFP64 underlying its role as a dependency in leukemia, we cannot entirely exclude the possibility that other target genes also contribute to its essential function. We detect more than 6,000 peaks of ZFP64 occupancy across the genome of MOLM-13 cells, however most of these regions only possess a single ZFP64 motif and exhibit markedly less ZFP64 enrichment than observed at MLL, as well as minimal changes in expression upon ZFP64 knockout. Based on our integrated ChIP-/RNA-seq analysis in two MLL-rearranged leukemia lines, we have identified six other transcriptional target genes under the control of ZFP64, including MEIS1 that encodes a leukemogenic TF (Wong et al., 2007). However, we only detect low levels of ZFP64 occupancy at the MEIS1 locus, and a modest change in MEIS1 expression upon ZFP64 knockout in the various cell lines examined throughout our study. Hence, it is unlikely that such a small effect on MEIS1 expression would be sufficient to influence the growth of MLLfusion leukemia cells. Nevertheless, important areas of future investigation will be to determine whether ZFP64 cooperates with other TFs/cofactors to carry out its transcriptional function in leukemia, how ZFP64 expression is regulated in hematopoietic cells, and to study the potential relevance of other ZFP64 target genes in other tissue contexts.
The evolutionary conservation of the high density of ZFP64 motifs at the MLL promoter suggests that this regulation has an important role in normal biological processes. The lack of ZFP64 motifs at the MLL homolog MLL2/KMT2B suggests that ZFP64 contributes to the specialization of these two paralogs by allowing for different patterns of expression. While a Zfp64 knockout mouse has yet to be reported, our findings suggest that such animals would phenocopy a loss of Mll, which would include homeotic malformations (Yu et al., 1995), defective fetal hematopoiesis and neurogenesis (Hess et al., 1997; Lim et al., 2009), as well as protection from leukemogenesis caused by expression of MLL fusion proteins from the endogenous Mll promoter (Corral et al., 1996).
A prevailing model of cis-element architecture at promoters and enhancers is the presence of diverse motifs within a short sequence interval, which enable cooperative functions among different TFs to specify a transcriptional state (Long et al., 2016; Spitz and Furlong, 2012). While this may reflect a general principle of most cis-regulatory elements, our study shows how homotypic clustering of TF motifs can create specialized promoter sequences with an augmented requirement for specific TFs for expression. In a prior study characterizing the transcription cofactor TRIM33, we identified a homotypic cluster of TF motifs as a mechanism that underlies the unique addiction of mouse lymphoblastic leukemia cells to this protein. In this study, we found that two enhancers in the mouse genome exhibit an exceptional density of PU.1 motifs, which are triggers for outlier levels of TRIM33 recruitment to cause repression of neighboring genes Bim and Atp1b3 (Wang et al., 2015a). In a recent study, it was found that DNA replication errors can produce tandem repeats to produce homotypic TF binding sites, which can promote the evolution of novel cis-regulatory elements (Balestrieri et al., 2018). Based on these findings, we propose that homotypic motif clustering as a general mechanism by which transcription factors and cofactors become endowed with high-precision regulatory functions during evolution.
While most transcriptional regulators occupy numerous sites across the genome, it has often been observed that the expression of cancer genes can be disproportionately sensitive to perturbations of the general transcriptional apparatus. Super-enhancers are one proposed mechanism underlying this phenomenon, in which clusters of enhancers with exceptional enrichment of transcriptional cofactors render nearby cancer genes hypersensitive to perturbation (Loven et al., 2013). While this model may apply to specific oncogenes (e.g. MYC), our study calls attention to an alternative mechanism by which oncogenes can become disproportionately suppressed by regulator perturbations. Since homotypic motif clusters are present at a significant fraction of cis-regulatory elements in the human genome (Gotea et al., 2010), this mechanism may have a broader significance as a source of transcriptional vulnerabilities in cancer and in other diseases.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Christopher Vakoc (vakoc@cshl.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse models
All animal procedures and studies were approved by the Cold Spring Harbor Laboratory Animal Care and Use committee in accordance to IACUC.
Cell lines
Leukemia cell lines MOLM-13, NOMO-1, THP-1, MV4–11, HEL, SET-2, U937, K562, RN2c, lung cancer cell lines NCI-H526, NCI-H211, NCI-H524, DMS114, NCI-H446, NCI-H82, pancreatic cell lines ASPC1, CFPAC-1, BXPC-3, MIAPACA-2, SUIT-2, HPAF-IIP, hM6.2D, hF3.2D, hT2.2D, PANC-1, CAPAN-2, sarcoma cell lines RH30 were cultured in RPMI 1640 supplemented with 10% FBS. NCI-H1048 was cultured in DMEM:F12 supplemented with 0.005 mg/mL Insulin, 0.01 mg/mL Transferrin, 30 nM Sodium selenite, 10 nM Hydrocortisone, 10 nM beta-estradiol, 4.5 mM L-glutamine and 5%FBS. OCI-AML3 was cultured in alpha-MEM with 20% FBS. Sarcoma cell lines RD, RH30, CTR, RH4, lung cancer cell line A549, HEK293T were cultured in DMEM supplemented with 10% FBS.
Mouse retroviral MLL-AF9/NrasG12D AML (RN2) cells were generated as described previously (Zuber et al., 2011). Briefly, fetal liver cells (HSPC) from mouse embryo D13.5-D15.5 were co-transduced by two retroviral vectors expressing MLL-AF9 and NrasG12D, followed by transplantation into irradiated recipient mice. Leukemia cells were harvested from moribund mice and cultured in RPMI with 10% FBS. Human retroviral MLL-AF9/NrasG12D AML cells were generated as described previously (Wei et al., 2008; Wunderlich et al., 2013). Briefly human cord blood CD34+ cells were transduced with a retroviral co-expressing MLL-AF9 and GFP followed by retroviral transduction with NrasG12D or FLT3ITD, and cultured in cytokine independent media. Human MLL-AF9/FLT3ITD and MLL-AF9/NrasG12D cells were cultured in IMDM with 20% FBS.
MLL-AF9 mouse AML cell lines were derived from MLL-AF9 knock-in mouse. Bone marrow cells from individual mice were plated in M3434 medium (Stem Cell Technologies, MethoCult GF M3434). A density of 10,000 cells/1ml medium was plated in each 35mm dish (Thermo Fisher, 174926). The primary cultured colonies and cells were counted and re-plating in M3434 medium, and re-plating occurred every 7 days. Three times after re-plating, 2,000 cells/1ml medium was plated in each 35mm dish for maintenance. Established clones were cultured in M3434 or IMDM medium supplemented with 10% FBS, 1×P/S, 10 ng/ml murine SCF, 10 ng/ml murine GM-CSF, 10 ng/murine IL-3 and 10 ng/ml murine IL-6 as previously described. All of the cell culture media was supplemented with Penicillin/streptomycin. All cell lines were cultured at 37°C with 5% CO2, and were periodically tested mycoplasma negative.
METHOD DETAILS
Plasmid construction and sgRNA cloning
All the Cas9-positive cancer cell lines in this study were derived by lentiviral transduction with a Cas9 expression vector (EFS-Cas9-P2A-Puro, Addgene: 108100). In this study, all the sgRNAs targeting human genes were cloned into either LRG2.1 (U6-sgRNA-GFP, Addgene: 108098) or LRCherry2.1 (U6-sgRNA-mCherry, Addgene: 108099) as indicated. All sgRNAs targeting mouse genes were cloned into LRG (Addgene:65656). Single sgRNAs were cloned by annealing two DNA oligos and T4 DNA ligation into a BsmB1-digested LRG2.1, LRCherry2.1, or LRG vector. To improve U6 promoter transcription efficiency, an additional 5’ G nucleotide was added to all sgRNA oligo designs that did not already start with a 5’ G.
For the dual CRISPR-based transcription activation and mutagenesis perturbation experiments, Streptococcus pyogenes (Sp) Cas9 system was used for transcription activation and Staphylococcus aureus (Sa) Cas9 system was used for target gene mutagenesis. The previous published three-vector CRISPR SAM system (dCAS-VP64_Blast, Addgene:61425; MS2-P65-HSF1_Hygro, Addgene: 61426; sgRNA(MS2)_zeo, Addgene:61427) was adapted for activation of the endogenous MLL gene (Konermann et al., 2015). Briefly, the hygro marker was replaced with a puromycin resistant gene and zeo marker was replaced with a neomycin resistant gene. To mutagenize ZFP64, an all-in-one SaCas9 and sgRNA expression vector was constructed (U6-sgRNA-EFS-SaCas9-P2A-GFP, will be available through Addgene). The SaCas9 cDNA and sgRNA cassette were derived from pX601 (Addgene: 61591).
For the cDNA overexpression experiments, a full length ZFP64 cDNA (DNASU, clone: HsCD00514448) was cloned into a lentiviral expression vector LentiV_Neo (Addgene: 108101). CRISPR sgRNA resistant synonymous mutations were introduced to ZFP64 cDNA by PCR mutagenesis. For the FLAG tag ChIP-seq experiment, ZFP64 cDNA was cloned into a pHAGE-puro vector (a gift from Zan Huang, Wuhan Univeristy, China) containing a 3X N terminal FLAG tag. For the domain mapping experiments, triple FLAG-tagged cDNA of ZFP64 was PCR amplified from pHAGE-puro vector, and then cloned into LentiV_neo. To make constructs without C-terminal transactivation domain (TAD) of ZFP64, Zinc finger domain ZF1~ZF11 was amplified from WT ZFP64 cDNA and cloned into LentiV neo vector with 3XFLAG tag fused to the C-terminal using In-Fusion following manufacturer’s instructions (Clontech: #638909), constructs will be made available through Addgene.
To map the ZFP64 transactivation domain, a series of ZFP64 deletion mutants were PCR amplified and inserted into plasmid pFN26 (BINB) hRluc neo Flexi ® (Promega #E1380) by In-Fusion method.
Sequences of ZFP64, MLL, and control sgRNAs are provided in Table S2.
Construction of domain-focused sgRNA pooled library
A gene list of transcription factors (TF) in the human genome was manually curated based on the presence of DNA binding domain(s). The TF DNA binding domain sequence information was retrieved from NCBI Conserved Domains Database. 6 sgRNAs were designed against individual DNA binding domains. The design principle of sgRNA was based on previous reports and the sgRNAs with the predicted high off-target effect were excluded (Hsu et al., 2013). All of the sgRNAs oligos including positive and negative control sgRNAs were synthesized in a pooled format (Twist Bioscience) and then amplified by PCR. PCR amplified products were cloned into BsmB1-digested LRG2.1 vector using Gibson Assembly kit (NEB#E2611). To verify the identity and relative representation of sgRNAs in the pooled plasmids, a deep-sequencing analysis was performed on a MiSeq instrument (Illumina) and confirmed that 100% of the designed sgRNAs were cloned in the LRG2.1 vector and the abundance of >95% of individual sgRNA constructs was within 5-fold of the mean (data not shown). This TF DNA binding domain CRISPR sgRNA pooled library will be available through Addgene.
Lentiviral transduction
Lentivirus was produced by transfecting HEK293T cells with helper plasmids (VSVG and psPAX2 (Addgene: #12260)) using Polyethylenimine (PEI 25000). Briefly, 10 μg of plasmid DNA, 5 μg of VSVG, 7.5 μg of psPAX2, and 40 μl of 1 mg/mL PEI were mixed, incubated and added to the 10 cm plate HEK293T cells, and media was replaced 6~8 hr post transfection, virus was collected 48 and 72 hr post transfection and pooled. Virus-containing supernatant was mixed with the indicated target cell lines supplied with 4 μg/mL polybrene, and then centrifuged at 1,700 rpm for 30 min at room temperature. Fresh media was changed 24 hr post-infection. Antibiotics (10 μg/mL blasticidin and/or 1 μg/mL puromycin and/or 1 mg/mL G418) were added 24 hr post-infection when selection was required.
Pooled CRISPR negative selection screen
CRISPR-based negative selection screenings were performed in various cancer cell lines with stable Cas9 expression (LentiV-Cas9-Puro vector, Addgene: 108100). Lentivirus of pooled sgRNA library targeting TF DNA binding domain were produced as described above. To ensure a single copy sgRNA transduction per cell, multiplicity of infection (MOI) was set to 0.3–0.4. To maintain the representation of sgRNAs during the screen, the number of sgRNA positive cells was kept at least 1000 times the sgRNA number in the library. Cells were harvested at day 3 post-infection and served as a reference representation of the pooled sgRNA library. Cells were cultured for 14 population doublings and harvested as the final time point. Genomic DNA was extracted using QIAamp DNA mini kit (QIAGEN) according to the manufacturer’s instructions.
To quantify the sgRNA abundance of reference and end time points, the sgRNA cassette was PCR amplified from genomic DNA using high-fidelity polymerase (Phusion master mix, catalog#F531S). The PCR product was end-repaired by T4 DNA polymerase (NEB B02025), DNA Polymerase I, Large (Klenow) Fragment (NEB M0210L), and T4 polynucleotide kinase (NEB M0201L). Next, a 3’ A-overhang was then added to the ends of blunted DNA fragments with Klenow Fragment (3’−5’ exo-) (NEB M0212L). The DNA fragments were ligated to diversity-increased custom barcodes with Quick ligation kit (NEB M2200L). Illumina paired-end sequencing adaptors were attached to the barcoded ligated products through PCR reaction with high-fidelity polymerase (Phusion master mix, catalog#F531S). The final product was quantified by Bioanalyzer Agilent DNA 1000 (Agilent 5067–1504) and pooled together in equal molar ratio and pair-end sequenced by using MiSeq (Illumina) with MiSeq Reagent Kit V3 150-cycle (Illumina).
The sequencing data was de-multiplexed and trimmed to contain only the sgRNA sequence cassettes. The read count of each individual sgRNA was calculated with no mismatches and compared to the sequence of reference sgRNA as described previously (Shi et al., 2015). Individual sgRNAs with the read count lower than 50 in the initial time point were discarded and remaining sgRNA counts normalized to total sample read counts. Average log2 fold change in abundance of all sgRNA against a given domain were calculated, as described previously (Wang et al., 2015b). MLLfusion specificity was determined by subtracting average of log2 fold-change in MLLfusion cell lines from average log2 fold-change in MLLWT leukemia cell lines. The DNA binding domain of transcription factor screen data from 33 cancer cell lines is provided in Table S1.
Competition-based assay to measure cell growth defects caused by sgRNAs
For the arrayed-format cellular proliferation competition assays, Cas9-expressing cell lines were infected with sgRNAs linked with either GFP or mCherry reporter (LRG, LRG2.1 or LRCherry2.1). The percentage of sgRNA positive (either GFP or mCherry positive) cell population was monitored over a time course using a Guava Easycyte HT instrument (Millipore). To assess the impact of individual sgRNA on cellular proliferation, final time point GFP% (or mCherry%) was divided by initial time point GFP% (or mCherry%) to calculate negatively selection fold-change of sgRNA positive cell populations.
FLAG-ZFP64 IP-MS analysis
100×106 HEK293T cell transfected with LentiV_neo or LentiV_neo_3XFLAG_ZFP64 were resuspended in 5 mL AFC buffer (10 mM TRIS-HCL, pH7.9, 420 mM NaCI, 0.1% NP40), supplemented with protease and phosphatase inhibitors. After 3 cycles of freeze-thaw steps (dry ice/ethanol and 37°C), cells were treated with 125 units of benzonase nuclease (Sigma, E1014) for 20 min at 4 °C with rotation. Cellular debris was removed by centrifugation at 13,000 rpm for 30 min at 4°C, followed by the addition of 3 volume of no salt AFC buffer. 100 μl of M2 beads (anti-flag, Sigma A2220) was added to the cell lysate and incubated at 4 °C for 3 hr. The lysate and beads were spin down at 2,000 rpm for 1 min at 4°C and washed 5 times with 1 mL low salt AFC buffer (10 mM TRIS-HCL, pH7.9, 150 mM NaCI, 0.1% NP40), Beads were pelleted down and resuspended in 100 μl low salt AFC with the addition of 3XFlag peptide (0.5 mg/mL, final conc), and then rotated 4°C for 2 hr. Aqueous phase was taken out and then precipitated using trichloroacetic acid (TCA), washed once with 10 % TCA and twice with ice acetone, and then followed by mass spectrometry analysis. All the mass spectrometry and data analysis were performed at Taplin Mass Spectrometry Facility at Harvard University.
Western blot
2×106 cells were resuspended in Lammli sample buffer (Biorad) containing 5% β-mercaptoethanol, and then denatured at 95 °C for 10 min. These whole cell extracts were loaded on NuPAGE 4–12% Bis-Tris protein Gels (Thermofisher) for series GAL4-ZFP64 mutant or on NuPAGE 3–8% Tris-Acetate protein Gels (Thermofisher) for detection of MLLWT or MLLfusion proteins. Running buffers were based on manufacturer instructions. Protein separation was electrophoresed for 150 V, 1.5 hr, and samples were transferred onto Nitrocellulose membrane using wet transfer at 90 V for 3 hr. Membrane was first blocked by 5% nonfat milk in TBST, and incubated with first antibody in 5% milk at 4 °C overnight. After incubation, membrane was washed for 3 times with TBST and followed by incubation with secondary antibody for 1 hr at room temperature, and then incubated with chemiluminescent HRP substrate (Thermofisher).
Chromatin immunoprecipitation (ChIP) assay
For ChIP experiments, 5×106 leukemia cells were cross-linked with 1% formaldehyde for 20 min at room temperature and quenched by the addition of 0.125 M glycine (final) for 10 min. Cells were pelleted and lysed with cell lysis buffer (10 mM Tris, pH8.0, 10 mM NaCl, 0.2% NP-40, protease inhibitor) at 4°C for 10 min, followed by centrifugation at 4,200 rpm for 30 sec and then lysed with 500 μL nulear lysis buffer (50 mM Tris, pH 8.0, 10 mM EDTA, 1% SDS, protease inhibitor) at 4°C for 10 min. Lysed cells were sonicated within nucluei lysis buffer using a Bioruptor (Diagenode) for 15 min at 4°C. Chromatin was pre-cleared by centrifugation at 14,000 rpm for 15 min at 4°C. Surpernatants containing chromatin were pre-incubated by adding 3.5 mL IP dilution buffer (20 mM Tris, pH 8.0, 2 mM EDTA, 150 mM NaCI, 1% Triton X-100, 0.01% SDS), 1.5 μg rabbit antibody (ZFP64, Proteintech, 17187–1-AP; FLAG, Sigma Aldrich F1048), and followed by addition of 20 μL Protein A or G magnetic beads (Invitrogen) for overnight incubation with rotation at 4°C.
The next day, immunocomplexes were spun down and washed once by IP Wash I buffer (20 mM Tris-CI pH 8.0, 2mM EDTA, 50 mM NaCI, 1% Triton X-100, 0.1% SDS), twice with High salt buffer (20 mM Tris-CI pH 8.0, 2 mM EDTA, 500 mM NaCI, 1% Triton X-100, 0.1% SDS), once with IP Wash II buffer (10 mM Tris-CI pH 8.0, 1 mM EDTA, 0.2 M LiCI, 1% NP-40, 1% Nadeoxycholate) and twice with TE pH 8.0. Immunocomplexes were then eluted by the addition of 200 μL elution buffer (50 mM Tris 8.0,10 mM EDTA, 1% SDS) at 65°C for 15 min with constant shaking. Eluted chromatins were reverse-crosslinked by the addition of RNase A (1μg/μL) and 0.25 M NaCI overnight at 65 °C within water bath and followed by the treatment of Proteinase K (0.2 mg/mL) for 2 hr at 42 °C and purification with QIAquick PCR purification kit within 60 μL distilled water.
ChIP-seq library preparation
ChIP-seq library construction was based on the manufacturer instruction (Illumina). Briefly, 50 μL ChIP DNA was first end-repaired, A-tailed and adaptor ligated to different barcodes. Agarose-gel purification was used to size-select (250 – 300 bp) adaptor-ligated ChIP-DNA, and followed by 15 cycles of PCR amplification. AMPure XP bead (Invitrogen) was used to clean-up amplified DNA. Quality of ChIP-seq library was checked by Bioanalyzer using High Sensitivity chip (Agilent). Equal molar ratio ChIP-seq library was mixed together and sequenced with Illumina NextSeq platform with single-end reads of 75 bases.
RNA-seq Library Preparation
For RNA-Seq, total RNA was extracted by using TRIzol reagent according to the standard protocol. Briefly, 1~5×106 leukemia cells were lysed in 1 mL TRIzol. For MOLM-13, and MV4–11 cells, samples were collected 4 days post infection of sgRNA lentivirus targeting ZFP64 and MLL. For NOMO-1 cells, samples for sgZFP64 were colleted 6 days post infection, and 4 days post infection of sgMLL. For THP1 cells, samples for sgZFP64 were collected 11 days post infection and 9 days post infection of sgMLL. For OCI-AML3 cells, samples were collected 9 days post infection of lentivirus targeting ZFP64 and MLL. Lysed samples were followed by the addition of chloroform and incubated at room temperature for 5 min, and then centrifuged for 15 min at 4 degrees. Aqueous phase was removed and mixed together with an equal volume of isopropanol and incubated for 10 min. RNA was pelleted by centrifugation at 4 degree for 10 min at 10,000xg, followed by a wash with 75% EtOH and dissolved in DEPC-treated water. RNA-seq libraries were prepared with TruSeq Sample Prep Kit V2 (Illumina) following the manufacturer’s protocol by using 2 μg RNA. Briefly, 2 μg RNA was poly-A selected and fragmented with fragmentation enzyme. First and second strand cDNA were synthesized according the standard protocol, followed by end-repair and PCR amplification of standard library construction protocol. Before sequencing, the quality of the library was checked by Bioanalyzer (Agilent) and multiple libraries with different barcodes were pooled together and sequenced by using Illumina HiSeq 2500 or NextSeq platform with single-end reads of 50 bases.
RT-PCR
RNA extraction was performed by using TRIzol reagent following manufacture’s recommendation. 2μg RNA was treated with DNaseI and reverse transcribed into cDNA using qScript cDNA SuperMix, then followed by RT-PCR with PCR mater mix containing SYBR green on ABI 7900HT fast real-time PCR machine.
In vivo transplantation of MOLM-13 cells into NSG mice
MOLM-13-Cas9 cells were first transduced with a Luciferase expressing cassette in Lenti-luciferase-P2A-Neo (Addgene #105621, Xu et al. 2018) vector, followed by G-418 (1mg/ml) selection and then viral transduction with LRG2.1-sgRNA-GFP vectors targeting the ZFP64 gene and Negative control (Neg1). Three replicates were performed for each sgRNA. On day 3 post infection with the sgRNA, the infection rate was checked by the percentage of GFP positive cells, and all samples had over 90% infection rate. 0.5 million cells were injected intravenous into sublethally irradiated (2.5 Gy) NSG mice (Jax 005557). To detect the disease progression, mice were imaged with IVIS Spectrum system (Caliper Life Sciences) on Day 12 and 16 post injection as previously described (Xu et al. 2018).
For FACS analysis, the animals were sacrificed on day 16 post injection, and bone marrow was flushed and collected. A single cell suspension was prepared and the percentage of GFP positive cells was measured using a Guava Easycyte HT instrument (Millipore).
Surface maker staining and flow cytometry
MOLM-13 cells were infected with sgRNAs and collected at day 8 post infection. 1 million cells were collected and incubated with APC-conjugated c-Kit or Mac-1 antibodies in FACS buffer (5% FBS, 0.05 % NaN3 in PBS) for 30 min at 4 degrees in the dark. Cells were washed for 3 times with FACS buffer and then analyzed on BD LSR2 flow cytometer. Cell surface maker staining was evaluated in GFP+ cell populations.
CRISPR scanning of ZFP64 motifs in the MLL promoter
To functionally evaluate ZFP64 binding sites of the MLL promoter, a pooled sgRNA library was constructed tiling MLL promoter region, which covered all six ZFP64 binding motifs. All possible sgRNAs were designed based on the presence of a PAM sequence of NGG that is recognized by the S. pyogenes Cas9 protein. The sgRNAs were synthesized individually, annealed to complementary sgRNAs, pooled in equal molar ratios, and ligated into BsmB1-digested LRG2.1 vectors. Pooled CRISPR negative selection genetic screening was performed similarly to the above-described screening. Cas9+ MOLM-13 and OCI-AML3 cells were infected with the pooled sgRNA library at low MOI. Day 3 and Day 21 post-infection samples were collected for Miseq quantification of sgRNA representation. The total read counts were normalized between samples. To quantify the negative selection effect of individual ZFP64 binding motifs, sgRNAs, that cut within +/− 12 nucleotides window of ChIP-seq derived ZFP64 binding motif, were grouped together. There were around 7–12 sgRNAs against each ZFP64 binding motif. All sgRNA sequences used in this study are in the Table S3.
Luciferase assays
Genomic DNA sequence upstream TSS of MLL (−783 bp) and MLL2 (−752 bp) were PCR amplified from genomic DNA and ligated into the plasmid pGL4.21 digested with EcoRV and HindIII. 400 ng MLL- or MLL2- pGL4.21 plasmid and 10 ng pGL4.74 control plasmid were co-transfected into K562 cells by Lipofectamine 2000 in a 96 well plate. 24 hr post transfection, equal volume Dual-Glo luciferase buffer and Dual-Glo Stop&Glo buffer were added sequentially and relative luciferase activity was measured according to manufacturer’s instructions.
To measure transcriptional activation activity of ZFP64 mutant, GAL4 DNA-binding domain (DBD) fused ZFP64 mutant were co-transfected with pGL4.35[luc2P/9XGAL4UAS/Hygro] vetor (Promega, #E1370) into HEK293T cells for 24 hr. Luciferase activity was measured as described above. Firefly luciferase activity was normalized to internal Renilla luciferase activity.
ChIP-Seq and RNA-Seq data analysis
For RNA-Seq analysis, Tophat2 software (Kim et al., 2013) was used to map raw reads to human (hg38) or mouse (mm10) genome using sensitive settings. Mapped reads were then analyzed using Cuffdiff (Trapnell et al., 2013) identifying differentially expressed coding genes, while structural RNAs were masked. Log2 fold-change was calculated from RPKM (reads per kilobase per million) of control (sgNeg) and biological replicates of sgRNAs targeting ZFP64 or MLL. During this step, genes with RPKM above 1 were considered expressed and were used for further analysis.
For ChIP-Seq analysis, raw reads were mapped to human (hg19) or mouse (mm10) genomes using Bowtie2 software (Langmead and Salzberg, 2012) using sensitive settings. Duplicate reads were removed prior to peak calling. Peaks were identified using MACS2 (Feng et al., 2012) software using 5% FDR cut off and broad peak option for histone- and Polymerase II or narrow peak option for transcription factor-ChIP-seq datasets. Histone modification marks and Pol II data were obtained from public GEO datasets, H3K27ac (GSM2944372), H3K4me1 (GSM2136939), H3K4me3 (GSM2136940), H3K9ac (GSM2136941), H3K79me2 (GSM1055774), Pol II (GSM2136943).
Sequencing depth normalized ChIP-seq pileup tracks were generated using the UCSC genome browser (Kent et al., 2002). Heatmaps density plot and metagene plots were generated using ±2 or 5 kb around the summit of the ZFP64 peaks with a binning size of 50 bps.
The high confidence peaks of ZFP64 were identified using the overlapping peaks of 5% FDR called peaks in two independent ChIP –seq datasets of each endogenous and FLAG-tagged ZFP64 ChIP-seq. Peaks with an enrichment below 10-fold or located at chrM were filtered out prior to overlapping. Overlapping peaks were identified using Bedtools intersect (Quinlan and Hall, 2010). AnnotatePeaks tool from HOMER suite was used to annotate peaks with the nearest expressed gene and to functional genomic features (Heinz et al., 2010). Normalized tag counts were calculated using the Bamliquidator package (https://github.com/BradnerLab/pipeline) without read extension.
ZFP64 de novo motif identification
For de novo motif identification, we performed endogenous ChIP using antibodies against ZFP64 followed by deep sequencing. The corresponding sequence of 400 bp around the summit of the top 1000 peaks with the highest ZFP64 occupancy was used for the MEME-ChIP software (Machanick and Bailey, 2011). The background model and shuffled sequences were generated using fasta-shuffle-letter with –kmer 2 option and fasta-get-markov –m 1 option. The motif width was limited to 30 bp and the number of generated motifs was limited to 100 and using the -anr option to allow any number of repetitions of motifs in the given sequences.
Transcription factor motif scanning
Genomic DNA and transcriptional start site annotation were downloaded from GENCODE: GRCh37 v19 for human (n=73,182), GRCm38 vM15 for mouse (n=51,884). Promoters were defined as 500 bp sequences upstream of annotated start sites of protein-coding transcripts. FIMO (Grant et al., 2011) was used to scan both strands of each promoter DNA sequence using a p-value cutoff of 10−4 and a background model matching the GC content of the region of the genome that was scanned. Analogous computations were performed for sequences defined by ChIP-Seq peaks. In case of H3K27ac peaks we used the ±500 bp sequence around the center of the bound region. For ZFP64 the high confidence peak regions were used. For the ortholog MLL promoter sequence analysis, 500 bp upstream the annotated TSS (Refseq or Ensemble) from the genomes hg38 (Human), rheMac8 (Rhesus), mm10 (MOUSE), rn6 (RAT), oryCun2 (RABBIT), susScr11 (PIG), galGal4 (CHICKEN), xenTro9 (XENOPUS), danRer10 (ZEBRAFISH), dm6 (DROSOPHILA) were extracted and analyzed for ZFP64 motif occurrences using FIMO as described above. In case of Drosophila, the TRX gene promoter was used.
We also scanned human promoter sequences using the non-redundant JASPER core vertebrate database (v2016) containing motifs of 519 transcription factors. In this analysis, the number of motif occurrences for each gene was quantified using only one promoter per gene (the one with the most putative sites). Cancer genes identified by (Lawrence et al., 2014) (n=260) were then investigated for exceptional motif count enrichment using a ranked promoter motif count list for each motif in the JASPAR database. Promoters for eight oncogenes are illustrated in Figure S4F. Overlapping motif were collapsed into one prior to generating the ranked count lists.
Our motif analysis was performed using custom Python scripts, which will be made publicly available at https://github.com/jbkinney/17_vakoc upon publication.
Recombinant protein expression and purification
E. coli Rosetta2 (DE3) pLysS cells transformed with ZFP64(ZF3–7)-pGEX6P plasmid were grown in LB media at 37 °C to an OD600 of ~0.5, then induced with isopropyl-β-D-thiogalactopyranoside and further grown at 25 °C for 18 h in the presence of 300 μM ZnSO4. Cell pellets were lysed in 50 mM Tris pH 7.5, 500 mM NaCl, 100 μM ZnCl2, 0.5 μM PMSF, 1 mM DTT, and the clarified lysate treated with protamine sulphate (1.5 mg mL−1) to remove contaminating nucleic acids. GST-ZFP64 (ZF3–7) was purified by affinity chromatography on Glutathione Sepharose® 4B resin (GE Healthcare Life Sciences), and eluted using 100 mM reduced glutathione in lysis buffer. The GST tag was cleaved using HRV-3C protease and ZFP64(ZF3–7) further purified by cation exchange on a UNO S1 column (Bio-Rad Laboratories) in 20 mM Tris pH 7.5, 1 mM DTT over a 50–1000 mM NaCl gradient. For EMSAs, ZFP64 (ZF3–7) was dialysed into 20 mM Tris pH 7.5, 150 mM NaCl, 1 mM DTT. Protein concentration was determined by UV absorbance at 280 nm.
Electrophoretic mobility shift assays
To construct motif F (wild-type and mutant), 5’ fluorescein-labelled (1WTfwd: TAGGCAGGTTCCGGGGCTT; 1MUTfwd: TAGGCAGGTTAAGTTTCTT) and unlabeled (1WTrev: AAGCCCCGGAACCTGCCTA; 1MUTrev: AAGAAACTTAACCTGCCTA) single-stranded DNA sequences were purchased as synthetic oligonucleotides from IDT (Integrated DNA Technologies). Double-stranded DNA probes for EMSAs were generated by mixing complementary oligonucleotides (fluorescein-labelled/unlabelled pair) in a 1:1 ratio, then heating at 95 °C for 5 min before annealing by slow cooling to room temperature. ZFP64 (ZF3–7) (0 –2.5 μM) was incubated with 20 nM dsDNA probe in gel shift buffer (10 mM MOPS pH 7, 50 mM KCl, 5 mM MgCl2, 10% glycerol [v/v]) on ice for 30 min, then electrophoresed on 8% polyacrylamide gels cast in 1× TB buffer (90 mM Tris, 90 mM boric acid, 5 mM MgCl2) at 250 V for 1.5 h in 0.5× TB buffer. Gels were imaged on a Typhoon FLA9000 scanner at 473 nm.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical significance was evaluated by p-value from unpaired Student t-test using Prism software.
DATA AND SOFTWARE AVAILABILITY
The ChIP-seq and RNA-seq data in this study is available in the Gene Expression Omnibus database https://www.ncbi.nlm.nih.gov/geo/ with accession number GSE115238.
Supplementary Material
Table S1, CRISPR screen data related to STAR Methods
Table S2, sgRNA and primer sequences related to STAR Methods
Table S3, sgRNA sequences related to STAR Methods
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-HSC70 (clone B-6) | Santa Cruz Biotechnology | Cat#Sc-7298;RRID:AB_637761 |
| Monoclonal ANTI-FALG® M2 antibody | Sigma-Aldrich | Cat# F1804, RRID:AB_262044 |
| Rabbit Anti-ZFP64 Polyclonal Antibody | Proteintech Group | Cat# 17187–1-AP, RRID:AB_2218826 |
| GAL4 (DBD) antibody | Santa Cruz Biotechnology | Cat# sc-577, RRID:AB_631554 |
| Anti-MLL1 (D2M7U) Rabbit Antibody | Cell Signaling Technology | Cat# 14689, RRID:AB_2688009 |
| Rabbit polyclonal anti-H3K27ac | Abcam | Cat# ab4729; RRID: AB_211829 |
| Donkey anti-rabbit IgG, HRP-linked | GE Healthcare | Cat# NA934; RRID: AB_772206 |
| Rabbit polyclonal anti-mouse IgG, HRP-linked | Dako | Cat#p0260; RRID: AB_2636929 |
| CD11b Monoclonal Antibody (M1/70), APC, | eBioscience | Cat#17-0112-81 |
| APC anti-human CD117 (c-kit) Antibody | Biolegend | Cat# 313205 |
| Bacterial and Virus Strains | ||
| E. coli Rosetta2 (DE3) pLysS | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Hexadimethrine bromide (polybrene) | Sigma-Aldrich | H9268 |
| Polyethylenimine, Linear, MW 25,000 (PEI 25000) | Polysciences | 23966-1 |
| Puromycin dihydrochloride | Sigma-Aldrich | P8833 |
| Geneticin Selective Antibiotic (G418 Sulfate) | Thermo Fisher Scientific | 10131035 |
| Agencourt AMPure XP | Beckman Coulter | A63880 |
| Dynabeads Protein A | Thermo Fisher Scientific | 10002D |
| Dynabeads Protein G | Thermo Fisher Scientific | 10004D |
| Formaldehyde, 37% solution | Avantor | 2106-01 |
| Glycine | Fisher Scientific | BP381-1 |
| Proteinase K | New England Biolabs | P8107S |
| Ribonuclease A from bovine pancreas | Sigma-Aldrich | R4875 |
| T4 DNA polymerase | New England Biolabs | M0203L |
| T4 polynucleotide kinase | New England Biolabs | M0201L |
| DNA Polymerase I, Large (Klenow) Fragment | New England Biolabs | M0210L |
| Agarose, Standard, Low Electroendosmosis (EEO) | New England Biolabs | M0212L |
| Agarose, Standard, Low Electroendosmosis (EEO) | Avantor | A426-07 |
| TRIzol Reagent | Thermo Fisher Scientific | 15596018 |
| SuperScript II Reverse Transcriptase | Thermo Fisher Scientific | 18064014 |
| Power SYBR Green Master Mix | Thermo Fisher Scientific | 4367659 |
| 2-Mercaptoethanol | Sigma-Aldrich | M6250 |
| 2XLaemmli Sample Buffer | Bio-Rad | 1610737 |
| Penicillin/Streptomycin | Thermo Fisher Scientific | 15140122 |
| 30% Acrylamide/Bis Solution, 37.5:1 | Bio-Rad | 1610158 |
| Critical Commercial Assays | ||
| MinElute Gel Extraction Kit | QIAGEN | 28604 |
| QIAquick PCR Purification Kit | QIAGEN | 28104 |
| TruSeq ChIP Sample Prep Kit | Illumina | IP-202-1012 |
| TruSeq RNA Sample Prep Kit v.2 | Illumina | RS-122-2001 |
| QIAamp DNA mini kit | QIAGEN | 51304 |
| In-Fusion HD Cloning Kit | Takara Bio | 638909 |
| Agilent High Sensitivity DNA Kit | Agilent | 5067-4626 |
| Dual-Glo® Luciferase Assay System | Promega | E2920 |
| Deposited Data | ||
| ChIP-seq and RNA-seq data | This study | GSE115238 |
| Human reference genome GRCh37/hg19 | Genome Reference Consortium | |
| Mouse reference genome GRCm38/mm10 | Genome Reference Consortium | |
| Experimental Models: Cell Lines | ||
| Human: MOLM-13 | DSMZ | ACC-554 |
| Human: NOMO-1 | DSMZ | ACC-542 |
| Human: THP-1 | ATCC | TIB-202 |
| Human: MV4-11 | ATCC | CRL-9591 |
| Human: K-562 | ATCC | CCL-243 |
| Human: SET-2 | DSMZ | ACC-608 |
| Human: OCI-AML3 | DSMZ | ACC-582 |
| Human: HEL | ATCC | TIB-180 |
| Human: U-937 | ATCC | CRL-1593.2 |
| Human: NCI-H526 | ATCC | CRL-5811 |
| Human: NCI-H1048 | ATCC | CRL-5853 |
| Human: NCI-H211 | ATCC | CRL-5824 |
| Human: NCI-H524 | ATCC | CRL-5831 |
| Human: DMS114 | ATCC | CRL-2066 |
| Human: NCI-H446 | ATCC | HTB-171 |
| Human: NCI-H82 | ATCC | HTB-175 |
| Human: A549 | ATCC | CCL-185 |
| Human: RH30 | DSMZ | ACC-489 |
| Human: RD | ATCC | CCL-136 |
| Human: CTR | Gift from Javed Khan | N/A |
| Human: RH4 | Gift from Javed Khan | N/A |
| Human: ASPC-1 | ATCC | CRL-1682 |
| Human: CFPAC-1 | ATCC | CRL-1918 |
| Human: SUIT-2 | JCRB | JCRB1094 |
| Human: BXPC-3 | ATCC | CRL-1687 |
| Human: MIAPACA-2 | ATCC | CRL-1420 |
| Human: HPAF-IIP | ATCC | CRL-1997 |
| Human: hM6.2D | Gift from David Tuveson | N/A |
| Human: hF3.2D | Gift from David Tuveson | N/A |
| Human: hT2.2D | Gift from David Tuveson | N/A |
| Human: PANC-1 | ATCC | CRL-1469 |
| Human: CAPAN-2 | ATCC | HTB-80 |
| Human: HEK293T | ATCC | CRL-3216 |
| Human: MLL-AF9/FLT3ITD | Wei et al., 2008 | N/A |
| Human: MLL-AF9/NrasG12D | Wei et al., 2008 | N/A |
| Mouse: Mll-AF9-KI | Gift from Gang Huang | N/A |
| Mouse: RN2 | Tarumoto et at., 2018 | N/A |
| Oligonucleotides | ||
| qPCR primers see Table S2 | This study | N/A |
| sgRNA sequence see Table S2 | This study | N/A |
| MLL promoter tiling sequence see Table S3 | This study | N/A |
| Recombinant DNA | ||
| LentiV_neo_empty | Tarumoto et at., 2018 | Addegene: 108101 |
| LentiV_neo_ZFP64_N_3XFLAG | This study | N/A |
| pHAGE_puro | This study | N/A |
| pHAGE-puro_ZFP64_N_3XFLAG | This study | N/A |
| LentiV_neo_ZFP64_CR | This study | N/A |
| LentiV_Cas9_puro | Tarumoto et at., 2018 | N/A |
| LRG(Lenti_sgRNA_EFS_GFP) | Tarumoto et at., 2018 | Addgene:65656 |
| LRG2.1T | Tarumoto et at., 2018 | Addgene:108098 |
| LRCherry2.1 | Tarumoto et at., 2018 | N/A |
| pGEX-6P | Gift from Joel P. Mackay | N/A |
| pGL4.21_MLL_promoter | This study | N/A |
| pGL4.21_MLL2_promoter | This study | N/A |
| Lenti_SaCRISPR_GFP | This study | N/A |
| Lenti_sgRNA_MS2_neo | This study | N/A |
| Lenti_dCas9_VP64_2A_Blast | This study | N/A |
| LentiV_ZFP64_ΔTAD | This study | N/A |
| Software and Algorithms | ||
| Tophat2 | Kim et al., 2013 | http://ccb.jhu.edu/software/tophat/index.shtml |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| MACS2 | Feng et al., 2012 | https://github.com/taoliu/MACS |
| BEDtools | Quinlan and Hall, 2010 | http://bedtools.readthedocs.io/en/latest/ |
| UCSC Genome Browser | UCSC | http://genome.ucsc.edu/ |
| HOMER v.4.9 | Heinz et al., 2010 | http://homer.ucsd.edu/homer/ |
| Cufflinks | Trapnell et al., 2013 | http://cole-trapnell-lab.github.io/cufflinks/cuffdiff/ |
| Others | ||
Highlights.
A CRISPR screen identifies ZFP64 as selectively essential in MLL leukemia
The essential function of ZFP64 in leukemia is to maintain MLL expression
The MLL promoter is the most enriched location of ZFP64 in the human genome
ZFP64 motif dominance underlies its exceptional specificity for MLL regulation
Significance.
Cancer cells are known to be hypersensitive to perturbations of the transcriptional regulatory machinery, an observation that has motivated the development of a class of therapies entering the clinic. However, the molecular mechanisms that underlie the addiction of cancer cells to specific transcriptional regulators remain poorly understood. In this study, we show how a homotypic cluster of transcription factor binding motifs within a single oncogene promoter can lead to a powerful addiction of leukemia cells to a transcription factor. This mechanism may have broader significance as a source of transcriptional vulnerabilities in cancer.
Acknowledgements
We would like to thank members of the Vakoc laboratory for helpful discussions and suggestions throughout the course of this study. We thank James C. Mulloy for retroviral human leukemia lines, the CSHL Cancer Center Shared Resources for deep-sequencing and bioinformatics supported by NCI Cancer Center Support grant 5P30CA045508. Additional funding for C.R.V was provided by the Forbeck Foundation, the Pershing Square Sohn Cancer Research Alliance, the Don Monti Foundation, National Institutes of Health grant NCI RO1 CA174793, NCI 5P01CA013106-Project 4, and a Leukemia & Lymphoma Society Scholar Award. Y.T. is supported by the Lauri Strauss Leukemia Foundation. J.P.M. is supported by a Senior Research Fellowship from the NHMRC. T.D.D.S. is supported by a grant from the State of New York (C150158).
Declaration of Interests
C.R.V. is an advisor to KSQ Therapeutics and receives research funding from Boehringer-Ingelheim.
References
- Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, and Korsmeyer SJ (2002). MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nature genetics 30, 41–47. [DOI] [PubMed] [Google Scholar]
- Ayton PM, Chen EH, and Cleary ML (2004). Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Molecular and cellular biology 24, 10470–10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagger FO, Sasivarevic D, Sohi SH, Laursen LG, Pundhir S, Sonderby CK, Winther O, Rapin N, and Porse BT (2016). BloodSpot: a database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic acids research 44, D917–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrieri C, Alfarano G, Milan M, Tosi V, Prosperini E, Nicoli P, Palamidessi A, Scita G, Diaferia GR, and Natoli G (2018). Co-optation of Tandem DNA Repeats for the Maintenance of Mesenchymal Identity. Cell. [DOI] [PubMed] [Google Scholar]
- Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, et al. (2011). MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer cell 20, 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F, Townsend EC, Karatas H, Xu J, Li L, Lee S, Liu L, Chen Y, Ouillette P, Zhu J, et al. (2014). Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Molecular cell 53, 247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Anastassiadis K, Kranz A, Stewart AF, Arndt K, Waskow C, Yokoyama A, Jones K, Neff T, Lee Y, et al. (2017). MLL2, Not MLL1, Plays a Major Role in Sustaining MLL-Rearranged Acute Myeloid Leukemia. Cancer cell 31, 755–770 e756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral J, Lavenir I, Impey H, Warren AJ, Forster A, Larson TA, Bell S, McKenzie AN, King G, and Rabbitts TH (1996). An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell 85, 853–861. [DOI] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, and Liu XS (2012). Identifying ChIP-seq enrichment using MACS. Nature protocols 7, 1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotea V, Visel A, Westlund JM, Nobrega MA, Pennacchio LA, and Ovcharenko I (2010). Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome research 20, 565–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, and Noble WS (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Lawton LN, Rozovskaia T, Frampton GM, Levine SS, Volkert TL, Croce CM, Nakamura T, Canaani E, and Young RA (2008). Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes & development 22, 3403–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess JL, Yu BD, Li B, Hanson R, and Korsmeyer SJ (1997). Defects in yolk sac hematopoiesis in Mll-null embryos. Blood 90, 1799–1806. [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nature biotechnology 31, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D (2002). The human genome browser at UCSC. Genome research 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. (2015). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, and Armstrong SA (2007). MLL translocations, histone modifications and leukaemia stem-cell development. Nature reviews Cancer 7, 823–833. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al. (2006). Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 442, 818–822. [DOI] [PubMed] [Google Scholar]
- Kuhn MW, Song E, Feng Z, Sinha A, Chen CW, Deshpande AJ, Cusan M, Farnoud N, Mupo A, Grove C, et al. (2016). Targeting Chromatin Regulators Inhibits Leukemogenic Gene Expression in NPM1 Mutant Leukemia. Cancer discovery 6, 1166–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, and Weirauch MT (2018). The Human Transcription Factors. Cell 172, 650–665. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nature methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, and Getz G (2014). Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, and Alvarez-Buylla A (2009). Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 458, 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, and Shilatifard A (2010). AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Molecular cell 37, 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Prescott SL, and Wysocka J (2016). Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell 167, 1170–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, and Young RA (2013). Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P, and Bailey TL (2011). MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiques-Diaz A, Spencer GJ, Lynch JT, Ciceri F, Williams EL, Amaral FMR, Wiseman DH, Harris WJ, Li Y, Sahoo S, et al. (2018). Enhancer Activation by Pharmacologic Displacement of LSD1 from GFI1 Induces Differentiation in Acute Myeloid Leukemia. Cell reports 22, 3641–3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathas S, Hinz M, Anagnostopoulos I, Krappmann D, Lietz A, Jundt F, Bommert K, Mechta-Grigoriou F, Stein H, Dorken B, et al. (2002). Aberrantly expressed c-Jun and JunB are a hallmark of Hodgkin lymphoma cells, stimulate proliferation and synergize with NF-kappa B. The EMBO journal 21, 4104–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA (2017). Mouse models of MLL leukemia: recapitulating the human disease. Blood 129, 2217–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Martin ME, Brock HW, Slany RK, and Hess JL (2005). Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer research 65, 11367–11374. [DOI] [PubMed] [Google Scholar]
- Mishra BP, Zaffuto KM, Artinger EL, Org T, Mikkola HK, Cheng C, Djabali M, and Ernst P (2014). The histone methyltransferase activity of MLL1 is dispensable for hematopoiesis and leukemogenesis. Cell reports 7, 1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson E, Hasemann MS, Willer A, Lauridsen FK, Rapin N, Jendholm J, and Porse BT (2014). Initiation of MLL-rearranged AML is dependent on C/EBPalpha. The Journal of experimental medicine 211, 5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persikov AV, and Singh M (2014). De novo prediction of DNA-binding specificities for Cys2His2 zinc finger proteins. Nucleic acids research 42, 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbitts TH (1994). Chromosomal translocations in human cancer. Nature 372, 143–149. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Tamamura Y, Katsube K, and Yamaguchi A (2008). Zfp64 participates in Notch signaling and regulates differentiation in mesenchymal cells. Journal of cell science 121, 1613–1623. [DOI] [PubMed] [Google Scholar]
- Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, and Vakoc CR (2015). Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nature biotechnology 33, 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, and Furlong EE (2012). Transcription factors: from enhancer binding to developmental control. Nature reviews Genetics 13, 613–626. [DOI] [PubMed] [Google Scholar]
- Tarumoto Y, Lu B, Somerville TDD, Huang YH, Milazzo JP, Wu XS, Klingbeil O, El Demerdash O, Shi J, and Vakoc CR (2018). LKB1, Salt-Inducible Kinases, and MEF2C Are Linked Dependencies in Acute Myeloid Leukemia. Molecular cell 69, 1017–1027 e1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel AT, Blessington P, Zou T, Feather D, Wu X, Yan J, Zhang H, Liu Z, Ernst P, Koretzky GA, et al. (2010). MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer cell 17, 148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, and Pachter L (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature biotechnology 31, 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Liu X, Liu Y, Zhang Q, Yao Z, Huang B, Zhang P, Li N, and Cao X (2013). Zinc finger protein 64 promotes Toll-like receptor-triggered proinflammatory and type I interferon production in macrophages by enhancing p65 subunit activation. The Journal of biological chemistry 288, 24600–24608. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang E, Kawaoka S, Roe JS, Shi J, Hohmann AF, Xu Y, Bhagwat AS, Suzuki Y, Kinney JB, and Vakoc CR (2015a). The transcriptional cofactor TRIM33 prevents apoptosis in B lymphoblastic leukemia by deactivating a single enhancer. eLife 4, e06377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, and Sabatini DM (2015b). Identification and characterization of essential genes in the human genome. Science 350, 1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Wunderlich M, Fox C, Alvarez S, Cigudosa JC, Wilhelm JS, Zheng Y, Cancelas JA, Gu Y, Jansen M, et al. (2008). Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia. Cancer cell 13, 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB (2002). Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science (New York, NY) 297, 63–64. [DOI] [PubMed] [Google Scholar]
- Wellbrock C, Rana S, Paterson H, Pickersgill H, Brummelkamp T, and Marais R (2008). Oncogenic BRAF regulates melanoma proliferation through the lineage specific factor MITF. PloS one 3, e2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong P, Iwasaki M, Somervaille TC, So CW, and Cleary ML (2007). Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes & development 21, 2762–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich M, Chou FS, Link KA, Mizukawa B, Perry RL, Carroll M, and Mulloy JC (2010). AML xenograft efficiency is significantly improved in NOD/SCID-IL2RG mice constitutively expressing human SCF, GM-CSF and IL-3. Leukemia 24, 1785–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich M, Mizukawa B, Chou FS, Sexton C, Shrestha M, Saunthararajah Y, and Mulloy JC (2013). AML cells are differentially sensitive to chemotherapy treatment in a human xenograft model. Blood 121, e90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Valerio DG, Eisold ME, Sinha A, Koche RP, Hu W, Chen CW, Chu SH, Brien GL, Park CY, et al. (2016). NUP98 Fusion Proteins Interact with the NSL and MLL1 Complexes to Drive Leukemogenesis. Cancer cell 30, 863–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M, Zhang H, Yang H, Koche R, Staber PB, Cusan M, Levantini E, Welner RS, Bach CS, Zhang J, et al. (2015). Hematopoietic Differentiation Is Required for Initiation of Acute Myeloid Leukemia. Cell stem cell 17, 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Lin M, Naresh A, Kitabayashi I, and Cleary ML (2010). A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer cell 17, 198–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, and Cleary ML (2005). The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell 123, 207–218. [DOI] [PubMed] [Google Scholar]
- Yu BD, Hess JL, Horning SE, Brown GA, and Korsmeyer SJ (1995). Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378, 505–508. [DOI] [PubMed] [Google Scholar]
- Zhou J, Wu J, Li B, Liu D, Yu J, Yan X, Zheng S, Wang J, Zhang L, Zhang L, et al. (2014). PU.1 is essential for MLL leukemia partially via crosstalk with the MEIS/HOX pathway. Leukemia 28, 1436–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, and Lowe SW (2011). Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nature biotechnology 29, 79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, CRISPR screen data related to STAR Methods
Table S2, sgRNA and primer sequences related to STAR Methods
Table S3, sgRNA sequences related to STAR Methods
Data Availability Statement
The ChIP-seq and RNA-seq data in this study is available in the Gene Expression Omnibus database https://www.ncbi.nlm.nih.gov/geo/ with accession number GSE115238.