Abstract
Interferon (IFN)-mediated antiviral responses are central to host defence against viral infection. Despite the existence of at least 20 IFNs, there are only three known cell surface receptors. IFN signalling and viral evasion mechanisms form an immensely complex network that differs across species. In this Review, we begin by highlighting some of the advances that have been made towards understanding the complexity of differential IFN signalling inputs and outputs that contribute to antiviral defences. Next, we explore some of the ways viruses can interfere with, or circumvent, these defences. Lastly, we address the largely under-reviewed impact of IFN signalling on host tropism, and we offer perspectives on the future of research into IFN signalling complexity and viral evasion across species.
Subject terms: Interferons, Viral immune evasion
This Review highlights some of the advances that have been made towards understanding the complexity of differential interferon (IFN) signalling inputs and outputs as well as some of the strategies viruses use to interfere with or circumvent IFN-induced antiviral responses.
Main
Across species, the innate immune response serves as the first line of defence against viral infection. Pattern recognition receptors (PRRs) within the host cell sense pathogen-associated molecular patterns, triggering a range of signalling cascades and subsequent gene activation1. For many viral infections, this results in the production and secretion of interferons (IFNs), which are critical for innate immunity and also have profound impacts on adaptive immune responses. IFN signalling in turn induces transcription of hundreds of IFN-stimulated genes (ISGs) that further the immune response against the viral infection.
Many viruses have evolved to avoid, subvert or directly interfere with the IFN signalling of their hosts. This is accomplished through numerous mechanisms extensively reviewed elsewhere2–7; however, some new discoveries are highlighted later in this Review. A given virus’ ability to antagonize and prevent the inputs and outputs of IFN signalling not only dictates the success of the infection, but is also a direct factor that contributes to the host and tissue range of a given virus.
Originally discovered in the 1950s and named for their ability to ‘interfere’ with viral replication, IFNs form a diverse family of cytokines8. Each class of IFN—type I, II and III— signals through a distinct heterodimeric receptor and induces gene expression through the Janus kinase–signal transducer and activator of transcription (JAK–STAT) signalling pathway (Fig. 1). In humans, type I IFNs are comprised of IFN-β, 13 subtypes of IFN-α, as well as IFN-ε and IFN-ω. A number of distantly related genes and pseudogenes have been identified in other species and are included in the type I classification. Three type III IFNs (IFN-λ1, IFN-λ2 and IFN-λ3) were simultaneously discovered by two groups in 2003 and shown to be structurally related to interleukin-10 (IL-10) (refs. 9,10). A fourth type III IFN, IFN-λ4, was identified in 2013 and was associated with impaired clearance of hepatitis C virus (HCV) infection11. The type II IFN class has only one member: IFN-γ. Given that type II fits the classic model of one ligand for one receptor, this Review will focus only on type I and III IFNs.
Fig. 1. Simplified schematic of type I and III IFN signalling cascades.
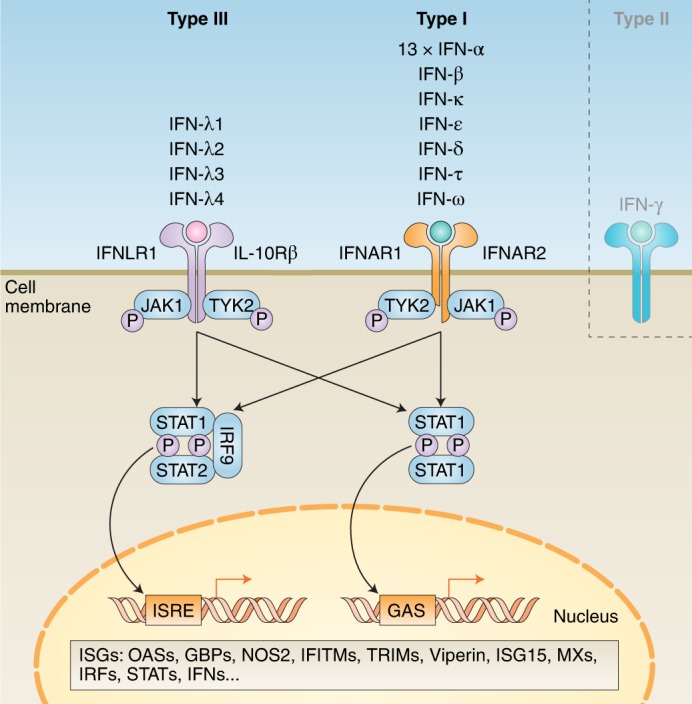
Within type I and III, there are multiple IFNs; within type II, there is only a single IFN. Each type has a distinct heterodimeric cell surface receptor: type I IFNs bind to the IFNAR receptor complex (composed of IFNAR1 and IFNAR2); type III IFNs bind to the IFNLR receptor complex (composed of INFLR1 and IL-10Rβ). Binding of an IFN to either receptor complex results in cross-phosphorylation of JAK1 and TYK2 on the cytoplasmic domains of the receptor subunits. This triggers phosphorylation of STAT1 and STAT2. Following phosphorylation, these STATs form various complexes that translocate into the nucleus, where they bind IFN-stimulated response elements (ISREs) or gamma-activated sequences (GASs) on the promoters of ISGs. Binding to these promoter elements results in the transcription of hundreds of genes involved in antiviral response, including ISGs, IFNs, IRFs and STATs. P, phosphate; OASs, oligoadenylate synthases; GBPs, guanylate-binding proteins; NOS2, nitric oxide synthase 2; IFITMs, IFN-induced transmembrane proteins; TRIMs, tripartate motif proteins.
IFNs vary considerably with regards to structure, receptor distribution and tissue-specific biological activities12, but are all capable of inducing an antiviral state. Type I IFN signalling within humans antagonizes innumerable viruses, such as influenza virus, West Nile virus (WNV), yellow fever virus (YFV), vesicular stomatitis virus (VSV), dengue virus (DENV), herpes simplex virus (HSV) and several hepatitis viruses13. As such, evading type I IFN signalling is crucial for viruses to successfully replicate within their hosts, as is demonstrated by many flaviviruses; Zika virus (ZIKV), Japanese encephalitis virus (JEV), WNV and DENV are all able to antagonize type I IFN signalling in human cells to promote replication of their genomes14. Deleting individual type I IFNs in vivo has revealed some of their specific roles during viral infection: within its extensive repertoire of immune functions, IFN-β is required for the production of IFN-α during vaccinia virus infection15 and IFN-ε protects the female reproductive tract against HSV (ref. 16). Less is known about type III IFNs, although studies have shown they are critical for antagonism of many viruses, particularly at epithelial surfaces17. However, across type I and III IFNs, there are vast differences with respect to their regulation and signalling outputs, which can dictate viral evasion strategies and ultimately impact host and tissue tropism.
Much remains unknown about how antiviral defences are encoded by IFN receptors, how viruses navigate these defences, and how differences in both of these areas contribute to viral susceptibility and permissivity across species. While many aspects of IFNs have been reviewed extensively elsewhere12,13,18, this Review will attempt to construct a holistic picture of the complicated relationship between IFN signalling and viral infection. To this end, we will progressively expand in scope with each section. We will begin at the level of individual IFN receptors, then broaden out to a discussion of IFN-mediated antiviral defences. We will then tackle some of the ways viruses can modulate or evade these defences. Lastly, we will consider how this intricate relationship between viruses and IFNs impacts host tropism.
Complex inputs
Despite the existence of approximately 20 IFNs, there are only three cell surface IFN receptors. The type I IFN receptor (IFNAR) is composed of subunits IFNAR1 and IFNAR2; the type II IFN receptor (IFNGR) is made up of subunits IFNGR1 and IFNGR2; and the type III IFN receptor (IFNLR) consists of subunits IFNLR1 and IL-10Rβ, the latter of which is shared by the IL-10 receptor. Of the three receptors, only IFNGR has one ligand: IFN-γ (ref. 19). In contrast, the type I and III receptors are shared by many IFNs. While the type I receptor is present on all nucleated cell types, the type III receptor is restricted mainly to epithelial cells, although some studies have observed that other cell types—such as epithelial cells20 and neutrophils21—are responsive to type III IFNs. Evidence suggests that type I and III IFNs can differentially engage their receptor to produce different signalling outcomes22,23.
How shared IFN receptors decipher the complexity of IFN signalling inputs remains elusive, yet it could be influenced by a variety of factors, such as ligand binding affinities, negative regulation, positive regulation and signal amplification, receptor expression and assembly, and biomolecular condensation (Fig. 2). These will be explored in detail in this section. Other factors that could not be extensively addressed here include post-translational modifications, epigenetics and epitranscriptomic regulation of IFN signalling components24,25. Among epigenetic modifications are STAT-mediated acetylation of histone H3 (ref. 26) and IFN-stimulated ubiquitination of histone 2B (ref. 27), both of which are associated with elevated expression of ISGs. Epitranscriptomic regulation includes RNA modifications by adenosine deaminases acting on RNA that inhibit IFN signalling28. Combinations of all of these factors may allow the type I and type III IFN receptors to process a dynamic range of signalling inputs during viral infection. Since many aspects of IFN signalling regulation have been reviewed elsewhere12,22,29–31, we will mainly highlight some of the emerging themes and new insights from the past few years.
Fig. 2. Factors that influence the decoding of IFN signalling inputs.
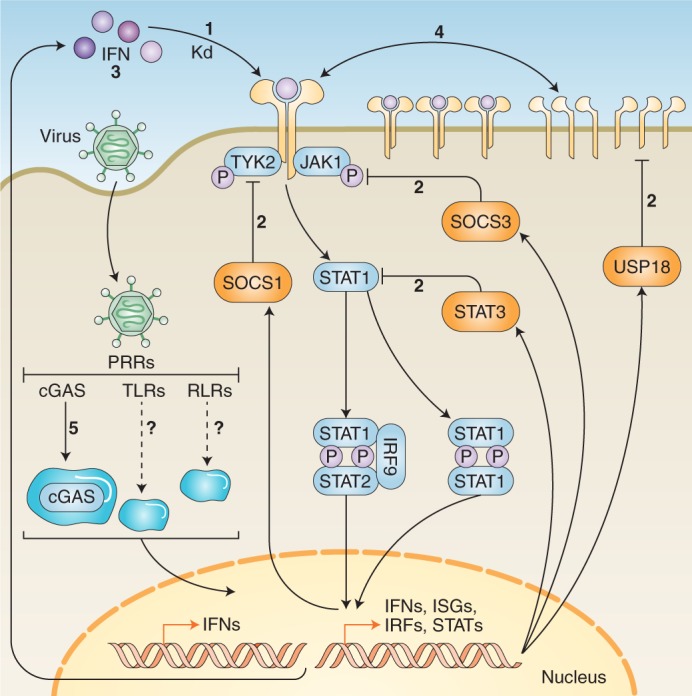
Various factors influence the decoding of IFN signalling inputs. (1) Receptor-binding affinities: each type I IFN binds to the IFNAR complex with varied affinities. These affinities correspond with differences in certain signalling outputs. As of yet, the binding affinities of type III IFNs have not been calculated. (2) Negative regulation: gene products induced by IFN signalling have the ability to negatively regulate the cascade at various stages; for example, SOCS1 and SOCS3, which interfere with TYK2 and JAK1, and STAT3, which can suppress STAT1 monomers, preventing homodimerization. (3) Positive regulation and signal amplification: factors such as STAT1, IRF9 and IRF7 can enhance IFN signal transduction. This leads to prolonged expression and secretion of antiviral cytokines and IFNs that can adjust the IFN cascade or lock the cell into an autocrine signalling loop. (4) Receptor expression and assembly: levels of receptor expression at the cell membrane contribute to differential signalling abilities of IFNs across cell types. Further, alterations to receptor assembly, such as the kinetics of subunit endocytosis, can influence signalling. (5) Biomolecular condensation: formation of cellular compartments caused by phase separation of biomolecules appears to influence various stages of IFN signalling; for example, phase separation of cGAS facilitates sensing of viral genetic material to induce expression of IFNs. Kd, dissociation constant.
An overarching model that has gained favourability is that many signalling differences result from changes in the stability of the ternary receptor complexes. This stability can be influenced by numerous factors, among which is ligand binding strength. Across type I IFNs, ligand binding affinities for IFNAR vary significantly, with IFN-β displaying the strongest affinity32. Differences in binding affinities appear to arise from the specific interactions each IFN makes with its receptor in addition to conserved ligand anchor points33. Type I IFNs that bind more strongly form a more stable ternary complex with the two receptor subunits, potentially resulting in increased binding duration34,35. Binding of type III IFNs with IFNLR forms a similar ternary complex, the stability of which increases when ligands are engineered to bind more strongly36. These studies suggest that stronger ligand binding affinities can stabilize the type I and type III receptors by increasing the duration of binding between the receptor subunits and ligand.
In contrast, negative regulatory mechanisms can obstruct the stability of the ternary receptor complexes and interfere with various stages of the IFN signalling cascade. For example, type I and type III IFN signalling induces expression of ubiquitin-specific protease 18 (USP18) (ref. 37), rendering the type I IFN receptor insensitive to IFN-α but not to IFN-β, thus implicating USP18 in the differential signalling activities of type I IFNs (ref. 38). Since overall IFN receptor expression remains unaltered, USP18 may participate in regulating receptor plasticity by preventing IFN-α (but not IFN-β) subtypes from recruiting IFNAR1 to the type I receptor complex39. Other well-characterized negative feedback mechanisms involve suppressor of cytokine signalling (SOCS) proteins, namely SOCS1 and SOCS3, whose targets include JAK1 (ref. 40) and tyrosine kinase 2 (TYK2) (ref. 41), which otherwise cross-phosphorylate to propagate the IFN signal relay and stabilize the receptor subunits. STAT proteins 1–6 have complicated roles in either propagating IFN signals or fine-tuning the cascade. For example, IFN-α/β-induced expression of STAT3 negatively regulates type I IFN signalling by sequestering STAT1 monomers42 or targeting transcription of IFN signalling components43. Strikingly, type III IFN signalling is negatively regulated by SOCS1, but not by SOCS3 or USP18 (ref. 44). These data indicate that negative regulatory mechanisms contribute to different signalling outcomes for type I IFNs, as well as the observed signalling differences between type I and III IFNs.
Conversely, positive regulatory mechanisms augment the IFN signalling cascade to maintain or amplify expression of ISGs or IFNs (refs. 30,45). Cascade components involved in amplifying IFN signals include STAT1 and IFN regulatory factor 9 (IRF9), which associate with STAT2 to form the ISGF3 transcription factor that binds to ISG promoter elements. Increased expression of these components in response to IFN-β prolongs expression of ISGs, resulting in enhanced viral resistance46. In addition to promoting the secretion of cytokines, positive feedback mechanisms can lock cells into an autocrine signalling loop that sustains IFN signal transduction47. Many positive regulatory mechanisms are cell type-specific, but certain factors such as IRF7 are thought to be common across many tissues48. Many details of positive regulation and signal amplification still need to be uncovered, as do the nuances of negative and positive regulation that keep IFN-mediated inflammation in check.
Upstream of many of these mechanisms, IFN receptors can decode signalling inputs through adjustments to receptor expression levels or assembly. It has been proposed that receptor expression levels contribute to the differential signalling observed by certain type I IFNs. For example, the potency of IFN-α surpasses that of IFN-β when human cells are engineered to overexpress IFNAR subunits49; however, the potency of IFN-β barely changes in response to receptor expression levels50. Beyond this, some models suggest that dynamic alterations to IFN receptor assembly may account for signalling differences. After formation of the ternary complex, receptor subunits are endocytosed. The kinetics of subunit endocytosis may factor into how signalling inputs are propagated22. Homodimerization of the IFNAR2 subunit may even be sufficient to induce certain signalling events51. However, to what extent receptor subunits can homodimerize and the role this plays in physiological conditions is still unknown. In all, it appears that adjusting the factors involved in receptor plasticity—expression, assembly and dimerization patterns—may fine-tune the signalling inputs of different IFNs.
Although somewhat distant from the other factors discussed, biomolecular condensates have been gaining attention as novel signalling structures that may guide the host response to viral infection. These are cellular compartments that lack a cell membrane and are typically formed by phase separation of polymeric biomolecules52. It was recently demonstrated that liquid phase separation of cyclic guanosine monophosphate (GMP)–adenosine monophosphate synthase (AMP) synthase (cGAS) plays a role in sensing pathogenic DNA upstream of IFN production53 (Fig. 2). Additionally, IFN-α signalling can prompt myxovirus resistance protein A (MxA) to form cytoplasmic, organelle-like structures adjacent to the cytoskeleton54. Since oligomerization of MxA is necessary for its antiviral activity against influenza virus55 and expression of MxA positively regulates IFN signalling in response to HCV infection56, biomolecular condensation of this ISG may facilitate IFN signalling during viral infection. Further studies should address the extent to which biomolecular condensation of ISGs and immune proteins in general influences IFN signalling dynamics.
Complex outputs
Complex IFN signalling inputs correspond with distinct transcriptional outputs and cellular responses during viral infection. Activation of type I and type III receptors triggers the transcription of hundreds of different ISGs as well as genes encoding other proteins, such as STATs 1–6. Signalling through IFN receptors also leads to the production of multiple other IFNs, which may vary in response to different viral infections57, although this phenomenon requires more study and characterization. Overall, the magnitude, kinetics and composition of IFN-stimulated transcriptional profiles appear to vary depending on which IFN initiated the signalling cascade58 (Fig. 3). As a result of these varied transcriptional profiles, IFNs induce different cellular responses, such as those associated with antiviral immunity, proliferation, apoptosis and clinical outcomes12. Often, differences in these transcriptional and biological outputs are both cell type- and IFN-specific. Understanding these outputs is imperative to relate differential IFN signalling to the antiviral responses of a cell.
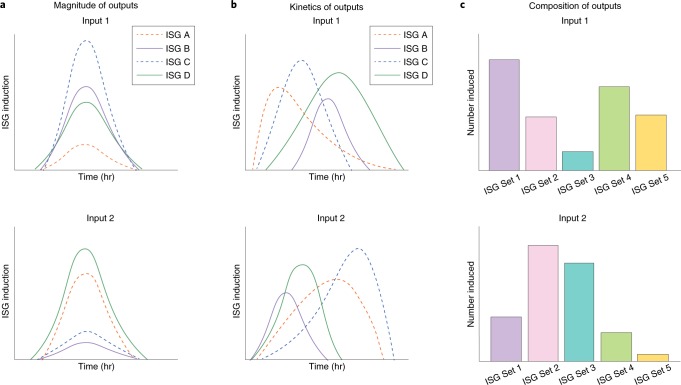
Fig. 3. Hypothetical models of different transcriptional outputs in response to viral infection.
During viral infection, differential IFN signalling inputs may manifest as differences in the magnitude of induced ISGs (a), kinetics of ISG transcription and degradation (b) or composition of ISGs expressed in the transcriptional profile (c). These variations, or a combination thereof, appear to play a significant role in cell-, virus- and species-specific antiviral responses.
As a group, ISGs can target most steps of viral replication, amplify IFN signalling cascades to strengthen the host response and exert combinatorial antiviral activities59. Mice deficient in certain ISG pathways still mount antiviral responses via other ISGs; such redundancy highlights the crucial role for ISG transcription during host defence60. The first ISGs discovered (now considered ‘classical ISGs’) were shown to induce potent antiviral responses by themselves (for example, Mx proteins61 or Protein kinase R (ref. 62)). Over time, however, the identification of less-potent ISGs indicates that weaker ISGs work together to temper the immune response depending on the type and magnitude of infection59. While much remains unknown about the roles of ISGs during viral infection and pathogenesis, large-scale ISG screens have identified and characterized hundreds of ISGs present during viral infection63,64, many with virus-, cell- and species-specific effects.
Unique ISG panels are induced in response to different viral infections57, resulting in specific antiviral responses. Some viruses are targeted primarily at a specific point during infection; for example, many ISGs that target HCV do so during translation of the entering genome63. Other viruses, such as DENV and WNV, are targeted by a number of ISGs at multiple stages of the viral replication cycle65. ISGs can exert virus-specific activities; for example, IFN-α-inducible protein 6 was found to selectively inhibit replication of certain flaviviridae family viruses by interfering with the formation of endoplasmic reticulum (ER) microenvironments66. Combinations of ISGs frequently increase defences against viruses. Additive effects were observed when pairs of ISGs were tested against viruses from different families, such as HCV, HIV-1 and YFV (ref. 63). ISG feedback loops also enhance or fine-tune antiviral effects by regulating expression of other ISGs or IFNs (ref. 45). In particularly striking cases, ISGs act in a manner that appears pro-viral on first glance. While some ISGs may indeed be co-opted by viruses to facilitate viral infection, others with curiously ‘pro-viral’ functions may in fact be working to fine-tune the IFN response against viral infection. For example, in humans (but not mice), ISG15 interacts with and stabilizes USP18, allowing for appropriate downregulation of IFN signalling67. Since ISG15 possesses well-documented antiviral properties68, these findings point to a species-specific ability of ISG15 to adjust the IFN signalling cascade during the later stages of viral infection in humans. Such studies indicate that ISGs can modulate the host’s antiviral response to fit the type of virus or stage of infection.
In early studies aiming to understand IFN signalling outputs, differential ISG outputs and cellular responses were linked with variations in the receptor-binding affinities of IFNs. Type I IFNs that bind more strongly to IFNAR, such as IFN-β, induce a subset of ISGs that are not expressed by the binding of weaker IFNs; furthermore, universally expressed ISGs are induced to a greater extent by IFN-β (refs. 64,69). A linear correlation exists between the binding affinities of type I IFNs and their anti-proliferative potency; however, the relationship between receptor-binding affinity and antiviral activity is less straightforward32. Although the signalling outputs of type III IFNs have been less thoroughly explored, an engineered IFN-λ that binds IFNLR with increased affinity leads to a corresponding increase in antiviral activity36. In contrast, increasing the binding affinity of a synthetic type I IFN does not increase antiviral activity compared with wild-type IFN, which may suggest that type I IFNs already exert their maximal antiviral potency36. Future studies should continue to explore how other factors contribute to the complexity of IFN-mediated transcriptional and antiviral signalling outputs.
An increasing number of reports have revealed instances where type I and III IFNs function together to coordinate viral clearance. Genetic disruption of type I and III signalling in mice results in pronounced viral neuroinvasion and overall hyper-susceptibility to infection by attenuated YFV (ref. 70). Hyper-susceptibility is also observed during influenza infection of mice deficient in type I and III IFN signalling; deficiency of either type alone does not abolish transcription of ISGs, which allows the type I and III IFN systems to compensate for the loss of the other71. Further research suggests that type III IFN signalling acts as the first line of defence in the lung epithelium during early influenza virus infection, and subsequent signalling by type I IFNs may offer enhanced protection during the later stages of viral spread72. Additionally, a comprehensive analysis of the antiviral landscape in mice infected with influenza virus found diverse antiviral gene signatures, indicating that the magnitude or number of rounds of influenza infection dictates the nature of IFN protection73. It thus appears that integration of the type I and III IFN systems, especially at barrier surfaces, can modulate or heighten a host’s antiviral response during infection by certain viruses.
While the antiviral activities of type I IFNs have been thoroughly explored13, those of type III are still being elucidated. Type III IFNs may hold therapeutic potential against viruses that preferentially infect cells of epithelial origin. Research in mice has demonstrated that IFN-λ signalling at the blood–brain barrier prevents neuroinvasion by WNV, and treatment with pegylated IFN-λ2 improves survival against viral challenge20. Similarly, intestinal epithelial cells infected with rotavirus respond more strongly to treatment with IFN-λ than type I IFNs in vivo74, and type III IFN signalling appears sufficient to restrict the spread of influenza virus in the respiratory tract75. IFN-λ also displays potent antiviral action against hepatitis viruses76, and promising clinical studies have tested pegylated IFN-λ against chronic HBV or HCV infection77,78. Counterintuitively, IFN-λ4 has been associated with lower viral clearance of HCV in humans79, possibly due to its ability to accentuate negative feedback mechanisms such as SOCS1 (ref. 80). However, certain IFN-λ4 genotypes may influence the efficacy of IFN therapy, as shown for HCV (refs. 81,82). Since IFNLR is expressed on fewer cell types than IFNAR, side effects from IFN-λ treatment may be more limited than those induced by treatment with type I IFNs (ref. 83). These studies and others indicate that type III IFNs can launch significant antiviral responses against a number of viruses and may be seriously considered as a clinical treatment; as such, type III IFNs will be an important area for future research.
Viral evasion of IFN-mediated defences
Viruses have evolved along with their hosts to evade these IFN systems, allowing them to propagate within their hosts and spread to neighbouring cells. Disruption of innate immune signalling both upstream (Table 1 and Fig. 4) and downstream (Table 2 and Fig. 5) of IFN induction is quite common among viruses. Many mechanisms behind such evasion have been extensively reviewed elsewhere2–7, but a few are briefly mentioned here: viruses can evade IFN-dependent innate immunity by hiding their genomes from detection, inhibit interactions with host inducers of the IFN response, regulate phosphorylation, ubiquitinylate related pathways of the host’s cellular machinery, cleave/ mark for degradation proteins vital to stimulating the IFN response, regulate host gene transcription and translation, regulate RNA processing and trafficking, and employ protein decoys. Rather than reiterate these evasion strategies, this section aims to highlight more recent discoveries relating to novel roles for organelle stress responses during viral infection and viral antagonism, both up- and downstream of host IFN signalling. These new discoveries broaden our understanding of host antagonism and, specifically, host tropism of RNA viruses.
Table 1.
Examples of RNA viral antagonism upstream of IFN induction
| Viral family | Virus | Proposed mechanism of evasion upstream of IFN signalling | References |
|---|---|---|---|
| Flaviviridae | DENV, WNV, ZIKV and JEV | Viral protease NS2B3 cleaves STING | 14, 91,118 |
| Flaviviridae | YFV | Viral protein NS4B interacts with STING | 124 |
| Flaviviridae | HCV | Viral protein NS4B interacts with and disrupts STING signalling complexes | 125,126 |
| Coronaviridae | HCoV-NL63 | Viral protein PLP2–TM interacts with and disrupts STING signalling complexes | 127,128 |
| Coronaviridae | PEDV | Viral protein PLP2 interacts with and disrupts polyubiquitination of STING | 129 |
| Coronaviridae | SARS-CoV | Viral protein Plpro–TM interacts with and disrupts STING signalling complexes | 127, 128,130 |
| Flaviviridae | HCV | Viral protease NS34A cleaves MAVS | 131,132 |
| Picornaviridae | HAV | Viral protease precursor 3ABC cleaves MAVS | 133 |
| Flaviviridae | DENV | Viral protease NS2B3 cleaves MAVS | 134 |
| Flaviviridae | HCV | Viral protease NS34A cleaves TRIF | 135 |
| Picornaviridae | HAV | Viral protease intermediate cleaves TRIF | 136 |
| Arteriviridae and Bunyaviridae | Arterivirus and nairovirus | Viral protease deubiquinates RIG-I | 137 |
| Arenaviridae | Arena virus synthetic molecules | Viral 5’ppp overhangs act as RIG-I decoy | 138 |
| Arenaviridae | GTOV, JUNV, MAVC and SABV | Viral Z protein binds to and inhibits RIG-I | 139 |
| Paramyxoviridae | hPIV5 | Viral V protein binds LGP2, cooperatively inhibiting induction by RIG-I ligands | 140 |
| Reoviridae | Synthetic reovirus RNA mimics | Synthetic particles trigger RIG-I; virus trims 5’ppp for innate immune evasion | 141 |
| Orthomyxoviridae | IAV | Viral protein NS1 binds RIG-I and prevents activation; RIG-I selectively evolved to distinguish self from non-self | 142,143 |
| Bornaviridae | BDV | Viral P protein antagonizes TBK1 | 144 |
| Bunyaviridae | SFTSV | Viral protein NSs sequesters TBK1/IKK-ε | 145 |
| Bunyaviridae | Hantavirus | Dephosphorylation of TBK1/IRF3 | 146–148 |
| Filoviridae | Ebola virus | Viral protein VP35 sequesters viral RNA | 149,150 |
| Togaviridae | Sindbis virus | Viral protein nsP2 downregulates IFN and minimizes virus visibility | 151 |
| Bunyaviridae | ANDV | Viral NP and GPC proteins inhibit IFN-β induction | 152 |
| Bunyaviridae | SNV | Viral GPC protein inhibits IFN-β induction | 152 |
| Bornaviridae | BDV | Evades detection by genome trimming | 153 |
GTOV, Guanarito virus; JUNV, Junin virus; MAVC, Machupo virus; SABV, Sabia virus; PLP2, papain-like protease 2; TM, transmembrane domain; VP35, virion protein 35; SFTSV, severe fever with thrombocytopenia syndrome virus; GPC, glycoprotein precursor; PEDV, porcine epidemic diarrhea virus; Plpro, papain-like protease; 3ABC, stable intermediate product of HAV polyprotein processing including viral proteins 3A, 3B and 3C; nsP2, non-structural protein 2; NP, nucleocapsid protein.
Fig. 4. Examples of RNA viral antagonism upstream of IFN induction.
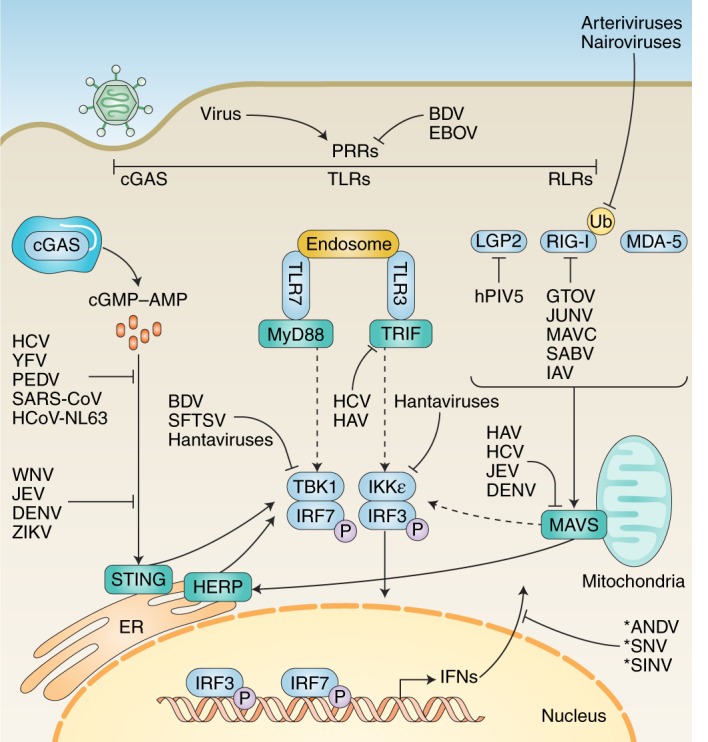
After entering a cell, viral genetic material may be recognized by one or more of the host’s PRRs. These include RLRs, Toll-like receptors (TLRs) and cGAS. Recognition by one or more of these PRRs triggers a signalling cascade that culminates in the transcription and subsequent generation of IFNs. Different viruses have evolved to antagonize these pathways at virtually all stages, indicated by blunt end arrows. Solid arrows indicate direct pathway connections. Dashed arrows indicate signal cascade components present in vivo but not shown for space concerns. Asterisks indicate viral antagonism by an unknown mechanism. See Table 1 for specific mechanisms at each stage. LGP2, laboratory of genetics and physiology protein 2; MDA-5, melanoma differentiation-associated protein 5; TRIF, TIR domain-containing adapter-inducing IFN-β; IKK-ε, IκB kinase-ε; HAV, hepatitis A virus; hCoV-NL63, human coronavirus NL63; PEDV, porcine epidemic diarrhea virus; GTOV, Guanarito virus; JUNV, Junin virus; MAVC, Machupo virus; SABV, Sabia virus; BDV, Borna disease virus; SFTSV, severe fever with thrombocytopenia syndrome virus; JEV, Japanese encephalitis virus; ANDV, Andes virus; SNV, Sin Nombre virus.
Table 2.
Examples of RNA viral antagonism downstream of IFN induction
| Viral family | Virus | Proposed mechanism of evasion downstream of IFN signalling | References |
|---|---|---|---|
| Paramyxoviridae | Hendra virus and Nipah virus | Prevents STAT protein nucleocytoplasmic trafficking by preventing STAT complex formation via viral N protein | 154 |
| Paramyxoviridae | hPIV1 | Viral C accessory protein prevents nuclear translocation of STAT proteins by an unknown mechanism | 107 |
| Paramyxoviridae | hPIV3 | Viral C accessory protein prevents phosphorylation of STAT proteins | 6 |
| Paramyxoviridae | RSV | Viral NS1 and NS2 proteins induce proteosomal degradation of STAT complexes | 155,156 |
| Paramyxoviridae | SeV | Viral C accessory protein associates with STAT 1, blocking STAT 1 and STAT2 activation | 109,110 |
| Paramyxoviridae | hPIV5 | Viral V protein induces STAT1 antagonism in a STAT2 dependent manner | 157,158 |
| Flaviviridae | JEV | Viral protein NS5 prevents tyrosine phosphorylation of TYK2 and STAT1 | 104 |
| Flaviviridae | WNV | Viral protein NS5 prevents accumulation of phosphorylated STAT1 | 101 |
| Paramyxoviridae | Hendra virus and Nipah virus | Viral V protein sequester STAT2 in the cytoplasm | 159–162 |
| Paramyxoviridae | hPIV2 | Viral V protein induces STAT2 degradation | 163 |
| Paramyxoviridae | Measles virus | Viral V protein sequesters STAT2 | 164 |
| Flaviviridae | DENV | Viral protein NS5 reduces STAT2 expression, mediates STAT2 binding and degradation, blocks STAT2 phosphorylation, and co-opts host UBR4 protein, mediating STAT2 degradation | 95–98 |
| Flaviviridae | ZIKV | Viral protein NS5 mediates STAT2 degradation in humans, but not in mice | 99,100 |
| Flaviviridae | JEV | Unknown viral protein blocks tyrosine phosphorylation, blunting JAK–STAT signalling | 103 |
| Flaviviridae | YFV | IFN-I activation of the JAK–STAT pathway allows viral protein NS5 to inhibit STAT signalling | 106 |
| Bunyaviridae | ANDV and SNV | Viral GPC protein inhibits JAK–STAT signal amplification | 152 |
| Flaviviridae | WNV and TBEV | Viral protein NS5 sequesters cellular prolidase, limiting expression of IFNAR1 cell surface expression | 102 |
| Paramyxoviridae | Measles virus | Viral V protein induces type I IFN in a STAT2-dependent and STAT1-independent manner, subverting antiviral IFN effects | 165 |
| Flaviviridae | JEV | Viral protein NS5 prevents IFN-β induced responses, such as ISG expression | 105 |
Fig. 5. Examples of RNA viral antagonism downstream of IFN induction.
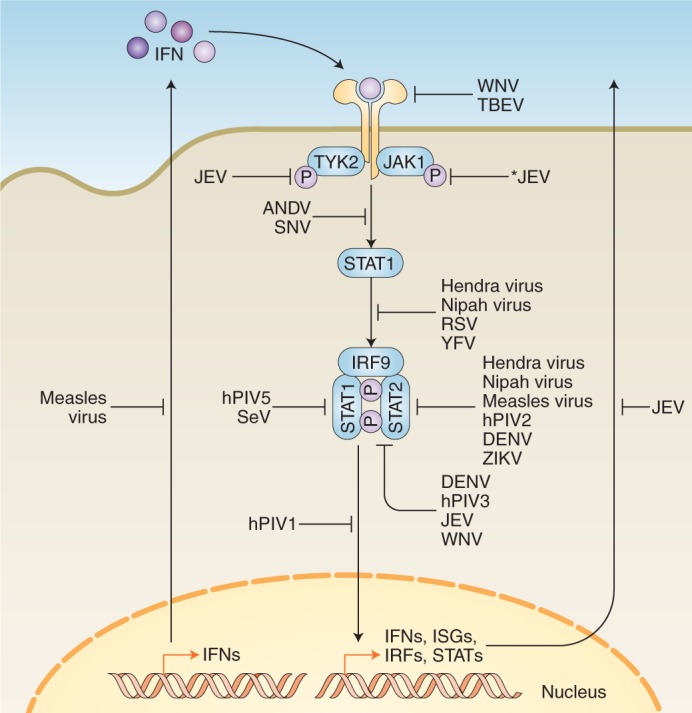
Following IFN binding to their receptor in an autocrine or paracrine manner, a signalling cascade takes place that results in the transcription and expression of ISGs. These ISGs are critical for establishing an antiviral state in the host and bystander cells. Different viruses have evolved to antagonize this pathway at virtually all stages, indicated by blunt end arrows. Solid arrows indicate direct pathway connections. The asterisk indicates viral antagonism by an unknown mechanism. See Table 2 for specific mechanisms at each stage.
The ER and mitochondria are two organelles that play important roles in detecting and responding to viral infection upstream of IFN induction. The organelle-damaging effects of certain viral infections signal to the host cell that something is amiss, prompting the cell to trigger antiviral responses through IFN signalling. In a healthy cell, ER stress induced by the unfolded protein response leads to ER-associated protein degradation84. Conversely, in a cell infected with influenza A virus (IAV), a potent antiviral response is triggered by the overproduction of viral haemagglutinin glycoproteins which stimulate the unfolded protein response85, offering a new perspective on the role of the ER during viral infection. Further, the homocysteine-induced ER protein (HERP), induced by the unfolded protein response and by ER stress, is implicated in antiviral immunity against IAV and several other RNA viruses. Mitochondrial antiviral signalling protein (MAVS), a vital host protein involved in the retinoic acid-inducible gene I (RIG-I) like receptor (RLR)–IFN signalling cascade, was shown to enhance HERP expression following viral infection86. HERP then binds TANK-binding kinase 1 (TBK1) to amplify the MAVS signalling cascade86, which, in turn, represses the replication of several RNA viruses, including Sendai virus (SeV), IAV, VSV and enterovirus 71 (EV71) (ref. 86) (Fig. 4). Similarly, viruses such as DENV and ZIKV can cause mitochondrial damage through elongation induced by viral non-structural proteins during infection87, but, in the past few years, several studies have come to conflicting conclusions regarding cellular defences against RNA virus-induced damage to this vital organelle, as outlined below.
It has been shown that IFNs can be induced in healthy cells when mitochondrial DNA (mtDNA) stress is induced by a deficiency in mtDNA binding protein88. This points to a role played by the fidelity and/or decompartmentalization of mtDNA in antiviral signalling. On the one hand, viruses from different viral families can prompt leakage of mtDNA into the cytosol, stimulating the DNA sensor cGAS, thereby elevating ISG expression through stimulator of IFN genes (STING)–IRF3 dependent signalling88,89. DENV (an RNA virus) has evolved to disrupt this canonical DNA sensing pathway by virally encoded, protease-mediated cleavage of STING (refs. 89–91). Additionally, the DENV non-structural protein NS4B causes mitochondrial elongation, compromising the integrity of ER-mitochondrial interfaces that are critical for RIG-I-mediated innate immune signalling87. Similarly, it was shown that other RNA viruses of diverse families, such as VSV, Sindbis virus, SeV, IAV and reovirus type 3 Dearing strain, all replicate at higher levels in cells expressing short-hairpin RNAs targeting STING (ref. 92). On the other hand, however, it is argued that STING exhibits an antiviral response through translational inhibition in an RLR-dependent manner and is not required for the sensing of leaked mtDNA due to organelle damage. This conclusion was drawn from findings that cells depleted of mtDNA are able to retain their ability to diminish viral RNA replication in a STING-dependent manner, and that STING-dependent translational inhibition does not depend on MAVS (ref. 92). These findings argue that RLR responses to RNA viral infections mediate IFN induction through MAVS and restrict viral translation through STING. Regardless of mechanism, RNA viruses must bypass these innate IFN signalling pathways to successfully and robustly replicate. More research is needed to understand the exact, and likely multifaceted, functions of STING during viral infection.
Similarly, it was recently discovered that like DNA sensors affecting RNA viral replication, the opposite is also true; several groups are now studying the mechanisms by which RIG-I is able to bind host RNA as a means to stimulate the innate immune response during viral infection. HSV-1, a DNA alphaherpes virus, was shown to induce the relocalization of host-derived 5S ribosomal RNA pseudogene 141 (RNA5SP141) from the nucleus to the cytoplasm, where it binds RIG-I and induces the type I IFN response93. This occurs due to viral induced shutoff of RNA5SP141-interacting proteins, thus freeing RNA5SP141 for interaction with RIG-I; silencing of RNA5SP141 was shown to dampen the antiviral response to not only HSV-1, but also to Epstein–Barr virus and IAV (ref. 93). Further, host RNAs were shown to facilitate the host immune response to Kaposi’s sarcoma-associated herpesvirus (KSHV), a gammaherpesvirus, which is capable of establishing latent and lysogenic infections. It was discovered that KSHV lytic reactivation from latency is restricted by RLRs through recognition of host-derived RNAs (ref. 94); this is accomplished by an infection-induced reduction in a cellular triphosphatase, leading to the accumulation of misprocessed, noncoding RNAs that are detected by RIG-I. These results, taken together with the above findings regarding cGAS–STING, demonstrate our recent advances in understanding the surprising interplay between the virus and host prior to IFN induction.
How RNA viruses antagonize antiviral responses downstream of IFN signalling appears to be largely restricted to a small subset of host proteins, albeit through a variety of viral proteins and mechanisms (Table 2 and Fig. 5). For example, STAT proteins serve as a key point of convergence within host defences. STAT2 is antagonized by the non-structural viral protein NS5 of numerous viruses, including DENV (refs. 95–98), ZIKV (refs. 99,100), WNV (refs. 101,102), JEV (refs. 103–105) and tick-borne encephalitis virus102. Notably, YFV also uses its NS5 protein to antagonize STAT2 and the STAT-signalling pathway, but this requires events upstream of IFN signalling to take place (specifically, phosphorylation of STAT1 and polyubiquitination of YFV NS5) (ref. 106). Less closely related viruses, such as human parainfluenza virus types 1 and 3 (hPIV1 and hPIV3) and SeV, also antagonize the STAT-signalling pathway by inhibiting STAT protein phosphorylation and/or STAT nuclear translocation, thereby blunting ISG induction107–110. Similarly, by mutating the critical residues of the STAT binding domain of rabies virus P protein, mice do not progress to a lethal neurological disease state, even following direct cranial inoculation by the mutant virus111, demonstrating that STAT inhibition is required for the lethality of the virus. Discoveries such as these highlight how certain viruses have evolved to counter IFN-mediated host defences, and allow us to look at how host and tissue tropism is dictated by differential regulation and antagonism of these host innate immune pathways.
Impact of IFN signalling complexity on host tropism
Determining the impact of IFN signalling on host tropism is vital to the pursuit of clinical interventions. However, gaining these insights comes with its fair share of challenges. Research conducted using different methods and bioinformatics analyses can be difficult to compare holistically and are typically carried out on a single-species level. This usually results in studies without sufficient power to understand and identify key evolutionary features, leaving it hard to draw meaningful conclusions from expanding data sets. The first piece of the puzzle is to understand how other species respond, or are immune to, varying infections that plague humans. Genomic- or transcriptomic-based studies are useful to investigate these differences, but require a well-annotated reference genome for the species in question as well as an ample number of samples from different donors to ensure the data represents the larger population. Identifying gene orthologues between species can be fruitless without a complete or near-complete reference genome for each species of interest. However, these complications pale in comparison to what we can learn about host–pathogen interactions from analysing other species, and we can be continually surprised by how other hosts combat, or even tolerate, viral infections.
Studies that analyse viral infection in multiple species using a single method are able to avoid the challenge of comparing results gathered from differing methods and bioinformatics analysis pipelines. One such study compared the type I ‘interferome’ of 10 species (human, rat, cow, sheep, pig, horse, dog, microbat, fruit bat and chicken), revealing a conserved core of 62 genes, some of which were not previously associated with response to type I IFN stimulation112. These conserved ISGs highlight the longstanding role of the innate immune system in responding to different pathogens. Additionally, ISG expression patterns were more similar in more closely related species than distant ones, while 14 conserved IFN repressing genes were upregulated in all species that target different points in the type I IFN pathway, demonstrating the importance of avoiding excess stimulation of these pathways112. While studies of this kind are necessary to draw comparisons across multiple species simultaneously, studies restricted to fewer species also have the potential to illuminate specific differences that confer resistance to, or tolerance of, different pathogens that already cross the species barrier.
Bats are a known reservoir for many pathogenic viruses that plague humans, such as Nipah virus, Hendra virus, severe acute respiratory syndrome coronavirus (SARS-CoV), Middle Eastern respiratory syndrome coronavirus and Marburg virus113,114. Originally, it was thought that certain species of bats become asymptomatically infected with these viruses due to enhanced potency of their antiviral response. A new hypothesis has recently emerged that immune responses in bats are differentially regulated compared to other species, leading to tolerance rather than clearance of infection. This differential immune regulation may occur because the metabolic demands of flight cause DNA damage, which releases host mtDNA into the cytoplasm. This is exemplified by Egyptian Rousette bats (Rousette aegyptiacus), in which the type I IFN subfamily IFN-ω is expanded to almost two dozen subtypes, whereas humans only have one115. Some bat species constitutively express type I IFNs (namely IFN-α) (ref. 116), but IFN-ω subfamily genes are only highly induced in R. aegyptiacus following infection with VSV. Additionally, bats have a high occurrence of point mutations in the innate immune signalling protein, STING. The Ser 358 residue of STING is conserved across all non-bat mammals117 and is a critical phosphorylation site for downstream IFN activation in these species. In bats, however, this residue can be mutated to asparagine, histidine or aspartic acid, preventing activation of cGAS–STING and thus resulting in a dampened innate immune response117. Taken together, these studies shed light on how bats evolved to differentially regulate innate immune signalling pathways, leading to infection tolerance and allowing them to be a reservoir for zoonotic pathogens. Notably, in other species such as mice, mutations in STING don’t impart a tolerance of viral infection, but rather prevent infection by some viruses altogether.
Mice do not support productive infection of many flaviviruses that affect humans, and one key mechanism behind this was recently elucidated. The viral protease NS2B3 in ZIKV, WNV and JEV, but not YFV, was found to mediate cleavage of human STING, increasing the permissiveness of mouse cells expressing human STING to ZIKV infection (including higher production of infectious particles and greater ZIKV RNA copy numbers)14; strikingly, murine STING is not cleavable by viral proteases in DENV (ref. 118) or ZIKV (ref. 14), thereby restricting the ability of these viruses to infect mice. Notably, STING-deficient mice are not more susceptible to ZIKV infection, pointing to other redundant antiviral pathways controlling ZIKV infection in vivo14. However, cGAS- or STING-deficient mice are more likely to progress to lethal infection by several DNA viruses119, demonstrating the vital role of this IFN signalling pathway’s ability to hinder viral replication following infection. Further, mouse STAT2, but not human STAT2, is known to restrict early DENV replication96, conferring additional resistance against DENV to mice. Similarly, the NS5 protein of ZIKV is able to bind and degrade human STAT2, but not mouse STAT2, preventing ZIKV from evading the innate immune response in mice99,100,120, and subsequently establishing a successful infection. Due to differing immune responses to differing infections, it can be hard to relevantly study these pathogens using mammalian model organisms. Recently, however, a large step forward for ZIKV was made by the generation of an immunocompetent mouse model for ZIKV infection. By garnering a key mutation in the NS4B gene of ZIKV to enhance replication in neuronal stem cells and the brain in mice, and replacing mouse STAT2 with human STAT2, an immunocompetent mouse model with which to study ZIKV was generated121, allowing for further research of this important human pathogen.
Future perspectives
Much remains to be learned about the complexity of IFN signalling during viral infection. Many factors have been proposed to explain how type I IFNs signal differently through their shared receptor. However, it is still unclear how many of these factors function individually as well as together. It is still relatively unknown as to what extent type III IFNs produce different signalling outputs and whether these outputs result from the same mechanisms that impact type I signalling. Further, given the complexity and vastness of ISG outputs uncovered by large-scale screening efforts, more work is needed to fully understand the transcriptional profiles associated with IFN signalling and their effect on viral antagonism. Future studies should continue to investigate the roles of upregulated ISGs and the antiviral consequences of downregulated genes in response to IFN signalling, which have been understudied so far.
As for viral evasion of host signalling pathways, new concepts and convergence points are starting to emerge. Fully elucidating the role of cGAS and STING in the detection of, and antagonism by, RNA viruses is a currently unfolding story. Likewise, the involvement of RNA sensors for fighting DNA viral infections is an intriguing new area of investigation. A better understanding of how certain species resist infection by pathogens that affect humans will further our efforts to develop interventions and treatments.
There is still much work to be done to understand viral species and tissue tropism. Ongoing difficulties associated with comparing genomic and transcriptomic studies across different species still pose a barrier to the field. Additional difficulties arise when developing appropriate models to study viral infections. Working with certain species may not be possible due to ethical or practical reasons, and cell models may not give us a biologically relevant picture of virus–host interactions. The use of induced pluripotent stem cells generated from naturally susceptible and permissive species may shed light on questions that cannot be answered using other models.
Efforts to understand host tropism, tissue tropism and viral pathogenicity may be aided by considering the evolution of host and viral factors. For example, in non-human primates, a conserved mutation in MAVS imparts an escape from hepaciviral protease antagonism, severely enhancing IFN response in the presence of HCV and other flaviviruses that infect primates, as demonstrated in cells expressing primate MAVS (ref. 122). Additionally, for duck Tembusu virus, a single point mutation in the envelope protein confers airborne transmissibility to the virus in the absence of its natural vector, the mosquito; this expands its tropism and ability to replicate in the lung tissue of its host123. Altogether, studies such as these demonstrate how small evolutionary changes can have drastic impacts on the viability of both the virus and host, and illustrate an important knowledge gap in the field that has yet to be filled.
Acknowledgements
We thank J. Gaska, F. Douam and other members of the Ploss lab, as well as A. Goglia, J. Toettcher and other members of the Toettcher lab at Princeton University for providing critical feedback on this manuscript. We apologize to all of our colleagues whose work we were unable to include or discuss due to space constraints. Work in the lab was supported by grants from the National Institutes of Health (R01 AI138797 and R01AI107301 both to A.P.), a Research Scholar Award from the American Cancer Society (RSG-15-048-01-MPC to A.P.), a Burroughs Wellcome Fund Award for Investigators in Pathogenesis (to A.P.), the New Jersey Health Foundation (to A.P.), a grant from the US Department of Defense (W81XWH18’10237 to A.P.) and funding from Princeton University. E.M. and R.L. were supported by the National Institute of General Medicine Sciences of the National Institutes of Health (grant no. T32GM007388). R.L. is a recipient of a predoctoral fellowship from the National Science Foundation.
Author contributions
E.V.M., R.A.L. and A.P. conceived and wrote this Review.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Emily V. Mesev, Robert A. LeDesma.
References
- 1.Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA. Pattern recognition receptors and the innate immune response to viral infection. Viruses. 2011;3:920–940. doi: 10.3390/v3060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia-Sastre A. Ten strategies of interferon evasion by viruses. Cell Host Microbe. 2017;22:176–184. doi: 10.1016/j.chom.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lei J, Hilgenfeld R. RNA-virus proteases counteracting host innate immunity. FEBS Lett. 2017;591:3190–3210. doi: 10.1002/1873-3468.12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miorin L, Maestre AM, Fernandez-Sesma A, Garcia-Sastre A. Antagonism of type I interferon by flaviviruses. Biochem. Bioph. Res. Co. 2017;492:587–596. doi: 10.1016/j.bbrc.2017.05.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Best SM. The many faces of the flavivirus NS5 protein in antagonism of type I interferon signaling. J. Virol. 2017;91:e01970–16. doi: 10.1128/JVI.01970-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers R, Takimoto T. Antagonism of innate immunity by paramyxovirus accessory proteins. Viruses. 2009;1:574–593. doi: 10.3390/v1030574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chowdhury FZ, Farrar JD. STAT2: A shape-shifting anti-viral super STAT. JAKSTAT. 2013;2:e23633. doi: 10.4161/jkst.23633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc. Roy. Soc. Lond. B Bio. 1957;147:258–267. doi: 10.1098/rspb.1957.0048. [DOI] [PubMed] [Google Scholar]
- 9.Kotenko SV, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 10.Sheppard P, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 11.O’Brien TR, Prokunina-Olsson L, Donnelly RP. IFN-lambda4: the paradoxical new member of the interferon lambda family. J. Interf. Cytok. Res. 2014;34:829–838. doi: 10.1089/jir.2013.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–372. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Teijaro JR. Type I interferons in viral control and immune regulation. Curr. Opin. Virol. 2016;16:31–40. doi: 10.1016/j.coviro.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding Q, et al. Species-specific disruption of STING-dependent antiviral cellular defenses by the Zika virus NS2B3 protease. Proc. Natl Acad. Sci. USA. 2018;115:E6310–E6318. doi: 10.1073/pnas.1803406115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deonarain R, et al. Impaired antiviral response and alpha/beta interferon induction in mice lacking beta interferon. J. Virol. 2000;74:3404–3409. doi: 10.1128/JVI.74.7.3404-3409.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fung KY, et al. Interferon-epsilon protects the female reproductive tract from viral and bacterial infection. Science. 2013;339:1088–1092. doi: 10.1126/science.1233321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donnelly RP, Kotenko SV. Interferon-lambda: a new addition to an old family. J. Interf. Cytok. Res. 2010;30:555–564. doi: 10.1089/jir.2010.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 19.Walter MR, et al. Crystal structure of a complex between interferon-gamma and its soluble high-affinity receptor. Nature. 1995;376:230–235. doi: 10.1038/376230a0. [DOI] [PubMed] [Google Scholar]
- 20.Lazear HM, et al. Interferon-lambda restricts West Nile virus neuroinvasion by tightening the blood-brain barrier. Sci. Transl. Med. 2015;7:284ra259. doi: 10.1126/scitranslmed.aaa4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Espinosa V, et al. Type III interferon is a critical regulator of innate antifungal immunity. Sci. Immunol. 2017;2:eaan5357. doi: 10.1126/sciimmunol.aan5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piehler J, Thomas C, Garcia KC, Schreiber G. Structural and dynamic determinants of type I interferon receptor assembly and their functional interpretation. Immunol. Rev. 2012;250:317–334. doi: 10.1111/imr.12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diegelmann J, et al. Comparative analysis of the lambda-interferons IL-28A and IL-29 regarding their transcriptome and their antiviral properties against hepatitis C virus. PLoS ONE. 2010;5:e15200. doi: 10.1371/journal.pone.0015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Licht K, Jantsch MF. Rapid and dynamic transcriptome regulation by RNA editing and RNA modifications. J. Cell Biol. 2016;213:15–22. doi: 10.1083/jcb.201511041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen K, Liu J, Cao X. Regulation of type I interferon signaling in immunity and inflammation: A comprehensive review. J. Autoimmun. 2017;83:1–11. doi: 10.1016/j.jaut.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 26.Paulson M, Press C, Smith E, Tanese N, Levy DE. IFN-Stimulated transcription through a TBP-free acetyltransferase complex escapes viral shutoff. Nat. Cell Biol. 2002;4:140–147. doi: 10.1038/ncb747. [DOI] [PubMed] [Google Scholar]
- 27.Fonseca GJ, et al. Adenovirus evasion of interferon-mediated innate immunity by direct antagonism of a cellular histone posttranslational modification. Cell Host Microbe. 2012;11:597–606. doi: 10.1016/j.chom.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Hartner JC, Walkley CR, Lu J, Orkin SH. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat. Immunol. 2009;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Weerd NA, Nguyen T. The interferons and their receptors—distribution and regulation. Immunol. Cell Biol. 2012;90:483–491. doi: 10.1038/icb.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schreiber G. The molecular basis for differential type I interferon signaling. J. Biol. Chem. 2017;292:7285–7294. doi: 10.1074/jbc.R116.774562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lavoie TB, et al. Binding and activity of all human alpha interferon subtypes. Cytokine. 2011;56:282–289. doi: 10.1016/j.cyto.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 33.Thomas C, et al. Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell. 2011;146:621–632. doi: 10.1016/j.cell.2011.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaitin DA, et al. Inquiring into the differential action of interferons (IFNs): an IFN-alpha2 mutant with enhanced affinity to IFNAR1 is functionally similar to IFN-beta. Mol. Cell Biol. 2006;26:1888–1897. doi: 10.1128/MCB.26.5.1888-1897.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalie E, Jaitin DA, Podoplelova Y, Piehler J, Schreiber G. The stability of the ternary interferon-receptor complex rather than the affinity to the individual subunits dictates differential biological activities. J. Biol. Chem. 2008;283:32925–32936. doi: 10.1074/jbc.M806019200. [DOI] [PubMed] [Google Scholar]
- 36.Mendoza JL, et al. The IFN-lambda-IFN-lambdaR1-IL-10Rbeta complex reveals structural features underlying type III IFN functional plasticity. Immunity. 2017;46:379–392. doi: 10.1016/j.immuni.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malakhova OA, et al. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006;25:2358–2367. doi: 10.1038/sj.emboj.7601149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Francois-Newton V, et al. USP18-based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon alpha response. PLoS ONE. 2011;6:e22200. doi: 10.1371/journal.pone.0022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilmes S, et al. Receptor dimerization dynamics as a regulatory valve for plasticity of type I interferon signaling. J. Cell Biol. 2015;209:579–593. doi: 10.1083/jcb.201412049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Babon JJ, et al. Suppression of cytokine signaling by SOCS3: characterization of the mode of inhibition and the basis of its specificity. Immunity. 2012;36:239–250. doi: 10.1016/j.immuni.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piganis RA, et al. Suppressor of cytokine signaling (SOCS) 1 inhibits type I interferon (IFN) signaling via the interferon alpha receptor (IFNAR1)-associated tyrosine kinase Tyk2. J. Biol. Chem. 2011;286:33811–33818. doi: 10.1074/jbc.M111.270207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ho HH, Ivashkiv LB. Role of STAT3 in type I interferon responses. Negative regulation of STAT1-dependent inflammatory gene activation. J. Biol. Chem. 2006;281:14111–14118. doi: 10.1074/jbc.M511797200. [DOI] [PubMed] [Google Scholar]
- 43.Lu L, et al. Gene regulation and suppression of type I interferon signaling by STAT3 in diffuse large B cell lymphoma. Proc. Natl Acad. Sci. USA. 2018;115:E498–E505. doi: 10.1073/pnas.1715118115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blumer T, Coto-Llerena M, Duong FHT, Heim MH. SOCS1 is an inducible negative regulator of interferon lambda (IFN-lambda)-induced gene expression in vivo. J. Biol. Chem. 2017;292:17928–17938. doi: 10.1074/jbc.M117.788877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michalska A, Blaszczyk K, Wesoly J, Bluyssen HAR. A positive feedback amplifier circuit that regulates interferon (IFN)-stimulated gene expression and controls type I and type II IFN responses. Front. Immunol. 2018;9:1135. doi: 10.3389/fimmu.2018.01135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheon H, et al. IFNβ-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J. 2013;32:2751–2763. doi: 10.1038/emboj.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kandhaya-Pillai R, et al. TNFα-senescence initiates a STAT-dependent positive feedback loop, leading to a sustained interferon signature, DNA damage, and cytokine secretion. Aging. 2017;9:2411–2435. doi: 10.18632/aging.101328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prakash A, Smith E, Lee CK, Levy DE. Tissue-specific positive feedback requirements for production of type I interferon following virus infection. J. Biol. Chem. 2005;280:18651–18657. doi: 10.1074/jbc.M501289200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moraga I, Harari D, Schreiber G, Uze G, Pellegrini S. Receptor density is key to the alpha2/beta interferon differential activities. Mol. Cell Biol. 2009;29:4778–4787. doi: 10.1128/MCB.01808-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Booy S, van Eijck CH, Dogan F, van Koetsveld PM, Hofland LJ. Influence of type-I Interferon receptor expression level on the response to type-I Interferons in human pancreatic cancer cells. J. Cell. Mol. Med. 2014;18:492–502. doi: 10.1111/jcmm.12200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pattyn E, et al. Dimerization of the interferon type I receptor IFNaR2–2 is sufficient for induction of interferon effector genes but not for full antiviral activity. J. Biol. Chem. 1999;274:34838–34845. doi: 10.1074/jbc.274.49.34838. [DOI] [PubMed] [Google Scholar]
- 52.Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Du M, Chen ZJ. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science. 2018;361:704–709. doi: 10.1126/science.aat1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davis D, Yuan H, Yang YM, Liang FX, Sehgal PB. Interferon-α-induced cytoplasmic MxA structures in hepatoma Huh7 and primary endothelial cells. Contemp. Oncol. (Pozn) 2018;22:86–94. doi: 10.5114/wo.2018.76149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao S, et al. Structure of myxovirus resistance protein a reveals intra- and intermolecular domain interactions required for the antiviral function. Immunity. 2011;35:514–525. doi: 10.1016/j.immuni.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 56.Shi X, Jiao B, Chen Y, Li S, Chen L. MxA is a positive regulator of type I IFN signaling in HCV infection. J. Med. Virol. 2017;89:2173–2180. doi: 10.1002/jmv.24867. [DOI] [PubMed] [Google Scholar]
- 57.Hillyer P, et al. Respiratory syncytial virus infection induces a subset of types I and III interferons in human dendritic cells. Virology. 2017;504:63–72. doi: 10.1016/j.virol.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou A, Paranjape JM, Der SD, Williams BR, Silverman RH. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology. 1999;258:435–440. doi: 10.1006/viro.1999.9738. [DOI] [PubMed] [Google Scholar]
- 61.Verhelst J, Hulpiau P, Saelens X. Mx proteins: antiviral gatekeepers that restrain the uninvited. Microbiol. Mol. Biol. R. 2013;77:551–566. doi: 10.1128/MMBR.00024-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dauber B, Wolff T. Activation of the antiviral kinase PKR and viral countermeasures. Viruses. 2009;1:523–544. doi: 10.3390/v1030523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schoggins JW, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl Acad. Sci. USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiang D, et al. Identification of five interferon-induced cellular proteins that inhibit west nile virus and dengue virus infections. J. Virol. 2010;84:8332–8341. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Richardson RB, et al. A CRISPR screen identifies IFI6 as an ER-resident interferon effector that blocks flavivirus replication. Nat. Microbiol. 2018;3:1214–1223. doi: 10.1038/s41564-018-0244-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Speer SD, et al. ISG15 deficiency and increased viral resistance in humans but not mice. Nat. Commun. 2016;7:11496. doi: 10.1038/ncomms11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morales DJ, Lenschow DJ. The antiviral activities of ISG15. J. Mol. Biol. 2013;425:4995–5008. doi: 10.1016/j.jmb.2013.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leaman DW, et al. Novel growth and death related interferon-stimulated genes (ISGs) in melanoma:greater potency of IFN-beta compared with IFN-alpha2. . J. Interf. Cytok. Res. 2003;23:745–756. doi: 10.1089/107999003772084860. [DOI] [PubMed] [Google Scholar]
- 70.Douam F, et al. Type III interferon-mediated signaling is critical for controlling live attenuated yellow fever virus infection in vivo. mBio. 2017;8:e00819–17. doi: 10.1128/mBio.00819-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Crotta S, et al. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog. 2013;9:e1003773. doi: 10.1371/journal.ppat.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Galani IE, et al. Interferon-λ mediates non-redundant front-line antiviral protection against influenza virus infection without compromising host fitness. Immunity. 2017;46:875–890. doi: 10.1016/j.immuni.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 73.Sjaastad LE, et al. Distinct antiviral signatures revealed by the magnitude and round of influenza virus replication in vivo. Proc. Natl Acad. Sci. USA. 2018;115:9610–9615. doi: 10.1073/pnas.1807516115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pott J, et al. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc. Natl Acad. Sci. USA. 2011;108:7944–7949. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Klinkhammer, J. et al. IFN-λ prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. eLife7, e33354 (2018). [DOI] [PMC free article] [PubMed]
- 76.Makjaroen J, et al. Comprehensive proteomics identification of IFN-λ3-regulated anti-viral proteins in HBV-transfected cells. Mol. Cell. Proteomics. 2018;17:2197–2215. doi: 10.1074/mcp.RA118.000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Muir AJ, et al. Phase 1b study of pegylated interferon lambda 1 with or without ribavirin in patients with chronic genotype 1 hepatitis C virus infection. Hepatology. 2010;52:822–832. doi: 10.1002/hep.23743. [DOI] [PubMed] [Google Scholar]
- 78.Phillips S, et al. Peg-interferon lambda treatment induces robust innate and adaptive immunity in chronic hepatitis B patients. Front. Immunol. 2017;8:621. doi: 10.3389/fimmu.2017.00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Prokunina-Olsson L, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 2013;45:164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Obajemu AA, et al. IFN-λ4 attenuates antiviral responses by enhancing negative regulation of IFN signaling. J. Immunol. 2017;199:3808–3820. doi: 10.4049/jimmunol.1700807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ramamurthy N, et al. Impact of interferon lamda 4 genotype on interferon-stimulated gene expression during DAA therapy for hepatitis C. Hepatology. 2018;68:3. doi: 10.1002/hep.29877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rosenberg BR, et al. Genetic variation at IFNL4 influences extrahepatic interferon-stimulated gene expression in chronic HCV patients. J. Infect. Dis. 2018;217:650–655. doi: 10.1093/infdis/jix593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Davidson S, et al. IFNλ is a potent anti-influenza therapeutic without the inflammatory side effects of IFNα treatment. EMBO Mol. Med. 2016;8:1099–1112. doi: 10.15252/emmm.201606413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Frabutt DA, Wang B, Riaz S, Schwartz RC, Zheng YH. Innate sensing of influenza A virus hemagglutinin glycoproteins by the host endoplasmic reticulum (ER) stress pathway triggers a potent antiviral response via ER-associated protein degradation. J. Virol. 2018;92:e01690–17. doi: 10.1128/JVI.01690-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ge M, et al. HERP binds TBK1 to activate innate immunity and repress virus replication in response to endoplasmic reticulum stress. J. Immunol. 2017;199:3280–3292. doi: 10.4049/jimmunol.1700376. [DOI] [PubMed] [Google Scholar]
- 87.Chatel-Chaix L, et al. Dengue virus perturbs mitochondrial morphodynamics to dampen innate immune responses. Cell Host Microbe. 2016;20:342–356. doi: 10.1016/j.chom.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.West AP, et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520:553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sun B, et al. Dengue virus activates cGAS through the release of mitochondrial DNA. Sci. Rep. 2017;7:3594. doi: 10.1038/s41598-017-03932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aguirre S, et al. Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat. Microbiol. 2017;2:17037. doi: 10.1038/nmicrobiol.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu CY, et al. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog. 2012;8:e1002780. doi: 10.1371/journal.ppat.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Franz KM, Neidermyer WJ, Tan YJ, Whelan SPJ, Kagan JC. STING-dependent translation inhibition restricts RNA virus replication. Proc. Natl Acad. Sci. USA. 2018;115:E2058–E2067. doi: 10.1073/pnas.1716937115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chiang JJ, et al. Viral unmasking of cellular 5S rRNA pseudogene transcripts induces RIG-I-mediated immunity. Nat. Immunol. 2018;19:53–62. doi: 10.1038/s41590-017-0005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao Y, Ye X, Dunker W, Song Y, Karijolich J. RIG-I like receptor sensing of host RNAs facilitates the cell-intrinsic immune response to KSHV infection. Nat. Commun. 2018;9:4841. doi: 10.1038/s41467-018-07314-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mazzon M, Jones M, Davidson A, Chain B, Jacobs M. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J. Infect. Dis. 2009;200:1261–1270. doi: 10.1086/605847. [DOI] [PubMed] [Google Scholar]
- 96.Ashour J, Laurent-Rolle M, Shi PY, Garcia-Sastre A. NS5 of dengue virus mediates STAT2 binding and degradation. J. Virol. 2009;83:5408–5418. doi: 10.1128/JVI.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jones M, et al. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J. Virol. 2005;79:5414–5420. doi: 10.1128/JVI.79.9.5414-5420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Morrison J, et al. Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLoS Pathog. 2013;9:e1003265. doi: 10.1371/journal.ppat.1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kumar A, et al. Zika virus inhibits type-I interferon production and downstream signaling. EMBO Rep. 2016;17:1766–1775. doi: 10.15252/embr.201642627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Grant A, et al. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe. 2016;19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Laurent-Rolle M, et al. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J. Virol. 2010;84:3503–3515. doi: 10.1128/JVI.01161-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lubick KJ, et al. Flavivirus antagonism of type I interferon signaling reveals prolidase as a regulator of IFNAR1 surface expression. Cell Host Microbe. 2015;18:61–74. doi: 10.1016/j.chom.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lin RJ, Liao CL, Lin E, Lin YL. Blocking of the alpha interferon-induced Jak-Stat signaling pathway by Japanese encephalitis virus infection. J. Virol. 2004;78:9285–9294. doi: 10.1128/JVI.78.17.9285-9294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lin RJ, Chang BL, Yu HP, Liao CL, Lin YL. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J. Virol. 2006;80:5908–5918. doi: 10.1128/JVI.02714-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang TC, et al. Proteomic analysis for Type I interferon antagonism of Japanese encephalitis virus NS5 protein. Proteomics. 2013;13:3442–3456. doi: 10.1002/pmic.201300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Laurent-Rolle M, et al. The interferon signaling antagonist function of yellow fever virus NS5 protein is activated by type I interferon. Cell Host Microbe. 2014;16:314–327. doi: 10.1016/j.chom.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bousse T, Chambers RL, Scroggs RA, Portner A, Takimoto T. Human parainfluenza virus type 1 but not Sendai virus replicates in human respiratory cells despite IFN treatment. Virus Res. 2006;121:23–32. doi: 10.1016/j.virusres.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 108.Chambers R, Takimoto T. Host specificity of the anti-interferon and anti-apoptosis activities of parainfluenza virus P/C gene products. J. Gen. Virol. 2009;90:1906–1915. doi: 10.1099/vir.0.011700-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Takeuchi K, et al. Sendai virus C protein physically associates with Stat1. Genes Cells. 2001;6:545–557. doi: 10.1046/j.1365-2443.2001.00442.x. [DOI] [PubMed] [Google Scholar]
- 110.Gotoh B, Takeuchi K, Komatsu T, Yokoo J. The STAT2 activation process is a crucial target of Sendai virus C protein for the blockade of alpha interferon signaling. J. Virol. 2003;77:3360–3370. doi: 10.1128/JVI.77.6.3360-3370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wiltzer L, et al. Interaction of rabies virus P-protein with STAT proteins is critical to lethal rabies disease. J. Infect. Dis. 2014;209:1744–1753. doi: 10.1093/infdis/jit829. [DOI] [PubMed] [Google Scholar]
- 112.Shaw AE, et al. Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type I interferon responses. PLoS Biol. 2017;15:e2004086. doi: 10.1371/journal.pbio.2004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Smith I, Wang LF. Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr. Opin. Virol. 2013;3:84–91. doi: 10.1016/j.coviro.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pavlovich SS, et al. The Egyptian rousette genome reveals unexpected features of bat antiviral immunity. Cell. 2018;173:1098–1110. doi: 10.1016/j.cell.2018.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhou P, et al. Contraction of the type I IFN locus and unusual constitutive expression of IFN-α in bats. Proc. Natl Acad. Sci. USA. 2016;113:2696–2701. doi: 10.1073/pnas.1518240113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xie J, et al. Dampened STING-dependent interferon activation in bats. Cell Host Microbe. 2018;23:297–301. doi: 10.1016/j.chom.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Aguirre S, et al. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 2012;8:e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ma Z, Damania B. The cGAS-STING defense pathway and its counteraction by viruses. Cell Host Microbe. 2016;19:150–158. doi: 10.1016/j.chom.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bowen JR, et al. Zika virus antagonizes type I interferon responses during infection of human dendritic cells. PLoS Pathog. 2017;13:e1006164. doi: 10.1371/journal.ppat.1006164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gorman MJ, et al. An immunocompetent mouse model of Zika virus infection. Cell Host Microbe. 2018;23:672–685. doi: 10.1016/j.chom.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Patel MR, Loo YM, Horner SM, Gale M, Jr., Malik HS. Convergent evolution of escape from hepaciviral antagonism in primates. PLoS Biol. 2012;10:e1001282. doi: 10.1371/journal.pbio.1001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yan D, et al. A single mutation at position 156 in the envelope protein of tembusu virus is responsible for virus tissue tropism and transmissibility in ducks. J. Virol. 2018;92:e00427–18. doi: 10.1128/JVI.00427-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nitta S, et al. Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type I interferon-dependent innate immunity. Hepatology. 2013;57:46–58. doi: 10.1002/hep.26017. [DOI] [PubMed] [Google Scholar]
- 126.Ding Q, et al. Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. J. Hepatol. 2013;59:52–58. doi: 10.1016/j.jhep.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 127.Sun L, et al. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS ONE. 2012;7:e30802. doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Clementz MA, et al. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 2010;84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xing Y, et al. The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. J. Gen. Virol. 2013;94:1554–1567. doi: 10.1099/vir.0.051169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen X, et al. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5:369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 132.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yang Y, et al. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc. Natl Acad. Sci. USA. 2007;104:7253–7258. doi: 10.1073/pnas.0611506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen S, Wu Z, Wang M, Cheng A. Innate immune evasion mediated by Flaviviridae non-structural proteins. Viruses. 2017;9:291. doi: 10.3390/v9100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li K, et al. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl Acad. Sci. USA. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Qu L, et al. Disruption of TLR3 signaling due to cleavage of TRIF by the hepatitis A virus protease-polymerase processing intermediate, 3CD. PLoS Pathog. 2011;7:e1002169. doi: 10.1371/journal.ppat.1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.van Kasteren PB, et al. Arterivirus and nairovirus ovarian tumor domain-containing Deubiquitinases target activated RIG-I to control innate immune signaling. J. Virol. 2012;86:773–785. doi: 10.1128/JVI.06277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Marq JB, Hausmann S, Veillard N, Kolakofsky D, Garcin D. Short double-stranded RNAs with an overhanging 5’ ppp-nucleotide, as found in arenavirus genomes, act as RIG-I decoys. J. Biol. Chem. 2011;286:6108–6116. doi: 10.1074/jbc.M110.186262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fan L, Briese T, Lipkin WI. Z proteins of New World arenaviruses bind RIG-I and interfere with type I interferon induction. J. Virol. 2010;84:1785–1791. doi: 10.1128/JVI.01362-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Childs K, Randall R, Goodbourn S. Paramyxovirus V proteins interact with the RNA Helicase LGP2 to inhibit RIG-I-dependent interferon induction. J. Virol. 2012;86:3411–3421. doi: 10.1128/JVI.06405-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Goubau D, et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5’-diphosphates. Nature. 2014;514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Davis WG, et al. The 3’ untranslated regions of influenza genomic sequences are 5’PPP-independent ligands for RIG.-I. PLoS ONE. 2012;7:e32661. doi: 10.1371/journal.pone.0032661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5’-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 144.Unterstab G, et al. Viral targeting of the interferon-{beta}-inducing Traf family member-associated NF-{kappa}B activator (TANK)-binding kinase-1. Proc. Natl Acad. Sci. USA. 2005;102:13640–13645. doi: 10.1073/pnas.0502883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ning YJ, et al. Viral suppression of innate immunity via spatial isolation of TBK1/IKKepsilon from mitochondrial antiviral platform. J. Mol. Cell Biol. 2014;6:324–337. doi: 10.1093/jmcb/mju015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Alff PJ, et al. The pathogenic NY-1 hantavirus G1 cytoplasmic tail inhibits RIG-I- and TBK-1-directed interferon responses. J. Virol. 2006;80:9676–9686. doi: 10.1128/JVI.00508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cimica V, Dalrymple NA, Roth E, Nasonov A, Mackow ER. An innate immunity-regulating virulence determinant is uniquely encoded by the Andes virus nucleocapsid protein. mBio. 2014;5:e01088–13. doi: 10.1128/mBio.01088-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lee MH, et al. RNA helicase retinoic acid-inducible gene I as a sensor of Hantaan virus replication. J. Gen. Virol. 2011;92:2191–2200. doi: 10.1099/vir.0.032367-0. [DOI] [PubMed] [Google Scholar]
- 149.Basler CF, et al. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl Acad. Sci. USA. 2000;97:12289–12294. doi: 10.1073/pnas.220398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cardenas WB, et al. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J. Virol. 2006;80:5168–5178. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Frolova EI, et al. Roles of nonstructural protein nsP2 and Alpha/Beta interferons in determining the outcome of Sindbis virus infection. J. Virol. 2002;76:11254–11264. doi: 10.1128/JVI.76.22.11254-11264.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Levine JR, et al. Antagonism of type I interferon responses by new world hantaviruses. J. Virol. 2010;84:11790–11801. doi: 10.1128/JVI.00916-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Schneider U, Martin A, Schwemmle M, Staeheli P. Genome trimming by Borna disease viruses: viral replication control or escape from cellular surveillance? Cell. Mol. Life Sci. 2007;64:1038–1042. doi: 10.1007/s00018-007-6545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sugai A, Sato H, Takayama I, Yoneda M, Kai C. Nipah and Hendra virus nucleoproteins inhibit nuclear accumulation of signal transducer and activator of transcription 1 (STAT1) and STAT2 by interfering with their complex formation. J. Virol. 2017;91:e01136–17. doi: 10.1128/JVI.01136-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Swedan S, Musiyenko A, Barik S. Respiratory syncytial virus nonstructural proteins decrease levels of multiple members of the cellular interferon pathways. J. Virol. 2009;83:9682–9693. doi: 10.1128/JVI.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ramaswamy M, et al. Respiratory syncytial virus nonstructural protein 2 specifically inhibits type I interferon signal transduction. Virology. 2006;344:328–339. doi: 10.1016/j.virol.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 157.Kraus TA, Garza L, Horvath CM. Enabled interferon signaling evasion in an immune-competent transgenic mouse model of parainfluenza virus 5 infection. Virology. 2008;371:196–205. doi: 10.1016/j.virol.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 158.Parisien JP, Lau JF, Horvath CM. STAT2 acts as a host range determinant for species-specific paramyxovirus interferon antagonism and simian virus 5 replication. J. Virol. 2002;76:6435–6441. doi: 10.1128/JVI.76.13.6435-6441.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rodriguez JJ, Horvath CM. Host evasion by emerging paramyxoviruses: Hendra virus and Nipah virus v proteins inhibit interferon signaling. Viral Immunol. 2004;17:210–219. doi: 10.1089/0882824041310568. [DOI] [PubMed] [Google Scholar]
- 160.Rodriguez JJ, Wang LF, Horvath CM. Hendra virus V protein inhibits interferon signaling by preventing STAT1 and STAT2 nuclear accumulation. J. Virol. 2003;77:11842–11845. doi: 10.1128/JVI.77.21.11842-11845.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rodriguez JJ, Cruz CD, Horvath CM. Identification of the nuclear export signal and STAT-binding domains of the Nipah virus V protein reveals mechanisms underlying interferon evasion. J. Virol. 2004;78:5358–5367. doi: 10.1128/JVI.78.10.5358-5367.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rodriguez JJ, Parisien JP, Horvath CM. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 2002;76:11476–11483. doi: 10.1128/JVI.76.22.11476-11483.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Parisien JP, et al. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology. 2001;283:230–239. doi: 10.1006/viro.2001.0856. [DOI] [PubMed] [Google Scholar]
- 164.Ramachandran A, Parisien JP, Horvath CM. STAT2 is a primary target for measles virus V protein-mediated alpha/beta interferon signaling inhibition. J. Virol. 2008;82:8330–8338. doi: 10.1128/JVI.00831-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hahm B, Trifilo MJ, Zuniga EI, Oldstone MB. Viruses evade the immune system through type I interferon-mediated STAT2-dependent, but STAT1-independent, signaling. Immunity. 2005;22:247–257. doi: 10.1016/j.immuni.2005.01.005. [DOI] [PubMed] [Google Scholar]