Abstract
A variety of amino acid substitutions in the NS3–4A protease of the hepatitis C virus (HCV) lead to protease inhibitor (PI) resistance. Many of these significantly impair the replication fitness of the resistant variants in a genotype- and subtype-dependent manner, a critical factor in determining the probability with which resistant variants will persist. However, the underlying molecular mechanisms are unknown. Here, we present a novel residue-interaction network approach to determine how near neighbor interactions of PI resistance mutations in NS3–4A can impact protease functional sites dependent on their genomic background. We constructed subtype-specific consensus residue networks for subtype 1a and 1b from protease structure ensembles combined with biological properties of protein residues and evolutionary amino acid conservation. By applying local and global network topology analysis and visual exploration, we characterize PI resistance-associated sites and outline differences in near neighbor interactions. We find local residue-interaction patterns and features at protease functional sites that are subtype specific. The noncovalent bonding patterns indicate higher fitness costs conferred by PI resistance mutations in a subtype 1b genomic background and explain the prevalence of Q80K and R155K in subtype 1a. Based on local residue interactions, we predict a subtype-specific role for the protease residue NS3–Q80 in molecular mechanisms related to the assembly of infectious virus particles that is supported by experimental data on the capacity of Q80K variants to replicate and produce infectious virus in subtype 1a and 1b cell culture.
Keywords: hepatitis C virus, subtype, viral variant fitness, residue networks, molecular determinants
INTRODUCTION
Graph-based approaches provide a convenient means for characterizing protein structure information (1). A comprehensive method for comparative analysis of molecular details from multiple protein structure models, however, is a difficult task. Here, we present a novel consensus residue network approach from ensembles of experimental protein structures that we applied to characterize near neighbor interactions of drug resistance mutations and their impact on the replicative fitness of hepatitis C virus (HCV) variants. Resistance-associated amino acid substitutions in drug target sites of direct antivirals frequently cause deficits in the replicative fitness of HCV resistance-associated viral variants (RAVs) (2). Interestingly, RAVs from patients that failed on inhibitors against the NS3–4A protease (PI) disappear significantly more rapidly in HCV subtype 1b-than 1a-infected patients (Table 1) (3). Moreover, some key resistance-associated amino acid substitutions are specifically observed in subtype 1a but not in 1b (Table 1). Such observations likely depend on the subtype-specific genomic background of the resistance mutations and are potentially explained by a genotype- and subtype-specific suite of resistant variants.
Table 1.
Subtype-specific RAV persistence.
| Median persistence (months) | ||||
|---|---|---|---|---|
| Residue | RAV | Subtype 1a | Subtype 1b | Reference |
| V36 | A | 2.8 | 3.6 | Sullivan et al. (3) |
| M | 9.3 | 6.3 | ||
| T54 | A | 2.3 | 2.8 | Sullivan et al. (3) |
| S | 11.5 | 1.8 | ||
| V55 | A | ND# | 57 | Susser et al. (57) |
| Q80 | K | 36 | 24 | Lenz et al. (24) |
| R155 | K | 9.8 | -* | Sullivan et al. (3) |
| A156 | S | 6.7 | 8.2 | Sullivan et al. (3) |
| T | 1.8 | 2.7 | ||
| D168 | A | 5 | ND# | Manns et al. (58) Krishnan et al. (59) |
| V | ND# | >9 | Zeuzem et al. (LEAGUE-1) (60) Lenz et al. (24) Krishnan et al. (59) |
|
| N | >6 | ND# | Buti et al. (C-SALVAGE) (61) | |
| I170 | V | ND# | ND# | |
Time to loss of NS3–4A protease RAVs reported in the literature as detected by population and/or clonal sequencing.
ND, no data available;
No RAVs observed.
The NS3–4A protease plays an essential role in the HCV replication cycle and is a prime target site for direct antiviral therapy. By proteolytically processing nonstructural proteins from the viral polyprotein, the protease is involved in RNA synthesis (4). Moreover, it modulates virus assembly by interference with the C-terminal RNA helicase domain of NS3 (4). An allosteric site in the domain interface regulates NS3 function via stabilizing conformational states (5). In addition, the protease blocks innate immune signalling of infected cells by cleavage of adaptor molecules, in particular Toll/interleukin-1 receptor (TIR) domain containing adapter inducing interferon–β TRIF (6) and mitochondrial antiviral signalling protein MAVS (also called Cardif, VISA, and IPS–1) (7, 8) (Fig 1). Binding of PIs to the protease active site and the related resistance-associated amino acid substitutions can interfere with these processes (9, 10). Clinical data suggest low fitness costs due to drug resistance mutations in a subtype 1a genomic background (11), yet there is little understanding of the underlying molecular mechanisms.
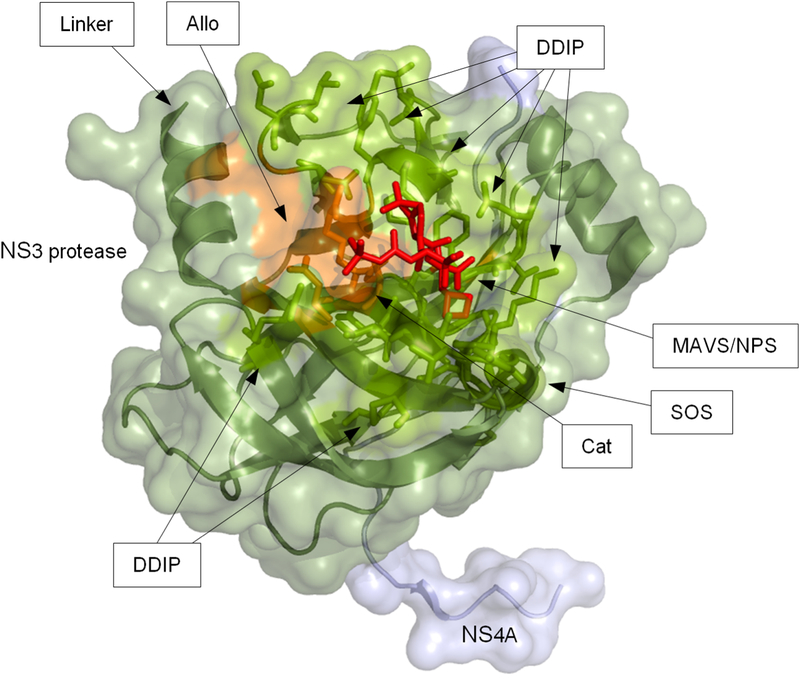
Fig 1. 3D protein structure of the NS3–4A protease domain.
The protein structure shows a subtype 1a NS3 protease domain from Protein Data Bank entry 2OC8 co-crystallized with NS4A (light blue) and a linear protease inhibitor (PI) (purple stick model). The protein backbone is given as ribbon model with transparent surface depiction. Residues where PI resistance-associated amino acid substitutions occur are highlighted as orange stick models; functionally important residues are shown as green stick models. Functionally important sites are indicated by arrows: allosteric site (Allo); protease active/catalytic site (Cat); domain-domain interaction sites (DDIP); linker region (Linker); MAVS binding site (MAVS); binding site for the natural protease substrate (NPS); SOS-binding site for TRIF (SOS).
We apply consensus residue networks to characterize drug resistance mutations in their subtype-specific protein structure environment to deduce biological phenotypes of drug-resistant viral variants dependent on their genomic background in different HCV subtypes.
RESULTS
Consensus residue network approach.
We made use of residue-interaction networks (RINs) for a subtype-specific characterization of PI resistance-associated sites and their potential impact on protein fold and function in NS3–4A (Fig 2). To compare available multiple structure models from the Protein Data Bank RCSB PDB, we constructed consensus residue networks from subtype 1a and 1b NS3–4A protease structure ensembles, enriched with information on molecular properties of amino acid residues, sequence conservation, and inter-subtype variability (see Materials and Methods). The networks provide information about the location and nature of resistance-associated sites and their neighboring residues in the NS3–4A protease structure to residues previously identified to pertain to different NS3 functional sites. Based on the network topology and the bonding patterns of resistance-associated sites and functional sites, we predict the subtype-specific impact of resistance-associated amino acid substitutions on the replicative fitness of the PI-resistant variants.
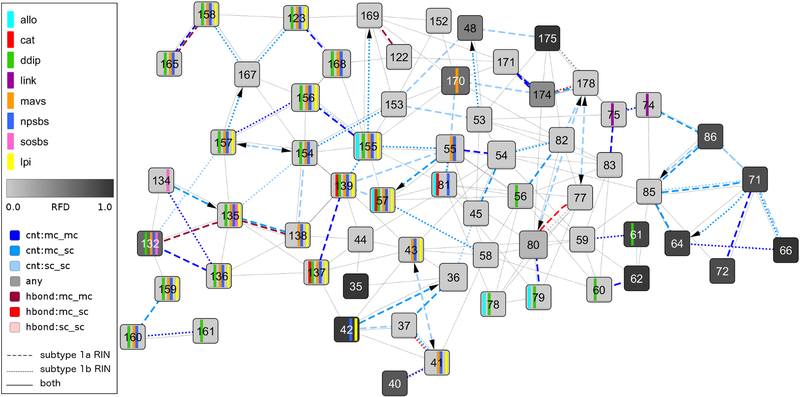
Fig 2. Network comparison view between subtype 1a and 1b consensus residue-interaction networks with focus on functional sites and protease inhibitor resistance-associated sites.
This comparison network view shows the noncovalent residue interactions between a subset of residues of the NS3–4A protease with focus on the differences between subtype 1a and 1b consensus residue-interaction networks. The view focuses on the protease inhibitor (PI) resistance-associated sites 36, 54, 55, 80, 155, 156, 168, and 170 and further includes their direct neighbors, all residues annotated as functionally important (see Materials and Methods), and residues that vary between subtype 1a and 1b. Functional residue annotations are shown as vertical bars (cyan – allosteric site; red - catalytic site; green - domain-domain interaction sites; violet - residues interacting with the linker region; orange - MAVS peptide; blue - natural peptide substrate; yellow - linear PI; purple - SOS-binding site of TRIF). The residue frequency distance is mapped to the node color using a white-to-grey gradient for small-to-large values. Solid edge lines represent noncovalent residue interactions present in both subtype consensus residue-interaction networks, dashed edge lines are residue interactions from the subtype 1a consensus residue-interaction network, and dotted edge lines correspond to residue interactions in the subtype 1b consensus residue-interaction network. Edges that represent contacts are colored in blue, hydrogen bonds in red, and overlaps in grey; a darker color indicates interaction between main chain atoms, while lighter color as well as an arrow stands for side chain atom interactions.
Global network topology and sequence analysis.
To compare the overall network topology between subtype 1a and 1b, we computed global topology measures for the subtype-specific consensus networks (Table 2). Each network comprises a total of 181 nodes, referring to residues of the NS3–4A protease. The network diameter is 12 edges in subtype 1a versus 11 edges in subtype 1b. The average number of neighbors, connected nodes and network density are similar between both subtypes (Table 2). Minor differences are observed in overall residue interactions between subtypes. Of the interaction edges, 86% in the subtype 1a consensus network can be mapped to 94% of the edges in the subtype 1b network. Hence, the residue interaction difference is only 14% (77 edges) for subtype 1a and 6% (29 edges) for subtype 1b. Despite this overall similarity, the consensus networks show subtype-specific differences in clustering coefficient, degree, and betweenness centrality; measures that reflect residue connectivity and control over neighboring residues (Table 2) (12–15). Although the subtype 1a network shows a higher clustering coefficient (3.4%) and higher average number of neighbors (6.5%), the average distance from resistance-associated sites to NS3–4A protease functional sites in subtype 1a is larger than in subtype 1b (2.5%) (Table 2, Suppl. Table 1 and 2).
Table 2.
Consensus residue-interaction network topology measures.
| Subtype | ||
|---|---|---|
| Topology measure | 1a | 1b |
| Radius | 6 | 6 |
| Diameter | 12 | 11 |
| Ave. path length | 4.777 | 4.785 |
| Ave. clustering coefficient | 0.422 | 0.408 |
| Ave. number of neighbors | 6.157 | 5.78 |
| Density | 0.035 | 0.034 |
| Connected nodes | 178 | 173 |
| Isolated nodes | 3 | 7 |
| Combi edges (unique*) | 548 (77) | 500 (29) |
| Common** combi edges | 85.9% | 94.2% |
| Contact edges (unique*) | 726 (184) | 676 (129) |
| H-bond edges (unique*) | 89 (30) | 74 (15) |
Edges unique to either the subtype 1a or 1b residue-interaction network;
Edges common to both subtype 1a and 1b residue-interaction networks.
We characterized the residue frequency distance (RFD) between subtypes to assess similarity between amino acid frequencies at equivalent sequence positions in the NS3–4A protease (Suppl. Table 3 and 4). Thereby, large RFD values (> 0.1) indicate a tendency to have a different amino acid distribution between subtypes (see Materials and Methods). From the 181 NS3–4A protease residues, only 19 residues (10.5%) show RFD values > 0.1, whereas the majority of residues (152 residues, 84%), comprising most of the PI resistance-associated sites, shows RFD values < 0.001, thus have the same or similar amino-acid profile in subtype 1a and 1b (Suppl. Table 1 and 2). A subtype-specific different amino acid profile at PI resistance-associated sites is observed at residue 80 and 170 (RFD > 0.1).
Local network topology at PI resistance-associated sites.
Next, we characterized the local residue neighborhood at PI resistance-associated sites to predict the potential impact of amino acid substitutions on the residue-bonding patterns and protease functional sites.
Network topology at V36.
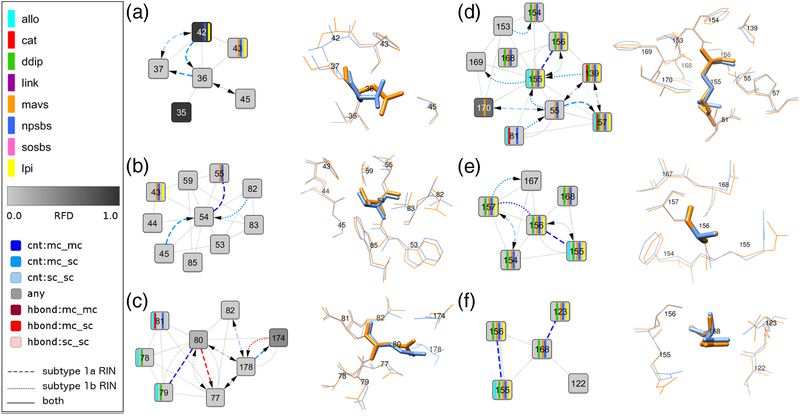
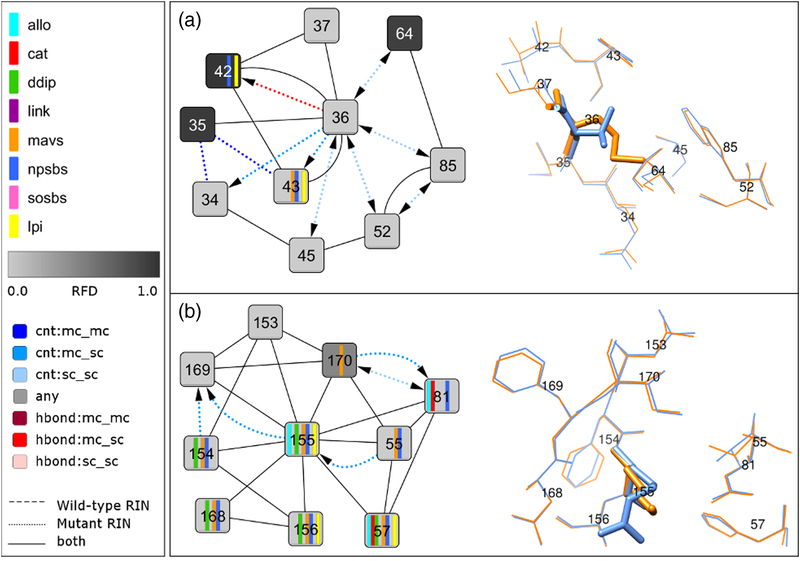
V36 is connected to five neighboring residues, 35, 37, 42, 43 and 45; 2/5 are protease functional sites (natural protease substrate-binding site) (Fig 3A). The clustering coefficient for V36 is higher in subtype 1a than 1b (0.333 versus 0.167) (Table 3, Suppl. Fig 1). The subtype 1a-bonding pattern for V36 shows specific interactions with residue 42 (natural protease substrate-binding site) that also has a different amino acid distribution between subtypes (RFD > 0.1). We identified an experimental protein structure comprising the amino acid substitution V36M in subtype 1a, PDB–2QV1, and constructed a comparison network from the subtype 1a–M36 mutant structure and the subtype 1a consensus network (Fig 4A). In the comparison network, M36 retains all seven noncovalent interactions with first neighbor residues, connecting to five additional residues through seven new side chain (sc) interactions; van-der-Waals (vdW) interactions (residues 34, 43, 45, 52, 64 and 85) and one H-bond interaction (residue 42). Although these changes amounts to a 100% gain of interactions for subtype 1a–M36 compared to wild type, only few protease functional sites are affected; 7/10 interactions occur with nonfunctional sites. As expected from the residue-network analysis, V36M in subtype 1a shows no significant deficit in its replicative fitness as suggested by the long term persistence of V36M-RAVs in subtype 1a-patients (Table 1).
Fig 3. Network comparison view and corresponding 3D structure close-up for the subtype-specific protease inhibitor resistance-associated sites: (A) 36, (B) 54, (C) 80, (D) 155, (E) 156, and (F) 168.
Each comparison network view shows the noncovalent residue interactions between the specified residue and its direct neighbors from the subtype 1a and 1b consensus residue-interaction networks. Node and edge colors as well as functional annotations are described in the graphical legend above and in the Fig 2 legend. The corresponding residues from the subtype 1a reference structure from Protein Data Bank entry 2OBQ and the subtype 1b reference structure from Protein Data Bank entry 1DXP are aligned and shown as blue and orange sticks, respectively.
Table 3.
Network topology of protease inhibitor resistance-associated sites.
| Topology measures | ||||||
|---|---|---|---|---|---|---|
| Class | Res | Subtype | Degree | Clust | Clos | Betw |
| L | 36 | 1a | 0.022 | 0.333 | 0.211 | 0.001 |
| 1b | 0.022 | 0.167 | 0.224 | 0.003 | ||
| 54 | 1a | 0.050 | 0.250 | 0.282 | 0.125 | |
| 1b | 0.050 | 0.250 | 0.269 | 0.100 | ||
| 55 | 1a | 0.050 | 0.278 | 0.262 | 0.052 | |
| 1b | 0.039 | 0.429 | 0.249 | 0.034 | ||
| 170 | 1a | 0.044 | 0.286 | 0.257 | 0.037 | |
| 1b | 0.034 | 0.400 | 0.245 | 0.022 | ||
| M* | 80 | 1a | 0.039 | 0.476 | 0.222 | 0.006 |
| 1b | 0.034 | 0.467 | 0.214 | 0.007 | ||
| B | 155 | 1a | 0.050 | 0.278 | 0.262 | 0.043 |
| 1b | 0.056 | 0.222 | 0.262 | 0.072 | ||
| 156 | 1a | 0.028 | 0.500 | 0.235 | 0.007 | |
| 1b | 0.028 | 0.500 | 0.229 | 0.008 | ||
| 168 | 1a | 0.022 | 0.333 | 0.222 | 0.004 | |
| 1b | 0.022 | 0.167 | 0.217 | 0.006 | ||
Class: class of compound (L – linear, M – macrocyclic, B – both) (22); Res: residue; Degree: degree centrality; Clust: clustering coefficient; Clos: closeness centrality; Betw: betweenness centrality. All values are rounded for better readability.
Exception: Sovaprevir (22)
Fig 4. Network comparison view between wild-type and mutant structure for V36M and R155K and corresponding 3D structure close-up.
(A) The comparison network view between the subtype 1a consensus residue-interaction network and the residue-interaction network of the V36M mutant structure from Protein Data Bank entry 2QV1 focuses on the direct noncovalent interactions of residue 36 with its neighbors. The corresponding residues in the structure are shown as blue and orange sticks, respectively. (B) The comparison network view between the subtype 1a consensus residue-interaction network and the residue-interaction network of the R155K mutant structure from Protein Data Bank entry 2OIN focuses on the direct noncovalent interactions of residue 155 with its neighbors. The corresponding residues in the structure are shown as blue and orange sticks, respectively.
Network topology at T54.
T54 is highly connected with neighboring residues with a node degree of 0.05 and a clustering coefficient of 0.25 in both subtypes (Table 3, Suppl. Fig 1). It shows direct interactions to nine neighboring residues, 43, 44, 45, 53, 55, 59, 82, 83 and 85 (Fig 3B); 2/9 residues are protease functional sites (natural protease-substrate binding sites). Among those sites, residue 55 is previously reported with a key role in structural rearrangements at the protease-helicase interface (16). The mechanism is known to involve a V55-neighboring surface loop in the NS3–4A protease structure (16). By using the consensus residue networks, we identified a unique 54–55 interaction in subtype 1a and a related subtype 1a-specific subnetwork of residue interactions comprising the loop residues 56, 57 and 58 (Suppl. Fig 2). Our network-based findings suggest a subtype-specific modulatory role of this loop structure on the protease-helicase interface in subtype 1a that likely explains the discrepancy in the persistence of T54S– RAVs between subtype 1a- and 1b-patients (Table 1).
Network topology at Q80.
Q80 is highly connected with neighboring nodes, showing a similar clustering coefficient of 0.476 and 0.467 in subtype 1a and 1b, respectively (Table 3, Suppl. Fig 1). In both subtypes, Q80 forms direct interactions with seven neighboring residues; 3/7 are protease functional sites (natural protease substrate-binding site, allosteric site, domain-domain interaction sites (DDIP)) (Fig 3C). Q80 has a distinct amino acid distribution between subtypes (RFD > 0.1) and a subtype 1a-specific bonding pattern with residues in the protease-helicase interface; residue 79 and 178 (DDIP, allosteric site, inter-domain linker). Moreover, Q80 interact indirectly via residue 178 with residues that neighbor the protease-helicase linker; residue 74 and 75 (Fig 2). Based on the network topology, we hypothesized a subtype-specific modulatory role for Q80 in the protease-helicase interaction and hence on RNA replication and assembly of infectious virus particles (17, 18). Molecular rearrangements in the protease-helicase interface due to Q80K are also supported from the local residue-bonding pattern at Q80. Different from the wild-type residue, the lysine sc in the mutant K80 is positively charged and provides for salt-bridge interactions with negatively charged neighboring residues. A potential K80 sc-interaction partner is D79, a domain-domain interaction site that pertain to the NS3 allosteric site (Fig 3C).
Network topology at R155.
R155 is a functionally important protease residue (natural protease substrate-binding site, allosteric site, DDIP). The subtype-specific network topology at R155 differs in betweenness centrality and node degree that are both higher in subtype 1b than 1a (Table 3, Suppl. Fig 1). Both measures indicate that R155 in the subtype 1b protease structure is more important for information spread and control over neighboring residues than in subtype 1a. R155 in both subtypes is connected to 10 neighboring residues; 8/10 interacting sites are functionally important (Fig 3D). In subtype 1b, a unique R155 sc-interaction with S139 of the protease catalytic triad suggests a critical impact of amino acid substitutions at R155 on the proteolytic activity. We identified an experimental protein structure comprising the amino acid substitution R155K in subtype 1a, PDB–2OIN. To characterize the structure impact of subtype 1a–R155K, we constructed a comparison network from the mutant structure and the subtype 1a-specific consensus network (Fig 4B) and quantified the fraction of changes in residue interactions. Interestingly, K155 in subtype 1a retains all 12 noncovalent interactions with first neighbors and only gains two additional interactions (17%). We find no direct impact on protease functional sites in subtype 1a–R155K.
Other resistance sites.
Only few clinical data are available on RAV persistence for amino acid substitutions at V55, A156, D168 and I170. These residues form dense bonding patterns with functionally important neighboring residues. For A156 and D168, we find only minor subtype-specific differences in the local residue-bonding patterns (Fig 3EF). In contrast, V55 and I170 form a unique sc interaction in subtype 1a (Suppl. Fig 3) and show subtype-specific differences in node degree, clustering coefficient and betweenness centrality (Table 3, Suppl. Fig 1).
Replicative fitness of Q80K in different genomic backgrounds.
Q80K is clinically most important and shows particularly strong subtype specificity. Based on our network analysis, we predicted subtype-specific structural rearrangements that impact RNA replication and virus assembly differentially in subtype 1a and 1b. To prove our in silico predictions and explore the mechanisms underlying the Q80K prevalence for subtype 1a, we applied reverse genetics in subtype-specific infectious HCV cell culture models. The amino acid substitution Q80K was created within the subtype 1a genomic background H77S.3 and the subtype 1b genomic background N.2, and their impact on replication of the viral RNA and production of infectious virus determined in RNA-transfected cells. Detailed information on experimental procedures is given in Materials and Methods. Both viruses are adapted to replicate in cell culture and as reported previously, the kinetics of RNA replication and virus production differ between H77S.3 and N.2 viruses (19).
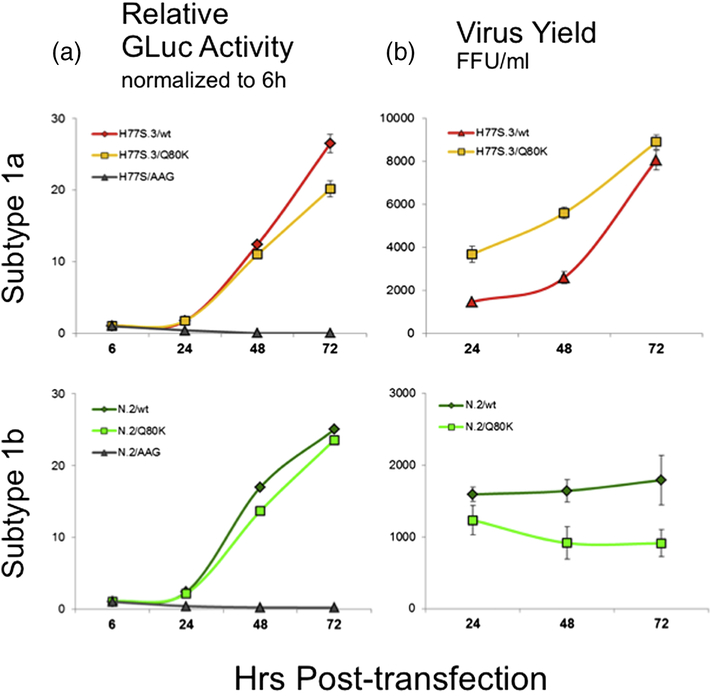
Replicative fitness: In both genomic backgrounds, Q80K showed only modest impairment of RNA replication capacity as measured by GLuc activity lower than the parental (wild type, WT) RNA. Compared to the parental H77S.3/GLuc2A and N.2/Gluc2A RNA, we observed a 20% reduction of replication in H77S.3 versus 6% reduction in N.2 at 72 hrs after transfection (Fig 5A).
Infectious virus yield: Yields of infectious virus generally correlate well with the RNA replication capacity (10). In a subtype 1a genomic background, the infectious virus production for H77S.3/Q80K was not reduced compared to the parental genome. Here, Q80K even produced slightly more infectious virus particles than WT at all times measured. In contrast, Q80K in the subtype 1b genetic background N.2 showed greater impairment in its ability to produce infectious virus than expected from reductions in RNA replication capacity. Virus production by N.2/Q80K was already lower at 24 hrs after transfection (23% compared to WT). By 72 hrs, virus production was almost 50% lower compared to parental N.2 (Fig 5B). As predicted from the residue-network analysis detailed above, the production of infectious virus showed distinct subtype-specific differences. The discordance between RNA replication capacity and yields of infectious virus for N.2/Q80K suggests a negative impact particularly on infectious virus production for Q80K in a subtype 1b genomic background.
Fig 5. Comparison of replication capacity and infectious virus yield of H77S.3 versus N.2 RNAs with the Q80K substitution in the NS3–4A protease.
(A) The effect of NS3-Q80K on the RNA replication capacity was measured by GLuc assay in the H77S.3/GLuc2A (left, upper panel) or the N.2/GLuc2A genetic background (left, lower panel). Results are normalized to GLuc activity at 6 hrs post transfection, and represent the mean ± standard deviation of triplicate samples. (B) The effect of NS3-Q80K on the infectious virus production from the H77S.3 (right, upper panel) or the N.2 background (right, lower panel) was measured by a focus forming unit (FFU) assay from cell culture supernatant fluids collected every 24 hr post transfection. Data represent the mean ± standard deviation from at least 3 independent experiments.
DISCUSSION
We made use of publicly-available sequence and structure data and present a graph-based approach to address subtype-specific differences in the replicative fitness of drug-resistant viral variants in HCV. Comparison views of subtype-specific consensus networks of the NS3–4A protease of HCV were constructed from protein structure ensembles and described as graphs that use common measures of node centrality. The consensus networks were characterized for their global network topology and local connectivity at PI resistance-associated sites. Different from coevolutionary analysis techniques that infer structural details of proteins based on primary sequence information (20, 21), we characterize bonding patterns in residue-interaction networks based on available experimental structure information from the Protein Data Bank. The most novel aspect is the idea of creating consensus residue networks that make multiple experimental structure information accessible and comparable to gain additional insights into the structural and functional roles of interacting residues.
In the NS3–4A protease of HCV, the overall network topology between subtype 1a and 1b is similar. In both consensus networks, the drug resistance-associated sites are highly connected with protease functional sites, however, we find that resistance-associated sites in the subtype 1a network are more distant from protease functional sites than in subtype 1b. This can explain lower replicative fitness costs of drug resistance mutations in a subtype 1a protease genomic background (as suggested by RAV persistence; Table 1). Although our approach is capable of detecting allosteric interactions as well as capturing effects of residues distant in the protein structure (Suppl. Fig 2), we find the local analysis of residue interactions most suitable to explain clinical observations in this particular application scenario. To focus on the specific PI resistance-associated sites, we characterized their local residue-interaction patterns with near neighbor residues. Here, the bonding patterns at major resistance-associated sites suggest a potential for better variant replicative fitness in a subtype 1a structure-specific context.
In particular, we identified specific molecular details that might explain the prevalence of Q80K and R155K for subtype 1a and higher fitness costs in subtype 1b. From pooled samples from 1797 patients in phase 3 clinical trials with a PI, Sullivan et al. observed no R155K–RAVs in subtype 1b but frequently observed R155K–RAVs in subtype 1a with long term persistence (Table 1) (3). Whereas a dense R155 residue-interaction pattern comprising multiple functional residues of the protease active site is observed in both subtypes, additional interactions to the protease catalytic triad are specific for subtype 1b. The unique noncovalent interactions suggest a larger subtype-specific structural impact on the protease catalytic site for R155K in subtype 1b. This notion is indirectly supported by experimental mutant structure data from R155K in subtype 1a. Here, the mutant structure revealed no direct impact on protease functional sites. Q80K is highly prevalent in subtype 1a (up to 47%) but rarely observed in subtype 1b (0.5%) (22). As a natural polymorphism in treatment-naïve patients, Q80K has been associated with a reduced treatment response to simeprevir (23, 24) and a median time until loss of 36 months in subtype 1a versus 24 months in subtype 1b (24). By exploratory visual analysis of the residue-interaction networks, we identified subtype-specific bonding patterns of Q80 with residues of the protease-helicase interface and indirect residue interactions with the protease-helicase linker that suggest a subtype-specific role of this residue in viral replicative fitness. Because the two enzymatic domains of NS3 are highly interdependent and sensitive to mutations in the domain interface (17, 18), we predicted subtype-specific molecular rearrangements due to Q80K that, by modulating NS3 helicase activity, can impair RNA replication and in addition can interfere with virus assembly. We tested our hypothesis using infectious cell culture models of subtype 1a and 1b. Whereas Q80K showed only modest impairment of RNA replication in both subtypes, we observed a pronounced drop of infectious virus yield that was specific for subtype 1b, whereas Q80K in subtype 1a even produced slightly higher amounts of infectious virus particles than the parental wild type virus. Given that high replicative fitness confers a general determinant of multidrug resistance (25), our data also potentially explain the lower sustained-virological response rates for voxilaprevir in subtype 1a-patients (26), although Q80K is reported to not confer changes to voxilaprevir susceptibility in vitro (27).
Collectively, we depicted differences in the network topology and local residue-interaction patterns of PI resistance-associated sites in the NS3–4A protease of HCV that are subtype specific and affect functionally important sites in the protease. Our results suggest that the subtype 1a NS3–4A genomic background provides for lower replicative fitness costs due to PI resistance mutations than the subtype 1b genomic background.
MATERIALS AND METHODS
Sequence data.
Sequences of subtype 1a and 1b proteases were downloaded from the euHCV database (28) on April 23, 2014 after filtering based on the following criteria: single protein, standard name ns3, confirmed subtype and published before 2010. Identical sequences were removed. This resulted in 163 sequences for subtype 1a and 178 sequences for subtype 1b. As suggested by Berger et al. (29), we also downloaded sequences with identifiers between KC123434 and KC127656 from the GenBank database (30) to have a better representation of the natural genetic diversity. After filtering out NS5 sequences, we retrieved 2111 subtype 1a and 1335 subtype 1b NS3 sequences (as of December 12, 2014) and generated multiple sequence alignments for each subtype using MUSCLE (31) with default settings as provided by the EMBL-EBI bioinformatics web framework (32). We computed the conservation as defined by Valdar (33) and the frequency of amino acid (AA) occurrence at each sequence position using the RINerator package (34). The residue frequency distance (RFD) between subtypes at each alignment position was computed using the cosine distance between the AA frequency vectors of each subtype. The cosine distance between two vectors is equal to one minus the cosine similarity, which is defined as the normalized dot product of the two vectors. Euclidean distance, cosine and correlation similarity (35) correlated highly for all measures. We chose the cosine distance because they were more distinctive and easier to interpret than those with a similarity measure. Since RFD values agree very well between the two sequence data sources, we only discuss the GenBank RFD. We chose a threshold of 0.1 to distinguish between scores that represent conservation (RFD < 0.1) and substitution tendency (RFD > 0.1) based on the distribution of scores for sequences from the euHCV and GenBank databases (Suppl. Fig 4). There is a clear separation of scores for the euHCV sequences between RFD of 0.45 and 0.016 with only one residue with a score between these two. In addition, the 19 residues with an RFD > 0.1 overlap between both sets of sequences, although they do not have exactly the same values (see Suppl. Table 3 for RFD scores of each residue).
Structure data.
The Protein Data Bank RCSB PDB (36) was used for a comprehensive analysis of available NS3–4A protease structure information using subtype 1a and 1b structure ensembles and residue-interaction networks (RINs). We identified all database entries of the NS3–4A protease (102 as of June 5, 2014) and selected representative protein structures based on the following criteria: unbound, without helicase domain, high resolution and R-free, no mutations, that is 3KF2 (37), 1A1R (38), 2OBQ (39) for subtype 1a and 1DXP (40), 1NS3 (41) for subtype 1b (Suppl. Table 5). Since the quality of 1NS3 in comparison to the other selected structures is poor, we made sure the consensus makes sense by comparing the number of edges between the consensus networks. For additional analysis, we selected 1CU1 (42), a subtype 1b 3D structure of the whole nonstructural protein NS3, as well as the subtype 1a structures 2QV1 (43) and 2OIN (44), which contain a V36M and a R155K amino acid substitution, respectively. For each structure, we used the longest resolved chain (chain A for 2OBQ, 1DXP, 1NS3, 1CU1 and chain B for 3KF2, 1A1R). We aligned the selected chains to create a residue mapping for each subtype using residue indices of 2OBQ and 1DXP for subtype 1a and 1b, respectively.
Residue interaction data.
For selected PDB structures, we created residue-interaction networks (RIN) using RINerator (34). Hydrogens were added to the 3D protein structures using the Reduce tool (45). Contacts on the van-der-Waals (vdW) surface of each atom were sampled by the Probe tool (46) and finally summarized into residue interactions by RINerator. Nodes in a RIN represent protein residues and edges indicate noncovalent residue interactions between residues; that is van-der-Waals contacts (cnt), hydrogen bonds (hbond), overlaps of van-der-Waals radii (ovl), and combined (any of the previous three). We also distinguish between main chain (mc) and side chain (sc) interactions. The RINs and additional data associated with them are visualized in Cytoscape using the RINalyzer and structureViz2 apps (47) (Suppl. Fig 5). We enriched the networks with sequence conservation, RFD, and functional site annotations. To define a representative RIN for each subtype, we combined noncovalent interactions from individual RINs from a subtype-specific structure ensemble into a single network and weighed the edges by the fraction of structures, in which they are present. For subtype 1a, the representative RIN contains edges with weights 0.33, 0.66 and 1.0, which corresponds to interactions in one, two or all three structures, while for subtype 1b, there are only weights of 0.5 and 1.0 for interactions in only one or both structures (Suppl. Fig 6). We created a consensus RIN for each subtype ensemble from all noncovalent interactions present in all individual RINs, which correspond to all edges with weight of 1.0.
Selection of functional sites.
Residues in the RIN previously identified to pertain to different NS3 function were grouped as follows:
Protease catalytic triad and oxyanion hole (residues 57, 81, 137, 139)
Putative domain-domain interaction sites (from PDB-1CU1, residues 56, 57, 60, 61, 78, 79, 123, 132, 135, 136, 137, 139, 154, 155, 156, 157, 158, 159, 160, 161, 165, 168, 186) (42)
Allosteric site (residues 57, 78, 79, 81, 155) (5)
Linker region (residues 182 to 195, but missing in the selected structures: residues 74, 75, 181 interact with them in the RIN) (17)
Binding site for the natural peptide (see e.g. PDB-1CU1, residues 41, 42, 43, 55, 57, 58, 81, 123, 132, 135, 136, 137, 138, 139, 154, 155, 156, 157, 158, 159, 160, 165, 168) (42)
MAVS binding specificity shows similar determinants as the protease natural substrate; MAVS binding sites (from PDB-3RC5, residues 41, 43, 55, 57, 123, 132, 135, 136, 137, 138, 139, 154, 155, 156, 157, 158, 159, 160, 161, 162, 168) (9, 10)
An eight-residue polyproline tract (SOS-binding motif) in TRIF is essential for binding to the protease (48); SOS-binding site for TRIF (residues 132, 134, 135) (48)
Linear PI binding site (from PDB-3SV6, residues 41, 42, 43, 57, 123, 135, 136, 137, 138, 139, 155, 156, 157, 158, 159) (49)
Binding sites for the natural peptide, MAVS and a linear PI were determined using PyMOL as all residues with an atom within 5Å of the respective ligand or product, respectively, in the bound NS3 structure (1CU1 (42), 3RC5 (50), 3SV6 (49), respectively). Domain-domain interaction sites (DDIP) contain the residues from the NS3–4A protease domain that are directly interacting noncovalently with residues of the NS3 helicase domain in the RIN of the NS3 protease-helicase complex structure from PDB–1CU1 (Suppl. Fig 7). According to Kohlway et al. (18), the NS3 inter-domain linker region comprises residues 182 to 195. Since these residues are missing from our selected subtype 1a and 1b structures, we identified all residues interacting with the linker region using the noncovalent interactions in the RIN of PDB–1CU1 (Suppl. Fig 8).
Topological network analysis.
We performed topological analysis on consensus and comparison RINs by computing topology measures using the NetworkAnalyzer plugin for Cytoscape (51) and the NetworkX Python package (52). Measures that describe the overall topology of a network are its radius and diameter, average shortest path length and network clustering coefficient. The length of a path between two nodes is the number of edges forming it with the length of the shortest path referred to as distance. Eccentricity is defined as the maximum distance between a given node and any other node in the network; largest and smallest eccentricities are called network diameter and radius, respectively. The average shortest path length is the average distance between two connected nodes. The network clustering coefficient is the average of the clustering coefficients for all nodes in the network.
For each residue, we calculated the following topology measures; for detailed definitions see (53):
Average distance to functional sites is the average shortest path length from a given node to any functional residue and gives a quantitative measure of how close a residue is to functionally important residues in NS3–4A.
Degree centrality is the fraction of all network nodes connected to a given node. This measure distinguishes between nodes with many neighbors and those that only have a few neighbors (for instance, nodes that represent residues in the protein inside have higher degree centrality than residues on the protein surface).
Clustering coefficient (density) measures the fraction of edges between direct neighbors of a given node relative to the total number of possible edges between all these direct neighbors and indicates how fully connected a group of nodes is.
Closeness centrality is defined as the reciprocal of the sum of the shortest path distances from a given node to all other nodes and is a measure of how fast information spreads from a given node to other reachable nodes (12–15).
Betweenness centrality is computed as the fraction of shortest paths that pass through a given node. This measure reflects the amount of control that a node exerts over interactions of other nodes in the network and favours nodes that join dense subnetworks, rather than nodes that lie inside such subnetworks or that lie in the periphery (12–15).
Edge fraction is the fraction of edges with a specific property that connect a given node to its direct neighbors, i.e., in the subtype comparison network this property describes the original network (subtype 1a, subtype 1b or both) to which the edges belong.
Cells and viruses.
Huh7.5 hepatoma cells (Apath LLC) (54) were maintained in DMEM with 10% FBS, 100U/ml Penicillin/Streptomycin, 1X non-essential amino acids, 1X sodium pyruvate and 1X Glutamax (all Gibco/Life Technologies). For virus production and GLuc assays, medium was supplemented with 1 μM vitamin E (α-tocopherol) (Sigma) and 50 mM HEPES buffer, pH7.4 (Cellgro) (19). The cell culture infectious subtype 1a HCV genome H77S.3, and its derivative H77S.3/GLuc2A, encoding a GLuc reporter gene, were both described in (10). The cell culture infectious subtype 1b HCV genome N.2, and its derivative N.2/GLuc2A, encoding a GLuc reporter gene, were both described in (55). The Q80K amino acid substitution was generated in H77S.3 and N.2 backgrounds using the Quikchange Site-Directed Mutagenesis Kit (Agilent).
Assays of viral fitness.
5×106 cells were electroporated with 10 μg HCV genomic RNA as described previously (56) and plated to 25 cm2 cell culture vessels for assays to assess intracellular viral RNA abundance (GLuc assay) or extracellular infectious virus production (FFU assay). For GLuc assays, cell culture medium was harvested and replaced at 6, 24, 48 and 72 hours after electroporation. Cell culture medium was analyzed for GLuc activity using the BioLux Gaussia substrate (New England BioLabs) on a Synergy 2 Multimode Plate Reader (Biotek). For FFU assay, cell culture medium was harvested and replaced at 24, 48 and 72 hours after electroporation. Infectious virus was measured as described previously (56).
Supplementary Material
Highlights.
A novel graph-based approach is applied to characterize multiple NS3–4A structure models.
Residue-interaction patterns at PI resistance sites are HCV subtype specific.
PI resistance sites are closer to NS3–4A protease functional sites in subtype 1b than 1a.
NS3-Q80K impairs infectious virus particle assembly specifically in subtype 1b.
This paper highlights subtype-specific molecular mechanisms in RAV persistence.
ACKNOWLEDGEMENT
SZ served as a consultant for AbbVie, Bristol-Myers Squibb, Gilead Sciences, Merck, and Janssen. All other authors declare no conflict of interest.
FUNDING INFORMATION
NTD and TL were supported by the Max Planck Society and the Cluster of Excellence for Multimodal Computing and Interaction (MMCI). FSD was supported by the Department of Innovation, Research, Development and Cooperatives, Autonomous Province of Bolzano-South Tyrol. MA was supported by BioTechMed-Graz. CW was supported by the German Research Foundation (WE 4388/6–1). TS and CW were supported by JSPS–DAAD under the Japan-Germany Research Cooperative Program. DRM was supported by National Institutes of Allergy and Infectious Diseases (R21-AI115207). CML and CW acknowledge support by the Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz (LOEWE) of the State of Hessen Research Center for Translational Medicine and Pharmacology TMP.
Abbreviation list:
- AA
amino acid
- allo
allosteric site
- cat
protease active/catalytic site
- cnt
van-der-Waals contacts
- DDIP
domain-domain interaction sites
- FFU
focus forming unit
- hbond
hydrogen bond
- HCV
hepatitis C virus
- mc
main chain
- link
inter-domain linker
- lpi
linear protease inhibitor
- MAVS
mitochondrial antiviral signalling protein
- NPS
natural protease substrate
- npsbs
natural protease substrate binding site
- NS
nonstructural protein
- ovl
overlaps of van-der-Waals radii
- PDB
Protein Data Bank
- PI
protease inhibitor
- RAV
resistance-associated amino acid variant
- RFD
residue frequency distance
- RIN
residue-interaction network
- RNA
ribonucleic acid
- sc
side chain
- sosbs
SOS binding site for TRIF
- TIR
Toll/interleukin-1 receptor
- TRIF
Toll/interleukin-1 receptor (TIR) domain containing adapter inducing interferon-β
- vdW
van-der-Waals interaction
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proofbefore it is published in its final citable form. Please note that during the productionprocess errors may be discovered which could affect the content, and all legal disclaimersthat apply to the journal pertain.
REFERENCES
- 1.Krishnan A, Zbilut JP, Tomita M, Giuliani A. 2008. Proteins as networks: usefulness of graph theory in protein science. Curr Protein Pept Sci 9:28–38. [DOI] [PubMed] [Google Scholar]
- 2.Welsch C 2014. Genetic barrier and variant fitness in hepatitis C as critical parameters for drug resistance development. Drug Discov Today Technol 11:19–25. [DOI] [PubMed] [Google Scholar]
- 3.Sullivan JC, De Meyer S, Bartels DJ, Dierynck I, Zhang EZ, Spanks J, Tigges AM, Ghys A, Dorrian J, Adda N, Martin EC, Beumont M, Jacobson IM, Sherman KE, Zeuzem S, Picchio G, Kieffer TL. 2013. Evolution of treatment-emergent resistant variants in telaprevir phase 3 clinical trials. Clin Infect Dis 57:221–9. [DOI] [PubMed] [Google Scholar]
- 4.Bartenschlager R, Lohmann V, Penin F. 2013. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat Rev Microbiol 11:482–96. [DOI] [PubMed] [Google Scholar]
- 5.Saalau-Bethell SM, Woodhead AJ, Chessari G, Carr MG, Coyle J, Graham B, Hiscock SD, Murray CW, Pathuri P, Rich SJ, Richardson CJ, Williams PA, Jhoti H. 2012. Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function. Nat Chem Biol 8:920–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M Jr, Lemon SM. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U A 102:2992–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–72. [DOI] [PubMed] [Google Scholar]
- 8.Welsch C, Haselow K, Gouttenoire J, Schneider M, Morikawa K, Martinez Y, Susser S, Sarrazin C, Zeuzem S, Antes I, Moradpour D, Lange CM. 2015. Hepatitis C virus variants resistant to macrocyclic NS3–4A inhibitors subvert IFN-beta induction by efficient MAVS cleavage. J Hepatol 62:779–84. [DOI] [PubMed] [Google Scholar]
- 9.McGivern DR, Masaki T, Lovell W, Hamlett C, Saalau-Bethell S, Graham B. 2015. Protease Inhibitors Block Multiple Functions of the NS3/4A Protease-Helicase during the Hepatitis C Virus Life Cycle. J Virol 89:5362–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimakami T, Welsch C, Yamane D, McGivern DR, Yi M, Zeuzem S, Lemon SM. 2011. Protease inhibitor-resistant hepatitis C virus mutants with reduced fitness from impaired production of infectious virus. Gastroenterology 140:667–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.EASL Recommendations on Treatment of Hepatitis C 2016. J Hepatol 66:153–194. [DOI] [PubMed] [Google Scholar]
- 12.Freeman LC. 1978. Centrality in social networks conceptual clarification. Soc Netw 1:215–239. [Google Scholar]
- 13.Koschützki D, Lehmann KA, Peeters L, Richter S, Tenfelde-Podehl D, Zlotowski O. 2005. Centrality Indices, p. 16–61. In Brandes U, Erlebach T (eds.), Network Analysis: Methodological Foundations. Springer; Berlin Heidelberg, Berlin, Heidelberg. [Google Scholar]
- 14.Borgatti SP. 1. Centrality and network flow. Soc Netw 27:55–71. [Google Scholar]
- 15.Barabasi AL, Oltvai ZN. 2004. Network biology: understanding the cell’s functional organization. Nat Rev Genet 5:101–13. [DOI] [PubMed] [Google Scholar]
- 16.Welsch C, Domingues FS, Susser S, Antes I, Hartmann C, Mayr G, Schlicker A, Sarrazin C, Albrecht M, Zeuzem S, Lengauer T. 2008. Molecular basis of telaprevir resistance due to V36 and T54 mutations in the NS3–4A protease of the hepatitis C virus. Genome Biol 9:R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beran RK, Pyle AM. Hepatitis C viral NS3–4A protease activity is enhanced by the NS3 helicase. J Biol Chem. 2008. October 31;283(44):29929–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohlway A, Pirakitikulr N, Ding SC, Yang F, Luo D, Lindenbach BD, Pyle AM. The linker region of NS3 plays a critical role in the replication and infectivity of hepatitis C virus. J Virol. 2014. September;88(18):10970–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamane D, McGivern DR, Wauthier E, Yi M, Madden VJ, Welsch C, Antes I, Wen Y, Chugh PE, McGee CE, Widman DG, Misumi I, Bandyopadhyay S, Kim S, Shimakami T, Oikawa T, Whitmire JK, Heise MT, Dittmer DP, Kao CC, Pitson SM, Merrill AHJ, Reid LM, Lemon SM. Regulation of the hepatitis C virus RNA replicase by endogenous lipid peroxidation. Nat Med. 2014. August;20(8):927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halabi N, Rivoire O, Leibler S, Ranganathan R. Protein sectors: evolutionary units of three-dimensional structure. Cell. 2009. August 21;138(4):774–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weigt M, White RA, Szurmant H, Hoch JA, Hwa T. Identification of direct residue contacts in protein-protein interaction by message passing. Proc Natl Acad Sci U S A. 2009. January 6;106(1):67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vermehren J, Welsch C, Sarrazin C. 2015. Protease inhibitor resistance, p. . In Gotte M, Berghuis A, Matlashewski G, Wainberg M, Sheppard D (eds.), Handbook of Antimicrobial Resistance. Springer, New York. [Google Scholar]
- 23.Sarrazin C, Lathouwers E, Peeters M, Daems B, Buelens A, Witek J, Wyckmans Y, Fevery B, Verbinnen T, Ghys A, Schlag M, Baldini A, De Meyer S, Lenz O. 2015. Prevalence of the hepatitis C virus NS3 polymorphism Q80K in genotype 1 patients in the European region. Antiviral Res 116:10–16. [DOI] [PubMed] [Google Scholar]
- 24.Lenz O, Verbinnen T, Fevery B, Tambuyzer L, Vijgen L, Peeters M, Buelens A, Ceulemans H, Beumont M, Picchio G, De Meyer S. 2015. Virology analyses of HCV isolates from genotype 1-infected patients treated with simeprevir plus peginterferon/ribavirin in Phase IIb/III studies. J Hepatol 62:1008–14. [DOI] [PubMed] [Google Scholar]
- 25.Sheldon J, Beach NM, Moreno E, Gallego I, Pineiro D, Martinez-Salas E, Gregori J, Quer J, Esteban JI, Rice CM, Domingo E, Perales C. 2014. Increased replicative fitness can lead to decreased drug sensitivity of hepatitis C virus. J Virol 88:12098–12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobson IM, Lawitz E, Gane EJ, Willems BE, Ruane PJ, Nahass RG, Borgia SM, Shafran SD, Workowski KA, Pearlman B, Hyland RH, Stamm LM, Svarovskaia E, Dvory-Sobol H, Zhu Y, Subramanian GM, Brainard DM, McHutchison JG, Brau N, Berg T, Agarwal K, Bhandari BR, Davis M, Feld JJ, Dore GJ, Stedman CAM, Thompson AJ, Asselah T, Roberts SK, Foster GR. 2017. Efficacy of 8 Weeks of Sofosbuvir, Velpatasvir, and Voxilaprevir in Patients With Chronic HCV Infection: 2 Phase 3 Randomized Trials. Gastroenterology 153:113–122. [DOI] [PubMed] [Google Scholar]
- 27.Taylor JG, Appleby T, Barauskas O, Chen X, Dvory-Sobol H, Gong R, Lee J, Nejati E, Schultz B, Wang Y, Yang C, Yu M, Zipfel S, Chan K. P0899 : Preclinical profile of the pan-genotypic HCV NS3/4A protease inhibitor GS-9857. J Hepatol 62:S681. [Google Scholar]
- 28.Combet C, Garnier N, Charavay C, Grando D, Crisan D, Lopez J, Dehne-Garcia A, Geourjon C, Bettler E, Hulo C, Le Mercier P, Bartenschlager R, Diepolder H, Moradpour D, Pawlotsky JM, Rice CM, Trepo C, Penin F, Deleage G. 2007. euHCVdb: the European hepatitis C virus database. Nucleic Acids Res 35:D363–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berger KL, Triki I, Cartier M, Marquis M, Massariol MJ, Bocher WO, Datsenko Y, Steinmann G, Scherer J, Stern JO, Kukolj G. 2014. Baseline hepatitis C virus (HCV) NS3 polymorphisms and their impact on treatment response in clinical studies of the HCV NS3 protease inhibitor faldaprevir. Antimicrob Agents Chemother 58:698–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2013. GenBank. Nucleic Acids Res 41:D36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W, Cowley A, Uludag M, Gur T, McWilliam H, Squizzato S, Park YM, Buso N, Lopez R. 2015. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res 43:W580–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valdar WSJ. 2002. Scoring residue conservation. Proteins 48:227–241. [DOI] [PubMed] [Google Scholar]
- 34.Doncheva NT, Klein K, Domingues FS, Albrecht M. 2011. Analyzing and visualizing residue networks of protein structures. Trends Biochem Sci 36:179–182. [DOI] [PubMed] [Google Scholar]
- 35.Deza MM, Deza E. 2009. Encyclopedia of Distances. Springer-Verlag, Berlin Heidelberg. [Google Scholar]
- 36.Rose PW, Bi C, Bluhm WF, Christie CH, Dimitropoulos D, Dutta S, others 2013. The RCSB Protein Data Bank: new resources for research and education. Nucleic Acids Res 41:D475–D482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cummings MD, Lindberg J, Lin TI, de Kock H, Lenz O, Lilja E, Fellander S, Baraznenok V, Nystrom S, Nilsson M, Vrang L, Edlund M, Rosenquist A, Samuelsson B, Raboisson P, Simmen K. 2010. Induced-fit binding of the macrocyclic noncovalent inhibitor TMC435 to its HCV NS3/NS4A protease target. Angew Chem Int Ed Engl 49:1652–5. [DOI] [PubMed] [Google Scholar]
- 38.Kim JL, Morgenstern KA, Lin C, Fox T, Dwyer MD, Landro JA, Chambers SP, Markland W, Lepre CA, O’Malley ET, Harbeson SL, Rice CM, Murcko MA, Caron PR, Thomson JA. 1996. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87:343–55. [DOI] [PubMed] [Google Scholar]
- 39.Prongay AJ, Guo Z, Yao N, Pichardo J, Fischmann T, Strickland C, Myers J Jr, Weber PC, Beyer BM, Ingram R, Hong Z, Prosise WW, Ramanathan L, Taremi SS, Yarosh-Tomaine T, Zhang R, Senior M, Yang RS, Malcolm B, Arasappan A, Bennett F, Bogen SL, Chen K, Jao E, Liu YT, Lovey RG, Saksena AK, Venkatraman S, Girijavallabhan V, Njoroge FG, Madison V. 2007. Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]- 3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization. J Med Chem 50:2310–8. [DOI] [PubMed] [Google Scholar]
- 40.Di Marco S, Rizzi M, Volpari C, Walsh MA, Narjes F, Colarusso S, De Francesco R, Matassa VG, Sollazzo M. 2000. Inhibition of the hepatitis C virus NS3/4A protease. The crystal structures of two protease-inhibitor complexes. J Biol Chem 275:7152–7. [DOI] [PubMed] [Google Scholar]
- 41.Yan Y, Li Y, Munshi S, Sardana V, Cole JL, Sardana M, Steinkuehler C, Tomei L, De Francesco R, Kuo LC, Chen Z. 1998. Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form. Protein Sci 7:837–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao N, Reichert P, Taremi SS, Prosise WW, Weber PC. 1999. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure 7:1353–63. [DOI] [PubMed] [Google Scholar]
- 43.Zhou Y, Bartels DJ, Hanzelka BL, Muh U, Wei Y, Chu HM, Tigges AM, Brennan DL, Rao BG, Swenson L, Kwong AD, Lin C. 2008. Phenotypic characterization of resistant Val36 variants of hepatitis C virus NS3–4A serine protease. Antimicrob Agents Chemother 52:110–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Y, Muh U, Hanzelka BL, Bartels DJ, Wei Y, Rao BG, Brennan DL, Tigges AM, Swenson L, Kwong AD, Lin C. 2007. Phenotypic and structural analyses of hepatitis C virus NS3 protease Arg155 variants: sensitivity to telaprevir (VX-950) and interferon alpha. J Biol Chem 282:22619–28. [DOI] [PubMed] [Google Scholar]
- 45.Word JM, Lovell SC, Richardson JS, Richardson DC. 1999. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J Mol Biol 285:1735–1747. [DOI] [PubMed] [Google Scholar]
- 46.Word JM, Lovell SC, LaBean TH, Taylor HC, Zalis ME, Presley BK, others 1999. Visualizing and quantifying molecular goodness-of-fit: small-probe contact dots with explicit hydrogen atoms. J Mol Biol 285:1711–1733. [DOI] [PubMed] [Google Scholar]
- 47.Doncheva NT, Klein K, Morris JH, Wybrow M, Domingues FS, Albrecht M. 2014. Integrative visual analysis of protein sequence mutations. BMC Proc 8:S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferreon JC, Ferreon AC, Li K, Lemon SM. 2005. Molecular determinants of TRIF proteolysis mediated by the hepatitis C virus NS3/4A protease. J Biol Chem 280:20483–92. [DOI] [PubMed] [Google Scholar]
- 49.Romano KP, Ali A, Aydin C, Soumana D, Ozen A, Deveau LM, Silver C, Cao H, Newton A, Petropoulos CJ, Huang W, Schiffer CA. 2012. The molecular basis of drug resistance against hepatitis C virus NS3/4A protease inhibitors. PLoS Pathog 8:e1002832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romano KP, Laine JM, Deveau LM, Cao H, Massi F, Schiffer CA. 2011. Molecular mechanisms of viral and host cell substrate recognition by hepatitis C virus NS3/4A protease. J Virol 85:6106–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Assenov Y, Ramirez F, Schelhorn S-E, Lengauer T, Albrecht M. 2008. Computing topological parameters of biological networks. Bioinformatics 24:282–284. [DOI] [PubMed] [Google Scholar]
- 52.Hagberg AA, Schult DA, Swart PJ. 2008. Exploring network structure, dynamics, and function using NetworkX, p. 11–15. In Proceedings of the 7th Python in Science Conference (SciPy2008). [Google Scholar]
- 53.Doncheva NT, Assenov Y, Domingues FS, Albrecht M. 2012. Topological analysis and interactive visualization of biological networks and protein structures. Nat Prot 7:670––685. [DOI] [PubMed] [Google Scholar]
- 54.Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol 76:13001–13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kannan RP, Hensley LL, Evers LE, Lemon SM, McGivern DR. 2011. Hepatitis C virus infection causes cell cycle arrest at the level of initiation of mitosis. J Virol 85:7989–8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McGivern DR, Masaki T, Williford S, Ingravallo P, Feng Z, Lahser F, Asante-Appiah E, Neddermann P, De Francesco R, Howe AY, Lemon SM. 2014. Kinetic analyses reveal potent and early blockade of hepatitis C virus assembly by NS5A inhibitors. Gastroenterology 147:453–462.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Susser S, Vermehren J, Forestier N, Welker MW, Grigorian N, Fuller C, Perner D, Zeuzem S, Sarrazin C. 2011. Analysis of long-term persistence of resistance mutations within the hepatitis C virus NS3 protease after treatment with telaprevir or boceprevir. J Clin Virol 52:321–7. [DOI] [PubMed] [Google Scholar]
- 58.Manns MP, Vierling JM, Bacon BR, Bruno S, Shibolet O, Baruch Y, Marcellin P, Caro L, Howe AY, Fandozzi C, Gress J, Gilbert CL, Shaw PM, Cooreman MP, Robertson MN, Hwang P, Dutko FJ, Wahl J, Mobashery N. 2014. The combination of MK-5172, peginterferon, and ribavirin is effective in treatment-naive patients with hepatitis C virus genotype 1 infection without cirrhosis. Gastroenterology 147:366–76.e6. [DOI] [PubMed] [Google Scholar]
- 59.Krishnan P, Schnell G, Tripathi R, Beyer J, Reisch T, Zhang X, Setze C, Rodrigues L Jr, Burroughs M, Redman R, Chayama K, Kumada H, Collins C, Pilot-Matias T. 2015. Analysis of Hepatitis C Virus Genotype 1b Resistance Variants in Japanese Patients Treated with Paritaprevir-Ritonavir and Ombitasvir. Antimicrob Agents Chemother 60:1106–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zeuzem S, Hezode C, Bronowicki JP, Loustaud-Ratti V, Gea F, Buti M, Olveira A, Banyai T, Al-Assi MT, Petersen J, Thabut D, Gadano A, Pruitt R, Makara M, Bourliere M, Pol S, Beumont-Mauviel M, Ouwerkerk-Mahadevan S, Picchio G, Bifano M, McPhee F, Boparai N, Cheung K, Hughes EA, Noviello S. 2016. Daclatasvir plus simeprevir with or without ribavirin for the treatment of chronic hepatitis C virus genotype 1 infection. J Hepatol 64:292–300. [DOI] [PubMed] [Google Scholar]
- 61.Buti M, Gordon SC, Zuckerman E, Lawitz E, Calleja JL, Hofer H, Gilbert C, Palcza J, Howe AY, DiNubile MJ, Robertson MN, Wahl J, Barr E, Forns X. 2016. Grazoprevir, Elbasvir, and Ribavirin for Chronic Hepatitis C Virus Genotype 1 Infection After Failure of Pegylated Interferon and Ribavirin With an Earlier-Generation Protease Inhibitor: Final 24-Week Results From C-SALVAGE. Clin Infect Dis 62:32–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.