Abstract
Short insertions and deletions (InDels) are a common type of mutation found in nature and a useful source of variation in protein engineering. InDel events have important consequences in protein evolution, often opening new pathways for adaptation. Yet much less is known about the effects of InDels compared to point mutations and amino acid substitutions. In particular, deep mutagenesis studies on the distribution of fitness effects of mutations have focused almost exclusively on amino acid substitutions. Here, we present a near-comprehensive analysis of the fitness effects of single amino acid InDels in TEM-1 β-lactamase. While we found InDels to be largely deleterious, partially overlapping deletion-tolerant and insertion-tolerant regions were observed throughout the protein, especially in unstructured regions and at the end of helices. The signal sequence of TEM-1 tolerated InDels more than the mature protein. Most regions of the protein tolerated insertions more than deletions, but a few regions tolerated deletions more than insertions. We examined the relationship between InDel tolerance and a variety of measures to help understand its origin. These measures included evolutionary variation in β-lactamases, secondary structure identity, tolerance to amino acid substitutions, solvent accessibility, and side-chain weighted contact number. We found secondary structure, weighted contact number, and evolutionary variation in class A beta-lactamases to be the somewhat predictive of InDel fitness effects.
Keywords: Fitness landscapes, InDels, antibiotic resistance protein, protein evolution
Introduction
Insertions and deletions (InDels) are an important source of genetic variation in nature. They occur nearly as frequently as point mutations in some genomes [1, 2]. For example, 15–21% of polymorphisms can be attributed to short InDels in the human genome [3]. Indels can result in dramatic effects on the properties of a protein and how it evolves [2, 4–6] and are known to be the cause of diseases such as cystic fibrosis and numerous types of cancer [7, 8].
The metaphor of an adaptive walk across a fitness landscape works well for substitution mutations [9]. Distance can be easily measured as the number of single substitutions necessary to convert one sequence to another. However, distance is not so cleanly defined for InDels because InDels add or subtract dimensions to a protein’s accessible sequence space. Once way to conceptualize InDels is that they represent a “leap” across sequence space rather than a step [10]. As such, InDels have the potential to open up new pathways for adaptation. For example, amino acid substitutions appear to be enriched around the site of InDel events in evolving proteins, either because InDel events make accumulation of substitution mutations near such sites more likely [10], or because substitutions makes InDel accumulation in their vicinity more likely via “neutral roaming” [2]. This enrichment of substitutions near InDels suggests that how the surrounding protein region changes during selection may be substantially impacted by InDels.
InDels also represent a potentially underutilized source of variation in protein engineering. Though routine engineering of backbone modifications has been challenging, InDels have long been recognized as important tools for altering protein structure and properties [2, 11]. Because insertions and deletions add or remove atoms from the polypeptide backbone, they can cause major structural modifications not available through substitutions alone. They may be particularly important when seeking to dramatically change active-site structure, as they have been found to propagate long-range effects on catalytic activity [11]. However, despite their importance in nature and the laboratory, InDels remain understudied compared to substitutions.
The fitness effects of point mutations and substitutions have been extensively studied in recent years [12, 13]. Previously, we comprehensively characterized the fitness effects of single amino acid substitutions in TEM-1 β-lactamase [14]. Other large-scale mutagenesis studies have been reported for over 14 proteins, characterizing the effects of single amino acid substitutions on function or fitness [13]. Such studies have advanced our understanding of the genetic code, protein structure, epistasis, and predictive models. However, we lack a similar systematic, large-scale analysis on the fitness effects of InDels.
Multiple studies have offered insight into the effects of deletions on a smaller scale. For example, a 2007 study of TEM-1 β-lactamase assayed 53 single amino acid deletions occurring throughout the protein, and found that 14 (26.4%) of the variants had a minimum inhibitory concentration (MIC) that was <1% of that conferred by TEM-1, while the remaining variants varied in activity, including four that retained wild-type levels, as measured by a MIC assay [15]. The majority of debilitating deletions occurred in secondary structure elements and buried/core residues. Similarly, a 2014 study on enhanced green fluorescent (EGFP) protein characterized the tolerance to 87 random single amino acid deletions throughout the protein [16]. They found that the majority of tolerated deletions occurred in loops, while the rest were found equally distributed in helices and β-strands, with the termini of β-strands being more tolerant than the middle. Computational analysis of the EGFP found that structural properties such as relative solvent accessibility and weighted contact number (WCN) can be used to predict tolerance to deletions to some extent [17].
Insertion studies are even more limited, generally examining only a few rationally chosen insertion sites in a protein. For example, 2006 study in TEM-1 assessed the impact of random peptide insertion into three loops and found that tolerance depended largely on the insertion site [15]. Based on their findings, they also suggested that tolerance to insertions was not well-correlated to tolerance to substitutions in the same region.
While these studies provide important insights into the effects of InDels, they are limited by their scale. Here, we present a near-comprehensive analysis of the fitness effects of single amino acid insertions and deletions in TEM-1 β-lactamase, a widely studied antibiotic resistance protein. We find that while InDels are largely deleterious compared to substitutions, partially overlapping regions of tolerance to insertions and deletions exist throughout the protein.
Results and Discussion
Fitness effects of single amino acid InDels
TEM-1 β-lactamase is a commonly studied protein and convenient model for protein evolution experiments. It confers high resistance to penicillin antibiotics, such as ampicillin, which can be used as a proxy for protein fitness [14, 18, 19]. We use our previously described band-pass, MIC-like approach for measuring antibiotic resistance in a high-throughput, high-resolution manner [14, 20, 21]. This method uses a synthetic biology approach to quantify Amp resistance as a proxy for fitness. Unlike growth competition experiments and standard MIC assays, the fitness measures are ampicillin concentration independent and low fitness values are as precisely measured as high fitness values.
We focused on in-frame insertions or deletions of three nucleotides. We did not study insertions or deletions that are one or two nucleotides in length, as such mutations are frame-shifting mutations with drastic changes to protein sequence and nearly always inactivate proteins, although such InDels can be bypassed by transcriptional or translational slippage to give full-length functional proteins [22]. We did not study three-nucleotide insertions or deletions that are out of frame, as they cause substitutions in the amino acid sequence in addition to the amino acid insertion or deletion. We wanted to be able to isolate the effect of the single amino acid insertion or deletion away from any substitution effects.
We used inverse-PCR to create a plasmid library designed to code for every possible single amino acid insertion (5,720 variants) and every possible single amino acid deletion (286 variants) in TEM-1. For insertions, we used degenerate primers in which the 5’-end of the forward primer had an additional (NNN) sequence. For deletions, we used primers in which the 5’-end of the forward primer had a 3 base pair deletion. We transformed SNO301 E. coli cells with each library of InDel alleles and plated on LB plates supplemented with tetracycline and 13 different Amp concentrations ranging from 0.25 μg/ml to 1024 μg/ml. Amp prevents growth if the Amp concentration is too high relative to the amount of Amp resistance conferred. Tetracycline prevents growth if the concentration of Amp is too low relative to the amount of Amp resistance conferred. This behavior is the key feature of the band-pass gene circuit in SNO301 cells. As a result, a particular allele will confer growth only in a narrow range of Amp concentrations (see Firnberg et al. [14] for a detailed explanation), and the higher the midpoint of that range, the fitter the allele. After incubation at 37°C overnight, we recovered the 13 sublibraries and performed deep-sequencing to determine how often each allele appeared on each plate. Sequencing reads of alleles containing synonymous codon insertions were grouped together, with the exception of the stop codons. The amber (UAG) stop codon exhibits nonsense suppression in SNO301 E. coli via the supE44 tRNA allele, which results in glutamine incorporation at UAG codons with variable efficiency depending on the nucleotides immediately flanking UAG [14]. To avoid convolution, we included only non-amber stop codons in our analysis. The reported fitness values are calculated as the Amp concentration at which the mutant allele appeared most frequently relative to the wildtype allele (see Material and Methods for a more detailed description).
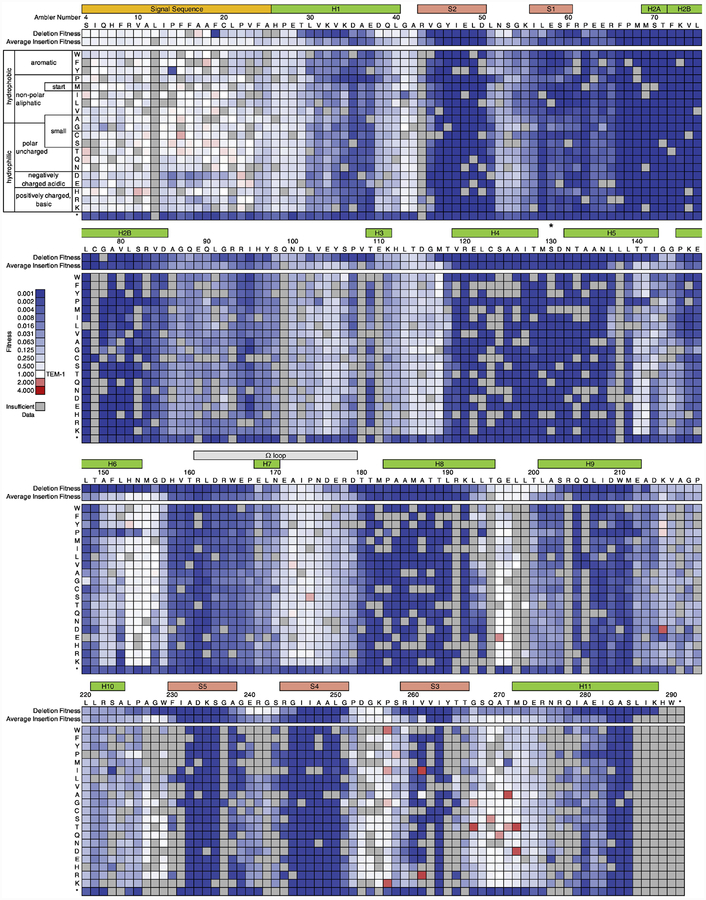
We obtained fitness values for 77.9% (4457/5720) of possible amino acid insertions and 97.9% (280/286) of possible amino acid deletions in TEM-1 (Fig. 1). As expected, we find that insertions and deletions are largely deleterious. Over half of insertions (51%) and deletions (59%) resulted in at least a 100-fold decrease in fitness relative to TEM-1. In contrast, only 9.8% of insertions and 11% of deletions retained 50% of wild-type fitness, though close to half (40.9%) of these were in the signal sequence, which is cleaved and not part of the mature protein. Though we measured 74 InDels alleles with fitness values nominally greater than 1, only 27 were statistically different than 1. However, we suspect many of these are not actually beneficial insertions. For example, the highest “significant” beneficial insertions cluster in the region 257–272. The insertions W and K at 257, I and R at 261, T at 267, A at 271, and T and D at 272 all have a peculiar distribution of sequencing counts that is defined by an abnormally high number of sequencing count on the plate with 512 μg/ml Amp with comparatively very few counts on 256 μg/ml and no counts on 1024 μg/ml. The abnormally high counts on just one plate suggests an artifact. For this reason, we focus our attention on broad patterns in our fitness effects rather than the fitness values associated with particular InDels.
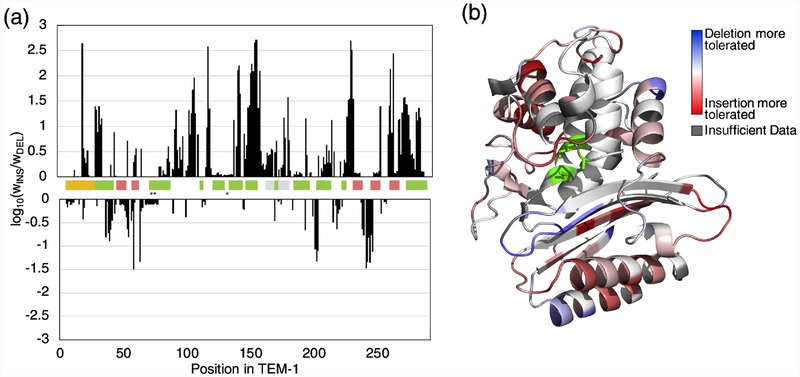
Fig. 1. The fitness effects of amino acid insertions and deletions in TEM-1.
The heat map indicates relative fitness values as calculated based on ampicillin resistance. Insertion position is defined by the new position of the inserted amino acid (e.g. an insertion denoted at position 50 was inserted between residues 49 and 50 in TEM-1). Ambler consensus numbering for beta-lactamases is used. The signal sequence (yellow), β helices (green), β strands (orange), Ω loop (grey), and active sites (*) are indicated. Tabulated sequencing counts and fitness data is provided as Supplementary Data S1 (insertions) and S2 (deletions).
Relationship between InDel fitness effects and TEM-1 sequence/structure
Visual examination of the heatmap depicting the fitness effects (Fig. 1) suggests a higher tolerance to InDels outside of secondary structures. It also appears that the fitness effect of an insertion depends more on the site of the insertion than on the amino acid identity. To examine this quantitatively, we looked at the distribution of mean fitness values per position and compared it to the distribution of mean fitness values grouped by amino acid (Supplementary Fig. 1). We found that the mean fitness effects per position have a wider distribution of values than the mean fitness effects grouped by the amino acid inserted (P=0.009, Brown-Forsythe test).
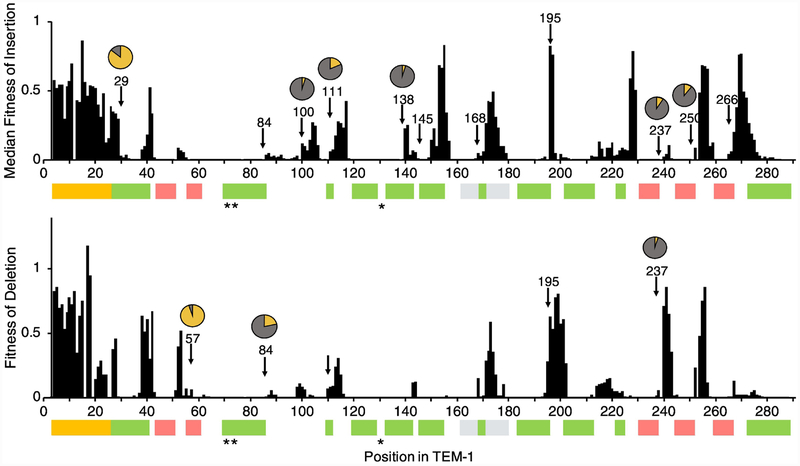
Examining the median fitness of alleles containing insertions and the fitness of alleles containing deletions across TEM-1, we observed “hot spots” of tolerance for InDels in TEM-1 (Fig. 2 and Fig. 3). The pattern suggests some correlation between where insertions and deletions are tolerated, and indicates higher tolerance in the signal sequence and in unstructured regions of the protein. Higher tolerance to InDels in loops compared to helices and strands is widely observed across many families of proteins [23]. Our results also agree with previous observations in TEM-1 in particular. For example, visual examination of Fig. 1 and Fig. 2 suggests a notable tolerance to insertions in the loop connecting the final β-strand to the C-terminal helix, which is a location previously found to be broadly tolerant to random sequences of insertions [15].
Fig. 2. Fitness of TEM-1 containing InDels as a function of primary sequence.
Median fitness values are presented for insertions. Arrows indicate positions at which other class A β-lactamases contain an insertion or deletion (based on a multiple sequence alignment of 156 class A β-lactamase and TEM-1). Pie charts indicate in yellow the fraction of sequences out of 156 that contain an insertion (top chart) or deletion (bottom chart) at that position. Pie charts are omitted for fractions less than 3%. The colored bars indicate the signal sequence (yellow), α helices (green), β strands (pink), Ωloop (grey), and active sites (*).
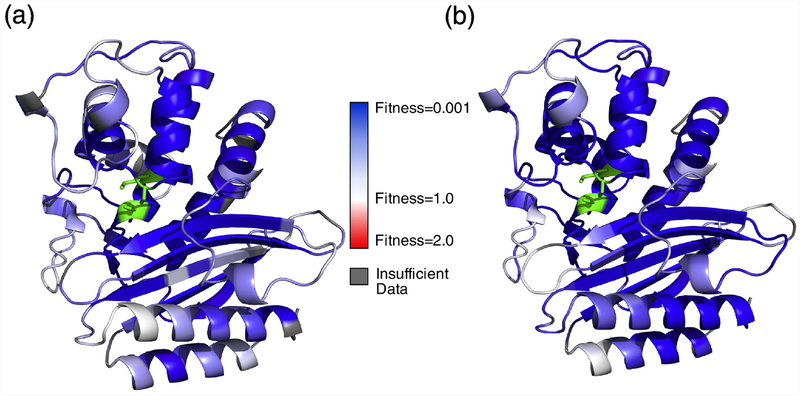
Fig. 3. InDel fitness mapped onto TEM-1 structure.
(a) TEM-1 secondary structure colored by median fitness of insertions. Positions for which we obtained fewer than 4 fitness values are indicated in grey. (b) TEM-1 secondary structure colored by fitness of deletions. In both figures, the active site residues are colored in green. No mean fitness values > 1 are observed.
We also examined the relationship between evolutionary variations in class A β-lactamases and InDel fitness effect in TEM-1. We aligned a published set of 157 class A β-lactamase sequences from different bacterial species (including TEM-1) [24] by progressive multiple alignment using a Gonnet scoring matrix in MATLAB. We chose this data set because it consists of close homologs of TEM-1 where there were enough examples of InDels, but the size of the InDels was general small (our analysis found 77% of the InDels were single amino acids and 94% were not more than 2 amino acids). We wanted to compare to homologs with small InDels, because our fitness measurements were of single amino acid InDels. We identified the positions at which these 157 sequences [24] contained an insertion or deletion relative to TEM-1. We find that these positions generally overlap insertion-tolerant regions in TEM-1, but several regions in TEM-1 that tolerate insertions and especially deletions are not observed in natural class A β-lactamases, at least in this dataset (Fig. 2).
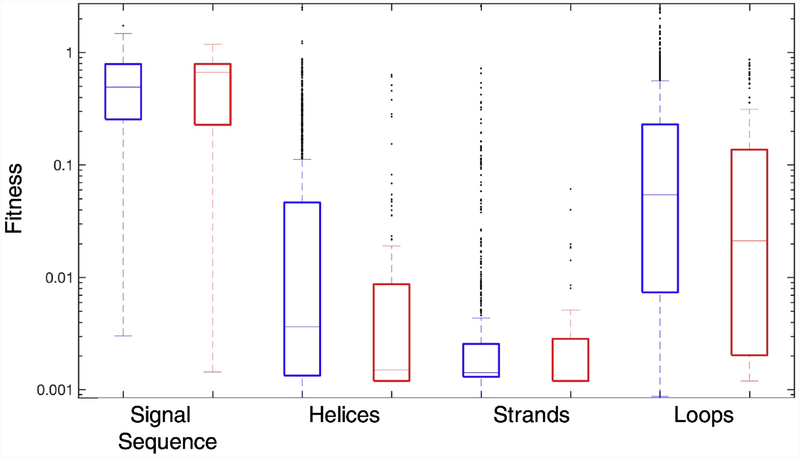
We find that the 23 amino acid signal sequence is the most InDel-tolerant region in TEM-1 (Fig. 4). This sequence directs TEM-1’s export to the periplasm via the Sec export pathway. The signal peptide is removed upon export to the periplasm and is not part of the mature protein. Presumably, mutations in the signal sequence affect fitness through changes of TEM-1’s export efficiency to the periplasm. The signal sequence of TEM-1 is also the most tolerant region to missense mutation [15]. This tolerance is consistent with the loose sequence constraints for Sec-dependent signal sequences and its lack of secondary structure elements [25].
Fig. 4. Relationship between InDel fitness and secondary structure.
Box plots of fitness values for insertions (blue) and deletions (red) are shown for the signal sequence and secondary structure elements. The central line indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles. The median fitness value for deletions in strands is at the 25th percentile, and therefore not visible on the plot. The whiskers extend to the most extreme data points not considered outliers, which are represented by circles. Outliers are defined as values more than 1.5 times the interquartile range away from the top or bottom of the box.
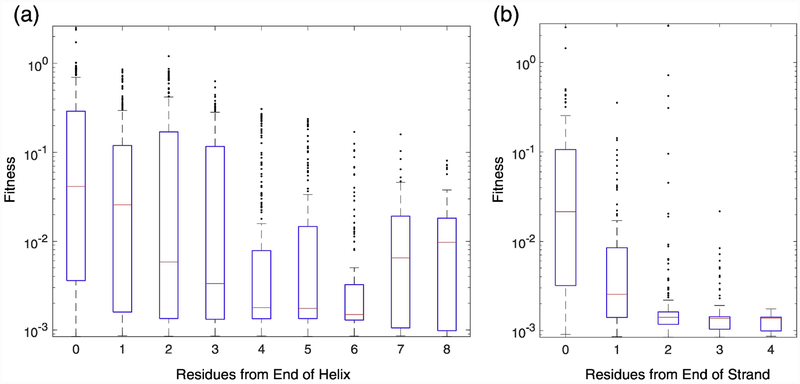
In the mature protein, helices and strands are the least tolerant to InDels. For both insertions and deletions, the mean fitness of mutant alleles in loop regions is higher than in secondary structure elements (P<0.0001 for insertions, P<0.001 for deletions, Student’s t-test). This is not surprising given that backbone modifications can cause structured regions to fold incorrectly and have dramatic effects on the protein [26]. However, we found some exceptions to this overall pattern. For example, the loop region between β-strand S1 and α-helix H2A, shows no tolerance for insertions or deletions. We also found that 2.9% (51/1765) of insertions in α-helices, often at the ends of the structure element, resulted in less than a 50% decrease in fitness. Tolerances to insertions was greatest at the ends of both α-helices and β-strands and decreased as one moved into the structural element (Fig. 5). The median fitness for an insertion immediately before or after an α-helix was 4.3- fold to 28-fold higher than the median values for insertions two to eight residues from the end (P< 2×10−5 for all pairwise comparisons, Mann-Whitney). When the inserted residue was one residue away from the end of the α-helix, the median fitness value was 2.6- to 17-fold higher than that for insertions located three to eight residues from the end (P< 0.0006 for all pairwise comparisons, Mann-Whitney). A notable exception to this trend was the 17- residue C-terminal β-helix H11, the longest α-helix in the protein, which showed significant tolerance for the first 11 positions before abruptly dropping to complete intolerance. The median fitness values for insertions immediately before or after a β-strand were 8- to 16- fold higher than the median values for insertions within a β-strand (P< 2×10−14 for all pairwise comparisons, Mann-Whitney). When the inserted residue was one residue away from the end of the β-strand, the median fitness value was 1.8- fold higher than those for insertions at other locations in the β-strand (P< 2×10−9 for all pairwise comparisons, Mann-Whitney)
Fig. 5. Relationship between insertion fitness and distance from the closest end of secondary structure element.
(a) Box plots of fitness values for insertions within helices. (b) Box plots of fitness values for insertions within strands. The number 0 corresponds to insertions immediately before or immediately after the structure element, the number 1 refers to insertions after the first or before the last residue in the structure element, and so on.
To more specifically examine the difference between tolerance to insertions versus deletions, we calculated the ratio of the mean fitness of alleles with insertions to the fitness of an allele with a deletion at each position across TEM-1 (Fig. 6). Overall, we find more regions where insertions are preferred over deletions, but a few regions where deletions are preferentially tolerated. For example, the C-terminal α-helix is dominated by preference to insertions, while the N-terminal α-helix contains several positions where deletions are relatively preferred. The extended loop between β-strands S5 and S4 (G238-S243) was the region that most preferred deletions over insertions (Fig. 6), Deletions relative to TEM-1 in this region are observed evolutionarily, though they are uncommon (Fig. 2).
Fig. 6. Differences in tolerance to insertions and deletions across TEM-1.
(a) The log10 of the ratio between mean fitness of insertions and the fitness of a deletion at each position across TEM-1. The colored bars indicate the signal sequence (yellow), αhelices (green), β strands (pink), Ω loop (grey), and active sites (*). (b) TEM-1 structure colored by the same ratio values. Blue indicates positions with higher tolerance to deletions, white indicates the same tolerance to both insertions and deletions, and red indicated higher tolerance to insertions.
We also examined the distribution of fitness effects of InDels compared to substitutions (measured in our previous study [14]). Unsurprisingly, we found InDels were more often deleterious than substitutions (Supplementary Fig. S2). The distributions of insertions and deletion fitness values were similar, and the mean fitness of alleles containing an insertion (0.14 ± 0.26) was not significantly different than the mean fitness of alleles containing a deletion (0.11 ± 0.23) (Supplementary Fig. S2).
To explore the comparison between insertions and deletions further, we examined the correlation between the mean fitness of alleles with an insertion at a given position and the fitness of an allele with a deletion at the corresponding position (Supplementary Fig. S3a) and found a weak correlation (R2=0.32). We also compared the mean fitness change of an insertion of a given amino acid against the mean fitness change of a deletion of the same amino acid and found almost no correlation (R2=0.07) (Supplementary Fig. S3c). This further indicates that the location of the InDel is more predictive than the identity of the amino acid inserted or deleted.
Next, we examined the correlation between fitness values when comparing insertions and substitutions. Specifically, we wondered if the fitness effect of an amino acid inserted before position N would correlate to the fitness effect of having position N mutated to the same amino acid. In this comparison, we included only fitness values of insertions at positions with a mean fitness ≥ 0.1. We do this to account for the predominance of insertions that result in complete loss of function. By excluding those positions, we instead ask the question: where insertions are tolerated to some degree, what is the correlation between the effects of insertions and substitutions? We find no significant correlation when we compare insertions to substitutions at the corresponding position (R2=0.07) (Supplementary Fig. S3b); however, the mean fitness change of an amino acid substitution is somewhat predictive of the mean fitness effect of the same amino acid insertion (R2=0.39) (Supplementary Fig. S3d). For example, the two least tolerated amino acid insertions (Pro and Trp) are also the least tolerated substitutions and the two most tolerated insertions (Ser and Thr) are among the most tolerated substitutions (Supplementary Fig. S3d).
We further explored TEM-1’s tolerance to insertions by determining the effective number of amino acid insertions at each position (). This measure is analogous to a measure of substitution tolerance (k*) that derives from information-theoretical entropy, which was originally proposed to quantify the variability at a given position in a set of aligned sequences [27]. As we showed previously, k* can be adapted to quantify the tolerance of substitutions based on measured fitness values [14]. For substitutions, a k* value of 1 indicates a position at which all missense mutations result in complete inactivation of the protein, and a k* value of 20 indicates that all amino acid substitutions result in the same fitness as wildtype. Here, we define a similar measure for insertions () which includes the possibility of no insertion (i.e. wild type) in the distribution of protein fitness values at each position (Eqs 1–4).
| (1) |
| (2) |
| (3) |
| (4) |
A value of 1 indicates a position at which no amino acid insertion is tolerated (i.e. the fitness values of all amino acid insertions are zero) and a value of 21 indicates a position at which all insertions retain wild-type fitness values.
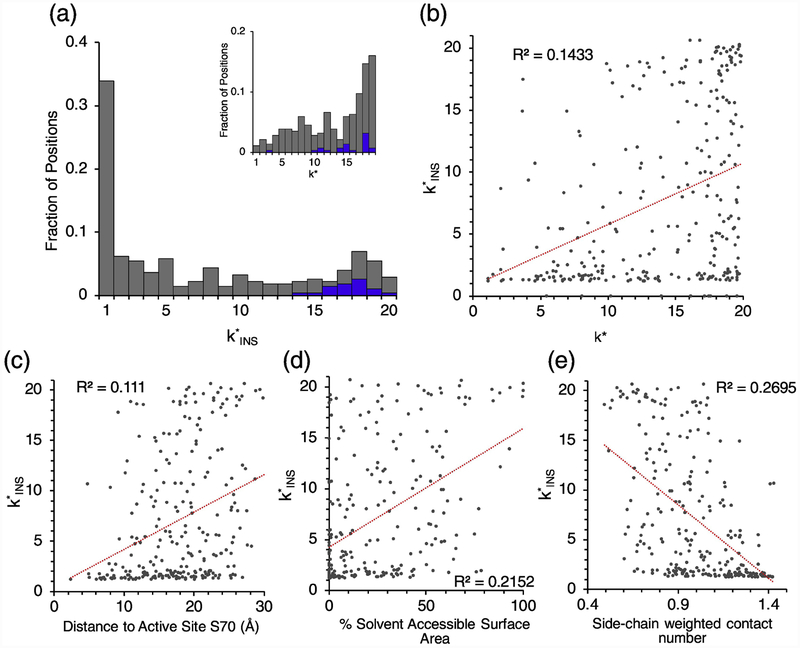
Over 30% of positions do not tolerate a single amino acid insertion of any kind () (Fig. 7a). The peak in the distribution of values between 17 and 20 indicates that there is a fraction of positions (19.3 %) for which most insertions are well-tolerated. However, there are no positions for which every inserted amino acid retains wildtype fitness ().
Fig. 7. Determinants of tolerance of TEM-1 to amino acid insertions.
(a) The distribution of values in TEM-1. values for the mature protein are colored in grey and values for the signal sequence are colored in blue. The inset shows the corresponding distribution of k* values for substitutions [14]. (b) Correlation of with k* of substitutions. [14] (c) Correlation of with distance from the active site. (d) Correlation of with percent solvent accessibly surface area. (e) Correlation of with side-chain weighted contact number (WCN).
All 23 positions in the signal sequence had values above 13, but five positions had a k* for substitutions less than 13 (Fig. 7a). In the entire protein, a position’s tolerance for insertion, as measured by , weakly correlated its tolerance for substitutions (Fig. 7b). We found that tolerance to insertions correlates weakly with distance from the active site (Fig. 7c). Positions less than 10 Å away from the active site are almost completely unaccepting of insertions. We observed a slightly stronger correlation between and percent solvent accessible surface area, with buried residues being less amenable to insertions (Fig. 7d). We found that side-chain weighted contact number (WCN), a measure of how densely packed a residue is [28], best predicts how well an insertion is tolerated (Fig. 7e), though the R2 is only 0.27. WCN was also the single best predictor of whether a deletion is tolerated in eGFP [17]. The lack of simple correlations between properties and insertion fitness were consistent with our expectation that the fitness effects of insertions have complex origins. Analogous correlations between these properties and substitution mutations were notably stronger, with ~2-fold higher R2 values [14].
Conclusions
Our analysis of InDels in TEM-1 provides the first systematic and near-comprehensive study of their fitness effect on a single protein and insight into a common yet understudied source of genetic variation. We found InDels to be largely deleterious, though regions of tolerance were observed, particularly in unstructured regions of the protein and at the ends of helices and strands. While regions of tolerance to insertions and deletions partially overlapped, we found that most regions of the protein tolerated insertions more than deletions. Of the measures we examined, we found secondary structure, weighted contact number, and evolutionary variation in class A beta-lactamases to be somewhat predictive of InDel fitness effects. A broader understanding the fitness effects of InDels and how they relate to structural properties should allow for more informed protein engineering strategies, more robust computational prediction of protein structure, and a deeper understanding of the role that different types of mutations play in protein evolution.
Materials and Methods
Insertion Library Creation
The TEM-1 gene was expressed on pSkunk2, a 4.36 kb plasmid containing spectinomycin resistance and the p15 origin of replication, under the IPTG-inducible tac promotor in E. coli. We used inverse PCR with oligo primers (IDT) designed to create every possible single amino acid insertion in TEM-1, using primers with a degenerate nucleotide (NNN) sequence on the 5’ end of the forward primer and a compatible reverse primer designed for each position. PCR products were visualized using gel electrophoresis, to confirm the creation of a linearized plasmid product at each of the 286 positions. We were unable to create a product for a small number of positions, despite troubleshooting efforts. We pooled the PCR products, creating a library for each third of the gene, to be compatible with Illumia MiSeq 2×300 bp sequencing. We isolated the ~4 kb band from an agarose electrophoresis gel for each third, phosphorylated the DNA at 37°C (NEB T4 PNK), and ligated it overnight at 16°C. NEB 5-alpha F’ lacIq E. coli were transformed with the ligation product and plated on LB-agar plates containing 50 μg/ml spectinomycin and 2% glucose. At least 500,000 transformants were obtained for each library (i.e. each third of the gene).
We recovered each library from the plate in LB media and isolated the plasmid library. We transformed electrocompetent SNO301 E. coli cells with each library and plated on LB-agar plates containing 50 μg/ml spectinomycin, 50 μg/ml chloramphenicol, and 2% glucose. At least 100,000 transformants were obtained from each third. We recovered each library from the plate in LB media and made glycerol stocks.
Deletion Library Creation
The deletion library was made in the same way as the insertion library with a few exceptions. The forward primer for inverse-PCR contained a 3-bp deletion on the 5’ end, to create a deletion at every position in TEM-1. The same reverse primers were used. The deletion library was not created in thirds, as it was subsequently sequenced using PacBio, which can accommodate longer reads.
Selection and Sequencing
High-throughput selection for resistance to ampicillin (Amp) was performed using a band-pass genetic circuit, described in previous work [14]. Briefly, E. coli SNO301 cells containing each library were plated on LB-agar plates containing 20 μg/ml tetracycline and 13 different Amp concentrations, ranging from 0.25 μg/ml to 1024 μg/ml, in 2-fold increments. Plates were incubated for 21 hours at 37°C. Each library was plated in triplicate on each Amp concentration and the CFUs from each plate were counted to determine the frequency of colonies appearing on each plate. Based on these counts, a proportional amount of DNA from each plate was deep sequenced. For the insertion library, barcoded amplicons were prepared by recovering the cells from each selection plate, isolating the plasmid DNA, and performing PCR with appropriate primers as described previously [14, 20]. Barcodes to identify each plate and adapters compatible with Illumina MiSeq platform were added in this PCR step. Amplicons were pooled and sequenced using Illumina MiSeq with 300 base pair, paired-end reads. For the deletion library, we recovered cells from each selection plate, isolated the plasmid DNA, linearized it with the SphI restriction enzyme, and separately sequenced each of the 13 linearized plasmid libraries using PacBio.
Data Analysis
The de-multiplexed MiSeq reads and the PacBio reads were analyzed using custom MATLAB scripts. For MiSeq reads, paired-end reads were trimmed and concatenated to yield full length reads. Each read was then aligned to TEM-1 using a Smith-Waterman algorithm with the lowest possible gap opening penalty of 1 and a gap extending penalty of 0.1. Reads with an alignment score lower than 100 were filtered out and only reads containing a single amino acid insertion (or deletion) were used for analysis. Fitness was calculated for each unique InDel mutant based on the counts from each plate (Amp concentration). For insertions, synonymous codons were grouped together and total counts were used to calculate the single amino acid fitness. Amber codons (UAG) were excluded from the stop codon analysis.
For each allele, counts were first adjusted based on the number of sequencing reads obtained from each plate relative to the CFUs observed on that plate, as described previously [20]. Detailed description of the fitness calculation can be found in our previous studies [14, 20], which we followed with just a few differences. For the insertion library, we excluded alleles with fewer than 20 counts and alleles with a maximum single plate count less than 1/3 the total count. For the deletion library, we excluded alleles with fewer than 5 counts. We used a lower cutoff for the deletion library because we used PacBio sequencing, which sequenced the entire TEM-1 gene. Thus, the sequencing data for deletions with low counts could not have artifactual data arising from TEM-1 base-substitution mutations arising spontaneously or during library creation, unlike the analogous data for the insertion library sequenced by MiSeq. Inspection of the sequencing counts per plate for the 17 deletions with between 5 and 20 counts supported the use of this lower threshold. Supplemental Data S1 and S2 contains sequencing counts and fitness values.
For each allele (i), the plate with the highest adjusted counts and the four plates on either side (i.e. two plates with higher Amp and two plates with lower Amp) were used to calculate an unnormalized fitness value, representing the midpoint resistance to Amp:
| (5) |
where ci, p is the adjusted count of allele i on plate p, and ap is the Amp concentration on plate p (in μ g/ml). The reported fitness values are normalized to wildtype TEM-1:
| (6) |
Wildtype fitness was calculated in the same way (i.e. using adjusted sequencing counts) and verified separately by plating wildtype in triplicate during the bandpass selection step. Both colony counts and sequencing counts revealed a midpoint Amp resistance of ~215 μg/ml.
Error in fitness () was estimated via Eqs 7 and 8, using our previously determined correlation between sequencing counts (ηi) and the standard deviation of the difference in fitness between synonymous alleles [14].
| (7) |
where ei, the upper-level estimate of the fraction error in fitness, is given by:
| (8) |
Fitness values were determined to be significantly different than 1 if they were greater or less than 1 by twice the error estimate.
Supplementary Material
Highlights.
Fitness effects of insertions and deletions (InDels) are understudied
Quantified fitness effect of 4,737 single amino acid InDels in TEM-1 β-lactamase Insertions are tolerated more than deletions except in a few regions
Relation between sequence/structural properties and fitness presented
Weighted contact number and evolutionary variation somewhat predictive of tolerance
Acknowledgements
This research was supported by the National Science Foundation (DEB-1353143, CBET-1402101, and MCB-1817646 to M.O.) and by the National Institutes of Health under a Ruth L. Kirschstein National Research Service Award (F31GM101941) to C.E.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Denver DR, Morris K, Lynch M, Thomas WK, High mutation rate and predominance of insertions in the caenorhabditis elegans nuclear genome, Nature 430 (2004) 679. [DOI] [PubMed] [Google Scholar]
- [2].Tóth-Petróczy Á, Tawfik DS, Protein insertions and deletions enabled by neutral roaming in sequence space, Mol. Biol. Evol 30 (2013) 761–771. [DOI] [PubMed] [Google Scholar]
- [3].Mullaney JM, Mills RE, Pittard WS, Devine SE, Small insertions and deletions (indels) in human genomes, Hum. Mol. Genet 19 (2010) R131–R136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hashimoto K, Panchenko AR, Mechanisms of protein oligomerization, the critical role of insertions and deletions in maintaining different oligomeric states, Proc. Natl. Acad. Sci. USA 107 (2010) 20352–20357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cooley RB, Arp DJ, Karplus PA, Evolutionary origin of a secondary structure: Π-helices as cryptic but widespread insertional variations of α-helices enhancing protein functionality, J. Mol. Biol 404 (2010) 232–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Britten RJ, Transposable element insertions have strongly affected human evolution, Proc. Natl. Acad. Sci. USA 107 (2010) 19945–19948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, La Starza R, Diverio D, Colombo E, Santucci A, Bigerna B, Pacini R, Pucciarini A, Liso A, Vignetti M, Fazi P, Meani N, Pettirossi V, Saglio G, Mandelli F, Lo-Coco F, Pelicci P-G, Martelli MF, Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype, N. Engl. J. Med 352 (2005) 254–266. [DOI] [PubMed] [Google Scholar]
- [8].Ye K, Wang J, Jayasinghe R, Lameijer E-W, McMichael JF, Ning J, McLellan MD, Xie M, Cao S, Yellapantula V, Huang K.-l., Scott A, Foltz S, Niu B, Johnson KJ, Moed M, Slagboom PE, Chen F, Wendl MC, Ding L, Systematic discovery of complex insertions and deletions in human cancers, Nature Med 22 (2015) 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kauffman S,Levin S, Towards a general theory of adaptive walks on rugged landscapes, J. Theor. Biol 128 (1987) 11–45. [DOI] [PubMed] [Google Scholar]
- [10].Leushkin EV, Bazykin GA, Kondrashov AS, Insertions and deletions trigger adaptive walks in drosophila proteins, Proc. Biol. Sci 279 (2012) 3075–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Shortle D, Sondek J, The emerging role of insertions and deletions in protein engineering, Curr. Opin. Biotechnol 6 (1995) 387–393. [DOI] [PubMed] [Google Scholar]
- [12].Gupta K,Varadarajan R, Insights into protein structure, stability and function from saturation mutagenesis, Curr. Opin. Struct. Biol 50 (2018) 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gray VE, Hause RJ, Fowler DM, Analysis of large-scale mutagenesis data to assess the impact of single amino acid substitutions, Genetics 207 (2017) 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Firnberg E, Labonte JW, Gray JJ, Ostermeier M, A comprehensive, high-resolution map of a gene’s fitness landscape, Mol. Biol. Evol 31 (2014) 1581–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mathonet P, Deherve J, Soumillion P, Fastrez J, Active tem-1 β-lactamase mutants with random peptides inserted in three contiguous surface loops, Protein Sci 15 (2009) 2323–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Arpino James A., Reddington Samuel C., Halliwell Lisa M., Rizkallah Pierre J., Jones DD, Random single amino acid deletion sampling unveils structural tolerance and the benefits of helical registry shift on gfp folding and structure, Structure 22 (2014) 889–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jackson EL, Spielman SJ,Wilke CO, Computational prediction of the tolerance to amino-acid deletion in green-fluorescent protein, PLoS ONE 12 (2017) e0164905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jacquier H, Birgy A, Le Nagard H, Mechulam Y, Schmitt E, Glodt J, Bercot B, Petit E, Poulain J, Barnaud G, Gros P-A, Tenaillon O, Capturing the mutational landscape of the beta-lactamase tem-1, Proc. Natl. Acad. Sci. USA 110 (2013) 13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Stiffler Michael A., Hekstra Doeke R., Ranganathan R, Evolvability as a function of purifying selection in tem-1 β-lactamase, Cell 160 (2015) 882–892. [DOI] [PubMed] [Google Scholar]
- [20].Steinberg B, Ostermeier M, Shifting fitness and epistatic landscapes reflect trade-offs along an evolutionary pathway, J. Mol. Biol 428 (2016) 2730–2743. [DOI] [PubMed] [Google Scholar]
- [21].Sohka T, Heins RA, Phelan RM, Greisler JM, Townsend CA, Ostermeier M, An externally tunable bacterial band-pass filter, Proc. Natl. Acad. Sci. USA 106 (2009) 10135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rockah-Shmuel L, Tóth-Petróczy Á, Sela A, Wurtzel O, Sorek R, Tawfik DS, Correlated occurrence and bypass of frame-shifting insertion-deletions (indels) to give functional proteins, PLOS Genetics 9 (2013) e1003882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pascarella S,Argos P, Analysis of insertions/deletions in protein structures, J. Mol. Biol 224 (1992) 461–471. [DOI] [PubMed] [Google Scholar]
- [24].Marciano DC, Brown NG, Palzkill T, Analysis of the plasticity of location of the arg244 positive charge within the active site of the tem-1 β-lactamase, Protein Sci 18 (2009) 2080–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Crane JM,Randall LL, The sec system: Protein export in escherichia coli, EcoSal Plus 7 (2017) 10.1128/ecosalplus.ESP-0002-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kim R,Guo J.-t., Systematic analysis of short internal indels and their impact on protein folding, BMC Struct. Biol 10 (2010) 24–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shenkin PS, Erman B, Mastrandrea LD, Information-theoretical entropy as a measure of sequence variability, Proteins 11 (1991) 297–313. [DOI] [PubMed] [Google Scholar]
- [28].Marcos ML,Echave J, Too packed to change: Side-chain packing and site-specific substitution rates in protein evolution, PeerJ 3 (2015) e911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.