Abstract
Characterization of proteins using NMR methods begins with assignment of resonances to specific residues. This is usually accomplished using sequential connectivities between nuclear pairs in proteins uniformly labeled with NMR active isotopes. This becomes impractical for larger proteins, and especially for proteins that are best expressed in mammalian cells, including glycoproteins. Here an alternate protocol for the assignment of NMR resonances of sparsely labeled proteins, namely ones labeled with a single amino acid type, or a limited subset of types, isotopically enriched with 15N or 13C, is described. The protocol is based on comparison of data collected using extensions of simple two-dimensional NMR experiments (correlated chemical shifts, nuclear Overhauser effects, residual dipolar couplings) to predictions from molecular dynamics trajectories that begin with known protein structures. Optimal pairing of predicted and experimental values is facilitated by a software package that employs a genetic algorithm, ASSIGN_SLP_MD. The approach is applied to the 36kDa luminal domain of the sialyltransferase, rST6Gal1, in which all phenylalanines are labeled with 15N, and the results are validated by elimination of resonances via single-point mutations of selected phenylalanines to tyrosines. Assignment allows the use of previously published paramagnetic relaxation enhancements to evaluate placement of a substrate analog in the active site of this protein. The protocol will open the way to structural characterization of the many glycosylated and other proteins that are best expressed in mammalian cells.
Keywords: mammalian cell culture, molecular dynamics, ligand docking, genetic algorithm, sialyltransferase
Graphical abstract
Introduction
NMR structural studies of uniformly 13C/15N labeled proteins larger than 40–60 kDa are challenging even when perdeuteration is used to enhance resolution and sensitivity. For glycosylated proteins, which are often expressed in mammalian cell culture to produce native- like glycosylation, perdeuteration is not possible; even structural studies of 20–30 kDa proteins are then challenging. Moreover, uniform isotopic labeling in mammalian cells with 13C and 15N can be costly as a mix of isotopically labeled amino acids, as opposed to isotopically labeled metabolic substrates, such as glucose and ammonium chloride, must be supplied. An economically viable alternative exists, namely sparse labeling using a single or small subset of isotopically labeled amino acids [1, 2]. Sparse labels can provide long range structural constraints through paramagnetic perturbations of resonance positions and intensities, as well as orientational constraints from residual dipolar couplings (RDCs) [3–5]. These constraints, along with chemical shift perturbation on interaction with other entities, can often be used to position ligands in binding sites and assemble proteins in multi-protein complexes [6, 7]. However, resonances must still be assigned to specific sites in proteins, and this must now be done without the aid of the triple resonance experiments usually applied to uniformly labeled proteins [8].
We recently introduced a strategy for resonance assignment of sparsely-labeled proteins that relies on acquisition of nuclear Overhauser effects (NOEs), RDCs and chemical shifts; all parameters measured directly from, or through modulation of, crosspeaks seen in basic two- dimensional heteronuclear single quantum coherence (HSQC) or multiple quantum coherence (HMQC) spectra. The strategy was implemented in a program package, ASSIGN_SLP, that employed a genetic algorithm to optimize pairing of specific spectral crosspeaks with specific protein sites using scores that compare experimental measurements of these parameters to predictions based on prior structural information, primarily from a single X-ray structure. The package was tested on a set of four small non-glycosylated proteins having known structures and crosspeak assignments, as well as a small glycoprotein [9]. It was subsequently applied to a larger non-glycosylated and perdeuterated protein, for which only the structure of isolated domains was known [10]. While the general approach showed success with smaller systems, it became clear that for larger systems, factors in addition to the technical aspects of associating predictions with experimental measurement would have to be considered. These include degeneracies in data that increase with the number of labeled sites, the greater probability of internal motion affecting observables and the more extensive spin-spin interactions that occur in larger proteins. Here, we introduce an approach that predicts parameters from molecular dynamics (MD) trajectories, as opposed to single structural snapshots from X-ray structures, to better account for effects internal motion and spin-spin interactions on predicted parameters. It also uses an improved procedure for identification of high-confidence assignments in the presence of data degeneracy. This approach, now embodied in a software package entitled ASSIGN_SLP_MD, proves useful in providing key assignments for a challenging 36 kDa glycoprotein, the luminal domain of rST6Gal1 (hereafter just rST6Gal1).
ST6Gal1 is a sialyltransferase that adds a sialic acid to the terminal galactose of N-linked glycans of many glycoproteins, and is therefore of importance in mammalian physiology [11]. The bond it forms is from the 2-carbon of sialic acid to the 6-oxygen of galactose, as opposed to the 3-oxygen of galactose. The specificity of the hemagglutinin of the avian influenza virus for the 2–3 linkage, found on glycans in the human gut, but seldom in the upper respiratory tract, is what restricts the transmission of bird flu to humans [12, 13]. Levels of 2–6 linked sialic acid also rise in certain types of cancer and there is significant effort devoted to understanding the possible role of sialylation in this disease [14, 15]. A decade ago we began an NMR-based structural study of rST6Gal1 [16]. At the time there were no crystal structures of ST6Gal1, or any of a close structural homolog. Using a sparse labeling approach in which all phenylalanines were labeled with 15N we demonstrated adequate resolution and sensitivity to detect HSQC crosspeaks from all 16 phenylalanine amide protons in the construct. Using a paramagnetic analog of the sialic acid donor (CMP-sialic acid), in which carboxy-TEMPO replaced the carboxyl-carrying sialic acid, we also showed that four of the 16 crosspeaks lost significant intensity. Based on an expected 1/r6 distance dependence of intensity loss, this number was deemed consistent with the number of phenylalanines in peptide segments believed to form the active site. However, in the absence of assignments we were unable to use the paramagnetic constraints to dock the donor analog in the active site of a homology model. In 2013 two X-ray structures appeared [17, 18], one of the rat enzyme on which our NMR work had been done [18]. With this structure in hand, along with previously collected RDC data, newly collected 1H-1H NOE data, and our new sparse label assignment strategy, we have proceeded with assignments of a new construct of rST6Gal1, isotopically labeled with 15N in all phenylalanines. A subset of the assignments are validated using a limited set of mutants in which single phenylalanines are changed to tyrosines, and then the assignments are used to place a sugar donor analog in the active site of rST6Gal1 in a manner consistent with paramagnetic perturbation data.
Results
Program development.
The ASSIGN_SLP_MD package is a collection of programs, primarily MATLAB scripts, that accepts as input a user-supplied MD trajectory, one or more files with experimental NOE peak lists (or NOE vectors derived from NOE strip plots), a file with 1H chemical shifts for labeled sites, a file with 15N or 13C chemical shifts for labeled sites and one or more files with RDC lists. Each of the files ends with a list of error estimates modified by weights for the specific data type. As success is very dependent on having adequate amounts of experimental data, it is recommended that at least one NOE file or one RDC file, in addition to chemical shifts, be present. Predicted data are appended to experimental files by scripts that call other programs to make these predictions. PPM [19] or SHIFTX2 [20] are used to predict chemical shifts averaged over frames of the trajectory. In the case of NOEs, a new version of our MD2NOE program, MD2NOE_Protein, is called; it uses the trajectory directly to make NOE predictions, taking into account the effects of internal motion and the extended interactions among multiple proton spins [21, 22]. In the case of RDCs, trajectories are used to calculate order parameters, which measure the amplitude of rapid variations in 1H-15N or 1H-13C bond orientations relative to the molecular frame, and produce coordinates for an average bond orientation; these in turn are used to adjust motionally-averaged experimental RDCs to a rigid equivalent and back-calculate predicted RDCs for each trial assignment using an algorithm similar to that in the REDCAT program [23]. A master script then calls a genetic algorithm that begins with a randomly generated set of assignments (each “gene” being a list of 16crosspeaks assigned to 16 different sites in our case). It calculates scores for each list based on an objective function that compares predicted and measured data, and it uses a series of runs with different crossover and mutation rates to mix assignments among the best scoring lists (genes) in an attempt to find an optimal assignment. Solutions with scores below a user-specified maximum are saved and latter analyzed by scripts that order output in terms of increasing scores and generate a heatmap showing the frequency of assignment of each crosspeak to each residue.
At the heart of the program is the objective function used in the genetic algorithm search for an optimal assignment. Initially this was defined as the sum of root-mean-square deviations (RMSDs) between measured and predicted values, divided by estimated errors (predicted plus observed standard deviations), for all data types except NOEs. The RMSDs minimize as agreement between measurements and predictions improves, as required for a well-behaved objective function. NOEs were treated differently because they are not represented by a single number, but by a series of intensities at the chemical shifts of NOE donating protons (actually a vector representation of a strip-plot from a 3D-NOESY spectrum). A Pearson correlation coefficient (R-value), which is a common way of assessing the similarity of two vectors was used to compare predicted and measured NOE vectors. The total NOE score, considering NOE vectors emanating from all crosspeaks, was then given as (1-R)2, as opposed to R2, divided by an estimated error, since R would go from 1 for perfect correlation to −1 for complete anti-correlation.
New additions.
The primary improvement in ASSIGN_SLP_MD comes from using, not just a single snap-shot of a protein structure as typically exists in a crystal structure, but from using long MD simulations (~1μs) to capture some of the effects of conformational averaging. This is not new in principle; MD simulations have been used previously to improve chemical shift prediction [19] and to provide order parameters which aid in interpretation of spin relaxation data [24], but they have not been used routinely. Until a few years ago a 1 μs MD run on a fully solvated protein, the size of rST6Gal1 would have been considered impractical. However, advances in computational hardware are now putting this timescale within reach of many laboratories. Our simulation of ST6Gal1 began with a crystal structure of the rat enzyme under conditions where neither donor nor acceptor was present (PDB ID 4MPS) [18]; these conditions match the conditions under which experimental data were collected. Unfortunately, this structure is missing a loop from 354–362 that contains two of the 16 phenylalanines. This loop was added directly from a structure of the homologous human protein in which the nucleoside portion of the donor was present (PDB ID 4JS1) [17]. The run required about two weeks on two GPUs running the PMEMD module of AMBER 14 [25]. Additional details are included in Materials and Methods.
The effect of using an MD simulation to improve prediction of RDCs is substantial. RDCs provide information on bond vector orientations (1H-15N bonds in our case) relative to a molecular alignment frame, but in the presence of internal motions that rapidly reorient these vectors, measured RDCs are reduced from their rigid limit (scaling by 1.0) to values scaled by the same order parameters that affect spin relaxation measurements. Dividing experimental RDCs by MD-derived order parameters scales values up to rigid equivalents that can easily be compared to predictions made during the genetic algorithm search. Order parameters for two of the residues within rST6Gal1, F132 and F356, are particularly small with values of 0.51 and 0.59 respectively.
While the use of the MD trajectory to better approximate RDC data proves valuable, the potential impact of using MD-based predictions is most dramatic in the case of NOEs. Our initial application to small proteins used an assumed 1/r6 distance dependence and distances extracted from crystal structures to predict NOE intensity contributions from each potential donating proton for each crosspeak. Chemicals shifts of donating protons were predicted byPPM_One [26] and predicted intensities were centered on these shifts, but spread over a region reflecting the uncertainty in prediction, to generate predicted NOE vectors [9]. For larger proteins and proteins having more internal motion, the 1/r6 assumption breaks down for two reasons. First, for internal motions that are fast compared to molecular tumbling, motional averaging depends on 1/r3 (plus an angular term), not 1/r6. Second, spin-diffusion effects, which are particularly prevalent in large proteins, make long-distance transfers by indirect mechanisms important. Direct calculation of correlation functions from an MD trajectory takes care of the former problem [27]. Use of a “complete” relaxation matrix takes care of the latter problem [28]. As a case in point, assuming a 1/r6 dependence, the ratio of NOEs for the amide proton of F208 in rST6Gal1 on inversion of neighboring proton HD1 on its phenyl ring and inversion of neighboring proton HB3 on its β-carbon would be 4.7. Using the program MD2NOE_Protein [21, 22], that incorporates both correlation function calculations and a “complete” relaxation matrix approach, one obtains a ratio of NOEs at a 40 ms mixing time of 0.9. We use this program to generate predicted NOE vectors in our new assignment strategy. For consistency with other terms in our objective function, NOEs are now scored as the RMSD of (1-R). There are, of course, other means of scoring agreement of predicted NOEs with experiment that may be considered in the future, including some based on Hausdorff distances which may be less sensitive to experimental outliers [29].
Weighting of various data types in our objective function have also changed. In our initial application various data types were simply weighted by the inverse of an estimated error. Dividing by estimated errors makes contributions of individual terms approach 1 when deviations approach estimated error. With standard estimates of error, a total score equal to the number of data types then provides a cutoff below which any total assignment should be considered acceptable. Because we are usually more interested in the confidence that can be placed in the assignment of a particular site to a crosspeak of interest (one perturbed on ligand binding, for example) than the assignment of all crosspeaks, we had suggested use of a confidence score based on the frequency of assignment of a crosspeak to one particular site within the set of all complete assignments deemed acceptable. We plan to keep this means of confidence assessment. However, there are factors, other than precision of measurements and predictions, that should be included in the weights used in the course of our genetic algorithm search. Factors that are not well represented in estimates of error include what we call “information content”. For example, degeneracies in RDC data may arise in certain proteins (an alpha-helical bundle) because the vectors connecting spin pairs (15N-1H amides) may be nearly parallel. This would reduce information content of the RDCs. Also, missing data allows interchange of assignments regardless of the precision of measurement. In this new version we have introduced an option that allows weighting by information content in addition to the inverse of an error estimate. In practice we define this as the variance in score relative to the square of the range of scores for each data type and provide MATLAB scripts that calculate weights for each data type. In addition, scaling by the ratio of the number of independent data points (measured values in most cases, but measured −5 in the case of RDCs where 5 order tensor elements must be determined from the measured values) to the number of sites to be assigned is included automatically as a part of the inverse of error estimates. This decreases the importance of data types when experimental data for particular sites are missing.
Establishing a confidence cutoff.
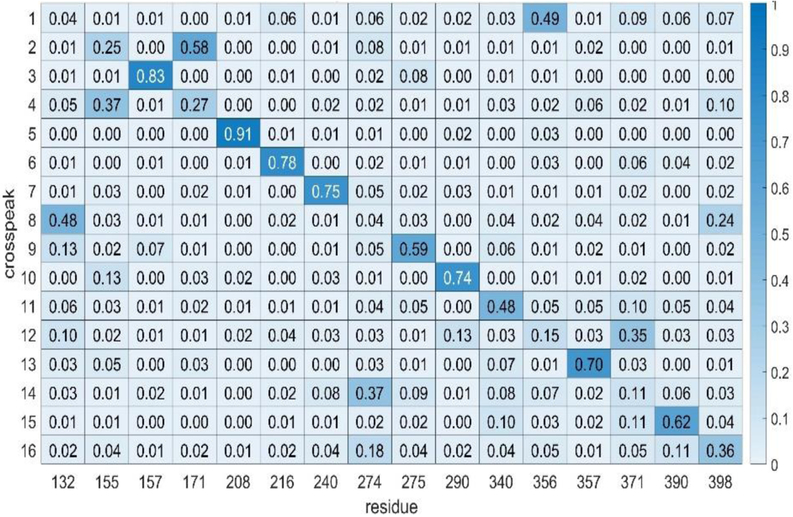
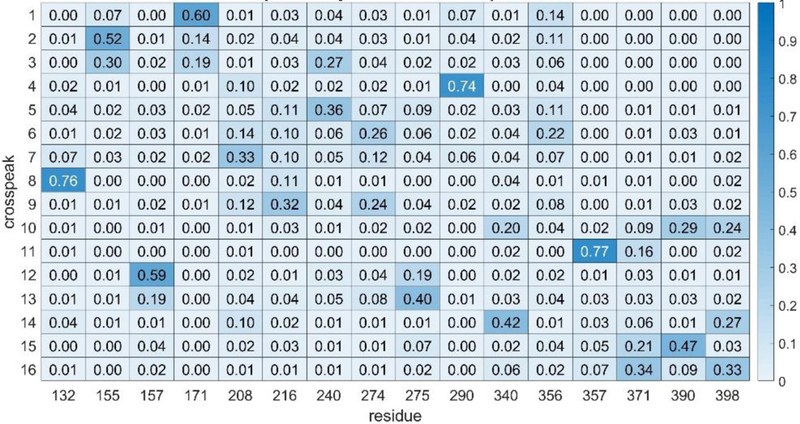
In using any assignment program, it is important to attach a level of confidence to the assignments made. This can be done by first testing the program on simulated experimental data that has been generated by adding random errors of a known magnitude to a predicted set. A predicted set appropriate for our eventual application to rST6Gal1 was generated using chemical shifts from a combination of PPM and SHIFTX2 calculations averaged over rST6Gal1 trajectory frames, using two RDC sets from application of the REDCAT program to a single frame from the trajectory, and using NOEs from the MD2NOE_Protein program as described above. A simulated experimental set was then generated by adding random errors to chemical shifts and RDCs, within limits that proved applicable to the actual experimental data (see section on application to experimental data below). For RDCs, 2 and 4 pieces of data, respectively, were also deleted from the two sets to mimic missing data in the actual experiments. NOE intensities were randomly varied within a ±25% limit and peak positions were varied within errors for shifts. Application of the program ASSIGN_SLP_MD gave a best solution with an unweighted best score of 4.1. Using an unweighted score cutoff of 5.0 (an average contribution of 1.0 for each of the 5 data types), the heatmap presented in Fig. 1 was produced. The numbers displayed in the heatmap are the fraction of time an assignment of a crosspeak to the same residue is made in a set of total assignments having scores between 4.1 and 5.0. The residues and calculated data were not scrambled, so the correct solutions occur on the diagonal. Note that most of the high fractions (darker blue) occur along the diagonal. If we choose a cutoff level of 0.50, we would identify 9 assignments as highly confident and there would be only one false positive. Hence, this cutoff can be associated with approximately a 90% confidence level. Using a less conservative approach in which errors are scaled down by 2/3, all 16 peaks have highly confident assignments and all 16 are assigned correctly.
Figure 1.
Heatmap showing ST6Gal1 phenylalanine assignments using simulated data. The numbers in each element are the fraction of times a crosspeak is assigned to a particular residue. Higher numbers are color-coded a darker blue and are taken to indicate a more confident assignment. All correct assignments should be on the diagonal in this case since the crosspeak order is was chosen to order with the residue order.
Application using experimental data.
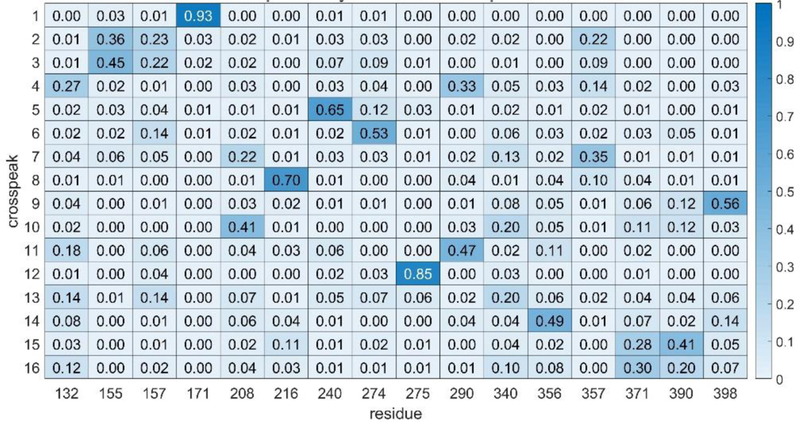
Experimental data on ST6Gal1 consisted of chemical shifts from an 800 MHz 1H-15N HSQC spectrum, NOEs from an 800 MHz NOESY-HSQC spectrum and two sets of 900 MHz RDC data, one using bacteriophage, and one using alkyl-ethylene-glycol (C12E5, PEG) bicelles to orient the protein. Errors for the chemical shifts are dominated by errors in predictions; these were initially set to two times the errors suggested by the authors of SHIFTX2. Errors in NOEs were taken from the noise level in experimental spectra and errors in RDCs were estimated based on line widths of spectra. Both the data and error estimates are detailed in Materials and Methods. Using these errors, the initial run of ASSIGN_SLP_MD failed to give any solutions with a score below the expected error-derived limit of 5.0. This is likely due to error contributions to data or simulations that are difficult to predict (for example, truncation of NOEs by exchange phenomena or failure to sample all conformers in the 1μs trajectory). We therefore raised all errors by 50% and repeated the run. The minimum solution then had a raw score of 4.4. A heatmap generated using all solutions below 5.0 is shown in Fig. 2. If we use the confidence cutoff of 0.50 suggested by our simulated data, we would confidently assign 6 of the 16crosspeaks.
Figure 2.
Heatmap showing ST6Gal1 phenylalanine assignments using experimental data. The 6 most confident assignments (fraction > 0.5) are shown in darker shades of blue These are now scattered throughout since we don’t know apriori how to order the crosspeaks.
Data not included in the objective function can in general be used to assess accuracy. Our prior work on rST6Gal1 had shown that addition of an analog of rST6Gal1’s nucleotide sugar donor that carries a paramagnetic TEMPO group causes paramagnetic relaxation enhancement (PRE) and intensity loss for four crosspeaks (6,7,10 and 14) [16]. Manually docking this donor analog into the active site of the 4MPS crystal structure and having the missing residues 356 and 357 modeled in from the 4JS1 structure results in a position for the TEMPO nitroxide group with the four closest phenylalanine amide protons (those of F208, F240, F356 and F357) at distances of 12, 16, 11 and 10 Å. Of this group, one assignment is at the edge of our confidence limit, F356 to crosspeak 14; this is in agreement with PRE data. While below our confidence limit, both F208 and F357 have their highest fraction of assignments to crosspeaks 10 and 7 respectively. This too is in agreement with PRE data. Based on frequency of assignment, peak 6 would be incorrectly assigned to F274, a residue far removed from the active site.
Validation.
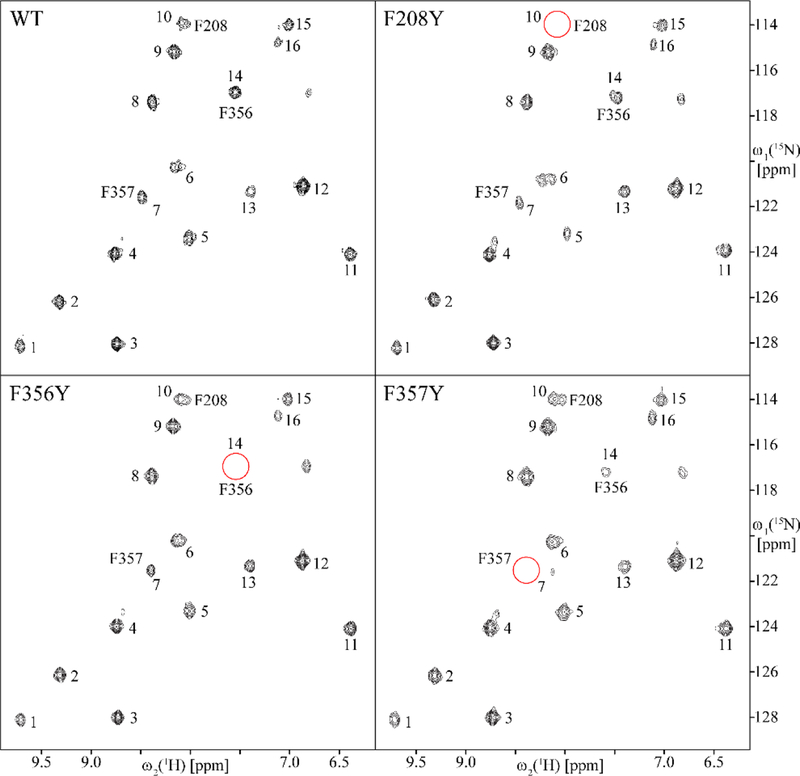
A more robust validation can be carried out by mutating phenylalanine residues to tyrosines, resulting in elimination of crosspeaks for the mutated residues. This was done for the three phenylalanines closest to the TEMPO group in the ST6Gal1 model, F208, F356, and F357. HSQC spectra for the 3 mutated proteins are shown in Fig. 3. Along with the HSQC spectrum of the wild type (WT) protein. In each of the spectra for mutated proteins one crosspeak is missing (red circles). This clearly assigns crosspeak 10 to F208 crosspeak 14 to F356 and crosspeak 7 to F357. The assignment made by ASSIGN_SLP_MD is therefore correct for our near-confident assignment (F356) as well as the highest fraction assignments of F357 and F208. While the mutational validation does not strictly overlap with our confident assignments (only the assignment of crosspeak 7 to F357 is close with a fraction of 0.49 as opposed to 0.50), the correlation of mutational assignments with the highest scoring assignment in each case adds confidence to our procedure.
Figure 3.
800 MHz 2D [15N, 1H] HSQC spectra of WT rST6Gal1 and single-point mutants F208Y, F356Y and F357Y One crosspeak disappears in each of the mutant spectra (red circles) identifying the crosspeak belonging to the mutated site.
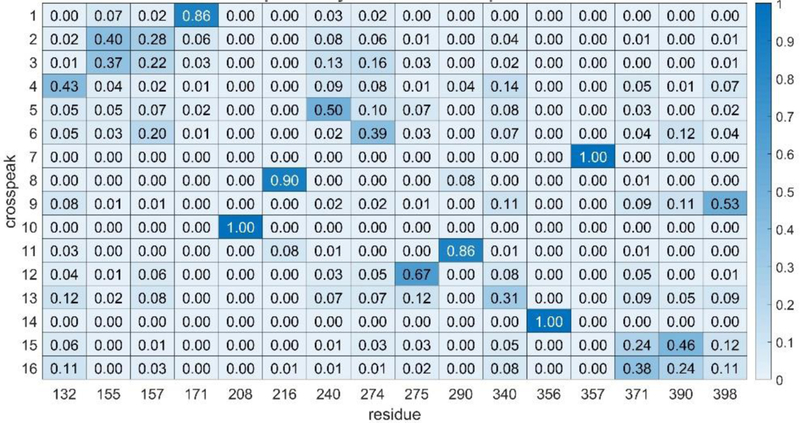
Mutational assignments can, of course be regarded as additional experimental data. These are easily incorporated into our assignment strategy through a penalty matrix (residue by crosspeak) that adds a zero score to our objective function for any assignment known to be correct and a high score (~10) for all other assignments. The results of applying this procedure using the 3 mutational assignments are shown in Fig. 4. There are now 9 assignments that we would regard as confident. There has been one notable removal of an assignment from the confident assignment list, that of crosspeak 6 to F274. Since crosspeak 6 is one of the crosspeaks showing intensity loss in the presence of a paramagnetically tagged donor analog, and F274 is not among the list of nearby residues, removal from this list is reassuring.
Figure 4.
Heatmap showing rST6Gal1 phenylalanine assignments using mutational constraints (F208 to 10, F356 to 14 and F357 to 7).
MD versus single frame analysis.
A remaining question is whether the use of an MD trajectory to simulate NMR data has made a significant difference in the quality of crosspeak assignments. To examine this, we used a single frame version of the ASSIGN_SLP_MD program which assumes a 1/r6 distance dependence to derive relative NOE intensities. Two single frames having the smallest RMSDs of backbone atom positions from the crystal structure (1.11 and 1.16 Å) were chosen from 500 samplings of the trajectory (the crystal structure could not be used directly because of the absence of the 354–362 loop). The procedures and errors used were identical to those used with the MD version of the program. The raw scores for the best solutions in the two frames were 4.3 and 4.2 respectively, not very different from those using the MD derived predictions. However, the fractions assigned to any particular pairing are generally lower and the number of high confidence scores are lower (4 and 6 in the 1.11Å and 1.16Å frames respectively). A heatmap for the 1.11Å frame produced using assignments with scores below 5.0 is presented in Fig. 5. There is some similarity to the MD-based assignment in Figs. 2 and 4. For example, crosspeak 1 is confidently assigned to F171, crosspeak 5 has its highest fraction of assignments to F240 and crosspeak 15 has its highest fraction of assignments to F390. However, neither of the single frame runs makes a highest fraction assignment consistent with any of the three mutationally validated assignments. Clearly, there is a substantial advantage in using MD simulations to improve predictions.
Figure 5.
Heatmap showing ST6Gal1 phenylalanine assignments using a single frame with a 1.10 Å RMSD of backbone atoms from those of the crystal structure, 4MPS.
Discussion
The data presented on the assignment of 1H-15N crosspeaks in HSQC spectra of the sparsely-labeled glycoprotein, rST6Gal1, suggests that similar assignments will be possible on a host of biomedically relevant proteins that are best expressed in mammalian, or other eukaryotic cells. Validation of assignments has confirmed an ability to set reasonable confidence limits on assignment so that, even when total assignments are not possible, a subset can be identified as trusted assignments. In many cases, some of these crosspeaks will be perturbed by ligand binding, leading to identification of residues involved in active sites of enzymes or binding pockets of receptors. For ST6Gal1, the peak at the edge of our confidence limit, (peak 14 assigned to F356) is perturbed by addition of a reaction product, cytidine monophosphate (CMP) that is known to inhibit sialylation activity [16]. In other cases, a sufficient number of trusted peaks may be perturbed by paramagnetic moieties, to allow use as constraints in ligand docking or refinement of protein structure.
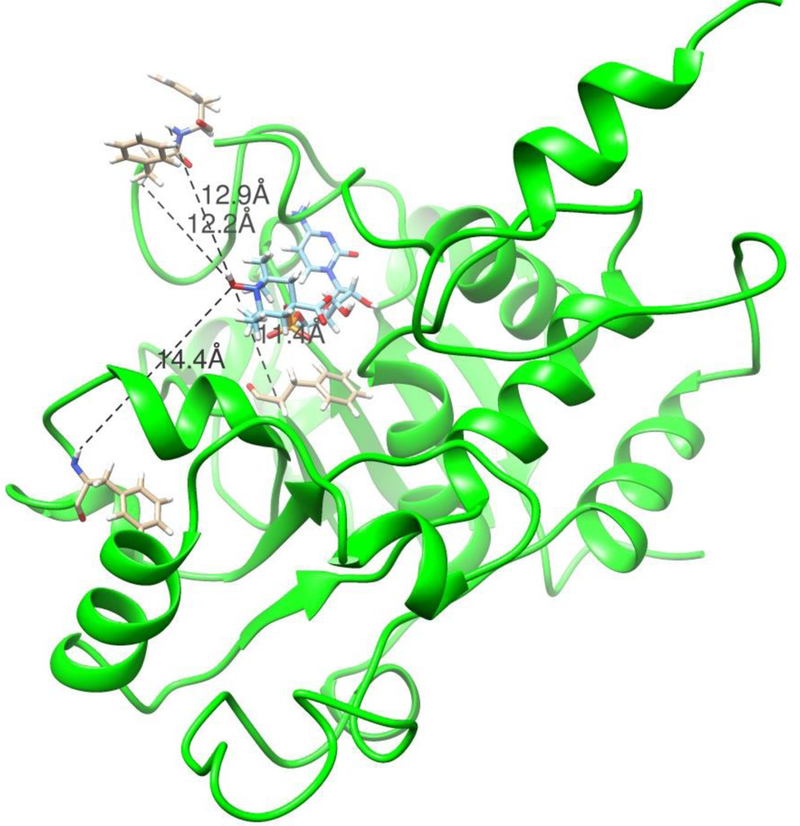
We can illustrate this latter case by using our previously published perturbations of crosspeaks by an analog of rST6Gal1’s sugar donor, sialylated cytidine monophosphate (NeuAc- CMP) [16]. The analog replaces the sialic acid with a carboxy-TEMPO group that retains the carboxyl group of sialic acid but replaces the six- membered ring of sialic acid with that of TEMPO. The TEMPO group carries a nitroxide oxygen with an unpaired electron distal from the phosphate ester connection to CMP. This oxygen is taken as the origin of paramagnetic perturbations. Prior estimates of distances between the oxygen and amide protons at sites associated with crosspeaks 6, 7, 10 and 14 were 14.3–15Å, 12.6–14.4Å, 10.6–12.9Å, and <17.0Å. The latter number is only an upper limit due to the low intensity of crosspeak 14 which is broadened significantly in the presence of CMP-TEMPO as well as CMP itself. To generate a model consistent with these distances, a structure taken from frame 100,000 of a rST6Gal1 trajectory (a stable point about 200 ns into the 1 μs simulation) was superimposed with the structure of hST6Gal1 determined with the reaction product (CMP) in place (4SJ2). Then the CMP moiety of our donor analog was superimposed with the CMP of 4SJ2, and the torsions of the two phosphate ester bonds plus the phosphate oxygen to TEMPO bond were adjusted to place the nitroxide oxygen within the above distance limits without introducing van der Waals clashes. The resulting structure is shown in Figure 6. The distances between nitroxide oxygen and the amide protons of F240 assigned to peak 6, F357 assigned to peak 7, F208 assigned to peak 10, and F356 assigned to peak 14 are 14.4Å, 12.9Å, 11.4Å and 12.2Å, respectively. The structure is chemically reasonable and places the carboxylated carbon of the analog in a position where SN2 attack by the O6 oxygen of a galactose-containing acceptor can approach.
Figure 6.
Model of rST6Gal1 with the donor analog, carboxy-TEMPO-CMP docked into the active site. Distances shown are those between the nitroxide oxygen of the TEMPO group and the amide protons of F240 (14.4), F357 (12.9), F208 (11.4) and F356 (12.2), respectively.
The use of an MD trajectory to improve prediction of NMR data has proven particularly useful. It provides one way of accounting for the averaging of parameters over the multiple conformational states sampled by a macromolecule. There are other ways of sampling conformational space; for example, by Monte Carlo based searches on the entire protein structure [30] or empirical representations of motions of individual peptide planes such as the Gaussian Axial Fluctuation models [31]. These may work equally well for observables such chemical shifts, which are primarily structure sensitive. However, NOEs are sensitive to both structure and dynamics. Among other factors their dependence on inter-proton distances, r, changes with the timescale of internal motion. When internal motions occur on timescales that are shorter than that of overall protein tumbling, the 1/r6 dependent term, often assumed in structure calculations, is replaced by a 1/r3 and angular dependent term that is best treated by directly calculating correlation functions from an MD trajectory [24]. This is the procedure we have adopted for calculation of the relaxation elements that are subsequently used in a complete relaxation matrix approach to NOE simulation. The MD approach may have its own limitations in terms of accuracy of time scale representations and practical limitations to the length of simulations, but forcefield improvements [32] and enhanced sampling methods [33] may address these limitations in the future. ST6Gal1 may not be representative of all proteins in the extent to which parameters are affected by internal motion. It has a loop containing two of the labeled phenylalanines that is not visible in the rat crystal structure. This loop is near the active site and clearly undergoes motion as evidenced by motional broadening of the phenylalanine resonances in the presence of CMP [16]. However, many enzymes share a tendency to have flexible regions as a part of their active site. Hence, the advantages of an MD-based analysis shown for ST6Gal1 may apply to certain subsets of proteins having high internal mobilities.
As for future applications, increasing the faction of confident assignments is clearly important. More precise experimental measurements and longer MD simulations will likely help. However, addition of other data types may be more important. We have already illustrated the impact of adding constraints from mutational studies. Other data types can also be added. Pseudo contact shifts (PCSs) share a functional form with RDCs [5, 34], as do PREs with NOEs, making addition straightforward. The procedure we have described for setting confidence limits, in which a simulated experimental data set is prepared by adding random errors to a predicted data set that has numbers of each data type equal to numbers of real experimental data and used in a search for best solutions (Fig. 1), provides a convenient means of assessing how much data is needed to reach assignment goals.
Applications to larger proteins are also of interest. Resolution of 1H-15N crosspeaks in the HSQC spectra shown here is certainly adequate to target proteins twice the size of rST6Gal1. However, sensitivity can be an issue. This will drop steeply for fully protonated glycoproteins of larger size. One encouraging prospect is the possibility of labeling with 13C methyl groups. Labeling all methyls in isoleucine, leucine and valine (ILV labeling) has provided a route to NMR characterization of large perdeuterated proteins expressed in bacterial cell cultures [35]. Assignment of methyl resonances in these instances presents challenges that parallel those for sparsely labeled glycoproteins. Alternative assignment strategies, similar in some respects to that described here, have been introduced recently [36–38]. Reliance on NOE data is one common aspect, but reduction in numbers of protonated sites by deuteration has allowed interpretation in terms of constraints on a very qualitative level. ASSIGN_SLP_MD is certainly applicable to data on ILV-labeled and perdeuterated proteins, and its use of MD trajectories to make interpretation of NOE data more quantitative may be particularly useful. A current limitation for many of these larger systems is the availability of appropriate crystal structures. This could be relaxed if an appropriate homology model could be selected. The minimum scores reached in making an assignment with ASSIGN_SLP_MD in many ways reflects the quality of the structural model used, and it may be possible to simultaneously obtain an assignment and select the best among several homology models. Comparison between predicted and measured chemical shifts from 13C-13C correlation spectra acquired by solids NMR have already been used to screen homology models [39], and with addition of more data types this may be possible with sparsely labeled samples as well.
Materials and Methods
Protein Expression, Mutagenesis and Purification.
Protein sample preparations used in collection of RDC data and PRE data were analogous to those described in a previous publication [16]. New samples were prepared for the collection of NOE data and validation by mutagenesis using modified methods for expression, labeling, and purification as described in the literature [40, 41]. Briefly, expression constructs encoding the luminal domain of rat ST6Gal1 (UniProt P13721, residues 103 to 403) in the pGEn2 vector were transiently transfected into HEK293S (GnTI−) cells [41] and metabolic labeling with [15N]-Phe was initiated 16 h after transfection by exchange of the culture medium for custom FreeStyle 293 expression medium (Thermo Fisher Scientific) depleted in Phe and supplemented with 150 mg/L [15N]-Phe 98% (Cambridge Isotope Laboratories, Andover, MA) and 2.2 mM valproic acid. The recombinant protein was harvested from the culture supernatant after 6 days of growth, purified by Ni2+-NTA chromatography, and concentrated to ~1 mg/mL. The resulting protein preparation was digested with recombinant TEV protease to cleave between ST6Gal1 and GFP, and recombinant endoglycosidase F1 (EndoF1) was used to cleave the glycans to single GlcNAc residues [41]. The preparation was then subjected to Ni2+-NTA chromatography a second time to remove the GFP fusion tag, TEV protease, and EndoF1, each of which contain a His tag [41]. The samples were further purified by Superdex75 chromatography (GE Healthcare Life Sciences) using a 20mM HEPES, pH 7.5, 250mMNaCl, and 60mM imidazole buffer. Peak fractions of ST6Gal1 were collected and concentrated to 20 mg/ml using an ultrafiltration pressure cell membrane. Exchange to NMR buffers (20mM Sodium Phosphate, pH 6.5, and 100mMNaCl for NOE and mutational studies) was accomplished usingCentricon centrifugal filtration units with a 10kDa cutoff. Site directed mutations of ST6Gal1 (F208Y, F357Y, and F357Y) were performed using the Q5 site-directed mutagenesis kit (New England Biolabs, Ipswich, MA) in the pGEn2- rST6Gal expression vector.
NMR data.
One bond 15N-1H RDCs were measured using the interleaved fHSQC and fHSQC-TROSY experiments collected at 25 C on a Varian Inova 900 MHz spectrometer equipped with a cryogenic triple resonance probe. Data were collected over a 24 h period with acquisition times of 30 and 80 ms for t1 and t2, respectively, and a 1.5 s recycle delay. NMR data were processed and analyzed using FELIX software. The rST6Gal1samples were in 10 mM phosphate, 200 mM NaCl, pH 6.8, with 10% 2H2O; partial alignment was obtained using PEG (3% C12E5) and pf1 phage (10mg/mL) media as previously described [4], giving deuterium splittings of the water resonance of 13 and 21 Hz respectively. Protein concentrations were at 350 and 400 μM for phage and PEG media respectively.
A 3D 15N-edited [1H, 1H] NOESY-HSQC spectrum of a 15N-Phe labeled WT rST6Gal1 sample was recorded on an 800 MHz Bruker AVANCE NEO spectrometer equipped with a 5mm cryogenic triple-resonance probe. The NMR sample contained 270 ul of 630 uM 15N-Phe WT rST6Gal1, 4 uM DSS, 0.02% sodium azide and 10% 2H2O in a Shigemi tube. NOE mixing time was set to 60 ms, and acquisition times t3,max(1H), t2,max(1H) and t3,max(15N) were set to 46 ms, 10ms and 10ms, respectively. Total acquisition time was 40 h, with a 1.1s recycle delay. A 2D [15N, 1H] HSQC was also recorded in 20 m with 1.0 s recycle delay and acquisition times t2,max(1H) and t1,max(15N) of 106 ms and 39 ms, respectively. Spectra were processed with TopSpin v3.5 (Bruker BioSpin) and analyzed with CARA v1.9.1.7.
Experimental NOE vectors were produced by averaging spectral intensity over an ellipse in the HSQC plane of NOE strip plots with dimensions ±0.03 ppm (1H) and ±0.65 ppm (15N). Diagonal peaks in all vectors, as well as the H2O resonance (4.79 ppm) in vectors 1 and 3, were removed by setting intensity within ±0.17 ppm of the corresponding signal to zero.
2D [15N, 1H] HSQC spectra of single-point rST6Gal1 tyrosine mutant samples were recorded on the same 800 MHz Bruker AVANCE NEO spectrometer but equipped with a 1.7 mm cryogenic triple-resonance probe. Samples consisted of 40 ul solutions of 330 uM F208Y, 580 uM F256Y, or 220 uM F357Y rST6Gal1 with 7.5uM DSS and 0.09% sodium azide in 10% 2H2O. Acquisition parameters were the same as for WT rST6Gal1, only the number of transients was adjusted.
MD Simulation and Docking.
The starting point for the MD simulation was the 4MPS crystal structure; the missing 354–362 segment was modeled in using the corresponding segment from the 4JS1 structure and minimized. The simulation was then carried out using the PMEMD module of the AMBER 14 package [25]. The ff14SB force field was used for protein residues and the GLYCAM_06j-1 force field [42] was used for the two GlcNAc residues attached to Asn residues at sites 146 and 158. A cubic box of TIP3 water extending a minimum of 8Å from the protein surface was used to solvate the protein. The system was first energy minimized by 50000 steps of minimization, then heated to 300 K in 2 fs steps over 1 ns. The 1 μs MD simulation was carried out using 2 NVIDIA GeForce GTX TITAN Black GPUs on a 4 GPU laboratory computer and required about 2 weeks. For use in NOE simulations, frames of the trajectory were aligned by minimizing deviations of backbone α-carbons using tools incpptraj, an AMBER 14 utility [43]. For chemical shift predictions by PPM and SHIFTX2 every 200th frame was extracted and saved as a model in PDB format, again using tools in cpptraj [43]. Graphic depictions of structures and docking of ligands was preformed using tools in the Chimera package [44].
ASSIGN_SLP_MD Package.
The ASSIGN_SLP_MD package, as implemented in this study, contained several modules that prepared input for the search module, executed the search and assembled output for presentation to the user. All are designed to operate under a LINUX operating system and execution with different input files can be facilitated by using bash scripts. Efforts are underway to integrate the separate modules of the program and develop a user interface. Up-to-date versions, as well as additional documentation, are available at http://tesla.ccrc.uga.edu/software/.
15N and 1H predicted chemical shifts for amide groups of the selected amino acid type were extracted from output of PPM [19] and SHIFTX2 [20] run on the PDB format trajectory by a MATLAB script called “Procedure for Spectra Generation”. The shifts were then appended to lists of experimental shifts, and a list of estimated error, as modified by weights, was added in separate input text files for 15N and 1H shifts.
NOE predicted peak lists for the 16 phenylalanine amide protons were prepared by a new version of the program, MD2NOE [21], written in C++ and called MD2NOE_Protein. Both predicted and experimental peak lists were converted to 512-point vectors containing gaussian lines of a user-specified width (0.2 Hz in this study) at the predicted or experimental chemical shift of donating protons and intensity as specified in the peak lists, again using the MATLAB script, “Procedure for Spectra Generation”. Such vectors correspond to a 1-dimensional slice along the indirect 1H frequency dimension of a NOESY spectrum. Diagonal NOE peaks were not included, but a “pseudo-peak” of intensity equal to the maximum NOE peak intensity averaged over all 16 vectors was added at the end of each vector. These were output as spreadsheets in csv format. Instead of experimental peak lists, NOE spectral intensities from 3D-15N-edited NOESY-HSQC spectra can also be converted to vectors with gaussian-broadened peaks with the same program. As in the case of 15N and 1H shifts, weighted errors are added to the end of each vector.
A MATLAB script entitled “Order Parameters” was used to calculate order parameters and average 1H-15N bond vectors for use in predicting RDCs for each trial assignment within the search module. Output was in the form of a text file with a line for each residue containing the six coordinate entries for the bonded pair and an order parameter. Experimental RDCs were provided in separate text files for each medium with an ordered list of weighted errors following the RDCs. Weights added to account for information content were calculated with separate MATLAB scripts for chemical shifts, RDCs, and NOEs; these generated distributions of scores by comparing each entry to every other entry and extracting a variance for the distribution, divided by the range of scores.
The search module, called ASSIGN_SLP_MD, is based on a genetic algorithm function call (ga) available as a part of the optimization toolbox of the MATLAB package. It reads in output from the various preparation modules and functions as described in our previous publication [9] except for the changes in score contributions to the objective function as described in the main text of this manuscript. Searches are repeated with 16 different combinations of mutation and crossover rates (2, 4, 6, 8 For each) to maximize the adequacy of the search. Every trial assignment with a score below a user specified level (raw score of 5 in the application presented) is saved in a text file along with the total score, and individual contributions from the various data types. Each search ceases when no improvement in score beyond the tolerance of 1e-4 is achieved or a maximum number of 500 iterations is reached.
The analysis module retrieves the output of the genetic algorithm search, orders the output by total score and eliminates all duplicates. A distribution of randomly generated scores is then calculated so that a mean and variance can be extracted, and Z-scores appended to each assignment in the ordered list. Heatmaps are then generated by considering the fraction of times the same crosspeak is assigned to the same residue within a set of all assignments having a score below a user specified limit. In the example presented, the limit selected to be an unweighted raw score equal to the number of experimental data types (5 in our example) or one unit above the minimum unweighted raw score when the this was greater than the number of data types; this resulted in inclusion of 1000 to 10,000 solutions in the sets discussed here.
Data Summary:
Data used in the application to rST6Gal1 are summarized in the following tables.
Table 1.
Non-NOE data used in rST6Gal115N-phenylalanine assignments.
| Data | Crosspeaks | Wta | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | ||
| 1H cs | 9.7 | 9.3 | 8.7 | 8.8 | 8.0 | 8.1 | 8.5 | 8.4 | 8.2 | 8.1 | 6.4 | 6.9 | 7.4 | 7.5 | 7.0 | 7.1 | 0.35 |
| error | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 | |
| 15N cs | 128 | 126 | 128 | 124 | 123 | 120 | 121 | 117 | 115 | 114 | 124 | 121 | 121 | 117 | 114 | 114 | 0.55 |
| error | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | |
| RDCpeg | −16 | 7 | −5 | 2 | −13 | 999 | 2 | 29 | −1 | 999 | 999 | −23 | −2 | 28 | 999 | 9 | 0.42 |
| error | 10 | 10 | 10 | 10 | 10 | 20 | 10 | 20 | 10 | 10 | 20 | 10 | |||||
| RDCpfl | 0 | −6 | −18 | −9 | −6 | −27 | −4 | 21 | −11 | 999 | 30 | −2 | −9 | 18 | −19 | 999 | 0.37 |
| error | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | |||
Weights (Wt) include an estimate of information content (variance/range2) and a penalty for missing data (#data/#sites for chemical shifts and ((#data-5)/#sites for RDCs). Errors for chemical shifts are 2x ShiftX2 estimates; RDC errors are approximately 20% of line widths.
Table 2.
NOE peak list representing data used in rST6Gal115N-phenylalanine assignments.a
| Data | Crosspeaks | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | |
| 1H cs | 2.4 | 2.8 | 2.5 | 1.5 | 2.1 | 0.8 | 3.0 | 1.8 | 1.6 | 9.4 | 1.1 | 2.1 | 4.0 | 1.6 | 4.0 | 3.2 |
| intensity | 17 | 21 | 18 | 18 | 18 | 12 | 22 | 12 | 17 | 4 | 12 | 10 | 23 | 16 | 13 | 11 |
| 1H cs | 2.8 | 3.1 | 2.9 | 1.7 | 2.3 | 2.7 | 4.7 | 2.4 | 3.0 | 1.6 | 2.0 | 6.8 | 3.0 | 8.3 | 4.2 | |
| intensity | 13 | 16 | 42 | 50 | 12 | 14 | 25 | 24 | 16 | 12 | 43 | 14 | 21 | 16 | 10 | |
| 1H cs | 3.7 | 4.7 | 3.3 | 2.8 | 2.9 | 4.5 | 3.5 | 3.2 | 3.4 | 3.2 | 3.2 | |||||
| intensity | 13 | 10 | 41 | 25 | 23 | 12 | 11 | 12 | 10 | 30 | 15 | |||||
| 1H cs | 6.1 | 3.6 | 3.3 | 3.0 | 7.7 | 4.2 | 3.6 | 4.2 | 4.0 | 4.1 | ||||||
| intensity | 28 | 10 | 18 | 14 | 12 | 39 | 14 | 47 | 55 | 17 | ||||||
| 1H cs | 8.0 | 7.4 | 4.4 | 4.1 | 5.1 | 3.8 | 6.1 | 4.6 | 4.3 | |||||||
| intensity | 22 | 30 | 120 | 20 | 20 | 23 | 10 | 20 | 22 | |||||||
| 1H cs | 4.4 | 4.3 | 7.3 | 4.6 | 6.6 | 4.7 | ||||||||||
| intensity | 120 | 12 | 22 | 35 | 43 | 25 | ||||||||||
| 1H cs | 4.8 | 6.3 | 8.1 | 7.0 | 7.0 | 7.7 | ||||||||||
| intensity | 30 | 12 | 26 | 19 | 17 | 18 | ||||||||||
| 1H cs | 7.1 | 7.8 | 9.4 | 8.4 | 7.4 | |||||||||||
| intensity | 19 | 11 | 40 | 24 | 44 | |||||||||||
| 1H cs | 8.3 | 9.5 | 8.9 | |||||||||||||
| intensity | 16 | 12 | 32 | |||||||||||||
NOE vectors used were a sum of gaussian peaks of width 0.4 ppm placed at chemical shifts and having intensities taken from the vectors emanating from crosspeaks in NOESY-HSQC spectra. Diagonal peaks and water peaks were removed and a peak of intensity equal to the average of the maximum peak in each vector was added to the end of experimental and predicted vectors to retain intensity sensitivity in R-factor calculations. Only points above 2 x noise = 5 are listed. Error was estimated comparing peak 10 to a vector having only a diagonal peak: (1-R) = 0.14. The NOE weight was 0.5.
Highlights:
A software package for NMR resonance assignment in sparsely-labeled proteins is described.
NMR resonances in a sparsely-labeled, mammalian-cell-expressed, protein are assigned.
Paramagnetic effects on assigned resonances facilitate docking of an enzyme substrate.
Funding
This work was supported by grants from the US National Institutes of Health, P41GM103390 and R01GM033225. Manuscript content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Moremen KW, Ramiah A, Stuart M, Steel J, Meng L, Forouhar F, et al. Expression system for structural and functional studies of human glycosylation enzymes. Nature Chemical Biology. 2018;14:156-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Prestegard JH, Agard DA, Moremen KW, Lavery LA, Morris LC, Pederson K. Sparse labeling of proteins: Structural characterization from long range constraints. Journal of Magnetic Resonance. 2014;241:32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chen K, Tjandra N. The Use of Residual Dipolar Coupling in Studying Proteins by NMR In: Zhu G, editor. Nmr of Proteins and Small Biomolecules2012. p. 47–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Prestegard JH, Bougault CM, Kishore AI. Residual dipolar couplings in structure determination of biomolecules. Chemical Reviews. 2004;104:3519–40. [DOI] [PubMed] [Google Scholar]
- [5].Nitsche C, Otting G. Pseudocontact shifts in biomolecular NMR using paramagnetic metal tags. Progress in Nuclear Magnetic Resonance Spectroscopy. 2017;98–99:20–49. [DOI] [PubMed] [Google Scholar]
- [6].Gobl C, Madl T, Simon B, Sattler M. NMR approaches for structural analysis of multidomain proteins and complexes in solution. Progress in Nuclear Magnetic Resonance Spectroscopy. 2014;80:26–63. [DOI] [PubMed] [Google Scholar]
- [7].Becker W, Bhattiprolu KC, Gubensak N, Zangger K. Investigating Protein-Ligand Interactions by Solution Nuclear Magnetic Resonance Spectroscopy. Chemphyschem. 2018;19:895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Frueh DP. Practical aspects of NMR signal assignment in larger and challenging proteins. Progress in Nuclear Magnetic Resonance Spectroscopy. 2014;78:47–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gao Q, Chalmers GR, Moremen KW, Prestegard JH. NMR assignments of sparsely labeled proteins using a genetic algorithm. Journal of Biomolecular Nmr. 2017;67:283–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pederson K, Chalmers GR, Gao Q, Elnatan D, Ramelot TA, Ma LC, et al. NMR characterization of HtpG, the E-coli Hsp90, using sparse labeling with C-13-methyl alanine. Journal of Biomolecular Nmr. 2017;68:225–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Varki A Biological roles of glycans. Glycobiology. 2017;27:3–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Schneider EK, Li J, Velkov T. A Portrait of the Sialyl Glycan Receptor Specificity of the H10 Influenza Virus Hemagglutinin-A Picture of an Avian Virus on the Verge of Becoming a Pandemic? Vaccines. 2017;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ji Y, White YJB, Hadden JA, Grant OC, Woods RJ. New insights into influenza A specificity: an evolution of paradigms. Current Opinion in Structural Biology. 2017;44:219–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Christiansen MN, Chik J, Lee L, Anugraham M, Abrahams JL, Packer NH. Cell surface protein glycosylation in cancer. Proteomics. 2014;14:525–46. [DOI] [PubMed] [Google Scholar]
- [15].Bhide GP, Colley KJ. Sialylation of N-glycans: mechanism, cellular compartmentalization and function. Histochemistry and Cell Biology. 2017;147:149–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Liu S, Venot A, Meng L, Tian F, Moremen KW, Boons GJ, et al. Spin-labeled analogs of CMP- NeuAc as NMR probes of the alpha-2,6-sialyltransferase ST6Gal I. Chemistry & Biology. 2007;14:409–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kuhn B, Benz J, Greif M, Engel AM, Sobek H, Rudolph MG. The structure of human alpha-2,6- sialyltransferase reveals the binding mode of complex glycans. Acta Crystallographica Section D- Biological Crystallography. 2013;69:1826–38. [DOI] [PubMed] [Google Scholar]
- [18].Meng L, Forouhar F, Thieker D, Gao ZW, Ramiah A, Moniz H, et al. Enzymatic Basis for N-Glycan Sialylation STRUCTURE OF RAT alpha 2,6-SIALYLTRANSFERASE (ST6GAL1) REVEALS CONSERVED AND UNIQUE FEATURES FOR GLYCAN SIALYLATION. Journal of Biological Chemistry. 2013;288:34680–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li DW, Bruschweiler R. PPM: a side-chain and backbone chemical shift predictor for the assessment of protein conformational ensembles. Journal of Biomolecular Nmr. 2012;54:257–65. [DOI] [PubMed] [Google Scholar]
- [20].Han B, Liu YF, Ginzinger SW, Wishart DS. SHIFTX2: significantly improved protein chemical shift prediction. Journal of Biomolecular Nmr. 2011;50:43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chalmers G, Glushka JN, Foley BL, Woods RJ, Prestegard JH. Direct NOE simulation from long MD trajectories. Journal of Magnetic Resonance. 2016;265:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sheikh MO, Thieker D, Chalmers G, Schafer CM, Ishihara M, Azadi P, et al. O-2 sensing-associated glycosylation exposes the F-box-combining site of the Dictyostelium Skp1 subunit in E3 ubiquitin ligases. Journal of Biological Chemistry. 2017;292:18897–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Valafar H, Prestegard JH. REDCAT: a residual dipolar coupling analysis tool. Journal of Magnetic Resonance. 2004;167:228–41. [DOI] [PubMed] [Google Scholar]
- [24].Gu Y, Li DW, Bruschweiler R. NMR Order Parameter Determination from Long Molecular Dynamics Trajectories for Objective Comparison with Experiment. Journal of Chemical Theory and Computation. 2014;10:2599–607. [DOI] [PubMed] [Google Scholar]
- [25].Case VB DA, Berryman JT, Betz RM, Cai Q, Cerutti DS, Cheatham TE III, Darden TA, Duke HG RE, Goetz AW, Gusarov S, Homeyer N, Janowski P, Kaus J, Kolossváry I, Kovalenko A,, Lee SL TS Luchko T, Luo R, Madej B, Merz KM, Paesani F, Roe DR, Roitberg A, Sagui C,, Salomon-Ferrer GS R, Simmerling CL, Smith W, Swails J, Walker RC, Wang J, Wolf RM, Kollman X WaPA. AMBER 14. In: University of California SF, editor.2014. [Google Scholar]
- [26].Li DW, Bruschweiler R. PPM_One: a static protein structure based chemical shift predictor. Journal of Biomolecular Nmr. 2015;62:403–9. [DOI] [PubMed] [Google Scholar]
- [27].Gu YN, Li DW, Bruschweiler R. Decoding the Mobility and Time Scales of Protein Loops. Journal of Chemical Theory and Computation. 2015;11:1308–14. [DOI] [PubMed] [Google Scholar]
- [28].Moseley HNB, Curto EV, Krishna NR. COMPLETE RELAXATION AND CONFORMATIONAL EXCHANGE MATRIX (CORCEMA) ANALYSIS OF NOESY SPECTRA OF INTERACTING SYSTEMS - 2-DIMENSIONAL TRANSFERRED NOESY. Journal of Magnetic Resonance Series B. 1995;108:243–61. [DOI] [PubMed] [Google Scholar]
- [29].Zeng J, Tripathy C, Zhou P, Donald BR. A HAUSDORFF-BASED NOE ASSIGNMENT ALGORITHM USING PROTEIN BACKBONE DETERMINED FROM RESIDUAL DIPOLAR COUPLINGS AND ROTAMER PATTERNS. Computational Systems Bioinformatics. p. 169–81. [PubMed] [Google Scholar]
- [30].Liwo A, Czaplewski C, Oldziej S, Scheraga HA. Computational techniques for efficient conformational sampling of proteins. Current Opinion in Structural Biology. 2008;18:134–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bouvignies G, Markwick PRL, Blackledge M. Characterization of protein dynamics fromresidual dipolar couplings using the three dimensional Gaussian axial fluctuation model. Proteins-Structure Function and Bioinformatics. 2008;71:353–63. [DOI] [PubMed] [Google Scholar]
- [32].Chen PC, Hologne M, Walker O, Hennig J. Ab lnitio Prediction of NMR Spin Relaxation Parameters from Molecular Dynamics Simulations. Journal of Chemical Theory and Computation. 2018;14:1009–19. [DOI] [PubMed] [Google Scholar]
- [33].Bernardi RC, Melo MCR, Schulten K. Enhanced sampling techniques in molecular dynamics simulations of biological systems. Biochimica Et Biophysica Acta-General Subjects. 2015;1850:872–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gao Q, Chen CY, Zong C, Wang S, Ramiah A, Prabhakar P, et al. Structural Aspects of Heparan Sulfate Binding to Robo1-Ig1–2. Acs Chemical Biology. 2016;11:3106–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Goto NK, Gardner KH, Mueller GA, Willis RC, Kay LE. A robust and cost-effective method for the production of Val, Leu, Ile (delta 1) methyl-protonated N-15-, C-13-, H-2-labeled proteins. Journal of Biomolecular Nmr. 1999;13:369–74. [DOI] [PubMed] [Google Scholar]
- [36].Kim J, Wang YJ, Li G, Veglia G. A Semiautomated Assignment Protocol for Methyl Group Side Chains in Large Proteins In: Kelman Z, editor. Isotope Labeling of Biomolecules - Applications2016. p. 35–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Schmidt E, Guntert P. A New Algorithm for Reliable and General NMR Resonance Assignment. Journal of the American Chemical Society. 2012;134:12817–29. [DOI] [PubMed] [Google Scholar]
- [38].Monneau YR, Rossi P, Bhaumik A, Huang CD, Jiang YJ, Saleh T, et al. Automatic methyl assignment in large proteins by the MAGIC algorithm. Journal of Biomolecular Nmr. 2017;69:215–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Courtney JM, Ye Q, Nesbitt AE, Tang M, Tuttle MD, Watt ED, et al. Experimental Protein Structure Verification by Scoring with a Single, Unassigned NMR Spectrum. Structure. 2015;23:1958–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gao Q, Chen CY, Zong C, Wang S, Ramiah A, Prabhakar P, et al. Structural Aspects of Heparan Sulfate Binding to Robo1-Ig1–2. ACS Chem Biol. 2016;11:3106–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Meng L, Forouhar F, Thieker D, Gao Z, Ramiah A, Moniz H, et al. Enzymatic basis for N-glycan sialylation: structure of rat alpha2,6-sialyltransferase (ST6GAL1) reveals conserved and unique features for glycan sialylation. J Biol Chem. 2013;288:34680–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kirschner KN, Yongye AB, Tschampel SM, Gonzalez-Outeirino J, Daniels CR, Foley BL, et al. GLYCAM06: A generalizable Biomolecular force field. Carbohydrates. Journal of Computational Chemistry. 2008;29:622–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Roe DR, Cheatham TE. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. Journal of Chemical Theory and Computation. 2013;9:3084–95. [DOI] [PubMed] [Google Scholar]
- [44].Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF chimera - A visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25:1605–12. [DOI] [PubMed] [Google Scholar]