Abstract
Ribosomal RNA (rRNA) biogenesis is a multistep process requiring several nuclear and cytoplasmic exonucleases. The exact processing steps for mammalian 5.8S rRNA remain obscure. Here, using loss-of-function approaches in mouse embryonic stem cells and deep sequencing of rRNA intermediates, we investigate at nucleotide resolution the requirements of exonucleases known to be involved in 5.8S maturation, and explore the role of the Perlman syndrome-associated 3’-5’ exonuclease Dis3l2 in rRNA processing. We uncover a novel cytoplasmic intermediate that we name ‘7SB’ rRNA that is generated through sequential processing by distinct exosome complexes. 7SB rRNA can be oligoadenylated by an unknown enzyme and/or oligouridylated by TUT4/7 and subsequently processed by Dis3l2 and Eri1. Moreover, exosome depletion triggers Dis3l2-mediated decay (DMD) as a surveillance pathway for rRNAs. Our data identify previously unknown 5.8S rRNA processing steps and provide nucleotide level insight into the exonuclease requirements for mammalian rRNA processing.
Keywords: Dis3l2, Ribosomal RNA, Exosome, TUTase, Uridylation, Eri1, Exosc3, Dis3, Exosc10
INTRODUCTION
Eukaryotic ribosomal RNA (rRNA) biogenesis is a complicated process that begins in the nucleolus. Three of the four rRNAs that make up the RNA constituents of the ribosome, 18S, 5.8S and 28S rRNAs, are transcribed by RNA Polymerase I (RNA Pol I) as a long polycistronic precursor, whereas 5S rRNA is transcribed by RNA Pol III1. A complex sequence of processing events progressively releases mature 18S, 5.8S, and 28S rRNAs from their primary transcript2–4. Specifically, the formation of 5.8S rRNA requires the combined action of both endo- and exoribonucleases. Although 5.8S rRNA processing has been extensively studied in budding yeast5 and mammals6, a precise molecular characterization of each rRNA precursor is still lacking.
The exosome complex is the major 3’-5’ ribonuclease acting in processing, degradation, and surveillance of various RNA species in eukaryotic cells7. The core exosome complex is comprised of multiple subunits including a 9-subunit catalytically inert ring8,9 with an associated Rrp44 (DIS3) subunit with both endo- and 3’-5’ exo-ribonucleolytic activity10, as well as another 3’-5’ exonuclease, Rrp6 (EXOSC10)11. The exosome complex was originally found to degrade pre-5.8S intermediates in yeast12,13 and subsequently many studies have pointed to its pivotal role in rRNA biogenesis in both yeast and mammals6,14.
Mammalian genomes encode three homologues of yeast Rrp44: Dis3, Dis3l1, and Dis3l2. Among those, Dis3 is considered the mammalian ortholog of yeast Rrp44. Dis3l1 is cytoplasmic and can also associate with the core exosome subunits, and while its molecular function is not well understood, it has been implicated in the degradation of poly(A)-tailed 28S rRNA intermediates15,16. Dis3l2 is a 3’-5’ exoribonuclease specifically localized in the cytoplasm that does not associate with the exosome complex17–19. Dis3l2 displays a preferential activity towards RNA species possessing a non-templated oligo-uridine 3’-end tail, which serves as a degradation signal. Dis3l2 was first identified as the effector of Lin28-dependent degradation of uridylated pre-let-7 microRNAs17,20–22. Mutations in human DIS3L2 have been linked to Perlman overgrowth syndrome and hyper susceptibility to Wilms tumors23. Recently, we performed a global identification of Dis3l2-bound RNA species in mESCs by RNA immunoprecipitation and sequencing (RIP-Seq) and found that the majority of Dis3l2 direct targets are noncoding RNAs (ncRNAs)24,25. This finding, as well as similar works from others established Dis3l2-Mediated Decay (DMD) pathway as a surveillance pathway for a wide variety of ncRNAs26–30. Following the initial observation that Dis3l2 binds to 5S rRNA24, and also given that Rrp44 (Dis3), the exosome-associated homolog of Dis3l2, participates in pre-rRNA processing10,15,31, we sought to determine a functional interplay between Dis3l2 and other known RNA exonucleases in rRNA processing.
RESULTS
Systematic analysis of rRNA uridylation in Dis3l2-depleted cells
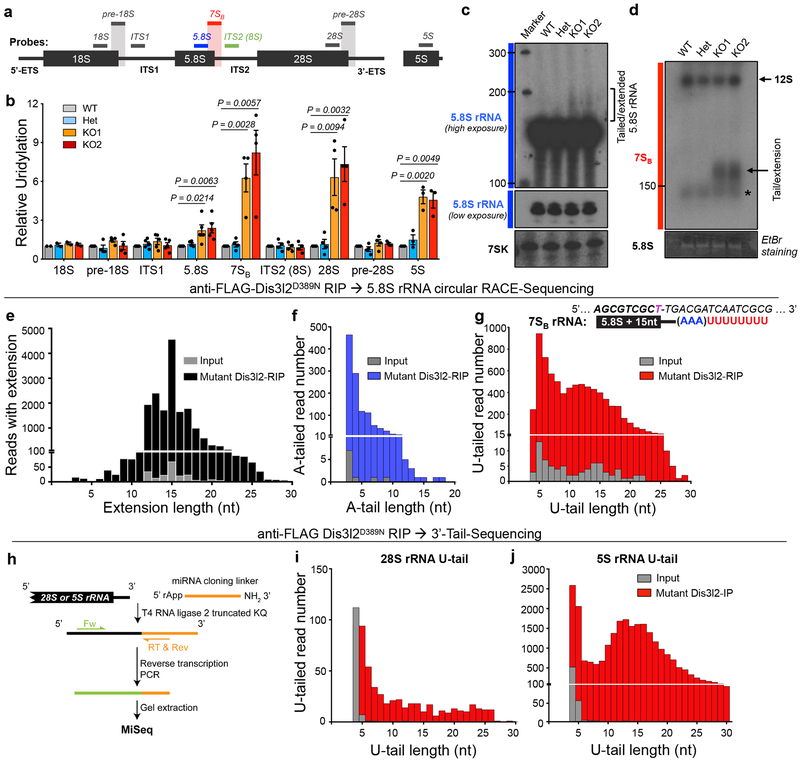
We previously performed a Dis3l2 RNA immunoprecipitation and high-throughput sequencing (RIP-Seq) to globally identify Dis3l2 target RNAs in mESCs24. Although this unbiased approach should have identified all RNA substrates of Dis3l2, we considered that certain RNAs, in particular rRNAs that are expressed as polycistronic transcripts and are present at numerous genomic loci in the mouse genome, might have been missed by our bioinformatics pipeline and alignment of the Dis3l2-bound transcript to the genome. Considering also our previous identification of 5S rRNA as a Dis3l2-bound RNA we set out to determine whether other rRNAs are also Dis3l2 targets. Since Dis3l2 preferentially targets U-tailed RNAs, we first measured the relative uridylation levels of the four rRNAs and their 3’-end elongated precursors in Dis3l2 knockout mESCs (Fig. 1a–d). We observed significant increases in relative uridylation of 5.8S, 28S, and 5S rRNAs, but not of 18S rRNA, in Dis3l2 knockout mESCs (Fig. 1b, Supplementary Fig. 1a). To specifically detect rRNA precursors, we used reverse primers and northern blotting probes that encompass the junctions between mature rRNAs and their 3’-end adjacent transcribed spacers (Fig. 1a). These primers detected precursor(s) of 18S rRNA (pre-18S), 5.8S rRNA (we called this 7SB for the reasons described later), 28S rRNA (pre-28S) as well as extended 18S (ITS1) and 5.8S (ITS2, i.e. 8S) rRNAs. Interestingly, among the precursors of the three potential Dis3l2 targets (5S, 5.8S, and 28S rRNAs), the only precursor showing marked increase in its relative uridylation was the short 5.8S rRNA precursor (7SB), but not the longer ITS2 (8S). The relative level of 7SB uridylation was even greater than 5.8S uridylation, suggesting specificity to 3’-elongated 5.8S rRNA (Fig. 1b). We focused further on this elongated 5.8S rRNA (7SB) and aimed to validate the presence of uridylated 5.8S rRNA in Dis3l2-depleted cells. Slow-migrating 5.8S rRNA species were observed in northern blot analysis of Dis3l2 knockout mESCs, but not in heterozygous or WT cells (Fig. 1c). More strikingly, probing with 7SB specific oligos reduced the background of mature 5.8S and showed even stronger signals in knockout samples that were undetected in controls (Fig. 1d). The size of observed band was greater than 150 nt and points to the 5.8S rRNA species with tails and/or genomic extensions. Combined with the increased uridylation levels detected by qRT-PCR, this signal most likely represents uridylated precursor(s) of 5.8S rRNA. Similar 7SB RNA species were found in stable knockdown Dis3l2 mESCs (Supplementary Fig. 1b,c). Thus, uridylated 5S, 5.8S, 7SB, and 28S rRNAs accumulate in Dis3l2-depleted mESCs.
Fig. 1. Analysis of rRNA uridylation in Dis3l2 depleted mESCs.
(a) Probing scheme for northern blot and qRT-PCR analyses in this study. Bars represent the reverse DNA oligos used as northern blot probes or qRT-PCR reverse primers. Internal probe to detect 5.8S rRNA (blue bar), and probe used to detect 3’-extended 5.8S species (7S) throughout the entire study encompassing 12 nt at 3’-end of 5.8S rRNA and 11 nt at 5’ end of ITS2 (red bar), as well as a probe used to detect 5.8S rRNA with longer extension (8S rRNA, green bar) are represented. Similar probes encompassing mature rRNA-adjacent spacer junctions were used to detect 3’-extended 18S and 28S (grey bars). (b) qRT-PCR analysis of relative uridylation for various rRNAs and their elongated species as depicted in Fig. 1a. Oligo-dA oligo was used to synthesize the cDNAs. Bars represent mean ± SEM. Individual P values are presented; two-tailed Student’s t-test (n=2 for 18S; n=3 for 5S; n=4 for pre-18S, 7SB, 8S, 28S and pre-28S; n=5 for ITS1, and 5.8S, representing biologically independent cell cultures). (c) Northern blotting of Dis3l2 control and knockout RNA samples with mature 5.8S rRNA and 7SK (as control) probes. (d) Northern blot analysis with 7S probe. Arrow points to the 7SB rRNA and the asterisk marks mature 5.8S rRNA (see main text). (e) Distribution of 5.8S rRNA genomic extension in FLAG-mutant Dis3l2 RIP (black bars) and input (grey bars). (f) Distribution of A-tail length in extended 5.8S rRNAs in RIP (blue bars) and input (grey bars). (g) Distribution of U-tail length in extended 5.8S rRNAs in RIP (red bars) and input (grey bars). The sequence of “7SB” rRNA with 15 nt extension in ITS2 after the canonical end of mature 5.8S rRNA is shown. (h) Schematic representation of 3’-ligation RACE procedure. 3’-end uridylation of 28S rRNA (i) and 5S rRNA (j) in input and FLAG-mutant Dis3l2-bound protein re-expressed in Dis3l2 knockout mESCs. No extension or adenylation was observed prior to the U-tail (data not shown). WT, wild type; Het, heterozygote; KO, knockout. Panels c and d are representative images from 2 and 3 repeats, respectively. Uncropped blot/gel images are shown in Supplementary Data Set 1. Source Date for panel b are available online.
To address whether Dis3l2 directly targets uridylated rRNAs, we asked if it physically associates with these rRNAs. Dis3l2 knockout mESCs were transfected with FLAG-tagged WT or catalytically mutant (D389N) Dis3l2 vectors. Dis3l2 ribonucleoproteins (RNPs) were UV-crosslinked and immunoprecipitated with anti-FLAG antibody and RNAs were isolated. qRT-PCR analysis showed a strong enrichment of uridylated 5.8S, 7SB, and 5S rRNAs in mutant FLAG-Dis3l2 RIP samples, but not in mock- or WT Dis3l2-RIPs (Supplementary Fig. 1d). Northern blot analysis with 7SB probe confirmed the specific binding of 7SB rRNA to mutant Dis3l2 (Supplementary Fig. 1e). Finally, Dis3l2 re-expression in knockout cells significantly reduced uridylated 7SB rRNA levels (Supplementary Fig. 1f,g). Therefore, Dis3l2 binds and processes uridylated pre-rRNA species.
3’-end sequencing identifies 7SB rRNA and reveals extensive modification of rRNAs
We next used rapid amplification of cDNA ends from circularized RNAs (cRACE)25 to precisely characterize the 3’-end of the 7SB rRNA in Dis3l2-depleted cells. To enrich for 7SB species over abundant mature 5.8S rRNA, a primer encompassing the 5.8S-ITS2 junction, similar to that used for detection of 7SB by northern blot (Fig. 1d), was used for the cDNA synthesis step (Supplementary Fig. 2a). By analyzing 7SB in input and especially in RIP samples, we observed many reads containing various genomic extensions at their 3’-ends corresponding to the genomic sequence in ITS2. The peak extension length was at 15 nt after the canonical 3’-end of mature 5.8S rRNA. The same trend was observed in input sample, although with lower frequency (Fig. 1e), highlighting the enrichment of the extended 5.8S rRNAs by Dis3l2. We noticed that extended reads occasionally contained a stretch of A-rich tails upstream of U-tails as well. To analyze the length of A-tailed reads, a stretch of three adenosines or more (≥AAA) was considered as an A-tail. Interestingly, A-tails mostly comprised stretches of 3–6 As, but longer A-tails were also detected at a lower frequency (Fig. 1f). To measure the number and the length of the U-tails, we considered only a stretch of 4 or more continuous uridines (≥UUUU) as a U-tail. Analyzing U-tail length showed a bimodal distribution with two peaks, one of ~5 Us and another of ~12–15 Us, especially in the RIP sample (Fig. 1g), similar to what was found previously at the 3’-end of Dis3l2-targeted Rmrp24. Notably, no other mixed tailings comprised of non-templated Gs or Cs32 were observed (data not shown). To gain insight to overall distribution of A-, U-, or both A- and U-tailed reads, the frequency of each category in extended reads in RIP sample was measured. Most of the reads contained only U-tails (70%) or mixed tails (A- followed by U-tails; 26%) after the 3’-end genomic extensions, whereas only a small fraction of extended reads (4%) contained A-tails, but no U-tail (Supplementary Fig. 2b). Noteworthy, the order of the tails occurrence was exclusively as 5.8S/ITS2 genomic extension/A-tail/U-tail (p value = 2.2*10−16; Wilcoxon t-test). The length of extension did not depend on the presence or absence of A- or U-tails. Moreover, the presence or absence of A-tail did not change the length of U-tails in extended reads (Supplementary Fig. 2c). Altogether, in Dis3l2 knockout mESCs, we detected accumulation of 5.8S rRNA species that is genomically extended 15 nt downstream into ITS2 (now known as 7SB), and is mostly oligouridylated with occasional oligoadenylation that typically precedes the Us. The size of extended and tailed 5.8S rRNA is on average between 170–200 nt that corresponds to the distinct 7SB band that we identified by northern blot analysis (Fig. 1d). The last known step of mammalian 5.8S rRNA maturation happens in the cytoplasm, where Eri1 removes the last 1–2 nt from the 3’-end of 6S rRNA33, whereas the last nuclear 5.8S rRNA precursor so far proposed in human is 5.8S with ~40 nt ITS2 extension, trimmed by the EXOSC10 exosome subunit14. However, no such intermediate has so far been described in mice. Based on its migration pattern seen by northern blot as well as the MiSeq data, the 7SB rRNA (5.8S+ 15 nt) that we found to be uridylated and accumulates in Dis3l2-depleted cells likely represents a novel intermediate that occurs between these two reported steps (i.e. upstream of cytoplasmic 6S rRNA). We therefore call these rRNA species “7SB rRNA”. Noteworthy, Dis3l2 loss did not affect steady state levels of 5.8S rRNA as measured by northern blotting (Fig. 1c, Fig. 2a,f, and Supplementary Fig. 1b), and qRT-PCR (Supplementary Fig. 2d). Depletion of human DIS3L2 protein in two different human cell lines similarly resulted in elevated levels of human 7SB rRNAs indicating that this pathway is conserved (Supplementary Fig. 2e).
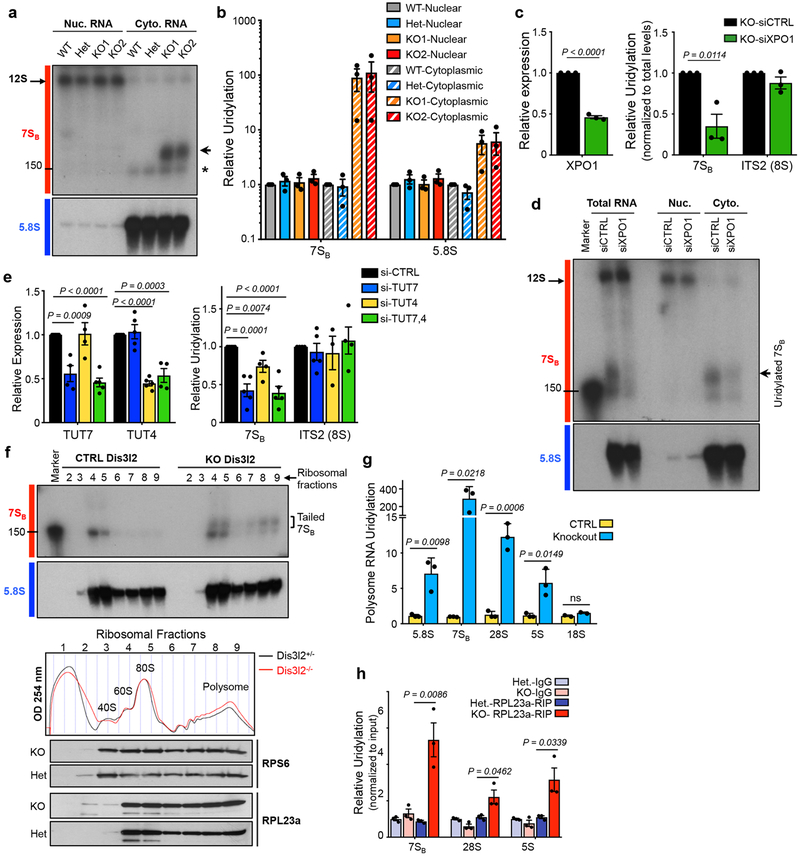
Fig. 2. 7SB rRNA export and cytoplasmic uridylation.
(a) Northern blot analysis of nuclear (Nuc.) and cytoplasmic (Cyto.) fractions with indicated probes. Arrow points to uridylated 7SB and asterisk marks 5.8S rRNA. (b) qRT-PCR analysis of relative uridylation of 7SB and 5.8S rRNAs in the cytoplasmic fraction of knockout mESCs as depicted in Fig. 1a. Bars represent mean ± SEM. Individual P values are presented; two-tailed Student’s t-test (n=3 biologically independent cell cultures). (c) qRT-PCR analysis of XPO1 expression (left panel) and relative uridylation levels of 7SB and ITS2 (8S) (right panel). Bars represent mean ± SEM. Individual P values are presented; two-tailed Student’s t-test (n=3 biologically independent cell cultures). (d) Northern blot analysis with 7SB and 5.8S rRNA probes in indicated fractions from Dis3l2 knockout cells. (e) Left panel: TUT7 and TUT4 levels measured by qRT-PCR in Dis3l2 knockout cells expressing individual siRNAs against TUT4 or TUT7, or a combination of both. Right panel: Relative uridylation of 7SB and ITS2 (8S) rRNAs measured by qRT-PCR after TUT4 and TUT7 knockdown. Bars represent mean ± SEM. Individual P values are presented; two-tailed Student’s t-test (n=4 biologically independent cell cultures). (f) Upper panels: Northern blot analysis of 7SB and 5.8S rRNAs in ribosomal fractions from lysates resolved through 10–50% sucrose gradients. Tailed 7SB in monosome and also in polysome fractions of knockout mESCs are marked by square bracket. Middle panel: Absorbance profiles at 254 nm (OD 254) recorded during fractionation through sucrose gradients. Ribosomal fractions: approximate positions of collected fractions. Lower panels: WB analysis of large (RPL23a) and small (RPS6) subunits protein components of ribosomes in corresponding ribosome fractions. (g) qRT-PCR analysis of uridylated rRNAs in the polysome fraction of Dis3l2 knockout cells. Bars represent mean ± SEM. Individual P values are presented; two-tailed Student’s t-test (n=3 biologically independent cell cultures). (h) Enrichment of uridylated rRNAs in RPL23a-immunoprecipitated samples in Dis3l2 knockout samples. Bars represent mean ± SEM. Individual P values are presented; two-tailed Student’s t-test (n=3 biologically independent cell cultures). To measure the uridylation levels, values were first normalized to Gapdh and then to the total levels of indicated transcript. All the other qRT-PCRs were normalized to Gapdh. WT, ; Het, heterozygote; KO, knockout. Panels a, d, and f are representative images from at least 2 repeats. Uncropped blot/gel images are shown in Supplementary Data Set 1. Source data for panels b, c, e-h are available online.
For the analysis of the 3’-ends of 5S and 28S rRNAs we used a 3’-ligation RACE and deep sequencing method25 since we could not obtain circularized RNAs for them (Fig. 1h). MiSeq analysis revealed 3’-end uridylation of Dis3l2-bound 28S rRNA (Fig. 1i), and 5S rRNA (Fig. 1j). Of note, no genomic extension or oligoadenylation was observed in input or Dis3l2-bound RIP species, which is consistent with qRT-PCR analysis using probes against extended transcripts (Fig. 1b). In conclusion, our deep sequencing analysis of the 3’-ends of rRNAs provides evidence that mature 5S and 28S can be oligouridylated and bound by Dis3l2, and furthermore we identify a putative new 5.8S rRNA intermediate (7SB rRNA) with oligo-A and/or oligo-U tails.
Pre-rRNA uridylation occurs in the cell cytoplasm and is catalyzed by TUT4/7
Dis3l2 is mainly localized in the cytoplasm17,18,20. To determine the subcellular localization of 7SB and Dis3l2 function during pre-rRNA processing, we analyzed the distribution of 7SB rRNA in nuclear and cytoplasmic RNA fractions. Northern blot analysis revealed that uridylated 7SB rRNAs accumulate exclusively in the cytoplasmic fractions of the knockout cells (Fig. 2a), while an earlier nuclear intermediate (12S pre-rRNAs) is unaffected by Dis3l2 deficiency. qRT-PCR analysis also confirmed the accumulation of uridylated 7SB and 5.8S in the cytoplasm of Dis3l2 knockout mESCs (Fig. 2b). Previously, we implicated two Terminal Uridylyl Transferase (TUTase) enzymes, TUT7 (Zcchc6) and TUT4 (Zcchc11), in the 3’-end terminal uridylation of Dis3l2 targets24,25,34–37. We next asked if these two TUTases are also involved in the uridylation of 7SB rRNA. In Dis3l2 knockout cells, downregulation of TUT7 and TUT4, individually and in combination, led to significant reduction of 7SB rRNA uridylation (Fig. 2e). Since XPO1 is involved in the nuclear export of pre-60S ribosomal particles containing pre-rRNAs, we tested whether XPO1 might be involved in the nuclear export of 7SB rRNA (likely as a part of pre-60S particles) as well. Relative uridylation of 7SB rRNA was significantly decreased in XPO1-depleted Dis3l2 knockout cells (Fig. 2c). Furthermore, northern blot analysis of RNA samples revealed decreased levels of uridylated 7SB in total and cytoplasmic samples due to XPO1 depletion (Fig. 2d). Therefore, 7SB rRNA uridylation requires XPO1-mediated export to the cell cytoplasm and is catalyzed by TUT4/7.
Uridylated rRNAs are incorporated into ribosomes in the absence of Dis3l2
A series of biochemical assays were performed to assess the physical association of uridylated rRNAs to ribosomes. First, northern blotting analysis of sucrose gradient fractionated lysates showed 7SB rRNA association with monosomes and translating polysomes38 from Dis3l2 knockout cells (Fig. 2f). Notably, Dis3l2 depletion did not cause an imbalanced 40S:60S ratio, which indicates that loss of rRNA processing did not prevent 60S subunit maturation (Fig. 2f, middle panel). Second, polysome fractions of Dis3l2-depleted cells also contained high levels of uridylated 5.8S, 7SB, 5S and 28S rRNAs (Fig. 2g). Third, immunoprecipitation of ribosome machinery in Dis3l2 knockout cells using a specific antibody against the large ribosomal subunit protein component RPL23a significantly enriched for uridylated rRNAs, verifying direct association of these rRNAs with ribosomes (Fig. 2h). The physiological consequence of Dis3l2 loss on protein synthesis will be discussed elsewhere. Nevertheless, these results established that in the absence of Dis3l2, at least a subset of ribosomes contain uridylated rRNAs with aberrant 3’-ends, with no gross defects in their overall biogenesis.
Dis3l2 functions downstream of the exosome in the rRNA processing pathway
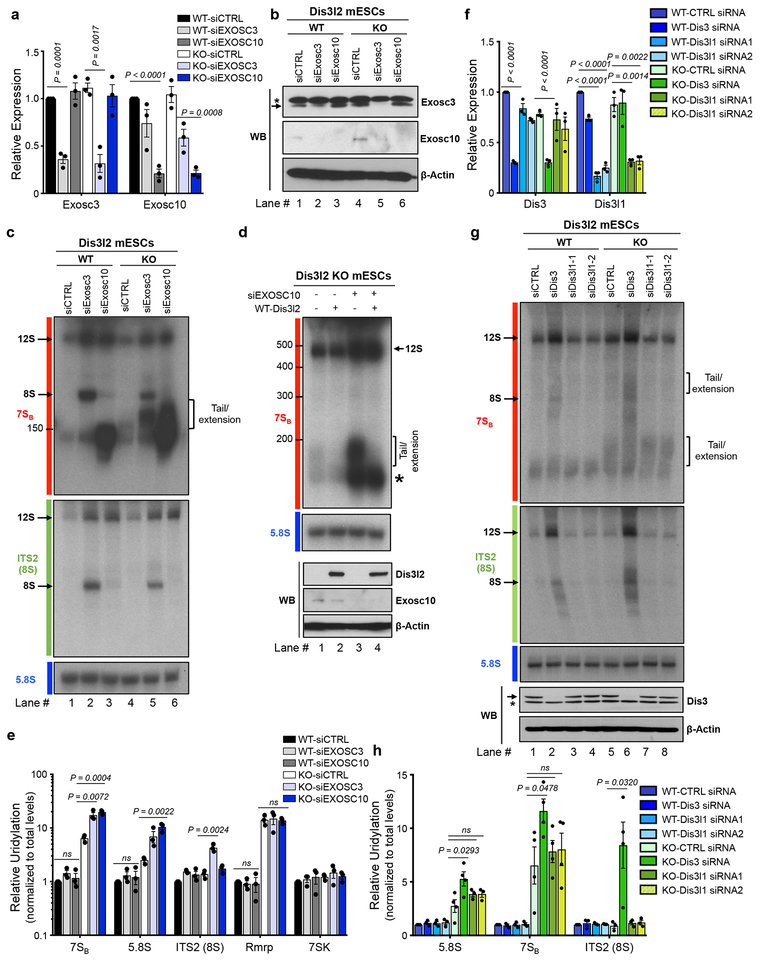
We next examined the role of individual exonucleases known to be involved in rRNA processing and their functional relationship with Dis3l2 in the context of 7SB rRNA processing. WT and Dis3l2 knockout ESCs were transfected with control (as mock), Exosc3- or Exosc10-targeting siRNAs (Fig. 3a,b). Northern blots revealed the accumulation of ~250 nt precursor in the Exosc3- but not the Exosc10-depleted WT cells that likely corresponds to known murine 8S pre-rRNA (Fig. 3c, upper panel, lane 2). The accumulation of 8S rRNA exclusively in siExosc3 knockdown samples was further confirmed by ITS2-specific probe (Fig. 3c, middle panel) located downstream of 7SB probe (Fig. 1a). Exosc10 knockdown had no effect on 8S rRNA processing but instead led to a strong accumulation of a band above 150 nt (Fig. 3c, upper panel, lane 3). This stabilization upon Exosc10 depletion resembles yeast 5.8S+30 nt and human 5.8S+ ~40 nt species, both detected after inhibiting Rrp6 or EXOSC10, respectively6,12,39. However, based on its migration pattern, the apparent length of its 3’-end extension is likely shorter than the yeast and human counterparts. Both Exosc3 and Exosc10 knockdowns increased the steady-state levels of a larger 5.8S precursor, likely corresponding to an upstream 12S intermediate. Dis3l2 deficiency had no effect on 12S or 8S rRNA accumulation upon Exosc3 or Exosc10 knockdown as revealed by northern blot analysis of Dis3l2 knockout cells treated with the same siRNAs (Fig. 3c, lanes 4–6). As confirmed by ITS2 probe (Fig. 3c) accumulation of 8S rRNA upon Exosc3 knockdown and 12S rRNA in both Exosc3 and Exosc10 knockdown were unaffected by Dis3l2 knockout. Notably, a slow migrating species [ranging up to 200 nt] was found to accumulate with the combined deficiency of Exosc10 and Dis3l2 (Fig. 3c, upper panel, lane 6). This band might contain both uridylated 7SB as well as a similarly sized putative Exosc10 substrate rRNA intermediate. These elongated species were also detected upon Exosc3 knockdown in Dis3l2 knockout ESCs. Indeed, Dis3l2 re-expression in the Exosc10-Dis3l2 (Fig. 3d, Supplementary Fig. 3a) and Exosc3-Dis3l2 co-depleted cells (Supplementary Fig. 3a) resulted in the specific loss of uridylated species. To further test whether the observed accumulation of elongated rRNA intermediates might represent U-tailed pre-rRNAs that escape Dis3l2-mediated processing we performed qRT-PCR and the results were normalized first to Gapdh, and then to total level of each transcript (Fig. 3e). In Dis3l2 WT cells, Exosc3 and Exosc10 knockdown had no effect on 7SB uridylation. However, in Dis3l2 knockout cells, depletion of either Exosc3 or Exosc10 resulted in an increased amount of uridylated 7SB compared to the Dis3l2 knockout cells transfected with control siRNA (Fig. 3e). The uridylation level of lncRNA Rmrp, a DMD target that we identified previously24, was unchanged upon exosome depletion, suggesting this functional relationship between Dis3l2 and the exosome complex is specific to a subset of Dis3l2 substrates including rRNAs (Fig. 3e). Altogether, our results suggest that in addition to its roles in 7SB rRNA processing, Dis3l2 might also be involved in the processing of rRNA biogenesis intermediates that accumulate upon exosome perturbation.
Fig. 3. Dis3l2-mediated rRNA processing and its relationship with the exosome.
qRT-PCR (a) and western blot (b) analysis of Exosc3 and Exosc10 expression in Dis3l2 wild type (WT) and knockout (KO) cells after siRNA transfection. In (a), bars represent mean ± SEM. Individual P values are presented; two-tailed Student’s t-test (n=3 biologically independent cell cultures). In (b), asterisk represent an unspecific band, whereas the arrow points to Exosc3 band. (c) Northern blot analysis of total RNAs from WT and Dis3l2 knockout ESCs depleted of Exosc3 or Exosc10 with 7SB, ITS2 and 5.8S rRNA probes. Arrows mark discrete bands of 12S and 8S rRNAs. The square bracket represents slow migrating tailed rRNAs. (d) Upper panel: Northern blot for 7SB rRNA in Dis3l2 knockout cells after Exosc10 knockdown and/or re-expression of WT Dis3l2. Lower panel: WB analysis of Exosc10 and Dis3l2 (FLAG-tagged) expression. (e) qRT-PCRs quantifying the relative uridylation of RNAs in samples treated as in (c); 7SK and Rmrp RNAs were assessed as negative controls. Bars represent mean ± SEM. Individual P values are presented; two-tailed Student’s t-test (n=3 biologically independent cell cultures). (f) qRT-PCR on control and Dis3- or Dis3l1-knockdown samples. Bars represent mean ± SEM. Individual P values are presented; two-tailed Student’s t-test (n=3 biologically independent cell cultures). (g) Upper and middle panels: Northern blot analysis of WT and Dis3l2 knockout ESCs after depletion of Dis3, or Dis3l1 using specific siRNAs with probes against 7SBor ITS2 (8S), respectively. Lower panel: Western blot analysis of Dis3 in respective samples. Asterisk represents an unspecific band. (h) qRT-PCR analysis of relative uridylation in respective RNA samples from (g). Bars represent mean ± SEM. Individual P values are presented; two-tailed Student’s t-test (n=4 biologically independent cell cultures).To measure relative uridylation levels, values were first normalized to Gapdh and then to the total levels of indicated transcript. WT, wild type Dis3l2; KO, knockout Dis3l2; ns, not significant. Panels b, c, d, and g are representative images from at least 3 repeats. Uncropped blot/gel images are shown in Supplementary Data Set 1. Source Data for panels a, e, f and h are available online.
Next, to examine the role of other Dis3l2 homologs in 7SB processing, we knocked down the expression of Dis3 and Dis3l1 (Fig. 3f–h). Similar to the effect of Exosc3 knockdown, perturbation of Dis3 expression caused the accumulation of 8S and 12S rRNA precursors assessed by 7SB (Fig. 3g, upper panel) and ITS2 (Fig. 3g, middle panel) probes. However, no such effects were observed upon downregulation of Dis3l1. qRT-PCR confirmed the elevated uridylation only in Dis3-depleted cells (Fig. 3h). Taken together, these results show that Dis3l2 acts downstream of the exosome and is primarily involved in the processing of 7SB and other 5.8S rRNA intermediates that accumulate when the exosome is perturbed.
Exosc3, Exosc10, and Dis3l2 rRNA substrates at single nucleotide resolution
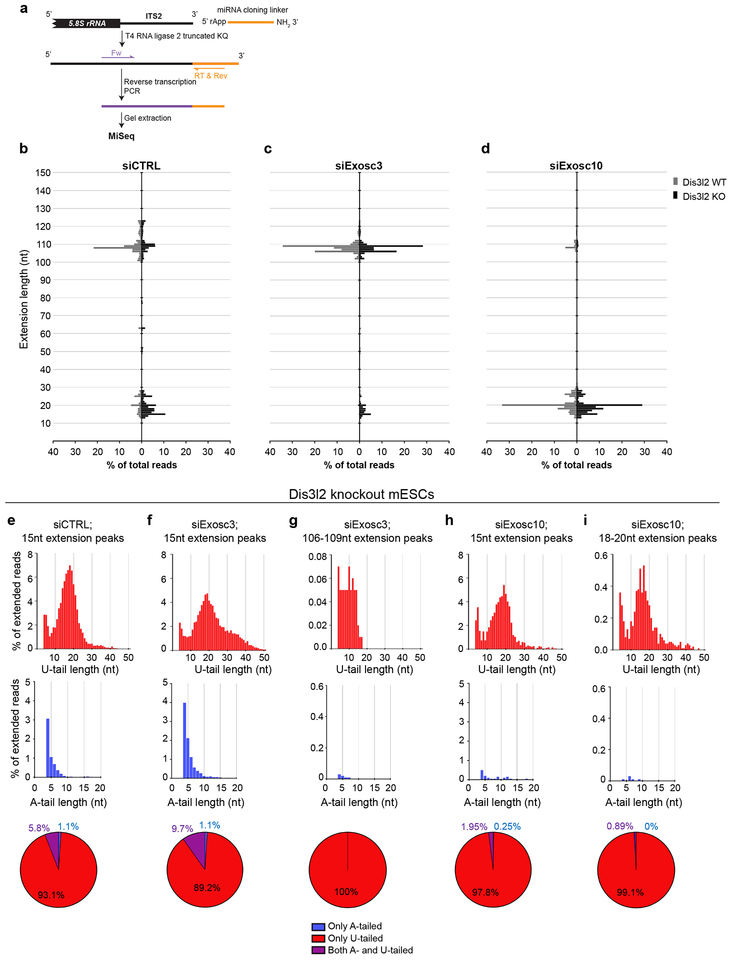
Given our finding that Exosc3 and Exosc10 impairment upregulates specific rRNA decay intermediates, with the latter closely resembling the size of 7SB, and aiming to further connect Dis3l2 to exosome activity in rRNA processing, RNA samples from Fig. 3c were subjected to 3’-ligation RACE followed by MiSeq analysis (Fig. 4a). The ITS2 genomic extension, A- and U-tails of rRNA intermediates were analyzed in WT and Dis3l2 knockout mESCs upon depletion of Exosc3 and Exosc10. In siCTRL-treated WT cells (Fig. 4b), several extended intermediates of 5.8S were detected (grey bars), whereas in knockout cells, 5.8S rRNAs with ~15 nt extension (ranging from ~10 to 20 nt) were dominant (black bars, up to 30% of the reads) as previously identified as 7SB (Fig. 1d). Similar to what we found by cRACE on the Dis3l2 RIP samples, we also found by this 3’-ligation RACE analysis of total RNA samples that 7SB is specifically oligoadenylated (blue bars) and/or oligouridylated (red bars) (Fig. 4e) with a similar length distribution to that we found with the alternative cRACE approach. In Exosc3-depleted WT cells (Fig. 4c), the prominent extension was 106–109 nt long (grey bars) that corresponds to the length of murine 8S rRNA. Similarly, in siExosc3-treated Dis3l2 knockout ESCs (black bars), beside 7SB (5.8S+15 nt) extension, an abundant 8S (5.8S+106–109 nt) was observed (Fig. 4c). This reveals that in knockout cells, both extended 5.8S rRNAs (7SB and 8S) are stabilized. In siExosc3-transfected WT cells, no U-tails were detected, whereas in Dis3l2 knockout cells U-tails were abundant both on the 7SB rRNA as well as the 8S rRNA (Fig. 4f, g). The longer U-tails on 7SB rRNA in siExosc3-treated Dis3l2 knockout cells probably corresponds to the slow-migrating RNA species (Fig. 3c) compared to the size of 7SB band with Dis3l2 depletion alone. Markedly, A-tailed reads (blue bars) were majorly evident in knockout cells but occurred almost exclusively on the 7SB rRNA (see below). Upon Exosc10 depletion (Fig. 4d) in WT cells, extensions of 13–30 nt (with a peak at 20 nt) corresponding to ITS2 (grey bars) were prominently stabilized. We hereafter refer to this pre-rRNA species as 7SA. Comparably, also in Dis3l2 knockout cells, similar extensions of around 20 nt (7SA) [together with a peak at 15 nt (7SB)] were highly stabilized (black bars) (Fig. 4d). In Dis3l2 knockout cells, while both the 7SB and the longer 7SA (5.8S+20 nt) and 8S (5.8S+106−109 nt) species were highly uridylated (red bars), only in siCTRL and siExosc3 samples the 7SB rRNA was found to be adenylated (Fig. 4e–i). This implies that Exosc10 is required for the processing and subsequent 7SB oligoadenylation. Altogether, these results provide a nucleotide resolution view of the 8S rRNA substrate of the exosome, identify a novel intermediate 7SA (5.8S+20nt) as a substrate of Exosc10, and find that while all intermediates can be detected as oligouridylated species in Dis3l2 knockout cells, only the 7SB rRNA is adenylated indicating that specific 7SB rRNA oligoadenylation is part of the 5.8S rRNA processing pathway.
Fig. 4. Deep sequencing analysis of the 3’-end of 5.8S rRNA species and their precursors following Dis3l2, Exosc3, and Exosc10 perturbation.
(a) Schematic representation of 3’-ligation RACE procedure. Dis3l2 wild type (WT) and knockout (KO) mESCs were transfected with siCTRL (b), siExosc3 (c) or siExosc10 (d) siRNAs. X-axis represents the percentage of the reads and Y-axis marks the nucleotide length. (e-i) Upper panels show the U-tail and middle panels show A-tails in reads with indicated genomic extensions in Dis3l2 knockout samples. Lower panels show the overall distribution of tailed reads. WT, wild type Dis3l2; KO, knockout Dis3l2.
Eri1 functions redundantly with Dis3l2 in 7SB rRNA processing
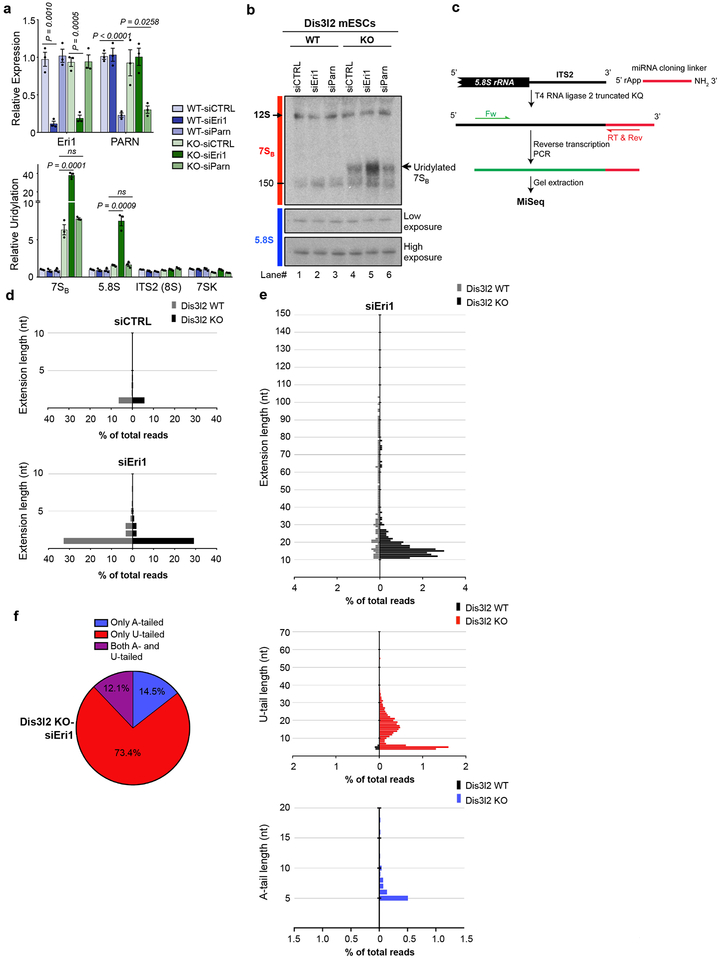
Eri1 is a cytoplasmic 3’-5’ exoribonuclease responsible for the final maturation of 5.8S rRNA33, as well as for degradation of oligouridylated histone mRNAs40. We asked whether Eri1 could also function in the processing of 7SB rRNA that is specifically stabilized in Dis3l2-depleted cells. Eri1 downregulation, but not of PARN (Poly(A)-Specific Ribonuclease), caused even stronger upregulation of 7SB uridylation in Dis3l2 knockout cells (Fig. 5a). This effect was specific to 7SB (and 5.8S), but not to 8S or 7SK RNAs. Northern blotting further confirmed a pronounced accumulation of 7SB in Dis3l2 knockout cells after Eri1 knockdown (Fig. 5b) with no effect on overall abundance of 5.8S rRNA. 3’-ligation RACE and MiSeq analysis (Fig. 5c) showed strong accumulation of 5.8S precursor with 1 nucleotide (a single U in the genomic-encoded ITS2) extension in both WT and knockout ESCs (Fig. 5d). This is consistent with the previous observation that Eri1 catalyzes the trimming of the last nucleotide(s) of the ITS2 and confirms 6S rRNA (5.8S+1 nt rRNA) as the substrate for Eri133. Furthermore, this shows that Dis3l2 is dispensable for Eri1 function on this substrate. However, a small portion of sequencing reads with longer extensions were also found in Dis3l2 knockout samples (Fig. 5e–f). Examination of these extensions upon Eri1 knockdown revealed a similar pattern to 7SB rRNA (10–20 nt extension with a peak at 15 nt) (Fig. 5e) that further implies Eri1 function in the processing of 7SB rRNA in the absence of Dis3l2. These extensions were also oligoadenylated and/or oligouridylated (Fig. 5e). Overall distribution of tails showed extensive uridylation as well as a considerable portion of A- or both A- and U-tailed reads (Fig. 5f). Altogether, these results certify the function of Eri1 in the trimming of the ITS2 last nucleotide and further suggest an extra role of Eri1 in redundant processing of 7SB rRNA in Dis3l2-depleted cells.
Fig. 5. Eri1 exonuclease functions in parallel to Dis3l2 to process uridylated 7SB rRNA.
(a) qRT-PCR analysis of Eri1 or PARN expression (upper panel) and relative uridylation of indicated RNAs (lower panel) in Dis3l2 WT or knockout ESCs. Bars represent mean ± SEM. Individual P values are presented; two-tailed Student’s t-test (n=3 biologically independent cell cultures). (b) Northern blot analysis of the samples from (a) using 7SB and 5.8S rRNA probes. Note to the similar size of tailed 7SB in siEri1 sample. (c) Schematic representation of 3’-ligation RACE procedure. Note that for the amplification of short tailed 5.8S rRNA species, an internal forward primer was used instead of the one used in Fig. 4. (d) The genomic extension of 5.8S rRNA upon Eri1 perturbation. (e) Tail analysis of low abundant extended (>10 nt) rRNA precursors after Eri1 knockdown.. (f) Overall distribution of different tails in Eri1-depleted Dis3l2 knockout cells showing A-, U-, and both A- and U-tailed reads. WT, wild type Dis3l2; KO, knockout Dis3l2; ns, not significant. Panel b shows representative images from at least 3 repeats. Uncropped blot/gel images are shown in Supplementary Data Set 1. Source Data for panel a are available online.
DISCUSSION
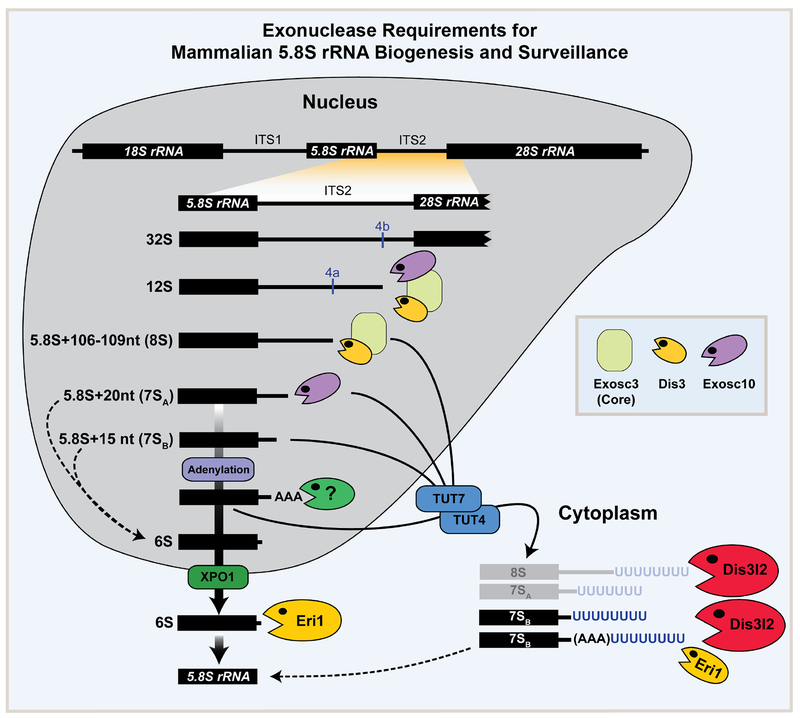
Based on extensive northern blot, qRT-PCR, and sequencing coupled to loss-of-function studies in mESCs we propose a model for the function of various exonucleases involved rRNA processing (Fig. 6). Accordingly, 1) The core exosome (Exosc3) together with its catalytic subunits Dis3 and Exosc10 process the 12S rRNA intermediate to the 8S intermediate with a 106–109 nt extension. 2) The 8S intermediate is processed by Dis3-Exosc3 to the 7SA (5.8S+20 nt) intermediate. 3) Exosc10 (independent of the exosome core and Dis3) trims the 7SA intermediate to the generate 7SB rRNA (5.8S+15 nt) intermediate. 4) 7SB can be adenylated (by an unknown enzyme) and is further processed by an unknown nuclease to generate 6S rRNA. 5) Finally, Eri1 trims the last nucleotide on the 6S rRNA to generate the mature 5.8S rRNA. 6) Failure of any of these processing steps results in oligouridylation by TUT4/7 and processing by Dis3l2 and Eri1 in the cytoplasm.
Fig. 6. Exonuclease requirements for mammalian 5.8S rRNA biogenesis and surveillance.
A model summarizing the function of various exoribonucleases in the processing of 5.8S rRNA intermediates. During step-wise maturation of 5.8S rRNA, exosome and Exosc10 are responsible for processing of the 8S rRNA and 7SA (5.8S+20nt) intermediates, respectively. 7SB rRNA (5.8S+15nt) which is a potential product of Exosc10 can be adenylated (by an unknown enzyme) and/or further processed by an unknown nuclease to the 6S pre-rRNA that processed by Eri1 to mature 5.8S rRNA in the cytoplasm (thick solid arrows). Failure of any of these processing steps results in oligouridylation by TUT4/7 and processing by Dis3l2 and Eri1 in the cytoplasm (arrows). Dashed lines represent possible alternative steps in the 5.8S rRNA processing pathway.
Until now, the only 5.8S precursors defined in mouse are the 12S and 8S pre-RNAs, possessing 3’-end extensions of ~794 nt and ~105 nt, respectively4,41–43. Recent studies highlight the existence of multiple yet uncharacterized processing intermediates in human14. However, the length of different 5.8S rRNA intermediates are mostly determined based on their migration pattern on northern blotting gels rather than by sequencing6,11,12,39. Our results add new details to the current understanding of the pathway. We provide nucleotide resolution of the 5.8S precursors and implicate different exosome subunits in their processing. We confirm that the length of murine 8S pre-rRNA is indeed 106–109 nt and show its stabilization upon the loss of Exosc3 core exosome component, as well as Dis3, but not Exosc10. Moreover, we identified a 7SA (5.8S+20nt) intermediate in murine cells that is a likely counterpart of yeast 5.8S+30 nt and human 5.8S+40 nt6,14. 5.8S+30 nt is a substrate of the yeast Exosc10 (Rrp6) that generates a product of 5.8S+~5–8 nt. We uncover an equivalent step in mice that involves the Exosc10-mediated processing of 7SA to produce the 7SB (5.8S+15 nt) intermediate product. While our model shows processing all the way to the 6S rRNA occurring in the nucleus (as is the case for yeast 5.8S rRNA biogenesis) it remains possible that some of the processing steps could take place in the cytoplasm. Moreover, we find oligoadenylated 7SB RNA and it is tempting to speculate that this might be required to stimulate the activity of the putative downstream nuclease to process 7SB to the 6S rRNA intermediate. In light of our findings, it will be of interest to identify the putative terminal transferase(s) that add the oligoA tail to the 7SB RNA. This would be analogous to the coordinated activity of the TRAMP (Trf4p/Air2p/Mtr4p Polyadenylation complex) and exosome complexes44 where the non-canonical polyA polymerase Trf4/5 in yeast, and TENT4B (PAPD5) in humans, facilitates RNA processing by the nuclear exosome complex44. These may include TENT2 (PAPD4)45, TENT4A (PAPD7) and/or TENT4B (PAPD5)32,46–48, etc. Since we do not see accumulation of 7SB in exosome-deficient cells it seems likely that a different nuclease is responsible for processing 7SB to 6S rRNA.
rRNAs have been co-immunoprecipitated with Dis3l2 protein in human26 and in fly cells28. Moreover, bacterial members of RNase II/RNB family have been implicated in rRNA processing49 suggesting the universal targeting of rRNAs by RNase II family enzymes like Dis3l2. We expand the repertoire of Dis3l2 targets in mammals to three (5.8S, 5S, and 28S) out of four rRNAs and identify uridylated 7SB pre-rRNA as a major Dis3l2 substrate. Whether Dis3l2 functions exclusively as a surveillance pathway for rRNA for unprocessed/aberrant intermediates or is also part of the 5.8S biogenesis mechanism requires further investigation (see below). Nevertheless, we test for the first time the Dis3l2-mediated decay (DMD) concept and showed that in fact Dis3l2 is responsible for elimination of aberrant rRNAs in the cytoplasm where it degrades unprocessed and thus aberrant 5.8S rRNA precursors downstream of Dis3-Exosc3 and Exosc10 (Fig. 6). SUSI-1, a Dis3l2 homolog in C. elegans has been recently identified to target erroneous, uridylated rRNAs, implying an evolutionary conserved role of Dis3l2 in the surveillance of impaired rRNAs50. Therefore, we propose that TUTase-Dis3l2 axis ensures the elimination of cytoplasmic aberrant rRNA intermediates.
The levels of 5.8S rRNA are unaffected in the Dis3l2 knockout mESCs even after multiple cell passages. This indicates that if Dis3l2 is involved in 5.8S production then it must function in a redundant pathway. In this regard, it is interesting that we find that Eri1 can also process oligouridylated 7SB in the cell cytoplasm. In several eukaryotes, including C. elegans, mouse, and human, 3’-5’ exonuclease activity of Eri1 was documented on different RNA species including replication-dependent histone mRNAs, siRNAs and 6S rRNA33,51–54. Here, we extend the Eri1 activity to 7SB rRNA in the absence of Dis3l2, where Eri1 can process oligouridylated 7SB. Similar to Dis3l2, Eri1 also has a preferential activity towards RNAs with oligouridyl tails33,40 and the last nucleotide in the 6S rRNA that Eri1 trims is a uridine. Moreover, Eri1 substrates share structural features like the presence of an RNA duplex followed by single-stranded extensions33,55, similar to what is seen in 3’-end of 7SB rRNA. While the exonuclease activities of both Dis3l2 and Eri1 may lead to complete degradation of 7SB rRNA (as an “off-pathway” dead-end intermediate), their processivity is likely halted when they reach to the annotated 3’-end of mature 5.8S rRNA that is protected by ribosomal proteins and/or the stem-like structure formed between the 3’-end of the mature 5.8S rRNA and the 5’-end of the mature 28S rRNA56,57. Accordingly, Dis3l2-Eri1 could redundantly be involved in the 5.8S rRNA “on-pathway” maturation step in the cytoplasm. Further investigation is needed to distinguish these alternative pathways and to determine the relative contribution of Dis3l2 and Eri1 to 5.8S rRNA maturation. Nevertheless, in the absence of Dis3l2 or Eri1 individually, the steady state levels of 5.8S rRNA are unchanged (our results and33), and even with loss of both enzymes i.e. when we knocked down Eri1 (albeit transiently) in the Dis3l2 knockout ESCs we see no decrease in 5.8S rRNA levels. Therefore we conclude that either there is further redundancy in this 5.8S rRNA biogenesis step or that the major role of Dis3l2-Eri1 is to degrade these unprocessed intermediate rRNAs. This study sheds new light on mammalian 5.8S rRNA processing, identifies 7SB as a novel rRNA intermediate, and reveals DMD as a cytoplasmic surveillance pathway, that ensures the accuracy of rRNAs biogenesis.
METHODS
ESC culture and transfection.
TC1 mouse embryonic stem cells (mESCs, a gift from Dr. F. W. Alt; Boston Children’s Hospital) have been tested for mycoplasma and cultured as previously described22. Dis3l2 knockout mES cells were generated previously using CRISPR/Cas9 gene editing24. For transient knockdown experiments, following ON-TARGET plus siRNAs (all Dharmacon) were used: Control siRNA pool (D-001810–10), siXPO1 pool (L-064569–00), siExosc3 pool (L-064537–01), siExosc10 pool (L-049286–00), siTUT7 (L-056770–01), and siTUT4 pool (L-065226–00). ESCs were reverse transfected using siRNAs and Lipofectamine RNAiMax (Invitrogen) complexes prepared in Opti-MEM (gibco) for 48–72 hours. In rescue experiments, 1 μg of WT Dis3l2 expressing vectors or empty vectors (as mock) were reverse transfected by Lipofectamine 2000 (Invitrogen) into ESCs for 48–72 hours. Dis3l2 stable knockdown ESCs (shDis3l2 line) were generated previously17. Cytoplasmic and nuclear fractions of ESCs were prepared using PARIS kit (Ambion) as previously described24.
RNA extraction and qRT-PCR.
ESCs were washed twice with PBS, lyzed in Trizol (Ambion) and RNAs were chloroform-isopropanol extracted and washed twice with 70% ethanol. 2 μg RNA was treated with RQ1 DNase for 30 minutes at 37 °C. Using random hexamers (to analyze relative expression) or oligo-dA12 oligonucleotides (to measure relative uridylation), cDNAs were made with SuperScript III reverse transcriptase (Invitrogen) and RNaseOUT (Invitrogen). List of primers and DNA oligos are provided in Supplemental Table 1. All the qRT-PCR experiments were normalization to Gapdh levels in the respective cDNA samples.
Western and Northern blotting.
Protein or RNA samples from ESCs were analyzed by Western or Northern blotting as previously described24. For Western blotting, following antibodies were used: Rabbit anti-Dis3l2 (Novus Biologicals); Rabbit anti-beta-Actin (abcam); Rabbit anti-TUT4 (Proteintech Group); mouse HRP-conjugated anti-FLAG M2 antibody (Sigma); Rabbit anti-Exosc3 (Bethyl); Rabbit anti-Exosc10 (abcam); Rabbit anti-RPL23a (Proteintech Group) and Rabbit anti-RPS6 (Cell Signaling, #2217). 7 μg of total or cytoplasmic RNAs, and 700 ng of the nuclear RNAs were used for Northern blotting. List of probes used in this study is provided Supplemental Table 1. Oligo DNA probes were 5’-labeled by32P-γ-ATP using T4 Polynucleotide Kinase.
RNA Immunoprecipitation.
mESCs were transfected with FLAG-WT Dis3l2, FLAG-mutant Dis3l217, or empty pFLAG-CMV2 (as mock) vectors. 48h after transfection, mESCs were UV-crosslinked, lyzed and then, RNA immunoprecipitation was performed using anti-FLAG M2 Affinity Gel beads (Sigma) as previously described24. For ribosome immunoprecipitation, the same procedure was taken and instead, rabbit anti-RPL23a antibody (Proteintech Group) was used.
cRACE and 3’-ligation RACE.
Input and FLAG-mutant Dis3l2 RIP samples were circularized with 10 units of T4 RNA ligase I, 10 mM ATP, 10% PEG 8000, in 1X T4 RNA ligase buffer for 2 hours at 37 °C and then the ligase was inactivated, as previously described24,25. After DNase treatment, circularized RNAs were reverse transcribed with 7SB specific reverse primer (Supplemental Table 1) and SuperScript III. cDNAs were amplified by divergent internal primers (see Supplementary Figure 2a and also Supplemental Table 1) and AccuPrime GC-rich DNA Polymerase (Invitrogen) to generated chimeric PCR products corresponding to 5’ and 3’-ends of 5.8S rRNA transcripts. PCR products were size selected on 2% agarose gel, purified and used in library preparation for MiSeq analysis using TruSeq Stranded mRNA Sample Preparation Kits (Illumina). For 3’-ligation RACE, 5 μg of total RNAs from Dis3l2 WT and knockout transfected with control siRNAs, siExosc3, or siExosc10 were ligated to miRNA cloning linker (Fig. 4a) as previously described24. Note that for 3’-ligation RACE analysis of Eri1-depleted cells, a different forward (Fw) primer was used (Fig. 5c). RT primer annealing to the linker was used to prepare chimeric cDNA, on which PCR was performed using forward and reverse (on linker) primers. After gel extraction of this PCR product, libraries were made and used for MiSeq.
Polysome profiling
Polysome fractionations were performed as described previously38. Briefly, four 15-cm plates of 80% confluent mESCs were lysed and layered onto 10%–50% sucrose gradient tube and centrifuged at 36,000 rpm in a Beckman SW-41Ti rotor for 2.5 hr at 4°C. Gradients were fractionated and monitored at absorbance 254 nm (Brandel). Collected fractions were then analyzed by western blotting and northern blotting.
Statistics.
All the experiments were performed more than three times. Quantitative data are presented as mean ± standard error of means (SEM). Student’s t-tests were used to analyze the significance of difference between different samples.
DATA AVAILABILITY
Raw sequencing data are deposited with GEO Series accession code GSE129734. Source data for figures 1b, 2b, c, e, g, h, 3a, e, f, h, and 5a are available with the paper online. Other data are available upon request.
Supplementary Material
Supplementary Data Set 1 Uncropped blot/gel images.
Supplementary Table 1 List of oligonucleotide primers and probes used in this study.
ACKNOWLEDGMENTS
This work was supported by grants to R.I.G. from the US National Institutes of Health (R01GM086386; R01CA211328), and the March of Dimes Foundation (FY15–3339). M.P. was supported by a research fellowship from the Manton Center for Orphan Disease Research.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Ciganda M & Williams N Eukaryotic 5S rRNA biogenesis. Wiley Interdiscip Rev RNA 2, 523–33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tomecki R, Sikorski PJ & Zakrzewska-Placzek M Comparison of preribosomal RNA processing pathways in yeast, plant and human cells - focus on coordinated action of endo- and exoribonucleases. FEBS Lett 591, 1801–1850 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Henras AK, Plisson-Chastang C, O’Donohue MF, Chakraborty A & Gleizes PE An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 6, 225–42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mullineux ST & Lafontaine DL Mapping the cleavage sites on mammalian pre-rRNAs: where do we stand? Biochimie 94, 1521–32 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Thomson E & Tollervey D The final step in 5.8S rRNA processing is cytoplasmic in Saccharomyces cerevisiae. Mol Cell Biol 30, 976–84 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tafforeau L et al. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol Cell 51, 539–51 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Chlebowski A, Lubas M, Jensen TH & Dziembowski A RNA decay machines: the exosome. Biochim Biophys Acta 1829, 552–60 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Liu Q, Greimann JC & Lima CD Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127, 1223–37 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Makino DL, Baumgartner M & Conti E Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature 495, 70–5 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Dziembowski A, Lorentzen E, Conti E & Seraphin B A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol 14, 15–22 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Allmang C et al. The yeast exosome and human PM-Scl are related complexes of 3’ --> 5’ exonucleases. Genes Dev 13, 2148–58 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell P, Petfalski E & Tollervey D The 3’ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev 10, 502–13 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Mitchell P, Petfalski E, Shevchenko A, Mann M & Tollervey D The exosome: a conserved eukaryotic RNA processing complex containing multiple 3’-->5’ exoribonucleases. Cell 91, 457–66 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Schillewaert S, Wacheul L, Lhomme F & Lafontaine DL The evolutionarily conserved protein Las1 is required for pre-rRNA processing at both ends of ITS2. Mol Cell Biol 32, 430–44 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomecki R et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J 29, 2342–57 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Staals RH et al. Dis3-like 1: a novel exoribonuclease associated with the human exosome. EMBO J 29, 2358–67 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang HM, Triboulet R, Thornton JE & Gregory RI A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature 497, 244–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lubas M et al. Exonuclease hDIS3L2 specifies an exosome-independent 3’-5’ degradation pathway of human cytoplasmic mRNA. EMBO J 32, 1855–68 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malecki M et al. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J 32, 1842–54 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ustianenko D et al. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA 19, 1632–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Triboulet R, Pirouz M & Gregory RI A Single Let-7 MicroRNA Bypasses LIN28-Mediated Repression. Cell Rep 13, 260–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pirouz M et al. Destabilization of pluripotency in the absence of Mad2l2. Cell Cycle 14, 1596–610 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Astuti D et al. Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat Genet 44, 277–84 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Pirouz M, Du P, Munafo M & Gregory RI Dis3l2-Mediated Decay Is a Quality Control Pathway for Noncoding RNAs. Cell Rep 16, 1861–1873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pirouz M, Ebrahimi AG & Gregory RI Unraveling 3’-end RNA uridylation at nucleotide resolution. Methods (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ustianenko D et al. TUT-DIS3L2 is a mammalian surveillance pathway for aberrant structured non-coding RNAs. EMBO J 35, 2179–2191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Towler BP, Jones CI, Harper KL, Waldron JA & Newbury SF A novel role for the 3’-5’ exoribonuclease Dis3L2 in controlling cell proliferation and tissue growth. RNA Biol 13, 1286–1299 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reimao-Pinto MM et al. Molecular basis for cytoplasmic RNA surveillance by uridylation-triggered decay in Drosophila. EMBO J 35, 2417–2434 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Labno A et al. Perlman syndrome nuclease DIS3L2 controls cytoplasmic non-coding RNAs and provides surveillance pathway for maturing snRNAs. Nucleic Acids Res 44, 10437–10453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eckwahl MJ, Sim S, Smith D, Telesnitsky A & Wolin SL A retrovirus packages nascent host noncoding RNAs from a novel surveillance pathway. Genes Dev 29, 646–57 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Preti M et al. Gradual processing of the ITS1 from the nucleolus to the cytoplasm during synthesis of the human 18S rRNA. Nucleic Acids Res 41, 4709–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim J et al. Mixed tailing by TENT4A and TENT4B shields mRNA from rapid deadenylation. Science 361, 701–704 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Ansel KM et al. Mouse Eri1 interacts with the ribosome and catalyzes 5.8S rRNA processing. Nat Struct Mol Biol 15, 523–30 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thornton JE, Chang HM, Piskounova E & Gregory RI Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA 18, 1875–85 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thornton JE et al. Selective microRNA uridylation by Zcchc6 (TUT7) and Zcchc11 (TUT4). Nucleic Acids Res 42, 11777–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagan JP, Piskounova E & Gregory RI Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol 16, 1021–5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heo I et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138, 696–708 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Lin S, Choe J, Du P, Triboulet R & Gregory RI The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell 62, 335–345 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allmang C et al. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J 18, 5399–410 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoefig KP et al. Eri1 degrades the stem-loop of oligouridylated histone mRNAs to induce replication-dependent decay. Nat Struct Mol Biol 20, 73–81 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Bowman LH, Goldman WE, Goldberg GI, Hebert MB & Schlessinger D Location of the initial cleavage sites in mouse pre-rRNA. Mol Cell Biol 3, 1501–10 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reddy R et al. The nucleotide sequence of 8 S RNA bound to preribosomal RNA of Novikoff hepatoma. The 5’-end of 8 S RNA is 5.8 S RNA. J Biol Chem 258, 584–9 (1983). [PubMed] [Google Scholar]
- 43.Michot B, Joseph N, Mazan S & Bachellerie JP Evolutionarily conserved structural features in the ITS2 of mammalian pre-rRNAs and potential interactions with the snoRNA U8 detected by comparative analysis of new mouse sequences. Nucleic Acids Res 27, 2271–82 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LaCava J et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121, 713–24 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Chung CZ, Jo DH & Heinemann IU Nucleotide specificity of the human terminal nucleotidyltransferase Gld2 (TUT2). RNA 22, 1239–49 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rammelt C, Bilen B, Zavolan M & Keller W PAPD5, a noncanonical poly(A) polymerase with an unusual RNA-binding motif. RNA 17, 1737–46 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shcherbik N, Wang M, Lapik YR, Srivastava L & Pestov DG Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep 11, 106–11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barandun J, Hunziker M & Klinge S Assembly and structure of the SSU processome-a nucleolar precursor of the small ribosomal subunit. Curr Opin Struct Biol 49, 85–93 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng ZF & Deutscher MP Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc Natl Acad Sci U S A 100, 6388–93 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou X et al. RdRP-synthesized antisense ribosomal siRNAs silence pre-rRNA via the nuclear RNAi pathway. Nat Struct Mol Biol 24, 258–269 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Dominski Z, Yang XC, Kaygun H, Dadlez M & Marzluff WFA 3’ exonuclease that specifically interacts with the 3’ end of histone mRNA. Mol Cell 12, 295–305 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Yang XC, Purdy M, Marzluff WF & Dominski Z Characterization of 3’hExo, a 3’ exonuclease specifically interacting with the 3’ end of histone mRNA. J Biol Chem 281, 30447–54 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Gabel HW & Ruvkun G The exonuclease ERI-1 has a conserved dual role in 5.8S rRNA processing and RNAi. Nat Struct Mol Biol 15, 531–3 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kennedy S, Wang D & Ruvkun G A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427, 645–9 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Tan D, Marzluff WF, Dominski Z & Tong L Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3’hExo ternary complex. Science 339, 318–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cote CA, Greer CL & Peculis BA Dynamic conformational model for the role of ITS2 in pre-rRNA processing in yeast. RNA 8, 786–97 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Joseph N, Krauskopf E, Vera MI & Michot B Ribosomal internal transcribed spacer 2 (ITS2) exhibits a common core of secondary structure in vertebrates and yeast. Nucleic Acids Res 27, 4533–40 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data Set 1 Uncropped blot/gel images.
Supplementary Table 1 List of oligonucleotide primers and probes used in this study.
Data Availability Statement
Raw sequencing data are deposited with GEO Series accession code GSE129734. Source data for figures 1b, 2b, c, e, g, h, 3a, e, f, h, and 5a are available with the paper online. Other data are available upon request.