Abstract
Autosomal dominant cerebellar ataxias (ADCAs) are a group of neurodegenerative disorders characterized by degeneration of the cerebellum and its connections. All ADCAs have progressive ataxia as their main clinical feature, frequently accompanied by dysarthria and oculomotor deficits. The most common spinocerebellar ataxias (SCAs) are 6 polyglutamine (polyQ) SCAs. These diseases are all caused by a CAG repeat expansion in the coding region of a gene. Currently, no curative treatment is available for any of the polyQ SCAs, but increasing knowledge on the genetics and the pathological mechanisms of these polyQ SCAs has provided promising therapeutic targets to potentially slow disease progression. Potential treatments can be divided into pharmacological and gene therapies that target the toxic downstream effects, gene therapies that target the polyQ SCA genes, and stem cell replacement therapies. Here, we will provide a review on the genetics, mechanisms, and therapeutic progress in polyglutamine spinocerebellar ataxias.
Key Words: Spinocerebellar ataxia, SCA, polyglutamine disorders, gene therapy, stem cell-based therapy, antisense oligonucleotides
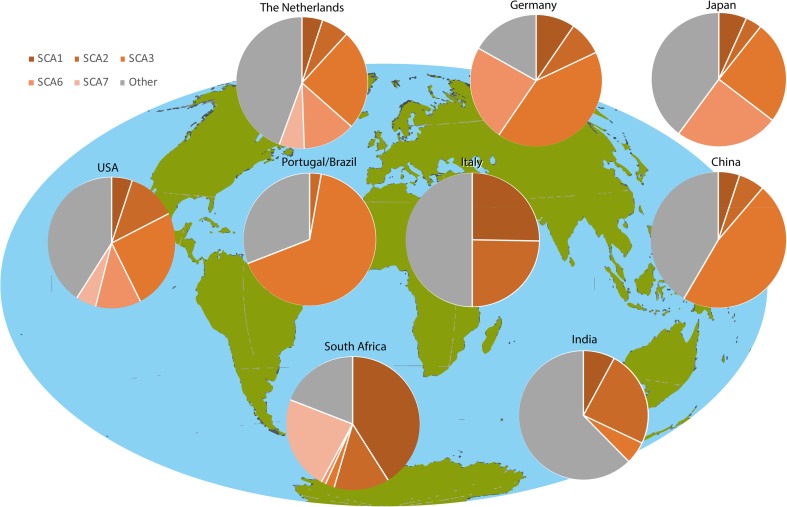
Autosomal dominant cerebellar ataxias (ADCAs) are a group of neurodegenerative disorders characterized by degeneration of the cerebellum and its connections [1, 2]. All ADCAs have progressive ataxia as their main clinical feature, frequently accompanied by dysarthria and oculomotor deficits. The ADCAs of which the causative gene has been identified belong to the spinocerebellar ataxias (SCAs). The SCAs are numbered in chronological order of discovered gene locus with SCA47 being the most recent SCA reported [3–5]. However, around 1 third of families with ADCA remain without a genetic diagnosis [6, 7]. Based on a meta-analysis of prevalence studies, the prevalence of SCAs is estimated to be approximately 1–5:100,000 [7, 8], but can vary widely between different geographical and ethnical groups (see Fig. 1) [17]. The highest prevalence rates in population-based studies were found in Portugal (5.6:100,000 [6]), in Norway (4.2:100,000 [19]), and in Japan (5.0:100,000 [20]).
Fig. 1.
Worldwide prevalence of polyglutamine spinocerebellar ataxias. In the ADCA families investigated, no SCA17 was identified. The Netherlands [8]; Germany [9]; Japan [10]; the USA [11]; Portugal/Brazil [12]; Italy [13]; China [14]; South Africa [15]; India [16]. Figure based on Schols et al. [17] and adapted from Bird [18] (GeneReviews)
The most common SCAs are 6 polyglutamine (polyQ) SCAs. These diseases are all caused by a CAG repeat expansion in the coding region of a gene. For each SCA, the causative gene is different (see Table 1) [21–41]. The expanded CAG repeat in the DNA is translated into an expanded repeat of glutamine amino acids (Qs) in the respective ataxin proteins, hence the term polyQ SCA. In general, an inverse correlation exists between the length of the expanded polyQ stretch and the age of symptom onset [42, 43]. Although the exact pathogenic mechanism for each polyQ SCA is not fully understood, the known mechanisms have been extensively reviewed previously (see review [44]). In short, data suggests that the disruption of the native protein functions and the toxic gain of function conveyed by the expanded polyQ stretch [45] are interrelated and disrupt several common cellular processes contributing to disease pathology. These disruptions include transcriptional deregulation, RNA toxicity, toxicity caused by repeat-associated non-ATG (RAN) translation peptides, dysregulation of the ubiquitin proteasome system, and autophagy [46]. In addition, the altered protein confirmation caused by the expanded polyQ sequence leads to the formation of large insoluble protein aggregates containing the expanded diseased protein mainly in neurons of the cerebellum. Whether these large aggregates, a hallmark of all polyQ SCAs, are neurotoxic or neuroprotective is still under debate [47]. However, for most polyQ SCAs, soluble mono- or oligomer fragments are thought to be the main toxic entity [48, 49]. Together, these disrupted cellular processes cause perturbations of the intracellular homeostasis and eventually lead to the death of neuronal cells, inducing the characteristic neurodegenerative symptoms of the polyQ SCAs. Despite their similarities, the different polyQ SCAs have distinct clinical and neuropathological features described in the sections below.
Table 1.
Characteristics of polyglutamine disease-associated genes
| Disease | PDAG | Locus | Protein | Repeat | CAG repeat number | ||
|---|---|---|---|---|---|---|---|
| Normal | Intermediate | Pathological | |||||
| SCA1 | ATXN1 | 6p22.3 | Ataxin-1 | (CAG)n(CAT)n(CAG)na | 6–35 (6–44a) | 36–38 | 39–91 (45–91a) |
| SCA2 | ATXN2 | 12q24.12 | Ataxin-2 | [(CAG)nCAA(CAG)n]nb | 14–31 | 32 | 33–500 |
| SCA3 | ATXN3 | 14q32.12 | Ataxin-3 | (CAG)2CAA AAG CAG CAA(CAG)n | 11–44 | 45–59 | 60–87 |
| SCA6 | CACNA1A | 19p13.13 | CACNA1A | (CAG)n | 4–18 | 19 | 20–33 |
| SCA7 | ATXN7 | 3p14.1 | Ataxin-7 | (CAG)n | 4–19 | 28–33 | 34–460 |
| SCA17 | TBP | 6q27 | TBP | [(CAG)n(CAA)n(CAG)n] | 25–40 | – | 41–66 |
CAG = cytosine-adenine-guanine; PDAG = polyglutamine disease-associated genes; CACNA1A = calcium channel, voltage-dependent P/Q type, α1A subunit; TBP = thymine-adenine-thymine-adenine (TATA) box-binding protein; SCA = spinocerebellar ataxia
aRange if CAT trinucleotide repeat interruptions are present
bCould be interrupted by 1-4 CAA trinucleotide repeats
SCA1
The prevalence of SCA1 worldwide is around 1 to 2 per 100,000 individuals [50]. A large variance exists in the number of individuals with ADCAs that are confirmed to be SCA1. The prevalence ranges from no cases reported in a study among Korean patients [51] to essentially 100% of all patients in an Eastern Siberian study, where SCA1 was the only type of ADCA identified in the ethnic Sakha population [52].
The main symptom in SCA1 is progressive cerebellar ataxia characterized by disturbances in balance and gait. Oculomotor movements are also affected [53, 54]. Furthermore, patients frequently suffer from pyramidal, extrapyramidal, and bulbar symptoms [1]. As the disease advances, muscle atrophy arises. Cognitive symptoms have been reported in the final stages of the disease with impaired executive function being the most common defect [55]. Disease onset is usually in the third or fourth decade of life and shows a more rapid disease progression than the other polyQ SCAs. Affected individuals eventually develop respiratory failure, which is the main cause of death [56]. Interval from onset to death varies from 10 to 30 years. Individuals with juvenile onset show a more rapid disease progression and more severe disease [17]. The neuropathology in SCA1 shows severe atrophy of the cerebellum and brainstem. In addition, degeneration of the frontal, temporal, and parietal cerebral lobes is seen with the basal ganglia, midbrain, and thalamus also affected [57].
SCA1 is caused by an expanded CAG repeat in the ATXN1 gene [21]. Normal alleles have 6 to 39 repeats, while pathological repeats have 39 to 91 repeats [21, 22]. Ataxin-1 is a protein of 98 kDa, and research has shown that this protein is required for cognitive function, motor coordination, as well as the processing of β-amyloid precursor protein (APP) [58–60]. Furthermore, ataxin-1 can act as a transcriptional repressor [61–63]. The ataxin-1 protein is involved in transcriptional regulation and RNA metabolism [64] and has a role in extracellular matrix (ECM) remodeling through formation of a transcriptional repressor complex with Capicua [65].
SCA2
Around 0.1 to 5.8 per 100,000 people suffer from SCA2 [66] with 6% to 33% of ADCA patients being diagnosed with SCA2. Lowest percentages were reported in Japan (5.9%) [67], while the highest percentages were reported in Korea (33%) [68].
SCA2 is characterized by progressive cerebellar ataxia, dysarthria, and oculomotor deficits, including abnormally slow saccades and nystagmus [69]. Patients can also suffer from intention and postural tremors, myoclonus, Parkinsonism, sleep disturbances, autonomic dysfunction, and initial hyperreflexia followed by peripheral neuropathy with hyporeflexia or areflexia [70]. In addition, different studies have reported late cognitive deficits [71] as well as psychiatric symptoms [72]. The onset of symptoms is usually in the fourth decade of life with a disease duration of 10 to 15 years [73]. Onset before the age of 20 years correlates with a more aggressive disease course [74]. SCA2 presents with atrophy of the cerebellum characterized by severe loss of cerebellar Purkinje cells (PCs) while the deep cerebellar nuclei (DCN) are relatively spared. Other major affected brain regions are the pons, medulla oblongata, and spinal cord. Pronounced neuronal loss is also seen in the cerebral cortex, basal forebrain, basal ganglia, and midbrain [57].
SCA2 is caused by an unstable expansion of a CAG/CAA repeat in the ATXN2 gene [75, 76]. Normal alleles have 14 to 31 repeats, while pathological repeats have 33 to 500 repeats [23–25, 75, 76]. The protein associated with SCA2 is the ataxin-2 protein. Ataxin-2 is a protein of 145 kDa with its main function in RNA metabolism, regulation of translation, stress granule formation, and P-body formation [77]. Of particular interest are the interactions of ATXN2 with polyadenylate-binding protein (PABP) and transactive response (TAR) DNA-binding protein 43 (TDP43), each of which also binds directly to RNA. One hypothesis is that the polyQ tract length in ataxin-2 impairs interactions with PABP [78] and TDP43, thereby contributing to SCA2 pathogenesis, as well as the risk of amyotrophic lateral sclerosis [79].
SCA3/Machado–Joseph Disease
SCA3, also known as Machado–Joseph disease (MJD), is, in many populations, the most common polyQ SCA with an estimated prevalence of 1 to 2 per 100,000 individuals with significant geographical and ethnic variations. Prevalence differs greatly per population investigated with percentages of SCA3 diagnosed patients ranging from 0 to 1% to 58% among ADCA patients. The lowest percentages were reported in Italy (approximately 1%) [13] and Finland (~0%) [80], while the highest percentages were seen in Taiwan (47.3%) [81] and Portugal (57.8%) [82].
General SCA3 symptoms include oculomotor symptoms, cranial nerve deficits, sleep disturbances, involuntary weight loss, and autonomic problems [83]. Furthermore, SCA3 patients are known to suffer from mild cognitive impairments [84]. Disease duration is reported to range between 6 and 29 years [85]. Clinically, SCA3 can be further divided into 4 subtypes [83, 86]. SCA3 type 1 has an early age of onset between 10 and 30 years and is characterized by pyramidal and extrapyramidal symptoms with cerebellar ataxia being less prominent. Type 2 is the most common SCA3 subtype with an age of onset between 20 and 50 years. This type involves distinct cerebellar ataxia, dysarthria, and pyramidal symptoms. Age of onset of type 3 is typically later (40–75 years). Symptoms include cerebellar ataxia, but additionally, peripheral neuropathy leading to muscle atrophy and areflexia can be present. Type 4 has a variable age of onset and is characterized by Parkinsonism [87].
SCA3 affects various different regions of the brain and spinal cord, although the cerebellar cortex and olivary nuclei are relatively spared. The brain weight of SCA3 patients is significantly reduced, and neuronal cell loss is most prominent in the cerebellar dentate nucleus and substantia nigra. Neurodegeneration can also be seen in the cerebral cortex, pons, medulla oblongata, thalamus, and basal ganglia. In the spinal cord, the dorsal root ganglia, dorsal nuclei, and anterior horn are affected [57, 88].
SCA3 is caused by a CAG trinucleotide repeat expansion in the ATXN3 gene [26]. Normal alleles range from 12 to 44 CAG repeats, while pathological alleles have 60 to 87 CAG repeats [26, 27]. The approximately 42 kDa SCA3-associated protein, the ataxin-3 protein (ATXN3), is ubiquitously expressed. This protein contains an N-terminal Josephin domain that displays ubiquitin protease activity and a C-terminal tail with several ubiquitin interacting motifs. ATXN3 functions mainly as a de-ubiquitinating enzyme [89].
SCA6
The overall prevalence of SCA6 is between 0.02 and 0.031 per 100,000 individuals [90]. Around 12% of the families with ADCAs are diagnosed with SCA6 with the lowest percentage reported in Germany (1%) [91] and the highest in Japan (31%) [92].
SCA6 is considered a pure cerebellar ataxia. Primary symptoms include slow progressive cerebellar ataxia, dysarthria, and nystagmus. In 40 to 50% of the patients, pyramidal symptoms such as hyperreflexia and extensor plantar responses have been noted. Furthermore, signs of basal ganglia involvement including dystonia and blepharospasm are reported in 25% of the cases. Cognitive function is usually intact. The mean age of onset of SCA6 is between 43 and 52 years (range 19–71 years). Although the disease causes a high morbidity, the lifespan of patients is usually unaltered [93, 94].
In SCA6, widespread neurodegeneration comparable to that of SCA1, SCA2, SCA3, and SCA7 is present, but less severe. Degeneration mainly occurs in the cerebellum. Particularly, the cerebellar Purkinje cells are affected [95]. Microscopic pathology is more widespread in the thalamus, midbrain, pons, and medulla oblongata [57].
SCA6 is caused by a CAG expansion in the CACNA1A gene [28]. Normal alleles range from 4 to 18 CAG repeats, while expanded repeats range from 20 to 33 CAGs [28–31]. CACNA1A encodes 2 structurally unrelated proteins with distinct functions: the α1A subunit of P/Q-type voltage-gated calcium channel and α1ACT, a newly recognized transcription factor, with the polyglutamine repeat at the C-terminal end [96]. The α1A subunit of P/Q-type voltage-gated calcium channel is highly expressed in the Purkinje cells [97].
SCA7
The prevalence of SCA7 is less than 1 per 100,000 individuals [11, 98]. Adult-onset SCA7 is characterized by progressive cerebellar ataxia, slowed ocular saccades, dysarthria, dysphagia, and pyramidal symptoms, such as hyperreflexia and spasticity [99]. Retinal degeneration is a distinctive feature of SCA7 with progressive cone-rod dystrophy leading to eventual blindness [100]. A decline in cognitive function and episodes of psychosis have also been reported [101]. The mean age of onset is about 32 years (range of 1–72 years) [102]. Clinical features differ between patients with an adult onset and patients with an onset in infancy or early childhood with the disease starting in childhood showing a more rapid and aggressive progression within a few years. When disease symptoms first appear later in life, the disease progresses more slowly and the degree of disability will vary accordingly [103].
In SCA7, extensive degeneration is observed in the retina. Furthermore, severe atrophy of the cerebellum and the brainstem is observed as well as neurodegeneration in the cerebral cortex, basal ganglia, thalamus, and midbrain [57].
SCA7 is caused by a CAG repeat expansion in the ATXN7 gene [32]. Normal SCA7 alleles range from 4 to 17 CAG repeats. Alleles bearing 28 to 33 repeats can give rise to disease alleles but are not associated with a SCA7 phenotype. Disease alleles with 34 to 460 CAG repeats have been reported [32–35]. The expanded polyQ protein in SCA7, the ataxin-7 protein (ATXN7), was found to associate with microtubules and to be involved in the cytoskeletal regulation [104]. Furthermore, this protein is an integral component of the mammalian SPT3-TAF9-ADA-GCN5 acetyltransferase (STAGA) transcription coactivator complex and mediates the direct interaction of STAGA with the CRX transactivator of photoreceptor genes [105].
SCA17
Worldwide, less than 100 families with SCA17 have been reported and the prevalence is less than 1 in 100,000 individuals [106, 107].
SCA17 is also known as Huntington disease-like 4, and symptoms of this disease indeed resemble Huntington disease (HD). SCA17 presents itself with cerebellar ataxia, pyramidal signs, and involuntary movements including chorea and dystonia. Parkinsonism has also been reported. Furthermore, SCA17 patients are known to suffer from psychiatric symptoms, as well as dementia [108, 109]. The mean age of onset is 35 years with a broad range of 3 to 75 years, and the mean disease duration of about 20 years [102, 110].
SCA17 shows a mild global atrophy of the brain. Neurodegeneration is seen in the cingulate and para-hippocampal gyri, striatum, ventral thalamic nuclei, cerebellar Purkinje cell layer and dentate nucleus, substantia nigra, and inferior olive [57].
SCA17 is caused by an expanded CAG repeat in the TATA-binding protein (TBP) gene [111]. The repeat in TBP can generally be divided into 5 regions including 2 polymorphic (CAG)n stretches: (CAG)3 (CAA)3′(CAG)n′CAA CAG CAA′ (CAG)n′CAA CAG where the CAA triplets also code for a glutamine amino acids [112]. Normal SCA17 alleles contain 25 to 40 repeats while alleles with 41 to 49 repeats have incomplete penetrance. Alleles with 50 or more repeats were reported to have a full penetrance [36–38]. The SCA17-associated protein TBP is an important component of the initiation complex of eukaryotic RNA polymerases [113]. TBP is 1 of several transcription factors that form a preinitiation complex with RNA polymerase II, and this step is essential for initiating transcription [112].
Therapeutic Progress
Currently, no curative treatment is available for any of the (polyQ) SCAs. However, several compounds and treatment strategies that aid in improving the quality of life exist. Readers are directed to more comprehensive reviews on the symptomatic treatment elsewhere [43, 114–117]. The aim of this review is to provide an overview of potential treatments for the polyQ SCAs that can be divided into pharmacological and gene therapies that target the toxic downstream effects, gene therapies that target the polyQ SCA genes, and stem cell replacement therapies.
Pharmacological Therapies
Increasing knowledge on the pathological pathways of the polyQ SCAs has provided promising therapeutic targets to potentially slow disease progression. Since the polyQ SCAs are rare, targeting a disease mechanism that is present in several polyQ SCAs would provide a therapeutic approach applicable to more than 1 disease. However, therapeutic targets that are more specific to particular polyQ SCAs are also discussed (Table 2).
Table 2.
Pharmacological compounds
| Compound | Mechanism | Model system | Outcome | Reference |
|---|---|---|---|---|
| Dantrolene | Stabilization of intracellular calcium signaling | MJD84.2 SCA3 mouse | Prevented neuronal cell loss and improved motor phenotype | [118] |
| Temsirolimus | Autophagy induction | SCA3 mouse line 70.61 | Reduces the number of aggregates in the mouse brain, decreases levels of cytosolic soluble mutant ataxin-3, and improves motor performance | [119] |
| Sodium butyrate | HDAC inhibitor, reversal of transcriptional downregulation | Ataxin-3-Q79 transgenic mice | Reversed histone hypoacetylation/transcriptional, ameliorated neurological phenotypes, and improved survival | [120] |
| H1152 | Rock inhibitor, ataxin-3 downregulation | Ataxin-3-Q79 transgenic mice | Reduction mutant ataxin-3 protein level in the brain, improved motor phenotype | [121] |
| Caffeine | Nonselective adenosine receptor antagonist | Lentiviral-induced SCA3 mice | Reduction in ataxin-3 inclusions, cell injury, and striatal degeneration | [122] |
| 17-DMAG | Hsp90 inhibitor but may act through autophagy induction | CMV MJD135 SCA3 mice | Delay in motor deficit progression and rescue coordination deficit | [123] |
| Lithium chloride | Autophagy induction | CMVMJD135 SCA3 mice | No overall beneficial effects | [124] |
| Citalopram | Serotonin reuptake inhibitor | CMVMJD135 SCA3 mice | Reduced ataxin-3 neuronal inclusions and ameliorated motor symptoms | [125] |
| Valproic acid | HDAC inhibitor | CMVMJD135 SCA3 mice | Limited effects on motor deficits; no effects on ataxin-3 inclusions | [126] |
| Combination of temsirolimus and lithium chloride | Autophagy induction | CMVMJD135 SCA3 mice | Deleterious effect; no improvement in neurological symptoms; induced neurotoxicity induced | [127] |
| Riluzole | Inhibition of glutamate release | Inducible SCA3 mouse | No improvement in motor deficits | [128] |
| Caloric restriction or resveratrol | SIRT1 activation, autophagy activation | Lentiviral-induced SCA3 mice | Amelioration of motor deficits and neuropathology | [129] |
| Lithium chloride | Induction autophagy + inhibition of GSK3β activity | SCA3 Drosophila | Prevented eye depigmentation, alleviated locomotor disability, and extended lifespan | [130] |
| Calpeptin | Inhibition of calpain cleavage | SCA3 iPSC model | Prevented ataxin-3 aggregate formation in neurons | [49] |
| Interferon-β | Induces expression PML protein, degrades mutant ataxin-7 | SCA7(266Q/5Q) knock-in mice | Reduction of mutant ataxin-7 in neuronal inclusions; improvement in motor coordination | [131] |
| Granulocyte–colony-stimulating factor | Upregulating chaperones and autophagy | SCA17 mice | Improved motor coordination; reduced cell loss | [132] |
| Lithium carbonate | Speculated to correct gene expression changes | SCA1 mice (154Q) | Slowed neurodegeneration and improved motor coordination but did not improve lifespan | [133] |
Protein Clearance Potentiation
Cellular homeostasis is maintained through a delicate balance between protein synthesis and protein degradation. Neurons are particularly dependent on efficient protein degradation mechanisms, 1 of which is autophagy [134]. Indeed, autophagy is essential for maintaining neuronal health, and autophagic dysfunction is involved in many neurodegenerative disorders [135] since loss of autophagy can result in disruption of neuronal function and neurodegeneration [136]. For the polyQ SCAs, the misfolded mutant polyQ proteins present a major challenge to cellular protein quality control systems. For this reason, enhancing autophagy is considered a viable therapeutic strategy, and a variety of compounds have been tested to this end for the SCAs as well as several other neurodegenerative diseases [137, 138].
Pharmacological Stimulation of Autophagy
One compound that has been extensively researched is lithium. Lithium has pleiotropic effects and can act on multiple pathways, but autophagic potentiation is 1 of the effects researched in the context of neurodegeneration. For instance, lithium chloride has been investigated in Alzheimer disease, amyotrophic lateral sclerosis, and Parkinson disease (PD) [139]. For SCA3, lithium chloride was tested in a Drosophila model, where the eye depigmentation and locomotor disability were successfully reduced. The observed effect was in part attributed to GSK3β/shaggy inhibition, which, in turn, perturbs the Wnt pathway [130]. However, the authors concluded that lithium clearly had additional beneficial effects that could not be solely explained by changes in this pathway [130]. In SCA1 knock-in (KI) mice, lithium treatment reduced neurodegeneration, improved motor symptoms, and restored cerebellar ascorbate levels [133, 140]. However, other studies have questioned the suitability of lithium for treatment of the polyQ SCAs. For instance, in SCA1 mice, life expectancy of mice was not improved despite improved motor function [133], suggesting that the underlying pathology was not sufficiently addressed through lithium treatment. Other research in SCA3 mice was unable to show any beneficial effect of lithium on a range of behavioral tests altogether, despite activation of autophagy [124]. Finally, a phase 2, double-blind, placebo-controlled trial testing lithium carbonate in 62 SCA3 patients was performed. The investigators concluded that lithium was well tolerated, but no effect on the Neurological Examination Score for Spinocerebellar Ataxia (NESSCA) was detected [141]. A clinical trial with lithium was also performed in 20 SCA2 patients, where only a significant effect on the Beck Depression Inventory could be established, but not on the Scale for the Assessment and Rating of Ataxia (SARA) score [142]. Based on these clinical trials, lithium treatment of the polyQ SCAs is currently not very well supported. More and larger clinical trials would be required to give a definitive decision on the suitability of lithium for SCA treatment [143], though major beneficial effects of lithium treatment for SCAs seem unlikely based on current knowledge.
Though lithium has thus far not been very successful as potential SCA treatment, potentiation of autophagy may still be a useful therapeutic strategy. Other compounds and means to stimulate autophagy have also been tested in preclinical SCA research. For instance, temsirolimus, through inhibition of mammalian target of rapamycin (mTOR), can potentiate autophagy and reduce toxicity in HD mouse models [144]. Temsirolimus was administered to SCA3 mice, where an improvement in motor phenotype was established and the number of aggregates in the mouse brain was also reduced [119]. However, when tested in combination with lithium chloride, no improvement in any parameter could be seen in SCA3 mice. In fact, only deleterious neurotoxic effects of the treatment were seen [127]. Whether the negative treatment effect was due to a toxic interactive effect of the 2 drugs or, potentially, an excessive degree of autophagic potentiation remains unclear [127]. To our knowledge, no clinical trials with temsirolimus have been performed for any of the SCAs.
Other Strategies to Induce Autophagy
Apart from the more traditional small molecule-based therapeutics, autophagy can be stimulated through other, more indirect methods. For instance, caloric restriction is a potent inducer of autophagy and is hypothesized to be a potential strategy to delay neurodegenerative disease progression [145]. Caloric restriction indeed resulted in marked improvement in motor phenotype and neuropathology when performed in SCA3 mice [129]. The authors also show that the disease alleviation is mediated by the SIRT1 protein, a deacetylase found to slow aging [129]. Trehalose, an alpha-linked disaccharide, has been shown to clear accumulated proteins by activating autophagy, primarily in an mTOR-independent manner [146]. In SCA17 cell models and a SCA17 mouse model, nuclear protein aggregation was significantly reduced after trehalose treatment. Furthermore, gait behavior and motor coordination were significantly improved in the trehalose-treated SCA17 mice [147]. Moreover, in SCA3 cell models, protein aggregation was significantly prohibited by trehalose analogues [148]. A phase 2, open-label clinical trial has being conducted in SCA3 patients, but no results have been published yet.
Lastly, Beclin-1, an important initiator protein of autophagy [149], was found to be reduced in a SCA3 rat model and brain material from patients [150]. Using lentiviral expression, the researchers overexpressed Beclin-1, resulting in autophagic flux. As a result, mutant ataxin-3 was cleared more efficiently and neuroprotective effects were established [150]. Research using the Hsp90 inhibitor 17-DMAG in the CMV MJD mouse model for SCA3 resulted in rescue of the neuropathology and accompanying alleviation of the motor deficits. Somewhat unexpectedly, researchers did not find evidence for induction of heat shock proteins but instead found induction of Beclin-1. It was hence speculated that the beneficial effect of 17-DMAG may occur through autophagic potentiation [123]. Similarly, granulocyte–colony-stimulating factor was assessed as a potential therapy in SCA17 mice. The treatment reduced insoluble mutant TBP protein, and reduced Purkinje cell loss, resulting in improvement of the motor phenotype. As Beclin-1 and other autophagic factors were increased, this suggests that the beneficial effect was mediated through autophagy [132].
Efforts have also been made to potentiate proteasomal degradation of mutant polyQ proteins, such as through treatment with the ROCK inhibitor H1152. In SCA3 mice, intraperitoneal injection of H1152 successfully improved the neurological phenotype of the mice through reduced mutant ataxin-3 protein levels in the cerebellum, pontine nuclei, and the spinal cord [121]. Taken together, many efforts have been made to stimulate autophagy and protein clearance in an effort to diminish toxicity of the mutant proteins underlying the SCAs. Though success has certainly been achieved in cell and animal models, translation toward a suitable clinical drug is still lacking. This is in part due to issues with the compounds tested so far that are, in many cases, not suitable for direct use in patients and are also not specific for the expanded polyQ proteins. Additionally, the models used in the study to test these compounds may overestimate the contribution of autophagy due to overexpression of the mutant proteins with very long repeat sizes.
Inhibiting Generation of Toxic Protein Fragments
For several of the polyQ disorders, a body of evidence exists supporting the notion that generation of intracellular toxic protein fragments is involved in the pathogenicity. The general understanding is that proteolytic cleavage of the mutant polyQ protein results in generation of short polyQ-containing protein fragments that result in a greater toxicity than the intact full-length protein. Evidence for this hypothesis has been found for HD [151–153], SCA3 [49, 154–158], SCA6 [159], and SCA7 [160–162].
A relation between proteolytic cleavage and generation of aggregates has been shown most convincingly for SCA3. Inhibition of the proteolytic enzyme calpain-2 in cell culture experiments was sufficient to prevent not only generation of the polyQ cleavage fragments but also the aggregation of mutant protein [163]. The cleavage of mutant ataxin-3 is likely to initiate the aggregation, after which both expanded and nonexpanded ataxin-3 are sequestered into the aggregates [164]. This observation was reproduced in SCA3 neurons derived from induced pluripotent stem cells (iPSCs), in which calpains were determined to be essential for ataxin-3 cleavage and subsequent induction of polyQ aggregates [49]. Although the aggregates are a hallmark of the diseases, they are not clearly correlated with cellular toxicity. Convincing evidence exists that at least the short polyQ-containing fragments generated through proteolytic cleavage are toxic [154]. For these reasons, preclinical research has been performed to find strategies capable of preventing generation of these toxic polyQ protein fragments. One obvious way to prevent generation of the toxic fragments is through inhibition of the proteolytic enzymes responsible for the cleavage of the polyQ-containing proteins.
In this context, the orally administered calpain inhibitor BDA-410 was tested during an 8-week period in a mouse model for SCA3. This treatment resulted in a 38% reduction in the number of mutant ataxin-3 inclusions in the striatum. Importantly, cerebellar cell loss was prevented, and an improvement in the motor phenotype of the mice could be achieved [158]. A second well-investigated way to inhibit calpains is by treatment with calpeptin, a cysteine protease inhibitor. In vitro experiments revealed that calpeptin treatment was able to prevent generation of ataxin-3 cleavage fragments and aggregates in iPSC-derived SCA3 neurons, proving the validity of this approach [49]. Furthermore, calpeptin proved protective in a SCA3 zebrafish model by induction of autophagy, which led to complete removal of the ataxin-3 protein [165]. Additionally, when overexpressing the endogenous calpain inhibitor calpastatin, ataxin-3 fragmentation was prevented and neuroprotective effects were established in a SCA3 mouse model [166]. This strategy was further supported by an additional study where knockout of calpastatin led to increased nuclear aggregates and aggravated neurodegeneration in SCA3 mice [156]. For HD, results from mouse models also indicated that inhibition of calpain cleavage can alleviate cellular toxicity of the polyQ protein and that this approach may thus constitute a viable therapy for polyQ disorders [167].
Despite these promising preclinical research efforts, no inhibitors preventing the formation of the toxic polyQ fragments have been assessed in clinical trials. One of the issues is that inhibiting a proteolytic enzyme will not only affect the targeted polyQ protein but will also affect the proteolytic cleavage of a wider range of proteins, and this could have unwanted side effects. Furthermore, for most of the polyQ proteins, the proteolytic enzymes that are involved in mediating toxicity for the various polyQ disorders are not conclusively identified [168]. For calpains, establishing isoform-specific inhibitors that are required to prevent side effects have been proven difficult when moving toward the clinic [169]. Similarly, caspase inhibitors have been investigated for clinical application, but many studies have been suspended due to side effects. As such, the path to caspase-targeted therapy is currently not clear [170]. Future developments may provide compounds capable of achieving isoform specificity for the various proteolytic enzymes, in turn providing better feasibility to the cleavage inhibition strategy as a potential treatment for the polyQ SCAs.
Correcting Transcriptional Dysregulation
One well-established effect of the mutant polyQ proteins is that these proteins lead to transcriptional dysregulation. Many of the polyQ-containing proteins function as transcription factors, and evidence for a regulatory role of the polyQ repeat in transcription factors has been found [171]. This implicates that abnormal elongation of the repeat hampers this function, leading to alterations in transcription of target genes. Also, co-aggregation of other transcription factors that harbor a polyQ stretch, such as CBP [172], could contribute to this transcriptional dysregulation. Indeed, ataxin-1 knockout mice revealed overlapping alterations in cerebellar gene expression changes with Atxn1 154Q SCA1 mice, indicative of a loss-of-function effect [59]. In contrast, a similar study comparing SCA3 transgenic mice with ataxin-3 knockout mice did not find an overlap in transcriptional alterations [173]. Especially, the shorter polyQ fragments generated through proteolytic cleavage are known to translocate to the nucleus, where these fragments may interfere with transcription [174]. For instance, polyQ bound to factors such as TAFII130 interferes with CREB-dependent transcription [175]. Additionally, expanded TBP showed enhanced interaction with general transcription factor IIB, leading to downregulation of neuroprotective factor heat shock protein HSPB1 in SCA17 [176]. As such, a range of compounds aimed at reversing the transcriptional abnormalities has been tested (reviewed in [177]).
Histone acetylation and deacetylation is a process critical for the regulation of gene expression. As such, a study with transgenic SCA3 mice revealed that inhibiting histone deacetylases (HDACs) through sodium butyrate treatment was able to reverse the pathogenic histone hypoacetylation and transcriptional downregulation in the cerebellum of the mice [120]. Additionally, the motor symptoms were successfully ameliorated through sodium butyrate treatment [120]. Similar observations were made in SCA3 cell models, where the HDAC inhibitors valproic acid [178] as well as divalproex sodium [179] and sodium valproate [180] attenuated cellular toxicity induced by mutant ataxin-3. For SCA7, the HDAC inhibitor trichostatin A was tested in astrocytes expressing mutant ataxin-7 and was able to partially restore aberrant transcription of reelin, a factor involved in synaptic plasticity [181]. Based on the promising effects in cell and animal models, a randomized, double-blind, placebo-controlled trial was performed in 36 SCA3 patients to test the efficacy of the HDAC inhibitor valproic acid. Daily dosing of valproic acid was performed during 12 weeks, and at the end of the study, a significant improvement in the SARA score was observed. Some adverse effects related to dizziness and appetite were noted, but the overall conclusion was that valproic acid is a potentially useful treatment for SCA3 and that further clinical research is warranted [182]. Recent preclinical research has, however, called into question the suitability of valproic acid, as this HDAC inhibitor did not improve motor phenotype or the ataxin-3 inclusions and astrogliosis in the CMV MJD135 mouse model of SCA3 [126]. Taken together, therapies aimed at correcting the transcriptional dysregulation in the polyQ SCAs are subject to several limitations. Firstly, it is not currently established to what extent the transcriptional dysregulation is a key component of polyQ toxicity and whether targeting this pathway is enough to achieve clinically meaningful effects. Secondly, the currently tested drugs have pleiotropic effects and potential long-term side effects have not yet been adequately established.
Other Pharmacological Targets
PolyQ disorders have a complex pathology where many cellular processes are suggested to be involved. As such, therapeutic interventions in cell and animal models targeting a range of different pathways have been suggested (reviewed in [183]). One approach has been to correct aberrant neuronal calcium signaling, a pathway suggested to be effected in multiple neurodegenerative diseases, including the polyQ disorders [184]. In SCA3, mutant ataxin-3 is able to associate with an intracellular calcium release channel. Treatment of transgenic SCA3 mice with dantrolene, a stabilizer of calcium signaling, resulted in improved motor performance and decreased neuronal cell loss [118]. However, in the treatment of the polyQ SCAs, few other studies have been performed regarding calcium signaling stabilization, and more evidence in favor of this therapeutic strategy is required before pursuing clinical trials.
Another interesting pharmacological approach for the treatment of polyQ SCAs has been resveratrol, a compound that is thought to activate SIRT1. Treatment of SCA3 mice with resveratrol improved the motor function [129]. Although the neuroprotective effect of SIRT1 activation is not fully understood, the neuroprotection could result from inhibition of neuro-inflammation and induction of autophagy [129]. Some clinical benefit of resveratrol has been observed in an open label study of Friedreich ataxia, indicating that further clinical research of resveratrol for treatment of SCA is warranted [185].
A third approach with some preclinical success is treatment with caffeine. In a SCA3 mouse model expressing mutant ataxin-3 in the striatum of 1 hemisphere, caffeine was tested by administration through the drinking water. The neuronal dysfunction and damage was successfully reduced in mice that received caffeine [122]. Later, the same research group tested caffeine in a different SCA3 mouse model, where alleviation of the behavioral deficits was achieved [186]. The mechanism is thought to involve adenosine receptor antagonism, as these receptors are involved in synaptic viability, neuroinflammation, and neuronal apoptosis [122].
Activation of serotonergic signaling has also been tested as potential treatment for SCA3. The serotonin reuptake inhibitor citalopram was recently identified in a small molecule screening as a compound capable of reducing aggregation of mutant ataxin-3 in a Caenorhabditis elegans model [125]. When tested in the CMV MJD135 mouse model of SCA3, citalopram strikingly reduced the motor symptoms, as well as the molecular hallmarks of SCA3, such as neuronal inclusions and astrogliosis in the mouse brain [125]. The mechanism behind this treatment may be the same as identified in HD models, where serotonin reuptake inhibition led to increased levels of brain-derived neurotrophic factor (BDNF) and enhanced neurogenesis [187].
Furthermore, the anti-glutamatergic compound riluzole was identified to improve ataxia symptoms in a randomized, double-blind, placebo-controlled pilot trial with 40 ataxia patients of different etiologies [188]. Given the proposed role of glutamatergic signaling for the induction of toxicity in SCA3 [49], riluzole was assessed in a SCA3 mouse model [128]. However, treatment with riluzole in the drinking water during 10 months did not provide an improvement in the rotarod performance, home cage activity, or body weight [128]. Surprisingly, riluzole-treated animals showed a higher number of damaged Purkinje neurons. These paradoxical results were hypothesized to be due to an excessive influx of Ca2+, in turn inducing increased ataxin-3 protein accumulates that are toxic to the Purkinje cells. Together, these results indicate that caution should be taken when further assessing clinical application of riluzole for the polyQ SCAs [128].
It was also shown that interferon-β treatment was capable of inducing clearance of ataxin-7 in a knock-in mouse model, resulting in improved motor function. Interferon-β induces expression of promyelocytic leukemia (PML) protein, resulting in formation of PML nuclear bodies and decreased neuronal inclusions of ataxin-7. It was therefore concluded that interferon-β is a promising molecule requiring further investigation into its suitability in treatment of SCA7 and the other polyQ disorders [131].
Gene Therapies
Knockdown or modification of the polyQ genes by RNA interference (microRNA (miRNA), short hairpin RNA (shRNA), and small interfering RNA (siRNA)), antisense oligonucleotides, or DNA editing techniques (clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated nuclease 9 (Cas9)) can reduce the levels of mutant expanded polyQ proteins, which will have an effect on all downstream pathological pathways (Fig. 2). Specifically targeting the disease-causing genes has the advantage of reducing the chance of nonspecific side effects but will require a unique approach for every disease. Here, we will provide an overview of the different gene therapies which are currently explored in the polyglutamine SCA field (Table 3).
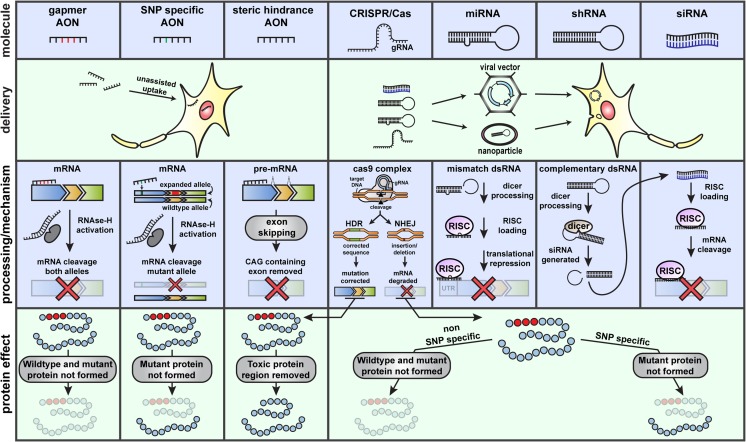
Fig. 2.
Potential genetic therapies for the polyQ SCAs. Several different nucleic acid-based molecules (top panel) are available to target the RNA or DNA of the polyQ-associated genes. The different therapeutic molecules differ in their chemical composition, delivery method, and functional mechanism. AONs can be delivered to the central nervous system as naked molecules, since their distribution, uptake, and stability in this context are excellent. CRISPR/Cas and double-stranded RNA molecules require supportive delivery methods, such as viral vectors or lipid nanoparticles. Assisted delivery of these types of molecules can be performed using different viruses, where nonintegrative gene therapy vectors based on AAV are usually preferred to avoid random integration and mutagenesis. AONs can be used to induce mRNA degradation (gapmer AONs), which activate RNase H due to formation of an RNA–DNA hybrid. Alternatively, fully 2′O-modified AONs do not activate RNase H and can be implemented to affect splicing and remove the CAG-containing exon. Downregulation of target transcripts can also be achieved through miRNA, shRNA, or siRNA. miRNA is generally designed with a mismatch, resulting in translational inhibition. shRNA and siRNA act through the same RISC pathway to degrade target mRNA. CRISPR/Cas is the most recent genetic therapy and the only strategy listed here that is able to target DNA. It can be used to inhibit expression by introducing insertions/deletions through nonhomologous end joining (NHEJ) or can introduce a corrected DNA sequence through homology-directed repair (HDR). In principle, all the mentioned molecules can be used to target SNPs associated with the pathogenic allele, resulting in downregulation or correction of the mutant allele. AON = antisense oligonucleotide, CRISPR/Cas = clustered regularly interspaced short palindromic repeats/CRISPR-associated nuclease, dsRNA = double-stranded RNA, gRNA = guide RNA, HDR = homology-directed repair, miRNA = microRNA, mRNA = messenger RNA, NHEJ = nonhomologous end joining, RISC = RNA-induced silencing complex, shRNA = short hairpin RNA, siRNA = small interfering RNA, SNP = single nucleotide polymorphism
Table 3.
Gene therapies
| SCA | Mechanism | Model system | Outcome | Reference |
|---|---|---|---|---|
| SCA1 | miRNA: miR-19, miR-101, and miR-130 | MCF7 and HEK293T cells | ± 60% reduction of ataxin-1 protein | [189] |
| SCA1 | miRNA: miR-144 and miR-101 | HEK293T cells | 20–30% reduction of ataxin-1 protein | [190] |
| SCA3 | miRNA: ban miRNA | Drosophila | Suppression of neurodegeneration | [191] |
| SCA3 | miRNA: miR-34 | Drosophila | Reduced inclusion formation; the protein retained greater solubility, and neural degeneration was suppressed | [192] |
| SCA3 | miRNA: mir-9, mir-181a, and mir-494 | SCA3 mice | Reduction of ATXN3 levels, aggregate counts, and neuronal dysfunction | [193] |
| SCA6 | miRNA: miR-3191-5p | SCA6 KI mice | Alleviation of motor deficits and Purkinje cell degeneration | [194] |
| SCA7 | miRNA: miR-124 | N2A cells and SCA7 mice | ± 80% reduction of ataxin-7 | [195] |
| SCA1 | shRNA: ataxin-1 downregulation | SCA1 mice | No efficiency data on RNA or protein level; improved motor coordination, restored cerebellar morphology, and resolved characteristic ataxin-1 inclusions in Purkinje cells | [196] |
| SCA1 | Artificial miRNA harboring a siRNA | SCA1 KI mice | 58–72% reduction of both ATXN1 and Atxn1; improvement of rotarod performance and neuropathology | [197, 198] |
| SCA1 | Artificial miRNA harboring a siRNA | B05 transgenic SCA1 mice | Improved behavior paradigms and neuropathology | [198, 199] |
| SCA1 | Artificial miRNA harboring a siRNA | Rhesus monkeys | ≥ 30% reduction of ATXN1 mRNA levels | [200] |
| SCA3 | Artificial miRNA harboring a siRNA | HEK293 cells and MJD84.2 SCA3 mice | ± 75% reduction of ataxin-3 levels (in vitro and in vivo); alleviation of nuclear accumulation of mutant ataxin-3 | [201] |
| SCA3 | Artificial miRNA harboring a siRNA | MJD84.2 SCA3 mice | Lifelong suppression of ATXN3 in the cerebellum; no mitigation of motor impairment and prolonged survival | [202] |
| SCA3 | siRNA | SCA3 mice | Reduction of both behavior deficits and neuropathology | [203] |
| SCA7 | Artificial miRNA harboring a siRNA | SCA7 mice | ≥ 50% reduction of mutant and wild-type ataxin-7 | [204] |
| SCA7 | Artificial miRNA harboring a siRNA | SCA7 mice | Improvement of ataxia phenotypes and a reduction in cerebellar molecular layer thickness and nuclear inclusions | [205] |
| SCA3 | siRNA: allele-specific downregulation | COS-7 and HeLa cells | > 90% reduction of mutant ataxin-3 and 25% reduction of WT ataxin-3 | [206] |
| SCA3 | siRNA: allele-specific downregulation | HEK293T cells | 96% reduction of mutant ataxin-3 and 6% reduction of WT ataxin-3 | [207] |
| SCA6 | siRNA: allele-specific downregulation | HEK293T cells | > 90% reduction of the mutant protein and no reduction of WT levels | [208] |
| SCA7 | siRNA: allele-specific downregulation | SCA7 patient-derived fibroblasts | More efficiently silencing of mutant transcript, but allele selectivity is lost at the highest dose of siRNA | [209] |
| SCA3 | shRNA: allele-specific downregulation | SCA3 rat model | Mitigated neuropathological abnormalities | [210] |
| SCA3 | shRNA: allele-specific downregulation | SCA3 mice | Alleviation of motor and neuropathological phenotypes | [211, 212] |
| SCA3 | AON: ataxin-3 downregulation | MJD84.2 SCA3 mice | 30% reduction mutant ataxin-3; over 75% reduction in ataxin-3 oligomers; strong improvement of motor phenotype | [213, 214] |
| SCA3 | AON: allele-specific downregulation by targeting CAG repeat | SCA3 fibroblasts | Complete downregulation of ataxin-3 protein, with preferential targeting of mutant protein | [215, 216] |
| SCA3 | AON and ss-siRNA: allele-specific downregulation by targeting CAG repeat | SCA3 fibroblasts | Complete downregulation of ataxin-3 protein, preferential targeting of mutant protein | [217] |
|
SCA1 SCA3 |
AON: allele-specific downregulation by targeting CAG repeat | Fibroblasts | Reduction of ATXN1 and ATXN3 mutant allele at RNA; other ataxin RNAs not tested | [218] |
| SCA2 | AON: ataxin-2 downregulation | ATXN2-Q127 and BAC-Q72 SCA2 mice | Up to 75% reduction of ataxin-2 protein in Purkinje cells of the mouse brain and significant improvement of motor phenotype | [219] |
| SCA7 | AON: allele-specific ataxin-7 downregulation | SCA7 fibroblasts | Mutant ataxin-7 reduced up to 50% and UCHL1 expression restored | [220] |
| SCA3 | CRISPR/Cas9 | Neurons derived from patient-specific iPSCs | Successful removal of polyQ-encoding region; the ubiquitin-binding capacity of ATXN3 was retained | [221] |
HEK = human embryonic kidney; MCF7 = Michigan Cancer Foundation-7; COS-7 = CV-1 in Origin Simian-7; miRNA = microRNA; siRNA = small interfering RNA; shRNA = short hairpin RNA; AON = antisense oligonucleotide
RNA Interference
RNA interference (RNAi) is a process of sequence-specific, post-transcriptional gene silencing initiated by double-stranded RNA that is homologous to the target gene, whereby RNA molecules inhibit gene expression or translation by targeting messenger RNA (mRNA) molecules. In 1998, Fire et al. [222] discovered that RNAi could be used to manipulate gene expression in C. elegans, and 3 years later, Elbashir et al. [223] extended this research to include mammalian cells. RNAi can be mediated through miRNA, siRNA, or shRNA. All types of RNAi can be used to achieve similar functional outcomes, but miRNAs, siRNAs, and shRNAs are functional different molecules (Fig. 2) [224–226]. Most of the protein-coding genes in the human genome are regulated by miRNAs, which can mediate both transcriptional gene activation (TGA) and transcriptional gene silencing (TGS) [227]. In short, after transcription, primary miRNA is cleaved by Drosha to form precursor miRNA (pre-miRNA). These pre-miRNAs are transported to the cytoplasm and processed by Dicer into a mature miRNA duplex. The miRNA antisense strand is loaded into the RNA-induced silencing complex (RISC). This active complex can scan for complementary mRNAs, which subsequently can be cleaved and degraded [228]. In case there is a single nucleotide mismatch in complementarity between the miRNA and its target, this will lead to translational repression [229] (Fig. 2). siRNAs are chemically synthesized double-stranded RNAs. Upon entering a cell, siRNAs are recognized by the RISC. Next, the siRNA is unwound to form single-stranded siRNA which is part of RISC, where it follows the same pathway as described before (Fig. 2). shRNAs are vector-based and can be delivered into mammalian cells through viral vectors or nanoparticles, allowing stable integration and long-term knockdown of the targeted gene. shRNAs consist of RNA sequences linked by a short loop. After transcription, the shRNA sequence is exported to the cytosol and recognized by Dicer. Dicer subsequently processes the shRNA into siRNA duplexes, and it follows a similar pathway as the chemically synthesized siRNAs [230] (Fig. 2).
miRNA-Based Therapies
In the polyQ SCA field, several miRNAs have been identified that regulate the polyQ genes causing SCA that could provide potential therapeutic targets [231]. By using miRNA target prediction databases, several evolutionarily conserved miRNA-binding sites in the 3′ UTR of the human ATXN1 gene were found [232]. These miRNAs were validated in MCF7 cells. Furthermore, inhibition of the miRNA-mediated post-transcriptional regulation of ATXN1 caused severe cytotoxicity in a SCA1 cell model [189]. Another SCA1 study found age-related differences in miRNA expression levels in the cortex and cerebellum of humans and nonhuman primates on a genome-wide scale. Moreover, this study found that miR-144 and miR-101 inhibition increased ATXN1 levels in a human cell model, concluding that activation of miRNA expression may protect from cytotoxicity caused by ATXN1 [190]. In Drosophila models, miRNA expression studies were performed for SCA1, SCA2, SCA3, and SCA7. Various miRNAs were found to be (almost) differentially expressed and could be potential targets for a miRNA-based therapy [191, 192, 233, 234]. In a SCA3 animal model and human neurons, 3 miRNAs were identified that interact with the ATXN3 3′ UTR and whose expression is dysregulated. Injecting lentiviral vectors encoding for these miRNAs in the striatum of 5-week-old SCA3 mice resulted in reduction of ataxin-3 levels and nuclear aggregates and improvement of neuronal dysfunction [193]. Similar results have been obtained with adeno-associated virus (AAV)-mediated delivery of miR-3191-5p in SCA6 mice. Viral delivery of this miRNA ameliorated behavioral deficits and Purkinje cell degeneration [194]. Finally, in a mouse neuroblastoma cell line and a SCA7 mouse model, it has been found that miR-124 mediates the interaction of lnc-SCA7 and Atxn7 transcripts. Delivery of miR124 mimics decreased the expression of Atxn7 [195]. Altogether, these findings suggest that dysregulation of miRNA pathways can be found in the polyQ SCAs and that restoring expression of these miRNAs can improve disease phenotypes. However, the fact that 1 miRNA can target many other transcripts [235] and cause unwanted off-target effects is a major drawback to take miRNA-based therapies to the clinic.
shRNA- and siRNA-Based Therapies
The first use of shRNA as gene-specific therapeutics in the polyQ SCA field was published in 2004. Upon intracerebellar injection of recombinant AAV vectors expressing shRNAs, cerebellar Purkinje cells were successfully transduced. No data on the percentage of reduction was shown, but treated animals showed improved motor function, restored cerebellar morphology, and resolved ataxin-1 inclusions in Purkinje cells in SCA1 mice [196]. Keiser et al. [197–199] took advantage of improvements in both expression systems and siRNA design and performed experiments in different SCA1 mouse models. Upon bilateral injection into the DCN, SCA1 mice showed improvement in both behavioral and neuropathological phenotypes [197–199]. Furthermore, after a single injection, adult rhesus macaque showed reduction of endogenous ATXN1 mRNA levels in the DCN, cerebellar cortex, inferior olive, and thalamus compared to the uninjected hemispheres. No clinical complications were observed, and quantitative and qualitative analyses suggest that this therapeutic intervention strategy and subsequent reduction of ATXN1 are well tolerated [200]. Altogether, these preclinical data are supportive of a clinical application of AAV-based siRNA therapy based on artificial miRNAs in SCA1 [196, 197, 199, 200]. For SCA3, AAV-mediated delivery of siRNAs targeting the 3′ UTR of ATXN3 to the cerebellum of a humanized SCA3 mouse model leads to gene silencing of human mutant ATXN3 [201, 202]. Short-term treatment cleared the nuclear accumulation of mutant ataxin-3 throughout the cerebellum [201], but long-term treatment did not reverse motor impairment or survival phenotypes [202]. These results suggest that targeting a large extent of the cerebellum may not be sufficient for an effective clinical trial in SCA3 patients [202]. Interestingly, Conceição et al. [203] used systemic administration of nonviral, stable nucleic acid lipid particles (SNALPs), incorporating a short peptide derived from rabies virus glycoprotein and encapsulating the siRNAs targeting the mutant ataxin-3. The authors reported efficient silencing of the mutant ataxin-3 and reduction of both behavior deficits and neuropathology in SCA3 mouse models. This study was the first to report that a noninvasive, systemic administration was beneficial in a preclinical study of a polyQ disorder [203]. A ≥ 50% reduction of both mutant and wild-type ataxin-7 is reported after viral siRNA delivery in a SCA7 mouse model. Although this mouse model expresses the mutant transgene in the retina, no obvious retinal phenotype is seen. Nevertheless, upon treatment, the normal retinal function is preserved and no toxicity has been reported [204]. Furthermore, this research group demonstrated a significant improvement of ataxia phenotypes and a reduction in cerebellar molecular layer thickness and nuclear inclusions [205], concluding that the reduction of ataxin-7 is well tolerated without any adverse toxicity [204, 205].
Above-described strategies are promising but have the disadvantage that both wild-type and mutant genes are simultaneously downregulated. A promising RNAi strategy to overcome this problem is to suppress the expression of only the expanded polyQ genes implicated in the polyQ SCAs. A selective reduction in the expression of the expanded genes can be achieved by exploiting differences in nucleotide sequence between the expanded polyQ alleles and the wild-type polyQ alleles. This strategy holds great promise for the treatment of polyQ SCAs, since the function of the wild-type allele is maintained [236]. In SCA3, a single nucleotide polymorphism (SNP) linked to the expanded polyQ allele [237] is used in this manner. Research demonstrated that in SCA3 cell models, allele-specific silencing of ATXN3 can be achieved by targeting this linked SNP by siRNAs [206, 207]. Studies in cell models for SCA6 and SCA7 showed that a similar siRNA approach was successful in the knockdown of these mutant transcripts. Moreover, there was only a modest reduction of the WT ataxin levels [208, 209]. Furthermore, allele-specific RNAi using shRNA showed improvement of motor and neuropathological deficits in SCA3 mouse and rat models [210–212]. Recently, the FDA approved the first-ever RNAi therapeutic, patisiran, for the treatment of polyneuropathy of hereditary transthyretin-mediated amyloidosis [238]. These results paved the way for the first clinical trial in polyQ SCA patients.
Antisense Oligonucleotides
One category of gene therapy that has made great progress over the last decades is antisense oligonucleotides (AONs). AONs are short, synthetic, single-stranded strings of nucleic acids of approximately 8 to 50 nucleotides in length [239]. AONs are designed to interfere with RNA through a variety of mechanisms, such as interfering with splicing, or downregulation through RNase H-mediated degradation of the target transcript (Fig. 2) [240]. For monogenetic disorders such as the polyQ SCAs, AONs are a promising therapeutic tool for a variety of reasons. Firstly, the genetic target for the polyQ SCAs is known, as the responsible gene and location of the toxic CAG repeat have been well described [40]. Secondly, recent years have shown great promise for AON therapies in application for neurodegenerative disorders. The first preclinical experiments using AONs to treat a neurodegenerative disorder were performed in 2006. AON treatment significantly slowed disease progression in a rat model of amyotrophic lateral sclerosis (ALS) caused by a SOD1 mutation [241], followed, 7 years later, by the first clinical trial of intrathecal delivery of an AON in SOD1 patients. No serious adverse events occurred in patients, and re-enrolment and retreatment were also well tolerated [242]. The successful delivery in the cerebrospinal fluid (CSF) and the outstanding safety profile paved the way for AONs as treatment for other neurodegenerative diseases. In 2009, experiments in an animal model for spinal muscular atrophy (SMA), a rare neuromuscular disorder characterized by a loss of motor neurons and progressive muscle wasting, show that AONs that abrogate aberrant splicing of SMN2 are promising compounds for treating SMA [243]. Most notably, this AON-based drug named Spinraza recently received FDA and EMA approval for treatment of SMA [244], following favorable outcomes in a phase 3 clinical trial where the treated children showed improved survival compared to placebo [245]. Upon intrathecal injection, AONs distribute well throughout the CNS and are taken up efficiently by cells of the brain. Also, the AONs remain present and stable in the tissue for months, allowing for infrequent dosing [246, 247]. In 2012, it has been demonstrated for the first time in a polyglutamine disease that infusion into the CSF of HD mouse models delays disease progression [248]. Preliminary results from a phase 1b/2a clinical trial following this preclinical research have already been released by Ionis Pharmaceuticals, with reports stating that the safety and tolerability profile are favorable and warrant further development [249]. Additionally, dose-dependent reduction in huntingtin protein levels in the CSF was reported [250]. Given that the similar approach and mechanism, it can be assumed that similar AONs for application for the polyQ SCAs can be equally well tolerated.
Given their favorable therapeutic properties, AONs clearly hold promise for the treatment of the polyQ SCAs. As such, several AON-based therapies are currently under research to this end, in particular for SCA3. The most straightforward approach is to target the mutant transcript for downregulation. This can be achieved either allele specifically for the transcript containing the expanded CAG repeat or nonallele specifically by targeting both mutant and wild-type transcripts.
Allele-Specific Downregulation
In order to specifically target the mutant allele, the CAG repeat can be directly targeted with AONs that preferentially bind to the longer repeat. Such AONs have been tested for SCA3, where peptide nucleic acids (PNAs) and locked nucleic acid (LNA) AONs were designed complementary to the CAG repeat and tested in different cell lines. Several of the tested AONs were indeed capable of downregulating mutant ataxin-3 at both transcript and protein levels [215]. The authors then tested a new range of AONs and showed several candidates capable of preferentially targeting the mutant ataxin-3 protein [216]. More recently, the same group has investigated a large range of new CAG repeat-targeting AONs with combinations of different chemical modifications and mismatches for downregulation of mutant ataxin-3. The authors found that subtle changes to the composition of the AON led to significant effects on efficacy and allele specificity. Through the screening of many candidates, the researchers identified several very promising AONs capable of potently downregulating mutant ataxin-3 in cells through CAG targeting [217]. Similar repeat-targeting AONs were tested in SCA7 cell lines, where ataxin-7 downregulation was achieved with varying efficiency based on the specific chemistry used [220]. To our knowledge, the CAG repeat-targeting AONs have not yet been tested in animal models of SCA, but positive results have been obtained where these types of AONs were injected in the brains of HD mice [251]. Specifically, CAG repeat-targeting AONs reduced mutant huntingtin (HTT) protein with 15 to 60% throughout the mouse brain, and corresponding improvements in motor tasks were reported [251]. These studies thus suggest good promise for these particular CAG repeat-targeting AONs for application in the polyQ SCAs, and indeed the other polyQ disorders [218].
Apart from targeting the CAG repeat, the mutant allele can contain SNPs that can be useful for specific targeting with AONs. The SNP targeting strategy is also largely investigated in the context of HD, where especially the newer AON chemistries show good potential of targeting the mutant allele [252, 253]. For instance for SCA3, a SNP that is associated with the mutant allele in 70% of SCA3 patients has been described [237] and has been proven as a viable target for targeting with shRNA [210]. A similar strategy has been described for SCA7 [209]. Despite these findings, no AON-mediated approach targeting the SNPs has been described for the polyQ SCAs to date.
Nonallele-Specific Downregulation
Specific downregulation of only the mutant allele may not be viable for all polyQ SCAs, and therefore, nonallele-specific downregulation, in which both alleles are targeted, may be a more achievable approach. An important consideration when pursuing this approach is whether prolonged efficient downregulation of the targeted protein will not have a detrimental effect, since most of the proteins that cause polyQ SCAs have important cellular function. A screening of AONs for downregulation of ATXN3 yielded several candidates capable of reducing expression with 50% [213]. Subsequent testing in SCA3 mice showed that the AON distributed well throughout the mouse brain and was capable of alleviating the motor symptoms in addition to preventing nuclear accumulation of mutant ataxin-3 for at least 14 weeks in the mouse brain [214]. Taking into account the level of ataxin-3 overexpression in the homozygous SCA3 mouse that was used, the lead AON is an exciting candidate for further clinical advancement [213]. Similarly, AONs have been designed to target ataxin-2 for downregulation. In a recent seminal study, again by screening a large number of different AON sequences, researchers were able to find an AON capable of potently downregulating ataxin-2 in cells. Intracerebroventricular injection of this AON in 2 different SCA2 mouse models yielded potent suppression of ATXN2 in the cerebellum. As a result, the motor phenotype of the SCA2 mice was markedly improved after AON injection [219]. Though the therapeutic AON candidates for the polyQ SCAs are currently still in preclinical phase, some speculation on the applicability of AONs for this purpose can already be made. For instance, an RNase H-based AON for downregulation of huntingtin is currently undergoing clinical trial. The AON is capable of downregulating both huntingtin alleles and has shown strong efficacy in preclinical research [248].
Exon Skipping
The polyQ SCAs are all caused by the same type of mutation, the expansion of a CAG repeat in the coding region of the responsible gene. For this reason, an interesting AON-based approach is to exclude the exon containing the CAG repeat from the transcript. This can be achieved through an approach termed exon skipping, where specific splicing signals are masked by AONs [254]. If removal of the exon in question does not result in a reading frame shift, the mRNA transcript is not degraded and can be translated in a normal fashion. This manner of splice modulation with AONs is a particularly useful strategy for application in the brain [239, 255]. Nonetheless, AON-mediated removal of the CAG repeat is not yet widely investigated for the polyQ disorders. This is in part due to the fact that the target gene needs to meet certain requirements. For instance, in ATXN2, the CAG repeat is located in the first exon and skipping of this exon would thus remove the start codon, resulting in downregulation of the transcript [23].
In this regard, ATXN3 has been most extensively investigated with regard to exon skipping. Firstly, Evers et al. [256] proved that AON mediated skipping of exon 9 and the CAG-containing exon 10 from the ATXN3 transcript led to formation of an internally truncated ataxin-3 protein lacking the polyQ repeat in SCA3 patient-derived fibroblasts. Moreover, the modified ataxin-3 protein was shown to retain its ubiquitin binding function [256]. More recent research from the same group demonstrates the possibility to skip only exon 10 from ATXN3 pre-mRNA, which then results in formation of a truncated ataxin-3 protein lacking the polyQ repeat and C-terminal region of the protein. This strategy was subsequently tested in a SCA3 mouse model, where AON treatment led to formation of the truncated ataxin-3 protein in all tested brain regions [257]. Interestingly, researchers serendipitously discovered that targeting the CAG repeat directly with 2′O-modified AONs also led to exclusion of ATXN3 exon 10, resulting in similar formation of a truncated ataxin-3 protein [217]. Targeting the mutant ATXN3 allele could be preferentially targeted for exon skipping by the CAG-targeting AONs, and this might be a very favorable therapeutic approach, since only the mutation could be specifically targeted for exclusion from the transcript.
Apart from skipping the CAG repeat-containing exon, AONs can also be implemented to skip other exons that have the potential to contribute to toxicity. For instance, proteolytic cleavage of the polyQ proteins can liberate shorter, more toxic, protein fragments for HD [258, 259] and SCA3 [154]. Hence, removing the protein region containing the proteolytic cleavage sites associated with the toxic fragments would, in theory, prevent toxicity of the mutant polyQ protein. For SCA3, the approach was concluded to be suboptimal due to inefficient exon skipping and is currently no longer pursued for clinical application [260]. The splice-modulating AONs, implemented for either exon skipping or inclusion, have been well investigated over the last years, especially for application in Duchene muscular dystrophy and SMA, where Spinraza is currently the first approved AON drug for clinical application in the brain.
CRISPR/Cas9
Advances in genome engineering techniques created the possibility to facilitate efficient genome engineering in eukaryotic cells [261–263]. The CRISPRs and Cas9 have been developed to target and edit specific genomic regions in (mammalian) cells [261, 263]. In short, guide RNAs (gRNAs) complementary to specific genomic sites are used to target Cas9 and induce double-stranded breaks (DSBs), which can be repaired through error-prone nonhomologous end joining (NHEJ) or the high-fidelity homology-directed repair (HDR) pathway. NHEJ leads to insertions or deletions (indels), leading to a frameshift and a premature stop codon. Therefore, this process can be used to make gene-specific knockouts. HDR generates defined gene modifications due to the introduction of a repair template [263].
Recently, Ouyang et al. [221] performed CRISPR/Cas9-mediated deletion of exon 10 containing the polyQ-encoding region in SCA3 patient-derived iPSCs. The corrected lines retained pluripotency and neural differentiation capacities. Moreover, the ubiquitin-binding capacity of ATXN3 was retained [221]. In a publication investigating HD, several allele-specific gRNAs for polyQ SCAs were proposed. In this study, a personalized, allele-specific CRISPR/Cas9 strategy was successfully tested for HD [264]. First, a genomic DNA locus was screened for SNPs that generate or eliminate protospacer adjacent motif (PAM) sequences. PAM sequences in the promoter region and transcription start site that are present on the mutant allele, but absent from the normal allele, were selected, and gRNAs were designed. Next, mutant alleles were inactivated in an allele-specific manner, resulting in the absence of mutant HTT RNA and HTT protein containing the expanded polyQ repeat. Based on the 1000 Genomes database, the authors proposed dozens of personalized, allele-specific gRNAs that can be tested in polyQ SCA preclinical studies [264].
CRISPR/Cas9 has the potential to become a great success in clinical trials for many diseases including the polyQ SCAs. However, off-target effects, which may cause genomic instability and disrupt functionality of nontargeted genes, are still a major concern [265]. Thus far, it was thought that CRISPR/Cas9 was reasonably specific. Recently, it has been shown that repair of double-strand breaks induced by CRISPR/Cas9 leads to large deletions and complex genomic rearrangements at the target sites [266]. This is an important issue that needs to be solved before clinical trials are started. Another point of concern is the delivery to the target cells. Many delivery strategies of the CRISPR/Cas9 system have been developed and are reviewed in Liu et al. [267]. So far, it is challenging to target the nondividing neurons in the human brain. Recently, Nishiyama et al. [268] showed that genome editing via HDR was successfully induced in mitotic and postmitotic cells in vitro and in vivo. The efficiency of HDR was with approximately 10% relatively low, but similar among different brain regions. This suggests that gene editing is applicable to practically any brain region and cell type at any given time point in life [268].
Stem Cell Therapies
Since the first stem cell-based clinical trials using fetal midbrain tissue to replace the dopamine neurons that are lost in PD [269, 270], stem cell replacement therapy has evolved and many more clinical trials were initiated [271]. Here, we give an overview of the current state of the stem cell-based therapies in the polyQ SCA field (Table 4).
Table 4.
Stem cell-based therapies
| SCA | Cell type | Delivery method | Mouse model | Results | Reference |
|---|---|---|---|---|---|
| SCA1 | ESC | Stereotaxic injection into the deep cerebellar nuclei | B05 transgenic SCA1 mice | Better performance on multiple behavioral tests of cerebellar function | [272] |
| SCA1 | Adult NPC | Stereotaxic injection into the cerebellar white matter | B05 transgenic SCA1 mice |
Only in mice with significant cell loss, grafted NPCs migrated into the cerebellar cortex. Improved motor skills, a significantly thicker molecular layer, more surviving PCs, and normalization of the PC basal membrane potential |
[273] |
| SCA3 | Cerebellar NSC | Transplantation into the cerebellum | SCA3/MJD transgenic mice | A significant and robust alleviation of the motor behavior impairments, which correlated with preservation from SCA3/MJD-associated neuropathology, namely reduction of Purkinje cell loss and reduction of cellular layer shrinkage and aggregates | [274] |
| SCA1 | MSC | Stereotaxic intrathecal injection | B05 transgenic SCA1 mice | Suppression atrophy of PC dendrites and better performance on rotarod | [275] |
| SCA1 | MSC | Stereotaxic intrathecal injection | SCA1-KI mice | Suppressing peripheral nervous system degeneration | [276] |
| SCA2 | MSC | Intravenous and intracranial transplantation | SCA2 transgenic mice |
Intracranial transplantation: no effect Intravenous: improved rotarod performance and delayed onset of motor function deterioration |
[277] |
| SCA3 | MSC | Single intracranial injection and repeated systemic administration | SCA3 transgenic mice |
Transplantation: only transient effects Periodic administration: Sustained motor behavior and neuropathology alleviation |
[278] |
SCA = spinocerebellar ataxia; ESC = embryonic stem cell; NPC = neural precursor cell; NSC = neural stem cell; MSC = mesenchymal stem cell
In the polyQ SCA field, the first preclinical experiment of embryonic cerebellar transplants into a transgenic mouse model of SCA1 has been published by Kaemmerer and Low [272] at the end of the previous millennium. Compared with sham-operated littermates, grafted mice functioned better on multiple cerebellar behavior tests. Improvements continued for approximately 3 months after the transplantation, after which a progressive decline in motor performance was observed. In most graft recipients, donor Purkinje cell survival was evident 20 weeks after surgery [272]. These first experiments show that transplants can survive and have behavioral benefits despite the ongoing pathological process in the diseased brain. Chintawar et al. [273] transplanted neural precursor cells (NPCs) derived from adult mice into the cerebellar white matter of SCA1 mice at different stages of the disease process. Only in mice with significant cell loss, grafted NPCs migrated into the cerebellar cortex. These animals showed behavioral benefits, a thicker molecular layer, and more surviving Purkinje cells compared to sham-treated controls. Despite these results, the grafted cells did not adopt the morphological characteristics of Purkinje cells and levels of neurotrophic factors were not increased, suggesting that the neuroprotective effect of grafted NPCs was mediated by direct contact with the host Purkinje cells [273]. Mendonça et al. [274] transplanted cerebellar neural stem cells into the cerebellum of adult SCA3 mice and found alleviation of the motor behavior impairments, which correlated with the prevention of SCA3 disease-associated neuropathology. Furthermore, a significant reduction of neuroinflammation and an increase of neurotrophic factors levels were found, suggesting that transplantation triggers important neuroprotective effects [274]. The increase in neurotrophic factors can be explained by the fact that a pool of multipotent, SOX2-positive cells remained in the graft [274].
Another cell type that is widely used in cell-based clinical trials is mesenchymal stromal cells (MSCs) [279–283]. MSCs are multipotent progenitor cells that can be isolated from multiple sources such as bone marrow [284], umbilical cord blood [285], or adipose tissue [286, 287]. Furthermore, MSCs have the ability to migrate and integrate into the endogenous neural network and subsequently produce various trophic factors involved in functional recovery, neuronal cell survival, and stimulation of endogenous regeneration [288]. In 2008, a protocol has been developed for inducing differentiation of MSCs toward neurotrophic factor (NTF)-secreting cells [289]. These cells can act as a protective agent in neurodegenerative diseases [290]. Chang et al. [277] showed that intravenous injection of MSCs in SCA2 mice improved rotarod performance and Purkinje cell survival, while intracranial transplantation failed to achieve these neuroprotective effects [277]. Intrathecal injection of MSCs and intravenous injection of MSC-conditioned medium ameliorated both cerebellar pathology and behavior phenotypes in different mouse models of SCA1 and SCA3 [275, 276, 278, 291]. The first phase 1/2 clinical trials to evaluate the safety, tolerability, and efficacy of intrathecal injection or intravenous administration of MSCs from healthy donors in polyQ SCA patients have been performed [292–295]. All these studies report that administration of MSCs is safe without any adverse events or rejection reactions. Administration of MSCs might delay the progression of neurologic deficits for polyQ SCAs, but a next, important step will be randomized, double-blind, placebo-controlled phase 2 trials.
The ground-breaking finding that somatic cells can be reprogrammed into iPSCs that can self-renew [296, 297] has enabled the development of an unlimited source of any type of human cells which can be used for disease modeling, drug discovery, and stem cell-based therapies (reviewed in [298]). All studies report a positive effect on both neuropathology and different behavior paradigms. So far, no iPSC replacement studies have been reported for the polyQ SCAs, but for HD, cell replacement therapy using iPSCs has been studied, mainly in rat models [299–301]. To date, patient-derived polyQ iPSCs are used for disease modeling and drug screens [49, 302–306]. Although the first preclinical experiments in HD and other neurodegenerative disease models are positive, an iPSC-based therapy for the polyQ SCAs is far from a clinical application.
Future Perspectives
A wide diversity of therapies for polyQ SCAs have been tested in preclinical models, and some of these therapies have even moved to the clinical trial stage. Most of these studies show promising results in both cell-based and animal models. In polyQ SCA mouse models, improvement of disease-associated neuropathology, most importantly the reduction of Purkinje cell loss and toxic aggregates, and alleviation of the behavior deficits have been achieved. So far however, these successes have not been translated to noteworthy breakthroughs in clinical trials. This may be in part due to the relatively small patient population and slow disease progression with a large variation in symptoms that is inherent to the SCAs. Nonetheless, there are plentiful compounds and treatment strategies being investigated that show promise for treatment of the SCAs.
Arguably, the most promising and elegant strategy to cure the polyQ SCAs is replacing the expanded CAG repeat by a control repeat length using CRISPR/Cas9. In this manner, the gain-of-function toxicity caused by the expanded allele is cleared and the repaired allele results in normal expression levels of the various ataxin proteins. To date, most CRISPR/Cas9 strategies are not immediately applicable for treating polyQ SCA patients. Various technical challenges, including minimizing off-target effects and optimizing delivery in all of the targets cells, need to be solved first. Therefore, the development of genome-editing therapies that induce HDR in the mutant allele only is crucial. Given the current challenges for clinical application of CRISPR/Cas9, RNA targeting therapies will be the solution for the near future. Exciting recent results from clinical trials in various neurodegenerative diseases have shown that RNA targeting compounds such as AONs can safely and effectively enter the cells of the CNS without inducing strong side effects.
The polyQ SCAs are a group of devastating progressive disease where historically, only symptomatic treatment has been available. With research progress in recent years, however, there is great promise that a therapy to delay disease progression or even to halt disease onset can be achieved. However, caution is still needed because progressing therapeutic approaches into approved drugs has been proven to be challenging.
Acknowledgments
This study was supported by the Dutch SCA1 Families Fund, the Dutch Brain Foundation (Nederlandse Hersenstichting) grant (HA2016-02-02), VENI grant (no. 91615080) from the Netherlands Organization of Scientific Research, and AFM Telethon (no. 20577).
Footnotes
Ronald A.M. Buijsen and Lodewijk J.A. Toonen should be regarded as joint first authors.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jacobi H, et al. The natural history of spinocerebellar ataxia type 1, 2, 3, and 6: a 2-year follow-up study. Neurology. 2011;77(11):1035–41. doi: 10.1212/WNL.0b013e31822e7ca0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dohlinger S, et al. Magnetic resonance imaging in spinocerebellar ataxias. Cerebellum. 2008;7(2):204–14. doi: 10.1007/s12311-008-0025-0. [DOI] [PubMed] [Google Scholar]
- 3.Gennarino VA, et al. A Mild PUM1 Mutation Is Associated with Adult-Onset Ataxia, whereas Haploinsufficiency Causes Developmental Delay and Seizures. Cell. 2018;172(5):924–936.e11. doi: 10.1016/j.cell.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paulson HL. The spinocerebellar ataxias. J Neuroophthalmol. 2009;29(3):227–37. doi: 10.1097/WNO0b013e3181b416de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matilla-Duenas A, et al. The spinocerebellar ataxias: clinical aspects and molecular genetics. Adv Exp Med Biol. 2012;724:351–74. doi: 10.1007/978-1-4614-0653-2_27. [DOI] [PubMed] [Google Scholar]
- 6.Coutinho P, et al. Hereditary ataxia and spastic paraplegia in Portugal: a population-based prevalence study. JAMA Neurol. 2013;70(6):746–55. doi: 10.1001/jamaneurol.2013.1707. [DOI] [PubMed] [Google Scholar]
- 7.Ruano L, et al. The global epidemiology of hereditary ataxia and spastic paraplegia: a systematic review of prevalence studies. Neuroepidemiology. 2014;42(3):174–83. doi: 10.1159/000358801. [DOI] [PubMed] [Google Scholar]
- 8.van de Warrenburg BP, et al. Spinocerebellar ataxias in the Netherlands: prevalence and age at onset variance analysis. Neurology. 2002;58(5):702–8. doi: 10.1212/wnl.58.5.702. [DOI] [PubMed] [Google Scholar]
- 9.Schols L, et al. Autosomal dominant cerebellar ataxia: phenotypic differences in genetically defined subtypes? Ann Neurol. 1997;42(6):924–32. doi: 10.1002/ana.410420615. [DOI] [PubMed] [Google Scholar]
- 10.Maruyama H, et al. Difference in disease-free survival curve and regional distribution according to subtype of spinocerebellar ataxia: a study of 1286 Japanese patients. Am J Med Genet. 2002;114(5):578–83. doi: 10.1002/ajmg.10514. [DOI] [PubMed] [Google Scholar]
- 11.Moseley ML, et al. Incidence of dominant spinocerebellar and Friedreich triplet repeats among 361 ataxia families. Neurology. 1998;51(6):1666–71. doi: 10.1212/wnl.51.6.1666. [DOI] [PubMed] [Google Scholar]
- 12.Silveira I, et al. Trinucleotide repeats in 202 families with ataxia: a small expanded (CAG)n allele at the SCA17 locus. Arch Neurol. 2002;59(4):623–9. doi: 10.1001/archneur.59.4.623. [DOI] [PubMed] [Google Scholar]
- 13.Brusco A, et al. Molecular genetics of hereditary spinocerebellar ataxia: mutation analysis of spinocerebellar ataxia genes and CAG/CTG repeat expansion detection in 225 Italian families. Arch Neurol. 2004;61(5):727–33. doi: 10.1001/archneur.61.5.727. [DOI] [PubMed] [Google Scholar]
- 14.Tang B, et al. Frequency of SCA1, SCA2, SCA3/MJD, SCA6, SCA7, and DRPLA CAG trinucleotide repeat expansion in patients with hereditary spinocerebellar ataxia from Chinese kindreds. Arch Neurol. 2000;57(4):540–4. doi: 10.1001/archneur.57.4.540. [DOI] [PubMed] [Google Scholar]
- 15.Bryer A, et al. The hereditary adult-onset ataxias in South Africa. J Neurol Sci. 2003;216(1):47–54. doi: 10.1016/s0022-510x(03)00209-0. [DOI] [PubMed] [Google Scholar]
- 16.Saleem Q, et al. Molecular analysis of autosomal dominant hereditary ataxias in the Indian population: high frequency of SCA2 and evidence for a common founder mutation. Hum Genet. 2000;106(2):179–87. doi: 10.1007/s004390051026. [DOI] [PubMed] [Google Scholar]
- 17.Schols L, et al. Autosomal dominant cerebellar ataxias: clinical features, genetics, and pathogenesis. Lancet Neurol. 2004;3(5):291–304. doi: 10.1016/S1474-4422(04)00737-9. [DOI] [PubMed] [Google Scholar]
- 18.Bird TD, et al. Hereditary Ataxia Overview. In: Adam MP, et al., editors. GeneReviews((R)) Seattle: University of Washington; 1993. [PubMed] [Google Scholar]
- 19.Erichsen AK, et al. Prevalence of hereditary ataxia and spastic paraplegia in southeast Norway: a population-based study. Brain. 2009;132(Pt 6):1577–88. doi: 10.1093/brain/awp056. [DOI] [PubMed] [Google Scholar]
- 20.Tsuji S, et al. Sporadic ataxias in Japan--a population-based epidemiological study. Cerebellum. 2008;7(2):189–97. doi: 10.1007/s12311-008-0028-x. [DOI] [PubMed] [Google Scholar]
- 21.Orr HT, et al. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet. 1993;4(3):221–6. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- 22.Zuhlke C, et al. Spinocerebellar ataxia type 1 (SCA1): phenotype-genotype correlation studies in intermediate alleles. Eur J Hum Genet. 2002;10(3):204–9. doi: 10.1038/sj.ejhg.5200788. [DOI] [PubMed] [Google Scholar]
- 23.Pulst SM, et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat Genet. 1996;14(3):269–76. doi: 10.1038/ng1196-269. [DOI] [PubMed] [Google Scholar]
- 24.Cancel G, et al. Molecular and clinical correlations in spinocerebellar ataxia 2: a study of 32 families. Hum Mol Genet. 1997;6(5):709–15. doi: 10.1093/hmg/6.5.709. [DOI] [PubMed] [Google Scholar]
- 25.Mao R, et al. Childhood-onset ataxia: testing for large CAG-repeats in SCA2 and SCA7. Am J Med Genet. 2002;110(4):338–45. doi: 10.1002/ajmg.10467. [DOI] [PubMed] [Google Scholar]
- 26.Kawaguchi Y, et al. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat Genet. 1994;8(3):221–8. doi: 10.1038/ng1194-221. [DOI] [PubMed] [Google Scholar]
- 27.Costa Mdo C, Paulson HL. Toward understanding Machado-Joseph disease. Prog Neurobiol. 2012;97(2):239–57. doi: 10.1016/j.pneurobio.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishikawa K, et al. Japanese families with autosomal dominant pure cerebellar ataxia map to chromosome 19p13.1-p13.2 and are strongly associated with mild CAG expansions in the spinocerebellar ataxia type 6 gene in chromosome 19p13.1. Am J Hum Genet. 1997;61(2):336–46. doi: 10.1086/514867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shizuka M, et al. Molecular analysis of a de novo mutation for spinocerebellar ataxia type 6 and (CAG)n repeat units in normal elder controls. J Neurol Sci. 1998;161(1):85–7. doi: 10.1016/s0022-510x(98)00270-6. [DOI] [PubMed] [Google Scholar]
- 30.Mariotti C, et al. Pathogenic effect of an intermediate-size SCA-6 allele (CAG)(19) in a homozygous patient. Neurology. 2001;57(8):1502–4. doi: 10.1212/wnl.57.8.1502. [DOI] [PubMed] [Google Scholar]
- 31.Yabe I, et al. SCA6 mutation analysis in a large cohort of the Japanese patients with late-onset pure cerebellar ataxia. J Neurol Sci. 1998;156(1):89–95. doi: 10.1016/s0022-510x(98)00009-4. [DOI] [PubMed] [Google Scholar]
- 32.David G, et al. Cloning of the SCA7 gene reveals a highly unstable CAG repeat expansion. Nat Genet. 1997;17(1):65–70. doi: 10.1038/ng0997-65. [DOI] [PubMed] [Google Scholar]
- 33.Nardacchione A, et al. Definition of the smallest pathological CAG expansion in SCA7. Clin Genet. 1999;56(3):232–4. doi: 10.1034/j.1399-0004.1999.560309.x. [DOI] [PubMed] [Google Scholar]
- 34.Lebre AS, Brice A. Spinocerebellar ataxia 7 (SCA7) Cytogenet Genome Res. 2003;100(1–4):154–63. doi: 10.1159/000072850. [DOI] [PubMed] [Google Scholar]
- 35.van de Warrenburg BP, et al. Striking anticipation in spinocerebellar ataxia type 7: the infantile phenotype. J Neurol. 2001;248(10):911–4. doi: 10.1007/s004150170082. [DOI] [PubMed] [Google Scholar]
- 36.Koide R, et al. A neurological disease caused by an expanded CAG trinucleotide repeat in the TATA-binding protein gene: a new polyglutamine disease? Hum Mol Genet. 1999;8(11):2047–53. doi: 10.1093/hmg/8.11.2047. [DOI] [PubMed] [Google Scholar]
- 37.Nanda A, et al. Case of spinocerebellar ataxia type 17 (SCA17) associated with only 41 repeats of the TATA-binding protein (TBP) gene. Mov Disord. 2007;22(3):436. doi: 10.1002/mds.21275. [DOI] [PubMed] [Google Scholar]
- 38.Maltecca F, et al. Intergenerational instability and marked anticipation in SCA-17. Neurology. 2003;61(10):1441–3. doi: 10.1212/01.wnl.0000094123.09098.a0. [DOI] [PubMed] [Google Scholar]
- 39.Bettencourt C, et al. DNA repair pathways underlie a common genetic mechanism modulating onset in polyglutamine diseases. Ann Neurol. 2016;79(6):983–90. doi: 10.1002/ana.24656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan HC, et al. Polyglutamine (PolyQ) diseases: genetics to treatments. Cell Transplant. 2014;23(4–5):441–58. doi: 10.3727/096368914X678454. [DOI] [PubMed] [Google Scholar]
- 41.Sequeiros J, Seneca S, Martindale J. Consensus and controversies in best practices for molecular genetic testing of spinocerebellar ataxias. Eur J Hum Genet. 2010;18(11):1188–95. doi: 10.1038/ejhg.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nethisinghe S, et al. PolyQ Tract Toxicity in SCA1 is Length Dependent in the Absence of CAG Repeat Interruption. Front Cell Neurosci. 2018;12:200. doi: 10.3389/fncel.2018.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saute JAM, Jardim LB. Machado Joseph disease: clinical and genetic aspects, and current treatment. Expert Opinion on Orphan Drugs. 2015;3(5):517–535. [Google Scholar]
- 44.Paulson HL, et al. Polyglutamine spinocerebellar ataxias—from genes to potential treatments. Nat Rev Neurosci. 2017;18(10):613–626. doi: 10.1038/nrn.2017.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kratter IH, Finkbeiner S. PolyQ disease: too many Qs, too much function? Neuron. 2010;67(6):897–9. doi: 10.1016/j.neuron.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evers MM, Toonen LJ, van Roon-Mom WM. Ataxin-3 protein and RNA toxicity in spinocerebellar ataxia type 3: current insights and emerging therapeutic strategies. Mol Neurobiol. 2014;49(3):1513–31. doi: 10.1007/s12035-013-8596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi T, Katada S, Onodera O. Polyglutamine diseases: where does toxicity come from? what is toxicity? where are we going? J Mol Cell Biol. 2010;2(4):180–91. doi: 10.1093/jmcb/mjq005. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi T, et al. Soluble polyglutamine oligomers formed prior to inclusion body formation are cytotoxic. Hum Mol Genet. 2008;17(3):345–56. doi: 10.1093/hmg/ddm311. [DOI] [PubMed] [Google Scholar]
- 49.Koch P, et al. Excitation-induced ataxin-3 aggregation in neurons from patients with Machado-Joseph disease. Nature. 2011;480(7378):543–6. doi: 10.1038/nature10671. [DOI] [PubMed] [Google Scholar]
- 50.Opal P, Ashizawa T, et al. Spinocerebellar Ataxia Type 1. In: Adam MP, et al., editors. GeneReviews((R)) Seattle: University of Washington; 1993. [PubMed] [Google Scholar]
- 51.Jin DK, et al. Frequency of spinocerebellar ataxia types 1,2,3,6,7 and dentatorubral pallidoluysian atrophy mutations in Korean patients with spinocerebellar ataxia. J Neurol. 1999;246(3):207–10. doi: 10.1007/s004150050335. [DOI] [PubMed] [Google Scholar]
- 52.Platonov FA, et al. Genetic fitness and selection intensity in a population affected with high-incidence spinocerebellar ataxia type 1. Neurogenetics. 2016;17(3):179–85. doi: 10.1007/s10048-016-0481-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferrarin M, et al. Procedure for the quantitative evaluation of motor disturbances in cerebellar ataxic patients. Med Biol Eng Comput. 2005;43(3):349–56. doi: 10.1007/BF02345812. [DOI] [PubMed] [Google Scholar]
- 54.Rivaud-Pechoux S, et al. Eye movement abnormalities correlate with genotype in autosomal dominant cerebellar ataxia type I. Ann Neurol. 1998;43(3):297–302. doi: 10.1002/ana.410430306. [DOI] [PubMed] [Google Scholar]
- 55.Burk K, et al. Cognitive deficits in spinocerebellar ataxia type 1, 2, and 3. J Neurol. 2003;250(2):207–11. doi: 10.1007/s00415-003-0976-5. [DOI] [PubMed] [Google Scholar]
- 56.Orengo, J.P., et al., Motor neuron degeneration correlates with respiratory dysfunction in SCA1. Disease Models & Mechanisms, 2018. 11(2): p. dmm032623. [DOI] [PMC free article] [PubMed]
- 57.Seidel K, et al. Brain pathology of spinocerebellar ataxias. Acta Neuropathol. 2012;124(1):1–21. doi: 10.1007/s00401-012-1000-x. [DOI] [PubMed] [Google Scholar]
- 58.Matilla A, et al. Mice lacking ataxin-1 display learning deficits and decreased hippocampal paired-pulse facilitation. J Neurosci. 1998;18(14):5508–16. doi: 10.1523/JNEUROSCI.18-14-05508.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crespo-Barreto J, et al. Partial loss of ataxin-1 function contributes to transcriptional dysregulation in spinocerebellar ataxia type 1 pathogenesis. PLoS Genet. 2010;6(7):e1001021. doi: 10.1371/journal.pgen.1001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang C, et al. Loss of function of ATXN1 increases amyloid beta-protein levels by potentiating beta-secretase processing of beta-amyloid precursor protein. J Biol Chem. 2010;285(12):8515–26. doi: 10.1074/jbc.M109.079079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsai CC, et al. Ataxin 1, a SCA1 neurodegenerative disorder protein, is functionally linked to the silencing mediator of retinoid and thyroid hormone receptors. Proc Natl Acad Sci U S A. 2004;101(12):4047–52. doi: 10.1073/pnas.0400615101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolger TA, et al. The neurodegenerative disease protein ataxin-1 antagonizes the neuronal survival function of myocyte enhancer factor-2. J Biol Chem. 2007;282(40):29186–92. doi: 10.1074/jbc.M704182200. [DOI] [PubMed] [Google Scholar]
- 63.Tong X, et al. Ataxin-1 and Brother of ataxin-1 are components of the Notch signalling pathway. EMBO Rep. 2011;12(5):428–35. doi: 10.1038/embor.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matilla-Duenas A, Goold R, Giunti P. Clinical, genetic, molecular, and pathophysiological insights into spinocerebellar ataxia type 1. Cerebellum. 2008;7(2):106–14. doi: 10.1007/s12311-008-0009-0. [DOI] [PubMed] [Google Scholar]
- 65.Lee Y, et al. ATXN1 protein family and CIC regulate extracellular matrix remodeling and lung alveolarization. Dev Cell. 2011;21(4):746–57. doi: 10.1016/j.devcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lorenzetti D, Bohlega S, Zoghbi HY. The expansion of the CAG repeat in ataxin-2 is a frequent cause of autosomal dominant spinocerebellar ataxia. Neurology. 1997;49(4):1009–13. doi: 10.1212/wnl.49.4.1009. [DOI] [PubMed] [Google Scholar]
- 67.Watanabe H, et al. Frequency analysis of autosomal dominant cerebellar ataxias in Japanese patients and clinical characterization of spinocerebellar ataxia type 6. Clin Genet. 1998;53(1):13–9. doi: 10.1034/j.1399-0004.1998.531530104.x. [DOI] [PubMed] [Google Scholar]
- 68.Lee WY, et al. Frequency analysis and clinical characterization of spinocerebellar ataxia types 1, 2, 3, 6, and 7 in Korean patients. Arch Neurol. 2003;60(6):858–63. doi: 10.1001/archneur.60.6.858. [DOI] [PubMed] [Google Scholar]
- 69.Geschwind DH, et al. The prevalence and wide clinical spectrum of the spinocerebellar ataxia type 2 trinucleotide repeat in patients with autosomal dominant cerebellar ataxia. Am J Hum Genet. 1997;60(4):842–50. [PMC free article] [PubMed] [Google Scholar]
- 70.Rub U, et al. Clinical features, neurogenetics and neuropathology of the polyglutamine spinocerebellar ataxias type 1, 2, 3, 6 and 7. Prog Neurobiol. 2013;104:38–66. doi: 10.1016/j.pneurobio.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 71.Burk K. Cognition in hereditary ataxia. Cerebellum. 2007;6(3):280–6. doi: 10.1080/14734220601115924. [DOI] [PubMed] [Google Scholar]
- 72.Storey E, et al. Spinocerebellar ataxia type 2: clinical features of a pedigree displaying prominent frontal-executive dysfunction. Arch Neurol. 1999;56(1):43–50. doi: 10.1001/archneur.56.1.43. [DOI] [PubMed] [Google Scholar]
- 73.Schols L, et al. Spinocerebellar ataxia type 2. Genotype and phenotype in German kindreds. Arch Neurol. 1997;54(9):1073–80. doi: 10.1001/archneur.1997.00550210011007. [DOI] [PubMed] [Google Scholar]
- 74.Moretti P, et al. Spinocerebellar ataxia type 2 (SCA2) presenting with ophthalmoplegia and developmental delay in infancy. Am J Med Genet A. 2004;124a(4):392–6. doi: 10.1002/ajmg.a.20428. [DOI] [PubMed] [Google Scholar]
- 75.Sanpei K, et al. Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique, DIRECT. Nat Genet. 1996;14(3):277–84. doi: 10.1038/ng1196-277. [DOI] [PubMed] [Google Scholar]
- 76.Imbert G, et al. Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat Genet. 1996;14(3):285–91. doi: 10.1038/ng1196-285. [DOI] [PubMed] [Google Scholar]
- 77.Kaehler C, et al. Ataxin-2-like is a regulator of stress granules and processing bodies. PLoS One. 2012;7(11):e50134. doi: 10.1371/journal.pone.0050134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Satterfield TF, Pallanck LJ. Ataxin-2 and its Drosophila homolog, ATX2, physically assemble with polyribosomes. Hum Mol Genet. 2006;15(16):2523–32. doi: 10.1093/hmg/ddl173. [DOI] [PubMed] [Google Scholar]
- 79.Elden AC, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466(7310):1069–75. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Juvonen V, et al. The occurrence of dominant spinocerebellar ataxias among 251 Finnish ataxia patients and the role of predisposing large normal alleles in a genetically isolated population. Acta Neurol Scand. 2005;111(3):154–62. doi: 10.1111/j.1600-0404.2005.00349.x. [DOI] [PubMed] [Google Scholar]
- 81.Soong BW, et al. Frequency analysis of autosomal dominant cerebellar ataxias in Taiwanese patients and clinical and molecular characterization of spinocerebellar ataxia type 6. Arch Neurol. 2001;58(7):1105–9. doi: 10.1001/archneur.58.7.1105. [DOI] [PubMed] [Google Scholar]
- 82.Vale J, et al. Autosomal dominant cerebellar ataxia: frequency analysis and clinical characterization of 45 families from Portugal. Eur J Neurol. 2010;17(1):124–8. doi: 10.1111/j.1468-1331.2009.02757.x. [DOI] [PubMed] [Google Scholar]
- 83.Paulson HL. Dominantly inherited ataxias: lessons learned from Machado-Joseph disease/spinocerebellar ataxia type 3. Semin Neurol. 2007;27(2):133–42. doi: 10.1055/s-2007-971172. [DOI] [PubMed] [Google Scholar]
- 84.Braga-Neto P, et al. Cerebellar cognitive affective syndrome in Machado Joseph disease: core clinical features. Cerebellum. 2012;11(2):549–56. doi: 10.1007/s12311-011-0318-6. [DOI] [PubMed] [Google Scholar]
- 85.Sequeiros J, Coutinho P. Epidemiology and clinical aspects of Machado-Joseph disease. Adv Neurol. 1993;61:139–53. [PubMed] [Google Scholar]
- 86.Rosenberg RN. Machado-Joseph disease: an autosomal dominant motor system degeneration. Mov Disord. 1992;7(3):193–203. doi: 10.1002/mds.870070302. [DOI] [PubMed] [Google Scholar]
- 87.Buhmann C, Bussopulos A, Oechsner M. Dopaminergic response in Parkinsonian phenotype of Machado-Joseph disease. Mov Disord. 2003;18(2):219–21. doi: 10.1002/mds.10322. [DOI] [PubMed] [Google Scholar]
- 88.Koeppen AH. The Neuropathology of Spinocerebellar Ataxia Type 3/Machado-Joseph Disease. Adv Exp Med Biol. 2018;1049:233–241. doi: 10.1007/978-3-319-71779-1_11. [DOI] [PubMed] [Google Scholar]
- 89.Doss-Pepe EW, et al. Ataxin-3 interactions with rad23 and valosin-containing protein and its associations with ubiquitin chains and the proteasome are consistent with a role in ubiquitin-mediated proteolysis. Mol Cell Biol. 2003;23(18):6469–83. doi: 10.1128/MCB.23.18.6469-6483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Geschwind DH, et al. Spinocerebellar ataxia type 6. Frequency of the mutation and genotype-phenotype correlations. Neurology. 1997;49(5):1247–51. doi: 10.1212/wnl.49.5.1247. [DOI] [PubMed] [Google Scholar]
- 91.Stevanin G, et al. Clinical and molecular features of spinocerebellar ataxia type 6. Neurology. 1997;49(5):1243–6. doi: 10.1212/wnl.49.5.1243. [DOI] [PubMed] [Google Scholar]
- 92.Ikeuchi T, et al. Spinocerebellar ataxia type 6: CAG repeat expansion in alpha1A voltage-dependent calcium channel gene and clinical variations in Japanese population. Ann Neurol. 1997;42(6):879–84. doi: 10.1002/ana.410420609. [DOI] [PubMed] [Google Scholar]
- 93.Globas C, et al. The cerebellum and cognition. Intellectual function in spinocerebellar ataxia type 6 (SCA6) J Neurol. 2003;250(12):1482–7. doi: 10.1007/s00415-003-0258-2. [DOI] [PubMed] [Google Scholar]
- 94.Gomez CM, et al. Spinocerebellar Ataxia Type 6. In: Adam MP, et al., editors. GeneReviews((R)) Seattle: University of Washington; 1993. [PubMed] [Google Scholar]
- 95.Sasaki H, et al. Neuropathological and molecular studies of spinocerebellar ataxia type 6 (SCA6) Acta Neuropathol. 1998;95(2):199–204. doi: 10.1007/s004010050787. [DOI] [PubMed] [Google Scholar]
- 96.Du X, et al. Second cistron in CACNA1A gene encodes a transcription factor mediating cerebellar development and SCA6. Cell. 2013;154(1):118–33. doi: 10.1016/j.cell.2013.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhuchenko O, et al. Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the alpha 1A-voltage-dependent calcium channel. Nat Genet. 1997;15(1):62–9. doi: 10.1038/ng0197-62. [DOI] [PubMed] [Google Scholar]
- 98.Filla A, et al. Relative frequencies of CAG expansions in spinocerebellar ataxia and dentatorubropallidoluysian atrophy in 116 Italian families. Eur Neurol. 2000;44(1):31–6. doi: 10.1159/000008189. [DOI] [PubMed] [Google Scholar]
- 99.Garden G, et al. Spinocerebellar Ataxia Type 7. In: Adam MP, et al., editors. GeneReviews((R)) Seattle: University of Washington; 1993. [Google Scholar]
- 100.Aleman TS, et al. Spinocerebellar ataxia type 7 (SCA7) shows a cone-rod dystrophy phenotype. Exp Eye Res. 2002;74(6):737–45. doi: 10.1006/exer.2002.1169. [DOI] [PubMed] [Google Scholar]
- 101.Benton CS, et al. Molecular and clinical studies in SCA-7 define a broad clinical spectrum and the infantile phenotype. Neurology. 1998;51(4):1081–6. doi: 10.1212/wnl.51.4.1081. [DOI] [PubMed] [Google Scholar]
- 102.Monin ML, et al. Survival and severity in dominant cerebellar ataxias. Ann Clin Transl Neurol. 2015;2(2):202–7. doi: 10.1002/acn3.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Giunti P, et al. Molecular and clinical study of 18 families with ADCA type II: evidence for genetic heterogeneity and de novo mutation. Am J Hum Genet. 1999;64(6):1594–603. doi: 10.1086/302406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nakamura Y, et al. Ataxin-7 associates with microtubules and stabilizes the cytoskeletal network. Hum Mol Genet. 2012;21(5):1099–110. doi: 10.1093/hmg/ddr539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Palhan VB, et al. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc Natl Acad Sci U S A. 2005;102(24):8472–7. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Craig K, et al. Minimum prevalence of spinocerebellar ataxia 17 in the north east of England. J Neurol Sci. 2005;239(1):105–9. doi: 10.1016/j.jns.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 107.Alendar A, et al. Spinocerebellar ataxia type 17 in the Yugoslav population. Acta Neurol Scand. 2004;109(3):185–7. doi: 10.1034/j.1600-0404.2003.00196.x. [DOI] [PubMed] [Google Scholar]
- 108.Toyoshima, Y., et al., SCA17 homozygote showing Huntington's disease-like phenotype. Ann Neurol, 2004. 55(2): p. 281-6. [DOI] [PubMed]
- 109.Toyoshima Y, et al. et al. Spinocerebellar Ataxia Type 17. In: Adam MP, et al.et al., editors. GeneReviews((R)) Seattle: University of Washington; 1993. [PubMed] [Google Scholar]
- 110.Stevanin G, Brice A. Spinocerebellar ataxia 17 (SCA17) and Huntington’s disease-like 4 (HDL4) Cerebellum. 2008;7(2):170–8. doi: 10.1007/s12311-008-0016-1. [DOI] [PubMed] [Google Scholar]
- 111.Nakamura K, et al. SCA17, a novel autosomal dominant cerebellar ataxia caused by an expanded polyglutamine in TATA-binding protein. Hum Mol Genet. 2001;10(14):1441–8. doi: 10.1093/hmg/10.14.1441. [DOI] [PubMed] [Google Scholar]
- 112.Gostout B, Liu Q, Sommer SS. “Cryptic” repeating triplets of purines and pyrimidines (cRRY(i)) are frequent and polymorphic: analysis of coding cRRY(i) in the proopiomelanocortin (POMC) and TATA-binding protein (TBP) genes. Am J Hum Genet. 1993;52(6):1182–90. [PMC free article] [PubMed] [Google Scholar]
- 113.van Roon-Mom WM, et al. TATA-binding protein in neurodegenerative disease. Neuroscience. 2005;133(4):863–72. doi: 10.1016/j.neuroscience.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 114.Zesiewicz TA, et al. Comprehensive systematic review summary: Treatment of cerebellar motor dysfunction and ataxia: Report of the Guideline Development, Dissemination, and Implementation Subcommittee of the American Academy of Neurology. Neurology. 2018;90(10):464–471. doi: 10.1212/WNL.0000000000005055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ilg W, et al. Consensus paper: management of degenerative cerebellar disorders. Cerebellum. 2014;13(2):248–68. doi: 10.1007/s12311-013-0531-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Perlman SL. Symptomatic and disease-modifying therapy for the progressive ataxias. Neurologist. 2004;10(5):275–89. doi: 10.1097/01.nrl.0000141651.35193.67. [DOI] [PubMed] [Google Scholar]
- 117.Sarva, H. and V.L. Shanker, Treatment Options in Degenerative Cerebellar Ataxia: A Systematic Review. Mov Disord Clin Pract, 2014. 1(4): p. 291–298. [DOI] [PMC free article] [PubMed]
- 118.Chen X, et al. Deranged calcium signaling and neurodegeneration in spinocerebellar ataxia type 3. J Neurosci. 2008;28(48):12713–24. doi: 10.1523/JNEUROSCI.3909-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Menzies FM, et al. Autophagy induction reduces mutant ataxin-3 levels and toxicity in a mouse model of spinocerebellar ataxia type 3. Brain. 2010;133(Pt 1):93–104. doi: 10.1093/brain/awp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chou AH, et al. HDAC inhibitor sodium butyrate reverses transcriptional downregulation and ameliorates ataxic symptoms in a transgenic mouse model of SCA3. Neurobiol Dis. 2011;41(2):481–8. doi: 10.1016/j.nbd.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 121.Wang HL, et al. H1152 promotes the degradation of polyglutamine-expanded ataxin-3 or ataxin-7 independently of its ROCK-inhibiting effect and ameliorates mutant ataxin-3-induced neurodegeneration in the SCA3 transgenic mouse. Neuropharmacology. 2013;70:1–11. doi: 10.1016/j.neuropharm.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 122.Goncalves N, et al. Caffeine and adenosine A(2A) receptor inactivation decrease striatal neuropathology in a lentiviral-based model of Machado-Joseph disease. Ann Neurol. 2013;73(5):655–66. doi: 10.1002/ana.23866. [DOI] [PubMed] [Google Scholar]
- 123.Silva-Fernandes A, et al. Chronic treatment with 17-DMAG improves balance and coordination in a new mouse model of Machado-Joseph disease. Neurotherapeutics. 2014;11(2):433–49. doi: 10.1007/s13311-013-0255-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Duarte-Silva S, et al. Lithium chloride therapy fails to improve motor function in a transgenic mouse model of Machado-Joseph disease. Cerebellum. 2014;13(6):713–27. doi: 10.1007/s12311-014-0589-9. [DOI] [PubMed] [Google Scholar]
- 125.Teixeira-Castro A, et al. Serotonergic signalling suppresses ataxin 3 aggregation and neurotoxicity in animal models of Machado-Joseph disease. Brain. 2015;138(Pt 11):3221–37. doi: 10.1093/brain/awv262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Esteves S, et al. Limited Effect of Chronic Valproic Acid Treatment in a Mouse Model of Machado-Joseph Disease. PLoS One. 2015;10(10):e0141610. doi: 10.1371/journal.pone.0141610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Duarte-Silva S, et al. Combined therapy with m-TOR-dependent and -independent autophagy inducers causes neurotoxicity in a mouse model of Machado-Joseph disease. Neuroscience. 2016;313:162–73. doi: 10.1016/j.neuroscience.2015.11.030. [DOI] [PubMed] [Google Scholar]
- 128.Schmidt J, et al. In vivo assessment of riluzole as a potential therapeutic drug for spinocerebellar ataxia type 3. J Neurochem. 2016;138(1):150–62. doi: 10.1111/jnc.13606. [DOI] [PubMed] [Google Scholar]
- 129.Cunha-Santos J, et al. Caloric restriction blocks neuropathology and motor deficits in Machado-Joseph disease mouse models through SIRT1 pathway. Nat Commun. 2016;7:11445. doi: 10.1038/ncomms11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jia DD, et al. Lithium chloride alleviates neurodegeneration partly by inhibiting activity of GSK3beta in a SCA3 Drosophila model. Cerebellum. 2013;12(6):892–901. doi: 10.1007/s12311-013-0498-3. [DOI] [PubMed] [Google Scholar]
- 131.Chort A, et al. Interferon beta induces clearance of mutant ataxin 7 and improves locomotion in SCA7 knock-in mice. Brain. 2013;136(Pt 6):1732–45. doi: 10.1093/brain/awt061. [DOI] [PubMed] [Google Scholar]
- 132.Chang YC, et al. Targeting the prodromal stage of spinocerebellar ataxia type 17 mice: G-CSF in the prevention of motor deficits via upregulating chaperone and autophagy levels. Brain Res. 2016;1639:132–48. doi: 10.1016/j.brainres.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 133.Watase K, et al. Lithium therapy improves neurological function and hippocampal dendritic arborization in a spinocerebellar ataxia type 1 mouse model. PLoS Med. 2007;4(5):e182. doi: 10.1371/journal.pmed.0040182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cortes CJ, La Spada AR. Autophagy in polyglutamine disease: Imposing order on disorder or contributing to the chaos? Mol Cell Neurosci. 2015;66(Pt A):53–61. doi: 10.1016/j.mcn.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci. 2010;13(7):805–11. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 137.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19(8):983–97. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 138.Towers CG, Thorburn A. Therapeutic Targeting of Autophagy. EBioMedicine. 2016;14:15–23. doi: 10.1016/j.ebiom.2016.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Forlenza OV, De-Paula VJ, Diniz BS. Neuroprotective effects of lithium: implications for the treatment of Alzheimer’s disease and related neurodegenerative disorders. ACS Chem Neurosci. 2014;5(6):443–50. doi: 10.1021/cn5000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Perroud B, et al. Pharmacometabolomic signature of ataxia SCA1 mouse model and lithium effects. PLoS One. 2013;8(8):e70610. doi: 10.1371/journal.pone.0070610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kieling C, et al. A neurological examination score for the assessment of spinocerebellar ataxia 3 (SCA3) Eur J Neurol. 2008;15(4):371–6. doi: 10.1111/j.1468-1331.2008.02078.x. [DOI] [PubMed] [Google Scholar]
- 142.Sacca F, et al. A randomized controlled pilot trial of lithium in spinocerebellar ataxia type 2. J Neurol. 2015;262(1):149–53. doi: 10.1007/s00415-014-7551-0. [DOI] [PubMed] [Google Scholar]
- 143.Saute JAM, Jardim LB. Planning Future Clinical Trials for Machado-Joseph Disease. Adv Exp Med Biol. 2018;1049:321–348. doi: 10.1007/978-3-319-71779-1_17. [DOI] [PubMed] [Google Scholar]
- 144.Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36(6):585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 145.Ntsapi C, Loos B. Caloric restriction and the precision-control of autophagy: A strategy for delaying neurodegenerative disease progression. Exp Gerontol. 2016;83:97–111. doi: 10.1016/j.exger.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 146.Aguib Y, et al. Autophagy induction by trehalose counteracts cellular prion infection. Autophagy. 2009;5(3):361–9. doi: 10.4161/auto.5.3.7662. [DOI] [PubMed] [Google Scholar]
- 147.Chen ZZ, et al. Trehalose attenuates the gait ataxia and gliosis of spinocerebellar ataxia type 17 mice. Neurochem Res. 2015;40(4):800–10. doi: 10.1007/s11064-015-1530-4. [DOI] [PubMed] [Google Scholar]
- 148.Lin CH, et al. Novel Lactulose and Melibiose Targeting Autophagy to Reduce PolyQ Aggregation in Cell Models of Spinocerebellar Ataxia 3. CNS Neurol Disord Drug Targets. 2016;15(3):351–9. doi: 10.2174/1871527314666150821101522. [DOI] [PubMed] [Google Scholar]
- 149.Kang R, et al. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18(4):571–80. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nascimento-Ferreira I, et al. Overexpression of the autophagic beclin-1 protein clears mutant ataxin-3 and alleviates Machado-Joseph disease. Brain. 2011;134(Pt 5):1400–15. doi: 10.1093/brain/awr047. [DOI] [PubMed] [Google Scholar]
- 151.Ona VO, et al. Inhibition of caspase-1 slows disease progression in a mouse model of Huntington’s disease. Nature. 1999;399(6733):263–7. doi: 10.1038/20446. [DOI] [PubMed] [Google Scholar]
- 152.Graham RK, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125(6):1179–91. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 153.Aharony I, et al. A Huntingtin-based peptide inhibitor of caspase-6 provides protection from mutant Huntingtin-induced motor and behavioral deficits. Hum Mol Genet. 2015;24(9):2604–14. doi: 10.1093/hmg/ddv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Goti D, et al. A mutant ataxin-3 putative-cleavage fragment in brains of Machado-Joseph disease patients and transgenic mice is cytotoxic above a critical concentration. J Neurosci. 2004;24(45):10266–79. doi: 10.1523/JNEUROSCI.2734-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hubener J, et al. N-terminal ataxin-3 causes neurological symptoms with inclusions, endoplasmic reticulum stress and ribosomal dislocation. Brain. 2011;134(Pt 7):1925–42. doi: 10.1093/brain/awr118. [DOI] [PubMed] [Google Scholar]
- 156.Hubener J, et al. Calpain-mediated ataxin-3 cleavage in the molecular pathogenesis of spinocerebellar ataxia type 3 (SCA3) Hum Mol Genet. 2013;22(3):508–18. doi: 10.1093/hmg/dds449. [DOI] [PubMed] [Google Scholar]
- 157.Jung J, et al. Preventing Ataxin-3 protein cleavage mitigates degeneration in a Drosophila model of SCA3. Hum Mol Genet. 2009;18(24):4843–52. doi: 10.1093/hmg/ddp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Simoes AT, et al. Calpain inhibition reduces ataxin-3 cleavage alleviating neuropathology and motor impairments in mouse models of Machado-Joseph disease. Hum Mol Genet. 2014;23(18):4932–44. doi: 10.1093/hmg/ddu209. [DOI] [PubMed] [Google Scholar]
- 159.Kordasiewicz HB, et al. C-termini of P/Q-type Ca2+ channel alpha1A subunits translocate to nuclei and promote polyglutamine-mediated toxicity. Hum Mol Genet. 2006;15(10):1587–99. doi: 10.1093/hmg/ddl080. [DOI] [PubMed] [Google Scholar]
- 160.Helmlinger D, et al. Hsp70 and Hsp40 chaperones do not modulate retinal phenotype in SCA7 mice. J Biol Chem. 2004;279(53):55969–77. doi: 10.1074/jbc.M409062200. [DOI] [PubMed] [Google Scholar]
- 161.Young JE, et al. Proteolytic cleavage of ataxin-7 by caspase-7 modulates cellular toxicity and transcriptional dysregulation. J Biol Chem. 2007;282(41):30150–60. doi: 10.1074/jbc.M705265200. [DOI] [PubMed] [Google Scholar]
- 162.Guyenet SJ, et al. Proteolytic cleavage of ataxin-7 promotes SCA7 retinal degeneration and neurological dysfunction. Hum Mol Genet. 2015;24(14):3908–17. doi: 10.1093/hmg/ddv121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Haacke A, Hartl FU, Breuer P. Calpain inhibition is sufficient to suppress aggregation of polyglutamine-expanded ataxin-3. J Biol Chem. 2007;282(26):18851–6. doi: 10.1074/jbc.M611914200. [DOI] [PubMed] [Google Scholar]
- 164.Haacke A, et al. Proteolytic cleavage of polyglutamine-expanded ataxin-3 is critical for aggregation and sequestration of non-expanded ataxin-3. Hum Mol Genet. 2006;15(4):555–68. doi: 10.1093/hmg/ddi472. [DOI] [PubMed] [Google Scholar]
- 165.Watchon M, et al. Calpain Inhibition Is Protective in Machado-Joseph Disease Zebrafish Due to Induction of Autophagy. J Neurosci. 2017;37(32):7782–7794. doi: 10.1523/JNEUROSCI.1142-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Simoes AT, et al. Calpastatin-mediated inhibition of calpains in the mouse brain prevents mutant ataxin 3 proteolysis, nuclear localization and aggregation, relieving Machado-Joseph disease. Brain. 2012;135(Pt 8):2428–39. doi: 10.1093/brain/aws177. [DOI] [PubMed] [Google Scholar]
- 167.Menzies FM, et al. Calpain inhibition mediates autophagy-dependent protection against polyglutamine toxicity. Cell Death Differ. 2015;22(3):433–44. doi: 10.1038/cdd.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Weber JJ, et al. From pathways to targets: understanding the mechanisms behind polyglutamine disease. Biomed Res Int. 2014;2014:701758. doi: 10.1155/2014/701758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Donkor IO. An updated patent review of calpain inhibitors (2012 - 2014) Expert Opin Ther Pat. 2015;25(1):17–31. doi: 10.1517/13543776.2014.982534. [DOI] [PubMed] [Google Scholar]
- 170.Kudelova J, et al. Pharmacological caspase inhibitors: research towards therapeutic perspectives. J Physiol Pharmacol. 2015;66(4):473–82. [PubMed] [Google Scholar]
- 171.Atanesyan L, et al. Polyglutamine tracts as modulators of transcriptional activation from yeast to mammals. Biol Chem. 2012;393(1–2):63–70. doi: 10.1515/BC-2011-252. [DOI] [PubMed] [Google Scholar]
- 172.Li F, et al. Ataxin-3 is a histone-binding protein with two independent transcriptional corepressor activities. J Biol Chem. 2002;277(47):45004–12. doi: 10.1074/jbc.M205259200. [DOI] [PubMed] [Google Scholar]
- 173.Ramani B, et al. Comparison of spinocerebellar ataxia type 3 mouse models identifies early gain-of-function, cell-autonomous transcriptional changes in oligodendrocytes. Hum Mol Genet. 2017;26(17):3362–3374. doi: 10.1093/hmg/ddx224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Havel LS, Li S, Li XJ. Nuclear accumulation of polyglutamine disease proteins and neuropathology. Mol Brain. 2009;2:21. doi: 10.1186/1756-6606-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Shimohata T, et al. Expanded polyglutamine stretches interact with TAFII130, interfering with CREB-dependent transcription. Nat Genet. 2000;26(1):29–36. doi: 10.1038/79139. [DOI] [PubMed] [Google Scholar]
- 176.Friedman MJ, et al. Polyglutamine domain modulates the TBP-TFIIB interaction: implications for its normal function and neurodegeneration. Nat Neurosci. 2007;10(12):1519–28. doi: 10.1038/nn2011. [DOI] [PubMed] [Google Scholar]
- 177.Xiang C, et al. Transcriptional Dysregulation and Post-translational Modifications in Polyglutamine Diseases: From Pathogenesis to Potential Therapeutic Strategies. Front Mol Neurosci. 2018;11:153. doi: 10.3389/fnmol.2018.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lin XP, et al. Valproic acid attenuates the suppression of acetyl histone H3 and CREB activity in an inducible cell model of Machado-Joseph disease. Int J Dev Neurosci. 2014;38:17–22. doi: 10.1016/j.ijdevneu.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 179.Wang ZJ, et al. Divalproex sodium modulates nuclear localization of ataxin-3 and prevents cellular toxicity caused by expanded ataxin-3. CNS Neurosci Ther. 2018;24(5):404–411. doi: 10.1111/cns.12795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yi J, et al. Sodium valproate alleviates neurodegeneration in SCA3/MJD via suppressing apoptosis and rescuing the hypoacetylation levels of histone H3 and H4. PLoS One. 2013;8(1):e54792. doi: 10.1371/journal.pone.0054792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.McCullough SD, et al. Reelin is a target of polyglutamine expanded ataxin-7 in human spinocerebellar ataxia type 7 (SCA7) astrocytes. Proc Natl Acad Sci U S A. 2012;109(52):21319–24. doi: 10.1073/pnas.1218331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lei LF, et al. Safety and efficacy of valproic acid treatment in SCA3/MJD patients. Parkinsonism Relat Disord. 2016;26:55–61. doi: 10.1016/j.parkreldis.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 183.Duarte-Silva S, Maciel P. Pharmacological Therapies for Machado-Joseph Disease. Adv Exp Med Biol. 2018;1049:369–394. doi: 10.1007/978-3-319-71779-1_19. [DOI] [PubMed] [Google Scholar]
- 184.Bezprozvanny I. Calcium signaling and neurodegenerative diseases. Trends Mol Med. 2009;15(3):89–100. doi: 10.1016/j.molmed.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yiu EM, et al. An open-label trial in Friedreich ataxia suggests clinical benefit with high-dose resveratrol, without effect on frataxin levels. J Neurol. 2015;262(5):1344–53. doi: 10.1007/s00415-015-7719-2. [DOI] [PubMed] [Google Scholar]
- 186.Goncalves N, et al. Caffeine alleviates progressive motor deficits in a transgenic mouse model of spinocerebellar ataxia. Ann Neurol. 2017;81(3):407–418. doi: 10.1002/ana.24867. [DOI] [PubMed] [Google Scholar]
- 187.Peng Q, et al. The antidepressant sertraline improves the phenotype, promotes neurogenesis and increases BDNF levels in the R6/2 Huntington’s disease mouse model. Exp Neurol. 2008;210(1):154–63. doi: 10.1016/j.expneurol.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ristori G, et al. Riluzole in cerebellar ataxia: a randomized, double-blind, placebo-controlled pilot trial. Neurology. 2010;74(10):839–45. doi: 10.1212/WNL.0b013e3181d31e23. [DOI] [PubMed] [Google Scholar]
- 189.Lee Y, et al. miR-19, miR-101 and miR-130 co-regulate ATXN1 levels to potentially modulate SCA1 pathogenesis. Nat Neurosci. 2008;11(10):1137–9. doi: 10.1038/nn.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Persengiev S, et al. Genome-wide analysis of miRNA expression reveals a potential role for miR-144 in brain aging and spinocerebellar ataxia pathogenesis. Neurobiol Aging. 2011;32(12):2316.e17–27. doi: 10.1016/j.neurobiolaging.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 191.Bilen J, et al. MicroRNA pathways modulate polyglutamine-induced neurodegeneration. Mol Cell. 2006;24(1):157–63. doi: 10.1016/j.molcel.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 192.Liu N, et al. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature. 2012;482(7386):519–23. doi: 10.1038/nature10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Carmona V, et al. Unravelling Endogenous MicroRNA System Dysfunction as a New Pathophysiological Mechanism in Machado-Joseph Disease. Mol Ther. 2017;25(4):1038–1055. doi: 10.1016/j.ymthe.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Miyazaki Y, et al. An miRNA-mediated therapy for SCA6 blocks IRES-driven translation of the CACNA1A second cistron. Sci Transl Med. 2016;8(347):347ra94. doi: 10.1126/scitranslmed.aaf5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Tan JY, et al. Cross-talking noncoding RNAs contribute to cell-specific neurodegeneration in SCA7. Nat Struct Mol Biol. 2014;21(11):955–961. doi: 10.1038/nsmb.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Xia H, et al. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat Med. 2004;10(8):816–20. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- 197.Keiser MS, Boudreau RL, Davidson BL. Broad therapeutic benefit after RNAi expression vector delivery to deep cerebellar nuclei: implications for spinocerebellar ataxia type 1 therapy. Mol Ther. 2014;22(3):588–595. doi: 10.1038/mt.2013.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Keiser MS, et al. RNAi prevents and reverses phenotypes induced by mutant human ataxin-1. Ann Neurol. 2016;80(5):754–765. doi: 10.1002/ana.24789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Keiser MS, et al. RNAi or overexpression: alternative therapies for Spinocerebellar Ataxia Type 1. Neurobiol Dis. 2013;56:6–13. doi: 10.1016/j.nbd.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Keiser MS, et al. Broad distribution of ataxin 1 silencing in rhesus cerebella for spinocerebellar ataxia type 1 therapy. Brain. 2015;138(Pt 12):3555–66. doi: 10.1093/brain/awv292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Rodriguez-Lebron E, et al. Silencing mutant ATXN3 expression resolves molecular phenotypes in SCA3 transgenic mice. Mol Ther. 2013;21(10):1909–18. doi: 10.1038/mt.2013.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Costa Mdo C, et al. Toward RNAi therapy for the polyglutamine disease Machado-Joseph disease. Mol Ther. 2013;21(10):1898–908. doi: 10.1038/mt.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Conceição M, et al. Intravenous administration of brain-targeted stable nucleic acid lipid particles alleviates Machado-Joseph disease neurological phenotype. Biomaterials. 2016;82:124–37. doi: 10.1016/j.biomaterials.2015.12.021. [DOI] [PubMed] [Google Scholar]
- 204.Ramachandran PS, et al. RNA interference-based therapy for spinocerebellar ataxia type 7 retinal degeneration. PLoS One. 2014;9(4):e95362. doi: 10.1371/journal.pone.0095362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ramachandran PS, et al. Nonallele specific silencing of ataxin-7 improves disease phenotypes in a mouse model of SCA7. Mol Ther. 2014;22(9):1635–42. doi: 10.1038/mt.2014.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Miller VM, et al. Allele-specific silencing of dominant disease genes. Proc Natl Acad Sci U S A. 2003;100(12):7195–200. doi: 10.1073/pnas.1231012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Li Y, et al. Sequence-dependent and independent inhibition specific for mutant ataxin-3 by small interfering RNA. Ann Neurol. 2004;56(1):124–9. doi: 10.1002/ana.20141. [DOI] [PubMed] [Google Scholar]
- 208.Kubodera T, et al. New RNAi strategy for selective suppression of a mutant allele in polyglutamine disease. Oligonucleotides. 2005;15(4):298–302. doi: 10.1089/oli.2005.15.298. [DOI] [PubMed] [Google Scholar]
- 209.Scholefield J, et al. Allele-specific silencing of mutant Ataxin-7 in SCA7 patient-derived fibroblasts. Eur J Hum Genet. 2014;22(12):1369–75. doi: 10.1038/ejhg.2014.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Alves S, et al. Allele-specific RNA silencing of mutant ataxin-3 mediates neuroprotection in a rat model of Machado-Joseph disease. PLoS One. 2008;3(10):e3341. doi: 10.1371/journal.pone.0003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Nobrega C, et al. RNA interference mitigates motor and neuropathological deficits in a cerebellar mouse model of Machado-Joseph disease. PLoS One. 2014;9(8):e100086. doi: 10.1371/journal.pone.0100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Nobrega C, et al. Silencing mutant ataxin-3 rescues motor deficits and neuropathology in Machado-Joseph disease transgenic mice. PLoS One. 2013;8(1):e52396. doi: 10.1371/journal.pone.0052396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Moore LR, et al. Evaluation of Antisense Oligonucleotides Targeting ATXN3 in SCA3 Mouse Models. Mol Ther Nucleic Acids. 2017;7:200–210. doi: 10.1016/j.omtn.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.McLoughlin, H.S., et al., Oligonucleotide therapy mitigates disease in spinocerebellar ataxia type 3 mice. Ann Neurol, 2018. 84(1): p. 64–77. [DOI] [PMC free article] [PubMed]
- 215.Hu J, et al. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat Biotechnol. 2009;27(5):478–84. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Hu J, et al. Allele-selective inhibition of ataxin-3 (ATX3) expression by antisense oligomers and duplex RNAs. Biol Chem. 2011;392(4):315–25. doi: 10.1515/BC.2011.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Liu J, et al. ss-siRNAs allele selectively inhibit ataxin-3 expression: multiple mechanisms for an alternative gene silencing strategy. Nucleic Acids Res. 2013;41(20):9570–83. doi: 10.1093/nar/gkt693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Evers MM, et al. Targeting several CAG expansion diseases by a single antisense oligonucleotide. PLoS One. 2011;6(9):e24308. doi: 10.1371/journal.pone.0024308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Scoles DR, et al. Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature. 2017;544(7650):362–366. doi: 10.1038/nature22044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Fiszer, A., et al., Mutant CAG Repeats Effectively Targeted by RNA Interference in SCA7 Cells. Genes, 2016. 7(12): p. 132. [DOI] [PMC free article] [PubMed]
- 221.Ouyang S, et al. CRISPR/Cas9-Targeted Deletion of Polyglutamine in Spinocerebellar Ataxia Type 3-Derived Induced Pluripotent Stem Cells. Stem Cells Dev. 2018;27(11):756–770. doi: 10.1089/scd.2017.0209. [DOI] [PubMed] [Google Scholar]
- 222.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 223.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–8. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 224.Rao DD, et al. siRNA vs. shRNA: similarities and differences. Adv Drug Deliv Rev. 2009;61(9):746–59. doi: 10.1016/j.addr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 225.Lam JK, et al. siRNA Versus miRNA as Therapeutics for Gene Silencing. Mol Ther Nucleic Acids. 2015;4:e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Torrecilla J, et al. Lipid nanoparticles as carriers for RNAi against viral infections: current status and future perspectives. Biomed Res Int. 2014;2014:161794. doi: 10.1155/2014/161794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Friedman RC, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9(9):1034–48. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18(5):504–11. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Moore CB, et al. Short hairpin RNA (shRNA): design, delivery, and assessment of gene knockdown. Methods Mol Biol. 2010;629:141–58. doi: 10.1007/978-1-60761-657-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Koscianska E, Krzyzosiak WJ. Current understanding of the role of microRNAs in spinocerebellar ataxias. Cerebellum Ataxias. 2014;1:7. doi: 10.1186/2053-8871-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 233.McCann C, et al. The Ataxin-2 protein is required for microRNA function and synapse-specific long-term olfactory habituation. Proc Natl Acad Sci U S A. 2011;108(36):E655–62. doi: 10.1073/pnas.1107198108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Reinhardt A, et al. Lack of miRNA Misregulation at Early Pathological Stages in Drosophila Neurodegenerative Disease Models. Front Genet. 2012;3:226. doi: 10.3389/fgene.2012.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Singh NK. miRNAs target databases: developmental methods and target identification techniques with functional annotations. Cell Mol Life Sci. 2017;74(12):2239–2261. doi: 10.1007/s00018-017-2469-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Rodriguez-Lebron E, Paulson HL. Allele-specific RNA interference for neurological disease. Gene Ther. 2006;13(6):576–81. doi: 10.1038/sj.gt.3302702. [DOI] [PubMed] [Google Scholar]
- 237.Gaspar C, et al. Ancestral origins of the Machado-Joseph disease mutation: a worldwide haplotype study. Am J Hum Genet. 2001;68(2):523–8. doi: 10.1086/318184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Adams D, et al. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N Engl J Med. 2018;379(1):11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 239.Evers MM, Toonen LJ, van Roon-Mom WM. Antisense oligonucleotides in therapy for neurodegenerative disorders. Adv Drug Deliv Rev. 2015;87:90–103. doi: 10.1016/j.addr.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 240.Rinaldi C, Wood MJA. Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nat Rev Neurol. 2017;14:9. doi: 10.1038/nrneurol.2017.148. [DOI] [PubMed] [Google Scholar]
- 241.Smith RA, et al. Antisense oligonucleotide therapy for neurodegenerative disease. J Clin Invest. 2006;116(8):2290–6. doi: 10.1172/JCI25424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Miller TM, et al. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurol. 2013;12(5):435–42. doi: 10.1016/S1474-4422(13)70061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Williams JH, et al. Oligonucleotide-mediated survival of motor neuron protein expression in CNS improves phenotype in a mouse model of spinal muscular atrophy. J Neurosci. 2009;29(24):7633–8. doi: 10.1523/JNEUROSCI.0950-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Shorrock HK, Gillingwater TH, Groen EJN. Overview of Current Drugs and Molecules in Development for Spinal Muscular Atrophy Therapy. Drugs. 2018;78(3):293–305. doi: 10.1007/s40265-018-0868-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Finkel RS, et al. Nusinersen versus Sham Control in Infantile-Onset Spinal Muscular Atrophy. N Engl J Med. 2017;377(18):1723–1732. doi: 10.1056/NEJMoa1702752. [DOI] [PubMed] [Google Scholar]
- 246.Geary RS, et al. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv Drug Deliv Rev. 2015;87:46–51. doi: 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 247.Schoch KM, Miller TM. Antisense Oligonucleotides: Translation from Mouse Models to Human Neurodegenerative Diseases. Neuron. 2017;94(6):1056–1070. doi: 10.1016/j.neuron.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Kordasiewicz HB, et al. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron. 2012;74(6):1031–44. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.van Roon-Mom WMC, Roos RAC, de Bot ST. Dose-Dependent Lowering of Mutant Huntingtin Using Antisense Oligonucleotides in Huntington Disease Patients. Nucleic Acid Ther. 2018;28(2):59–62. doi: 10.1089/nat.2018.0720. [DOI] [PubMed] [Google Scholar]
- 250.Tabrizi, S., et al., Effects of IONIS-HTTRx in Patients with Early Huntington’s Disease, Results of the First HTT-Lowering Drug Trial (CT.002). Neurology, 2018. 90(15 Supplement): p. CT.002.
- 251.Datson NA, et al. The expanded CAG repeat in the huntingtin gene as target for therapeutic RNA modulation throughout the HD mouse brain. PLoS One. 2017;12(2):e0171127. doi: 10.1371/journal.pone.0171127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Carroll JB, et al. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene / allele-specific silencing of mutant huntingtin. Mol Ther. 2011;19(12):2178–85. doi: 10.1038/mt.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Ostergaard ME, et al. Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res. 2013;41(21):9634–50. doi: 10.1093/nar/gkt725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Aartsma-Rus A, van Ommen GJ. Antisense-mediated exon skipping: a versatile tool with therapeutic and research applications. RNA. 2007;13(10):1609–24. doi: 10.1261/rna.653607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Zalachoras I, et al. Antisense-mediated RNA targeting: versatile and expedient genetic manipulation in the brain. Front Mol Neurosci. 2011;4:10. doi: 10.3389/fnmol.2011.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Evers MM, et al. Ataxin-3 protein modification as a treatment strategy for spinocerebellar ataxia type 3: removal of the CAG containing exon. Neurobiol Dis. 2013;58:49–56. doi: 10.1016/j.nbd.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 257.Toonen LJA, et al. Antisense Oligonucleotide-Mediated Removal of the Polyglutamine Repeat in Spinocerebellar Ataxia Type 3 Mice. Mol Ther Nucleic Acids. 2017;8:232–242. doi: 10.1016/j.omtn.2017.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Cooper JK, et al. Truncated N-terminal fragments of huntingtin with expanded glutamine repeats form nuclear and cytoplasmic aggregates in cell culture. Hum Mol Genet. 1998;7(5):783–90. doi: 10.1093/hmg/7.5.783. [DOI] [PubMed] [Google Scholar]
- 259.Schilling G, et al. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum Mol Genet. 1999;8(3):397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- 260.Toonen LJ, et al. Antisense oligonucleotide-mediated exon skipping as a strategy to reduce proteolytic cleavage of ataxin-3. Sci Rep. 2016;6:35200. doi: 10.1038/srep35200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Shin JW, et al. Permanent inactivation of Huntington’s disease mutation by personalized allele-specific CRISPR/Cas9. Hum Mol Genet. 2016;25(20):4566–4576. doi: 10.1093/hmg/ddw286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Zhang XH, et al. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol Ther Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36(8):765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Liu C, et al. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J Control Release. 2017;266:17–26. doi: 10.1016/j.jconrel.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Nishiyama J, Mikuni T, Yasuda R. Virus-Mediated Genome Editing via Homology-Directed Repair in Mitotic and Postmitotic Cells in Mammalian Brain. Neuron. 2017;96(4):755–768.e5. doi: 10.1016/j.neuron.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Lindvall O, et al. Grafts of fetal dopamine neurons survive and improve motor function in Parkinson’s disease. Science. 1990;247(4942):574–7. doi: 10.1126/science.2105529. [DOI] [PubMed] [Google Scholar]
- 270.Lindvall O, et al. Human fetal dopamine neurons grafted into the striatum in two patients with severe Parkinson’s disease. A detailed account of methodology and a 6-month follow-up. Arch Neurol. 1989;46(6):615–31. doi: 10.1001/archneur.1989.00520420033021. [DOI] [PubMed] [Google Scholar]
- 271.Kopyov OV, et al. Safety of intrastriatal neurotransplantation for Huntington’s disease patients. Exp Neurol. 1998;149(1):97–108. doi: 10.1006/exnr.1997.6685. [DOI] [PubMed] [Google Scholar]
- 272.Kaemmerer WF, Low WC. Cerebellar allografts survive and transiently alleviate ataxia in a transgenic model of spinocerebellar ataxia type-1. Exp Neurol. 1999;158(2):301–11. doi: 10.1006/exnr.1999.7099. [DOI] [PubMed] [Google Scholar]
- 273.Chintawar S, et al. Grafting neural precursor cells promotes functional recovery in an SCA1 mouse model. J Neurosci. 2009;29(42):13126–35. doi: 10.1523/JNEUROSCI.0647-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Mendonca LS, et al. Transplantation of cerebellar neural stem cells improves motor coordination and neuropathology in Machado-Joseph disease mice. Brain. 2015;138(Pt 2):320–35. doi: 10.1093/brain/awu352. [DOI] [PubMed] [Google Scholar]
- 275.Matsuura S, et al. Mesenchymal stem cells ameliorate cerebellar pathology in a mouse model of spinocerebellar ataxia type 1. Cerebellum. 2014;13(3):323–30. doi: 10.1007/s12311-013-0536-1. [DOI] [PubMed] [Google Scholar]
- 276.Mieda T, et al. Mesenchymal stem cells attenuate peripheral neuronal degeneration in spinocerebellar ataxia type 1 knockin mice. J Neurosci Res. 2016;94(3):246–52. doi: 10.1002/jnr.23698. [DOI] [PubMed] [Google Scholar]
- 277.Chang YK, et al. Mesenchymal stem cell transplantation ameliorates motor function deterioration of spinocerebellar ataxia by rescuing cerebellar Purkinje cells. J Biomed Sci. 2011;18:54. doi: 10.1186/1423-0127-18-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Oliveira Miranda, C., et al., Repeated Mesenchymal Stromal Cell Treatment Sustainably Alleviates Machado-Joseph Disease. Mol Ther, 2018. 26(9): p. 2131–2151. [DOI] [PMC free article] [PubMed]
- 279.Olson SD, et al. Genetically engineered mesenchymal stem cells as a proposed therapeutic for Huntington’s disease. Mol Neurobiol. 2012;45(1):87–98. doi: 10.1007/s12035-011-8219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Bonab MM, et al. Autologous mesenchymal stem cell therapy in progressive multiple sclerosis: an open label study. Curr Stem Cell Res Ther. 2012;7(6):407–14. doi: 10.2174/157488812804484648. [DOI] [PubMed] [Google Scholar]
- 281.Liu J, et al. Clinical analysis of the treatment of spinal cord injury with umbilical cord mesenchymal stem cells. Cytotherapy. 2013;15(2):185–91. doi: 10.1016/j.jcyt.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 282.Hare JM, et al. A randomized, double-blind, placebo-controlled, dose-escalation study of intravenous adult human mesenchymal stem cells (prochymal) after acute myocardial infarction. J Am Coll Cardiol. 2009;54(24):2277–86. doi: 10.1016/j.jacc.2009.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Newman RE, et al. Treatment of inflammatory diseases with mesenchymal stem cells. Inflamm Allergy Drug Targets. 2009;8(2):110–23. doi: 10.2174/187152809788462635. [DOI] [PubMed] [Google Scholar]
- 284.Baddoo M, et al. Characterization of mesenchymal stem cells isolated from murine bone marrow by negative selection. J Cell Biochem. 2003;89(6):1235–49. doi: 10.1002/jcb.10594. [DOI] [PubMed] [Google Scholar]
- 285.Lee OK, et al. Isolation of multipotent mesenchymal stem cells from umbilical cord blood. Blood. 2004;103(5):1669–75. doi: 10.1182/blood-2003-05-1670. [DOI] [PubMed] [Google Scholar]
- 286.Gimble J, Guilak F. Adipose-derived adult stem cells: isolation, characterization, and differentiation potential. Cytotherapy. 2003;5(5):362–9. doi: 10.1080/14653240310003026. [DOI] [PubMed] [Google Scholar]
- 287.Kern S, et al. Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem Cells. 2006;24(5):1294–301. doi: 10.1634/stemcells.2005-0342. [DOI] [PubMed] [Google Scholar]
- 288.Kan I, Melamed E, Offen D. Autotransplantation of bone marrow-derived stem cells as a therapy for neurodegenerative diseases. Handb Exp Pharmacol. 2007;180:219–42. doi: 10.1007/978-3-540-68976-8_10. [DOI] [PubMed] [Google Scholar]
- 289.Sadan O, et al. Migration of neurotrophic factors-secreting mesenchymal stem cells toward a quinolinic acid lesion as viewed by magnetic resonance imaging. Stem Cells. 2008;26(10):2542–51. doi: 10.1634/stemcells.2008-0240. [DOI] [PubMed] [Google Scholar]
- 290.Nakamura K, et al. Mesenchymal stem cells as a potential therapeutic tool for spinocerebellar ataxia. Cerebellum. 2015;14(2):165–70. doi: 10.1007/s12311-014-0604-1. [DOI] [PubMed] [Google Scholar]
- 291.Suto N, et al. Morphological and Functional Attenuation of Degeneration of Peripheral Neurons by Mesenchymal Stem Cell-Conditioned Medium in Spinocerebellar Ataxia Type 1-Knock-in Mice. CNS Neurosci Ther. 2016;22(8):670–6. doi: 10.1111/cns.12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Tsai YA, et al. Treatment of Spinocerebellar Ataxia With Mesenchymal Stem Cells: A Phase I/IIa Clinical Study. Cell Transplant. 2017;26(3):503–512. doi: 10.3727/096368916X694373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Dongmei H, et al. Clinical analysis of the treatment of spinocerebellar ataxia and multiple system atrophy-cerebellar type with umbilical cord mesenchymal stromal cells. Cytotherapy. 2011;13(8):913–7. doi: 10.3109/14653249.2011.579958. [DOI] [PubMed] [Google Scholar]
- 294.Miao X, Wu X, Shi W. Umbilical cord mesenchymal stem cells in neurological disorders: A clinical study. Indian J Biochem Biophys. 2015;52(2):140–6. [PubMed] [Google Scholar]
- 295.Jin JL, et al. Safety and efficacy of umbilical cord mesenchymal stem cell therapy in hereditary spinocerebellar ataxia. Curr Neurovasc Res. 2013;10(1):11–20. doi: 10.2174/156720213804805936. [DOI] [PubMed] [Google Scholar]
- 296.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 297.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 298.Ross CA, Akimov SS. Human-induced pluripotent stem cells: potential for neurodegenerative diseases. Hum Mol Genet. 2014;23(R1):R17–26. doi: 10.1093/hmg/ddu204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Delli Carri A, et al. Developmentally coordinated extrinsic signals drive human pluripotent stem cell differentiation toward authentic DARPP-32+ medium-sized spiny neurons. Development. 2013;140(2):301–12. doi: 10.1242/dev.084608. [DOI] [PubMed] [Google Scholar]
- 300.Fink KD, et al. Intrastriatal transplantation of adenovirus-generated induced pluripotent stem cells for treating neuropathological and functional deficits in a rodent model of Huntington’s disease. Stem Cells Transl Med. 2014;3(5):620–31. doi: 10.5966/sctm.2013-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Mu S, et al. Transplantation of induced pluripotent stem cells improves functional recovery in Huntington’s disease rat model. PLoS One. 2014;9(7):e101185. doi: 10.1371/journal.pone.0101185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Buijsen RAM, et al. Generation of 3 spinocerebellar ataxia type 1 (SCA1) patient-derived induced pluripotent stem cell lines LUMCi002-A, B, and C and 2 unaffected sibling control induced pluripotent stem cell lines LUMCi003-A and B. Stem Cell Res. 2018;29:125–128. doi: 10.1016/j.scr.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 303.Xia G, et al. Generation of human-induced pluripotent stem cells to model spinocerebellar ataxia type 2 in vitro. J Mol Neurosci. 2013;51(2):237–48. doi: 10.1007/s12031-012-9930-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Ou Z, et al. Autophagy Promoted the Degradation of Mutant ATXN3 in Neurally Differentiated Spinocerebellar Ataxia-3 Human Induced Pluripotent Stem Cells. Biomed Res Int. 2016;2016:6701793. doi: 10.1155/2016/6701793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Ishida Y, et al. Vulnerability of Purkinje Cells Generated from Spinocerebellar Ataxia Type 6 Patient-Derived iPSCs. Cell Rep. 2016;17(6):1482–1490. doi: 10.1016/j.celrep.2016.10.026. [DOI] [PubMed] [Google Scholar]
- 306.Luo Y, et al. Generation of induced pluripotent stem cells from skin fibroblasts of a patient with olivopontocerebellar atrophy. Tohoku J Exp Med. 2012;226(2):151–9. doi: 10.1620/tjem.226.151. [DOI] [PubMed] [Google Scholar]