Abstract
Codon optimization by synonymous substitution is widely used for recombinant protein expression. Recent studies have investigated sequence features for codon optimization based on large-scale expression analyses. However, these studies have been limited to common host organisms such as Escherichia coli. Here, we develop a codon optimization method for Rhodococcus erythropolis, a gram-positive GC-rich actinobacterium attracting attention as an alternative host organism. We evaluate the recombinant protein expression of 204 genes in R. erythropolis with the same plasmid vector. The statistical analysis of these expression data reveals that the mRNA folding energy at 5’ regions as well as the codon frequency are important sequence features for codon optimization. Intriguingly, other sequence features such as the codon repetition rate show a different tendency from the previous study on E. coli. We optimize the coding sequences of 12 genes regarding these sequence features, and confirm that 9 of them (75%) achieve increased expression levels compared with wild-type sequences. Especially, for 5 genes whose expression levels for wild-type sequences are small or not detectable, all of them are improved by optimized sequences. These results demonstrate the effectiveness of our codon optimization method in R. erythropolis, and possibly in other actinobacteria.
Subject terms: Expression systems, Applied microbiology
Introduction
Recombinant protein expression using bacterial and other host organisms is a fundamental technology for protein production1. A key step in recombinant protein expression is codon optimization where a coding sequence for a protein of interest is designed by synonymous substitution aiming to increase its expression level2.
Current approaches to codon optimization are based on sequence features considered to influence protein expression levels3–6. For example, a conventional approach is to substitute rare codons by frequent codons according to the genomic codon usage in a host organism. The basis of this approach is that endogenous genes whose coding sequences consist of frequent codons have high protein expression levels, and thus recombinant protein expression is also considered to be improved by increasing the codon frequency. Another approach is to introduce synonymous substitution computationally predicted to destabilize mRNA secondary structures. Since stable mRNA secondary structures may inhibit translation, this approach is considered to improve recombinant protein expression by enhancing translational efficiency. The association between these sequence features and protein expression levels has been indicated by omics analyses of endogenous genes (e.g.7). On the other hand, the direct evidence of their influences in recombinant protein expression has been shown using a relatively small number of genes (e.g.8).
Recently, large-scale analyses of recombinant protein expression have revealed sequence features for codon optimization in unprecedented detail9,10. In these studies, the recombinant protein expression of thousands of genes is evaluated in a systematic way using the same host organism and the same plasmid vector. Then, the influence of various sequence features on protein expression levels is investigated by statistical analyses. This strategy has provided new insights into conventionally-used sequence features such as the effect of the codon frequency depending on sequence positions (e.g. 5′ regions or others). In addition, a variety of new sequence features have been shown to be important including the use of specific di-codons and the repeated occurrence of codons in neighboring positions. However, such studies have been so far limited to common host organisms such as Escherichia coli, presenting a question whether these sequence features are also effective for codon optimization in less-studied host organisms.
Rhodococcus erythropolis, a gram-positive GC-rich actinobacterium, has been used as a host organism for recombinant protein expression and for the heterologous production of antimicrobial compounds11–15. R. erythropolis grows and produces recombinant proteins at a wide temperature range from 4 to 35 °C, and has different intracellular milieu compared to other host organisms such as E. coli (a gram-negative bacterium) and Bacillus and Lactococcus species (gram-positive bacteria with moderate GC contents). Due to these characteristics, R. erythropolis can produce recombinant proteins that are difficult to be expressed in E. coli. R. erythropolis has also been shown to produce the bacterial lipoglycoproteins from Mycobacterium tuberculosis, which cannot be expressed in E. coli for their post-translational modifications16. Based upon these performances, R. erythropolis is recognized as an alternative next-generation host microorganism.
Here, we develop a codon optimization method based on the statistical analysis of the recombinant protein expression data of 204 genes using R. erythropolis with the same plasmid vector. The statistical analysis reveals the mRNA folding energy at 5′ regions and the codon frequency as the most important sequence features for codon optimization. Interestingly, other sequence features including the codon repetition rate show a tendency different from E. coli, suggesting the species specificity of their influence on protein expression levels. We design the coding sequences of selected genes based on the optimization of these sequence features, and demonstrate that most of them become to show increased expression levels compared to wild-type sequences.
Results
Recombinant protein expression dataset in R. erythropolis
To develop a codon optimization method for R. erythropolis, we sought to investigate sequence features that influence protein expression levels. For this purpose, we evaluated the recombinant protein expression of 204 genes in R. erythropolis (Supplementary Data S1). These genes were selected from Streptomyces coelicolor, and were heterologously expressed in R. erythropolis using the pTip plasmid vector12. This expression system allowed us to evaluate the recombinant protein expression of various genes under the transcription from the same promoter, thereby focusing on their difference in translational efficiency. Based on the visual inspection of SDS-PAGE gels (Supplementary Fig. S1), protein expression levels were measured by integer scores: 1 (low or not detected), 2 (medium), and 3 (high). Note that such a discrete scoring scheme has also been employed in the previous study on E. coli9.
Statistical analysis of sequence features influencing protein expression levels
We used our data to analyze the influence of sequence features on protein expression levels. We considered various sequence features including the measures of the codon frequency such as the codon adaptation index (CAI)17 and the tRNA adaptation index (tAI)18 and the measures of the repeated occurrence of codons such as the codon repetition rate9 and the amino acid repetition rate9. In addition, the stability of mRNA secondary structures at 5′ regions was measured by the folding energy (ΔGUH) predicted by EnsembleEnergy program in RNAStructure package19. For computing ΔGUH, 5′ regions were defined as the 5′ untranslated region (UTR) in the pTip plasmid vector plus 33 nucleotides at the head of coding sequences. Calculated feature values are summarized in Supplementary Data S1. For each type of sequence feature, a polyserial correlation coefficient20 was evaluated between feature values and protein expression levels (Fig. 1a). The positive correlation coefficients were detected for CAI, tAI, and ΔGUH, which is consistent with the previous report on E. coli9. Intriguingly, the results for the codon repetition rate and the amino acid repetition rate showed a tendency different from E. coli. While these sequence features have been reported to be negatively correlated with protein expression levels in E. coli, our results in R. erythropolis showed positive correlation coefficients, implicating the species-specific influence of these sequence features. Supplementary Fig. S2 summarizes the comparison between our results on R. erythropolis and the previous study on E. coli. CAI and ΔGUH are important factors not only in R. erythropolis but also in E. coli. On the other hand, the contributions of the codon repetition rate and the amino acid repetition rate are larger in E. coli compared with R. erythropolis.
Figure 1.
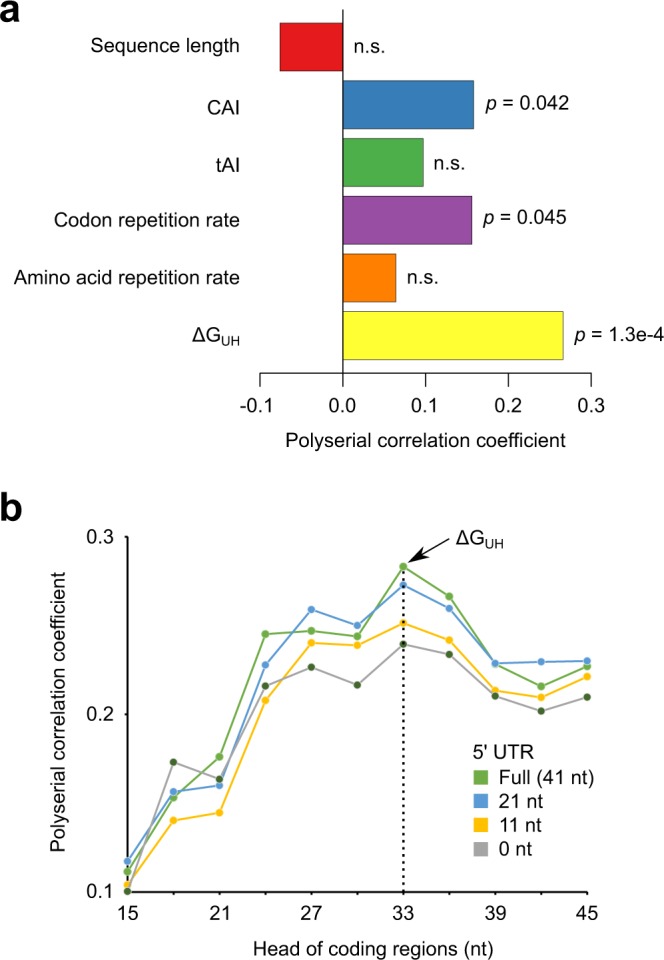
Influence of sequence features on protein expression levels. (a) For each type of sequence feature, a polyserial correlation coefficient between feature values and expression levels is shown with its p-value. The ΔGUH gave the largest correlation coefficient, meaning that higher ΔGUH (weaker mRNA secondary structures at 5′ regions) associate with higher expression levels. n.s.: p > 0.05. (b) The polyserial correlation coefficients for mRNA folding energies are shown using the different lengths of subsequences in 5′ UTR and the head coding region. The largest correlation coefficient was obtained when the full-length 5′ UTR and 33 head nucleotides were used, which is equivalent to ΔGUH in (a).
Among the sequence features considered, ΔGUH showed the largest correlation coefficient, suggesting that higher mRNA folding energies (i.e. weaker mRNA secondary structures) at 5′ regions lead to increased protein expression levels. The influence of the mRNA folding energy was further investigated by changing the definition of 5′ regions (Fig. 1b). The correlation coefficient was the maximum for ΔGUH with the full-length 5′ UTR plus 33 head nucleotides, whereas the use of extended or truncated 5′ regions did not achieve larger correlation coefficients. These results motivated us to develop a codon optimization method based on ΔGUH. In addition to ΔGUH, CAI showed the second-largest correlation coefficient, suggesting that the use of frequent codons is also effective for increasing protein expression levels.
H-method: codon optimization based on mRNA folding energy
We first devised a codon optimization method based solely on ΔGUH, which we named “H-method”. For a given protein, H-method computationally generates coding sequences for all possible synonymous variants regarding to 33 head nucleotides. H-method then calculates ΔGUH for each synonymous variant, and proposes the coding sequence with the highest ΔGUH.
We note that H-method introduces mutations only to 33 head nucleotides (i.e. 11 codons) with downstream nucleotides unmodified. Therefore, the number of generated synonymous variants can be kept relatively small, which allows us to calculate ΔGUH for all possible synonymous variants regarding 33 head nucleotides. Such an exhaustive computation is not feasible when the entire coding sequence is mutated, since the number of possible synonymous variants increases exponentially with the sequence length. The merits of focusing on head nucleotides are not only computational costs but also experimental costs. If codon optimization modifies the entire coding sequence, we need to use full-length gene synthesis that requires a relatively high experimental cost. In contrast, the modification of head nucleotides can be performed by primer-based mutagenesis that is much cheaper than full-length gene synthesis (Methods). As will be shown later, this allows us to test the effectiveness of our method using a large number of sequences. Such a low experimental cost is important for facilitating a wide applicability of codon optimization.
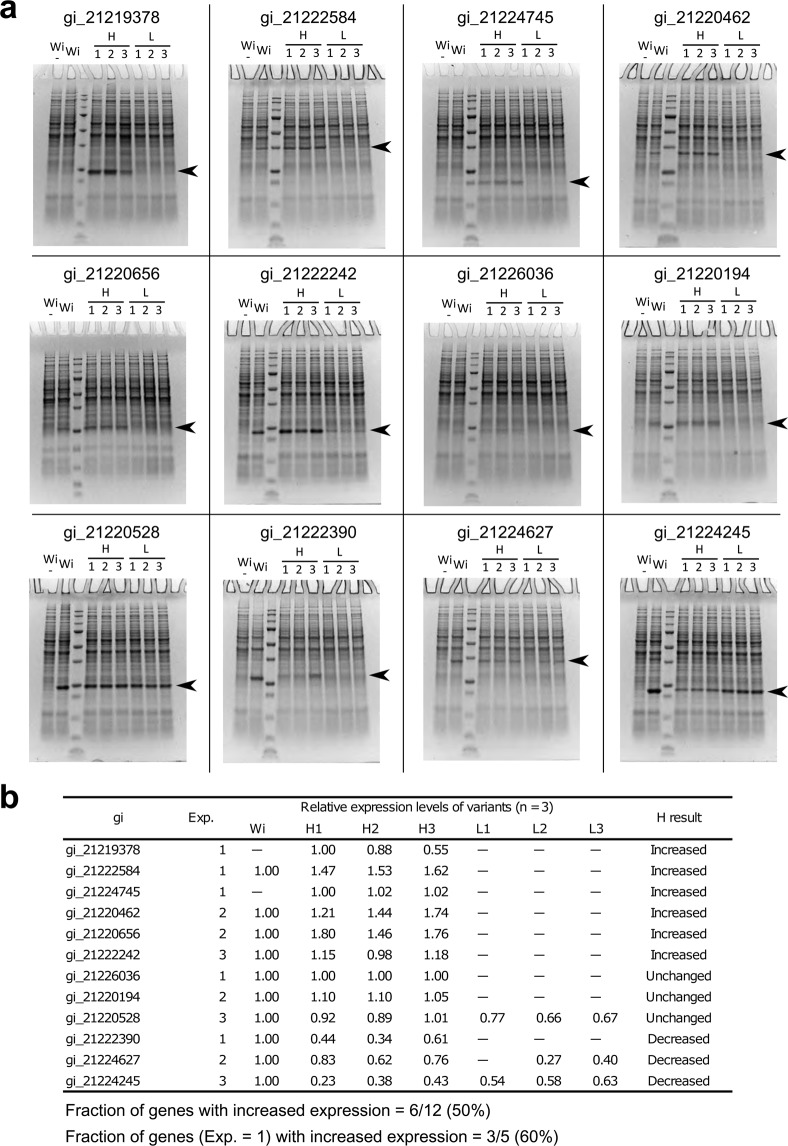
To test the effectiveness of H-method, we design the coding sequences of genes using H-method, and compared protein expression levels between the optimized sequences and the wild-type sequences. We selected 12 genes so that their protein expression levels before codon optimization vary: 5 genes with the score of 1 (low or not detected), 4 genes with the score of 2 (medium), and 3 genes with the score of 3 (high). For each gene, we designed the 3 sequences with the first-to-third highest ΔGUH using H-method (H1-3). For comparison, we also designed the 3 sequences with the first-to-third lowest ΔGUH (i.e. deoptimized sequences; L1-3). These sequences were transformed into R. erythropolis using the pTip plasmid vector (Fig. 2a), and the protein expression levels were measured using three biological replicates (Fig. 2b). In summary, the optimized sequences showed increased expression levels compared with the wild-type sequences for 6 out of the 12 genes. However, for the remaining 6 genes, the improvement in protein expression was not observed. These results demonstrated the effectiveness of H-method, while indicating its limitation in the success rate. The effect of ΔGUH in codon optimization was also supported by the deoptimized sequences whose protein expression levels were substantially decreased from the wild-type sequences for all of the 12 genes.
Figure 2.
Codon optimization by H-method. (a) For each of 12 genes, the recombinant protein expression using different coding sequences is shown by SDS-PAGE. The expected positions of the recombinant proteins are indicated by black arrows. Wi: wild-type sequence. H1-3: optimized sequences with the first-to-third highest ΔGUH. L1-3: deoptimized sequences with the first-to-third lowest ΔGUH. W-: negative control where the wild-type sequence was used but the induction of expression was not performed. Note that although multiple gels are presented, the comparison of bands is performed only within each gel, but not between different gels. No gels are cropped and regrouped for comparison. For confirmation, raw image data are available as Supplementary Data S5. (b) Expression levels for the optimized sequences relative to the wild-type sequence. For gi_21219378 and gi_21224745, expression levels relative to H1 are shown since the expression for the wild-type sequences was not detected. Each expression level is the average of three biological replicates from individual recombinant clones. —: expression not detected. Also shown is the summary of the results for the H-series sequences. Increased: more than 1.1-fold increase. Decreased: less than 0.9-fold decrease. Unchanged: not changed by means of the threshold of the 1.1-fold or 0.9-fold change. Exp.: integer expression score for the wild-type sequence in our expression dataset (Supplementary Data S1).
For investigating the cause that H-method could not improve protein expression, we recall that CAI showed the second-largest correlation coefficient next to ΔGUH (Fig. 1a). Therefore, we hypothesized that the mutations introduced by H-method might decrease CAI, thereby leading to reduced protein expression levels. To test this hypothesis, we calculated CAI for the coding sequences designed by H-method focusing only on 33 head nucleotides (CAIH). CAIH was compared between the two groups of genes whose protein expression levels were decreased or not (Fig. 3a). We also measured the number of rare codons used in 33 head nucleotides (Fig. 3b; Supplementary Data S2) where rare codons were defined as TTA, ATA, and AGA whose normalized codon frequencies are less than 0.1 (Methods). Indeed, the coding sequences designed by H-method showed smaller CAIH and more rare codons for the genes with decreased protein expression levels compared to the other genes. These results suggested that a more powerful codon optimization method can be developed by combining both ΔGUH and CAIH.
Figure 3.
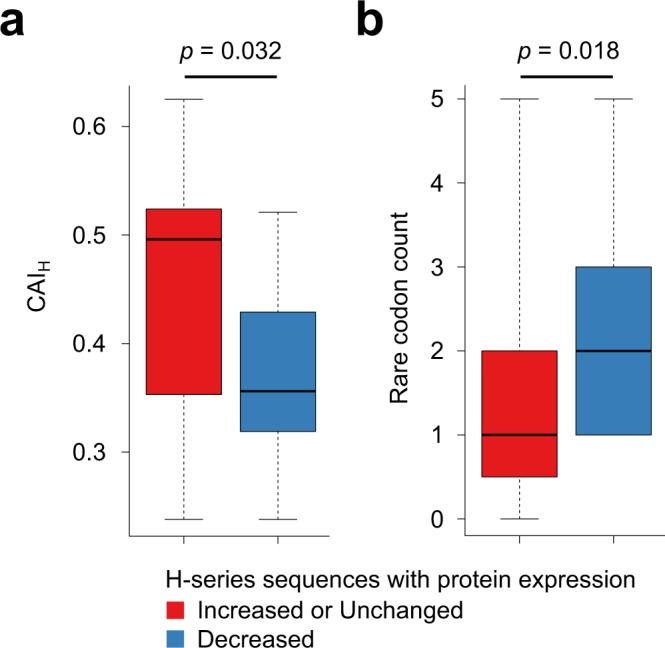
Failure of expression improvement by H-method is associated with decreased codon frequencies. For the H-series sequences, CAIH (a) and the rare codon count (b) are compared between genes whose expression levels were decreased by H-method (indicated as “Decreased” in Fig. 2b) or not (indicated as “Increased” or “Unchanged” in Fig. 2b). P-values were calculated by Mann-Whitney U test.
C-method: codon optimization combining mRNA folding energy and codon frequency
To improve the success rate of our codon optimization method, we next devised an approach combining both ΔGUH and CAIH, which we named “C-method”. Similar to H-method, C-method proposes the coding sequence with the highest ΔGUH from synonymous variants regarding 33 head nucleotides. However, generated synonymous variants are restricted so that they have CAIH larger than a user-specified threshold and contain no rare codon. This approach enables us to design coding sequences with high ΔGUH while controlling CAIH (Supplementary Fig. S3).
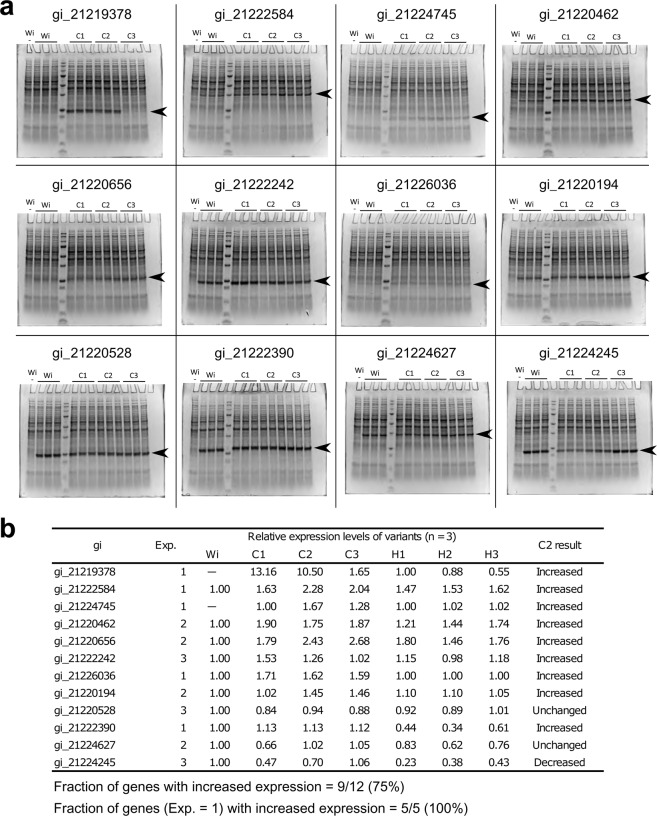
We tested the effectiveness of C-method by an experiment similar to that for H-method (Fig. 4a). For each gene, the 3 sequences were designed by C-method using the different thresholds for CAIH: 0.60 (C1), 0.75 (C2), and 0.90 (C3). C-method achieved the success rate better than H-method (Fig. 4b). For the C2 sequences, protein expression levels were increased for 9 out of the 12 genes (75%). Strikingly, when focusing on the 5 genes whose protein expression levels for the wild-type sequences were low or not detectable, all of them were improved by optimized sequences. For the C1 and C3 sequences, 8 out of the 12 genes were improved in each case, showing the success rate slightly worse than the C2 sequences. However, the improvement was still observed for all of the 5 genes with poor wild-type expression levels. These results demonstrated the effectiveness of C-method for improved expression of recombinant proteins in R. erythropolis.
Figure 4.
Codon optimization by C-method. (a) For each of 12 genes, the recombinant protein expression using different coding sequences is shown by SDS-PAGE. The expected positions of the recombinant proteins are indicated by black arrows. Wi: wild-type sequence. C1-3: optimized sequences with the highest ΔGUH satisfying the CAIH thresholds of 0.60 (C1), 0.75 (C2), and 0.90 (C3). H1-3: optimized sequences with the first-to-third highest ΔGUH without the restriction of CAIH. W-: negative control where the wild-type sequence was used but the induction of expression was not performed. Note that although multiple gels are presented, the comparison of bands is performed only within each gel, but not between different gels. No gels are cropped and regrouped for comparison. For confirmation, raw image data are available as Supplementary Data S5. (b) Expression levels for the optimized sequences relative to the wild-type sequence. For gi_21219378 and gi_21224745, expression levels relative to H1 are shown since the expression for the wild-type sequences was not detected as indicated by “—”. Each expression level is the average of three biological replicates from individual recombinant clones. Increased: more than 1.1-fold increase. Decreased: less than 0.9-fold decrease. Unchanged: not changed by means of the threshold of the 1.1-fold or 0.9-fold change. Exp.: integer expression score for the wild-type sequence in our expression dataset (Supplementary Data S1).
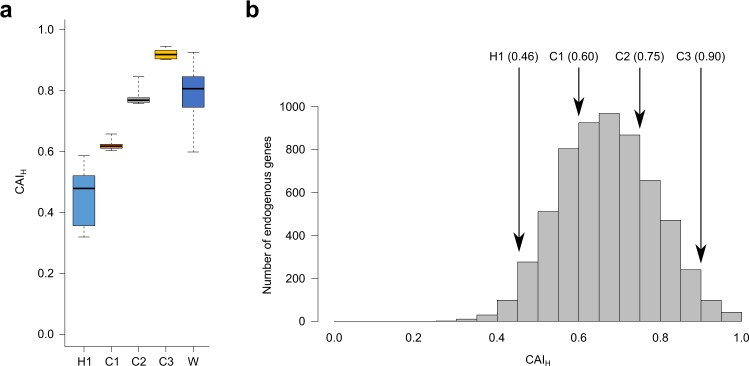
Since C-method uses a CAIH threshold as a parameter, we need the criteria for selecting the CAIH threshold when applying C-method to other genes and/or host organisms. For this purpose, we conducted the following analyses. First, we compared CAIH of the C1, C2, and C3 sequences with the corresponding wild-type sequences before codon optimization (Fig. 5a). We found that the C2 sequences had on average a similar level of CAIH to the wild-type sequences while the C1 and C3 sequences were at the low and high extremes, respectively, in the wild-type distribution. This result suggests a criterion that an appropriate CAIH threshold can be determined according to CAIH of the wild-type sequences of genes to be expressed. Second, we compared the CAIH thresholds of C1, C2, and C3 with the all endogenous genes in R. erythropolis (Fig. 5b). We found that C2 was moderately higher from the median of the endogenous gene distribution whereas C1 and C3 were lower and much higher than the median, respectively. This result suggests another criterion that CAIH thresholds should be higher from the median of the endogenous gene distribution while avoiding extremely high values. Taken together, we recommend that the CAIH threshold is selected so that it does not largely deviate from those of the wild-type sequences and the endogenous genes. We also note that although C2 was the best performing in our verification experiment, C1 and C3 achieved the similar performance to C2 (Fig. 4b). Their success rates were comparable: C2 (9 out of the 12 genes) versus C1 and C3 (8 out of the 12 genes). In addition, all of the 5 genes with poor wild-type expression levels were improved not only by C2, but also by C1 and C3. Therefore, the exact value of the CAIH threshold may not drastically affect the performance of C-method as long as the above criteria are roughly satisfied.
Figure 5.
Selecting CAIH thresholds for C-method. (a) CAIH of the sequences designed by C-method are compared with the wild-type sequences. C1-3: sequences designed with the CAIH thresholds of 0.60 (C1), 0.75 (C2), and 0.90 (C3). W: wild-type sequences. H1: sequences designed by H-method. (b) CAIH thresholds are compared with the distribution of CAIH computed from the all endogenous genes in R. erythropolis. For H1, the mean CAIH of designed sequences (0.46) is shown.
Discussion
We developed a codon optimization method to be used in R. erythropolis, an attractive host organism for recombinant protein expression. The method was developed based on the statistical analysis of our recombinant protein expression data from 204 genes. The resulting method named C-method was used to optimize the coding sequences of selected genes, achieving to increase protein expression levels compared with wild-type sequences. Our method will be a useful tool for improved expression of recombinant proteins in R. erythropolis, and possibly in other actinobacteria.
In the statistical analysis of recombinant protein expression data, our results on R. erythropolis were partly consistent with the previous study on E. coli (Fig. 1a; Supplementary Fig. S2). On the other hand, we observed the species-specific influence regarding the codon repetition rate and the amino acid repetition rate. Although the interpretation of this species specificity is not straightforward, we provide possible molecular mechanisms underlying this phenomenon. First, several studies reported an effect called codon order where the repeated use of the same type of codon increases translational efficiency21,22. These studies proposed a model that a ribosome can “recycle” a tRNA when scanning from a codon to the next codon if these neighboring codons correspond to the same type of tRNA, thereby enhancing translational efficiency. According to this model, codon repetition rates are expected to show positive correlation with protein expression levels. Second, some amino acid repeats such as proline-rich stretches are known to decrease translational efficiency by inducing ribosome stalling. (See the recent review23). This suggests that amino acid repetition rates negatively correlate with protein expression levels. Finally, and most importantly, the codon repetition rate and the amino acid repetition rate are not independent since codon repeats are necessarily translated into amino acid repeats. Therefore, the joint effect of these sequence features can be complicated. For example, increased codon repetition rates, which themselves imply higher protein expression levels, may result in lower protein expression levels due to the counter effect of increased amino acid repetition rates. In summary, we speculate that the species-specific influence of these sequence features may reflect a different balance between their effects in R. erythropolis and E. coli due to e.g. differences in the structures of ribosomal machineries and/or the copy numbers of tRNA genes. While the dissection of such possible causes cannot be performed in the present study, our results will contribute as an example showing the species-specific influence of sequence features on protein expression levels.
In the development of codon optimization methods, H-method based solely on the mRNA folding energy was first devised (Fig. 2), and then improved to C-method by incorporating the codon frequency (Fig. 4). This was motivated by the feedback from our experimental data that genes whose protein expression levels could not be increased by H-method contained rare codons (Fig. 3). Such a feedback strategy may also be useful for developing a codon optimization method for host organisms other than R. erythropolis, including bacteria, fungi, insects and mammals1.
In the verification experiment of C-method (Fig. 4), 9 of 12 tested genes (75%) achieved increased expression levels. Especially for 5 genes whose expression levels for wild-type sequences were small or not detectable, all of them were improved by codon optimization. On the other hand, the improvement was not observed for the remaining 3 genes. We note that the expression levels of these genes were relatively high even with wild-type sequences before codon optimization. Their expression scores in our dataset (Supplementary Data S1) were 3 (high) for 2 genes and 2 (medium) for one gene. Thus, one possibility is that the expression of these genes is already at near-optimal levels, and is difficult to be further improved by codon optimization.
We note that our method currently has the limitations as summarized below. First, since C-method only considers ΔGUH and CAIH, the improvement of protein expression may be hindered by the involvement of other sequence features. For example, the modulation of CAIH in C-method only considers 33 head nucleotides, which suggests a possibility that the improvement is hindered by the poor CAI in downstream nucleotides. We confirmed that this possibility did not apply to the genes used in the verification experiment of C-method (Supplementary Fig. S4). The overall CAI of the failed genes did not show substantial differences from that of the succeeded genes. Nevertheless, the success rate of our method may be improved by incorporating new sequence features in addition to ΔGUH and CAIH. Recently, Cambray et al. has conducted an expression analysis in E. coli using 244,000 genes24. Even with such a large-scale analysis, they have reported that sequence features (similar to those used in our study) only explain approximately 30% of variations in protein expression levels, suggesting the existence of unknown sequence features. To explore such new sequence features, larger-scale analysis of recombinant protein expression in R. erythropolis as well as in other host organisms will be necessary. Second, our method was developed based upon sequence features influencing translational efficiency. Thus, the improvement of protein expression will be limited when the expression is hindered by factors other than translational efficiency. These factors include the membrane sorting, the S-S bond formation, and the toxicity of proteins to be expressed. We note that all proteins used in this study were cytoplasmic proteins not including transmembrane proteins nor proteins with S-S bonds (Supplementary Data S1). While this choice of proteins enabled us to develop and evaluate our method focusing on translational efficiency, the improvement based on the other factors was not addressed in the present study.
In the present study, we addressed the use of codon optimization methods mainly for increasing expression levels. On the other hand, there is also some need for decreasing expression levels, which includes weakening unnecessary fluxes in a metabolic pathway to improve the production of objective metabolites25. In this regard, our method may also be useful since the L-series sequences designed by the deoptimization of the mRNA folding energy succeeded to decrease expression levels for most of the tested genes (Fig. 2). Another issue not addressed in this study is a fine tuning of expression levels, that is, not only to simply maximize or minimize expression levels, but to regulate expression at a desired level26. The design of coding sequences for that purpose may be possible by choosing synonymous variants with moderate values of sequence features, rather than those with the maximum or the minimum value. These points should be addressed as a future direction.
Methods
Recombinant protein expression dataset
Gene expression
A total of 204 genes (listed in Supplementary Data S1) from S. coelicolor A3(2) was cloned into the inducible expression vector pTip-QC212 at the NdeI restriction site (CATATG) in order to arrange the start codon at the same position. For genes whose start codon is other than ATG, the start codon was replaced with ATG. Each of the plasmid vector was introduced into the host strain, R. erythropolis L-8827. The recombinant strains were precultured in LB liquid media containing 34 µg/ml chloramphenicol at 28 °C for overnight. 2 ml of the preculture was transferred to 20 ml of the same fresh media containing 0.5 µg/ml thiostrepton and cultured with 120 rpm agitation for 16 hours. The cells were harvested and washed twice with 100 mM sodium phosphate buffer. The cell pellets were then resuspended with the same buffer containing 8 M urea and cell disruption was performed by glass beads using the Multi-beads shocker instrument (Yasui Kikai, Japan). The supernatant of the disrupted sample was used as the denatured crude cell extract.
Protein expression level measurement
20 µg of the crude cell extract protein from each of the recombinants was analyzed by SDS-PAGE and the protein was visualized by Coomassie Brilliant Blue staining. The expression level of each recombinant protein was manually scored and classified into 1-3 (1, low or not detected; 2, medium; 3, high) with visual inspection by bare eye (Supplementary Fig. S1; Supplementary Data S1).
Sequence features and statistical analysis
CAI and tAI were calculated as previously described17,18 where the genomic codon usage in R. erythropolis was obtained from Codon Usage Database28 with the accession number 234621. For tAI, the weight matrix (also known as the codon-tRNA interaction matrix) was calculated by the method proposed in the previous study29. Briefly, we adjusted the weight matrix considering the copy numbers of tRNA genes and the codon usage bias in the R. erythropolis genome. The resulting weight matrix is available at the authors’ GitHub web site30. The codon repetition rate9 was calculated by where di is the distance from each codon position to the next occurring position of the same type of codon, and L is the sequence length. The amino acid repetition rate9 was calculated similarly except that the repetition was counted for the type of encoded amino acids rather than the type of codons. ΔGUH was calculated by EnsembleEnergy program version 5.8.1 in RNAstructure package19. For statistical analysis, polyserial correlation coefficients20 and their p-values were calculated using polycor package version 0.7.9 in R software31. CAIH was calculated similarly to CAI focusing on 33 head nucleotides (i.e. 11 codons). Rare codons were defined as codons with normalized codon frequencies less than 0.1. For each type of codon c, a normalized codon frequency was defined as wc = fc/maxs fs where f. is a genomic codon frequency and s is a synonymous codon encoding the same amino acid as c.
Codon optimization methods
The algorithms of H-method and C-method are described in Results. The software is available at the authors’ GitHub web site30. Coding sequences designed by our method are shown in Supplementary Data S3.
Verification experiments
PCR amplification and cloning of the codon-optimized genes: The nucleotide sequences of the codon-optimized genes were listed in Supplementary Data S3. The designed genes were amplified by PCR using the wild-type genes as the templates and the forward primers containing the codon-altered nucleotide sequences. The both forward and reverse PCR primers also contained overhang sequences of the cloning vector, pTip-QC2, to combine them by In-Fusion cloning (TAKARA, Japan). The PCR primers used in this study were listed in Supplementary Data S4. The resultant plasmids were introduced into the host strain, and the protein expression experiment was performed as described above except for the culture volume of 11 ml and 10 µg of the crude extract protein used for the SDS-PAGE analysis.
Measurement of protein expression levels
The SDS-PAGE gels were optically analyzed by the image analyzer WSE-6100 LuminoGraph (ATTO, Japan). Quantitative analysis of the recombinant protein was performed by the ImageSaver6 / CS Analyzer software (ATTO, Japan). The expression level of each mutant relative to the wild type was determined by the average value of three independent recombinant clones.
Supplementary information
Acknowledgements
This work was supported by New Energy and Industrial Technology Development Organization (NEDO) and National Project on Protein Structural and Functional Analyses (“Protein 3000” Project). Y. S. is supported by JSPS KAKENHI (17H06410). W. K. is supported by JSPS KAKENHI (17H05457).
Author Contributions
Y.S., N.T., T. Kumagai and T. Kameda conducted the computational analysis, and developed the codon optimization method. W.K. and T.T. conducted the protein expression experiment. Y.S. and W.K. wrote the paper. T.T. and T. Kameda conceived of the study. Y.N., K.T. and Y.Y. participated in the data interpretation. All authors read and approved the final manuscript. In this research work, we used the supercomputer of ACCMS, Kyoto University.
Data Availability
All data in this study are available in the main text or in Supplementary Material. The program implementation of our codon optimization methods is available in the authors’ GitHub web site30.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yutaka Saito and Wataru Kitagawa contributed equally.
Contributor Information
Tomohiro Tamura, Email: t-tamura@aist.go.jp.
Tomoshi Kameda, Email: kameda-tomoshi@aist.go.jp.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44500-z.
References
- 1.Structural Genomics Consortium, China Structural Genomics Consortium, Northeast Structural Genomics Consortium, Gräslund, S., Nordlund, P. et al. Protein production and purification. Nat. Methods. 5, 135–46 (2008). [DOI] [PMC free article] [PubMed]
- 2.Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–53. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Gustafsson C, et al. Engineering genes for predictable protein expression. Protein Expr. Purif. 2012;83:37–46. doi: 10.1016/j.pep.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quax TE, Claassens NJ, Söll D, van der Oost J. Codon bias as a means to fine-tune gene expression. Mol. Cell. 2015;59:149–61. doi: 10.1016/j.molcel.2015.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parret AH, Besir H, Meijers R. Critical reflections on synthetic gene design for recombinant protein expression. Curr. Opin. Struct. Biol. 2016;38:155–62. doi: 10.1016/j.sbi.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Brule CE, Grayhack EJ. Synonymous codons: choose wisely for expression. Trends Genet. 2017;33:283–297. doi: 10.1016/j.tig.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuller T, Waldman YY, Kupiec M, Ruppin E. Translation efficiency is determined by both codon bias and folding energy. Proc. Natl. Acad. Sci. USA. 2010;107:3645–50. doi: 10.1073/pnas.0909910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welch M, Govindarajan S, Ness JE, Villalobos A, Gurney A, et al. Design parameters to control synthetic gene expression in Escherichia coli. PLoS One. 2009;4:e7002. doi: 10.1371/journal.pone.0007002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boël G, et al. Codon influence on protein expression in E. coli correlates with mRNA levels. Nature. 2016;529:358–363. doi: 10.1038/nature16509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodman DB, Church GM, Kosuri S. Causes and effects of N-terminal codon bias in bacterial genes. Science. 2013;342:475–9. doi: 10.1126/science.1241934. [DOI] [PubMed] [Google Scholar]
- 11.Nakashima N, Tamura T. A novel system for expressing recombinant proteins over a wide temperature range from 4 to 35 degrees C. Biotechnol. Bioeng. 2004;86:136–48. doi: 10.1002/bit.20024. [DOI] [PubMed] [Google Scholar]
- 12.Nakashima N, Tamura T. Isolation and characterization of a rolling-circle-type plasmid from Rhodococcus erythropolis and application of the plasmid to multiple-recombinant-protein expression. Appl. Environ. Microbiol. 2004;70:5557–68. doi: 10.1128/AEM.70.9.5557-5568.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasuga K, et al. Heterologous production of kasugamycin, an aminoglycoside antibiotic from Streptomyces kasugaensis, in Streptomyces lividans and Rhodococcus erythropolis L-88 by constitutive expression of the biosynthetic gene cluster. Appl. Microbiol. Biotechnol. 2017;101:4259–4268. doi: 10.1007/s00253-017-8189-5. [DOI] [PubMed] [Google Scholar]
- 14.Kitagawa W, Mitsuhashi S, Hata M, Tamura T. Identification of a novel bacteriocin-like protein and structural gene from Rhodococcus erythropolis JCM 2895, using suppression-subtractive hybridization. J. Antibiot. (Tokyo) 2018;71:872–879. doi: 10.1038/s41429-018-0078-3. [DOI] [PubMed] [Google Scholar]
- 15.Kitagawa W, et al. Cloning and heterologous expression of the aurachin RE biosynthesis gene cluster afford a new cytochrome P450 for quinoline N-hydroxylation. Chembiochem. 2013;14:1085–93. doi: 10.1002/cbic.201300167. [DOI] [PubMed] [Google Scholar]
- 16.Vallecillo AJ, Parada C, Morales P, Espitia C. Rhodococcus erythropolis as a host for expression, secretion and glycosylation of Mycobacterium tuberculosis proteins. Microb. Cell Fact. 2017;16:12. doi: 10.1186/s12934-017-0628-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharp PM, Li WH. The codon Adaptation Index - a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–95. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuller T, et al. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–54. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 19.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olsson U, Drasgow F, Dorans NJ. The polyserial correlation coefficient. Psychometrika. 1982;47:337–47. doi: 10.1007/BF02294164. [DOI] [Google Scholar]
- 21.Cannarozzi G, et al. A role for codon order in translation dynamics. Cell. 2010;141:355–67. doi: 10.1016/j.cell.2010.02.036. [DOI] [PubMed] [Google Scholar]
- 22.Shao ZQ, Zhang YM, Feng XY, Wang B, Chen JQ. Synonymous codon ordering: a subtle but prevalent strategy of bacteria to improve translational efficiency. PLoS One. 2012;7:e33547. doi: 10.1371/journal.pone.0033547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodnina MV. The ribosome in action: Tuning of translational efficiency and protein folding. Protein Sci. 2016;25:1390–406. doi: 10.1002/pro.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cambray G, Guimaraes JC, Arkin AP. Evaluation of 244,000 synthetic sequences reveals design principles to optimize translation in Escherichia coli. Nat. Biotechnol. 2018;36:1005–1015. doi: 10.1038/nbt.4238. [DOI] [PubMed] [Google Scholar]
- 25.Woolston BM, Edgar S, Stephanopoulos G. Metabolic engineering: past and future. Annu. Rev. Chem. Biomol. Eng. 2013;4:259–88. doi: 10.1146/annurev-chembioeng-061312-103312. [DOI] [PubMed] [Google Scholar]
- 26.Marschall L, Sagmeister P, Herwig C. Tunable recombinant protein expression in E. coli: enabler for continuous processing? Appl. Microbiol. Biotechnol. 2016;100:5719–28. doi: 10.1007/s00253-016-7550-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitani Y, Meng X, Kamagata Y, Tamura T. Characterization of LtsA from Rhodococcus erythropolis, an enzyme with glutamine amidotransferase activity. J. Bacteriol. 2005;187:2582–91. doi: 10.1128/JB.187.8.2582-2591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sabi R, Tuller T. Modelling the efficiency of codon-tRNA interactions based on codon usage bias. DNA Res. 2014;21:511–26. doi: 10.1093/dnares/dsu017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.GitHub, https://github.com/yutaka-saito/codon_optimization.
- 31.polycor: Polychoric and polyserial correlations, https://cran.r-project.org/web/packages/polycor/index.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data in this study are available in the main text or in Supplementary Material. The program implementation of our codon optimization methods is available in the authors’ GitHub web site30.