Abstract
Purpose
Recently, a new Bayesian Penalized Likelihood (BPL) Reconstruction Algorithm was introduced by GE Healthcare, Q.Clear; it promises to provide better PET image resolution compared to the widely used Ordered Subset Expectation Maximization (OSEM). The aim of this study is to compare the performance of these two algorithms on several types of findings, in terms of image quality, lesion detectability, sensitivity, and specificity.
Methods
Between September 6th 2017 and July 31st 2018, 663 whole body 18F-FDG PET/CT scans were performed at the Nuclear Medicine Department of S. Martino Hospital (Belluno, Italy). Based on the availability of clinical/radiological follow-up data, 240 scans were retrospectively reviewed. For each scan, a hypermetabolic finding was selected, reporting both for OSEM and Q.Clear: SUVmax and SUVmean values of the finding, the liver and the background close to the finding; size of the finding; percentage variations of SUVmax and SUVmean. Each finding was subsequently correlated with clinical and radiological follow-up, to define its benign/malignant nature.
Results
Overall, Q.Clear improved the SUV values in each scan, especially in small findings (< 10 mm), high SUVmax values (≥ 10), and medium/low backgrounds. Furthermore, Q.Clear amplifies the signal of hypermetabolic findings without modifying the background signal, which leads to an increase in signal-to-noise ratio, improving overall image quality. Finally, Q.Clear did not affect PET sensitivity or specificity, in terms of number of reported findings and characterization of their nature.
Conclusions
Q.Clear is an iterative algorithm that improves significantly the quality of PET images compared to OSEM, increasing the SUVmax of findings (in particular for small findings) and the signal-to-noise ratio. However, due to the intrinsic characteristics of this algorithm, it will be necessary to adapt and/or modify the current interpretative criteria based of quantitative evaluation, to avoid an overestimation of the disease burden.
Keywords: Q.Clear, OSEM, 18FDG-PET, Reconstruction algorithm, BPL reconstruction algorithm, PET/CT
Introduction
Positron emission tomography (PET/CT) is nuclear medicine functional imaging technique that allows to evaluate in vivo some metabolic processes involved in oncological, inflammatory-infectious, and neurodegenerative pathologies.
The system is able to define the bio-distribution of a radioactive tracer, thanks to the detection of pairs of gamma rays generated by a positron-emitting radionuclide (usually fluorine-18).
However, PET/CT presents some limitations, such as spatial resolution, related on one hand to the physics of the β + decay, on the other hand to the technologies used for the detection of incident γ rays [1].
Over the past years, there have been introduced many reconstruction algorithms, in order to reduce the errors and artifacts, which affect the quality of images and the accuracy and reproducibility of the measurement of the standardized uptake value (SUV). Currently, the Ordered Subset Expectation Maximization (OSEM) is the most used algorithm worldwide [2–5]. However, a new Bayesian Penalized Likelihood (BPL) Reconstruction Algorithm was recently introduced by GE Healthcare, named Q.Clear, which promises to provide a better image resolution [6].
Ordered Subset Expectation Maximization
Ordered Subset Expectation Maximization (OSEM) is an accelerated variant of expectation maximization (EM), and nowadays, it is the most widely used algorithm for PET image reconstruction. This algorithm is characterized by the fact that the projection data are divided into subgroups (subsets) that are analyzed sequentially during each iteration. The number of subsets determines the acceleration factor. However, since the background noise increases with the number of iterations, the OSEM algorithm is prematurely stopped before the noise becomes excessive. Finally, a post-processing image filter is subsequently applied to further improve the quality [7–9].
In general, OSEM generates higher-quality images than analytical algorithms; however, it has to be kept in mind that there is a trade-off between the number of subsets and the quality of the image: when the number of subsets is high, each subset contains a smaller amount of tomographic and statistical information, so the noise and artifacts can increase [7].
Therefore, due to the non-uniform and object-dependent convergence, the relationship between resolution and noise obtained by prematurely stopping the algorithm is not always optimal and it is difficult to determine the most suitable number of iterations/subsets and parameters of the post-processing filters for a heterogeneous patient population.
Bayesian Penalized Likelihood Reconstruction Algorithm
In order to achieve complete convergence of images during their reconstruction, numerous iterations are required; however, as the number of iterations increases, the background noise increases too. On the other hand, the accuracy and quality of PET images is reduced because the full convergence is not reached, as the reconstruction of the images is limited to reduce noise.
Recently, GE Healthcare (Milwaukee, USA) has introduced a new iterative image reconstruction algorithm, a Bayesian Penalized Likelihood (BPL) Reconstruction Algorithm named Q.Clear.
Q.Clear takes into account all aspects of the image reconstruction process, including PSF modeling and an innovative penalty factor. This parameter is a function given by the difference in values between adjacent voxels and their sum; it acts as a term of noise suppression, allowing the algorithm to achieve full convergence [10–22]. This term leads to a greater uniformity in regions with low concentration of radiopharmaceutical activity and less uniformity in areas of higher concentration (or near the margins of findings with higher concentration). In other words, at each iteration, the voxels, with a smaller variation between neighboring voxels, are slightly favored compared to the noisier ones. The SUV values obtained with the Q.Clear (called Q.SUV) are therefore much more accurate and images’ resolution is better than those obtained with OSEM, thanks to its intrinsic characteristics and to the association with Point Spread Function (PSF) modeling and Time of Flight (TOF).
This penalty function is controlled by a penalty factor called β, which is the only variable of the algorithm that can be modified by the operator. Beyond the penalty factor, the other modifiable parameter is TOF, which can be removed during image reconstruction; however, it is not possible to modify the PSF modeling associated with the Q.Clear (SharpIR).
The primary aim of this study was to evaluate the efficiency and the diagnostic performance of Q.Clear compared to OSEM on a wide range of findings, characterized by different sizes, locations, and activity concentrations. All findings were subsequently correlated with clinical and radiological follow-up to determine their benign or malignant nature.The secondary aim was to further compare these two algorithm, modifying some parameters (such as TOF and PSF), in order to evaluate the impact of each parameter on the images’ rendering.
Materials and Methods
This retrospective observational study was performed according to the declaration of Helsinki and to national regulations. All the patients provided informed consent for participation and anonymous publication of data. The approval of the Ethics Committee is not required for retrospective observational studies at the hospital where the present study was conducted (S. Martino Hospital, Belluno, Italy).
Between the 6th September 2017 and the 31st July 2018, 663 whole body PET/CT scans (both oncological and non-oncological) were performed on 625 patients at the Nuclear Medicine Department of the S. Martino Hospital (Belluno, Italy). Based on the availability of clinical and radiological follow-up data, 240 scans of 204 patients were retrospectively reviewed. The patients’ characteristics are reported in Table 1.
Table 1.
Patients’ characteristics
| Patients 204 (♀ 86, ♂ 118) | Average | Range |
|---|---|---|
| Age (years) | 64.8 | 25–91 |
| BMI (kg/cm2) | 26.3 | 14.7–38.9 |
| Glycemia (mg/ml) | 103.5 | 52–232 |
| Injected dose (MBq) | 296 | 133–494 |
| Uptake time (min) | 59 | 51–100 |
|
Clinical indications Oncological: 199 Non-oncological: 5 |
||
| Follow-up (months) | 5.3 | 3–12 |
Clinical indications were lung cancer (67), breast cancer (63), lymphoproliferative diseases (39), head-neck tumors (9), sarcoma (8), colorectal cancer (5), melanoma (5), gynecological cancer (3), endocarditis (2), sarcoidosis (2), and FUO (1).
Technical Specifications of Tomograph and Radiopharmaceutical
The tomograph is a GE Digital Ready PET/CT equipped with 3 rings of LYSO crystals (therefore equipped with TOF). The radiopharmaceutical used was 18F-FDG. This radiotracer is not produced on site, but comes from the cyclotron of Castelfranco Veneto Hospital (Italy). The average activity administered was 3.8 MBq/kg.
Image Acquisition Protocol
All scans were performed according to the European (EANM) and national (AIMN) guidelines related to 18F-FDG studies. Intravenous administration of approximately 3.8 MBq/kg of 18F-FDG was performed, with an average uptake time of approximately 59 min. The CT scan features were 120 kV, 150 mA, DFOV 70 cm, Q.AC WIDEVIEW reconstruction. The PET protocols were diversified according to BMI, adapting the Z-axis filter and the acquisition time per bed (1.30–3.30 min). The β value was 350 for each scan, while OSEM protocol was set to 4 iterations/16 subsets.
Image Analysis and Validation Criteria
For each scan, a hypermetabolic finding was selected, for a total of 240 findings. In order to analyze every possible scenario, in each scan, the choice of the finding has been modified, in terms of type, location, size, and uptake. For each scan, the following were reported:
SUVmax and SUVmean values of the finding (both with OSEM + PSF + TOF and Q.Clear + TOF);
SUVmax and SUVmean values of the liver (both with OSEM + PSF + TOF and Q.Clear + TOF) with a standard VOI of 35 mm3;
SUVmax values of the background close to the selected finding, obtained through a circular ROI drawn around the finding, thanks to which it was possible to define three categories: (a) low background, with SUVmax background < to the SUVmax of the blood pool (measured on the aortic arch), (b) medium background, with SUVmax background ≈ to the blood pool SUVmax, (c) high background, with SUVmax background > to the blood pool SUVmax;
the size of the finding, defining three categories: ≤ 10 mm, > 10 mm, and ≥ 20 mm.
Then, for each finding and for the liver, the percentage variations of SUVmax and SUVmean between OSEM + PSF + TOF and Q.Clear + TOF were calculated.
Each finding was subsequently correlated with clinical and radiological follow-up, to define its benign or malignant nature.
Furthermore, 15 out of 240 scans were consecutively selected, on which other reconstructions were carried out, specifically:
OSEM + PSF without TOF;
OSEM + TOF without PSF;
OSEM without TOF and without PSF;
Q.Clear without TOF (it is not possible to exclude the PSF).
On these subgroups, the percentage variations of SUVmax and SUVmean of the findings and the liver were analyzed, in order to evaluate the effect of TOF and PSF on the qualitative output of the images for both algorithms.
Finally, each scan was visually evaluated with both reconstructions (OSEM + PSF + TOF and Q.Clear + TOF), in order to highlight any differences in the number of hypermetabolic findings. A Nuclear Medical Physician with a 4-year experience reviewed all scans.
Statistics
The results were analyzed using the Sign test, the Wilcoxon test, and the Kolmogorov-Smirnov test, considering p values < 0.05 as statistically significant.
Results
The general characteristics of the findings are summarized in Table 2.
Table 2.
General characteristics of the findings
| Number | Size (mm) | |
|---|---|---|
| Lymph nodes | 91 | 32 ≤ 10, 45 > 10, 14 ≥ 20 |
| Pulmonary nodules | 46 | 15 ≤ 10, 11 > 10, 20 ≥ 20 |
| Hepatic findings | 20 | 6 ≤ 10, 6 > 10, 8 ≥ 20 |
| Bone findings | 23 | 5 ≤ 10, 12 > 10, 6 ≥ 20 |
| Soft tissue | 11 | 3 ≤ 10, 5 > 10, 3 ≥ 20 |
| Others (brain, colon, heart, esophagus, spleen, retroperitoneal fibrosis, pleura, muscle, kidney, adrenal, stomach, thyroid, testicles, uterus, vessels) | 49 | 17 ≤ 10, 20 > 10, 12 ≥ 20 |
Of these, 89 findings presented low background values, 121 medium background, 20 high background; the average values of SUVmax were Q.Clear 11.2 (range: 1.7–43.1) and OSEM 8.9 (range: 1.5–36.8).
The main results obtained from the analysis of the images with the two reconstruction algorithms are reported in Table 3.
Table 3.
Main results of the comparison between OSEM and Q.Clear algorithms
| Number | Average Var % SUVmax | Standard deviation | Average Var % SUVmean | Standard deviation | P values | |
|---|---|---|---|---|---|---|
| Total findings | 240 | 18.8 | 12.7 | 19.6 | 14.1 | p < 0.001 |
| Size | ||||||
| ≤ 10 mm | 60 | 25.2 | 14.8 | 25.5 | 18.2 | p < 0.001 |
| > 10 mm | 91 | 20.2 | 11.9 | 22.2 | 12.8 | p < 0.01 |
| ≥ 20 mm | 89 | 12.9 | 9 | 12.9 | 8.8 | p < 0.1 |
| Background | ||||||
| Low | 89 | 18.3 | 12.4 | 19,4 | 12,7 | p < 0.05 |
| Medium | 121 | 21.2 | 12.8 | 21.8 | 15.2 | p < 0.005 |
| High | 30 | 10.6 | 9 | 11.2 | 10.3 | p > 0.1 |
| SUVmax | ||||||
| < 5 | 37 | 11.5 | 8.6 | 8.5 | 10.4 | p < 0.001 |
| ≥ 5 | 96 | 17.3 | 11.2 | 12.3 | 19.3 | p < 0.001 |
| ≥ 10 | 107 | 22.6 | 13.8 | 23.6 | 14.8 | p < 0.001 |
The average percentage variations of SUVmax and SUVmean of all the findings were, respectively, 18.8 and 19.6, with a significant gain in terms of SUV thanks to Q.Clear. Looking at the results reported in Table 3, Q.Clear is clearly more efficient than OSEM + PSF + TOF in all the subgroups to determine SUVmax and SUVmean values (p < 0.001).
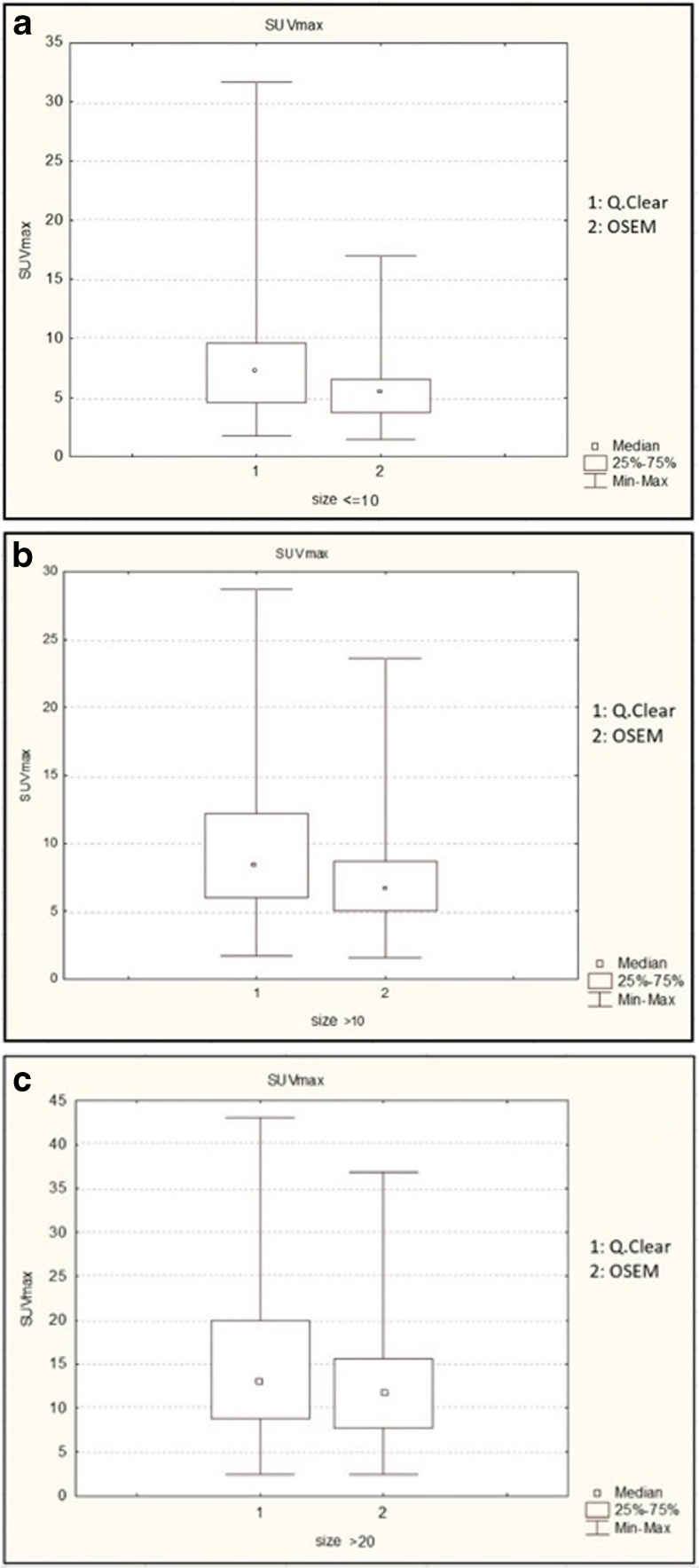
It is also possible to observe an average percentage variation of SUVmax and SUVmean higher for findings ≤ 10 mm (p < 0.001) and > 10 mm (p < 0.01), while there are no statistically significant differences (p > 0.1) for findings > 20 mm (see Graph 1a–c).
Graph 1.
a SUVmax variation based on the size of the findings, ≤ 10 mm. b SUVmax variation based on the size of the findings, > 10 mm. c SUVmax variation based on the size of the findings, ≥ 20 mm
As for the background, compared to OSEM, Q.Clear + TOF showed a statistically significant SUV variation in those findings with medium (e.g., mediastinal lymph nodes, p < 0.005) and low backgrounds (p < 0.05), while there were not significant differences for those with high background (e.g., a lesion localized in the liver parenchyma, p > 0.1). Furthermore, the mean percentage variations of SUVmax and SUVmean of the liver relative to all the scans examined are, respectively, − 0.1 and 0.2, therefore not significant (p > 0.1).
The correlation between findings and clinical and radiological follow-up data did not show a significant difference between OSEM + PSF + TOF and Q.Clear + TOF in the determination of the benign or malignant nature of the finding itself. Furthermore, Q.Clear + TOF did not increase the number of findings, which are quantitatively comparable to those reported with OSEM + PSF + TOF reconstruction.
The general characteristics of the 15 findings, on which further reconstructions were carried out (OSEM ± PSF ± TOF and Q.Clear-TOF), were as follows:
3 pulmonary nodules, 6 lymph nodes, 2 bones, 2 soft tissues, 1 breast, and 1 thyroid;
5 ≥ 20 mm, 6 > 10 mm, and 4 ≤ 10 mm;
Q. Clear SUVmax: 8.6 (range: 4.7–14), OSEM SUVmax: 7 (range: 4.3–10.4).
The statistical analysis of these image reconstruction protocols showed a greater impact of TOF on SUV values compared to PSF (p < 0.0003). However, the PSF showed a significant impact too, even though lower than TOF (p < 0.002).
Discussion
Currently, the Ordered Subset Expectation Maximization (OSEM) is the most used algorithm worldwide in PET image reconstruction. However, a new Bayesian Penalized Likelihood (BPL) Reconstruction Algorithm was recently introduced by GE Healthcare, named Q.Clear, which promises to provide a better image resolution.
In literature, some phantom and clinical studies have been already conducted in order to evaluate the performance of Q.Clear compared to OSEM [10–24]. In regard to phantom studies, Reynés-Llompart G. et al. showed that as the β increases, the ability to quantify the signal (measured as contrast recovery) increases, while the noise is reduced, leading to an overall increase in detectability of the findings (in terms of signal-to-noise ratio). Both these parameters improve with the increase of β up to a plateau (above 500 and below 150). In general, the optimal values of β reported are between 300 and 400, based on the signal-to-noise ratio, the size of the finding, and the acquisition time.
Teoh EJ et al. analyzed the diagnostic performance of these two algorithms on small pulmonary nodules; mediastinal lymph nodes in non-small cell lung cancer (NSCLC); hepatic metastases from colorectal cancer. In all three cases, Q.Clear demonstrated an increase in the signal-to-noise and signal-to-background ratio compared to reconstruction obtained with OSEM. Moreover, Q.Clear provided a more accurate report on the metabolic activity of the findings. However, it did not significantly increase the accuracy in the differential diagnosis between benign and malignant lesions.
Since a high BMI corresponds to an increase in noise, with a consequent reduction in image quality, Chilcott AK et al. analyzed the efficiency of Q.Clear over a wide range of patients’ weights. Their study showed that the liver’s signal-to-noise ratio improved significantly with Q.Clear compared to OSEM. Furthermore, it demonstrated that the quadratic dose of the radiopharmaceutical based on weight (commonly used in patients with high BMI) is no longer necessary with Q.Clear. Similar results were obtained by Ahn S. et al. and Vallot D. et al.
Howard BA et al. evaluated the efficiency of Q.Clear on a cohort of 29 patients with pulmonary nodules smaller than 8 mm. In this small population, Q.Clear demonstrated a significant increase in SUVmax and lesion detectability, but only with low values of penalty factor (β recommended: 150). In this regard, it is important to remember that, in order to obtain an optimal image quality with Q.Clear, it is necessary to pay attention on the type of radiopharmaceutical used, as well as on the anatomical region under examination. For example, Reynés-Llompart G. et al. reported an optimal β value between 300 and 400 for whole-body scan and between 150 and 250 for the cerebral acquisitions with 18F-FDG. Rowley LM et al. reported that the optimal β value is 4000 for the acquisitions obtained with 90Y, while Ter Voert EEGW et al. demonstrated that for 68Ga-PSMA, the optimal β values are between 400 and 550. Finally, Teoh EJ et al. analyzed the scans of 15 patients studied with 18F-Fluciclovine (FACBC), suggesting an optimal β value of 300 for this radiotracer.
Despite the limitations of this study, mainly related to the great variability of the selected findings and enrolled patients (corresponding to a physiological biodistribution diversity of the radiotracer), it was possible to make some important considerations. The results of this study confirmed the data available in literature. In particular, Q.Clear proved to increase the SUV values, especially for small findings (< 10 mm) and high SUVmax values (≥ 10) in scans reconstructed with TOF. Regarding the relationship between the finding and its background, Q.Clear appeared more efficient in findings with medium and low backgrounds (such as mediastinal lymph nodes and pulmonary nodules). The lack of significance in the mean percentage variation of SUVmax and SUVmean of the liver showed that Q.Clear amplifies the signal of hypermetabolic findings without modifying the background signal. This leads to an increase in the signal-to-noise ratio, improving overall image quality. Furthermore, according to the literature data, Q.Clear did not increase PET sensitivity or specificity, in terms of number of reported findings and the characterization of their nature.
However, the increase in SUVmax/SUVmean and in signal-to-noise ratio, obtained thanks to Q.Clear, opens up a relevant matter. To date, in some pathologies, visual assessment criteria are implemented in order to categorize the malignant/benign nature of a finding and to quantify the response to therapy. For example, the Deauville Five-point scale visually compares the uptake of lymphomas’ lesions with those of the mediastinum and liver, in order to assess response to therapy [25, 26]. Since Q.Clear increases the SUVmax of the hypermetabolic findings, without significantly affecting the background values (which leads to a higher signal-to-noise ratio, as already reported in several studies), it is possible that these interpretation criteria could no longer be used with this reconstruction algorithm, as they could overestimate the overall tumor burden.
Therefore, it is essential to
conduct further studies to investigate the impact of Q.Clear on current interpretation criteria and eventually adapt those criteria, to correctly characterize the findings;
always specify in the final report the type of reconstruction algorithm used, in order to allow a correct comparison between exams conducted in different Nuclear Medicine Departments.
Conclusion
In conclusion, Q.Clear is an algorithm that improves the quality of PET images compared to OSEM, increasing the SUVmax of findings (in particular for small findings) and improving the signal-to-noise ratio, without affecting PET sensitivity and the specificity. However, due to the intrinsic characteristics of this algorithm, further studies are needed to investigate the impact of Q.Clear on current interpretation criteria and eventually adapt and/or modify those criteria, in order to avoid an overestimation of the disease burden.
Acknowledgements
None.
Compliance with Ethical Standards
Conflict of Interest
Antonella Matti, Giacomo Maria Lima, Cinzia Pettinato, Francesca Pietrobon, Felice Martinelli, and Stefano Fanti declare that they have no conflict of interest. There is no source of funding.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed Consent
The approval of the Ethics Committee is not required for retrospective observational studies at the hospital where the present study was conducted (S. Martino Hospital, Belluno, Italy). Informed consent was obtained from all individual participants included in the study.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Antonella Matti, Email: antomatti@hotmail.com.
Giacomo Maria Lima, Email: giacomo.maria.lima@gmail.com.
Cinzia Pettinato, Email: cinzia.pettinato@aosp.bo.it.
Francesca Pietrobon, Email: francesca.pietrobon@ulss.belluno.it.
Felice Martinelli, Email: felice.martinelli@aulss1.veneto.it.
Stefano Fanti, Email: stefano.fanti@aosp.bo.it.
References
- 1.Volterrani D, Erba PA, Mariani G. Fondamenti di medicina nucleare. Tecniche e Applicazioni. 1st ed: Springer Verlag; 2010.
- 2.Tarantola G, Zito F, Gerundini P. PET Instrumentation and Reconstuction Algorthms in whole-body applications. J Nucl Med. 2003;44:756–769. [PubMed] [Google Scholar]
- 3.Berg E, Cherry SR. Innovations in instrumentation for positron emission tomography. Semin Nucl Med. 2018. 10.1053/j.semnuclmed.2018.02.006. [DOI] [PMC free article] [PubMed]
- 4.Slomka PJ, Pan T, Germano G. Recent advances and future Progress in PET instrumentation. Semin Nucl Med. 2016;46:5–19. doi: 10.1053/j.semnuclmed.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Iriarte A, Marabini R, Matej S, Sorzano COS, Lewitt RM. System models for PET statistical iterative reconstruction: a review. Comput Med Imaging Graph. 2015. 10.1016/j.compmedimag.2015.12.003. [DOI] [PubMed]
- 6.GE Healthcare, PET/CT Millennium specifics, http://www3.gehealthcare.co.uk/en-gb/products/categories/molecular_imaging/pet-ct/discovery_mi. Accessed 21 Nov 2018.
- 7.Van der Vos CS, Koopman D, Rijnsdorp S, Arends AJ, Boellaard R, Van Dalen JA, et al. Quantification, improvement, and harmonization of small lesion detection with state-of-the-art PET. Eur J Nucl Med Mol Imaging. 2017;44:4–16. doi: 10.1007/s00259-017-3727-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vandenberghe S, Mikhaylova E, D’Hoe E, Mollet P, Karp JS. Recent developments in time-of-flight PET. EJNMMI Phys. 2016;3:3. doi: 10.1186/s40658-016-0138-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Surti S, Karp JS. Advances in time-of-flight PET. Phys Med. 2016;32:12–22. doi: 10.1016/j.ejmp.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teoh EJ, McGowan RD, Macpherson RE, Bradley KM, Gleeson FV. Phantom and clinical evaluation of the Bayesian penalized likelihood reconstruction algorithm Q.Clear on an LYSO PET/CT system. J Nucl Med. 2015;56:1447–1452. doi: 10.2967/jnumed.115.159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reynés-Llompart G, Gámez-Cenzano C, Vercher-Conejero JL, Sabaté-Llobera A, Calvo-Malvar N, Martí-Climent JM. Phantom, clinical, and texture indices evaluation and optimization of a penalized-likelihood image reconstruction method (Q.Clear) on a BGO PET/CT scanner. Med Phys. 2018;45:3214–3222. doi: 10.1002/mp.12986. [DOI] [PubMed] [Google Scholar]
- 12.Teoh EJ, McGowan RD, Bradley KM, Belcher E, Black E, Gleeson FV. Novel penalised likelihood reconstruction of PET in the assessment of histologically verified small pulmonary nodules. Eur J Radiol. 2016;26:576–584. doi: 10.1007/s00330-015-3832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teoh EJ, McGowan RD, Bradley KM, Belcher E, Black E, Moore A, et al. 18F-FDG PET/CT assessment of histopathologically confirmed mediastinal lymph nodes in non-small cell lung cancer using a penalised likelihood reconstruction. Eur J Radiol. 2016;26:4098–5006. doi: 10.1007/s00330-016-4253-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parvizi N, Franklin JM, McGowan RD, Teoh EJ, Bradley KM, Gleeson FV. Does a novel penalized likelihood reconstruction of 18F-FDG PET-CT improve signal-to-background in colorectal liver metastases? Eur J Radiol. 2015;84:1873–1878. doi: 10.1016/j.ejrad.2015.06.025. [DOI] [PubMed] [Google Scholar]
- 15.Ahn S, Ross SG, Asma E, Miao J, Jin X, Cheng L, et al. Quantitative comparison of OSEM and penalized likelihood image reconstruction using relative difference penalties for clinical PET. Phys Med Biol. 2015;60:5733–5751. doi: 10.1088/0031-9155/60/15/5733. [DOI] [PubMed] [Google Scholar]
- 16.Chilcott AK, Bradley KM, McGowan RD. Effect of a Bayesian penalized likelihood PET reconstruction compared with ordered subset expectation maximization on clinical image quality over a wide range of patient weights. AJR Am J Roentgenol. 2018;210:153–157. doi: 10.2214/AJR.17.18060. [DOI] [PubMed] [Google Scholar]
- 17.O’ Doherty J, McGowan RD, Abreu C, Barrington S. Effect of Bayesian penalized likelihood reconstruction on [13N]-NH3 rest perfusion quantification. J Nucl Cardiol. 2016;24:282–290. doi: 10.1007/s12350-016-0554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teoh EJ, McGowan RD, Schuster DM, Tsakok MT, Gleeson FV, Bradley KM. Bayesian penalised likelihood reconstruction (Q.Clear) of 18F-fluciclovine PET for imaging of recurrent prostate cancer: semi-quantitative and clinical evaluation. Br J Radiol. 2018;91:20170727. doi: 10.1259/bjr.20170727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowley LM, Bradley KM, Boardman P, Hallam A, McGowan DR. Optimization of image reconstruction for 90Y selective internal radiotherapy on a lutetium yttrium Orthosilicate PET/CT system using a Bayesian penalized likelihood reconstruction algorithm. J Nucl Med. 2017;58:658–664. doi: 10.2967/jnumed.116.176552. [DOI] [PubMed] [Google Scholar]
- 20.Ter Voert EEGW, Muehlematter UJ, Delso G, Pizzuto DA, Műller J, Nagel HW, et al. Quantitative performance and optimal regularization parameter in block sequential regularized expectation maximization recontructions in clinical 68Ga-PSMA PET/MR. EJNMMI Res. 2018;8:70. doi: 10.1186/s13550-018-0414-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howard BA, Morgan R, Thorpe MP, Turkington TG, Oldan J, James OG, et al. Comparison of Bayesian penalized likelihood reconstruction versus OS-EM for characterization of small pulmonary nodules in oncologic PET/CT. Ann Nucl Med. 2017;31:623–628. doi: 10.1007/s12149-017-1192-1. [DOI] [PubMed] [Google Scholar]
- 22.Vallot D, Caselles O, Chaltiel L, Fernandez A, Gabianche E, Dierickx L, et al. A clinical evaluation of the impact of the Bayesian penalized likelihood reconstruction algorithm on PET FDG metrics. Nucl Med Commun. 2017;38:979–984. doi: 10.1097/MNM.0000000000000729. [DOI] [PubMed] [Google Scholar]
- 23.Teoh EJ, McGowan RD, Macpherson RE, Bradley KM, Gleeson FV. Phantom and clinical evaluation of the Bayesian penalized likelihood reconstruction algorithm Q.Clear on an LYSO PET/CT system. J Nucl Med. 2015;56:1447–1452. doi: 10.2967/jnumed.115.159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reynés-Llompart G, Gámez-Cenzano C, Vercher-Conejero JL, Sabaté-Llobera A, Calvo-Malvar N, Martí-Climent JM. Phantom, clinical, and texture indices evaluation and optimization of a penalized-likelihood image reconstruction method (Q.Clear) on a BGO PET/CT scanner. Med Phys. 2018;45:3214–3222. doi: 10.1002/mp.12986. [DOI] [PubMed] [Google Scholar]
- 25.Barrington SF, Kluge R. FDG PET for therapy monitoring in Hodgkin and non-Hodgkin lymphomas. Eur J Nucl Med Mol Imaging. 2017;44:97–110. doi: 10.1007/s00259-017-3690-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheson BD, Fisher RI, Barrington SF, Cavalli F, Schwartz LH, Zucca E, et al. Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: the Lugano classification. J Clin Oncol. 2014;32:3059–3068. doi: 10.1200/JCO.2013.54.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]