Abstract
We introduce a method for registration and visualization of correlative super-resolution microscopy images from different microscopy techniques. We established an automated registration procedure based on the generalized Hough transform. We developed a software tool to apply this algorithm and visualize correlated images from structured illumination microscopy (SIM) and direct stochastic optical reconstruction microscopy (dSTORM). To demonstrate the potential of this super-resolution correlator, we visualize the distribution of the presynaptic protein bassoon in the active zones of synapses in the molecular layer of the mouse cerebellum. First, a multiple labeled sample is imaged by SIM, followed by imaging of one of the fluorescent labels by dSTORM. To avoid the use of artificial fiducial markers, we used the signal of Alexa Fluor 647 recorded in switching buffer on the two microscopes for image superposition. We recorded multicolor SIM images in 20-μm thick brain slices to identify synapses in the dendritic system of Purkinje cells and put higher-resolved dSTORM images of the synaptic distribution of bassoon in registry.
Significance
We introduce a method for automated registration and visualization of correlative super-resolution microscopy images from different microscopy techniques. The tool is applicable to correlative analysis of images acquired on different microscopes. For example, overview images from multicolor structured illumination microscopy that provide contextual information in a large field-of-view can be used to identify regions of interest that have been better resolved by single-molecule localization microscopy. Registering the two images will help to identify structures within a cell or tissue environment and provide highly resolved information at the same time.
Main Text
Single-molecule localization microscopy (SMLM) has been established as a powerful tool to visualize cellular structures with a spatial resolution approaching the molecular level (1, 2). SMLM delivers super-resolved images of cellular structures along with single-molecule information, even providing estimates for absolute numbers of molecules that are present in subcellular compartments or protein complexes (2). However, data analysis within a cellular context (e.g., the allocation of pre- and postsynaptic protein localizations to special synapses and neurons in brain slices) remains challenging. Often, different molecular components (e.g., a synaptic marker and a receptor of interest) need to be imaged independently and correlated thereafter to get molecular scale information on signal identity and relative positions. It can be very helpful to identify regions of interest (as for instance a synapse) through a large field-of-view, multicolor image (e.g., recorded by correlative structured illumination microscopy (SIM)) and correlate this with a higher resolution but smaller field-of-view image (e.g., SMLM). Depending on the fluorescence signals and the imaged structures, manual alignment might be feasible. In most cases, however, it requires a large amount of try-and-error alignment procedures and thus an automated registration procedure reduces data analysis effort and time and increases accuracy and thus ensures reproducibility and performance.
Problems arise from the fact that multicolor SMLM experiments suffer from different photophysical characteristics and associated photoswitching and photoactivation rates of different fluorescent proteins and organic dyes (2). So far, only a few dye pairs have been successfully used for multicolor SMLM (3, 4, 5, 6, 7). For quantitative direct stochastic optical reconstruction microscopy (dSTORM) with organic dyes, only the carbocyanine dye Cy5 and its structural analog Alexa Fluor 647 provide reliable results (4, 8). Although other fluorophores are used in combination with Alexa Fluor 647, the resolution and reliability in the photophysical properties is superior for Alexa Fluor 647 as compared to all other dyes (see Supporting Material in (8)).
Recently, sequential SMLM techniques have been introduced for multitarget SMLM using the same dye in subsequent labeling and imaging steps (9, 10, 11). However, the techniques are time consuming and less suited for whole cell or tissue imaging.
On the other hand, SMLM can be correlated with electron microscopy (EM). Correlative light and EM (12, 13, 14) can be used to visualize the distribution of specifically targeted proteins or organelles in cells and tissue. Here, EM enables a structural overview with nanometer resolution, whereas fluorescence microscopy allows specific molecular labeling with a higher affinity (4, 5, 15, 16, 17, 18). Importantly, fluorescence imaging enables higher labeling efficiencies than immunogold EM using fluorophore-tagged antibodies facilitating structure determination by SMLM. Thus, localization microscopy and EM are complementary methods that can be combined advantageously to determine molecular positions in the context of the cellular ultrastructure. However, correlative light and EM approaches remain challenging because of the vastly different requirements for the sample preparation of SMLM and EM.
Technically less demanding is to correlate confocal or widefield fluorescence imaging and SMLM enabling quantitative super-resolved imaging of proteins in the context of other subcellular structures imaged at diffraction-limited spatial resolution (5, 19, 20). Also, SIM and SMLM can serve as complementary approaches with SIM being superior in terms of speed, field-of-view, multicolor imaging, fluorophore availability, and photophysical requirements but SMLM excelling at resolution and quantification (21, 22, 23).
Here, we present an optimized workflow for correlative three-dimensional (3D)-SIM (24) and 3D-dSTORM (25), together with a new algorithm for automated registration of a pixel-based fluorescence image and localization data. The optimized workflow enables dSTORM visualization of the distribution of labeled molecules in cells and tissue in the context of other cellular markers imaged with SIM resolution (about twofold improved). To avoid the use of artificial fiducial markers, we aimed at imaging the same dye first by SIM and then by dSTORM. Fiducial markers in general are helpful for registration and drift correction. However, it can also be challenging to introduce artificial fiducials in sample tissue because they tend to accumulate on cell membranes or sample dish surfaces. Fiducials with an emission wavelength different to the imaged fluorophores require registration of spectrally distinct channels. Therefore, we focused on demonstrating a registration procedure without fiducials. We tested different buffer and embedding conditions and identified a switching buffer containing 100 mM mercaptoethylamine and PBS (pH 7.4) without an oxygen scavenger as a sufficient buffer to preserve the fluorescence signal of Alexa Fluor 647 in both imaging modes. Because efficient photoswitching of organic dyes to off states requires irradiation intensities in the kW/cm2 range (7, 25, 26), Alexa Fluor 647 does not show pronounced blinking in SIM experiments using irradiation intensities of ∼100 W/cm2. Hence, the main effect of the switching buffer in SIM experiments is minimization of photobleaching. Correspondingly, the fluorescence signal and localizations of Alexa Fluor 647 in the SIM and SMLM image modes, respectively, can be used to overlay the images.
We present registration of images from SMLM and alternative widefield microscopy based on the GHT. To align super-resolution images from different microscopy modalities, we established suitable image representations to compute overlay characteristics and search for optimal alignment of selected image regions (workflow outlined in Fig. 1). We will demonstrate the procedure with dSTORM and SIM images, noting that other image-based techniques can be used instead.
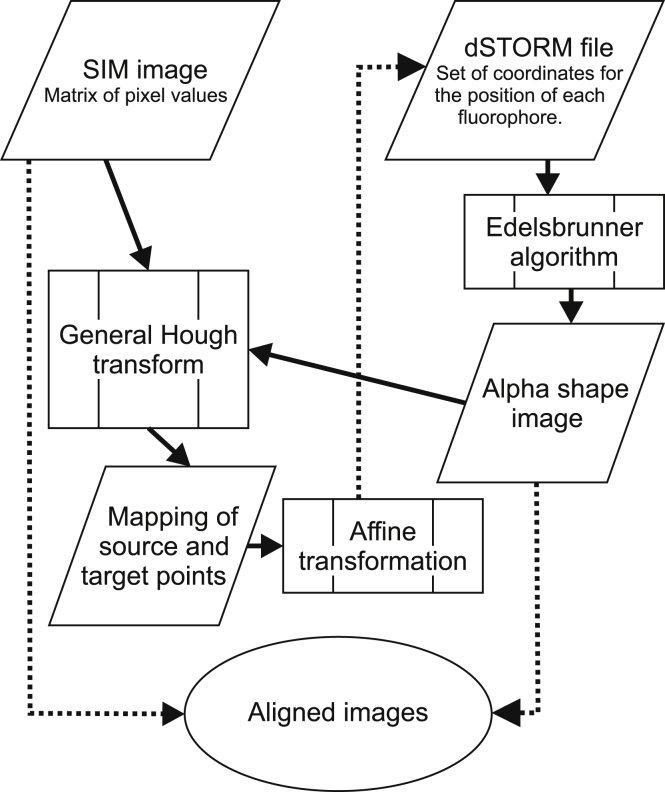
Figure 1.
Workflow of the registration and visualization process. SIM and dSTORM files serve as an initial input and are processed to yield an affine transformation (solid lines). This intermediate result is then applied to the initial dSTORM coordinates. For visualization and further analysis, the aligned images are rendered as described (dotted line).
The experimental workflow (illustrated in Fig. 2) typically provides SIM images of larger regions for the field-of-view (target) and dSTORM images of smaller regions (source) such that the source image overlaps with a subregion in the target image. For automated alignment, we need to identify a mapping from the dSTORM (source) space to the SIM (target) space in the form of an affine transformation that brings the source features in registry with the target features. However, finding a subset of corresponding points in both images is challenging because of the differences in field-of-view, imaging aberrations, resolution, and signal-to-noise for different microscope modalities. We found a robust solution to this problem by representing dSTORM data (i.e., the individual localizations represented by two-dimensional (2D) or 3D points) as α-shapes that are rendered with SIM pixel size. The α-shapes represent a concave hull of the point data that depends on a single parameter (α value). The α-shapes can be computed from a Delaunay triangulation of point data according to the Edelsbrunner algorithm (27). Other representations as, for instance, being derived from a Voronoi tessellation (being dual to the Delaunay triangulation) are feasible.
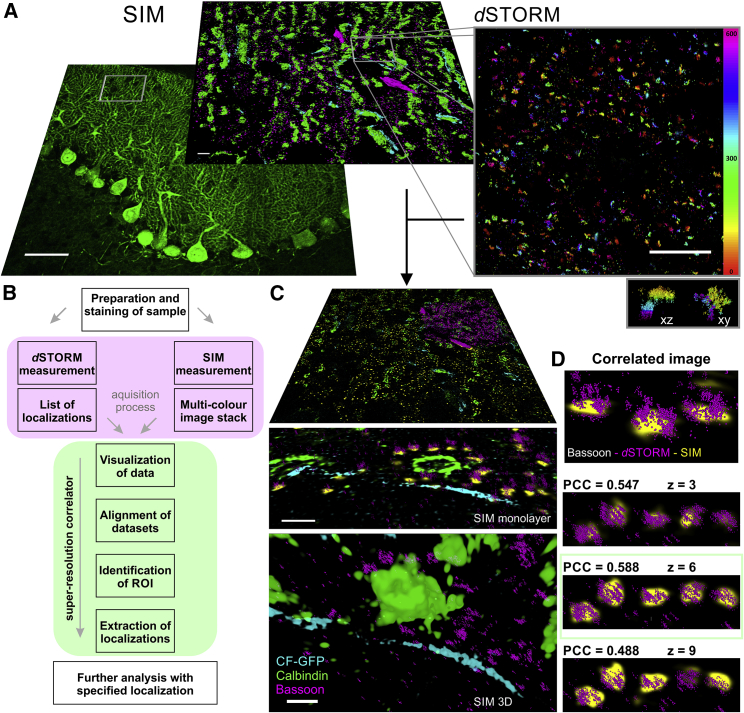
Figure 2.
Principle of the super-resolution correlation procedure, including imaging on two microscopes, registration, and visualization. (A) Shown is a three-color 3D-SIM (left) at a different magnification (10× and 63×) and 3D-dSTORM (right) image of a selected area in the 20-μm thick brain slice. Climbing fibers (CF) are labeled by GFP (cyan), calbindin in Purkinje cells are labeled by Alexa Fluor 568 (green), and bassoon are labeled by Alexa Fluor 647 (magenta). The 3D bassoon distribution is color coded over an axial range of 600 nm. The inset (right lower corner) shows the x-z and x-y image of two synapses. Scale bars, 50 μm (10×, SIM), 5 μm (63×, SIM and dSTORM). (B) Schematic workflow of the super-resolution correlator software is shown. (C) Functions of the super-resolution correlator are shown. Localization- and pixel-based information are displayed in 3D. Scale bars, 1 μm. (D) Selected planes are aligned, and the quality of alignment is monitored by Pearson correlation coefficient analysis. The upper image shows the 3D visualization of the combined registered images. Images and localization data of interest can be exported.
The rendered dSTORM representation is then partitioned into several smaller, equally sized segments (typically 16) that are all aligned with the SIM image using an adaptation of the generalized Hough transform (GHT). The GHT is a robust way to detect arbitrary shapes that correlate in different images (28). The general procedure consists of the following computation steps (details in the Supporting Materials and Methods). First, edges are detected for both source and target image using a Canny edge detection algorithm. Second, a gradient representation is computed for both images using a convolution with 2D-Sobel filter kernels in x and y direction. All nonzero pixels in the edge image of the source data are then defined as edge points and represented as a vector relative to a predefined arbitrary origin. This list of edge point vectors is grouped by the orientation angle of the corresponding gradient vector defining the so-called r-table. In a similar way, edge and gradient images are computed for the target image. In addition, an accumulator array with the same size as the target image is created and initially filled with zeros. Then, for each pixel in the target gradient image, the corresponding slice of the r-table is selected. For each vector in each r-table slice, the position of the corresponding source pixel is shifted by the current vector, and the accumulator array is incremented by one at the resulting point. The resulting accumulator array is a rating of the likelihood for each pixel of the target image to be an origin point of the segment.
Because the standard GHT did not provide sufficient matching accuracy for a proper alignment of dSTORM and SIM data, a weighting scheme was developed following the suggestions by Ballard et al. (28). It was noticed that, depending on the imaged structure, the local density of identified edges might strongly vary. By not taking the edge density into account, a random feature overlay between source and target structures will influence the GHT algorithm and favor random correspondence in dense regions over true correspondence in sparse regions (Fig. S3). We modified the GHT by introducing a normalization procedure. A performant solution was found by thresholding the accumulator array and scaling the accumulator array values with the number of target edge points in a local region of a size that corresponds to the segment size. This weighted GHT is repeated for each segment, and the optimal transformation is identified from the maximal weighted accumulator array values.
We accounted for differences in orientation between source and target images by rotating each segment by a discrete set of angles φ (typically with −20° < φ < 20° with Δφ = 1°) and repeating the weighted GHT. We compared all segment transformations and identified those that are similar for at least three segments of a given source image. This grid consistency of the segment transformations was used to reject possible mismatches by an error handling algorithm (see Supporting Materials and Methods and Fig. S4 for an error estimate of a false positive match in multiple segment transformations). If grid consistency was assured, we constructed the total affine transformation for registering the complete source image with the target image. From the group of points (source segments and the corresponding target points) that represent valid segment transformations and that were identified through the GHT algorithm, the optimal affine transformation (including translation, rotation, and shear) was constructed by optimizing transformation parameters.
The quality of registration was estimated by computing the Pearson correlation coefficient (29) from the registered source and the target image. The Pearson correlation image allows comparing the registration under various parameters and in comparison to manual registration and confirmed a superior performance of the automated procedure based on the weighted GHT on all the presented images (see Supporting Materials and Methods). The precision for 2D registration depends strongly on the image features (Fig. S5). In addition, it is limited by the pixel size of the target image or the SIM image resolution. In all our presented example data, the SIM resolution is the limiting factor for the alignment precision to be on the order of 100 nm.
We increased computation performance by implementing the GHT for graphical processing unit-based computation using a Python-based (using scipy libraries) cuda-accelerated implementation (30) (for details see Supporting Materials and Methods). A Delaunay triangulation was constructed. The Edelsbrunner algorithm (27) was implemented in python to compute 2D α-shapes. Here, performance was enhanced by using parallel computation in PyCUDA. The Python implementation was compared with a Python binding of CGAL:α_shape_2 (31) (CGAL being a state-of-the-art C++ library) to confirm that both implementations yield identical results for the same α and with Boundary set to REGULAR in the CGAL implementation. The runtime of GHT was significantly improved by outsourcing the critical computation step of incrementing the accumulator array to the graphical processing unit.
To perform the superposition of the two images, we developed a super-resolution correlator software that is based on vividSTORM, an open-source software for correlative confocal and SMLM (19). The super-resolution correlator is a Python-based program enabling the registration and high-performance visualization of big localization data files. It implements visualization of 3D localization-based dSTORM data and pixel-based SIM information and enables fast image rendering and alignment (Fig. 1 B). The quality of the registration can be monitored by quantifying the co-localization of the dSTORM image and the chosen SIM z-layer using Pearson coefficient (Fig. 1 C).
To demonstrate the universal applicability of the registration procedure, we imaged the distribution of the presynaptic protein bassoon in active zones affiliated to Purkinje cells and climbing fibers in 20-μm thick cryosectioned brain slices. Bassoon was marked by immunolabeling using a secondary Alexa Fluor 647 antibody. For visualization of Purkinje cells, dendritic spines were identified by calbindin staining immunolabeled using secondary Alexa Fluor 568 antibodies. Climbing fibers were identified by transgenic GFP expression. Three-color 3D-SIM images were acquired on an ELYRA S.1 using a 63× objective at an axial step size of 150 nm (Fig. 1 A). To identify and select the region of interest in the brain slice, a fluorescence image with a 10× objective was acquired first at a low irradiation intensity. After the SIM measurement, the sample was moved to a widefield microscope for biplane temporal radial-aperture-based intensity estimation (TRABI) 3D-dSTORM (32) (Supporting Materials and Methods). The intensity was increased to 4–5 kW/cm2 to induce the photoswitching required for the dSTORM imaging of bassoon (Fig. 1 A).
In summary, we presented a registration algorithm that performs well on different super-resolution imaging modalities. We demonstrated the performance with SIM and dSTORM images of bassoon signals recorded from Purkinje cells in thin brain slices. The experiment illustrates the advantage of using SIM to gain a large field-of-view overview together with additional information about the SMLM-recorded region from additional color channels, albeit at a lower resolution. From the overview, it is possible to identify the overall brain region as well as individual cells within a specific tissue structure. The SMLM image, on the other hand, gives us the ability to analyze the bassoon distribution pattern in a specific cell at the highest resolution. Registration and visualization was carried out with the newly developed super-resolution correlator software, which visualizes 3D-dSTORM data sets, containing a million localizations, in large 3D-SIM image areas acquired on two different microscopes.
The open-source programming code together with test data is available for the registration algorithm at https://github.com/super-resolution/Impro (https://doi.org/10.5281/zenodo.2275436) and for the super-resolution correlator at https://github.com/super-resolution/Super-resolution-correlator (https://doi.org/10.5281/zenodo.2275730).
Author Contributions
S.A., S.D., and M.S. conceived the project and designed and managed the study. S.A., S.R., and S.D. planned and outlined the software. S.R. developed the software. S.A. performed the experiments. All authors wrote the manuscript.
Acknowledgments
We thank J. Eilers and Ch. Pätz (Institute for Physiology, University of Leipzig) for provision of perfused mouse brain, A.-L. Sirén and B. Gado (Experimental Neurosurgery, University Clinic of Würzburg) for assistance in cryoslicing, and C. Franke (Department of Biotechnology and Biophysics, University of Würzburg) for the assistance in TRABI analysis of biplane 3D dSTORM data.
The work was supported financially by the Deutsche Forschungsgemeinschaft (German Research Foundation) – CRC-TRR 166 - A04 and CRC-TRR 166 - A02 to M.S. (project A04) and to S.D. (project B02).
Editor: Amy Palmer.
Footnotes
Sebastian Reinhard and Sarah Aufmkolk contributed equally to this work.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.04.029.
Contributor Information
Markus Sauer, Email: m.sauer@uni-wuerzburg.de.
Sören Doose, Email: soeren.doose@uni-wuerzburg.de.
Supporting Citations
References (33, 34, 35) appear in the Supporting Material.
Supporting Material
References
- 1.Patterson G., Davidson M., Lippincott-Schwartz J. Superresolution imaging using single-molecule localization. Annu. Rev. Phys. Chem. 2010;61:345–367. doi: 10.1146/annurev.physchem.012809.103444. [DOI] [PMC free article] [PubMed] [Google Scholar]; Patterson, G., M. Davidson, …, J. Lippincott-Schwartz. 2010. Superresolution imaging using single-molecule localization. Annu. Rev. Phys. Chem. 61:345-367. [DOI] [PMC free article] [PubMed]
- 2.Sauer M., Heilemann M. Single-molecule localization microscopy in eukaryotes. Chem. Rev. 2017;117:7478–7509. doi: 10.1021/acs.chemrev.6b00667. [DOI] [PubMed] [Google Scholar]; Sauer, M., and M. Heilemann. 2017. Single-molecule localization microscopy in eukaryotes. Chem. Rev. 117:7478-7509. [DOI] [PubMed]
- 3.Winterflood C.M., Platonova E., Ewers H. Dual-color 3D superresolution microscopy by combined spectral-demixing and biplane imaging. Biophys. J. 2015;109:3–6. doi: 10.1016/j.bpj.2015.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]; Winterflood, C. M., E. Platonova, …, H. Ewers. 2015. Dual-color 3D superresolution microscopy by combined spectral-demixing and biplane imaging. Biophys. J. 109:3-6. [DOI] [PMC free article] [PubMed]
- 4.Löschberger A., van de Linde S., Sauer M. Super-resolution imaging visualizes the eightfold symmetry of gp210 proteins around the nuclear pore complex and resolves the central channel with nanometer resolution. J. Cell Sci. 2012;125:570–575. doi: 10.1242/jcs.098822. [DOI] [PubMed] [Google Scholar]; Loschberger, A., S. van de Linde, …, M. Sauer. 2012. Super-resolution imaging visualizes the eightfold symmetry of gp210 proteins around the nuclear pore complex and resolves the central channel with nanometer resolution. J. Cell Sci. 125:570-575. [DOI] [PubMed]
- 5.Flottmann B., Gunkel M., Erfle H. Correlative light microscopy for high-content screening. Biotechniques. 2013;55:243–252. doi: 10.2144/000114099. [DOI] [PubMed] [Google Scholar]; Flottmann, B., M. Gunkel, …, H. Erfle. 2013. Correlative light microscopy for high-content screening. Biotechniques. 55:243-252. [DOI] [PubMed]
- 6.Gould T.J., Verkhusha V.V., Hess S.T. Imaging biological structures with fluorescence photoactivation localization microscopy. Nat. Protoc. 2009;4:291–308. doi: 10.1038/nprot.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gould, T. J., V. V. Verkhusha, and S. T. Hess. 2009. Imaging biological structures with fluorescence photoactivation localization microscopy. Nat. Protoc. 4:291-308. [DOI] [PMC free article] [PubMed]
- 7.van de Linde S., Löschberger A., Sauer M. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 2011;6:991–1009. doi: 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]; van de Linde, S., A. Loschberger, …, M. Sauer. 2011. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 6:991-1009. [DOI] [PubMed]
- 8.Ehmann N., van de Linde S., Kittel R.J. Quantitative super-resolution imaging of Bruchpilot distinguishes active zone states. Nat. Commun. 2014;5:4650. doi: 10.1038/ncomms5650. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ehmann, N., S. van de Linde, …, R. J. Kittel. 2014. Quantitative super-resolution imaging of Bruchpilot distinguishes active zone states. Nat. Commun. 5:4650. [DOI] [PMC free article] [PubMed]
- 9.Valley C.C., Liu S., Lidke K.A. Sequential superresolution imaging of multiple targets using a single fluorophore. PLoS One. 2015;10:e0123941. doi: 10.1371/journal.pone.0123941. [DOI] [PMC free article] [PubMed] [Google Scholar]; Valley, C. C., S. Liu, …, K. A. Lidke. 2015. Sequential superresolution imaging of multiple targets using a single fluorophore. PLoS One. 10:e0123941. [DOI] [PMC free article] [PubMed]
- 10.Yi J., Manna A., Samelson L.E. madSTORM: a superresolution technique for large-scale multiplexing at single-molecule accuracy. Mol. Biol. Cell. 2016;27:3591–3600. doi: 10.1091/mbc.E16-05-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yi, J., A. Manna, …, L. E. Samelson. 2016. madSTORM: a superresolution technique for large-scale multiplexing at single-molecule accuracy. Mol. Biol. Cell. 27:3591-3600. [DOI] [PMC free article] [PubMed]
- 11.Schnitzbauer J., Strauss M.T., Jungmann R. Super-resolution microscopy with DNA-PAINT. Nat. Protoc. 2017;12:1198–1228. doi: 10.1038/nprot.2017.024. [DOI] [PubMed] [Google Scholar]; Schnitzbauer, J., M. T. Strauss, …, R. Jungmann. 2017. Super-resolution microscopy with DNA-PAINT. Nat. Protoc. 12:1198-1228. [DOI] [PubMed]
- 12.Müller-Reichert T., Srayko M., McDonald K. Correlative light and electron microscopy of early Caenorhabditis elegans embryos in mitosis. Methods Cell Biol. 2007;79:101–119. doi: 10.1016/S0091-679X(06)79004-5. [DOI] [PubMed] [Google Scholar]; Muller-Reichert, T., M. Srayko, …, K. McDonald. 2007. Correlative light and electron microscopy of early Caenorhabditis elegans embryos in mitosis. Methods Cell Biol. 79:101-119. [DOI] [PubMed]
- 13.Hauser M., Wojcik M., Xu K. Correlative super-resolution microscopy: new dimensions and new opportunities. Chem. Rev. 2017;117:7428–7456. doi: 10.1021/acs.chemrev.6b00604. [DOI] [PubMed] [Google Scholar]; Hauser, M., M. Wojcik, …, K. Xu. 2017. Correlative super-resolution microscopy: new dimensions and new opportunities. Chem. Rev. 117:7428-7456. [DOI] [PubMed]
- 14.Ando T., Bhamidimarri S.P., Zifarelli G. The 2018 correlative microscopy techniques roadmap. J. Phys. D Appl. Phys. 2018;51:443001. doi: 10.1088/1361-6463/aad055. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ando, T., S. P. Bhamidimarri, …, G. Zifarelli. 2018. The 2018 correlative microscopy techniques roadmap. J. Phys. D Appl. Phys. 51:443001. [DOI] [PMC free article] [PubMed]
- 15.Watanabe S., Punge A., Jorgensen E.M. Protein localization in electron micrographs using fluorescence nanoscopy. Nat. Methods. 2011;8:80–84. doi: 10.1038/nmeth.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]; Watanabe, S., A. Punge, …, E. M. Jorgensen. 2011. Protein localization in electron micrographs using fluorescence nanoscopy. Nat. Methods. 8:80-84. [DOI] [PMC free article] [PubMed]
- 16.Kopek B.G., Shtengel G., Hess H.F. Correlative 3D superresolution fluorescence and electron microscopy reveal the relationship of mitochondrial nucleoids to membranes. Proc. Natl. Acad. Sci. USA. 2012;109:6136–6141. doi: 10.1073/pnas.1121558109. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kopek, B. G., G. Shtengel, …, H. F. Hess. 2012. Correlative 3D superresolution fluorescence and electron microscopy reveal the relationship of mitochondrial nucleoids to membranes. Proc. Natl. Acad. Sci. USA. 109:6136-6141. [DOI] [PMC free article] [PubMed]
- 17.Sochacki K.A., Shtengel G., Taraska J.W. Correlative super-resolution fluorescence and metal-replica transmission electron microscopy. Nat. Methods. 2014;11:305–308. doi: 10.1038/nmeth.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sochacki, K. A., G. Shtengel, …, J. W. Taraska. 2014. Correlative super-resolution fluorescence and metal-replica transmission electron microscopy. Nat. Methods. 11:305-308. [DOI] [PMC free article] [PubMed]
- 18.Markert S.M., Britz S., Stigloher C. Filling the gap: adding super-resolution to array tomography for correlated ultrastructural and molecular identification of electrical synapses at the C. elegans connectome. Neurophotonics. 2016;3:041802. doi: 10.1117/1.NPh.3.4.041802. [DOI] [PMC free article] [PubMed] [Google Scholar]; Markert, S. M., S. Britz, …, C. Stigloher. 2016. Filling the gap: adding super-resolution to array tomography for correlated ultrastructural and molecular identification of electrical synapses at the C. elegans connectome. Neurophotonics. 3:041802. [DOI] [PMC free article] [PubMed]
- 19.Barna L., Dudok B., Katona I. Correlated confocal and super-resolution imaging by VividSTORM. Nat. Protoc. 2016;11:163–183. doi: 10.1038/nprot.2016.002. [DOI] [PubMed] [Google Scholar]; Barna, L., B. Dudok, …, I. Katona. 2016. Correlated confocal and super-resolution imaging by VividSTORM. Nat. Protoc. 11:163-183. [DOI] [PubMed]
- 20.Soeller C., Hou Y., Crossman D. Correlative single-molecule localization microscopy and confocal microscopy. Methods Mol. Biol. 2017;1663:205–217. doi: 10.1007/978-1-4939-7265-4_17. [DOI] [PubMed] [Google Scholar]; Soeller, C., Y. Hou, …, D. Crossman. 2017. Correlative single-molecule localization microscopy and confocal microscopy. Methods Mol. Biol. 1663:205-217. [DOI] [PubMed]
- 21.Pinnington S.J.L., Marshall J.F., Wheeler A.P. Correlative 3D structured illumination microscopy and single-molecule localization microscopy for imaging cancer invasion. Methods Mol. Biol. 2018;1764:253–265. doi: 10.1007/978-1-4939-7759-8_15. [DOI] [PubMed] [Google Scholar]; Pinnington, S. J. L., J. F. Marshall, and A. P. Wheeler. 2018. Correlative 3D structured illumination microscopy and single-molecule localization microscopy for imaging cancer invasion. Methods Mol. Biol. 1764:253-265. [DOI] [PubMed]
- 22.Hamel V., Guichard P., Gönczy P. Correlative multicolor 3D SIM and STORM microscopy. Biomed. Opt. Express. 2014;5:3326–3336. doi: 10.1364/BOE.5.003326. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hamel, V., P. Guichard, …, P. Gonczy. 2014. Correlative multicolor 3D SIM and STORM microscopy. Biomed. Opt. Express. 5:3326-3336. [DOI] [PMC free article] [PubMed]
- 23.Mönkemöller V., Øie C., McCourt P. Multimodal super-resolution optical microscopy visualizes the close connection between membrane and the cytoskeleton in liver sinusoidal endothelial cell fenestrations. Sci. Rep. 2015;5:16279. doi: 10.1038/srep16279. [DOI] [PMC free article] [PubMed] [Google Scholar]; Monkemoller, V., C. Oie, …, P. McCourt. 2015. Multimodal super-resolution optical microscopy visualizes the close connection between membrane and the cytoskeleton in liver sinusoidal endothelial cell fenestrations. Sci. Rep. 5:16279. [DOI] [PMC free article] [PubMed]
- 24.Gustafsson M.G., Shao L., Sedat J.W. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 2008;94:4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gustafsson, M. G., L. Shao, …, J. W. Sedat. 2008. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94:4957-4970. [DOI] [PMC free article] [PubMed]
- 25.Heilemann M., van de Linde S., Sauer M. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int.Engl. 2008;47:6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]; Heilemann, M., S. van de Linde, …, M. Sauer. 2008. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. Engl. 47:6172-6176. [DOI] [PubMed]
- 26.Burgert A., Letschert S., Sauer M. Artifacts in single-molecule localization microscopy. Histochem. Cell Biol. 2015;144:123–131. doi: 10.1007/s00418-015-1340-4. [DOI] [PubMed] [Google Scholar]; Burgert, A., S. Letschert, …, M. Sauer. 2015. Artifacts in single-molecule localization microscopy. Histochem. Cell Biol. 144:123-131. [DOI] [PubMed]
- 27.Edelsbrunner H. Springer; New York: 2014. A Short Course in Computational Geometry and Topology. [Google Scholar]; Edelsbrunner, H. 2014. A Short Course in Computational Geometry and Topology: Springer, New York.
- 28.Ballard D.H. Generalizing the Hough transform to detect arbitrary shapes. Pattern Recognit. 1981;13:111–122. [Google Scholar]; Ballard, D. H. 1981. Generalizing the Hough transform to detect arbitrary shapes. Pattern Recognit. 13:111-122.
- 29.Ruszczycki B., Bernas T. Quality of biological images, reconstructed using localization microscopy data. Bioinformatics. 2018;34:845–852. doi: 10.1093/bioinformatics/btx597. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ruszczycki, B., and T. Bernas. 2018. Quality of biological images, reconstructed using localization microscopy data. Bioinformatics. 34:845-852. [DOI] [PMC free article] [PubMed]
- 30.Jones, E., and E. Oliphant. 2001. SciPy: Open Source Scientific Tools for Python. http://www.scipy.org.
- 31.Da T.K.F. CGAL User and Reference Manual. 14 edition. Vol. 4. CGAL Editorial Board; 2019. 2D Alpha Shapes. [Google Scholar]; Da, T.K.F. 2019. 2D Alpha Shapes. In CGAL User and Reference Manual. CGAL Editorial Board, 4.14 edition.
- 32.Franke C., Sauer M., van de Linde S. Photometry unlocks 3D information from 2D localization microscopy data. Nat. Methods. 2017;14:41–44. doi: 10.1038/nmeth.4073. [DOI] [PubMed] [Google Scholar]; Franke, C., M. Sauer, and S. van de Linde. 2017. Photometry unlocks 3D information from 2D localization microscopy data. Nat. Methods. 14:41-44. [DOI] [PubMed]
- 33.Dunn K.W., Kamocka M.M., McDonald J.H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 2011;300:C723–C742. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dunn, K. W., M. M. Kamocka, and J. H. McDonald. 2011. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 300:C723-C742. [DOI] [PMC free article] [PubMed]
- 34.Wolter S., Löschberger A., Sauer M. rapidSTORM: accurate, fast open-source software for localization microscopy. Nat. Methods. 2012;9:1040–1041. doi: 10.1038/nmeth.2224. [DOI] [PubMed] [Google Scholar]; Wolter, S., A. Loschberger, …, M. Sauer. 2012. rapidSTORM: accurate, fast open-source software for localization microscopy. Nat. Methods. 9:1040-1041. [DOI] [PubMed]
- 35.Heilemann M., Margeat E., Tinnefeld P. Carbocyanine dyes as efficient reversible single-molecule optical switch. J. Am. Chem. Soc. 2005;127:3801–3806. doi: 10.1021/ja044686x. [DOI] [PubMed] [Google Scholar]; Heilemann, M., E. Margeat, …, P. Tinnefeld. 2005. Carbocyanine dyes as efficient reversible single-molecule optical switch. J. Am. Chem. Soc. 127:3801-3806. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.