Abstract
UNCoordinated-6 (UNC-6) was the first member of the netrin family to be discovered in Caenorhabditis elegans. With homology to human netrin-1, it is a key signaling molecule involved in directing axon migration in nematodes. Similar to netrin-1, UNC-6 interacts with multiple receptors (UNC-5 and UNC-40, specifically) to guide axon migration in development. As a result of the distinct evolutionary path of UNC-6 compared to vertebrate netrins, we decided to employ an integrated approach to study its solution behavior and compare it to the high-resolution structure we previously published on vertebrate netrins. Dynamic light scattering and analytical ultracentrifugation on UNC-6 (with and without its C-domain) solubilized in a low-ionic strength buffer suggested that UNC-6 forms high-order oligomers. An increase in the buffer ionic strength resulted in a more homogeneous preparation of UNC-6, that was used for subsequent solution x-ray scattering experiments. Our biophysical analysis of UNC-6 ΔC solubilized in a high-ionic strength buffer suggested that it maintains a similar head-to-stalk arrangement as netrins −1 and −4. This phenomenon is thought to play a role in the signaling behavior of UNC-6 and its ability to move throughout the extracellular matrix.
Introduction
Netrins are a class of proteins involved in axon guidance and therefore suitably named after the Sanskrit word “netr,” which means “one who guides.” In mammals, the family of netrins is composed of both secreted (netrin-1, -2, -3, -4, and -5) and membrane-bound proteins through glycosylphosphatidylinositol linkages (netrin-G1 and -G2) (1). Structurally, they all comprise an N-terminal domain (VI) followed by several epidermal growth factor-like domains (V-1, V-2, etc.) and a positively charged C-terminal domain (2). The N-terminal domains (V and VI) of netrin-1, -2, -3, and -5 are homologous to the domains present in laminin γ1, whereas the N-terminal domains of netrin-4, -G1, and -G2 are homologous to the domains present in laminin β1. The basic C-terminal domain (C-domain) (also known as the netrin-like domain) is not homologous to any laminin domains but exhibits sequence similarity to tissue inhibitor of metalloproteinases (3).
Netrin-1 is by far the most studied protein in the netrin family since its discovery in 1994. It was originally discovered for its role in guidance of commissural axons in the development of the vertebrate central nervous system (4). Since then, netrin-1 has been found to play pivotal roles in cell migration as well as in angiogenesis and morphogenesis of organs (5). Netrin-1 is an extracellular matrix protein secreted from floor plate cells and neural progenitor cells (4, 6). High levels of netrin-1 produced from neural progenitor cells extend from the ventricular zone to the basal lamina to attract commissural axons of the embryonic brain and spinal column. The role of netrin-1 secreted from floor plate cells is still unclear. It is hypothesized that as the commissural axons grow, they create a netrin-1–filled pathway for other axons to follow (6). As a result of the captured netrin-1, the axon growth pathway is netrin-1–driven through attraction (via Deleted in Colorectal Cancer or Neogenin receptors) or repulsion (UNCoordinated-5 [UNC-5]) of its receptors (3).
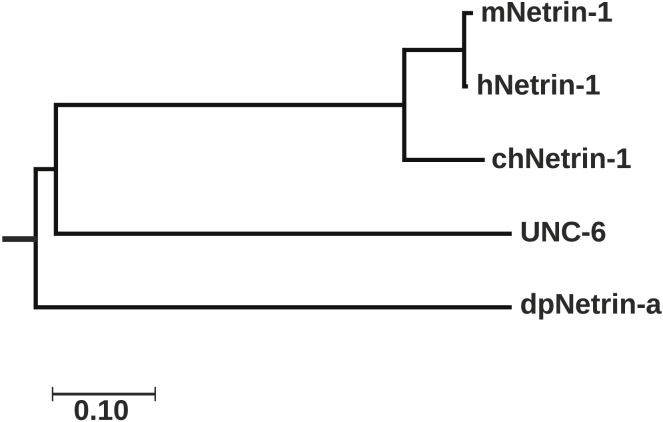
UNC-6 was the first reported member of the netrin family from Caenorhabditis elegans (3). It was identified in 1990 to be present as a gradient along the entire length of the nematode body wall guiding diverse migrations (7). Specifically, UNC-6 was found to be a signaling molecule that guides the circumferential migration of pioneer axons and mesodermal cells on the nematode body wall through interactions with its receptors. Interactions with UNC-40 attract ventrally directed axons and interactions with UNC-5 repulse dorsally directed axons (8, 9). In vertebrates UNC-40 is closely related to Deleted in Colorectal Cancer and Neogenin, whereas UNC-5 has been further characterized to have four orthologs UNC-5A, UNC-5B, UNC-5C, and UNC-5D. Upon sequence alignment with other vertebrate and invertebrate netrins, UNC-6 only has 40–50% similarity to netrins from human, mouse, chicken or drosophila (Fig. S1). Phylogenetic analysis shown in Fig. 1 depicts that netrin from invertebrates branch off quickly from vertebrates suggesting that UNC-6 might possess different properties than vertebrate netrin-1.
Figure 1.
Phylogenetic tree of netrins from different species with the highest log likelihood tree shown. The tree is drawn to scale with branch lengths measured in the number of substitutions per site. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA7 (46) and inferred using the Maximum Likelihood method based on JTT matrix-based model (14).
From the netrin family, we have already determined the high resolution crystal structure of netrin-1 ΔC (10) and netrin-4 ΔC (11). Both proteins form a head-to-stalk arrangement ranging from 150 to 175 Å for netrin-1 ΔC and -4 ΔC, respectively, with a globular N-terminal domain and a protein stalk formed from the laminin-type epidermal growth factor-like (LE) domains. The laminin N-terminal (LN) domain forms a β-sandwich jelly roll motif with single Ca2+ binding motif. The LE domains adopt irregular coil structures in a linear extended structure. Both proteins are stabilized through multiple conserved disulfide bridges and have electron density for three N-linked glycosylation sites. However, the positioning of the N-glycosylation differs between the two netrins. In netrin-1 ΔC, the N-glycosylation sites are displayed on the dorsal face of the protein, which is not true for netrin-4 ΔC. Additionally, they also differ because of the extensive contact between the N-terminal domain of netrin-4 and its LE1 domain, which may limit the rotational freedom of the LN domain when compared to other netrins (11).
In this study, we are the first to investigate the solution structure and hydrodynamics of C. elegans UNC-6 using an integrated approach involving dynamic light scattering (DLS), analytical ultracentrifuge (AUC), and small angle x-ray scattering (SAXS) techniques. We determined that its oligomeric state in solution depends on the ionic strength, with low-ionic strength conditions inducing aggregation of UNC-6 (with and without the C-domain), especially at high concentrations. However, in high-ionic strength conditions, we found UNC-6 (with and without the C-domain) to have a more homogeneous solution behavior across all concentrations of protein. This property could aid in its ability to move throughout the extracellular matrix (12).
Materials and Methods
Phylogenetic analysis
Sequence alignment of the Mus musculus Netrin-1 (mNetrin-1; Uniprot ID: O09118), Homo sapiens Netrin-1 (hNetrin-1, Uniprot ID: O95631), Gallus gallus Netrin-1 (chNetrin-1, Uniprot ID: Q90922), C. elegans UNC-6 (Uniprot ID: P34710), Drosophila melanogaster Netrin-a (dpNetrin-a, Uniprot ID: Q24567) was performed by using Clustal W (13). From this alignment, evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix-based model (14) using the MEGA-X program (15). Initial tree(s), which were used for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model before selecting the topology with superior log likelihood.
Protein expression and purification
C. elegans UNC-6 without its C-terminal domain (UNC-6 ΔC, residues 18–487, Uniprot ID: P34710) and full-length UNC-6 (UNC-6 FL, residues 18–612, Uniprot ID: P34710) both containing a thrombin cleavable C-terminal (UNC-6 ΔC) or N-terminal (UNC-6 FL) double Strep II-tag were expressed using the inducible Sleeping Beauty transposon system in human embryonic kidney 293T cells (16). Cells were grown to confluency in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum before being transferred into TripleFlask cell culture vessels (Nunclon; Thermo Fisher Scientific, Waltham, MA) for expression. Protein expression was induced with 0.5 μg/mL doxycycline after cells reached confluency and spent media were collected following 0.2 μm filtration four times every 2 days. With UNC-6 FL only, 1 IU/mL heparin sodium salt (Pan Reac AppliChem, 200 IU/mg) was added to the media from the time of induction onwards. Collections were applied to a 4 mL Strep-Tactin column (IBA Lifesciences, Göttingen, Germany) equilibrated with 50 mM tris, pH 8, 200 mM NaCl (I = 234 mM). After washing the bound protein with a high-ionic strength buffer (50 mM tris, pH 8, 1000 mM NaCl [I = 1035 mM]), both UNC-6 versions were eluted with 50 mM tris, pH 8, 200 mM (I = 234 mM) containing 0.053% (w/v) d-desthiobiotin. UNC-6 ΔC was incubated with 0.5 U thrombin/mg protein in combination with dialysis into 50 mM tris, pH 8.0, 1000 mM NaCl, 2.5 mM CaCl2 (I = 1042 mM) overnight to remove the Strep-tag. The final purification step involved passing cleaved UNC-6 ΔC over a size exclusion chromatography (SEC) Superdex 200 column (GE Healthcare, Fairfield, CT) using the ÄKTA FPLC system in 50 mM tris, pH 8.0, 1000 mM NaCl (I = 1035 mM). After tag-cleavage and purification, an N-terminal sequence extension before residue 18 and a C-terminal extension after residue 487 (DYKDDDDKGSLVPR) remained on the protein in comparison to the UniProt sequence. UNC-6 FL was polished on a Superdex 200 increase column in 50 mM tris, pH 7.5, 500 mM NaCl and 500 mM (NH4)2SO4, before dialyzing it into the analysis buffer (50 mM tris, pH 7.5, 200 mM or 1000 mM NaCl). No thrombin cleavage was performed. Compared to the UniProt sequence, the N-terminus contained the following extension:
APLESWSHPQFEKGGGSGGGSGGGSWSHPQFEKSGLVPRGSAS.
Hydrodynamic characterization
The solution behavior of UNC-6 was analyzed using DLS and sedimentation velocity (SV) AUC in 50 mM tris, pH 7.5, 200 mM NaCl (I = 244 mM, low-ionic strength) and 50 mM tris, pH 7.5, 1000 mM NaCl (I = 1044 mM, high-ionic strength) buffers. For DLS analysis, SEC-purified samples from peak 1 (13 mL in the case of UNC-6 ΔC and 12 mL in the case of UNC-6 FL, Fig. S2 A and C) were concentrated and subjected to 0.1 μm filtration followed by equilibration at 20.0°C for 5 min in the Nano-S Zetasizer (Malvern, Toronto, ON, Canada). The hydrodynamic radius (Rh) distributions were obtained for each buffer condition at concentrations between 0.5 and 8.0 mg/mL (UNC-6 ΔC) and 0.3–0.9 mg/mL (UNC-6 FL) with methods described previously (17, 18, 19, 20). SV experiments were performed as previously described (18, 20, 21) using a ProteomeLab XL-I AUC and an An50Ti 8-cell rotor (Beckman Coulter, Mississauga, ON, Canada). Standard 12 mm double-sector cells were filled with 400 μL of buffer and 400 μL of sample with a concentration range of 0.19–1.90 mg/mL (UNC-6 ΔC) and 0.31–0.88 mg/mL (UNC-6 FL) for each buffer condition. After a temperature equilibration of at least 2 h at rest and under vacuum, the sedimenting samples were measured at rotors speeds of first 30,000 rpm and in a second experiment at 25,000 rpm for 24 h. Both absorbance and interference data were collected. Two-dimensional distributions c(s, fr) of sedimentation coefficient s and frictional ratio fr were calculated using the SEDFIT program as previously described (21, 22, 23, 24). The c(s, fr) distribution deconvolutes diffusion (in form of fr) and sedimentation (as sedimentation constant s) and provides a detailed map of all populations of particles in solution. Since both diffusion and sedimentation constants are obtained, the distribution can be converted to show the molecular mass distribution c(s, M). The c(s, fr) distribution can be collapsed along the fr axis to obtain the one-dimensional c(s, ∗) distribution, which looks similar to the traditional c(s) distribution but without the incorrect assumption of a common fr value for all species. After calculating the map, species analysis (23, 25) was performed to obtain more accurate values for M and s for each population. For the stationary species (monomer), the values of multiple concentrations were linearly extrapolated to infinite dilution. Values for larger particles were averaged. All obtained parameters were corrected to standard solvent conditions (pure water at 20°C).
SAXS
Synchrotron HPLC-SAXS data was collected at the Diamond Light Source (Didcot, UK) using the B21 beamline in-line with an Agilent 1200 (Agilent Technologies, Stockport, UK) HPLC system connected to a temperature-controlled quartz cell capillary. Data were collected for UNC-6 ΔC in low- (2.01 mg/mL protein concentration) and high-ionic (9.2 mg/mL protein concentration) strength buffer using the Superdex 200 SEC column. Each frame was exposed for 3 s, and the frames in the sample peak region were integrated and buffer subtracted using the program scÅtter (26) and further processed using the GNOM program (27) to obtain radius of gyration (Rg) and maximum particle dimension (Dmax), as described previously (17, 20, 21). The ab initio structures for UNC-6 ΔC in high-ionic strength buffer were calculated using the program DAMMIN (28), and each model was verified for quality by the goodness of fit parameter (χ value). The ab initio models were then rotated and averaged using the program DAMAVER (29), resulting in the final low-resolution model.
Calculation of hydrodynamic parameters from ab initio models
The hydrodynamic properties of UNC-6 ΔC were calculated for each model using the program HYDROPRO (30), as described previously (18, 31, 32) (Table 1). The density (1.040120 g/mL) and viscosity (0.011131 Poise) of the high-ionic strength buffer were calculated using SEDNTERP (24, 33). The molecular mass (Mw) and partial specific volume for UNC-6 ΔC (58.65 kDa, 0.70662 cm3/g) and UNC-6 FL (76.49 kDa, 0.71101 cm3/g) were calculated based on amino acid sequence and glycans present using SEDNTERP (24, 33) (Tables S1 B, S2 B and S3 B).
Table 1.
Analysis of SAXS Data for UNC-6 ΔC in High-Ionic Strength Buffer
| Methods | Parameters | Values |
|---|---|---|
| Guinier | Rg (nm) | 4.28 ± 0.03 |
| q.Rg | 0.46–1.29 | |
| I(0) | 0.0140 ± 0.0001 | |
| Points used | 11–81 | |
| GNOM | Rg (nm) | 4.41 ± 0.01 |
| I(0) | 0.0130 ± 0.0001 | |
| Dmax (nm) | 14.3 | |
| Points used | 26–1374 | |
| DAMMIN Ab initio modeling | Models calculated | 12 |
| Χˆ2 | 1.4 | |
| NSD | 0.82 | |
| error NSD | 0.01 | |
| HYDROPRO | Rg (nm) | 4.44 ± 0.04 |
| Dmax (nm) | 15.6 ± 0.01 | |
| Rh (nm) | 4.26 ± 0.02 | |
| s20,w (S) | 2.65 ± 0.01 | |
| Experimental | Rh(nm) - SEC | 4.7 ± 0.2 |
| Rh (nm) - AUC | 4.44 [3.14–7.61] | |
| s20,w (S) - AUC | 3.3 ± 0.7 |
Figure preparation
The figures were prepared using the computer software GUSSI (34) (http://biophysics.swmed.edu/MBR/software.html), QTIPLOT (https://www.qtiplot.com), PRO FIT (https://www.quansoft.com), and Matplotlib (35).
Results
Evolutionary and domain analysis of netrins
Phylogeny analysis using the maximum likelihood method suggested that netrins from vertebrates are more closely related than netrins from invertebrates. The branch length in Fig. 1 represents the number of amino acid substitutions (i.e. length of time) that has occurred to reach the current protein sequence. Starting at the root, dpNetrin-a branches off from the other netrin species followed by UNC-6. Generations later, chNetrin-1 branches off from the vertebrate netrins with mNetrin-1 and hNetrin-1 being the most closely related (99.2% identical).
Macromolecular state of UNC-6 as a function of ionic strength
UNC-6 was purified using affinity chromatography followed by dialysis in buffers containing low- and high-ionic strengths (244 and 1044 mM, respectively). The isoelectric point calculated with ProtParam (36) tool using the protein sequence is 8.5 for the ΔC mutant and 8.7 for the FL version; therefore, we chose to work in 50 mM tris/tris-HCl, pH 7.5 containing 200 or 100 mM NaCl, respectively, to keep the protein soluble. In order to study the macromolecular state of UNC-6 ΔC in solution, it was first analyzed using SEC in the appropriate buffer conditions (low- and high-ionic strengths, Fig. S2, A and B). The SEC elution profile for UNC-6 ΔC in low-ionic strength buffer (Fig. S2 A, magenta) exhibits a comparably broad distribution eluting from ∼12 to ∼15 mL (single peak 1 at 13.09 mL). In contrast, UNC-6 ΔC elutes with a major peak (peak 1) at 12.94 mL and a minor peak at 10.71 mL in high-ionic strength buffer (Fig. S2 A, light blue, peak 2). The elution peaks correspond to hydrodynamic radii Rh of 4.7 ± 0.2 nm (peak 1 in high-ionic strength), 4.6 ± 0.2 nm (peak 1 in low-ionic strength), and 6.5 ± 0.3 nm (peak 2 in high-ionic strength) as derived from our calibration with protein standards of known Rh (Fig. S2 B; Table S1). UNC-6 ΔC recovery from the FPLC run on the Superdex 200 column was significantly lower in low-ionic strength buffer (∼40%) compared to high-ionic strength buffer (60–70%). Nevertheless, the purified peaks under both conditions (peak 1) were concentrated to further study the homogeneity using DLS. The SEC elution profile of UNC-6 FL in high-ionic strength looks very similar to the one of UNC-6 ΔC, eluting with a major peak 1 at 12.00 mL and a minor peak at 9.63 mL, which corresponds to hydrodynamic radii of 5.1 ± 0.2 nm (peak 1) and 7.6 ± 0.3 nm (peak 2) (Fig. S2, C and D). Due to its strong tendency to aggregate in low-ionic strength, we did not perform SEC in this buffer.
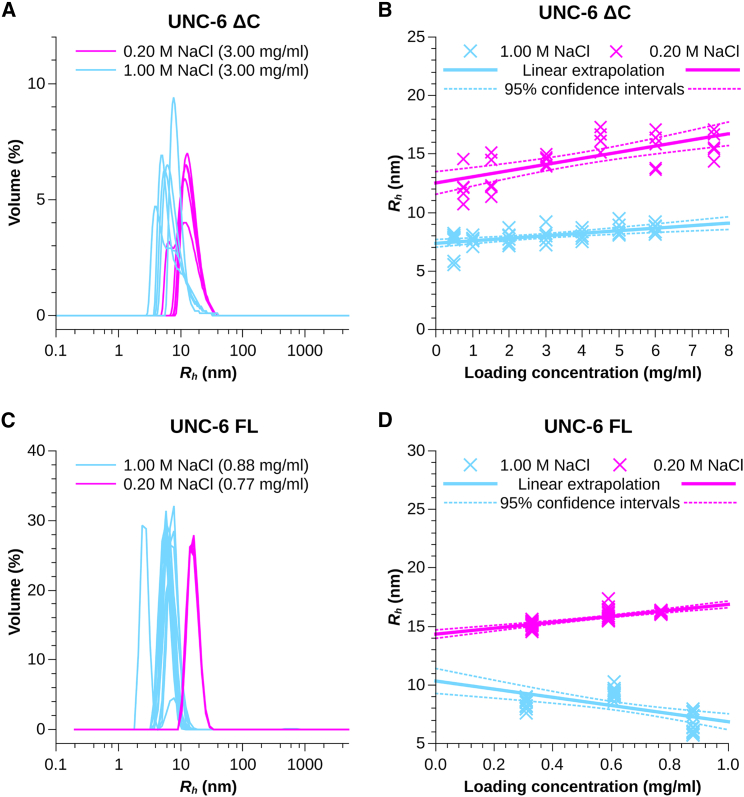
We observed that the UNC-6 ΔC preparation in high-ionic strength buffer could be concentrated up to 6 mg/mL without affecting its Rh, which was not the case for the preparation in low-ionic strength buffer. Although the protein samples used had been previously purified by SEC, further concentration was required for the DLS experiment which induced additional aggregation. The heterogeneity of the volume weighted Rh distributions in Fig. 2 A suggests that more than one species is present and the obtained Rh is too large for a monomer. DLS analysis of UNC-6 ΔC prepared in low-ionic strength buffer suggested much higher values of Rh compared to the ones obtained under high-ionic strength conditions. Clearly, at low-ionic strength, the protein assumes higher oligomeric states of self-association with an Rh of 12.5 ± 1.0 nm, which is further confirmed by a strong increase of Rh (up to 17 nm) with increasing protein concentration (Fig. 2 B). Extrapolation of the results obtained at individual concentrations to infinite dilution provided an Rh of 7.4 ± 0.4 nm for UNC-6 ΔC in high-ionic strength buffer (Fig. 2 B ).
Figure 2.
(A) Volume weighted hydrodynamic radii Rh distributions of UNC-6 ΔC at 3.0 mg/mL in low- (magenta) and high-ionic (light blue) strength. At least five measurements are shown. The larger size of UNC-6 ΔC in 0.20 M NaCl is obvious. The heterogeneity of the distributions suggests the presence of multiple species. (B) Extrapolation (solid line) of the hydrodynamic radii obtained at UNC-6 ΔC concentrations from 0.5 to 7.5 mg/mL to infinite dilution. The obtained Rh are 7.4 ± 0.4 nm in high-ionic strength (light blue) and 12.5 ± 1.0 nm at low ionic-strength (magenta). 95% confidence intervals of the extrapolation are given by stippled lines. The peak position of each measured size distribution is indicated by an Andrew cross. (C) Volume weighted hydrodynamic radii Rh distributions of UNC-6 FL at 0.8 mg/mL in low-ionic (magenta) and high-ionic (light blue) strength. At least 10 measurements are shown. UNC-6 FL also has a larger Rh in 0.20 M NaCl than in 1.00 M NaCl. The heterogeneity of the distributions suggests the presence of multiple species in high-ionic strength. At low-ionic strength, the distribution is surprisingly homogeneous, suggesting their monodispersed nature. However, this finding is not supported by our SV results. (D) Extrapolation (solid line) of the hydrodynamic radii obtained at UNC-6 ΔC concentrations from 0.3 to 0.9 mg/mL to infinite dilution. The obtained Rh are 10.3 ± 1.1 nm in high-ionic strength (light blue) and 14.4 ± 0.4 nm at low-ionic strength (magenta), reflecting the larger size of the FL protein variant. 95% confidence intervals of the extrapolation are given by stippled lines. The peak position of each measured size distribution is indicated by an Andrew cross. To see this figure in color, go online.
The DLS results for UNC-6 FL are similar to that of UNC-6 ΔC, but we could only explore the concentrations range of 0.3–0.9 mg/mL because we observed that it aggregates in low-ionic strength buffer (stronger than the UNC-6 ΔC) and the solubility is limited to ∼1 mg/mL. However, in high-ionic strength buffer conditions, we were able to attain concentrations up to 5 mg/mL without any sign of precipitation or light scattering in the absorbance spectrum. In high-ionic strength, the volume-weighted Rh distributions are heterogenous like UNC-6 ΔC, suggesting the presence of more than one species (Fig. 2 C). However, in low-ionic strength, the Rh distributions presented a single peak, suggesting a homogeneous preparation. The Rh extrapolated to infinite dilution yielded values of 10.3 ± 1.1 nm in high-ionic strength and 14.4 ± 0.4 nm in low-ionic strength buffers (Fig. 2 D). However, DLS is very limited in resolution and gives a disproportionate large weight to larger species, because they dominate the light scattering. The results can therefore be misleading.
To obtain more detailed insight into the hydrodynamics of UNC-6, we performed SV-AUC using loading concentrations from 0.19 to 1.90 mg/mL of UNC-6 ΔC and 0.31–0.88 mg/mL of UNC-6 FL in both, low- and high-ionic strength buffers. We applied the 2-dimensional c(s, fr) analysis (37) to the data, which produces an accurate map of all sedimenting particles separated by the sedimentation coefficient (s) on the abscissa and the frictional coefficient (fr) on the ordinate. Figs. 3 and S3 show the c(s, M) distributions obtained from the interference and absorbance data, respectively, of both versions of UNC-6 in 0.20 M NaCl and 1.00 M NaCl close to 1 mg/mL loading concentration. Fig. S4 and S5 show the one-dimensional c(s, ∗) distributions of all measured loading concentrations obtained by collapsing the c(s, fr) distribution along the fr-axis to a single dimension. The complete analysis of each individual data sets and each loading concentration for both absorbance and interference optics measured at a rotor speed of 30,000 rpm can be found in Fig. S9 (UNC-6 ΔC in 0.20 M NaCl), Fig. S10 (UNC-6 ΔC in 1.00 M NaCl), Fig. S11 (UNC-6 FL in 0.20 M NaCl), and Fig. S12 (UNC-6 FL in 1.00 M NaCl). The fit to the data together with the residuals are shown there as well. To save space, we did not include the data collected at 25,000 rpm in the supporting material, but the results are identical.
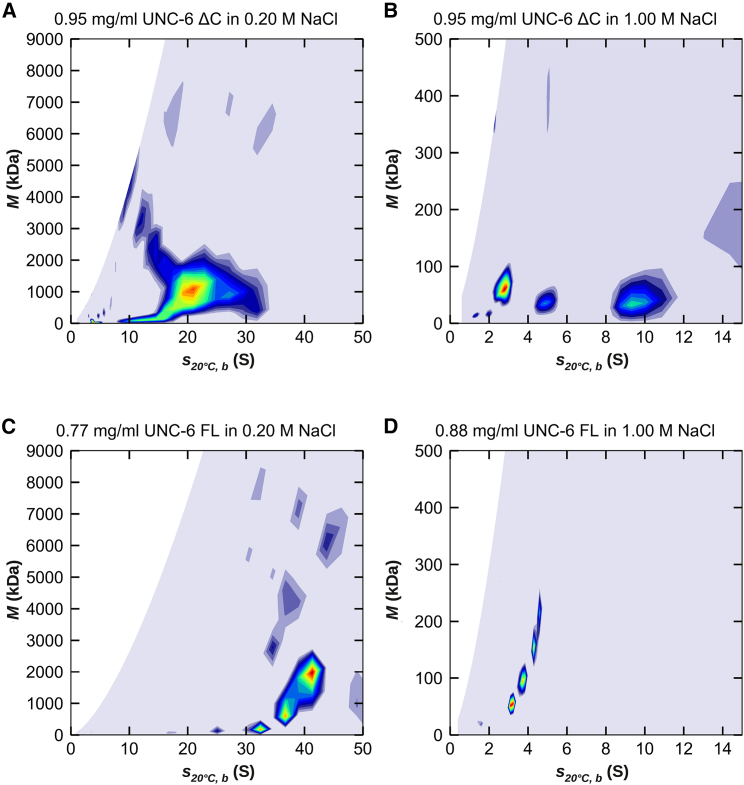
Figure 3.
Two-dimensional c(s, M) distributions obtained from the interference data of (A) C. elegans UNC-6 ΔC in low-ionic strength buffer, (B) C. elegans UNC-6 ΔC in high-ionic strength buffer, (C) C. elegans UNC-6 FL in low-ionic strength buffer, and (D) C. elegans UNC-6 FL in high-ionic strength buffer at concentrations close to 1 mg/mL. In low-ionic strength, the protein forms assemblies of high molecular mass in the range of 1–2 MDa. At high-ionic strength, the formation of these assemblies is suppressed, and the distributions suggest populations of monomers, dimers, and multimers in the case of the ΔC truncation and a monomer/dimer equilibrium (plus a small amount of a larger species) in the case of the FL version. Interestingly, in low-ionic strength, the assemblies of the FL protein have sedimentation coefficients twice as large as the truncated protein; however, their masses are similar. This suggests that the shapes of the assemblies differ between the two protein versions. To see this figure in color, go online.
The difference in the behavior of the protein in the two buffers is striking. In low-ionic strength, we observe a large population of assembled material with molecular masses of 1–2 MDa at all investigated concentrations (Figs. 3, A and C and S3, A and C). In high-ionic strength, the formation of the assemblies is suppressed, and we observe only two major populations of particle sizes (Figs. 3, B and D and S3, B and D). For UNC-6 FL, the suppression is complete, and we were able to fit the two major populations and one small third population with the pure noninteracting species model in SEDPHAT (Fig. S8; Table S3). The first population has a sedimentation coefficient of 3.8 ± 0.6 S and a molecular mass of 70 ± 10 kDa (Fig. 4, C and F; Fig. S8, A and D), which can be attributed to a UNC-6 FL monomer (sequence mass 76.49 kDa). Sedimentation coefficient and mass remain stationary or slightly decrease with increasing loading concentration. This stationary behavior is commonly observed for the smallest species in a self-interacting system. The second population corresponds to an effective (or apparent) particle that has contributions from real monomers and dimers undergoing active self-interaction during the time scale of the sedimentation experiment (24 h). Its sedimentation coefficient and molecular mass is therefore intermediate between monomers and dimers with increasing contribution of the dimer as the loading concentration increased (Fig. S8, B and E). We obtained a sedimentation coefficient of 5.0 ± 0.3 S and a molecular mass of 106 ± 12 kDa. We averaged the values from all loading concentrations, and they underestimate the true values of the dimer. The third population is very small (Fig. S8 G) and contributes less than 3% of the total signal in the investigated concentration range. Its hydrodynamic parameters are therefore not very well defined (Fig. S8, C and F), and its contribution to the effective particle is negligible.
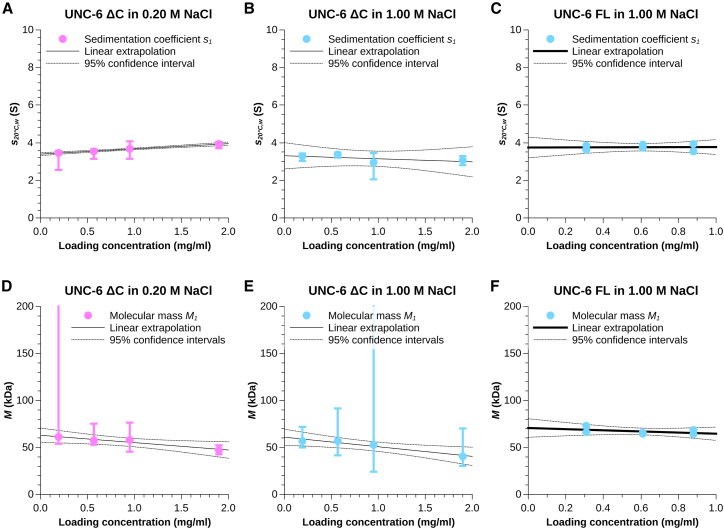
Figure 4.
(A and D) Sedimentation coefficients (A) and molecular masses (D) of monomeric UNC-6 ΔC (species 1) in low-ionic strength buffer as determined by separately fitting a hybrid model of a continuous c(s) distribution and two discrete species to the SV data measured at each protein loading concentration. (B and E) Sedimentation coefficients (B) and molecular masses (E) of monomeric UNC-6 ΔC (species 1) in high-ionic strength buffer as determined by separately fitting a noninteracting discrete species model with two discrete species to the SV data measured at each protein loading concentration. (C and F) Sedimentation coefficients (C) and molecular masses (F) of monomeric UNC-6 FL (species 1) in high-ionic strength buffer as determined by separately fitting a noninteracting discrete species model with three discrete species to the SV data measured at each protein loading concentration. Vertical error bars indicate the 95.4% confidence interval of the respective parameter at each loading concentration. The parameters were extrapolated (without weighting) to infinite dilution (black line) to account for protein-buffer interactions. The 95.4% confidence intervals of the extrapolation are shown by black dotted lines. To see this figure in color, go online.
In the case of UNC-6 ΔC, we also observe two major populations in 1.00 M NaCl (Fig. 3 B; Fig. S10) and we could robustly fit those with the pure noninteracting species model in SEDPHAT. The first population is again the monomeric species (Fig. 4, B and E) with a sedimentation coefficient of 3.3 ± 0.7 nm and a molecular mass of 61 ± 9 kDa (sequence mass 58.65 kDa, Table S2). The second species is again an effective particle containing contributions from self-interacting real particles. It is apparent, however, especially at higher concentrations, that the presence of 1.00 M NaCl does not completely suppress higher oligomeric states and various populations of them are still present in a significant amount. Their contribution prevented a meaningful interpretation of the sedimentation coefficient and molecular mass of the effective particle (Fig. S7, C and D).
In low-ionic strength, UNC-6 ΔC still has a stable pool of monomers in solution (Fig. S9). We fitted these together with a second species using the Hybrid Continuous Distribution and Discrete Species Model SEDPHAT, where we modeled the high mass assemblies using the continuous distribution (Fig. 4, A and B; Figs. S6 and S9). We obtained values of 3.4 ± 0.1 nm for the sedimentation coefficient of the monomer and 63 ± 8 kDa for its mass, matching those obtained at high-ionic strength. The second species was required to obtain a robust fit and is an effective particle but cannot be meaningfully interpreted due to the presence of the high-mass assemblies.
Solution structure of UNC-6 ΔC
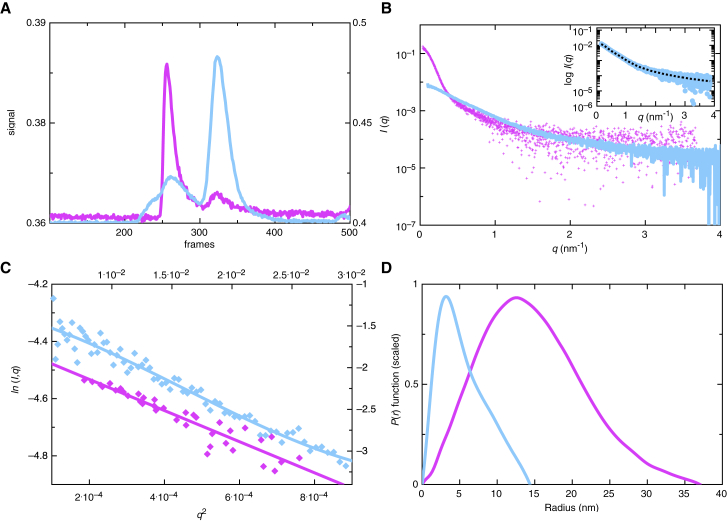
We employed the method of SAXS to study the low-resolution structure of UNC-6 ΔC. Here, we prepared UNC-6 ΔC in low- and high-ionic strength buffers as described above, followed by synchrotron data collection using an HPLC-SAXS (Fig. 5). This method was necessary to separate out the larger aggregates in the protein sample and obtain accurate solution information. Data were collected for UNC-6 ΔC in low- and high-ionic strength buffers (2 mg/mL) in which the sample peaks were integrated and buffer subtracted before further processing. The Kratky analysis for UNC-6 ΔC solubilized in both buffer systems suggested that both proteins are well-folded. The P(r) distribution of UNC-6 ΔC differed with the ionic strength of the buffer. In low-ionic strength, the P(r) distribution peaked at ∼15 nm and had an extended tail up to a Dmax of 37 nm while at high-ionic strength the P(r) distribution peaked at ∼4 nm and contained an extended tail with a Dmax of 14.3 nm (Fig. 5 D). Both P(r) distributions are indicative of elongated proteins, with the Dmax varying depending on the ionic strength of the buffer.
Figure 5.
SAXS data for UNC-6 ΔC in low- and high-ionic strength buffer (magenta and light blue, respectively). (A) Light scattering profile of UNC-6 ΔC after passing through the Superdex 200 column. (B) Raw scattering data from the protein peak chosen at 250–300 frames for UNC-6 ΔC in the low-ionic strength buffer and 300–350 frames for UNC-6 ΔC in high-ionic strength. Inset to (B) displays the DAMMIN fit to the raw scattering data in high-ionic strength buffer (C) Guinier plots show that the scattering data fit to a line. (D) Pair-distance distribution P(r) plot of averaged SAXS data suggest that UNC-6 ΔC has a defined shape with an extended tail that reaches to 14.3 nm in high-ionic strength buffer and 37 nm in low-ionic strength buffer. To see this figure in color, go online.
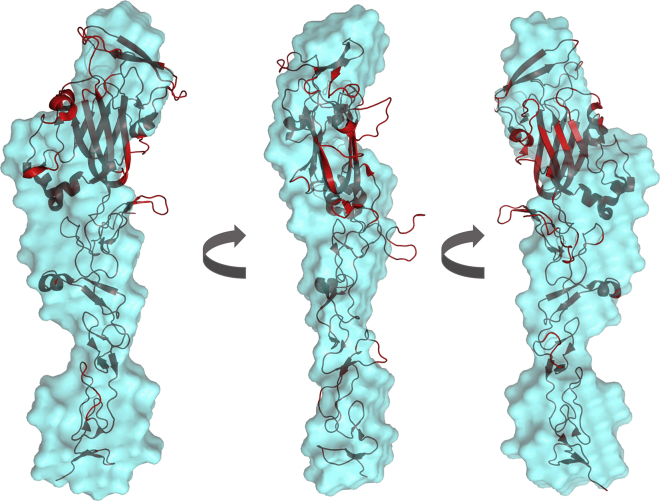
Using DAMMIN analysis of UNC-6 ΔC in high-ionic strength (28), 12 models were generated and averaged to obtain a low-resolution structure (Fig. 6) with a χ2 of 1.4 and a normalized spatial discrepancy (NSD) of 0.82 ± 0.01 (Table 1). This indicates that the individually calculated models agree well with the data and to each other, respectively. Through this low-resolution model we observed that in high-ionic strength buffer, UNC-6 ΔC is an elongated molecule that is monomeric in solution with an Rg = 4.41 ± 0.01 nm. Superimposing the low-resolution UNC-6 ΔC structure with a high-resolution structure of murine Netrin-1 ΔC (PDB: 4OVE) (10) (Fig. 6), we are able to visualize that UNC-6ΔC has a similar overall shape to that of Netrin-1 ΔC.
Figure 6.
Ab initio model of UNC-6 ΔC in high-ionic strength buffer (light blue) compared to crystal structure of murine Netrin-1 ΔC (PDB: 4OVE) (red) shows that monomeric UNC-6 ΔC has a similar shape in solution as murine Netrin-1 ΔC. To see this figure in color, go online.
Discussion
UNC-6 has a distinct evolutionary path, being the first of the entire family of netrins to be discovered in 1990 (3). The LN and LE domains of UNC-6 are somewhat conserved (52–58% identical) among other netrins, specifically drosophila, human, mouse, and chicken, whereas the C-domain (Netrin-like) is more diverse (25–31% identical). This information alone suggests that it is the N-terminal portion (LN and LE domains) that is necessary for the functional role of netrins. This has been experimentally confirmed, determining that the N-terminal domain plays a role of axon guidance in the aforementioned species. A recent study discussed that it is the truncated Netrin-1 ΔC, which is the active protein in cells, modulating vascular permeability in conjunction with the UNC-5B receptor (38). In addition to axon guidance, UNC-6 has been found to not only guide axons but also identified to play a role in cell invasion of basement membranes. Its additional roles include aiding in distal tip migration and neuronal regionalization, neuromuscular junction formation, and polarization of neuron cells (39).
A detailed investigation of the function of each domain in UNC-6 determined that the axon guidance roles of the nematode are domain-specific. Specifically, the VI, V-2, and V-3 domains are involved in dorsal cell and axon guidance, but only the VI domain is additionally involved in ventral cell and axon guidance. In terms of neuronal cell migration, the VI and V-3 domain are essential for its function; however, the V-2 and C-domain only play an influential role in migration (40). However, as we have elucidated to earlier, the C-domain of UNC-6 is much more diverse compared to the other netrins, but the actual function of this domain remains to be determined. Experiments involving UNC-6 conclude that the C-domain is involved in preventing axon-guidance branching (40). More specifically, a study determined that the calcium/calmodulin protein-kinase and diacylglycerol-dependent axon branching is inhibited by the C-domain of UNC-6 (41). Other hypotheses suggest that the basic C-domain is required to tether UNC-6 to other molecules for display on the cell surface or other matrix sites (42). Furthermore, it is believed that the positive charges in this domain are required for its interaction with heparin sulfates (43). Due to the ∼70% sequence difference in the C-domain of UNC-6 with respect to other species, its shape and overall function could be completely species specific.
In our study, we have deduced that the ionic strength of the buffer conditions dramatically affects the solution behavior of UNC-6 (both in presence and absence of the C-domain). At low-ionic strength, the protein forms high molecular weight assemblies with a mass of 1–2 MDa as evidenced by the c(s, M) distributions in Fig. 3. These aggregates are partially (UNC-6 ΔC) or totally (UNC-6 FL) suppressed in high-ionic strength. This is further corroborated by DLS where we noticed a larger Rh of UNC-6 in low-ionic strength buffer than in high-ionic strength buffer. Also the SEC-SAXS data demonstrated greater than a 50% increase in the Dmax of UNC-6 ΔC in low-ionic strength compared to high-ionic strength. There is still a significant pool of monomeric UNC-6 ΔC at both low- and high-ionic strength (Tables S1 and S2), and the monomer maintains a similar head-to-stalk arrangement of its domains as previously published Netrin-1 ΔC and Netrin-4 ΔC crystal structures (10, 11, 44, 45). UNC-6 FL is clearly a monomer/dimer equilibrium in high-ionic strength at the investigated concentration range (0.3–0.9 mg/mL) with a tiny population (<3%) of a larger species. Our data are compatible with a pool of 27–63% monomers (Table S3). It is probable that the UNC-6 ΔC version also forms a certain amount of dimeric species at this ionic strength, but due to the still significant presence of populations with larger masses, we could not determine this with absolute certainty.
In a low-ionic strength environment, both UNC-6 versions are present in an aggregated state, and we have shown that they disassemble in the case of UNC-6 FL into monomers and dimers or in the case of UNC6 ΔC into monomers and undetermined larger species, most likely also a dimer, when the ionic strength of the environment increases. The kinetics of the self-association are fast enough that we could observe it during the time course of the sedimentation experiment (24 h) but slow enough that we could isolate the monomeric species by SEC during the SEC-SAXS experiment. Studies of the extracellular matrix have determined that high-ionic strength contributes to the release of molecules, which are trapped or stored in their hydrogel, allowing them to form electrostatically neutral complexes for transport (12). These transport mediators include heparin, which binds to UNC-6 (presumably the C-domain), allowing it to be carried to commissural axons to guide in their growth (43). Furthermore, the ability of UNC-6 to change its aggregation state could play a major role in its bifunctional axon guidance activity, with some receptors having a higher binding affinity when UNC-6 is in a monomeric state rather than aggregated. With this information, we begin to understand that UNC-6 behavior in solution is not simple and has an added layer of complexity when it comes to understanding its signaling pathway.
Conclusions
Our work demonstrates the effect that ionic strength has on the solution behavior of C. elegans UNC-6. AUC and DLS determined that UNC-6 solubilized in low-ionic strength buffer is present in an aggregated state, whereas in a high-ionic strength buffer it disassembles into monomers and dimers. SAXS data confirm this aggregated nature of UNC-6 ΔC while also concluding that in high-ionic strength buffer it maintains a similar head-to-stalk arrangement as Netrin-1 and Netrin-4. This unique behavior of UNC-6 allows it to be transported through the extracellular matrix and could play a major role in its bifunctional axon guidance activity.
Author Contributions
M.K. and R.R. prepared the UNC-6 ΔC expression vector and optimized protein expression. R.R. and M. K. created the UNC-6 FL expression vector. R. R. performed protein expression and purification. N.K. prepared all UNC-6 ΔC samples and performed the corresponding AUC/DLS experiments. M.M. prepared all UNC-6 FL samples, collected the corresponding AUC/DLS data and analyzed and interpreted all AUC data. T.R.P. collected and processed SAXS data. N.K., M.M., T.R.P., and J.S. wrote the manuscript. J.S. and T.R.P. were both main supervisors of this work.
Acknowledgments
N.K. was supported by a University of Manitoba Faculty of Science scholarship. R.R. was supported by a Mildred-Scheel Postdoctoral Fellowship grant of the German Cancer Aid (Deutsche Krebshilfe). J.S. was funded by the Natural Sciences and Engineering Research Council (NSERC) Discovery Grant (342077 2012), NSERC Research Tool and Infrastructure support (345517 07), and Canada Research Chair program. T.R.P. acknowledges the Canada Research Chair program. We are grateful to the DIAMOND Light Source and their staff for Bio-SAXS (B21) beamtime allocation.
Editor: Andreas Engel.
Footnotes
Natalie Krahn, Markus Meier, and Raphael Reuten contributed equally to this work.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.04.033.
Contributor Information
Joerg Stetefeld, Email: jorg.stetefeld@umanitoba.ca.
Trushar R. Patel, Email: trushar.patel@uleth.ca.
Supporting Material
References
- 1.Moore S.W., Tessier-Lavigne M., Kennedy T.E. Netrins and their receptors. In: Bagnard D., editor. Axon Growth and Guidance. Landes Bioscience and Springer Science; 2007. pp. 17–26. [Google Scholar]; Moore, S. W., M. Tessier-Lavigne, and T. E. Kennedy. 2007. Netrins and their receptors. In Axon Growth and Guidance, D. Bagnard, ed. (Landes Bioscience and Springer Science), pp. 17-26.
- 2.Patel T.R., Reuten R., Stetefeld J. Determination of a molecular shape for netrin-4 from hydrodynamic and small angle X-ray scattering measurements. Matrix Biol. 2012;31:135–140. doi: 10.1016/j.matbio.2011.11.004. [DOI] [PubMed] [Google Scholar]; Patel, T. R., R. Reuten, …, J. Stetefeld. 2012. Determination of a molecular shape for netrin-4 from hydrodynamic and small angle X-ray scattering measurements. Matrix Biol. 31:135-140. [DOI] [PubMed]
- 3.Lai Wing Sun K., Correia J.P., Kennedy T.E. Netrins: versatile extracellular cues with diverse functions. Development. 2011;138:2153–2169. doi: 10.1242/dev.044529. [DOI] [PubMed] [Google Scholar]; Lai Wing Sun, K., J. P. Correia, and T. E. Kennedy. 2011. Netrins: versatile extracellular cues with diverse functions. Development. 138:2153-2169. [DOI] [PubMed]
- 4.Serafini T., Colamarino S.A., Tessier-Lavigne M. Netrin-1 is required for commissural axon guidance in the developing vertebrate nervous system. Cell. 1996;87:1001–1014. doi: 10.1016/s0092-8674(00)81795-x. [DOI] [PubMed] [Google Scholar]; Serafini, T., S. A. Colamarino, …, M. Tessier-Lavigne. 1996. Netrin-1 is required for commissural axon guidance in the developing vertebrate nervous system. Cell. 87:1001-1014. [DOI] [PubMed]
- 5.Bradford D., Cole S.J., Cooper H.M. Netrin-1: diversity in development. Int. J. Biochem. Cell Biol. 2009;41:487–493. doi: 10.1016/j.biocel.2008.03.014. [DOI] [PubMed] [Google Scholar]; Bradford, D., S. J. Cole, and H. M. Cooper. 2009. Netrin-1: diversity in development. Int. J. Biochem. Cell Biol. 41:487-493. [DOI] [PubMed]
- 6.Dominici C., Moreno-Bravo J.A., Chédotal A. Floor-plate-derived netrin-1 is dispensable for commissural axon guidance. Nature. 2017;545:350–354. doi: 10.1038/nature22331. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dominici, C., J. A. Moreno-Bravo, …, A. Chedotal. 2017. Floor-plate-derived netrin-1 is dispensable for commissural axon guidance. Nature. 545:350-354. [DOI] [PMC free article] [PubMed]
- 7.Ishii N., Wadsworth W.G., Hedgecock E.M. UNC-6, a laminin-related protein, guides cell and pioneer axon migrations in C. elegans. Neuron. 1992;9:873–881. doi: 10.1016/0896-6273(92)90240-e. [DOI] [PubMed] [Google Scholar]; Ishii, N., W. G. Wadsworth, …, E. M. Hedgecock. 1992. UNC-6, a laminin-related protein, guides cell and pioneer axon migrations in C. elegans. Neuron. 9:873-881. [DOI] [PubMed]
- 8.Hedgecock E.M., Culotti J.G., Hall D.H. The unc-5, unc-6, and unc-40 genes guide circumferential migrations of pioneer axons and mesodermal cells on the epidermis in C. elegans. Neuron. 1990;4:61–85. doi: 10.1016/0896-6273(90)90444-k. [DOI] [PubMed] [Google Scholar]; Hedgecock, E. M., J. G. Culotti, and D. H. Hall. 1990. The unc-5, unc-6, and unc-40 genes guide circumferential migrations of pioneer axons and mesodermal cells on the epidermis in C. elegans. Neuron. 4:61-85. [DOI] [PubMed]
- 9.Wadsworth W.G., Hedgecock E.M. Hierarchical guidance cues in the developing nervous system of C. elegans. BioEssays. 1996;18:355–362. doi: 10.1002/bies.950180505. [DOI] [PubMed] [Google Scholar]; Wadsworth, W. G., and E. M. Hedgecock. 1996. Hierarchical guidance cues in the developing nervous system of C. elegans. BioEssays. 18:355-362. [DOI] [PubMed]
- 10.Grandin M., Meier M., Stetefeld J. Structural decoding of the Netrin-1–UNC5 interaction and its therapeutical implications in cancers. Cancer Cell. 2016;29:173–185. doi: 10.1016/j.ccell.2016.01.001. [DOI] [PubMed] [Google Scholar]; Grandin, M., M. Meier, …, J. Stetefeld. 2016. Structural decoding of the Netrin-1-UNC5 interaction and its therapeutical implications in cancers. Cancer Cell. 29:173-185. [DOI] [PubMed]
- 11.Reuten R., Patel T.R., Koch M. Structural decoding of netrin-4 reveals a regulatory function towards mature basement membranes. Nat. Commun. 2016;7:13515. doi: 10.1038/ncomms13515. [DOI] [PMC free article] [PubMed] [Google Scholar]; Reuten, R., T. R. Patel, …, M. Koch. 2016. Structural decoding of netrin-4 reveals a regulatory function towards mature basement membranes. Nat. Commun. 7:13515. [DOI] [PMC free article] [PubMed]
- 12.Lieleg O., Baumgärtel R.M., Bausch A.R. Selective filtering of particles by the extracellular matrix: an electrostatic bandpass. Biophys. J. 2009;97:1569–1577. doi: 10.1016/j.bpj.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lieleg, O., R. M. Baumgartel, and A. R. Bausch. 2009. Selective filtering of particles by the extracellular matrix: an electrostatic bandpass. Biophys. J. 97:1569-1577. [DOI] [PMC free article] [PubMed]
- 13.Larkin M.A., Blackshields G., Higgins D.G. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]; Larkin, M. A., G. Blackshields, …, D. G. Higgins. 2007. Clustal W and clustal X version 2.0. Bioinformatics. 23:2947-2948. [DOI] [PubMed]
- 14.Jones D.T., Taylor W.R., Thornton J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]; Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275-282. [DOI] [PubMed]
- 15.Kumar S., Stecher G., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kumar, S., G. Stecher, …, K. Tamura. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35:1547-1549. [DOI] [PMC free article] [PubMed]
- 16.Kowarz E., Löscher D., Marschalek R. Optimized sleeping beauty transposons rapidly generate stable transgenic cell lines. Biotechnol. J. 2015;10:647–653. doi: 10.1002/biot.201400821. [DOI] [PubMed] [Google Scholar]; Kowarz, E., D. Loscher, and R. Marschalek. 2015. Optimized sleeping beauty transposons rapidly generate stable transgenic cell lines. Biotechnol. J. 10:647-653. [DOI] [PubMed]
- 17.Patel T.R., Nikodemus D., Stetefeld J. Biophysical analysis of a lethal laminin alpha-1 mutation reveals altered self-interaction. Matrix Biol. 2016;49:93–105. doi: 10.1016/j.matbio.2015.06.005. [DOI] [PubMed] [Google Scholar]; Patel, T. R., D. Nikodemus, …, J. Stetefeld. 2016. Biophysical analysis of a lethal laminin alpha-1 mutation reveals altered self-interaction. Matrix Biol. 49:93-105. [DOI] [PubMed]
- 18.Meier M., Patel T.R., McKenna S.A. Binding of G-quadruplexes to the N-terminal recognition domain of the RNA helicase associated with AU-rich element (RHAU) J. Biol. Chem. 2013;288:35014–35027. doi: 10.1074/jbc.M113.512970. [DOI] [PMC free article] [PubMed] [Google Scholar]; Meier, M., T. R. Patel, …, S. A. McKenna. 2013. Binding of G-quadruplexes to the N-terminal recognition domain of the RNA helicase associated with AU-rich element (RHAU). J. Biol. Chem. 288:35014-35027. [DOI] [PMC free article] [PubMed]
- 19.Stetefeld J., McKenna S.A., Patel T.R. Dynamic light scattering: a practical guide and applications in biomedical sciences. Biophys. Rev. 2016;8:409–427. doi: 10.1007/s12551-016-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stetefeld, J., S. A. McKenna, and T. R. Patel. 2016. Dynamic light scattering: a practical guide and applications in biomedical sciences. Biophys. Rev. 8:409-427. [DOI] [PMC free article] [PubMed]
- 20.Krahn N., Meier M., Stetefeld J. Nanoscale assembly of high-mobility group AT-hook 2 protein with DNA replication fork. Biophys. J. 2017;113:2609–2620. doi: 10.1016/j.bpj.2017.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]; Krahn, N., M. Meier, …, J. Stetefeld. 2017. Nanoscale assembly of high-mobility group AT-hook 2 protein with DNA replication fork. Biophys. J. 113:2609-2620. [DOI] [PMC free article] [PubMed]
- 21.Meier M., Moya-Torres A., Stetefeld J. Structure and hydrodynamics of a DNA G-quadruplex with a cytosine bulge. Nucleic Acids Res. 2018;46:5319–5331. doi: 10.1093/nar/gky307. [DOI] [PMC free article] [PubMed] [Google Scholar]; Meier, M., A. Moya-Torres, …, J. Stetefeld. 2018. Structure and hydrodynamics of a DNA G-quadruplex with a cytosine bulge. Nucleic Acids Res. 46:5319-5331. [DOI] [PMC free article] [PubMed]
- 22.Dam J., Schuck P. Calculating sedimentation coefficient distributions by direct modeling of sedimentation velocity concentration profiles. Methods Enzymol. 2004;384:185–212. doi: 10.1016/S0076-6879(04)84012-6. [DOI] [PubMed] [Google Scholar]; Dam, J., and P. Schuck. 2004. Calculating sedimentation coefficient distributions by direct modeling of sedimentation velocity concentration profiles. Methods Enzymol. 384:185-212. [DOI] [PubMed]
- 23.Schuck P. Sedimentation analysis of noninteracting and self-associating solutes using numerical solutions to the Lamm equation. Biophys. J. 1998;75:1503–1512. doi: 10.1016/S0006-3495(98)74069-X. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schuck, P. 1998. Sedimentation analysis of noninteracting and self-associating solutes using numerical solutions to the Lamm equation. Biophys. J. 75:1503-1512. [DOI] [PMC free article] [PubMed]
- 24.Laue T.M., Shah B.D., Pelletier S.L. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S.E., Horton J.C., editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]; Laue, T. M., B. D. Shah, …, S. L. Pelletier. 1992. Computer-aided interpretation of analytical sedimentation data for proteins. In Analytical Ultracentrifugation in Biochemistry and Polymer Science, S. E. Harding and J. C. Horton, eds. (Royal Society of Chemistry), pp. 90-125.
- 25.Brown P.H., Schuck P. A new adaptive grid-size algorithm for the simulation of sedimentation velocity profiles in analytical ultracentrifugation. Comput. Phys. Commun. 2008;178:105–120. doi: 10.1016/j.cpc.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; Brown, P. H., and P. Schuck. 2008. A new adaptive grid-size algorithm for the simulation of sedimentation velocity profiles in analytical ultracentrifugation. Comput. Phys. Commun. 178:105-120. [DOI] [PMC free article] [PubMed]
- 26.Förster S., Apostol L., Bras W. Scatter: software for the analysis of nano- and mesoscale small-angle scattering. J. Appl. Cryst. 2010;43:639–646. [Google Scholar]; Forster, S., L. Apostol, and W. Bras. 2010. Scatter: software for the analysis of nano- and mesoscale small-angle scattering. J. Appl. Cryst. 43:639-646.
- 27.Svergun D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 1992;25:495–503. [Google Scholar]; Svergun, D. I. 1992. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 25:495-503.
- 28.Franke D., Svergun D.I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]; Franke, D., and D. I. Svergun. 2009. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 42:342-346. [DOI] [PMC free article] [PubMed]
- 29.Volkov V.V., Svergun D.I. Small-angle scattering techniques. J. Appl. Cryst. 2003;36:860–864. [Google Scholar]; Volkov, V. V., and D. I. Svergun. 2003. Small-angle scattering techniques. J. Appl. Cryst. 36:860-864.
- 30.Ortega A., Amorós D., García de la Torre J. Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys. J. 2011;101:892–898. doi: 10.1016/j.bpj.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ortega, A., D. Amoros, and J. Garcia de la Torre. 2011. Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys. J. 101:892-898. [DOI] [PMC free article] [PubMed]
- 31.Deo S., Patel T.R., McKenna S.A. Activation of 2′ 5′-oligoadenylate synthetase by stem loops at the 5′-end of the West Nile virus genome. PLoS One. 2014;9:e92545. doi: 10.1371/journal.pone.0092545. [DOI] [PMC free article] [PubMed] [Google Scholar]; Deo, S., T. R. Patel, …, S. A. McKenna. 2014. Activation of 2′ 5′-oligoadenylate synthetase by stem loops at the 5′-end of the West Nile virus genome. PLoS One. 9:e92545. [DOI] [PMC free article] [PubMed]
- 32.Dzananovic E., Patel T.R., McKenna S.A. Recognition of viral RNA stem-loops by the tandem double-stranded RNA binding domains of PKR. RNA. 2013;19:333–344. doi: 10.1261/rna.035931.112. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dzananovic, E., T. R. Patel, …, S. A. McKenna. 2013. Recognition of viral RNA stem-loops by the tandem double-stranded RNA binding domains of PKR. RNA. 19:333-344. [DOI] [PMC free article] [PubMed]
- 33.Tucker H., Wright A., Philo J. University of New Hampshire; Durham, New Hampshire: 2013. Sedimentation interpretation program. [Google Scholar]; Tucker, H., A. Wright, …, J. Philo. 2013. Sedimentation interpretation program: University of New Hampshire, Durham, New Hampshire.
- 34.Brautigam C.A. Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol. 2015;562:109–133. doi: 10.1016/bs.mie.2015.05.001. [DOI] [PubMed] [Google Scholar]; Brautigam, C. A. 2015. Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol. 562:109-133. [DOI] [PubMed]
- 35.Hunter J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. [Google Scholar]; Hunter, J. D. 2007. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9:90-95.
- 36.Gasteiger E., Hoogland C., Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker J.M., editor. The Proteomics Protocols Handbook. Humana Press; 2005. pp. 571–607. [Google Scholar]; Gasteiger, E., C. Hoogland, …, A. Bairoch. 2005. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook, J. M. Walker, ed. (Humana Press), pp. 571-607.
- 37.Brown P.H., Schuck P. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys. J. 2006;90:4651–4661. doi: 10.1529/biophysj.106.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]; Brown, P. H., and P. Schuck. 2006. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys. J. 90:4651-4661. [DOI] [PMC free article] [PubMed]
- 38.Miloudi K., Binet F., Sapieha P. Truncated netrin-1 contributes to pathological vascular permeability in diabetic retinopathy. J. Clin. Invest. 2016;126:3006–3022. doi: 10.1172/JCI84767. [DOI] [PMC free article] [PubMed] [Google Scholar]; Miloudi, K., F. Binet, …, P. Sapieha. 2016. Truncated netrin-1 contributes to pathological vascular permeability in diabetic retinopathy. J. Clin. Invest. 126:3006-3022. [DOI] [PMC free article] [PubMed]
- 39.Ziel J.W., Sherwood D.R. Roles for netrin signaling outside of axon guidance: a view from the worm. Dev. Dyn. 2010;239:1296–1305. doi: 10.1002/dvdy.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ziel, J. W., and D. R. Sherwood. 2010. Roles for netrin signaling outside of axon guidance: a view from the worm. Dev. Dyn. 239:1296-1305. [DOI] [PMC free article] [PubMed]
- 40.Lim Y.S., Wadsworth W.G. Identification of domains of netrin UNC-6 that mediate attractive and repulsive guidance and responses from cells and growth cones. J. Neurosci. 2002;22:7080–7087. doi: 10.1523/JNEUROSCI.22-16-07080.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lim, Y. S., and W. G. Wadsworth. 2002. Identification of domains of netrin UNC-6 that mediate attractive and repulsive guidance and responses from cells and growth cones. J. Neurosci. 22:7080-7087. [DOI] [PMC free article] [PubMed]
- 41.Wang Q., Wadsworth W.G. The C domain of netrin UNC-6 silences calcium/calmodulin-dependent protein kinase- and diacylglycerol-dependent axon branching in Caenorhabditis elegans. J. Neurosci. 2002;22:2274–2282. doi: 10.1523/JNEUROSCI.22-06-02274.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang, Q., and W. G. Wadsworth. 2002. The C domain of netrin UNC-6 silences calcium/calmodulin-dependent protein kinase- and diacylglycerol-dependent axon branching in Caenorhabditis elegans. J. Neurosci. 22:2274-2282. [DOI] [PMC free article] [PubMed]
- 42.Wadsworth W.G., Bhatt H., Hedgecock E.M. Neuroglia and pioneer neurons express UNC-6 to provide global and local netrin cues for guiding migrations in C. elegans. Neuron. 1996;16:35–46. doi: 10.1016/s0896-6273(00)80021-5. [DOI] [PubMed] [Google Scholar]; Wadsworth, W. G., H. Bhatt, and E. M. Hedgecock. 1996. Neuroglia and pioneer neurons express UNC-6 to provide global and local netrin cues for guiding migrations in C. elegans. Neuron. 16:35-46. [DOI] [PubMed]
- 43.Kappler J., Franken S., Koch K.W. Glycosaminoglycan-binding properties and secondary structure of the C-terminus of netrin-1. Biochem. Biophys. Res. Commun. 2000;271:287–291. doi: 10.1006/bbrc.2000.2583. [DOI] [PubMed] [Google Scholar]; Kappler, J., S. Franken, …, K. W. Koch. 2000. Glycosaminoglycan-binding properties and secondary structure of the C-terminus of netrin-1. Biochem. Biophys. Res. Commun. 271:287-291. [DOI] [PubMed]
- 44.Xu K., Wu Z., Nikolov D.B. Neural migration. Structures of netrin-1 bound to two receptors provide insight into its axon guidance mechanism. Science. 2014;344:1275–1279. doi: 10.1126/science.1255149. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xu, K., Z. Wu, …, D. B. Nikolov. 2014. Neural migration. Structures of netrin-1 bound to two receptors provide insight into its axon guidance mechanism. Science. 344:1275-1279. [DOI] [PMC free article] [PubMed]
- 45.Finci L.I., Krüger N., Meijers R. The crystal structure of netrin-1 in complex with DCC reveals the bifunctionality of netrin-1 as a guidance cue. Neuron. 2014;83:839–849. doi: 10.1016/j.neuron.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; Finci, L. I., N. Kruger, …, R. Meijers. 2014. The crystal structure of netrin-1 in complex with DCC reveals the bifunctionality of netrin-1 as a guidance cue. Neuron. 83:839-849. [DOI] [PMC free article] [PubMed]
- 46.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kumar, S., G. Stecher, and K. Tamura. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33:1870-1874. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.