Abstract
Small extracellular vesicles (EVs) are 50–200 nm sized mediators in intercellular communication that reflect both physiological and pathophysiological changes of their parental cells. Thus, EVs hold great potential for biomarker detection. However, reliable purification methods for the downstream screening of the microRNA (miRNA) cargo carried within urinary EVs by small RNA sequencing have yet to be established. To address this knowledge gap, RNA extracted from human urinary EVs obtained by five different urinary EV purification methods (spin column chromatography, immunoaffinity, membrane affinity, precipitation and ultracentrifugation combined with density gradient) was analyzed by small RNA sequencing. Urinary EVs were further characterized by nanoparticle tracking analysis, Western blot analysis and transmission electron microscopy. Comprehensive EV characterization established significant method-dependent differences in size and concentration as well as variances in protein composition of isolated vesicles. Even though all purification methods captured enough total RNA to allow small RNA sequencing, method-dependent differences were also observed with respect to library sizes, mapping distributions, number of miRNA reads and diversity of transcripts. Whereas EVs obtained by immunoaffinity yielded the purest subset of small EVs, highly comparable with results attained by ultracentrifugation combined with density gradient, precipitation and membrane affinity, sample purification by spin column chromatography indicated a tendency to isolate different subtypes of small EVs, which might also carry a distinct subset of miRNAs. Based on our results, different EV purification methods seem to preferentially isolate different subtypes of EVs with varying efficiencies. As a consequence, sequencing experiments and resulting miRNA profiles were also affected. Hence, the selection of a specific EV isolation method has to satisfy the respective research question and should be well considered. In strict adherence with the MISEV (minimal information for studies of extracellular vesicles) guidelines, the importance of a combined evaluation of biophysical and proteomic EV characteristics alongside transcriptomic results was clearly demonstrated in this present study.
Abbreviations: A, spin column chromatography; Ago2, argonaute-2 protein; ANOVA, analysis of variance; B, immunoaffinity; C, membrane affinity; D, precipitation; DGE, differential gene expression; DTT, dithiothreitol; E, ultracentrifugation combined with density gradient; EV(s), extracellular vesicle(s); FM, fluorescent mode; mIgG, murine immunoglobulin G; miRNA, microRNA; MISEV, minimal information for studies of extracellular vesicles; mRNA, messenger RNA; nm, nanometer(s); nt, nucleotide(s); NTA, nanoparticle tracking analysis; PC, principal component; RIN, RNA integrity number; rRNA, ribosomal RNA; RNA-Seq, RNA sequencing; SM, scattering mode; snRNA, small nuclear RNA; snoRNA, small nucleolar RNA; TEM, transmission electron microscopy; tRNA, transfer RNA; UCrea, urinary creatinine; uEVs, urinary extracellular vesicles
Keywords: Extracellular vesicles, Small RNA sequencing, microRNA, Human, Urine, Biomarker
1. Introduction
Urine presents a highly preferred biofluid in disease detection due to its non-invasive accessibility, allowing fast and easy sampling, that can even be carried out by patients themselves. Due to its broad variety in proteins, metabolites, cells and cellular contents not only from the urogenital tract, but also from glomerular filtration of plasma, urine offers great potential for comprehensive analyses of an individual’s physical and pathological condition [1]. For some time, urine proteomics and transcriptomics have already been investigated for prostate or bladder cancer identification [[2], [3], [4]]. However, a consistent approach for sample collection and handling across all clinical disciplines prior to any downstream “-omics” analysis, and transcriptomics in particular, has yet to be established. Additionally, many pathological conditions remain unnoticed until their significant manifestation, due to the lack of trustable and precise biomarkers for early disease detection and prevention.
Besides total urine analysis, more and more research points towards urinary extracellular vesicles (uEVs) as promising cellular candidates for reliable biomarker identification. EVs are secreted by all cell types as important mediators in intercellular communication and are thought to reflect physiological as well as pathophysiological changes of their secreting tissues [5,6]. With a size ranging from 50 up to 200 nanometers (nm), the bilayered exosomes putatively represent the smallest and best described type of EVs [5,7,8]. By now, those small EVs were already investigated by numerous research groups from various biological fluids, with the major focus on blood derived compartments [[9], [10], [11], [12]], saliva [13], milk [14,15], cerebrospinal fluid [16], semen [17], ascites [18], and urine [19,20]. Previous studies focusing on urine demonstrated intense uEV secretion from all parts of the nephron and collecting duct, whose content is characteristic for the urinary tract, including kidney, ureter and bladder [[21], [22], [23]]. Since Valadi et al. discovered in 2007 that particularly small EVs contain functional RNAs including microRNA (miRNA) [24], small EV-miRNA has come to the fore. As part of the non-coding RNA family with an average size of 22 nucleotides (nt), miRNAs rank amongst the most fundamental post-transcriptional regulators, mediating gene silencing by inhibition or degradation of complementary messenger RNA (mRNA) [25,26]. Contrary to free urinary miRNAs, small EV-miRNAs are protected against prevalent RNA-hydrolyzing enzymes by the vesicular membrane, but also when bound to argonaute-2 proteins (Ago2) and lipoproteins [[27], [28], [29], [30]]. Driven by their non-invasive accessibility and exceptional stability as well as recent advances in nucleic acid quantification, both EV and non EV derived urinary miRNA profiles have emerged as promising discriminators for disease progression of bladder cancer [31], chronic kidney disease [32], type 1 diabetes [[33], [34], [35]], lupus nephritis [36] and primary focal segmental glomerulosclerosis [37], exemplarily. Despite the multitude of uEV research, no concordance exists on the best possible isolation and, at the same time, an extensive characterization of the isolated vesicles is only poorly reported. According to EV-TRACK [38], a crowdsourcing platform providing higher transparency on EV-related experiments, staggering 34 different strategies for EV isolation from urine have already been applied in more than 130 experiments, emphasizing the pressing need for more standardized protocols.
Indeed, more and more researchers realized the necessity of comprehensive sample characterization and proper uEV purification to provide high reproducibility and comparability. At present, the MISEV guidelines are state of the art for reliable and valid EV research [39]. Based on best sample knowledge, appropriate purification methods suitable for any downstream analyses can be applied. With ultracentrifugation being the gold standard for a long time, closer examinations of the different received compartments during centrifugation as well as performance optimization via combined application of different methods were attempted [20,40,41]. Additionally, various new isolation kits have been developed and commercialized [42]. Moreover, to improve the quality and understanding of the purified material, many efforts comparing major purification strategies based on EV morphology, proteome and nucleic acid content were made, outlining prevailing advantages and pitfalls depending on the respective sample type, purification strategy and downstream analyses [[43], [44], [45], [46], [47], [48]].
In terms of valuable biomarker discovery, either related to EVs or not, small RNA sequencing (RNA-Seq) has been evidenced as the most promising tool, especially by detecting both novel and known miRNAs. Although small RNA-Seq results have been shown to be strongly dependent on the chosen isolation strategy of miRNA originating from serum EVs [11,49], an equivalent screening of isolation-dependent effects on urinary derived EV-miRNA is still absent. Up to now, it has not been ascertained adequately whether the application of small RNA-Seq is possible for uEV derived miRNA, and which uEV purification strategy would deliver the most reproducible and reliable results. To this end, we aimed to test for general feasibility of small RNA-Seq of uEV-miRNA by applying five different EV isolation strategies and evaluating their success in small RNA-Seq. In addition, uEVs were characterized according to MISEV guidelines [39]. Particularly, particle concentration, morphology and protein content of purified uEVs were analyzed to provide sound sample knowledge, thus, contributing to forward-looking urinary biomarker research.
2. Methods
2.1. Urine sampling and processing
Spontaneously voided urine from six healthy male and non-smoking subjects aged between 18 and 36 was collected into sterile containers. Ten ml urine were aliquoted instantly and saved for the determination of urinary creatinine concentration. The remaining urine was kept at 4 °C in the dark for no longer than eight hours. Then, to sediment cells, urine was centrifuged at 4 °C at 300 x g for 10 min. The supernatant was centrifuged another 20 min. at 4 °C at 2,000 x g to pellet cell debris and larger particles. The resulting cell-free urine was stored at 80 °C until further processing. Written informed consent has been obtained from every participant as part of an ethically approved study (test number 359-14) with consistent compliance with the Declaration of Helsinki [50].
2.2. Urinary creatinine determination
The urinary creatinine level was determined by an external laboratory based on the Jaffé method [51], which is a kineticcolour reaction test happening in an alkaline medium in presence of picric acid (AU 5800, Beckman Coulter). As it has been shown that urinary creatinine is strongly correlated with uEV number and urine dilution conditions, and, thus, is highly suitable for normalization purposes, it has been applied to account for individual differences [52,53].
2.3. Extracellular vesicle purification
Frozen urine samples were slowly thawed over-night at 4 °C and vortexed vigorously. Five different strategies were utilized to purify small EVs from cell-free urine (see Table 1).
Table 1.
Overview of applied strategies to purify small extracellular vesicles from urine.
| Purification type: | Based on: | Manufacturer: |
|---|---|---|
| A | Spin column chromatography | Urine Exosome Purification and RNA Isolation Midi Kit (Norgen Biotek, Norway) |
| B | Immunoaffinity | Exosome Isolation Kit Pan, human (Miltenyi Biotec, Germany) |
| C | Membrane affinity | exoRNeasy Serum/Plasma Midi Kit (Qiagen, Germany) |
| D | Precipitation | miRCURY Exosome Isolation Kit – Cells, urine and CSF (Exiqon, Denmark) |
| E | Ultracentrifugation with density gradient | Optima LE-80 K (Beckman Coulter, USA) and OptiPrep (Merck, Germany) |
Since every purification strategy is optimized for different input volumes, Amicon Ultra-15 Centrifugal Filter Devices (100 K, Ultracel membrane of regenerated cellulose, Merck, Germany) were used to concentrate 10 ml cell-free urine, each, to the recommended input volume according to the manufacturer’s instructions. The concentration and purification process was conducted in duplicate from which one was used for subsequent EV characterization via nanoparticle tracking analysis (NTA), Western blot analysis and transmission electron microscopy (TEM), and the other one used as starting material for RNA isolation and small RNA-Seq.
EVs from concentrated urine were isolated consistent with the provided manuals (A to D). In terms of ultracentrifugation (E), a modified protocol already published by Greening et al. [54] was applied. In brief, 4 ml concentrated cell-free urine were centrifuged at 4 °C for 30 min. at 17,000 x g (k-factor: 987.4) in a SW60Ti swinging-bucket rotor (Beckman Coulter Optima LE-80 K, USA). The recovered supernatant was transferred into a new centrifugal tube and centrifuged another two hours at 4 °C at 100,000 x g (k-factor: 167.9) in a SW60Ti swinging-bucket rotor. The resulting pellet was resuspended in 750 μl 1x PBS and loaded on top of a discontinuous 5–40 % OptiPrep density gradient. Prepared as such, an over-night centrifugation for 18 h was conducted at 4 °C at 100,000 x g (k-factor: 390.8) using the SW40Ti swinging-bucket rotor. Eleven fractions of 900 μl, each, were recovered from top-to-bottom and washed once in 1x PBS at 4 °C for one hour at 100,000 x g (k-factor: 277.5). After discarding the supernatant, the obtained pellet was retained in 100 μl 1x PBS. A separate control density gradient loaded with double-distilled water was prepared and centrifuged alongside samples. The thereby obtained fractions were applied to a refractometer for density calculation.
Purified uEVs from all five purification methods (A–E) were stored at −80 °C until RNA isolation and subsequent small RNA-Seq. Samples for uEV characterization were vacuum-evaporated to a final volume of around 45 μl. The actual volume was determined to calculate initial uEV concentrations in urine based on NTA results. uEV samples were then stored at -80 °C until further processing.
2.4. Nanoparticle tracking analysis
Particle concentration as well as size distribution of uEV preparations were measured using the ZetaView PMX110 (Particle Metrix, Germany) and the corresponding software, version 8.04.02 SP2. After instrument calibration, the temperature was clamped at 24 °C, and the pre-acquisition parameters to measure scattered light (SM) were set to a shutter of 70, frame rate of 30 and a sensitivity of 80% at high resolution. Post-acquisition parameters were adjusted to a minimum brightness of 25, size range of 5–1,000 nm and a trace length of 15. Appropriate sample dilution in 1x PBS was evaluated before every measurement. To discriminate between biological and non-biological particles, such as the microbeads contained within the immunoaffinitybased EV purification, all samples were mixed with 5 μg/ml CellMask Orange Plasma Membrane Stain (Invitrogen, USA), incubated at 37 °C for 30 min. and additionally measured in the fluorescence mode (FM) of the instrument. Besides adjusting sensitivity to 95%, all software settings were kept the same for scatter and fluorescence analyses. To account for the sensitivity adjustment in the fluorescent mode, a correcting calibration factor was introduced according to the manufacturer’s recommendations. uEV concentration was re-calculated according to Eitan et al. [55] accounting for sample dilution, EV sample volume and starting urine volume. The re-calculated particle concentrations were normalized to urinary creatinine (UCrea) [52]. Using GraphPad Prism, version 7.04, one-way analysis of variance (ANOVA) with Tukey post-hoc test was performed to determine significance of results and adjust for multiple comparisons. Adjusted p values < 0.05 were treated as significant.
2.5. Western blotting
For Western blotting, thawed EV samples were lysed in 1x RIPA buffer supplemented with protease inhibitors for 15 min. on ice supported by three 1 min bouts of ultrasonication. Depending on the investigated proteins, samples were analyzed in either reducing (Laemmli buffer + β-mercaptoethanol; 10 min. at 95 °C) or non-reducing (Laemmli buffer, 5 min. at 95 °C) conditions. 20 μl of protein lysate, each, were loaded onto SDS-PAGE gels (NuPAGE Bis-Tris,4–12 %; Invitrogen, USA). Whole cell lysates from HEK293 (8 μg; OriGene Technologies, Inc, USA) and HeLa (10 μg; OriGene Technologies, Inc., USA) cells were loaded as positive controls. The separated proteins were transferred to a nitrocellulose membrane with a 0.45 μm pore size (GE Healthcare Life Sciences, USA). After blocking the membrane in blocking buffer, containing 1% skim milk powder in 1x TBST, for one hour at room temperature, it was incubated over-night with the primary antibody at 4 °C, followed by another incubation with the secondary antibody for one hour at room temperature. Afterwards, the membrane was incubated with Clarity Western ECL Substrate (Bio-Rad, UK) for five minutes at room temperature allowing for chemiluminescent signal capturing using the Fusion FX6 system (Vilber, France).
The following primary antibodies were used, diluted in blocking buffer: goat anti-Calnexin (WA-AF1179a, Biomol, 1:2,500), mouse anti-Uromodulin (Clone B-2, sc-271022, Santa Cruz, 1:100), rabbit anti-Alix (ab186728, Abcam, 1:1,000), rabbit anti-EPCAM (ab32392, Abcam, 1:1,000), rabbit anti-Syntenin (Clone EPR8102, ab133267, Abcam, 1:5,000), mouse anti-TSG101 (Clone 4A10, ab83, Abcam, 1:800), mouse anti-CD63 (Clone TS63, ab59479, Abcam, 1:500), mouse anti-CD81 (Clone M38, ab79559, Abcam, 1:833), and rabbit anti-CD9 (ab92726, Abcam, 1:1,000). All proteins except CD63 and CD81 were investigated under reducing conditions. The secondary antibodies conjugated to horseradish peroxidase were purchased from Abcam and diluted in blocking buffer (1:6,666): goat anti-Mouse (ab97040), goat anti-Rabbit (ab97080), and rabbit anti-Goat (ab97105).
2.6. Transmission electron microscopy
Five μl of each uEV preparation were left to adhere onto formvar carbon coated grids (Nickel Grid 200 mesh; Electron Microscopy Sciences, USA) for five minutes followed by five minutes of negative staining with 2% aqueous uranyl acetate. Excess liquids were blotted. Total grid preparation was performed at room temperature. Images were acquired of air-dried grids on the same day at 80 kV using the Zeiss EM 900 instrument (Zeiss, Germany).
We have submitted all relevant data of our uEV experiments to the EV-TRACK knowledgebase (EV-TRACK ID: EV190007) [38].
2.7. Extracellular RNA isolation
The miRNeasy Mini Kit (Qiagen, Germany) was used to extract total RNA from all uEV samples. Due to different uEV elution volumes, the amount of used QIAzol lysis reagent was adapted according to the manufacturer’s recommendations. To achieve a higher RNA yield, the first eluate of 30 μl was applied to the membrane a second time. Isolated RNA was quality controlled and quantified by capillary gel electrophoresis using the RNA 6000 Pico Kit (Agilent Technologies, Germany) and the Bioanalyzer 2100 (Agilent Technologies). Total RNA was stored at -80 °C until library preparation.
2.8. Small RNA library preparation and sequencing reaction
After complete vacuum evaporation, total RNA samples were resuspended in 12 μl nuclease-free water. The manufacturer’s instructions provided with the NEBNext Multiplex Small RNA Library Prep Set for Illumina (New England BioLabs, USA) were followed to prepare small RNA libraries. Recommended adjustments for minimal RNA input amount were carried out additionally. The MinElute PCR Purification Kit (Qiagen, Germany) was utilized to purify resulting PCR products prior to cDNA library evaluation via capillary gel electrophoresis using the DNA 1000 Kit and the Bioanalyzer 2100 (Agilent Technologies) according to the manual. For size selection at a miRNA-specific length of 130–150 base pairs, barcoded cDNA libraries were pooled to 5 ng cDNA input and fractionated on a 4% agarose gel (MetaPhor, USA). Bands appropriate in size were cut and cleaned up using the MinElute Gel Extraction Kit (QIAGEN GmbH, Germany). Correct size and purity of the cut cDNA band were analyzed via capillary gel electrophoresis using the Bioanalyzer DNA High Sensitivity Kit (Agilent Technologies) before sequencing via 50 cycles of single-end sequencing-by-synthesis reactions using the HiSeq Rapid SBS Kit v2 (Illumina, USA) on the HiSeq 2500 instrument (Illumina, USA).
2.9. Small RNA-Seq data analysis
The FastQC software (Babraham Bioinformatics, UK, Version 0.11.7) was applied to check for the overall sequencing success, such as the per base sequence quality indicated by the Quality Phred Score and sequence length distribution, based on the imported raw sequencing data [56]. In the following, 3′ adapter sequences added during cDNA preparation were trimmed using Btrim [57] and reads without detectable adapter (“No_Adapter”) were excluded. To avoid bias by false positive mappings introduced by reads shorter than 16 nucleotides (“Short”), corresponding reads were rejected, as well. Remaining reads were mapped to sequences obtained from RNAcentral, v9 [58] and those reads which belonged to other RNA classes (“rRNA”, “snRNA”, “snoRNA”, and “tRNA”) were omitted in ensuing processing. Further, thereby filtered reads were aligned to human precursor miRNA sequences obtained from miRBase, release 22, allowing for one mismatch during alignment using Bowtie [59,60]. Residual reads were considered as unmapped (“Unmapped”). By summing up the hits per miRNA sequence, a read count table was generated which was loaded into R, version 3.5.1 [61]. Processed by the Bioconductor package DESeq2 (version 1.20.0) [[62], [63], [64]], only miRNAs with more than 20 DESeq2-normalized reads were treated as valid. Exploratory data analysis was visualized by Venn diagrams, principal component analysis, hierarchical clustering and heatmap based on Euclidean distances. DESeq2 internal differential gene expression (DGE) analysis was conducted to determine significant differences in miRNA detection dependent on the applied EV purification method. A Benjamini-Hochberg adjusted p value < 0.05 and a log2 fold change > |1| was set as level of significance. Raw data of trimmed sequence reads obtained by small RNA-Seq are accessible in the European Nucleotide Archive (accession number PRJEB30403) (http://www.ebi.ac.uk/ena/data/view/PRJEB30403).
3. Results
3.1. Quantitative characterization of purified urinary extracellular vesicles
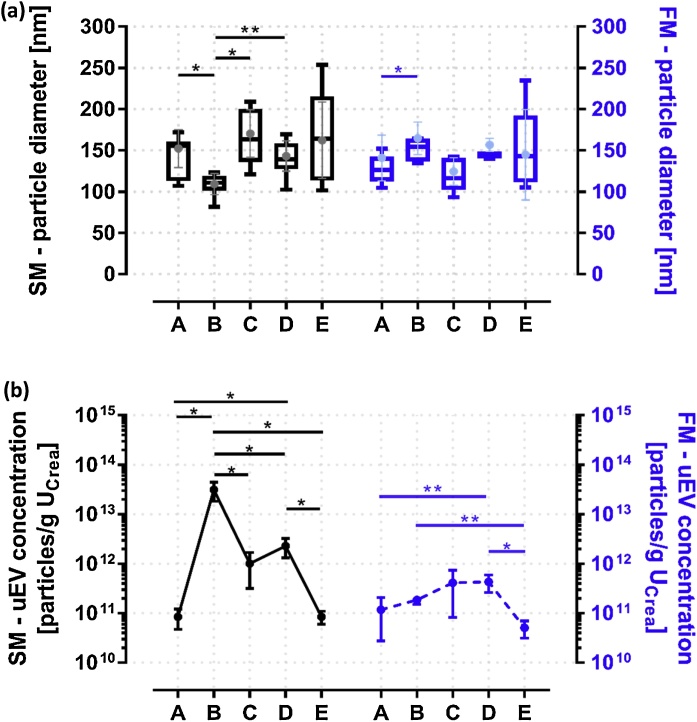
In order to evaluate purification-specific differences, protein composition as well as size and concentration of uEVs were examined. For the quantification and size determination of purified uEV samples, NTA was performed in two different experiments (Fig. 1). First, all available particles were measured in the scattering mode (SM). Second, only particles stained with the membrane dye were analyzed in the fluorescence mode (FM). For the latter approach, any autofluorescence was excluded beforehand. Overall, the mean and modal particle diameters overlapped broadly (Fig. 1a). The widest size distribution, ranging from nearly 100 to 250 nm, was observed in uEVs obtained by ultracentrifugation combined with density gradient (E). While most of these uEVs seemed to have a size around 160 nm in scattering and 145 nm in fluorescence measurements, a wide size dispersion was still apparent. Particle diameters ranging from 100 to 150 nm, with an average size of 125 nm in FM, were detected in uEVs obtained by spin column chromatography (A). The lowest particle diameters with sizes ranging from 80 to 120 nm were identified in SM for uEV samples purified by immunoaffinity (B), whereas an average size of 150 nm was ascertained in FM. This finding is mainly due to the unavoidable measurement of kit-derived magnetic beads that appear with a size of 30 to 80 nm according to the manufacturer, resulting in significant differences in size compared to competitors A (SM: p = 0.018), C (SM: p = 0.015) and D (SM: p = 0.006). uEVs purified by membrane affinity (C) emerged with a broad size distribution ranging from 120 to 220 nm, while the measured average size in FM was 120 nm. The size distribution of uEV samples derived from precipitation (D) was narrower; however, an averaged size of 145 nm was detected in both FM and SM.
Fig. 1.
Results of size and concentration measurements of purified uEVs using nanoparticle tracking analysis (NTA) in scattering mode (SM, black) and in fluorescence mode (FM, blue): spin column chromatography (A), immunoaffinity (B), precipitation (C), membrane affinity (D) and ultracentrifugation combined with density gradient (E). (n = 6; p value, adjusted for multiple comparisons: * < 0.05, ** < 0.01) (a) Illustration of the mean (box-whisker-plot: whiskers indicating minimum and maximum values) and modal (dot-plot: mean ± SD) particle diameter in nanometer (nm). (b) Detected particle concentration corrected for sample dilution and normalized to urinary creatinine (UCrea); left and right Y axes log10 transformed (mean ± SD).
Regarding concentration measurements of uEVs (Fig. 1b), exceedingly few particles were detected in uEVs derived from E with an average of 8.4E + 10 and 5.1E + 10 particles/g UCrea, which was significantly less compared to B (SM: p = 0.012; FM: p = 0.003) and D (SM: p = 0.013, FM: p = 0.012). Similar amounts of particles were detected in uEVs purified by A, which yielded an average of 8.5E + 10 and 1.2E + 11 particles/g UCrea, which was still significantly less than in D (SM: p = 0.013; FM: p = 0.008). The highest particle concentration was observed in SM in samples purified by B (3.1E + 13 particles/g UCrea). However, this is again based on the presence of kit-derived magnetic beads, resulting in significant differences in particle concentration when comparing B to all other purification strategies (0.02 > p > 0.01). In FM, by contrast, concentrations of uEVs isolated by B dropped to 1.9E + 11 particles/g UCrea, which was still significantly higher than in E (p = 0.003). uEV samples purified by C emerged with a particle concentration of 1.0E + 12 particles/g UCrea in SM, while the FM measurement resulted in 2.9E + 11 particles/g UCrea. Slightly higher particle concentrations were measured for uEVs derived from D, with 2.3E + 12 particles/g UCrea (SM) and 4.3E + 11 particles/g UCrea (FM).
3.2. Qualitative characterization of purified urinary extracellular vesicles
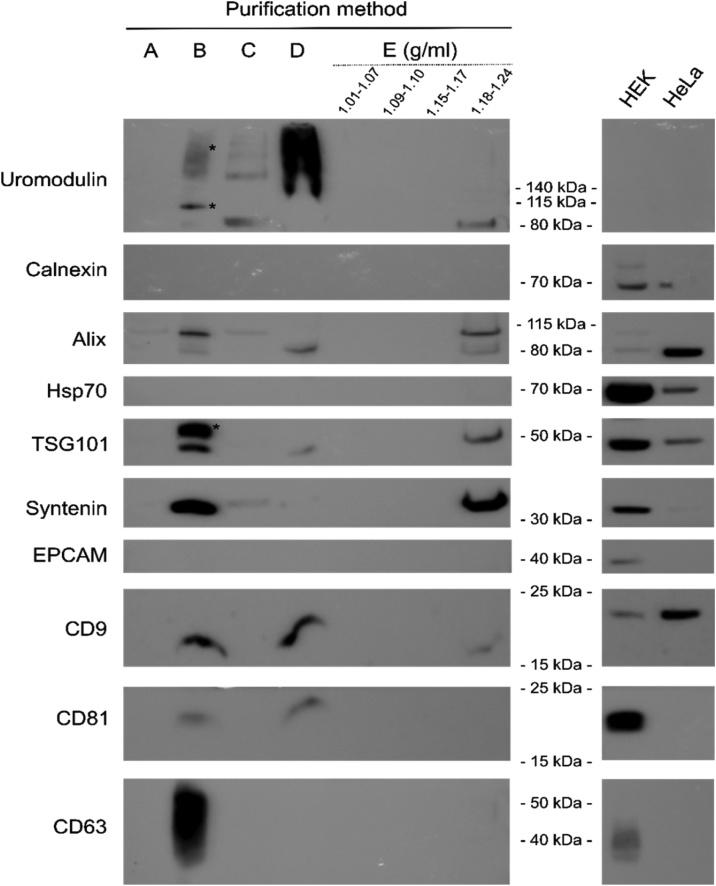
As specified in the MISEV guidelines [39], the presence of EV-enriched and contaminating protein markers was assessed by Western blot analysis (Fig. 2). All purified uEV samples appeared free from cellular contamination, as indicated by the absence of calnexin, which originates from the endoplasmatic reticulum. However, uromodulin, a very dominant 85 kDa protein in urine [65], was present in some samples, more precisely in protein lysates obtained from uEV samples purified by C and from the 1.18–1.24 g/ml fractions obtained by E. Unspecific bands in uEVs purified by B, as indicated with asterisks, corresponded to kit-derived murine immunoglobulin G (mIgG), which was confirmed by an individual mIgG staining (supplemental Figure A.1). An intense smearing above 115 kDa could be detected in uEV lysates obtained by D, pointing towards substantial contamination with a uromodulin isoform at higher molecular weight. Hsp70 and EPCAM were detected in none of the uEV lysates. Despite a hardly noticeably detection of Alix (around 100 kDa) in protein lysates purified based on A and C, a more visible band at 80 kDa was detected in D, and even sharper bands were identified for both isoforms in B and E (1.18–1.24 g/ml fraction). Clearly visible bands at 46 kDa, belonging to TSG101, were detected in B and E (1.18–1.24 g/ml fraction). Signal intensities for TSG101 were lower in D, while the purification methods A and C did not show any specific protein signal. Syntenin appeared with strong signals in B and E (1.18–1.24 g/ml fraction) (32 kDa), especially compared to C, where a band was barely identifiable. However, it was not substantiated neither in A nor in D. Regarding the tetraspanins CD9 and CD81, both were identified in B and D (around 20 kDa). CD9 was additionally detected in E (1.18–1.24 g/ml fraction). However, CD63 was solely confirmed in B with a characteristic smearing between 30 and 60 kDa.
Fig. 2.
Western blot analysis of EV-specific protein markers in protein lysates obtained from five different purification types: spin column chromatography (A), immunoaffinity (B), precipitation (C), membrane affinity (D) and ultracentrifugation combined with density gradient (E). Uromodulin and calnexin indicate uEV contamination with non-EV structures. The other proteins are used as positive marker proteins. Asterisks indicate bands related to murine immunoglobin G. HEK and HeLa cell lysates were used as positive controls. The blot is representative of one sample with a urinary creatinine content of 1.04 g/l.
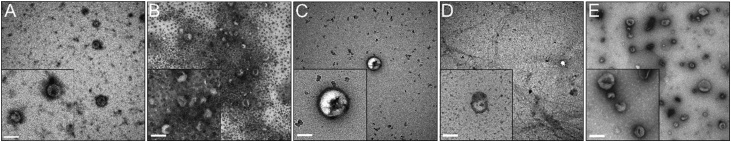
Further, when investigating uEV morphology by TEM, the results of nanoparticle tracking and Western blot analysis could be reinforced (Fig. 3). All of the applied uEV purification methods appeared with high image qualities and EV typical “cup-shape” morphologies, which result from desiccation during grid preparation [66]. Since TEM size estimations are performed on desiccated uEVs, the corresponding sizes around 100 nm are smaller compared to the hydrodynamic sizes estimated in NTA. The broad size distribution, which was recognized in NTA in uEVs purified by E, was also noticed in TEM. Moreover, the huge sample contamination with uromodulin in D could be ascertained likewise, as indicated by the filamentous network [67]. Even though high-contrasting beads in B increase the overall background, a large number of small uEVs could be clearly detected. The lowest number of uEVs was detected in TEM preparations of C and D.
Fig. 3.
Results of transmission electron microscopy (TEM). Urinary EV (uEV) samples were obtained by five different purification methods: spin column chromatography (A), immunoaffinity (B), precipitation (C), membrane affinity (D) and ultracentrifugation combined with density gradient (E). Images represent wide field and clos-ups (boxes at the lower left) of operator selected locations. Scale bar is the same for all images and represent 100 nm.
3.3. Characterization of urinary extracellular vesicle derived RNA in small RNA sequencing
The uEV characterization was complemented by vesicle content analysis. First, total RNA was extracted from uEVs and analyzed using capillary gel electrophoresis (Supplementary Fig. A.2). Since 18S and 28S ribosomal RNA are not extensively present in EV-derived RNA [68], the RNA integrity numbers (RIN) generated by the Bioanalyzer’s software are mainly used to assess the degree of cellular RNA contamination, which was largely inapplicable in this case (RIN 2.5 ± 0.3). Total RNA yields, also listed in Table 2, were highest in uEVs obtained by D (542.0 ± 292.9 pg/ml urine), followed by C (457.0 ± 2116.8 pg/ml urine) and E (331.0 ± 233.0 pg/ml urine). The lowest RNA yields were detected for A (289.5 ± 134.0 pg/ml urine) and B (277.0 ± 90.0 pg/ml urine). When normalizing total RNA yields to urinary creatinine, accounting for every individual’s urinary composition, the order of the different purification methods was similar, however, overall deviation was reduced due to this normalization.
Table 2.
Performance indicators for RNA extraction and small RNA-Seq.
| A | B | C | D | E | |
|---|---|---|---|---|---|
| Total RNA [pg/ml urine] | |||||
| MEAN | 289.5 | 277.0 | 457.0 | 542.0 | 331.0 |
| SD | 134.0 | 90.9 | 216.8 | 292.9 | 233.0 |
| Total RNA [ng/g UCrea] | |||||
| MEAN | 456.3 | 450.7 | 697.5 | 852.6 | 523.0 |
| SD | 86.3 | 72.6 | 162.2 | 289.7 | 281.3 |
| Total readcounts [million reads] | |||||
| MEAN | 9.01 | 10.8 | 13.6 | 14.1 | 11.0 |
| SD | 1.79 | 2.90 | 2.80 | 3.81 | 1.49 |
| miRNA mapped readcounts [%] | |||||
| MEAN | 2.9 | 5.0 | 3.4 | 5.8 | 3.7 |
| SD | 0.6 | 2.2 | 1.1 | 2.3 | 0.7 |
| Individually detected canonical miRNAs | |||||
| Absolute number | 102 | 119 | 114 | 119 | 124 |
Spin column chromatography (A), immunoaffinity (B), precipitation (C), membrane affinity (D) and ultracentrifugation combined with density gradient (E).
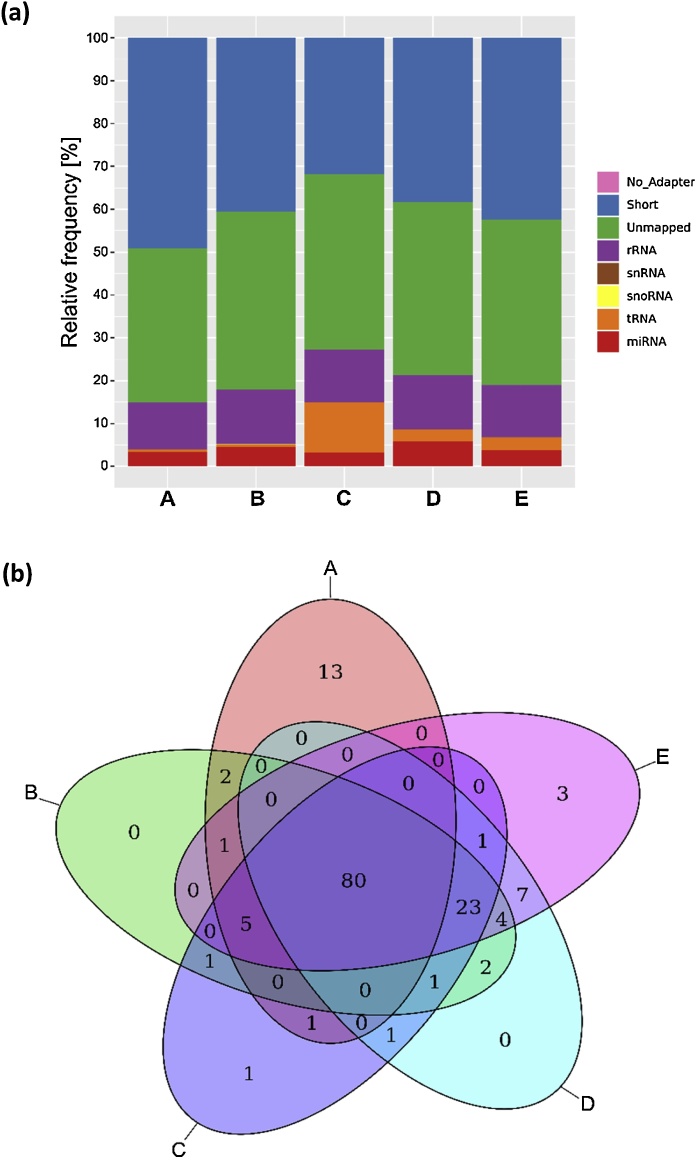
In the following, small RNA-Seq was conducted to analyze the miRNA cargo of purified uEVs using a normalized input of 5 ng of purified cDNA libraries. The electropherogram of the size-selected cDNA library is depicted in Supplemental Fig. A.3. Quality control of sequencing results revealed a mean phred score of 39.5, indicating a base call accuracy of almost 99.99%. The obtained length distribution (Supplemental Fig. A.4) emerged with a single peak at 22 nt. Though there were no significant variances in total library sizes and the percentage of mapped miRNA reads, clear differences in small RNA-Seq performance were still noticeable. Library sizes ranged from 9.01E + 6 ± 1.79E + 6 reads (A) to 1.40E + 7 ± 3.81E + 6 reads (D), and miRNA-mapped reads ranged from 2.9 ± 0.6% (A) to 5.8 ± 2.3% (D) (Table 2). For all uEV isolation methods, the majority of reads (>70%) remained unmapped or was shorter than 16 nt and hence excluded from further alignments (Fig. 4a). About 10% of total reads related to ribosomal RNA (rRNA), and an even smaller fraction to transfer RNA (tRNA).
Fig. 4.
Assignment of small RNA sequencing results to different RNA species (n = 6): spin column chromatography (A), immunoaffinity (B), precipitation (C), membrane affinity (D) and ultracentrifugation combined with density gradient (E). (a) Mean relative frequency of reads that mapped to microRNA (miRNA), transfer RNA (tRNA), small nucleolar RNA (snoRNA), small nuclear RNA (snRNA), ribosomal RNA (rRNA), remained unmapped, were shorter than 16 nucleotides (short) or did not show any adapter). (b) Overlap of distinct miRNA species detected in sequencing libraries from each isolation method, data are filtered for miRNAs with more than 20 reads per group.
Reads that mapped to miRNAs were further examined for their diversity, revealing the detection of between 102 (A) and 124 (E) distinct miRNAs for all methods (Table 2). Fig. 4b depicts the attributions of identified miRNAs to each uEV isolation method, with an overlap of 80 shared miRNAs between all uEV purification strategies. Thirteen % (13/102) of the detected miRNAs were exclusively found in uEVs purified by A. Another 23 distinct miRNAs were found to be shared by the purification methods B to E, but not by A. The strongest overlaps of identified miRNAs were detected for D and E, sharing 93% (115/124) as well as B and E, sharing 91% (113/124) of detected miRNAs. An extensive list of all miRNAs detected with a mean expression of more than 20 reads per group can be found in the supplemental Table A.1. The top 10 of highest expressed miRNAs shared between all uEV purification strategies are displayed in Table 3. The number of miRNA reads strongly depended on the chosen uEV purification method as illustrated by the percentage deviation from the miRNA reads’ average calculated based on all methods.
Table 3.
Top 10 expressed miRNAs identified by small RNA sequencing in every uEV isolation method.
| MEAN [reads] | A [%] | B [%] | C [%] | D [%] | E [%] | |
|---|---|---|---|---|---|---|
| miR-451a | 18,970 | +110 (24) | −3 (44) | −9 (25) | −65 (21) | −32 (30) |
| miR-148a-3p | 14,326 | −91 (4) | +112 (118) | +10 (76) | +11 (29) | −42 (25) |
| miR-486-5p | 12,702 | +110 (28) | −4 (46) | −10 (27) | −64 (22) | −31 (32) |
| miR-26a-5p | 6,547 | −82 (4) | −13 (32) | +2 (52) | +51 (27) | +42 (66) |
| miR-92a-3p | 6,021 | +100 (27) | −3 (37) | −13 (22) | −57 (19) | −28 (26) |
| miR-30d-5p | 5,251 | −78 (2) | +28 (55) | −4 (23) | +54 (36) | 0 (31) |
| let-7a-5p | 3,947 | −79 (4) | +8 (47) | +1 (30) | +49 (32) | +21 (61) |
| miR-191-5p | 3,011 | +95 (24) | −6 (40) | −11 (18) | −56 (16) | −21 (24) |
| let-7i-5p | 2,802 | −9 (10) | −9 (15) | −9 (23) | +4 (14) | +23 (33) |
| miR-30c-5p | 2,127 | −66 (2) | +4 (32) | +30 (45) | +28 (21) | +4 (43) |
Spin column chromatography (A), immunoaffinity (B), precipitation (C), membrane affinity (D) and ultracentrifugation combined with density gradient (E). miRNAs are ranked by their mean normalized reads over all groups. Method-specific variances in miRNA detection are indicated by their percentage deviations from the average miRNA-specific reads with standard deviations in brackets.
3.4. Substantial differences in miRNA profiles of disparately purified urinary extracellular vesicles
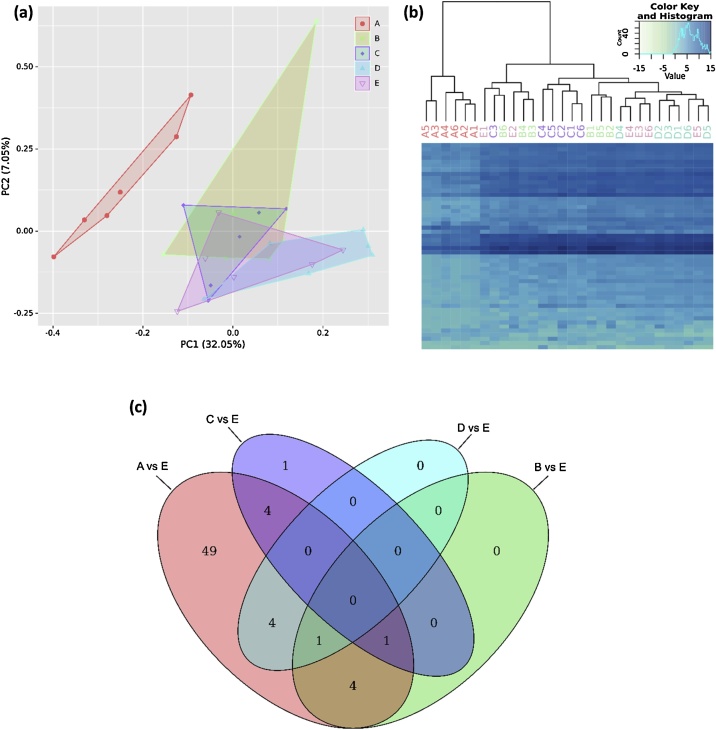
Despite a large overlap of detected miRNAs, a clear separation of the miRNA profile in uEVs purified by A from miRNA profiles of all other methods was observed in principal component analysis (Fig. 5a). miRNA profiles of uEVs isolated byB–E, however, were highly comparable. Similar findings were observed in hierarchical clustering analysis, which confirmed the definite separation of A from all other groups B–E (Fig. 5b). As evidenced in differential gene expression (DGE) analysis, the partition of A was based on 63 significantly differentially expressed miRNAs when compared to the commonly used purification method (E) (Fig. 5c). All 63 miRNAs significantly different between A and E are listed in Supplementary Table A.2. Analysis of the miRNA profiles of the other purification methods also revealed some significantly differentially expressed miRNAs, in total 15 (Fig. 5c), however, these did not induce any purification-specific clustering (Fig. 5a and b).
Fig. 5.
Exploratory data analysis of differences in urinary miRNAs derived from distinct uEV purification strategies (n = 6): spin column chromatography (A), immunoaffinity (B), precipitation (C), membrane affinity (D) and ultracentrifugation combined with density gradient (E). (a) Principal component analysis of the complete miRNA gene set showing the first and second principal component (PC) of explained variance. (b) Heatmap and hierarchical cluster analysis indicated strong separation of A from other methods. (c) Venn diagram of significantly differentially detected miRNAs compared to E. Results were determined by DESeq2-based differential gene expression analysis and filtered for adjusted p value < 0.05 and log2 fold change > |1|.
4. Discussion
Urine contains a large number of EVs, which originate from different cell types and have proven to be remarkably stable [23,52]. Particularly small uEVs carry a majority of urinary miRNAs and assure an even greater protection of urinary miRNAs compared to cells [20,22,69,70]. On this basis, uEV-derived miRNA has become a promising candidate in current biomarker research, whereby untargeted approaches, such as small RNA-Seq, provide the greatest possibilities and are frequently used for deep miRNA profiling and even identification of novel miRNAs. Particularly diagnostics dealing with disease identification and progression monitoring, such as bladder or prostate cancer, could benefit from non-invasive biomarkers based on uEV-derived miRNA. However, sample processing is crucial, as it heavily influences the sequencing output and can thereby evoke misleading statements based on flawed data. Although the dependency of sequencing results on the chosen isolation method has already been demonstrated for serum samples [11], the influence and suitability of different uEV purification methods has not yet been examined. We therefore aimed to identify the most appropriate EV purification method for ensuing small RNA-Seq analysis. At the same time, we comprehensively characterized uEV samples by NTA, TEM and Western blot analysis in order to facilitate the proper interpretation of results.
By performing NTA, we observed significant differences in particle diameters and concentrations depending on the chosen EV purification method, indicating that each of the applied methods seems to preferably isolate different vesicular subpopulations with distinct properties and efficiencies. This might mainly be based on the heterogeneity of uEVs with respect to their size, solubility, density, surface proteins and cargo. It has additionally to be considered that the measurement in scattering mode (SM) does not discriminate between membrane-enclosed and non-enclosed particles, hence introducing a presumable overestimation of the actual uEV proportion of totally detected particles in SM. Indeed, when analyzing only membrane-stained particles in fluorescence mode (FM), we identified drops in particle concentrations to a varying extent combined with perceptible shifts in particle diameters. Those findings point towards unavoidable co-isolation of contaminating structures without membranes, mainly occurring in uEV samples purified by immunoaffinity (B), membrane affinity (C) and precipitation (D). Contaminants observed in B are principally introduced by the bead-based purification method itself and do not hinder downstream analyses. Methods C and D, however, are well known to unspecifically co-purify proteins and protein complexes [20,[71], [72], [73]], which we could confirm by the drop in particle concentration in FM-NTA and the dominant uromodulin detection in Western blot analysis and TEM. Moreover, without additional cleanup, such heavy protein impurities could mask the detection of EV-specific protein markers such as CD63 and syntenin in Western blot analysis [11]. Even if precipitation-based purification methods can be performed in an expeditious manner, its major drawback of co-precipitates has to be well-considered in biomarker research, since a definite assignment of analyzed miRNAs to small uEVs would be hard to establish. Co-purification of uromodulin was also detected in uEVs obtained by ultracentrifugation combined with density gradient (E), albeit less pronounced. However, we did not observe extensive differences in vesicle diameter and concentration of uEV samples in SM and FM, which stands in contrast to reports of others, who identified heavy contaminations in ultracentrifugation-based preparations by NTA [20]. Nevertheless, the detection of the lowest uEV concentration in E could still result from uromodulin-caused vesicle aggregates, which would pellet at low-speed centrifugation and lead to vesicle loss [74]. Previously suggested sample treatment with dithiothreitol (DTT) was intended to alleviate uromodulin polymerization, but it was shown to be of limited usefulness or even completely unable to increase vesicle yield [65,69]. Besides, a large proportion of vesicles might have gotten lost in the discarded supernatants of the first centrifugation steps, as already reported elsewhere [20,40]. Associated therewith, huge technical biases are introduced during decanting and resuspension steps, which are part of the complex differential centrifugation procedure, as well as during high-speed centrifugations, which generate artifacts by EV clumping and vesicle rupture [20,75,76]. Moreover, the low scalable ultracentrifugation still requires a lot of time and cost-intensive equipment that might not be available in every laboratory, thus emphasizing the growing need for more efficient and cleaner EV purification methods, whilst allowing for comparability to previously conducted experiments.
In fact, two of the methods utilized here, spin column chromatography (A) and immunoaffinity (B), were free from calnexin and uromodulin contamination as illustrated by Western blot analysis. However, A did also not show any of the EV-specific marker proteins. According to Webber and Clayton, who suggested a high ratio of particles to protein content as a measure for high EV purity [77], that accounts for co-purified proteins, method A would reveal highly pure uEVs. But in the present case, this ratio might be delusive, since A seemed to rather purify EV quantities below the limit of detection, or vesicles with the complete absence of EV-specific protein markers. Furthermore, additional protein was introduced to the samples via bead-bound antibodies in B, which artificially increased the samples’ protein content and erroneously lowered the EV purity ratio. This ambiguity emphasizes the conflated evaluation of proteomic and transcriptomic observations, especially when aiming at identifying the carrier of detected miRNAs.
With respect to transcriptomics, both A and B revealed only minor amounts of total RNA compared to C or D, which was most likely related to the differences in particle concentration. When accounting for these variances by using the same cDNA input amount for subsequent small RNA-Seq, total readcounts were higher in less EV-specific purification methods (C, D and E). Besides, the majority of generated reads in all of the applied methods appeared with less than 16 nt or remained unmapped, raising the question where they might derive from and whether they are of small uEV origin. It is already known that library preparation bares a huge risk for introduction of artifacts, such as adapter dimers [78], but sequences of RNA breakdown and exogenous origin, such as contaminants and bacteria, could also contribute to the high number of unmapped and short reads [[79], [80], [81]]. The relative mapping distributions, however, pointed towards method-dependent preferences in capturing different RNA species as suggested by a larger proportion of mapped reads to transfer RNA (tRNA) in C compared to the other methods. Though this phenomenon was already observed by others [11], the underlying reason has not yet been clarified. One attempt at explanation could be the incidence of co-purified urinary high-density lipoproteins, which carry considerable amounts of tRNA [73,82,83]. Nevertheless, when focusing on miRNA contents, the highest percentage was observed in uEVs isolated by D. Based on our protein analyses, which insinuated heavy co-purification, it is suspected that parts of the detected miRNAs might belong to extracellular RNA other than EV-derived. However, due to the high occurrence of nuclease activities in urine, which makes nuclease treatment redundant, the presence of RNA other than uEV-related is highly unlikely [84], even though it cannot be excluded completely, since extracellular RNA could be also protected by carrier proteins and lipids [[28], [29], [30]]. The second highest percentage of miRNA mapping reads was detected for uEVs obtained by B, though a lower RNA yield was obtained, most probably based on the positive selection of only CD63+/CD81+/CD9+-vesicles. Thus, the present results indicate that predominantly CD63+/CD81+/CD9+-vesicles contain large amounts of uEV-specific miRNAs, as the EV-specificity of immunoaffinity-based methods was confirmed by Western blot analysis and has already been established by others [85,86]. The broad overlap of most of the individually detected miRNAs in B, D and E suggests that D and E also purify vesicles with high amounts of uEV-specific miRNAs, albeit with a lower degree of purity. However, even highly abundant miRNAs were still affected by method-dependent variations indicated by huge deviations in the number of miRNA-specific reads. Especially in biomarker discovery, this bares great risk for false negative or positive results. Overall, purification methods B–E still resulted in comparable miRNA profiles without prevalent significant differences, as also indicated by principal component analysis and hierarchical clustering. However, purification method A appeared to be clearly separated from the others, which we confirmed by detecting a large number of significantly differentially expressed miRNAs. The combined results of uEV characterization and small RNA-Seq results, hence, imply the isolation of a different subtype of small uEVs that also comprise miRNAs. Based on their size and the absence of detectable tetraspanins, especially CD63, these vesicles might be ectosomes [87], whereas the characteristics of the small uEVs purified by B–E are suspected to represent exosomes. However, since the investigation of the underlying biogenesis was not within the scope of this project, the unambiguous assignment to specific EV classes is not possible and care should be taken in naming the purified vesicles.
5. Conclusion
To conclude, proper selection of uEV purification methods depending on the research question is highly recommend. In the present study, we could confirm the general feasibility of small RNA-Seq for all the applied small uEV purification methods. However, minor and major deviations were observed, which have to be considered. The purest vesicles might be obtained by the positive selection via immunoaffinity (B), which enriched enough Alix+/Hsp70-/TSG101+/Syntenin+/EPCAM-/CD63+/CD81+/CD9+-uEVs to perform small RNA-Seq and indicated a high content of miRNAs. Whenever less pure vesicle preparations without further clean-up are sufficient, e.g. in some fields of biomarker research, precipitation-based (D), membrane affinity-based (C) purification methods and ultracentrifugation combined with density gradient (E) still emerge with highly comparable results. Special care should be taken when purifying vesicles by spin column chromatography (A), as a larger diversity of uEV subtypes seems to be obtained which provide a miRNA profile significantly different from those from more homogenous EV preparations. This again emphasizes the need for a combined evaluation of purified uEVs and the corresponding small RNA-Seq results to ensure a high degree of reproducibility and comparability.
Conflict of interest
The authors declare that there are no conflicts of interest.
Funding
This project was supported by intramural funding from the Technical University of Munich (MP).
Contributions
VM and MP designed the study setup. VM and GW acquired samples and data. VM performed the experiments and data analysis. All authors contributed to the manuscript and approved the final version to be submitted.
Author agreement
All authors have read and approved the final version of the manuscript being submitted. They warrant that the article is the authors' original work, hasn't received prior publication and isn't under consideration for publication elsewhere.
Acknowledgements
We gratefully acknowledge Dominik Buschmann for excellent assistance by critically proof reading the manuscript. We further thank Benedikt Kirchner for his support in processing small RNA sequencing data.
Handled by Jim Huggett
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.bdq.2019.100089.
Contributor Information
Veronika Mussack, Email: mussack@wzw.tum.de.
Georg Wittmann, Email: georg.wittmann@med.uni-muenchen.de.
Michael W. Pfaffl, Email: michael.pfaffl@wzw.tum.de.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Baig A. 2011. Biochemical Composition of Normal Urine. [Google Scholar]
- 2.Tolle A., Jung M., Rabenhorst S., Kilic E., Jung K., Weikert S. Identification of microRNAs in blood and urine as tumour markers for the detection of urinary bladder cancer. Oncol. Rep. 2013;30(4):1949–1956. doi: 10.3892/or.2013.2621. [DOI] [PubMed] [Google Scholar]
- 3.Sapre N., Macintyre G., Clarkson M., Naeem H., Cmero M., Kowalczyk A. A urinary microRNA signature can predict the presence of bladder urothelial carcinoma in patients undergoing surveillance. Br. J. Cancer. 2016;114(4):454–462. doi: 10.1038/bjc.2015.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davalieva K., Kiprijanovska S., Komina S., Petrusevska G., Zografska N.C., Polenakovic M. Proteomics analysis of urine reveals acute phase response proteins as candidate diagnostic biomarkers for prostate cancer. Proteome Sci. 2015;13(1):2. doi: 10.1186/s12953-014-0059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raposo G., Nijman H.W., Stoorvogel W., Liejendekker R., Harding C.V., Melief C.J., Geuze H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996;183(3):1161–1172. doi: 10.1084/jem.183.3.1161. https://www.ncbi.nlm.nih.gov/pubmed/8642258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skog J., Wurdinger T., van Rijn S., Meijer D.H., Gainche L., Sena-Esteves M. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Niel G., Porto-Carreiro I., Simoes S., Raposo G. Exosomes: a common pathway for a specialized function. J. Biochem. 2006;140(1):13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- 8.van Balkom B.W., Pisitkun T., Verhaar M.C., Knepper M.A. Exosomes and the kidney: prospects for diagnosis and therapy of renal diseases. Kidney Int. 2011;80(11):1138–1145. doi: 10.1038/ki.2011.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rekker K., Saare M., Roost A.M., Kubo A.L., Zarovni N., Chiesi A. Comparison of serum exosome isolation methods for microRNA profiling. Clin. Biochem. 2014;47(1-2):135–138. doi: 10.1016/j.clinbiochem.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 10.Muller L., Hong C.S., Stolz D.B., Watkins S.C., Whiteside T.L. Isolation of biologically-active exosomes from human plasma. J. Immunol. Methods. 2014;411:55–65. doi: 10.1016/j.jim.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buschmann D., Kirchner B., Hermann S., Marte M., Wurmser C., Brandes F. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J. Extracell. Vesicles. 2018;7(1):1481321. doi: 10.1080/20013078.2018.1481321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reithmair M., Buschmann D., Marte M., Kirchner B., Hagl D., Kaufmann I. Cellular and extracellular miRNAs are blood-compartment-specific diagnostic targets in sepsis. J. Cell. Mol. Med. 2017 doi: 10.1111/jcmm.13162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zlotogorski-Hurvitz A., Dayan D., Chaushu G., Korvala J., Salo T., Sormunen R., Vered M. Human saliva-derived exosomes: comparing methods of isolation. J. Histochem. Cytochem. 2015;63(3):181–189. doi: 10.1369/0022155414564219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lasser C., Alikhani V.S., Ekstrom K., Eldh M., Paredes P.T., Bossios A. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J. Transl. Med. 2011;9:9. doi: 10.1186/1479-5876-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zempleni J., Aguilar-Lozano A., Sadri M., Sukreet S., Manca S., Wu D. Biological activities of extracellular vesicles and their cargos from bovine and human milk in humans and implications for infants. J. Nutr. 2017;147(1):3–10. doi: 10.3945/jn.116.238949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stuendl A., Kunadt M., Kruse N., Bartels C., Moebius W., Danzer K.M. Induction of alpha-synuclein aggregate formation by CSF exosomes from patients with Parkinson’s disease and dementia with Lewy bodies. Brain. 2016;139(Pt 2):481–494. doi: 10.1093/brain/awv346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang C., Guo W.B., Zhang W.S., Bian J., Yang J.K., Qi T. Extraction and identification of semen-derived exosomes using PEG6000. Nan Fang Yi Ke Da Xue Xue Bao. 2016;36(11):1531–1535. https://www.ncbi.nlm.nih.gov/pubmed/27881345 [PubMed] [Google Scholar]
- 18.Peng P., Yan Y., Keng S. Exosomes in the ascites of ovarian cancer patients: origin and effects on anti-tumor immunity. Oncol. Rep. 2011;25(3):749–762. doi: 10.3892/or.2010.1119. [DOI] [PubMed] [Google Scholar]
- 19.Perakis S., Speicher M.R. Emerging concepts in liquid biopsies. BMC Med. 2017;15(1):75. doi: 10.1186/s12916-017-0840-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gheinani A.H., Vogeli M., Baumgartner U., Vassella E., Draeger A., Burkhard F.C., Monastyrskaya K. Improved isolation strategies to increase the yield and purity of human urinary exosomes for biomarker discovery. Sci. Rep. 2018;8(1):3945. doi: 10.1038/s41598-018-22142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pisitkun T., Shen R.F., Knepper M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. U. S. A. 2004;101(36):13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miranda K.C., Bond D.T., McKee M., Skog J., Paunescu T.G., Da Silva N. Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int. 2010;78(2):191–199. doi: 10.1038/ki.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dhondt B., Van Deun J., Vermaerke S., de Marco A., Lumen N., De Wever O., Hendrix A. Urinary extracellular vesicle biomarkers in urological cancers: from discovery towards clinical implementation. Int. J. Biochem. Cell Biol. 2018;99:236–256. doi: 10.1016/j.biocel.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J.J., Lotvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 25.Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. https://www.ncbi.nlm.nih.gov/pubmed/14744438 [DOI] [PubMed] [Google Scholar]
- 27.Delic D., Eisele C., Schmid R., Baum P., Wiech F., Gerl M. Urinary exosomal miRNA signature in type II diabetic nephropathy patients. PLoS One. 2016;11(3) doi: 10.1371/journal.pone.0150154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arroyo J.D., Chevillet J.R., Kroh E.M., Ruf I.K., Pritchard C.C., Gibson D.F. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U. S. A. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turchinovich A., Weiz L., Langheinz A., Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39(16):7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner J., Riwanto M., Besler C., Knau A., Fichtlscherer S., Roxe T. Characterization of levels and cellular transfer of circulating lipoprotein-bound microRNAs. Arterioscler. Thromb. Vasc. Biol. 2013;33(6):1392–1400. doi: 10.1161/ATVBAHA.112.300741. [DOI] [PubMed] [Google Scholar]
- 31.Long J.D., Sullivan T.B., Humphrey J., Logvinenko T., Summerhayes K.A., Kozinn S. A non-invasive miRNA based assay to detect bladder cancer in cell-free urine. Am. J. Transl. Res. 2015;7(11):2500–2509. https://www.ncbi.nlm.nih.gov/pubmed/26807194 [PMC free article] [PubMed] [Google Scholar]
- 32.Khurana R., Ranches G., Schafferer S., Lukasser M., Rudnicki M., Mayer G., Huttenhofer A. Identification of urinary exosomal non-coding RNAs as novel biomarkers in Chronic Kidney Disease. RNA. 2016;23(2):142–152. doi: 10.1261/rna.058834.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Argyropoulos C., Wang K., McClarty S., Huang D., Bernardo J., Ellis D. Urinary microRNA profiling in the nephropathy of type 1 diabetes. PLoS One. 2013;8(1) doi: 10.1371/journal.pone.0054662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Argyropoulos C., Wang K., Bernardo J., Ellis D., Orchard T., Galas D., Johnson J.P. Urinary MicroRNA Profiling Predicts the Development of Microalbuminuria in Patients with Type 1 Diabetes. J. Clin. Med. 2015;4(7):1498–1517. doi: 10.3390/jcm4071498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barutta F., Tricarico M., Corbelli A., Annaratone L., Pinach S., Grimaldi S. Urinary exosomal microRNAs in incipient diabetic nephropathy. PLoS One. 2013;8(11) doi: 10.1371/journal.pone.0073798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sole C., Cortes-Hernandez J., Felip M.L., Vidal M., Ordi-Ros J. miR-29c in urinary exosomes as predictor of early renal fibrosis in lupus nephritis. Nephrol. Dial. Transplant. 2015;30(9):1488–1496. doi: 10.1093/ndt/gfv128. [DOI] [PubMed] [Google Scholar]
- 37.Huang Z., Zhang Y., Zhou J., Zhang Y. Urinary exosomal miR-193a can be a potential biomarker for the diagnosis of primary focal segmental glomerulosclerosis in children. Biomed Res. Int. 2017;2017 doi: 10.1155/2017/7298160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Deun J., Mestdagh P., Agostinis P., Akay O., Anand S., Anckaert J. EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. Methods. 2017;14(3):228–232. doi: 10.1038/nmeth.4185. [DOI] [PubMed] [Google Scholar]
- 39.Théry C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2019;8(1):1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Musante L., Tataruch-Weinert D., Kerjaschki D., Henry M., Meleady P., Holthofer H. Residual urinary extracellular vesicles in ultracentrifugation supernatants after hydrostatic filtration dialysis enrichment. J. Extracell. Vesicles. 2017;6(1):1267896. doi: 10.1080/20013078.2016.1267896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alvarez M.L., Khosroheidari M., Kanchi Ravi R., DiStefano J.K. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012;82(9):1024–1032. doi: 10.1038/ki.2012.256. [DOI] [PubMed] [Google Scholar]
- 42.Markowska A., Pendergrast R.S., Pendergrast J.S., Pendergrast P.S. A novel method for the isolation of extracellular vesicles and RNA from urine. J. Circ. Biomark. 2017;6 doi: 10.1177/1849454417712666. 1849454417712666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Royo F., Zuniga-Garcia P., Sanchez-Mosquera P., Egia A., Perez A., Loizaga A. Different EV enrichment methods suitable for clinical settings yield different subpopulations of urinary extracellular vesicles from human samples. J. Extracell. Vesicles. 2016;5:29497. doi: 10.3402/jev.v5.29497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Royo F., Diwan I., Tackett M.R., Zuniga P., Sanchez-Mosquera P., Loizaga-Iriarte A. Comparative miRNA analysis of urine extracellular vesicles isolated through five different methods. Cancers (Basel) 2016;8(12) doi: 10.3390/cancers8120112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crossland R.E., Norden J., Bibby L.A., Davis J., Dickinson A.M. Evaluation of optimal extracellular vesicle small RNA isolation and qRT-PCR normalisation for serum and urine. J. Immunol. Methods. 2016;429:39–49. doi: 10.1016/j.jim.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 46.Lozano-Ramos I., Bancu I., Oliveira-Tercero A., Armengol M.P., Menezes-Neto A., Del Portillo H.A. Size-exclusion chromatography-based enrichment of extracellular vesicles from urine samples. J. Extracell. Vesicles. 2015;4:27369. doi: 10.3402/jev.v4.27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Deun J., Mestdagh P., Sormunen R., Cocquyt V., Vermaelen K., Vandesompele J. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles. 2014;3 doi: 10.3402/jev.v3.24858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Channavajjhala S.K., Rossato M., Morandini F., Castagna A., Pizzolo F., Bazzoni F., Olivieri O. Optimizing the purification and analysis of miRNAs from urinary exosomes. Clin. Chem. Lab. Med. 2014;52(3):345–354. doi: 10.1515/cclm-2013-0562. [DOI] [PubMed] [Google Scholar]
- 49.Guo Y., Vickers K., Xiong Y., Zhao S., Sheng Q., Zhang P. Comprehensive evaluation of extracellular small RNA isolation methods from serum in high throughput sequencing. BMC Genomics. 2017;18(1):50. doi: 10.1186/s12864-016-3470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.World Medical A. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 51.Delanghe J.R., Speeckaert M.M. Creatinine determination according to Jaffe-what does it stand for? NDT Plus. 2011;4(2):83–86. doi: 10.1093/ndtplus/sfq211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou H., Yuen P.S., Pisitkun T., Gonzales P.A., Yasuda H., Dear J.W. Collection, storage, preservation, and normalization of human urinary exosomes for biomarker discovery. Kidney Int. 2006;69(8):1471–1476. doi: 10.1038/sj.ki.5000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.ISEV 2018 abstract book. J. Extracell. Vesicles. 2018;7(Suppl. 1):1461450. [Google Scholar]
- 54.Greening D.W., Xu R., Ji H., Tauro B.J., Simpson R.J. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol. Biol. 2015;1295:179–209. doi: 10.1007/978-1-4939-2550-6_15. [DOI] [PubMed] [Google Scholar]
- 55.Eitan E., Green J., Bodogai M., Mode N.A., Baek R., Jorgensen M.M. Age-Related Changes in Plasma Extracellular Vesicle Characteristics and Internalization by Leukocytes. Sci. Rep. 2017;7(1):1342. doi: 10.1038/s41598-017-01386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andrews S. 2010. FastQC: A Quality Control Tool for High Throughput Sequence Data. [Google Scholar]
- 57.Kong Y. Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics. 2011;98(2):152–153. doi: 10.1016/j.ygeno.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 58.RNAcentral Consortium RNAcentral: a comprehensive database of non-coding RNA sequences. Nucleic Acids Res. 2017;45(D1):D128–D134. doi: 10.1093/nar/gkw1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2018. R: A Language and Environment for Statistical Computing.https://www.R-project.org/ [Google Scholar]
- 62.Love M.I. 2016. Differential Analysis of Count Data – The DESeq2 Package. [Google Scholar]
- 63.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fernandez-Llama P., Khositseth S., Gonzales P.A., Star R.A., Pisitkun T., Knepper M.A. Tamm-Horsfall protein and urinary exosome isolation. Kidney Int. 2010;77(8):736–742. doi: 10.1038/ki.2009.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chernyshev V.S., Rachamadugu R., Tseng Y.H., Belnap D.M., Jia Y., Branch K.J. Size and shape characterization of hydrated and desiccated exosomes. Anal. Bioanal. Chem. 2015;407(12):3285–3301. doi: 10.1007/s00216-015-8535-3. [DOI] [PubMed] [Google Scholar]
- 67.Darisipudi M.N., Thomasova D., Mulay S.R., Brech D., Noessner E., Liapis H., Anders H.J. Uromodulin triggers IL-1beta-dependent innate immunity via the NLRP3 inflammasome. J. Am. Soc. Nephrol. 2012;23(11):1783–1789. doi: 10.1681/ASN.2012040338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hill A.F., Pegtel D.M., Lambertz U., Leonardi T., O’Driscoll L., Pluchino S. ISEV position paper: extracellular vesicle RNA analysis and bioinformatics. J. Extracell. Vesicles. 2013;2 doi: 10.3402/jev.v2i0.22859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li M., Zeringer E., Barta T., Schageman J., Cheng A., Vlassov A.V. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369(1652) doi: 10.1098/rstb.2013.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schageman J., Zeringer E., Li M., Barta T., Lea K., Gu J. The complete exosome workflow solution: from isolation to characterization of RNA cargo. Biomed Res. Int. 2013;2013 doi: 10.1155/2013/253957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Witwer K.W., Buzas E.I., Bemis L.T., Bora A., Lasser C., Lotvall J. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zarovni N., Corrado A., Guazzi P., Zocco D., Lari E., Radano G. Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods. 2015;87:46–58. doi: 10.1016/j.ymeth.2015.05.028. [DOI] [PubMed] [Google Scholar]
- 73.Stranska R., Gysbrechts L., Wouters J., Vermeersch P., Bloch K., Dierickx D. Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J. Transl. Med. 2018;16(1):1. doi: 10.1186/s12967-017-1374-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hogan M.C., Manganelli L., Woollard J.R., Masyuk A.I., Masyuk T.V., Tammachote R. Characterization of PKD protein-positive exosome-like vesicles. J. Am. Soc. Nephrol. 2009;20(2):278–288. doi: 10.1681/ASN.2008060564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Linares R., Tan S., Gounou C., Arraud N., Brisson A.R. High-speed centrifugation induces aggregation of extracellular vesicles. J. Extracell. Vesicles. 2015;4:29509. doi: 10.3402/jev.v4.29509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Erdbrugger U., Rudy C.K., Etter M.E., Dryden K.A., Yeager M., Klibanov A.L., Lannigan J. Imaging flow cytometry elucidates limitations of microparticle analysis by conventional flow cytometry. Cytometry A. 2014;85(9):756–770. doi: 10.1002/cyto.a.22494. [DOI] [PubMed] [Google Scholar]
- 77.Webber J., Clayton A. How pure are your vesicles? J. Extracell. Vesicles. 2013;2 doi: 10.3402/jev.v2i0.19861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baran-Gale J., Kurtz C.L., Erdos M.R., Sison C., Young A., Fannin E.E. Addressing Bias in small RNA library preparation for sequencing: a new protocol recovers MicroRNAs that evade capture by current methods. Front. Genet. 2015;6:352. doi: 10.3389/fgene.2015.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mateescu B., Kowal E.J., van Balkom B.W., Bartel S., Bhattacharyya S.N., Buzas E.I. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. J. Extracell. Vesicles. 2017;6(1):1286095. doi: 10.1080/20013078.2017.1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ghosal A. Importance of secreted bacterial RNA in bacterial-host interactions in the gut. Microb. Pathog. 2017;104:161–163. doi: 10.1016/j.micpath.2017.01.032. [DOI] [PubMed] [Google Scholar]
- 81.Heintz-Buschart A., Yusuf D., Kaysen A., Etheridge A., Fritz J.V., May P. Small RNA profiling of low biomass samples: identification and removal of contaminants. BMC Biol. 2018;16(1):52. doi: 10.1186/s12915-018-0522-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Allen R.M., Zhao S., Ramirez Solano M.A., Zhu W., Michell D.L., Wang Y. Bioinformatic analysis of endogenous and exogenous small RNAs on lipoproteins. J. Extracell. Vesicles. 2018;7(1):1506198. doi: 10.1080/20013078.2018.1506198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Michell D.L., Allen R.M., Landstreet S.R., Zhao S., Toth C.L., Sheng Q., Vickers K.C. Isolation of high-density lipoproteins for non-coding small RNA quantification. J. Vis. Exp. 2016;117 doi: 10.3791/54488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Blank A., Dekker C.A. Ribonucleases of human serum, urine, cerebrospinal fluid, and leukocytes. Activity staining following electrophoresis in sodium dodecyl sulfate-polyacrylamide gels. Biochemistry. 1981;20(8):2261–2267. doi: 10.1021/bi00511a030. https://www.ncbi.nlm.nih.gov/pubmed/7236597 [DOI] [PubMed] [Google Scholar]
- 85.Tauro B.J., Greening D.W., Mathias R.A., Ji H., Mathivanan S., Scott A.M., Simpson R.J. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56(2):293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 86.Rana S., Yue S., Stadel D., Zoller M. Toward tailored exosomes: the exosomal tetraspanin web contributes to target cell selection. Int. J. Biochem. Cell Biol. 2012;44(9):1574–1584. doi: 10.1016/j.biocel.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 87.Cocucci E., Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25(6):364–372. doi: 10.1016/j.tcb.2015.01.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.