Abstract
Transforming growth factor β (TGF-β) drives epithelial-mesenchymal transition (EMT), playing vital roles in cancer metastasis. The crosstalk between microRNAs (miRNAs) and TGF-β are frequently observed and involved in TGF-β-induced EMT. Here, we determine that miR-577 is significantly upregulated in gastric cancer (GC). miR-577 expression is positively correlated with GC metastasis status and poor patient prognosis. Functional assays demonstrate that miR-577 promotes metastasis and chemoresistance by inducing EMT and stemness-like properties. Moreover, TGF-β promotes the expression of miR-577, and miR-577 participates TGF-β-mediated cancer metastasis. Mechanistically, TGF-β activates miR-577 via NF-κB-mediated transcription, and miR-577 enhances TGF-β signaling by targeting the serum deprivation protein response (SDPR), which directly interacts with ERK to inactivate the ERK-NF-κB pathway, hence forming a feedback loop to drive tumor metastasis. A plausible mechanism of EMT induction by the TGF-β network is elucidated. Our findings suggest that the TGF-β-miR-577-SDPR axis may be a potential prognostic marker and therapeutic target against cancer metastasis in GC.
Keywords: TGF-β, gastric cancer, metastasis, miR-577-SDPR axis, feedback loop
Luo et al. find that mir-577 can be as a prognostic marker and therapeutic target against cancer metastasis in gastric cancer. TGF-β activates miR-577 via NF-κB-mediated transcription, and miR-577 enhances TGF-β signaling by forming a feedback loop with SDPR-ERK-NF-κB pathway.
Introduction
The incidence and mortality of gastric cancer (GC) are the factors for its being listed among the top three of all malignant tumors, with invasion and metastasis being important factors for poor prognosis.1, 2 Due to the lack of early symptoms, GC metastasis has occurred in most patients by the time of their diagnoses.3, 4 Thus, it is critical to explore the molecular mechanisms of GC metastasis and to identify new prognostic markers and therapeutic targets.
Epithelial-mesenchymal transition (EMT) is an evolutionarily conserved program for cellular development. However, in pathological conditions such as cancer, EMT can drive tumor cell metastasis. EMT allows tumor cells to reprogram the polarized epithelial cells into a mesenchymal phenotype and acquire enhanced movement capacity, invasiveness, and resistance to apoptosis.5 Transforming growth factor beta (TGF-β) is the most important inducer of EMT, and studies on its complicated molecular mechanisms have only just begun. Our previous study demonstrated that LIM and SH3 protein 1 (LASP1) is a downstream effector of TGF-β and activates the Smad pathway by modulating expression of the calcium-binding protein S100A4.6 However, additional downstream molecules and pathways, which may serve as specific targets for managing GC metastasis, still need to be explored.
MicroRNAs (miRNAs) are members of the non-coding RNA family and regulate the expression of target genes at the post-transcriptional level. Accumulating evidence has demonstrated that miRNAs can affect biological behaviors such as cell development, proliferation, differentiation, apoptosis, and EMT.6 Previous studies have defined miR-338-3p as an EMT-specific suppresser miRNA in GC that acts by repressing the expression of the metastasis-associated in colon cancer-1 (MACC1) gene, which has been identified as a metastasis-associated protein in GC.3, 7, 8 Recent studies have found that miRNA can be involved in TGF-β-induced EMT as a downstream effector.9, 10 Our previous study illustrated that miR-187 is induced by TGF-β and suppresses SOX4, NT5E, and PTK6 proteins to inhibit the Smad pathway.11 Recently, it was demonstrated that miR-577 is a tumor-associated gene and that its expression is highly heterogeneous in different cancers. miR-577 functions as a tumor suppressor in papillary thyroid cancer (PTC) by targeting SphK2.12 In addition, miR-577 suppresses cell growth and enhanced 5-fluorouracil (5-FU) sensitivity though targeting HSP27 in colorectal cancer.13 However, a recent study specified that miR-577 promotes esophageal squamous cell carcinoma progression by inactivation of the p53 pathway and the Rb pathway.14 Currently, the expression and molecular function of miR-577 in GC remain largely unknown.
Suppressed serum deprivation protein response (SDPR), also known as Cavin-2, is a member of the cavin family and plays an important role in the formation of caveolae.15 SDPR was first identified as a substrate for protein kinase C (PKC) isoforms and recruits polymerase I and transcript release factor (PTRF) to caveolae.16 Recently, downregulation of SDPR was found to occur in breast cancer, kidney cancer, prostate cancer, and hepatocellular carcinoma.17, 18 In breast cancer, hypermethylation of the SDPR promoter region leads to a low expression of SDPR, and the lower expression of SDPR promoted cancer metastasis by relieving the inhibition of the extracellular-signal-regulated kinase (ERK) pathway and the nuclear factor κB (NF-κB) pathway.19 Another study found that SDPR suppression activates the TGF-β pathway and contributes to EMT in breast cancer cells.20 Moreover, SDPR was also a potential prognostic and diagnostic indicator in hepatocellular carcinoma.17 Nevertheless, the role of SDPR in GC is still obscure.
Here, we reported a crosstalk between TGF-β and miR-577. We found that miR-577 was induced by TGF-β through NF-κB-mediated transcriptional activation. The expression of miR-577 targeted SDPR mRNA and repressed the SDPR-ERK-NF-κB pathway in GC cells. By forming a positive-feedback loop, the miR-577-SDPR axis enhanced the aggressive effects of TGF-β on GC cells. These findings demonstrate that miR-577-SDPR axis exerts oncogenic and metastatic effects in GC and may serve as a potential target for therapeutic applications in the clinic.
Results
miR-577 Is Upregulated in GC and Associated with a Poor Prognosis for GC Patients
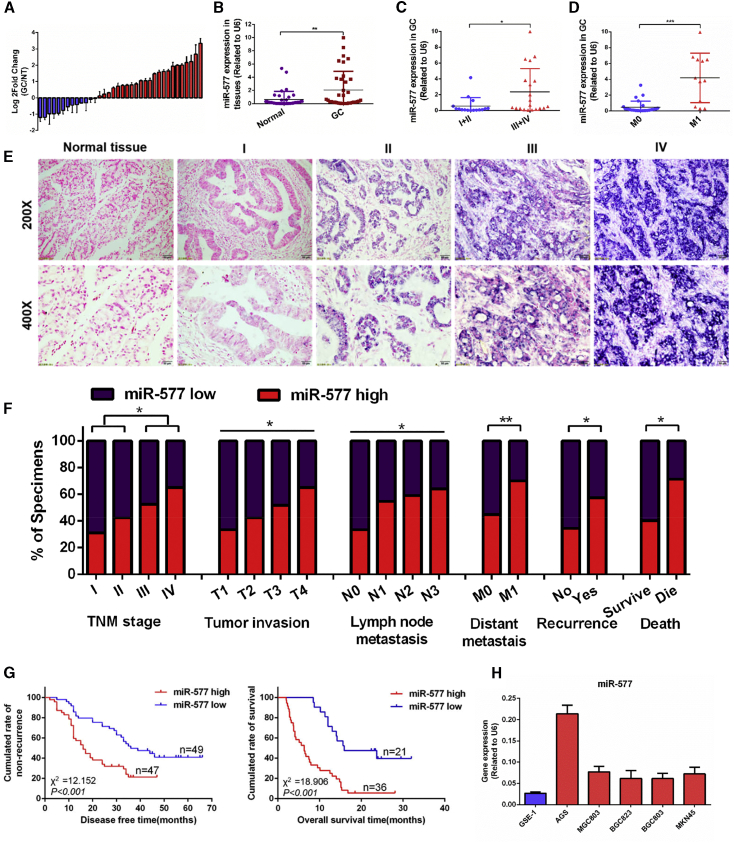
The Cancer Genome Atlas (TCGA) microarray21 analysis indicated that miR-577 was upregulated in GC (p < 0.001; Figure S1). The real-time qPCR was used to measure the expression of miR-577 transcripts in GC tissues and cell lines. The upregulated miR-577 was observed from 24 of all 36 GC samples when compared to the matched normal tissues (Figure 1A). The expression of miR-577 in GC tissues was significantly higher than the paired normal tissues (p < 0.01; Figure 1B). Meanwhile, high miR-577 expression was associated with progressed tumor node metastasis (TNM) stage (p < 0.05; Figure 1C) and the occurrence of metastasis (p < 0.001; Figure 1D). In addition, high expression of miR-577 was found in all five GC cell lines compared with the immortalized gastric mucosal epithelial cell line GSE-1 (Figure 1H). The expression level of miR-577 was also evaluated using in situ hybridization (ISH) by analyzing a large cohort of 153 archived paraffin-embedded GC specimens and normal tissues. miR-577 was found to be highly expressed in GC, and its expression increased relative to the progression of the tumor stage (Figure 1E). It showed a significant correlation between miR-577 expression and clinical variables, including TNM stage (p < 0.05), tumor invasion (p < 0.05), lymph node metastasis (p < 0.05), distant metastasis (p < 0.01), recurrence (p < 0.05), and OS (p < 0.05), while age, gender, and tumor differentiation were not correlated with miR-577 expression (Figure 1F; Table S1). Kaplan-Meier survival analysis revealed that GC patients with high miR-577 expression had worse disease-free survival (DFS) in stage I–III patients and worse overall survival in stage-IV patients (p < 0.001 and p < 0.001, respectively; Figure 1G). Univariate survival analysis showed that high miR-577 expression was associated with the shorter OS (p < 0.001, hazard ratio [HR] = 2.473; Table 1). Furthermore, multivariate survival analysis indicated that the expression of miR-577, T classification, and age were independent predictors for prognosis in GC patients (Table 1).
Figure 1.
miR-577 Is Upregulated in GC and Associated with Poor Prognosis
(A) qRT-PCR analysis of miR-577 expression in 36 pairs of GC specimens and normal tissues. miR-577 was normalized to endogenous U6 RNA and expressed relative to their respective match normal tissues. (B) The expression of miR-577 in 36 pairs of GC specimens and normal tissues. **p < 0.01. (C) The miR-577 expression in TNM stage I and stage II GC tissues and stage III and stage IV GC tissues. *p < 0.05. (D) The miR-577 expression in GC tissues with or without metastasis. ***p < 0.001. (E) In situ hybridization (ISH) analysis of miR-577 expression in 153 human normal gastric tissues and GC specimens from TNM stage I–IV patients. (F) Frequency of low and high miR-577 expressions categorized by TNM stage, tumor invasion, lymph node metastasis, distant metastasis, recurrence, and death (p = 0.022, p = 0.019, p = 0.027, p = 0.002, p = 0.029, and p = 0.030, respectively). Patients were separated into high- and/or low-expression groups by the expression score of the miR-577. *p < 0.05; **p < 0.01. (G) Retrospective analysis of Kaplan-Meier plots for miR-577 expression in association with disease-free survival and overall survival. (H) qRT-PCR analysis of miR-577 expression in GC cell lines and an immortalized human gastric cell line. Data represent mean ± SD.
Table 1.
Univariate and Multivariate Analyses of Individual Parameters for Correlations with Overall Survival Rate: Cox Proportional Hazards Model
| Variables | Univariate |
Multivariate |
||||
|---|---|---|---|---|---|---|
| HR | CI (95%) | p Value | HR | CI (95%) | p Value | |
| Age (≥55/< 55) | 0.574 | 0.434–0.759 | 0.000* | 0.738 | 0.547–0.995 | 0.046* |
| Gender (male/female) | 0.985 | 0.693–1.401 | 0.308 | 1.137 | 0.781–1.655 | 0.504 |
| T classification (III + IV/I + II) | 3.935 | 2.041–7.585 | 0.000* | 3.172 | 1.607–3.552 | 0.001* |
| N classification (N1–3/N0) | 1.538 | 1.027–2.302 | 0.037* | 1.44 | 0.953–2.176 | 0.083 |
| M classification (M1/M0) | 1.841 | 1.275–2.657 | 0.01* | 1.496 | 0.979–2.285 | 0.062 |
| Differentiation (moderate/well) | 0.895 | 0.488–1.640 | 0.719 | 1.207 | 0.644–2.262 | 0.558 |
| Differentiation (poor/well) | 1.094 | 0.614–1.949 | 0.76 | 1.67 | 0.905–3.085 | 0.101 |
| miR-577 expression (high/low) | 2.473 | 1.707–3.580 | 0.000* | 2.389 | 1.607–3.552 | 0.000* |
HR; hazard ratio; CI; confidence interval. *Statistically significant (p < 0.05).
miR-577 Promotes GC Metastasis and Chemoresistance In Vivo and In Vitro
MGC803 cells and MKN45 cells with a moderate expression level of miR-577 were chosen for further investigation. We transfected miR-577 AgomiR to overexpress miR-577 and miR-577 AntagomiR to inhibit miR-577. At the same time, MGC803 cells were chosen for the lentiviral-mediated stable overexpression of miR-577, and MKN45 cells were used for lentiviral-mediated stable inhibition of miR-577. According to qRT-PCR, transfection efficiencies were all effective (p < 0.001, p < 0.001, p < 0.001, and p < 0.01 in Figures S2A–S2D, respectively). 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assays, colony formation assays, and EdU cell proliferation assays revealed that miR-577 had no effect on GC cell proliferation in vitro (Figures S2E–S2G). For in vivo analysis, we constructed the subcutaneous-tumor mouse model and found that miR-577 overexpression or suppression showed no impacts on tumor weights, volumes, tumor signals, or the Ki-67 index (Figures S2H–S2K).
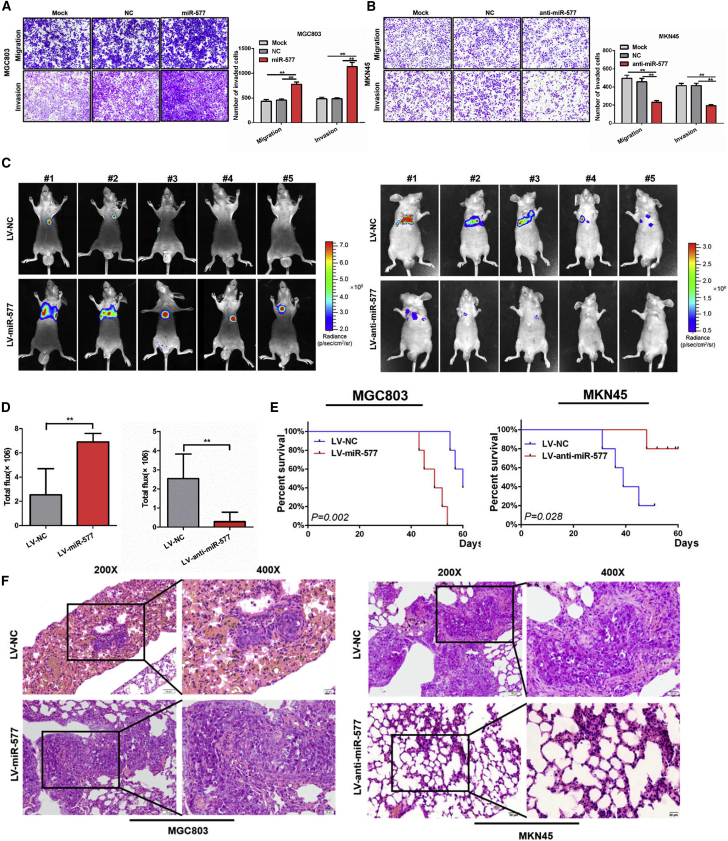
We then assessed the metastatic potential of miR-577. Results from Transwell assays showed that the overexpression of miR-577 significantly enhanced cell migration and invasiveness, while this effect was abolished when treated with the miR-577 antagonist AntagomiR (p < 0.01; Figures 2A and 2B). Subsequently, to observe the effect of miR-577 on lung colonization, cancer cells were injected into the tail vein of nude mice. Higher metastasis signals and shorter survival time were found in the miR-577 overexpressed group compared with the control group, while miR-577 suppression in MKN45 cells led to the opposite effects (Figures 2C–2E). We also found that more and larger tumor nodules were formed in the LV-miR-577 group compared with the lentivirus of negative control (LV-NC) group. In contrast, miR-577 inhibition reduced the number of lung metastases compared with that in the control group (Figure 2F).
Figure 2.
miR-577 Promotes GC Metastasis In Vitro and In Vivo
(A and B) Effect of miR-577 AgomiR or AntagomiR on the migration and invasiveness of MGC803 (A) and MKN45 (B) cells based on Transwell assays. **p < 0.01. Magnification, 100×. (C) Fluorescence images of lung homing potential of miR-577-expressing or silencing group mice and control group mice. (D) Luciferase signals of lung metastasis nudes in miR-577-expressing or silencing group mice and control group mice; the data were expressed as mean ± SD. **p < 0.01. (E) Kaplan–Meier survival curves and univariate analyses (log rank) for the mice with miR-577-expressing or silencing cells versus control cells were performed. (F) Representative image of H&E staining of lung metastasis in miR-577-expressing or silencing group mice and control group mice.
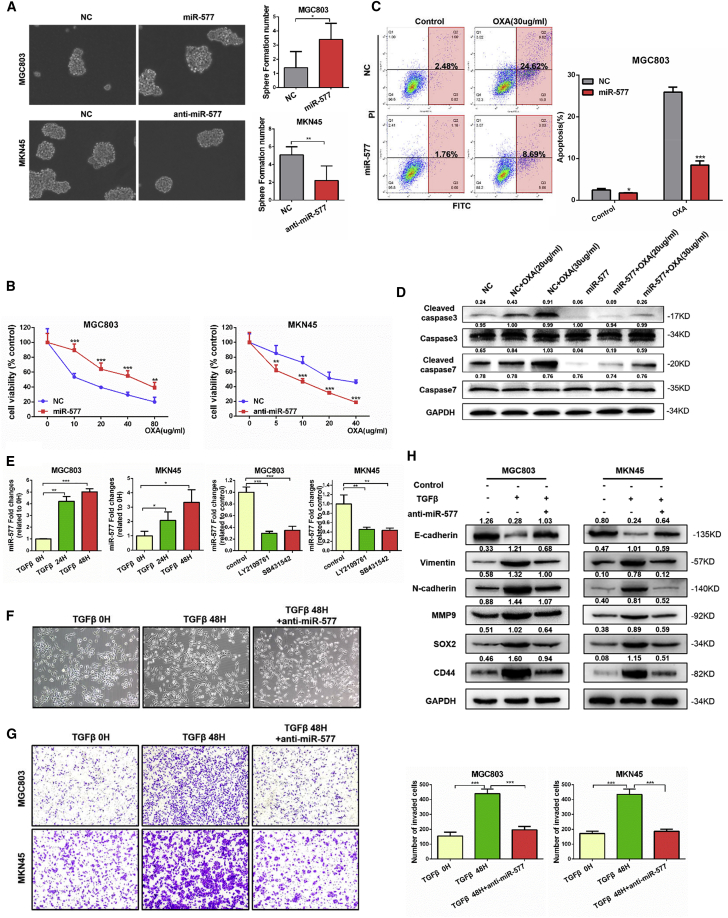
To further assess the effect of miR-577 on tumor stemness, tumorsphere formation assays were performed. As shown in Figure 3A, the overexpression of miR-577 in MGC803 cells induced the sphere-forming ability, while miR-577 silencing reduced sphere formation. Since cancer stemness is associated with chemoresistance,22 we performed MTT assays, and the results indicated that overexpression of miR-577 led to chemoresistance to oxaliplatin, while suppression of miR-577 increased chemosensitivity (Figure 3B). Consistent with the results from the MTT assays, flow cytometry analysis showed that oxaliplatin-induced apoptosis was reduced following the overexpression of miR-577 (Figure 3C). Further, we carried out immunoblot assays to detect apoptosis markers and found the suppressed effect on oxaliplatin-induced apoptosis from miR-577 (Figure 3D). Collectively, these results suggested that miR-577 promoted cancer cell migration, invasion, lung colonization, and chemoresistance but without affecting the proliferation of GC cells.
Figure 3.
miR-577 Is Essential for TGF-β-Mediated EMT and Stemness in GC
(A) Effect of miR-577 on tumorsphere formation in serum-free stem cell medium. Magnification 100×. Student’s t test. *p < 0.05; **p < 0.01. (B) Dose-response curves of cells transfected with miR-577 AgomiR or AntagomiR after treatment with oxaliplatin for 24 h. Parametric generalized linear model with random effects. (C) Effects of miR-577 on cell apoptosis analyzed by flow cytometry with or without oxaliplatin treatment (30 μg/mL). *p < 0.05; ***p < 0.001. (D) Western blot of endogenous caspase-3, cleaved caspase-3, caspase-7, and cleaved caspase-7 protein expression levels in MGC803 cells transfected with negative control (NC) or miR-577 AgomiR treated with oxaliplatin (20 μg/mL or 30 μg/mL) or not, as indicated. GAPDH served as a loading control. (E) qRT-PCR analysis of miR-577 expression after GC cells were treated with TGF-β (20 ng/mL) for 24 and 48 h and TGF-β inhibitors LY2109761 (10 μM) and SB431542 (10 μM) for 24 h; transcript levels were normalized to U6 expression. (F–H) Effects on the morphology of MGC803 (F), GC cell migration ability (G), and the expression of EMT and stemness markers (H) mentioned earlier were detected following transfection of miR-577 AntagomiR into GC cells treated with TGF-β (20 ng/mL) for 48 h. Data represent mean ± SD. GAPDH served as a loading control. ***p < 0.001. (F and G) Magnification, 100×.
miR-577 Is Essential for TGF-β-Mediated GC EMT and Stemness
Under inverted microscopy, we found that cells overexpressing miR-577 displayed a morphology typical of aggressive cells, presenting a spindle-shaped morphology (Figure S3A). This finding suggested that miR-577 might promote EMT in GC. To determine whether miR-577 contributed to the EMT process, we evaluated EMT and cancer stemness markers. Immunoblot assays revealed that miR-577 suppressed the expression of epithelial markers (E-cadherin) and stimulated the expression of mesenchymal markers (vimentin, N-cadherin, and MMP9) and stemness markers (SOX2 and CD44). In contrast, suppression of miR-577 inhibited the EMT process in MKN45 cells (Figure S3B). Similarly, immunofluorescence analysis showed that miR-577 decreased the fluorescence intensity of the epithelial marker E-cadherin and increased the fluorescence intensity of the mesenchymal marker vimentin (Figure S3C).
The tumor microenvironment promotes the development of EMT through multiple growth factors such as epidermal growth factor (EGF), hepatocyte growth factor (HGF), and TGF-β5. To identify the potential upstream effectors of miR-577, we stimulated GC cell lines MGC803 and MKN45 with EGF, HGF, and TGF-β. Treatment with EGF and HGF resulted in no significant change of miR-577 (Figure S3D). However, the expression of miR-577 was induced at the presence of TGF-β in a time-dependent manner, while cells treated with TGF-β inhibitors LY2109761 (10 μM) and SB431542 (10 μM) for 24 h significantly decreased miR-577 expression (Figure 3E). As TGF-β was the major inducer of EMT, TGF-β-treated cells were induced to display a spindle-shaped morphology (Figure S3E). Furthermore, Transwell assays showed that TGF-β stimulation dramatically enhanced GC cell migration (Figure S3F). As expected, the protein levels of the mesenchymal markers and stemness markers were also upregulated in TGF-β-treated cells compared to levels in control cells, whereas the epithelial marker E-cadherin was downregulated (Figure S3G).
It has been demonstrated that TGF-β is able to regulate the expression of miR-577, suggesting that miR-577 may be a downstream target of TGF-β. Therefore, we treated MGC803 and MKN45 cells with miR-577 antagonistic oligonucleotide in combination with TGF-β. Suppression of miR-577 reversed the morphological shift observed in GC cells induced by TGF-β (Figure 3F). Similarly, the enhancement of cell migration under TGF-β treatment was also abrogated by the silencing of miR-577 (Figure 3G). Western blot analysis showed that, under the conditions of miR-577 silence, the expression of epithelial markers was recovered in cells in response to TGF-β treatment, while the expression levels of the mesenchymal markers and stemness markers were reduced (Figure 3H), suggesting that miR-577 was essential for TGF-β-induced EMT and stemness in GC.
TGF-β Regulates miR-577 Expression by Activating NF-κB
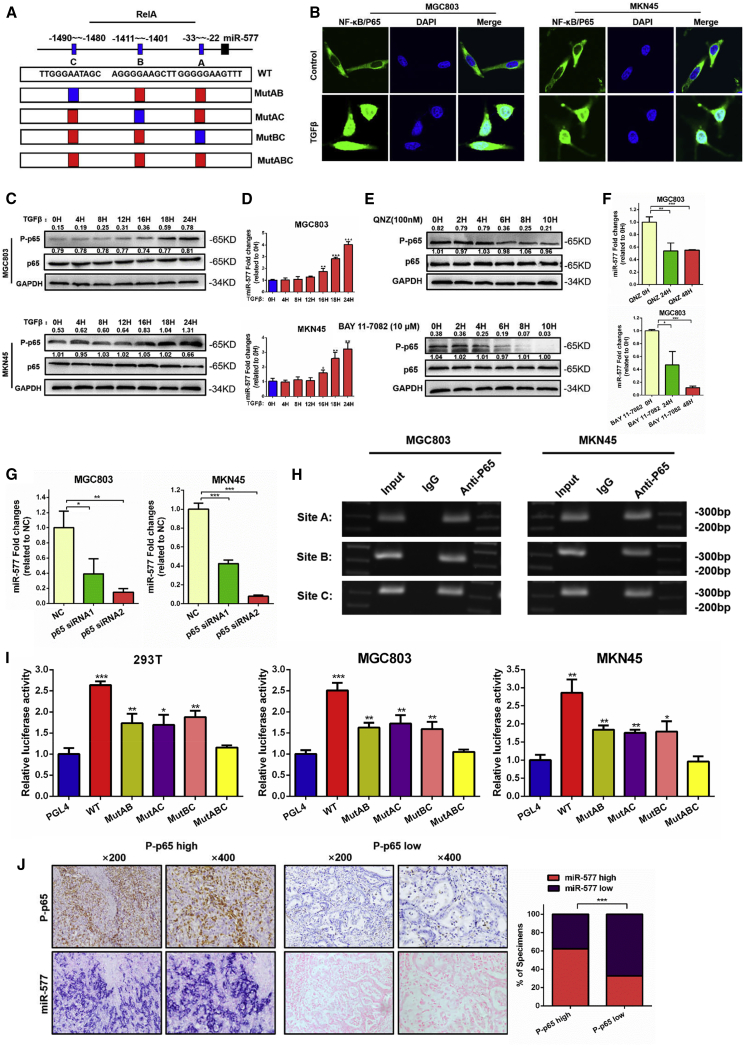
Since TGF-β can upregulate miR-577 expression, we speculated that TGF-β might regulate miR-577 transcription. To clarify the transcriptional mechanisms of miR-577, we analyzed the University of California Santa Cruz (UCSC) genomic database and the transcription factor prediction database PROMO. We determined that NF-κB p65, also known as RelA, was possibly the transcription factor of miR-577 and that there were three potential binding sites. These three transcription-factor binding sites (TFBSs) were termed A, B, and C according to their relative positions (Figure 4A). It has been reported that TGF-β contributes to the activation of the NF-κB pathway and assists p65 entering into the nucleus.23, 24 Immunofluorescence was used to examine p65 localization. As we suspected, the nuclear localization of p65 significantly increased following TGF-β treatment for 24 h (Figure 4B). Western blot analysis revealed that the expression of phosphorylated p65 (p-p65) was upregulated in conjunction with prolonged TGF-β treatment time (Figure 4C). Moreover, the expression of miR-577 was increased, accompanied with the activation of p-p65 (Figure 4D). To further confirm the regulation between the NF-κB pathway and miR-577, we treated cells with NF-κB pathway inhibitors QNZ and BAY 11-7082. QNZ was a specific NF-κB inhibitor and directly inhibited phosphorylation of NF-κB.25 BAY 11-7082 inhibited phosphorylation of IkappaB kinase (IKBα) to suppress the NF-κB pathway.26 Results from immunoblot assays demonstrated that the phosphorylation of p65 was suppressed (Figure 4E). We also observed a time-dependent decrease of miR-577, accompanied with the change in p-p65, after treatment with NF-κB pathway inhibitors (Figure 4F). We also designed specific small interfering RNAs (siRNAs) targeting p65. Western blot analysis showed that both siRNAs effectively suppressed the expression of p65 protein (Figure S4A). Quantitative analyses of miR-577 expression using qRT-PCR were consistent with the p65 change (Figure 4G). Together, these findings indicated that p65 might be the transcription factor of miR-577 regulation and was involved in the TGF-β-mediated upregulation of miR-577.
Figure 4.
miR-577 Is Modulated by TGF-β through Activation of NF-κB (p65)
(A) Schematic representation of the promoter regions of miR-577 with the putative RelA (p65) transcription factor-binding sites (A, B, and C) and the concrete structure of the wild-type (WT) and mutant (MutAB, MutAC, MutBC, and MutABC) luciferase reporters driven by the promoter. (B) Immunofluorescence assays of p65 protein cellular localization in MGC803 and MKN45 cells treated with TGF-β, as indicated. Representative figures are shown. Magnification, 180×. (C) Expression levels of p-p65 and p65 proteins in MGC803 and MKN45 cells treated with TGF-β were detected by western blot. GAPDH served as a loading control. (D) qRT-PCR analysis of miR-577 expression in MGC803 and MKN45 cells treated with TGF-β for different hours; transcript levels were normalized to U6 expression. *p < 0.05; **p < 0.01; ***p < 0.001. (E) The effects of NF-κB pathway inhibitors (QNZ and BAY 11-7082) on the expression of p-p65 and p65 were analyzed by western blot. (F) qRT-PCR analysis was performed for detecting miR-577 expression in MGC803 cells treated with QNZ and BAY 11-7082 for 24 h or 48 h. *p < 0.05; **p < 0.01; ***p < 0.001. (G) Expression of miR-577 in MGC803 and MKN45 cells transfected with si-p65 (#1–2) were analyzed by qRT-PCR. *p < 0.05; **p < 0.01; ***p < 0.001. (H) Amplification of p65-binding sites A, B, and C after ChIP analysis using anti-p65 antibody is indicated in the PCR gel. The gel figures are accompanied by the locations of molecular weight markers. (I) Relative luciferase activity of the indicated promoter vectors in 293T, MGC803, and MKN45 cells transfected with p65 plasmids. *p < 0.05; **p < 0.01; ***p < 0.001. (J) Representative images of successive IHC and ISH staining of p-p65 and miR-577 expression in GC tumor specimens. Bars on the right represent the percentage of specimens; ***p < 0.001.
Furthermore, to confirm the binding of p65 to the miR-577 promoter, chromatin immunoprecipitation (ChIP) assays were performed, and the results demonstrated that the p65 protein was recruited to all three binding sites in the miR-577 promoter (Figure 4H). We then constructed luciferase reporter vectors WT, MUTAB, MUTAC, MUTBC, and MUTABC based on the three TFBSs, and they are described in the Materials and Methods. Luciferase activity was detected following co-transfection with a p65-expressing plasmid. The results confirmed that TFBS A, B, and C all enhanced luciferase activity, indicating that all three sites were functional sites in p65 regulating miR-577 expression (Figure 4I). Notably, we detected the expression of p-p65 in 153 tissue specimens and found that miR-577 expression was positively associated with the expression of p-p65 (Figure 4J).
Based on the aforementioned experimental results, it was determined that miR-577 is downstream of TGF-β in the signaling pathway. TGF-β activated p65 to enter into the cell nucleus to transcriptionally regulate miR-577, ultimately leading to the induction of EMT and stemness in GC.
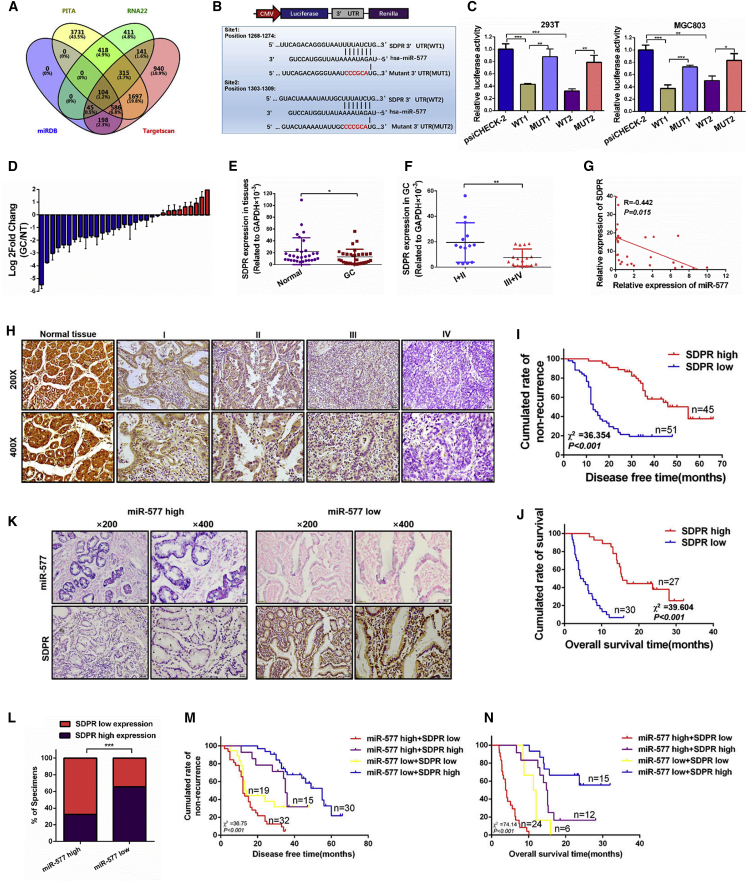
SDPR Is the Target of miR-577 in GC
Since the function of miRNAs is dependent on the regulation of target genes, it is important to identify the target genes of miR-577 in GC. Potential targets were selected from an analysis of four prediction databases (Targetscan, RNA22, PITA, and miRDB); the findings are shown in Figure 5A. Among them, seven candidate genes were chosen for further verification. The qRT-PCR results showed that four target genes were decreased after miR-577 transfection, among which SDPR presented the most dramatic change (Figure S4B). Accordingly, we chose SDPR for further exploration.
Figure 5.
SDPR Is a Metastasis Suppressor Downstream of miR-577 in GC
(A) Bioinformatics prediction of potential miR-577 targets by four common miRNA databases (TargetScan, PITA, RNA22, and miRDB). (B) Potential miR-577 binding sequences in the 3′ UTR of SDPR mRNAs and the structure of the WT (WT1 and WT2) and mutant (Mut1 and Mut2) luciferase reporters. (C) Relative luciferase activity of the indicated 3′UTR vectors in 293T and MGC803 cells co-transfected with miR-577 AgomiR and SDPR plasmids. (D) qRT-PCR analysis of SDPR expression in 30 pairs of GC specimens and normal tissues, which are normalized to an endogenous GAPDH RNA control. Data represent the mean ± SD. (E and F) The SDPR expression in 30 pairs of GC specimens and normal tissues (E) and in GC tissues in the TNM stages I and II or stages III and IV (F). *p < 0.05; **p < 0.01. (G) Correlation between miR-577 and SDPR mRNA expression levels. Two-tailed Spearman’s correlation analysis. Data represent mean ± SD; p = 0.015. (H) Immunohistochemistry (IHC) analysis of SDPR expression in 153 human normal gastric tissues and GC specimens from TNM stage I–IV patients. (I and J) Retrospective analysis of Kaplan-Meier plots for SDPR expression and the association with disease-free survival (I) and overall survival (J). ***p < 0.001. (K and L) Representative images of successive IHC and ISH staining of miR-577 and SDPR expression in GC tumor specimens (K). Bars represent the percentage of specimens (L). ***p < 0.001. (M and N) Retrospective analysis of Kaplan-Meier plots of the co-expression of SDPR and miR-577 in association with disease-free survival (M) and overall survival (N). ***p < 0.001.
Luciferase reporter assays were performed to determine whether miR-577 was able to directly target the 3′ UTR of SDPR. We constructed the wild-type 3′ UTR fragments WT1 and WT2 and the mutant 3′ UTR fragments MUT1 and MUT2 according to two binding sites (Figure 5B). These vectors and miR-577 AgomiR were co-transfected into 293T cells and MGC803 cells. As shown in Figure 5C, the luciferase activities were decreased for both WT vectors; of note, mutants of the binding site restored the luciferase signal intensity, thereby confirming the direct interaction between miR-577 and the SDPR 3′ UTR. Meanwhile, overexpression of miR-577 decreased SDPR protein expression. Conversely, miR-577 silence elevated the SDPR levels (Figure S4C). Our studies confirmed that miR-577 was downstream of TGF-β; accordingly, TGF-β stimulation caused a time-dependent reduction of SDPR expression at the protein level (Figure S4D).
SDPR is reported to serve as a tumor-suppressive gene. Analysis of the Oncomine database indicated that the downregulation of SDPR occurs in multiple tumor types, including GC (Figures S4E and S4F). The KMPlot database27 was used to analyze the survival value of SDPR. According to Kaplan-Meier survival analyses, high expression levels of SDPR were correlated with benign DFS and overall survival (OS) in GC patients, which implied that SDPR is a tumor suppressor gene (Figure S4G). To further determine whether these findings would be supported in clinical samples, we performed qRT-PCR analysis on 30 pairs of GC specimens. Consistent with the database predictions, SDPR expression was downregulated in 21 of 30 paired GC specimens (Figure 5D), and SDPR expression was lower in the tumor samples than in the matched normal tissues (Figure 5E). In addition, as shown in Figure 5F, the expression of SDPR was lower in tumor tissues at advanced TNM stages (III and IV) than in those at early TNM stages (I and II). Correlation analysis revealed that the expression level of SDPR was negatively correlated with miR-577 expression (Figure 5G). We also performed an analysis on 153 GC specimens using immunohistochemistry and found that the expression of SDPR in GC tumor tissue was lower than that in the adjacent normal tissues and that the expression of SDPR decreased as the tumor stage increased (Figure 5H). Statistical analysis showed that the expression of SDPR was associated with TNM stages (p < 0.001), tumor invasion (p < 0.001), lymph node metastasis (p < 0.001), and distant metastasis (p < 0.001) in patients (Figure S4H; Table S1). Prognostic analysis showed that high SDPR expression was associated with better DFS for patients in TNM stages I–III and better OS for patients in TNM stage IV (Figure 5I and 5J). Notably, in GC specimens expressing high levels of miR-577, the SDPR expression levels were low, while the expression levels of SDPR in tissues with low expression of miR-577 were strongly positive (p < 0.001; Figure 5K and 5L). Moreover, patients with high miR-577 expression and low SDPR expression had the worst prognosis, and patients with low miR-577 expression and high SDPR expression had the best outcome (Figure 5M and 5N). Taken together, these findings demonstrated that SDPR was the target gene of miR-577. In addition, SDPR was downregulated in GC and associated with a better prognosis.
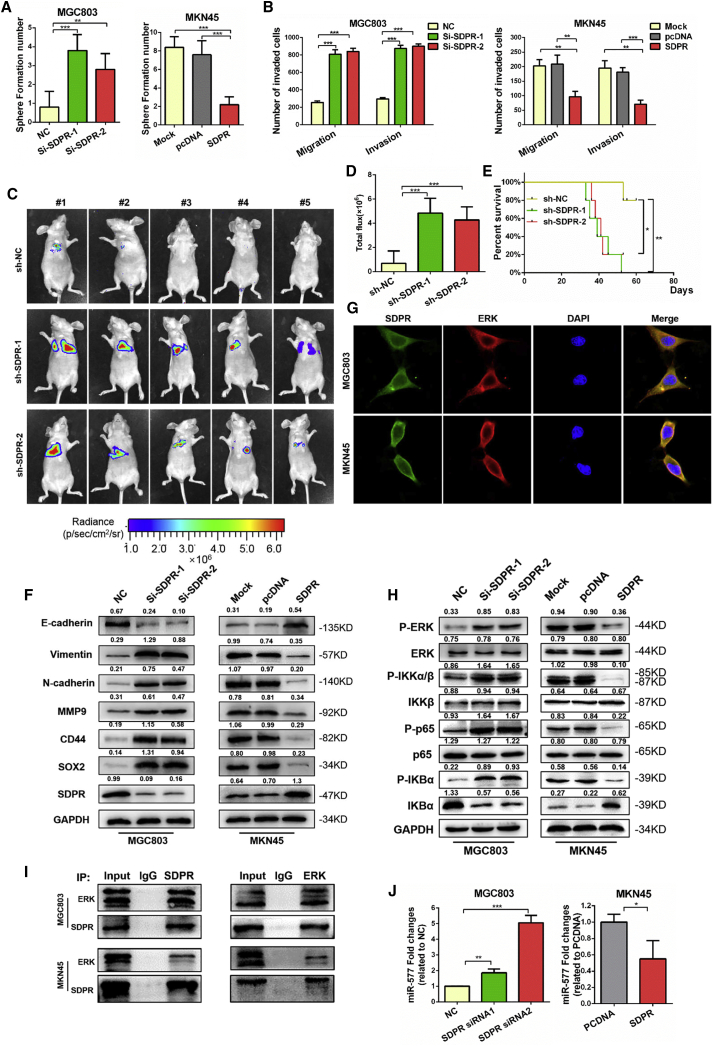
SDPR Suppresses GC Metastasis by Directly Inhibiting the ERK-NF-κB Signaling Pathway
Currently, the biological function of SDPR in GC has not yet been reported. To investigate the role of SDPR in GC metastasis, we suppressed SDPR expression in MGC803 cells using siRNAs and induced the overexpression of SDPR in MKN45 cells by transfecting SDPR-expressing plasmid. The success of knockdown and overexpression of SDPR were confirmed by immunoblot assays (Figure S5A), and siRNA1 and siRNA2 were chosen for further studies. Tumorsphere-formation assays and Transwell assays showed that SDPR- knockdown cells exhibited reduced sphere formation, migration, and invasion capacities, whereas the opposite results were observed in SDPR-overexpressing cells (Figures 6A and 6B; Figures S5D and S5E). To evaluate the in vivo effect of SDPR on GC cell metastasis, we established that MGC803 stably suppressed SDPR cells by lentivirus infection (Figure S5B). MGC803 stable SDPR knockdown cells and control cells were injected into the tail vein of mice, and bioluminescence images showed that SDPR knockdown enhanced the metastasis signals and metastasis tumor nodules in lung and reduced the survival time of mice (Figures 6C–6E; Figure S5F). Through the performance of functional experiments, we provide supportive evidence that SDPR is a tumor suppressor gene in GC.
Figure 6.
Downregulation of SDPR Induces GC EMT and Stemness through the ERK-NF-κB Signaling Pathway
(A) Tumorsphere-formation assays were performed to detect the effect of SDPR on sphere formation ability. The number of spheres were counted under a microscope in five randomly selected fields. **p < 0.01; ***p < 0.001. (B) The ability of cell migration and invasiveness tested using Transwell chamber migration assays. The cells were counted under a microscope in five randomly selected fields. **p < 0.01; ***p < 0.001. (C) Fluorescence images of mice after tail-vein injections with SDPR-silencing cells and control cells. (D) Luciferase signals of lung metastasis nudes in SDPR-silencing group mice and control group mice; the data were expressed as mean ± SD. ***p < 0.001. (E) Kaplan-Meier survival curves and univariate analyses (log rank) for the mice after injections with SDPR-silencing cells and control cells. *p < 0.05; **p < 0.01. (F) Western blot of EMT markers (E-cadherin, vimentin, and N-cadherin), stemness markers (CD44 and SOX2), and SDPR protein expression levels in MGC803 cells transfected with small interfering RNA targeting SDPR (si-SDPR) (#1–2) and MKN45 cells transfected with SDPR plasmid. (G) Immunofluorescence microscopy colocalization analysis of SDPR and ERK in MGC803 and MKN45 cells. Magnification, 180×. (H) P-ERK, ERK, P-IKKα-β, IKKβ, P-IKBα, IKBα, p-p65, and p65 were quantitated by western blot after transfection with NC and si-SDPR (#1–2), pcDNA, and SDPR, as indicated. GAPDH served as a loading control. (I) Lysates from MGC803 and MKN45 cells were immunoprecipitated using anti-SDPR or anti-ERK antibodies and immunoblotted with the indicated antibodies. Twenty percent of the lysate used for immunoprecipitation was loaded as the input control. (J) qRT-PCR analysis was performed for miR-577 expression in MGC803 cells transfected with pcDNA or SDPR plasmid and MKN45 cells transfected with si-SDPR (#1–2). Endogenous U6 RNA was used as a control. *p < 0.05; **p < 0.01; ***p < 0.001.
Previous studies revealed that miR-577 promotes GC EMT; therefore, we investigated the effect of SDPR on GC EMT. After transfection of the SDPR-expressing plasmid into cells, the SDPR protein level was upregulated in conjunction with the reduction in expression of EMT markers. The opposing changes in EMT markers were observed when SDPR was knocked down (Figure 6F). Immunofluorescence analysis showed that SDPR silence decreased the fluorescence intensity of E-cadherin and increased the fluorescence intensity of vimentin (Figure S5C). These data supported the conclusion that SDPR inhibited the EMT process in GC cells.
It has been reported that SDPR inhibits ERK and the NF-κB pathway in breast cancer.19 Moreover, ERK can promote the phosphorylation of IKKα-β and activate the NF-κB pathway.24, 28, 29 Therefore, we speculated that SDPR might inhibit EMT by directly interacting with the ERK protein and thereby inhibit the ERK-NF-κB pathway. To test our hypothesis, we analyzed the protein localization of ERK and SDPR using immunofluorescent labeling and found a strong co-localization of ERK and SDPR (Figure 6G). As the SDPR protein level was effectively suppressed, there was a corresponding boost in expression of the ERK-NF-κB pathway markers, including P-ERK, P-IKKα-β, P-IKBα, and p-p65. In contrast, overexpression of SDPR reduced the expression of the ERK-NF-κB pathway (Figure 6H).
Using co-immunoprecipitation techniques, we also confirmed the direct binding between ERK and SDPR in both MGC803 and MKN45 cells (Figure 6I). As confirmed by earlier experiments in our study, NF-κB was a transcription factor that regulated miR-577 expression. Additionally, miR-577 was upstream of SDPR, and SDPR inhibited the ERK-NF-κB pathway. Taken together, these findings implied the existence of a positive-feedback loop in the regulation of this signaling pathway. Thus, we sought to examine the effect of SDPR on the expression level of miR-577, using qRT-PCR analysis. Knockdown of SDPR resulted in miR-577 being significantly upregulated. Conversely, miR-577 expression was inhibited following the overexpression of SDPR in MKN45 cells (Figure 6J).
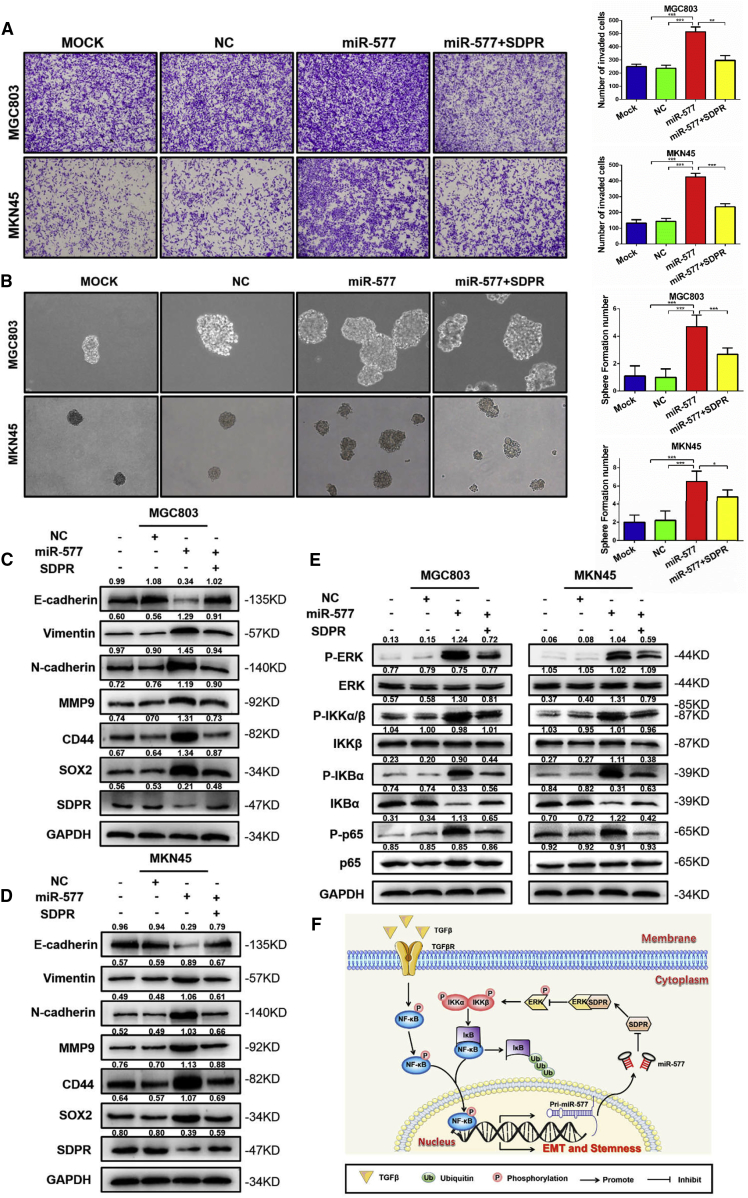
Restoring SDPR Expression Reverses EMT and Stemness Induced by miR-577 in GC
To demonstrate that SDPR suppression mediated the role of miR-577 in GC cells, we restored SDPR expression in cells overexpressing miR-577. While miR-577 overexpression resulted in an increased migration phenotype, restoring SDPR treatment abolished miR-577-induced cancer cell migration (Figure 7A). Tumorsphere-formation assays demonstrated that the number of pluripotent spheres increased following the overexpression of miR-577, whereas a reduction of sphere numbers was observed when SDPR expression was rescued (Figure 7B). In addition to the observations of biological phenotype, reversed changes of EMT and stemness markers were observed following SDPR restoration in both MGC803 cells and MKN45 cells (Figure 7C and 7D). Since previous experiments in our study confirmed that SDPR mediated the ERK-NF-κB pathway, we also evaluated the expression of ERK-NF-κB-related markers by western blot analysis. Consistent results were detected in the ERK-NF-κB pathway (Figure 7E). These findings strongly supported the conclusion that SDPR was an important mediator of miR-577-induced EMT and stemness in GC cells.
Figure 7.
SDPR Plays a Crucial Role in TGF-β-miR-577-Induced EMT and Stemness of GC Cells
(A) The ability of MGC803 and MKN45 cell migration and invasiveness after transfection with NC, MOCK, miR-577 AgomiR, and miR-577 AgomiR + SDPR plasmids were detected by Transwell chamber migration assays. Bars on the right represent the number of invaded cells; the cells were counted under a microscope in five randomly selected fields. **p < 0.01; ***p < 0.001. Magnification, 100×. (B) Tumorsphere-formation assays were performed to detect the sphere formation ability. Bars on the right represent the number of spheres; the number of spheres were counted under a microscope in five randomly selected fields. *p < 0.05; ***p < 0.001. Magnification 100×. (C and D) Western blot of EMT markers (vimentin, N-cadherin, and MMP9), stemness markers (CD44 and SOX2) proteins expression levels in MGC803 (C) and MKN45 (D) cells transfected with NC, MOCK, miR-577 AgomiR, miR-577 AgomiR + SDPR plasmids, as indicated. (E) Western blot of the proteins expression levels of ERK/NF-κB signaling pathway (P-ERK, ERK, P-IKKα/β, IKKβ, P-IKBα, IKBα, P-p65, and p65) in MGC803 and MKN45 cells transfected with NC, MOCK, miR-577 AgomiR, miR-577 AgomiR + SDPR plasmids, as indicated. TGF-β induced miR-577 expression by NF-κB-mediated transcription, while SDPR as a direct target was a metastasis suppressor gene of miR-577 and inhibited the ERK-NF-κB pathway, which formed a positive-feedback loop to promote GC metastasis.
Discussion
Our present results demonstrated that the overexpression of miR-577 contributed to TGF-β-induced EMT and stemness by forming a positive-feedback loop, which played a vital role in GC development and sustaining an aggressive phenotype. The present study highlighted the oncogenic role of miR-577 in GC and revealed a relevant role for TGF-β during EMT. These findings may provide insight for the development of potential targets for managing GC.
Ectopic expression of miR-577 has been reported in multiple types of cancer, including esophageal squamous cell carcinoma,14 glioblastoma,30 colorectal cancer,13 and osteosarcoma.31 Available data suggest that miR-577 promotes or suppresses cancer progression in a cancer-specific manner. For instance, Zhang et al. found that miR-577 inhibits glioma cell proliferation.30 In osteosarcoma, miR-577 acts as an endogenous competitive RNA for the long non-coding RNA (lncRNA) SNHG1 and assists in the regulation of cell proliferation, invasion, and EMT.31 However, in esophageal squamous cell carcinoma (ESCC), miR-577 overexpression promotes G1-S phase transition in vitro and accelerates cell proliferation.14 Currently, the regulatory effects of miR-577 in GC remain unknown. Although miR-577 promotes or inhibits proliferation capacities in multiple types of cancer, we failed to observe any significant effect on the ability of GC cell proliferation from either our in vitro or in vivo studies. Interestingly, miR-577 contributed to increased migration, invasion, and lung colonization abilities and induced chemoresistance to oxaliplatin in GC. In this study, miR-577 was determined for the first time to be an oncogenic gene in GC development.
Since miR-577 was frequently associated with cancer cell migration, invasion, and chemosensitivity in GC, this strongly suggested that miR-577 may be related to patient outcome in cancer. The present study analyzed the expression of miR-577 in clinical samples. Statistical analysis suggested that miR-577 was correlated with GC metastasis, and Kaplan-Meier survival analysis found that high expression of miR-577 was associated with a worse clinical prognosis in GC patients. Intriguingly, results from the in vivo experiments were consistent with this finding in that miR-577 overexpression in the mice lead to a reduced survival. These findings may suggest that GC patients with high miR-577 expression should receive more aggressive clinical intervention in the treatment of their cancer.
The relationship between miR-577 and GC metastasis remains unclear. Recently, EMT has been identified as a critical step during cancer progression and metastasis. EMT generates significant changes in tumor cells and includes the loss of tight junctions and cell polarity and the gain of invasive forces to break through the surrounding tissues of the tumor.32 In previous studies, we demonstrated that several miRNAs act as key regulators of EMT and modulate cancer metastasis.11, 33 In the present study, we observed that miR-577 was able to promote a mesenchymal morphology. Conversely, miR-577 antagonism promoted the retention of an epithelial morphology. Additional studies confirmed miR-577-induced changes in EMT-related markers. Consistent with EMT being closely associated with cancer stemness,5 we also found that miR-577 enhanced the expression of stemness markers CD44 and SOX2. These results strongly suggested that miR-577 plays an essential role in the EMT process.
TGF-β is the most important inducer of EMT by regulating the Smad pathway and non-Smad pathway. In the classical pathway, TGF-β binds with its receptor, TGF-βR1/2, and stimulates phosphorylation of Smad2/3. The phosphorylated Smad2/3 forms a protein complex with Smad4 and is translocated to the nucleus, leading to the induced transcription of EMT-related genes. Moreover, TGF-β can also activate Smad-independent pathways such as phosphatidylinositol 3-kinase (PI3K)/serine/threonine kinase (AKT), mitogen-activated protein kinase (MAPK), and the NF-κB pathway.34 Among these, the NF-κB pathway is frequently activated in cancers and widely regulates cancer cell proliferation, survival, angiogenesis, metastasis, and therapeutic resistance.35 The NF-κB family contains NF-κB1 (p50), NF-κB2 (p52), RELA (p65), RELB, and c-REL. In the canonical pathway, the IκB kinase (IKK) complex, especially IKKβ, is phosphorylated by growth factor or signaling transduction. The activated IKK complex then phosphorylates the negative regulator of NF-κB, inhibitor of the kappa B-α (IκBα) protein, which leads to the degradation of IκBα through ubiquitination and proteasome-mediated means. The degradation of IκBα results in the release of the p65-p50 heterodimer, which mediates the NF-κB complex being translocated into the nucleus.35 Since TGF-β is able to activate the kinase TGF-β-activated kinase 1 (TAK1), and TAK1 is essential for IKK complex phosphorylation,23, 34 NF-κB is involved in this TGF-β-mediated biological function. In the present study, miR-577 was initially identified as the downstream target of TGF-β. TGF-β stimulation activated NF-κB and translocated it from the cytoplasm to the nucleus in order to activate the transcription of miR-577. Notably, a positive correlation between miR-577 and P-NF-κB was also found in clinical tissues. Moreover, suppressing the expression of miR-577 was able to neutralize the TGF-β-mediated invasion and EMT, which may prove to be a novel target for managing GC metastasis.
Generally, miRNAs exert their biological functions by inhibiting target-gene expression post-transcriptionally. Recently, some target genes of miR-577 have been identified. In glioblastoma (GBM), the Wnt signaling pathway genes lipoprotein receptor-related protein 6 (LRP6) and β-catenin are reported to be target genes of miR-577.30 In colorectal carcinoma (CRC), heat shock protein 27 (HSP27) has been identified as a target gene of miR-577.13 In our study, the SDPR protein was identified as a new target of miR-577. miR-577 overexpression suppressed SDPR expression, while knockdown of miR-577 improved SDPR expression. Furthermore, TGF-β treatment also suppressed SDPR expression, suggesting that SDPR may be required for TGF-β function.
Our study demonstrated that SDPR was downregulated in GC, both in fresh tissues and clinical specimens, and that the levels are associated with cancer metastasis. Survival analysis showed that patients with a high level of expression of SDPR had a better relative prognosis, which is highly consistent with the findings in hepatocellular carcinoma (HCC),17 suggesting a potential prognostic value and clinical significance for SDPR in cancers. With SDPR being the target of miR-577, there was a negative correlation between miR-577 levels and SDPR expression in both fresh tissues and clinical specimens. Functional assays confirmed the comprehensive effects of SDPR in suppressing GC progression and that restored SDPR expression offset the miR-577-derived activation of EMT and cancer metastasis. Nevertheless, the molecular mechanism of SDPR in GC remains unclear. Ozturk et al.19 found that SDPR directly interacts with ERK by binding phosphatidylserine (PS) and inhibits the ERK and NF-κB pathways. Another study also found that SDPR suppresses the ERK pathway and induces cell-cycle arrest in oral cancer.36 The ERK pathway is the continuation of the RAS-RAF-MEK pathway and plays an important role in cell migration, angiogenesis, proliferation, differentiation, and cell survival.37 More importantly, as a protein kinase, ERK may phosphorylate the IKK complex, thus activating the NF-κB pathway.24, 28, 29 Therefore, we hypothesized that SDPR may interact with ERK and suppress the ERK-NF-κB pathway to block EMT and the metastasis of GC cells. Here, the interaction between SDPR and ERK was confirmed by immunofluorescence and co-immunoprecipitation analyses. The regulation of the ERK-NF-κB pathway was also verified by immunoblotting. Previous studies have found that NF-κB is a transcription factor of miR-577. We also found that SDPR could regulate the expression of miR-577, thus forming a positive-feedback loop.
In summary, this study has refined the understanding of the signaling network of TGF-β-induced EMT; successfully identified a new downstream target, miR-577; and confirmed that TGF-β regulated the expression of miR-577 through activating NF-κB. The overexpression of miR-577 suppressed SDPR expression and released the SDPR-ERK protein complex to activate the ERK-NF-κB pathway, which induced EMT and formed a positive-feedback loop to upregulate the transcription of miR-577 (Figure 7F). Consequently, miR-577 and SDPR may serve as new therapeutic targets and prognostic markers for GC patients.
Materials and Methods
Cell Culture
Five human GC cell lines (AGS, BGC823, MKN28, MKN45, and MGC803), cells from an immortalized human gastric epithelial cell line (GES-1), and HEK293T cells were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and authenticated using the short tandem repeat (STR) method. GC cell lines and GES-1 cells were cultured in RPMI-1640 medium (Hyclone, Logan, UT, USA) with 10% fetal bovine serum (FBS) (GIBCO BRL, Invitrogen, Grand Island, NY, USA), while HEK293T cells were cultured in DMEM (Invitrogen, Grand Island, NY, USA) with 10% FBS. All cell lines were incubated at an atmosphere of 5% CO2 at 37°C.
Tumor Tissue Samples
36 fresh gastric cancer specimens and the paired adjacent noncancerous tissues from Nanfang Hospital, Southern Medical University, were preserved in liquid nitrogen immediately after resection. 153 paraffin-embedded GC specimens and the paired adjacent noncancerous specimens were obtained from Nanfang Hospital as well. Each specimen was attached to a confirmed pathological diagnosis. None of the patients had received any chemoradiotherapy before surgery. All experiments in this study were endorsed by the Ethics Committee of Southern Medical University and complied with the Declaration of Helsinki. The Ethics Committee of Southern Medical University specifically approved that no informed consent was required, because data were analyzed anonymously.
Construction of Plasmid, siRNA, and Cell Transfection
The sequences of SDPR and p65 or RelA were constructed into the pcDNA3.1 plasmid vector. siRNAs for p65 (siRNAs #1–2) and SDPR (siRNAs #1–3) were synthesized by GenePharma (Suzhou, China). All siRNA sequences were listed in Table S2. To generate the promoter vector of miR-577, a wild-type fragment containing 3 binding sties of p65 was introduced into PGL4 luciferase reporter vector. In addition, p65-binding site mutation vectors were constructed. The PGL4-derived vectors and p65 plasmids were cotransfected into 293T or MGC803 and MKN45 cells using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA). To confirm the direct binding of the miR-577 and SDPR 3′ UTRs, two wild-type SDPR 3′ UTR fragments and two mutant fragments were introduced into psiCHECK2 luciferase reporter vector. These psiCHECK2-derived vectors and miR-577 AgomiR were cotransfected into 293T and MGC803 cells. After 48 h, the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) was used to measure the luciferase activity according to the manufacturer’s protocols.
Real-Time qPCR and Western Blot
The real-time qPCR and western blot assays were conducted according to a previously described method.3, 11 All the primers designed for qPCR were listed in Table S3, and final data were analyzed with the 2−ΔΔCt method. The details of antibodies for the western blot are listed in Table S4. Further details are provided in the Supplemental Materials and Methods.
MTT, EdU Cell Proliferation Assays and Colony Formation Assays
MTT, Edu, and colony formation assays were performed according to a previously described method.7 Further details are provided in the Supplemental Materials and Methods.
Cell Migration and Invasion Assays
The cell migration and invasion assays were performed according to previously described methods.8 Further details are provided in the Supplemental Materials and Methods.
Tumorsphere-Formation Assays
After being resuspended, 1,000 cells were seeded into ultra-low-attachment 6-well plates (Corning, Corning, NY, USA) in conditions with 2 mL serum-free DMEM/F-12 medium (GIBCO, Rockville, MD, USA) supplemented with 20 ng/mL basic fibroblast growth factor (bFGF), 10 ng/mL EGF (Peprotech, Rocky Hill, NJ, USA), and 2% B27 (Life Technologies, Grand Island, NY, USA; catalog no. 17504044) for 14 days. Morphological examinations were performed by collecting pictures taken under the inverted microscope (IX71, Olympus, Tokyo, Japan).
Flow Cytometry Analysis
Flow cytometry analyses (BD Biosciences, San Jose, CA, USA) were performed to measure the cell apoptosis percentage with an Annexin-PI kit (Keygen Biotech, Nanjing, China). After cells were incubated in the culture medium with a concentration of 30 μg/mL oxaliplatin treatment, or not, for 24 h, they were resuspended and collected to detect an apoptosis percentage according to the Annexin-PI kit instructions.
ISH and IHC Analysis
The ISH and IHC were conducted according to previously described methods.11, 33 A specific probe (5′- CAGGTACCAATATTTTATCT -3′) of miR-577 was designed for ISH. Scoring was measured by three pathologists separately as: 0 = no staining, 1 = weak staining, 2 = moderate staining, and 3 = strong staining. According to the staining score, 0–1 was considered as low expression and 2–3 was considered as high expression.
The information of antibodies was listed in Table S4. Further details are provided in the Supplemental Materials and Methods.
ChIP and CoIP
For the ChIP assay: After cells were cross-linked and lysed, DNA fragments were sonicated on ice to lengths between 200 and 1,000 bp. Anti-p65 antibody (catalog no. 26156, 1:50, Cell Signaling Technology) and control immunoglobulin G were used to precipitate protein-DNA complexes. The detail processes were performed according to the instructions of the Pierce Agarose ChIP Kit (Thermo Scientific, Waltham, MA, USA; catalog no. 26156). Finally, PCR was done to examine the putative p65-binding sites in the miR-577 promoter with its specific primers (Table S5). For the co-immunoprecipitation (coIP) assay: About 80%–90% adherent MGC803 and MKN45 cells were extracted by cell lysis buffer for IP. Then coIP assays were performed with the Dynabeads Protein A Immunoprecipitation Kit (Thermo Scientific, Waltham, MA, USA; catalog no. 10006D) according to the manufacturer’s protocols. Immunoprecipitated proteins (SDPR and ERK) were proved by western blot.
Mouse Xenograft Model
For in vivo tumor growth assay, 4-week-old BALB/c nude mice were subcutaneously injected with stable overexpressing or silencing cells and control cells (1 × 107 cells per mouse). Tumor volumes were measured with a precise caliper every 3 days from day 3 to day 28 after injection. After 28 days, mice with general anesthesia were intraperitoneally injected with luciferin (150 mg/kg per mouse) to detect tumor size using the CRi Maestro In Vivo Imaging System (FX PRO, Bruker, Billerica, MA, USA). Then, mice were sacrificed, subcutaneous tumors were fixed, and IHC staining was performed to detect Ki-67 expression in two groups. For an in vivo metastasis assay, a total of 1 × 106 cells were injected into the tail veins of 6-week-old BALB/c nude mice. After 42 days, mice after general anesthesia were intraperitoneally injected with luciferin (150 mg/kg per mouse) to detect tumor systemic metastasis through the CRi Maestro In Vivo Imaging System. Endpoint time for observation was 60 days after injection, or mice died naturally. Then lung tissues were fixed, and H&E staining was used to detect the metastatic tumors, which were quantified by counting metastatic lesions in each section. Images were taken by an upright microscope (Olympus DP72, Tokyo, Japan). All animal experiments involved were approved by the Guangdong Provincial Bureau of Science, and all animals were ethically and humanely treated.
Bioinformatics
The expression profiling data of differential miRNAs in 42 paired GC patient tissues were analyzed by TCGA STAD (Stomach Adenocarcinoma). The expression of miRNAs was analyzed between GC tissues and paired normal tissues; miRNAs with the number of changes above 30 in 42 GC tissues and a total fold change (Fold) above 2 were selected. Statistical significance was established at p < 0.001. To search transcription factors that can regulate miR-577, PROMO was used to analyze promoter region binding sites. Targetscan, RNA22, PITA, and miRDB were used to predict the potential miR-577 targets, which were under consideration when they were positive in four analyses mentioned earlier. Furthermore, the expression of SDPR and its prognosis role in GC were confirmed by Oncomine and Kaplan-Meier Plotter. All the databases are listed in Table S6.
Statistical Analysis
Data were analyzed using SPSS v20.0 software (Abbott Laboratories, Chicago, IL, USA) and GraphPad Prism software 6.0 (GraphPad Software, San Diego, CA, USA). Results were presented as the mean ± SD. Statistical analyses were performed using the Student’s t test, one-way ANOVA, or a Mann-Whitney U test for nonparametric data. Kaplan-Meier plots were used to estimate the prognostic relevance of miR-577-SDPR in univariate analysis. Univariate and multivariate Cox proportional hazards models were applied to determine the effects of variables on survival. Pearson’s chi-square test was used to analyze the associations between miR-577 expression and SDPR expression. Statistical significance was established at p < 0.05.
Author Contributions
W.L. and N.H. led the study design and prepared the manuscript; Y. Luo and J.W. performed the experiments; X.L. performed statistical analysis; J.W. and J.Z. performed data analysis and interpretation; Q.W. and X.R. provided data collection; and J.R., Y. Liao and J.B. contributed to the editing of the manuscript.
Conflicts of Interest
The authors declare no competing financial interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (nos. 81472314, 81602705, and 81772580).
Footnotes
Supplemental Information includes five figures, six tables, and Supplemental Materials and Methods and can be found with this article online at https://doi.org/10.1016/j.ymthe.2019.02.002.
Contributor Information
Na Huang, Email: lala343@126.com.
Wangjun Liao, Email: nfyyliaowj@163.com.
Supplemental Information
References
- 1.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Van Cutsem E., Sagaert X., Topal B., Haustermans K., Prenen H. Gastric cancer. Lancet. 2016;388:2654–2664. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 3.Davidson M., Okines A.F., Starling N. Current and future therapies for advanced gastric cancer. Clin. Colorectal Cancer. 2015;14:239–250. doi: 10.1016/j.clcc.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Digklia A., Wagner A.D. Advanced gastric cancer: current treatment landscape and future perspectives. World J. Gastroenterol. 2016;22:2403–2414. doi: 10.3748/wjg.v22.i8.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mittal V. Epithelial mesenchymal transition in tumor metastasis. Annu. Rev. Pathol. 2018;13:395–412. doi: 10.1146/annurev-pathol-020117-043854. [DOI] [PubMed] [Google Scholar]
- 6.Wang H., Shi J., Luo Y., Liao Q., Niu Y., Zhang F., Shao Z., Ding Y., Zhao L. LIM and SH3 protein 1 induces TGFβ-mediated epithelial-mesenchymal transition in human colorectal cancer by regulating S100A4 expression. Clin. Cancer Res. 2014;20:5835–5847. doi: 10.1158/1078-0432.CCR-14-0485. [DOI] [PubMed] [Google Scholar]
- 7.Xia J., Huang N., Huang H., Sun L., Dong S., Su J., Zhang J., Wang L., Lin L., Shi M. Voltage-gated sodium channel Nav 1.7 promotes gastric cancer progression through MACC1-mediated upregulation of NHE1. Int. J. Cancer. 2016;139:2553–2569. doi: 10.1002/ijc.30381. [DOI] [PubMed] [Google Scholar]
- 8.Wang L., Wu Y., Lin L., Liu P., Huang H., Liao W., Zheng D., Zuo Q., Sun L., Huang N. Metastasis-associated in colon cancer-1 upregulation predicts a poor prognosis of gastric cancer, and promotes tumor cell proliferation and invasion. Int. J. Cancer. 2013;133:1419–1430. doi: 10.1002/ijc.28140. [DOI] [PubMed] [Google Scholar]
- 9.Yu J., Lei R., Zhuang X., Li X., Li G., Lev S., Segura M.F., Zhang X., Hu G. MicroRNA-182 targets SMAD7 to potentiate TGFβ-induced epithelial-mesenchymal transition and metastasis of cancer cells. Nat. Commun. 2016;7:13884. doi: 10.1038/ncomms13884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bu P., Wang L., Chen K.Y., Rakhilin N., Sun J., Closa A., Tung K.L., King S., Varanko A.K., Xu Y. miR-1269 promotes metastasis and forms a positive feedback loop with TGF-β. Nat. Commun. 2015;6:6879. doi: 10.1038/ncomms7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang F., Luo Y., Shao Z., Xu L., Liu X., Niu Y., Shi J., Sun X., Liu Y., Ding Y., Zhao L. MicroRNA-187, a downstream effector of TGFβ pathway, suppresses Smad-mediated epithelial-mesenchymal transition in colorectal cancer. Cancer Lett. 2016;373:203–213. doi: 10.1016/j.canlet.2016.01.037. [DOI] [PubMed] [Google Scholar]
- 12.Xue K.C., Hu D.D., Zhao L., Li N., Shen H.Y. MiR-577 inhibits papillary thyroid carcinoma cell proliferation, migration and invasion by targeting SphK2. Eur. Rev. Med. Pharmacol. Sci. 2017;21:3794–3800. [PubMed] [Google Scholar]
- 13.Jiang H., Ju H., Zhang L., Lu H., Jie K. MicroRNA-577 suppresses tumor growth and enhances chemosensitivity in colorectal cancer. J. Biochem. Mol. Toxicol. 2017 doi: 10.1002/jbt.21888. Published online February 2, 2017. [DOI] [PubMed] [Google Scholar]
- 14.Yuan X., He J., Sun F., Gu J. Effects and interactions of miR-577 and TSGA10 in regulating esophageal squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2013;6:2651–2667. [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen C.G., Bright N.A., Howard G., Nichols B.J. SDPR induces membrane curvature and functions in the formation of caveolae. Nat. Cell Biol. 2009;11:807–814. doi: 10.1038/ncb1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aboulaich N., Vainonen J.P., Strålfors P., Vener A.V. Vectorial proteomics reveal targeting, phosphorylation and specific fragmentation of polymerase I and transcript release factor (PTRF) at the surface of caveolae in human adipocytes. Biochem. J. 2004;383:237–248. doi: 10.1042/BJ20040647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jing W., Luo P., Zhu M., Ai Q., Chai H., Tu J. Prognostic and diagnostic significance of SDPR-Cavin-2 in hepatocellular carcinoma. Cell. Physiol. Biochem. 2016;39:950–960. doi: 10.1159/000447803. [DOI] [PubMed] [Google Scholar]
- 18.Li X., Jia Z., Shen Y., Ichikawa H., Jarvik J., Nagele R.G., Goldberg G.S. Coordinate suppression of Sdpr and Fhl1 expression in tumors of the breast, kidney, and prostate. Cancer Sci. 2008;99:1326–1333. doi: 10.1111/j.1349-7006.2008.00816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozturk S., Papageorgis P., Wong C.K., Lambert A.W., Abdolmaleky H.M., Thiagalingam A., Cohen H.T., Thiagalingam S. SDPR functions as a metastasis suppressor in breast cancer by promoting apoptosis. Proc. Natl. Acad. Sci. USA. 2016;113:638–643. doi: 10.1073/pnas.1514663113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian Y., Yu Y., Hou L.K., Chi J.R., Mao J.F., Xia L., Wang X., Wang P., Cao X.C. Serum deprivation response inhibits breast cancer progression by blocking transforming growth factor-β signaling. Cancer Sci. 2016;107:274–280. doi: 10.1111/cas.12879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J., Lichtenberg T., Hoadley K.A., Poisson L.M., Lazar A.J., Cherniack A.D., Kovatich A.J., Benz C.C., Levine D.A., Lee A.V. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–416.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Batlle E., Clevers H. Cancer stem cells revisited. Nat. Med. 2017;23:1124–1134. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 23.Freudlsperger C., Bian Y., Contag Wise S., Burnett J., Coupar J., Yang X., Chen Z., Van Waes C. TGF-β and NF-κB signal pathway cross-talk is mediated through TAK1 and SMAD7 in a subset of head and neck cancers. Oncogene. 2013;32:1549–1559. doi: 10.1038/onc.2012.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han D., Wu G., Chang C., Zhu F., Xiao Y., Li Q., Zhang T., Zhang L. Disulfiram inhibits TGF-β-induced epithelial-mesenchymal transition and stem-like features in breast cancer via ERK/NF-κB/Snail pathway. Oncotarget. 2015;6:40907–40919. doi: 10.18632/oncotarget.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tobe M., Isobe Y., Tomizawa H., Nagasaki T., Takahashi H., Fukazawa T., Hayashi H. Discovery of quinazolines as a novel structural class of potent inhibitors of NF-kappa B activation. Bioorg. Med. Chem. 2003;11:383–391. doi: 10.1016/s0968-0896(02)00440-6. [DOI] [PubMed] [Google Scholar]
- 26.Miyamoto R., Ito T., Nomura S., Amakawa R., Amuro H., Katashiba Y., Ogata M., Murakami N., Shimamoto K., Yamazaki C. Inhibitor of IkappaB kinase activity, BAY 11-7082, interferes with interferon regulatory factor 7 nuclear translocation and type I interferon production by plasmacytoid dendritic cells. Arthritis Res. Ther. 2010;12:R87. doi: 10.1186/ar3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szász A.M., Lánczky A., Nagy Á., Förster S., Hark K., Green J.E., Boussioutas A., Busuttil R., Szabó A., Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ling M., Li Y., Xu Y., Pang Y., Shen L., Jiang R., Zhao Y., Yang X., Zhang J., Zhou J. Regulation of miRNA-21 by reactive oxygen species-activated ERK/NF-κB in arsenite-induced cell transformation. Free Radic. Biol. Med. 2012;52:1508–1518. doi: 10.1016/j.freeradbiomed.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 29.Chen B.C., Chang Y.S., Kang J.C., Hsu M.J., Sheu J.R., Chen T.L., Teng C.M., Lin C.H. Peptidoglycan induces nuclear factor-kappaB activation and cyclooxygenase-2 expression via Ras, Raf-1, and ERK in RAW 264.7 macrophages. J. Biol. Chem. 2004;279:20889–20897. doi: 10.1074/jbc.M311279200. [DOI] [PubMed] [Google Scholar]
- 30.Zhang W., Shen C., Li C., Yang G., Liu H., Chen X., Zhu D., Zou H., Zhen Y., Zhang D., Zhao S. miR-577 inhibits glioblastoma tumor growth via the Wnt signaling pathway. Mol. Carcinog. 2016;55:575–585. doi: 10.1002/mc.22304. [DOI] [PubMed] [Google Scholar]
- 31.Jiang Z., Jiang C., Fang J. Up-regulated lnc-SNHG1 contributes to osteosarcoma progression through sequestration of miR-577 and activation of WNT2B/Wnt/β-catenin pathway. Biochem. Biophys. Res. Commun. 2018;495:238–245. doi: 10.1016/j.bbrc.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 32.Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang N., Wu Z., Lin L., Zhou M., Wang L., Ma H., Xia J., Bin J., Liao Y., Liao W. MiR-338-3p inhibits epithelial-mesenchymal transition in gastric cancer cells by targeting ZEB2 and MACC1/Met/Akt signaling. Oncotarget. 2015;6:15222–15234. doi: 10.18632/oncotarget.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neuzillet C., Tijeras-Raballand A., Cohen R., Cros J., Faivre S., Raymond E., de Gramont A. Targeting the TGFβ pathway for cancer therapy. Pharmacol. Ther. 2015;147:22–31. doi: 10.1016/j.pharmthera.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Li F., Zhang J., Arfuso F., Chinnathambi A., Zayed M.E., Alharbi S.A., Kumar A.P., Ahn K.S., Sethi G. NF-κB in cancer therapy. Arch. Toxicol. 2015;89:711–731. doi: 10.1007/s00204-015-1470-4. [DOI] [PubMed] [Google Scholar]
- 36.Unozawa M., Kasamatsu A., Higo M., Fukumoto C., Koyama T., Sakazume T., Nakashima D., Ogawara K., Yokoe H., Shiiba M. Cavin-2 in oral cancer: a potential predictor for tumor progression. Mol. Carcinog. 2016;55:1037–1047. doi: 10.1002/mc.22347. [DOI] [PubMed] [Google Scholar]
- 37.Fey D., Matallanas D., Rauch J., Rukhlenko O.S., Kholodenko B.N. The complexities and versatility of the RAS-to-ERK signalling system in normal and cancer cells. Semin. Cell Dev. Biol. 2016;58:96–107. doi: 10.1016/j.semcdb.2016.06.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.