The Shigella bacterium is one of the most significant causes of waterborne and foodborne bacterial dysentery. A lytic bacteriophage infecting Shigella flexneri was isolated from wastewater in Japan.
ABSTRACT
The Shigella bacterium is one of the most significant causes of waterborne and foodborne bacterial dysentery. A lytic bacteriophage infecting Shigella flexneri was isolated from wastewater in Japan. We report here the complete genome sequence of this bacteriophage, revealing that it belongs to the Myoviridae family and possesses linear genomic DNA.
ANNOUNCEMENT
The Shigella bacterium is one of the most significant causes of waterborne and foodborne bacterial dysentery in the world (1). Among the four species of the genus Shigella, S. flexneri is most commonly associated with shigellosis outbreaks in the developing world (2). Bacteriophages have been proposed as a means for treating bacterial disease (bacteriophage-based therapy) (3–5), detecting and typing bacteria (6), and decontaminating surfaces and water (7, 8). We report here the complete genome sequence of a novel bacteriophage, SfΦ01, that infects S. flexneri and was isolated from wastewater in Japan.
Bacteriophage SfΦ01 was isolated from municipal wastewater by serial plaque purification using S. flexneri (strain identifier, RIMD 3102037) as a host bacterium grown in R2A agar and incubated overnight at 37°C. Spot tests using other bacterial strains (all grown in R2A agar at 37°C) demonstrated that bacteriophage SfΦ01 is capable of infecting Shigella sonnei (RIMD 3104005) and Escherichia coli O1:K1:H7 (JCM1649) as well. Replication of bacteriophage SfΦ01 in these bacterial strains was confirmed by an increase in bacteriophage SfΦ01 genome copy numbers after infection (data not shown). However, this bacteriophage did not infect other types of E. coli, such as the E. coli K12 (MG1655), O26:H11 (RIMD 05091992), O111, and O157:H7 Sakai (RIM0509952) strains. Electron micrographs of bacteriophage particles showed that SfΦ01 had an icosahedral head with a contractile tail (Fig. 1), which morphologically resembled bacteriophages belonging to the family Myoviridae (in the order Caudovirales) (9).
FIG 1.
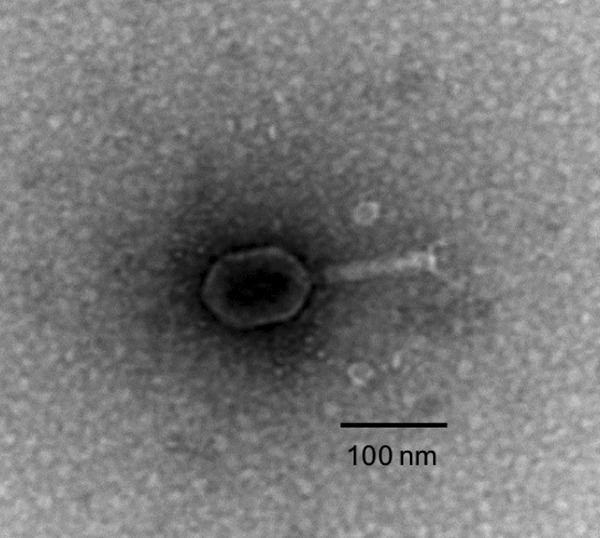
Transmission electron microscope image of bacteriophage SfΦ01 taken at ×100,000 magnification.
For genomic DNA extraction, bacteriophage SfΦ01 was inoculated to S. flexneri grown in R2A medium and incubated overnight at 37°C. Bacterial cells were removed by centrifugation and filtration with a 0.45-μm-pore-size filter. Bacteriophage particles were concentrated using the Centricon Plus-70 filter (Merck Millipore), and 160 μl of bacteriophage concentrate was mixed with 20 μl of DNase I (Promega) to digest free DNA. Bacteriophage genomic DNA was extracted from the resultant sample using the PowerBiofilm DNA isolation kit (Mo Bio Laboratories). Sequencing libraries were prepared using the TruSeq PCR-free library prep kit (Illumina) with an insert fragment size of ca. 350 bp and paired-end sequenced by using the MiSeq platform (Illumina) with v2 chemistry (250 cycles). The sequencing reads (541,594 reads each for forward and reverse sequencing reactions) were assembled de novo by using the SPAdes v. 3.12 program (10). The genome assembly depth (coverage) was 1,618. Genes were predicted by using PHANOTATE (11) and translated and annotated by using in-house perl scripts and a BLASTP algorithm against the nonredundant (nr) database. Default parameters were used for all software tools.
The sequencing results revealed that the genome of bacteriophage SfΦ01 is double-stranded linear DNA with a size of 168,000 bp and a G+C content of 35.29% and containing 288 protein-coding sequences (CDSs). A BLASTn search of the complete genome of SfΦ01 showed the highest identity of 95.58% with Shigella phage Sf21 (GenBank accession number MF327007), which belongs to the family Myoviridae and possesses a linear genome. The present study provides the complete genome sequence information of a novel bacteriophage, SfΦ01, infecting Shigella spp.
Data availability.
The complete genome sequence of bacteriophage SfΦ01 has been deposited in the NCBI database under the GenBank accession number LC465543. Raw data corresponding to the bacteriophage SfΦ01 genome were deposited in the DDBJ DRA database under the SRA accession number DRR175055.
ACKNOWLEDGMENTS
We thank Reiko Hirano at Hokkaido University for her technical assistance.
This work was partly supported by the Japan Society for the Promotion of Science (JSPS) through a Grant-in-Aid for Challenging Research (Exploratory) (17K1889507).
This work was done using computing resources at the Minnesota Supercomputing Institute.
REFERENCES
- 1.Niyogi SK. 2005. Shigellosis. J Microbiol 43:133–143. [PubMed] [Google Scholar]
- 2.Anderson M, Sansonetti PJ, Marteyn BS. 2016. Shigella diversity and changing landscape: insights for the twenty-first century. Front Cell Infect Microbiol 6:1–9. doi: 10.3389/fcimb.2016.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kortright KE, Chan BK, Koff JL, Turner PE. 2019. Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 25:219–232. doi: 10.1016/j.chom.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 4.Doss J, Culbertson K, Hahn D, Camacho J, Barekzi N. 2017. A review of phage therapy against bacterial pathogens of aquatic and terrestrial organisms. Viruses 9:50. doi: 10.3390/v9030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cisek AA, Dąbrowska I, Gregorczyk KP, Wyżewski Z. 2017. Phage therapy in bacterial infections treatment: one hundred years after the discovery of bacteriophages. Curr Microbiol 74:277–283. doi: 10.1007/s00284-016-1166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagens S, Loessner MJ. 2007. Application of bacteriophages for detection and control of foodborne pathogens. Appl Microbiol Biotechnol 76:513–519. doi: 10.1007/s00253-007-1031-8. [DOI] [PubMed] [Google Scholar]
- 7.Woolston J, Parks AR, Abuladze T, Anderson B, Li M, Carter C, Hanna LF, Heyse S, Charbonneau D, Sulakvelidze A. 2013. Bacteriophages lytic for Salmonella rapidly reduce Salmonella contamination on glass and stainless steel surfaces. Bacteriophage 3:e25697. doi: 10.4161/bact.25697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jun JW, Giri SS, Kim HJ, Yun SK, Chi C, Chai JY, Lee BC, Park SC. 2016. Bacteriophage application to control the contaminated water with Shigella. Sci Rep 6:1–7. doi: 10.1038/srep22636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ackermann HW. 2007. 5500 phages examined in the electron microscope. Arch Virol 152:227–243. doi: 10.1007/s00705-006-0849-1. [DOI] [PubMed] [Google Scholar]
- 10.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McNair K, Zhou C, Souza B, Edwards R. 2018. THEA: a novel approach to gene identification in phage genomes. bioRxiv. 10.1101/265983. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequence of bacteriophage SfΦ01 has been deposited in the NCBI database under the GenBank accession number LC465543. Raw data corresponding to the bacteriophage SfΦ01 genome were deposited in the DDBJ DRA database under the SRA accession number DRR175055.


