Abstract
Neuroimaging investigations have revealed interindividual variations in anatomy, metabolism, activity, and connectivity of specific cortical association areas through which years of education (YoE), as a common proxy of cognitive reserve, may operate in the face of age- or pathology-associated brain changes. However, the associated molecular properties of YoE-related brain regions and the biological pathways involved remain poorly understood. In the present study we first identified brain areas that showed an association between cortical thickness and YoE among 122 cognitively healthy older human individuals (87 female). We subsequently characterized molecular properties of these regions by studying brain-wide microarray measurements of regional gene expression. In accordance with previous studies, we observed that YoE were associated with higher cortical thickness in medial prefrontal, anterior cingulate, and orbitofrontal areas. Compared with the rest of the cortex, these regions exhibited a distinct gene expression profile characterized by relative upregulation of gene sets implicated in ionotropic and metabotropic neurotransmission as well as activation of immune response. Our genome-wide expression profile analysis of YoE-related brain regions points to distinct molecular pathways that may underlie a higher capacity for plastic changes in response to lifetime intellectual enrichment and potentially also a higher resilience to age-related pathologic brain changes.
SIGNIFICANCE STATEMENT We combined a neuroimaging-based analysis with a transcriptome-wide gene expression approach to investigate the molecular-functional properties of cortical regions associated with educational attainment, as a commonly used proxy for cognitive reserve, in older individuals. The strongest association with education was observed in specific areas of the medial prefrontal cortex, and these areas exhibited a distinct gene expression profile characterized by relative upregulation of gene sets implicated in neurotransmission and immune responses. These findings complement previous neuroimaging studies in the field and point to novel biological pathways that may mediate the beneficial effects of high educational attainment on adaptability to cope with, or prevent, age-related brain changes. The identified genes and pathways now warrant further exploration in mechanistic studies.
Keywords: cognitive reserve, cortical thickness, gene expression, immune response, synaptic transmission
Introduction
The concept of cognitive reserve (CR), most commonly estimated through the use of “proxies” such as years of education (YoE; Nucci et al., 2012), addresses interindividual differences in the adaptability and susceptibility of cognitive abilities or day-to-day function to brain aging, pathology, or insult (Stern et al., 2018b). Neuroimaging studies of cognitively normal older individuals have revealed positive associations between CR proxies and measures of brain integrity (i.e., regional volumes or cortical thickness; for review, see Bartrés-Faz and Arenaza-Urquijo, 2011). These associations are primarily observed in limbic, paralimbic, and heteromodal cortical association areas, particularly within the prefrontal cortex, and have typically been interpreted as reflecting a higher capacity for plastic change, which may increase the adaptability of higher-educated individuals to age- or disease-related brain changes (Valenzuela et al., 2008; Foubert-Samier et al., 2012; Arenaza-Urquijo et al., 2013, 2017; Lee et al., 2016; Vaqué-Alcázar et al., 2017). This interpretation is corroborated by functional imaging studies that have demonstrated differential patterns of brain activation in these latter regions that are suggestive of greater neural efficiency and capacity in higher-educated individuals (Bartrés-Faz et al., 2009; Solé-Padullés et al., 2009; Fernández-Cabello et al., 2016; Stern et al., 2018a) and may reflect greater capacity for plastic change associated to the CR concept (Bartrés-Faz and Arenaza-Urquijo, 2011).
The involvement of distinct brain areas in mediating the positive effects of CR is likely to be related to their specific anatomo-functional properties. For example, although the prefrontal cortex is highly affected by the aging process in terms of brain atrophy (Hedden and Gabrieli, 2004; Fjell et al., 2014), the functional reorganization capacities of both its lateral and medial sections have been related to maintenance of cognitive function (Cabeza et al., 2002; Davis et al., 2008). Also, parts of the lateral prefrontal cortex exert a regulatory role on the functional organization of other networks strongly involved in cognitive processes (Chen et al., 2013; Spreng et al., 2013; Franzmeier et al., 2017, 2018). Within the medial parts of the frontal lobe, the anterior cingulate cortex (ACC) has been one of the most frequently involved brain areas in previous CR studies (see references in the previous paragraph), and this paralimbic region is known to exert an integrative role between limbic and associative cortices, promoting goal-directed behaviors and executive cognitive processes (Devinsky et al., 1995; Carter et al., 1999; Ridderinkhof et al., 2004). At the cellular level, both the ACC region and the dorsolateral prefrontal cortex (Fajardo et al., 2008) contain a characteristic type of large spindle-shaped neuron called von Economo neurons (Allman et al., 2011; Butti et al., 2013) that have been linked to superior memory capacity in old age (Gefen et al., 2015).
Although previous neuroimaging investigations identified heteromodal and paralimbic brain regions and associated networks as likely substrates of the “neural implementation” of CR, our understanding of the associated biological pathways through which they may operate to sustain the mechanisms predicated by the CR theory (i.e., efficiency, capacity, flexibility, and compensation; Stern et al., 2018b) are poorly understood. Former investigations have shown that differential gene expression profiles across brain regions are tightly associated with differences in morphometric (Romero-Garcia et al., 2018b; Seidlitz et al., 2018; Shin et al., 2018) and functional (Hawrylycz et al., 2015; Richiardi et al., 2015; Wang et al., 2015) brain tissue characteristics, as well as differential vulnerability to disease (Romme et al., 2017; Grothe et al., 2018; Romero-Garcia et al., 2018a). Here we aimed to further delineate molecular-functional properties of selective cortical regions associated to YoE among cognitively healthy older individuals. First, through an magnetic resonance image (MRI)-based cortical thickness analysis, we identified brain regions associated to high versus low educational attainment. Subsequently we investigated the transcriptional architecture of these regions using brain-wide regional gene expression data (Hawrylycz et al., 2012) in combination with gene set enrichment analysis (Subramanian et al., 2005; Grothe et al., 2018).
Materials and Methods
Participants.
One hundred and twenty-two normal functioning older human participants (87 female, 35 male, mean age 68.2 years) were enrolled from the Fundació Institut Català de l'Envelliment. All volunteers had normal cognitive function, with Mini-Mental State Examination test (MMSE) scores ≥25 and performances ≥1.5 SD according to normative scores in a neuropsychological assessment (i.e., they did not fulfill cognitive criteria for mild cognitive impairment; Petersen and Morris, 2005). The study was approved by the local ethics committee and was conducted in accordance with the Helsinki Declaration. Written informed consent was obtained from each participant before enrollment in the study.
Experimental design.
For each subject we recorded the total years of completed formal education and stratified the sample according to high or low education using a cutoff point of 15 years of education as in our previous report (Vaqué-Alcázar et al., 2017). We used this cutoff on the basis of a recent study examining which kind of information derived from educational measures was most closely related to a lower risk of dementia and concluding that the stratification of subjects into high versus low educational groups (i.e., tertiary vs non-tertiary education) showed the strongest associations (Then et al., 2016). A further reason to use this dichotomous stratification of our sample rather than a continuous approach was based on the frequency distribution of educational years. Hence, in accordance with the National Spanish education stages at the time our older participants completed the education, educational years were bimodally distributed, with a concentration of cases ∼8 years of completed education and a second major grouping at 15 years. Using the dichotomous classification with a cutoff point of 15 years, 48 participants were classified as high-educated and 74 as low-educated (see group comparisons in Results).
Neuropsychological assessment.
Applying a similar approach used in previous studies (Vidal-Piñeiro et al., 2014; Vaqué-Alcázar et al., 2017) we used principal components analysis (PCA) based on the dataset of the current study to create a composite scale representing separate declarative memory and “frontal lobe function” domains. The following cognitive tests were used to estimate the memory factor: Rey Auditory Verbal Learning Test total learning and delayed recall (RAVLT-total and RAVLT-delayed, N = 94 cases) and Buschke test (total and delayed scores, N = 28 cases). Previously, the total learning and delayed scores from both memory tests were transformed to a comparable metric by the percentage of maximum possible (“POMP”) method (Moeller, 2015). On the other hand, the tests contributing to the frontal lobe function domain included the Trail Making Tests (TMT B-A), the Symbol Digit Modalities Test, and the phonemic (sum of letters F, A, S, 1 min each, N = 94; and letters P, M, R, N = 28) and semantic (animals, 1 min) fluency tests. Both factors were calculated in a way that higher scores on the factor indicated better cognitive performances.
MRI acquisition.
MRI for each participant were acquired in a Siemens Magnetom Trio Tim syngo 3-T system. High-resolution T1-weighted structural images were obtained with a magnetization-prepared rapid acquisition gradient echo 3-dimensional protocol (repetition time = 2300 ms, echo time = 3 ms, inversion time = 900 ms, field-of-view = 244 mm, and 1 mm isotropic voxel).
Cortical thickness analyses.
Cortical surface reconstruction and calculation of cortical thickness (CTh) from the structural T1-weighted images were performed using FreeSurfer v5.1 software package (http://surfer.nmr.mgh.harvard.edu). In summary, the procedures performed by the main FreeSurfer pipeline include removal of non-brain data, intensity normalization (Sled et al., 1998), tessellation of the gray matter/white matter boundary, automated topology correction (Ségonne et al., 2007), and accurate surface deformation to identify tissue borders (Dale et al., 1999). CTh is then calculated as the distance between the white and gray matter surfaces at each vertex of the reconstructed cortical mantle (Fischl and Dale, 2000). Individual results were inspected visually to ensure accuracy of registration, skull stripping, segmentation, and cortical surface reconstruction. Manual editing of the reconstructed surfaces was required in 15 subjects and the threshold used for brain extraction was changed in 31 other subjects. The CTh maps were smoothed using a Gaussian kernel of 15 mm full-width at half-maximum. We then evaluated the statistical analysis using voxelwise general linear models (GLMs) to study group differences in CTh between high and low YoE subjects, adjusted by age and gender (Vaqué-Alcázar et al., 2017). Image was corrected for familywise error using a Monte Carlo null-Z simulation, with 10,000 repetitions. Both the initial vertex and the cluster thresholds were set to p < 0.05.
Statistical analysis of group differences in sociodemographic and cognitive characteristics.
Data analyses were performed using SPSS v24 (IBM). Demographic and cognitive data were described as mean ± SD. For quantitative data, we evaluated differences between groups using two-sample t tests. For categorical variables, we assessed χ2 tests. In addition, when the normal distribution assumptions were not met as indicated by the Shapiro–Wilk test, the Mann–Whitney's U test was used. Finally, because age and gender showed differences between the YoE groups (see results section) the MMSE scores and the final cognitive factors obtained from the PCA analyses (see Neuropsychological assessment) were regressed by age and gender. Because there were differences in the type of memory and verbal fluency tests administered between individuals, this “group test” variable was also introduced as a regressor to obtain the final memory and frontal lobe domain values. For all the analyses results were considered statistically significant at p < 0.05.
Cortical gene expression data.
The transcriptome data from the Allen Institute Human Brain Atlas (http://human.brain-map.org/; Hawrylycz et al., 2012, 2015) was used in the present study. In brief, complete microarray gene expression datasets from a total of 3702 regional tissue samples from postmortem brain tissue of six individuals were mapped to anatomical brain locations in corresponding structural MRI scans; these steps were performed by the Allen Institute (Shen et al., 2012). The produced raw expression values were then mean averaged across probes to obtain single expression values for each gene, resulting in 20,737 gene expression values per tissue sample. From the 3702 brain samples available in the atlas, we first discarded samples not belonging to the cortex according to their anatomical annotation (N = 1752). Further, because of the lack of right hemisphere gene expression data for most of the donors (Hawrylycz et al., 2012), we only focused on the left hemisphere in further analyses, leaving a total of 1452 cortical tissue samples. Next, for each donor the individual tissue sample coordinates were spatially mapped to a cortical surface reconstruction obtained by processing the donors' MRI images in FreeSurfer (data available from Romero-Garcia et al., 2018b). The YoE-related region-of-interest (ROI) together with a reference ROI covering the rest of the cortex were then mapped from the FreeSurfer template space used for cortical thickness analysis to the individual surface reconstruction of each donor's brain. The surface-based ROIs of each donor's brain were then transformed into volumetric parcellations and extended 2 mm into the subjacent white matter to account for possible registration misalignments. Individual tissue samples were then mapped to their nearest point on the cortex, resulting in a total of 44 tissue samples being mapped to the YoE-related ROI and 1356 samples to the cortical reference region. Fifty-two of the 1452 initial cortical samples (3.6%) were discarded because their distance to the nearest cortical voxel was larger than 2 mm. Finally, for each ROI the expression values were averaged across all tissue samples to obtain median expression values for each gene (French and Paus, 2015; Romero-Garcia et al., 2018b).
Gene expression profile of YoE-related brain regions and gene set enrichment analysis.
After identification of brain regions with higher cortical thickness in the higher-education group (YoE-related areas; see Results), we quantified the difference in gene expression between these areas and the background expression levels in the rest of the cortex, calculating a delta-score for each gene (Freer et al., 2016):
here, for each gene (g) the median gene expression within the YoE-related brain area (E′g,YoE) was contrasted to the median expression in the rest of the cortex (E′g,rest). In an additional sensitivity analysis we calculated delta scores using an alternative reference region that excluded all cortical regions with positive (i.e., high > low YoE) β estimates in the GLM assessing cortical thickness differences between high and low YoE groups, regardless of their statistical significance. This spatially reduced reference region covered a total of 814 tissue samples with gene expression data (compared with the 1354 samples in the original reference region covering the entire rest of the cortex). The rationale behind this sensitivity analysis is that a reference region based on the entire rest of the cortex may include gene expression signal from some regions that show marginal, subthreshold associations with YoE, thus possibly blurring gene expression differences of YoE-related areas with cortical background levels.
The resulting delta scores were then ranked in descending order, where the top (positive values) and bottom parts (negative values) of the ranked list contain the genes with relative overexpression or underexpression, respectively, in YoE-related areas compared with the rest of the cortex. Instead of examining single genes at the extremes of the ranked list, we then used gene set enrichment analysis (GSEA; http://software.broadinstitute.org/gsea/index.jsp, software version 3.0; Subramanian et al., 2005) to more broadly explore differentially expressed functional pathways. Because to our knowledge this represents the first study using the present approach in this specific field, we used the GSEA approach to characterize molecular properties of the YoE-related brain regions in an exploratory manner. Instead of focusing on one or a few predefined genes based on a priori hypotheses, the enrichment analysis helps to uncover general trends in differential gene expression in a more comprehensive manner, while extracting meaningful and interpretable information from the high-throughput microarray data. To identify functional gene sets that are differentially expressed in YoE-related regions in reference to cortical background levels, GSEA determines whether the genes of prespecified functional gene sets, derived from curated gene set databases (see next paragraph), cluster toward one of the extremes (top or bottom) of the ranked list. Using this location information an enrichment score (ES) is calculated for each gene set, reflecting the degree of clustering of the gene set's genes toward the top (positively enriched/overexpressed) or bottom (negatively enriched/underexpressed) of the ranked list. A gene set nominal p value is then created by comparing the ES with a null-distribution obtained after permuting 1000 times the gene set's gene positions in the list and recomputing a new ES at each permutation. To adjust the estimated significance level and account for the independent testing of multiple gene sets, the ES are normalized by the size of the set, obtaining a normalized ES (NES). We then controlled the proportion of false-positives by calculating the false discovery rate (FDR) for every NES (Subramanian et al., 2005).
To increase the power and coverage of our analyses, we included gene sets from multiple independent sources. The gene sets are defined by the common implication of genes in particular biological states or processes, and are retrieved from a reviewed, curated, and annotated repository [the Molecular Signatures Database (MSigDB) v6.0; http://software.broadinstitute.org/gsea/msigdb/index.jsp; Liberzon et al., 2011, 2015]. In the present study, we explored a total of 5429 gene sets, including 497 curated and peer-reviewed gene sets of functional pathways derived from the Reactome database (http://reactome.org/), 177 from the Kyoto Encyclopedia of Genes and Genomes (https://www.genome.jp/kegg/), 50 from the MSigDB hallmark collection (Liberzon et al., 2015), 147 from the BioCarta repository (http://cgap.nci.nih.gov/Pathways/BioCarta_Pathways), and 4558 gene sets that group genes annotated by the same gene ontology term (http://www.geneontology.org/).
Given that statistical inference in GSEA is based on complete functional gene sets, not all of the genes included in a significant gene set are necessarily differentially expressed. A so called leading-edge subset of genes can be identified that primarily accounts for a gene set's enrichment score, and a leading-edge analysis then aims to find commonalities among the identified gene sets by clustering the respective leading-edge subsets (Subramanian et al., 2005). To account for the existing redundancy between the different gene sets and to facilitate interpretation of the enrichment results, functionally coherent gene sets were detected using a clusterization algorithm (Merico et al., 2010; Isserlin et al., 2014), and organized into a network layout (“enrichment map”). In this network, nodes represent gene sets, and their edges or connections represent the genetic overlap (i.e., number of overlapping genes) between pairs of nodes. Each resulting cluster in the network indicates functionally related gene sets implicated in the same overarching biological pathway. The network was created with Enrichment Map v2.0.1 (http://www.baderlab.org/Software/EnrichmentMap) using the GSEA leading-edge subsets as input (FDR p < 0.05; combined Jaccard and overlap coefficients cutoff = 0.5; Fig. 1).
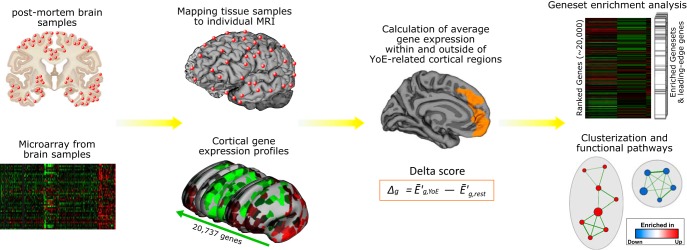
Figure 1.
Enrichment analysis workflow. Outline of the processing of gene expression data from regional probe level to enrichment map for the identification of functionally coherent clusters of gene sets. First, regions associated with YoE in healthy older adults are identified. Then, individual tissue sample coordinates are assigned to the nearest point of the reconstructed MRI brain surface, and for each gene the median expression levels within and outside YoE-related cortical areas are calculated and subtracted (delta score). Finally, genes are ranked according to their delta score (differential expression within YoE-related areas), and the ranked list is submitted to GSEA to search for significantly over- or under-represented gene sets in YoE-related areas. Identified gene sets are then clustered and organized into a network layout to identify groupings of functionally related gene sets into overarching functional pathways.
Results
Sociodemographic and cognitive differences related to YoE
Table 1 depicts the comparison of high and low YoE groups in terms of sociodemographic and cognitive characteristics. Between-group comparisons revealed that the low-educated individuals were significantly older and there were more low-educated women. There were no age and sex-adjusted MMSE differences between groups, although high YoE older adults outperformed those with low YoE in the composite scores for memory and “frontal lobe” domains. When comparisons within the specific cognitive domains were retested adding the MMSE as a covariate, differences were still observed indicating that the cognitive advantage of higher-educated elders in memory and frontal lobe function is not explained by global cognitive difference.
Table 1.
Demographic and cognitive differences between groups
| High YoE (N = 48) | Low YoE (N = 74) | Group differences | |
|---|---|---|---|
| Age, y | 67.2 ± 3.2 | 68.9 ± 3.6 | U = 2.850 |
| p = 0.005a* | |||
| Gender, F/M | 24/24 | 63/11 | χ2 = 17.568 |
| p < 0.001b* | |||
| YoE | 15.7 ± 1.7 | 9.0 ± 2.6 | U = 9.480 |
| p < 0.001a* | |||
| MMSE | 29.6 ± 0.7 | 29.4 ± 1 | U = 0.656 |
| p = 0.512a | |||
| Memory domain | 0.39 ± 0.94 | −0.25 ± 0.96 | t = 3.122 |
| p = 0.002c* | |||
| Frontal lobe domain | 0.55 ± 0.66 | −0.30 ± 1.02 | t = 4.427 |
| p < 0.001c* |
Data are presented as mean ± SD. F/M, Female/male. MMSE represent direct values, Memory and Frontal lobe scores were composite factors calculated using PCA. Statistical comparisons reflect the results after these variables were further adjusted by age, gender, and for the test group variable (see Materials and Methods).[1]
ap values were obtained by Mann–Whitney's U test pairwise comparisons (nonparametric data).
bp values were obtained by χ2 test (categorical data).
cp values were obtained by two-sample t test.
*Significant differences.
Identification of YoE-related cortical areas
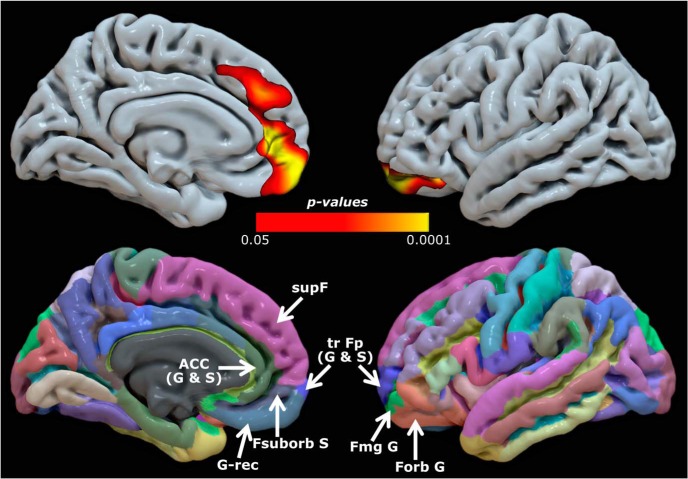
The vertex-wise CTh analysis identified a large cluster where healthy older adults in the high YoE group exhibited greater CTh than those in the low YoE group (left hemisphere only). The cluster was restricted to areas within the frontal lobe, including parts of the dorsomedial prefrontal cortex (medial parts of the superior frontal gyrus) overlapping with Brodmann areas (BAs) 8, 9, and 32, the ACC (BA 24), frontal pole (BA 10), and orbitofrontal cortex (BA 11; Fig. 2). No brain areas reached significant differences in the opposite direction (i.e., greater thickness in low compared with high YoE groups).
Figure 2.
Top, Regions showing greater cortical thickness in healthy older adults with high YoE compared with low YoE individuals. For anatomical reference, overlapping anatomical regions as delineated in the Desikan–Killiany atlas are shown (bottom). G, Gyrus; S, sulcus; trFp, transverse frontopolar; Fmg, fronto-marginal; Forb, frontal orbital (including H-shaped); G-rec, gyrus rectus; Fsuborb, frontal suborbital; supF, superior frontal.
A distinct gene expression profile characteristic of YoE-related areas
GSEA analysis identified 11 gene sets that were positively enriched (FDR < 0.05) in YoE-related areas compared with cortical background levels (Table 2), and the respective enrichment signals were driven by a total of 130 unique leading-edge genes (Table 3).
Table 2.
GSEA
| Gene set | NES | FDR q-val | Size | GSEA ID | URL |
|---|---|---|---|---|---|
| 1. Ionotropic neurotransmission | |||||
| GO_EXTRACELLULAR_LIGAND_GATED_ION_CHANNEL_ACTIVITY | 2.159 | 0.012 | 25 | M18911 | http://amigo.geneontology.org/amigo/term/GO:0005230 |
| GO_EXCITATORY_EXTRACELLULAR_LIGAND_GATED_ION_CHANNEL_ACTIVITY | 2.204 | 0.011 | 28 | M18473 | http://amigo.geneontology.org/amigo/term/GO:0005231 |
| GO_TRANSMITTER_GATED_CHANNEL_ACTIVITY | 1.999 | 0.048 | 29 | M17983 | http://amigo.geneontology.org/amigo/term/GO:0022835 |
| GO_NEUROTRANSMITTER_RECEPTOR_ACTIVITY | 2.049 | 0.031 | 79 | M18363 | http://amigo.geneontology.org/amigo/term/GO:0030594 |
| 2. NEUROPEPTIDE SIGNALING | |||||
| GO_NEUROPEPTIDE_HORMONE_ACTIVITY | 2.057 | 0.033 | 68 | M18214 | http://amigo.geneontology.org/amigo/term/GO:0005184 |
| GO_NEUROPEPTIDE_RECEPTOR_BINDING | 2.120 | 0.012 | 51 | M18007 | http://amigo.geneontology.org/amigo/term/GO:0071855 |
| 3. Transmembrane signaling | |||||
| GO_RESPONSE_TO_AMMONIUM_ION | 2.037 | 0.033 | 26 | M10335 | http://amigo.geneontology.org/amigo/term/GO:0060359 |
| 4. Purinergic signaling | |||||
| GO_PURINERGIC_RECEPTOR_SIGNALING_PATHWAY | 1.991 | 0.049 | 28 | M16435 | http://amigo.geneontology.org/amigo/term/GO:0035587 |
| 5. Immune response | |||||
| GO_MYELOID_DENDRITIC_CELL_ACTIVATION | 2.124 | 0.014 | 55 | M16283 | http://amigo.geneontology.org/amigo/term/GO:0001773 |
| GO_MACROPHAGE_ACTIVATION | 2.133 | 0.016 | 75 | M13410 | http://amigo.geneontology.org/amigo/term/GO:0042116 |
| 6. Vesicle trafficking | |||||
| GO_CLATHRIN_COATED_VESICLE_MEMBRANE | 2.013 | 0.043 | 31 | M17518 | http://amigo.geneontology.org/amigo/term/GO:0030665 |
GSEA results identifying 11 gene sets belonging to 6 larger functional clusters.
Table 3.
Leading-edge genes of positively enriched gene sets in education-related cortical areas
| Gene | Rank in gene list | Delta-score | Functional group |
|---|---|---|---|
| CHRNA1 | 2073 | 0.055 | Ionotropic neurotransmission |
| GABRB3 | 4013 | 0.034 | Ionotropic neurotransmission |
| GABRB1 | 1966 | 0.057 | Ionotropic neurotransmission |
| CHRNA2 | 383 | 0.112 | Ionotropic neurotransmission |
| CHRNA4 | 3822 | 0.035 | Ionotropic neurotransmission |
| GRIK5 | 3748 | 0.036 | Ionotropic neurotransmission |
| CHRNA6 | 30 | 0.228 | Ionotropic neurotransmission |
| GRIK3 | 2188 | 0.053 | Ionotropic neurotransmission |
| GRIK4 | 1311 | 0.069 | Ionotropic neurotransmission |
| GRIK1 | 725 | 0.090 | Ionotropic neurotransmission |
| GRIK2 | 3892 | 0.035 | Ionotropic neurotransmission |
| CHRND | 2309 | 0.051 | Ionotropic neurotransmission |
| GLRA2 | 2534 | 0.049 | Ionotropic neurotransmission |
| GLRA3 | 193 | 0.139 | Ionotropic neurotransmission |
| PTK2B | 1126 | 0.074 | Ionotropic neurotransmission |
| GABRD | 3157 | 0.042 | Ionotropic neurotransmission |
| GRIA4 | 3367 | 0.040 | Ionotropic neurotransmission |
| GABRQ | 1422 | 0.067 | Ionotropic neurotransmission |
| CHRNB2 | 3933 | 0.035 | Ionotropic neurotransmission |
| GRID2 | 3549 | 0.038 | Ionotropic neurotransmission |
| CHRNB3 | 172 | 0.145 | Ionotropic neurotransmission |
| GABRA5 | 2148 | 0.054 | Ionotropic neurotransmission |
| GABRA3 | 2268 | 0.052 | Ionotropic neurotransmission |
| HTR3A | 1185 | 0.072 | Ionotropic neurotransmission |
| HTR3B | 107 | 0.166 | Ionotropic neurotransmission |
| GRIN2C | 886 | 0.083 | Ionotropic neurotransmission |
| GRIN2B | 1247 | 0.071 | Ionotropic neurotransmission |
| GRIN2D | 3196 | 0.041 | Ionotropic neurotransmission |
| GRIN3A | 293 | 0.122 | Ionotropic neurotransmission |
| GRIA3 | 4508 | 0.030 | Ionotropic neurotransmission |
| CHRM3 | 1533 | 0.064 | Ionotropic neurotransmission |
| CHRM1 | 695 | 0.092 | Ionotropic neurotransmission |
| CHRM5 | 841 | 0.084 | Ionotropic neurotransmission |
| HRH3 | 1832 | 0.059 | Ionotropic neurotransmission |
| DRD4 | 679 | 0.092 | Ionotropic neurotransmission |
| UCN | 2726 | 0.046 | Neuropeptide signaling |
| GRP | 61 | 0.194 | Neuropeptide signaling |
| CCK | 327 | 0.118 | Neuropeptide signaling |
| TRH | 1531 | 0.064 | Neuropeptide signaling |
| ADCYAP1 | 723 | 0.090 | Neuropeptide signaling |
| CORT | 2852 | 0.045 | Neuropeptide signaling |
| GAL | 13 | 0.300 | Neuropeptide signaling |
| NPY | 1125 | 0.074 | Neuropeptide signaling |
| PNOC | 98 | 0.170 | Neuropeptide signaling |
| NPPA | 1561 | 0.064 | Neuropeptide signaling |
| CRH | 308 | 0.120 | Neuropeptide signaling |
| VIP | 1573 | 0.064 | Neuropeptide signaling |
| HCRT | 2355 | 0.051 | Neuropeptide signaling |
| EDN1 | 1281 | 0.070 | Neuropeptide signaling |
| CCKBR | 2641 | 0.048 | Neuropeptide signaling |
| GHRH | 847 | 0.084 | Neuropeptide signaling |
| NMU | 27 | 0.235 | Neuropeptide signaling |
| SHANK1 | 746 | 0.089 | Neuropeptide signaling |
| KCNC2 | 1181 | 0.072 | Transmembrane signaling |
| SLC34A1 | 187 | 0.140 | Transmembrane signaling |
| OPRM1 | 255 | 0.127 | Transmembrane signaling |
| ASS1 | 1852 | 0.058 | Transmembrane signaling |
| PPP1R9B | 3268 | 0.041 | Transmembrane signaling |
| CCNA2 | 976 | 0.079 | Transmembrane signaling |
| CRHBP | 1198 | 0.072 | Transmembrane signaling |
| HRH1 | 674 | 0.092 | Transmembrane signaling |
| GNA15 | 3229 | 0.041 | Transmembrane signaling |
| GNAQ | 2989 | 0.043 | Transmembrane signaling |
| GNB1 | 1626 | 0.062 | Transmembrane signaling |
| RGS10 | 910 | 0.082 | Transmembrane signaling |
| RGS8 | 1677 | 0.061 | Transmembrane signaling |
| CDK5R1 | 2769 | 0.046 | Transmembrane signaling |
| P2RY12 | 95 | 0.172 | Purinergic signaling |
| P2RX7 | 1776 | 0.060 | Purinergic signaling |
| P2RY13 | 33 | 0.224 | Purinergic signaling |
| P2RX6 | 263 | 0.126 | Purinergic signaling |
| P2RY6 | 309 | 0.120 | Purinergic signaling |
| P2RX5 | 1255 | 0.071 | Purinergic signaling |
| P2RY11 | 1570 | 0.064 | Purinergic signaling |
| GPR34 | 271 | 0.124 | Purinergic signaling |
| ADORA3 | 155 | 0.148 | Purinergic signaling |
| PTAFR | 961 | 0.080 | Purinergic signaling |
| GNAI2 | 1428 | 0.067 | Purinergic signaling |
| PYCARD | 517 | 0.101 | Immune response |
| BATF3 | 985 | 0.079 | Immune response |
| TGFB1 | 564 | 0.098 | Immune response |
| PYDC1 | 505 | 0.102 | Immune response |
| DOCK2 | 461 | 0.105 | Immune response |
| DHRS2 | 122 | 0.160 | Immune response |
| TM7SF4 | 253 | 0.127 | Immune response |
| CX3CR1 | 269 | 0.125 | Immune response |
| ZAP70 | 545 | 0.099 | Immune response |
| TYROBP | 653 | 0.093 | Immune response |
| SYK | 295 | 0.121 | Immune response |
| CD93 | 777 | 0.088 | Immune response |
| SLC11A1 | 950 | 0.080 | Immune response |
| TLR7 | 1188 | 0.072 | Immune response |
| AIF1 | 96 | 0.171 | Immune response |
| CLU | 1748 | 0.060 | Immune response |
| TLR3 | 994 | 0.078 | Immune response |
| CX3CL1 | 1772 | 0.060 | Immune response |
| SNCA | 1060 | 0.076 | Immune response |
| RAB3A | 2097 | 0.055 | Vesicle trafficking |
| TYRP1 | 328 | 0.118 | Vesicle trafficking |
| CLTA | 3693 | 0.037 | Vesicle trafficking |
| AP2A1 | 1624 | 0.062 | Vesicle trafficking |
| KIAA1199 | 3293 | 0.041 | Vesicle trafficking |
| AP2A2 | 2058 | 0.055 | Vesicle trafficking |
| NCALD | 511 | 0.101 | Vesicle trafficking |
| NRGN | 229 | 0.132 | Vesicle trafficking |
| CLTCL1 | 693 | 0.092 | Vesicle trafficking |
| AP2S1 | 666 | 0.093 | Vesicle trafficking |
| SLC17A7 | 2179 | 0.053 | Vesicle trafficking |
| FCGR1A | 1250 | 0.071 | Vesicle trafficking |
| FCGR1B | 257 | 0.127 | Vesicle trafficking |
| HLA-DQA2 | 2184 | 0.053 | Vesicle trafficking |
| LDLR | 3312 | 0.040 | Vesicle trafficking |
| AP1M2 | 1516 | 0.065 | Vesicle trafficking |
| AP2M1 | 2364 | 0.051 | Vesicle trafficking |
| HLA-DQA1 | 4017 | 0.034 | Vesicle trafficking |
| AP1M1 | 2776 | 0.046 | Vesicle trafficking |
| HLA-DPA1 | 4290 | 0.032 | Vesicle trafficking |
| CD74 | 708 | 0.091 | Vesicle trafficking |
| HLA-DRB5 | 3488 | 0.039 | Vesicle trafficking |
| HLA-DRB4 | 8 | 0.312 | Vesicle trafficking |
| DBNL | 764 | 0.088 | Vesicle trafficking |
| FZD5 | 4176 | 0.033 | Vesicle trafficking |
| SLC32A1 | 4411 | 0.031 | Vesicle trafficking |
| WNT5A | 4491 | 0.031 | Vesicle trafficking |
| AP1B1 | 3964 | 0.035 | Vesicle trafficking |
| AP3B2 | 3468 | 0.039 | Vesicle trafficking |
| HLA-DPB1 | 2465 | 0.050 | Vesicle trafficking |
| HLA-DRA | 62 | 0.193 | Vesicle trafficking |
| CD9 | 3327 | 0.040 | Vesicle trafficking |
| HLA-DQB1 | 239 | 0.130 | Vesicle trafficking |
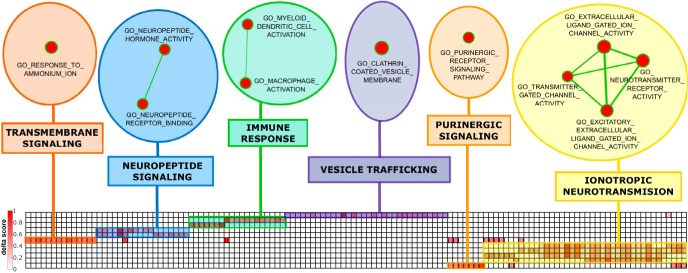
No significantly negatively enriched gene sets were identified. Network organization of the gene sets clustered by their leading-edge subsets (enrichment map) evidenced strongly overlapping leading-edge genes among some of the gene sets, resulting in a total of six different gene sets/clusters (Fig. 3). The largest cluster included four gene sets implicated in biological events related to ionotropic neurotransmission, and the large majority of the upregulated leading-edge genes corresponded to genes coding for components of several different ligand-gated ion channels, including those receptive for glutamate (kainate, AMPA, and NMDA receptors), GABA (GABAA), glycine, acetylcholine (nicotinic receptors), and serotonin (5-HT3). Other clusters related to neurotransmission included gene sets implicated in neuropeptide (2 gene sets) and G-protein-coupled transmembrane signaling (1 gene set), as well as purinergic signaling (1 gene set). A different category of overexpressed gene sets was represented by a cluster related to immune response (2 gene sets), including several leading-edge genes related to pathogen recognition (e.g., TLR family), cytokine signaling, and immune activation (e.g., TGFB1, AIF1, PYCARD), as well as a closely related gene set implicated in vesicle trafficking, which mainly contained leading-edge genes coding for major histocompatibility complex (MHC) molecules and adaptor complex proteins (AP1–3). Complementary GSEA analysis of delta scores calculated with the alternative reference region revealed identical results.
Figure 3.
Heat map of the clustered leading edge genes of the positively enriched gene sets. Rows correspond to the gene sets listed in Table 2 and columns correspond to their respective leading-edge gene as detailed in Table 3. Color coding from white to dark red reflects degree of relative upregulation (delta score). The upper part shows the enrichment map of the clustered gene sets, containing six different clusters representing the overarching functional pathways. Node size represents the gene set size and line thickness shows the degree of overlap (shared genes) between the two gene sets it connects.
Discussion
The observed association between YoE and higher cortical thickness in medial prefrontal, ACC, and orbitofrontal cortices provides further converging evidence to other neuroimaging studies conducted within the CR model (Arenaza-Urquijo et al., 2013; Kim et al., 2015; Lee et al., 2016; Boller et al., 2017; Vaqué-Alcázar et al., 2017). The relevance of distinct frontal lobe areas as mediators of CR mechanisms is probably related to their key role in cognitive processes such as working memory (Owen et al., 2005; Barbey et al., 2011), cognitive conflict monitoring (Jahn et al., 2016), or decision-making (Shenhav et al., 2013). We further observed that these cortical areas exhibited a distinct gene expression profile characterized by relative upregulation of gene sets implicated in neurotransmission and immune response.
Our pathway analyses identified several gene sets involved in cell communication and neurotransmission processes. The majority of the upregulated leading-edge genes encode for major ionotropic glutamate receptors (i.e., GRIK, GRIA, and GRIN gene families), nicotinic acetylcholine receptors (CHRN gene family), as well as GABAA receptor subunits (GABR gene family), suggesting an important role for these brain areas in fast excitatory and inhibitory signaling. This molecular characteristic corresponds to the well described role of these medial prefrontal areas as highly active, functionally connected network hubs subserving transmodal information integration (Buckner et al., 2009; Sepulcre et al., 2012; Braga et al., 2013). In addition, neurotransmitter signaling through calcium-permeable ion channels, particularly glutamatergic AMPA and NMDA receptors as well as nicotinic acetylcholine receptors, is considered to be one of the most important molecular features of postsynaptic plasticity (Cull-Candy et al., 2006; Anggono and Huganir, 2012; Pankratov and Lalo, 2014).
In addition to ionotropic neurotransmission, we also identified several other upregulated gene set families involved in G-protein-coupled transmembrane signaling through neuropeptide and purinergic receptor binding. Beyond their role in homeostatic and neuroendocrine processes, neuropeptide/G-coupled receptor signaling is increasingly recognized as a mediator of activity-dependent refinement of local brain circuits and plasticity (McClard and Arenkiel, 2018), including important roles for learning and memory (Gøtzsche and Woldbye, 2016). Postmortem autopsy studies (Riudavets et al., 2007; Iacono et al., 2009), revealed that subjects with considerable AD pathology but preserved cognition exhibited neuronal hypertrophy in specific brain areas including the ACC. Such changes were interpreted as evidence of morphological plasticity in response to the incipient neuropathological changes of AD, suggesting that modulation of synaptic transmission and associated plasticity may reflect a neurobiological mechanism of resilience to cope with pathology and maintain cognition, which is the core concept of the CR theory.
A second category of gene sets that were found to be upregulated in the examined YoE-related areas was related to immune processes. This may reflect a particular sensitivity of the identified cortical regions for microglial activation (e.g., MHC-II molecules are frequently used as markers of microglial activation; Hopperton et al., 2018). In this sense, imaging-based PET studies have demonstrated region-specific activation of microglia in normal aging, including the frontal lobe and ACC regions (Schuitemaker et al., 2012). Although excessive microglial hyperactivation seems to contribute to the susceptibility of cognitive deficits (Kohman, 2012), transiently activated microglia also play an important role in the clearance and degradation of misfolded protein aggregates associated with neurodegenerative disease, such as the extracellular Aβ fibrils characteristic for Alzheimer's disease (Cai et al., 2014; Cho et al., 2014). Recent PET imaging studies have indicated that high education and lifelong intellectual enrichment may result in attenuation of age-related Aβ aggregation in medial prefrontal cortical regions such as the ACC (Landau et al., 2012) and orbitofrontal areas (Arenaza-Urquijo et al., 2017), particularly in those individuals at risk for AD (Wirth et al., 2014). Thus, together with these previous findings our present results could indicate that such education-related Aβ suppression effects may be primarily observed in these brain areas partially because they are characterized by a high expression of microglial activity genes.
Regarding the characteristic overexpression of both microglial activation and neurotransmission pathways, it is interesting to note that purinergic signaling has been implicated in excitatory neurotransmission as well as in neuronal–glial communication, inflammation regulation, and phagocytotic activity of microglia (North and Verkhratsky, 2006), including clearance of Aβ fibrils (Kim et al., 2012; Erb et al., 2018).
In summary, our results suggest that the gene expression profile and enriched biological pathways of the prefrontal regions identified here may entail fundamental mechanisms to account for why these brain areas represent key regions mediating the positive physiological and cognitive effects of high educational attainment. One possibility is that because of their particular molecular characteristics, lifetime exposure to CR proxies such as YoE may lead to specific changes in the anatomy and activity of these regions over other cortical areas, possibly explaining the frequently reported increases of gray matter or cortical thickness measures in these areas in elders with high CR ratings. On the other hand, enrichment for biological pathways implicated in immune response and microglial activity would align with evidence for a protective effect of lifetime exposure to enriched cognitive activity against pathology accumulation in these regions (Landau et al., 2012; Arenaza-Urquijo et al., 2017).
The present study is not without limitations. A first constraint refers to the lack of biomarker information to characterize our sample of older adults. According to population-based studies (Jansen et al., 2015; Toledo et al., 2015) ∼20% of individuals with normal cognition and a mean age comparable to our older adult individuals are likely to harbor significant brain pathology (i.e., amyloid-β or tau deposition). Furthermore, because our main inclusion criterion was normal cognitive function, these percentages may be expected to be even higher among highly educated elders, as preserved cognition in the face of brain pathology is likely to be more frequent among high CR elders. Therefore, our current approach does not allow discerning the degree to which the detected associations between education and cortical thickness may be specific to normal aging or confounded by coexisting brain pathology.
A further restriction of our study is that the cross-sectional structural MRI analysis does not provide specific information regarding the possible mechanisms underlying the observed education/cortical thickness relationship. Thus, these relations could either reflect a higher capacity for plastic brain changes related to efficiency or compensation mechanisms of CR (for review, see Bartrés-Faz and Arenaza-Urquijo, 2011), or they could reflect brain maintenance associated to less age-related cortical thickness loss (Nyberg et al., 2012). Although CR and brain maintenance are two interrelated and not mutually exclusive concepts (Stern et al., 2018b), both being putatively related to brain plasticity mechanisms (Bartrés-Faz and Arenaza-Urquijo, 2011; Cabeza et al., 2018), disentangling their relative contributions would typically require a longitudinal study design (or comparison with young brains' characteristics; Nyberg et al., 2012; Cabeza et al., 2018). The detailed characterization of neuronal systems underlying CR and maintenance effects is an ongoing and highly active area of research (Stern et al., 2018a). In future studies the inclusion of other CR proxies or lifestyle-based measures (IQ, occupation, leisure-time, physical activity), cognitively impaired individuals (e.g., patients with AD or MCI), and functional imaging modalities, together using distinct approaches such as the residual (van Loenhoud et al., 2017) or those based on physiological age estimations (Steffener et al., 2016) will help to further characterize detailed CR-related brain networks, possibly also disentangling regionally diverging implications in compensation/efficiency and maintenance mechanisms.
Finally, our study is limited by the available data for estimating region-specific cortical gene expression profiles. Although the used gene expression data from the Allen Brain Atlas provides the anatomically most comprehensive transcriptome data for the human brain available to date, the data were derived from only six brain donors. Furthermore, this gene expression dataset was obtained from postmortem specimens of relatively young subjects (mean age 42.5 at death; Hawrylycz et al., 2012, 2015; Shen et al., 2012) with no educational background information provided. We acknowledge that these aspects may pose a bias when interpreting the findings based on brain regions identified from older participants, given that cortical gene expression values are known to change across the lifespan (Naumova et al. 2013).
In conclusion, to our knowledge, this study represents the first attempt to characterize the transcriptome-wide gene expression profile of YoE-related brain areas. Our results point to a distinctive enrichment with certain biologic pathways that may equip these regions with a higher capacity for plastic change in response to lifetime intellectual enrichment and potentially also a higher resilience to age-related pathologic brain changes. Further research is needed to advance our mechanistic understanding of the biological pathways through which cognitive and brain reserve operate to preserve cognition and day-to-day functionality among older adults.
Footnotes
This work was supported by a Spanish Ministry of Economy and Competitiveness (MINECO) grant to D-BF [grant number PSI2015-64227-R (AEI/FEDER, UE)]; by the Walnuts and Healthy Aging study (http://www.clinicaltrials.gov; Grant NCT01634841) funded by the California Walnut Commission, Sacramento, California; by a Grant from the Alzheimer Forschung Initiative (16037) to M.J.G.; and by the German Research Foundation; (Grants SFB-1193 and SFB/CRC-TR-128) to G.G.-E.
The authors declare no competing financial interests.
References
- Allman JM, Tetreault NA, Hakeem AY, Manaye KF, Semendeferi K, Erwin JM, Park S, Goubert V, Hof PR (2011) The von economo neurons in the frontoinsular and anterior cingulate cortex. Ann N Y Acad Sci 1225:59–71. 10.1111/j.1749-6632.2011.06011.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anggono V, Huganir RL (2012) Regulation of AMPA receptor trafficking and synaptic plasticity. Curr Opin Neurobiol 22:461–469. 10.1016/j.conb.2011.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenaza-Urquijo EM, Molinuevo JL, Sala-Llonch R, Solé-Padulles C, Balasa M, Bosch B, Olives J, Antonell A, Lladó A, Sánchez-Valle R, Rami L, Bartrés-Faz D (2013) Cognitive reserve proxies relate to gray matter loss in cognitively healthy elderly with abnormal cerebrospinal fluid amyloid-beta levels. J Alzheimers Dis 35:715–726. 10.3233/JAD-121906 [DOI] [PubMed] [Google Scholar]
- Arenaza-Urquijo EM, Bejanin A, Gonneaud J, Wirth M, La Joie R, Mutlu J, Gaubert M, Landeau B, de la Sayette V, Eustache F, Chételat G (2017) Association between educational attainment and amyloid deposition across the spectrum from normal cognition to dementia: neuroimaging evidence for protection and compensation. Neurobiol Aging 59:72–79. 10.1016/j.neurobiolaging.2017.06.016 [DOI] [PubMed] [Google Scholar]
- Barbey AK, Koenigs M, Grafman J (2011) Orbitofrontal contributions to human working memory. Cereb Cortex 21:789–795. 10.1093/cercor/bhq153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartrés-Faz D, Arenaza-Urquijo EM (2011) Structural and functional imaging correlates of cognitive and brain reserve hypotheses in healthy and pathological aging. Brain Topogr 24:340–357. 10.1007/s10548-011-0195-9 [DOI] [PubMed] [Google Scholar]
- Bartrés-Faz D, Solé-Padullés C, Junqué C, Rami L, Bosch B, Bargalló N, Falcón C, Sánchez-Valle R, Molinuevo JL (2009) Interactions of cognitive reserve with regional brain anatomy and brain function during a working memory task in healthy elders. Biol Psychol 80:256–259. 10.1016/j.biopsycho.2008.10.005 [DOI] [PubMed] [Google Scholar]
- Boller B, Mellah S, Ducharme-Laliberté G, Belleville S (2017) Relationships between years of education, regional grey matter volumes, and working memory-related brain activity in healthy older adults. Brain Imaging Behav 11:304–317. 10.1007/s11682-016-9621-7 [DOI] [PubMed] [Google Scholar]
- Braga RM, Sharp DJ, Leeson C, Wise RJ, Leech R (2013) Echoes of the brain within default mode, association, and heteromodal cortices. J Neurosci 33:14031–14039. 10.1523/JNEUROSCI.0570-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, Andrews-Hanna JR, Sperling RA, Johnson KA (2009) Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer's disease. J Neurosci 29:1860–1873. 10.1523/JNEUROSCI.5062-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butti C, Santos M, Uppal N, Hof PR (2013) Von economo neurons: clinical and evolutionary perspectives. Cortex 49:312–326. 10.1016/j.cortex.2011.10.004 [DOI] [PubMed] [Google Scholar]
- Cabeza R, Anderson ND, Locantore JK, McIntosh AR (2002) Aging gracefully: compensatory brain activity in high-performing older adults. Neuroimage 17:1394–1402. 10.1006/nimg.2002.1280 [DOI] [PubMed] [Google Scholar]
- Cabeza R, Albert M, Belleville S, Craik FIM, Duarte A, Grady CL, Lindenberger U, Nyberg L, Park DC, Reuter-Lorenz PA, Rugg MD, Steffener J, Rajah MN (2018) Maintenance, reserve and compensation: the cognitive neuroscience of healthy ageing. Nat Rev Neurosci 19:701–710. 10.1038/s41583-018-0068-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Hussain MD, Yan LJ (2014) Microglia, neuroinflammation, and beta-amyloid protein in Alzheimer's disease. Int J Neurosci 124:307–321. 10.3109/00207454.2013.833510 [DOI] [PubMed] [Google Scholar]
- Carter CS, Botvinick MM, Cohen JD (1999) The contribution of the anterior cingulate cortex to executive processes in cognition. Rev Neurosci 10:49–57. [DOI] [PubMed] [Google Scholar]
- Chen AC, Oathes DJ, Chang C, Bradley T, Zhou ZW, Williams LM, Glover GH, Deisseroth K, Etkin A (2013) Causal interactions between fronto-parietal central executive and default-mode networks in humans. Proc Natl Acad Sci U S A 110:19944–19949. 10.1073/pnas.1311772110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MH, Cho K, Kang HJ, Jeon EY, Kim HS, Kwon HJ, Kim HM, Kim DH, Yoon SY (2014) Autophagy in microglia degrades extracellular β-amyloid fibrils and regulates the NLRP3 inflammasome. Autophagy 10:1761–1775. 10.4161/auto.29647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cull-Candy S, Kelly L, Farrant M (2006) Regulation of Ca2+-permeable AMPA receptors: synaptic plasticity and beyond. Curr Opin Neurobiol 16:288–297. 10.1016/j.conb.2006.05.012 [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI (1999) Cortical surface-based analysis: I. Segmentation and surface reconstruction. Neuroimage 9:179–194. 10.1006/nimg.1998.0395 [DOI] [PubMed] [Google Scholar]
- Davis SW, Dennis NA, Daselaar SM, Fleck MS, Cabeza R (2008) Que PASA? The posterior-anterior shift in aging. Cereb Cortex 18:1201–1209. 10.1093/cercor/bhm155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devinsky O, Morrell MJ, Vogt BA (1995) Contributions of anterior cingulate cortex to behaviour. Brain 118:279–306. 10.1093/brain/118.1.279 [DOI] [PubMed] [Google Scholar]
- Erb L, Woods LT, Khalafalla MG, Weisman GA (2018) Purinergic signaling in Alzheimer's disease. Brain Res Bull. Advance online publication. Retrieved November 22, 2018. doi: 10.1016/j.brainresbull.2018.10.014. [DOI] [PubMed] [Google Scholar]
- Fajardo C, Escobar MI, Buriticá E, Arteaga G, Umbarila J, Casanova MF, Pimienta H (2008) Von economo neurons are present in the dorsolateral (dysgranular) prefrontal cortex of humans. Neurosci Lett 435:215–218. 10.1016/j.neulet.2008.02.048 [DOI] [PubMed] [Google Scholar]
- Fernández-Cabello S, Valls-Pedret C, Schurz M, Vidal-Piñeiro D, Sala-Llonch R, Bargallo N, Ros E, Bartrés-Faz D (2016) White matter hyperintensities and cognitive reserve during a working memory task: a functional magnetic resonance imaging study in cognitively normal older adults. Neurobiol Aging 48:23–33. 10.1016/j.neurobiolaging.2016.08.008 [DOI] [PubMed] [Google Scholar]
- Fischl B, Dale AM (2000) Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A 97:11050–11055. 10.1073/pnas.200033797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell AM, McEvoy L, Holland D, Dale AM, Walhovd KB; Alzheimer's Disease Neuroimaging Initiative (2014) What is normal in normal aging? Effects of aging, amyloid and Alzheimer's disease on the cerebral cortex and the hippocampus. Prog Neurobiol 117:20–40. 10.1016/j.pneurobio.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foubert-Samier A, Catheline G, Amieva H, Dilharreguy B, Helmer C, Allard M, Dartigues JF (2012) Education, occupation, leisure activities, and brain reserve: a population-based study. Neurobiol Aging 33:423.e15–25. 10.1016/j.neurobiolaging.2010.09.023 [DOI] [PubMed] [Google Scholar]
- Franzmeier N, Duering M, Weiner M, Dichgans M, Ewers M; Alzheimer's Disease Neuroimaging Initiative (2017) Left frontal cortex connectivity underlies cognitive reserve in prodromal Alzheimer disease. Neurology 88:1054–1061. 10.1212/WNL.0000000000003711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmeier N, Hartmann J, Taylor ANW, Araque-Caballero MÁ, Simon-Vermot L, Kambeitz-Ilankovic L, Bürger K, Catak C, Janowitz D, Müller C, Ertl-Wagner B, Stahl R, Dichgans M, Duering M, Ewers M (2018) The left frontal cortex supports reserve in aging by enhancing functional network efficiency. Alzheimers Res Ther 10:28. 10.1186/s13195-018-0358-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freer R, Sormanni P, Vecchi G, Ciryam P, Dobson CM, Vendruscolo M (2016) A protein homeostasis signature in healthy brains recapitulates tissue vulnerability to Alzheimer's disease. Sci Adv 2:e1600947. 10.1126/sciadv.1600947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French L, Paus T (2015) A FreeSurfer view of the cortical transcriptome generated from the allen human brain atlas. Front Neurosci 9:323. 10.3389/fnins.2015.00323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gefen T, Peterson M, Papastefan ST, Martersteck A, Whitney K, Rademaker A, Bigio EH, Weintraub S, Rogalski E, Mesulam MM, Geula C (2015) Morphometric and histologic substrates of cingulate integrity in elders with exceptional memory capacity. J Neurosci 35:1781–1791. 10.1523/JNEUROSCI.2998-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gøtzsche CR, Woldbye DP (2016) The role of NPY in learning and memory. Neuropeptides 55:79–89. 10.1016/j.npep.2015.09.010 [DOI] [PubMed] [Google Scholar]
- Grothe MJ, Sepulcre J, Gonzalez-Escamilla G, Jelistratova I, Schöll M, Hansson O, Teipel SJ (2018) Molecular properties underlying regional vulnerability to Alzheimer's disease pathology. Brain 141:2755–2771. 10.1093/brain/awy189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, Abajian C, Beckmann CF, Bernard A, Bertagnolli D, Boe AF, Cartagena PM, Chakravarty MM, Chapin M, Chong J, Dalley RA, et al. (2012) An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489:391–399. 10.1038/nature11405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz M, Miller JA, Menon V, Feng D, Dolbeare T, Guillozet-Bongaarts AL, Jegga AG, Aronow BJ, Lee CK, Bernard A, Glasser MF, Dierker DL, Menche J, Szafer A, Collman F, Grange P, Berman KA, Mihalas S, Yao Z, Stewart L, et al. (2015) Canonical genetic signatures of the adult human brain. Nat Neurosci 18:1832–1844. 10.1038/nn.4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden T, Gabrieli JD (2004) Insights into the ageing mind: a view from cognitive neuroscience. Nat Rev Neurosci 5:87–96. 10.1038/nrn1323 [DOI] [PubMed] [Google Scholar]
- Hopperton KE, Mohammad D, Trépanier MO, Giuliano V, Bazinet RP (2018) Markers of microglia in post-mortem brain samples from patients with Alzheimer's disease: a systematic review. Mol Psychiatry 23:177–198. 10.1038/mp.2017.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono D, Markesbery WR, Gross M, Pletnikova O, Rudow G, Zandi P, Troncoso JC (2009) The nun study clinically silent AD, neuronal hypertrophy, and linguistic skills in early life. Neurology 73:665–673. 10.1212/WNL.0b013e3181b01077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isserlin R, Merico D, Voisin V, Bader GD (2014) Enrichment map: a cytoscape app to visualize and explore OMICs pathway enrichment results. F1000Research 3:141. 10.12688/f1000research.4536.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn A, Nee DE, Alexander WH, Brown JW (2016) Distinct regions within medial prefrontal cortex process pain and cognition. J Neurosci 36:12385–12392. 10.1523/JNEUROSCI.2180-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen WJ, Ossenkoppele R, Knol DL, Tijms BM, Scheltens P, Verhey FR, Visser PJ; Amyloid Biomarker Study Group (2015) Prevalence of cerebral amyloid pathology in persons without dementia: a meta-analysis. JAMA 313:1924–1938. 10.1001/jama.2015.4668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Ajit D, Peterson TS, Wang Y, Camden JM, Gibson Wood W, Sun GY, Erb L, Petris M, Weisman GA (2012) Nucleotides released from Abeta(1)(-)(4)(2)-treated microglial cells increase cell migration and Abeta(1)(-)(4)(2) uptake through P2Y(2) receptor activation. J Neurochem 121:228–238. 10.1111/j.1471-4159.2012.07700.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JP, Seo SW, Shin HY, Ye BS, Yang JJ, Kim C, Kang M, Jeon S, Kim HJ, Cho H, Kim JH, Lee JM, Kim ST, Na DL, Guallar E (2015) Effects of education on aging-related cortical thinning among cognitively normal individuals. Neurology 85:806–812. 10.1212/WNL.0000000000001884 [DOI] [PubMed] [Google Scholar]
- Kohman RA. (2012) Aging microglia: relevance to cognition and neural plasticity. Methods Mol Biol 934:193–218. 10.1007/978-1-62703-071-7_11 [DOI] [PubMed] [Google Scholar]
- Landau SM, Marks SM, Mormino EC, Rabinovici GD, Oh H, O'Neil JP, Wilson RS, Jagust WJ (2012) Association of lifetime cognitive engagement and low β-amyloid deposition. Arch Neurol 69:623–629. 10.1001/archneurol.2011.2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Shin HY, Kim HJ, Jang YK, Jung NY, Lee J, Kim YJ, Chun P, Yang JJ, Lee JM, Kang M, Park KC, Na DL, Seo SW (2016) Combined effects of physical exercise and education on age-related cortical thinning in cognitively normal individuals. Sci Rep 6:24284. 10.1038/srep24284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP (2011) Molecular signatures database (MSigDB) 3.0. Bioinformatics 27:1739–1740. 10.1093/bioinformatics/btr260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P (2015) The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst 1:417–425. 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClard CK, Arenkiel BR (2018) Neuropeptide signaling networks and brain circuit plasticity. J Exp Neurosci 12:1179069518779207. 10.1177/1179069518779207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD (2010) Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PloS One 5:e13984. 10.1371/journal.pone.0013984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller J. (2015) A word on standardization in longitudinal studies: don't. Front Psychol 6:1389. 10.3389/fpsyg.2015.01389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova OY, Lee M, Rychkov SY, Vlasova NV, Grigorenko EL (2013) Gene expression in the human brain: the current state of the study of specificity and spatiotemporal dynamics. Child Dev 84:76–88. 10.1111/cdev.12014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North RA, Verkhratsky A (2006) Purinergic transmission in the central nervous system. Pflugers Arch 452:479–485. 10.1007/s00424-006-0060-y [DOI] [PubMed] [Google Scholar]
- Nucci M, Mapelli D, Mondini S (2012) Cognitive reserve index questionnaire (CRIq): a new instrument for measuring cognitive reserve. Aging Clin Exp Res 24:218–226. 10.3275/7800 [DOI] [PubMed] [Google Scholar]
- Nyberg L, Lövdén M, Riklund K, Lindenberger U, Bäckman L (2012) Memory aging and brain maintenance. Trends Cogn Sci 16:292–305. 10.1016/j.tics.2012.04.005 [DOI] [PubMed] [Google Scholar]
- Owen AM, McMillan KM, Laird AR, Bullmore E (2005) N-back working memory paradigm: a meta-analysis of normative functional neuroimaging studies. Hum Brain Mapp 25:46–59. 10.1002/hbm.20131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratov Y, Lalo U (2014) Calcium permeability of ligand-gated Ca2+ channels. Eur J Pharmacol 739:60–73. 10.1016/j.ejphar.2013.11.017 [DOI] [PubMed] [Google Scholar]
- Petersen RC, Morris JC (2005) Mild cognitive impairment as a clinical entity and treatment target. Arch Neurol 62:1160–1163; discussion 1167. 10.1001/archneur.62.7.1160 [DOI] [PubMed] [Google Scholar]
- Richiardi J, Altmann A, Milazzo AC, Chang C, Chakravarty MM, Banaschewski T, Barker GJ, Bokde AL, Bromberg U, Büchel C, Conrod P, Fauth-Bühler M, Flor H, Frouin V, Gallinat J, Garavan H, Gowland P, Heinz A, Lemaître H, Mann KF, et al. (2015) BRAIN NETWORKS. correlated gene expression supports synchronous activity in brain networks. Science 348:1241–1244. 10.1126/science.1255905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridderinkhof KR, Ullsperger M, Crone EA, Nieuwenhuis S (2004) The role of the medial frontal cortex in cognitive control. Science 306:443–447. 10.1126/science.1100301 [DOI] [PubMed] [Google Scholar]
- Riudavets MA, Iacono D, Resnick SM, O'Brien R, Zonderman AB, Martin LJ, Rudow G, Pletnikova O, Troncoso JC (2007) Resistance to Alzheimer's pathology is associated with nuclear hypertrophy in neurons. Neurobiol Aging 28:1484–1492. 10.1016/j.neurobiolaging.2007.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Garcia R, Warrier V, Bullmore ET, Baron-Cohen S, Bethlehem RAI (2018a) Synaptic and transcriptionally downregulated genes are associated with cortical thickness differences in autism. Mol Psychiatry. Advance online publication. Retrieved February 26, 2018. doi: 10.1038/s41380-018-0023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Garcia R, Whitaker KJ, Váša F, Seidlitz J, Shinn M, Fonagy P, Dolan RJ, Jones PB, Goodyer IM, Bullmore ET, Vértes PE (2018b) Structural covariance networks are coupled to expression of genes enriched in supragranular layers of the human cortex. Neuroimage 171:256–267. 10.1016/j.neuroimage.2017.12.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romme IA, de Reus MA, Ophoff RA, Kahn RS, van den Heuvel MP (2017) Connectome disconnectivity and cortical gene expression in patients with schizophrenia. Biol Psychiatry 81:495–502. 10.1016/j.biopsych.2016.07.012 [DOI] [PubMed] [Google Scholar]
- Schuitemaker A, van der Doef TF, Boellaard R, van der Flier WM, Yaqub M, Windhorst AD, Barkhof F, Jonker C, Kloet RW, Lammertsma AA, Scheltens P, van Berckel BN (2012) Microglial activation in healthy aging. Neurobiol Aging 33:1067–1072. 10.1016/j.neurobiolaging.2010.09.016 [DOI] [PubMed] [Google Scholar]
- Ségonne F, Pacheco J, Fischl B (2007) Geometrically accurate topology-correction of cortical surfaces using nonseparating loops. IEEE Trans Med Imaging 26:518–529. 10.1109/TMI.2006.887364 [DOI] [PubMed] [Google Scholar]
- Seidlitz J, Váša F, Shinn M, Romero-Garcia R, Whitaker KJ, Vértes PE, Wagstyl K, Kirkpatrick Reardon P, Clasen L, Liu S, Messinger A, Leopold DA, Fonagy P, Dolan RJ, Jones PB, Goodyer IM, Goodyer IM, Raznahan A, Bullmore ET (2018) Morphometric similarity networks detect microscale cortical organization and predict inter-individual cognitive variation. Neuron 97:231–247.e7. 10.1016/j.neuron.2017.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulcre J, Sabuncu MR, Yeo TB, Liu H, Johnson KA (2012) Stepwise connectivity of the modal cortex reveals the multimodal organization of the human brain. J Neurosci 32:10649–10661. 10.1523/JNEUROSCI.0759-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen EH, Overly CC, Jones AR (2012) The allen human brain atlas: comprehensive gene expression mapping of the human brain. Trends Neurosci 35:711–714. 10.1016/j.tins.2012.09.005 [DOI] [PubMed] [Google Scholar]
- Shenhav A, Botvinick MM, Cohen JD (2013) The expected value of control: an integrative theory of anterior cingulate cortex function. Neuron 79:217–240. 10.1016/j.neuron.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, French L, Xu T, Leonard G, Perron M, Pike GB, Richer L, Veillette S, Pausova Z, Paus T (2018) Cell-specific gene-expression profiles and cortical thickness in the human brain. Cereb Cortex 28:3267–3277. 10.1093/cercor/bhx197 [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC (1998) A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 17:87–97. 10.1109/42.668698 [DOI] [PubMed] [Google Scholar]
- Solé-Padullés C, Bartrés-Faz D, Junqué C, Vendrell P, Rami L, Clemente IC, Bosch B, Villar A, Bargalló N, Jurado MA, Barrios M, Molinuevo JL (2009) Brain structure and function related to cognitive reserve variables in normal aging, mild cognitive impairment and Alzheimer's disease. Neurobiol Aging 30:1114–1124. 10.1016/j.neurobiolaging.2007.10.008 [DOI] [PubMed] [Google Scholar]
- Spreng RN, Sepulcre J, Turner GR, Stevens WD, Schacter DL (2013) Intrinsic architecture underlying the relations among the default, dorsal attention, and frontoparietal control networks of the human brain. J Cogn Neurosci 25:74–86. 10.1162/jocn_a_00281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffener J, Habeck C, O'Shea D, Razlighi Q, Bherer L, Stern Y (2016) Differences between chronological and brain age are related to education and self-reported physical activity. Neurobiol Aging 40:138–144. 10.1016/j.neurobiolaging.2016.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern Y, Gazes Y, Razlighi Q, Steffener J, Habeck C (2018a) A task-invariant cognitive reserve network. Neuroimage 178:36–45. 10.1016/j.neuroimage.2018.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern Y, Arenaza-Urquijo EM, Bartres-Faz D, Belleville S, Cantilon M, Chetelat G, Ewers M, Franzmeier N, Kempermann G, Kremen WS, Okonkwo O, Scarmeas N, Soldan A, Udeh-Momoh C, Valenzuela M, Vemuri P, Vuoksimaa E; Reserve, Resilience and Protective Factors PIA Empirical Definitions and Conceptual Frameworks Workgroup (2018b) Whitepaper: defining and investigating cognitive reserve, brain reserve, and brain maintenance. Alzheimer's Dement. Advance online publication. Retrieved September 14, 2018. doi: 10.1016/j.jalz.2018.07.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Then FS, Luck T, Angermeyer MC, Riedel-Heller SG (2016) Education as protector against dementia, but what exactly do we mean by education? Age Ageing 45:523–528. 10.1093/ageing/afw049 [DOI] [PubMed] [Google Scholar]
- Toledo JB, Zetterberg H, van Harten AC, Glodzik L, Martinez-Lage P, Bocchio-Chiavetto L, Rami L, Hansson O, Sperling R, Engelborghs S, Osorio RS, Vanderstichele H, Vandijck M, Hampel H, Teipl S, Moghekar A, Albert M, Hu WT, Monge Argilés JA, Gorostidi A, et al. (2015) Alzheimer's disease cerebrospinal fluid biomarker in cognitively normal subjects. Brain 138:2701–2715. 10.1093/brain/awv199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela MJ, Sachdev P, Wen W, Chen X, Brodaty H (2008) Lifespan mental activity predicts diminished rate of hippocampal atrophy. PloS One 3:e2598. 10.1371/journal.pone.0002598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loenhoud AC, Wink AM, Groot C, Verfaillie SCJ, Twisk J, Barkhof F, van Berckel B, Scheltens P, van der Flier WM, Ossenkoppele R (2017) A neuroimaging approach to capture cognitive reserve: application to Alzheimer's disease. Hum Brain Mapp 38:4703–4715. 10.1002/hbm.23695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaqué-Alcázar L, Sala-Llonch R, Valls-Pedret C, Vidal-Piñeiro D, Fernández-Cabello S, Bargalló N, Ros E, Bartrés-Faz D (2017) Differential age-related gray and white matter impact mediates educational influence on elders' cognition. Brain Imaging Behav 11:318–332. 10.1007/s11682-016-9584-8 [DOI] [PubMed] [Google Scholar]
- Vidal-Piñeiro D, Valls-Pedret C, Fernández-Cabello S, Arenaza-Urquijo EM, Sala-Llonch R, Solana E, Bargalló N, Junqué C, Ros E, Bartrés-Faz D (2014) Decreased default mode network connectivity correlates with age-associated structural and cognitive changes. Front Aging Neurosci 6:256. 10.3389/fnagi.2014.00256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GZ, Belgard TG, Mao D, Chen L, Berto S, Preuss TM, Lu H, Geschwind DH, Konopka G (2015) Correspondence between resting-state activity and brain gene expression. Neuron 88:659–666. 10.1016/j.neuron.2015.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth M, Villeneuve S, La Joie R, Marks SM, Jagust WJ (2014) Gene-environment interactions: lifetime cognitive activity, APOE genotype, and β-amyloid burden. J Neurosci 34:8612–8617. 10.1523/jneurosci.4612-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]