Abstract
Pollination services provided by the honey bee, Apis mellifera (Hymenoptera: Apidae, Linnaeus, 1758) have broad economic impacts and are necessary for production of a diversity of important crops. Hives may be transported multiple times per year to provide pollination. To test how temperature may contribute to transportation stress, temperature sensors were placed in hives in different locations and orientations on the trailer during shipping. Colony size prior to shipping significantly contributed to loss of population immediately after shipping which contributed to colony failure with smaller colonies more likely to fail and fail faster. Colony size also affects thermoregulation and temperature stress. Internal hive temperature varies significantly based on location and orientation. While colonies near the front and rear of the trailer and those oriented toward the center aisle had significantly different average internal temperatures, colony size best predicts loss of thermoregulation. Additionally, we profiled gene expression at departure, on arrival, and after a recovery period to identify transcriptional responses to transportation. Functional and enrichment analysis identified increased methylation and decreased ribosomal and protein-folding activity. Pheromone and odorant-binding transcripts were up-regulated after transportation. After recovery, transcripts associated with defense response, immune activity, and heat shock decreased, while production of antibiotic peptides increased. We conclude that hives experience considerable temperature stress possibly caused by turbulent airflow in exposed locations. Transportation stress should be considered an important component of annual colony losses which can be mitigated with improved management strategies.
Keywords: thermoregulation, temperature stress, Apis mellifera, pollinator, gene expression
The use and management of the honey bee, Apis mellifera (Hymenoptera: Apidae, Linnaeus, 1758) has changed in recent decades to an increasing focus on transportation of hives for pollination services during intensive short-term pollination events due to rapid growth in production of animal-pollinated crops driving growth in the industry. Since 1960 honey production has tracked global population growth, approximately doubling over the following 50 yr. The production of animal-pollinated crops experienced a fourfold increase over the same time period and as a result crop production is growing much faster than the pollination service industry (Aizen and Harder 2009). While animal-pollinated crops represent a minority of agricultural output in tonnage (3–8%) due to the prevalence of wind-pollinated grains, insect-pollinated fruits, vegetables, and oil crops are a primary source of agricultural diversity (84%) (Southwick and Southwick 1992, Klein et al. 2007). The economic value by weight of insect-pollinated crops overtakes other crops with a valuation of 9.5% of global agricultural output (Gallai et al. 2009). Intensive regional production requires coordinated pollination during brief periods when plants are in bloom, prior to which there is limited forage to attract natural pollinators. For these reasons the transportation of hives to provide pollination is necessary to maintain output of the majority of crop species with a large impact on agricultural markets and food security (Gallai et al. 2009).
In the United States, large numbers of hives are transported to multiple locations throughout the country by truck to pollinate seasonal fields and orchards. During transportation, colonies are challenged by a variety of stressors. The condition of a hive prior to transportation is often locally acclimated to ecological conditions which often differ greatly from those of the destination. They are moved between locations at interstate highway speeds and deployed in fields and orchards prior to the bloom. Changes in temperature, day length, and nutrient supplementation that bees experience after transportation can increase foraging activity and brood production earlier than would have occurred before relocation and in agricultural environments prior to floral bloom with low availability of resources (Avitabile 1978, Fewell and Winston 1992). Relocating hives also causes some loss of foragers which typically require time and an obstruction or landmark to reorient them to the new location (Free 1958, Capaldi and Dyer 1999).
Abiotic stress during hive relocation for pollination events and overwintering has received some attention, though many investigations in to the causes of annual colony losses have focused on parasites and pathogens, some of which have been identified as predictive markers (vanEngelsdorp and Meixner 2010, vanEngelsdorp et al. 2010, Dainat et al. 2012, Kulhanek et al. 2017). Transportation has been described as a likely contributor to colony loss but the focus has been on changing forage quality and consistency, not stress endured during transportation (Oldroyd 2007). Transportation stress has received less attention because of the difficulty of collecting data during shipping. Even though transportation lasts only a few days, colonies experience confinement, increased variation in temperature, air pressure, and vibration. During shipping, colonies experience a rapid progression of changing elevation and latitude. Proper ventilation is a primary concern because poorly ventilated colonies often die from overheating. The consequences of low-temperature stress are less obvious. A healthy colony will maintain an internal temperature between 32 and 35°C which is necessary for the development of brood, flight, and efficient worker activity (Bernd 1979, Fahrenholz et al. 1989). A colony may experience extended periods of sub-lethal chill stress and loss of thermoregulation (LT) that affects long-term colony survival without proximate mortality by inducing developmental defects in new brood (Tautz et al. 2003, Groh et al. 2004, Jones et al. 2005). Colonies have many possible locations on the trailer and may be oriented inward toward a center aisle or outward toward the road which may affect airflow, especially at interstate highway speeds. Hives in locations exposed to turbulent airflow or near areas of aerodynamic drag such as the tractor-trailer gap and back of the trailer may also experience greater temperature variation.
The effects of temperature variation during shipping on long-term colony survival are unknown. Our objective was to monitor temperature during shipping to determine how much variation occurs among colonies on the same truck as they are transported across the United States from North Dakota to California to provide pollination services. We hypothesize that during transportation colonies experience cold temperature stress which causes internal hive temperature to vary outside of normal thermoregulation. Colony size may affect thermoregulation with larger colonies possessing both a greater thermal mass and ability to produce metabolic heat. Additionally, we hypothesize that shipping will produce a transcriptional response in genes and pathways that respond to temperature stress and confinement. The physiological mechanisms and transcriptional regulation in insects associated with cold stress, chill injury, and repair are well-understood (Clark and Worland 2008, MacMillan and Sinclair 2011, Xu et al. 2017). There is some evidence in A. mellifera that confinement is a source of stress affecting juvenile hormone (JH) levels in workers that is normally associated with worker age and function (Lin et al. 2004, Herb et al. 2012). In this study, we identify hives with different orientations and locations on the trailer and equip them with internal and external temperature sensors to monitor variation in temperature during shipping. We also perform gene expression analysis on A. mellifera workers using Illumina mRNA sequencing prior to transport from bee yards in North Dakota, on arrival in California, and 3 wk after departure to profile the expression of transcripts associated with temperature and other physiological stressors. We identify gene functional categories, enriched pathways and gene ontology (GO) terms after transportation to determine whether pathways associated with physiological, immunological, and abiotic stress are correlated with transport conditions.
Materials and Methods
Transportation and Hive Management
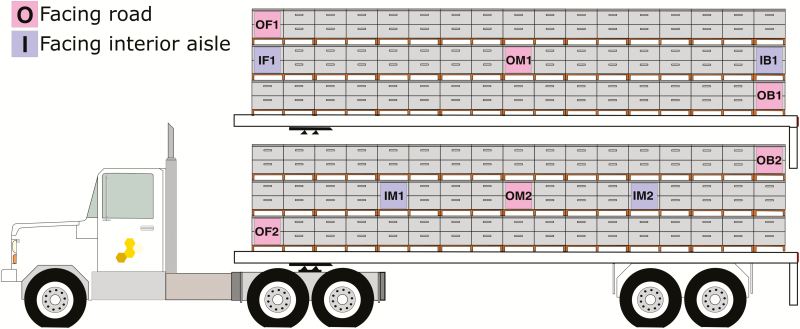
Hives were provided by AgPollen, LLC. Hives were prepared for departure on 10 October 2012. Four hundred eight hives were placed on pallets, loaded on a flatbed trailer, and covered by a net. Each pallet had four hives, columns of three pallets formed two rows on the bed of the trailer with an aisle between pallets in the center. Ten of the 408 hives were selected for monitoring, and these represented different locations and orientations on the flatbed trailer. Hives are identified by their orientation, facing Outward or Inward (O, I), and their location, Front, Middle, or Back (F, M, B), on the trailer (Fig. 1). The truck departed Towner, ND (48.361755, −100.402613) on 10 October 2012 and arrived in Sheep Ranch, CA (38.209894, −120.464346) on 14 October 2012. Hive strength was measured by visual inspection of frames to determine the number of active, colonized frames per hive. Hive strength was assessed on departure (day 1, October 10), and on days 6, 26, 119, 146, and 170 (Fig. 2). Colonies received supplemental protein patties (454 g, 15.7% protein by weight) on days 10, 33, 87, 107, and 123 and 3.8 liters of supplemental high fructose corn syrup on days 10 and 123. Hives received antifungal treatments (fumagillin, 9 g per hive) on days 26 and 40. For a full calendar of hive management see Supp. Table S1.
Fig. 1.
Position and orientation of hives during transport. Hives were organized on pallets with two facing out and two facing toward the interior center aisle, stacked three pallets high. Hives with temperature sensors are labeled by orientation and location. Orientation (I, O) of hives is indicated by ‘I’ for hives facing the central aisle and ‘O’ for hives facing outward. Position (F, M, B) indicates front, middle, or back location of hives on the trailer.
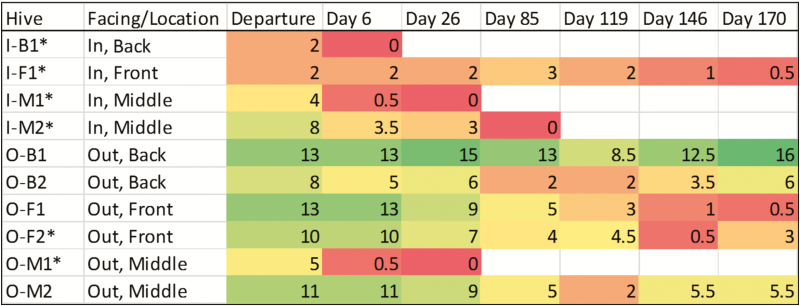
Fig. 2.
Assessment of colony strength. Colony strength was assessed by visual count of active colonized frames within each hive. Colony strength was assessed prior to departure, after arrival, and at semi-regular intervals. Colonies that lost thermoregulation during shipping are indicated by an asterisk (*). Colonies with fewer active frames experience disproportionate loss during shipping, while larger colonies maintain stable populations (Z = −2.191, P < 0.05). Departure strength, arrival strength, and the loss of thermoregulation during shipping all significantly interact to affect colony survival, with smaller colonies failing faster than larger colonies (Z = −4.366, P < 0.0001).
Measurement and Analysis of Temperature During Transportation
During preparation for shipping 14 iButton data logging temperature sensors (Maxim Integrated, San Jose, CA) were placed in each hive. Sensors were located in the top, middle, and bottom corners of the hive and the center frame of each brood box to allow continuous measurement of colony temperature and to compensate for colony movement and position in the hive. Sensors were programmed to record temperature at 1-h intervals. External sensors were placed on each pallet to record external temperature during transport. After transport, sensors and corresponding temperature data were recovered. Temperatures were downloaded from iButtons and analyzed using R (v3.4.2) (R Core Team 2017). Data from populated frames were analyzed from 12 a.m. on the 10 of October to 11 p.m. October 14th. Temperatures were compared between hives, orientation of hives, and the position of hives within the truck. After analyzing the distributions of temperatures and positions of each hive, the assumptions for homogenous variance were not met. Thus, hive temperatures and positions were analyzed using a nonparametric analysis of variance (ANOVA).
Hive strength assessments were performed at regular intervals. This was used to determine the number of days each hive survived, with the final strength assessment, 170 d, as the maximum value. The effect of colony strength prior to departure on colony strength after arrival was determined using ANOVA. Colony survival was assessed via generalized linear model (GLM) in R (R Core Team 2017). Colony survival time was used as the dependent variable. The number of active frames at departure, active frames after arrival, and the number of hours below a thermoregulation threshold of 32°C as predictors. Interactions between all three predictors were included in the model to determine the effect of hive strength prior to shipping on the number of frames remaining and the ability to thermoregulate during shipping. A mixed effects model was used to determine whether location and orientation on the truck influenced LT using the R package nlme (v3.1–137) (Pinheiro et al. 2018). The fixed effects were orientation (in or out), location on the truck (front, middle, back), and the number of frames the hive had at the start of transportation. Hive identification number was included as a random effect to account for the repeated measure of temperature during each day of transport.
RNA Collection and Sequencing
Nurse bees were collected at three time points: prior to, immediately after, and 3 wk after transport. Nurse bees were identified by location in the hive and inability to fly by removing a brood frame, shaking bees into a plastic tub and agitating the remaining bees to remove those able to fly. Three adult workers were collected from three hives totaling nine replicate bees per time point. mRNA was extracted from whole adults using the Tri-zol protocol and quality assessment and quantification was performed by Nanodrop and Qubit and stored at −80°C prior to sequencing. Illumina sequencing was performed by Georgia Genomics Facility which assessed quality using a Bioanalyzer. Libraries were generated from 27 samples which include three time points and 9 replicates per time point. Sequencing consisted of two NextSeq flowcells. Quality assessment of sequence reads was done using FastQC (v0.11.7) (Andrews 2010). Overrepresented sequences and Illumina sequencing artifacts and remaining adaptor sequences were removed using the BBDuk functions of the BBMap software package (v38.18) (Bushnell 2014). Illumina data is archived at the NCBI Sequence Read Archive indexed under BioProject PRJNA495845.
Differential Gene Expression and Enrichment Analysis
Reads were mapped and annotated to the A. mellifera genome (Release v4.5, accession number PRJNA10625) hosted by NCBI (Honeybee Genome Sequencing Consortium 2006) using Hisat2 (v2.0.5) (Kim et al. 2015). Gene expression analysis was done using Cufflinks (v2.2.1) to perform a three-way comparison of nine replicates collected at each time point (Trapnell et al. 2012). Transcripts and isoforms were annotated using to the A. mellifera genome. Differential expression and data exploration were performed using the cummeRbund R package (v2.22.0) (Goff et al. 2012). Overall variation among replicates was assessed by plotting density and the distribution of FPKM values among replicates. Variation among replicates across treatments was assessed by clustering using Jensen-Shannon distance in R (Goff et al. 2012, R Core Team 2017).
Significant differentially expressed genes (Supp. Tables S3–S8) were uploaded to the Database for Annotation, Visualization, and Integrated Discovery (DAVID v6.8) ID Conversion and Functional Annotation tools (Huang et al. 2007, 2009a, 2009b). GO, protein domain, and pathway enrichment were performed using a count threshold of 2 record matches, an EASE threshold of 0.05, P-value correction using Benjamini–Hochberg. The fold-enrichment values for significantly enriched gene sets were also obtained using this analysis. Enriched protein domains were identified using InterPro functional annotation tools in DAVID. Proteins were classified using UniProt keywords. DAVID was also used to visualize enriched pathways, gene function classifications, and generate KEGG pathway assignments (Kanehisa and Goto 2000).
Results
Effects of Hive Strength and Position on Thermoregulation
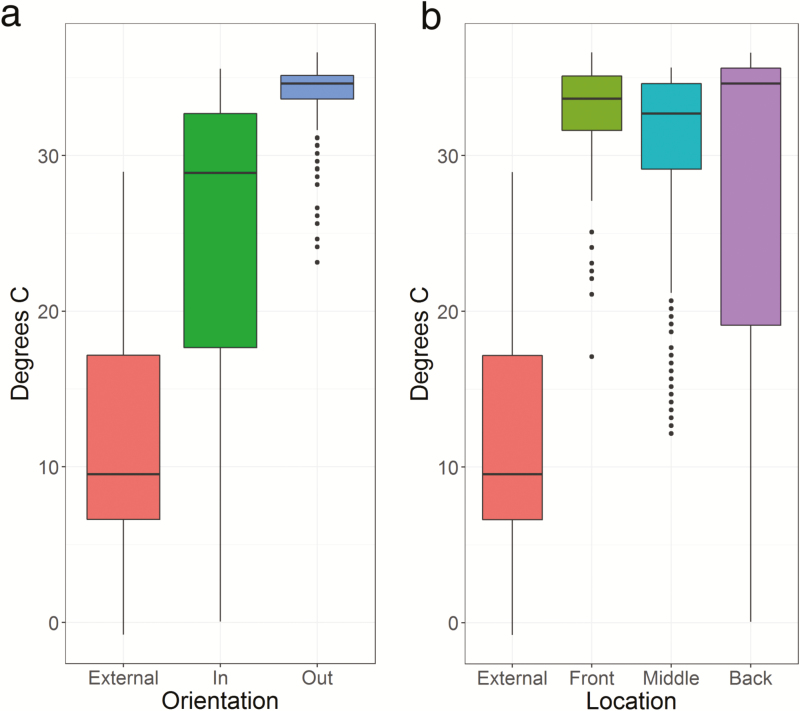
Internal hive temperature was recorded every hour during shipping with the goal of determining whether they experience thermal stress (Fig. 3). The average temperature among the 10 hives was significantly different (Supp. Fig. S1; nonparametric ANOVA: F9, χ2 = 766.53, P < 0.001). Pairwise comparisons determined that hives were significantly different from each other except hives O-M1 and I-F1 (P = 0.235), hives O-F2 and IM-2 (P = 0.054), and hives O-F1 and O-B2 (P = 0.169). There was a significant difference in average temperature depending on whether the hive was oriented facing the outside or inside of the truck (nonparametric ANOVA: F2, χ2 = 27.74, P < 0.001), with hives facing the inside having a lower average temperature than those facing outside (Fig. 4a). Average temperature of hives located in the middle of the truck were significantly different than those in the front (P < 0.001) and back (P < 0.001). Average temperatures of hives in the front and back were not significantly different (P = 0.99). Average temperatures of hives oriented inward toward the center aisle were significantly different than those oriented outward (nonparametric ANOVA: F1, χ2 = 418.06, P < 0.001).
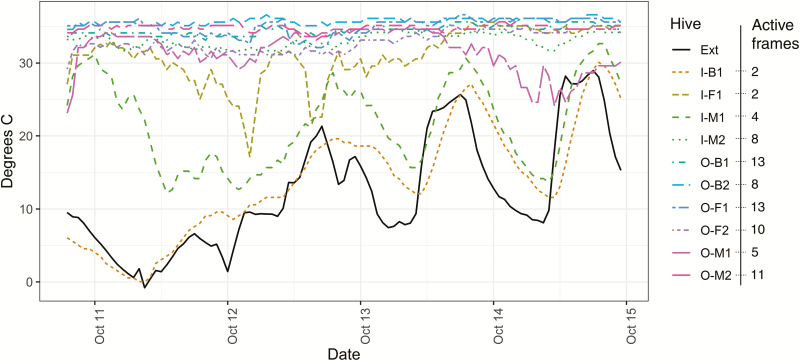
Fig. 3.
Internal temperature of hives and number of colonized frames during transportation from North Dakota to California. Hive temperature during transportation was recorded hourly. Orientation (I, O) of hives is indicated by ‘I’ for hives facing the central aisle and ‘O’ for hives facing outward. Position (F, M, B) indicates front, middle, or back location of hives on the trailer. The number of actively colonized frames is indicated for each hive. External temperature is shown in black. Hive orientation had a significant effect on thermoregulation, with hives facing inward more likely to lose thermoregulation (F = 10.523, P = 0.023). Colony size at departure, indicated as the number of active frames, did not significantly affect thermoregulation (F = 4.266, P = 0.094) nor did hive location (F = 0.024, P = 0.976).
Fig. 4.
Hive temperature by location and orientation. Internal hive temperature was measured at 1 h intervals throughout transportation. A sensor outside the hive recorded external temperature. Hives facing the center aisle (In) have significantly greater variation in mean temperature and difficulty maintaining thermoregulation (F = 10.523, P = 0.023). Those facing the road (Out) have significantly less variation in mean temperature (a). While mean temperature varies by location on the trailer (b), location was not a significant predictor of loss of thermoregulation (F = 0.024, P = 0.976).
After transportation hives remained in bee yards at Sheep Ranch, CA from the initial arrival, October 14 through February 4, totaling 114 d before they were relocated to almond orchards to provide pollination. During this time 4 of 10 hives failed before they were used for pollination, corresponding to three hives facing inward and one outward during transport. At the final hive strength assessment, two additional hives were near-failing and considered a loss (Fig. 2, Table 1).
Table 1.
Hive strength and hours below 32°C
| Hive | Active frames | Mean temperature | Hours 32–36°C | Hours < 32°C | ||
|---|---|---|---|---|---|---|
| Departure | Arrival | Final | ||||
| IB1 | 2 | 0 | 0 | 13.9 | 0 | 101 |
| IF1 | 2 | 2 | 0.5 | 31.0 | 43 | 58 |
| IM1 | 4 | 0.5 | 0 | 21.8 | 2 | 99 |
| IM2 | 8 | 3.5 | 0 | 33.1 | 85 | 16 |
| OB1 | 13 | 13 | 16 | 34.5 | 101 | 0 |
| OB2 | 8 | 5 | 6 | 35.7 | 101 | 0 |
| OF1 | 13 | 13 | 0.5 | 35.0 | 101 | 0 |
| OF2 | 10 | 10 | 3 | 32.9 | 72 | 29 |
| OM1 | 5 | 0.5 | 0 | 31.6 | 57 | 44 |
| OM2 | 11 | 11 | 5.5 | 34.8 | 101 | 0 |
Hive strength is measured in the number of active, colonized frames. Internal hive temperature is shown as the number of hours within or below normal thermoregulation range.
To determine the effects of shipping on colony survival we developed a generalized linear model in which colony survival time was the dependent variable and colony strength (measured by the number of colonized frames) before shipping, after shipping, and temperature stress (measured by hours below 32°C) during shipping were the predictor variables (Table 2). Arrival strength (AS) significantly contributed to colony survival (Z = 3.006, P < 0.01). Departure strength (DS) and LT, are both significant interaction effects that influence AS (Z = −2.191, P < 0.05; Z = 5.469, P < 0.0001). The interaction between all three predictor variables, DS, AS, and LT, also significantly effects colony survival time (Z = −4.366, P < 0.0001). Colony strength prior to departure significantly affects colony strength after arrival with smaller hives experiencing greater losses of colonized frames (ANOVA: F1,8 = 64.68, P < 0.0001). We found no evidence of overheating with 36.62°C the highest recorded temperature over the duration of the study.
Table 2.
Effect of initial colony size on loss after shipping and colony survival
| Term | Estimate | SE | Z | P | LRT | P |
|---|---|---|---|---|---|---|
| Intercept | 3.619 | 0.969 | 3.734 | <0.001* | 148.64 | <0.0001* |
| DS | 0.108 | 0.138 | 0.782 | 0.43 | ||
| AS | 0.231 | 0.077 | 3.006 | <0.01* | ||
| LT | −0.008 | 0.011 | −0.741 | 0.46 | ||
| DS * AS | −0.017 | 0.008 | −2.191 | <0.05* | ||
| DS * LT | −0.003 | 0.002 | −1.313 | 0.19 | ||
| AS * LT | 0.018 | 0.003 | 5.469 | <0.0001* | ||
| DS * AS * LT | −0.001 | 0 | −4.366 | <0.0001* |
GLM of effects on colony survival time with DS, AS, and hours <32°C indicating LT as predictor variables. Values represent results of generalized linear models with Poisson error.
We assessed whether location and orientation on the truck influenced whether a hive lost the ability to thermoregulate during shipping using a mixed effects model. An ANOVA on the model showed that orientation was a significant predictor of the LT (F = 10.523, P = 0.0228) but location on the truck (F = 0.024, P = 0.9762) and number of frames at shipping (F = 4.266, P = 0.0938) were not significant. These results indicate that hives that face inward are likely to lose thermoregulation during shipping regardless of where they are located on the truck and hive strength at the start of transport.
Gene Expression and Enrichment Analysis
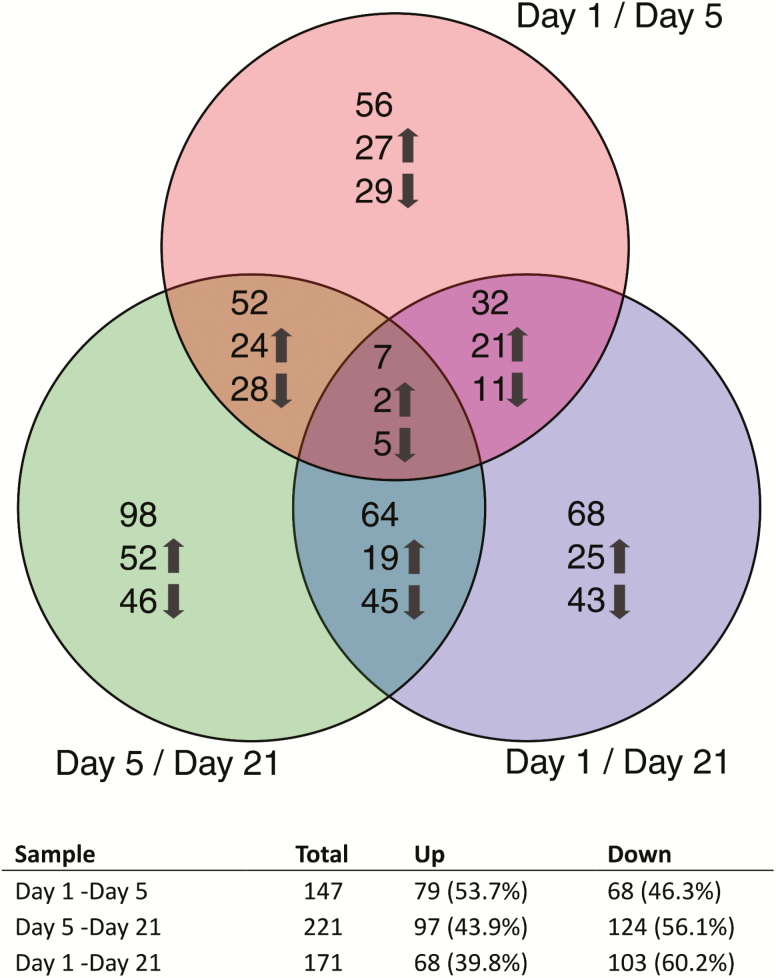
Sequencing generated 873.7 million 100 base pair paired-end reads averaging 16.7 million reads per sample. An average of approximately 83.1% of reads across samples mapped to the A. mellifera genome (v4.5, accession number PRJNA10625). ANOVA of FPKM values across replicates did not show a significant difference in the distribution of values within samples (F = 0.020, P = 0.887) or between samples (F = 0.792, P = 0.428). Clustering of replicates by Jensen-Shannon distance show T1 prior to transport has slightly higher variation among individual bees than the post-transport time points, T2 and T3 (Supp. Fig. S2). Gene expression of pairwise comparisons between day 1 (prior to departure), day 5 (after arrival), and day 21 (after the recovery period) identified significant differentially expressed transcripts and the direction and magnitude of expression (Fig. 5). The number of differentially expressed transcripts that are unique to one comparison range from 56 to 98 (38.1–44.3% by comparison). The approximate 60% of remaining transcripts appear as significant in multiple comparisons indicating dynamic gene expression across time points with seven transcripts appearing in all comparisons. While the number of significant transcripts appear evenly distributed between comparisons, the direction of expression skewed downward over time with a 60.2% majority of transcripts down-regulated on day 21 versus day 1 (Fig. 5).
Fig. 5.
Expression profile of transcripts at sampling intervals before shipping, immediately after arrival, and after a recovery period. RNA was extracted from workers from 9 hives at three intervals, immediately prior to departure from the Towner, North Dakota bee yard (day 1, October 9), after arrival in Sheep Ranch, California (day 5, October 15), and after a recovery period in Sheep Ranch, California (day 21, November 5). Discrepancy in the total number of up or down-regulated genes shown in the Venn diagram reflects change in the direction of expression of genes shared between comparisons.
Enrichment analysis using DAVID identified enriched functional groups, KEGG pathways, GO terms, and enriched InterPro protein domains (Tables 3, 4, 5). The set of frequently occurring GO terms for all differentially expressed transcripts are shown in Supp. Fig. S3–S5 (n > 1 GO terms for Biological process and Cellular component, n > 2 GO terms for Molecular Function).
Table 3.
KEGG pathway enrichment
| Comparison | Term | n | P | FE |
|---|---|---|---|---|
| Day 1–day 5 up-regulated | ||||
| Metabolic pathways | 5 | 0.039 | 2.60 | |
| Day 1–day 21 up-regulated | ||||
| Metabolic pathways | 10 | 0.001 | 2.60 | |
| Tyrosine metabolism | 3 | 0.002 | 35.40 | |
| Biosynthesis of amino acids | 3 | 0.03 | 9.83 | |
| Ubiquinone and other terpenoid-quinone biosynthesis | 2 | 0.046 | 39.33 | |
| Biosynthesis of antibiotics | 4 | 0.046 | 4.34 | |
| Day 1–day 21 down-regulated | ||||
| Protein processing in endoplasmic reticulum | 4 | 0.039 | 4.83 |
Pathway enrichment of significant differentially expressed genes used a P-value cutoff of P < 0.05. N = the number of genes per term, fold enrichment is indicated by FE.
Table 4.
GO term enrichment
| Comparison | Category | Term | n | P | FE |
|---|---|---|---|---|---|
| Day 1–day 5 up-regulated | |||||
| GO CC | Nucleosome | 5 | <0.0001 | 28.7 | |
| GO CC | Extracellular region | 4 | 0.038 | 5.1 | |
| GO MF | Odorant binding | 6 | 0.002 | 6.1 | |
| GO MF | DNA binding | 6 | 0.038 | 3.1 | |
| Day 5–day 21 down-regulated | |||||
| GO BP | Chitin catabolic process | 3 | 0.003 | 36.4 | |
| GO BP | Carbohydrate metabolic process | 4 | 0.023 | 6.1 | |
| GO BP | Defense response | 2 | 0.023 | 80.9 | |
| GO BP | Nucleotide catabolic process | 2 | 0.023 | 80.9 | |
| GO BP | Phospholipid metabolic process | 2 | 0.039 | 48.6 | |
| GO BP | Arachidonic acid secretion | 2 | 0.039 | 48.6 | |
| GO CC | Extracellular region | 12 | <0.0001 | 11.2 | |
| GO MF | Chitinase activity | 3 | 0.005 | 27.7 | |
| GO MF | Odorant binding | 5 | 0.033 | 4 | |
| Day 1–day 21 up-regulated | |||||
| GO BP | Nucleosome assembly | 5 | <0.0001 | 55.6 | |
| GO BP | DNA-templated transcription, initiation | 3 | 0.004 | 30.8 | |
| GO BP | Aromatic amino acid family metabolic process | 2 | 0.035 | 53.4 | |
| GO CC | Nucleosome | 9 | <0.0001 | 47.8 | |
| GO CC | Nucleus | 9 | 0.01 | 2.7 | |
| GO MF | DNA binding | 9 | 0.001 | 3.9 | |
| Day 1–day 21 down-regulated | |||||
| GO BP | Chitin metabolic process | 4 | 0.002 | 14.2 | |
| GO BP | Innate immune response | 3 | 0.004 | 29.1 | |
| GO BP | Defense response | 2 | 0.027 | 71.2 | |
| GO BP | Carbohydrate metabolic process | 4 | 0.032 | 5.4 | |
| GO BP | Phospholipid metabolic process | 2 | 0.044 | 42.7 | |
| GO BP | Arachidonic acid secretion | 2 | 0.044 | 42.7 | |
| GO CC | Extracellular region | 13 | <0.0001 | 11.8 | |
| GO MF | Chitin binding | 5 | <0.0001 | 13.2 | |
| GO MF | Fatty-acyl-COA reductase (alcohol-forming) activity | 3 | 0.003 | 34.6 |
GO term enrichment of significant differentially expressed genes used a P-value cutoff of P < 0.05. N = the number of genes per term, fold enrichment is indicated by FE.
Table 5.
Interpro protein domain enrichment
| Term | n | P | FE | |
|---|---|---|---|---|
| Day 1–day 5 up-regulated | ||||
| Pheromone/general odorant-binding protein | 5 | <0.0001 | 32.3 | |
| Histone-fold | 4 | 0.002 | 16.7 | |
| Histone core | 3 | 0.005 | 26.7 | |
| Haemolymph juvenile hormone binding | 2 | 0.042 | 20.3 | |
| Day 5–day 21 up-regulated | ||||
| Histone-fold | 3 | 0.031 | 10.7 | |
| Mab-21 domain | 2 | 0.032 | 60.5 | |
| Zinc finger, RanBP2-type | 2 | 0.041 | 26.9 | |
| Day 5–day 21 down-regulated | ||||
| Pheromone/general odorant-binding protein | 5 | <0.0001 | 25.4 | |
| Glycoside hydrolase, family 18, catalytic domain | 3 | 0.003 | 33.5 | |
| Chitinase II | 3 | 0.003 | 33.5 | |
| Glycoside hydrolase, superfamily | 4 | 0.006 | 10.6 | |
| Cytochrome b5, heme-binding site | 2 | 0.035 | 55.8 | |
| Glycoside hydrolase, catalytic domain | 3 | 0.036 | 9.9 | |
| Chitin binding domain | 3 | 0.038 | 9.6 | |
| Phospholipase A2 domain | 2 | 0.043 | 44.7 | |
| Day 1–day 21 up-regulated | ||||
| Histone-fold | 7 | <0.0001 | 30.5 | |
| Histone core | 4 | <0.0001 | 37 | |
| Histone H4 | 3 | <0.0001 | 111.1 | |
| Histone H4, conserved site | 3 | <0.0001 | 111.1 | |
| TATA box binding protein associated factor (TAF) | 3 | 0.001 | 74.1 | |
| Histone H5 | 2 | 0.013 | 148.1 | |
| Pyridoxal phosphate-dependent transferase, major region, subdomain 2 | 3 | 0.022 | 12.7 | |
| Pyridoxal phosphate-dependent transferase, major region, subdomain 1 | 3 | 0.027 | 11.4 | |
| Pyridoxal phosphate-dependent transferase | 3 | 0.03 | 10.8 | |
| Histone H1/H5 | 2 | 0.033 | 59.2 | |
| Histone H2B | 2 | 0.033 | 59.2 | |
| Haemolymph juvenile hormone binding | 2 | 0.049 | 21.2 | |
| Day 1–day 21 down-regulated | ||||
| Chitin binding domain | 5 | <0.0001 | 17 | |
| Fatty acyl-CoA reductase | 3 | 0.002 | 44.6 | |
| Male sterility, NAD-binding | 3 | 0.002 | 44.6 | |
| Heat shock protein Hsp90, N-terminal | 2 | 0.033 | 59.5 | |
| Heat shock protein Hsp90 | 2 | 0.033 | 59.5 | |
| Cytochrome b5, heme-binding site | 2 | 0.033 | 59.5 | |
| Insect cuticle protein | 3 | 0.039 | 9.4 | |
| Phospholipase A2 domain | 2 | 0.041 | 47.6 | |
| Glycoside hydrolase, superfamily | 3 | 0.047 | 8.5 | |
| Aquaporin-like | 2 | 0.049 | 39.7 | |
| Lipase, GDSL | 2 | 0.049 | 39.7 |
InterPro protein domain enrichment of significant differentially expressed genes used a P-value cutoff of P < 0.05. N = the number of genes per term, fold enrichment is indicated by FE.
From day 1 to day 5 enriched GO terms associated with changes to gene expression through modification of histones, nucleosome methylation activity, and DNA binding are up-regulated while those associated with translation are down-regulated. Six genes associated with pheromone activity and odorant binding are up-regulated. Pathway enrichment analysis shows up-regulation of metabolic pathways (generic). At day 5 proteins associated with pheromone binding, JH binding, and modification of histones were significantly enriched.
From day 5 to day 21 no enriched GO terms are associated with up-regulated genes. Terms associated with metabolic processes are down-regulated, notably chitinases, those associated with chitin catabolism, and carbohydrate metabolism. Pheromone and odorant enrichment which was up-regulated on day 5 is down-regulated on day 21. Genes associated with arachidonic acid metabolism are down-regulated. Enriched protein domains that are up-regulated from day 5 to 21 include histone folding, Mab-21 domain, and zinc finger protein. Down-regulated protein domains include pheromones and odorants, and numerous metabolic enzymes. Up-regulated transcripts associated with transport proteins showed significant enrichment including transmembrane transport proteins and RanBP2, a member of the RAS superfamily, which mediates transport of proteins across the nuclear membrane. Up-regulation of transcripts and enrichment of proteins associated with histone activity is maintained.
By comparing the first and last time points, day 1 and day 21, we identified additional enriched terms associated with stress which are down-regulated by day 21 including innate immune response, defense response to bacterium, response to stress, defense response, and protein folding related to a decrease in Hsp90, Hsp83, Hsp70, and DNAJ activity. GO terms associated with metabolism, nucleosome activity, and DNA binding are maintained from day 5 to day 21. Protein possessing in the endoplasmic reticulum is down-regulated. Enriched protein domains that are up-regulated include multiple histone and DNA binding proteins, pyridoxal phosphate transferase, and JH binding proteins. Down-regulated protein domains include Hsp90 and multiple metabolic enzymes. Genes that are down-regulated after the recovery period on day 21 are involved in immune response, venom components, and protein-folding chaperones associated with stress response, while production of antibiotic peptides is up-regulated (Table 6).
Table 6.
Transcripts associated with stress down-regulated after recovery
| Category | Name | Symbol | Log FC |
|---|---|---|---|
| Immune response | Defensin 2 | Def2 | −4.428 |
| Hymenoptaecin | LOC406142 | −5.350 | |
| Peptidoglycan recognition protein S2 | Pgrp-s2 | −2.020 | |
| Biosynthesis of antibiotics | |||
| Kynurenine/alpha-aminoadipate aminotransferase | LOC724239 | 2.726 | |
| Multifunctional protein ADE2 | LOC551966 | 0.794 | |
| Probable phosphoserine aminotransferase | LOC412670 | 0.752 | |
| Tyrosine aminotransferase | LOC725204 | 0.788 | |
| Defense/venom component | |||
| Hyaluronoglucosaminidase | LOC406146 | −1.477 | |
| Secapin | LOC406145 | −2.752 | |
| Venom allergen Api m 6 | LOC678674 | −2.081 | |
| Venom carboxylesterase-6 | Est-6 | −1.499 | |
| Phospholipase A2 | Pla2 | −4.462 | |
| Stress response | |||
| Heat shock protein Hsp70Ab-like | LOC410620 | −1.724 | |
| Heat shock protein 83 | LOC411700 | −1.411 | |
| Heat shock protein 90 | Hsp90 | −1.324 | |
| dnaJ protein homolog 1 | LOC411071 | −0.978 |
Differentially expressed genes used a P-value cutoff of P < 0.05. Log fold change of expression between time points is indicated by Log FC.
Discussion
A. mellifera and pollination services are essential for improving the productivity and economic viability of many crops. Insect-pollinated crops in the United States are numerous and highly diverse. Many require intensive local pollination during brief periods when they are in bloom. By transporting colonies, pollination service providers are able to pollinate crops that bloom at different times in different climates. Transportation helps to make the industry economically viable and sustainable but presents a number of challenges that contribute to colony loss. Increasingly, bees are transported to multiple locations for short-term pollination events. The effects of transportation are an understudied area of pollinator management with a potentially large impact on colony health and survival and we focus specifically on colonies that are being actively transported as a source of stress that impacts the colony. It is unknown what percentage of hives relocated for pollination services fail during or shortly after transportation. Additionally, little is known about the effects of transportation on colonies that do not result in complete loss shortly after relocation but may increase susceptibility to factors that contribute to long-term failure. Considering that hives may be relocated multiple times in a season these effects may accumulate if bees are not provided adequate forage and recovery time between moves. Confounding factors such as disease, a high parasite load, or small colony size may compromise health and may cause an inability to thermoregulate during shipping or recover after relocation.
Effects of Hive Strength and Location on Internal Temperature
During transportation, A. mellifera are impacted by abiotic stressors including variable temperatures, airflow, humidity, and air pressure as they are moved across latitude and elevation. The data presented here were collected during a 4-d relocation of colonies by AgPollen in October from North Dakota bee yards to California. Our study used an industry standard layout of hives during transport, with four hives on pallets and three stacked pallets. Pollination service providers favor increased airflow to prevent extreme temperatures and suffocation. For this reason, hives are oriented so the entrance faces either outward or toward a narrow central aisle to provide adequate airflow. Our first objective was to test whether transportation is a significant source of temperature stress and whether colony size significantly affects internal temperature stability. While in transit the external temperature ranged from −1.68°C to 38.26°C with a mean temperature of 12.52°C (Supp. Table 2). Healthy colonies thermoregulate between 32 and 35°C, temperature variation outside this range negatively impacts brood development and worker efficiency (Bernd 1979, Fahrenholz et al. 1989). Our analysis shows that 6 of 10 hives experienced a significant LT indicated by internal hive temperature falling below 32°C for more than 1 h. Four hives recovered, while two were unable to recover thermoregulation (Fig. 3). Colony size affects thermal stability. Smaller colonies have significantly more internal temperature variation and are more likely to lose thermoregulation. Colonies with low population are also at the greatest risk of failure after shipping. Our analysis shows colony strength at departure significantly affects strength at arrival. Additionally, our model shows significant interactions between colony strength at departure, strength after arrival, and LT which affect colony survival (Table 2). Colonies with small populations are more likely to lose population after shipping. These colonies are significantly more likely to fail, and fail faster than robust colonies. Further investigation of the effects of hive strength on survival after shipping are needed. This would allow the pollination service industry to develop a predictive model of colony loss at different populations. This could be incorporated into management strategies to determine a minimum population threshold, below which transporting colonies is not economically viable.
There is some evidence that hive position during shipping resulted in different average hive temperatures although further investigation is needed to determine whether this effect is caused by hive location and orientation or if it is caused by variation in colony size which we show here to be the greatest predictor of thermal stability during transportation and long-term survival. Colonies facing the central aisle experienced significantly different average temperatures but also had lower average populations (Fig. 4a). Hive temperature in the front and back significantly differ from those in the middle, but these locations were not significantly more likely to lose thermoregulation (Fig. 4b). It is possible that turbulent airflow is responsible for temperature variation. The front and back of the trailer are regions of high turbulence caused by the aerodynamic drag of the tractor-trailer gap and the trailer draft respectively. The center aisle may also generate air turbulence responsible for temperature variation in inward-facing hives, however additional data from hives of similar strength are needed to test that hypothesis.
The effects of temperature stress during shipping have the potential to influence long-term colony survival. While LT has been shown to cause mortality, it is also associated with sublethal physiological effects. Adults that receive chill injuries may have impaired performance. The effects of short-term exposure to temperatures slightly below 32°C on larva cause developmental abnormalities. This condition results in workers that present a variety of permanent neurological deficiencies related to memory that reduce foraging performance and are unable to navigate or communicate effectively (Tautz et al. 2003, Jones et al. 2005). The sublethal effects caused by chilled brood have the potential to impact both long-term colony survival and the performance of adults providing pollination services to agriculture (Jones et al. 2005, Khoury et al. 2011). A robust, healthy colony can tolerate most challenges through intensive brood production and attrition of sick or parasitized bees, an investment in ineffective workers may exacerbate factors that lead to colony failure such as nutrient resource shortfalls, reduced ability to purge infections, through slower brood production, and inability to restore robust population.
Effects of Transportation on Gene Expression
Our second objective was to profile changes in gene expression associated with stress of transportation and relocation of colonies by assessing gene expression in A. mellifera workers immediately before and after transportation and again after a short recovery period. We hypothesized that during transportation colonies encounter diverse stressors that produce a transcriptional response that is detectable by profiling gene expression before and after relocation. We predicted that immediately after transportation the transcriptomic response would be most pronounced and that after a recovery period differentially expressed transcripts would return to a baseline established by samples collected prior to shipping. Although we are interested in identifying transcription profiles that are specific to an abiotic stress response in individual bees, relocating a colony causes a significant behavioral disruption in a complex social organism. During this time workers are unable to leave the hive to forage, and may be limited in their ability to perform hygienic activities and respond to pheromone cues which would likely produce a transcriptional response.
Differential expression analysis identified GO terms across all time points associated with changes to metabolic activity, regulation of transcription and translation through histone modification and DNA-binding activity, and pheromone activity. Our analysis of enriched GO terms and InterPro protein domains between time points suggests an increase in DNA methylation activity and a reduction in protein synthesis. After the recovery period several protein-folding chaperones, Hsp90, Hsp83, Hsp70, co-chaperone DNAJ and genes involved in ribosomal activity are down-regulated. This may indicate an epigenetic response immediately after transportation (Table 4). DNA methylation has been identified as a common mechanism for regulating behavior in social insects. In A. mellifera DNA methylation is not only implicated in caste determination but specific methylation patterns are associated with worker subcaste jobs and behaviors (Lyko et al. 2010, Herb et al. 2012). Workers that are unable to leave the hive may receive environmental cues that cause them to revert to other subcastes through epigenetic activity. While subcaste reassignment is reversible, it requires workers to encounter environmental cues that produce the methylation profile associated with a subcaste. Therefore, transported colonies may have an unbalanced distribution of worker subcastes which is resolved over time but reduces the ability of the colony to buffer stress. Characterizing the methylation profiles of workers may identify changes to subcaste assignment caused by confinement. After transportation, there is an increase in JH binding activity. The analysis we performed indicates that JH receptor activity is significantly differentially expressed and has a fold-enrichment score of 21.2. JH titers naturally increase as workers age from nurses to foragers with high JH titers associated with older foragers (Rutz et al. 1976, Jassim et al. 2000). JH has been shown to respond to both cold stress and confinement. Confinement produces a strong response in both foragers and nurses even though nurses do not leave the hive (Lin et al. 2004). JH titers in foragers that revert to nurses in experimentally manipulated hives decrease significantly and are similar to those of other nurses (Robinson et al. 1992). Considering that subcastes have distinct methylation profiles, and JH titers vary significantly and reversibly with subcaste, and that JH has been shown to respond to both cold stress and confinement, it is possible that confinement has induced an epigenetic change in worker subcastes. It is important to note that nurse bees were sampled randomly and the age at the time of sampling is unknown. While differentially expressed transcripts are identified across multiple replicates, the average age of the sampled nurse bees may vary between time points which would affect JH levels. Further investigation of the interaction between JH titers and methylation profiles using a cohort of known-aged bees may reveal a mechanism that produces reverted nurses during confinement. The magnitude and duration of change in subcaste assignment caused by confinement may produce a maladaptive distribution of subcaste demographics that impact forager efficiency.
Immediately after transportation colonies also show GO term and protein domain enrichment of transcripts associated with pheromones and odorant binding which decrease after the recovery period (Tables 4 and 5). Enriched GO terms associated with metabolic pathways are down-regulated after transportation and remain down-regulated after the recovery period. The Drosophila homolog yellow was also down-regulated after the recovery period. In A. mellifera, yellow has a high degree of sequence homology with the family of major royal jelly proteins secreted by nurse bees as part of the specialized diet fed to larva for the development of new queens (Schmitzová et al. 1998, Han et al. 2002).
By comparing the first and last time point several things specific to stress response stand out. Transcripts mapped to enriched GO terms for innate immune response, defense response to bacterium were down-regulated while we identified an increase in transcripts for production of antibiotics (Table 6). Many of these transcripts were not differentially expressed from departure to arrival but change after the recovery period indicating that colonies may have been experiencing an immune challenge prior to transportation.
Conclusions
In this study, we identified one of many potential stressors that occur during transport. Cold stress occurs over multiple days and impacted colony survival after transportation. By measuring the interior and exterior temperature of the hive in multiple locations we identified significant differences in the ability of colonies to thermoregulate. Hive strength was the greatest predictor of thermal stability during transportation, loss of population after arrival, and long-term colony survival. Hive location and orientation may affect internal temperature variation, although further data collection is needed to determine whether this effect persists after accounting for variation in hive strength. The impact of other transport-related stressors such as vibration, exhaust, changes in humidity and barometric pressure, and confinement remain unexplored. Gene expression analysis indicated that hives may experience confinement stress during transport that affects worker subcastes. Three weeks after transportation, genes associated with immune responses were still down-regulated compared to before transportation indicating a long recovery period. Relocating hives for pollination services is the primary economic focus of many large beekeeping operations. Identifying sources of stress during transportation will inform management practices that decrease colony loss and improve pollination.
Availability of Data and Materials
The data supporting the conclusions of this article are included within the article (and its additional files) as Supp. Tables S1–S8. Sequence reads associated with the mRNA sequencing analysis are archived at NCBI under BioProject PRJNA495845.
Supplementary Material
Acknowledgments
We would like to thank David Moreland of AgPollen, LLC for providing hives, transportation, additional resources, and field support for this research. This work was also supported by the US Department of Agriculture Agricultural Research Service, Insect Genetics and Biochemistry unit. Additional support was provided by North Dakota State University College of Science and Mathematics, the Department of Biological Sciences (NSF-1557940), and the Department of Biological Sciences Shockey-Scoby Graduate Fellowship. J.R., G.Y., S.P., and D.M. designed the research plan. J.R. and G.Y. obtained the animals in collaboration with S.P. at AgPollen. Tissue collection and sensor placement were performed by S.P. at AgPollen and G.Y. at the USDA ARS Red River Valley Agricultural Research Center. Gene expression analysis was performed by D.M. Statistical analysis of temperature in transit was performed by E.W., D.M., and J.B. All authors helped write and edit the manuscript. J.R. and G.Y. provided funding for the research. All authors contributed to and approved the content of the final manuscript. The authors declare that they have no competing interests.
References Cited
- Aizen M. A., and Harder L. D.. . 2009. The global stock of domesticated honey bees is growing slower than agricultural demand for pollination. Curr. Biol. 19: 915–918. [DOI] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data.http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Avitabile A. 1978. Brood rearing in honeybee colonies from late autumn to early spring. J. Apic. Res. 17: 69–73. [Google Scholar]
- Bernd H. 1979. Thermoregulation of African and European honeybees during foraging, attack, and hive exits and returns. J. Exp. Biol. 80: 217–229. [Google Scholar]
- Bushnell B. 2014. BBMap: a fast, accurate, splice-aware aligner (No. LBNL-7065E). Lawrence Berkeley National Lab (LBNL), Berkeley, CA. [Google Scholar]
- Capaldi E. A., and Dyer F. C.. . 1999. The role of orientation flights on homing performance in honeybees. J. Exp. Biol. 202: 1655–1666. [DOI] [PubMed] [Google Scholar]
- Clark M. S., and Worland M. R.. . 2008. How insects survive the cold: molecular mechanisms-a review. J. Comp. Physiol. B 178: 917–933. [DOI] [PubMed] [Google Scholar]
- Dainat B., Evans J. D., Chen Y. P., Gauthier L., and Neumann P.. 2012. Predictive markers of honey bee colony collapse. PLoS One 7: e32151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Engelsdorp D., and Meixner M. D.. . 2010. A historical review of managed honey bee populations in Europe and the United States and the factors that may affect them. J. Invertebr. Pathol. 103 (Suppl 1): S80–S95. [DOI] [PubMed] [Google Scholar]
- van Engelsdorp D., Hayes J., Underwood R. M., and Pettis J. S.. . 2010. A survey of honey bee colony losses in the United States, fall 2008 to spring 2009. J. Apic. Res. 49: 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenholz L., Lamprecht I., and Schricker B.. . 1989. Thermal investigations of a honey bee colony: thermoregulation of the hive during summer and winter and heat production of members of different bee castes. J. Comp. Physiol. B 159: 551–560. [Google Scholar]
- Fewell J. H., and Winston M. L.. . 1992. Colony state and regulation of pollen foraging in the honey bee, Apis mellifera L. Behav. Ecol. Sociobiol. 30: 387–393. [Google Scholar]
- Free J. B. 1958. The Behaviour of Honeybees when their Hive is Moved to a New Site. Bee World. 39: 109–115. [Google Scholar]
- Gallai N., Salles J.-M., Settele J., and Vaissière B. E.. . 2009. Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol. Econ. 68: 810–821. [Google Scholar]
- Goff L., Trapnell C., and Kelley D.. . 2012. cummeRbund: analysis, exploration, manipulation, and visualization of Cufflinks high-throughput sequencing data. R Package Version. 2. [Google Scholar]
- Groh C., Tautz J., and Rössler W.. 2004. Synaptic organization in the adult honey bee brain is influenced by brood-temperature control during pupal development. Proc. Natl. Acad. Sci. U. S. A. 101: 4268–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q., Fang J., Ding H., Johnson J. K., Christensen B. M., and Li J.. 2002. Identification of Drosophila melanogaster yellow-f and yellow-f2 proteins as dopachrome-conversion enzymes. Biochem. J. 368: 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herb B. R., Wolschin F., Hansen K. D., Aryee M. J., Langmead B., Irizarry R., Amdam G. V., and Feinberg A. P.. 2012. Reversible switching between epigenetic states in honeybee behavioral subcastes. Nat. Neurosci. 15: 1371–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honeybee Genome Sequencing Consortium 2006. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 443: 931–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Tan Q., Collins J. R., Alvord W. G., Roayaei J., Stephens R., Baseler M. W., Lane H. C., and Lempicki R. A.. 2007. The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 8: R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. A. W., Sherman B. T., and Lempicki R. A.. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. A. W., Sherman B. T., and Lempicki R. A.. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Jassim O., Huang Z. Y., and Robinson G. E.. 2000. Juvenile hormone profiles of worker honey bees, Apis mellifera, during normal and accelerated behavioural development. J. Insect Physiol. 46: 243–249. [DOI] [PubMed] [Google Scholar]
- Jones J. C., Helliwell P., Beekman M., Maleszka R., and Oldroyd B. P.. 2005. The effects of rearing temperature on developmental stability and learning and memory in the honey bee, Apis mellifera. J. Comp. Physiol. A. Neuroethol. Sens. Neural. Behav. Physiol. 191: 1121–1129. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., and Goto S.. . 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury D. S., Myerscough M. R., and Barron A. B.. 2011. A quantitative model of honey bee colony population dynamics. PLoS One 6: e18491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., and Salzberg S. L.. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A. M., Vaissière B. E., Cane J. H., Steffan-Dewenter I., Cunningham S. A., Kremen C., and Tscharntke T.. 2007. Importance of pollinators in changing landscapes for world crops. Proc. Biol. Sci. 274: 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulhanek K., Steinhauer N., Rennich K., Caron D. M., Sagili R. R., Pettis J. S., Ellis J. D., Wilson M. E., Wilkes J. T., Tarpy D. R., . et al. 2017. A national survey of managed honey bee 2015–2016 annual colony losses in the USA. J. Apic. Res. 56: 328–340. [Google Scholar]
- Lin H., Dusset C., and Huang Z. Y.. . 2004. Short-term changes in juvenile hormone titers in honey bee workers due to stress. Apidologie. 35: 319–327. [Google Scholar]
- Lyko F., Foret S., Kucharski R., Wolf S., Falckenhayn C., and Maleszka R.. 2010. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 8: e1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macmillan H. A., and Sinclair B. J.. . 2011. Mechanisms underlying insect chill-coma. J. Insect Physiol. 57: 12–20. [DOI] [PubMed] [Google Scholar]
- Oldroyd B. P. 2007. What’s killing American honey bees? PLoS Biol. 5: e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro J., Bates D., DebRoy S., and Sarkar D.. . 2018. R Core Team nlme: linear and nonlinear mixed effects modelsR package version 3.1–137.https://CRAN.R-Proj.OrgpackageNlme.
- R Core Team 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Robinson G. E., Page R. E., Strambi C., and Strambi A.. . 1992. Colony integration in honey bees: mechanisms of behavioral reversion. Ethology. 90: 336–348. [Google Scholar]
- Rutz W., Gerig L., Wille H., and Lüscher M.. . 1976. The function of juvenile hormone in adult worker honeybees, Apis mellifera. J. Insect Physiol. 22: 1485–1491. [Google Scholar]
- Schmitzová J., Klaudiny J., Albert S., Schröder W., Schreckengost W., Hanes J., Júdová J., and Simúth J.. 1998. A family of major royal jelly proteins of the honeybee Apis mellifera L. Cell. Mol. Life Sci. 54: 1020–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwick E. E., and Southwick L.. . 1992. Estimating the economic value of honey bees (Hymenoptera: Apidae) as agricultural pollinators in the United States. J. Econ. Entomol. 85: 621–633. [Google Scholar]
- Tautz J., Maier S., Groh C., Rossler W., and Brockmann A.. 2003. Behavioral performance in adult honey bees is influenced by the temperature experienced during their pupal development. Proc. Natl. Acad. Sci. U. S. A. 100: 7343–7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L., and Pachter L.. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Niu Q., Zhao H., Du Y., and Jiang Y.. 2017. Transcriptomic analysis to uncover genes affecting cold resistance in the Chinese honey bee (Apis cerana cerana). PLoS One 12: e0179922. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the conclusions of this article are included within the article (and its additional files) as Supp. Tables S1–S8. Sequence reads associated with the mRNA sequencing analysis are archived at NCBI under BioProject PRJNA495845.