Abstract
The repair of DNA damage requires the ordered recruitment of many different proteins that are responsible for signaling and subsequent repair. A powerful and widely used tool for studying the orchestrated accumulation of these proteins at damage sites is laser microirradiation in live cells, followed by monitoring the accumulation of the fluorescently labeled protein in question. Despite the widespread use of this approach, there exists no rigorous method for characterizing the recruitment process quantitatively. Here, we introduce a diffusion model that explicitly accounts for the unique sizes and shapes of individual nuclei and uses two variables: Deff, the effective coefficient of diffusion, and F, the fraction of mobile protein that accumulates at sites of DNA damage. Our model quantitatively describes the accumulation of three test proteins, poly-ADP-ribose polymerases 1 and 2 (PARP1/2) and histone PARylation factor 1. Deff for PARP1, as derived by our approach, is 6× greater than for PARP2 and in agreement with previous literature reports using fluorescence correlation spectroscopy and fluorescence recovery after photobleaching. Our data indicate that histone PARylation factor 1 arrives at sites of DNA damage independently of either PARP. Importantly, our model, which can be applied to existing data, allows for the direct comparison of the coefficient of diffusion for any DNA repair protein between different cell types, obtained in different laboratories and by different methods, and also allows for the interrogation of cell-to-cell variability.
Significance
We present a new approach to mechanistically and quantitatively describe the accumulation of DNA repair proteins at sites of DNA damage. Here, we demonstrate that our quantitation of fluorescence accumulation after DNA damage method accommodates the unique topology of each nucleus to provide physically meaningful (and mathematically robust) information on the mechanism of transport. For the first time, direct comparisons of rates of accumulation at sites of DNA damage can be made between different cells, different cell types, different methods (i.e., fluorescence recovery after photobleaching or single-molecule detection), and different laboratories.
Introduction
Genomic DNA is continuously subjected to endogenous and exogenous insults from free radicals, ionizing radiation, and DNA-modifying chemicals. All of these, either directly or during the repair process, cause single-strand and double-strand breaks (SSBs and DSBs). Without proper DNA damage detection and repair, the resulting genomic instabilities can lead to premature aging, sensitivity toward radiation damage, and cancer. Although specific repair pathways exist for different types of DNA lesions, the sequential accumulation and binding of many signaling and repair proteins at damage sites is conserved (1). Thus, a quantitative understanding of protein accumulation, order of accumulation, and variability due to type and amount of damage, cell type, etc., is not only critical for establishing a fundamental framework of DNA repair pathways but also of clinical relevance because mutations or misregulation in DNA damage detection and repair pathways are strongly associated with or even cause many cancers (2).
Poly(ADP-ribose) polymerase 1 (PARP1) and the less abundant PARP2 are two proteins with partially overlapping functions; the presence of each protein is essential for the DNA damage repair response to both SSBs and DSBs (3, 4, 5, 6, 7, 8, 9). These two known first responders to DNA damage (10) are members of the larger family of diphtheria-toxin-like ADP-ribosyltransferases, and both are enzymatically activated upon binding to DNA strand breaks. When active, they use NAD+ to polymerize long chains of poly(ADP)-ribose (PAR) onto themselves and other nuclear acceptor proteins such as histones and DNA repair proteins. These PAR chains recruit a cohort of repair proteins that contain PAR-binding motifs (11, 12, 13), thus leading to the activation of DNA repair. Recently, a newly discovered protein, histone PARylation factor 1 (HPF1), has been shown to direct modifications after DNA damage onto serine (instead of glutamate and aspartate) residues, thereby facilitating the PARylation of histone tails by both PARP1 and PARP2 (14, 15, 16).
There are many different genetic, biochemical, cellular, and animal-model methods to investigate DNA repair pathways. Laser microirradiation is a particularly powerful and accessible method to study these processes in living cells, with >100 published applications before 2017 (17) and at least 15 more since then. In this approach, cells are first transiently transfected with a fluorescently tagged protein of interest. Local DNA damage is induced in the transfected cells with a short-wavelength (355–405 nm) laser, and the time-dependent accumulation of the tagged proteins of interest at the site of laser-induced DNA damage is monitored by fluorescence microscopy. This unique combination of biophysical manipulation and cell biology has yielded compelling data on the order and kinetics for the recruitment of many different DNA repair proteins such as PARP1, ataxia-telangiectasia mutated kinase, Ku80, p21, and even phospholipids at sites of DNA damage (18, 19, 20, 21, 22, 23, 24, 25, 26, 27).
Although robust analytical methods have been developed for quantitation of fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP) (28, 29, 30), analysis of laser microirradiation experiments typically is limited to reports of appearance at sites of damage, often expressed as the time to half-maximal accumulation (t1/2) (26). Although this approach is simple, it provides no insight into the mechanism (e.g., simple, facilitated, or anomalous diffusion) or rate of protein movement (i.e., diffusion coefficients). Furthermore, t1/2 analyses typically rely on averaged data from many individual cells, despite significant cell-to-cell variability in the amounts and rates of protein accumulation. Thus, t1/2 analyses do not allow for quantitative comparisons with other methods that monitor protein movement such as FRAP, fluorescence correlation spectroscopy (FCS), or single-molecule tracking. Of additional concern, t1/2 analyses ignore variation in nuclei size and shape, which, as we show here, leads to qualitatively and quantitatively incorrect conclusions.
Some analyses of laser microirradiation data rely on quantitative fitting using single (17) or multiple first-order rate constants (18). However, these exponential fits are not based on a physical model for protein accumulation and thus suffer from the same deficiencies as the t1/2 analyses while also failing to account for the unique size and shape of each nucleus. Additionally, using multiple exponentials for fitting a single accumulation curve is most likely overfitting and might yield values that cannot be compared meaningfully with other experiments.
Here, we develop a quantitative Monte Carlo model to account for protein accumulation at sites of DNA damage, which assumes that the movement of PARPs and HPF1, like most nuclear proteins (28, 31), can be described by simple diffusion. We employ the model to explicitly account for the unique size and shape of each nucleus and demonstrate that the traditional approaches of fitting accumulation kinetics (t1/2 and exponential fitting) can lead to erroneous results because of nucleus shape and size variability. Experimentally, we benchmark our model, quantitation of fluorescence accumulation after DNA damage (Q-FADD), by determining effective diffusion constants (Deff) for both PARP1 and PARP2, which are in good agreement with previously reported values for PARP1 (32). We then apply our approach to HPF1, which has recently been shown to act as an essential modulator of both PARP1 and PARP2 (14, 15, 16). Analysis of the accumulation kinetics with Q-FADD reveals that all three proteins diffuse independently to sites of DNA damage.
Materials and Methods
Expression plasmids, cell culture, and transient transfection
Mammalian expression plasmid (pEGFP-C3) encoding full-length GFP-tagged human PARP1 and wild-type mouse embryo fibroblasts (MEFs) were a kind gift from Dr. Françoise Dantzer (University of Strasbourg, Strasbourg, France). The expression plasmid for GFP-PARP2 was generated by subcloning human PARP2 cDNA into pEGFP-C3 between the restriction sites SalI and BamHI. dsRed-HPF1 construct was kindly provided by Dr. Shan Zha (Columbia University, New York, NY). HPF1 cDNA was amplified from this construct and was introduced into pEGFP-C3 by Gibson assembly. PARP1 chromobody (Chb-PARP1) was purchased from ChromoTek GmbH (Planegg-Martinsried, Germany).
MEF cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 50 μg/mL of gentamicin and 10% fetal bovine serum. HeLa-CCL2 cells were purchased from ATCC (Manassas, VA) and were cultured in Dulbecco’s modified Eagle’s medium supplemented with 1× penicillin/streptomycin and 10% fetal bovine serum. For the laser microirradiation experiments, cells were grown on CELLview slides (Greiner Bio-One, Monroe, NC) and were transfected with jetPEI (Polyplus-transfection, Illkirch-Graffenstaden, France) according to the manufacturer’s instructions. Briefly, 20,000 cells were plated and transfected 24 h later with 250 ng of DNA. Cells were sensitized with Hoechst 33342 (Invitrogen, Carlsbad, CA) (10 μg/mL) for 10 min before the start of the experiment.
Data collection
Live-cell imaging and laser microirradiation experiments were performed on a Nikon A1R laser scanning confocal microscope (Nikon, Tokyo, Japan) equipped with a 405 nm diode laser and an argon-ion laser for 488 nm excitation (Biofrontiers Institute, University of Colorado, Boulder, CO). Cells were imaged using a 100 × 1.45 NA oil immersion objective. A stage-top incubator was used to maintain proper environmental conditions of 37°C and 5% CO2, and for each data collection, a cell was positioned so that its nucleus was at the center of the acquisition field of view. To induce DNA damage (including SSBs, DSBs, and pyrimidine dimers (33)), the 405 nm laser was used at ∼1.7 mW to irradiate a predetermined rectangular region of interest (ROI) across the nucleus for 1 s. Excitation of the GFP fluorophore, using the 488 nm laser lines, was used to monitor the accumulation of PARP in the ROI. Six preirradiation and 150 postirradiation frames were recorded at 2 s time intervals. Images were collected at a frame size of 512 × 512 as per Nyquist sampling (1.2 AiryUnits). The resulting time lapse video was saved as an ND2 file (proprietary Nikon format), and the region of irradiation was saved as a TIF image.
Data processing
Automated analysis of the fluorescent image was carried out using a custom code in MATLAB (The MathWorks, Natick, MA), which is described here (https://www.openmicroscopy.org/bio-formats/). First, the digital ND2 image (16 bit) was imported into MATLAB using the Bioformats Image toolbox (The Open Microscopy Environment). The region of the image corresponding to the nucleus was segmented using an intensity threshold from either the EGFP or Hoechst channel as converted to grayscale. The mean intensity Imean in a 20-pixel radius circle around the center of the image, i.e., the center of the nucleus (Video S1. Accumulation of GFP-PARP1 at Sites of Laser-Induced DNA Damage, Video S2. Accumulation of GFP-PARP2 at Sites of Laser-Induced DNA Damage, Document S2. Article plus Supporting Material a), was used to determine the threshold level (0.5 × Imean). Applying this grayscale threshold to the image generated a binary mask of the nucleus (Fig. S1 b). Objects in the mask less than 500 pixels in area were removed, and gaps in the mask were filled in. Finally, a disk-shaped structuring element (radius = 3 pixels) was used to perform morphological dilation to smooth the edges and generate the final nuclear mask (Document S1. Figs. S1–S10, Video S1. Accumulation of GFP-PARP1 at Sites of Laser-Induced DNA Damage, Video S2. Accumulation of GFP-PARP2 at Sites of Laser-Induced DNA Damage, Document S2. Article plus Supporting Material c). This binary mask is simply used to identify the pixels in the image that belong to the nucleus and those that belong to the background. The mask has a value of “true” for the nucleus, “false” for the background. This mask is then used in following algorithms for computations such as calculating the mean intensities. If multiple nuclei were present in the field of view, the nucleus closest to the center of the image was selected for further analysis.
To measure the fluorescence intensity across the nucleus as identified above, we divided the cell vertically into a number of ROIs. The initial ROI (ROI0) was generated from the TIF image of the irradiation region and was vertically expanded by 10 pixels to avoid edge effects. Depending on the height of the nucleus, additional ROIs were then generated by translating the original ROI above and below its original position (Fig. S2). The total intensity of the nucleus within each ROI was measured for each frame of the video. These intensity measurements were corrected for both background fluorescence and photobleaching. To determine the background fluorescence level, the image background was segmented using an intensity threshold. An intensity histogram of the image, smoothed by a 3-pixel moving window to remove spurious peaks, was used to set the threshold level. The first peak of the histogram, corresponding to the darkest grayscale values, was identified, and the grayscale value at which the counts dropped to 1/e of the peak height was used to generate a mask of the background. The background intensity was then calculated as the mean intensity of the background mask, as shown in Fig. S3. This process was repeated for the first six preirradiation frames, and the background correction level was calculated as the mean of these six values. This correction level was then subtracted for each frame of the video. A separate correction level was calculated for each fluorescence channel in the video. To correct for photobleaching, the total intensity of the nucleus in each frame was normalized to the total intensity of the nucleus in the first frame after applying the background correction. The correction for photobleaching was minimal (<10%). For each ND2 image file, the code generated a CSV file containing the time-series measurements of the intensity within each ROI. The cell-based data are displayed as normalized fluorescence values compared to the initial fluorescence value within ROI0 (averaged over the first six frames). The ROI and the nuclear mask were also exported as a text file to serve as inputs for the simulation of diffusion.
Simulation of diffusion
Protein diffusion was modeled via standard Monte Carlo approaches on a two-dimensional grid using a custom code in the Mathematica environment. Because the image plane of the confocal microscope is ∼1 μm deep, the data we have collected integrate over this depth, allowing us to make the assumption of a two-dimensional plane with net flux of zero. We note that Monte Carlo simulations have been used to analyze FRAP data that account for differing shapes of illumination but not individual nuclear shapes (34, 35). The region confined by the nuclear mask served as the active region of the simulation. 12,000 sample points were randomly generated at uniform density within the nuclear mask and allowed to propagate in a random walk. The step size for the random walk is constant, set to the pixel dimension of the experimental image. The direction is chosen randomly, with a 50/50 probability of going left or right and a 50/50 probability of going up or down. Control studies were performed with more sample points (up to 16,000); however, results did not differ beyond a negligible reduction in noise observed for the accumulation and depletion kinetics. At each time step of the trajectory, a pseudorandom number generator was employed to determine step interval in the x (left, right) and y (up, down) directions. Under these conditions, the diffusion coefficient, D is given by
where Δx is the step size and Δt is the time interval. The grid size of the simulation was chosen to match the pixel resolution of the experimental microscope images (0.08678 μm/pixel). The time step was typically fixed at 0.16 s; however, variation of this parameter within a factor of five had no effect on the results of the simulation. At each time step (n), the position of each point at step (n + 1) was calculated and verified to lie within the nuclear boundary. If the new (n + 1) position was determined to escape the nuclear region, reflecting boundary conditions were enforced by reverting the (n + 1) position to the position at step (n). Trapping in ROI0 was similarly enforced. At each step (n), the point position was checked to determine whether it lay within the trapping region. If so, position at step (n + 1) was set to the position at step (n). The trajectories of each of the 12,000 points were stored in memory for postrun analysis. Accumulation and depletion kinetics were determined by summing the total number of points in each ROI at each time step from the stored trajectories. The experimental data were fitted empirically using r-squared coefficients between the simulated curves and the experimental data. Systematic testing of ranges of Deff and F indicate that there is only one best solution for each nucleus. We processed the accumulation kinetics for all the nuclei shown in Figs. S4–S7 and only analyzed selected depletion kinetics from regions adjacent to the DNA damage site. (Note that the depletion kinetics are accurately modeled only when the ROI areas are located entirely inside the nucleus.) The software for processing of individual nuclei in MATLAB and for deriving Deff and F in Mathematica are publicly available at https://biof-git.colorado.edu/biofrontiers-imaging/luger-DNA-damage.
Results
Laser microirradiation and accumulation of PARPs at sites of DNA damage
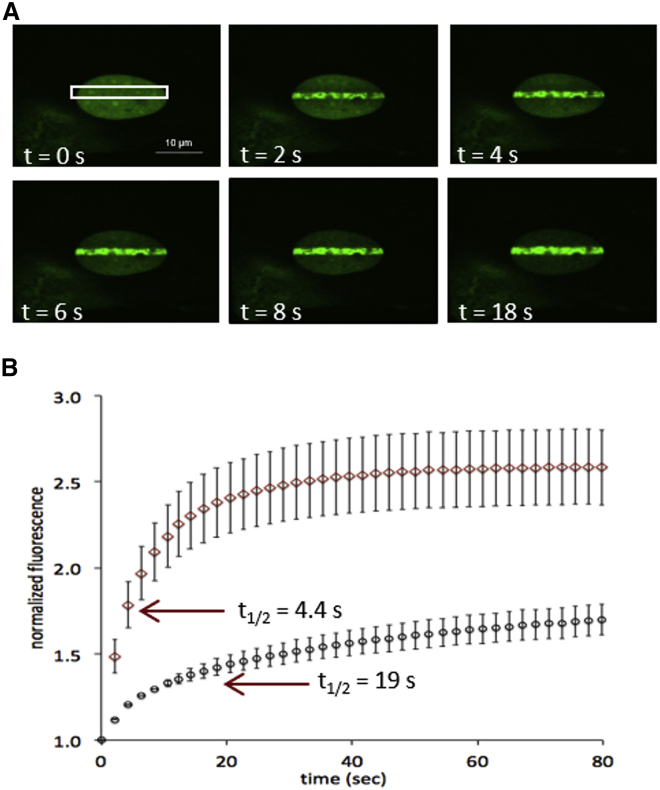
To develop and test our method for analyzing protein accumulation at sites of DNA damage, we first needed to generate a sufficiently large data set for which we had all the necessary raw data, including the time-dependent intensities for the fluorescent and Hoechst channels, as well as individual nuclear envelopes for each cell. We transfected either GFP-PARP1 or GFP-PARP2 into MEFs, where both proteins were easily visualized in nuclei by fluorescence microscopy (e.g., Fig. 1 A). We next monitored the recruitment of GFP-PARP1 and GFP-PARP2 to sites of laser irradiation (see Fig. 1 A; representative videos can be found as Video S1 and S2). This method of laser irradiation is known to cause a variety of DNA alterations, including SSBs, DSBs, and pyrimidine dimers (33), and is thus expected to recruit both PARP1 and PARP2, which are both known to bind and respond to SSBs and DSBs (6, 7). In agreement with previous reports (24, 36), we saw a rapid increase in fluorescence intensity of PARP1 and PARP2 at the site of DNA damage by factors of 1.2–5.5, reaching maximal intensities within 60–200 s (Fig. 1 B). As noted previously (24), PARP1 accumulates more rapidly than PARP2, as measured by t1/2 of data averaged from all the nuclei (Fig. 1 B). For both PARP1 and PARP2, we observed significant depletion of fluorescence from sites outside the region of DNA damage (Fig. 1 A). As reported previously, both PARP1 and PARP2 are observed to deplete from the site of damage when the nuclei are monitored for much longer time courses (data not shown) (24, 36). We thus demonstrate that we can generate microirradiation data that is qualitatively similar to previously published reports.
Figure 1.
PARP1 accumulates faster than PARP2 at sites of DNA damage after laser microirradiation: (A) video snapshots showing GFP-PARP1 accumulating at a site of DNA damage (white box) in an MEF nucleus. (B) Accumulation of GFP-PARP1 (diamonds) and GFP-PARP2 (circles) in MEF cells is shown as the average ± standard error of the mean (SEM) of 28 and 19 nuclei, respectively. Accumulation of PARP1 and PARP2 for individual nuclei are shown in Figs. S4 and S5, respectively.
Modeling of diffusion
In the data collected for the nuclei above (see also Figs. S4–S9), we noted significant cell-to-cell variations in apparent t1/2 and amount of accumulation, as well as in nuclear size and shape. Because there may be unknown factors that contribute to cell-to-cell variability, we ruled out any method that involved averaging of data from multiple nuclei. Also, we envisioned that our method should yield numerical values with a physical meaning for comparison with data obtained in other laboratories and by other methods. We therefore ruled out fitting our data from individual nuclei to single or multiple exponentials, as this approach has no physical basis. Instead, we developed a method based on the physics of diffusion because the movement of many nuclear proteins, even those known to bind tightly to chromatin, has been well-described by diffusion (28, 34, 37, 38). Although a continuum diffusion approach could, in principle, be used to model experimental results (as is the standard for FRAP (28)), the boundary conditions for each uniquely shaped nucleus would either need to be ignored (see also below) or would have to be parameterized, which is challenging.
We therefore used a Monte Carlo approach to model protein movement in the nucleus and accumulation at sites of DNA damage. This method provides the ability to simulate a discrete diffusional process while incorporating an experimentally accurate nuclear profile into the simulation. In these simulations, the effective diffusion coefficient (Deff) of the particles is parameterized with the particle step frequency (1/Δt) and the particle step size (Δx). The trajectories of the particles as they undergo a random two-dimensional walk are confined to the experimentally accurate nuclear boundary and are trapped at the defined site of laser damage. Because we see no significant depletion from the irradiated area of PARP1, PARP2, or HPF1 over the time course of observation and analysis (1–2 min), we make the simplest assumption that these proteins are trapped for the full time course of our simulation. F is defined as the fraction of mobile particles, all of which will accumulate within the trap by the end of the simulation. It is important to note that Deff is the effective diffusion coefficient that comprises the fast binding and release of the protein from DNA and, in general, produces a slower diffusion coefficient than expected from the molecule’s size (28). That is, this analysis does not reveal the thermodynamic or kinetic parameters of the DNA-protein interaction but does reveal the mechanism, rate, and amount of protein that accumulates at sites of DNA damage.
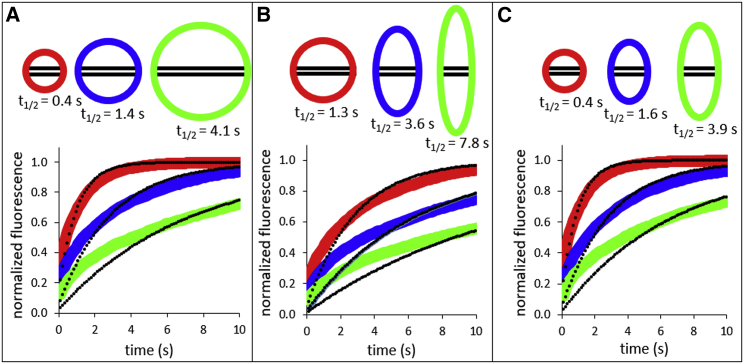
Nuclei in living cells are highly variable in both size and shape (see, e.g., Figs. S4–S9). To highlight the importance of accounting for both the size and shape of a nucleus to correctly determine Deff, we modeled a series of simple test cases, which are presented in Fig. 2. In the simplest example, we model diffusion using the same Deff in three different circular nuclei of varying diameter (Fig. 2 A). Apparent accumulation as measured by t1/2 is slowest in the largest circle, consistent with the intuition that the average molecule must traverse a longer distance to be caught in the trap. Next, we keep both Deff and the area of each nucleus constant but vary the ellipticity (Fig. 2 B). As measured by t1/2, the accumulation of particles takes longer in more elongated nuclei, again consistent with the intuition that the average molecule must traverse a longer distance to be caught in the trap. In a final example, which most realistically mimics the diversity we observe in real nuclei, we allow both the size and ellipticity to vary while holding constant the size of the trap and Deff (Fig. 2 C). Again, particles appear to accumulate more slowly in the more elliptical and larger nuclei. We emphasize that in all three examples, Deff is constant, yet the t1/2 parameters vary by as much as a factor of 10 solely because of the differences in size and shape of the nuclei. From these simulations, we infer that the opposite must also be true, namely that similar t1/2 values can be found for different nuclei whose Deff differs by a factor of 10. If we perform simulations with circular instead of rectangular damage sites, the differences in t1/2 introduced by varying nuclear sizes and shapes are still apparent unless the damage sites are so small that the data for protein accumulation become noisy. These results highlight the importance of explicitly accounting for the varying size and shape of individual nuclei in the analysis of experimental microirradiation data when the damaged area is greater than ∼2% of the total nuclear area. The overwhelming majority of published data fall into this category.
Figure 2.
Varying nucleus size and shape affect t1/2 at constant values of Deff in a simulation: each panel shows a different scenario of size and/or shape variation of the nucleus along with the simulated accumulation of GFP-PARP1 in the region of laser damage (box, with unchanged height) and the apparent t1/2 assuming a Deff = 4.53 μm2/s and F = 1. The colors of the nuclei correspond to the data in the graph. (A) Three different circular nuclei with radii of 60, 100, and 150 pixels are shown. (B) Three different nuclei of varying ellipticity (100 × 100, 70 × 143, 50 × 200 pixels) but with the same overall area are shown. (C) Three different nuclei of varying size and ellipticity (60 × 60, 60 × 100, 60 × 150 pixels) are shown but, as in (A) and (B), with the same trap size. The black dots superimposed on each curve indicate the best fit to a first-order exponential and demonstrate that size and shape also influence the quality of this fit, which may lead to arbitrary application of multiexponential fitting for larger or more elongated nuclei.
Analysis of the simulated accumulation kinetics using simple rate models leads to similarly misleading conclusions. For example, fits to the accumulation kinetics in Fig. 2 using a single exponential model (black dots) yield slower apparent derived rates for larger or more elongated nuclei compared to smaller and rounder nuclei, despite identical diffusion coefficients. Furthermore, as can be seen by comparing the single exponential fits in Fig. 2 C, larger or more elongated nuclei are fitted more poorly to a single exponential than smaller rounder ones, thus potentially leading to the different numbers of exponentials for the same protein purely because of differences in nucleus shape. Efforts to obtain better fits have in the past led to overparameterization (18). Also, as noted above, there is no physical basis for fitting microirradiation data using exponentials. Thus, unless applied only as a comparative tool in the same nucleus, analysis of protein accumulation at sites of DNA damage by either t1/2 measurements or exponential models will lead to specious results, which are both qualitatively and quantitatively incorrect.
Testing the diffusion model for PARP1, PARP2, and HPF1 recruitment
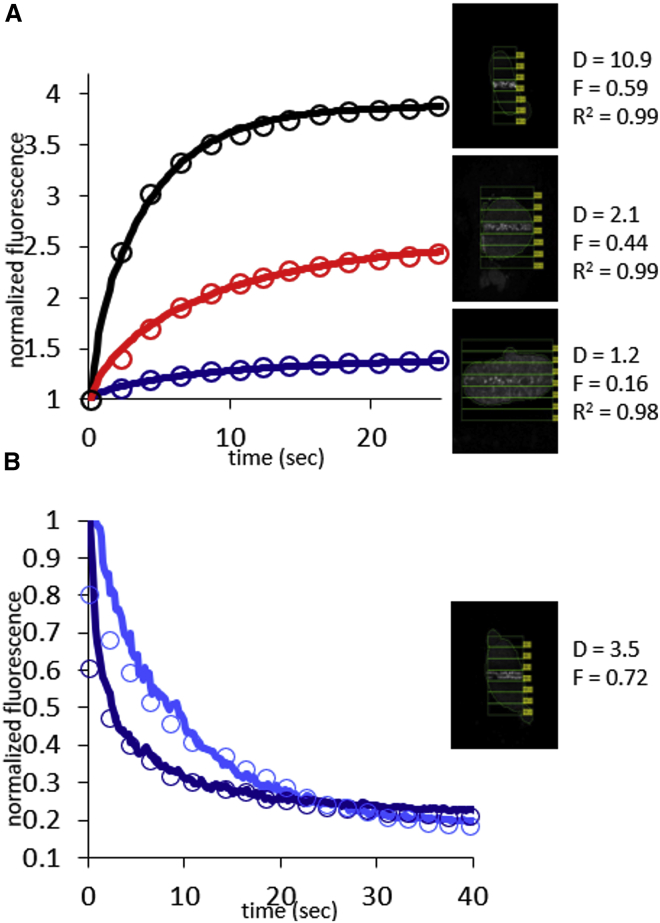
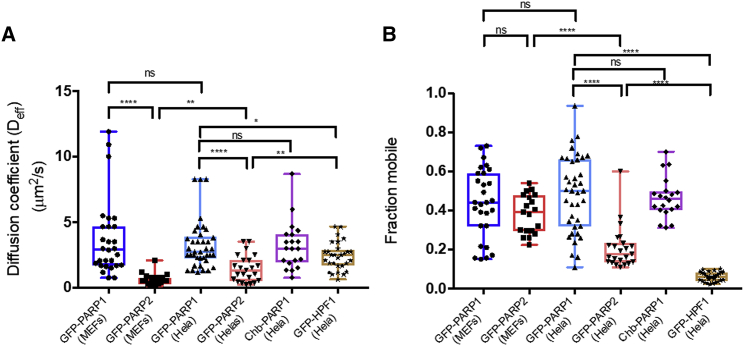
To quantify the difference in rates of accumulation between PARP1 and PARP2, we used our model of diffusion to simulate the accumulation of PARPs in the region of DNA damage in MEF cells, comparing the modeled curves with actual experimental data. For both PARP1 and PARP2, two parameters were sufficient to generate curves that fit the data very well (as judged by r-squared coefficient >0.96), namely Deff, the effective coefficient of diffusion, and F, the fraction of mobile protein. Errors in both Deff and F of 5–20%, depending on the nucleus, can be estimated from empirical fitting with the criterion that r-squared remains >0.96. Three example data sets for PARP1 are shown in Fig. 3 A. In addition to the accumulation kinetics, our model also accurately describes the depletion of PARP1 from regions adjacent to the damage site using the same parameter values Deff and F. As one can intuit, we find that depletion of PARP1 occurs more quickly at sites closer to the damage site, although all proteins in the nucleus move with the same Deff (Fig. 3 B). In Figs. S4 and S5, we show the raw data, respective fits, and variability in size and shape for 28 nuclei accumulating GFP-PARP1 and 19 nuclei accumulating GFP-PARP2 at sites of DNA damage in MEF cells. We summarize the derived values of Deff and F in box-and-whisker plots (Fig. 4) and in Table 1.
Figure 3.
Appearance of fluorescence at sites of DNA damage and disappearance of fluorescence from the nucleus can both be modeled by simple diffusion. (A) Overlay of experimental data from Q-FADD for GFP-PARP1 and simulation to determine Deff for GFP-PARP1 in MEF cells is shown for three representative nuclei (see also Fig. S4). (B) Depletion of PARP1 from two different sites within one nucleus can be quantitatively described using the same Deff (dark blue = ROI-1, adjacent to site of damage; light blue = ROI-2, adjacent to ROI-1) (see also Fig. S2).
Figure 4.
Q-FADD reveals statistically significant differences between PARP1, PARP2, and HPF1 recruitment. Box-and-whisker plots for Deff (A) and F (B) as determined by simulation and matching to Q-FADD with GFP-PARP1, GFP-PARP2, and GFP-HPF1 are given. Data are shown for transient transfections of GFP-PARP1, GFP-PARP2, and GFP-HPF1 in MEF and/or HeLa cells and detection of endogenous PARP1 using chromobodies (Chb). Statistical differences between pairs of samples were evaluated using the unpaired t-test with p < 0.05 (∗) and p < 0.001 (∗∗∗∗).
Table 1.
Summary of Parameters Derived from Diffusion Analysis of Q-FADD Data
| GFP-PARP1 | GFP-PARP2 | GFP-PARP1 | GFP-PARP2 | Chb-PARP1 | GFP-HPF1 | |
|---|---|---|---|---|---|---|
| Cell type | MEFs | MEFs | HeLa | HeLa | HeLa | HeLa |
| n | 28 | 19 | 38 | 23 | 20 | 29 |
| Deff (μm2/s) ± SEM | 3.7 ± 0.6 | 0.62 ± 0.1 | 3.2 ± 0.3 | 1.4 ± 0.2 | 3.2 ± 0.4 | 2.4 ± 0.2 |
| F ± SEM | 0.44 ± 0.03 | 0.38 ± 0.02 | 0.48 ± 0.03 | 0.21 ± 0.02 | 0.46 ± 0.02 | 0.059 ± 0.0044 |
Deff and F values represent the mean and SEM for the indicated number of nuclei (n). The corresponding box-and-whisker plots are shown in Fig. 4, along with a statistical evaluation of differences.
Statistical analysis demonstrates that PARP2 has a sixfold slower diffusion coefficient than PARP1 in MEF cells (3.7 vs. 0.62 μm2/s; Fig. 4; Table 1). Test simulations using a “leaky” trap, in which we allow proteins to escape again, show that this difference cannot be reproduced by assuming, based on experimental observations (39), that PARP2 has a weaker affinity to DNA in the damaged site than PARP1. In fact, the opposite behavior is observed in simulations, with accumulation to maximum occurring faster when trapped particles are allowed to become free with finite probability. Recall that Deff describes the net movement of proteins from throughout the nucleus to the sites of DNA damage and thus encompasses both free (unbound) diffusion as well as the kinetics of PARP binding to DNA and other proteins. Although at present we cannot account for the smaller Deff for PARP2 versus PARP1, the ability to model different mechanisms (e.g., leaky traps, nonanomalous diffusion) is one advantage of the Q-FADD approach.
We next investigated the effects of cell type on the accumulation of PARP1 and PARP2 at sites of DNA damage, comparing MEFs and HeLa cells. Not surprisingly, there are some cell-type-specific variations, particularly for PARP2. Despite these variations, Deff for PARP1 is significantly higher than the Deff for PARP2 in both HeLa and MEF cells (Fig. 4; Table 1).
To test the effects of endogenous versus exogenous PARP1 expression, we needed to find a way to also measure the accumulation of endogenous (untagged) PARP1. Chromobodies are small functional antibodies that are tagged with a chromophore and are readily transfected into cells (40). Chromobodies to human PARP1 were used to monitor endogenous PARP1 accumulation in HeLa cells to compare with accumulation of transiently transfected GFP-PARP1. Subsequent analysis yielded statistically identical values for both Deff and F as confirmed by unpaired Student’s t-test (Fig. 4; Fig. S8; and Table 1). These results demonstrate that transfection of GFP-tagged PARP1 does not significantly affect its accumulation at sites of DNA damage in the nucleus, as has been previously observed for other proteins (41).
Finally, given the recent discovery that HPF1 redirects PARP-mediated PARylation onto histones, specifically targeting serine residues on histone tails (14, 15, 16), we asked whether HPF1 travels to sites of DNA damage with PARP1 or PARP2 or rather is recruited independently. Interestingly, HPF1 appears to diffuse more slowly than PARP1 but still faster than PARP2 (Fig. 4; Fig. S9; and Table 1). The independent arrival of HPF1 at sites of DNA damage is consistent with our finding that there is no interaction between PARP1 (or PARP2) and HPF1 in vitro in the absence of DNA binding (J.R., unpublished data). We also observe that the fraction of HPF1 accumulation is much lower than either PARP1 or PARP2, as measured by F. Thus, even under conditions of transient transfection, the ratio of PARP1 to HPF1 at sites of DNA damage must favor PARP1 by a large margin. These results appear to reflect the situation in untransfected cells, where the concentration of HPF1 (0.1 μM) is much lower than that of PARP1 (2 μM) (14). Given the slower diffusion coefficient and much lower accumulation at sites of DNA damage of HPF1 compared to PARP1, there exists a potential dilemma that HPF1 cannot physically visit all sites fast enough to modify PARP1 activity and to ensure that all PARylation ends up on serine residues.
Correlation analysis
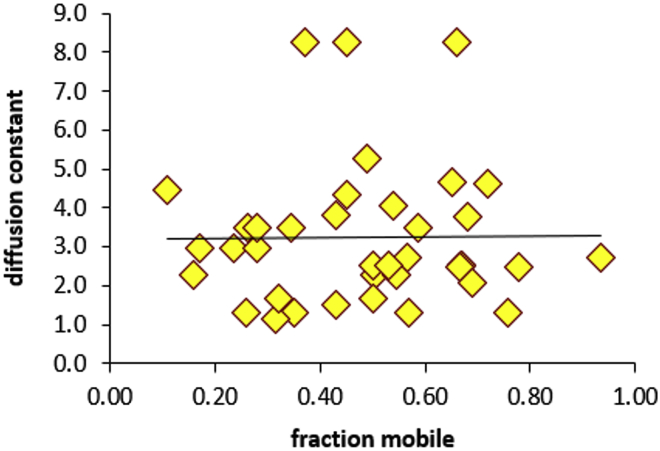
Our extensive description of the variability between different nuclei in Deff and F for PARP1 and PARP2 is novel, to our knowledge, in the analysis of protein accumulation by laser microirradiation, in which data are generally presented as averages of many different nuclei. We observed that both Deff and F varied significantly for different nuclei, despite accounting for the different sizes and shapes of the nuclei (note the whiskers in Fig. 4). Using our largest data set of 38 nuclei of GFP-PARP1 in HeLa cells, we found no correlation between Deff and the fraction of mobile protein, F (Fig. 5). In analysis of diffusion by FRAP, F is typically interpreted as the ratio of the effective rates of protein binding and release from stationary sites (i.e., DNA) (28). If this assumption were to hold for PARP1, we would expect a direct correlation between Deff and F because a larger F would imply less binding of PARP1 to DNA during its travel from its initial position to the damage site and thus faster arrival and a larger observed Deff. The fact that we do not observe a correlation between Deff and F strongly suggests that this assumption is incorrect, at least for PARP1 and PARP2 (data for PARP2 not shown). The lack of correlation and the variability in both Deff and F for PARP1 and PARP2 hint at a yet unidentified cause for the cell-to-cell variability that requires further investigation. For example, one might expect to see variable rates of diffusion depending on cell-cycle phase or levels of DNA damage, and this is an area of future studies, now made possible with our novel, to our knowledge, method of quantitation. We also saw no correlation of either Deff or F with the level of overexpression of GFP-PARP1 as measured by initial total fluorescence intensity of the entire nucleus (Fig. S10).
Figure 5.
Deff and F are not correlated. Deff is plotted versus F for the accumulation kinetics of 38 HeLa nuclei transfected with GFP-PARP1 and analyzed by Q-FADD. A similar lack of correlation was seen for GFP-PARP2 (data not shown).
Discussion
For PARP1, PARP2, and HPF1, accumulation at sites of DNA damage can be described by simple diffusion. Our results imply that movement of these three proteins is neither facilitated (i.e., actively transported by directed motion) nor anomalous. Our results are in agreement with the finding that many other proteins move by diffusion, as determined by FRAP, FCS, or single-molecule tracking (28, 34, 37, 38). With our new, to our knowledge, method of analysis, it is now possible to correlate microirradiation data with these other methods for any protein of interest.
The mean Deff of GFP-PARP1 is very fast (3.7 μm2/s, Table 1), only ∼9-fold slower than the theoretical limit of 31.5 μm2/s, assuming a spherical protein of 143 kDa (r = 3.45 nm) and a viscosity of the nuclear milieu of 2 × 10−3 N s/m2, twofold as viscous as pure water (42). In fact, the Deff of PARP1 is among the fastest known of any nuclear protein whose Deff has been determined by FRAP, FLIP, FCS, or single-molecule tracking (28, 38, 43). Our derived Deff for PARP1 is consistent with the values of 4.6 and 2.9 μm2/s as determined by FCS and FRAP, respectively (32), wherein it was also demonstrated that the diffusion of PARP1 was essentially not anomalous (anomaly factor = 0.89). The high Deff for PARP1 is surprising because PARP1 is known to bind tightly to damaged and undamaged DNA (44, 45) and in light of the fact that PARP1 has many other roles that involve binding to chromatin or other proteins (46). The fast diffusion of PARP1 is consistent with our recent demonstration of PARP1 release from DNA being facilitated by interaction with an additional strand of undamaged DNA (monkey-bar mechanism). The high DNA concentration in the nucleus promotes rapid binding and release of PARP1, leading to its rapid movement through the nucleus (47).
Although both PARP2 and HPF1 have a lower molecular weight than PARP1 and are therefore expected to move more rapidly, both proteins diffuse more slowly than PARP1. In fact, PARP2 diffuses 60-fold slower than its theoretical limit of 36.4 μm2/s. Although the reason for the slower diffusion of PARP2 is unknown at this time, the application of Q-FADD now allows for quantitative investigations using mutants of PARP2 and/or knockdown of potential binding partners that might increase its effective size and slow its rate of diffusion. We speculate that PARP2, which has fewer DNA-binding domains than PARP1, may not use the highly effective monkey-bar mechanism used by PARP1 in moving from one piece of DNA to the next (47). We noted that the accumulation kinetics of PARP2 do not show a lag phase, a characteristic feature that has been previously attributed to sequential accumulation (18). It is therefore unlikely that PARP2 recruitment depends on the accumulation of some other protein. Elegant experiments have recently demonstrated that PARP2 is recruited to PAR chains generated by PARP1 at sites of DNA damage (48). However, because of the exceedingly fast accumulation of PARP1, it is likely that any characteristic lag of PARP2 accumulation due to this dependence is undetectable.
Finally, our observation that HPF1 does not travel with either PARP1 or PARP2 and accumulates to much lower levels raises a number of interesting questions about the observed predominance of PARylation of serine residues after DNA damage in vivo (14). First, assuming that PARP1 arrives first and immediately becomes activated by binding to damaged DNA, how does it not cause massive amounts of PARylation of glutamate and aspartate residues on itself before modulation of its activity by the later-arriving HPF1? Are these initial labile modifications transferred in a secondary reaction? Second, because HPF1 levels are ∼20-fold lower than PARP1 in the cell (16) and we observe >7-fold lower accumulation for HPF1 compared to PARP1, is there an as-yet unknown assistant to HPF1, as suggested previously (49)?
In summary, we have developed a powerful and, to our knowledge, new method that combines the cell-based technique of laser-induced microirradiation with Monte Carlo simulation to derive the diffusion coefficient Deff and the fraction accumulation F for quantifying proteins at sites of DNA damage. Q-FADD makes it possible to compare protein movement derived from different methods such as FRAP, FCS, or single-molecule tracking. Our method can also be used to quantitatively probe interesting biological questions regarding the function of proteins recruited to sites of DNA damage. Because monitoring accumulation of fluorescent proteins after laser microirradiation is a relatively simple technique available to many researchers with standard cell culture and microscopy facilities and because it can be readily applied to already collected data, we anticipate that Q-FADD will find wide application in the field of DNA repair biology.
Author Contributions
J.M. performed all the cell biology experiments. A.J. helped with designing constructs. J.W.T. and E.M.G. developed the software. J.M. and J.R. did all the data analysis and manuscript preparation. J.D. provided microscopy support. K.L. provided overall guidance and funding. All authors have read and accepted the submission file.
Acknowledgments
We thank Françoise Dantzer (University of Strasbourg) for GFP-PARP clones. The imaging work was performed at the BioFrontiers Institute Advanced Light Microscopy Core. We also thank Samuel Bowerman for help with correlation plots.
Laser scanning confocal microscopy was performed on a Nikon A1R microscope supported by NIST-CU Cooperative Agreement award number 70NANB15H226. J.M., J.R., A.J., and K.L. are supported by the National Institutes of Health-National Cancer Institute R01 CA218255, the University of Colorado Cancer Center (Pilot Funding Grant ST63501792), and the Howard Hughes Medical Institute. J.W.T. and J.D. are funded by the NIST-CU Cooperative Agreement award number 70NANB15H226 (2015–2020).
Editor: Toshio Tsukiyama.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.04.032.
Supporting Material
References
- 1.Dexheimer T.S. DNA repair pathways and mechanisms. In: Mathews L.A., Cabarcas S.M., Hurt E., editors. DNA Repair of Cancer Stem Cells. Springer; 2013. pp. 19–32. [Google Scholar]; Dexheimer, T. S. 2013. DNA repair pathways and mechanisms. In DNA Repair of Cancer Stem Cells, L. A. Mathews, S. M. Cabarcas, E. Hurt, eds. (Springer), pp. 19-32.
- 2.Khanna K.K., Shiloh Y. Springer; Netherlands, Dordrecht: 2009. The DNA Damage Response: Implications on Cancer Formation and Treatment. [Google Scholar]; Khanna, K. K. and Y. Shiloh. 2009. The DNA Damage Response: Implications on Cancer Formation and Treatment: Springer Netherlands, Dordrecht.
- 3.Bai P. Biology of poly(ADP-ribose) polymerases: the factotums of cell maintenance. Mol. Cell. 2015;58:947–958. doi: 10.1016/j.molcel.2015.01.034. [DOI] [PubMed] [Google Scholar]; Bai, P. 2015. Biology of poly(ADP-ribose) polymerases: the factotums of cell maintenance. Mol. Cell. 58:947-958. [DOI] [PubMed]
- 4.Bock F.J., Chang P. New directions in PARP biology. FEBS J. 2016;283:4017–4031. doi: 10.1111/febs.13737. [DOI] [PubMed] [Google Scholar]; Bock, F. J., and P. Chang. 2016. New directions in PARP biology. FEBS J. 283:4017-4031. [DOI] [PubMed]
- 5.Morales J., Li L., Boothman D.A. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit. Rev. Eukaryot. Gene Expr. 2014;24:15–28. doi: 10.1615/critreveukaryotgeneexpr.2013006875. [DOI] [PMC free article] [PubMed] [Google Scholar]; Morales, J., L. Li, …, D. A. Boothman. 2014. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit. Rev. Eukaryot. Gene Expr. 24:15-28. [DOI] [PMC free article] [PubMed]
- 6.De Vos M., Schreiber V., Dantzer F. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem. Pharmacol. 2012;84:137–146. doi: 10.1016/j.bcp.2012.03.018. [DOI] [PubMed] [Google Scholar]; De Vos, M., V. Schreiber, and F. Dantzer. 2012. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem. Pharmacol. 84:137-146. [DOI] [PubMed]
- 7.Beck C., Robert I., Dantzer F. Poly(ADP-ribose) polymerases in double-strand break repair: focus on PARP1, PARP2 and PARP3. Exp. Cell Res. 2014;329:18–25. doi: 10.1016/j.yexcr.2014.07.003. [DOI] [PubMed] [Google Scholar]; Beck, C., I. Robert, …, F. Dantzer. 2014. Poly(ADP-ribose) polymerases in double-strand break repair: focus on PARP1, PARP2 and PARP3. Exp. Cell Res. 329:18-25. [DOI] [PubMed]
- 8.Daniels C.M., Ong S.E., Leung A.K. The promise of proteomics for the study of ADP-ribosylation. Mol. Cell. 2015;58:911–924. doi: 10.1016/j.molcel.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; Daniels, C. M., S. E. Ong, and A. K. Leung. 2015. The promise of proteomics for the study of ADP-ribosylation. Mol. Cell. 58:911-924. [DOI] [PMC free article] [PubMed]
- 9.Mashimo M., Kato J., Moss J. Structure and function of the ARH family of ADP-ribosyl-acceptor hydrolases. DNA Repair (Amst.) 2014;23:88–94. doi: 10.1016/j.dnarep.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mashimo, M., J. Kato, and J. Moss. 2014. Structure and function of the ARH family of ADP-ribosyl-acceptor hydrolases. DNA Repair (Amst.). 23:88-94. [DOI] [PMC free article] [PubMed]
- 10.Yang G., Liu C., Yu X. Super-resolution imaging identifies PARP1 and the Ku complex acting as DNA double-strand break sensors. Nucleic Acids Res. 2018;46:3446–3457. doi: 10.1093/nar/gky088. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yang, G., C. Liu, …, X. Yu. 2018. Super-resolution imaging identifies PARP1 and the Ku complex acting as DNA double-strand break sensors. Nucleic Acids Res. 46:3446-3457. [DOI] [PMC free article] [PubMed]
- 11.Liu C., Vyas A., Yu X. The role of poly ADP-ribosylation in the first wave of DNA damage response. Nucleic Acids Res. 2017;45:8129–8141. doi: 10.1093/nar/gkx565. [DOI] [PMC free article] [PubMed] [Google Scholar]; Liu, C., A. Vyas, …, X. Yu. 2017. The role of poly ADP-ribosylation in the first wave of DNA damage response. Nucleic Acids Res. 45:8129-8141. [DOI] [PMC free article] [PubMed]
- 12.Gagné J.P., Isabelle M., Poirier G.G. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gagne, J. P., M. Isabelle, …, G. G. Poirier. 2008. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 36:6959-6976. [DOI] [PMC free article] [PubMed]
- 13.Teloni F., Altmeyer M. Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res. 2016;44:993–1006. doi: 10.1093/nar/gkv1383. [DOI] [PMC free article] [PubMed] [Google Scholar]; Teloni, F., and M. Altmeyer. 2016. Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res. 44:993-1006. [DOI] [PMC free article] [PubMed]
- 14.Palazzo L., Leidecker O., Ahel I. Serine is the major residue for ADP-ribosylation upon DNA damage. eLife. 2018;7:34334. doi: 10.7554/eLife.34334. [DOI] [PMC free article] [PubMed] [Google Scholar]; Palazzo, L., O. Leidecker, …, I. Ahel. 2018. Serine is the major residue for ADP-ribosylation upon DNA damage. eLife. 7:34334. [DOI] [PMC free article] [PubMed]
- 15.Bonfiglio J.J., Fontana P., Matic I. Serine ADP-ribosylation depends on HPF1. Mol. Cell. 2017;65:932–940.e6. doi: 10.1016/j.molcel.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bonfiglio, J. J., P. Fontana, …, I. Matic. 2017. Serine ADP-ribosylation depends on HPF1. Mol. Cell. 65:932-940.e6. [DOI] [PMC free article] [PubMed]
- 16.Gibbs-Seymour I., Fontana P., Ahel I. HPF1/C4orf27 is a PARP-1-interacting protein that regulates PARP-1 ADP-ribosylation activity. Mol. Cell. 2016;62:432–442. doi: 10.1016/j.molcel.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gibbs-Seymour, I., P. Fontana, …, I. Ahel. 2016. HPF1/C4orf27 is a PARP-1-interacting protein that regulates PARP-1 ADP-ribosylation activity. Mol. Cell. 62:432-442. [DOI] [PMC free article] [PubMed]
- 17.Kochan J.A., Desclos E.C.B., Krawczyk P.M. Meta-analysis of DNA double-strand break response kinetics. Nucleic Acids Res. 2017;45:12625–12637. doi: 10.1093/nar/gkx1128. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kochan, J. A., E. C. B. Desclos, …, P. M. Krawczyk. 2017. Meta-analysis of DNA double-strand break response kinetics. Nucleic Acids Res. 45:12625-12637. [DOI] [PMC free article] [PubMed]
- 18.Aleksandrov R., Dotchev A., Stoynov S.S. Protein dynamics in complex DNA lesions. Mol. Cell. 2018;69:1046–1061.e5. doi: 10.1016/j.molcel.2018.02.016. [DOI] [PubMed] [Google Scholar]; Aleksandrov, R., A. Dotchev, …, S. S. Stoynov. 2018. Protein dynamics in complex DNA lesions. Mol. Cell. 69:1046-1061.e5. [DOI] [PubMed]
- 19.Haince J.F., McDonald D., Poirier G.G. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]; Haince, J. F., D. McDonald, …, G. G. Poirier. 2008. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 283:1197-1208. [DOI] [PubMed]
- 20.Wang Y.H., Hariharan A., Sheetz M.P. DNA damage causes rapid accumulation of phosphoinositides for ATR signaling. Nat. Commun. 2017;8:2118. doi: 10.1038/s41467-017-01805-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang, Y. H., A. Hariharan, …, M. P. Sheetz. 2017. DNA damage causes rapid accumulation of phosphoinositides for ATR signaling. Nat. Commun. 8:2118. [DOI] [PMC free article] [PubMed]
- 21.Hartlerode A.J., Morgan M.J., Ferguson D.O. Recruitment and activation of the ATM kinase in the absence of DNA-damage sensors. Nat. Struct. Mol. Biol. 2015;22:736–743. doi: 10.1038/nsmb.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hartlerode, A. J., M. J. Morgan, …, D. O. Ferguson. 2015. Recruitment and activation of the ATM kinase in the absence of DNA-damage sensors. Nat. Struct. Mol. Biol. 22:736-743. [DOI] [PMC free article] [PubMed]
- 22.Koike M., Koike A. Accumulation of Ku80 proteins at DNA double-strand breaks in living cells. Exp. Cell Res. 2008;314:1061–1070. doi: 10.1016/j.yexcr.2007.11.014. [DOI] [PubMed] [Google Scholar]; Koike, M., and A. Koike. 2008. Accumulation of Ku80 proteins at DNA double-strand breaks in living cells. Exp. Cell Res. 314:1061-1070. [DOI] [PubMed]
- 23.Koike M., Yutoku Y., Koike A. Accumulation of p21 proteins at DNA damage sites independent of p53 and core NHEJ factors following irradiation. Biochem. Biophys. Res. Commun. 2011;412:39–43. doi: 10.1016/j.bbrc.2011.07.032. [DOI] [PubMed] [Google Scholar]; Koike, M., Y. Yutoku, and A. Koike. 2011. Accumulation of p21 proteins at DNA damage sites independent of p53 and core NHEJ factors following irradiation. Biochem. Biophys. Res. Commun. 412:39-43. [DOI] [PubMed]
- 24.Mortusewicz O., Amé J.C., Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mortusewicz, O., J. C. Ame, …, H. Leonhardt. 2007. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 35:7665-7675. [DOI] [PMC free article] [PubMed]
- 25.Mortusewicz O., Leonhardt H. XRCC1 and PCNA are loading platforms with distinct kinetic properties and different capacities to respond to multiple DNA lesions. BMC Mol. Biol. 2007;8:81. doi: 10.1186/1471-2199-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mortusewicz, O., and H. Leonhardt. 2007. XRCC1 and PCNA are loading platforms with distinct kinetic properties and different capacities to respond to multiple DNA lesions. BMC Mol. Biol. 8:81. [DOI] [PMC free article] [PubMed]
- 26.Mortusewicz O., Leonhardt H., Cardoso M.C. Spatiotemporal dynamics of regulatory protein recruitment at DNA damage sites. J. Cell. Biochem. 2008;104:1562–1569. doi: 10.1002/jcb.21751. [DOI] [PubMed] [Google Scholar]; Mortusewicz, O., H. Leonhardt, and M. C. Cardoso. 2008. Spatiotemporal dynamics of regulatory protein recruitment at DNA damage sites. J. Cell. Biochem. 104:1562-1569. [DOI] [PubMed]
- 27.Izhar L., Adamson B., Elledge S.J. A systematic analysis of factors localized to damaged chromatin reveals PARP-dependent recruitment of transcription factors. Cell Reports. 2015;11:1486–1500. doi: 10.1016/j.celrep.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]; Izhar, L., B. Adamson, …, S. J. Elledge. 2015. A systematic analysis of factors localized to damaged chromatin reveals PARP-dependent recruitment of transcription factors. Cell Reports. 11:1486-1500. [DOI] [PMC free article] [PubMed]
- 28.Mazza D., Abernathy A., McNally J.G. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res. 2012;40:e119. doi: 10.1093/nar/gks701. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mazza, D., A. Abernathy, …, J. G. McNally. 2012. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res. 40:e119. [DOI] [PMC free article] [PubMed]
- 29.Mueller F., Wach P., McNally J.G. Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching. Biophys. J. 2008;94:3323–3339. doi: 10.1529/biophysj.107.123182. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mueller, F., P. Wach, and J. G. McNally. 2008. Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching. Biophys. J. 94:3323-3339. [DOI] [PMC free article] [PubMed]
- 30.Kühn T., Ihalainen T.O., Timonen J. Protein diffusion in mammalian cell cytoplasm. PLoS One. 2011;6:e22962. doi: 10.1371/journal.pone.0022962. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kuhn, T., T. O. Ihalainen, …, J. Timonen. 2011. Protein diffusion in mammalian cell cytoplasm. PLoS One. 6:e22962. [DOI] [PMC free article] [PubMed]
- 31.Bénichou O., Chevalier C., Voituriez R. Facilitated diffusion of proteins on chromatin. Phys. Rev. Lett. 2011;106:038102. doi: 10.1103/PhysRevLett.106.038102. [DOI] [PubMed] [Google Scholar]; Benichou, O., C. Chevalier, …, R. Voituriez. 2011. Facilitated diffusion of proteins on chromatin. Phys. Rev. Lett. 106:038102. [DOI] [PubMed]
- 32.Kozlowski M. Ludwig Maximilians Universität; 2014. The molecular mechanism of PARP1 activation and its downstream roles in ALC1-regulated transcription. Doctoral Thesis. [Google Scholar]; Kozlowski, M. 2014. The molecular mechanism of PARP1 activation and its downstream roles in ALC1-regulated transcription. Doctoral Thesis (Ludwig Maximilians Universitat).
- 33.Dinant C., de Jager M., Vermeulen W. Activation of multiple DNA repair pathways by sub-nuclear damage induction methods. J. Cell Sci. 2007;120:2731–2740. doi: 10.1242/jcs.004523. [DOI] [PubMed] [Google Scholar]; Dinant, C., M. de Jager, …, W. Vermeulen. 2007. Activation of multiple DNA repair pathways by sub-nuclear damage induction methods. J. Cell Sci. 120:2731-2740. [DOI] [PubMed]
- 34.Blumenthal D., Goldstien L., Gheber L.A. Universal approach to FRAP analysis of arbitrary bleaching patterns. Sci. Rep. 2015;5:11655. doi: 10.1038/srep11655. [DOI] [PMC free article] [PubMed] [Google Scholar]; Blumenthal, D., L. Goldstien, …, L. A. Gheber. 2015. Universal approach to FRAP analysis of arbitrary bleaching patterns. Sci. Rep. 5:11655. [DOI] [PMC free article] [PubMed]
- 35.Bläßle A., Soh G., Müller P. Quantitative diffusion measurements using the open-source software PyFRAP. Nat. Commun. 2018;9:1582. doi: 10.1038/s41467-018-03975-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Blaßle, A., G. Soh, …, P. Muller. 2018. Quantitative diffusion measurements using the open-source software PyFRAP. Nat. Commun. 9:1582. [DOI] [PMC free article] [PubMed]
- 36.Riccio A.A., Cingolani G., Pascal J.M. PARP-2 domain requirements for DNA damage-dependent activation and localization to sites of DNA damage. Nucleic Acids Res. 2016;44:1691–1702. doi: 10.1093/nar/gkv1376. [DOI] [PMC free article] [PubMed] [Google Scholar]; Riccio, A. A., G. Cingolani, and J. M. Pascal. 2016. PARP-2 domain requirements for DNA damage-dependent activation and localization to sites of DNA damage. Nucleic Acids Res. 44:1691-1702. [DOI] [PMC free article] [PubMed]
- 37.Sprague B.L., Pego R.L., McNally J.G. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys. J. 2004;86:3473–3495. doi: 10.1529/biophysj.103.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sprague, B. L., R. L. Pego, …, J. G. McNally. 2004. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys. J. 86:3473-3495. [DOI] [PMC free article] [PubMed]
- 38.Normanno D., Boudarène L., Dahan M. Probing the target search of DNA-binding proteins in mammalian cells using TetR as model searcher. Nat. Commun. 2015;6:7357. doi: 10.1038/ncomms8357. [DOI] [PMC free article] [PubMed] [Google Scholar]; Normanno, D., L. Boudarene, …, M. Dahan. 2015. Probing the target search of DNA-binding proteins in mammalian cells using TetR as model searcher. Nat. Commun. 6:7357. [DOI] [PMC free article] [PubMed]
- 39.Langelier M.-F., Riccio A.A., Pascal J.M. PARP-2 and PARP-3 are selectively activated by 5′ phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1. Nucleic Acids Res. 2014;42:7762–7775. doi: 10.1093/nar/gku474. [DOI] [PMC free article] [PubMed] [Google Scholar]; Langelier, M.-F., A. A. Riccio, and J. M. Pascal. 2014. PARP-2 and PARP-3 are selectively activated by 5′ phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1. Nucleic Acids Res. 42:7762-7775. [DOI] [PMC free article] [PubMed]
- 40.Buchfellner A., Yurlova L., Rothbauer U. A new nanobody-based biosensor to study endogenous PARP1 in vitro and in live human cells. PLoS One. 2016;11:e0151041. doi: 10.1371/journal.pone.0151041. [DOI] [PMC free article] [PubMed] [Google Scholar]; Buchfellner, A., L. Yurlova, …, U. Rothbauer. 2016. A new nanobody-based biosensor to study endogenous PARP1 in vitro and in live human cells. PLoS One. 11:e0151041. [DOI] [PMC free article] [PubMed]
- 41.Stadler C., Rexhepaj E., Lundberg E. Immunofluorescence and fluorescent-protein tagging show high correlation for protein localization in mammalian cells. Nat. Methods. 2013;10:315–323. doi: 10.1038/nmeth.2377. [DOI] [PubMed] [Google Scholar]; Stadler, C., E. Rexhepaj, …, E. Lundberg. 2013. Immunofluorescence and fluorescent-protein tagging show high correlation for protein localization in mammalian cells. Nat. Methods. 10:315-323. [DOI] [PubMed]
- 42.Erickson H.P. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol. Proced. Online. 2009;11:32–51. doi: 10.1007/s12575-009-9008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Erickson, H. P. 2009. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol. Proced. Online. 11:32-51. [DOI] [PMC free article] [PubMed]
- 43.Phair R.D., Scaffidi P., Misteli T. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Phair, R. D., P. Scaffidi, …, T. Misteli. 2004. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 24:6393-6402. [DOI] [PMC free article] [PubMed]
- 44.Kim M.Y., Mauro S., Kraus W.L. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]; Kim, M. Y., S. Mauro, …, W. L. Kraus. 2004. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 119:803-814. [DOI] [PubMed]
- 45.Muthurajan U.M., Hepler M.R., Luger K. Automodification switches PARP-1 function from chromatin architectural protein to histone chaperone. Proc. Natl. Acad. Sci. USA. 2014;111:12752–12757. doi: 10.1073/pnas.1405005111. [DOI] [PMC free article] [PubMed] [Google Scholar]; Muthurajan, U. M., M. R. Hepler, …, K. Luger. 2014. Automodification switches PARP-1 function from chromatin architectural protein to histone chaperone. Proc. Natl. Acad. Sci. USA. 111:12752-12757. [DOI] [PMC free article] [PubMed]
- 46.Kim M.Y., Zhang T., Kraus W.L. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]; Kim, M. Y., T. Zhang, and W. L. Kraus. 2005. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 19:1951-1967. [DOI] [PubMed]
- 47.Rudolph J., Mahadevan J., Luger K. Poly(ADP-ribose) polymerase 1 searches DNA via a ‘monkey bar’ mechanism. eLife. 2018;7:e37818. doi: 10.7554/eLife.37818. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rudolph, J., J. Mahadevan, …, K. Luger. 2018. Poly(ADP-ribose) polymerase 1 searches DNA via a ‘monkey bar’ mechanism. eLife. 7:e37818. [DOI] [PMC free article] [PubMed]
- 48.Chen Q., Kassab M.A., Yu X. PARP2 mediates branched poly ADP-ribosylation in response to DNA damage. Nat. Commun. 2018;9:3233. doi: 10.1038/s41467-018-05588-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chen, Q., M. A. Kassab, …, X. Yu. 2018. PARP2 mediates branched poly ADP-ribosylation in response to DNA damage. Nat. Commun. 9:3233. [DOI] [PMC free article] [PubMed]
- 49.Crawford K., Bonfiglio J.J., Ahel I. Specificity of reversible ADP-ribosylation and regulation of cellular processes. Crit. Rev. Biochem. Mol. Biol. 2018;53:64–82. doi: 10.1080/10409238.2017.1394265. [DOI] [PubMed] [Google Scholar]; Crawford, K., J. J. Bonfiglio, …, I. Ahel. 2018. Specificity of reversible ADP-ribosylation and regulation of cellular processes. Crit. Rev. Biochem. Mol. Biol. 53:64-82. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.