Abstract
Fabry disease, a rare, X-linked lysosomal storage disease, arises from deficiency of the lysosomal hydrolase, α-galactosidase A (GLA) which disrupts the catabolism of globo-series glycosphingolipids (GSLs). One potential link between GLA deficiency and vascular dysfunction may be changes in endothelial nitric oxide synthase (eNOS) function. GLA-deficient EA.hy926 cells were obtained by siRNA knockdown of GLA expression and by mutation of GLA with CRISPR/Cas9 gene editing to investigate the effects of GLA deficiency on eNOS. As previously observed with siRNA knockdown of GLA, globotriaosylceramide (Gb3) accumulated in EA.hy926 cells. In contrast, Gb3 did not accumulate in CRISPR/Cas9 gene edited GLA-deficient cells, but instead, globotetraosylceramide (Gb4). However, in both the siRNA and CRISPR/Cas9 models globotriaosylsphingosine (lyso-Gb3) was elevated. As was previously observed with siRNA knockdown of GLA expression, CRISPR/Cas9 GLA-deficient cells had lower eNOS activity. Restoring GLA activity in GLA-deficient cells with exogenous GLA treatment improved eNOS activity. In contrast, treating cells with the glucosylceramide synthase inhibitor, eliglustat, decreased NOS activity. These results suggest that eNOS uncoupling is due to GLA deficiency, and not necessarily due to elevated Gb3 per se. It was observed that lyso-Gb3 inhibits eNOS activity.
Keywords: Endothelium, endothelial nitric oxide synthase, globotriaosylceramide, globotriaosylsphingosine, Fabry disease, alpha-galactosidase A, lyso-globotriaosylceramide
1. Introduction
Fabry disease is an X-linked lysosomal storage disorder caused by mutations in the GLA gene. Fabry disease affects multiple organs and gives rise to a variety of complications, including kidney failure, neuropathy, and cardiovascular and cerebrovascular events (Eng, Fletcher et al. 2007). Deficient α-galactosidase A (GLA) activity results in elevated levels of glycosphingolipids (GSLs) with terminal α−1,4-galactose groups. These globo-series GSLs accumulate in various cells and tissues, but are particularly prominent in vascular tissues and more specifically in endothelial cells (Linhart and Elliott 2007). In clinical and laboratory studies the primary GSL observed to accumulate in tissues is globotriaosylceramide (Gb3), a primary substrate for GLA. However, plasma Gb3 is not a reliable biomarker for individual symptoms (Vedder, Linthorst et al. 2007). Globotriaosylsphingosine (lyso-Gb3), the deacylated metabolite of Gb3, may better correlate with symptoms, especially in symptomatic heterozygous females with the disease (Aerts, Groener et al. 2008).
Impaired endothelial nitric oxide synthase (eNOS) activity has been observed in several experimental models of Fabry disease. This impairment is manifest as both decreased NO availability and eNOS uncoupling (Eitzman, Bodary et al. 2003, Bodary, Shen et al. 2005, Park, Whitesall et al. 2008, Shu, Park et al. 2009, Shu, Vivekanandan-Giri et al. 2014). Decreased NO bioavailability is associated with the loss of normal vasodilation in arterial vessels. eNOS uncoupling occurs when there is improper flow of electrons from the reductase domain of eNOS, to the oxygenase domain, which results in formation of O2− instead of NO (Vasquez-Vivar, Kalyanaraman et al. 1998, Xia, Tsai et al. 1998). O2− can then react with NO to produce a reactive nitrogen species, peroxynitrite (ONOO−). ONOO− can then rapidly oxidize a cofactor of eNOS, tetrahydrobiopterin (BH4), to the inactive form, dihydrobiopterin (BH2), which causes further uncoupling (Milstien and Katusic 1999). A biomarker for ONOO−, 3-nitrotyrosine, which results from protein nitrosylation (Heinecke 2002), was found to be elevated in the plasma of Fabry patients (Shu, Vivekanandan-Giri et al. 2014). The mechanism by which GLA deficiency causes eNOS uncoupling has not been well elucidated.
Previous in vitro models of Fabry disease our group has utilized mouse aortic endothelial cells (MAECs) from Gla-knockout mice, and the human endothelial cell line, EA.hy926 with anti-GLA siRNA treatment (Shu and Shayman 2007, Shu, Vivekanandan-Giri et al. 2014). Both models have provided insight into how GLA-deficiency affects GSL accumulation in the cell and affects eNOS function. However, each has its limitations. MAECs are challenging to maintain in culture, and in anti-GLA siRNA-treated EA.hy926 cells the GLA depletion is not sustained for longer than the duration of the siRNA treatment (3–6 days). In this study, cell lines with a permanent absence of GLA protein expression were developed to be able to study eNOS uncoupling in the setting of GLA deficiency. These cells were obtained by using the CRISPR/Cas9 method of gene editing.
We report the surprising finding that CRISPR/Cas9 gene edited EA.hy926 cells displaying a permanent absence of GLA expression did not accumulate Gb3 in contrast to EA.hy926 cells treated with anti-GLA siRNA. However, both models of GLA-deficient EA.hy926 cells showed loss of eNOS activity.
2. Material and methods
2.1. Cell culture
EA.hy926 cells (ATCC, Manassas, VA) were maintained at 37 °C with 95/5 O2/CO2, in DMEM/F-12, GlutaMAX (Thermo Fisher Scientific, Waltham, MA) supplemented with heat-inactivated 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin, which will be later referred to as “growth medium.”
2.2. CRISPR/Cas9 plasmid generation and delivery
In order to disrupt GLA expression in EA.hy926 cells, single-stranded guide RNA was designed to target Exon 1 of Gla using the Optimized Design Tool (crispr.mit.edu). The forward primer was ATTGGCAAGGACGCCTACCA (GTTTT), and the reverse primer was TGGTAGGCGTCCTTGCCAAT (CGGTG). The parentheses indicate sequence complementary to the 3’ overhang sequence in the CRISPR nuclease vector. Forward and reverse single-stranded oligonucleotides were annealed to generate a double-stranded oligonucleotide. The oligonucleotide was cloned into the GeneArt CRISPR nuclease vector (Thermo Fisher Scientific), which expresses the orange florescence protein (OFP) reporter. The vector was transformed into One Shot TOP10 E. coli cells (Thermo Fisher Scientific). Bacteria were grown on LB agar plates with ampicillin and incubated at 37 °C overnight. Clones were selected and grown in LB broth overnight. Plasmids were purified and sequenced with the U6 forward primer to verify the presence and proper orientation of double-stranded oligonucleotide.
EA.hy926 cells were plated at 600,000 cells/well on six-well dishes. The following day, 2 µg of CRISPR plasmid DNA was transfected using Lipofectamine 3000 (Thermo Fisher Scientific), and incubated with DMEM-F12 GlutaMAX™ supplemented with 10% FBS for 24 hours. Cells were FACS sorted on a BD FACSAria™ II flow cytometer (BD Biosciences, San Jose, CA) with BD FACSDiva 8.0 software. OFP positive cells were collected and plated onto 150 mm dishes at low density in normal growth conditions. Cells were allowed to grow for several days, and then cloning cylinders were used to isolate individual colonies. Cells were grown in the same media conditions described for EA.hy926 cells.
2.3. siRNA silencing of GLA in EA.hy926 cells
The transfection of EA.hy926 cells was performed as previously described, but with minor modification (Shu, Vivekanandan-Giri et al. 2014). For cells that were to be harvested after a single transfection, the following protocol was observed. One day before transfection, 0.8 × 106 cells were seeded onto 150 mm cell culture dishes with growth medium. At the time of transfection (day 0), growth medium was replaced with Opti-MEM-I (Thermo Fisher Scientific). Next, anti-GLA siRNA or scrambled-siRNA control (scr-siRNA) was mixed with Lipofectamine RNAiMax (Thermo Fisher Scientific) according to the manufacturer’s protocol. After 8 hours, the medium was replaced with growth medium. Cells were harvested on day 1 or 3 post-transfection.
For cells that were to be harvested after a double transfection, the following protocol was observed. One day before transfection, 0.7 × 106 cells were seeded onto 150 mm cell culture dishes with growth medium. The same 3-day transfection protocol was followed as described above, but instead of harvesting the cells on day 3, cells were transfected once more with the same treatment, and later harvested on day 6.
2.4. Western blotting
Colonies that were identified as having a mutation were grown to confluency on 100 mm dishes. Cells were harvested with 0.05% trypsin-EDTA and lysed in a buffer consisting of 25 mM Tris-HCl, 137 mM NaCl, 2 mM EDTA, 2 mM Na3VO4, 20 mM NaF, 1% Triton X-100, 10% glycerol, and protease inhibitor (P8340, Sigma). Cells were probe sonicated on ice and centrifuged at 10,000 x g for 10 minutes at 4 °C. Supernatants were saved and total cellular protein determined with bicinchoninic acid (BCA) assay using bovine serum album (BSA) as a standard (Pierce BCA Protein Assay Kit, Thermo Fisher Scientific). Thirty μg of protein were denatured with 2-mercaptoethanol in Laemmli buffer, heated at 95 °C for 10 minutes, and then separated on NuPAGE 4–12% SDS-PAGE (Thermo Fisher Scientific). Proteins were transferred to nitrocellulose membranes, blocked for 1 hour with 5% nonfat dry milk-TBST (tris-buffered saline, tween-20), then incubated with primary antibody against GLA (1:1000 in 5% milk-TBST, EPR5829, (ab129173) Abcam, Cambridge MA), eNOS (1:1000 in 5% nonfat dry milk-TBST, M221 (ab76198) Abcam), or GAPDH (1:1000 in 1% BSA-TBST, #MAB374, EMD Millipore, Billerica, MA) overnight at 4 °C. After washing with TBST, membranes were incubated with anti-mouse (for eNOS and GAPDH detection) or anti-rabbit (for GLA detection) secondary antibody at (1:15,000 in 1% BSA-TBST). Membranes were washed with TBST and incubated with enhanced chemiluminescence (Pierce ECL Western Blotting Substrate, Thermo Fisher Scientific). Protein bands were detected with film exposure. The film was scanned and protein bands quantified with ImageJ Software.
2.5. Lipid extraction
Cells were grown to confluency on 150 mm cell culture dishes and harvested by scraping into 700 μL PBS. The cell suspension was transferred into 16 mm × 100 mm glass tubes, and 1 mL chloroform and 2.4 mL methanol were added. After water bath sonication, cell protein was precipitated by centrifuging at 2400 × g for 30 minutes. The supernatant was transferred to a new glass tube and 4.5 mL chloroform and 1.2 mL 0.9% NaCl were added. The sample was centrifuged at 900 × g for 5 minutes. The upper aqueous phase was discarded, and the lower organic phase was washed twice with 2 mL methanol and 0.8 mL 0.9% NaCl. The lower phase was extracted using a 1 mL glass syringe (Hamilton, Reno, NV) and transferred to a new glass tube. Lipids were dried under a stream of nitrogen.
2.6. Total phospholipid assay
Total phospholipid was determined as follows (Ames 1966). Dried lipids were suspended in 500 μL of chloroform/methanol (2:1, v/v). Thirty μL of sample was transferred to 13 × 100 mm glass tube, and 30 μL of 10% Mg(NO3)2.6H2O in 95% EtOH was added. The mixture was dried under a stream of nitrogen and ashed for 30 seconds over a flame. After cooling to room temperature, 300 μl of 0.5 M HCl was added and incubated at room temperature for 30 minutes. Then, 700 µL of (1:6, v/v) 10% ascorbic acid: 0.42% (NH4)2Mb·4H2O was added and samples and standards were incubated for 45 minutes at 45 °C. Samples were cooled to room temperature and absorbance measured at 820 nm. Samples were compared to standards of 0, 5, 10, 20, 30, 40 µL of 1 mM KH2PO4, that were prepared simultaneously. Total phospholipid was calculated according to the standard curve. The volume according to 100 nmol of total phospholipid was transferred to a new glass tube and dried under a stream of nitrogen.
2.7. Alkaline methanolysis and acid hydrolysis
Two mL chloroform and 1 mL 0.21 M NaOH in 100% methanol were added to the dried lipids. Samples were incubated at room temperature for 1 hour. The reaction was terminated by addition of 0.8 mL 0.25 M HCl and centrifuged at 900 × g for 5 minutes. The upper aqueous phase was removed and lower organic phase was transferred with a 1 mL Hamilton syringe to a new 16 mm × 125 mm glass tube. Four mL methanol and 1.6 mL 0.05 M HCl/25 mM HgCl2 were added. The mixture was incubated at 37 °C for 15 minutes. Two mL of chloroform and 1.6 mL of water was added. Samples were centrifuged at 900 × g for 5 minutes. The upper aqueous phase was removed and 2 mL methanol and 1 mL 30 mM EDTA pH 8.0 were added to the lower organic phase. Samples were centrifuged and the upper phase was removed again. The lower organic phase was washed twice with 2 mL methanol and 1.6 mL water. The lower layer was transferred with a 1 mL glass syringe to a new 12 × 100 mm glass tube and dried under a stream of nitrogen.
2.8. Thin layer chromatography – LacCer, Gb3, and Gb4 measurements
One hundred nmol of total phospholipid was applied to a silica high performance TLC plate (Sigma-Aldrich). The plate was first developed in a solvent system consisting of chloroform/methanol (98:2, v/v), and air dried. The plate was then developed in a solvent system consisting of chloroform/methanol/acetic acid/water (61/31/5/3, v/v/v/v) and air dried. Plates were sprayed with 1% orcinol in 11% H2SO4 and charred at 130 °C for 5 minutes. Plates were scanned and densitometry measured using ImageJ software. Lipids were quantified by running glucosylceramide (GlcCer), lactosylceramide (LacCer), globotriaosylceramide (Gb3), and globotetraosylceramide (Gb4) standards on each plate (Matreya LLC, State College, PA).
2.9. Exogenous GLA treatment
Cells (1.0 × 106) were seeded on 150 mm cell culture dishes with growth medium. The following day, growth medium was replaced with fresh media, and 3 U of α-galactosidase A (Green coffee bean extract, Sigma), was added. Cells grew for 3 days and were harvested for lipid analysis with TLC. This dose was chosen based on a previous study using 10 μg/mL GLA to treat cells (Shu and Shayman 2007).
2.10. Eliglustat treatment
Cells (1.0 × 106) were seeded on 150 mm cell culture dishes with DMEM/F12 GlutaMAX supplemented with 5% FBS. The following day, medium was replaced with DMEM/F12 GlutaMAX supplemented with 2% FBS. Cells were treated with either PBS vehicle control, 20 or 200 nM eliglustat. Cells were harvested 3 days later for lipid analysis with TLC.
2.11. NOS activity measurement
Endothelial nitric oxide synthase (eNOS) activity in EA.hy926 cell lysates was analyzed by using a NOS activity assay kit (Cayman Chemical, Ann Arbor, MI) according to the manufacturer’s instructions. This assay measures the biochemical conversion of L-arginine to L-citrulline by NOS. Cells were harvested with 0.05% trypsin-EDTA and washed with PBS two times. Cell pellets were lysed in the provided homogenization buffer (final concentration 1 mM EDTA and 1 mM EGTA in 25 mM Tris-HCl (pH 7.4) buffer). After brief sonication, samples were centrifuged at 12,000 × g for 5 minutes, and supernatants transferred to new tubes. Cell lysates (5 µL) were incubated at room temperature for 3 hours with 1 µCi [3H]L-arginine (PerkinElmer), 100 nM calmodulin, and the provided reaction buffer (final concentration 1 mM NADPH, 600 µM CaCl2, 25 mM Tris-HCl (pH 7.4), 3 µM tetrahydrobiopterin, 1 µM flavin adenine dinucleotide, and 1 µM flavin adenine mononucleotide in 50 µL). The reaction was stopped by adding 400 µL of 5 mM EDTA in 50 mM HEPES (pH 5.5) buffer. The provided resin was added to each sample to remove [3H]L-arginine. Radioactivity due to [3H]L-citrulline was measured as counts per minute (cpm) using a scintillation counter. Cpm were also measured in samples with no cell lysate as a background control. NOS activity of each sample was calculated by subtracting background cpm from the cpm for each sample, and then normalizing to total protein in each 5 µL sample, determined by the BCA protein assay. In some experiments glucosylsphingosine (GlcSph) or globotriaosylsphingosine (lyso-Gb3) (Matreya) were added to each reaction for a final concentration of 0 – 60 μM.
2.12. Lyso-Gb3 measurements
To measure lyso-Gb3 in EA.hy926-CRISPR/Cas9 clonal cell lines, 0.7 × 106 cells were seeded on 100 mm dishes. The following day, media was replaced with fresh growth media. Three days later, media was collected and cells harvested with 0.05% trypsin-EDTA. Cells and media were collected the same way in siRNA-treated EA.hy926 cells with the transfection method described above. Cell pellets were sonicated on ice in 200–250 μL PBS, centrifuged at 12,000 × g, and supernatant collected. BCA protein assay was performed. Lyso-Gb3 was measured with as previously described (Gold, Mirzaian et al. 2013). Results were normalized to protein concentration.
2.13. Statistical analysis
Data were analyzed by using Prism 7.0 software (GraphPad Software Inc., La Jolla, CA). Significance was determined by two-way analysis of variance (ANOVA) with Dunnett’s or Sidak’s multiple comparisons test. Values are shown as mean ± SEM. Differences between groups were considered statistically significant at a P-value of <0.05.
3. Results
3.1. GLA-deficient cells accumulate Gb4
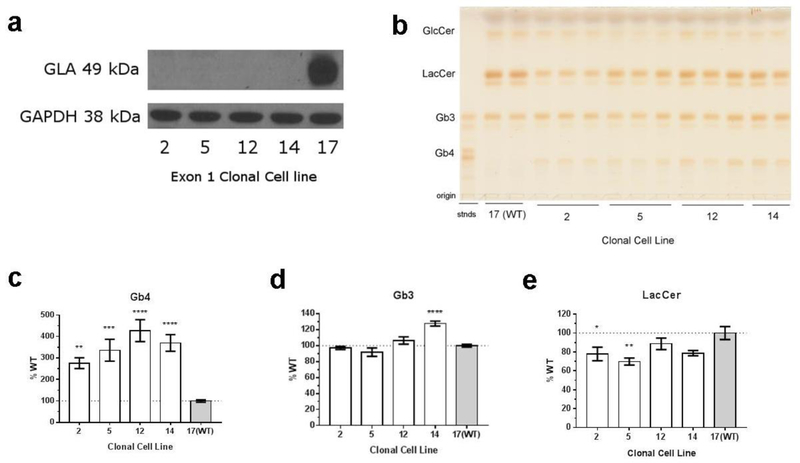
Gla-deficient clonal cell lines were obtained by CRISPR/Cas9 gene editing, and the absence of Gla expression was confirmed by Western blot (figure 1a) and GLA enzyme assay (figure S.1). DNA sequencing confirmed a single base pair deletion in the targeted region of Gla (figure S.2). To determine the effect of GLA deficiency on globo-series GSL accumulation, whole cell lipids were isolated and separated by thin layer chromatography (figure 1b). Globo-series GSLs was detected by staining with 1% orcinol (w/v) solution in 11% H2SO4 (v/v). Cell lysates from GLA-deficient cells have a 2.5 to 4-fold higher amount of Gb4 content over the WT cell line (figure 1c). Only one clonal cell line had an increase in Gb3, the primary substrate for GLA (figure 1d). This is in contrast to a previous study that used EA.hy926 cells treated with anti-GLA siRNA for 3 or 6 days, which resulted in Gb3 accumulation (Shu, Vivekanandan-Giri et al. 2014). In light of the prevalence of Gb4 instead of Gb3 in the CRISPR/Cas9 GLA-deficient EA.hy926 cells, Gb4 content in anti-Gla siRNA – treated EA.hy926 cells was also measured. Anti-GLA siRNA treatment completely knocked down Gla expression (figure S.3), and increased Gb4 by 35% compared to scrambled siRNA treatment (figure S.4). LacCer accumulation is also different between the CRISPR/Cas9 and siRNA models. CRISPR/Cas9 Gla-deficient cells have a decrease in LacCer (figure 1e), whereas cells treated with anti-Gla siRNA have an increase in LacCer content (figure S.4).
Figure 1.
(a) CRISPR/Cas9-modified clones 2, 5, 12, and 14 do not express Gla. Clone 17 represents the WT control used for all studies. (b) A representative TLC plate charred with 1% (wt/vol) orcinol in 11% (v/v) H2SO4 is shown. Quantification of (c) Gb4, (d) Gb3, (e) LacCer compared to WT control (100%, dotted line). Abbreviations: glucosylceramide (GlcCer), lactosylceramide (LacCer), globotriaosylceramide (Gb3), globotetraosylceramide (Gb4), standards (stnds). n=10 ****P<0.0001, ***P<0.001, **P<0.01, *P<0.05
3.2. Lyso-Gb3 is elevated in both GLA-siRNA knockdown and CRISPR/Cas9 GLA-deficient cells
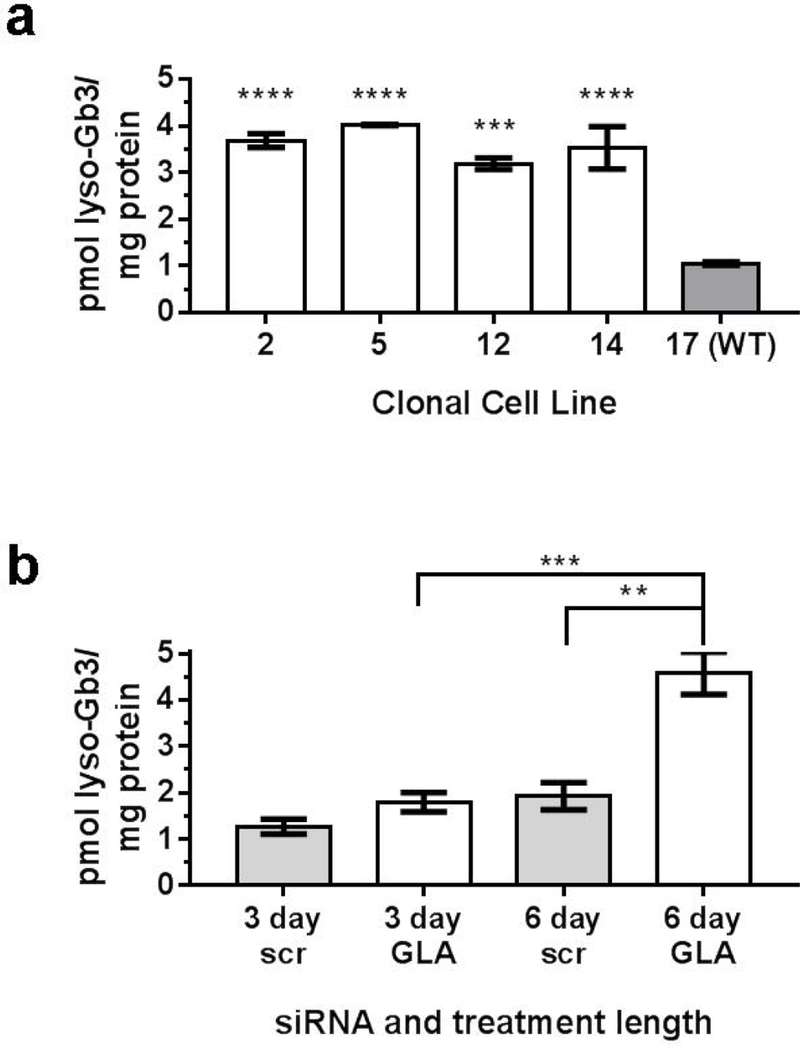
Because the correlation between GLA deficiency and Gb3 or Gb4 content differed between the CRISPR/Cas9 and siRNA models of GLA in EA.hy926 cells, the levels of lyso-Gb3 in the two models were assessed. Cell lysates from CRISPR/Cas9 GLA-deficient clonal cell lines had a 3 to 4-fold increase of lyso-Gb3 compared to WT control (figure 2a). Similarly, 6-day treatment of anti-GLA siRNA in EA.hy926 cells caused a significant increase of lyso-Gb3 compared to scr-siRNA control and 3-day anti-GLA siRNA treated cells (figure 2b). In contrast to LacCer, Gb3, and Gb4, lyso-Gb3 is a marker that is consistently elevated in both siRNA and CRISPR/Cas9 models of GLA deficiency in EA.hy926 cells.
Figure 2.
(a) Lyso-Gb3 is elevated in CRISPR/Cas9 GLA-deficient cells compared to clone 17 (WT). (b) Lyso-Gb3 is elevated in 6-day anti-GLA siRNA treated cells compared to 3-day anti-GLA siRNA treatment and 6-day scrambled-siRNA (scr) treatment. n=3. ****P<0.0001, ***P<0.001, **P<0.01.
3.3. Decreased NO production and eNOS expression in GLA-deficient cells
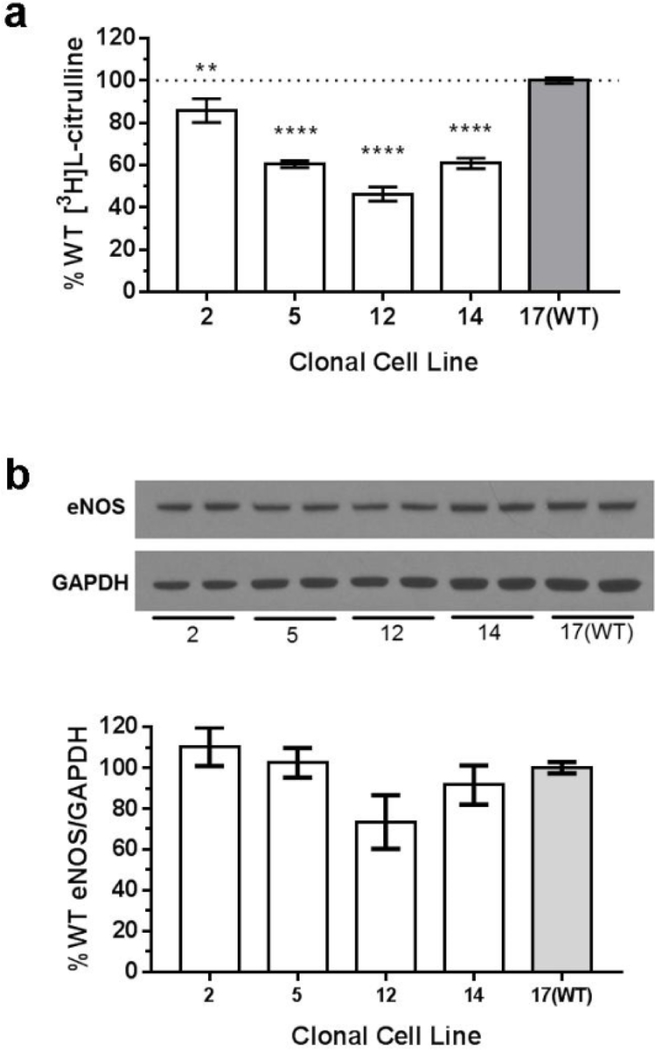
Three of the four GLA-deficient cell lines had 40–50% of NOS activity compared to WT cells (figure 3a). One GLA-deficient cell line, clone 2, had a 14% reduction of NOS activity compared to WT. To verify [3H]L-citrulline that was measured in the NOS activity assay originated from NOS activity, reactions were repeated with the addition of a competitive NOS inhibitor, NG-nitro-L-arginine (L-NNA) to the NOS reaction mixture. There was no detectable [3H]L-citrulline above background with L-NNA (data not shown).
Figure 3.
(a) NO production was significantly lower in GLA-deficient cell lines, 5, 12, and 14, when compared to the WT control 17. n=18. (b) GLA-deficient cells have less eNOS protein expression. A representative Western blot from CRISPR/Cas9 clonal cell lines is shown. GLA-deficiency causes a decrease in eNOS expression in clone 12, but is not statistically significant (p=0.08). Clone 2, n=3; clones 5, 12, and 14, n=4; clone 17(WT), n=6. **P<0.01, ****P<0.0001 compared to 17(WT)
A possible reason for decreased NOS activity is reduced eNOS expression. A Western blot was performed to detect eNOS expression in WT and GLA-deficient cells (figure 3b). Although there is a trend of decreased eNOS expression in GLA-deficient cells that correlates with decreased NOS activity, the change was not statistically significant (p=0.08 for clone 12 compared with 17(WT)).
3.4. Two methods of reducing neutral GSLs have different effects on NOS activity
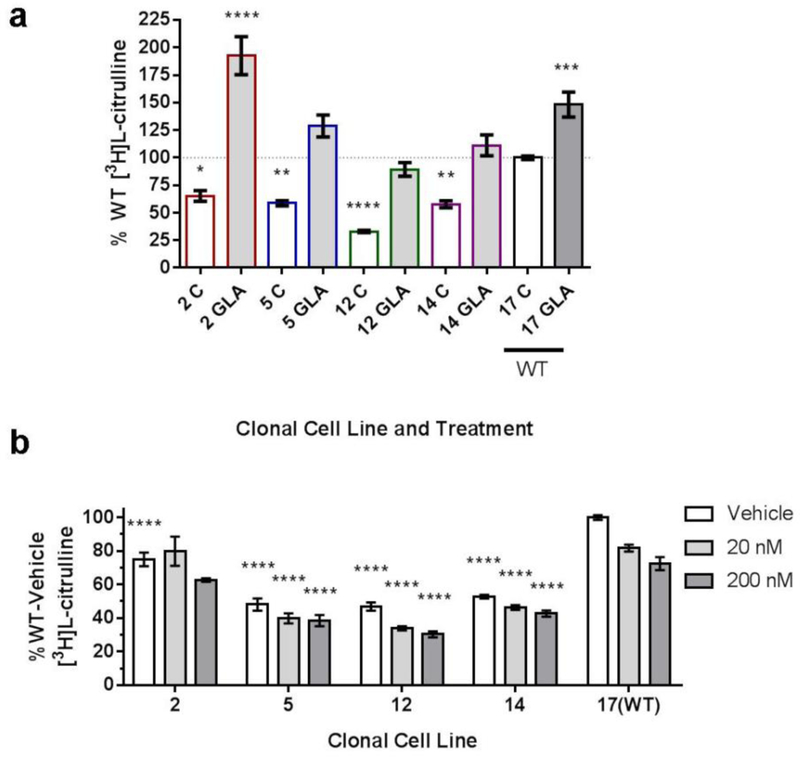
Two pharmacological approaches were used to determine if diminished NOS activity is due to the absence of functional GLA or accumulation of globo-series GSLs. Cells were treated for three days with either exogenous GLA, which primarily decreases Gb3 levels (figure S.5), or eliglustat (20 or 200 nM), which decreases globo-series GSLs (figure S.6) due to the inhibition of glucosylceramide synthase. GLA enzyme treatment increased NOS activity in all cells (figure 4a), indicating a direct correlation between the presence of the functional GLA enzyme and functioning NOS. In contrast, eliglustat treatment decreased NOS activity in a dose-dependent manner (figure 4b), suggesting that decreasing all globo-series GSLs decreases NOS activity. The level of lyso-Gb3 in cell lysates after treatment with eliglustat or exogenous GLA was not measured.
Figure 4.
(a) GLA-deficient cell lines (2, 5, 12, 14) and the WT cell line (17) were treated with exogenous GLA (shaded bars), resulting in increased NO production. Dotted line indicates WT control (C) normalized to 100%. n=8. Significance shown compared to 17(WT) control. (b) GLA-deficient cell lines (2, 5, 12, 14) and the WT cell line (17) were treated with eliglustat (20 or 200 nM) for 3 days, resulting in decreased NO production. Results are reported as %17(WT)-vehicle. n=5. Significance shown compared to 17(WT) each treatment. ****P<0.0001, ***P<0.001, **P<0.01, *P<0.05
3.5. Lyso-Gb3 inhibits NOS activity
We previously determined that GLA-deficient EA.hy926 cells have elevated globotriaosylsphingosine (lyso-Gb3) compared to WT cells (figure 2). GLA-deficient cells were also shown to have decreased NOS activity (figure 3a). To determine if there is a correlation between presence of lyso-Gb3 and NOS activity, lyso-Gb3 (250 or 500 nM) was added to cell culture media for three days, and NOS activity was measured in cell lysates. The concentrations used are within the range reported to increase smooth muscle cell proliferation (Aerts, Groener et al. 2008). However, there was no change in NOS activity compared to cells treated with DMSO or glucosylsphingosine (GlcSph) (data not shown).
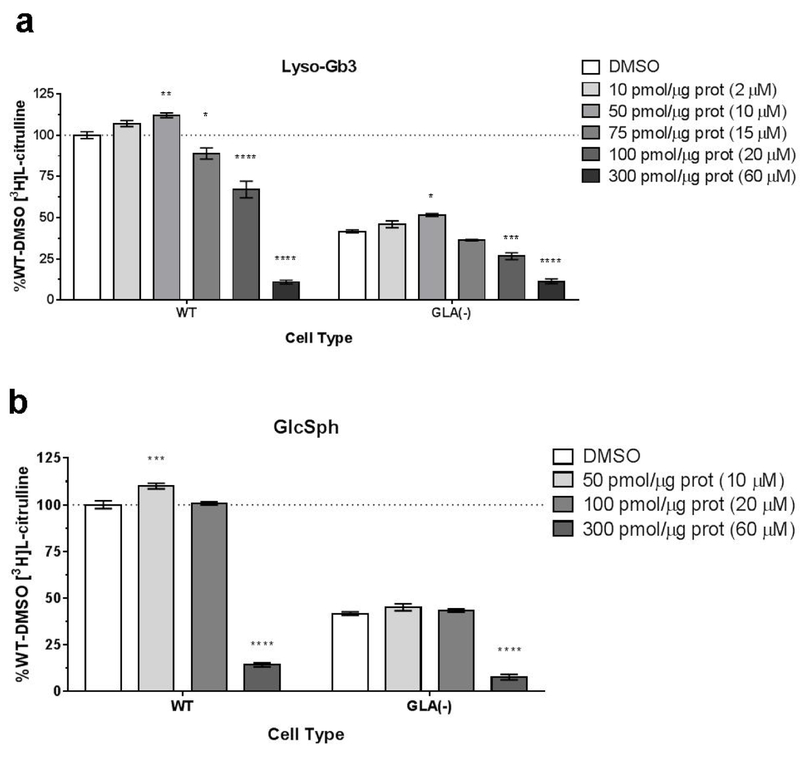
To directly assess the effect of lyso-Gb3 on eNOS, lyso-Gb3 or GlcSph were added to the NOS reaction mixture to achieve a final concentration of 0 – 60 μM lyso-Gb3 or GlcSph in each reaction volume. NOS activity was measured in the GLA-deficient clonal cell line 12, and wild type cell line 17. Lyso-Gb3 inhibited the NOS reaction in a dose-dependent manner (figure 5a). GlcSph also decreases NOS activity, but at a higher dose (figure 5b). These effects on NOS activity were observed in both WT and GLA-deficient cells.
Figure 5.
The NOS reaction was performed with different amounts of lyso-Gb3 in the reaction mixture. Each reaction contained 10 µg of protein from WT or GLA (-) lysates. (A) Lyso-Gb3 was added to the NOS reaction mixture. (B) GlcSph was added to the NOS reaction mixture. Results are reported as %WT-DMSO (dotted line). n=3. ****P<0.0001, ***P<0.001, **P<0.01, *P<0.05 compared to DMSO each cell type.
4. Discussion
In this study, we developed cell lines that are GLA-deficient to investigate the effect of specific changes in globo-series GSLs on eNOS activity. In contrast to other in vitro models of Fabry disease, these cells do not accumulate Gb3, the primary substrate for GLA, but still exhibit less NOS activity. Consistent with other Fabry disease models and clinical data, these cells had elevated lyso-Gb3 content. Furthermore, two pharmacological approaches of decreasing globo-series GSLs have opposite effects on NOS activity, suggesting restoring GLA function is better for improving NO production and reversing eNOS uncoupling than is reducing GSL accumulation. The relative changes in the globo-series glycolipids and their association with eNOS activity is shown in table 1.
Table 1.
Comparative changes in globo series glycosphingolipids and eNOS activity with siRNA and CRISPR/Cas9 GLA deficiency.
| Cell Model | Gb3 | Gb4 | lyso-Gb3 | eNOS activity |
|---|---|---|---|---|
| siRNA | ↑↑↑ | ↑ | ↑↑↑ | ↓↓↓ |
| CRISPR/Cas9 | -↑ | ↑↑↑ | ↑↑↑ | ↓↓↓ |
Patients with Fabry disease have an increased incidence of vasculopathy compared to the non-Fabry population (Sims, Politei et al. 2009). GLA deficiency causes accumulation of globo series GSLs, most notably in vascular endothelial cells (Desnick, Ioannou et al. 2001). Past studies indicate that defects of vascular dysfunction can be localized to the endothelium, and eNOS uncoupling may be the basis for impaired vascular function (Shu, Park et al. 2009, Shu, Vivekanandan-Giri et al. 2014). In the microvasculature, eNOS uncoupling may be a factor in gastrointestinal complications seen in patients with Fabry disease (Kang, Shu et al. 2014). To further investigate biochemical changes in endothelial cells due to GLA deficiency, we used the CRISPR/Cas9 method of gene editing to obtain permanent cell lines that were GLA deficient.
Previously, our group used primary cells isolated from mouse aortas of GLA knockout mice (mouse aortic endothelial cells, MAECs), which were useful for characterizing lipid changes and efficacy of GlcCer synthase inhibition for decreasing accumulating Gb3 (Shu, Murphy et al. 2005). However, this model was not sustainable for studying long-term consequences of treating cells with different agents since these cells could only be maintained in cell culture for a limited time (3 to 6 days). The anti-GLA siRNA EA.hy926 cell model was useful for making initial observations of how GLA-deficiency affects NOS activity, even when GLA was absent for only a few days, and for identifying 3-nitrotyrosine (a marker of eNOS uncoupling (Heinecke 2002)) as a biomarker for GLA deficiency (Shu, Vivekanandan-Giri et al. 2014). However, similar to MAECs, this in vitro model is not sustainable because a single siRNA treatment lasts for only three days. There can also be off-target effects of the siRNA.
An advantage of the CRISPR/Cas9 GLA-deficient cell model is that they can be continually passaged and retain the GLA-deficient characteristic with no additional treatment. To account for off-target effects of the initial CRISPR/Cas9 transfection, multiple single cell colonies were isolated, grown, and analyzed. While the measurements of Gb3, Gb4, and NOS activity were variable (figures 1 and 3), the lyso-Gb3 levels were consistently elevated in all four GLA-deficient clonal cell lines (figure 2). Potential off-target effects of the CRISPR/Cas9 transfection are unknown, but may have contributed to this observation. Moreover, the differences seen between the CRISPR/Cas9 and siRNA models of GLA depletion may be due to adaptive changes taking place in the CRISPR/Cas9 treated cells because they are continually passaged and have been without GLA expression for a longer period time than the cells treated with siRNA. The variation in Gb3 accumulation could be due to changes in Gb3 synthesis, or degradation to lyso-Gb3.
Despite differences of Gb3 and Gb4 accumulation, both the CRISPR/Cas9 and siRNA models of GLA deficiency in EA.hy926 cells have decreased NOS activity (figure 3 and (Shu, Vivekanandan-Giri et al. 2014)) and elevated lyso-Gb3 (figure 2). Lyso-Gb3, a deacylated form of Gb3, has emerged as a biomarker for Fabry disease (Aerts, Groener et al. 2008). Lyso-Gb3 is elevated in plasma from Fabry patients and in GLA-deficient mice. Lyso-Gb3 levels go down post-enzyme replacement therapy (ERT) (van Breemen, Rombach et al. 2011). Furthermore, lyso-Gb3 is useful for monitoring Fabry disease in females. Heterozygous females have normal circulating levels of GLA and Gb3, but may still experience symptoms of the disease. Symptomatic heterozygous females have elevated plasma lyso-Gb3, while lyso-Gb3 cannot be detected in asymptomatic heterozygous females (Aerts, Groener et al. 2008, Rombach, Dekker et al. 2010, Togawa, Kodama et al. 2010, Smid, van der Tol et al. 2015). In our study, Gb3 was not consistently elevated in all GLA-deficient clonal cell lines, but lyso-Gb3 was.
Lyso-lipids found to be elevated in other lipid storage diseases, for example, Krabbe disease (galactosylsphingosine), and Gaucher disease (glucosylsphingosine), have been shown to have toxic effects (Tanaka and Webster 1993, Schueler, Kolter et al. 2003). Lyso-Gb3 has also been shown to be involved in the pathogenesis of Fabry disease. Lyso-Gb3, but not Gb3 or lactosylsphingosine, promotes smooth muscle cell proliferation, which may contribute to the increased intima-media thickening seen in patients (Aerts, Groener et al. 2008). Lyso-Gb3 induces podocyte injury, which is an early feature of Fabry nephropathy (Sanchez-Nino, Sanz et al. 2011). In the present study, when cells were treated with physiologically relevant doses of lyso-Gb3, there were no observed changes in NOS activity in cell lysates. It is unknown if lyso-Gb3 in the cell culture medium was able to get into cells and affect intracellular lyso-Gb3 content. However, lyso-Gb3 was found to inhibit NOS when added to cell lysates (figure 5), but further work needs to be done to determine the mechanism of interaction. The concentration of lyso-Gb3 that inhibits NOS in this in vitro experiment is higher than that observed in the plasma of Fabry patients. Considering that eNOS is localized to the caveolae in the plasma membrane (Shaul, Smart et al. 1996), further studies need to be done to assess if lyso-Gb3 is also found at the plasma membrane, and if the concentrations of lyso-Gb3 used in these experiments are physiologically relevant.
The primary treatment method for Fabry disease is enzyme replacement therapy (ERT) with recombinant GLA. Substrate deprivation is an alternative approach to reducing accumulation of globo-series GSLs, where a small molecule inhibitor of glucosylceramide (GlcCer) synthase prevents the de novo synthesis of downstream GSLs, including LacCer, Gb3, and Gb4 (Abe, Arend et al. 2000, Abe, Gregory et al. 2000). A previous in vitro experiment performed with MAECs from Gla-knockout mice reported that treating cells with recombinant GLA or a GlcCer synthase inhibitor reduced the accumulation of Gb3 (Shu, Murphy et al. 2005). Similarly, treating CRISPR/Cas9 GLA-deficient EA.hy926 cells with exogenous GLA or the GlcCer inhibitor, eliglustat, decreased the accumulation of globo-series GSLs (figs. S.5 and S.6). Eliglustat treatment decreases NOS activity, even in WT cells (figure 4b). These results are in agreement with a previous study where treatment of MAECs from Gla-deficient mice with the GlcCer synthase inhibitor, D-threo-ethylenedioxyphenyl-2-palmitoylamino-3-pyrrolidinopropanol (P4), did not improve NOS activity, although Gb3 was cleared from cells (Shu, Park et al. 2009). In contrast, treating EA.hy926 cells with exogenous GLA increased NOS activity, in both GLA-deficient and WT cells (figure 4A). The opposite effects of two methods of reducing GSLs on NOS activity suggests that restoring GLA activity, rather than decreasing GSL content, may be more important for improving NOS activity.
5. Conclusion
eNOS uncoupling, observed in in vivo models of Fabry disease, is also observed in the CRISPR/Cas9 GLA-deficient EA.hy926 GLA-deficient endothelial cells. However, eNOS uncoupling tracks with increased lyso-Gb3 and is not necessarily the result of elevated Gb3 per se.
Supplementary Material
Acknowledgments
This work was supported by NIH grant RO1 DK 055823.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe A, Arend LJ, Lee L, Lingwood C, Brady RO and Shayman JA (2000). “Glycosphingolipid depletion in fabry disease lymphoblasts with potent inhibitors of glucosylceramide synthase.” Kidney Int 57(2): 446–454. [DOI] [PubMed] [Google Scholar]
- Abe A, Gregory S, Lee L, Killen PD, Brady RO, Kulkarni A and Shayman JA (2000). “Reduction of globotriaosylceramide in Fabry disease mice by substrate deprivation.” J Clin Invest 105(11): 1563–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aerts JM, Groener JE, Kuiper S, Donker-Koopman WE, Strijland A, Ottenhoff R, van Roomen C, Mirzaian M, Wijburg FA, Linthorst GE, Vedder AC, Rombach SM, Cox-Brinkman J, Somerharju P, Boot RG, Hollak CE, Brady RO and Poorthuis BJ (2008). “Elevated globotriaosylsphingosine is a hallmark of Fabry disease.” Proc Natl Acad Sci U S A 105(8): 2812–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames BN (1966). “[10] Assay of inorganic phosphate, total phosphate and phosphatases.” Methods in Enzymology 8: 115–118. [Google Scholar]
- Bodary PF, Shen Y, Vargas FB, Bi X, Ostenso KA, Gu S, Shayman JA and Eitzman DT (2005). “Alpha-galactosidase A deficiency accelerates atherosclerosis in mice with apolipoprotein E deficiency.” Circulation 111(5): 629–632. [DOI] [PubMed] [Google Scholar]
- Desnick RJ, Ioannou YA and Eng CM (2001). “α-Galactosidase A deficiency: Fabry disease.” The Metabolic and Molecular Bases of Inherited Disease(136): 3733–3774. [Google Scholar]
- Eitzman DT, Bodary PF, Shen Y, Khairallah CG, Wild SR, Abe A, Shaffer-Hartman J and Shayman JA (2003). “Fabry disease in mice is associated with age-dependent susceptibility to vascular thrombosis.” J Am Soc Nephrol 14(2): 298–302. [DOI] [PubMed] [Google Scholar]
- Eng CM, Fletcher J, Wilcox WR, Waldek S, Scott CR, Sillence DO, Breunig F, Charrow J, Germain DP, Nicholls K and Banikazemi M (2007). “Fabry disease: baseline medical characteristics of a cohort of 1765 males and females in the Fabry Registry.” J Inherit Metab Dis 30(2): 184–192 [DOI] [PubMed] [Google Scholar]
- Gold H, Mirzaian M, Dekker N, Joao Ferraz M, Lugtenburg J, Codee JD, van der Marel GA, Overkleeft HS, Linthorst GE, Groener JE, Aerts JM and Poorthuis BJ (2013). “Quantification of globotriaosylsphingosine in plasma and urine of fabry patients by stable isotope ultraperformance liquid chromatography-tandem mass spectrometry.” Clin Chem 59(3): 547–556. [DOI] [PubMed] [Google Scholar]
- Heinecke JW (2002). “Oxidized amino acids: culprits in human atherosclerosis and indicators of oxidative stress.” Free Radic Biol Med 32(11): 1090–1101. [DOI] [PubMed] [Google Scholar]
- Linhart A and Elliott PM (2007). “The heart in Anderson-Fabry disease and other lysosomal storage disorders.” Heart 93(4): 528–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstien S and Katusic Z (1999). “Oxidation of tetrahydrobiopterin by peroxynitrite: implications for vascular endothelial function.” Biochem Biophys Res Commun 263(3): 681–684. [DOI] [PubMed] [Google Scholar]
- Kang JJ, Shu L, Park JL, Shayman JA and Bodary PF (2014). “Endothelial nitric oxide synthase uncoupling and microvascular dysfunction in the mesentery of mice deficient in alpha-galactosidase A.” Am J Physiol Gastrointest Liver Physiol 306(2): G140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JL, Whitesall SE, D’Alecy LG, Shu L and Shayman JA (2008). “Vascular dysfunction in the alpha-galactosidase A-knockout mouse is an endothelial cell-, plasma membrane-based defect.” Clin Exp Pharmacol Physiol 35(10): 1156–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombach SM, Dekker N, Bouwman MG, Linthorst GE, Zwinderman AH, Wijburg FA, Kuiper S, Vd Bergh Weerman MA, Groener JE, Poorthuis BJ, Hollak CE and Aerts JM (2010). “Plasma globotriaosylsphingosine: diagnostic value and relation to clinical manifestations of Fabry disease.” Biochim Biophys Acta 1802(9): 741–748. [DOI] [PubMed] [Google Scholar]
- Sanchez-Nino MD, Sanz AB, Carrasco S, Saleem MA, Mathieson PW, Valdivielso JM, Ruiz-Ortega M, Egido J and Ortiz A (2011). “Globotriaosylsphingosine actions on human glomerular podocytes: implications for Fabry nephropathy.” Nephrol Dial Transplant 26(6): 1797–1802. [DOI] [PubMed] [Google Scholar]
- Schueler UH, Kolter T, Kaneski CR, Blusztajn JK, Herkenham M, Sandhoff K and Brady RO (2003). “Toxicity of glucosylsphingosine (glucopsychosine) to cultured neuronal cells: a model system for assessing neuronal damage in Gaucher disease type 2 and 3.” Neurobiol Dis 14(3): 595–601. [DOI] [PubMed] [Google Scholar]
- Shaul PW, Smart J, Robinson LJ, German Z (1996). “Acylation targets endothelial nitric-oxide synthase to plasmalemmal caveolae.” J Biol Chem 271(11): 6518–6522 [DOI] [PubMed] [Google Scholar]
- Shu L, Murphy HS, Cooling L and Shayman JA (2005). “An in vitro model of Fabry disease.” J Am Soc Nephrol 16(9): 2636–2645. [DOI] [PubMed] [Google Scholar]
- Shu L, Park JL, Byun J, Pennathur S, Kollmeyer J and Shayman JA (2009). “Decreased nitric oxide bioavailability in a mouse model of Fabry disease.” J Am Soc Nephrol 20(9): 1975–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu L and Shayman JA (2007). “Caveolin-associated accumulation of globotriaosylceramide in the vascular endothelium of alpha-galactosidase A null mice.” J Biol Chem 282(29): 20960–20967. [DOI] [PubMed] [Google Scholar]
- Shu L, Vivekanandan-Giri A, Pennathur S, Smid BE, Aerts JM, Hollak CE and Shayman JA (2014). “Establishing 3-nitrotyrosine as a biomarker for the vasculopathy of Fabry disease.” Kidney Int 86(1): 58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims K, Politei J, Banikazemi M and Lee P (2009). “Stroke in Fabry Disease Frequently Occurs Before Diagnosis and in the Absence of Other Clinical Events: Natural History Data From the Fabry Registry.” Stroke 40(3): 788–794. [DOI] [PubMed] [Google Scholar]
- Smid BE, van der Tol L, Biegstraaten M, Linthorst GE, Hollak CE and Poorthuis B (2015). “Plasma globotriaosylsphingosine in relation to phenotypes of Fabry disease.” J Med Genet 52(4): 262–268. [DOI] [PubMed] [Google Scholar]
- Tanaka K and Webster HD (1993). “Effects of psychosine (galactosylsphingosine) on the survival and the fine structure of cultured Schwann cells.” J Neuropathol Exp Neurol 52(5): 490–498. [DOI] [PubMed] [Google Scholar]
- Togawa T, Kodama T, Suzuki T, Sugawara K, Tsukimura T, Ohashi T, Ishige N, Suzuki K, Kitagawa T and Sakuraba H (2010). “Plasma globotriaosylsphingosine as a biomarker of Fabry disease.” Mol Genet Metab 100(3): 257–261. [DOI] [PubMed] [Google Scholar]
- van Breemen MJ, Rombach SM, Dekker N, Poorthuis BJ, Linthorst GE, Zwinderman AH, Breunig F, Wanner C, Aerts JM and Hollak CE (2011). “Reduction of elevated plasma globotriaosylsphingosine in patients with classic Fabry disease following enzyme replacement therapy.” Biochim Biophys Acta 1812(1): 70–76. [DOI] [PubMed] [Google Scholar]
- Vasquez-Vivar J, Kalyanaraman B, Martasek P, Hogg N, Masters BS, Karoui H, Tordo P and Pritchard KA Jr. (1998). “Superoxide generation by endothelial nitric oxide synthase: the influence of cofactors.” Proc Natl Acad Sci U S A 95(16): 9220–9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedder AC, Linthorst GE, van Breemen MJ, Groener JE, Bemelman FJ, Strijland A, Mannens MM, Aerts JM and Hollak CE (2007). “The Dutch Fabry cohort: diversity of clinical manifestations and Gb3 levels.” J Inherit Metab Dis 30(1): 68–78. [DOI] [PubMed] [Google Scholar]
- Xia Y, Tsai AL, Berka V and Zweier JL (1998). “Superoxide generation from endothelial nitric-oxide synthase. A Ca2+/calmodulin-dependent and tetrahydrobiopterin regulatory process.” J Biol Chem 273(40): 25804–25808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.