Abstract
Background
Adherent junction associated protein 1 (AJAP1), a typical molecule of adherent junctions, has been found to be a tumor suppressor in many cancer types. Aberrant activation of β-catenin has been demonstrated to be associated with malignant biological properties of tumors including breast cancer. This study aimed to investigate the function and mechanism of AJAP1-mediated β-catenin activity of breast cancer lines in vitro and in breast cancer patients.
Methods
AJAP1 and β-catenin expressions in breast cancer tissues and cell lines were detected by immunohistochemistry, western blotting and qRT-PCR. The EGF/EGFR axis-mediated AJAP1 attenuated β-catenin nuclear location was measured by western blotting, immunofluorescence assay, co-immunoprecipitation, luciferase assay and ubiquitination assays. Furthermore, the function of AJAP1 and β-catenin regulated breast cancer progression was explored both in vivo and in vitro.
Results
It was found that AJAP1 had a high negative correlation with β-catenin nuclear expression and was a novel tumor suppressor in breast cancer. AJAP1 loss can mediate β-catenin accumulated in cytoplasm and then transferred it to the nucleus, activating β-catenin transcriptional activity and downstream genes. Additionally, β-catenin can reverse the invasion, proliferation ability and tumorigenicity of the depletion of AJAP1 caused both in vivo and in vitro. Besides, EGF/EGFR also involved in the process of AJAP1-depiction induced β-catenin transactivation to the nucleus. More importantly, EGFR depletion/AJAP1 knocked down promoted the progression of breast cancer by regulating the activity of β-catenin nuclear transactivation.
Conclusion
This study demonstrated that AJAP1 acted as a putative tumor suppressor while β-catenin nuclear localization positively fed back on EGF/EGFR-attenuated AJAP1 expression in breast cancer, which might be beneficial to develop new therapeutic targets for decreasing nuclear β-catenin-mediated malignancy in breast cancer.
Electronic supplementary material
The online version of this article (10.1186/s13046-019-1252-6) contains supplementary material, which is available to authorized users.
Keywords: AJAP1, β-Catenin, Nuclear location, EGF, EGFR, Tumor progression
Background
Breast cancer, a biologically and molecularly heterogeneous disease derived from epithelial cells, has been one of the most common malignancies in women worldwide for many years [1–3]. As fundamental components of epithelial cells, adherent junctions (AJs) have been proven to play important roles in cancer progression [4–10]. However, data on AJs in breast cancer is still scarce.
Adherens junctions-associated protein 1(AJAP1), also called Shrew-1, was initially discovered as a novel transmembrane protein of AJs in epithelial cells [11]. Some studies then verified that AJAP1 was a promising tumor candidate gene in glioma [12, 13], hepatocellular carcinoma [14–16], esophagus carcinoma [17] and oligodendrogliomas [18]. However, its role in breast cancer has not been fully elucidated.
In addition, previous reports showed that 50% of breast cancer cases have Wnt signaling abnormal activation and low rates of somatic mutations [19–21]. Additionally, abnormal activation of Wnt signaling often led to β-catenin nuclear accumulation [22–25]. Nuclear β-catenin can function as a transcriptional co-activator of the TCF/LEF complex, resulting in a series of changes in proliferation, invasion and metastasis. Moreover, β-catenin has been implicated in the transduction of mechanical signals from junctions to the nucleus [26].
In this study, the roles of AJAP1 and β-catenin in breast cancer were explored. Immunohistochemistry assay showed that AJAP1 depletion was positively related with β-catenin nuclear expression and poor prognosis of patients. Besides, AJAP1 was a putative tumor suppressor that suppressed the growth, migration, invasion of breast cancer and cell cycle by mediating the nuclear β-catenin activity. More importantly, β-catenin localization and tumor progression also positively fed back on EGF/EGFR-attenuated AJAP1 expression. In summary, these findings might be beneficial in developing new therapeutic targets for decreasing nuclear β-catenin-mediated malignancy in breast cancer.
Materials and methods
Patients and breast cancer samples
283 cases of paraffin-embedded breast cancer patients’ specimen and 25 pairs of fresh tumor tissues were randomly selected at Cancer Hospital of Tianjin Medical University. The patients received treatments from January 1, 2006 to December 31, 2006. None of the patients underwent chemotherapy or radiotherapy before surgery. The patient clinical pathologic features are showed in Additional file 1: Table S1. All cases had decent follow-up and reliable clinical data. Besides, this study followed the Declaration of Helsinki, and the patients provided written informed consents.
Immunohistochemistry (IHC) and evaluation
All paraffinized tissue blocks were cut at 4 μm thicknesses and detected by the SP immunochemistry kit (Zhongshan Golden Bridge Biotechnology, Beijing, China). IHC assay was conducted as previously described [27]. The rabbit monoclonal anti-human AJAP1 antibody (Bioss, China) at 1:100 dilution or the mouse monoclonal anti-human β-catenin antibody (CTNNB1, Boster) at 1:200 dilution was used for IHC. Two senior pathologists (Yun Niu and Shuhua Lv) evaluated the score without any knowledge of the clinicopathological outcomes of the patients. The percentage of positivity of the tumor was scored as “0” (no tumor cells), “1” (1–25%), “2” (26–50%), “3” (51–75%), and “4” (> 75%). The staining intensity of the positive tumor cells was scored as “0” (no staining), “1” (weak staining), “2” (moderate staining), and “3” (strong staining). As for AJAP1, the multiplier of the scoring of (0–3) for low expression and (4–12) for high expression were used. The IHC staining results of β-catenin were evaluated independently according to the subcellular localization of the nucleus and membrane. A positive/abnormal nuclear expression was defined as over 10% of the nuclear-stained tumor cells. An abnormal membranous expression demonstrated either no immunoreactivity in the membrane or less than 10% of the cancer cells with a positive membranous staining [28, 29].
Cell line and culture
All cell lines were purchased from the American Type Culture Collection (ATCC, USA). T47D, SK-BR-3, MCF-7 and MDA-MB-231 were cultured with the 1640 (Gibco, USA), MCF10A was cultured in DMEM-F12 (Gibco, USA) media. MDA-MB-453 was cultured in L-15 media. All media contained 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin and cell culture carried out at 37 °C in a 5% CO2 incubator.
Immunofluorescence
The cells were grown in 24 -well plates and then stimulated with 50 ng/ml EGF for 24 h. After 1 day, the cells were fixed, rehydrated, and incubated with β-catenin (Santa Cruz Biotech, China), AJAP1 (Bioss, China), and fluorescein-labeled secondary antibodies. The nuclei were stained with DAPI for 10 min. The slides were examined through a fluorescent confocal microscope.
Western blot
Cellular protein was extracted by RIPA buffer containing PMSF. The Nuclear and Cytoplasmic Isolation Kit (KeyGEN, China) was used for the cytoplasmic and nuclear protein extraction according to the manufacturer’s instructions. The proteins were separated on 10% SDS-PAGE gels, transferred onto PVDF membranes, blocked using 5% skim milk, and incubated with primary antibodies overnight at 4 °C. The protein bands were detected by the ECL detection Kit (Solarbio, China).
Plasmids and lentiviral transfection
The related siRNAs and control plasmids were showed in Additional file 2: Table S2. The viruses were packaged in 293 T cells according to the manufacturer’s instructions. The infected cells were selected with 2 μg/mL puromycin.
Quantitative reverse-transcription polymerase chain reaction (qRT- PCR) assay
Total RNAs were extracted using Trizol reagent (Takara, Japan). Reverse transcription was conducted by the SuperScript RT kit (Takara, Japan). The qRT-PCR assay was performed with SYBR Green PCR kit (Takara, Japan). Primers used are listed in Additional file 2: Table S2. GAPDH was used as the internal control. The relative expression levels were calculated by the 2-ΔΔCt method. All experiments were assayed in triplicate.
Co-immunoprecipitation (co-IP)
The co-IP of AJAP1 and β-catenin was performed using the Pierce Co-IP kit (Thermo Scientific, USA) according to the manufacturer’s protocol as described previously [30]. The protein quantity was then detected by western blotting.
Luciferase assays
5 × 104 cells were seeded in 24-well plates and cultured for 24 h at a density of 70–80%. The cells were transferred with 2 μg of β-catenin-responsive firefly luciferase reporter plasmid TOP-FLASH or the negative control FOP-FLASH (Merck-Millipore) using fugene6 (Invitrogen, USA) according to the manufacturer’s instructions. The cells were subjected to luciferase reporter assay 24 h after transfection according the Dual Luciferase Reporter Assay System (Promega, Madison, WI). The relative Renilla luciferase activity was normalized to that of firefly luciferase activity. Each experiment was measured in triplicate.
Migration and invasion assay
2 × 104 cells were seeded in the upper chamber with RMPI 1640 medium without FBS and a medium containing 10% FBS at 37 °C in the lower chamber. After culturing at 37 °C for 24 h, the cells in the upper chamber were harvested and then stained according to the three-step set (Thermo Scientific, USA) protocol. Scratches were made when the cells reached a 95% confluence in each well using a 10 μL plastic pipette tip. The scratches were measured and photographed. Each experiment was conducted at least three times.
MTT and colony-formation assay
2 × 103 cells were seeded in 96-well plates and the activities of the viable cells were measured at different time points using a microplate reader. In the colony formation assay, 8 × 102 cells were cultured in 6-well plates for 2 weeks. The colonies were stained with crystal violet and counted.
Subcutaneous xenograft assay
Female BALB/c nude mice (4–6 week) were injected with 5 × 106 treated cells and kept in the Tianjin Medical University Cancer Institute and Hospital SPF’s animal feeding center. The tumors were measured as described previously [31]. All animals were sacrificed after 60 days. All experimental procedures were approved by the International Animal Care and Use Committee of Tianjin Medical University Cancer Institute and Hospital.
Statistical analyses
Statistical significance was determined by SPSS version 24.0. Values are shown as mean ± SD. Multiple groups were compared by one-way analysis of variance followed by Dunnett’s t-test. p < 0.05 was considered statistically significant.
Results
AJAP1 was low expressed in breast cancer and inhibited the proliferation of breast cells
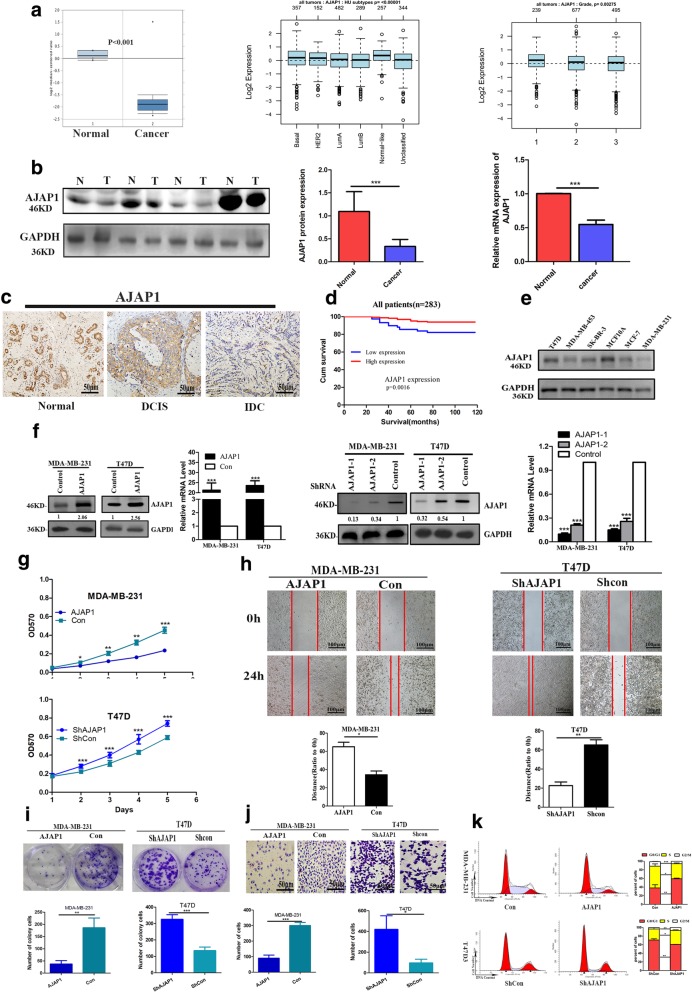
To determine the expression profile of AJAP1 in breast cancer tissues, breast cancer datasets from Oncomine and GOBO datasets were analyzed in this study. The dataset from Finak (GOBO)and Oncomine showed that AJAP1 was downregulated in the breast cancer tissues compared with the normal tissues (Fig. 1a left panel). In addition, GOBO dataset demonstrated that the expression of AJAP1 was related with cancer subtypes (Fig. 1a middle panel, p = 0.00402) and grades (Fig. 1a right panel, p = 0.00275). These conclusions were confirmed by Western blotting and qRT-PCR of 25 pairs of breast cancer and corresponding normal tissues (Fig. 1b).
Fig. 1.
AJAP1 was low expressed in breast cancer and inhibited the proliferation of breast cells. a Dataset from Oncomine and GOBO on AJAP1 in tumors tissues versus non-tumor tissues (n = 59), different subtypes of breast cancer(n = 1881) and grades(n = 1411). b AJAP1 protein and mRNA levels in 25 pairs of tumor tissues versus non-normal tissue. c Representative IHC image of AJAP1 expression location in different stages of breast cancer (normal lobules, Ductal carcinoma in situ, invasive cancer). d overall survival of AJAP1 in 283 cases of breast cancer. e AJAP1 protein levels in breast cancer cell lines. f confirmation of upregulation and downregulation of AJAP1 expression by Western blot and qRT-PCR (g-k) effects of AJAP1 knockdown and overexpressed on in vitro proliferation (g, i), invade (h) migration(j) and cell cycle progression (k) Data were shown as mean ± SD *p < 0.05; **p < 0.01; ***p < 0.001
Furthermore, 283 breast cancer formalin-fixed, paraffin-embedded tumor slides with complete clinicopathological characteristics and follow-up data were utilized to analyze the expression of AJAP1. In this work, AJAP1 was mainly located in the cytoplasm and was downregulated in breast cancer. Moreover, its expression was decreased in the progression of breast cancer (Fig. 1c). As shown in Table 1, low AJAP1 expression was significantly associated with advanced histological grade and higher lymph node involvement. In addition, Cox proportional hazards regression analyses (Additional file 3: Table S3) demonstrated that AJAP1 was an independent prognostic marker for overall survival (p = 0.024). Meanwhile, Kaplan-Meier analysis showed that patients with low expression of AJAP1 exhibited poor survival (Fig. 1d).
Table 1.
The clinicopathologic characteristics of AJAP1 expression in 283 Breast cancer patients
| Parameters | Cases | AJAP1 | p | |
|---|---|---|---|---|
| Low expression (N, %) | High expression (N, %) | |||
| Age(yr) | 118(41.7) | 165(58.3) | ||
| ≤50 | 130 | 49(41.5) | 81(49.1) | 0.208 |
| > 50 | 153 | 69(58,5) | 84(50.9) | |
| Menopausal status | ||||
| Premenopausal | 156 | 60(50.8) | 96(58.2) | 0.221 |
| Postmenopausal | 127 | 58(49.2) | 69(41.8) | |
| Family history | ||||
| No | 194 | 85(72.0) | 109(66.1) | 0.286 |
| Yes | 89 | 33(28.0) | 56(33.9) | |
| Tumor size | ||||
| T1 | 103 | 42(35.6) | 68(41.2) | 0.615 |
| T2 | 148 | 66(55.9) | 83(50.3) | |
| T3 | 32 | 10(8.5) | 14(8.5)) | |
| Histological grade | ||||
| 1 | 44 | 9(7.6) | 35(21.2) | < 0.0001* |
| 2 | 153 | 59(50.0) | 94(57.0) | |
| 3 | 86 | 50(42.4) | 36(21.8) | |
| LN involvement | ||||
| 0 | 180 | 65(55.1) | 115(69.7) | 0.010* |
| 1–3 | 43 | 19(16.1) | 24(14.5) | |
| 4–9 | 41 | 20(16.9) | 21(12.7) | |
| ≥ 10 | 19 | 14(11.9) | 5(3.0) | |
| ER | ||||
| Negative | 111 | 50(42.4) | 61(37) | 0.359 |
| Positive | 172 | 68(57.6) | 104(63) | |
| PR | ||||
| Negative | 142 | 83(41.5) | 82(49.7) | 0.849 |
| Positive | 141 | 35(42.2) | 83(50.3) | |
| Her-2 | ||||
| Negative | 200 | 83(41.5) | 117(58.5) | 0.917 |
| Positive | 83 | 35(42.2) | 48(57.8) | |
| Ki67 | ||||
| < 20 | 83 | 31(26.3) | 52(31.5) | 0.339 |
| ≥ 20 | 200 | 87(73.7) | 113(68.5) | |
| P53 | ||||
| Negative | 143 | 59(50) | 84(50.9) | 0.880 |
| Positive | 140 | 59(50) | 81(49.1) | |
*significantly different
To better explore the role of AJAP1 in breast cancer, AJAP1 expression in different wild types of breast cancer cell lines were detected. T47D and MDA-MB-231 cells showed typical high and low expression of AJAP1, respectively (Fig. 1e). T47D cells with knocking down of AJAP1 expression and MDA-MB-231 cells with overexpression of AJAP1 were then generated (Fig. 1f) to evaluate the proliferation, migration and invasion of breast cancer by MTT, Transwell, colony-formation and wound-healing assays. These results showed that AJAP1-depletion in T47D cells promoted the capacity of proliferation (Fig. 1g, Fig. 1i), migration (Fig. 1h) and invasion (Fig. 1j). In contrast, AJAP1 overexpressed in MDA-MB-231 cells showed the opposite results. Furthermore, flow cytometry (Fig. 1k) results showed that downregulation of AJAP1 in T47D cells significantly reduced the population of G0/G1 cells but increased S/G2/M cells. On the contrary, AJAP1-overexpressed MDA-MB-231 cells showed the opposite results. In conclusion, AJAP1 was a putative tumor suppressor in breast cancer.
Different expression patterns of AJAP1 and β-catenin in breast cancer patients
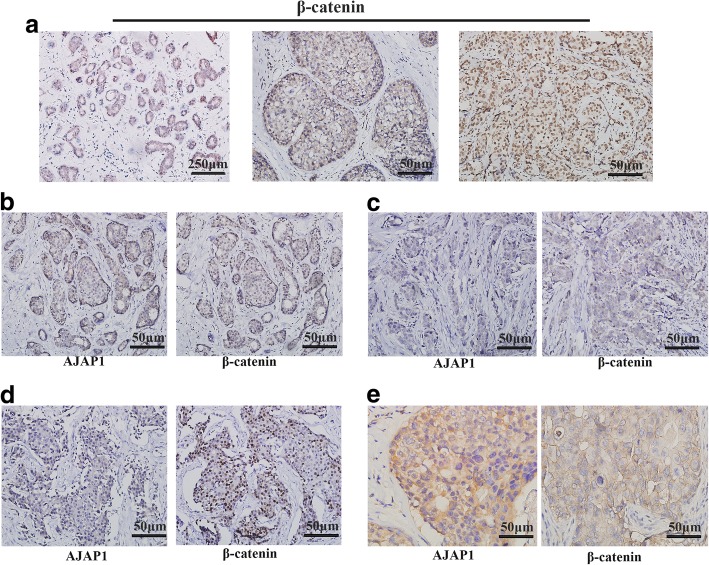
The 283 breast cancer cases were then used to explore the relationship between β-catenin expression and clinicopathological parameters. Figure 2a represents the typical progression of breast cancer. It was observed that β-catenin presented a strong uniform membrane in the luminal cells of para-carcinoma normal tissues (Fig. 2a, left). In the high-grade ductal carcinoma in situ and invasive cancer, β-catenin demonstrated different expression patterns of cytoplasmic (Fig. 2a, middle) and nuclear expression (Fig. 2a, right). The different expression patterns of AJAP1 and β-catenin in different cases were illustrated in Fig. 2b–e. The abnormal membranous expression of β-catenin and the positive nuclear/cytoplasmic expression of β-catenin were detected in 43.5% (123/283) and 78.1% (221/283) of the breast cancer tissues, respectively. As shown in Table 2, the β-catenin expression in membrane staining was correlated with histological grade (p = 0.005) and lymph node (LN) involvement (p = 0.018) in breast cancer patients. Likewise, the expression of β-catenin in the nucleus and cytoplasm was related to histological grade (p < 0.0001) and LN involvement (p = 0.010) in breast cancer patients.
Fig. 2.
Immunohistochemical expression of β-catenin and AJAP1 in breast tissue slides. a β-catenin expression in normal breast cancer tissues, ductal carcinoma in situ and invasive ductal carcinoma (magnification, × 40, × 200 and × 200). b-e Different expression patterns of AJAP1 and β-catenin in breast cancer tissues. Micrographs showing low expression (c, d) and high expression (b, e) AJAP1 cytoplasm (b, c, d) and membrane (e) expression and negative (c) and positive (b, d, e) β-catenin membrane(e), cytoplasm (b, c) and nuclear expression(d) through the immunohistochemical staining of breast cancer specimens (magnification, × 200)
Table 2.
Correlation between the clinicopathologic characteristics and membranous, cytoplasmic and nuclear expression of β-catenin in 283 breast cancer patients
| Parameters | Cases | β-catenin(M) | p | β-catenin(C/N) | p | ||
|---|---|---|---|---|---|---|---|
| Normal (N, %) | Abnormal (N, %) | Negative(N, %) | Positive (N, %) | ||||
| Age(yr) | 160(56.5) | 123(43.5) | 62(21.9) | 221(78.1) | |||
| ≤50 | 130 | 79(49.4) | 51(41.5) | 0.186 | 32(51.6) | 123(55.7) | 0.310 |
| > 50 | 153 | 81(50.6) | 72(58.5) | 30(48.4) | 98(44.3) | ||
| Menopausal status | |||||||
| Premenopausal | 156 | 90(56.3) | 66(53.7) | 0.664 | 37(59.7) | 119(53.8) | 0.415 |
| Postmenopausal | 127 | 70(43.8) | 57(46.3) | 25(40.3) | 102(46.2) | ||
| Family history | |||||||
| No | 194 | 111(69.4) | 83(67.5) | 0.734 | 69(31.2) | 152(68.8) | 0.877 |
| Yes | 89 | 49(30.6) | 40(32.5) | 20(32.3) | 69(31.2) | ||
| Tumor size | |||||||
| T1 | 110 | 66(41.3) | 44(35.8) | 0.428 | 32(51.6) | 78(35.3) | 0.055 |
| T2 | 149 | 83(51.9) | 66(53.7) | 27(43.5) | 122(55.2) | ||
| T3 | 24 | 11(6.9) | 13(10.6) | 3(4.8) | 21(9.5) | ||
| Histological grade | |||||||
| 1 | 44 | 33(20.6) | 11(8.9) | 0.005* | 16(25.8) | 28(12.7) | 0.029* |
| 2 | 153 | 88(55.0) | 65(52.8) | 32(51.6) | 121(54.8) | ||
| 3 | 86 | 39(24.4) | 47(38.2) | 14(22.6) | 72(32.6) | ||
| LN involvement | |||||||
| 0 | 180 | 114(71.3) | 66(53.7) | 0.018* | 49(79.0) | 131(59.3) | 0.028* |
| 1–3 | 43 | 19(11.9) | 24(19.5) | 7(11.3) | 36(16.3) | ||
| 4–9 | 41 | 20(12.5) | 21(17.1) | 5(8.1) | 36(16.3) | ||
| ≥ 10 | 19 | 7(4.4) | 12(9.8) | 1(1.6) | 18(8.1) | ||
| ER | |||||||
| Negative | 111 | 68(42.5) | 43(35.0) | 0.198 | 27(43.5) | 84(38.0) | 0.430 |
| Positive | 172 | 92(57.5) | 80(65.0) | 35(56.5) | 137(62.0) | ||
| PR | |||||||
| Negative | 142 | 87(54.4) | 55(44.7) | 0.107 | 35(56.5) | 107(48.4) | 0.263 |
| Positive | 141 | 73(45.6) | 68(55.3) | 27(43.5) | 114(51.6) | ||
| Her-2 | |||||||
| Negative | 200 | 109(54.5) | 91(45.5) | 0.283 | 44(22.0) | 156(78.0) | 0.953 |
| Positive | 83 | 51(61.4) | 32(38.6) | 18 (21.7) | 65 (78.3) | ||
| Ki67 | |||||||
| < 20 | 83 | 54 (33.8) | 29 (23.6) | 0.062 | 19 (30.6) | 64 (29.0) | 0.797 |
| ≥ 20 | 200 | 106 (66.3) | 94 (76.4) | 43 (69.4) | 157 (71.0) | ||
| P53 | |||||||
| Negative | 143 | 80(50.0) | 63(51.2) | 0.839 | 27(43.5) | 116(52.5) | 0.213 |
| Positive | 140 | 80(50.0) | 60(48.8) | 35(56.5) | 105(47.5) | ||
*significantly different
There were different expression forms of AJAP1 and β-catenin observed in Fig. 2b–e, thus the correlation between the expressions of β-catenin and AJAP1 was analyzed. Table 3 showed that the low expression of AJAP1 was linked to abnormal membrane expression (p < 0.0001) and cytoplasmic/nuclear staining of β-catenin (p = 0.001). Moreover, the membrane staining of β-catenin had a significant difference with the cytoplasmic/nuclear expression (Table 4, p < 0.001).
Table 3.
Correlation between β-catenin and AJAP1 expression
| AJAP1 | p | ||||
|---|---|---|---|---|---|
| Low expression | High expression | ||||
| Membrane | 160 | Normal | 49(41.5) | 111(67.3) | < 0.0001* |
| 123 | Abnormal | 69(58.5) | 54(32.7) | ||
| Cytoplasmic/Nuclear | 62 | Negative | 14(11.9) | 48(29.1) | 0.001* |
| 221 | Positive | 104(88.1) | 117(70.9) | ||
Table 4.
Correlation between membrane and nuclear of β-catenin expression
| Cytoplasmic/Nuclear | p | ||||
|---|---|---|---|---|---|
| Negative | Positive | ||||
| Membrane | 160 | Normal | 57(91.9) | 103(46.6) | < 0.0001* |
| 123 | Abnormal | 5(8.1) | 118(53.4) | ||
Correlation between AJAP1 and β-catenin expressions with patient outcome
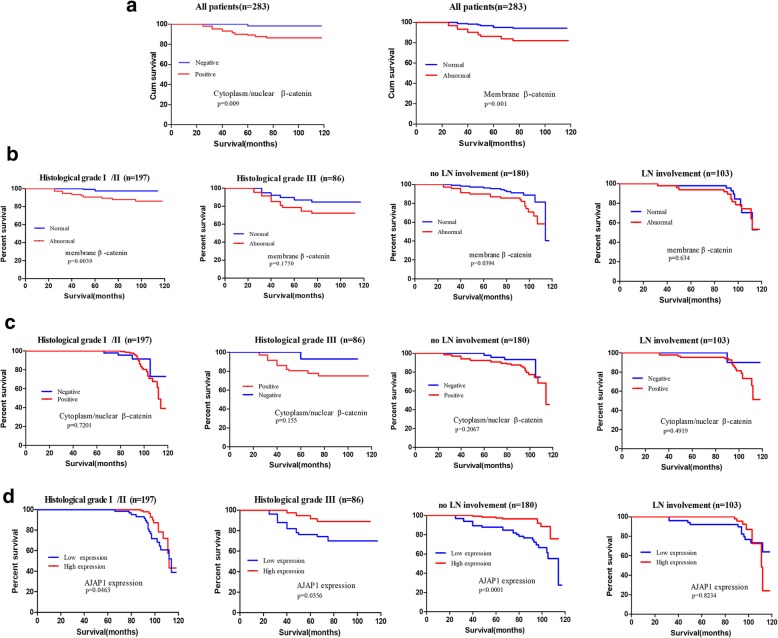
As shown in Fig. 3a, abnormal membrane expression, cytoplasm and nuclear staining of β-catenin characterized a poor prognosis, while univariate and multivariate analyses showed that the location of β-catenin was not an independent prognostic factor for OS (Additional file 3: Table S3).
Fig. 3.
The effect of different expression locations of β-catenin and AJAP1 expression on overall survival was demonstrated in breast cancer patients. All breast cancer patients were divided into different groups according to histological grade and LN involvement. The Kaplan-Meier survival plot was evaluated, and log-rank tests were utilized to analyze the prognostic value of AJAP1 and β-catenin expressions in all patients (a) and subgroups (b-d)
Many studies hold different perspectives on the prognosis value of β-catenin. The membrane expression of β-catenin seemed to predict OS better in the I/II histological grade (p = 0.0039) and no LN involvement (p = 0.0394) as shown in Fig. 3b. However, the cytoplasmic or nuclear expression of β-catenin did not show a significant difference in the different risks of subgroups in breast cancer patients (Fig. 3c).
Patients with a low expression of AJAP1 predicted poor OS in all the subgroups except the patients without involvement of LN (Fig. 3d, p = 0.8234). In summary, these results indicated that AJAP1 expression could be a prognostic marker in the different risks of subgroups of breast cancer patients, while β-catenin didn’t have the same function.
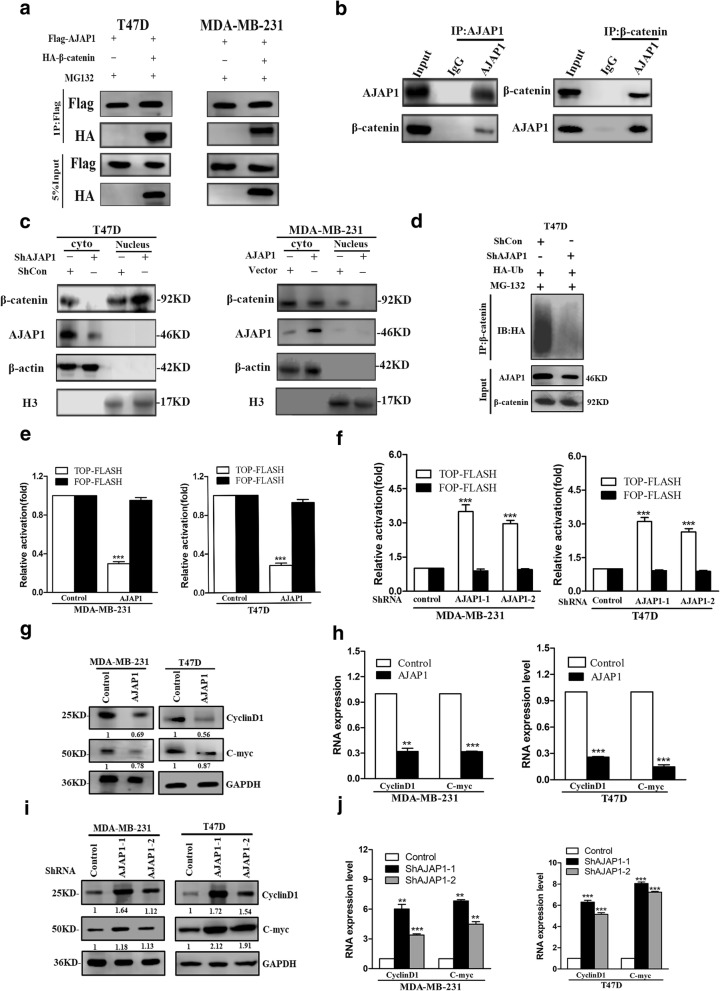
AJAP1 inhibited nuclear accumulation of β-catenin via interaction with β-catenin binding sites in breast cancer cell
To further explore how AJAP1 mediated β-catenin, the relationship and potential mechanism between AJAP1 and the different locations of β-catenin in breast cancer were investigated. Firstly, it was found that AJAP1 and β-catenin interacted in T47D and MDA-MB-231 cells (Fig. 4a). Co-IP assay was also conducted to confirm this theory (Fig. 4b). Loss of AJAP1 reduced the β-catenin cytoplasm expression but increased its nuclear location in T47D. In contrast, upregulated AJAP1 in MDA-MB-231 cells showed opposite results (Fig. 4c). To determine the exact path of β-catenin degradation mediated by AJAP1, the ubiquitination level of β-catenin in AJAP1-deficient T47D cells was examined. As shown in Fig. 4d, the ubiquitination of β-catenin was reduced in the AJAP1 knockdown cells, demonstrating that AJAP1-depletion activated Wnt signaling which caused β-catenin degradation.
Fig. 4.
AJAP1 interacted with β-catenin and mediated its nuclear location and transcriptional activity. a Co-IP assay results showed that AJAP1 formed a complex with β-catenin in T47D and MDA-MB-231 cells. b Co-IP assays showed that endogenous protein of AJAP1 interacted with β-catenin. c Western blot results showed that AJAP1 depletion increased nuclear translocation of β-catenin and overexpression of AJAP1 had the opposite function. d AJAP1-depleted stable cell lines were transfected with HA-Ub and then treated with MG132 as demonstrated. Cell lysates were subjected to β-catenin and showed that AJAP1-depletion inhibited the ubiquitination level of β-catenin. e AJAP1 reduced the β-catenin/TCF/LEF-mediated transcription activity in MDA-MB-231 and T47D cells with and without the upregulation of AJAP1. f AJAP1-depletion in MDA-MB-231 and T47D cells increased the β-catenin/TCF/LEF-mediated transcription activity. g-h overexpressed AJAP1 can reduce the protein (g) and mRNA(h) levels of β-catenin downstream genes as C-myc and CyclinD1 in MDA-MB-231 and T47D cells by western blot and RT-PCR. i-j AJAP1 knockdown can increase the protein (i) and mRNA(j) levels of β-catenin downstream genes, such as C-myc and CyclinD1 in MDA-MB-231 and T47D cells by western blot and qRT-PCR. Data were shown as mean ± SD. Each experiment was conducted in triplicate. **p < 0.01, ***p < 0.001
AJAP1 regulated the transcriptional activity of β-catenin and its downstream gene expression
Above experiments showed that the knockdown of AJAP1 influenced the location of β-catenin, however it is unclear whether the transcriptional activity of β-catenin was also changed. TCF/LEF-responsive luciferase (TOP-FlASH) and non-responsive (FOP-FLASH) reporters were used to evaluate the effect of AJAP1 on the β-catenin transcriptional activity. As shown in Fig. 4e, upregulation of AJAP1 led to a lower transcription activity from the β-catenin/TCF/LEF-dependent promoter (TOP-FLASH) but had little effect on FOP-FLASH. Conversely, downregulation of AJAP1 obviously increased the transcription activity of the β-catenin/TCF/LEF-dependent promoter (Fig. 4f). To further examine the influence of knockdown or overexpression of AJAP1 on β-catenin’s downstream gene expression, the expressions of C-myc and CyclinD1 were analyzed by Western blotting and qRT-PCR. Overexpression of AJAP1 in MDA-MB-231 and T47D cells decreased C-myc and CyclinD1 expression both in protein and mRNA levels (Fig. 4g and h). On the contrary, knockdown of AJAP1 showed the opposite results (Fig. 4i and j). These findings showed that AJAP1 depletion increased β-catenin nuclear expression, activated the transcriptional activity of β-catenin and promoted its expressions of downstream genes (C-myc and CyclinD1).
AJAP1 regulated breast cancer tumorigenesis via mediating β-catenin expression
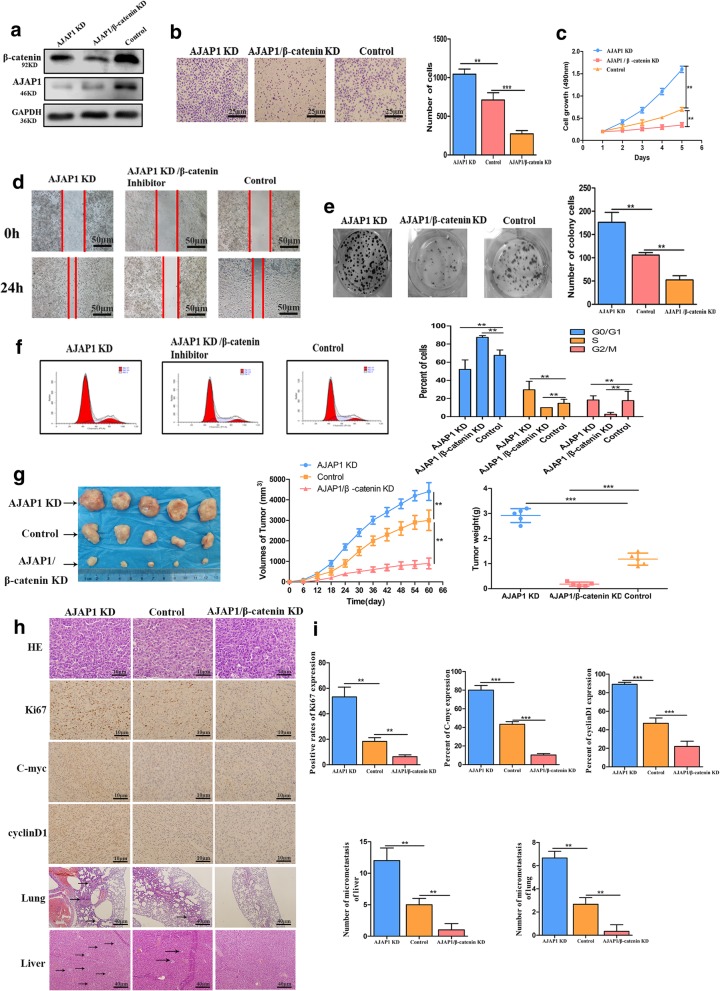
The above results indicated that AJAP1 inhibited cell invasion, proliferation and migration ability in breast cancer. Then we explored whether the function of AJAP1 in breast cancer progression relied on mediation of β-catenin expression. For this purpose, three groups of T47D cells were constructed: depletion of AJAP1 (AJAP1 KD), AJAP1 knockdown and β-catenin knockdown (AJAP1/β-catenin KD), and the control. Figure 5a shows the different protein levels of AJAP1 and β-catenin in the three groups. Transwell assay and wound healing were used to examine the migratory and invasion abilities of these cells. As shown in Fig. 5b and Fig. 5d, AJAP1/β-catenin KD group suppressed the migratory and invasion abilities in comparison with the AJAP1 KD and control groups. Colony formation and MTT assays were performed on the three groups and the results showed that the proliferation ability of the AJAP1/β-catenin KD group was also lower than AJAP1 KD and control groups (Fig. 5c and Fig. 5e). The cell cycle results demonstrated that the AJAP1/β-catenin KD group had a higher number of G0/G1 cells but a lower number of S/G2/M cells than the other two groups (Fig. 5f). This phenomenon indicated that inhibited β-catenin expression reversed the effect caused by the depletion of AJAP1.To better analyze the significance of β-catenin in the tumorigenicity of AJAP1-mediated tumor progression, stable T47D cells were successfully established in the AJAP1 KD, AJAP1/β-catenin KD, and control groups. The AJAP1/β-catenin KD group had the lowest tumor capacity and tumor growth rate among the three groups (Fig. 5g).
Fig. 5.
AJAP1 regulated tumor progression by regulating β-catenin in breast cancer a Validation of protein levels of three groups after transfected with related shRNA by Western Blot b、d Results of the transwell assay (b) and wound-healing assay(d) showed that AJAP1/β-catenin KD inhibited invasion and migration abilities of T47D cell, indicating that β-catenin can reverse the effect of AJAP1-depletion caused (Transwell assay: original magnification, × 200). c、e MTT(c) and Colony-forming assay(e) were conducted to determine the clone-initiating and proliferation ability among AJAP1 KD, AJAP1/β-catenin KD and control in T47D cells. f Flow cytometry results of AJAP1 KD, AJAP1/β-catenin KD and control in T47D cells. g Representative images of tumor isolated from three groups (left panel) as demonstrated and tumor growth (middle panel), tumor weight representative graph(right) h H&E staining of tumors isolated from three experimental groups (magnification, × 400), IHC staining of Ki67, C-myc and CyclinD1 in slides removed from the three groups (magnification, × 400) and typical images of micrometastasis of the lung and liver (magnification, × 100). i Corresponding analysis of the positive rates of Ki67, C-myc and CyclinD1 expression in three groups(top), the number of liver and lung micrometastasis in three related groups. Data were shown as mean ± SD. Each experiment was conducted in triplicate. **p < 0.01, ***p < 0.001
The expression levels of ki67, C-myc, CyclinD1, lung and liver metastasis situation were evaluated in the mouse xenografts (Fig. 5h and i). It is clear that AJAP1/β-catenin KD group shows the lower expression of Ki67, less metastasis in lung and liver compared with the other two groups. This group also shows the lowest C-myc and CyclinD1 expressions (Fig. 5h and i). Collectively, AJAP1 inhibited breast cancer growth by regulating β-catenin transcriptional activity through suppressing C-myc and CyclinD1 expression both in vivo and in vitro.
β-Catenin nuclear localization negatively mediated by EGF/EGFR attenuate AJAP1 expression
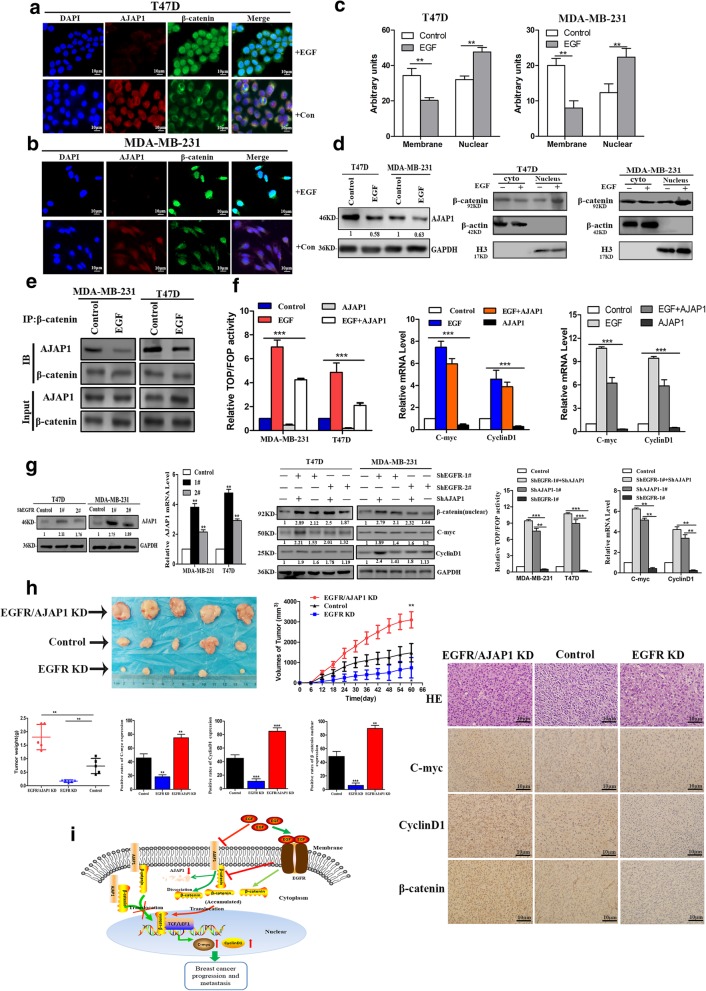
The above experiments verified β-catenin as the downstream target of AJAP1 in controlling breast cancer tumorigenesis and metastasis. However, it is necessary to further investigate which molecule controls the AJAP1 expression or mediate the conjunction between AJAP1 and β-catenin. Using the String online prediction software and the reported literature (Additional file 4: Figure S1), it was presumed that EGFR or EGF could participate in the AJAP1-β-catenin axis modulation process. To verify this assumption, EGF was added to T47D and MDA-MB-231 cells to observe the locational changes of β-catenin and AJAP1 through immunofluorescence assay. Consistently, Fig. 6a-c showed that EGF led the β-catenin expression transfer from the membrane/cytoplasm to the nucleus. Besides, the expression of AJAP1 was reduced but didn’t show locational changes. Western blot validated the same results in the MDA-MB-231 and T47D cell lines (Fig. 6d). In the subsequent co-IP assay, Fig. 6e presented that interaction between AJAP1 and β-catenin was obviously deceased by EGF. However, the total β-catenin of the cells did not change significantly. These experiments confirmed that β-catenin transferred from the cytoplasm to the nucleus. Subsequently, transcriptional activity of β-catenin and its downstream genes, such as C-myc and CyclinD1, were explored via luciferase assay and qRT-PCR. Figure 6f shows that the overexpression of AJAP1 with EGF reduced the relative TOP/FOP activity and the C-myc 、CyclinD1 mRNA levels in comparison with the group treated EGF alone. Taken together, EGF promoted the transfer of β-catenin from the membrane/cytoplasm to the nucleus and activated its transcriptional activity by inhibiting the expression of AJAP1.
Fig. 6.
EGF and EGFR regulated the AJAP1-mediated β-catenin nuclear translocation. a-b The changes of AJAP1 and β-catenin were observed through immunofluorescence assay of both cells with EGF stimulation (100 ng/ml). (magnification, × 400) c Statistical analysis of the membrane and nuclear expressions of β-catenin with EGF treatment by Image J. d protein changes of AJAP1 and β-catenin expression in T47D and MDA-MB-231 cells under EGF stimulation e Immunoprecipitation assays were conducted to evaluate the interaction between AJAP1 and β-catenin under EGF treatment. f Relative luciferase activities of TOP/FOP dual-luciferase reporter performed to observe the effects of EGF (100 ng/ml) on both cell lines (left) qRT-PCR was performed to assess the effects of EGF stimulation on the β-catenin downstream genes C-myc and CyclinD1 in the experimental groups. g Western blot and qRT-PCR were conducted to examine the AJAP1 expression in EGFR-depleted without EGF stimulating cells (first and second panel). The results showed that EGFR-depletion in AJAP1 knocked down cells promote the β-catenin transfer to the nucleus and its transcription activity (third and fourth panels) and increased the downstream gene levels (third and last panels). h the results of tumor growth, H&E and immunohistochemistry staining of C-myc,CyclinD1, and β-catenin nuclear expression in vivo assays (magnification, × 400). Data were expressed as mean ± SD. **p < 0.01, ***p < 0.001 i A mechanism model showed that the β-Catenin nuclear localization positively fed back on the EGF/EGFR attenuated AJAP1 expression in breast cancer to promote breast cancer progression and metastasis
EGF is considered as a ligand part of EGFR, and previous studies have demonstrated that its activation could stabilize and enhance the β-catenin nuclear accumulation in cancer [32–34]. Thus, investigations were carried out on whether EGFR had the same function as EGF and whether it participated in the process of AJAP1-mediated β-catenin expression. Cells without EGF stimulation were used to observe above changes. As shown in the first and second panels of Fig. 6g, EGFR depletion obviously inhibited the expression of AJAP1, reduced the expression of β-catenin and its downstream genes (C-myc and CyclinD1). However, knockdown AJAP1 and EGFR group also enhanced these effects and increased the relative β-catenin TOP/FOP activity.
Overall, these results showed that EGFR and EGF could inhibit the expression of AJAP1 and promot the β-catenin nuclear localization in the breast cancer cell lines.
AJAP1 knockdown inhibited tumorigenicity of EGFR-depletion groups in vivo
Numerous reports showed that overexpression of EGFR decreased OS and DFS in women with early breast cancer [35, 36]. Therefore, it also seemed to be a major driving factor of malignancy in breast cancer [37–39]. Experiments were carried out to study whether AJAP1 had the same effect on EGFR in vivo. As shown in Fig. 6h, AJAP1/EGFR KD group improved the tumor growth and weight, β-catenin nuclear expression and the expressions of C-myc and CyclinD1 compared with the other two groups isolated from the breast cancer cell-inoculated nude mice.
Figure 6i showed that AJAP1 could interact with β-catenin and the loss of AJAP1 could promote breast cancer progression and metastasis by stimulating β-catenin nuclear translocation and TCF/LEF1 transcriptional activity, increasing C-myc and CyclinD1 expression. Furthermore, EGF/EGFR could inhibit the expression of AJAP1 and negatively control the location and the activity of β-catenin. EGF reduced the AJAP1–β-catenin complex joint efficiency by reducing AJAP1 expression and dissociating β-catenin from the junctions. Further, dissociative β-catenin could accumulate in the cytoplasm and then move to the nucleus, which aggravates the breast cancer malignancy. Collectively, EGF/EGFR/AJAP1 axis controled breast cancer tumor progression and metastasis by mediating the β-catenin nuclear transactivation.
Discussion
Adherens junction (AJs) are multi-protein complexes which play vital roles in many behaviors of the cell like adhesion, transfer signals for inhibition of cell growth, resistance to apoptosis and regulation of cell shape and polarity [40–42]. Besides, AJs core structural component is a complex of cadherin and catenin proteins [40]. β-catenin can bind to the cadherins in AJs. The novel molecule of AJs, AJAP1, was found to be a tumor suppressor in glioma, hepatocellular carcinoma, esophagus carcinoma, oligodendrogliomas, and cervical cancer [12–14, 17, 43–45]. This study demonstrated that AJAP1 was a putative suppressor in breast cancer and it can interact with β-catenin in the cytoplasm and membrane. AJAP1 knocked down contributed to breast cancer malignancy through promoting accumulation of aberrant β-catenin nuclear expression, which indicated that increased expression of AJAP1 in higher grades of breast cancer could be beneficial to abrogate the β-catenin driven malignancy. Besides, the results of AJAP1 and β-catenin expression in 283 cases of breast cancer patients and overall survival further confirmed this theory.
As for β-catenin stabilization, recent researches [32, 34, 46, 47] have revealed that the accumulation of β-catenin nuclear expression activated many oncogenes about cellular proliferation. It is noteworthy that β-catenin is an important molecule which is involved in many metastasis signals. Thus, inhibiting nuclear accumulation of β-catenin might be an effective strategy for prohibiting malignancy of tumor cells. Here, a new mechanism for β-catenin nuclear location was identified. AJAP1 depletion promoted β-catenin nuclear translocation, affected the transcription activity of β-catenin and its downstream genes like C-myc and CyclinD1 expression. Likewise, AJAP1 regulated breast cancer tumorigenesis via mediating the β-catenin activity, indicating an effective way to prevent the tumor progression.
Increasing evidence demonstrated that EGF affected the β-catenin transactivation in breast cancer [33, 46, 48]. As expected, it was found that EGF-stimulated cells accelerated β-catenin nuclear transaction, decreased AJAP1 expression and activated the C-myc and CycliD1 expression. Previous studies revealed that EGF activated EGFR and led to the activation of downstream signaling pathways, such as PI3K/AKT and MAPK/ERK, which are critical in metastasis and tumor progression [49–51]. And now EGFR mutant also occurred in breast cancer. A great deal of studies show that EGFR-target medicine made a huge breakthrough in treating cancer especially lung cancer. Above achievements inspired us to use these target medicine to treat breast cancer with high EGFR mutant rates. Besides, our results showed that EGFR had similar function on β-catenin as previously illustrated [32, 34]. Additionally, Chao Yang et al. [13] revealed that EGFR/EGFRvIII inhibited AJAP1 in the cytoskeleton remodel of glioma cell. However, our study explored and validated that EGFR also can inhibit AJAP1 expression and β-catenin activity in breast cancer cells. Knockdown of EGFR and AJAP1 promoted tumor growth and β-catenin nuclear expression. Collectively, EGFR acted as an important role activating β-catenin nuclear expression. These results implied that the combination of EGFR targets and AJAP1-enhanced medicine might improve the prognosis of breast cancer patients. However, further researches on different β-catenin phosphorylation spots and mutants mediated by AJAP1 need to be further conducted. Likewise, these discoveries inspire more studies to explore the role of AJAP1 in EGFR-target medicine like Gefitinib in future studies.
To the best of our knowledge, this is the first study to reveal that the aberrant nuclear localization of β-catenin in breast cancer tissues, which is associated with the expression level of AJAP1. This work clarified one new feed loop for EGF-EGFR-AJAP1-controlled nuclear location and transcription activity of β-catenin. This change affected breast cancer progression and metastasis. Given the inhibition effect of AJAP1 on breast cancer both in vivo and in vitro, new medicine targets need to be developed. Determining whether this inducer for AJAP1 has the same effect as other adherens junction-associated proteins is also significant. If so, then many new anticancer occurrences may bring new hope to breast cancer patients.
Conclusion
We reported a novel EGF/EGFR axis that negatively fed back on the AJAP1-mediated β-catenin expression pathway and it accelerated breast cancer progression. This research provided novel findings demonstrating that the combination of the inhibitor of EGFR and the AJAP1 inducer may be beneficial to breast cancer prognosis.
Additional files
Table S1. Clinicopathological parameters of breast cancer patients. (DOC 54 kb)
Table S2. Primers of real-time PCR and oligos for siRNAs or shRNAs in this study. (XLSX 9 kb)
Table S3. Cox proportional hazard regression model analysis. (DOC 36 kb)
Figure S1. Predicted molecules that may be associated with AJAP1 and β-catenin using STRING software. (TIF 3124 kb)
Acknowledgements
This study was funded by National Natural Science Foundation of China (81172532,81470119,81602340 of Shuling Wang) and the Tianjin Municipal Science and the Technology Commission Research Fund (15JCYBJC27800).
Abbreviations
- AJAP1
Adherens junctions associated protein 1
- AJs
Adherent junctions
- EGF
Epidermal Growth Factor
- EGFR
Epidermal Growth Factor Receptor
- LN
Lymph node
- qRT-PCR
quantitative reverse-transcription polymerase chain reaction
Authors’ contributions
YN and CX conceived and designed the experiments. CX, LC performed most of the experiments. CX, LC, SLW performed the statistical analysis. GMX, FL and DNX cultured the cells and conducted the stable cell lines. GMX, JL, QXM and JJ conducted some animal experiments. CX and YN wrote the manuscript. YN, SHL evaluated the scores of breast cancer tissues. Eventually, all authors read and approved the final manuscript.
Funding
This study was funded by National Natural Science Foundation of China (81172532,81470119,81602340 of Shuling Wang) and the Tianjin Municipal Science and the Technology Commission Research Fund (15JCYBJC27800).
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was approved by the Tianjin Medical University Cancer Institute and Hospital, China, and has been performed in accordance with the ethical standards laid down in the 1964 Helsinki Declaration and its later amendments, For the in vivo tumor growth study,20 nude mice were used and comply with the ARRIVE guidelines and were carried out in accordance with the U.K. Animals (Scientific Procedures) Act, 1986 and associated guidelines, EU Directive 2010/63/EU for animal experiments.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Cong Xu, Email: Congxu2016@163.com.
Fang Liu, Email: 825243165@qq.com.
Guomin Xiang, Email: 928020622@qq.com.
Lu Cao, Email: 897242711@qq.com.
Shuling Wang, Email: wangshuling006@163.com.
Jing Liu, Email: 18202508979@163.com.
Qingxiang Meng, Email: M18522258305@126.com.
Danni Xu, Email: janedan21@126.com.
Shuhua Lv, Email: Shuhualv2005@126.com.
Jiao Jiao, Email: 1181414165@qq.com.
Yun Niu, Email: yunniu0823@126.com.
References
- 1.Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389:1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Vasioukhin V. Adherens junctions and cancer. Subcell Biochem. 2012;60:379–414. doi: 10.1007/978-94-007-4186-7_16. [DOI] [PubMed] [Google Scholar]
- 5.Tepass U. Adherens junctions: new insight into assembly, modulation and function. Bioessays. 2002;24:690–695. doi: 10.1002/bies.10129. [DOI] [PubMed] [Google Scholar]
- 6.Brooke MA, Nitoiu D, Kelsell DP. Cell-cell connectivity: desmosomes and disease. J Pathol. 2012;226:158–171. doi: 10.1002/path.3027. [DOI] [PubMed] [Google Scholar]
- 7.Wang YC, Khan Z, Kaschube M, Wieschaus EF. Differential positioning of adherens junctions is associated with initiation of epithelial folding. Nature. 2012;484:390–393. doi: 10.1038/nature10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baum B, Georgiou M. Dynamics of adherens junctions in epithelial establishment, maintenance, and remodeling. J Cell Biol. 2011;192:907–917. doi: 10.1083/jcb.201009141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mège René Marc, Ishiyama Noboru. Integration of Cadherin Adhesion and Cytoskeleton at Adherens Junctions. Cold Spring Harbor Perspectives in Biology. 2017;9(5):a028738. doi: 10.1101/cshperspect.a028738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McEwen AE, Escobar DE, Gottardi CJ. Signaling from the adherens junction. Subcell Biochem. 2012;60:171–196. doi: 10.1007/978-94-007-4186-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bharti S, Handrow-Metzmacher H, Zickenheiner S, Zeitvogel A, Baumann R, Starzinski-Powitz A. Novel membrane protein shrew-1 targets to cadherin-mediated junctions in polarized epithelial cells. Mol Biol Cell. 2004;15:397–406. doi: 10.1091/mbc.e03-05-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di C, Mladkova N, Lin J, et al. AJAP1 expression modulates glioma cell motility and correlates with tumor growth and survival. Int J Oncol. 2018;52(1):47–54. doi: 10.3892/ijo.2017.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang C, Li YS, Wang QX, Huang K, Wei JW, Wang YF, Zhou JH, Yi KK, Zhang KL, Zhou BC, et al. EGFR/EGFRvIII remodels the cytoskeleton via epigenetic silencing of AJAP1 in glioma cells. Cancer Lett. 2017;403:119–127. doi: 10.1016/j.canlet.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Han J, Xie C, Pei T, Wang J, Lan Y, Huang K, Cui Y, Wang F, Zhang J, Pan S, et al. Deregulated AJAP1/beta-catenin/ZEB1 signaling promotes hepatocellular carcinoma carcinogenesis and metastasis. Cell Death Dis. 2017;8:e2736. doi: 10.1038/cddis.2017.126. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Qu W, Wen X, Su K, Gou W. MiR-552 promotes the proliferation, migration and EMT of hepatocellular carcinoma cells by inhibiting AJAP1 expression. J Cell Mol Med. 2019;23:1541–1552. doi: 10.1111/jcmm.14062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ezaka K, Kanda M, Sugimoto H, Shimizu D, Oya H, Nomoto S, Sueoka S, Tanaka Y, Takami H, Hashimoto R, et al. Reduced expression of Adherens junctions associated protein 1 predicts recurrence of hepatocellular carcinoma after curative hepatectomy. Ann Surg Oncol. 2015;22(Suppl 3):S1499–S1507. doi: 10.1245/s10434-015-4695-9. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka H, Kanda M, Koike M, Iwata N, Shimizu D, Ezaka K, Sueoka S, Tanaka Y, Takami H, Hashimoto R, et al. Adherens junctions associated protein 1 serves as a predictor of recurrence of squamous cell carcinoma of the esophagus. Int J Oncol. 2015;47:1811–1818. doi: 10.3892/ijo.2015.3167. [DOI] [PubMed] [Google Scholar]
- 18.McDonald JM, Dunlap S, Cogdell D, Dunmire V, Wei Q, Starzinski-Powitz A, Sawaya R, Bruner J, Fuller GN, Aldape K, Zhang W. The SHREW1 gene, frequently deleted in oligodendrogliomas, functions to inhibit cell adhesion and migration. Cancer Biol Ther. 2006;5:300–304. doi: 10.4161/cbt.5.3.2391. [DOI] [PubMed] [Google Scholar]
- 19.Martin GS. Cell signaling and cancer. Cancer Cell. 2003;4:167–174. doi: 10.1016/S1535-6108(03)00216-2. [DOI] [PubMed] [Google Scholar]
- 20.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461–1473. doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howe LR, Brown AM. Wnt signaling and breast cancer. Cancer Biol Ther. 2004;3:36–41. doi: 10.4161/cbt.3.1.561. [DOI] [PubMed] [Google Scholar]
- 22.Geyer FC, Lacroix-Triki M, Savage K, Arnedos M, Lambros MB, MacKay A, Natrajan R, Reis-Filho JS. Beta-catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Mod Pathol. 2011;24:209–231. doi: 10.1038/modpathol.2010.205. [DOI] [PubMed] [Google Scholar]
- 23.Khramtsov AI, Khramtsova GF, Tretiakova M, Huo D, Olopade OI, Goss KH. Wnt/beta-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am J Pathol. 2010;176:2911–2920. doi: 10.2353/ajpath.2010.091125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Zhang H, Hou J, et al. Clinical implications of β-catenin protein expression in breast cancer. Int J Clin Exp Pathol. 2015;8:14989–14994. [PMC free article] [PubMed] [Google Scholar]
- 25.Cuello-Carrion FD, Shortrede JE, Alvarez-Olmedo D, Cayado-Gutierrez N, Castro GN, Zoppino FC, Guerrero M, Martinis E, Wuilloud R, Gomez NN, et al. HER2 and beta-catenin protein location: importance in the prognosis of breast cancer patients and their correlation when breast cancer cells suffer stressful situations. Clin Exp Metastasis. 2015;32:151–168. doi: 10.1007/s10585-015-9694-5. [DOI] [PubMed] [Google Scholar]
- 26.Farge E. Mechanical induction of twist in the Drosophila foregut/stomodeal primordium. Curr Biol. 2003;13:1365–1377. doi: 10.1016/S0960-9822(03)00576-1. [DOI] [PubMed] [Google Scholar]
- 27.Cao L, Xu C, Xiang G, Liu F, Liu X, Li C, Liu J, Meng Q, Jiao J, Niu Y. AR-PDEF pathway promotes tumour proliferation and upregulates MYC-mediated gene transcription by promoting MAD1 degradation in ER-negative breast cancer. Mol Cancer. 2018;17:136. doi: 10.1186/s12943-018-0883-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun L, Chen F, Shi W, Qi L, Zhao Z, Zhang J. Prognostic impact of TAZ and beta-catenin expression in adenocarcinoma of the esophagogastric junction. Diagn Pathol. 2014;9:125. doi: 10.1186/1746-1596-9-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woo DK, Kim HS, Lee HS, Kang YH, Yang HK, Kim WH. Altered expression and mutation of beta-catenin gene in gastric carcinomas and cell lines. Int J Cancer. 2001;95:108–113. doi: 10.1002/1097-0215(20010320)95:2<108::AID-IJC1019>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 30.Liu X, Feng C, Liu J, Cao L, Xiang G, Liu F, Wang S, Jiao J, Niu Y. Androgen receptor and heat shock protein 27 co-regulate the malignant potential of molecular apocrine breast cancer. J Exp Clin Cancer Res. 2018;37:90. doi: 10.1186/s13046-018-0762-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naito S, von Eschenbach AC, Giavazzi R, Fidler IJ. Growth and metastasis of tumor cells isolated from a human renal cell carcinoma implanted into different organs of nude mice. Cancer Res. 1986;46:4109–4115. [PubMed] [Google Scholar]
- 32.Li W, Hou JZ, Niu J, Xi ZQ, Ma C, Sun H, Wang CJ, Fang D, Li Q, Xie SQ. Akt1 inhibition promotes breast cancer metastasis through EGFR-mediated beta-catenin nuclear accumulation. Cell Commun Signal. 2018;16:82. doi: 10.1186/s12964-018-0295-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, Gao X, Aldape K, Lu Z. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee CH, Hung HW, Hung PH, Shieh YS. Epidermal growth factor receptor regulates beta-catenin location, stability, and transcriptional activity in oral cancer. Mol Cancer. 2010;9:64. doi: 10.1186/1476-4598-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez-Conchas GA, Rodriguez-Romo L, Hernandez-Barajas D, Gonzalez-Guerrero JF, Rodriguez-Fernandez IA, Verdines-Perez A, Templeton AJ, Ocana A, Seruga B, Tannock IF, et al. Epidermal growth factor receptor overexpression and outcomes in early breast cancer: a systematic review and a meta-analysis. Cancer Treat Rev. 2018;62:1–8. doi: 10.1016/j.ctrv.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 36.Saxena R, Dwivedi A. ErbB family receptor inhibitors as therapeutic agents in breast cancer: current status and future clinical perspective. Med Res Rev. 2012;32:166–215. doi: 10.1002/med.20209. [DOI] [PubMed] [Google Scholar]
- 37.O'Donovan N, Crown J. EGFR and HER-2 antagonists in breast cancer. Anticancer Res. 2007;27:1285–1294. [PubMed] [Google Scholar]
- 38.Teng YH, Tan WJ, Thike AA, Cheok PY, Tse GM, Wong NS, Yip GW, Bay BH, Tan PH. Mutations in the epidermal growth factor receptor (EGFR) gene in triple negative breast cancer: possible implications for targeted therapy. Breast Cancer Res. 2011;13:R35. doi: 10.1186/bcr2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams CB, Soloff AC, Ethier SP, Yeh ES. Perspectives on epidermal growth factor receptor regulation in triple-negative breast Cancer: Ligand-Mediated Mechanisms of Receptor Regulation and Potential for Clinical Targeting. Adv Cancer Res. 2015;127:253–281. doi: 10.1016/bs.acr.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 40.Rudini N, Dejana E. Adherens junctions. Curr Biol. 2008;18:R1080–R1082. doi: 10.1016/j.cub.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 41.Meng W, Takeichi M. Adherens junction: molecular architecture and regulation. Cold Spring Harb Perspect Biol. 2009;1:a002899. doi: 10.1101/cshperspect.a002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brüser Lena, Bogdan Sven. Adherens Junctions on the Move—Membrane Trafficking of E-Cadherin. Cold Spring Harbor Perspectives in Biology. 2017;9(3):a029140. doi: 10.1101/cshperspect.a029140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han L, Zhang KL, Zhang JX, Zeng L, Di CH, Fee BE, Rivas M, Bao ZS, Jiang T, Bigner D, et al. AJAP1 is dysregulated at an early stage of gliomagenesis and suppresses invasion through cytoskeleton reorganization. CNS Neurosci Ther. 2014;20:429–437. doi: 10.1111/cns.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng L, Kang C, Di C, Fee BE, Rivas M, Lin J, Adamson DC. The adherens junction-associated protein 1 is a negative transcriptional regulator of MAGEA2, which potentiates temozolomide-induced apoptosis in GBM. Int J Oncol. 2014;44:1243–1251. doi: 10.3892/ijo.2014.2277. [DOI] [PubMed] [Google Scholar]
- 45.Chen Yu-Chih, Huang Rui-Lan, Huang Yung-Kai, Liao Yu-Ping, Su Po-Hsuan, Wang Hui-Chen, Chang Cheng-Chang, Lin Ya-Wen, Yu Mu-Hsien, Chu Tang-Yuan, Lai Hung-Cheng. Methylomics analysis identifies epigenetically silenced genes and implies an activation of β-catenin signaling in cervical cancer. International Journal of Cancer. 2013;135(1):117–127. doi: 10.1002/ijc.28658. [DOI] [PubMed] [Google Scholar]
- 46.Liu L, Yang Y, Liu S, Tao T, Cai J, Wu J, Guan H, Zhu X, He Z, Li J, et al. EGF-induced nuclear localization of SHCBP1 activates beta-catenin signaling and promotes cancer progression. Oncogene. 2019;38:747–764. doi: 10.1038/s41388-018-0473-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paul D, Islam S, Manne RK, Dinesh US, Malonia SK, Maity B, Boppana R, Rapole S, Shetty PK, Santra MK. F-box protein FBXO16 functions as a tumor suppressor by attenuating nuclear beta-catenin function. J Pathol. 2019. 10.1002/path.5252. [DOI] [PMC free article] [PubMed]
- 48.Ji H, Wang J, Nika H, Hawke D, Keezer S, Ge Q, Fang B, Fang X, Fang D, Litchfield DW, et al. EGF-induced ERK activation promotes CK2-mediated disassociation of α-catenin from β-catenin and transactivation of β-catenin. Mol Cell. 2009;36:547–559. doi: 10.1016/j.molcel.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gan Y, Shi C, Inge L, Hibner M, Balducci J, Huang Y. Differential roles of ERK and Akt pathways in regulation of EGFR-mediated signaling and motility in prostate cancer cells. Oncogene. 2010;29:4947–4958. doi: 10.1038/onc.2010.240. [DOI] [PubMed] [Google Scholar]
- 50.Gabitova L, Gorin A, Astsaturov I. Molecular pathways: sterols and receptor signaling in cancer. Clin Cancer Res. 2014;20:28–34. doi: 10.1158/1078-0432.CCR-13-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Claperon A, Mergey M, Nguyen Ho-Bouldoires TH, Vignjevic D, Wendum D, Chretien Y, Merabtene F, Frazao A, Paradis V, Housset C, et al. EGF/EGFR axis contributes to the progression of cholangiocarcinoma through the induction of an epithelial-mesenchymal transition. J Hepatol. 2014;61:325–332. doi: 10.1016/j.jhep.2014.03.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Clinicopathological parameters of breast cancer patients. (DOC 54 kb)
Table S2. Primers of real-time PCR and oligos for siRNAs or shRNAs in this study. (XLSX 9 kb)
Table S3. Cox proportional hazard regression model analysis. (DOC 36 kb)
Figure S1. Predicted molecules that may be associated with AJAP1 and β-catenin using STRING software. (TIF 3124 kb)
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.