Abstract
Cytokinins (CKs) are a group of mobile adenine derivatives that act as chemical signals regulating a variety of biological processes implicated in plant development and stress responses. Their synthesis, homeostasis, and signaling perception evoke complicated intracellular traffic, intercellular movement, and in short- and long-distance translocation. Over nearly two decades, subsets of membrane transporters have been recognized and implicated in the transport of CKs as well as the related adenylates. In this review, we aim to recapitulate the key progresses in exploration of the transporter proteins involved in cytokinin traffic and translocation, discuss their functional implications in the cytokinin-mediated paracrine and long-distance communication, and highlight some knowledge gaps and open issues toward comprehensively understanding the molecular mechanism of membrane transporters in controlling spatiotemporal distribution of cytokinin species.
Keywords: cytokinin, ATP-binding cassette transporter, purine permease, equilibrative nucleoside transporter, isopentenyl adenine, trans-zeatin
Introduction
Cytokinins (CKs) are the mobile adenine derivatives that carry N6-linked isopentenyl or aromatic side chains. They serve as hormonal signals functioning in a myriad of biological processes, such as cell division and differentiation, seed germination, apical dominance, leaf senescence, root growth, branching and nodulation, nutrient homeostasis, and stress responses (Mok and Mok, 2001; Sakakibara, 2006; Werner and Schmulling, 2009; Kieber and Schaller, 2018). The majority of naturally occurring CKs in planta are the N6-isopentenyl conjugated adenine derivatives, in addition to a small amount of N6-aromatic CK species. The isoprenoid CKs chemically differ in their side chain structures, including the hydroxylation at the side chain terminus, the stereo-isomeric position, and the hydrocarbon chain saturation status. Correspondingly, they are classified into four main categories: N6-(Δ2-isopentenyl) adenine (iP), trans-zeatin (tZ), cis-zeatin (cZ), and dihydrozeatin (DHZ) (Sakakibara, 2006). Structural variations of the side chain may confer substantial differences in chemical properties of CK species, especially for their lipophilicity/hydrophobicity. Predictably, the iP-type CKs that lack hydroxyl moiety at their N6-linked side chains would possess the highest hydrophobicity compared to the others.
The isoprenoid CKs are generated through N-prenylation of adenosine 5′-phosphates (AMP, ADP, or ATP) or tRNAs, which is catalyzed by adenosine 5′-phosphate isopentenyltransferase (IPT) (Kakimoto, 2001; Takei et al., 2001, 2004a; Miyawaki et al., 2006). The resulting product isopentenyl adenosine 5′-phosphates, i.e., iP nucleotides, are then converted to tZ derivatives by trans-hydroxylases, the cytochrome P450 enzymes CYP735A1 and CYP735A2 (Takei et al., 2004b). Both iP- and tZ- nucleotides can be activated by phosphoribohydrolase LONELY GUY (LOG), yielding free CK bases (Kurakawa et al., 2007; Ueda et al., 2007). Alternatively, the formation of free CK bases may potentially undergo a two-step sequential dephosphorylation and deribosylation process. However, only a nucleoside N-ribohydrolase that mediates deribosylation reaction has been recognized so far (Jung et al., 2009; Kopecna et al., 2013). The enzymes involved in the two-step conversion in planta remain to be further identified.
Cytokinins are synthesized in a number of different cell types in both roots and shoots, and cross-talk with other phytohormones, particularly auxins, to regulate plant growth and development (Sakakibara, 2006). As chemical signals, CKs mediate both local and long-distance communications, and are transported either in short distance among neighboring cells or as acropetal and basipetal messengers translocated in long distance between roots and shoots. In Arabidopsis, tZ- and iP-type of CK species are confirmed as the active forms in respect to their specific recognition by three sensor histidine kinases, AHK2, AHK3, and CRE1/AHK4 (Inoue et al., 2001; Kakimoto, 2003; Romanov et al., 2006; Lomin et al., 2015). The tZ-type CKs are mainly synthesized in roots and transported apoplastically to shoots, which promote the growth of the above-ground parts of the plant (Beveridge et al., 1997; Hirose et al., 2008). In contrast, the iP- and cZ type CKs are the major forms found in phloem and are translocated rootward to transmit messages from shoots to roots (Corbesier et al., 2003; Hirose et al., 2008). The shoot-borne iP-type CKs have been suggested to serve as a signal of nitrogen satiety, regulating root architecture, suppressing nitrogen uptake in the root, and/or modulating nodulation (Sasaki et al., 2014). In addition to nutritional signaling, the shoot-derived iP-type CKs also regulate root development by modulating polar auxin transport and vascular patterning in the root meristem (Bishopp et al., 2011). Interestingly, although the iP-type CKs are considered to be synthesized throughout the whole plant body of Arabidopsis (Miyawaki et al., 2004, 2006; Takei et al., 2004a), a couple of recent studies reveal that iP ribosides can be predominantly synthesized in the roots of young tobacco and Arabidopsis seedlings (Zd’arska et al., 2013; Gelova et al., 2018), which implicate that a shootward transport of iPR via phloem could also be possible, although its underlying mechanism and related physiological functions remain to be explored.
As mobile signals, CKs biosynthesis, metabolism, distribution and perception evoke considerable intra- and inter-cellular movement and translocation. However, compared with the knowledge on transport and distribution of other plant growth and development related phytohormones, such as auxin, our understanding on molecular mechanisms of CK transport is just emerging. During the last two decades, three types of membrane transporters have been recognized and implicated in the CK transmembrane transport and intercellular translocation (Figure 1 and Table 1). These proteins include the subsets of purine permeases (PUPs) and equilibrative nucleoside transporters (ENTs), which act as influx carriers and are implicated in the transport of CK nucleobases or nucleosides, respectively (Figure 1 and Table 1), and the ATP-binding cassette transporter G subfamily member, ABCG14 in Arabidopsis, that acts as an efflux pump involved in long-distance acropetal translocation of the root-born CKs (Figure 1 and Table 1). Over the years, several excellent review articles have summarized the progress in understanding CK transport and in characterizing the related transporters (Hirose et al., 2008; Kudo et al., 2010; Borghi et al., 2015; Lacombe and Achard, 2016; Duran-Medina et al., 2017; Kang et al., 2017; Ko and Helariutta, 2017; Park et al., 2017). In this article, we aim to recapitulate the advances in identifying and characterizing the membrane transporters involved in short- and long-distance translocation of CKs as well as the related metabolites, to discuss their implicated functions in CK biosynthesis, homeostasis, and signal perception, and to tentatively point out some gaps and open questions toward fully understanding molecular mechanism of CK transport.
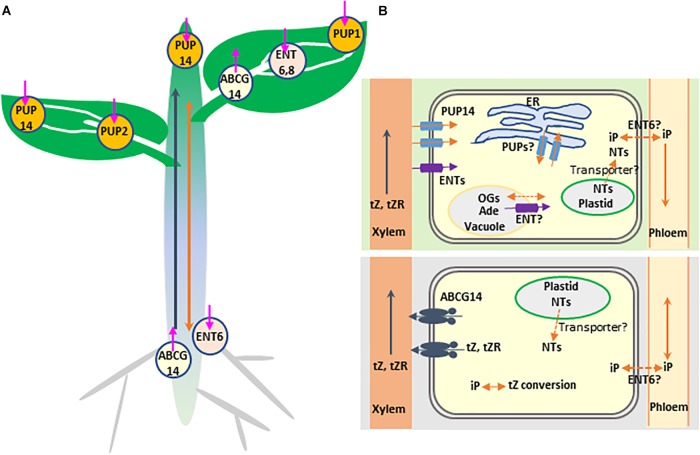
FIGURE 1.
Schematic representation of transporter-mediated long- and short-distance movement of cytokinins in Arabidopsis thaliana. (A) Involvement of transporters in cytokinin distribution. The determined or the speculated functional sites of cytokinin transporters, based on their expression patterns (see Table 1 for further details), are mapped on the different tissues. The three types of transporters are indicated with distinctly colored circles. The orientation of the transport is indicated by an arrow pointing inside (influx) or outside (efflux) the circle symbol. Long-distance transport of tZ-type cytokinins from roots to photosynthetic tissues through xylem and the rootward and the hypothetic shootward translocation of iP-type cytokinins through phloem are indicated with blue and orange arrows, respectively. (B) Involvement of transporters in the inter-and intra-cellular movements of cytokinins in root (bottom) and shoot (top) cells. The plastid-synthesized iP nucleotides (NTs) may be delivered to the cytoplasm, nucleus, or even endoplasmic reticulum (ER) via transporter activity. ABCG14 as an efflux pump transports tZ-type cytokinins from cytoplasm to apoplast (xylem vessel). PUP14 and ENTs act as importers to take up apoplastic free bases or nucleosides of cytokinins, respectively, into cytoplasm. The ER localized PUP members may involve in sequestering cytokinin species to or out ER lumen. ENT transporter perhaps delivers vacuolar adenosine (Ade) to cytosol; meanwhile, the unidentified transporters might be responsible for the vacuolar sequestration of reversible cytokinin glucosides (OGs). Finally, transport of iP-type cytokinin species in or out of the cells of their synthesis and the signaling target sites as well as phloem may also require transporter proteins; ENT6 was considered as one of the candidates.
Table 1.
The identified and/or putative cytokinin transporters∗.
| Transporter family | Name | Substrates (organism for detection) | Physiological functions in plant | Subcellular localization | Expression pattern | References |
|---|---|---|---|---|---|---|
| ENT | AtENT1 | Adenosine in yeast | Unknown | Plasma membrane or vacuolar membrane | Li and Wang, 2000; Sun et al., 2005; Jaquinod et al., 2007 | |
| AtENT8/SOI33 | iPR in seedlings | Reduced sensitivity of soi33-1 to iPR; Overexpression increases sensitivity to iPR | Unknown | Seedling, hypocotyls, flowers, midribs, abscission zones, siliques | Sun et al., 2005 | |
| AtENT3 | iPR in seedlings; adenosine in yeast; nucleoside in Xenopus oocytes | Reduced uptake efficiency of nucleoside-type CK (iPR), seedlings | Unknown | Unknown | Li et al., 2003; Sun et al., 2005 | |
| AtENT6 | iPR, tZR and adenosine in yeast | Unknown | Plasma membrane | Vascular tissues of root, leaf and flower, and cotyledons, stomata | Wormit et al., 2004; Hirose et al., 2008 | |
| OsENT2 | iPR, tZR, adenosine and uridine in yeast | Unknown | Unknown | Vascular tissues of leaf and root | Hirose et al., 2005 | |
| PUP | AtPUP1 | Hypoxanthine, zeatin, kinetin, caffeine in yeast (proton-gradient dependent symporter) | Unknown | Unknown | Leaf mesophyll cells, stems, flowers, siliques, and hydathodes, stigma | Gillissen et al., 2000; Bürkle et al., 2003 |
| AtPUP2 | Adenine, tZ, cZ, iPA, kinetin, BAP and tZR in yeast (proton-gradient dependent symporter) | Unknown | Unknown | Vascular tissues of leaf, stem, particularly in phloem, pollen | Bürkle et al., 2003 | |
| AtPUP14 | tZ, iPA, BA in protoplast and microsomes of plant. (ATP- dependent) | CK signaling, morphogenesis in embryos, roots and the shoot apical meristem | Plasma membrane | Various organ and tissues, including seedlings, embryos and mesophyll cells | Zürcher et al., 2016 | |
| OsPUP7 | Caffeine in yeast | Plant height, seed size and flowering time. Higher iP and iPR levels in mutant plant | Endoplasmic reticulum | Vascular bundle of roots, culms and leaves, hull vein and flower, stigma, style, stamens | Qi and Xiong, 2013; Kirschner et al., 2018; Xiao et al., 2018 | |
| OsPUP4/BG3 | Unknown | Seed size | Plasma membrane | Parenchyma cells near vascular tissue | Xiao et al., 2018 | |
| ABCG | AtABCG14 | tZ-type CKs | Reduced root-to-shoot translocation in mutant. Shoot and root growth, nitrogen signaling | Plasma membrane | Vascular tissue in root (mainly) and shoot, leaf midribs, veins, mature anthers | Ko et al., 2014; Zhang et al., 2014; Poitout et al., 2018 |
∗iPA or iP, isopentenyladenine; iPR, isopentenyladenosine; BAP or BA, benzylaminopurine or benzyladenine; tZ, trans-zeatin; tZR, trans-zeatin riboside.
Transport Proteins Potentially Involved in Intracellular Traffic of Cytokinin
Subcellular compartmentation is a key feature of eukaryotic cells to effectively organize their metabolic and signaling processes (Skalicky et al., 2018). At the site of synthesis, the formation, conversion and storage of CKs take place in different compartments of the cell. Correspondingly the key enzymes/proteins involved in the processes are located within different places (Skalicky et al., 2018). IPTs catalyze N-prenylation of adenine ribotides, which diverts both the plastidial and cytosolic sources of terpenoid building blocks, the five carbon units, isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) to CK synthesis (Sakakibara, 2006). Among several IPTs identified in Arabidopsis, AtIPT1, 3, and 5 were proved the major players for iP- and tZ-type CKs formation. Their fluorescently tagged proteins were found to primarily localize to the chloroplasts of mesophyll cells (Kasahara et al., 2004). Interestingly, besides its plastidial localization, AtIPT3 can be farnesylated, which directs it to the nucleus. The subcellular localization of farnesylated and non-farnesylated protein is closely correlated with either iP-type or tZ-type CK biosynthesis (Galichet et al., 2008). By contrast, AtIPT2 and 9 catalyze isopentenylation of tRNA to provide a source for cZ-type CKs (Miyawaki et al., 2006). The AtIPT2-GFP fusion appeared within cytoplasm, indicating that AtIPT2 utilizes isoprenoid precursors synthetized via the mevalonate pathway in cytosol for cZ-type CK synthesis. Additionally, AtIPT7-GFP was observed in mitochondria (Kasahara et al., 2004). Different subcellular localization of IPTs implicates multiple sites occurrence of CK species.
After formation of iP nucleotides, their subsequent hydroxylation is catalyzed by cytochrome P450 monooxygenases CYP735A1 and CYP735A2, which yields tZ-type CKs (Takei et al., 2004b). Cytochrome P450s are the integral endoplasmic reticulum (ER)-resident proteins. Therefore, the hydroxylation reaction of CKs must take place on the surface of ER. In addition, the resulting CK nucleotides need to be dephosphoribosylated to become active free-base forms (Kurakawa et al., 2007). The responsible enzyme LOGs have been demonstrated to predominantly present in the nucleus and cytosol (Kuroha et al., 2009). Therefore, the translocation of iP nucleotides from plastid to cytoplasm, to nucleus, or even to ER is conceivable (Figure 1). However, until now there is no specific study elucidating how the CK biosynthetic intermediates are intracellularly sequestrated, and whether the specific transporters are required. An Arabidopsis plastidial membrane-localized nucleotide uniport carrier, Brittle 1, has been found to transport adenylates such as AMP, ADP, and ATP into the cytosol (Kirchberger et al., 2008). The gene encoding Brittle 1 primarily expressed in the root tips and the maturating or germinating pollens. Down-regulation of Brittle 1 gene expression substantially retarded plant growth; but feeding adenosine complemented the growth defect of the mutant (Kirchberger et al., 2008). Giving the structural similarity of prenylated adenosine-5′-phosphate to the reported substrates of Brittle 1, it remains interesting to examine whether such a plastidial purine nucleotide exporter would have overlapped functions in delivering CK biosynthetic intermediates from plastid to cytoplasm.
The free CK bases within the cell can be reversibly glycosylated at their hydroxyl moieties of the N6-side chains (for tZ, cZ, and DHZ), or irreversibly glycosylated at N7- or N9- positions by cytosolic localized glycosyltransferases and sequestered into the vacuole for storage. The vacuolar stored CK glucosides can also be reconverted back to the active CK species via β-glucosidases (Brzobohaty et al., 1993) and released to cytosol. In addition, the free CK bases can also be reversibly inactivated into their nucleotide forms by enzymes in common with purine metabolism, such as adenine phosphoribosyltransferases, which appear to be cytosolic and act antagonistically to LOGs (Moffatt et al., 1991; Zhang et al., 2013). Such a complexity in subcellular compartmentation of CK de novo synthesis, bioconversion, storage and inactivation implicates the presence of a sophisticated intracellular CK transport network. Members of equilibrative nucleoside transporter (ENT) family have been demonstrated with broad substrate specificities, transporting both purine and pyrimidine nucleosides (Mohlmann et al., 2001; Wormit et al., 2004; Table 1). Eight members (AtENT1-8) are found in Arabidopsis and four (OsENT1-4) in rice (Li et al., 2003; Hirose et al., 2004, 2005). Among them AtENT1 was the first member with demonstrated adenosine transport activity by a complementation assay with Arabidopsis cDNAs in yeast cells deficient in synthesizing adenine (Mohlmann et al., 2001). Instead of the previously reported plasma membrane localization (Li and Wang, 2000), AtENT1 was re-verified to present on vacuolar membrane (Jaquinod et al., 2007; Bernard et al., 2011; Schulze et al., 2012). Overexpression of AtENT1 reduced vacuolar contents of adenosine and 2’,3’-adenosine monophosphate, the RNA degradation products (Bernard et al., 2011). Conversely, down-regulation of AtENT1 by RNAi resulted in a significant increase in vacuolar accumulation of both metabolites. Furthermore, upregulating AtENT1-mediated vacuolar export activity concurred with the increase of the cytosolic adenosine kinase activity (Bernard et al., 2011). These observations suggest that AtENT1 might participate in the export of vacuolar nucleosides and/or RNA breakdown products to cytosol, and the exported nucleosides may re-enter the salvage pathways of adenylates with actions of nucleoside kinases, hydrolases, and/or phosphoribosyltransferases (Bernard et al., 2011). Therefore, expectedly AtENT1 activity might connect to the cytosolic CK biosynthesis and CK homeostasis. Although it was reported that the null mutant line of AtENT1 exhibited no substantial effect on the CK response (Sun et al., 2005), it is worthy to re-examine the functions of this transporter in CK metabolism and homeostasis by using more sensitive CK signaling read-out reporters.
Transporters Responsible for Cytokinin Long Distance Translocation
After synthesis, CKs undergo intercellular movement and long-distance translocation from the sites of their synthesis to the target cells. Consistently, CKs are detected in the transduction systems phloem and xylem sap. In xylem sap, tZ, and tZR account for about 15 and 80% of CK species, respectively (Beveridge et al., 1997; Hirose et al., 2008; Kuroha et al., 2009; Osugi et al., 2017); whereas in phloem sap, the major forms are the iP- and cZ-type CKs (Corbesier et al., 2003; Hirose et al., 2008). The compartmentation of tZ- and iP-type of CKs in distinct vascular tissues suggest that plant has elaborated sophisticated selective transport mechanisms for distributing different types of CKs.
Translocation of the root-born tZ-type of CKs from roots to shoots requires an ABC transporter (Figure 1). A few years ago, we adopted a reverse genetics approach to systematically characterize the functions of ABCG subfamily proteins. We noticed that disruption of ABCG14 in Arabidopsis caused obvious morphological alterations, such as smaller inflorescences, smaller rosettes, slender stems with reduced number of vascular bundles, severely retarded primary root growth, and differential responses to the exogenously supplied phytohormones. Such developmental defects resembled those of the cytokinin biosynthetic mutants defective in CK signaling (Zhang et al., 2014). AtABCG14 expressed primarily in the pericycle and stele of roots. Profiling CKs in both roots and shoots of Arabidopsis seedling deficient in abcg14 revealed a clear overaccumulation of the root-derived tZ- and DHZ-type CKs in the roots and reduced content in the shoots, indicating that disruption of AtABCG14 substantially inhibits the translocation and distribution of the root-synthesized CKs to the shoots. Interestingly, in contrast to the decline of both tZ- and DHZ-type CKs in the shoots, the iP-type CKs levels were slightly increased in both the shoots and roots of abcg14, so was the cZ-type CKs (Zhang et al., 2014). These data point out that (1) AtABCG14 participates in the long-distance translocation of the root-derived CKs. (2) AtABCG14 possesses profound substrate specificity and is able to discriminate the subtle structural variation of the isopentenyl side chain of CK species. (3) An intrinsic CK homeostatic mechanism occurs in plant cells that compensates the perturbation of CK allocation. Furthermore, by employing radiolabeled tZ in an in planta feeding and in the detached leaf assay with ABCG14 overexpressing plants, we demonstrated that ABCG14, as a plasma membrane-localized protein, functioned as an efflux pump (Zhang et al., 2014). The data affirm that ABCG14 is a CK exporter responsible for loading the root-born tZ- and DHZ-type CKs into xylem vessels from xylary parenchyma cells. Coincidently, a couple of months later, Ko et al. (2014) reported a very similar study and finding, with additional reciprocal grafting experiments confirming the roles of AtABCG14 in the long-distance translocation of the root-synthesized CKs to the shoots (Ko et al., 2014). ABC transporter is an ATP-dependent primary transporter. Its activity consumes ATP yielding AMP, which directly links to the purine nucleotide metabolism. Therefore, it might have metabolic and regulatory advantages for plant to adopt this type of transporter to deliver CKs species in terms of the potential co-sharing and recycling of purine nucleotides, and more effective coordination and organization of energy flow, CKs synthesis and transport processes.
When AtABCG14::GUS was employed to examine gene expression, we noticed that in adult plant, AtABCG14 was not only expressed in the roots but also in young rosette leaves, especially in the leaf midribs, veins and the nearby leaf cells. In addition, AtABCG14 was also detected in mature anthers, and in siliques (Zhang et al., 2014; Table 1). The broad expression pattern of AtABCG14 hints its possibility of more comprehensive functions than the involvement in xylem loading of root CKs. Speculatively, AtABCG14 may also reside on the cell membrane of the lignifying yet alive vascular cells near midribs or veins of young aerial tissues, potentially as an efflux pump responsible for the redistribution of CK species that are accumulated in those alive xylary cells (Figure 1), although currently it remains unclear how the CK species from xylem sap enrich in those cells. Alternatively, when AtABCG14 expresses in the leaf vein neighboring cells, it may also be possible to switch its transport direction to function as an importer taking up the xylem sap CKs to the cells. Although it is quite a rare case and mechanistically unclear, some ABC transporters seem pretty versatile in their flux pump direction. For example, AtABCB4, the transporter involved in auxin distribution, exhibited a substrate concentration-dependent switch from auxin influx to efflux when it was heterologously expressed in tobacco BY-2 cells, yeast, or HeLa cells (Terasaka et al., 2005; Yang and Murphy, 2009; Kubes et al., 2012). It is unknown whether such a unique behavior occurs in planta. Clearly, more refined experimental evidences are required for corroborating the postulated dual roles of AtABCG14 in CK translocation and cellular distribution.
While ABCG14 mediates acropetal transport of the root-born tZs to the shoots, systemic transport of the shoot-derived CKs (primarily iP- and cZ-types) takes place in phloem and likely involves cell to cell translocation through symplastic connection (Figure 1). Plasmodesmata connects neighboring cells and forms a highway for the movement of endogenous CKs as well as other photosynthetic assimilates from the site of synthesis to phloem and from phloem to the target cells (Romanov et al., 2018). This symplastic process appears to have no requirement for the involvement of particular transporters. However, previous studies revealed that AtENT6 in Arabidopsis and OsENT2 in rice were likely involved in mediating the transport of adenosine and other nucleosides, including iP riboside (iPR) (Hirose et al., 2005, 2008; Table 1). Comparably, the affinity of ENTs to iPR was much higher than that to tZR (Hirose et al., 2005, 2008). Moreover, AtENT6 predominantly expressed in the vascular tissues of rosette leaves, flowers, cotyledons and roots (Hirose et al., 2008), while OsENT2 expressed primarily in rice leaf vascular bundles and phloem tissues (Hirose et al., 2005; Table 1). These data led to the speculation that ENTs may be responsible at least in part for the selective transport of iP nucleosides in vascular tissues (Sakakibara, 2006). Considering the hydrophobic characteristics of iP species, another possibility is that the concentration-dependent free diffusion might also contribute to iPs’ spatial distribution and movement. This notion seems to be supported with the observation that the iP-type CKs abundantly presented in xylem sap when both CYP735A1 and CYP735A2 genes were knocked-out (Ko et al., 2014). Presumably disruption of two CYP735A genes depleted conversion of iP to tZ, consequently the overaccumulation of iP species in the xylem parenchyma cells resulted in their passive release into apoplastic space (xylem vessel). Alternatively, it is also possible for the existence of a low affinity exporter for iP-type species in root xylary cells. When high concentration of iPs accumulate, the transporter turns on. Molecular mechanism for iP systemic translocation needs to be further clarified.
Transporters Involved in CK Signal Perception
The transported CKs, when reaching the target cells, will eventually meet their receptors either at the cell surface or inside the cells to trigger signaling cascade. The activity of CK transporters guides cellular and subcellular localization of CKs and affects ligand-receptor interactions. Fragmentary evidences indicate that both PUP and ENT family transporters participate in CK uptake, therefore, directly or indirectly affect CK cellular or subcellular distribution and signal perception. PUPs are a class of small, integral membrane proteins that exhibit influx activity to a broad range of nucleobases (Girke et al., 2014; Table 1). Similar to the discovery of ENTs, the recognition of PUPs also came from complementation assay by adopting a purine transport-deficient yeast mutant transformed with an Arabidopsis cDNA library. In the subsequent competition assays, besides adenine and other nucleobases, it was found that AtPUP1 also took up CKs kinetin and zeatin, meanwhile showed a lower activity to their ribosides (Gillissen et al., 2000). This discovery eventually led to the recognition of 23 PUP family members in Arabidopsis (Schwacke et al., 2003; Zürcher et al., 2016). So far AtPUP1, AtPUP2, and AtPUP14 have been demonstrated with direct CK uptake activity in yeast expression system, Arabidopsis mesophyll protoplasts, and/or microsomes derived from Nicotiana benthamiana transfected with PUP genes (Bürkle et al., 2003; Zürcher et al., 2016; Table 1). Interestingly, AtPUP1 and AtPUP2 behaved as the proton-gradient dependent symporter. Protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP) and H+-ATPase inhibitor significantly depleted their uptake activity to the radiolabeled tZ (Bürkle et al., 2003; Cedzich et al., 2008); whereas the activity of AtPUP14 is seemingly independent of proton-gradient potential but requests the presence of ATP (Zürcher et al., 2016). This phenomenon suggests that PUP members differ substantially in their biochemical properties. Moreover, PUP members in planta exhibit substantially different gene expression patterns, which infer their potential multifaceted physiological roles. For instance, AtPUP1 is mainly expressed in the epithem of hydathodes and at the stigma surface of siliques (Figure 1), implicating its potential role in the retrieval of nucleobase derivatives in its expressing tissues (Bürkle et al., 2003). AtPUP14 prevailed in all the examined tissues including seedlings, embryos and mesophyll protoplasts, where it displays the highest expression level among all other family members (Zürcher et al., 2016). In heart stage embryos, the expression of AtPUP14 was inversely correlated to the cytokinin signaling readout. The reciprocal correlation led to a conclusion that AtPUP14, as a principal plasma membrane-resident protein, is able to import the active CK nucleobases into the cells consequently depleting apoplastic CK pools and inhibiting perception of CK signals by plasma membrane-localized receptors, by which, AtPUP14 modulates spatiotemporal CK sink patterns and influences plant morphogenesis (Zürcher and Muller, 2016; Zürcher et al., 2016). This hypothesis, however, evoked substantial argument in respect to whether the CK signaling initiation site occurs at plasma membrane or ER, since so far the experimentally defined CK receptors in different plant species appeared as the ER-localization (Romanov et al., 2006; Lomin et al., 2011, 2018). In addition, there are also some concerns about the binding affinity of PUPs versus the CK receptors to the ligands. Even if CK sensors do localize on the plant membrane, unless they have substantially lower ligand binding affinity than PUPs, it is not easy to explain the complete inhibition of CK signaling by the activity of AtPUP14 (Duran-Medina et al., 2017; Kang et al., 2017; Romanov et al., 2018). With these concerns, Romanov et al. (2018) argued that AtPUP14 or other PUP family members, instead of as the PM-localized proteins, might function at the ER membrane, where they pump CKs from ER lumen to the cytosol, thereby modulating CK signaling. Recently a rice PUP member OsPUP7 has been proved to localize at the ER and be involved in CK transport, although its flux pump direction remains ambiguous (Xiao et al., 2018), which seemingly aligns with Romanov et al’s argument.
In favor of CK signal perception primarily at the ER, Romanov et al proposed a model of multiple sites cellular CK signaling initiation and perception (Romanov et al., 2018), which integrates the importer activities of both PUP and ENT family proteins and differentiates the fate of tZ and tZR. The root-derived tZ that is transported through xylem to the particular region or cell types of the shoot directly interacts with the hypothetic plasma membrane-localized CK sensors, thereby, triggering hormonal signaling at the surface of cells. The established apoplastic CK pool can also be either degraded by the extracellular cytokinin oxidases, or transformed into the corresponding riboside by extracellular purine nucleoside phosphorylases, or imported into the cytoplasm by PUP transporters. The imported CK bases then could be either degraded or transformed into the inactive tZRP, and re-enter into the intracellular CK pool. On the other hand, the xylem-delivered tZR is taken up into the cells of the shoot via nucleoside importer ENTs. The imported tZRs in the cytosol could further undergo metabolic conversion by tZR kinase into tZRP, and by the activity of LOG enzymes release the biologically active form, free base of tZ.
While this multiple-site CK signal perception model integrates well with the experimental evidences on the ER-localization of CK sensors, the proposed transporter components call for more solid experimental confirmation. Primarily, the transport efficiency and specificity of ENTs to tZR is a matter of concern. The ENTs identified from Arabidopsis and rice exhibit quite broad substrate specificity and are apparently not optimal for the transport of tZR, since the affinity of the transporters to tZR (Km = 630∼660 μM) is substantially lower than that to iPR (Km = 17∼32 μM) or adenosine (Km = 3∼91 μM) (Hirose et al., 2005, 2008). Although it was reported that mutation of AtENT8 has suppressed the phenotypes of a plant with overproduction of CKs, and a loss-of-function of AtENT3 presented reduced CK uptake (Sun et al., 2005), compelling biological evidence for ENTs’ involvement in the tZR uptake is still missing.
Future Perspectives
Short- and/or long-distance translocation of CKs are important components in CK biosynthesis, salvage, homeostasis, and signaling processes. Elucidating molecular mechanism of CK transport and identifying the related molecular factors of CK translocation have substantially deepened our understandings of the physiological roles that CKs play with in plant growth and development, and in plant-environmental interactions. Grafting experiments between abcg14 mutant and WT have revealed that root-to-shoot transport of CKs is crucially required for the regulation of shoot growth and development (Ko et al., 2014). A recent study through disturbing root CK synthesis and translocation discovered that both tZ and tZR act as systemic signals traveling from root to shoot through xylem, differentially influencing leaf size and/or vegetative meristem activity (Osugi et al., 2017). Moreover, the long-distance translocated CKs also function as second messengers to signal the level of nutrient availability, particularly the soil nitrate level, to the shoot apical meristem, thereby mediating the organogenesis activity of shoot apical meristem in respect to the availability of soil mineral nutrients (Landrein et al., 2018). Aided with split-root system to uncouple local and systemic signaling, analysis of mutants ipt3,5,7 and abcg14 deficient in the root CK synthesis and translocation revealed that accumulation of the root-born tZ in shoots is an integral component of systemic signaling network of the root-shoot-root, which mediates the molecular and physiological responses of plants to nitrate heterogeneity in their roots, and controls gene expression in roots and shoots (Poitout et al., 2018). These studies exemplify the importance of unveiling CKs transport in understanding the physiological functions of this group of mobile hormonal molecules.
On the other hand, although a few transporter proteins have been implicated or partially evidenced in CK signal transport and distribution, the biological relevance of many putative CK transporter members require more detail and systematic exploration. PUP as well as ENT proteins are encoded by a multigene family in planta. The potential functional redundancy and their substrate promiscuity pose significant challenges for pinpointing their biochemical and biological functions in CK synthesis, intracellular traffic, cellular distribution and signal perception. To unequivocally elucidate the functions of ENTs and PUPs, it requires the integrated biochemical and biophysical approaches to precisely determine their influx or efflux carrier properties, substrate preferences, and subcellular localizations. Meanwhile, establishing higher order of mutant lines via conventional genetic cross and/or by CRISPR-Cas9 gene editing, together with the employment of high sensitive CK signal reporter system such as TCSn::GFP (Zürcher et al., 2013; Kirschner et al., 2018) could facilitate the determination and discrimination of biological roles of both PUP and ENT members in CK metabolism and signaling.
Recent study revealed that tZ and tZR transported through xylem flow exert distinct physiological roles for leaf development and apical meristem activity. The ratio of tZ and tZR in xylem flow and their delivery rate are modulated in response to the environmental conditions, which render different physiological consequence of shoot growth (Osugi et al., 2017). Currently it remains unclear if transporter proteins are involved in determining the ratio and delivery rate of root-born CK species in xylem flow. In the future, besides further tackling down additional physiological roles of ABCG14 in aerial tissues pertaining to the CK distribution, it is also intriguing to explore whether ABCG14, the primary transporter for xylem loading of tZ and tZR in root, coordinates with particular PUPs and/or ENTs in shoot to fine-tune the status and flow rate of different CK species in xylem sap and their differential cellular distribution, in response to plant growth and development cues and environmental stimuli.
Author Contributions
C-JL wrote the manuscript. C-JL, YZ, and KZ prepared the data. All authors have read, edited, and agreed to the content.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was part of the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by the United States Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the United States Department of Energy. C-JL was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences under contract number DE-SC0012704 -specifically through the Physical Biosciences program of the Chemical Sciences, Geosciences and Biosciences Division. The ABCG14 related work was supported by the United States National Science Foundation grant MCB-1051675 to C-JL. KZ was supported by the National Natural Science Foundation of China (31470370).
References
- Bernard C., Traub M., Kunz H. H., Hach S., Trentmann O., Mohlmann T. (2011). Equilibrative nucleoside transporter 1 (ENT1) is critical for pollen germination and vegetative growth in Arabidopsis. J. Exp. Bot. 62 4627–4637. 10.1093/jxb/err183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge C. A., Murfet I. C., Kerhoas L., Sotta B., Miginiac E., Rameau C. (1997). The shoot controls zeatin riboside export from pea roots. evidence from the branching mutant rms4. Plant J. 11 339–345. 10.1046/j.1365-313x.1997.11020339.x [DOI] [Google Scholar]
- Bishopp A., Lehesranta S., Vaten A., Help H., El-Showk S., Scheres B., et al. (2011). Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Curr. Biol. 21 927–932. 10.1016/j.cub.2011.04.049 [DOI] [PubMed] [Google Scholar]
- Borghi L., Kang J., Ko D., Lee Y., Martinoia E. (2015). The role of ABCG-type ABC transporters in phytohormone transport. Biochem. Soc. Trans. 43 924–930. 10.1042/BST20150106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzobohaty B., Moore I., Kristoffersen P., Bako L., Campos N., Schell J., et al. (1993). Release of active cytokinin by a β-glucosidase localized to the maize root-meristem. Science 262 1051–1054. 10.1126/science.8235622 [DOI] [PubMed] [Google Scholar]
- Bürkle L., Cedzich A., Dopke C., Stransky H., Okumoto S., Gillissen B., et al. (2003). Transport of cytokinins mediated by purine transporters of the PUP family expressed in phloem, hydathodes, and pollen of Arabidopsis. Plant J. 34 13–26. 10.1046/j.1365-313x.2003.01700.x [DOI] [PubMed] [Google Scholar]
- Cedzich A., Stransky H., Schulz B., Frommer W. B. (2008). Characterization of cytokinin and adenine transport in Arabidopsis cell cultures. Plant Physiol. 148 1857–1867. 10.1104/pp.108.128454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L., Prinsen E., Jacqmard A., Lejeune P., Van Onckelen H., Perilleux C., et al. (2003). Cytokinin levels in leaves, leaf exudate and shoot apical meristem of Arabidopsis thaliana during floral transition. J. Exp. Bot. 54 2511–2517. 10.1093/jxb/erg276 [DOI] [PubMed] [Google Scholar]
- Duran-Medina Y., Diaz-Ramirez D., Marsch-Martinez N. (2017). Cytokinins on the move. Front. Plant Sci. 8:146. 10.3389/fpls.2017.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galichet A., Hoyerova K., Kaminek M., Gruissem W. (2008). Farnesylation directs AtIPT3 subcellular localization and modulates cytokinin biosynthesis in Arabidopsis. Plant Physiol. 146 1155–1164. 10.1104/pp.107.107425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelova Z., ten Hoopen P., Novak O., Motyka V., Pernisova M., Dabravolski S., et al. (2018). Antibody-mediated modulation of cytokinins in tobacco: organ-specific changes in cytokinin homeostasis. J. Exp. Bot. 69 441–454. 10.1093/jxb/erx426 [DOI] [PubMed] [Google Scholar]
- Gillissen B., Burkle L., Andre B., Kuhn C., Rentsch D., Brandl B., et al. (2000). A new family of high-affinity transporters for adenine, cytosine, and purine derivatives in Arabidopsis. Plant Cell 12 291–300. 10.1105/tpc.12.2.291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke C., Daumann M., Niopek-Witz S., Mohlmann T. (2014). Nucleobase and nucleoside transport and integration into plant metabolism. Front. Plant Sci. 5:443. 10.3389/fpls.2014.00443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N., Makita N., Yamaya T., Sakakibara H. (2004). Characterization of the nucleoside transporters in rice: a possible role as cytokinin transporters. Plant Cell Physiol. 45 S40–S40. [Google Scholar]
- Hirose N., Makita N., Yamaya T., Sakakibara H. (2005). Functional characterization and expression analysis of a gene, OsENT2, encoding an equilibrative nucleoside transporter in rice suggest a function in cytokinin transport. Plant Physiol. 138 196–206. 10.1104/pp.105.060137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N., Takei K., Kuroha T., Kamada-Nobusada T., Hayashi H., Sakakibara H. (2008). Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 59 75–83. 10.1093/jxb/erm157 [DOI] [PubMed] [Google Scholar]
- Inoue T., Higuchi M., Hashimoto Y., Seki M., Kobayashi M., Kato T., et al. (2001). Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409 1060–1063. 10.1038/35059117 [DOI] [PubMed] [Google Scholar]
- Jaquinod M., Villiers F., Kieffer-Jaquinod S., Hugouvieu V., Bruley C., Garin J., et al. (2007). A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol. Cell Proteomics 6 394–412. 10.1074/mcp.m600250-mcp200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung B., Florchinger M., Kunz H. H., Traub M., Wartenberg R., Jeblick W., et al. (2009). Uridine-ribohydrolase is a key regulator in the uridine degradation pathway of Arabidopsis. Plant Cell 21 876–891. 10.1105/tpc.108.062612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto T. (2001). Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate : ATP/ADP isopentenyltransferases. Plant Cell Physiol. 42 677–685. 10.1093/pcp/pce112 [DOI] [PubMed] [Google Scholar]
- Kakimoto T. (2003). Perception and signal transduction of cytokinins. Ann. Rev. Plant Biol. 54 605–627. [DOI] [PubMed] [Google Scholar]
- Kang J., Lee Y., Sakakibara H., Martinoia E. (2017). Cytokinin transporters: go and stop in signaling. Trends Plant Sci. 22 455–461. 10.1016/j.tplants.2017.03.003 [DOI] [PubMed] [Google Scholar]
- Kasahara H., Takei K., Ueda N., Hishiyama S., Yamaya T., Kamiya Y., et al. (2004). Distinct isoprenoid origins of cis- and trans-zeatin biosyntheses in Arabidopsis. J. Biol. Chem. 279 14049–14054. 10.1074/jbc.m314195200 [DOI] [PubMed] [Google Scholar]
- Kieber J. J., Schaller G. E. (2018). Cytokinin signaling in plant development. Development 145:dev149344. 10.1242/dev.149344 [DOI] [PubMed] [Google Scholar]
- Kirchberger S., Tjaden J., Neuhaus H. E. (2008). Characterization of the Arabidopsis Brittle1 transport protein and impact of reduced activity on plant metabolism. Plant J. 56 51–63. 10.1111/j.1365-313X.2008.03583.x [DOI] [PubMed] [Google Scholar]
- Kirschner G. K., Stahl Y., Imani J., von Korff M., Simon R. (2018). Fluorescent reporter lines for auxin and cytokinin signalling in barley (Hordeum vulgare). PLoS One 13:e0196086. 10.1371/journal.pone.0196086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko D., Helariutta Y. (2017). Shoot-root communication in flowering plants. Curr. Biol. 27 R973–R978. 10.1016/j.cub.2017.06.054 [DOI] [PubMed] [Google Scholar]
- Ko D., Kang J., Kiba T., Park J., Kojima M., Do J., et al. (2014). Arabidopsis ABCG14 is essential for the root-to-shoot translocation of cytokinin. Proc. Natl. Acad. Sci. U.S.A. 111 7150–7155. 10.1073/pnas.1321519111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecna M., Blaschke H., Kopecny D., Vigouroux A., Koncitikova R., Novak O., et al. (2013). Structure and function of nucleoside hydrolases from Physcomitrella patens and maize catalyzing the hydrolysis of purine, pyrimidine, and cytokinin ribosides. Plant Physiol. 163 1568–1583. 10.1104/pp.113.228775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubes M., Yang H. B., Richter G. L., Cheng Y., Mlodzinska E., Wang X., et al. (2012). The Arabidopsis concentration-dependent influx/efflux transporter ABCB4 regulates cellular auxin levels in the root epidermis. Plant J. 69 640–654. 10.1111/j.1365-313X.2011.04818.x [DOI] [PubMed] [Google Scholar]
- Kudo T., Kiba T., Sakakibara H. (2010). Metabolism and long-distance translocation of cytokinins. J. Integr. Plant Biol. 52 53–60. 10.1111/j.1744-7909.2010.00898.x [DOI] [PubMed] [Google Scholar]
- Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., et al. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445 652–655. 10.1038/nature05504 [DOI] [PubMed] [Google Scholar]
- Kuroha T., Tokunaga H., Kojima M., Ueda N., Ishida T., Nagawa S., et al. (2009). Functional analyses of lonely guy cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 21 3152–3169. 10.1105/tpc.109.068676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe B., Achard P. (2016). Long-distance transport of phytohormones through the plant vascular system. Curr. Opin. Plant Biol. 34 1–8. 10.1016/j.pbi.2016.06.007 [DOI] [PubMed] [Google Scholar]
- Landrein B., Formosa-Jordan P., Malivert A., Schuster C., Melnyk C. W., Yang W., et al. (2018). Nitrate modulates stem cell dynamics in Arabidopsis shoot meristems through cytokinins. Proc. Natl. Acad. Sci. U.S.A. 115 1382–1387. 10.1073/pnas.1718670115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Liu K., Baldwin S. A., Wang D. (2003). Equilibrative nucleoside transporters of Arabidopsis thaliana. cDNA cloning, expression pattern, and analysis of transport activities. J. Biol. Chem. 278 35732–35742. 10.1074/jbc.m304768200 [DOI] [PubMed] [Google Scholar]
- Li J., Wang D. W. (2000). Cloning and in vitro expression of the cDNA encoding a putative nucleoside transporter from Arabidopsis thaliana. Plant Sci. 157 23–32. 10.1016/s0168-9452(00)00261-2 [DOI] [PubMed] [Google Scholar]
- Lomin S. N., Krivosheev D. M., Steklov M. Y., Arkhipov D. V., Osolodkin D. I., Schmulling T., et al. (2015). Plant membrane assays with cytokinin receptors underpin the unique role of free cytokinin bases as biologically active ligands. J. Exp. Bot. 66 1851–1863. 10.1093/jxb/eru522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomin S. N., Myakushina Y. A., Arkhipov D. V., Leonova O. G., Popenko V. I., Schmulling T., et al. (2018). Studies of cytokinin receptor-phosphotransmitter interaction provide evidences for the initiation of cytokinin signalling in the endoplasmic reticulum. Funct. Plant Biol. 45 192–202. [DOI] [PubMed] [Google Scholar]
- Lomin S. N., Yonekura-Sakakibara K., Romanov G. A., Sakakibara H. (2011). Ligand-binding properties and subcellular localization of maize cytokinin receptors. J. Exp. Bot. 62 5149–5159. 10.1093/jxb/err220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K., Matsumoto-Kitano M., Kakimoto T. (2004). Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 37 128–138. 10.1046/j.1365-313x.2003.01945.x [DOI] [PubMed] [Google Scholar]
- Miyawaki K., Tarkowski P., Matsumoto-Kitano M., Kato T., Sato S., Tarkowska D., et al. (2006). Roles of Arabidopsis ATP/ADP isopentenyltransferases and trna isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 103 16598–16603. 10.1073/pnas.0603522103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt B., Pethe C., Laloue M. (1991). Metabolism of benzyladenine is impaired in a mutant of Arabidopsis thaliana lacking adenine phosphoribosyltransferase activity. Plant Physiol. 95 900–908. 10.1104/pp.95.3.900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohlmann T., Mezher Z., Schwerdtfeger G., Neuhaus H. E. (2001). Characterisation of a concentrative type of adenosine transporter from Arabidopsis thaliana (AtENT1). Febs Lett. 509 370–374. 10.1016/s0014-5793(01)03195-7 [DOI] [PubMed] [Google Scholar]
- Mok D. W. S., Mok M. C. (2001). Cytokinin metabolism and action. Annu. Rev. Plant Phys. 52 89–118. [DOI] [PubMed] [Google Scholar]
- Osugi A., Kojima M., Takebayashi Y., Ueda N., Kiba T., Sakakibara H. (2017). Systemic transport of trans-zeatin and its precursor have differing roles in Arabidopsis shoots. Nat. Plants 3:17112. 10.1038/nplants.2017.112 [DOI] [PubMed] [Google Scholar]
- Park J., Lee Y., Martinoia E., Geisler M. (2017). Plant hormone transporters: what we know and what we would like to know. BMC Biol. 15:93. 10.1186/s12915-017-0443-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poitout A., Crabos A., Petrik I., Novak O., Krouk G., Lacombe B., et al. (2018). Responses to systemic nitrogen signaling in Arabidopsis roots involve trans-zeatin in shoots. Plant Cell 30 1243–1257. 10.1105/tpc.18.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z., Xiong L. (2013). Characterization of a purine permease family gene OsPUP7 involved in growth and development control in rice. J. Integr. Plant Biol. 55 1119–1135. 10.1111/jipb.12101 [DOI] [PubMed] [Google Scholar]
- Romanov G. A., Lomin S. N., Schmulling T. (2006). Biochemical characteristics and ligand-binding properties of Arabidopsis cytokinin receptor AHK3 compared to CRE1/AHK4 as revealed by a direct binding assay. J. Exp. Bot. 57 4051–4058. 10.1093/jxb/erl179 [DOI] [PubMed] [Google Scholar]
- Romanov G. A., Lomin S. N., Schmulling T. (2018). Cytokinin signaling: from the ER or from the PM? That is the question! New Phytol. 218 41–53. 10.1111/nph.14991 [DOI] [PubMed] [Google Scholar]
- Sakakibara H. (2006). Cytokinins: activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57 431–449. 10.1146/annurev.arplant.57.032905.105231 [DOI] [PubMed] [Google Scholar]
- Sasaki T., Suzaki T., Soyano T., Kojima M., Sakakibara H., Kawaguchi M. (2014). Shoot-derived cytokinins systemically regulate root nodulation. Nat. Commun. 5:4983. 10.1038/ncomms5983 [DOI] [PubMed] [Google Scholar]
- Schulze W. X., Schneider T., Starck S., Martinoia E., Trentmann O. (2012). Cold acclimation induces changes in Arabidopsis tonoplast protein abundance and activity and alters phosphorylation of tonoplast monosaccharide transporters. Plant J. 69 529–541. 10.1111/j.1365-313X.2011.04812.x [DOI] [PubMed] [Google Scholar]
- Schwacke R., Schneider A., van der Graaff E., Fischer K., Catoni E., Desimone M., et al. (2003). Aramemnon, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131 16–26. 10.1104/pp.011577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalicky V., Kubes M., Napier R., Novak O. (2018). Auxins and cytokininsthe role of subcellular organization on homeostasis. Int. J. Mol. Sci. 19:3115. 10.3390/ijms19103115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. P., Hirose N., Wang X., Wen P., Xue L., Sakakibara H., et al. (2005). Arabidopsis SOI33/AtENT8 gene encodes a putative equilibrative nucleoside transporter that is involved in cytokinin transport in planta. J. Integr. Plant Biol. 47 588–603. 10.1111/j.1744-7909.2005.00104.x [DOI] [Google Scholar]
- Takei K., Sakakibara H., Sugiyama T. (2001). Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J. Biol. Chem. 276 26405–26410. 10.1074/jbc.m102130200 [DOI] [PubMed] [Google Scholar]
- Takei K., Yamaya T., Sakakibara H. (2004b). Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-zeatin. J. Biol. Chem. 279 41866–41872. 10.1074/jbc.m406337200 [DOI] [PubMed] [Google Scholar]
- Takei K., Ueda N., Aoki K., Kuromori T., Hirayama T., Shinozaki K., et al. (2004a). AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol. 45 1053–1062. 10.1093/pcp/pch119 [DOI] [PubMed] [Google Scholar]
- Terasaka K., Blakeslee J. J., Titapiwatanakun B., Peer W. A., Bandyopadhyay A., Makam S. N., et al. (2005). PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. Plant Cell 17 2922–2939. 10.1105/tpc.105.035816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda N., Kojima M., Kurakawa T., Maekawa M., Kobayashi K., Nagato Y., et al. (2007). LOG encodes a novel cytokinin activating enzyme that directly controls meristern activity in rice. Plant Cell Physiol. 48 S118–S118. [Google Scholar]
- Werner T., Schmulling T. (2009). Cytokinin action in plant development. Curr. Opin. Plant Biol. 12 527–538. 10.1016/j.pbi.2009.07.002 [DOI] [PubMed] [Google Scholar]
- Wormit A., Traub M., Florchinger M., Neuhaus H. E., Mohlmann T. (2004). Characterization of three novel members of the Arahidopsis thaliana equilibrative nucleoside transporter (ENT) family. Biochem. J. 383 19–26. 10.1042/bj20040389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Liu D., Zhang G., Gao S., Liu L., Xu F., et al. (2018). Big grain3, encoding a purine permease, regulates grain size via modulating cytokinin transport in rice. J. Integr. Plant Biol. 61 581–597. 10.1111/jipb.12727 [DOI] [PubMed] [Google Scholar]
- Yang H., Murphy A. S. (2009). Functional expression and characterization of Arabidopsis ABCB, AUX1 and PIN auxin transporters in Schizosaccharomyces pombe. Plant J. 59 179–191. 10.1111/j.1365-313X.2009.03856.x [DOI] [PubMed] [Google Scholar]
- Zd’arska M., Zatloukalova P., Benitez M., Sedo O., Potesil D., Novak O., et al. (2013). Proteome analysis in Arabidopsis reveals shoot- and root-specific targets of cytokinin action and differential regulation of hormonal homeostasis. Plant Physiol. 161 918–930. 10.1104/pp.112.202853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. W., Novak O. R., Wei Z. Y., Gou M. Y., Zhang X. B., Yu Y., et al. (2014). Arabidopsis ABCG14 protein controls the acropetal translocation of root-synthesized cytokinins. Nat. Commun. 5:3274. 10.1038/ncomms4274 [DOI] [PubMed] [Google Scholar]
- Zhang X. Y., Chen Y. T., Lin X., Hong X. Y., Zhu Y., Li W. Y., et al. (2013). Adenine phosphoribosyl transferase 1 is a key enzyme catalyzing cytokinin conversion from nucleobases to nucleotides in Arabidopsis. Mol. Plant 6 1661–1672. 10.1093/mp/sst071 [DOI] [PubMed] [Google Scholar]
- Zürcher E., Liu J. C., di Donato M., Geisler M., Muller B. (2016). Plant development regulated by cytokinin sinks. Science 353 1027–1030. 10.1126/science.aaf7254 [DOI] [PubMed] [Google Scholar]
- Zürcher E., Muller B. (2016). Cytokinin synthesis, signaling, and function-advances and new insights. Int. Rev. Cell. Mol. Biol. 324 1–38. 10.1016/bs.ircmb.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Zürcher E., Tavor-Deslex D., Lituiev D., Enkerli K., Tarr P. T., Muller B. (2013). A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol. 161 1066–1075. 10.1104/pp.112.211763 [DOI] [PMC free article] [PubMed] [Google Scholar]