Abstract
The ability to monitor specific molecules in real-time directly in a flowing sample stream and in a manner that does not adulterate that stream could greatly augment quality control in, for example, food processing and pharmaceutical manufacturing. Because they are continuous, reagentless, and able to work directly in complex samples, electrochemical DNA-based (E-DNA) sensors, a modular and, thus, general sensing platform, are promising candidates to fill this role. In support, we describe here an E-DNA sensor supporting the continuous, real-time measurement of melamine in flowing milk. Using target-driven DNA triplex formation to generate an electrochemical output, the sensor responds to rising and falling melamine concentration in seconds without contaminating the product stream. The continuous, autonomous, real-time operation of sensors such as this could provide unprecedented safety, convenience, and cost-effectiveness relative to the batch processes historically employed in molecular quality control.
Graphical Abstract
The ability to monitor toxic substances, such as illegal additives, pesticide residues, or reaction intermediates in a flowing sample stream continuously, in real time, and without adulterating that sample stream could greatly augment quality control in food processing and the pharmaceutical industry.1,2 For example, in milk processing (as in much of the rest of the food processing industry), whole milk collected from farms is pumped into storage silos where it undergoes pasteurization, homogenization, separation, and further processing. Quality control in such processes has historically been relegated to batch-based laboratory procedures,3,4 which are so slow that they do not return an answer regarding a feedstock until after the feedstock has already entered the processing chain. In contrast, an ability to perform immediate, real-time measurements directly in crude foodstuffs would provide a means of identifying and rejecting contaminated feed stocks before they enter the processing chain, improving both safety and cost-effectiveness.
Motivated by the above arguments we describe here a sensor supporting high frequency, real-time molecular measurements in a flowing stream of foodstuffs using the detection of melamine in whole milk as our test bed. This compound, a triazine heterocyclic organic compound with particularly high nitrogen content (66% by mass), has been unethically used to increase the apparent protein content of foods despite the fact that exposure to this compound can lead to fatal kidney and tissue damage; melamine adulteration in milk products has caused thousands of renal failure cases and several infant deaths in China.5,6 For this reason, melamine levels are strictly regulated, with the legal safety limits being 2.5 ppm in the USA/EU and 1 ppm in China.7
Melamine interacts with nucleobase thymine (T) via the formation of complementary hydrogen bonds to form a DNA triplex (Figure 1),8,9 an effect that provides melamine recognition in a variety of previously reported sensors for the detection of this important adulterant.10–14 A colorimetric melamine-detecting approach, for example, employs this triplex motif to release the thymine-rich DNA sequence from the surface of gold nanoparticles, inducing their aggregation and an associated change in their absorption.10 A previously described fluorescent assay similarly employs a DNA triplex as melamine recognition element in a “molecular beacon” format by introducing an abasic site into the triplex stem,12,13 which provides a hydrophobic vacancy for targeting melamine through hydrogen bonding interaction. Although these approaches are sensitive and selective, they require washing steps, sample pretreatment, reagent additions, and bulky cumbersome laboratory equipment, and thus, they are not practical for point-of-use application,10,15–17 much less for, as shown here, continuous, seconds-resolved monitoring of a flowing, complex feedstock.
Figure 1.
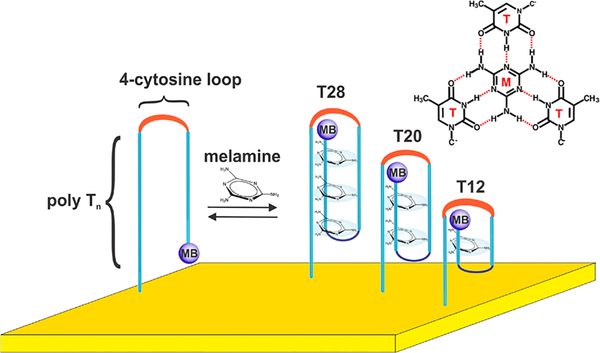
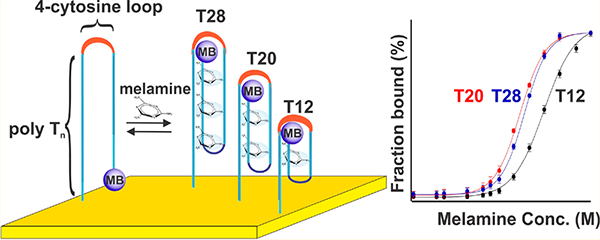
An electrochemical DNA-based (E-DNA) sensor supporting continuous, reagentless detection in a complex sample stream. Here, we have fabricated and validated sensors for the continuous monitoring of melamine (M) using a family of redox-reporter (methylene blue, MB) modified, thiol-anchored DNA sequences comprised of two polythymine segments connected by a four-cytosine loop: TnC4Tn, with the length of the polythymine varying from 4 to 12 bases. By tuning the length of polythymine segments, we expect that we could tune the extent of cooperativity.
In response to the inability of optical approaches to support continuous monitoring in raw food stocks, here we explore adaption of electrochemical DNA-based (E-DNA) sensors to the problem of continuous, real-time melamine measurements. E-DNA sensors are comprised of an electrode-bound, redox reporter-modified DNA that undergoes a binding-induced conformational change (Figure 1). This conformational change alters the ability of the redox reporter to approach the electrode, producing a target-dependent change in current when the sensor is interrogated via square wave voltammetry. This signaling mechanism renders E-DNA sensors single-step, reagentless, selective, and rapidly reversible, attributes that support continuous, real-time measurements without altering the sample stream.18–22 Together, these attributes suggest that the approach could prove a valuable tool for performing real-time molecular quality control, an application we explore here.
Guided by the variety of thymine-rich, melamine recognizing DNA elements previously described in the literature,23,24 we explored here E-DNA sensors employing a family of sequences comprised of two polythymine segments connected by a four-cytosine loop: TnC4Tn, with the length of the polythymine varying from 4 to 12 bases (Figure 1, Table S1). Specifically, we characterized three constructs, T12, T20, and T28, containing a total of 12, 20, or 28 nucleobases, respectively. These two polythymine segments of each construct presumably facilitate melamine binding via the formation of a triplex structure based on hydrogen-bond motif between melamine and thymine.10 Such binding should induce a conformational change in the DNA strand from disordered to a well-defined folded conformation exposing multiple melamine binding sites. This binding-induced conformational change reduces the ability of the attached redox reporter to approach the electrode surface, slowing electron transfer and causing a decrease square-wave voltammetric peak current (Figure 2).
Figure 2.
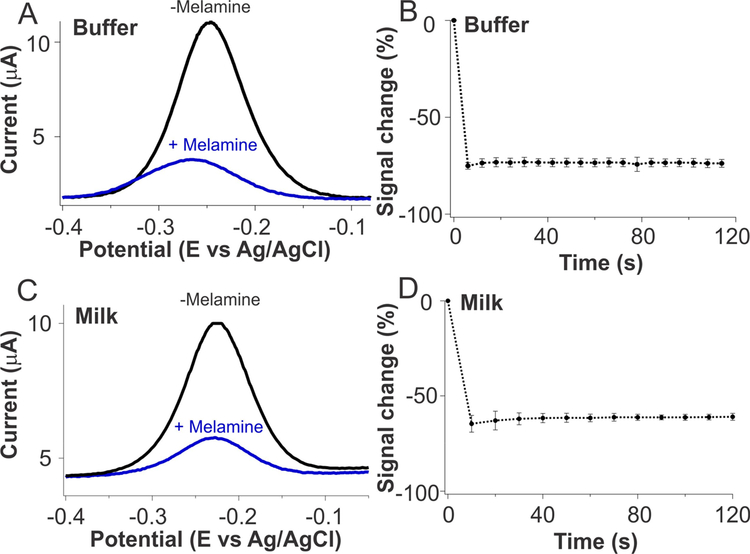
Sensors for the continuous, high frequency measurement of melamine in simple buffer solutions and in whole milk. (Top) When challenged with 500 μM melamine in a simple buffered solution (A), the T20 sensor’s signaling current changes dramatically and (B) the binding event is effectively complete within the few seconds required to perform the first square-wave scan. (Bottom) When challenged with the same amount of melamine in undiluted whole milk, we observe (C) a significant signal change and (D) rapid binding event. Of note, sensors built from the other two T-rich sequences we have explored exhibit similar binding kinetics (Figure S1). The error bars here and in the following figures reflect the standard deviation of measurements derived from at least three independently fabricated sensors, illustrating the magnitude of the sensor-to-sensor variation.
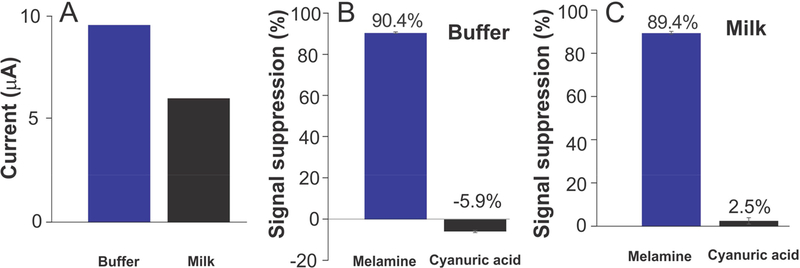
Our melamine sensors rapidly and specifically detect their target in both simple buffer solution and undiluted whole milk. The T20 sensor, for example, responds rapidly and strongly to its target; a challenge with 500 μM melamine in buffer produces a signal decrease of 75% (Figure 2A), with all of this occurring within the few seconds required to perform a square-wave scan (Figure 2B), a time resolution more than sufficient for the proposed quality control applications. As expected for sensors in this class,21 the melamine sensor performs well when challenged directly in complex media, including, for example, undiluted whole milk, with little loss in performance relative to simpler sample matrices (Figures 2C,D and 3A). Finally, the sensor is specific, and it can easily distinguish between melamine and its molecular analog, cyanuric acid, in both simple buffer solution and whole milk (Figure 3B, C). Attempts to determine its specificity relative against the related compounds ammeline and ammelide were precluded by their poor water solubility; this poor solubility, however, also precludes their use as adulterants.
Figure 3.
The sensors are selective and specific. For example, the T20 sensor shown here (A) exhibits little loss in signal when transferred from simple buffer solution to undiluted whole milk and (B, C) performs well when challenged in this complex sample matrix. Specifically, the signal gain observed in undiluted whole milk is within the error of that seen in simple buffer, when challenging the sensors with 1 mM melamine. The sensor is also specific, responding to melamine but not (<5% signal change, which is within normal sensor-to-sensor variation for these hand-fabricated devices) to its molecular analogue, cyanuric acid. Of note, sensors built from the other two T-rich sequences we have explored exhibit similar selectivity (Figure S1).
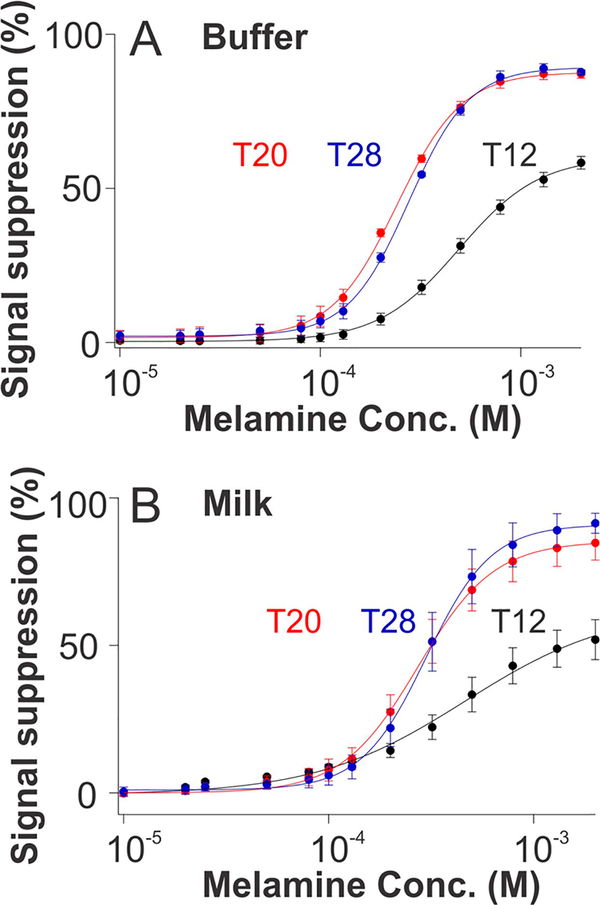
Although sensors employing all three of our thymine-rich constructs we have characterized exhibit a similar binding kinetics and binding specificity, their signal gains (relative change in signal upon the addition of saturating target) differ rather significantly. Specifically, the gains of T20 or T28 sensors are twice that of the T12 sensor within their common detection range, as seen in both buffer and whole milk (Figure 4). Given this, the three sensors achieve different detection limits. Specifically, sensors based on T20 and T28 constructs readily detected 80 μM melamine (10 ppm) in buffer at a signal change of 5% (Figure 4). T12 sensors, in contrast, perform less well, achieving a detection limit of 150 μM melamine (~19 ppm). When interrogated in whole milk, T12 sensor exhibits an improved detection limit of 20 μM, while the performance of T20 and T28 sensors do not change. The latter detection limit, corresponding to 2.5 ppm, is the legal safety limit regulated by the FDA in consumer products. The concentration of melamine in adulterated feed stocks, in contrast, is much higher than this: given the unethical purpose for which melamine is employed, it is generally present in concentrations of hundreds to thousands of parts-per-million (ppm) in order to, as criminally and dangerously intended, significantly and falsely raise the apparent protein content of adulterated feedstocks.7
Figure 4.
The sensors are quantitative. The response curves of sensors fabricated using T12, T20, and T28 in (A) simple buffer and (B) undiluted whole milk. Due to their thymine rich nature, these constructs are cooperative, steepening their binding curves. They are less cooperative, however, when challenged in milk rather than in a simpler sample matrix. The signals produced at lower concentrations (below 50 μM) are presented in Figure S3.
Given the presence of multiple binding sites, the thymine-rich recognition elements we employ here bind melamine in a cooperative manner,25–27 with the extent of cooperativity varying with the length of thymine segments (Figure 4). That is, the binding curves of sensors fabricated using all three constructs are well fitted by the Hill equation (Figures 4, S2, and S3) with Hill coefficients, nH, of greater than unity
where K1/2 is the target concentration at which one-half of the receptor binding sites are occupied. In a simple buffer solution, the shortest construct, T12, achieved a Hill coefficient of 1.9 ± 0.1 (97% confidence intervals based on estimated fitting errors) and K1/2 of order 490 ± 10 μM. Given the sequences of these constructs (three thymine bases bind one copy of melamine), it is reasonable to assume that the T12 construct, containing a total of 8 thymines, binds a maximum of two copies of melamine, thus accounting for the Hill coefficient approaching its “ideal” value of 2 for a two-site receptor.
As mentioned above, while the T20 and T28 constructs contain 4 or 6 potential binding sites, they exhibit Hill coefficients of only 2.7 ± 0.1 and 2.9 ± 0.1, respectively (Table S2). This less than ideal cooperativity, however, is associated with ~2-fold better affinity (K1/2 = 241 ± 3 and 276 ± 4 μM, respectively) than that of the shorter construct. This presumably reflects the thermodynamic trade off between cooperativity and affinity in which lower (than ideal, which would be 4 or 6 here for T20 and T28, respectively) Hill coefficients are associated with higher affinity (than would be expected if the Hill coefficients of these constructs were closer to ideal).25,28 Cooperative binding such as this “steepens” the binding curve and improves the sensor’s ability to measure small changes in the concentration of its target by narrowing its useful dynamic range (defined here as the ratio between the target concentration at which fraction bound is 90% to that at which it is 10%) of 9.7-, 5.2-, and 4.6-fold for T12, T20, and T28, respectively.
The melamine sensors are less cooperative when interrogated in whole milk, presumably due to the different ionic composition of this sample matrix. For example, the T12 construct loses its cooperativity compared to that in buffer, exhibiting a Hill coefficient of 1.2 ± 0.2. This reduced cooperativity is, however, associated with improved affinity, achieving a slightly lower K1/2 (463 ± 10 μM in milk versus 490 ± 10 μM in simple buffer). The decreased cooperativity also broadens the sensor’s useful dynamic range, expanding it to 41-fold in milk. The T20 and T28 sensors likewise exhibit reduced cooperativity in milk but to less of an extent than was true for the T12 construct, achieving improved K1/2 and dynamic ranges of 6.7- and 4.8-fold, respectively.
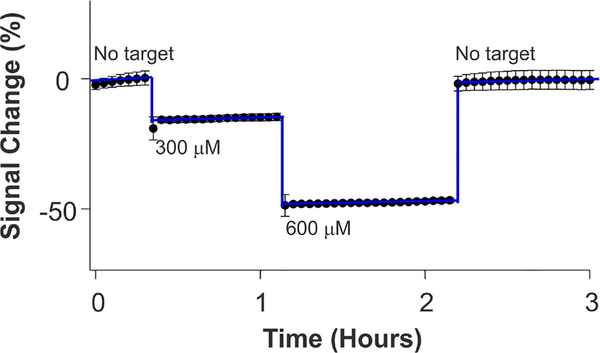
Because E-DNA sensors are rapid, reagentless, single-step, and reversible, they easily support continuous, real-time measurements in a flowing sample stream. To see this, we fabricated sensors employing the T20 construct and challenged them in flowing, undiluted whole milk in which we varied the concentration of melamine (Figure 5). As expected, the sensors respond rapidly and correspondingly to the concentration of spiked melamine, with the signal returning back to the baseline (within 2%) when the sensor is returned to melamine-free milk.
Figure 5.
Continuous, real-time molecular monitoring in a flowing sample stream. We challenged a melamine-detecting sensor in undiluted, flowing whole milk to demonstrate the real-time measurement of this target. Specifically, we fabricated these sensors with a T20 construct and challenged them against varying pulses of the target in flowing whole milk over the course of several hours. As expected, our sensors responded correspondingly and rapidly to the spiked concentrations (300 and 600 μM, respectively), and the signal returned back to the baseline (within 2%) when the sensor is returned to melamine-free milk.
Here, we describe a proof-of-principle study of the ability of E-DNA sensors to perform the continuous, autonomous, real-time monitoring of specific molecular analytes directly in a flowing sample stream. Specifically, we have demonstrated the ability of sensors in this class to monitor the adulterant melamine rapidly and sensitively in flowing, undiluted whole milk, achieving detection limits in line with current regulations without the need for batch processing or the addition of reagents That latter observation should render it possible to place sensors directly in a feedstock without fear of contaminating it or otherwise altering it, an attribute not seen in other approaches to melamine detection (e.g., ref 29).
When coupled with their continuous nature, reasonably long shelf life,30 and the fact that they do not require the addition of reagents (thus leaving the food uncontaminated), the ability of E-DNA sensors to perform well in complex sample matrices suggests that they could have utility in performing quality control and safety monitoring in the food chain, ranging from allergens,31 toxic pollutants (such as heavy metals), illegally used additives (such as, here, melamine in milk), and toxins (such as mycotoxins).22,32–34 They could also be valuable in the monitoring of other aqueous manufacturing processes, such as those employed in the pharmaceutical industry. More broadly, the continuous, autonomous, real-time operation of sensors in this (modular, versatile) class could provide unprecedented safety, convenience, and cost-effectiveness relative to the batch processes historically employed in molecular quality control.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant R01EB022015 (K.W.P) and by the Fundamental Research Funds (No. CUG170665) for the Central Universities, China University of Geosciences (Wuhan) (H.L.). H.L. acknowledges partial support from an “Early Postdoc Mobility fellowship” from the Swiss National Science Foundation. The authors also appreciate helpful discussions with Dr. Gabriel Ortega.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.anal-chem.8b01993.
Detailed description of the experimental procedures, DNA sequences employed, binding properties, binding kinetic plots, fraction bound curves, and signal response of sensors (PDF)
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Bagheri H; Asgharinezhad AA; Ebrahimzadeh H Food Anal. Methods 2016, 9, 876. [Google Scholar]
- (2).Chen X; Dong F; Xu J; Liu X; Wu X; Zheng YJ Agric. Food Chem. 2016, 64, 8935. [DOI] [PubMed] [Google Scholar]
- (3).https://www.fda.gov/ICECI/Inspections/InspectionGuides/ucm074974.htm#MILK%20AND%20MILK (accessed 2017).
- (4).https://library.e.abb.com/public/e3bec3791905b20cc1257106002e54d1/AG_BAT_2.pdf (accessed 2006).
- (5).Mauer LJ; Chernyshova AA; Hiatt A; Deering A; Davis RJ J. Agric. Food Chem. 2009, 57, 3974. [DOI] [PubMed] [Google Scholar]
- (6).Yang S; Ding J; Zheng J; Hu B; Li J; Chen H; Zhou Z; Qiao X Anal. Chem 2009, 81, 2426. [DOI] [PubMed] [Google Scholar]
- (7).https://www.federalregister.gov/documents/2008/11/13/E8-26869/interim-safety-and-risk-assessment-of-melamine-and-its-analogues-in-food-for-humans-availability (accessed 2008).
- (8).Sherrington DC; Taskinen KA Chem. Soc. Rev 2001, 30, 83. [Google Scholar]
- (9).Huang H; Li L; Zhou G; Liu Z; Ma Q; Feng Y; Zeng G; Tinnefeld P; He Z Talanta 2011, 85, 1013. [DOI] [PubMed] [Google Scholar]
- (10).Ai K; Liu Y; Lu LJ Am. Chem. Soc 2009, 131, 9496. [DOI] [PubMed] [Google Scholar]
- (11).Fodey TL; Thompson CS; Traynor IM; Haughey SA; Ken-nedy DG; Crooks SR Anal. Chem 2011, 83, 5012. [DOI] [PubMed] [Google Scholar]
- (12).Wang Y; Zhang J; Zhu L; Lu L; Feng C; Wang F; Xu Z; Zhang W Analyst 2015, 140, 7508. [DOI] [PubMed] [Google Scholar]
- (13).Wang Y; Sun Q; Zhu L; Zhang J; Wang F; Lu L; Yu H; Xu Z; Zhang W Chem. Commun 2015, 51, 7958. [DOI] [PubMed] [Google Scholar]
- (14).Wang Q; Gao F; Ni J; Liao X; Zhang X; Lin Z Sci. Rep 2016, 6, 22441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Gu C; Xiang Y; Guo H; Shi H Analyst 2016, 141, 4511. [DOI] [PubMed] [Google Scholar]
- (16).Yun W; Li H; Chen S; Tu D; Xie W; Huang Y Eur. Food Res. Technol 2014, 238, 989. [Google Scholar]
- (17).Han S; Zhu S; Liu Z; Hu L; Parveen S; Xu G Biosens. Bioelectron 2012, 36, 267. [DOI] [PubMed] [Google Scholar]
- (18).Lubin AA; Plaxco KW Acc. Chem. Res 2010, 43, 496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Li H; Dauphin-Ducharme P; Arroyo-Curras N; Tran CH; Vieira PA; Li S; Shin C; Somerson J; Kippin TE; Plaxco KW Angew. Chem., Int. Ed 2017, 56, 7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Li H; Arroyo-Currás N; Kang D; Ricci F; Plaxco KW J. Am. Chem. Soc 2016, 138, 15809. [DOI] [PubMed] [Google Scholar]
- (21).Plaxco KW; Soh HT Trends Biotechnol. 2011, 29, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Somerson J; Plaxco KW Molecules 2018, 23, 912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zeng Y; Pratumyot Y; Piao X; Bong DJ Am. Chem. Soc 2012, 134, 832. [DOI] [PubMed] [Google Scholar]
- (24).Piao X; Xia X; Bong D Biochemistry 2013, 52, 6313. [DOI] [PubMed] [Google Scholar]
- (25).Simon AJ; Vallée-Bélisle A; Ricci F; Plaxco KW Proc. Natl. Acad. Sci. U. S. A 2014, 111, 15048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Macazo FC; Karpel RL; White RJ Langmuir 2015, 31, 868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Ricci F; Valleé-Beĺisle A; Simon AJ; Porchetta A; Plaxco KW Acc. Chem. Res 2016, 49, 1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Simon A; Vallée-Bélisle A; Ricci F; Watkins HM; Plaxco KW Angew. Chem., Int. Ed 2014, 53, 9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Gu C; Lan T; Shi H; Lu Y Anal. Chem 2015, 87, 7676. [DOI] [PubMed] [Google Scholar]
- (30).Lai RY; Seferos DS; Heeger AJ; Bazan GC; Plaxco KW Langmuir 2006, 22, 10796. [DOI] [PubMed] [Google Scholar]
- (31).Knoedler M; Reisenhauer K; SCHIEBER A; Carle RJ Agric. Food Chem. 2009, 57, 3639. [DOI] [PubMed] [Google Scholar]
- (32).Cuccioloni M; Mozzicafreddo M; Barocci S; Ciuti F; Pecorelli I; Eleuteri AM; Spina M; Fioretti E; Angeletti M Anal. Chem 2008, 80, 9250. [DOI] [PubMed] [Google Scholar]
- (33).Bianco M; Sonato A; De Girolamo A; Pascale M; Romanato F; Rinaldi R; Arima V Sens. Actuators, B 2017, 241, 314. [Google Scholar]
- (34).McCullum C; Tchounwou P; Ding L-S; Liao X; Liu Y-MJ Agric. Food Chem. 2014, 62, 4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.