Abstract
Objective
Spinocerebellar ataxia type 2 (SCA2) is a neurodegenerative disease caused by a CAG repeat expansion in the gene ataxin-2 (ATXN2). ATXN2 intermediate-length CAG expansions were identified as a risk factor for amyotrophic lateral sclerosis (ALS). The ATXN2 CAG repeat is translated into polyglutamine, and SCA2 pathogenesis has been thought to derive from ATXN2 protein containing an expanded polyglutamine tract. However, recent evidence of bidirectional transcription at multiple CAG/CTG disease loci has led us to test whether additional mechanisms of pathogenesis may contribute to SCA2.
Methods
In this work, using human postmortem tissue, various cell models, and animal models, we provide the first evidence that an antisense transcript at the SCA2 locus contributes to SCA2 pathogenesis.
Results
We demonstrate the expression of a transcript, containing the repeat as a CUG tract, derived from a gene (ATXN2-AS) directly antisense to ATXN2. ATXN2-AS transcripts with normal and expanded CUG repeats are expressed in human postmortem SCA2 brains, human SCA2 fibroblasts, induced SCA2 pluripotent stem cells, SCA2 neural stem cells, and lymphoblastoid lines containing an expanded ATXN2 allele associated with ALS. ATXN2-AS transcripts with a CUG repeat expansion are toxic in an SCA2 cell model and form RNA foci in SCA2 cerebellar Purkinje cells. Finally, we detected missplicing of amyloid beta precursor protein and N-methyl-D-aspartate receptor 1 in SCA2 brains, consistent with findings in other diseases characterized by RNA-mediated pathogenesis.
Interpretation
These results suggest that ATXN2-AS has a role in SCA2 and possibly ALS pathogenesis, and may therefore provide a novel therapeutic target for these diseases.
Spinocerebellar ataxia type 2 (SCA2) is an autosomal dominant disorder characterized by near universal limb and gait ataxia, dysarthria, and abnormal eye movements, frequently accompanied by neuropathy, chorea, and dystonia, and occasionally accompanied by pyramidal signs.1,2 Neuropathologically, SCA2 presents a complicated picture, with marked loss of cerebellar Purkinje cells, as well as loss of cerebellar granule cells, motor neurons, and neurons in the substaniatia nigra, inferior olive, and pontocerebellar nuclei, and atrophy of white matter tracts.3–6
Genetically, SCA2 is caused by a CAG repeat mutation in the first exon of ATXN2, which is translated into an expanded polyglutamine (polyQ) tract.7 The normal ATXN2 allele contains 15 to 32 CAG repeats, whereas the disease allele has 33 to 64 triplets,8 with rare cases of expansions of >200 triplets.9 Normal SCA2 alleles contain CAA interruptions, whereas the majority of expanded alleles are uninterrupted. The most common configuration for the normal allele is (CAG)8CAA(-CAG)4CAA(CAG)8,10 whereas the most common SCA2 pathogenic allele contains 37 uninterrupted CAG trip-lets.11 The prevalence of SCA2 is 1–2/100,000, similar to that of another form of autosomal dominant ataxia, SCA1.12 However, significant geographic and ethnic variations exist, with the prevalence reaching 41/100,000 in the Holguin region of Cuba due to a possible founder effect.3 Recently, intermediate-length ATXN2 repeat expansions (27–33 triplets), mostly interrupted by 1 to 3 CAA triplets, have been associated with an increased risk for amyotrophic lateral sclerosis (ALS).13
ATXN2 protein is widely expressed, localizes to the endoplasmic reticulum and Golgi apparatus, and plays roles in a number of cellular pathways, including mRNA maturation, translation, and endocytosis.14,15 ATXN2 knockout mice have adult onset obesity, indicating that ATXN2 may also play a role in energy metabolism.16 Multiple cell and mouse models of SCA2 suggest that SCA2 arises, at least in part, by a gain of toxic function of mutant ATXN2 protein.14,17–19 Recent work has demonstrated that ATXN2 is an RNA-binding protein,20 and that in ALS, the RNA-dependent interaction between mutant ATXN2 protein and TDP-43 is involved in the disease pathogenesis.13
Natural antisense transcripts (NATs) that at least partially overlap with the gene on the opposite strand appear to contribute to the pathogenesis of a number of diseases caused by trinucleotide repeat expansions, including SCA8,21 fragile X syndrome, fragile X–associated tremor/ataxia syndrome,22 Huntington disease (HD),23 SCA7,24 and Huntington disease-like 2 (HDL2).25,26
Here we report that the ATXN2 locus is bidirectionally transcribed in SCA2 brain and cells, as well as in ALS tissue with an ATXN2 expansion. The antisense transcript ATXN2-AS with a CUG repeat expansion is neurotoxic, and therefore may contribute to SCA2 pathology. This finding suggests that ATXN2-AS is a potential therapeutic target in SCA2 and ALS.
Materials and Methods
Brain Tissue
Five human SCA2 and 4 control brains were obtained from Johns Hopkins University Brain Bank (Baltimore, MD), University of Maryland Brain Bank (Baltimore, MD), and the hereditary ataxia tissue repository at the Veterans Affairs Medical Center (Albany, NY). Postmortem interval varied from 6 to 24 hours, and expanded allele lengths ranged from 37 to 44 triplets. Control brains were matched to the SCA2 brains for age and postmortem interval. Clinical information for control and SCA2 patients is included in the Table 1. Sample 2LC, the normal human cerebral cortex RNA (Catalog No. 636561; Lot No. 1010040) pooled from 5 control males (aged 20–44 years), was purchased from Clontech (Mountain View, CA). Bacterial artificial chromosome (BAC)-SCA2 mice were maintained in the FVB background and bred and maintained according to the University of Utah Institutional Animal Care and Use Committee Protocol, in accordance with National Institutes of Health guidelines.17 Six-month-old BAC-SCA2 mice were deeply anesthetized, and cerebella were collected. Tissues were kept at −80 °C until the time of processing.
Table 1.
Control and Patient Brain Information
| Disease | Case ID | ATXN2 Alleles | Age of Death, yr | Age of Onset, yr | Gender | PMI, h | Brain Region |
|
|---|---|---|---|---|---|---|---|---|
| Cortex | Cerebellum | |||||||
| Control | 201 | 22/22 | 62 | N/A | M | 14 | + | |
| 219 | 22/22 | 35 | N/A | F | 8 | + | ||
| 249 | 22/22 | 49 | N/A | M | 12 | + | ||
| 221 | 22/22 | 45 | N/A | F | 20 | + | ||
| 2LC | Normal | 20–44 | N/A | M | N/A | + | ||
| SCA2 | K1 | 22/37 | 72 | 40 | F | 6 | + | + |
| H1 | 22/38 | 74 | 60 | F | 19 | + | + | |
| K2 | 22/41 | 49 | 26 | M | 24 | + | + | |
| K3 | 22/41 | 55 | 35 | F | 24 | + | + | |
| M1 | 22/44 | 43 | 30 | M | 23 | + | + | |
F = female; M = male; N/A = not applicable/not available PMI = postmortem interval; SCA2 = spinocerebellar ataxia type 2.
Cell Culture
Published control and patient-derived SCA2 fibroblasts, induced pluripotent stem cells (iPSCs), and neural stem cells (NSCs) differentiated from iPSCs were cultured as previously described.27 Human lymphoblastoid cell lines were obtained from patients with ALS (Coriell, Camden, NJ) or controls, and cultured as previously described.13 SK-N-MC neuroblastoma cells (ATCC, Manassas, VA) were maintained in Dulbecco modified Eagle medium with high glucose, supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin/amphotericin B (Sigma, St Louis, MO). SK-N-MC cells were screened regularly for mycoplasma contamination using Hoechst DNA staining method (Sigma). Primary mouse cortical neurons were prepared as previously described.28 Cells were transfected using Lipofectamine 2000 (Life Technologies, Grand Island, NY) according to the manufacturer’s protocol.
RNA Extraction and Strand-Specific Reverse Transcription Polymerase Chain Reaction
Human control RNA from different brain regions and peripheral tissues were obtained from Clontech. RNA from human postmortem control and SCA2 cortices and cerebella, patient cells, mouse brains, and transfected SK-N-MC cells were extracted by TRIZOL (Life Technologies), further purified (RNeasy Mini Kit; Qiagen, Valencia, CA), and cleaned of genomic DNA (Ambion Turbo DNA-free kit; Life Technologies). To examine ATXN2-AS transcript expression in human tissue or cells by strand-specific reverse transcription polymerase chain reaction (SS-RT-PCR), 500ng of RNA was reverse transcribed using the LK-F1 primer (SuperScript III First-Strand Synthesis System; Life Technologies). A first round of PCR used the linkered (LK) primer and a R1 primer for 25 cycles, followed by a second round of PCR using the LK primer and a nested R2 primer for 30 additional cycles. To examine ATXN2 transcript expression by SS-RT-PCR, LK-R1 primer was used during the reverse transcription. LK primer and F1 primer were used for the first round of PCR for 25 cycles, and F1 and R1 primers for the second round of PCR for 30 cycles. To examine the ATXN2-AS transcript expression in BAC-SCA2 mice by SS-RT-PCR, cDNA was made from 500ng of mouse cerebellum RNA and LK-F2 primer, and LK and R3 primers were used for PCR for 35 cycles. To validate the expression of ATXN2-AS_v1 and _v2, (or ATXN2-AS_v3 and _v4) in SCA2 iPSCs and control human brains by SS-RT-PCR, LK-R6 (or LK-R7) primer was used during the reverse transcription. LK primer and R2 primer were used for the first round of PCR for 25 cycles, and then LK primer and R3 primer for the second round of 30 cycles. The PCR products were resolved on 1.5% or 2% agasrose gels. Primer sequences are listed in Supplementary Table 1. Primer locations are shown in Figures 1A and 2C, E.
FIGURE 1:
Bidirectional transcription across the CAG/CTG repeat region at the ATXN2 locus. (A) Location of primers for the detection of ATXN2 and ATXN2-AS by strand-specific reverse transcription polymerase chain reaction (SS-RT-PCR). Dark arrows indicate primers used for SS-RT-PCR of human tissue and cells, and gray arrows indicate SS-RT-PCR primers for BAC-SCA2 mice. (B) Expression of ATXN2 and ATXN2-AS in multiple control human peripheral tissue and brain regions. Primers used to amplify transcript spanning the CAG/CTG repeat region are indicated as dark arrows in A. (C–E) SS-RT-PCR detected both normal and expanded alleles of ATXN2 and ATXN2-AS, in human spinocerebellar ataxia type 2 (SCA2) cerebellum (C), cortex (D), fibroblasts (FB), neural stem cells (NSCs), and induced pluripotent stem cells (iPSCs; E), whereas only expression from the normal alleles was detected in the controls. (F, G) Presence of the ATXN2-AS in the cerebellum (F) and cortex (G) of the bacterial artificial chromosome (BAC)-SCA2 mouse model, as detected by SS-RT-PCT using primers flanking the repeat (gray arrows in A). Deidentified ID numbers for brains and cells are included above each panel. PCRs using control (Ctl) or SCA2 genomic DNAs (gDNA), or water (no template), were used as positive and negative controls, respectively. No reverse transcription (−RT) control is a mock reverse transcription containing all the RT-PCR reagents, except the reverse transcriptase. For B–G, each experiment was repeated at least 3 times using distinct RNA preparations of each brain; representative gel images are shown. WT = wild type.
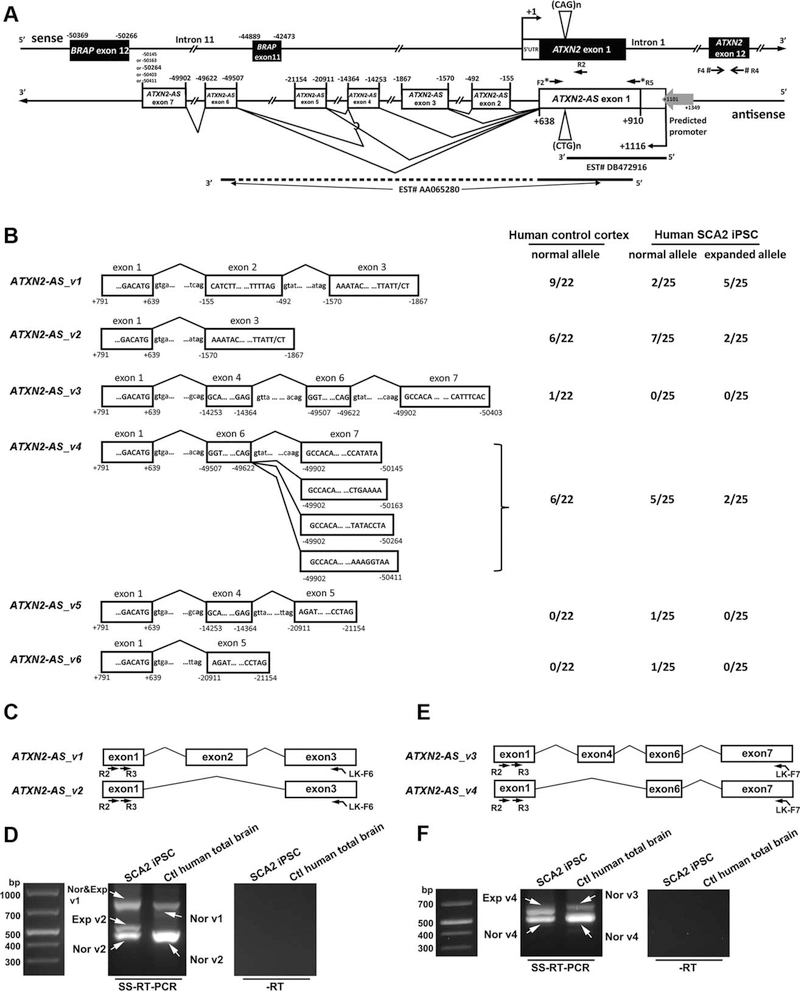
FIGURE 2:
Characterization and expression of ATXN2-AS. (A) Genomic structure of ATXN2-AS. Numbers are relative to the transcription start site of ATXN2. Quantitative polymerase chain reaction (PCR) primers for ATXN2 (F4/R4) and ATXN2-AS (F2/R5) are indicated by pound signs and asterisks, respectively. A putative promoter region for ATXN2-AS was predicted by the Proscan V1.7 software. R2 primer was used to characterize the 3′ ends of ATXN2-AS splice variants. (B) Characterized 3′ ends of ATXN2-AS in control human cortex and human spinocerebellar ataxia type 2 (SCA2) induced pluripotent stem cells (iPSCs). For each transcript variant bearing a normal or an expanded CUG repeat, the number of clones in which the transcript has been identified is shown per a total number of clones examined. A total of 22 and 25 clones were analyzed for human control cortex and SCA2 iPSCs, respectively. Consensus splicing donor/acceptor sites are shown in bold. (C–F) ATXN2-AS transcript is processed and represents a contiguous molecule. In both SCA2 iPSCs and control (Ctl) human brains, nested strand-specific reverse transcription PCRs (SS-RT-PCRs) amplified the ATXN2-AS_v1 and ATXN2-AS_v2 transcript from exon 1 to exon 3 (C, D), as well as the ATXN2-AS_v3 and ATXN2-AS_v4 transcript from exon 1 to exon 7 (E, F). Primer locations are indicated in C and E. Bands indicated by arrows (D, F) were confirmed by sequencing. For C-F, each experiment was repeated 3 times using different RNA preparations; representative gel images are shown. Exp = expanded; Nor = normal; −RT = no reverse transcription.
3′ Rapid Amplification of the cDNA End
Five milligrams of total RNA from human control frontal cortex (Clontech) or human SCA2 iPSCs was reverse transcribed using GeneRacer oligo-dT primer and SuperScript III First-Strand Synthesis System (Life Technologies). PCR was performed using Generacer-3′ and R2 primers with the touchdown PCR protocol, as previously described.23 The PCR products were cloned into pCR4-TOPO or pCR2.1-TOPO vector (Life Technologies) and sequenced.
Real-Time Quantitative PCR
To measure ATXN2 and ATXN2-AS expression levels by quantitative PCR (qPCR), 1mg of human cerebellum RNA was reverse transcribed using the ImProm-II Reverse Transcription System and random hexamer primers (Promega, Madison, WI). iTaq universal SYBR green super mix (Bio-Rad, Hercules, CA) was used for qPCR. Primer sequences are listed in Supplementary Table 1. To calculate the absolute ratio of ATXN2-AS to ATXN2, a sense–antisense (S-AS) plasmid was constructed by cloning both ATXN2-AS and ATXN2 amplicons into the same pCR2.1-TOPO vector (Life Technologies). Serial dilutions of the S-AS plasmid were used to construct a standard curve for both ATXN2-AS and ATXN2 amplification. ACTB was used as an internal control. qPCR was performed using an ABI 7900HT detection system (Applied Biosystems, Foster City, CA).
Each qPCR experiment involving human brains was performed 3 times. In each separate experiment, fresh RNA was extracted from each brain, converted to cDNA, and assayed in triplicate for ATXN2, ATXN2-AS, and ACTB expression. Each set of triplicates was averaged, and ATXN2 and ATXN2-AS means for each brain were normalized to the mean ACTB of a particular brain sample to yield normalized ATXN2/ACTB and ATXN2-AS/ACTB expression. The means of normalized ATXN2/ACTB and ATXN2-AS/ACTB for each brain from 3 separate experiments were then determined, and the data were grouped into control (n=3) and SCA2 (n=5) brains and subjected to final statistical analysis.
Splicing Analysis
To examine amyloid beta precursor protein (APP) exon 7 and N-methyl-D-aspartate receptor 1 (NMDA-R1) exon 5 splicing in human control and SCA2 brains, and iPSC or NSC lines, PCRs were performed on human cerebellum, iPSC or NSC cDNA, using FailSafe 2X buffer J (Epicentre, Madison, WI), Taq recombinant DNA polymerase (Life Technologies), and specific primers. Primer sequences were as previously described.29 PCR band intensities were measured using ImageJ software. The brain splicing experiments were performed 4 times for each brain (n=3 for control and n=5 for SCA2) and analyzed as described for brain qPCR experiments. Cell experiments to examine the relative APP exon 7 inclusion in control versus SCA2 cells were preformed similary, except that only 1 line was used for each condition.
Fluorescent In Situ Hybridization
The presence of foci containing expanded ATXN2-AS in human tissue or transfected cells was detected using a 5′ Texas Red–labeled CAG riboprobe (IDT, Coralville, IA), as previously described.30 Fluorescent in situ hybridization (FISH) with DNaseor RNase treatment was performed as previously described.31
Caspase 3/7 Assay
Cell viability was assessed by measuring caspase 3/7 activity (Caspase-Glo 3/7 Assay; Promega) in a 96-well format at 72 hours post-transfection as previously described.30 Each experiment was performed 4 separate times by transfecting constructs into SK-N-MC cells with a different passage number. In each experiment, independent constructs were transfected in quadruplicates (4 technical replicates). For each quadruplicate, the mean value, normalized to control, was determined. For each construct, the mean values from 4 separate experiments were averaged to provide a final outcome.
Nuclear Condensation Assay
Viability of transfected primary mouse cortical neurons was assessed by measuring the Hoechst staining intensity 48 hours post-transfection as previously described.28
cDNA Constructs
An ATXN2 fragment containing the region 21 bp upstream of the CAG repeat and 105 bp downstream from the repeat was PCR-amplified and cloned into pcDNA3.1(−) myc-his A vector (Life Technologies) at the EcoRV site. DNA extracted from human SCA2 postmortem brain with 22 or 44 CAG repeats served as a template for the PCR. ATXN2 inserts were flipped to obtain ATXN2-AS constructs with 22 and 44 CTG repeats. To obtain ATXN2-AS-(CTG)43M constructs, the CTG repeat region was replaced by a (CTG/TTG)43 repeat. A (CTG)32M fragment was synthesized by BioMatik (Wilmington, DE) and cloned into pcDNA3.1(−) myc-his A vector. CTG/TTG composition of the repeat region in ATXN2-AS constructs is shown in Figures 3D and 4B. The JPH3-(CTG)55 construct was made by subcloning the NT-55 insert30 into pcDNA3.1(−)myc-his A vector at the Nhe1 and Not1 sites. To examine RAN translation, ATXN2-AS-(CTG)n inserts were PCR-amplified and cloned into the Xhol and Xbal sites of A8(*KKQEXP)-3Tf1 vector, a kind gift from Dr Laura P. W Ranum,32 to produce ATXN2-AS-(CTG)n-3T plasmids. eEF1A1-Myc-Flag encoding myc- and flag-tagged eEF1A1 was from Origene (Rockville, MD), and hemagglutinin (HA)-tagged DISC1 was a kind gift from Dr Mikhail Pletnikov.33 The green fluorescent protein (GFP)-tagged MBNL1 (GFP-MBNL1) plasmid was a kind gift from Dr Charles A. Thornton.31,34
FIGURE 3:
Expression of ATXN2-AS in postmortem spinocerebellar ataxia type 2 (SCA2) cerebella. (A, B) Validation of quantitative polymerase chain reaction (qPCR) primers for ATXN2 and ATXN2-AS by strand-specific reverse transcription PCR (SS-RT-PCR). SS-RT-PCRs suggest that exon 12 (for ATXN2, A) and exon 1/intron 1 junction (for ATXN2-AS, B) regions are unidirectionally transcribed. Each experiment was repeated 3 times; representative gel images are shown. (C, D) Expression level of ATXN2 (C) and ATXN2-AS (D) as determined by qPCR and normalized to ACTB in control (Ctl) and SCA2 human cerebella. Three control and 5 SCA2 brains were included. The ATXN2/ACTB or ATXN2-AS/ACTB ratio in control was normalized to 1. (E) Expression level of ATXN2-AS relative to ATXN2 in human control and SCA2 cerebella. The ATXN2/ACTB ratio in control was normalized to 100. Data are expressed as mean ± standard deviation for control (n = 3) and SCA2 (n = 5) brains. n.s. = not significant by Mann–Whitney test; −RT = no reverse transcription.
FIGURE 4:
The expATXN2-AS transcript in spinocerebellar ataxia type 2 is toxic to neuronal-like SK-N-MC neuroblastoma cells. (A) ATXN2-AS-(CTG)44, but not ATXN2-AS-(CTG)22, is toxic to SK-N-MC cells, as indicated by a caspase 3/7 activity assay performed 72 hours after transfection. ATXN2-AS-(CTG)44 toxicity is higher than that of ATXN2-Q44 encoding a truncated ATXN2 protein with 44 glutamines. JPH3-(CUG)55 is included as a positive control.30 The caspase 3/7 activity in empty vector transfected SK-N-MC cells was normalized to 100. Data are expressed as mean ± standard deviation (SD) from 4 independent samples per condition (n =4 ). *p < 0.05, n.s.=not significant by Kruskal–Wallis test. (B) Schematic presentation of the construct and primer locations is shown. (C) Comparable expression levels of exogenous ATXN2, ATXN2-AS, and JPH3 RNAs were confirmed by reverse transcription polymerase chain reaction (RT-PCR). ACTB transcript was used as a loading control. The expression ratio of pcDNA3.1 A(−) to ACTB in vector-transfected cells was normalized to 1. Data are expressed as mean ± SD from 3 independent samples per condition (n = 3), and representative gel images are shown. (D) Schematic presentation of CTG/TTG composition in the ATXN2-AS-(CTG)n constructs used in E and F. Filled circles represent CTG repeats, and open circles represent TTG interruptions. (E) ATXN2-AS-(CTG)44, but not ATXN2-AS-(CTG)22 or ATXN2-AS-(CTG)43M, is toxic to primary mouse cortical neurons, as indicated by a nuclear condensation assay performed 48 hours after transfection. Data are expressed as mean ± SD from 8 independent samples per condition (n = 8). In each sample, 4,000 neurons per condition were analyzed. **p < 0.01 and ***p < 0.001 by Kruskal–Wallis test. (F) expATXN2-AS toxicity depends on the repeat’s ability to form toxic hairpin structures. Interrupted ATXN2-AS-(CTG)43M construct is much less toxic than the ATXN2-AS-(CTG)44 construct with a pure repeat. The caspase 3/7 activity in empty vector transfected SK-N-MC cells was normalized to 100. Data are expressed as mean ± SD from 4 independent samples per condition (n = 4). *p < 0.05, **p < 0.01 by Kruskal–Wallis test. (G, H) Comparable expression levels of exogenous ATXN2-AS transcripts were confirmed by RT-PCR. ACTB transcript was used as a loading control. Data are expressed as mean ± SD from 3 independent samples per condition (n = 3); representative gel images are shown.
Statistics
Power calculation was based on preliminary results (not included in the final analysis) using 2-sample Mann–Whitney tests (PASS 11; NCSS Statistical Software, Kaysville, UT). Samples of 3 and 5 for each control and SCA2 group for the brain experiments (Figs 5C–E and 6B, C) are capable of detecting an effect size as low as 3.26 (statistical power=0.80, alpha=0.05), and samples of 4 for each condition for the cell experiments (see Figs 3A, F, 4C, and 6E, F) are capable of detecting an effect size as low as 3.07 (statistical power=0.80. alpha=0.05). The small number of available brains (only 5 SCA2 postmortem brains were available to this study) was recognized as limiting the power to demonstrate the statistical significance of small differences in the data. The results were analyzed using nonparametric methods. The 2-tailed Mann–Whitney test was used for 2-group comparison, whereas the Kruskal–Wallis test, followed by Dunn multiple comparison test, was used for multiple-group comparison. Statistical significance was set at p<0.05.
FIGURE 5:
The expATXN2-AS transcript is expressed in amyotrophic lateral sclerosis (ALS) and triggers toxicity in neuronal-like SK-N-MC neuroblastoma cells. (A) Strand-specific reverse transcription polymerase chain reaction (SS-RT-PCR) detected the expression of ATXN2 and ATXN2-AS from both normal and expanded alleles in human ALS lymphoblastoid cells associated with the ATXN2 mutation. Gel images were representative of n = 3 independent experiments. (B) Schematic presentation of CTG/TTG composition in the ATXN2-AS constructs used in C. Filled circles represent CTG repeats, and open circles represent TTG interruptions. (C) ATXN2-AS-(CTG)32M is approximately 10% more toxic than ATXN2-AS-(CTG)22. The caspase 3/7 activity in empty vector transfected SK-N-MC cells was normalized to 100. Data are expressed as mean ± standard deviation (SD) from 4 independent samples per condition (n = 4). **p<0.01, n.s. = not significant by Kruskal–Wallis test. (D, E) Comparable expression levels of exogenous ATXN2-AS transcripts were confirmed by RT-PCR. ACTB transcript was used as a loading control. The expression ratio of pcDNA3.1 A(−) to ACTB in vector-transfected cells was normalized to 1. Data are expressed as mean ± SD from 3 independent samples per condition (n = 3); representative gel images are shown. Ctl = control; −RT = no reverse transcription.
FIGURE 6:
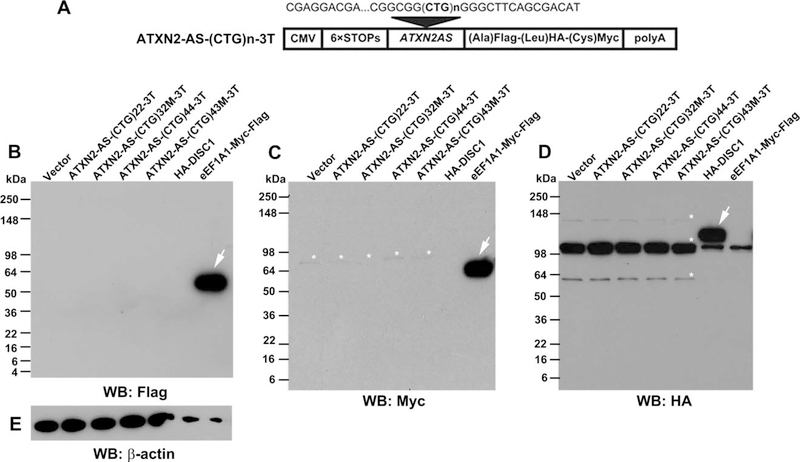
Non-ATG initiated (RAN) translation does not contribute to the toxicity of expATXN2-AS in SK-N-MC cells. (A) Schematic presentation of ATXN2-AS-(CTG)n-3T constructs with the 6XSTOP cassette and 3 tags (Flag, hemagglutinin [HA], and Myc) in 3 open reading frames. (B–E) SK-N-MC cells were transfected with ATXN2-AS-(CTG)n-3T constructs, and the presence of RAN translation was assessed by Western blot (WB) 72 hours post-transfection. pcDNA3.1 empty vector was used as a negative control. eEF1A1-Myc-Flag and HA-DISC1 plasmids were used as positive controls for antibodies used in the experiment. β-Actin was used as a loading control. Arrows point to positive control bands, and asterisks indicate nonspecific bands. Note that positive controls were purposefully underloaded. Each Western blot was repeated 3 times using different protein extracts; representative gel blots are shown.
Results
Bidirectional Transcription at the ATXN2 Locus
Modeled on our previous work on HDL2,25,26,30 a disease in which a relatively short repeat expansion triggers a devastating phenotype, probably through a combination of loss-of-function, RNA-mediated, and protein-mediated toxicity, we speculated that SCA2 may similarly involve complex mechanisms of pathogenesis. We therefore tested the possibility that an ATXN2 antisense (ATXN2-AS) transcript with an expanded CUG repeat is expressed at the ATXN2 locus. Using RNA from human control brain, we performed SS-RT-PCR35 with LK primers flanking the repeat in ATXN2 exon 1 (see Fig 1A), and detected the expression of an ATXN2-AS transcript in multiple brain regions and peripheral tissues (see Fig 1B). The identity of the transcript was confirmed by sequencing (data not included).
Expression of the Expanded ATXN2-AS (expATXN2-AS) Transcript in SCA2
In myotonic dystrophy type 1 (DM1), HDL2, SCA7, and HD human tissue, antisense transcripts derived from the normal allele have been detected, but not from the expanded allele,23,24,26,35 whereas both alleles yield antisense transcripts in SCA8.36 SS-RT-PCR demonstrated expression of both ATXN2-AS with a normal CUG repeat length (nATXN2-AS) and ATXN2-AS transcript with an expanded CUG repeat (expATXN2-AS) in human SCA2 postmortem cerebellum and cortex (see Fig 1). Moreover, both nATXN2-AS and expATXN2-AS were expressed in patient-derived SCA2 fibroblasts, as well as in an SCA2 iPSC line27 and SCA2 NSCs. In contrast, only nATXN2-AS was expressed in the corresponding control cells. Expression of expATXN2-AS in the human SCA2 iPSC line was confirmed by sequencing of the SS-RT-PCR product (data not included). Finally, SS-RT-PCR using primers specific for the human ATXN2-AS sequence detected the ATXN2-AS transcript in the cortices and cerebella of SCA2 BAC mice17 expressing either 22 or 72 CAG/CUG repeats.
Characterization of ATXN2-AS
Bioinformatic analysis indicates that the known ATXN2-AS region (+797 to+639) includes several expressed sequence tags (ESTs; AA065279, AA065280, and DB472916; see Fig 2). The ATXN2-AS transcript overlaps at least partially with intron 1 of ATXN2 in both human control and SCA2 cerebella, as detected by SS-RT-PCR against that region (+911 to+639). EST DB472916 (+1,119 to+661) may contain the 5′ end of ATXN2-AS transcript, as a functional promoter region (+1,349 to+1,101) has been predicted immediately upstream of the ATXN2-AS exon 1 by the Proscan V1.7 program. The characterized ATXN2-AS exon 1 (+911 to+639) lacks ATG start codons upstream of the CTG repeat, suggesting that the repeat in ATXN2-AS may not be translated. Additional bioinformatic analysis using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) reveals that the human genomic DNA sequence encoding the so far known ATXN2-AS exon 1 (+911 to+639) is 97.6% identical to green monkey, 96.7% to chimpanzee, 95.6% to rhesus, and 70.3% to mouse. To identify the 3′ end of ATXN2-AS, total RNA from human control cortex or SCA2 iPSCs were reverse transcribed using an oligo-dT primer with an attached linker sequence. Sequence analysis of the 3′ rapid amplification of the cDNA end (RACE) PCR products revealed 6 splice variants (see Fig 2B and Supplementary Table 2). The exon-intron structure in relationship to ATXN2 is depicted in Figure 2A. In human SCA2 iPSCs, 3′ alternative splicing is associated with both normal and expanded alleles of ATXN2-AS. ATXN2-AS exons 6 and 7 overlap with intron 11 of BRCA1-associated protein (BRAP), a gene upstream from ATXN2. SS-RT-PCR amplified the ATXN2-AS transcript from exon 1 to exon 3, and from exon 1 to exon 7, indicating that the ATXN2-AS is indeed processed and reflects a continuous transcript. In contrast to the characterization of the 3′ end of ATXN2-AS, characterization of the 5′ end has proven to be difficult. Our capacity to identify the 3′ ends of ATXN2-AS transcripts confirms that RNA quality was not an issue. We suspect that a very high G/C content led to our inability to detect 5′ ends of the SCA2 transcript, as high G/C content interferes with RACE. We confirmed this by examining DB472916, a potential 5′ end of the ATXN2-AS transcript in GenBank; its regional G/C content is up to 87%.
ATXN2-AS Expression in Patient Cerebella
NATs are common throughout the human genome; however, they are usually expressed at much lower levels than their sense counterparts.37 We therefore examined the expression level of ATXN2-AS relative to ATXN2 in human postmortem control and SCA2 cerebella. qPCR primers for ATXN2 resided in exon 12, whereas the primers for ATXN2-AS resided in exonl/intronl (see Fig 2A). SS-RT-PCRs were performed to confirm that qPCR primer pairs specifically amplify the transcript expression of either the ATXN2 or the ATXN2-AS strand (see Fig 5). Interestingly, qPCR revealed that the expression levels of both ATXN2 and ATXN2-AS (relative to that of ACTB) are slightly but not significantly higher in SCA2 than in control cerebella, subject to the caveat that the study was only powered to detect a difference of 0.75 or larger (power=0.80, alpha=0.05, estimated standard deviation=0.23 and effect size=3.26). The ratio of ATXN2-AS to ATXN2 (ATXN2-AS/ATXN2) is similar in control (14.3±1.6%) and SCA2 cerebella (13.6 6 1.3%). The expanded allele lengths in the human SCA2 postmortem brains ranged from 37 to 44 triplets (see Table 1), and the ATXN2-AS/ATXN2 ratio in SCA2 does not seem to vary by the repeat length.
expATXN2-AS Triggers Toxicity in Neuronal-like and Neuronal Cells
Transcripts containing expanded CUG repeat contribute to toxicity in DM1 and HDL2.30,38 We therefore examined whether expATXN2-AS with a CUG repeat expansion in the typical range of adult onset SCA2 is toxic to neuronal-like cells. ATXN2-AS-(CTG)n constructs expressing truncated ATXN2-AS transcripts with either 22 or 44 CUG triplets were transfected into neuronallike SK-N-MC neuroblastoma cells, which express endogenous ATXN2-AS and ATXN2 at a ratio similar to that observed in human brains (data not included). The toxicity of exogenous ATXN2-AS was compared to that of the corresponding exogenous sense ATXN2 fragment of a similar length, which is translated into a truncated ATXN2 protein with either 22 or 44 glutamines, respectively. As a positive control, we used a construct containing a fragment of junctophilin-3 (JPH3) expressing a transcript with 55 CUG triplets (JPH3-[CTG]55), previously associated with HDL2 pathogenesis.30 ATXN2-Q22 (expressing a truncated ATXN2 protein with 22 glutamines) and ATXN2-AS-(CTG)22 (expressing a truncated ATXN2-AS transcript with 22 CUG triplets) do not appear to be toxic, compared with the empty vector control. The truncated ATXN2 construct expressing a protein with 44 glutamines (ATXN2-Q44) is not stastically more toxic (20%, p>0.05) than a normal length glutamine repeat (ATXN2-Q22). However, ATXN2-AS-(CTG)44, expressing an ATXN2-AS transcript with 44 CUG triplets, triggered significant toxicity that was similar to that of the JPH3-(CTG)55 transcript and nearly twice that of ATXN2-Q44 (see Fig 3). Similar results were obtained using tetrazolium dye MTT 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide activity to assay for cell viability (data not shown). Equal expression of all the transcripts in SK-N-MC cells was confirmed by semiquantitative RT-PCR. To determine whether the toxicity of antisense transcripts with an expanded CUG repeat that we observed in neuroblastoma cell lines extended to neurons, we transfected ATXN2-AS-(CTG)44 and ATXN2-AS-(CTG)22 into primary mouse cortical neurons and measured toxicity by determining the extent of nuclear condensation. The expanded repeat increased toxicity by approximately 40%.
It has been suggested that the CUG transcript toxicity is dependent on the structure formed by the repeats.39,40 CUG repeats form hairpin structures, the stems of which sequester RNA-binding proteins (RBPs; ie, MBNL1); however, UUG repeats do not form hairpins or any higher-order structure.39 To examine whether preventing the formation of hairpin structures in expATXN2-AS ameliorates its neurotoxicity, we replaced the CTG repeat region in the ATXN2-AS-(CTG)44 construct with a synthetic fragment of 43 heavily interrupted CTG/TTG triplets, to obtain the ATXN2-AS-(CTG)43M construct (see Fig 3). Inserting interruptions abolished expATXN2-AS toxicity in mouse primary neurons and in SK-N-MC cells. Equal expression of the overexpressed transcripts in SK-N-MC cells was confirmed by semiquantitative RT-PCR. This suggests that expATXN2-AS toxicity is dependent on the ability of the repeat to form hairpin structures.
expATXN2-AS Is Expressed in Lymphoblastoid Cells from ALS Patients and Triggers Toxicity
ATXN2 intermediate-length polyQ expansions have recently been identified as a risk factor for ALS.13 Given our evidence of the neurotoxicity of ATXN2-AS with repeats typical of adult onset SCA2, we explored the potential relevance of ATXN2-AS to ALS. We detected ATXN2-AS expression from both normal and expanded alleles in 3 human ALS lymphoblastoid lines that have intermediate CAG expansions in ATXN2 (see Fig 4). The expanded ATXN2 allele lengths in ALS lymphoblastoid lines examined ranged from 31 to 32 triplets. Expression of expATXN2-AS in ALS lymphoblastoid cells was confirmed by sequencing (data not included). We next tested whether the expATXN2-AS transcript associated with ALS is toxic. A representative repeat from the expanded allele in ALS patients with 30 CTG triplets interrupted by 2 TTG triplets was cloned to make the ATXN2-AS-(CTG)32M construct. The configuration of the repeat in ATXN2-AS-(CTG)32M is (CTG)9TTG(CTG)8TTG(CTG)13. Caspase 3/7 activity assay suggested that the ATXN2-AS-(CTG)32M is approximately 10% more toxic to SK-N-MC cells than the control ATXN2-AS-(CTG)22 construct, although less toxic than ATXN2-AS-(CTG)44. Equal expression of the transcripts was confirmed by semiquantitative RT-PCR. Therefore, our data support the idea that expATXN2-AS may possibly contribute to the ALS phenotype, interestingly analogous to the potential contribution of an antisense transcript at mutant C9ORF72 allele to ALS pathogenesis.41
Non-ATG-Initiated Translation Products Do Not Contribute to Toxicity of expATXN2-AS in SK-N-MC Cells
The ATXN2-AS-(CTG)n construct naturally lacks ATG start codons from 5′ to the repeat. However, in the absence of an ATG start codon, an expanded repeat can be translated by non–ATG-initiated (RAN) translation.32 To confirm that neurotoxicity of expATXN2-AS is derived from the transcript itself, ATXN2-AS-(CTG)n fragments were cloned into a previously described 3-tagged vector (Fig 7A), so that translation from any of the 3 open reading frames (ORFs) across the repeat is readily detectable.32 Western blot analysis confirmed that no RAN products from ATXN2-AS fragment are translated in the SK-N-MC cells that overexpress these constructs (see Fig 7B–D). This supports the idea that the toxicity detected following overexpression of ATXN2-AS-(CUG)44 and ATXN2-AS-(CUG)32M transcripts is solely triggered by toxic RNA and does not involve a contributory toxicity of noncanonically translated proteins in our cell model. However, we cannot exclude the possibility that, in other cell types and in the context of full-length ATXN2-AS in SCA2 brains, such noncanonical proteins are translated from ATXN2-AS and trigger an additional mechanism of neurotoxicity in SCA2.
FIGURE 7:
expATXN2-AS RNA foci in spinocerebellar ataxia type 2 (SCA2). (A–F) Exogenous expATXN2-AS transcripts form nuclear CUG RNA foci in SK-N-MC neuroblastoma cells. ATXN2-AS-(CUG)32M (C) and ATXN2-AS-(CUG)44 (D), but not vector control (Ctl; A) or ATXN2-AS-(CUG)22 (B) transcripts formed foci when overexpressed in SK-N-MC cells. The ATXN2-AS-(CUG)44 RNA foci were resistant to DNase treatment (E), but were susceptible to RNase treatment (F). (G, H) expATXN2-AS transcript forms RNA foci in cerebellar Purkinje cells of human SCA2 brains (H), but not in control ones (G). (I–J) expATXN2-AS transcripts form nuclear CUG RNA foci in cerebellar Purkinje cells of bacterial artificial chromosome (BAC)-SCA2 Q72 mice (J), but not in wild-type (WT) mice (I). (K–V) MBNL1 is colocalized with expATXN2-AS RNA foci in SK-N-MC neuroblastoma cells. The ATXN2-AS-(CUG)32M (O-R) and ATXN2-AS-(CUG)44 (S-V) RNA foci sequester exogenous green fluorescent protein (GFP)-MBNL1 in SK-N-MC cells, whereas GFP-MBNL1 showed diffused nuclear localization in ATXN2-AS-(CTG)22-overexpressing cells (K–N). Scale bars = 5 mm. Arrows point to RNA foci, and asterisks indicate cerebellar Purkinje cells. Each experiment was repeated 3 times; representative images are shown. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
CUG RNA Foci and Splicing Abnormalities in SCA2 Patient Brains
In DM1, HDL2, and SCA8, expanded CUG repeat-containing transcripts aggregate into RNA foci.29,30,36 Although CUG RNA foci were not formed in SK-N-MC neuroblastoma cells transfected with control vector or ATXN2-AS-(CTG)22, using FISH with a CAG riboprobe, we detected CUG RNA aggregates in SK-N-MC neuroblastoma cells transfected with ATXN2-AS-(CTG)44or -(CTG)32M (Fig 8). The expATXN2-AS foci were resistant to DNase treatment, but susceptible to RNase treatment. Of the 5 human postmortem SCA2 brains available for this study, CUG RNA foci were detected in approximately 2% of cerebellar Purkinje cells in 2 brains that had 41 and 44 triplets, respectively, for the expanded alleles. In addition, CUG RNA foci were also detected in cerebellar Purkinje cells of BAC-SCA2 Q72 mice.17 In DM1 and HDL2 brains, the splicing factor MBNL1 localizes to CUG RNA foci, leading to missplicing of a set of MBNL1-regulated genes, including APP and NMDA-R1.29,30 Similarly, although exogenously expressed GFP-MBNL1 is diffusely localized to the nuclei of SK-N-MC neuroblastoma cells transfected with ATXN2-AS-(CTG)22, GFP-MBNL1 is colocalized with RNA foci containing expATXN2-AS in SK-N-MC cells. Finally, consistent with the possibility that sequestration of MBNL1 into expATXN2-AS foci may trigger missplicing in SCA2, we found that APP exon 7 and NMDA-R1 exon 5 were abnormally spliced in human postmortem SCA2 cerebella, compared with the controls (see Fig 6). As quantified in Figure 6B and C, SCA2 is associated with an increase in APP exon 7 inclusion, as well as a decrease in NMDA-R1 exon 5 inclusion, which resembles the pattern of missplicing in Alzheimer disease (AD).42,43 In addition, although APP exon 7 inclusion is similar in a control and an SCA2 iPSC line, following differentiation exon 7 inclusion decreases in control NSCs, but remains unchanged in SCA2 NSCs.
FIGURE 8:
Missplicing in spinocerebellar ataxia type 2 (SCA2). (A–C) Reverse transcription polymerase chain reaction suggests that amyloid beta precursor protein (APP) exon 7 and N-methyl-D-aspartate receptor 1 (NMDA-R1) exon 5 are misspliced in human SCA2 cerebella compared with control (A), as quantified in B and C. ACTB was used as a loading control. Three control and 5 SCA2 brains were included. Data are expressed as mean ± standard deviation (SD) from control (Ctl; n = 3) and SCA2 (n = 5) brains. *p<0.05 by Mann–Whitney test. (D) APP exon 7 splicing patterns in control or SCA2 induced pluripotent stem cells (iPSCs) and neural stem cells (NSCs). APP exon 7 is spliced similarly in SCA2 and control iPSCs, but is misspliced in human SCA2 NSCs compared to control, as quantified in E and F, respectively. ACTB was used as a loading control. One control iPSC line and 1 SCA2 iPSC line were differentiated into control NSCs and SCA2 NSCs, respectively. Data are expressed as mean ± SD from 4 technical replicates per condition; representative gel images are shown.
Discussion
We demonstrate that both normal and expanded CUG repeat-containing ATXN2-AS transcripts are expressed in human postmortem SCA2 brains, SCA2 fibroblasts, SCA2 iPSCs and NSCs, and a BAC-SCA2 mouse model,17 as well as in ALS lymphoblastoid cell lines carrying an intermediate length expansion in ATXN2. Nontranslatable expATXN2-AS with a CUG repeat size in the typical range for adult onset SCA2 and ALS is toxic to neuronal-like SK-N-MC neuroblastoma cells. Furthermore, we show that the expATXN2-AS transcripts form foci that sequester the splicing factor MBNL1. Finally, APP exon 7 and NMDA-R1 exon 5 are abnormally spliced in SCA2, similar to abnormalities observed in AD. We conclude that the expATXN2-AS transcripts may play an important role in the pathogenesis of SCA2, as well as ALS.
In DM1, HD, and HDL2 normal, but not expanded, antisense transcripts are expressed at the respective disease loci in human tissue.23,26,35 In distinction, our data demonstrate that in SCA2 the antisense allele with the expanded repeat is also expressed at significant levels. Thus far, SCA8 is the only other repeat expansion disorder to exhibit a similar phenomenon. Whether mutant antisense transcripts are expressed at other repeat expansion loci, including the most intensively studied dominant spinocerebellar ataxias, SCA1 and SCA3, remains to be determined.
It is estimated that at least 40% of gene loci are transcribed bidirectionally. Yet, to date, only a few NATs have been characterized, and most are expressed at much lower levels than their sense counterparts.44,45 However, even at low levels of expression NATs play important roles, for example, in regulating sense expression. For instance, HTT-AS, a transcript identified antisense to HTT at the HD locus, is expressed 100 times less than its sense counterpart. Overexpression of HTT-AS decreases the expression of HTT in a cell model, supporting the idea that upregulation of HTT-AS may serve as an endogenous strategy to suppress mutant HTT expression.23 We have found that ATXN2-AS is expressed at at least 12% of the level of ATXN2, 10 times higher than the ratio of expression of HTTAS to HTT. Hence, we hypothesized that the expression of ATXN2-AS was sufficient to directly contribute to SCA2 pathogenesis. In support of this possibility, we have shown that, when expressed at equal levels in a cell model, a nontranslatable ATXN2-AS-(CTG)44 construct triggered much higher toxicity than a ATXN2-Q44 construct that expresses both mutant truncated ATXN2 transcript and mutant truncated ATXN2 protein (see Fig 3A). In addition, RNA expressed from ATXN2-AS-(CTG)44 construct is toxic to primary mouse cortical neurons (see Fig 3E). Moreover, even the ATXN2-AS containing a CUG repeat with 2 interruptions, which is associated with ALS, may be slightly toxic to neuronal-like cells, although we did not find that the toxicity reached statistical significance (see Fig 4C). Although our models do not allow us to reach a conclusion about the relative toxicity of ATXN2-AS transcript with a CUG expansion in comparison to the toxicity of mutant ATXN2 protein, the data suggest that mutant ATXN2-AS may significantly contribute to SCA2 pathogenesis and may contribute to ALS pathogenesis.
Although overexpressed mutant ATXN2-AS fragments readily aggregate into RNA foci in SK-N-MC cells (see Fig 8C, D), RNA foci in Purkinje neurons in SCA2 human brain were uncommon (2%), and present in only 2 (of 5) brains (see Fig 8G, H). We suspect this likely underestimates the actual frequency of RNA foci in Purkinje cells. It is possible that our in situ approach is less effective in detecting RNA foci in tissue than in our cell models. In addition, Purkinje cells containing foci may have been lost at an earlier stage of disease. Also, levels of lipofuscin in Purkinje neurons may have obscured foci. Although we have previously used treatment with Sudan black B30 to decrease lipofuscin autofluorescence in human HDL2 tissue,30 this treatment also significantly decreases the fluorescent signal from the in situ probes. In the future, alternative approaches may be necessary to more accurately determine the frequency of ATXN2-AS RNA foci in SCA2 brains. It should be noted, however, that the role of RNA foci in neurotoxicity remains controversial46,47; it is not fully established whether the foci themselves are protective or neurotoxic. Recent work describing a transgenic BAC mouse model expressing expanded C9orf72 (expC9orf72) and exhibiting widespread RNA foci, but lacking behavioral abnormalities and neurodegeneration, even at advanced ages, suggests that RNA foci are not sufficient to trigger toxicity in ALS.48 Interestingly, protein inclusions are also rare in SCA2 brains and may not be an essential component of the pathogenesis of SCA2 49 or of other polyglutamine diseases.50 Similarly, although RNA foci are found in all CAG/CTG repeat diseases associated with RNA neurotoxicity,30,51–53 we predict that soluble RNA and/or RNA foci precursors rather than fully formed RNA foci are the essential feature of CUG repeat neurotoxicity.
NATs frequently regulate the expression of the gene on the sense strand.23,37 Our preliminary data suggest that overexpression of an ATXN2-AS fragment in trans does not have a substantial effect on the expression of endogenous ATXN2 transcript or protein in SK-N-MC neuroblastoma cells (data not shown). Whether ATXN2-AS affects the expression of ATXN2 or ATXN2 in cis remains to be examined. In addition, ATXN2-AS may affect the expression or splicing of BRAP on the sense strand, upstream from the ATXN2 locus (see Fig 2A). Although additional experiments are required to investigate the regulatory role of ATXN2-AS, ATXN2-AS may also serve other cellular functions, the disruption of which by the CUG expansion triggers pathogenic mechanisms that contribute to SCA2 and ALS. In a number of CAG/CUG repeat diseases, mutant transcripts aberrantly interact with RBPs involved in a range of cellular processes, including rRNA processing and splicing. Consistent with these findings, we demonstrate that APP and NMDA-R1 are misspliced in SCA2. The splicing of these two genes is, as mentioned above, thought to be under the control of MBNL1 that is sequestered into RNA foci in brain or model systems of several CAG/CUG repeat expansion diseases,30,31,36,51,52 and in our ATXN2-AS cell model (see Fig 8M–V). The APP splicing in SCA2 cerebellum shifts toward exon 7 inclusion, whereas that of the NMDA-R1 shifts toward exon 5 exclusion (see Fig 6A–C). Interestingly, this appears to be opposite to the splicing changes observed in DM1 and HDL2 cortex.29,30 We speculate that this is due to the cell-type specificity of missplicing (neurons in SCA2 vs glia in DM and HDL2), which may be dependent on the CUG repeat flanking sequence. Future studies examining the effect of expATXN2-AS on APP and NMDA-R1 splicing in cultured differentiated neurons and glia will help address this question. It is intriguing that the APP and NMDA-R1 splicing patterns in SCA2 resemble those in AD,42,43 suggesting the possibility of a shared pathogenic mechanism.
Our results provide further evidence that the pathogeneses of CAG/CTG repeat diseases is unlikely to be fully explained by the “single gene with a strong effect” model of pathogenesis, as bidirectionally expressed genes may each lead to a smaller, but additive or interacting, effect and thereby contribute to pathogenesis. This is of relevance to the development of therapy, as it suggests that targeting sense proteins/transcripts may not always be sufficient to achieve significant therapeutic benefits. Hence, suppressing the expression of ATXN2-AS may offer an additional or an alternative therapeutic approach to SCA2, and perhaps aid treatment of ALS associated with intermediate expansions in ATXN2. Because SCA2 belongs to a large group of diseases that are caused by a CAG/CTG repeat expansion, the data from this study may apply to other CAG/CTG repeat diseases involving additive or synergistic mechanisms of protein and RNA neurotoxicity.
Supplementary Material
Acknowledgment
This work was supported by the NIH National Center for Research Resources, NIH National Center for Advancing Translational Sciences (1UL1TR001079), and NIH National Institute of Neurological Disorders and Stroke (NS064138).
We thank Drs L. P. L. Ranum, M. Pletnikov, and C. A. Thornton for the kind gift of the A8(*KKQEXP)-3Tf1, HA-tagged DISC1, and GFP-MBNL1 constructs, respectively; Dr A. D. Gitler for the ALS lymphoblastoid genotyping data; and K. A. Carson for her advice on the statistical analysis.
Footnotes
Potential Conflicts of Interest
Nothing to report.
References
- 1.Geschwind DH, Perlman S, Figueroa CP, et al. The prevalence and wide clinical spectrum of the spinocerebellar ataxia type 2 trinucleotide repeat in patients with autosomal dominant cerebellar ataxia. Am J Hum Genet 1997;60:842–850. [PMC free article] [PubMed] [Google Scholar]
- 2.Cancel G, Durr A, Didierjean O, et al. Molecular and clinical correlations in spinocerebellar ataxia 2: a study of 32 families. Hum Mol Genet 1997;6:709–715. [DOI] [PubMed] [Google Scholar]
- 3.Orozco G, Estrada R, Perry TL, et al. Dominantly inherited olivopontocerebellar atrophy from eastern Cuba. Clinical, neuropathological, and biochemical findings. J Neurol Sci 1989;93:37–50. [DOI] [PubMed] [Google Scholar]
- 4.Rub U, Del Turco D, Del Tredici K, et al. Thalamic involvement in a spinocerebellar ataxia type 2 (SCA2) and a spinocerebellar ataxia type 3 (SCA3) patient, and its clinical relevance. Brain 2003; 126(pt 10):2257–2272. [DOI] [PubMed] [Google Scholar]
- 5.Rub U, Burk K, Schols L, et al. Damage to the reticulotegmental nucleus of the pons in spinocerebellar ataxia type 1, 2, and 3. Neurology 2004;63:1258–1263. [DOI] [PubMed] [Google Scholar]
- 6.Rub U, Del Turco D, Burk K, et al. Extended pathoanatomical studies point to a consistent affection of the thalamus in spinocerebellar ataxia type 2. Neuropathol Appl Neurobiol 2005;31:127–140. [DOI] [PubMed] [Google Scholar]
- 7.Pulst SM. Spinocerebellar ataxia type 2 In: Pagon RA, Adam MP, Ardinger HH, et al. , eds. GeneReviews. Seattle, WA: University of Washington, 1993. [PubMed] [Google Scholar]
- 8.Rub U, Farrag K, Seidel K, et al. Involvement of the cholinergic basal forebrain nuclei in spinocerebellar ataxia type 2 (SCA2). Neuropathol Appl Neurobiol 2013;39:634–643. [DOI] [PubMed] [Google Scholar]
- 9.Moretti P, Blazo M, Garcia L, et al. Spinocerebellar ataxia type 2 (SCA2) presenting with ophthalmoplegia and developmental delay in infancy. Am J Med Genet A 2004;124A:392–396. [DOI] [PubMed] [Google Scholar]
- 10.Laffita-Mesa JM, Velazquez-Perez LC, Santos Falcon N, et al. Unexpanded and intermediate CAG polymorphisms at the SCA2 locus (ATXN2) in the Cuban population: evidence about the origin of expanded SCA2 alleles. Eur J Hum Genet 2012;20:41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pulst SM, Nechiporuk A, Nechiporuk T, et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat Genet 1996;14:269–276. [DOI] [PubMed] [Google Scholar]
- 12.Subramony SH, Ashizawa T. Spinocerebellar ataxia type 1 In: Pagon RA, Adam MP, Ardinger HH, et al. , eds. GeneReviews. Seattle, WA: University of Washington, 1993. [Google Scholar]
- 13.Elden AC, Kim HJ, Hart MP, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 2010;466:1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huynh DP, Yang HT, Vakharia H, et al. Expansion of the polyQ repeat in ataxin-2 alters its Golgi localization, disrupts the Golgi complex and causes cell death. Hum Mol Genet 2003;12:1485–1496. [DOI] [PubMed] [Google Scholar]
- 15.Magana JJ, Velazquez-Perez L, Cisneros B. Spinocerebellar ataxia type 2: clinical presentation, molecular mechanisms, and therapeutic perspectives. Mol Neurobiol 2013;47:90–104. [DOI] [PubMed] [Google Scholar]
- 16.Kiehl TR, Nechiporuk A, Figueroa KP, et al. Generation and characterization of Sca2 (ataxin-2) knockout mice. Biochem Biophys Res Commun 2006;339:17–24. [DOI] [PubMed] [Google Scholar]
- 17.Dansithong W, Paul S, Figueroa KP, et al. Ataxin-2 regulates RGS8 translation in a new BAC-SCA2 transgenic mouse model. PLoS Genet 2015;11 :e1005182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Damrath E, Heck MV, Gispert S, et al. ATXN2-CAG42 sequesters PABPC1 into insolubility and induces FBXW8 in cerebellum of old ataxic knock-in mice. PLoS Genet 2012;8:e1002920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen ST, Meera P, Otis TS, Pulst SM. Changes in Purkinje cell firing and gene expression precede behavioral pathology in a mouse model of SCA2. Hum Mol Genet 2013;22:271–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yokoshi M, Li Q, Yamamoto M, et al. Direct binding of ataxin-2 to distinct elements in 3’ UTRs promotes mRNA stability and protein expression. Mol Cell 2014;55:186–198. [DOI] [PubMed] [Google Scholar]
- 21.Moseley ML, Zu T, Ikeda Y, et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet 2006;38:758–769. [DOI] [PubMed] [Google Scholar]
- 22.Ladd PD, Smith LE, Rabaia NA, et al. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet 2007;16:3174–3187. [DOI] [PubMed] [Google Scholar]
- 23.Chung DW, Rudnicki DD, Yu L, Margolis RL. A natural antisense transcript at the Huntington’s disease repeat locus regulates HTT expression. Hum Mol Genet 2011;20:3467–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sopher BL, Ladd PD, Pineda VV, et al. CTCF regulates ataxin-7 expression through promotion of a convergently transcribed, antisense noncoding RNA. Neuron 2011;70:1071–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilburn B, Rudnicki DD, Zhao J, et al. An antisense CAG repeat transcript at JPH3 locus mediates expanded polyglutamine protein toxicity in Huntington’s disease-like 2 mice. Neuron 2011;70:427–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seixas AI, Holmes SE, Takeshima H, et al. Loss of junctophilin-3 contributes to Huntington disease-like 2 pathogenesis. Ann Neurol 2012;71:245–257. [DOI] [PubMed] [Google Scholar]
- 27.Xia G, Santostefano K, Hamazaki T, et al. Generation of human-induced pluripotent stem cells to model spinocerebellar ataxia type 2 in vitro. J Mol Neurosci 2013;51:237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watkin EE, Arbez N, Waldron-Roby E, et al. Phosphorylation of mutant huntingtin at serine 116 modulates neuronal toxicity. PloS One 2014;9:e88284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang H, Mankodi A, Swanson MS, et al. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum Mol Genet 2004;13:3079–3088. [DOI] [PubMed] [Google Scholar]
- 30.Rudnicki DD, Holmes SE, Lin MW, et al. Huntington’s disease--like 2 is associated with CUG repeat-containing RNA foci. Ann Neurol 2007;61:272–282. [DOI] [PubMed] [Google Scholar]
- 31.Sun X, Li PP, Zhu S, et al. Nuclear retention of full-length HTT RNA is mediated by splicing factors MBNL1 and U2AF65. Sci Rep 2015;5:12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zu T, Gibbens B, Doty NS, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A 2011;108:260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abazyan B, Nomura J, Kannan G, et al. Prenatal interaction of mutant DISC1 and immune activation produces adult psychopathology. Biol Psychiatry 2010;68:1172–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin X, Miller JW, Mankodi A, et al. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet 2006;15:2087–2097. [DOI] [PubMed] [Google Scholar]
- 35.Cho DH, Thienes CP, Mahoney SE, et al. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell 2005;20:483–489. [DOI] [PubMed] [Google Scholar]
- 36.Daughters RS, Tuttle DL, Gao W, et al. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet 2009;5:e1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pelechano V, Steinmetz LM. Gene regulation by antisense transcription. Nat Rev Genet 2013;14:880–893. [DOI] [PubMed] [Google Scholar]
- 38.Ranum LP, Day JW. Pathogenic RNA repeats: an expanding role in genetic disease. Trends Genet 2004;20:506–512. [DOI] [PubMed] [Google Scholar]
- 39.Krzyzosiak WJ, Sobczak K, Wojciechowska M, et al. Triplet repeat RNA structure and its role as pathogenic agent and therapeutic target. Nucleic Acids Res 2012;40:11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nalavade R, Griesche N, Ryan DP, et al. Mechanisms of RNA-induced toxicityin CAG repeatdisorders. Cell Death Dis2013;4:e752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zu T, Liu Y, Banez-Coronel M, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U SA 2013;110:E4968–E4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka S, Shiojiri S, Takahashi Y, et al. Tissue-specific expression of three types of beta-protein precursor mRNA: enhancement of protease inhibitor-harboring types in Alzheimer’s disease brain. Biochem Biophys Res Commun 1989;165:1406–1414. [DOI] [PubMed] [Google Scholar]
- 43.Hynd MR, Scott HL, Dodd PR. Glutamate(NMDA) receptor NR1 subunit mRNA expression in Alzheimer’s disease. J Neurochem 2001;78:175–182. [DOI] [PubMed] [Google Scholar]
- 44.Zucchelli S, Fasolo F, Russo R, et al. SINEUPs are modular antisense long non-coding RNAs that increase synthesis of target proteins in cells. Front Cell Neurosci 2015;9:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oeder S, Mages J, Flicek P, Lang R. Uncovering information on expression of natural antisense transcripts in Affymetrix MOE430 datasets. BMC Genomics 2007;8:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Houseley JM, Wang Z, Brock GJ, et al. Myotonic dystrophy associated expanded CUG repeat muscleblind positive ribonuclear foci are not toxic to Drosophila. Hum Mol Genet 2005;14:873–883 [DOI] [PubMed] [Google Scholar]
- 47.Sicot G, Gomes-Pereira M. RNA toxicity in human disease and animal models: from the uncovering of a new mechanism to the development of promising therapies. Biochim Biophys Acta 2013; 1832:1390–1409. [DOI] [PubMed] [Google Scholar]
- 48.O’Rourke JG, Bogdanik L, Muhammad AK, et al. C9orf72 BAC transgenic mice display typical pathologic features of ALS/FTD. Neuron 2015;88:892–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huynh DP, Figueroa K, Hoang N, Pulst SM. Nuclear localization or inclusion body formation of ataxin-2 are not necessary for SCA2 pathogenesis in mouse or human. Nat Genet 2000;26: 44–50. [DOI] [PubMed] [Google Scholar]
- 50.Arrasate M, Finkbeiner S. Protein aggregates in Huntington’s disease. Exp Neurol 2012;238:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ranum LP, Day JW. Myotonic dystrophy: RNA pathogenesis comes into focus. Am J Hum Genet 2004;74:793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li LB, Yu Z, Teng X, Bonini NM. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature 2008;453:1107–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mykowska A, Sobczak K, Wojciechowska M, et al. CAG repeats mimic CUG repeats in the misregulation of alternative splicing. Nucleic Acids Res 2011;39:8938–8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.