Abstract
The discovery of fosfomycin more than 40 years ago was an important milestone in antibiotic therapy. The antibiotic’s usefulness, alone or in combination, for treating infections caused by multidrug-resistant microorganisms is clearer than ever. Both the European Medicines Agency and the US Food and Drug Administration have open processes for reviewing the accumulated information on the use of fosfomycin and the information from new clinical trials on this compound. The agencies’ objectives are to establish common usage criteria for Europe and authorize the sale of fosfomycin in the US, respectively. Fosfomycin’s single mechanism of action results in no cross-resistance with other antibiotics. However, various fosfomycin-resistance mechanisms have been described, the most important of which, from the epidemiological standpoint, is enzymatic inactivation, which is essentially associated with a gene carrying a fosA3-harboring plasmid. Fosfomycin has been found more frequently in Asia in extended-spectrum beta-lactamase-producing and carbapenemase-producing Enterobacterales. Although fosfomycin presents lower intrinsic activity against Pseudomonas aeruginosa compared with that presented against Escherichia coli, fosfomycin’s activity has been demonstrated in biofilms, especially in combination with aminoglycosides. The current positioning of fosfomycin in the therapeutic arsenal for the treatment of infections caused by multidrug-resistant microorganisms requires new efforts to deepen our understanding of this compound, including those related to the laboratory methods employed in the antimicrobial susceptibility testing study.
Keywords: Fosfomycin, Mechanisms of resistance, Susceptibility testing study, Biofilms, Antimicrobial combinations
BACKGROUND
Fosfomycin, a bactericidal antibiotic produced by, among others, Streptomyces fradiae, was discovered by a Spanish team from the Spanish Penicillin and Antibiotics Company (Compañía Española de Penicilina y Antibióticos) in 1969. Since then, fosfomycin has been employed in numerous countries for various indications, both in its intravenous (disodium salt) and oral formulations (calcium salt or trometamol). In recent years, the use of fosfomycin has increased spectacularly due to the considerable incidence of multidrug-resistant microorganisms for which fosfomycin constitutes, alone or in combination, a treatment alternative [1,2]. Due to the considerable usage differences worldwide, the need to establish common criteria and the need to expand the knowledge on this antibiotic, the European Medicines Agency has opened a process that seeks to collect evidence supporting fosfomycin’s indications and authorize and harmonize its usage criteria in Europe (https://www.ema.europa.eu/en/medicines/human/referrals/fosfomycin-containing-medicinal-products). Moreover, the US Food and Drug Administration included fosfomycin (according to the laboratory that conducts clinical trials of this antibiotic) in the list of drugs with antimicrobial activity (qualified infectious disease product), which facilitates a priority review of the results of the clinical trials and an accelerated registration process (https://www.nabriva.com/pipeline-research).
The implementation of epidemiological surveillance studies that include fosfomycin, the new clinical trials of this antimicrobial, as well as the pharmacokinetics-pharmacodynamics (PK-PD) studies necessary to support its formulation and to understand the significance of the possible development of resistances have deepened our microbiological understanding of this drug. The aim of this article is to review this new evidence from a microbiological standpoint that supports its clinical use.
MECHANISM OF ACTION AND PHARMACODYNAMICS OF FOSFOMYCIN
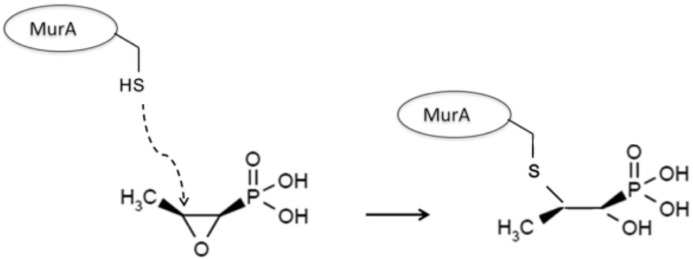
Fosfomycin has a single mechanism of action: blocking the first step of peptidoglycan synthesis. The transport of fosfomycin to the interior of the bacteria is performed through permeases, such as the glycerol-3-phosphate transporter (GlpT) and glucose-6-phosphate [G6P] transporter (UhpT). While GlpT maintains baseline activity without being induced, UhpT lacks activity in the absence of G6P. Once inside the bacterial cell, fosfomycin inhibits the UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) enzyme, responsible for catalyzing the formation of N-acetylmuramic acid (precursor of peptidoglycan) through the binding of N-acetylglucosamine and phosphoenolpyruvate. Fosfomycin is an analog of phosphoenolpyruvate, with an epoxide ring (essential in its mechanism of action) and a phosphonic group. Fosfomycin binds covalently with MurA, inhibiting the latter and thereby causing lysis of the bacterial cells (figure 1).
Figure 1.
Mechanism of action of fosfomycin
Fosfomycin is therefore a bactericidal compound that acts on bacteria in the growth phase. The fact that Gram-positive and Gram-negative bacteria require the formation of N-acetylmuramic acid for the synthesis of peptidoglycan means that fosfomycin’s spectrum of action is very broad. Likewise, there is no possibility of crossed resistances with this compound. Fosfomycin has therefore been employed for treating infections by multidrug-resistant pathogens such as methicillin-resistant Staphylococcus aureus (MRSA), methicillin-resistant coagulase-negative staphylococci (MRCNS), vancomycin-resistant enterococci (VRE), penicillin-resistant Streptococcus pneumoniae, extended-spectrum beta-lactamase (ESBL)-producing Enterobacterales, carbapenemase-producing Enterobacterales (CPE) and multidrug-resistant Pseudomonas aeruginosa [3].
In terms of its physical-chemical properties, fosfomycin is a low-molecular-weight, water-soluble compound with low plasma protein binding that disseminates easily to most tissues and to the interstitial fluid. Studies have shown that fosfomycin penetrates and reaches relevant concentrations in inflamed tissues, aqueous and vitreous humor, bones and lungs [4]. Likewise, fosfomycin actively accesses the interior of polymorphonuclear leukocytes. The compound is excreted almost exclusively in urine in a nonmetabolized form [5].
The PK-PD parameter associated with the compound’s bacteriological activity is not clearly defined and appears to depend on the microorganism. Recent studies have established that the PK-PD parameter that best predicts fosfomycin activity in Gram-negative bacilli (P. aeruginosa, Escherichia coli and Proteus spp.) is area under the curve (AUC)/minimum inhibitory concentration (MIC) [6, 7], while in S. aureus and enterococcus, fosfomycin has a time-dependent (T>MIC) behavior [8]. A study also demonstrated a high postantibiotic effect, even at subinhibitory concentrations [9].
Various studies have been published that have sought to elucidate the PK-PD parameter that determines fosfomycin activity in P. aeruginosa, with a number of conflicting results. A study using a murine model observed that AUC/MIC is the parameter that best fits fosfomycin activity [6], while another study showed that the antibiotic is time-dependent [10]. Bilal et al. determined that the PK-PD parameter that determines the total bactericidal activity of fosfomycin in P. aeruginosa is AUC/MIC, while T>MIC is related to resistance suppression [11].
MECHANISMS OF FOSFOMYCIN RESISTANCE
Fosfomycin resistance can be produced by 3 separate mechanisms: 1) transport impairment, 2) impairment of the target of action and 3) enzymatic inactivation (table 1) [5, 12, 13]. The first of these mechanisms is produced by mutants in chromosomal genes of the transporters GlpT and UhpT or in their regulator genes, impeding fosfomycin from reaching its location of action. This mechanism has been essentially described in E. coli and P. aeruginosa isolates. In Acinetobacter baumannii, it has been shown that mutants in the chromosomal gene abrp (essential for the bacteria’s growth and involved in wall patency) determine the resistance to fosfomycin, tetracyclines and chloramphenicol.
Table 1.
Mechanisms of fosfomycin resistance
| Process | Resistance mechanism | Microorganism | Localization |
|---|---|---|---|
| Transport reduction | Mutants in transporter genes glpT or uhpT Mutants in regulator genes of glpT or uhpT Mutants in cyaA and ptsI (regulate cAMP for glpT) Mutants in abrp |
Escherichia coli Escherichia coli Escherichia coli Acinetobater baumannii |
Crom Crom Crom Crom |
| Change in target or expression | Mutants in murA Increased murA expression Alternative pathways to MurA in peptidoglycan synthesis Limited participation of MurA in the biological cycle |
Mycobaterium tuberculosisa, Vibrio fischeria, Escherichia coli Escherichia coli Pseudomonas aeruginosab,c, Pseudomonas putidab Chlamydia trachomatisa |
Crom Crom Crom |
| Inactivation | Inactivation by metalloenzymes by incorporating: -glutathione (FosA, FosA2, FosA3, FosA4, FosA5, FosA6, etc.) -Bacillithiol or l-cysteine (FosB) -water (FosX) Phosphorylation of the phosphonate group by kinases and formation of: -diphosphates and triphosphates (FomA and FomB) -monophosphate (FosC) (FosC2) |
Enterobacteralesc, Pseudomonas sppb,c Acinetobacter spp. Staphylococcus spp., Enterococcus spp. Bacillus subtillisa Listeria monocytogenesa Streptomyces spp. Pseudomonas syringae Escherichia coli |
Crom / Pl Crom Crom / Pl Crom Crom Crom Crom Pl |
Intrinsic resistance
Reduced susceptibility
Some species of Enterobacterales (Serratia marcescens, Klebsiella spp., Enterobacter spp, Kluyvera georgiana, etc. have homologous chromosomal fosA genes and can present reduced fosfomycin susceptibility)
Crom: chromosome; Pl: plasmid
The target of action can be altered intrinsically or by murA gene mutants that affect the structure of MurA, with fosfomycin incapable of acting as a substrate. Mycobacterium tuberculosis naturally presents MurA with an aspartate residue instead of cysteine in position 117 and is incapable of interacting with fosfomycin, thereby resulting in its intrinsic resistance. Mutants with an altered active center of MurA are found relatively frequently in E. coli. The overproduction of MurA also results in insufficient inhibition by fosfomycin, with the microorganism non-susceptible to the action of this antibiotic. In some microorganisms such as P. aeruginosa and Pseudomonas putida, alternative metabolic pathways independent of MurA have been described in the synthesis of the peptidoglycan that explain the low fosfomycin susceptibility presented by these microorganisms. The lack of susceptibility of Chlamydia trachomatis to this antibiotic is due to the lack of importance of MurA in its biological cycle.
However, the mechanism that has attracted the most attention due to its greater epidemiological importance is fosfomycin inactivation, which can be caused by metalloenzymes that efficiently impare this antibiotic, blocking its inhibitory action on MurA. Various metalloenzymes have been described, including FosX and FosA, which inactivate fosfomycin by opening the epoxide ring by incorporating a water and glutathione molecule, respectively. FosB, another metalloenzyme, inactivates fosfomycin by adding a cysteine or bacillithiol molecule, the latter of which is used by Gram-positive microorganisms (Firmicutes) that do not produce glutathione. The incorporation of fosA in plasmids and their transformation in E. coli raises the MIC values of fosfomycin.
FosX has been found in environmental microorganisms with intrinsic fosfomycin resistance such as Mesorhizobium loti and Desulfitobacterium hafniense and in pathogens such as Listeria monocytogenes, Brucella melitensis and Clostridium botulinum. FosA and FosB have an approximate amino acid sequence homology of 48%, and their corresponding genes have been found in the case of fosB in plasmids and in the chromosomes of Gram-positive microorganisms (Staphylococcus epidermidis and Bacillus subtilis) and occasionally associated with plasmids in Enterobacterales [14]. The fosA gene and its various homologous genes, such as fosA2, fosA3, fosA4, fosA5 and fosA6, have been associated with plasmids in isolates of ESBL-producing E. coli and in carbapenemase-producing Klebsiella pneumoniae. For Klebsiella spp., Enterobacter spp., Serratia marcescens, Kluyvera spp. and P. aeruginosa, fosA variants have been identified in their chromosome, with differing sequences but preserving the active center, which could explain the low fosfomycin activity (modal MIC, 4-64 mg/L) in these species when compared with that presented against E. coli (modal MIC, 2-4 mg/L) (https://mic.eucast.org/Eucast2/). It has been shown that the deletion of these chromosomal genes reduces the MIC values of fosfomycin and that its insertion into a plasmid and transformation in E. coli confers an increase in MIC values.
Studies have also described kinases (FomA and FomB) that phosphorylate the phosphonate group of fosfomycin, forming diphosphate and triphosphate compounds that lack antimicrobial activity. Another reported kinase is FosC, a homologous phosphotransferase of FomA, which in Pseudomonas syringae (another microorganism able to synthesize fosfomycin) converts fosfomycin to fosfomycin monophosphate, which is non-susceptible to MurA.
MICROBIOLOGICAL CONSEQUENCES AND CLINICAL SIGNIFICANCE OF THE DEVELOPMENT OF FOSFOMYCIN RESISTANCE DEVELOPMENT
Despite the considerable ease with which fosfomycin-resistant mutants can be obtained, the clinical repercussion of such mutants has not been sufficiently tested [13]. In some cases, the mechanisms of fosfomycin resistance reduce the fitness of the bacteria that present fosfomycin resistance, and in numerous occasions reduce the bacterial virulence. Such is the case for some mutants in genes that participate in fosfomycin transport, such as cysA or pstI, which, in E. coli, reduce the formation of pili that limit its virulent nature by reducing its ability to adhere to epithelial cells and synthetic materials such as catheters. Lower fitness has also been observed in isolates with MurA overproduction, and its relationship with clinical failure has not been demonstrated. A noteworthy example is that of L. monocytogenes, which, in vitro, is considered inherently fosfomycin-resistant, not only because it has FosX, which inactivates fosfomycin but also because it is unable of transporting fosfomycin and accessing its location of action. However, in vivo, L. monocytogenes expresses a permease (Hpt) of G6P, which facilitates the entry of the antibiotic and its susceptibility.
The phenomenon of heteroresistance has been reported in various microorganisms, such as E. coli, A. baumannii, P. aeruginosa and even S. pneumoniae, which indicates the presence of bacterial subpopulations with lower fosfomycin susceptibility. This phenomenon would partly explain the high frequency of mutation for fosfomycin. Resistant mutants can be obtained in up to 40% of E. coli isolates at a rate of 10-7-10-5. These mutants present MCIs of 32-64 mg/L, with occasional mutants in genes glpT and uhpT. Their in vitro stability in laboratory media and urine is low, and the typical MIC values can be recovered in successive passages (2-4 mg/L). In approximately 1% of isolates, resistant mutants can be obtained at a lower rate (10-11-10-7) by deletions or insertions in genes uhpT and uhpA. These mutants present high MICs (512-1,024 mg/L) and lower growth stability than the isogenic strains but greater than that of the glpT and uhpT mutants [15-17]. These mutants are obtained more frequently in hypermutator strains. However, in all cases, their lower fitness, absence of stability and lower likelihood of selection in acidic environments (e.g., in urine) would also explain the low in vivo repercussion of fosfomycin resistance observed in vitro [18]. It should be noted that the high concentrations that fosfomycin reaches in some locations, such as urine, and its good penetration in biofilms minimize the possible selection of these mutants. This fact has been demonstrated in in vitro models in which the mutant selection window (the concentration range in which resistant mutants would be selected) has been able to be defined. This selection window can be avoided with therapeutic regimens higher than 4 g/8 h [19].
A recent meta-analysis estimated that the risk of selecting resistant mutants during fosfomycin monotherapy in various types of infections (urinary, respiratory, bacteremia, central nervous system and bone) with the involvement of various microorganisms was 3.4% [20]. Resistant mutants were obtained at a higher rate in Klebsiella spp., Proteus spp., Enterobacter spp. and P. aeruginosa, the latter of which can reach 20%. This fact could be due to fosfomycin’s lower intrinsic activity than that it presents against E. coli, which would facilitate its entry into the selection window and justify the administration of fosfomycin in combination with other antimicrobials for infections caused by P. aeruginosa. Additionally, a fitness cost associated with fosfomycin resistance in isolates of fosfomycin-resistant P. aeruginosa has not been demonstrated, which could reinforce the need for combined therapy in infections caused by this pathogen. These combinations would reduce the selection window in which resistant mutants could be selected [21].
Regardless of the mechanisms detailed earlier, the most important from the epidemiological and clinical standpoint is the enzymatic inactivation associated with fos genes. The most important of these genes due to its greater dispersion, plasmid characteristics and presence in ESBL-producing and carbapenemase-producing Enterobacterales is fosA3 [14]. Initially described in 2010, fosA3 has been found more frequently in Asia, in human and animal isolates, although it is also present in Europe [22, 23]. The rate of fosA3 varies according to the studied collection but can be present in 90% of fosfomycin-resistant isolates (3-15% of all isolates) that produce ESBL or carbapenemase.
Recently, the origin of the fosA3 gene in Kluyvera georgina has been confirmed. Its integration into plasmids of various incompatibility groups could be related with composite transposons with the insertion sequence IS26 [24].
FOSFOMYCIN SUSCEPTIBILITY TESTING STUDY IN THE LABORATORY, CLINICAL AND EPIDEMIOLOGICAL BREAKPOINTS
The study of in vitro fosfomycin susceptibility has always been a challenge in the laboratory due to the lack of unanimous criteria on how it should be conducted for all microorganisms involved in infections for which fosfomycin is indicated. In addition, not all microorganisms currently have interpretive breakpoints (table 2). This situation could change due to the growing interest in this antimicrobial and the need to study it against multidrug-resistant microorganisms in which fosfomycin represents a treatment option.
Table 2.
Clinical breakpoints for intrepreting fosfomycin susceptibility testing results
| EUCAST | CLSI | |||||||
|---|---|---|---|---|---|---|---|---|
| MIC (mg/L) | Inhibition zone (mm) | MIC (mg/L) | Inhibition zone (mm) | |||||
| ≤S | >R | ≥S | <R | ≤S | ≥R | ≥S | ≤R | |
| Enterobacterales | 32a | 32a | 24a | 24a | 64b | 256b | 16b | 12b |
| Pseudomonas spp. | 128c | 128c | 12c | 12c | - | - | - | - |
| Enterococcus spp. | - | - | - | - | 64d | 256d | 16d | 12d |
| Staphylococcus spp. | 32e | 32e | - | - | - | - | - | - |
| Streptococcus pneumoniae | IE | IE | IE | IE | - | - | - | - |
| Haemophilus influenzae | IE | IE | IE | IE | - | - | - | - |
| Moraxella catarrhalis | IE | IE | IE | IE | - | - | - | - |
EUCAST, European Antimicrobial Suceptiblity Testing Committee; CLSI, Clinical and Laboratory Standards Institute; IE: insufficient evidence to establish breakpoint values.
Intravenous and oral use (uncomplicated UTI)
E. coli isolates from the urinary tract
Epidemiological cutoff values (ECOFF) use in combination with other antimicrobials
E. faecalis isolates from the urinary tract
Intravenous use
To date, the reference method recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and the Clinical and Laboratory Standards Institute (CLSI) for the study of fosfomycin susceptibility is agar dilution, adding G6P to the medium (25 mg/L). The justification for this recommendation is that fosfomycin uses 2 types of transporters to penetrate bacterial cells. The first transporter, which has constitutive expression, uses glycerol 3-phosphate. This transporter reduces its activity in culture media that contain glucose or phosphate, which occurs with Mueller-Hinton agar, increasing fosfomycin’s MIC values compared with other culture media. The second transporter is induced by the presence of G6P; therefore, when G6P is added to the medium, fosfomycin enters the bacteria more effectively, and its MIC values are drastically reduced. The addition of 25 mg/L of G6P mimics the physiological situation of bacteria at the site of the infection; the MIC values would therefore approach the theoretical values. An increase in the amount of G6P above 25 mg/L in the medium has little effect on the MIC values.
Some microorganisms, such as P. aeruginosa, lack a G6P-dependent transporter and only present the glycerol 3-phosphate-dependent transporter. In this case, the addition of G6P to the medium does not change the MIC values [25]. It has recently been shown that fosfomycin activity is increased (lower MIC values) in this microorganism when studied in conditions with limited oxygen availability. This is explained by higher expression of the glycerol-3-phosphate-dependent transporter GlpT, which would resemble that of growth conditions in biofilms and would explain the strong fosfomycin activity against P. aeruginosa when grown in these conditions [26].
Although broth microdilution is not recommended for the study of fosfomycin susceptibility testing, a number of authors have demonstrated in P. aeruginosa the equivalence of agar dilution and broth microdilution [25]. In Enterobacterales, there is a very low correlation between the various methods, including the automatic systems and agar dilution, and are therefore not recommended for the susceptibility study [27, 28].
In the diffusion methods, G6P is added to the disc or to the gradient strips. The disc load recommended by EUCAST and CLSI is 200 μg with 50 μg of G6P. The reading of inhibition zone or ellipses is usually problematic because colonies can appear inside the inhibition zone in up to 41% of E. coli isolates. EUCAST has standardized its reading for E. coli, proposed that colonies considered susceptible within the inhibition zone should be ignored and has planned to offer recommendations for K. pneumoniae and P. aeruginosa. Using whole genome sequencing, Lucas et al. [17] recently studied the colonies observed inside the inhibition zone, estimating that only 0.8% of cases were considered resistant when re-examined by disc diffusion. These colonies are mutants whose resistance is due to deletions or nonsense mutants in the uhpT gene associated with G6P-dependent fosfomycin transport.
To facilitate reading the inhibition zones or ellipses, reducing the standard inoculum from 1.5x108 to 1.5x106 colony-forming units/mL has been proposed for P. aeruginosa [29]. This reduction decreases the presence of inner colonies and improves the correlation with the agar dilution MIC values to better define the wild-type population [MIC less than or equal to the epidemiological cutoff (ECOFF), 128 mg/L]. This approach should also be explored in Enterobacterales.
NEW DATA FROM EPIDEMIOLOGICAL SURVEILLANCE STUDIES
The revaluation of fosfomycin in recent years is due to the scarcity of new antibiotic options and the increased incidence of infections by multidrug-resistant microorganisms. Fosfomycin’s unique mechanism of action results in no crossed resistances with other antibiotics. Fosfomycin is therefore situated as one of the few therapeutic options for infections by multidrug-resistant microorganisms. The latest studies that detail fosfomycin activity in pathogens with various mechanisms of resistance are listed in table 3.
Table 3.
Fosfomycin activity in pathogens with various resistance mechanisms
| Author, date of publication | Microorganism, resistance, (n) | % Fosfomycin susceptibility | Other susceptibility data | Methodology (Breakpoints) | Source of isolate | Country | Ref. |
|---|---|---|---|---|---|---|---|
| Flamm, R., 2018 | E. coli (22)/ESBL K. pneumoniae (21) | 81.8%/91.7% | MIC50, MIC90= 0.5, 2 mg/L / MIC50, MIC90= 4, 8 mg/L | Agar dilution (CLSI) | SENTRY study | USA | (30) |
| E. coli (11)/Carbapenemase K. pneumoniae (12) | 92% | MIC50, MIC90= 8, 64 mg/L / MIC50, MIC90= 1, >256 mg/L | |||||
| Falagas, M.,2009 | MDR/XDR Enterobacterales (152) | 98% | Gradient strips (CLSI) | Clinical isolates | Greece | (31) | |
| Bouxom, H., 2018 | ESBL E. coli and K. pneumoniae (100) | 92.7% | Agar dilution (EUCAST) | Urinary-bacteremia isolates | France | (35) | |
| Bi, W. 2017 | ESBL E. coli (356) | 92,7% | MIC50, MIC90= 0.5, 32 mg/L | Agar dilution (CLSI) | Urinary isolates | China | (34) |
| Mezzatesta ML., 2017 | ESBL E. coli (24)/KPC K. pneumoniae (53) | 100%/78% | MIC50, MIC90= 0.5, 1 mg/L / MIC50, MIC90= 32, 128 mg/L | Agar dilution/microdilution/gradient diffusion (CLSI) | Urinary isolates | Italy | (32) |
| Flamm, R., 2018 | P. aeruginosa not susceptible to CAZ-AVI (21) | 85.7% | MIC50, MIC90= 32, 128 mg/L | Agar dilution (CLSI) | SENTRY study | USA | (30) |
| P. aeruginosa not susceptible to MER (20) | 75% | MIC50, MIC90= 32, 128 mg/L | |||||
| Walsh C., 2015 | MDR and non-MDR P. aeruginosa (64) | 61% | MIC50, MIC90= 64, 512 mg/L | Agar dilution/microdilution (CLSI) | Cystic fibrosis, bacteremia | Australia | (10) |
| Perdigao-Neto LV., 2014 | MDR P. aeruginosa (15) | 7% | Agar dilution/microdilution (CLSI) | Urinary, bacteremia and respiratory isolates | Brazil | (38) | |
| Flamm, R., 2018 | MRSA (101) | 100% | MIC50, MIC90= 4, 8 mg/L | Agar dilution (CLSI) | SENTRY study | USA | (30) |
| Falagas M., 2010 | MRSA (130) | 99.2% | Disc diffusion (200) (CLSI) | Nonurinary | Greece | (40) | |
| Lu CL., 2011 | MRSA (100) | 89% | Agar dilution (NE) | Clinical isolates | Taiwan | (41) | |
| López Díaz MC., 2017 | MRSA (55) | 43.6% | MIC50, MIC90= 128, 512 mg/L | Agar dilution (NE) | Clinical isolates | Spain | (42) |
| Wu D., 2018 | MRSA (293) | 46.8% | Agar dilution (CLSI) | Urinary, bacteremia and respiratory isolates | China | (43) | |
| Guo Y., 2017 | VRE (890) | 85.1% susceptible 13.4% intermediate |
Agar dilution (CLSI) | Rectal swabs | USA | (44) | |
| Tang HJ., 2013 | VR E. faecium (19) VR E. faecalis (21) |
30% 44% |
MIC50, MIC90=128 mg/L | Agar dilution (CLSI) | Clinical isolates | Taiwan | (45) |
CAZ/AVI, ceftazidime/avibactam; CLSI, Clinical and Laboratory Standards Institute; ESBL, extended-spectrum beta-lactamase; EUCAST, European Committee on Antimicrobial Susceptibility Testing; KPC, Klebsiella pneumoniae carbapenemase; MER, meropenem; MIC, minimum inhibitory concentration; MDR, multidrug-resistant; MRSA, methicillin-resistant S. aureus; NS, not specified; VR, vancomycin-resistant; VRE, vancomycin-resistant enterococcus; XDR, extremely drug-resistant
Enterobacterales with extended-spectrum beta-lactamase and carbapenemases. According to various in vitro susceptibility studies, fosfomycin maintains its activity against ESBL-producing and carbapenemase-producing Enterobacterales. It has been reported fosfomycin susceptibility rates of more than 80% against these microorganisms. The authors of a recent article that described fosfomycin activity clinical isolates from the US observed 100% (43/43 isolates) susceptibility to fosfomycin in ESBL-producing E. coli and K. pneumoniae (MIC50/MIC90 of 0.5/2 mg/L and 4/8 mg/L, respectively). In terms of CPE, a susceptibility of 81.8% (MIC50/90 of 1/>256 mg/L) was observed for E. coli isolates and 91.7% (MIC50/90 of 8/64 mg/L) for K. pneumoniae [30]. A susceptibility of 94.9% was observed in CPE from Greece [31], while 78% was observed in K. pneumoniae with Klebsiella pneumoniae carbapenemase (KPC) from Italy [32].
A review by Falagas et al. [33] that collected in vitro data calculated a fosfomycin susceptibility of 96.8% and 81.3% for ESBL-producing E. coli and K. pneumoniae, respectively. In China, a susceptibility of 92.7% was observed in E. coli with ESBL from urinary infections. The resistance in most isolates was associated with a plasmid that carries the blafosA and blaCTX-M genes [34]
In a study that compared the antibiotic susceptibility of fosfomycin with that of other noncarbapenem antibiotics in Enterobacterales with ESBL, 98% of the isolates were fosfomycin-susceptible, while 100% were ceftazidime-avibactam-susceptible, 97% were susceptible to amikacin and piperacillin-tazobactam, and 96% were nitrofurantoin-susceptible [35].
Although these data demonstrated high susceptibility rates in this type of microorganism, an increase in fosfomycin-resistant isolate was reported in Spain during a 4-year period, which was attributed to the increased use of this antibiotic in community-acquired urinary tract infections and to the dispersion of epidemic clones [36]. Likewise, PD studies conducted using time-kill curves and in vitro models of emergence of resistant mutants in enterobacteria with ESBL and/ or carbapenemases showed not only the bactericidal activity of fosfomycin but also a regrowth of resistant subpopulations that varied according to the species and isolate [37].
Multidrug-resistant Pseudomonas aeruginosa. Fosfomycin activity against P. aeruginosa is controversial due to the mutation freequency rate at which resistant mutants emerge. There is considerable heterogeneity in the in vitro susceptibility results, often due to the method employed for reading the susceptibility. In a study conducted in Australia, 61% of multidrug-resistant and nonmultidrug-resistant P. aeruginosa isolates were susceptible to fosfomycin (considering the MIC breakpoint as ≤64 mg/L), with a similar MIC distribution in the 2 groups [10]. In P. aeruginosa isolates not susceptible to ceftazidime-avibactam and not susceptible to meropenem, a fosfomycin susceptibility of 85.7% and 75%, respectively, was observed [30]. Much lower susceptibility rates (7%) were observed by Perdigao-Net et al. in Brazil [38].
A review of fosfomycin activity against nonfermenting Gram-negative bacilli collected 19 studies that measured a susceptibility rate in multidrug-resistant P. aeruginosa of 30.2%, with a considerable variety of methods employed and different mean susceptibility rates for each of them [39]: microdilution, 91.1% (mean 58.1%, range 0-100%, SD ±45%); agar, 90% (mean 70%, range 0-100%, SD ±41%); disc diffusion, 56.3% (mean 51%, range 0-100%, SD ±35%) and MIC gradient test, 11.1% (mean 28.6%, range 0-93.3%, SD ±35%). Given that agar dilution is the reference method for fosfomycin susceptibility testing, our group has proposed an alternative procedure for implementing the diffusion methods, in which the 0.5 McFarland inoculum is diluted 100 times, which significantly improves the correlation with the reference method [29].
Methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus. While a number of studies have observed good fosfomycin activity in methicillin-susceptible S. aureus (MSSA) and in MRSA, with susceptibility rates of up to 99.2% [30, 40, 41], other studies have reported susceptibility readings of less than 50% in MRSA [42], with differences according to the clonal lineage [43]. Likewise, data on fosfomycin activity against Enterococcus vary according to the study. Thus, more than 80% of vancomycin-resistant Enterococcus faecium have preserved fosfomycin susceptibility [44] versus 30% reported in other studies [45].
ANTIMICROBIAL ACTIVITY IN BIOFILMS
Fosfomycin has shown a high rate of penetration in mature biofilms of P. aeruginosa [46]. Likewise, the anaerobic environment present in the interior of these structures favors the expression of the fosfomycin transporter GlpT. A larger quantity of antibiotic therefore penetrates the interior of the bacteria [26]. There are several in vitro and animal model studies that have shown that fosfomycin combined with various antibiotics has the capacity to eradicate or reduce the biofilms of Gram-positive and Gram-negative bacteria. An example of this is the published studies on MRSA biofilms, in which good results have been obtained with fosfomycin combined with vancomycin [47], rifampicin [48], linezolid, minocycline, vancomycin or teicoplanin [49, 50] or with Enterococcus faecalis in monotherapy and in combination with gentamicin [8]. Likewise, synergy has been demonstrated against P. aeruginosa biofilms in combination with tobramycin, enhancing the penetration of this antibiotic to the cell’s interior [51-53].
FOSFOMYCIN ACTIVITY IN COMBINATION WITH OTHER ANTIMICROBIALS
One of the main problems presented by fosfomycin is the high rate at which resistant mutants emerge during the treatment, which, coupled with the lack of crossed resistances and antagonism with other families, means that fosfomycin is administered in most cases in combination with other antimicrobials. There are numerous in vitro studies that have sought to elucidate the effect of the combinations, against both Gram-negative bacilli and Gram-positive microorganisms.
Combinations against Gram-negative bacteria. Fosfomycin is one of the few alternatives (along with aminoglycosides and colistin) that present MICs within the susceptibility range in CPE. Therefore, the activity of the combinations of these antibiotics has been studied. The effect of the combination of fosfomycin and amikacin or colistin against KPC-2-producing K. pneumoniae was determined in a PK-PD model. A lower resistance rate was observed with the use of the fosfomycin-colistin combination than when colistin was employed in monotherapy [54]. This synergistic effect appears to be due to the fact that colistin facilitates the entry of fosfomycin into the bacteria’s interior, thereby increasing fosfomycin’s concentration in the active site. The effect of the fosfomycin-colistin combination appears to depend on the type of strain studied. Thus, the bactericidal effect was not boosted with the combination in colistin-heteroresistant or colistin-resistant strains [55, 56]. In vitro synergy with imipenem, ertapenem and tigecycline was also demonstrated in time-kill curves and checkerboard models in KPC-producing K. pneumoniae [57].
An interesting combination is the one with phosphonoformic acid (foscarnet) derivatives, an antiviral drug that also possess activity as inhibitor of the FosA enzyme which hydrolyze fosfomycin in Gram-negative microorganisms. Fosfomycin activity is thereby increased in bacteria such as P. aeruginosa, K. pneumoniae and Enterobacter cloacae, which have this enzyme encoded in their chromosome [58] .
The combination of temocillin and fosfomycin has also been shown to be synergistic in vitro and in vivo and prevents the emergence of resistant mutants when used against E. coli with KPC carbapenemases and even OXA-48, which confer resistance to temocillin [59].
In P. aeruginosa, there is an alternative pathway bound to the recycling of the peptidoglycan, which prevents its de novo synthesis. This fact could explain the lower fosfomycin activity in this microorganism. Peptidoglycan recycling inhibitors have been shown to increase fosfomycin susceptibility [60].
In terms of beta-lactam antibiotics, ceftolozane-tazobactam in combination with fosfomycin has demonstrated in vitro synergy, which could be useful for treating infections caused by multidrug-resistant P. aeruginosa [61]. Likewise, the combination with meropenem in a model of hollow-fiber infection increased the bactericidal effect and prevented the emergence of resistant mutants [62].
Combinations against Gram-positive bacteria. The combination of fosfomycin and daptomycin is one of the most studied strategies against Gram-positive bacteria. In a recent review that collected cases of infection caused by Gram-positive microorganisms treated with different fosfomycin combinations and the results of time-kill curves in MRSA and MSSA, the combination with daptomycin was shown to be the most effective [63]. An animal model of MRSA endocarditis showed the bactericidal and synergistic action of this combination, where the proportion of sterile vegetations and the bacterial inoculum in the vegetations were also improved [64]. Likewise, daptomycin combined with fosfomycin showed synergy in vitro and in a PK-PD model in VRE [65]. In MRSA with intermediate susceptibility to glycopeptides, the combination with imipenem or ceftriaxone was synergistic in an animal model and in time-kill curves [66]. Using time-kill curves and checkerboard assays, in vitro synergy has also been demonstrated against MRSA for fosfomycin combined with linezolid [67], rifampicin, tigecycline [68], acid fusidic [69] or quinupristin-dalfopristin [70].
CONCLUSIONS
The microbiological understanding and clinical use of fosfomycin has increased in recent years. However, various aspects still need to be defined, such as those related to its in vitro susceptibility study and the PK-PD parameters that best predict its clinical efficacy. Despite this need and the introduction of new antimicrobials with activity against multidrug-resistant microorganisms, the empiric and targeted use of fosfomycin (alone or in combination with other antimicrobials) has increased. It is therefore essential to have fosfomycin in countries with the highest resistance rates, as supported by surveillance studies on resistance and the clinical guidelines.
CONFLICTS OF INTEREST
RC has participated in training activities organized by ERN, Pfizer and MDS.
ACKNOWLEDGMENTS
MDA is funded by the iABC (reference 115721-2) of the Innovative Medicines Initiative of the European Commission.
REFERENCES
- 1.Falagas ME, Vouloumanou EK, Samonis G, Vardakas KZ. Fosfomycin. Clin Microbiol Rev 2016;29:321–47. doi: 10.1128/CMR.00068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Candel FJ, Cantón R. Current approach to fosfomycin: From bench to bedside. Enferm Infecc Microbiol Clin 2018. doi: 10.1016/j.ijggc.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Falagas ME, Giannopoulou KP, Kokolakis GN, Rafailidis PI. Fosfomycin: use beyond urinary tract and gastrointestinal infections. Clin Infect Dis 2008;46:1069–77. doi: 10.1086/527442. [DOI] [PubMed] [Google Scholar]
- 4.Roussos N, Karageorgopoulos DE, Samonis G, Falagas ME. Clinical significance of the pharmacokinetic and pharmacodynamic characteristics of fosfomycin for the treatment of patients with systemic infections. Int J Antimicrob Agents 2009;34:506–15. doi: 10.1016/j.ijantimicag.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 5.Dijkmans AC, Zacarías NVO, Burggraaf J, Mouton JW, Wilms E, van Nieuwkoop C, et al. Fosfomycin: pharmacological, clinical and future perspectives. Antibiotics 2017;6:24. doi: 10.3390/antibiotics6040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lepak AJ, Zhao M, VanScoy B, Taylor DS, Ellis-Grosse E, Ambrose PG, et al. In Vivo pharmacokinetics and pharmacodynamics of ZTI-01 (fosfomycin for injection) in the neutropenic murine thigh infection model against Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa. Antimicrob Agents Chemother 2017;61:1–11. doi: 10.1128/AAC.00476-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Docobo-Pérez F, Drusano GL, Johnson A, Goodwin J, Whalley S, Ramos-Martín V, et al. Pharmacodynamics of fosfomycin: Insights into clinical use for antimicrobial resistance. Antimicrob Agents Chemother 2015;59:5602–10. doi: 10.1128/AAC.00752-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oliva A, Furustrand Tafin U, Maiolo EM, Jeddari S, Bétrisey B, Trampuza A. Activities of fosfomycin and rifampin on planktonic and adherent Enterococcus faecalis strains in an experimental foreign-body infection model. Antimicrob Agents Chemother 2014;58:1284–93. doi: 10.1128/AAC.02583-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazzei T, Cassetta MI, Fallani S, Arrigucci S, Novelli A. Pharmacokinetic and pharmacodynamic aspects of antimicrobial agents for the treatment of uncomplicated urinary tract infections. Int J Antimicrob Agents 2006;28:35–41. doi: 10.1016/j.ijantimicag.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 10.Walsh CC, McIntosh MP, Peleg AY, Kirkpatrick CM, Bergen PJ. In vitro pharmacodynamics of fosfomycin against clinical isolates of Pseudomonas aeruginosa. J Antimicrob Chemother 2015;70:3042–50. doi: 10.1093/jac/dkv221. [DOI] [PubMed] [Google Scholar]
- 11.Louie A, Maynard M, Duncanson B, Nole J, Vicchiarelli M, Drusano GL. Determination of the dynamically linked indices of fosfomycin for Pseudomonas aeruginosa in the hollow fiber infection model. Antimicrob Agents Chemother 2018; 62:1–9. doi: 10.1128/AAC.02627-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castañeda-García A, Blázquez J, Rodríguez-Rojas A. Molecular mechanisms and clinical impact of acquired and intrinsic fosfomycin resistance. Antibiotics 2013; 2:217–36. doi: 10.3390/antibiotics2020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falagas ME, Athanasaki F, Voulgaris GL, Triarides NA, Vardakas KZ. Resistance to fosfomycin: mechanisms, frequency and clinical consequences. Int J Antimicrob Agents 2018. doi: 10.1103/PhysRevB.83.075123. [DOI] [PubMed] [Google Scholar]
- 14.Yang TY, Lu PL, Tseng SP. Update on fosfomycin-modified genes in Enterobacteralesceae. J Microbiol Immunol Infect 2017. doi: 10.1016/j.jmii.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 15.Nilsson AI, Otto B, Aspevall O, Kahlmeter G, Andersson DI. Biological costs and mechanisms of fosfomycin resistance in Escherichia coli. Antimicrob Agents Chemother 2003; 47:2850–8. doi: 10.1128/AAC.47.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ballestero-Téllez M, Docobo-Pérez F, Portillo-Calderón I, Rodríguez-Martínez JM, Racero L, Ramos-Guelfo MS, et al. Molecular insights into fosfomycin resistance in Escherichia coli. J Antimicrob Chemother 2017;72: 1303–9. doi: 10.1093/jac/dkw573. [DOI] [PubMed] [Google Scholar]
- 17.Lucas AE, Ito R, Mustapha MM, McElheny CL, Mettus RT, Bowler SL, et al. Frequency and mechanisms of spontaneous fosfomycin nonsusceptibility observed upon disk diffusion testing of Escherichia coli. J Clin Microbiol 2018;56:1–7. doi: 10.1128/JCM.01368-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karageorgopoulos DE, Wang R, Yu X-H, Falagas ME. FosfomycIn: evaluation of the published evidence on the emergence of antimicrobial resistance in Gram-negative pathogens. J Antimicrob Chemother 2012;67:255–68. doi: 10.1093/jac/dkr466. [DOI] [PubMed] [Google Scholar]
- 19.VanScoy B, McCauley J, Bhavnani SM, Ellis-Grosse EJ, Ambrose PG. Relationship between fosfomycin exposure and amplification of Escherichia coli subpopulations with reduced susceptibility in a hollow-fiber infection model. Antimicrob Agents Chemother 2016;60:5141–5. doi: 10.1128/AAC.00355-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grabein B, Graninger W, Rodríguez Baño J, Dinh A, Liesenfeld DB. Intravenous fosfomycin—back to the future. Systematic review and meta-analysis of the clinical literature. Clin Microbiol Infect 2017;23:363–72. doi: 10.1016/j.cmi.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Díez-Aguilar M, Morosini MI, Tedim AP, Rodríguez I, Aktaş Z, Cantón R. Antimicrobial activity of fosfomycin-tobramycin combination against Pseudomonas aeruginosa isolates assessed by time-kill assays and mutant prevention concentrations. Antimicrob Agents Chemother 2015;59. doi: 10.1128/AAC.00822-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, Wang D, Ding Y, Zhang L, Li X. Molecular epidemiology of plasmid-mediated fosfomycin resistance gene determinants in Klebsiella pneumoniae Carbapenemase-Producing Klebsiella pneumoniae Isolates in China. Microb Drug Resist 2018:mdr.2018.0137. doi: 10.1089/mdr.2018.0137. [DOI] [PubMed] [Google Scholar]
- 23.Benzerara Y, Gallah S, Hommeril B, Genel N, Decré D, Rottman M, et al. Emergence of plasmid-mediated fosfomycin-resistance genes among Escherichia coli isolates, France. Emerg Infect Dis 2017;23:1564–7. doi: 10.3201/eid2309.170560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito R, Pacey MP, Mettus RT, Sluis-Cremer N, Doi Y. Origin of the plasmid-mediated fosfomycin resistance gene fosA3. J Antimicrob Chemother 2018;73:373–6. doi: 10.1093/jac/dkx389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Díez-Aguilar M, Morosini M-I, Del Campo R, García-Castillo M, Zamora J, Cantóna R. In Vitro activity of fosfomycin against a collection of clinical Pseudomonas aeruginosa isolates from 16 Spanish hospitals: Establishing the validity of standard broth microdilution as susceptibility testing method. Antimicrob Agents Chemother 2013;57. doi: 10.1128/AAC.00589-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirakawa H, Kurabayashi K, Tanimoto K, Tomita H. Oxygen limitation enhances the antimicrobial activity of fosfomycin in Pseudomonas aeruginosa following overexpression of glpT Which encodes glycerol-3-phosphate/fosfomycin symporter. Front Microbiol 2018;9:1950. doi: 10.3389/fmicb.2018.01950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Camarlinghi G, Parisio EM, Antonelli A, Nardone M, Coppi M, Giani T, et al. Discrepancies in fosfomycin susceptibility testing of KPC-producing Klebsiella pneumoniae with various commercial methods. Diagn Microbiol Infect Dis 2018:2018–20. doi: 10.1016/j.diagmicrobio.2018.07.014. [DOI] [PubMed] [Google Scholar]
- 28.van Mens SP, ten Doesschate T, Kluytmans-van den Bergh MFQ, Mouton JW, Rossen JWA, Verhulst C, et al. Fosfomycin Etest for Enterobacteralesceae: Interobserver and interlaboratory agreement. Int J Antimicrob Agents 2018;52:678–81. doi: 10.1016/j.ijantimicag.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 29.Díez-Aguilar M, Martínez-García L, Cantón R, Morosini MI. Is a new standard needed for diffusion methods for in vitro susceptibility testing of fosfomycin against Pseudomonas aeruginosa? Antimicrob Agents Chemother 2016;60. doi: 10.1128/AAC.02237-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flamm RK, Rhomberg PR, Watters A, Sweeney K, Ellis-Grosse EJ, Shortridge D. Activity of fosfomycin when tested against US contemporary bacterial isolates. Diagn Microbiol Infect Dis 2018. doi: 10.1016/j.diagmicrobio.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 31.Falagas ME, Kastoris AC, Karageorgopoulos DE, Rafailidis PI. Fosfomycin for the treatment of infections caused by multidrug-resistant non-fermenting Gram-negative bacilli: a systematic review of microbiological, animal and clinical studies. Int J Antimicrob Agents 2009;34:111–20. doi: 10.1016/j.ijantimicag.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 32.Mezzatesta ML, La Rosa G, Maugeri G, Zingali T, Caio C, Novelli A, et al. In vitro activity of fosfomycin trometamol and other oral anti-biotics against multidrug-resistant uropathogens. Int J Antimicrob Agents 2017;49:763–6. doi: 10.1016/j.ijantimicag.2017.01.020. [DOI] [PubMed] [Google Scholar]
- 33.Falagas ME, Kastoris AC, Kapaskelis AM, Karageorgopoulos DE. Fosfomycin for the treatment of multidrug-resistant, including extended-spectrum β-lactamase producing, Enterobacteralesceae infections: a systematic review. Lancet Infect Dis 2010;10:43–50. doi: 10.1016/S1473-3099(09)70325-1. [DOI] [PubMed] [Google Scholar]
- 34.Bi W, Li B, Song J, Hong Y, Zhang X, Liu H, et al. Antimicrobial susceptibility and mechanisms of fosfomycin resistance in extended-spectrum β-lactamase-producing Escherichia coli strains from urinary tract infections in Wenzhou, China. Int J Antimicrob Agents 2017;50:29–34. doi: 10.1016/j.ijantimicag.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 35.Bouxom H, Fournier D, Bouiller K, Hocquet D, Bertrand X. Which non-carbapenem antibiotics are active against extended-spectrum β-lactamase-producing Enterobacteralesceae? Int J Antimicrob Agents 2018;52:100–3. doi: 10.1016/j.ijantimicag.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 36.Oteo J, Bautista V, Lara N, Cuevas O, Arroyo M, Fernández S, et al. Parallel increase in community use of fosfomycin and resistance to fosfomycin in extended-spectrum β-lactamase (ESBL)-producing Escherichia coli. J Antimicrob Chemother 2010;65:2459–63. doi: 10.1093/jac/dkq346. [DOI] [PubMed] [Google Scholar]
- 37.Fransen F, Hermans K, Melchers MJB, Lagarde CCM, Meletiadis J, Mouton JW. Pharmacodynamics of fosfomycin against ESBL- and/ or carbapenemase-producing Enterobacteralesceae. J Antimicrob Chemother 2017;72:3374–81. doi: 10.1093/jac/dkx328. [DOI] [PubMed] [Google Scholar]
- 38.Perdigão-Neto L V., Oliveira MS, Rizek CF, Carrilho CMDM, Costa SF, Levin AS. Susceptibility of multiresistant gram-negative bacteria to fosfomycin and performance of different susceptibility testing methods. Antimicrob Agents Chemother 2014;58:1763–7. doi: 10.1128/AAC.02048-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bilal H, Peleg AY, McIntosh MP, Styles IK, Hirsch EB, Landersdorfer CB, et al. comment on: Elucidation of the pharmacokinetic/pharmacodynamic determinants of fosfomycin activity against Pseudomonas aeruginosa using a dynamic in vitro model. J Antimicrob Chemother 2018;73:1570–8. doi: 10.1093/jac/dky045. [DOI] [PubMed] [Google Scholar]
- 40.Falagas ME, Maraki S, Karageorgopoulos DE, Kastoris AC, Kapaskelis A, Samonis G. Antimicrobial susceptibility of Gram-positive non-urinary isolates to fosfomycin. Int J Antimicrob Agents 2010;35:497–9. doi: 10.1016/j.ijantimicag.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 41.Lu C-L, Liu C-Y, Huang Y-T, Liao C-H, Teng L-J, Turnidge JD, et al. Antimicrobial susceptibilities of commonly encountered bacterial isolates to fosfomycin determined by agar dilution and disk diffusion methods. Antimicrob Agents Chemother 2011;55:4295–301. doi: 10.1128/AAC.00349-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.López Díaz MC, Ríos E, Rodríguez-Avial I, Simaluiza RJ, Picazo JJ, Culebras E. In-vitro activity of several antimicrobial agents against methicillin-resistant Staphylococcus aureus (MRSA) isolates expressing aminoglycoside-modifying enzymes: potency of plazomicin alone and in combination with other agents. Int J Antimicrob Agents 2017;50:191–6. doi: 10.1016/j.ijantimicag.2017.01.039. [DOI] [PubMed] [Google Scholar]
- 43.Wu D, Chen Y, Sun L, Qu T, Wang H, Yu Y. Prevalence of fosfomycin resistance in methicillin-resistant Staphylococcus aureus isolated from patients in a university hospital in China, 2013-2015. Jpn J Infect Dis 2018:312–4. doi: 10.7883/yoken.JJID.2018.013. [DOI] [PubMed] [Google Scholar]
- 44.Guo Y, Tomich AD, McElheny CL, Cooper VS, Tait-Kamradt A, Wang M, et al. High-level fosfomycin resistance in vancomycin-resistant Enterococcus faecium. Emerg Infect Dis 2017;23:1902–4. doi: 10.3201/eid2311.171130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang HJ, Chen CC, Zhang CC, Su BA, Li CM, Weng TC, et al. In vitro efficacy of fosfomycin-based combinations against clinical vancomycin-resistant Enterococcus isolates. Diagn Microbiol Infect Dis 2013;77:254–7. doi: 10.1016/j.diagmicrobio.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 46.Rodríguez-Martínez J, Ballesta S, Pascual A. Activity and penetration of fosfomycin, ciprofloxacin, amoxicillin/clavulanic acid and co-trimoxazole in Escherichia coli and Pseudomonas aeruginosa biofilm. Int J Antimicrob Agents. 2007;30(4):366-8. Doi: 10.1016/j.ijantimicag.2007.05.005 [DOI] [PubMed] [Google Scholar]
- 47.Shi J, Mao NF, Wang L, Zhang HB, Chen Q, Liu H, et al. Efficacy of combined vancomycin and fosfomycin against methicillin-resistant Staphylococcus aureus in biofilms in vivo. PLoS One 2014;9:1–14. doi: 10.1371/journal.pone.0113133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mihailescu R, Tafin UF, Corvec S, Oliva A, Betrisey B, Borens O, et al. High activity of fosfomycin and rifampin against methicillin-resistant Staphylococcus aureus biofilm in vitro and in an experimental foreign-body infection model. Antimicrob Agents Chemother 2014;58:2547–53. doi: 10.1128/AAC.02420-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang HJ, Chen CC, Cheng KC, Toh HS, Su BA, Chiang SR, et al. In vitro efficacy of fosfomycin-containing regimens against methicillin-resistant Staphylococcus aureus in biofilms. J Antimicrob Chemother 2012;67:944–50. doi: 10.1093/jac/dkr535. [DOI] [PubMed] [Google Scholar]
- 50.Chai D, Liu X, Wang R, Bai Y, Cai Y. Efficacy of linezolid and fosfomycin in catheter-related biofilm infection caused by methicillin-resistant Staphylococcus aureus. Biomed Res Int 2016;2016. doi: 10.1155/2016/6413982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Díez-Aguilar M, Morosini MI, Köksal E, Oliver A, Ekkelenkamp M, Cantón R. Use of Calgary and microfluidic BioFlux systems to test the activity of fosfomycin and tobramycin alone and in combination against cystic fibrosis Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 2018;62. doi: 10.1128/AAC.01650-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anderson GG, Kenney TF, Macleod DL, Henig NR, O’Toole G a. Eradication of Pseudomonas aeruginosa biofilms on cultured airway cells by a fosfomycin/tobramycin antibiotic combination. Pathog Dis 2013;67:39–45. doi: 10.1111/2049-632X.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Field TR, White A, Elborn JS, Tunney MM. Effect of oxygen limitation on the in vitro antimicrobial susceptibility of clinical isolates of Pseudomonas aeruginosa grown planktonically and as biofilms. Eur J Clin Microbiol Infect Dis 2005;24:677–87. doi: 10.1007/s10096-005-0031-9. [DOI] [PubMed] [Google Scholar]
- 54.Yu W, Zhou K, Guo L, Ji J, Niu T, Xiao T, et al. In vitro pharmacokinetics/pharmacodynamics evaluation of fosfomycin combined with amikacin or colistin against KPC2-producing Klebsiella pneumoniae. Front Cell Infect Microbiol 2017;7. doi: 10.3389/fcimb.2017.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J, He JT, Bai Y, Wang R, Cai Y. Synergistic activity of colistin/fosfomycin combination against carbapenemase-producing Klebsiella pneumoniae in an in vitro pharmacokinetic/pharmacodynamic model. Biomed Res Int 2018;2018. doi: 10.1155/2018/5720417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao M, Bulman ZP, Lenhard JR, Satlin MJ, Kreiswirth BN, Walsh TJ, et al. Pharmacodynamics of colistin and fosfomycin: A “treasure trove” combination combats KPC-producing Klebsiella pneumoniae. J Antimicrob Chemother 2017;72:1985–90. doi: 10.1093/jac/dkx070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu W, Shen P, Bao Z, Zhou K, Zheng B, Ji J, et al. In vitro antibacterial activity of fosfomycin combined with other antimicrobials against KPC-producing Klebsiella pneumoniae. Int J Antimicrob Agents 2017;50:237–41. doi: 10.1016/j.ijantimicag.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 58.Ito R, Tomich AD, McElheny CL, Mettus RT, Sluis-Cremer N, Doi Y. Inhibition of fosfomycin resistance protein fosa by phosphonoformate (foscarnet) in multidrug-resistant gram-negative pathogens Ryota. Antimicrob Agents Chemother 2017;61. doi: 10.1128/AAC.01424-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berleur M, Guérin F, Massias L, Chau F, Poujade J, Cattoir V, et al. Activity of fosfomycin alone or combined with temocillin in vitro and in a murine model of peritonitis due to KPC-3- or OXA-48-producing Escherichia coli. J Antimicrob Chemother 2018. doi: 10.1093/jac/dky283. [DOI] [PubMed] [Google Scholar]
- 60.Hamou-Segarra M, Zamorano L, Vadlamani G, Chu M, Sanchez-Diener I, Juan C, et al. Synergistic activity of fosfomycin, β-lactams and peptidoglycan recycling inhibition against Pseudomonas aeruginosa. J Antimicrob Chemother 2017;72:448–54. doi: 10.1093/jac/dkw456. [DOI] [PubMed] [Google Scholar]
- 61.Monogue ML, Nicolau DP. Antibacterial activity of ceftolozane/tazobactam alone and in combination with other antimicrobial agents against MDR Pseudomonas aeruginosa. J Antimicrob Chemother 2018;73:942–52. doi: 10.1093/jac/dkx483. [DOI] [PubMed] [Google Scholar]
- 62.Drusano GL, Neely M, Yamada W, Duncanson B, Brown D, Maynard M, et al. The combination of fosfomycin plus meropenem is synergistic for Pseudomonas aeruginosa PA01 in a Hollow Fiber Infection Model (HFIM). Antimicrob Agents Chemother 2018;7060:1–32. doi: 10.1128/AAC.00183-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Coronado-Álvarez NM, Parra D, Parra-Ruiz J. Clinical efficacy of fosfomycin combinations against a variety of gram-positive cocci. Enferm Infecc Microbiol Clin 2018. doi: 10.1016/j.eimc.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 64.García-De-La-Mària C, Gasch O, García-Gonzalez J, Soy D, Shaw E, Ambrosioni J. The combination of daptomycin and fosfomycin has of experimental endocarditis. Antimicrob Agents Chemother 2018;62:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Snyder ADH, Werth BJ, Nonejuie P, McRoberts JP, Pogliano J, Sakoulas G, et al. Fosfomycin enhances the activity of Daptomycin against Vancomycin-Resistant enterococci in an in Vitro pharmacokinetic-pharmacodynamic model. Antimicrob Agents Chemother 2016;60:5716–23. doi: 10.1128/AAC.00687-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Del Río A, García-de-la-Mària C, Entenza JM, Gasch O, Armero Y, Soy D, et al. Fosfomycin plus β-lactams as synergistic bactericidal combinations for experimental endocarditis due to methicillin-resistant and glycopeptide-intermediate Staphylococcus aureus. Antimicrob Agents Chemother 2016;60:478–86. doi: 10.1128/AAC.02139-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu-hong Y, Falagas ME, Dong W, Karageorgopoulos DE, De-feng L, Rui W. In vitro activity of fosfomycin in combination with linezolid against clinical isolates of methicillin-resistant Staphylococcus aureus. J Antibiot (Tokyo) 2014;67:369–71. doi: 10.1038/ja.2014.5. [DOI] [PubMed] [Google Scholar]
- 68.Simonetti O, Morroni G, Ghiselli R, Orlando F, Brenciani A, Xhuvelaj L, et al. In vitro and in vivo activity of fosfomycin alone and in combination with rifampin and tigecycline against Gram-positive cocci isolated from surgical wound infections. J Med Microbiol 2018;67:139–43. doi: 10.1099/jmm.0.000649. [DOI] [PubMed] [Google Scholar]
- 69.Yu X-H, Song X-J, Cai Y, Liang B-B, Lin D-F, Wang R. In vitro activity of two old antibiotics against clinical isolates of methicillin-resistant Staphylococcus aureus. J Antibiot (Tokyo) 2010;63:657–9. doi: 10.1038/ja.2010.105. [DOI] [PubMed] [Google Scholar]
- 70.Duez J-M, Adochitei A, Péchinot A, Siebor E, Sixt N, Neuwirth C. In vitro combinations of five intravenous antibiotics with dalfopristin-quinupristin against Staphylococcus aureus in a 3-Dimensional Model. J Chemother 2013;20:684–9. doi: 10.1179/joc.2008.20.6.684. [DOI] [PubMed] [Google Scholar]