Abstract
Candida auris is an emerging multidrug-resistant yeast with high mortality. We report the sentinel C. auris case on the United States West Coast in a patient who relocated from India. We identified close phylogenetic relatedness to the South Asia clade and ERG11 Y132F and FKS1 S639Y mutations potentially explaining antifungal resistance.
Keywords: Candida auris, emerging infection, antimicrobial resistance, echinocandin resistance, metagenomic
Introduction
Candida auris is an emerging fungal pathogen with high minimum inhibitory concentrations (MICs) for many antifungals. Since identification in 2009, it is increasingly recognized as an important cause of invasive disease and nosocomial outbreaks, with high-associated in-hospital mortality of 40–72%.1 Genomic evaluation of strains from multiple geographic regions suggests simultaneous emergence of distinct geographic clades on three continents, as opposed to dissemination from a single source.1 This observation suggests that environmental factors such as increased antifungal use may have contributed to C. auris emergence.1
In addition to high fluconazole MICs, C. auris isolates also frequently have high MICs for other antifungals, including amphotericin and less frequently echinocandins.1 Despite the alarming frequency of elevated antifungal MICs in C. auris, the underlying mechanisms and alleles associated with this resistance have not been fully characterized. In C. auris as well as in other Candida species, mutations in ERG11 (ergosterol synthetase), FKS1 (1,3 beta-d-glucan synthetase), and FUR1 (uracil phosphoribosyltransferase) have been associated with resistance to fluconazole, echinocandins, and flucytosine, respectively.1,2 Previous studies suggest that mutations in these genes can arise in the setting of systemic antifungal therapy.3
Despite first appearing in the eastern United States in 2013, C. auris had not been detected on the United States West Coast.2 In this study, we report the identification of C. auris in this region, which was unusual in that it did not establish endemicity, and use whole-genome sequencing (WGS) to identify strain origin and evaluate genetic mechanisms of antifungal resistance.
Methods
Case description
An elderly man with metastatic rectal cancer relocated from India to California. He had received chemotherapy and radiation while in India and had so undergone intraabdominal surgeries complicated by sepsis. In the year following his move, he required multiple admissions to the University of California, San Francisco (UCSF) Medical Center for management of his malignancy and for secondary infections with carbapenem-resistant Enterobacteriaceae (CRE), for which he was placed in contact isolation. During his initial multi-month admission, two cultures from his urostomy grew 10,000 colony-forming units of a non-Candida albicans yeast that was not further speciated due to unclear clinical significance. In the course of his care, he was treated with echinocandins with prophylactic intent. Several months after initial admission, he was transitioned to palliative care. Three days before death, a nephrostomy culture returned positive for yeast, which was ultimately speciated as C. auris.
Clinical microbiology and antifungal susceptibility testing
Urine collected from the patient's nephrostomy tube into a sterile container underwent quantitative culture for bacteria and yeast using standard culture methods. Species identification was made using matrix-assisted laser desorption ionization-time of flight mass spectrometry (Brucker Diagnostics), which returned a score value of 2.14, and was additionally confirmed by the California Department of Public Health. Antifungal susceptibility testing was performed using Sensititre YeastOne MIC plates (Trek Diagnostic Systems, Inc.), which has >95% agreement with the Clinical Laboratory Standards Institute reference method.4
Whole-genome sequencing
DNA was extracted from the cultured C. auris isolate using the Zymo ZR Bacterial/Fungal DNA kit. Library preparation was completed with the New England Biolabs NEBNext Ultra II DNA library prep kit and WGS was performed using an Illumina NextSeq. The same DNA also underwent library prep using the Oxford Nanopore Rapid Low Input by PCR Barcoding Kit and WGS on a MinION instrument.
Genome assembly, phylogenetic analyses, and antifungal resistance gene analysis
Raw Illumina sequencing reads were quality filtered using PriceSeqFilter5 and then parsed with Nanopore reads for hybrid de novo assembly using DBG2OLC.6 Reference-based whole-genome phylogenetic analysis constructed from core genome single-nucleotide polymorphisms (SNPs) was carried out with the Northern Arizona SNP Pipeline7 using Pakistan strain B8441 as the reference genome and incorporating genomes from Lockhart et al.1 as well as C. auris isolate 16B15b containing the FKS1 S639P mutation identified by Rhodes et al.3 RAxML-ng8 was used to build maximum likelihood phylogenetic trees as detailed in Supplementary Methods (see Supplementary Tables S2–S4). To identify genetic mutations associated with fluconazole or echinocandin resistance, Illumina sequences were aligned against ERG11 (GenBank KY410388.1) and FKS1 (GenBank XM_018312471.1) using BowTie2.9 Mutations were confirmed by ERG11 and FKS1 PCR followed by Sanger Sequencing (Supplementary Table S1) following previously described methods.10
Results
Assembly and phylogenetic characteristics
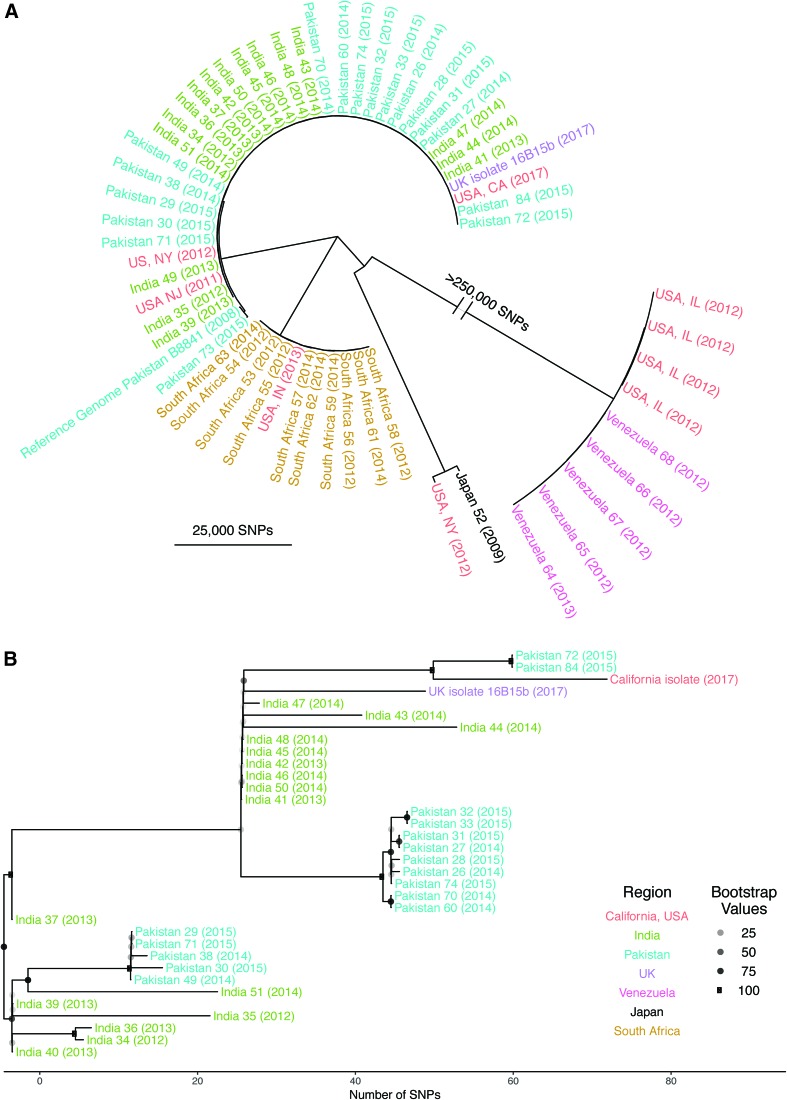
De novo hybrid assembly of Illumina and Oxford Nanopore reads produced a total of 33 contigs spanning 12 Mb, characterized by 44.9% GC content, consistent with prior estimates.1,3 Whole-genome phylogenetic analysis based on a core genome of 208,384 SNPs placed this isolate within the South Asia clade (Fig. 1A). On average, 56 SNPs separated this isolate from others from the South Asia clade (Fig. 1B).
FIG. 1.
(A) Phylogenetic assessment based on core genome single-nucleotide polymorphisms demonstrated the four known geographic clades1 and placed the California isolate within the South Asia clade. (B) Detailed phylogenetic tree describing the South Asia clade, including the California isolate and United Kingdom outbreak isolate16B15b,3 which both harbored the FKS1 S639P mutation. Color images are available online.
Phenotypic and genotypic assessment of antifungal resistance
The California isolate demonstrated low MICs to amphotericin (1 μg/mL), flucytosine (0.5 μg/mL), and voriconazole (0.032 μg/mL). The isolate had an elevated fluconazole MIC of 32 μg/mL. Assessment of this isolate's ERG11 (encoding ergosterol synthetase) allele revealed the well-characterized Y132F substitution in the azole resistance hotspot region.1,3 Unlike most C. auris strains, this California isolate also exhibited a high caspofungin MIC of 8 μg/mL. Interrogation of FKS1 [encoding (1,3)-β-d-glucan synthetase] revealed a S639Y mutation in the echinocandin resistance hotspot 1 region (Supplementary Table S1).3,10
Discussion
C. auris emerges on the West Coast of the United States
In this study, we report the first case of C. auris on the United States West Coast, a region that had no previous reports of the pathogen despite emergence in New York in 2013. The patient's history of health care exposure in India combined with the clustering of his C. auris isolate with the South Asia clade by WGS phylogenetic analysis suggests that he acquired C. auris abroad before hospitalization in California. This finding supports current guidance from the United States Centers for Disease Control and Prevention to speciate all Candida in high-risk patients, including those from regions of high C. auris prevalence, to allow for early implementation of infection control measures.1,11 Following identification of C. auris, enhanced infection control measures were implemented at UCSF, including surface disinfection, a unit-level point prevalence survey, and prospective surveillance. No additional cases of C. auris at our medical center have been identified in over a year. This case represents an unusual interruption in spread and prolonged health care environmental contamination that has been characteristic of detection of health care-associated C. auris. Early implementation of contact precautions for CRE may have contributed to curbing transmission of C. auris in this case.
This isolate had a high fluconazole MIC with an observed ERG11 Y132F mutation.1,3 The California C. auris isolate also demonstrated a high echinocandin MIC, which is observed in <10% of C. auris strains.1 It is possible that this patient's prophylactic treatment with echinocandins could have selected for resistance as observed in this isolate. This C. auris isolate also had a FKS1 hotspot-1 region mutation, which has been associated with echinocandin resistance in multiple other Candida species.3,10 The identified FKS1 S639 substitutions of nonpolar residues (Y, F, P) has also been identified in other C. auris strains with high echinocandin MIC values, suggesting a key role for this amino acid in echinocandin resistance.3,10
Further study is needed to estimate the prevalence and duration of colonization by this emerging pathogen. Future work using WGS is needed to clarify the origins of C. auris, transmission patterns, and mechanisms of resistance to prevent and manage this emerging fungal pathogen of global significance.
Data Availability
Raw sequences are available via BioProject ID PRJNA480539.
Supplementary Material
Acknowledgments
This work was supported by the National Center for Advancing Translational Sciences (grant number TL1 TR002382 to M.H.W.), the National Institute for Allergy and Infectious Disease (UM1AI104681 to M.H.W., P01AI091575 to C.L.), the National Heart, Lung, and Blood Institute (NHLBI K23HL138461-01A1 to C.L.), and the Chan Zuckerberg Biohub (J.L.D.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Funding was provided by NCATS TL1 TR002382, NIAID UM1AI104681(M.H.W.). NHLBI K12HL119997, Nina Ireland Foundation, Marcus Foundation (C.L.). NIAID P01AI091575 and the Chan Zuckerberg Biohub (E.D.C. and J.L.D.).
Disclosure Statement
No competing financial interests exist.
Supplementary Material
References
- 1. Lockhart S.R.,. Etienne K.A, Vallabhaneni S., Farooqi J., Chowdhary A., Govender N.P, Colombo A.L, Calvo B., Cuomo C.A, Desjardins C.A, Berkow E.L, Castanheira M., Magobo R.E, Jabeen K., Asghar R.J, Meis J.F, Jackson B., Chiller T., and Litvintseva A.P. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 64:134–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vallabhaneni S., Kallen A., Tsay S., Chow N., Welsh R., Kerins J., Kemble S.K, Pacilli M., Black S.R, Landon E., Ridgway J., Palmore T.N, Zelzany A., Adams E.H, Quinn M., Chaturvedi S., Greenko J., Fernandez R., Southwick K., Furuya E.Y, Calfee D.P, Hamula C., Patel G., Barrett P., Lafaro P., Berkow E.L, Moulton-Meissner H., Noble-Wang J., Fagan R.P, Jackson B.R, Lockhart S.R, Litvintseva A.P, and Chiller T.M. 2016. Investigation of the first seven reported cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus—United States, May 2013–August 2016. MMWR Morb. Mortal. Wkly. Rep. 65:1234–1237 [DOI] [PubMed] [Google Scholar]
- 3. Rhodes J., Abdolrasouli A., Farrer R.A, Cuomo C.A, Aanensen D.M, Armstrong-James D., Fisher M.C, and Schelenz S. 2018. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg. Microbes Infect. 7:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cuenca-Estrella M., Gomez-Lopez A., Alastruey-Izquierdo A., Bernal-Martinez L., Cuesta I., Buitrago M.J, and Rodriguez-Tudela J.L. 2010. Comparison of the Vitek 2 antifungal susceptibility system with the Clinical and Laboratory Standards Institute (CLSI) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) broth microdilution reference methods and with the Sensititre Ye. J. Clin. Microbiol. 48:1782–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ruby J.G.,. Bellare P., and Derisi J.L. 2013. PRICE: software for the targeted assembly of components of (Meta) genomic sequence data. G3 (Bethesda) 3:865–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ye C., Hill C.M, Wu S., Ruan J., and Ma Z.S. 2016. DBG2OLC: Efficient assembly of large genomes using long erroneous reads of the third generation sequencing technologies. Sci. Rep. 6:31900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roe C., Smith D.E, Williamson C.H.D, Aziz M., Keim P., Hepp C.M, Driebe E.M, Lemmer D., Travis J., Hicks N.D, Schupp J.M, Wagner D.M, Engelthaler D.M, Gillece J.D, Sahl J.W, and Drees K.P. 2016. NASP: an accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb. Genomics 2:e000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kozlov A. 2018. amkozlov/raxml-ng: RAxML-NG v0.6.0 BETA (Version 0.6.0). Zenodo. 10.5281/zenodo.1291478 [DOI]
- 9. Langmead B., and Salzberg S.L. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9:357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chowdhary A., Prakash A., Sharma C., Kordalewska M., Kumar A., Sarma S., Tarai B., Singh A., Upadhyaya G., Upadhyay S., Yadav P., Singh P.K, Khillan V., Sachdeva N., Perlin D.S, and Meis J.F. 2018. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J. Antimicrob. Chemother. 73:891–899 [DOI] [PubMed] [Google Scholar]
- 11. Chowdhary A., Sharma C., and Meis J.F. 2017. Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 13:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequences are available via BioProject ID PRJNA480539.