Preface
The development of mature blood cells from haematopoietic stem cells (HSCs) has long served as a paradigm for stem cell research, with the haematopoietic differentiation tree used widely as a model for maintenance of hierarchically organised tissues. Recent results and new technologies have challenged the demarcations between stem and progenitor populations, the timing of cell fate choices and the contribution of stem and multipotent progenitor cells to steady-state blood maintenance. These evolving views of haematopoiesis have broad implications for our understanding of adult stem cell function, as well as the development of new therapies for malignant and non-malignant haematopoietic diseases.
Introduction
When Ernst Haeckel first used the word stem cell (Stammzelle) in 1868, as a Darwinist he used it to refer to the primordial unicellular organism from which all multicellular life descended 1. This stem cell therefore sat at the root of a branching family tree, incidentally called a stem tree in German (Stammbaum, e.g. a tree that shows where things stem from). Haeckel’s biogenetic law (ontogeny recapitulates phylogeny) shortly thereafter prompted him to use the stem cell term also to describe the fertilised egg. Histopathologists subsequently applied this stem cell concept to normal and leukaemic haematopoiesis, putting forward the concept of a common progenitor of red and white blood cells 2 as well as a common precursor of myeloid and lymphoid leukaemic cells 3. From the very beginning, the stem cell concept has thus been framed into a tree-like model, where multipotent stem cells give rise to their progeny through an ordered series of branching steps.
The first in vivo assay for stem cell function was based on the rescue of lethal irradiation through bone marrow transplantation 4, followed by the first estimation of stem cell numbers by counting haematopoietic colonies in the spleens of transplanted mice (spleen colony forming unit assay, CFU-S). This not only provided an estimate of CFU-S frequency at 1 in 10,000 bone marrow cells 5, but also delivered the first definitive proof for in vivo multipotent progenitor cell function based on tracking cytogenetic abnormalities within individual CFU-S colonies 6. Fluorescence activated cell sorting (FACS) subsequently facilitated the purification of transplantable hematopoietic stem cells (HSCs), with a landmark 1988 publication7 demonstrating the utility of positive and negative selection. HSCs have historically been defined on the basis of two essential properties: self-renewal and multipotency. Operationally this is tested via transplantation experiments. In contrast, progenitors are defined by the absence of extended self-renewal and a restricted lineage differentiation capacity (most often bi- or unilineage), so that they are usually lost within the first 2–3 weeks after transplantation 8.
Characterisation of progenitor populations downstream of the HSC resulted around the year 2000 in a model of the haematopoietic differentiation tree still shown in many textbooks today (Fig. 1A). In this model, the first branch point segregates lymphoid potential from all other lineages (myeloid, erythroid and megakaryocytic), followed by a number of further branching steps on either side of the tree progressing from multi- to bi- and finally unipotent progenitor cells. The subsequent introduction of additional surface markers suggested several modifications of this classical tree, including lymphoid and myeloid fates remaining associated until further down the tree 8–10, early megakaryocyte branching 11,12 as well as subdivision of the multipotent progenitor compartment into distinct subpopulations13,14 (Fig. 1B). Moreover, the picture is further complicated because the HSC pool itself is functionally and molecularly heterogeneous11,12,15–20. These studies are most advanced in the murine system, where we now have what at first glance appears to be a bewildering number of different structures for the haematopoietic tree. While it is likely that all these structures capture true aspects of HSC differentiation, collectively they would be difficult to squeeze into a single, rigid branching tree. New ways of not only thinking about, but also graphically representing the process of HSC differentiation are thus required. Below we will illustrate how new technologies are challenging the classical view of the hematopoietic hierarchy as a highly compartmentalised and stable structure. The emerging picture is one of a collection of heterogeneous populations organised hierarchically, with gradual progression from one to the next, and which remains highly flexible to meet the changing needs of blood demand.
Fig.1: Timeline of hierarchical models of hematopoiesis.
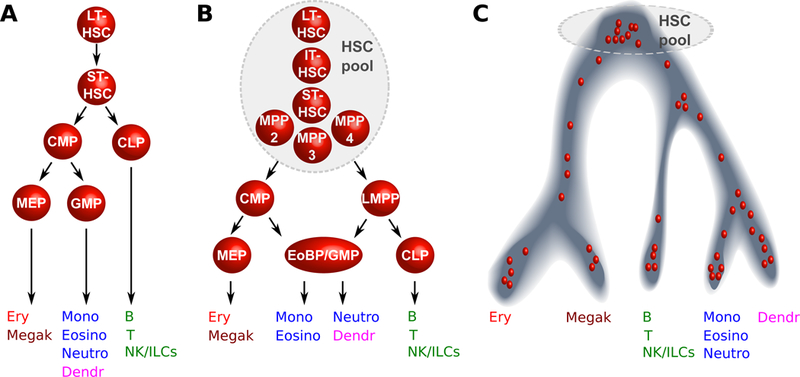
A. Visualisation based on state-of-the-art around the year 2000: HSCs are represented as a homogeneous population downstream of which the first lineage bifurcation separates the myeloid and lymphoid branches via the CMP and CLP populations; B. During the years 2005 to 2015 this visualisation incorporates new findings: the HSC pool is now accepted to be more heterogeneous both in terms of self-renewal (vertical axis) and differentiation properties (horizontal axis), the myeloid and lymphoid branches remain associated further down in the hierarchy via the LMPP population, the GMP compartment is shown to be fairly heterogeneous 141. C. From 2016 onwards, single cell transcriptomics snapshots indicate a continuum of differentiation. Each red dot represents a single cell and its localisation along a differentiation trajectory.
Stem cells and progenitors boundaries
With self-renewal and multipotency at the heart of what defines an HSC, much research has been invested into understanding the underlying cellular and molecular processes.
Defining the HSC state
At the cellular level, switching off self-renewal coincides with turning on lineage programs. It thus seemed plausible that this would also be true at the molecular level, and the concept of multilineage priming was proposed early on as a possible underlying molecular mechanism by which HSCs maintain multipotentiality21 (Box 1). However the advent of genomic technologies 14,22–25 coupled with mouse genetics studies, has demonstrated that the HSC transcriptional programme is defined by a collection of unique metabolic and cellular properties, which are not intuitively linked directly with multipotency. Approximately 70% of all expression changes between HSCs and early progenitors occur independently of lineage choice 24, with a similar dichotomy also at the levels of methylation26–29 and chromatin accessibility30. Correspondingly, HSCs reside in a quiescent 19,20,31,32, autophagy-dependent 33,34 and glycolytic 35,36 state marked by low mitochondrial activity 37,38 and tightly controlled levels of protein synthesis, below those of most other haematopoietic cell types 39. Stem-cell specific stress response and quality control mechanisms allow preservation of the integrity of the HSC compartment after exposure to DNA40,41 and protein damage 42,43 or metabolic stress 33,34. In contrast, progenitors are highly proliferative and metabolically active cells dependent on oxidative metabolism and mitochondrial function.
BOX1: Mechanistic underpinnings of cell fate choice.
Cell fate decisions entail a choice between alternative gene expression programs, executed by transcriptional and epigenetic regulators. Transcription factors (TFs) bind specific sequence motifs within promoter, enhancer and silencer regions. Cell type specific expression is achieved through combinatorial TF interactions, which form key building blocks of wider regulatory networks. TF complexes recruit epigenetic regulators to modulate the activation status of a gene locus, which can be transmitted to subsequent cell generations as so-called “epigenetic memory”. Importantly, a progenitor cell can become primed by opening up regulatory elements associated with genes that drive differentiation down a specific mature lineage143.
Multipotent cells are believed to exhibit multilineage priming, which entails simultaneous, low-level activation of expression programs for alternate lineages. Lineage choice then constitutes one program “winning out” while the alternative program is extinguished. Cross-antagonism between pairs of lineage-determining TFs represents an attractive mechanistic model, which initially focussed on the erythroid regulator Gatal and the myeloid regulator Pu.1. More recent single cell time-lapse imaging however questioned whether an erythroid /myeloid fate choice is indeed driven primarily by Gata1/Pu.1 cross-antagonism144. Evidence for lineage priming and its resolution by TF cross-antagonism has been reported at single cell resolution for other TF pairs, such as the neutrophil/macrophage fate choice controlled by Gfi 1/Irf8 87.
Both instructive and stochastic models have been proposed as mechanisms triggering the upregulation of one lineage program over another. Stochastic here generally refers to random, unequal distribution of molecules following cell division. Instructive models posit that low-level expression of a cytokine receptor is sufficient for a cell to be responsive to external signals, as shown for a number of myeloid cytokines 145. Autoregulation and feed forward loops also play important roles in fate choice decisions. For example, Gata1 positive autoregulation stabilizes erythroid fate 146, Pu.1 positive autoregulation stabilizes myeloid identity 147, and the highly connected triad of Gata2, Tal1/Scl and Fli1 is thought to stabilize the stem/progenitor state 148,149. In feed forward loops, an upstream regulator induces its target directly as well as through an intermediate regulator 150. Feed-forward motifs can filter out transient signals, and when coupled with autoregulation, generate forward momentum.
Importantly, so-called HSC specific characteristics are not absolute: HSCs occasionally divide during homeostasis and get activated in response to stress, and therefore transiently pass through a proliferative state. It is also worth noting that early lymphoid progenitors (mouse LMPP9 or human MLP8) share common transcription (TF) networks with HSCs24, which in the former push towards B cell differentiation24 while in the latter inhibit self-renewal44,45. While the induction of lineage-specific transcriptional programs may occur largely independently of the loss of stem cell characteristics, regulators such as Runx1 can be involved in both 46 suggesting that much remains to be learned about how lineage decisions are coordinated with changes of cellular state.
HSC self-renewal and cell cycle interplay
Whether the variability in HSC outputs is an intrinsic system property, a reflection of stochastic behaviour, environmental influences, technically related variability or a combination of all of these, has been and still is a subject of debate. Nonetheless, in the past 10–15 years, this heterogeneity in behaviour has been significantly formalised, and it is now accepted that HSCs are heterogeneous in terms of durability of engraftment upon transplantation, cell cycle properties and differentiation (Table 1).
Table 1: summary of phenotypic or functionally defined HSC subsets reported to date.
Listed are subtypes of HSCs defined based on combinations of cell surface markers and/or function. A global interpretation of the literature on HSC biology is complicated by the fact that each study usually uses only one of the classification schemes below, so the extent of overlap between HSC subsets remains to be clarified. N.A.: not assessed.
| Species | Name | Cell surface phenotype |
Self-renewal | Cell cycle properties |
Differentiation | Refs |
|---|---|---|---|---|---|---|
| Mouse | LT-HSC |
Lin- Sca1+ cKit+ CD34− CD150+ CD135− CD48− ± EPCR+ ± Rholo |
High | 17,47,48,140 | ||
| IT-HSC |
Lin− Sca1+ cKit+ CD34lo CD135− Rholo CD49bhi |
Intermediate | Short G0 exit |
16 | ||
| ST- HSC/MPP1 |
Lin− Sca1+ cKit+ CD135−CD150− CD48− |
Low | ||||
| α | N.A. | High | N.A. | Ly-deficient | 15,107 | |
| β | N.A. | High | N.A. | Balanced | ||
| γ | N.A. | Intermediate | N.A. | My-deficient | ||
| δ | N.A. | Low | N.A. | My-deficient | ||
| MPP2 |
Lin− Sca1+ cKit+ o CD135−CD150+ CD48+ |
Low | similar | Ly-deficient | 13,14 | |
| MPP3 |
Lin− Sca1+ cKit+ o CD135−CD150+ CD48+ |
Low | Balanced | |||
| MPP4 |
Lin− Sca1+ cKit+ o CD135+CD150− CD48+ |
Low | Ly-biased | |||
| d-HSC | N.A. | Higher | dormant | 19,20,51-53 | ||
| a-HSC | N.A. | Lower | activated | |||
| Human | LT-HSC |
Lin− CD34+ CD38− CD45RA− CD49f+ CD90+ ± Rholo |
High | Long G0 exit |
50,54 | |
| ST- HSC/MPP |
Lin− CD34+ CD38− CD45RA− CD49f+ CD90+ |
Low | Short G0 exit |
|||
| CD34− LT- HSC |
Lin−CD34− CD38−CD93hi |
High | Highly quiescent |
66 |
HSCs first of all differ in the degree to which they self-renew, equating to the number of symmetric divisions that an HSC can make over its lifetime. Operationally, the field now widely accepts that, both in mouse and human, HSC that repopulate in transplantation assays for more than 16 weeks in a primary transplantation and at least in a second round of transplantation are considered Long-Term (LT-) HSCs 17,47–50. |f cells can produce all differentiated cell types and engraft transiently in primary (and in some cases secondary) transplants, they are referred to as Intermediate (IT-) HSCs16, Short-Term (ST-) HSCs or MultiPotent Progenitors (MPPs) 13,14,47 depending on the length and robustness of the graft produced.
Heterogeneity in self-renewal capacity appears to be directly correlated to the time HSCs spend in quiescence. Label retaining studies have demonstrated that the most dormant HSCs (which retain the label over months at steady-state) display the most robust and longest repopulation capacity 19,20,51–53. Two cell cycle parameters are inversely correlated with repopulation capacity: the frequency of division (length of interval between divisions), but also the time a single HSC takes to exit quiescence in vitro 16,54. Of note, the level of pre-existent CDK6 mRNA and protein in quiescent HSCs directly determines the kinetics of quiescence exit54, and may thus serve as a marker of the quiescent state32,54 (less CDK6 corresponding to higher dormancy). Dormancy is also associated with high levels of vitamin A metabolism via retinoic acid signalling32, high levels of p5 732,55,low protein synthesis32, low Myc activity32,56, and may exist as a continuum of quiescent states between the most dormant HSCs and their activated counterparts 32. While dormant HSC forced into activation by stress signals can return to dormancy19,51,Bernitz et al. estimated that 4 divisions in adulthood are sufficient for irreversible loss of self-renewal 53.
A number of label-retaining assays have been developed to allow isolation of HSCs based on their division history19,20,32,51–53,57. One major limitation of all label-retaining studies to date is that the rate of symmetric divisions is inferred or indirectly measured at the bulk level. However, direct experimental measurement of asymmetric versus symmetric label-diluting divisions will be required to understand the dynamics at play within the HSC compartment. Novel integrative tools, most likely at the single cell level, will have to be developed to address this challenge. Nonetheless, it is already apparent that distinct cell cycle properties within the HSC pool are intimately linked to HSC function. Coupled with a wide range in division frequencies (depending on the HSC subset, from once a month to twice a year in mouse19,51,53,58), this means that there will be substantial variation in the contribution of distinct HSC subsets to blood formation.
Heterogeneity in HSC lineage output
The capacity to give rise to all differentiated blood cell types is a fundamental aspect of what constitutes an HSC. It is however now accepted that there are distinct differentiation behaviours within the HSC and MPP13,14 compartments (Table 1, Fig. 1B). Using limiting dilution analysis and single cell transplantation, the Müller-Sieburg and Eaves groups described HSCs that differ in their relative myeloid and lymphoid output 15,59. More recently the Jacobsen and Nakauchi groups identified HSCs that predominantly differentiate towards megakaryocytes and platelets (platelet- biased) 11,12. There are however limitations to single cell transplantation, in particular that very low contributions to certain lineages may be missed. Moreover, a cell with unilineage readout may have the potential to give rise to other lineages in a durable fashion in other conditions, and both platelet-biased HSC and long-lived platelet progenitors 11,60,61 may coexist and be difficult to distinguish.
Importantly, HSC heterogeneity is not simply stochastic as it can be propagated through serial transplantation15,18,62,63, indicating intrinsic programming, the molecular basis for which however remains unclear. As discussed below, these findings have important conceptual implications, because they question at what cellular stage lineage choices occur. Recent evidence suggests that the overall picture may even be more complex, with distinct metabolic needs 64 and clonal expansion capacity of HSCs. There are examples where HSCs with high clonal expansion capacity generate subsets with lower output 65, but also cases in which HSCs with very modest clonal expansion in the first transplantation, generate the most robust grafts upon serial transplantation 66,67.
Rethinking blood lineages relationships
A number of recent studies at the single cell level have questioned the routes by which lineage differentiation occurs.
Single cell assays to study potential
Cell fate decisions (Box 1) are executed at the level of individual single cells. To understand their regulation, it is therefore imperative that both the biological assays testing their cellular function as well as the biochemical assays examining their molecular profiles are performed at single cell resolution. While most advanced in the mouse, in vivo transplantation assays have been and remain fundamental for our understanding of HSC biology, as the only assay that can test for HSC self-renewal. Because of the suboptimal support of human cells from the mouse microenvironment, xenotransplants cannot robustly read-out all possible differentiation routes, particularly not at single cell level. The last 10 years have therefore seen a collective effort in defining the lineage potential of single human progenitor cells by using highly defined in vitro models that can support the differentiation of most mature blood cell types. Work from the Dick and Vyas groups, together with studies in mice from the Jacobsen group described that the first restriction in lineage potential does not segregate lymphoid and myeloid potential as postulated by the CMP-CLP model (Fig. 1A), but that these potentials remain coupled in the LMPP9,10 and MLP8 compartments (Fig. 1B). Many other subpopulations, most with either bi- or unilineage capacity, have since been found both in the lympho-myeloid branch 68 and the myelo-erythroid-megakaryocytic branch 69–73. Altogether it seems that few single cells read-out as multipotential within the progenitor compartment. There are however some caveats with such interpretations, because strong instructive signals provided by the in vitro cultures, or potentially high stress levels that HSC are exposed to during single cell transplants may promote unilineage output. Moreover, a given cell may be bipotential based on its molecular state, but if it makes a lineage choice before dividing, it will read out as unipotent in functional assays. Nonetheless the evidence to date suggests that lineage choice occurs earlier than previously thought, and as recently shown for dendritic cells 74, likely already within the phenotypic HSC populations.
Single-cell transcriptional landscapes
Single cell expression analysis of RNA was first reported over 25 years ago75, but remained low throughput both in number of genes and cells until microfluidic approaches were introduced. These quickly prompted studies reporting the expression of dozens of genes in hundreds of single HSCs and progenitor cells (HSPCs)76, and provided new insights into core regulatory circuits 77, novel progenitor populations 70,71, cellular hierarchies in normal and transformed haematopoiesis 78, dissociation between self-renewal potential and activation of lineage programs 79, and the molecular overlap between HSC populations purified with four different cell sorting strategies 80. However, it is not practical to assay more than 200 genes per single cell with PCR based methods, and these genes need to be predefined thus limiting the scope for novel discoveries. A real breakthrough was therefore provided by technical innovations, which now make it possible to perform transcriptome-wide RNA-Seq in thousands of single cells.
Following a landmark paper reporting the transcriptomes for more than 2600 murine single myelo-erythroid progenitor cells 81, a subsequent report of 1600 transcriptomes ranging from true long-term HSCs to progenitors of all major lineages focused on generic stem cell functions such as metabolic and cell cycle status82. A number of algorithms have been developed based on the idea that single cell transcriptomes represent snapshots of single cells as they traverse differentiation landscapes (Fig. 1C). A recent study on human bone marrow haematopoiesis comprehensively sampled the HSPC compartment88. Computational predictions suggestive of early lineage restriction were underpinned by in vitro single cell culture assays, which lead the authors to propose a model whereby acquisition of lineage-specific fates is a continuous process, and unilineage-restricted cells emerge directly from a continuum of low-primed undifferentiated haematopoietic stem and progenitor cells, without any major transition through multi- and bipotent stages. This is supported by other studies 69,81,85, which highlighted the abundance of unipotent progenitors within compartments that, at the population level, are multipotent.
There are however caveats with this new model and its heavy reliance on scRNA-Seq. A major conundrum here is that surface marker combinations can readily split the HSPC compartment into functionally distinct subpopulations, including within the space proposed to be a continuum of low-primed cells when analysed by scRNA-Seq. One possible explanation is that there is a decoupling between steady state mRNA and protein expression levels. The counter argument here is that multi-omics analysis of highly purified bulk HSPC populations has shown good concordance between mRNA and protein for the majority of genes14. It is possible also that functional heterogeneity of HSCs is primarily determined at the epigenetic level. Future measurements of chromatin accessibility and histone marks ideally at single cell resolution may reveal such mechanisms. A third possible explanation is that even though purifying cells based on protein markers followed by functional assays suggests clean splits into distinct cell types with different biological functions, the changes in biological functionality are in reality much more gradual, where there is no binary biological difference between for example LT-HSCs and MPPs, but instead any individual cell sits somewhere along a continuous spectrum. A fourth possible explanation is that current scRNA-Seq methods are not effective at distinguishing between closely related cell types, because many of the shared biological processes (e.g. cell cycle, metabolism, motility) generate substantial heterogeneity, which may exceed the number of differentially expressed genes between closely related stages of haematopoietic maturation. The degree to which early haematopoiesis is characterised by a continuum versus distinct populations therefore remains a question requiring further investigation.
New representations of haematopoiesis
An immediate challenge for the field is how all the recent findings can be reconciled into new graphical models describing the hierarchical organisation of haematopoiesis. It seems clear that a tree where a circle depicts each successive restriction in potential and each circle is connected to a few others by arrows is an over-simplification. First, the circle is intuitively viewed as a homogeneous set of cells with specific characteristics, a vision incompatible with the large degree of heterogeneity observed experimentally. Second, these trees indicate a restricted set of possible transitions between circles, which likely underestimates the possible differentiation journeys in vivo. Because cell surface markers can highly enrich for particular behaviours (differentiation, self-renewal or proliferative output), a model in which all lineages branch out directly from the HSC compartment also seems somewhat unrealistic (Fig. 1C). We thus propose here an alternative visualisation (Fig. 2A), in which trajectories of differentiation are mapped over the areas that have long been represented as circles, highlighting both the diversity in possible routes and the prevalence of early lineage choice. In addition, it is important to remember that one HSC will produce a very large number of progeny, which increases exponentially with each division, an element so far ignored in graphical representations of hematopoiesis (including Fig. 2A). Divisional histories are difficult to measure, and are likely to be heterogeneous, but should nevertheless be incorporated in future experimental and computational analyses, which could result in new graphical representations of the blood system (Fig. 2B).
Fig.2: Trajectory based visualizations of the hematopoietic hierarchy.
A. 2D visualisation of early hematopoiesis. Continuous lines: trajectories of differentiation for different types of single cells present in the phenotypic HSC compartment (grey shaded area). Along these trajectories, cells and their progeny pass through progenitor compartments commonly defined by specific combinations of cell surface markers (shaded areas). Horizontal lines represent snapshots of the lineage potential of the cells present in each phenotypic compartment (single colour circles: unilineage cells, 2 colours circles: bi-lineage cells, 3 colours circles: tri-lineage cells, black circles: multipotent cells). In most progenitor compartments the number of unilineage cells outnumbers that of bi- or tri-lineage ones. The figure illustrates differentiation trajectories reported in the literature to date, but their proportions may not reflect the in vivo situation. B. 3D visualisation of the progeny of one single HSC. Red, blue and green respectively represent the erythroid, myeloid and lymphoid lineage. Cell history, division and progenitor expansion should all be considered when modelling the differentiation journey of one HSC and all its progeny. In an adult human, there are an estimated 3000–10000 HSCs, which most likely divide only from once every 3 months to once every 3 years 58. Humans produce an estimated 1.4×1014 mature blood cells/year142. The amplification from a few thousand HSCs therefore is staggering and must include a strong contribution from a transient-amplifying compartment. Also, because there are many more terminally differentiated erythroid cells than myeloid cells, and even less lymphoid cells, all with different turn-over rates, the flux into each compartment must be highly regulated.
Making blood at steady state and under stress
In addition to defining the routes of lineage differentiation, another important question is to understand the quantitative contributions of HSCs and progenitors to daily and emergency haematopoiesis.
Studying unperturbed haematopoiesis
While the haematopoietic differentiation tree is widely used as a paradigm for how a hierarchically-organized tissue is maintained, it is important to remember that this tree was largely derived from experiments that measure cell potential in colony or transplantation assays, rather than cell fate during steady state differentiation. However, if a single cell gives rise to two lineages in a colony assay, this does not prove that the same cell, when left alone in an unperturbed bone marrow environment, would have done the same. To understand the dynamics of blood formation, cells can be individually tagged (by retro- or lentiviral insertions, barcodes etc), and transplanted into recipient mice to measure the contribution of each clone over time. Progressively more sensitive methods have been used in mouse, primates and humans 89–93, collectively supporting the lineage biases and/or restrictions observed in single cell transplants. Importantly, only a limited number of HSC produce the vast majority of the differentiated cells in a transplantation setting, consistent with studies of the divisional history of HSC 19,20,51–53,57, which demonstrate that only the very rare most dormant HSC can provide life-long reconstitution after transplant.
Substantial excitement was raised by new technologies to assess the lineage output of individual stem and progenitor cells in unperturbed haematopoiesis 94. Doxycycline induced mobilization of a sleeping beauty transposon in stem/progenitor cells was employed to generate unique integration events, which serve as barcodes then tracked by sequencing. Consequently, comparing barcodes in mature lineages following a pulse-chase label allows reconstruction of single cell behaviours in native, unperturbed haematopoiesis. In contrast to transplantation approaches, analysis of unperturbed haematopoiesis suggested that (i) MPPs contribute predominantly to the myeloid lineage during steady state, and (ii) cells functioning as HSCs in transplantation do not play a significant role in steady state haematopoiesis, which instead seems to be driven almost entirely by cells within the MPP compartment.
An alternative genetic fate mapping system based on Cre-loxP induced recombination of a transgenic barcode cassette recently achieved temporally-controlled barcode induction in single cells, and demonstrated that when HSCs are labelled at the fetal liver stage, their descendants in the adult will mostly contribute to multiple lineages 95. However, megakaryocytic fate was not analysed, and when the analysis was repeated in adult bone marrow, few barcodes were detected in HSCs as well as mature progeny. Given that each fetal liver HSC divides and therefore give rises to multiple HSCs in the adult, conclusions about the lineage contribution of individual adult HSCs therefore remained preliminary. Another study from the Camargo group96 addressed this issue more comprehensively by carrying out a 30-week pulse-chase experiment in adult mice with the sleeping beauty barcode system. 133 barcodes were detected in HSCs and at least one of four mature lineages (megakaryocyte, erythroid, granulocyte, B-cell). Interestingly, more than half of these 133 HSC barcodes were present only in megakaryocytes, and only a minority of the remaining barcodes were present in more than one mature lineage. Coupled with analysis at shorter pulse-chase intervals and comprehensive single cell RNA-Seq analysis, this study therefore concluded that during homeostatic unperturbed haematopoiesis, (i) megakaryocytes can arise independently from other lineages, and (ii) the phenotypic LT-HSC population as defined by transplantation assays actively contributes to megakaryocyte output.
In the HSC compartment, a transposon tag may often be present in just one or two cells, because HSC clones will rarely amplify during unperturbed haematopoiesis, thus making barcode detection less reliable. It is therefore noteworthy that both the Rodewald and Reizis groups 97,98 found that the HSC compartment contributed more to multilineage blood production than what was estimated by the transposon approach. Busch et al. also estimated the kinetics with which cells transit through their differentiation trajectories. Interestingly, flux into the lymphoid branch is 180 fold less than in the myeloid branch. Flux into the erythroid lineage was not assessed, but is likely to be even higher than in the myeloid (Fig. 2b). Flux analysis also found substantial self-renewal capacity in the ST-HSC/MPP compartment, consistent with a recent report of long-term normal haematopoiesis in mice where the HSPC compartment was ablated by 90% 99.
Future approaches are likely to employ barcodes that are expressed under a strong promoter, and therefore can be detected reliably by scRNA-Seq. This would afford true single cell resolution for the analysis of clonal relationships and single cell transcriptomes, offering the exciting possibility of defining the native hierarchy agnostic of sorting strategies that were developed using transplantation. Another pertinent question is to what extent laboratory mice kept under sterile, pathogen-free conditions are a suitable model for human haematopoiesis, which is constantly challenged by exposure to infectious agents, and has to function over a much longer lifespan. Long-term follow-up of autologous transplant patients has already revealed previously unknown functional aspects of human haematopoiesis, such as the number, stability and dynamics of individual HSCs over many years 100. Background somatic mutations represent unique barcodes that can be exploited to reconstruct clonal lineage relationships 101,102 and may thus represent another approach to investigate the dynamics of single human HSPCs over extended periods of time.
Hematopoiesis is flexible in space and time
Blood production needs to have the flexibility to adapt to drastic changes of demand, with evidence accumulating that HSC properties and differentiation journeys can adapt. As reviewed elsewhere103, haematopoietic development in the embryo is complex, with a series of transient haematopoietic waves across several organs (Fig. 3). Importantly, fetal and adult HSC have fundamentally different regulation and behaviours (reviewed in 104). In mice, there is a switch from a proliferative to a quiescent state between 3 and 4 weeks of age, which coincides with decrease in self-renewal 105. In humans, the timing of the fetal to adult switch somewhat differs. When compared to adult bone marrow, human HSCs from neonatal cord blood have increased proliferative potential (as expected from mouse studies), but their cell cycle properties already resemble the adult configuration 54. Consistently, telomere length in granulocytes, a surrogate for HSC division rate, rapidly declines during the first year of human life 106. There is evidence also that terminally differentiated cells are produced differently during fetal and adult hematopoiesis, because many more single progenitor cells from human fetal liver produce 2 lineages or more than from adult bone marrow 69. Interestingly, the relative proportion of HSC subsets also change over time with balanced HSC predominating in fetal liver, while lymphoid-deficient (also called myeloid-biased) HSC accumulate during ageing 107. The effects of ageing on HSC and more generally blood production are numerous and have been reviewed elsewhere108. Changes in both the composition of the HSC pool107,109,110 as well as the molecular circuitry of individual HSCs 34,111–113 can be due to either extrinsic or intrinsic properties, including alterations in the microenvironment, the proliferative history and accumulation of mutations.
Fig.3: The composition of the HSPC compartment changes in space and time.
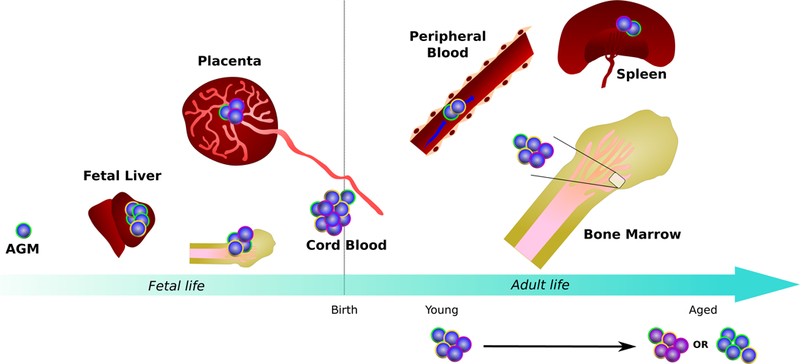
HSPCs are found in many organs in the body across a lifetime. Cells of different colours represent distinct HSPC subsets. To date it is unclear whether all HSPC subsets and differentiation trajectories are present in the same proportions in each of the organs. Current evidence suggests that age-related changes result from a combination of shifts in the composition of the HSPC pool, as well as phenotypic changes in particular cell types driven by intrinsic genetic and epigenetic changes as well as systemic alterations of the microenvironment. AGM: Aorta Gonad Mesonephros.
The HSC niche is a highly complex ecosystem that sustains HSC function, in particular promoting survival and long-term maintenance of the HSC pool 114. If and how niche interactions shape the activity of distinct HSC subsets and the differentiation journeys of HSCs remains unclear. Single cell transplants and clonal tracking have shown that clones display stereotypical behaviour over serial rounds of transplantation, arguing that their characteristic outputs are not extensively niche-dependent 15,59,62,63. However, it is possible that distinct HSC subsets may have different niche preferences. Furthermore, even though the vast majority of HSC are located in the bone marrow in adults, a small percentage of HSC are released in the blood with circadian-clock controlled patterns115, and HSCs can be found also in the spleen116 and lungs117 (Fig. 3). The role of these extra-medullary niches and whether they host specific subsets of HSC or influence their differentiation or clonal expansion capacity will have to be explored at single cell resolution. Finally, many types of stress directly affect HSC function: DNA damage 40,41, inflammation 118, acute or chronic infection119,120, psychosocial stress 121, metabolic stress33 and obesity 122. For most of these processes, insights have been gained on the molecular mechanisms driving the changes in HSC or progenitor cell function. Interestingly though, there are also examples showing that, at the cellular level, not all HSC or progenitor subsets equally respond to these stresses. For example, in emergency myelopoiesis, MPP2 and MPP3 drive enhanced production of GMPs 13,123, which reorganise themselves spatially and activate a self-renewal network 123. We are thus only beginning to understand how stress responses can reshape the relative abundance and possibly differentiation trajectories of different HSPC subsets, and less still is known about the cellular and molecular control/feed-back mechanisms that maintain and/or re-establish homeostasis.
Implications for Human Disease
Our understanding of haematopoiesis is currently undergoing a number of shifts. These include: (i) demarcations between stem and progenitor cells that previously were considered rather rigid are becoming increasingly blurred, (ii) cell fate choices upstream of the classically defined bi-/oligopotent progenitor cells may be more prevalent than previously thought, (iii) the loss of key stem cell characteristics may be largely decoupled from the initiation of specific lineage differentiation programmes, and (iv) measuring cellular fates in vivo without the need for highly disruptive transplantation procedures has highlighted a previously underappreciated importance of ST-HSC/MPPs in unperturbed haematopoiesis. These and other revisions of our understanding of haematopoiesis as a stem cell developmental system have major implications for the diagnosis, prognosis and treatment of haematological diseases.
Haematology as a clinical discipline has a long track record of being an early adopter of the latest technological developments, which recently included the first successful gene therapy trials 124,125 as well as some of the first comprehensive cancer genome studies 126. With respect to haematopoietic malignancies, much current research focuses on identifying the cell of origin, which acquired the first somatic mutation within the multistep progression towards a full-blown malignancy. It is widely accepted now that stem and progenitor cells play a major role in the development of myeloid malignancies (CML, AML) and myeloproliferative neoplasms. More recent evidence also implicates HSCs and lymphoid progenitors in the early stages of hairy cell leukemia 127 and lymphoma 128. It is beyond the scope of this review to list the effects of each of the driver mutations on HSC and progenitor cell function, but it is worth noting that each one of them will reshape the balance of differentiation trajectories and generate complex clonal dynamic patterns. Since cellular context influences the potential impact of leukaemogenic mutations, the newly recognized fluidity of cellular states within the HSPC compartment implies greater disease heterogeneity between patients, because even when two patients carry identical founder mutations, the likelihood that they arose in identical cellular states is small. Moreover, the malignant transformation is likely to open up new molecular states/trajectories. In AML for example, leukemic stem cells are defined by a chimeric transcriptional state 10,129, and single cell proteomic approaches have demonstrated the existence of distinct differentiation trajectories for malignant cells 130.
Given the increasing recognition of discrepancies between the clonal behaviour of native unperturbed HSPCs versus transplantation, xenotransplantation of human leukaemic cells into immune-compromised mice will share similar limitations, where exposing leukaemic cells to a transplantation assay may induce cellular behaviour that would never occur in a human patient. Exciting prospects may be offered here by new models that may permit transplantation without irradiation131 and the use of ossicles templated with human bone marrow stromal cells 132,133. It nevertheless seems imperative to invigorate research efforts that directly track disease in human patients. This will require careful design of patient cohorts and new screening technologies, including multi-omics single cell technologies to define the cellular state as well as the mutation burden of individual cells, as demonstrated recently for BCR-ABL 134.
Bone marrow transplantation is likely to remain an important curative therapy for many leukaemia patients, and with progress in the cell and gene therapy field, may find much wider applicability. Since suitable donor material for bone marrow transplantation remains rate limiting, one promising avenue here is to produce HSCs from other cell types by borrowing regulatory processes known to be important during developmental haematopoiesis, as illustrated by the recent generation of stem cells with long-term engrafting capability from human pluripotent and mouse adult endothelial cells 135,136. A better understanding of the HSC state and cell fate decision making will provide new avenues to develop protocols for in vitro amplification of HSCs 137–139, and also facilitate optimization of protocols for robust genome engineering of HSCs with vast potential for treating common diseases ranging from inherited red blood cell to autoimmune disorders. Expansion of clinical applications will greatly benefit from a better understanding of the self-renewal and differentiation potential of individual HSPCs, coupled with robust prediction algorithms from molecular profiling data to evaluate the efficacy of cell therapy products. Haematopoiesis is therefore well positioned to lead the way in the gene and cell therapy arena, and we may not be too far away from a future where haematopoiesis will be firmly established not just as a stem cell, but also as a therapeutic paradigm.
ACKNOWLEDGMENTS
We thank Dr David Kent for critical reading of the manuscript. E.L. is supported by a Sir Henry Dale fellowship from the Wellcome Trust (WT)/Royal Society. Research in the Laurenti and Gottgens laboratories is supported by the WT, CRUK, Bloodwise, MRC, BBSRC, NIH-NIDDK, and core support grants by the WT and MRC to the WT-MRC Cambridge Stem Cell Institute.
REFERENCES
- 1.Haeckel E Natürliche Schöpfungsgeschichte. (Georg Reimer, 1868). [Google Scholar]
- 2.Pappenheim A Ueber Entwickelung und Ausbildung der Erythroblasten. Virchows Arch Pathol Anat 145, 587–643 (1896). [Google Scholar]
- 3.Pappenheim A Zwei Fälle akuter grosslymphozytärer Leukämie. Fol Haematol 4, 301–308 (1907). [Google Scholar]
- 4.Jacobson LO, Simmons EL, Marks EK & Eldredge JH Recovery from Radiation Injury. Science 113, 510–511 (1951). [DOI] [PubMed] [Google Scholar]
- 5.Till JE & McCulloch EA A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat. Res. 14, 213–222 (1961). [PubMed] [Google Scholar]
- 6.Becker AJ, McCULLOCH EA & Till JE Cytological demonstration of the clonal nature of spleen colonies derived from transplanted mouse marrow cells. Nature 197, 452–454 (1963). [DOI] [PubMed] [Google Scholar]
- 7.Spangrude GJ, Heimfeld S & Weissman IL Purification and characterization of mouse hematopoietic stem cells. Science 241, 58–62 (1988). [DOI] [PubMed] [Google Scholar]
- 8.Doulatov S et al. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat. Immunol. 11, 585–593 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Adolfsson J et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell 121, 295–306 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Goardon N et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell 19, 138–152 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Sanjuan-Pla A et al. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature 502, 232–236 (2013). First evidence of an HSC differentiation bias along the megakaryocyte-platelet lineage. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto R et al. Clonal Analysis Unveils Self-Renewing Lineage-Restricted Progenitors Generated Directly from Hematopoietic Stem Cells. Cell 154, 1112–1126 (2013). Comprehensive single cell transplantation study demonstrating that single cells can repopulate long-term after transplantation without contributing to all blood lineages. [DOI] [PubMed] [Google Scholar]
- 13.Pietras EM et al. Functionally Distinct Subsets of Lineage-Biased Multipotent Progenitors Control Blood Production in Normal and Regenerative Conditions. Cell Stem Cell 17, 35–46 (2015). Evidence of lineage-biases in MPP populations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cabezas-Wallscheid N et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell 15, 507–522 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Dykstra B et al. Long-Term Propagation of Distinct Hematopoietic Differentiation Programs In Vivo. Cell Stem Cell 1, 218–229 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Benveniste P et al. Intermediate-Term Hematopoietic Stem Cells with Extended but Time-Limited Reconstitution Potential. Cell Stem Cell 6, 48–58 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Kiel MJ et al. SLAM Family Receptors Distinguish Hematopoietic Stem and Progenitor Cells and Reveal Endothelial Niches for Stem Cells. Cell 121, 1109–1121 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Muller-Sieburg CE, Cho RH, Thoman M, Adkins B & Sieburg HB Deterministic regulation of hematopoietic stem cell self-renewal and differentiation. Blood 100, 1302–1309 (2002). [PubMed] [Google Scholar]
- 19.Wilson A et al. Hematopoietic Stem Cells Reversibly Switch from Dormancy to Self-Renewal during Homeostasis and Repair. Cell 135, 1118–1129 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Foudi A et al. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat. Biotechnol. 27, 84–90 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyamoto T et al. Myeloid or Lymphoid Promiscuity as a Critical Step in Hematopoietic Lineage Commitment. Dev. Cell 3, 137–147 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Chambers SM et al. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell Stem Cell 1, 578–591 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novershtern N et al. Densely Interconnected Transcriptional Circuits Control Cell States in Human Hematopoiesis. Cell 144, 296–309 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laurenti E et al. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nat. Immunol. 14, 756–763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L et al. Transcriptional diversity during lineage commitment of human blood progenitors. Science 345, 1251033 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bock C et al. DNA Methylation Dynamics during In Vivo Differentiation of Blood and Skin Stem Cells. Mol. Cell 47, 633–647 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farlik M et al. DNA Methylation Dynamics of Human Hematopoietic Stem Cell Differentiation. Cell Stem Cell 19, 808–822 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bocker MT et al. Genome-wide promoter DNA methylation dynamics of human hematopoietic progenitor cells during differentiation and aging. Blood 117, e182–e189 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Ji H et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature 467, 338–342 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corces MR et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 48, 1193–1203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheshier SH, Morrison SJ, Liao X & Weissman IL In vivo proliferation and cell cycle kinetics of long-term self-renewing hematopoietic stem cells. Proc. Natl. Acad. Sci. 96, 3120–3125 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cabezas-Wallscheid N et al. Vitamin A-Retinoic Acid Signaling Regulates Hematopoietic Stem Cell Dormancy. Cell 169, 807–823.e19 (2017). Comprehensive study describing the molecular circuitry that maintains dormant HSCs. [DOI] [PubMed] [Google Scholar]
- 33.Warr MR et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature 494, 323–327 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ho TT et al. Autophagy maintains the metabolism and function of young and old stem cells. Nature 543, 205–210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simsek T et al. The Distinct Metabolic Profile of Hematopoietic Stem Cells Reflects Their Location in a Hypoxic Niche. Cell Stem Cell 7, 380–390 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takubo K et al. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 12, 49–61 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vannini N et al. Specification of haematopoietic stem cell fate via modulation of mitochondrial activity. Nat. Commun. 7, 13125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ito K et al. Self-renewal of a purified Tie2+ hematopoietic stem cell population relies on mitochondrial clearance. Science aaf5530 (2016). doi: 10.1126/science.aaf5530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Signer RAJ, Magee JA, Salic A & Morrison SJ Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509, 49–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohrin M et al. Hematopoietic Stem Cell Quiescence Promotes Error-Prone DNA Repair and Mutagenesis. Cell Stem Cell 7, 174–185 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milyavsky M et al. A Distinctive DNA Damage Response in Human Hematopoietic Stem Cells Reveals an Apoptosis-Independent Role for p53 in Self-Renewal. Cell Stem Cell 7, 186–197 (2010). [DOI] [PubMed] [Google Scholar]
- 42.van Galen P et al. The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature 510, 268–272 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Mohrin M et al. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 347, 1374–1377 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Galen P et al. Reduced Lymphoid Lineage Priming Promotes Human Hematopoietic Stem Cell Expansion. Cell Stem Cell 14, 94–106 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Beer PA et al. A Dominant-Negative Isoform of IKAROS Expands Primitive Normal Human Hematopoietic Cells. Stem Cell Rep. 3, 841–857 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cai X et al. Runx1 Deficiency Decreases Ribosome Biogenesis and Confers Stress Resistance to Hematopoietic Stem and Progenitor Cells. Cell Stem Cell 17, 165–177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oguro H, Ding L & Morrison SJ SLAM Family Markers Resolve Functionally Distinct Subpopulations of Hematopoietic Stem Cells and Multipotent Progenitors. Cell Stem Cell 13, 102–116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kent DG et al. Prospective isolation and molecular characterization of hematopoietic stem cells with durable self-renewal potential. Blood 113, 6342–6350 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Majeti R, Park CY & Weissman IL Identification of a Hierarchy of Multipotent Hematopoietic Progenitors in Human Cord Blood. Cell Stem Cell 1, 635–645 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Notta F et al. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science 333, 218–221 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Takizawa H, Regoes RR, Boddupalli CS, Bonhoeffer S & Manz MG Dynamic variation in cycling of hematopoietic stem cells in steady state and inflammation. J. Exp. Med. 208, 273–284 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qiu J, Papatsenko D, Niu X, Schaniel C & Moore K Divisional History and Hematopoietic Stem Cell Function during Homeostasis. Stem Cell Rep. 2, 473–490 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bernitz JM, Kim HS, MacArthur B, Sieburg H & Moore K Hematopoietic Stem Cells Count and Remember Self-Renewal Divisions. Cell 167, 1296–1309.e10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laurenti E et al. CDK6 Levels Regulate Quiescence Exit in Human Hematopoietic Stem Cells. Cell Stem Cell 16, 302–313 (2015). Demonstration of the molecular mechanism by which distinct quiescence exit kinetics are differentialy maintained in distinct HSC subsets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zou P et al. p57Kip2 and p27Kip1 Cooperate to Maintain Hematopoietic Stem Cell Quiescence through Interactions with Hsc70. Cell Stem Cell 9, 247–261 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Laurenti E et al. Hematopoietic Stem Cell Function and Survival Depend on c-Myc and N-Myc Activity. Cell Stem Cell 3, 611–624 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Säwén P et al. Mitotic History Reveals Distinct Stem Cell Populations and Their Contributions to Hematopoiesis. Cell Rep. 14, 2809–2818 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Catlin SN, Busque L, Gale RE, Guttorp P & Abkowitz JL The replication rate of human hematopoietic stem cells in vivo. Blood 117, 4460–4466 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muller-Sieburg CE, Cho RH, Karlsson L, Huang J-F & Sieburg HB Myeloid-biased hematopoietic stem cells have extensive self-renewal capacity but generate diminished lymphoid progeny with impaired IL-7 responsiveness. Blood 103, 4111–4118 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Haas S et al. Inflammation-Induced Emergency Megakaryopoiesis Driven by Hematopoietic Stem Cell-like Megakaryocyte Progenitors. Cell Stem Cell 17, 422–434 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Roch A, Trachsel V & Lutolf MP Brief Report: Single-Cell Analysis Reveals Cell Division-Independent Emergence of Megakaryocytes From Phenotypic Hematopoietic Stem Cells. STEM CELLS 33, 3152–3157 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Yu VWC et al. Epigenetic Memory Underlies Cell-Autonomous Heterogeneous Behavior of Hematopoietic Stem Cells. Cell 167, 1310–1322.e17 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Challen GA, Boles NC, Chambers SM & Goodell MA Distinct Hematopoietic Stem Cell Subtypes Are Differentially Regulated by TGF-β1. Cell Stem Cell 6, 265–278 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luchsinger LL, de Almeida MJ, Corrigan DJ, Mumau M & Snoeck H-W Mitofusin 2 maintains haematopoietic stem cells with extensive lymphoid potential. Nature 529, 528–531 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grinenko T et al. Clonal expansion capacity defines two consecutive developmental stages of long-term hematopoietic stem cells. J. Exp. Med. jem.20131115 (2014). doi: 10.1084/jem.20131115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anjos-Afonso F et al. CD34–Cells at the Apex of the Human Hematopoietic Stem Cell Hierarchy Have Distinctive Cellular and Molecular Signatures. Cell Stem Cell 13, 161–174 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Morita Y, Ema H & Nakauchi H Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J. Exp. Med. 207, 1173–1182 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kohn LA et al. Lymphoid priming in human bone marrow begins before expression of CD10 with upregulation of L-selectin. Nat. Immunol. (2012). doi: 10.1038/ni.2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Notta F et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science 351, aab2116 (2016). Study demonstrating that the structure of the hematopoietic hierarchy and the differentiation journeys downstream of HSCs change over a human lifetime. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Psaila B et al. Single-cell profiling of human megakaryocyte-erythroid progenitors identifies distinct megakaryocyte and erythroid differentiation pathways. Genome Biol. 17, 83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miyawaki K et al. Identification of unipotent megakaryocyte progenitors in human hematopoiesis. Blood blood-2016–09-741611 (2017). doi: 10.1182/blood-2016-09-741611 [DOI] [PubMed] [Google Scholar]
- 72.Sanada C et al. Adult human megakaryocyte-erythroid progenitors are in the CD34+CD38mid fraction. Blood 128, 923–933 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pronk CJH et al. Elucidation of the Phenotypic, Functional, and Molecular Topography of a Myeloerythroid Progenitor Cell Hierarchy. Cell Stem Cell 1, 428–442 (2007). [DOI] [PubMed] [Google Scholar]
- 74.Lee J et al. Lineage specification of human dendritic cells is marked by IRF8 expression in hematopoietic stem cells and multipotent progenitors. Nat. Immunol. 18, 877–888 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brady Gerard, Barbara Mary & Iscove Norman. Representative in Vitro cDNA Amplification From Individual Hemopoietic Cells and Colonies. METHODS Mol. Cell. Biol. 17–25 (1990). [Google Scholar]
- 76.Warren L, Bryder D, Weissman IL & Quake SR Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc. Natl. Acad. Sci. U. S. A. 103, 17807–17812 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moignard V et al. Characterisation of transcriptional networks in blood stem and progenitor cells using high-throughput single cell gene expression analysis. Nat. Cell Biol. 15, 363–372 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guo G et al. Mapping Cellular Hierarchy by Single-Cell Analysis of the Cell Surface Repertoire. Cell Stem Cell 13, 492–505 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pina C et al. Inferring rules of lineage commitment in haematopoiesis. Nat. Cell Biol. 14, 287–294 (2012). [DOI] [PubMed] [Google Scholar]
- 80.Wilson NK et al. Combined Single-Cell Functional and Gene Expression Analysis Resolves Heterogeneity within Stem Cell Populations. Cell Stem Cell 16, 712–724 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paul F et al. Transcriptional Heterogeneity and Lineage Commitment in Myeloid Progenitors. Cell 163, 1663–1677 (2015). Comprehensive single cell RNA-seq study of the myelo-erythroid progenitor compartment. [DOI] [PubMed] [Google Scholar]
- 82.Nestorowa S et al. A single cell resolution map of mouse haematopoietic stem and progenitor cell differentiation. Blood blood-2016–05-716480 (2016). doi: 10.1182/blood-2016-05-716480 Study providing the first description of the whole hematopoietic hierarchy by single cell RNA-seq. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trapnell C et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Haghverdi L, Buettner F & Theis FJ Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinforma. Oxf. Engl. 31, 2989–2998 (2015). [DOI] [PubMed] [Google Scholar]
- 85.Grün D et al. De Novo Prediction of Stem Cell Identity using Single-Cell Transcriptome Data. Cell Stem Cell 19, 266–277 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Setty M et al. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat. Biotechnol. 34, 637–645 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Olsson A et al. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature advance online publication, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Velten L et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat. Cell Biol. 19, 271–281 (2017). Study coupling single cell RNA-seq and index sorting to delineate differentiation journeys of human HSCs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Naik SH et al. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature 496, 229–232 (2013). [DOI] [PubMed] [Google Scholar]
- 90.Lu R, Neff NF, Quake SR & Weissman IL Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat. Biotechnol. 29, 928–933 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cheung AMS et al. Analysis of the clonal growth and differentiation dynamics of primitive barcoded human cord blood cells in NSG mice. Blood 122, 3129–3137 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dick JE, Magli MC, Huszar D, Phillips RA & Bernstein A Introduction of a selectable gene into primitive stem cells capable of long-term reconstitution of the hemopoietic system of W/Wv mice. Cell 42, 71–79 (1985). [DOI] [PubMed] [Google Scholar]
- 93.Lemischka IR, Raulet DH & Mulligan RC Developmental potential and dynamic behavior of hematopoietic stem cells. Cell 45, 917–927 (1986). [DOI] [PubMed] [Google Scholar]
- 94.Sun J et al. Clonal dynamics of native haematopoiesis. Nature 514, 322–327 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pei W et al. Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature advance online publication, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rodriguez-Fraticelli AE et al. Clonal analysis of lineage fate in unperturbed hematopoiesis. Nature in press, Clonal tracking analysis of unperturbed hematopoiesis reveals large contribution of single phenotypic HSC to the production of megakaryocytes and platelets at homeostatis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sawai CM et al. Hematopoietic Stem Cells Are the Major Source of Multilineage Hematopoiesis in Adult Animals. Immunity 45, 597–609 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Busch K et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature 518, 542–546 (2015). Clonal tracking analysis coupled with mathematical modeling defined the flux of stem cells into the different lineage branches under unperturbed and transplantation conditions. [DOI] [PubMed] [Google Scholar]
- 99.Schoedel KB et al. The bulk of the hematopoietic stem cell population is dispensable for murine steady-state and stress hematopoiesis. Blood blood-2016–03-706010 (2016). doi: 10.1182/blood-2016-03-706010 [DOI] [PubMed] [Google Scholar]
- 100.Biasco L et al. In Vivo Tracking of Human Hematopoiesis Reveals Patterns of Clonal Dynamics during Early and Steady-State Reconstitution Phases. Cell Stem Cell 19, 107–119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Behjati S et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature 513, 422–425 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Biezuner T et al. A generic, cost-effective, and scalable cell lineage analysis platform. Genome Res. (2016). doi: 10.1101/gr.202903.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ivanovs A et al. Human haematopoietic stem cell development: from the embryo to the dish. Development 144, 2323–2337 (2017). [DOI] [PubMed] [Google Scholar]
- 104.Copley MR & Eaves CJ Developmental changes in hematopoietic stem cell properties. Exp. Mol. Med. 45, e55 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bowie MB et al. Identification of a new intrinsically timed developmental checkpoint that reprograms key hematopoietic stem cell properties. Proc. Natl. Acad. Sci. 104, 5878–5882 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rufer N et al. Telomere Fluorescence Measurements in Granulocytes and T Lymphocyte Subsets Point to a High Turnover of Hematopoietic Stem Cells and Memory T Cells in Early Childhood. J. Exp. Med. 190, 157–168 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Benz C et al. Hematopoietic Stem Cell Subtypes Expand Differentially during Development and Display Distinct Lymphopoietic Programs. Cell Stem Cell 10, 273–283 (2012). Evidence that differentiation biases of HSC change over a lifetime. [DOI] [PubMed] [Google Scholar]
- 108.Geiger H, de Haan G & Florian MC The ageing haematopoietic stem cell compartment. Nat. Rev. Immunol. 13, 376–389 (2013). [DOI] [PubMed] [Google Scholar]
- 109.Young K et al. Progressive alterations in multipotent hematopoietic progenitors underlie lymphoid cell loss in aging. J. Exp. Med. jem.20160168 (2016). doi: 10.1084/jem.20160168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Grover A et al. Single-cell RNA sequencing reveals molecular and functional platelet bias of aged haematopoietic stem cells. Nat. Commun. 7, 11075 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Beerman I & Rossi DJ Epigenetic Control of Stem Cell Potential during Homeostasis, Aging, and Disease. Cell Stem Cell 16, 613–625 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Flach J et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature 512, 198–202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sun D et al. Epigenomic Profiling of Young and Aged HSCs Reveals Concerted Changes during Aging that Reinforce Self-Renewal. Cell Stem Cell 14, 673–688 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Baryawno N, Severe N & Scadden DT Hematopoiesis: Reconciling Historic Controversies about the Niche. Cell Stem Cell 20, 590–592 (2017). [DOI] [PubMed] [Google Scholar]
- 115.Méndez-Ferrer S, Lucas D, Battista M & Frenette PS Haematopoietic stem cell release is regulated by circadian oscillations. Nature 452, 442–447 (2008). [DOI] [PubMed] [Google Scholar]
- 116.Inra CN et al. A perisinusoidal niche for extramedullary haematopoiesis in the spleen. Nature advance online publication, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lefrangais E et al. The lung is a site of platelet biogenesis and a reservoir for haematopoietic progenitors. Nature 544, 105–109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Takizawa H, Boettcher S & Manz MG Demand-adapted regulation of early hematopoiesis in infection and inflammation. Blood 119, 2991–3002 (2012). [DOI] [PubMed] [Google Scholar]
- 119.Matatall KA et al. Chronic Infection Depletes Hematopoietic Stem Cells through Stress-Induced Terminal Differentiation. Cell Rep. 17, 2584–2595 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hirche C et al. Systemic Virus Infections Differentially Modulate Cell Cycle State and Functionality of Long-Term Hematopoietic Stem Cells In Vivo. Cell Rep. 19, 2345–2356 (2017). [DOI] [PubMed] [Google Scholar]
- 121.Heidt T et al. Chronic variable stress activates hematopoietic stem cells. Nat. Med. advance online publication, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ambrosi TH et al. Adipocyte Accumulation in the Bone Marrow during Obesity and Aging Impairs Stem Cell-Based Hematopoietic and Bone Regeneration. Cell Stem Cell 0, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hérault A et al. Myeloid progenitor cluster formation drives emergency and leukaemic myelopoiesis. Nature advance online publication, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Naldini L Gene therapy returns to centre stage. Nature 526, 351–360 (2015). [DOI] [PubMed] [Google Scholar]
- 125.Cavazzana M, Ribeil J-A, Lagresle-Peyrou C & André-Schmutz I Hematopoietic stem cell gene therapy: the point of view of the diseased bone marrow. Stem Cells Dev.(2016). doi: 10.1089/scd.2016.0230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Steensma DP The beginning of the end of the beginning in cancer genomics. N. Engl. J. Med. 368, 2138–2140 (2013). [DOI] [PubMed] [Google Scholar]
- 127.Chung SS et al. Hematopoietic Stem Cell Origin of BRAFV600E Mutations in Hairy Cell Leukemia. Sci. Transl. Med. 6, 238ra71–238ra71 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Horton SJ et al. Early loss of Crebbp confers malignant stem cell properties on lymphoid progenitors. Nat. Cell Biol. advance online publication, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Eppert K et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 17, 1086–1093 (2011). [DOI] [PubMed] [Google Scholar]
- 130.Levine JH et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 162, 184–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cosgun KN et al. Kit Regulates HSC Engraftment across the Human-Mouse Species Barrier. Cell Stem Cell 15, 227–238 (2014). [DOI] [PubMed] [Google Scholar]
- 132.Reinisch A et al. A humanized bone marrow ossicle xenotransplantation model enables improved engraftment of healthy and leukemic human hematopoietic cells. Nat. Med. 22, 812–821 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sontakke P et al. Modeling BCR-ABL and MLL-AF9 leukemia in a human bone marrow-like scaffold-based xenograft model. Leukemia 30, 2064–2073 (2016). [DOI] [PubMed] [Google Scholar]
- 134.Giustacchini A et al. Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia. Nat. Med. advance online publication, (2017). [DOI] [PubMed] [Google Scholar]
- 135.Sugimura R et al. Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature 545, 432–438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lis R et al. Conversion of adult endothelium to immunocompetent haematopoietic stem cells. Nature 545, 439–445 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wagner JE et al. Phase I/II Trial of StemRegenin-1 Expanded Umbilical Cord Blood Hematopoietic Stem Cells Supports Testing as a Stand-Alone Graft. Cell Stem Cell 18, 144–155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Boitano AE et al. Aryl Hydrocarbon Receptor Antagonists Promote the Expansion of Human Hematopoietic Stem Cells. Science 329, 1345–1348 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fares I et al. Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self-renewal. Science 345, 1509–1512 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Goodell MA, Brose K, Paradis G, Conner AS & Mulligan RC Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J. Exp. Med. 183, 1797–1806 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gorgens A et al. Revision of the Human Hematopoietic Tree: Granulocyte Subtypes Derive from Distinct Hematopoietic Lineages. Cell Rep. doi: 10.1016/j.celrep.2013.04.025 [DOI] [PubMed] [Google Scholar]
- 142.Dancey JT, Deubelbeiss KA, Harker LA & Finch CA Neutrophil kinetics in man. J. Clin. Invest. 58, 705–715 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hoogenkamp M et al. Early chromatin unfolding by RUNX1: a molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood 114, 299–309 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hoppe PS et al. Early myeloid lineage choice is not initiated by random PU.1 to GATA1 protein ratios. Nature 535, 299–302 (2016). [DOI] [PubMed] [Google Scholar]
- 145.Rieger MA, Hoppe PS, Smejkal BM, Eitelhuber AC & Schroeder T Hematopoietic Cytokines Can Instruct Lineage Choice. Science 325, 217–218 (2009). [DOI] [PubMed] [Google Scholar]
- 146.Nishikawa K et al. Self-association of Gata1 enhances transcriptional activity in vivo in zebra fish embryos. Mol. Cell. Biol. 23, 8295–8305 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Okuno Y et al. Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Mol. Cell. Biol. 25, 2832–2845 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pimanda JE et al. Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc. Natl. Acad. Sci. U. S. A. 104, 17692–17697 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Narula J, Smith AM, Gottgens B & Igoshin OA Modeling reveals bistability and low-pass filtering in the network module determining blood stem cell fate. PLoS Comput. Biol. 6, e1000771 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Swiers G, Patient R & Loose M Genetic regulatory networks programming hematopoietic stem cells and erythroid lineage specification. Dev. Biol. 294, 525–540 (2006). [DOI] [PubMed] [Google Scholar]


