Abstract
Heterologous expression of natural product biosynthetic gene clusters (BGCs) is a robust approach not only to decipher biosynthetic logic behind natural product (NP) biosynthesis, but also to discover new chemicals from uncharacterized BGCs. This approach largely relies on techniques used for cloning large BGCs into suitable expression vectors. Recently, several whole-pathway direct cloning approaches, including full-length RecE-mediated recombination in Escherichia coli, Cas9-assisted in vitro assembly, and transformation-associated recombination (TAR) in Saccharomyces cerevisiae, have been developed to accelerate BGC isolation. In this chapter, we summarize a protocol for TAR cloning large NP BGCs, detailing the process of choosing TAR plasmids, designing pathway-specific TAR vectors, generating yeast spheroplasts, performing yeast transformation, and heterologously expressing BGCs in various host strains. We believe that the established platforms can accelerate the process of discovering new NPs, understanding NP biosynthetic logic, and engineering biosynthetic pathways.
Keywords: Biosynthesis, Heterologous expression, Natural products, Yeast homologous recombination
1. Introduction
Genetic elements directly associated with the production of a natural product (NP) are generally clustered along a contiguous region of the microbial chromosome (Medema et al., 2015). These biosynthetic gene clusters (BGCs) code for proteins associated with NP biosynthesis, regulation, transport, and resistance. Recent advances in genome sequencing technologies and bioinformatics tools for NP BGC predication have led to the rapid accumulation of a large number of orphan or cryptic NP biosynthetic pathways in publicly available databases (Medema & Fischbach, 2015), providing unprecedented opportunities to (1) convert this genetic potential into chemical reality for the discovery of new molecules and (2) identify new biosynthetic capabilities and new enzymes associated with microbial metabolism. Genetic manipulation of native producing strains is a conventional strategy for exploring new BGCs and deciphering their biosynthetic mechanisms. However, this strategy relies heavily on the ability to genetically manipulate strains of interest by homologous recombination or transposon mutagenesis, which is often time consuming and not applicable to genera without established genetic approaches or for uncultured microbes. In contrast, expression of BGCs in a well-investigated and genetically amenable host is often more facile and broadly applicable (Kim, Moore, & Yoon, 2015; Luo, Enghiad, & Zhao, 2016). Heterologous expression requires (1) cloning of entire BGCs into suitable expression vectors and (2) accessibility of various heterologous hosts. Traditionally, BGC cloning involves the construction of large random libraries of genomic DNA fragments that are then screened for sequences of interest. This process is both inefficient and laborious, particularly given current accessibility of microbial genome sequences. Recently, several targeted approaches, including full-length RecE-mediated recombination (Fu et al., 2012) and oriT-directed capture (Kvitko, McMillan, & Schweizer, 2013) in Escherichia coli, Gibson assembly (Kvitko et al., 2013) or Golden-gate assembly (Liang, Liu, Low, Ang, & Zhao, 2017) based DNA ligation in a test tube, transformation-associated recombination (TAR) cloning and assembly in the yeast Saccharomyces cerevisiae (Kim et al., 2010; Yamanaka et al., 2014), have been developed for direct cloning of large BGCs. These methods have greatly accelerated the process of NP discovery and biosynthetic research.
TAR cloning, which leverages high rates of homologous recombination in S. cerevisiae, is arguably the most robust method for selective isolation of large chromosomal fragments. This method has enabled the direct cloning of complex chromosomal fragments up to 300kb into yeast artificial chromosome (YAC) vectors, with positive clone rates up to 32% (Kouprina & Larionov, 2006). The highly recombinant nature of S. cerevisiae was first reported in the early 1980s (Orr-Weaver, Szostak, & Rothstein, 1981), but the use of TAR for selective isolation of large genomic regions from complex genomic DNA was not established until the late 1990s by a group of scientists at the National Institutes of Health (Kouprina et al., 1998; Larionov et al., 1996; Larionov, Kouprina, Solomon, Barrett, & Resnick, 1997). Because early applications of this method involved the cloning of mammalian DNA, which possess yeast origin-like replicons (ARS elements) at a frequency of approximately one per 20–30 kb, propagation of TAR-based vectors in yeast cells relied on the acquisition of an ARS element from the targeted chromosomal DNA. Later, a modified TAR system was established by integrating an ARS element into the TAR vector backbone (Noskov et al., 2003), enabling the isolation of the genomic regions that do not possess ARS-like sequences, such as centromere and telomere regions or bacterial genomic DNA. Co-introduction of the counter-selectable marker URA3 alongside the ARS element helps to counteract high levels of vector re-circularization made possible by non-homologous end joining (NHEJ). For more details regarding the principles, development, and current applications of TAR cloning, please see other thorough review articles (Kouprina & Larionov, 2003, 2006, 2016).
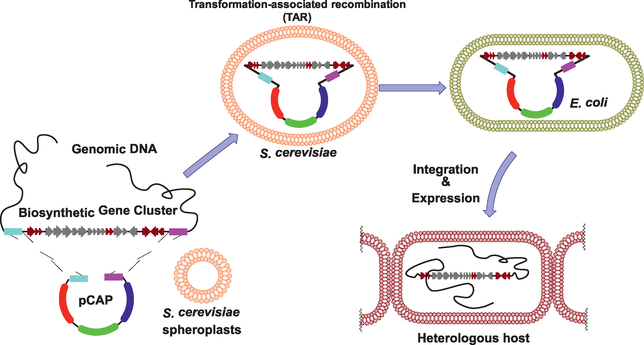
The first application of TAR cloning for NP research was the assembly of large NP BGCs from soil-derived eDNA cosmidlibraries (Kim et al., 2010). Later, our group developed a TAR-based genetic platform for direct cloning, genetic manipulation, and heterologous expression of large BGCs to identify new NPs and investigate their biosynthesis (Yamanaka et al., 2014). This platform was initially designed specifically for the study of large NP BGCs from Actinobacteria, an important source of bioactive NPs. Based on this platform, we successfully expressed a large nonribosomal peptide synthetase BGC from a marine actinomycete in the model expression host Streptomyces coelicolor, resulting in the discovery of a group of new lipopeptide antibiotics called the taromycins. Following this successful application, we have improved the approach by streamlining the process of capture vector construction and introducing the URA3 counter-selectable marker to improve cloning efficiency (Tang et al., 2015). Furthermore, we have expanded the range of heterologous hosts compatible with our platform by constructing two new TAR vectors, which are compatible with Gram-positive Bacillus subtilis (Li et al., 2015) and various Gram-negative Proteobacteria (i.e., Pseudomonas putida and Agrobacterium tumefaciens) (Zhang, Tang, Zhang, Nguyen, & Moore, 2017). Here, we summarize a protocol detailing all elements of our TAR platform, including choosing TAR plasmids, designing pathway-specific TAR vectors, TAR cloning in yeast, and heterologous expression of large BGCs in various host strains (Fig. 1).
Fig. 1.
Scheme for TAR cloning and heterologous expression of biosynthetic gene clusters using pCAP vectors.
2. Vector design
We have published three categories of TAR vectors for direct capture of large BGCs in yeast and heterologous expression in various host organisms. All vectors consist of elements for maintenance and selection in yeast, E. coli, and heterologous host organisms (Addgene plasmid numbers are provided in parenthesis).
2.1. pCAP01 (plasmid #59981)/pCAP03 (plasmid #69862)
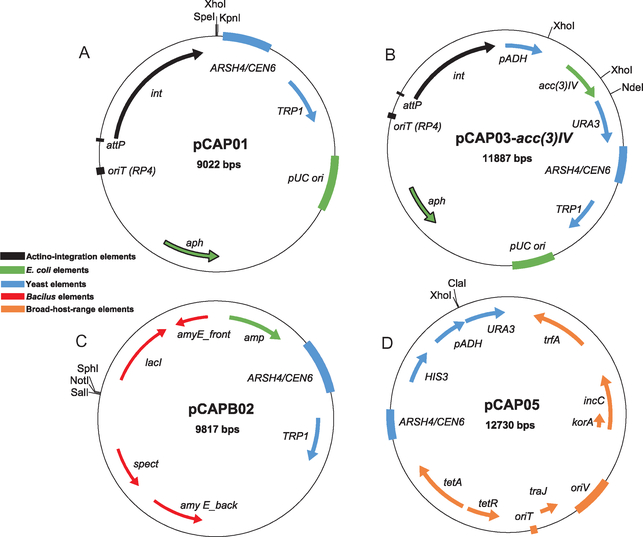
pCAP01 is a yeast-E. coli shuttle vector that can be integrated into the genome of Actinobacterial hosts. It is composed of a low copy-number yeast origin of replication (CEN6/ARSH4 from pRS314), a high copy-number E. coli origin of replication (pUC ori from SuperCos1), and a ΦC31 cassette for chromosomal integration in diverse Streptomyces species (int-attP-oriT-aph elements from pSET152) (Fig. 2A). For yeast selection, the TRP1 auxotrophic marker is used for selection of transformants on tryptophan-deficient media. Additionally, the antibiotic resistance marker neoR/kanR (neomycin transferase) is used for kanamycin selection in both E. coli and Actinobacteria. This marker can also be selected for using neomycin for hosts that possess the apramycin resistance gene (aac(3)IV) or are otherwise kanamycin resistant. This vector is maintained as a single copy yeast artificial chromosome in S. cerevisiae to avoid multiple copies of recombinants in a single yeast colony. This feature also stabilizes large constructs, and we have experimentally confirmed that DNA inserts up to 80kb can be maintained in yeast.
Fig. 2.
Vector maps for (A) pCAP01, (B) pCAP03, (C) pCAPB02, and (D) pCAP05.
Because the pCAP01 vector backbone contains a yeast origin of replication, we observed high rates of self-recircularization of empty TAR vectors by NHEJ, greatly reducing the efficiency of successful BGC cloning. To counteract this, pCAP03 was constructed through the addition of the ura3 gene under the control of the strong promoter of the Schizosaccharomyces pombe ADH1 gene (pADH1) into the SpeI and KpnI restriction sites of pCAP01 (Fig. 2B). pADH1 can tolerate an insertion of up to 130bp between the TATA box and the transcription initiation site and still act upon the downstream ura3 gene. Thus, vector-recircularization can be effectively selected against in the presence of 5-fluoroorotic acid (5-FOA). Additionally, the acc(3)IV cassette was cloned into pCAP03 to obtain pCAP03-acc(3)IV, which can be efficiently digested with XhoI and NdeI and agarose gel purified to generate ready-to-use pCAP03 (RTU-pCAP03).
2.2. pCAPB02 (plasmid #65628)
In collaboration with Dr. Pei-Yuan Qian and his group at Hong Kong University of Science and Technology, we initially built a self-replicating plasmid (pCAPB01) from pCAP01 by replacing the Streptomyces integration elements with Bacillus elements from plasmid pBU4. However, instability was observed following transfer of pCAPB01-sert (containing the 38 kb surfactin BGC from B. subtilis 1779) into the host organism B. subtilis JH642, resulting in no production of surfactins in the host. To overcome this issue, we constructed the integration vector pCAPB02 (Fig. 2C), which consists of yeast elements from pCAP01, E. coli elements from pBR322, and a homologous recombination-based integration system for B. subtilis from pDR111. pCAPB02 can be selected for in E. coli and B. subtilis with ampicillin and spectinomycin, respectively. The BGC insertion site is flanked by the 5′ and 3′ gene fragments of the B. subtilis amyE gene, enabling double-crossover recombination of BGCs into the chromosome of various B. subtilis hosts, as amyE is conserved across many B. subtilis strains.
2.3. pCAP05 (plasmid #102920)
pCAP05 (Fig. 2D) was constructed by combining yeast elements for cloning with self-replicating Gram-negative broad-host-range elements for heterologous expression. Broad-host-range elements were amplified from plasmid pRK442(H), a broad-host-range vector of the RK2 family. The RK2 replicon and its derivatives, which belong to the incompatibility group P plasmids, have been shown to replicate at low copy number (<30 in E. coli) in a wide range of Gram-negative bacteria using the oriV origin of replication. The plasmid copy number and host range are controlled by the essential trfA gene. HIS3 was chosen as the yeast selective marker to make pCAP05 orthogonal to pCAP01/03 and pCAPB02 vectors, which use TRP1 for yeast selection. Additionally, the counter-selective system from pCAP03 was introduced into pCAP05, which allows for the use of short capture arm sequences and improves cloning efficiency. pCAP05 can be selected for in E. coli and bacterial hosts using tetracycline. Although we imagine that pCAP05 can be transferred to a broad range of Gram-negative host organisms, we have thus far only validated this vector system in two proteobacterial expression hosts beyond E. coli—P. putida KT2440 and A. tumefaciens LBA4404.
3. TAR cloning
3.1. Yeast strains
Highly transformable yeast strains S. cerevisiae VL6–48 (ATCC MYA-3666: MATα, his3-D200, trp1-Δ1, ura3–52, lys2, ade2–101, met14, psi + cir°) and S. cerevisiae VL6–48N (MATα, his3-D200, trp1-Δ1, ura3-Δ1, lys2, ade2–101, met14, psi + cir°) (Larionov et al., 1997) were chosen as hosts for TAR cloning experiments. S. cerevisiae VL6–48N is more suitable for counter selection using ura3, as ura3 has been completely deleted in this strain. The yeast cells were grown in liquid or solid yeast extract-peptone-dextrose medium (YPD: 2% d-glucose, 1% yeast extract, and 2% peptone with or without agar), supplemented with 100mg/L adenine. Adenine is not necessary for yeast growth but is recommended for propagating yeast for generating spheroplasts. Yeast cells turn red if adenine is not added to YPD agar plates.
3.2. Solutions
All solutions can be sterilized by autoclave unless otherwise specified.
10 × nitrogen bases (100 mL): 1.7 g of Yeast Nitrogen Base without amino acids and ammonium sulfate (Sigma Y1251); 1.9 g of Yeast Synthetic Drop-out Medium Supplements without tryptophan or histidine; 5g of ammonium sulfate. Dissolve above components in 100mL warmed up Milli-Q water and filter through a 0.22μm filter to make 10 × stock solution (store at 4 °C).
100 × adenine (50mL): 0.5 g adenine, 3.7mL of 1 MHCl, and Milli-Q water up to 50mL. The solution can be sterilized by filtration through a 0.22μm filter to make 100 × stock solution.
Top selective agar (50mL, for 6 transformations): sorbitol, 9.1 g, 1.1 M; dextrose, 1.1 g, 2.2% (w/v); agar, 1.0 g. Before using, microwave to melt and add 10 × nitrogen bases and 100 × adenine.
Bottom selective agar (500 mL): sorbitol, 91g, 1.1 M; dextrose, 11g, 2.2% (w/v); agar, 15g. Before using, microwave and add 10 × nitrogen bases and 100 × adenine. For counter selection, add 100 × 5-fluoroorotic acid (5-FOA) stock solution (100mg/mL in DMSO) before pouring plates.
Dextrose, 40% (w/v).
1 M Sorbitol (500mL): 91 g sorbitol in 500mL Milli-Q water (store at 4 °C).
Milli-Q water, 200mL.
HEPES buffer, 1 M and pH 7.5.
EDTA solution, 0.5 M, adjust the pH to 8.0 with NaOH.
100mM CaCl2 solution: 1.1 g CaCl2 in 100mL Milli-Q water.
SPE (50mL) solution: 500 μL HEPES buffer (8), 1mL EDTA solution (9), 9.1 g sorbitol, Milli-Q water to 50mL. Store up to 2 months at room temperature after autoclaving.
SOS medium (50 mL): 3.25mL 100mM CaCl2 solution (10), 0.125 g yeast extract, 9.1 g sorbitol, 0.5 g peptone, Milli-Q water to 50mL. Store up to 2 months at room temperature after autoclaving.
Tris buffer, 1 M and pH 7.5.
STC solution (50 mL): 500 μL 1 M tris buffer (13), 5mL 100mM CaCl2 solution (10), 9.1 g sorbitol, Milli-Q water to 50mL. Store up to 2 months at room temperature after autoclaving.
PEG8000 solution (10 mL): 100 μL 1 M tris buffer (13), 1mL 100mM CaCl2 solution (10), 2.0 g PEG8000, Milli-Q water to 10mL. Filter and store up to 4 weeks at room temperature.
Zymolyase 20T solution: 10mg/mL zymolyase-20T (MP Biomedicals), 25% (w/v) glycerol, 50mM Tris-HCl (pH 7.5). No sterilization required. Dispense into 500μL aliquots and store up to 2 years at −20 °C.
3.3. Construction of pathway-specific capture vectors
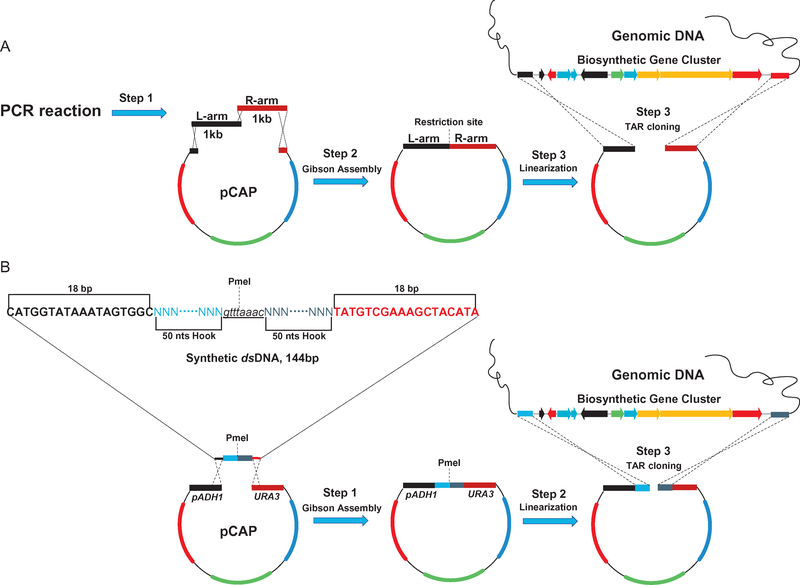
There are currently two ways to construct pathway-specific capture vectors (Fig. 3). The first approach, which applies to pCAP01 and pCAPB02, requires (1) the amplification of two 1kb homologous arms that flank the upstream and downstream regions of the BGC of interest, and (2) the insertion of the two 1kb sequences into a TAR vector by traditional cloning methods (digestion and ligation) or Gibson Assembly (Fig. 3A). Constructs prepared in this manner do not utilize the pADH-ura3 counter selection system, which we introduced to pCAP03 and pCAP05 to counteract high levels of plasmid recircularization in yeast due to NHEJ, resulting in capture rates below 2%. For second generation capture vectors pCAP03 and pCAP05, a 144bp dsDNA fragment, which contains two 18bp overlapping fragments with the vector backbone for Gibson Assembly, two 50bp pathway-specific capture hooks for recombination, and an 8 bp PmeI blunt end restriction site for linearization, has been designed as shown in Fig. 3B. This approach simplifies the procedure for constructing pathway-specific cloning vectors. While use of the counter selection system significantly decreases the percentage of yeast transformants containing empty vectors, it also dramatically reduces the total number of yeast transformants and may necessitate preparation of additional transformation plates. Additionally, there should be no ATG start codons within the homology arm sequences, which could cause mis-translation of the ura3 counterselection gene. Finally, homology arm sequences, no matter their length, should be designed as closely as possible to restriction or fragmentation sites used for preparation of the genomic DNA (gDNA) (see below for more details).
Fig. 3.
Two methods for generating pathway-specific capture vectors involving (A) PCR amplification and assembly of two 1kb homology arms or (B) assembly of synthetic ds-DNA fragment containing two 50bp homology arms.
3.4. Preparation of genomic DNA
gDNA must be appropriately fragmented before being transformed into yeast spheroplasts. Previous studies have demonstrated that homologous recombination combining two linear pieces of DNA into a yeast artificial chromosome is much more efficient if the homologous arm sequences are closer to the ends of the targeted DNA fragment (Noskov et al., 2003). Ideally, site specific double-stand breaks (DSBs) can be introduced directly upstream and downstream of BGCs of interest by restriction enzyme digestion, and homology arm sequences should be designed as close to these restriction sites as possible. However, this approach can be limited due to the lack of appropriate restriction sites directly beyond and not within BGCs of interest.
Alternatively, gDNA can be randomly fragmented by shearing with a needle or pipette tip. Recently, Dr. Natalay Kouprina and colleagues have successfully introduced DSBs to the ends of targeted gDNA fragments using the CRISPR-Cas9 endonuclease in vitro, resulting in high efficiency DNA capture, up to 32% for large fragments isolated from complex gDNA (Lee, Larionov, & Kouprina, 2015). However, in vitro CRISPR/Cas9-mediated DNA digestion necessitates several troubleshooting steps and thus may not be an attractive approach if commercial restriction enzymes can be used. The quality of the gDNA also greatly affects recombination efficiency. For isolation of gDNA from a single organism in pure culture, standard gDNA isolation procedures or commercial isolation kits are generally adequate, but gDNA should be checked by Nanodrop, Qubit, and agarose gel to confirm integrity. Finally, if the target of interest is low in quantity (such as from metagenomic DNA), PCR and/or genomic library preparation are encouraged for enrichment.
3.5. Preparation of yeast spheroplasts and transformation
The protocol for the preparation of yeast spheroplasts and transformation is outlined in the original TAR cloning protocol that was published elsewhere (Noskov et al., 2011). Although the authors of this protocol suggest using mid-log phase yeast cells (OD600 = 3.0–5.0) to generate spheroplasts, our protocol instead calls for early-log phase yeast cells (OD600 around 1.0) osmotically stabilized overnight in 1 M ice-cold sorbitol. Our slightly modified 3-day protocol is detailed below:
Day 1
Inoculate a large single colony of S. cerevisiae VL6–48 or S. cerevisiae VL6–48N from a YPD agar plate supplemented with adenine into 10 mL YPD liquid medium (in a 50mL falcon tube) with adenine overnight at 30 °C with shaking at 220 rpm. Although colonies grown on YPD plates can be kept at 4 °C for up to a week, we suggest growing fresh cells on YPD plates directly from a glycerol stock.
Day 2
Inoculate 0.5–1mL overnight culture into 50 mL YPD media containing adenine (in a 250mL-flask). Grow for 5–8h at 30 °C shaking at 220 rpm to OD600 of ~0.7–1.0. This will give you enough early-log phase cells for 10 transformations.
Put the flask on ice for 10min and then centrifuge the cells in a 50mL falcon tube for 3min at 1800 × g, 4 °C. The cells must be well pelleted; if they are not, re-streak fresh yeast and start the protocol again.
Decant the supernatant and resuspend the pellets in 50mL ice-cold sterile water by vortexing. Centrifuge for 3min at 1800 × g, 4 °C. Again, the cells must be well pelleted.
Decant the water and resuspend the cells in 50 mL of 1 M ice-cold sorbitol by vortexing. Leave the cells at 4 °C overnight for osmotic stabilization.
Day 3
Invert the tube several times and then centrifuge for 3 min at 1800 × g, 4 °C. The cells must be well pelleted.
Remove the supernatant completely by standing the tube upside down on a paper towel and resuspend the cells in 20mL SPE solution (room temperature) by vortexing. Add 40 μL of 2-mercaptoethanol (2-ME) and invert to mix. Then add 80 μL zymolyase-20T solution and invert to mix. The activity of the zymolyase enzyme may not be equivalent from brand to brand and even from batch to batch. Therefore, we suggest carefully monitoring the cells over the course of the incubation time.
Incubate the cell suspension in a 30 °C incubator, inverting the tube gently every 5min or using a shaker set at slow speed <25rpm.
After 20min, sample zymolyase treated cells by diluting five times with 1 M sorbitol and 2% SDS. Mix 200 μL cell suspension with 800 μL 1 M sorbitol and another 200 μL of cells with 800 μL 2% SDS. Monitor the progress of spheroplast formation by taking absorbance measurements of the two cell suspensions at 600 nm. Spheroplasts are determined to be ready when the absorbance of the 1 M sorbitol cell suspension is 5- to 20-fold higher than the cell suspension diluted in 2% SDS (when 80–95% of cells have been converted to spheroplasts). This process often takes about 40 min. If the fold change is not great enough, keep incubating for an additional 10 min and measure the OD difference again. The total incubation time, however, should not be longer than 50 min, as the OD of cells suspended in 1 M sorbitol will decrease over time, indicating that the cells have been over-digested and lysed. The level of spheroplast formation can greatly affect transformation efficiency (insufficiently digested cells do not take up large DNA fragments). You can use a large construct (any pCAP captured gene cluster) as a control to measure spheroplast quality.
Add 1 M sorbitol (4°C) up to 50mL and gently invert to mix. Harvest the cells by centrifuging for 10min at 600 × g, 4 °C.
Decant the supernatant and stand the tube upside down on a paper towel carefully (the cell pellet is not tightly packed) to remove the supernatant completely.
Gently resuspend the yeast spheroplasts in 20 mL of 1 M sorbitol (4 °C) by pipetting up and down with a 25-mL serological pipette. It generally takes 2 min to resuspend the spheroplasts completely (spheroplasts are extremely unstable, do not vortex to resuspend). Add ice-cold 1 M sorbitol to a total volume of 50mL and invert to mix.
Harvest the cells by centrifuging for 10 min at 600 × g, 4 °C. Remove supernatant completely by standing the tube upside down on a paper towel.
Resuspend the spheroplasts in up to 2 mL STC solution (room temperature) by pipetting up and down with a 10-mL serological pipette (it takes about 2 min to resuspend the spheroplasts completely). The volume of STC can be adjusted depending on cell density and how many transformations will be made (200 μL per transformation).
Incubate the spheroplasts for 10min at room temperature (22°C).
Mix 200 μL spheroplast solution with no more than 40 μL DNA solution (0.2–0.5 μg linearized vector and 2–3 μg gDNA) in a 1.5 mL microcentrifuge tube gently (wide-bore tip is recommended).
Incubate cell mixture for 10min at room temperature.
Add 0.8mL 20% PEG8000 solution and mix by inverting the tube 10 times.
Incubate the tube for 20min at room temperature.
Microcentrifuge the cells for 10min at 700 × g, 4°C.
Remove the supernatant with 1-mL pipette.
Add 800 μL SOS solution and resuspend the cells by pipetting with 1-mL wide-bore pipette tip.
Incubate the tube for 30–40min at 30 °C.
Add a cell suspension into 8mL melted top agar prepared in 15mL tube (keep at 50–55 °C) and invert a few times to mix. Immediately pour cells onto selective bottom agar plate.
Incubate the plates for 4–7 days at 30 °C.
Use circular capture vector (~0.1 μg) as a control to determine the quality of the spheroplast cells. Cells harboring an empty pCAP vector can form visible colonies in ~3 days, but cells carrying a recombined vector generally grow much slower.
3.6. Screening of candidate colonies
3.6.1. Picking colonies
After 4–7 days incubation, yeast transformants form visible colonies on the selective agar plate. Normally, you should observe dozens to several hundred colonies on a regular selection plate from one transformation. We suggest patching at least 100 colonies for initially screening; however, more colonies would need to be collected if no positive hit could be detected from the first screening. For the experiments using the counter selection marker ura3, dozens of colonies or even less are commonly found on one selection plate with 5-FOA. In this case, you can pick all colonies from the plate to screen.
Pick colonies from two to three plates with sterile toothpicks and transfer them onto new selective agar plates.
Incubate new plates for 2 days at 30 °C.
Make three replica plates by using filter paper or sterile velvet.
Incubate replica plates for 1–2 days at 30 °C.
3.6.2. PCR screening
DNA isolated from yeast colonies is generally harder to use as a template for PCR (especially for colony PCR) in comparison with gDNA. Therefore, primers and PCR conditions must be optimized using gDNA (giving bold and clear PCR products). Multiplex PCR is not recommended for screening.
Scoop cells from one replica plate with loop or 200-μL pipette tip, and resuspend into 200 μL of solution 1 (10% sucrose, 50mM Tris-HCl (pH 8.0), 10mM EDTA) containing 2-ME (1/1000 dilution) and 0.2 mg/mL zymolyase 20T.
Incubate and shake for 2h at 37 °C.
Option 1 (colony PCR, quick method)
Boil for 5 min (thermocycler can be used, 98 °C/5min).
Use 0.2–0.5μL as template for 50μL scale PCR reaction.
Option 2 (plasmid isolation and PCR, reliable method)
Mix the suspension with 400μL solution 2 (NaOH 0.2 M and 1% SDS (w/v)) by inversion. The solution should become clear.
Add 300μL solution 3 (KAc, 3 M, pH 4.8). Mix well by inversion and centrifuge for 10min at maximum speed.
Transfer clear supernatant into a fresh 1.5mL microcentrifuge tube. Precipitate the DNA by addition of 1 volume isopropanol (IPA) and centrifugation (maximum speed, 10min). A white DNA pellet should be visible on the bottom of the tube.
Discard the supernatant. Dissolve the DNA pellet in 200μL 10mM Tris- HCl (pH 8.0).
Use 0.2–0.5μL as a template for 50μL scale PCR reactions.
3.6.3. DNA extraction from PCR positive yeast clones
Pick up PCR-confirmed clone and inoculate into 10mL yeast selective liquid medium.
Incubate for 20–30h at 30 °C with shaking at 220rpm.
Resuspend cells in 200μL of solution 1 (10% sucrose, 50mM Tris-HCl (pH 8.0), 10mM EDTA) containing 2-ME (1/500 dilution) and 0.5mg/mL zymolyase.
Incubate and shake for 2h at 37 °C.
Mix the suspension with 400μL solution 2 (NaOH 0.2 M and 1% SDS (w/v)) by inversion. Solution should become clear.
Add 300μL solution 3 (KAc, 3 M, pH 4.8) and centrifuge for 10min at maximum speed.
Remove residual proteins by phenol-chloroform denaturation, and then precipitate and clean up DNA using IPA and 70% ethanol.
Dissolve DNA in 50μL of 10mM Tris-HCl (pH 8.0). Use 1–2μL of the DNA solution for E. coli transformation.
Stability issues are often observed in E. coli when the size of inserts is larger than 50kb. For propagation of large constructs, use of a recA− and endA− strain of E. coli such as Stbl4 (Invitrogen, Genotype: mcrA Δ(mcrBC-hsdRMS-mrr) recA1 endA1 gyrA96 gal – thi-1 supE44 λ – relA1 Δ (lac-proAB)/F′ proAB + lacIqZΔM15 Tn10 (TetR)) is recommended to avoid unwanted rearrangement of the cloned cluster. In recA+ and/or endA+ E. coli strains such as BW25113 (for X-red recombination) and ET12567 (used for conjugal DNA transfer), stability issues tend to arise more frequently. Cultivating E. coli at lower temperature (30 °C) might help to overcome these stability issues.
4. Heterologous hosts
Following the successful capture of BGCs of interest, constructs can be transferred to suitable heterologous hosts that can be analyzed for the production of associated metabolites. pCAP01/pCAP03 can be introduced to Gram-positive actinobacterial host organisms (Streptomyces species and an engineered strain of Salinispora tropica) by E. coli-mediated conjugation. pCAPB02 can be introduced to Gram-positive B. subtilis strains by natural competence transformation. Finally, pCAP05 can be introduced to Gramnegative proteobacterial hosts (P. putida and A. tumefaciens) by electrical transformation.
4.1. Gram-positive Actinobacteria: Streptomyces and Salinispora
pCAP01/pCAP03 can be directly introduced into most Streptomyces hosts by conjugation, since these vectors possess proper conjugative elements such as oriT (RK2). Inclusion of the phage ΦC31 attachment site attP in these vectors enables the genomic integration of whole constructs into any host that possesses the chromosomal attachment site attB, for example many established Streptomyces hosts. We recently engineered the marine actinomycete S. tropica to serve as apCAP01/pCAP03-compatible heterologous host by introducing the phage ΦC31 attachment site attB into the S. tropica CNB-440 genome. If a host carries a methyl-sensing restriction system (as is the case for S. coelicolor and Streptomyces avermitilis), it is necessary to utilize non-methylating E. coli strains such as E. coli ET12567 as the donor strain for conjugation. If the target host does not carry a methyl-sensing restriction system (as is the case for Streptomyces lividans and S. tropica), common E. coli strains such as DH5α can be used instead. Exconjugants can be selected for on MS agar plates using kanamycin or neomycin (50 μg/mL).
Prepare competent cells of E. coli ET12567 grown at 37 °C in LB containing chloramphenicol (25μg/mL) to maintain selection for the dam mutation.
Transform competent cells with pCAP-BGC and select for the incoming plasmid using kanamycin (50μg/mL) and chloramphenicol (25μg/mL).
Inoculate an E. coli ET12567/pCAP-BGC colony and E. coli ET12567/pUB307 into 10 mL LB medium (containing 25 μg/mL chloramphenicol and 50μg/mL kanamycin). Grow overnight at 37 °C.
Inoculate 250–500μL overnight culture into 10mL fresh LB plus antibiotics as above and grow for about 4h at 37 °C to an OD600 of 0.4–0.6. Normally, E. coli ET12567/pUB307 should grow faster than E. coli ET/pCAP-BGC.5.
Wash the cells twice with 10mL LB to remove antibiotics that might inhibit Streptomyces or Salinispora acceptor strains and resuspend E. coli ET12567/pUB307 in 0.5–1 mL LB and E. coli ET12567/pCAP-BGC in 0.2–0.4mL LB.
While washing the E. coli cells, add 10–20μL Streptomyces spore stock to 200μL 2 × YT broth (Thermo Fisher Scientific) or 10–20μL Salinispora spore stock to 200 μL Marine Broth (BD Difco) for each conjugation. Heat shock at 50 °C for 10min and then allow to cool at room temperature.
Mix 0.1mL of each E. coli cell suspension and 0.2mL heat-shocked spores.
Plate out Streptomyces and E. coli mixture on MS agar + 10 mM MgCl2 (for Streptomyces) or Salinispora and E. coli mixture on A1 agar (10 g soluble starch, 4 g yeast extract, 2 g peptone, 16g agar, 1L natural or artificial seawater) and incubate at 30 °C for 16–20h.
Overlay the plate with 1.5mL water containing 0.5 mg nalidixic acid (20 μL of25mg/mL stock; selectively kills E. coli) and 2–4 mg kanamycin (40–80μL of 50mg/mL stock). Use a spreader to lightly distribute the antibiotic solution evenly. Continue incubation at 30 °C for 5–7 days.
Pick a single colony and streak it on a MS agar plate (Streptomyces) or A1 agar plate (Salinispora) containing nalidixic acid (25 μg/mL) and kanamycin (50 μg/mL) for single colony growth and removal of E. coli contamination.
Prepare a spore suspension for two to three independent colonies.
4.2. Gram-positive Firmicutes: B. subtilis
pCAPB02-based vectors can be readily transformed into B. subtilis hosts by natural competence transformation (Albano, Hahn, & Dubnau, 1987). The region between amyE_front and amyE_back (containing the BGC of interest and the spectinomycin resistance gene) can be integrated into the amyE gene (encoding an alpha-amylase conserved in B. subtilis strains) by doublecrossover recombination at high frequency. Transformants can be selected for on LB agar plates using spectinomycin (100μg/mL).
Inoculate a single colony of B. subtilis from a LB plate into 10mL LB liquid medium (in a 50mL falcon tube) overnight at 30 °C with shaking at 220rpm.
Inoculate 100 μL overnight culture into 10 mL fresh LB and grow for about 3h at 37 °C to an OD600 of ~1. In the meantime, make MD media (see below).
Inoculate 20mL MD media with 1mL of LB B. subtilis culture.
Grow for about 4h at 37 °C at 220rpm.
Add 50% glycerol to the competent cells to get a final concentration of 20% glycerol. Divide into 1mL aliquots and freeze at 80 °C.
Transformation: Add DNA (5–10μL plasmid) to 200μL competent cells and incubate for 30min at 37 °C at 220rpm.
Plate out appropriate dilutions on plates containing the appropriate antibiotics.
4.2.1. Solutions
10 × Phosphate citrate (1000 mL): 107g K2HPO4, 60g KH2PO4, 10g sodium citrate·2H2O
MD media (50 mL): 45.6mL 10 × phosphate citrate, 2.0mL glucose (50%, w/v), 500 μL L-tryptophan (10mg/mL), 250 μL ferric ammonium citrate (2.2mg/mL), 1.25mL potassium aspartate (100mg/mL), 150μL 1 M MgSO4, and 250μL phenylalanine (10mg/mL).
4.3. Gram-negative Proteobacteria: Pseudomonas and Agrobacterium
pCAP05-based vectors are broad-host range vectors that are maintained as self-replicating plasmids in a broad range of Gram-negative bacteria. pCAP05 has been successfully transferred to two proteobacterial expression hosts beyond E. coli—P. putida KT2440 and A. tumefaciens LBA4404—using standard electrical transformation. pCAP05 can also be transferred by conjugation (see vector map details in Fig. 1D) and selected for using tetracycline.
For preparation of electrocompetent P. putida or A. tumefaciens, grow strains overnight in LB or YM medium (0.04% yeast extract, 1.0% mannitol, 1.7mM NaCl, 08mM MgSO4·7H2O, 2.2mM K2HPO4·3H2O), respectively, at 30 °C.
In the morning, inoculate 5–10mL of P. putida or A. tumefaciens precultures into 50 mL of fresh LB or YM media, respectively. Grow at 30 °C for approximately 4h or until cultures reach an OD600 of ~1.0–3.0 for P. putida or 0.5 for A. tumefaciens. P. putida will grow quickly; A. tumefaciens will grow more slowly.
Place cultures on ice for 10–15min. Wash P. putida with ice-cold 10% glycerol twice and resuspend cells in a small volume of glycerol. Wash A. tumefaciens with ice-cold 1mM HEPES buffer (pH 7.5) twice and cells resuspend in a small volume of 1mM HEPES-10% glycerol. Cells can be aliquoted and stored at −80 °C for future use.
Add 0.1–1μg of pCAP05-BGC DNA to cells and electroporate at 2500V in a 2-mm cuvette.
Recover P. putida for 2h at 30 °C in LB medium before plating on LB agar supplemented with 15 μg/mL tetracycline. Recover A. tumefaciens for 3 h at 30 °C in YM medium before plating on YM agar supplemented with 5μg/mL tetracycline.
For future heterologous expression experiments, both P. putida and A. tumefaciens can be grown on LB medium.
Because pCAP05 exists as a self-replicating plasmid in the host, we have observed occasional genetic instability in host organisms when carrying large and repetitive DNA sequences. Thus, we are currently developing and refining methods for facile and site-specific genomic integration of BGCs into Gram-negative hosts.
5. Conclusions
Heterologous expression of whole NP BGCs offers several advantages for discovery of new NPs and understanding their biosynthetic logic. First, successful expression of a genomic region containing a predicted BGC clearly defines all specialized genes that are essential for NP production. Second, genetic manipulation of cloned BGCs leveraging established E. coli or yeast recombineering strategies is comparatively faster, easier, and more broadly applicable than genetic manipulation of wild-type strains. Third, working in a sequenced model host organism provides the opportunity to rationally manipulate NP production by metabolic engineering. Thus, we believe our established platforms for cloning and heterologous expression provide a routine approach not only for the discovery of metabolites from “orphan” or “cryptic” BGCs but also to probe natural product biosynthetic logic and engineer biosynthetic pathways. We summarize per Table 1 a list of BGCs that have been successfully cloned and expressed using the pCAP vector systems. We believe that lessons learned through developing and optimizing these platforms will continue to be broadly useful to the natural products research community in future studies of microbial BGCs.
Table 1.
List of selected NPs cloned and expressed using the pCAP vector platforms.
| NP | BGC family | Native host | Size (kb) | Vector | Expression host | References |
|---|---|---|---|---|---|---|
| Taromycin | NRPS | Saccharomonospora sp. CNQ-490 | 67 | pCAPOl | Streptomyces coelicolor M1146 | Yamanaka et al. (2014); Reynolds et al. (2018) |
| Marinopyrrole | PKS-NRPS | Streptomyces sp. CNQ418 | 30 | pCAPOl | S. coelicolor M512 | Yamanaka et al. (2014) |
| Enterocin | Type II PKS | Salinispora pacifica CNT-150 | 18 | pCAPOl | S. coelicolor M1146 | Bonet, Teufel, Crusemann, Ziemert, and Moore (2015) |
| Alterochromide | NRPS-FAS | Pseudoalteromonas piscicida JCM 20779 | 34 | pCAPOl (capture) pETDuet-1 (expression) |
E. coli BL21(DE3) | Ross, Gulland, Dorrestein, and Moore (2015) |
| Ammosamide | Alkaloid | Streptomyces sp. CNR-698 | 37.5 | pCAPOl | S. coelicolor M1146 | Jordan and Moore (2016) |
| Cosmomycin | Type II PKS | Streptomyces sp. CNT-302 | 54 | pCAPOl | S. coelicolor M152 | Larson, Crusemann, and Moore (2017) |
| Salinamide | PKS-NRPS | Streptomyces sp. CNB-091 | 48.2 | pCAPOl | S. coelicolor M1146 | Ray, Yamanaka, and Moore (2016) |
| Jomthonic acid | PKS-NRPS | S. caniferus GUA-06–05–006A | 35 | pCAPOl |
S. coelicolor M1152 Streptomyces albus J1074 |
Garcia-Salcedo et al. (2018) |
| Chuangxinmycin | Alkaloid | Actinoplanes tsinanensis CPCC 200056 | unknown | pCAPOl | S coelicolor M1146 | Shi et al. (2018) |
| Nataxazole | Alkaloid | Streptomyces sp. Tü 6176 | unknown | pCAPOl | S. albus JT46 | Cano-Prieto et al. (2015) |
| Demethylchlortetracycline | Type II PKS | S. aureofaciens DM-1 | 43.5 | pCAPOl | Streptomyces aureofaciens DM-1 | Wu, Huang, Min, and Hu (2017) |
| Precolibactins | PKS-NRPS | Escherichia coli CFT073 | 51 | pCAPOl | Escherichia coli DH10B | Li et al. (2016) |
| Thiolactomycin | PKS-NRPS | Sa. pacifica CNS863 | 22 | pCAP03 | S. coelicolor M1152 and Salinispora tropica CNB4401 | Tang et al. (2015); Zhang, Moore, and Tang (2018) |
| Thiotetroamide | PKS-NRPS | S. afghaniensis NRRL 5621 | 33 | pCAP03 | S. coelicolor M1152 | Tang et al. (2015) |
| Scleric acid | NRPS | S. sclerotialus NRRL ISP-5269 | 33 | pCAP03 | S. coelicolor M1152 | Alberti et al. (2019) |
| Amicoumacin | PKS-NRPS | Bacillus subtilis 1779 | 47.4 | pCAPB02 | Bacillus subtilis JH642 | Li et al. (2015) |
| Plipastatin | NRPS | B. amyloliquefaciens HYM12 | 39 | pCAPB02 | B. subtilis 1A751 | Hu et al. (2018) |
| Violacein | Alkaloid | P. luteoviolacea 2tal6 | 9 | pCAP05 |
Pseudomonas putida KT2440 Agrobacterium tumefaciens LBA4404 |
Zhang et al. (2017) |
NRPS, nonribosomonal peptide synthetase; PKS, polyketide synthase; FAS, fatty acid synthase.
Acknowledgments
We kindly thank all past and current members of the Moore lab for providing their valuable experiences and discussion in developing the TAR-based platforms. We acknowledge Dr. Pei-Yuan Qian and his group for joint efforts in developing pCAPB plasmids. Finally, we thank Dr. Vladimir Larionov and Dr. Natalay Kouprina of the National Cancer Institute (NCI) at the National Institutes of Health for sharing their TAR expertise and yeast materials with our group. The research reported has been supported by a graduate fellowship from the National Science Foundation to J.J.Z. and National Institutes of Health grants F31-AI129299 to J.J.Z. and R01-GM085770 and R01-AI117712 to B.S.M.
References
- Albano M, Hahn J, & Dubnau D (1987). Expression of competence genes in Bacillus subtilis. Journal of Bacteriology, 169, 3110–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti F, Leng DJ, Wilkening I, Song L, Tosin M, & Corre C (2019). Triggering the expression of a silent gene cluster from genetically intractable bacteria results in scleric acid discovery. Chemical Science, 10, 453–463. 10.1039/C8SC03814G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonet B, Teufel R, Crusemann M, Ziemert N, & Moore BS (2015). Direct capture and heterologous expression of Salinispora natural product genes for the biosynthesis of enterocin. Journal of Natural Products, 78, 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Prieto C, Garcia-Salcedo R, Sanchez-Hidalgo M, Brana AF, Fiedler HP, Mendez C, et al. (2015). Genome mining of sp. Tü 6176: Characterization of the nataxazole biosynthesis pathway. Chembiochem, 16, 1461–1473. [DOI] [PubMed] [Google Scholar]
- Fu J, Bian X, Hu S, Wang H, Huang F, Seibert PM, et al. (2012). Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nature Biotechnology, 30, 440–446. [DOI] [PubMed] [Google Scholar]
- Garcia-Salcedo R, Alvarez-Alvarez R, Olano C, Canedo L, Brana AF, Mendez C, et al. (2018). Characterization of the jomthonic acids biosynthesis pathway and isolation of novel analogues in Streptomyces caniferus GUA-06–05-006A. Marine Drugs, 16, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Nan F, Maina SW, Guo J, Wu S, & Xin Z (2018). Clone of plipastatin biosynthetic gene cluster by transformation-associated recombination technique and high efficient expression in model organism Bacillus subtilis. Journal of Biotechnology, 288, 1–8. [DOI] [PubMed] [Google Scholar]
- Jordan PA, & Moore BS (2016). Biosynthetic pathway connects cryptic ribosomally synthesized posttranslationally modified peptide genes with pyrroloquinoline alkaloids. Cell Chemical Biology, 23, 1504–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Feng Z, Bauer JD, Kallifidas D, Calle PY, & Brady SF (2010). Cloning large natural product gene clusters from the environment: Piecing environmental DNA gene clusters back together with TAR. Biopolymers, 93, 833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Moore BS, & Yoon YJ (2015). Reinvigorating natural product combinatorial biosynthesis with synthetic biology. Nature Chemical Biology, 11, 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Annab L, Graves J, Afshari C, Barrett JC, Resnick MA, et al. (1998). Functional copies of a human gene can be directly isolated by transformation-associated recombination cloning with a small 3′ end target sequence. Proceedings of the National Academy of Sciences of the United States of America, 95, 4469–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, & Larionov V (2003). Exploiting the yeast Saccharomyces cerevisiae for the study of the organization and evolution of complex genomes. FEMS Microbiology Reviews, 27, 629–649. [DOI] [PubMed] [Google Scholar]
- Kouprina N, & Larionov V (2006). TAR cloning: Insights into gene function, long-range haplotypes and genome structure and evolution. Nature Reviews. Genetics, 7, 805–812. [DOI] [PubMed] [Google Scholar]
- Kouprina N, & Larionov V (2016). Transformation-associated recombination (TAR) cloning for genomics studies and synthetic biology. Chromosoma, 125, 621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitko BH, McMillan IA, & Schweizer HP (2013). An improved method for oriT-directed cloning and functionalization of large bacterial genomic regions. Applied and Environmental Microbiology, 79, 4869–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larionov V, Kouprina N, Graves J, Chen XN, Korenberg JR, & Resnick MA (1996). Specific cloning of human DNA as yeast artificial chromosomes by transformation-associated recombination. Proceedings of the National Academy of Sciences of the United States of America, 93, 491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larionov V, Kouprina N, Solomon G, Barrett JC, & Resnick MA (1997). Direct isolation of human BRCA2 gene by transformation-associated recombination in yeast. Proceedings of the National Academy of Sciences of the United States of America, 94, 7384–7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson CB, Crusemann M, & Moore BS (2017). PCR-independent method of transformation-associated recombination reveals the cosmomycin biosynthetic gene cluster in an ocean streptomycete. Journal of Natural Products, 80, 1200–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NC, Larionov V, & Kouprina N (2015). Highly efficient CRISPR/Cas9-mediated TAR cloning of genes and chromosomal loci from complex genomes in yeast. Nucleic Acids Research, 43, e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZR, Li J, Gu JP, Lai JY, Duggan BM, Zhang WP, et al. (2016). Divergent biosynthesis yields a cytotoxic aminomalonate-containing precolibactin. Nature Chemical Biology, 12, 773–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li Z, Yamanaka K, Xu Y, Zhang W, Vlamakis H, et al. (2015). Directed natural product biosynthesis gene cluster capture and expression in the model bacterium Bacillus subtilis. Scientific Reports, 5, 9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Liu Z, Low XZ, Ang EL, & Zhao H (2017). Twin-primer non-enzymatic DNA assembly: An efficient and accurate multi-part DNA assembly method. Nucleic Acids Research, 45, e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Enghiad B, & Zhao H (2016). New tools for reconstruction and heterologous expression of natural product biosynthetic gene clusters. Natural Product Reports, 33, 174–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, & Fischbach MA (2015). Computational approaches to natural product discovery. Nature Chemical Biology, 11, 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Kottmann R, Yilmaz P, Cummings M, Biggins JB, Blin K, et al. (2015). Minimum information about a biosynthetic gene cluster. Nature Chemical Biology, 11, 625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov VN, Chuang RY, Gibson DG, Leem SH, Larionov V, & Kouprina N (2011). Isolation of circular yeast artificial chromosomes for synthetic biology and functional genomics studies. Nature Protocols, 6, 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov VN, Kouprina N, Leem SH, Ouspenski I, Barrett JC, & Larionov V (2003). A general cloning system to selectively isolate any eukaryotic or prokaryotic genomic region in yeast. BMC Genomics, 4, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver TL, Szostak JW, & Rothstein RJ (1981). Yeast transformation: A model system for the study of recombination. Proceedings of the National Academy of Sciences of the United States of America, 78, 6354–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray L, Yamanaka K, & Moore BS (2016). A peptidyl-transesterifying type I thioesterase in salinamide biosynthesis. Angewandte Chemie (International Ed. in English), 55, 364–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds KA, Luhavaya H, Li J, Dahesh S, Nizet V, Yamanaka K, et al. (2018). Isolation and structure elucidation of lipopeptide antibiotic taromycin B from the activated taromycin biosynthetic gene cluster. Journal of Antibiotics (Tokyo), 71, 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AC, Gulland LE, Dorrestein PC, & Moore BS (2015). Targeted capture and heterologous expression of the Pseudoalteromonas alterochromide gene cluster in Escherichia coli represents a promising natural product exploratory platform. ACS Synthetic Biology, 4, 414–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Jiang Z, Li X, Zuo L, Lei X, Yu L, et al. (2018). Biosynthesis of antibiotic chuangxinmycin from Actinoplanes tsinanensis. Acta Pharmaceutica Sinica B, 8, 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Li J, Millan-Aguinaga N, Zhang JJ, O’Neill EC, Ugalde JA, et al. (2015). Identification of thiotetronic acid antibiotic biosynthetic pathways by target-directed genome mining. ACS Chemical Biology, 10, 2841–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Huang H, Min T, & Hu H (2017). TAR cloning and integrated overexpression of 6-demethylchlortetracycline biosynthetic gene cluster in Streptomyces aureofaciens. Acta Biochimica et Biophysica Sinica Shanghai, 49, 1129–1134. [DOI] [PubMed] [Google Scholar]
- Yamanaka K, Reynolds KA, Kersten RD, Ryan KS, Gonzalez DJ, Nizet V, et al. (2014). Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proceedings of the National Academy of Sciences of the United States of America, 111, 1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JJ, Moore BS, & Tang X (2018). Engineering Salinispora tropica for heterologous expression of natural product biosynthetic gene clusters. Applied Microbiology and Biotechnology, 102, 8437–8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JJ, Tang X, Zhang M, Nguyen D, & Moore BS (2017). Broad-host-range expression reveals native and host regulatory elements that influence heterologous antibiotic production in Gram-negative bacteria. MBio, 8, e01291–17. [DOI] [PMC free article] [PubMed] [Google Scholar]