Abstract
Chronic lymphocytic leukemia (CLL) patients progressively develop an immunosuppressive state. CLL patients have more plasma inerleukin-10 (IL-10), an anti-inflammatory cytokine, than healthy controls. In vitro human CLL cells produce IL-10 in response to B-cell receptor (BCR) cross-linking. We used the transgenic Eµ-TCL1 mouse CLL model to study the role of IL-10 in CLL associated immunosuppression. Eµ-TCL mice spontaneously develop CLL due to a B-cell specific expression of the oncogene, TCL1. Eµ-TCL1 mouse CLL cells constitutively produce IL-10, which is further enhanced by BCR cross-linking, CLL-derived IL-10 did not directly affect survival of murine or human CLL cells in vitro. We tested the hypothesis that the CLL-derived IL-10 has a critical role in CLL disease in part by suppressing the host immune response to the CLL cells. In IL-10R−/− mice, wherein the host immune cells are unresponsive to IL-10 mediated suppressive effects, there was a significant reduction in CLL cell growth compared to WT mice. IL-10 reduced the generation of effector CD4 and CD8 T-cells. We also found that activation of BCR signaling regulated the production of IL-10 by both murine and human CLL cells. We identified the transcription factor, Sp1, as a novel regulator of IL-10 production by CLL cells and that it is regulated by BCR signaling via the Syk/MAPK pathway. Our results suggest that incorporation of IL-10 blocking agents may enhance current therapeutic regimens for CLL by potentiating host anti-tumor immune response.
Introduction
Chronic lymphocytic leukemia (CLL), the most common human leukemia in Western world, is defined by accumulation of malignant CD5+CD19+ B-lymphocytes in the peripheral blood (PB), bone marrow (BM), and secondary lymphoid organs (1). CLL cells express B-cell receptor (BCR) as well as CD5, a molecule characteristic of T-lymphocytes, similar to the B-1 B-cell subset (2, 3). Systemic immunosuppression is associated with a more aggressive CLL disease resulting in infections and secondary cancers such as melanoma, head and neck cancer and lymphoblastic leukemia (4–6).
Many mechanisms of immunosuppression have been described in CLL. Interestingly, like normal B-1 cells, CLL cells themselves constitutively produce interleukin-10 (IL-10), an immunosuppressive factor (7, 8). IL-10, a T helper 2 (Th2) type cytokine, was identified by its ability to suppress macrophage cytokine production (9, 10). In addition, IL-10 regulates T-cell responses indirectly through its effects on antigen presenting cells (APCs), inhibiting their MHC class II expression and limiting their production of proinflammatory mediators (11). IL-10 can also directly inhibit T-cell proliferation and production of cytokine such as IL-2 and IFN-γ (12, 13). IL-10 from CLL cells was shown to inhibit macrophage TNF production, but its importance for CLL disease was not tested (14). In this study, we investigated the direct effects IL-10 has on T-cell responses in CLL using the Eµ-TCL1 mice. These mice serve as an excellent model of CLL as they overexpress T-cell leukemia oncogene-1 (TCL1) under the control of the B-cell-specific µ-enhancer and the IGHV promoter leading to a progressive CLL-like disease at 8–11 months of age (15). Here we showed that IL-10 failed to affect the survival of CLL cells in vitro, but that it played an important role in inhibiting T-cell anti-tumor immunity in vivo. This could play a significant role in affecting anti-tumor responses to current immunotherapy strategies. For example, the efficacy of CAR T-cell therapies is much less (26% complete response) for CLL than other B-cell leukemias (78% complete response) (16), which could be due to the inhibition of CD19-specific CAR T-cells by IL-10. Another study implicated IL-10 as one of the adaptive resistance mechanisms that undermine the efficacy of antibodies to programmed cell death protein-1 (PD-1) monotherapies in ovarian cancer (17). Indeed, the combination of PD-1 blockade and IL-10 neutralization improved survival and delayed tumor growth in tumor bearing mice, which was in part a result of increased T- and B-cell responses (17).
Despite several studies on molecular basis of IL-10 expression by various cell types (18), the mechanisms involved in IL-10 production by CLL cells remain understudied. BCR derived signals are crucial for CLL survival. Here, we found that BCR dependent constitutive activation of Src, Btk and Syk family kinases is required for constitutive IL-10 production by murine and human CLL cells. In addition, our results indicate a novel role for the transcription factor specific protein 1 (Sp1) in BCR dependent production of IL-10 by CLL cells.
Materials and Methods
Mice and cells
C57BL/6J, B6.129S2-IL10rbtm1Agt/J (IL-10R−/−), B6.129S7-Rag1tm1Mom/J (B6.Rag1−/−), and NOD.Cg-PrkdcscidIL2rgtm1Wjl/SzJ (NSG) mice were obtained from The Jackson Laboratory (Bar Harbor, ME). Eµ-TCL1 mice on C57/BL6J background were provided by Dr. John Byrd (Ohio State University, OH) and bred in house. The University of Kentucky IACUC protocol number for this study is 2011-0904. Cohorts of Eµ-TCL1 mice were maintained and monitored regularly for CLL cells in blood by flow cytometry. Mice were bled by submandibular bleeding using a lancet. Blood was collected in K2 EDTA Microtainer tubes (BD #365974, San Diego, CA). Most Eµ-TCL1 mice developed CLL between 6–9 months of age (at least 30% CD5+CD19+ B cells in the blood). Mice were euthanized when CLL cells were 80–90% or when their body condition score (BCS) was ≤2 (19). In addition to using CLL cells from primary Eµ-TCL1 mice, we adoptively transfer CLL cells from spleens of euthanized Eµ-TCL1 mice with 80–90% CLL into syngeneic mice, which lead to a reliable and consistent development of the disease in the recipient mice within weeks of injection. Actual kinetics of disease development in the adoptive transfer model vary depending on the individual Eµ-TCL1 mouse donor of the CLL cells used for the transfer. Hence in any given adoptive transfer experiment CLL cells from a single Eµ-TCL1 donor mouse were used. Spleens from mice with CLL lost their follicular architecture and had proliferative centers shown by histology as suggested by Bichi et al (15). Normal splenic cells were isolated from spleens of C57BL/6J mice and CD19+ microbeads were used to isolate B-cells. The human CLL cell line, MEC1 was cultured in Iscove's Modified Dulbecco's Medium (ThermoFisher Scientific, Waltham, MA) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biological Systems).
Human Samples
Patients with CLL were recruited from the University of Kentucky Markey Cancer Center (Table I). All patients gave informed consent according to protocols approved by the University of Kentucky Institutional Review Board. For healthy controls, leukopak units were obtained from the Kentucky blood bank. Normal human plasma samples were provided by the Sanders Brown Center on Aging at the University of Kentucky. B-cells were enriched using Ficoll-Paque PLUS density gradients (GE HealthCare, Pittsburgh, PA) and the Miltenyi Microbeads (San Diego, CA) as described previously by Ramsey et al (20). CLL preparations were always >90% CD5+CD19+ B-cells.
Table I.
Properties of the CLL patients’ donor pool
| Patient# | Age | Sex | WBC (K/µL) |
%CD5+ CD19+ |
CD38 Status |
Treatment | IGHV mutation |
|---|---|---|---|---|---|---|---|
| 1 | 82 | F | 20.7 | 90.54 | Negative | No | M-CLL |
| 2 | 56 | F | ND | 82.35 | ND | No | U-CLL |
| 3 | 36 | M | 38.3 | 88.08 | Negative | No | M-CLL |
| 4 | 46 | M | 41 | 44.01 | Negative | No | M-CLL |
| 5 | 52 | M | 15.2 | 81.37 | ND | Yes | M-CLL |
| 6 | 69 | M | 29 | 83.88 | Positive | No | M-CLL |
| 7 | 57 | M | 40 | 50.33 | ND | Yes | U-CLL |
| 8 | 62 | F | 83.7 | 10.55 | ND | No | U-CLL |
| 9 | 69 | M | 30.2 | 95.71 | ND | No | M-CLL |
| 10 | 70 | M | 17.4 | 80.45 | ND | No | M-CLL |
| 11 | 63 | M | 34.8 | 96.97 | ND | No | M-CLL |
| 12 | 53 | M | 12.1 | 84.73 | Positive | Yes | U-CLL |
| 13 | 55 | M | 6 | 13.64 | ND | Yes | M-CLL |
| 14 | 76 | F | 36.6 | 37.23 | ND | Yes | M-CLL |
| 15 | 80 | M | 142 | 97.73 | ND | Yes | U-CLL |
ND= not done
Reagents
PMA, Ionomycin, LPS and Thiazolyl Blue Tetrazolium Bromide (MTT) were purchased from Millipore Sigma-Aldrich (St. Louis, MO). Purified anti-mouse IL-10 and anti-mouse IL-10R antibodies were from BD PharMingen (San Diego, CA). AffiniPure F(ab')2 Fragment Goat anti-Mouse IgM and AffiniPure F(ab')2 Fragment Goat anti-human IgM were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA). Magentic beads to isolate mouse and human B cells, mouse CD8a and CD4+ T cells were from Miltenyi Biotech. Polybrene was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Dasatinib was manufactured by Bristol-Myers Squibb Company (Seattle, WA). Syk inhibitor IV (Bay 61-3606) was from EMD Millipore Calbiochem (Billerica, MA). Mithramycin A obtained from Enzo Life Sciences (Farmingdale, NY). Ibrutinib and ERK1/2 inhibitor were from APExBIO (Houston, TX). Fluorochrome-conjugated antibodies to mouse CD5 (53-7.3), CD19 (6D5), CD45 (30-F11), CD11b (M1/70), CD4 (GK1.5), CD8 (53-6.7), IL-10R (1B1.3a), IFN-γ (XMG1.2), and IL-10 (JESS-16E3) were purchased from BioLegend (San Diego, CA). Mouse Anti-CD90.2 (Thy1.2) (30-H12) was purchased from BD PharMingen. Antibodies to human CD5 (L17F12), CD19 (HIB19) and CD45 (2D1). Western Blot antibodies to P-Syk, total Syk, P-p38, total p38, P-ERK1/2, P-STAT3, total STAT3 and GAPDH were obtained from Cell Signaling Technology (Danvers, Massachusetts). Antibodies to IL-10, SP1, Lyn, and total ERK1, peroxidase coupled goat anti-rabbit and anti-mouse Ig secondary antibodies were acquired from Santa Cruz Biotechnology (Santa Cruz, CA).
CLL and T cell adoptive transfer
Adoptive transfers were performed by transferring 1–10×106 Eµ-TCL1 splenic cells into C57BL/6J, IL-10R−/−, BL/6.Rag−/− or NSG mice (without any preconditioning) intravenously via retro-orbital injections. CLL disease was monitored by periodic submandibular bleeding and CD5+CD19+ cells were quantified by flow cytometry (21). For T-cell adoptive transfer experiments. CD8+ T cells were purified as noted above. Refer to Supp. Figure 1 for additional experimental schematics.
Enzyme-linked immunosorbent assay (ELISA)
For cytokine analysis, normal B-1, murine CLL and human CLL cells were cultured in triplicate (2×106 cells/mL) for 24 hours in 96-well plate with various stimulants or treatments. Cells were removed by centrifugation and supernatants were assayed immediately or stored at −80°C. Plasma from mice was obtained by centrifuging blood collected in EDTA tubes at the time of euthanization by cardiac puncture and the samples were stored at −80°C. IL-10 levels in supernatants or plasma were quantified using IL-10 OptEIA ELISA kit (BD#555252). IFN-γ levels were measured using IFN-γ OptEIA ELISA kit (BD#555138). Human plasma or secreted IL-10 levels were quantified using IL-10 ELISA MAX set (BioLegend #430601). Limit of IL-10 detection is 8 pg/ml.
In vitro cell survival, proliferation and differentiation assays
Eµ-TCL1 CD5+CD19+ splenic cells were stimulated with LPS in the presence or absence of anti-IL-10R or anti-IL-10 antibodies for 48 hours and cell survival was determined by MTT assay (22). For T-cell proliferation and differentiation studies, CD8+ cells were purified using Miltenyi Microbeads according to the methods described by Keir et al (23), and cultured (1–2×105 cells) with irradiated (25Gy) Eµ-TCL1 cells (2×105 cells) for 72 hours. The cultures were pulsed with 1.0µCi of tritiated thymidine for four hours. The incorporated radioactivity was measured using a Matrix 96 β-counter. For cytokine analysis, cells were cultured in triplicate (2×105 cells) for 24 hours with or without various treatments. Secreted IL-10 and IFN-γ levels were quantified using ELISA.
Immunofluorescence analysis and cell sorting
Single-cell lymphocyte suspensions from mouse tissues were prepared by pressing tissues through a 40µm strainer using the plunger end of a syringe in Hank’s buffered salt solution (HBSS) (Millipore Sigma-Aldrich). Tibiae and femora were flushed with a 26G syringe in HBSS. Peritoneal cells were obtained by peritoneal lavage with HBSS. Cells were resuspended in RPMI 1640 medium (Corning, New York, NY) supplemented with 10% FBS. For multi-color immunofluorescence analysis, single-cell suspensions of mononuclear cells were first incubated with normal rat IgG (10µg/106 cells) at 4°C for 15 min to block Fcγ receptors. The cells were then labeled with fluorochrome conjugated anti-mouse antibodies for 30 minutes on ice. Stained cells were analyzed using Becton Dickinson LSRII flow cytometer and CellQuest Pro software. Anti-CD19, anti-CD11b and anti-CD5 were used to identify and sort B-1a (CD19+CD5+CD11b+), B-1b (CD19+CD5-CD11b+), B-2 (CD19+CD5-CD11b-) cells from the peritoneum of C57BL/6J mice using iCyt Synergy sorter system from (Sony Biotechnology, San Jose, CA). Anti-CD19 and anti-CD90.2 antibodies were used to identify and sort T-cells (CD19-CD90.2+). Intracellular staining of IL-10 and IFN-γ was performed according to Biolegend protocol after a short-term stimulation with PMA and Ionomycin for 4 hours.
Immunoblotting
Cells were lysed in Cell Signaling lysis buffer containing protease and phosphatase inhibitors. Proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and processed for Western blotting. The blots were developed with HyGLO chemiluminescence reagent and exposed to HyBlot CL autoradiography film (Denville Scientific, Holliston, MA). Band densitometry analysis was performed using the NIH ImageJ program. Phospho-protein levels were normalized to total protein. Total protein levels were normalized to either GAPDH or β-actin expression.
Short hairpin RNA (shRNA) sequence and cell infection
Lyn shRNAs in the pLKO.1 lentiviral vector were validated and selected by the RNAi Consortium and glycerol stocks were purchased from ThermoFisher Scientific. MEC1 cells were seeded at a density of 1.5×106 cells/ml in a 6-well assay plate, infected with 25–50µl of concentrated shRNA lentiviral supernatants with the addition of polybrene (10µg/ml) and centrifuging for 90 minutes at 2800rpm and 10°C. Virus and cells were incubated for 24hrs at 37°C and then fresh media was replenished. Puromycin antibiotic selection began at day 3 and remained in culture for the entire period of experimentation after proper titration. Lyn shRNA Clone# TRCN0000010101 is represented in this paper.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was isolated from Eµ-TCL1 cells using TRI reagent (Sigma-Aldrich). RNA was used to make cDNA using qScript cDNA SuperMix (Quanta Biosceince, Gaithersburg, MD) according to the manufacturer’s protocol. iTaq Universal SYBR Green Supermix (BIO-RAD, Hercules, CA) was used to perform the RT-PCR reaction. RT-PCR was performed on StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA). Primer sequences used were as follows: IL-10 forward, 5’-ACTGGCATGAGGATCAGCAG-3’; IL-10 reverse, 5’-AGAAATCGATGACAGCGCCT-3’; SP1 forward, 5’-TGCCACCATGAGCGACCAAGATCA-3’; SP1 reverse, 5’-TGCTGCTGCTTCGAGTCTGAGAAA-3’; 18S forward, 5’-CGCCGCTAGAGGTGAAATTCT-3’; 18S reverse, 5’-CGAACCTCCGACTTTCGTTCT-3’. The 18S–specific primers were used for loading control. All primers were obtained from IDT technologies (Coralville, IA).
Chromatin Immunoprecipitation (ChIP) for qChIP analysis
ChIP analysis was performed using Cell Signaling SimpleChIP Enzymatic Chromatin IP kit following manufacturer’s protocol. Proteins linked to the DNA were immunoprecipitated with anti-Sp1 antibody. For qChIP, RT-PCR was performed on the eluted DNA using SYBR Green Reaction Mix. Specific primers used to amplify a region of the IL-10 promoter described to contain an Sp1 binding site (24) were as follows; forward, 5’-GCAGAAGTTCATTCCGACCA-3’; reverse, 5’-GGCTCCTCCTCCCTCTTCTA-3’.
Human CLL IGHV Sequencing
For each patient, total RNA from PBMCs was reverse transcribed and immunoglobulin sequences were amplified from cDNA with IGH family specific forward primers and isotype specific reverse primers, as previously described (25, 26). The resulting PCR products were unidirectionally Sanger sequenced and aligned via the online IG BLAST tool. Sequence reads covered the entire variable domain and included the first 38–124 nucleotides of the constant region. For each patient only one productive sequence was obtained, and thus was termed the major clone. Patients with ≥98% sequence homology to the germline variable domain were categorized as unmutated, as others have done previously (25–28).
Tissue Histology and Disease Scoring
Colons were dissected from euthanized mice and fixed in 10%-buffered formalin (Fisher Scientific #SF93-4) (Fair Lawn, NJ), embedded in paraffin, and stained with hematoxylin and eosin (H&E) by the University of Kentucky histology services. Histological scoring was based on the method previously described (29).
Statistical analysis
GraphPad Prism 7 was used for statistical analyses. Statistical significance of differences between groups was evaluated by Student’s t test and p-values <0.05 were considered significant. Normality testing was conducted and wherever appropriate nonparametric tests were employed.
Results
Blocking of IL-10 signaling does not affect in vitro survival of the Eµ-TCL1 CLL cells
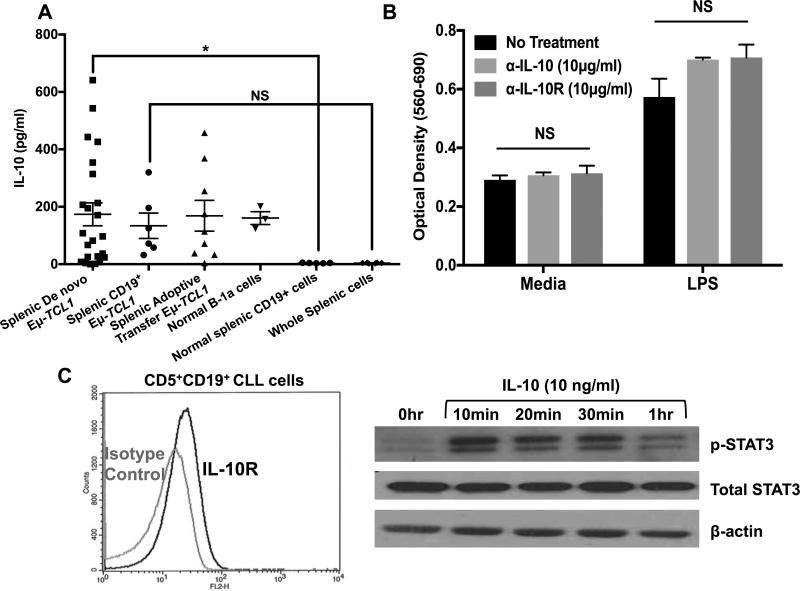
Eµ-TCL1 CD5+CD19+ CLL cells secrete IL-10 constitutively; similar to the peritoneal CD5+CD11b+ B-1 cells, although the amount produced is variable (Fig. 1A) (30). This constitutive production is a property of CLL B-cells since highly purified CD19+ CLL B-cells and unpurified Eµ-TCL1 splenic cells from mice with de novo disease or from adoptive transfer models produced similar amounts of IL-10, whereas normal spleen cells do not produce IL-10 (Fig. 1A). Neutralization of CLL-induced IL-10 using anti-IL-10 antibodies or anti-IL-10 receptor (IL-10R) antibodies did not affect the survival of Eµ-TCL1 cells with or without TLR4 stimulation (~1000pg/ml IL-10 produced upon LPS treatment) (Fig. 1B) in agreement with our previous observations (30). Flow cytometry analysis confirmed the presence of IL-10R on the surface of Eµ-TCL1 splenic CLL cells (Fig. 1C left) and it appeared to be functional since treatment of CLL cells with exogenous IL-10 induced phosphorylation of signal transducer and activator of transcription 3 (STAT3) transcription factor, a key-signaling molecule downstream of IL-10R (Fig. 1C right). Thus, Eµ-TCL1 cells appear to be distinct from normal B-1 cells in not responding to IL-10-mediated suppressive effects in vitro.
Figure 1. Constitutive IL-10 production by CLL cells and its role in CLL cell survival.
A) CLL cells were harvested from spleens of Eµ-TCL1 (n=23) or adoptive transfer mice (n=9). B-1a cells were isolated from the peritoneal cavity (PC) (n=3) and normal CD19+ cells were isolated from spleens of C57BL/6J mice (n=5). Purified CD19+ Eµ-TCL1 cells were cultured for 24 hours and IL-10 was measured in the supernatants by ELISA. Bars represent mean±SE. B) Splenic Eµ-TCL1 cells were cultured with αIL-10 or αIL-10R antibodies ±LPS (5µg/ml) for 48 hours. Survival was measured by MTT. Values represent mean±SD of triplicate cultures. C) Flow cytometric analysis of IL-10R expression by Eµ-TCL1 cells (left) and protein lysates of Eµ-TCL1 CLL cells stimulated with exogenous IL-10 were analyzed for STAT3 activation by immunoblot (right). β-actin is used for loading control. *p< 0.05, NS; not significant.
CLL cell growth is reduced in IL-10R−/− mice
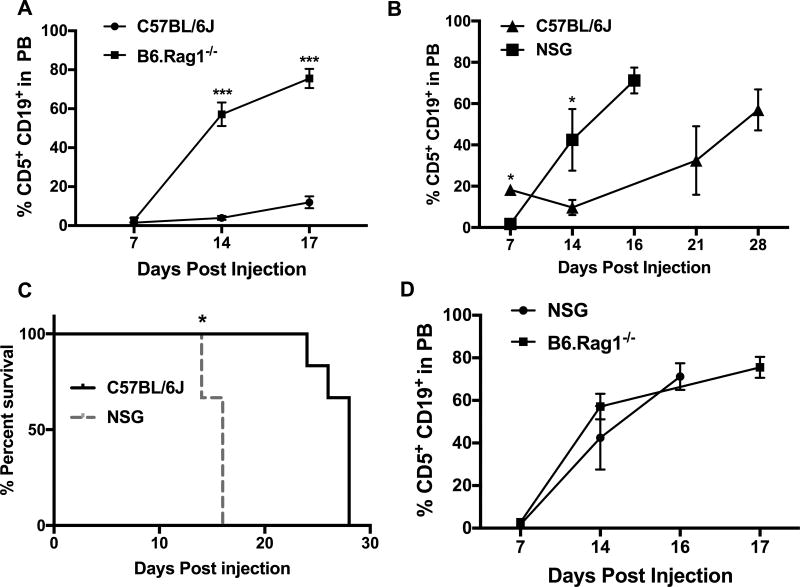
The lack of direct effects of CLL-derived IL-10 on their survival led us to hypothesize that IL-10 may contribute to immunosuppression in CLL by affecting adaptive immune responses to CLL. First, we evaluated if there is an adaptive tumor immune response against CLL in the Eµ-TCL1 mouse model, utilizing an adoptive transfer model by injecting CLL cells from the de novo Eµ-TCL1 mouse into syngeneic C57BL/6J wild type (WT) or immunodeficient mice (NSG or B6.Rag1−/−). NSG mice lack mature B and T-cells, as well as natural killer (NK) cells, while B6.Rag1−/− mice do not have an adaptive immune response due to a total lack of B and T-cells. CLL cells in B6.Rag1−/− mice were detectable at an earlier time point than WT mice and had to be euthanized at day 17 due to poor body condition with an average of 75% CD5+CD19+ cells in PB (Fig. 2A), whereas WT mice had only ~10% CD5+CD19+ cells in PB at this time (Fig. 2A). Similarly, in NSG mice, CLL cells grew at a faster rate than WT mice (Fig. 2B). Overall, C57BL/6J mice survived longer than NSG mice (Fig. 2C). Despite the fact that NSG mice were on a NOD background no significant difference was observed between CLL growth kinetics in NSG and B6.Rag1−/− mice (Fig. 2D).
Figure 2. Lack of B, T and NK cells leads to an acceleration of CLL growth kinetics.
A–D) Eµ-TCL1 cells (4×106) were adoptively transferred into WT, B6.Rag1−/− or NSG mice by retro-orbital injection. Leukemic status is determined by weekly submandibular bleeding. Graph shows the % CD5+CD19+ cells in the peripheral blood at indicated time points (A, B and D). Kaplan-Meier blot represents the survival of C57BL/6J and NSG mice during the course of the experiment (n=6) (C). * p< 0.05, *** p< 0.001 determined by Student’s t-test for panels A, B and D and by Log-rank (Mantel-Cox) test for panel C. Similar results were obtained in another experiment.
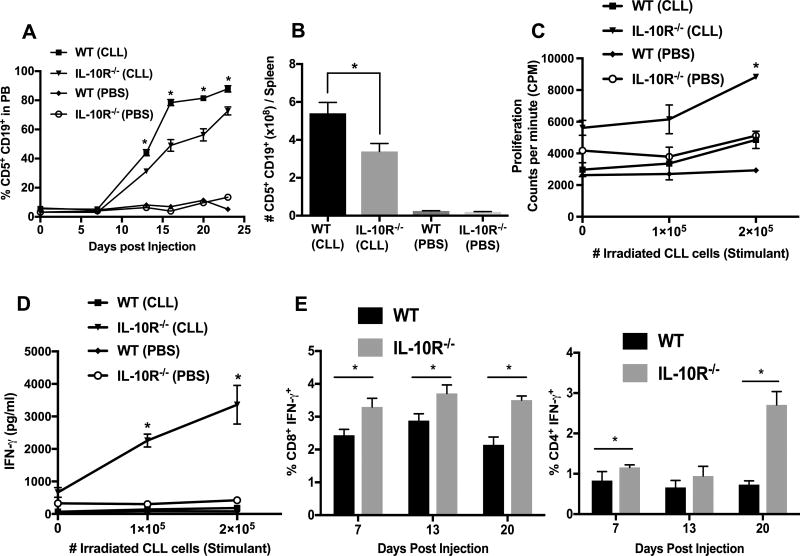
The difference in CLL growth kinetics between immunodeficient and C57BL/6J mice could be due to adaptive immunity and/or other differences in the microenvironment. Hence we investigated if murine CLL-derived IL-10 could inhibit anti-CLL immune responses. CLL cells were injected into WT and C57BL/6.IL-10R−/− mice. We hypothesized that the lack of IL-10R will eliminate any inhibitory effects of IL-10 in the CLL microenvironment of the recipient mice including its effects on T-cells allowing more effective anti-CLL immune response and hence a delay in CLL development. Accordingly, we found that CLL cells grew at a slower rate in IL-10R−/− mice than in WT mice (Fig. 3A). After euthanization, CLL tumor burden in spleen, BM, and peritoneal cavity (PC) was higher in WT mice than in IL-10R−/− mice (Fig. 3B and Supp. Fig. 2A,B). There was no significant difference in plasma IL-10 levels between WT and IL-10R−/− recipient mice at the end of the experiment (Supp. Fig. 2C). We used 3-weeks-old mice as recipients, since IL-10R−/− mice are known to develop colitis by 12 weeks of age. To be certain that the death of recipient mice in this experiment is not due to colitis we performed colon histology. Colon sections from both WT and IL-10R−/− recipients at the time of euthanization showed no significance difference in inflammation by histopathology (Supp. Fig. 2D).
Figure 3. CLL cell growth is reduced and T-cell function is enhanced in IL-10R−/− mice.
A–B) Eµ-TCL1 CLL cells (4×106) were intravenously transferred into WT and IL-10R−/− mice (n=5). Leukemia status was monitored by weekly bleeding and is shown as CD5+CD19+ cells after gating on CD45+ cells in PB (A) and tumor burden was expressed as total number of CD5+CD19+ cells per spleen (B). Values represent arithmetic mean±SD (n=5). C–D) Magnetic bead purified CD8+ cells (105) from WT and IL-10R−/− mice 20-days-post CLL injection were cultured with irradiated CLL cells for 72 hours. Proliferation, measured by 3[H] incorporation (C), and IFN-γ secretion by CD8+ T-cells (D) are shown. Values represent mean±SD of triplicate cultures. E) Intracellular staining of IFN-γ was performed after 4 hours of stimulation with PMA and Ionomycin. Levels of cytoplasmic IFN-γ+ CD8+(left) and CD4+(right) T-cells from WT and IL-10R−/− mice at different times post-CLL injection. Values represent arithmetic mean±SE (n=4). *p< 0.05 comparing wild type and IL-10R−/− groups.
Decrease in T-cell function in wild type compared to IL-10R−/− mice injected with CLL
To determine if the reduced CLL growth in IL-10R−/− mice is due to the inhibitory effects of IL-10 on host anti-CLL immune response, we investigated the function of T-cells in the adoptive transfer recipients. In agreement with such a notion, upon restimulation with CLL cells, CD8+ T-cells from IL-10R−/− recipients proliferated better than CD8+ T-cells from WT mice at day-20 post-injection (Fig. 3C). Similar results were found at other time points (data not shown). Also CD8+ T-cells isolated from IL-10R−/− recipients secreted more IFN-γ than CD8+ T-cells from WT recipients (Fig. 3D). In addition, intracellular staining of IFN-γ was performed after 4 hour stimulation with PMA and ionomycin. Cytoplasmic IFN-γ+ CD4+ and CD8+ T-cells were higher in IL-10R−/− than WT mice injected with CLL cells (Fig. 3E). These data suggest a role for IL-10 in inhibiting T-cell responses against Eµ-TCL1 CLL cells in vivo.
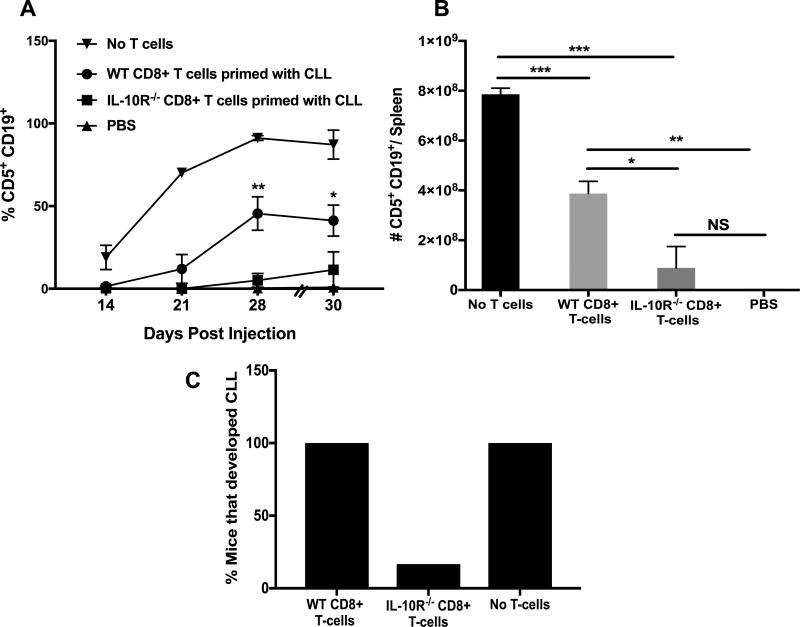
IL-10R−/− T-cells controlled CLL growth better than WT T-cells
To determine if the functional differences between WT and IL-10R−/− T-cells generated after CLL injection affects their ability to control CLL growth in vivo, we performed adoptive transfer experiments using NSG mice as recipients (Supp. Fig. 1A). Overall, injection of primed WT or IL-10R−/− T-cells along with CLL cells delayed onset of disease in PB by a significant amount of time (Supp. Fig. 3A). At day-65, when recipients were euthanized 60% of WT T cell recipients but none of the IL-10R−/− T cell recipients developed CLL disease (Sup. Fig. 3B). Thus IL-10R−/− T-cells that cannot respond to IL-10 were more effective in controlling CLL disease than IL-10 responsive wild type T-cells. To determine if the anti-CLL response of T cells is due to CD8+ T-cells, we injected purified CD8+ T-cells from WT and IL-10R−/− mice primed with CLL along with CLL cells at a T-cell:CLL-cell ratio of 1:32 into NSG mice (Supp. Fig. 1B). Interestingly, CD8+ T-cells were sufficient to delay CLL growth and CD8+ T-cells from IL-10R−/− mice were better at controlling disease development than CD8+ T-cells from WT mice (Fig. 4A). Upon euthanization, spleens of mice receiving IL-10R−/− CD8+ T-cells had fewer CLL cells than mice receiving WT CD8+ T-cells; however, no significant difference was observed between groups receiving CD8+ T-cells from IL-10R−/− mice and PBS control mice (Fig. 4B). At day-30 post-injection, 100% of the mice that received no T-cells or WT CD8+ T-cells but only 16.7% of IL-10R−/− CD8 T-cell recipients developed disease (Fig. 4C).
Figure 4. Adoptive transfer of CLL primed CD8+ T cells effectively delayed CLL growth.
A) Purified CD8+ cells from WT and IL-10R−/− mice primed with CLL cells 14-days before were injected into NSG mice along with CLL cells at a T-cell:CLL ratio of 1:32. Leukemic status was monitored by CD5+CD19+ cell accumulation in PB. Values represent mean±SE (n=6). B) CD5+CD19+ cell numbers in the spleens of recipients at the time of euthanization. C) Percentage of mice that developed CLL from each group. *p< 0.05, p**< 0.01, p***< 0.001, NS; not significant comparing disease in recipients of wild type and IL-10R−/− CD8+ T cells.
Novel role of BCR-signaling and Sp1 in constitutive and induced IL-10 production by CLL cells
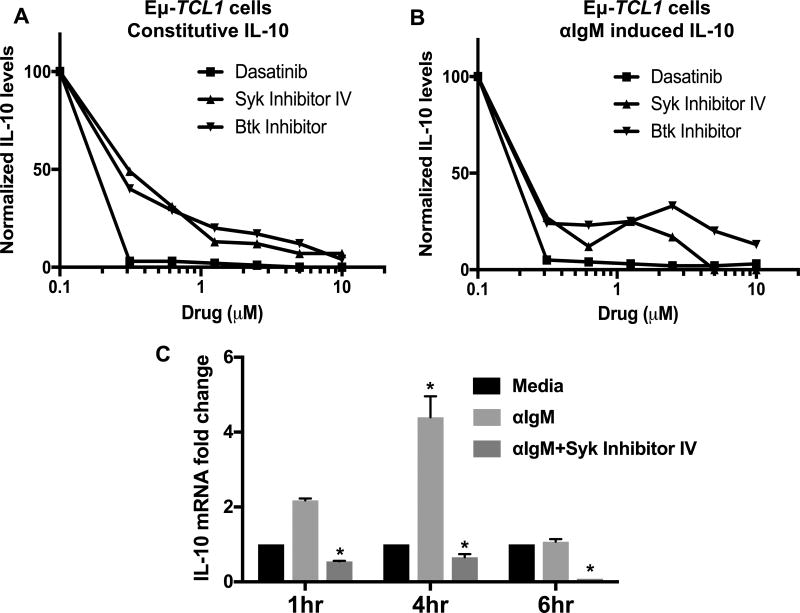
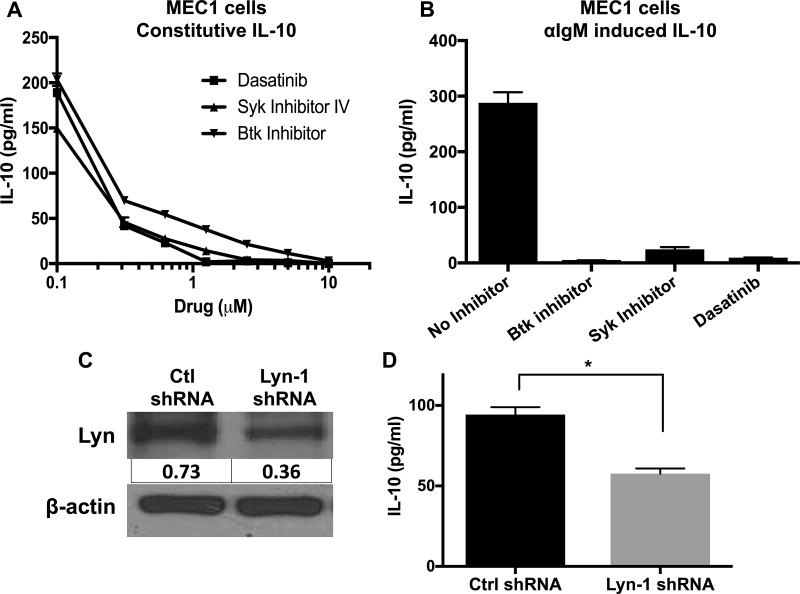
Establishing a role for IL-10 in CLL growth led us to investigate the possible mechanisms by which IL-10 is produced by CLL cells. Previously, we reported our preliminary studies about the novel role of BCR-signaling in IL-10 production by normal B-1 and malignant Eµ-TCL1 CLL cells (30). Inhibition of Src, Btk or Syk family kinases that are essential for signal transduction via BCR reduced both constitutive and anti-IgM induced IL-10 production by Eµ-TCL1 cells in a dose dependent manner (IC50≈0.3µM) (Fig. 5A,B). BCR cross-linking increases IL-10 mRNA levels, which were reduced by the Syk inhibitor (Fig. 5C), suggesting that IL-10 production is controlled at the transcript level. Intracellular staining for IL-10 showed that 60–80% of CLL cells were IL-10 producers, which was reduced by Syk inhibition (Supp. Fig. 4A–B). MEC1, a human CLL cell line, also secretes IL-10 constitutively, which is diminished by inhibition of Src family kinase (SFK), Syk or Btk (Fig. 6A). Moreover, BCR cross-linking in MEC1 cells increases IL-10 production, which is also diminished by the BCR inhibitors (Fig. 6B). In addition, we used a short hairpin RNA (shRNA) to knock down Lyn, an SFK, and one of the earliest enzymes activated with BCR cross-linking. MEC1 cells with 50% lyn knock-down produced less IL-10 than control shRNA treated cells (Fig. 6C,D).
Figure 5. The role of BCR signaling in IL-10 production by Eµ TCL1 CLL cells.
A–B) Eµ-TCL1 cells were cultured without (A) or with (B) αIgM ±indicated doses of dasatinib (an SFK inhibitor), Syk inhibitor-IV or Btk inhibitor (Ibrutinib). IL-10 levels in supernatants were measured after 24 hours (at which time the cell viability was 75–80%). Values are normalized to the no drug control. IL-10 range for untreated Eµ-TCL1 cells is 193–349pg/ml. C) IL-10 mRNA levels (normalized to mouse 18S RNA) are determined by qRT-PCR in Eµ-TCL1 CLL cells treated with or without αIgM ±Syk inhibitor-IV. Values represent mean±SD of triplicates. Results are representative of 3–8 experiments. *p< 0.05 comparing untreated to αIgM±Syk inhibitor treated cells.
Figure 6. The role of BCR signaling in IL-10 production by MEC1 cells.
A) MEC1 cells were cultured without (A) or with (B) αIgM ± dasatinib, Syk inhibitor-IV or Btk inhibitor. IL-10 levels in supernatants were measured after 24 hours. For (B) 2µM of indicated drugs is used. C) Immunoblot showing a reduction in Lyn in MEC1 cells expressing Lyn specific shRNA. Lyn protein values were normalized to β-actin. D) IL-10 levels were measured in the supernatants of MEC1 cells expressing either control or Lyn shRNA. Values represent mean±SD of triplicates. Results are representative of 2–5 experiments. *p< 0.05.
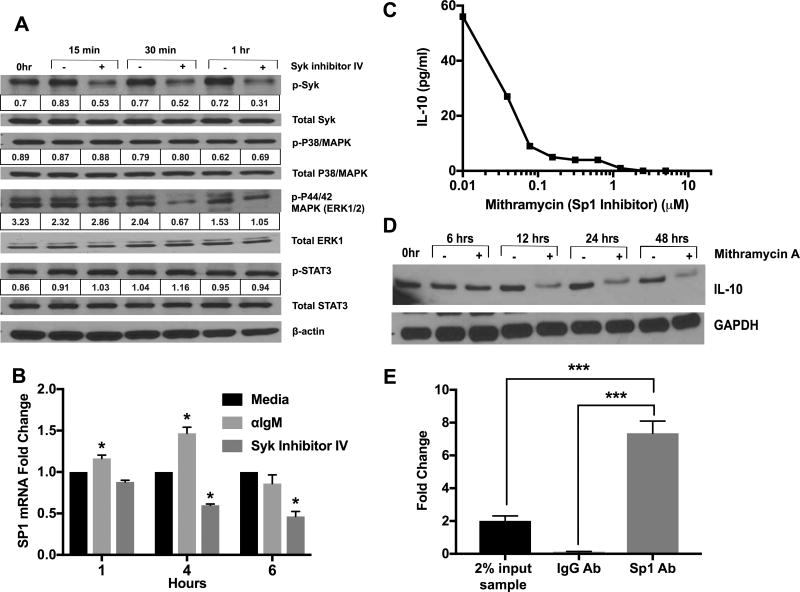
Importance of p38-MAPK and STAT3 was tested since they are known to be involved in IL-10 production by myeloid cells. Surprisingly, phosphorylation of the p38-MAPK or STAT3 was not affected by Syk inhibition (Fig. 7A). On the other hand, phosphorylation of extracellular signal-regulated kinase 1/2 (ERK1/2) was reduced after Syk inhibition (Fig. 7A). Similar results are obtained with ibrutinib (data not shown).
Figure 7. IL-10 production by Eµ TCL1 CLL cells is dependent on ERK1/2 MAPK and the transcription factor Sp1 but not on p38MAPK or STAT3.
A) Eµ-TCL1 cells were treated with Syk inhibitor-IV (5µM) for indicated time periods. Levels of key molecules downstream of BCR signaling were quantified by Immunoblotting. Band densitometry analysis was performed using the NIH ImageJ program. Phospho-protein levels were normalized to total protein. Results are representative of three experiments. B) Sp1 mRNA levels were quantified by qRT-PCR after treatment of Eµ-TCL1 cells with αIgM ±Syk inhibitor-IV. Fold change was normalized to the no-treatment group. Values represent mean±SD of triplicate determinations. C) Eµ-TCL1 cells were treated with various doses of mithramycin A for 24 hours and IL-10 in the supernatant was quantified by ELISA. D) Immunoblot analysis of IL-10 protein levels in CLL cells after treatment with mithramycin A (5µM). E) ChIP with anti-Sp1 or control IgG was carried out as described in the Methods. qRT-PCR was performed on the ChIP DNA product using primers specific for the consensus Sp1 binding site sequence in the IL-10 promoter. Results are calculated using the Fold Enrichment Method. *p< 0.05, ***p<0.001 comparing untreated to αIgM±Syk inhibitor treated cells.
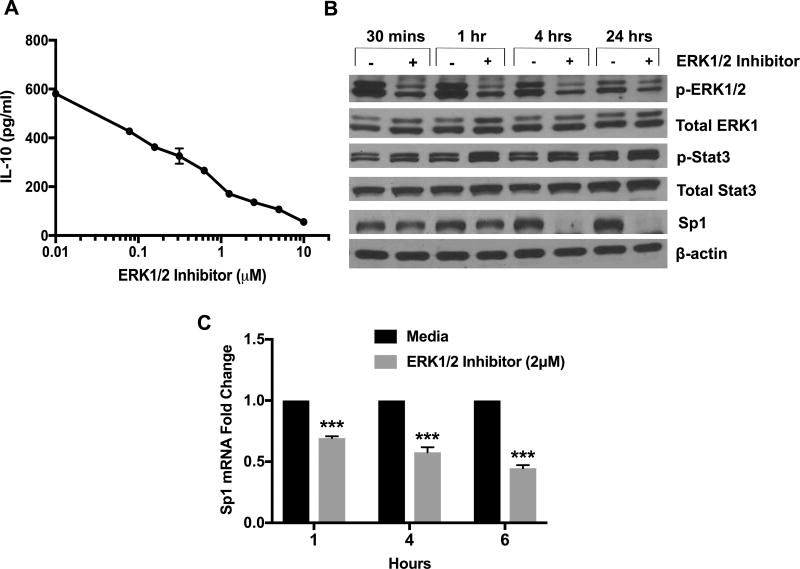
In order to find a possible downstream transcription factor, which is involved in BCR induced IL-10 production, we tested the transcript levels of multiple transcription factors (SMAD4, GATA3, CREB, ATF1 and Sp1) known to be required for IL-10 transcription in various immune cells (Supp. Fig, 4C) (18). Of these Sp1 was the only transcription factor enhanced by BCR signaling and reduced by Syk inhibition (Fig. 7B). Mithramycin A, a well-known inhibitor of Sp1 (31), reduced IL-10 protein levels in a dose dependent manner (Fig. 7C,D). Sp1 is proposed to bind and transactivate IL10 gene in macrophages and T-cells (32). To further establish its role in IL-10 transcription in Eµ-TCL1 cells, we utilized chromatin immunoprecipitation (ChIP) to test if Sp1 binds to the IL-10 promoter in CLL cells. RT-PCR of the ChIP product revealed 8-fold enrichment in binding of Sp1 to IL-10 promoter over the input sample (Fig. 7E). Taken together, these results indicate that IL-10 production by murine CD5+CD19+ CLL cells is dependent on the transcription factor Sp1. Since ERK1/2 activation is regulated by BCR signaling (Fig. 7A), we hypothesized that ERK1/2 activation is the link between BCR and Sp1, based on previous studies (33). Accordingly, treatment of CLL cells with SCH772984, a specific ERK1/2 inhibitor decreased IL-10 in a dose-dependent manner (Fig. 8A). Interestingly, Sp1 protein and transcript levels were reduced upon ERK1/2 inhibition (Fig. 8B,C). Also, consistent with data using Syk inhibitor, ERK1/2 inhibitor did not decrease activation of STAT3 (Fig. 8B). This suggests that IL-10 production by CLL cells is regulated by ERK1/2 dependent activation of Sp1.
Figure 8. Effect of ERK1/2 inhibition on Sp1 and IL-10 levels.
Eµ-TCL1 CLL cells were cultured with the ERK1/2 inhibitor (SCH772984) (2µM). A) IL-10 levels in 24-hour supernatants were measured by ELISA. Values represent mean±SD of triplicates. B) Levels of p-ERK1/2, total ERK1, p-STAT3, total STAT3 and Sp1 were quantified by immunoblot analysis. Results are representative of three experiments. C) Sp1 mRNA levels were quantified by qRT-PCR. Fold change was normalized to the no-treatment group. Values represent mean±SD of triplicates ***p<0.001 comparing groups with and without ERK1/2 inhibitor.
BCR signaling regulates IL-10 production by human CLL cells
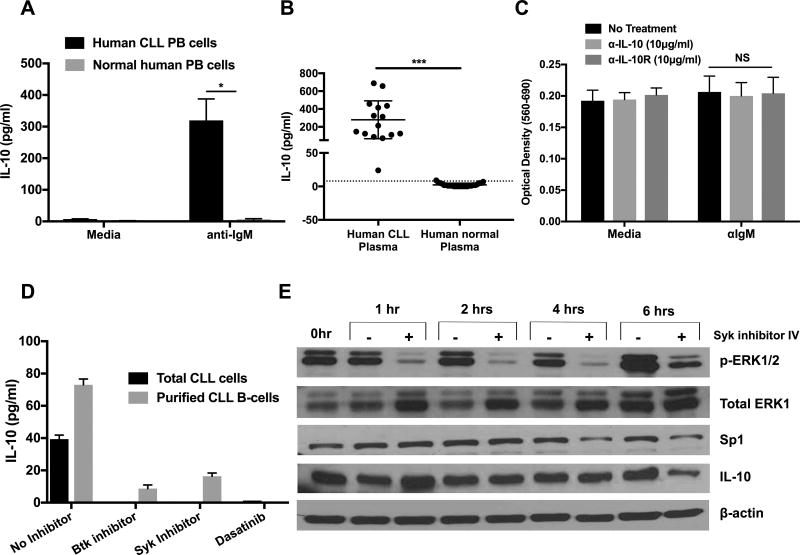
Human CLL peripheral blood mononuclear cells (PBMCs) produced a significant amount of IL-10 only after BCR cross-linking in comparison to normal human PBMCs, with very little constitutive production (Fig. 9A). There was a significant amount of IL-10 in the plasma of CLL patients in comparison to age-matched controls (Fig. 9B), suggesting that in vivo CLL cells may be receiving activation signals via BCR. Neutralization of anti-IgM induced-IL-10 did not affect the survival of human CLL cells (Fig. 9C). Inhibition of Src, Syk family kinases, Btk and ERK1/2 led to the complete abrogation of anti-IgM induced IL-10 production by human CLL cells (Fig. 9D). To confirm that CLL cells were the source of IL-10, we show that B-cells purified from the PBMCs of CLL patients secrete IL-10, which was also diminished by BCR inhibitors (Fig. 9D). Finally, inhibition of BCR signaling in human CLL reduced ERK1/2 activation, Sp1 and IL-10 levels (Fig. 9E).
Figure 9. Human CLL cells utilize BCR signaling for IL-10 production.
A–B) IL-10 levels in 24-hour supernatants of human CLL cells ±αIgM (A) or in the plasma of human CLL patients (n=15) and normal donors (n=28) (B). C) Human CLL cells were cultured with αIL-10 or αIL-10R antibodies ±αIgM for 48 hours. Survival of CLL cells was measured by MTT. D) Human total CLL cells or purified CLL B-cells were stimulated with αIgM±inhibitors of Btk, Syk, SFK or ERK1/2 for 24 hours (2µM). IL-10 levels in the supernatants were measured by ELISA. Human B-cell CLL cells were purified by negative selection (MojoSort Human B cells (CD43−) Isolation Kit from Biolegend. E) Human CLL cells were treated with Syk inhibitor-IV (2µM). Protein levels indicated were quantified by immunoblot analysis. For (A, C and D), values represent mean±SD of triplicate cultures. *p< 0.05, ***p< 0.0001, NS; not significant.
Discussion
A variety of functional defects in T-cells from patients with CLL as well as in Eµ-TCL1 mice with CLL have been reported including, reduction in activation-induced CD40L expression, immunologic synapse formation with APCs and alteration in gene expression profiles (34, 35). Here, for the first time, we show a clear link between CLL-derived IL-10 and dysfunction of T-cell responses to CLL cells. We demonstrated that T-cells from IL-10R−/− mice primed with CLL proliferate and differentiate better than T-cells isolated from mice with intact IL-10 signaling. Furthermore, primed T-cells and purified CD8+ T-cells reduced incidence and progression of the disease in immunodeficient mice injected with CLL cells. T-cells from IL-10R−/− mice were better at controlling CLL growth.
IL-10 has been found to have a dual function of immune stimulation and immune suppression in different types of cancer (36), which explains the complexity of studying the effects of IL-10 on cancer. For example, previous reports have shown contradictory results regarding the requirement of IL-10 for the survival and proliferation of CLL cells. Fluckiger et al reported that exogenous IL-10 inhibited the proliferation of human CLL cells and decreased the survival of CLL cells in culture by inducing apoptosis (37), similar to normal B-1 cell regulation by IL-10, as we showed previously that normal B-1 cell-derived IL-10 inhibited their proliferation responses to TLR or BCR ligation (8, 30). However, Kitabayashi et al reported that IL-10 prevents apoptotic cell death of CLL cells (38). In our study both mouse and human CLL cells appear to be unique in that their survival/proliferation is not suppressed directly by IL-10 in vitro but that IL-10 affects CLL growth indirectly by suppressing anti-CLL T-cells.
The question then remains of why T-cells in the de novo CLL disease do not control disease development. T-cells isolated from CLL patients have higher expression of checkpoint molecules such as cytotoxic T-lymphocyte-associated protein-4 and PD-1 (39, 40). This possibility of multiple pathways of immunosuppression in CLL raises the need for combination therapy to target multiple modulators of immune suppression. The combination of different immunotherapies is widely studied and is demonstrating tremendous promise for the treatment of diverse tumor types. In an interesting study, it was found that PD-1high tumor antigen specific CD8+ T-cells in patients with advanced melanoma upregulate IL-10R expression (41). When IL-10 directly acts on these CD8+ T-cells, it limits their proliferation and survival (41). The authors found that PD-1 blockade increases the expression of IL-10R by CD8+ T cells, increasing their sensitivity to the immunosuppressive effects of endogenous IL-10 (41). However when they used combination therapy of IL-10 and PD-1 inhibition, IL-10 blockade strengthened the effects of PD-1 blockade in expanding tumor specific CD8+ T-cells (41). Another study found that treatment of ovarian tumor-bearing mice with anti-PD-1 antibody caused as increase in IL-10 levels, which contributed to the resistance seen with anti-PD-1 monotherapy (17). When the combination of PD-1 blockade and IL-10 neutralization was used, improved survival and delayed tumor growth were seen in these mice (17).
Future studies will need to identify the CLL antigens recognized by the T-cells, which could be utilized in developing new chimeric antigen receptor T-cell therapy for CLL. A novel category of tumor-associated T-cell antigens were identified in a recent study that analyzed the landscape of naturally presented HLA class I and II ligands of primary CLL (42). Specific expression of these HLA ligands exclusively in CLL patients correlated with the frequencies of immune recognition by patient T-cells (42). Interestingly, patients displaying immune responses to multiple antigens exhibited better survival than those with responses to one or fewer antigens (42). Variable efficacy of CAR-T cells in CLL could be due to this suppression by CLL derived IL-10, which can be targeted to make CAR T-cells more effective.
Understanding the specific molecular events that regulate the production of IL-10 will help in designing new strategies of immune intervention. It was found that BAFF stimulation via TACI receptor enhanced IL-10 production by leukemic B-cells in CLL patients and Eµ-TCL1 mice (43). Another study demonstrated an epigenetic control of IL-10 production, in which differential IL10 gene methylation was responsible for the variability of IL-10 production by human CLL cells (44). Our studies demonstrated a novel role of BCR signaling in the constitutive production of IL-10. A novel role for Sp1 in BCR signaling dependent IL-10 production by CLL cells is described here. Sp1 bound to the IL-10 promoter in CLL cells and inhibition of Sp1 led to a decrease in IL-10 production. Sp1 activity required activation of ERK1/2, which is consistent with regulation of Sp1 by ERK1/2 in other cell types (45, 46). Our results introduce a new rationale to therapeutics targeting the ERK-Sp1 pathway.
In conclusion, we demonstrated that BCR signaling, a key mechanism for the survival of normal and CLL B-cells, leads to IL-10 production by CLL B-cells. The CLL-derived IL-10 suppresses anti-CLL immune response. Hence inhibition of IL-10 production by small molecules such as mithramycin or its analogues with less toxicity could be used in combination with current CLL therapies such as BCR signaling inhibitors and combination chemotherapy as well as the new therapeutic modalities like checkpoint inhibitor or CAR-T cells to induce better anti-CLL immunity.
Supplementary Material
Acknowledgments
We are thankful to Dr. Greg Bauman and Jennifer Strange for their help with flow cytometry and Beth Gachuki for her technical support. We thank Dr. Donna Wilcock for help with normal human plasma samples.
This work was supported by NIH grants (CA 165469), the “Flow Cytometry” and the “Biospecimen Procurement and Translational Pathology” shared resource facilities of the Markey Cancer Center (P30CA177558), and the Alzheimer’s Disease Center grant (P30-AG028383) at the University of Kentucky.
Footnotes
The authors declare no competing financial interests.
References
- 1.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352:804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 2.Hayakawa K, Formica AM, Brill-Dashoff J, Shinton SA, Ichikawa D, Zhou Y, Morse HC, 3rd, Hardy RR. Early generated B1 B cells with restricted BCRs become chronic lymphocytic leukemia with continued c-Myc and low Bmf expression. J Exp Med. 2016;213:3007–3024. doi: 10.1084/jem.20160712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayakawa K, Formica AM, Colombo MJ, Ichikawa D, Shinton SA, Brill-Dashoff J, Hardy RR. B cells generated by B-1 development can progress to chronic lymphocytic leukemia. Ann N Y Acad Sci. 2015;1362:250–255. doi: 10.1111/nyas.12768. [DOI] [PubMed] [Google Scholar]
- 4.Morrison VA. Infectious complications of chronic lymphocytic leukaemia: pathogenesis, spectrum of infection, preventive approaches. Best Pract Res Clin Haematol. 2010;23:145–153. doi: 10.1016/j.beha.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Nosari A. Infectious complications in chronic lymphocytic leukemia. Mediterr J Hematol Infect Dis. 2012;4:e2012070. doi: 10.4084/MJHID.2012.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Royle JA, Baade PD, Joske D, Girschik J, Fritschi L. Second cancer incidence and cancer mortality among chronic lymphocytic leukaemia patients: a population-based study. Br J Cancer. 2011;105:1076–1081. doi: 10.1038/bjc.2011.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Garra A, Chang R, Go N, Hastings R, Haughton G, Howard M. Ly-1 B (B-1) cells are the main source of B cell-derived interleukin 10. Eur J Immunol. 1992;22:711–717. doi: 10.1002/eji.1830220314. [DOI] [PubMed] [Google Scholar]
- 8.Sindhava V, Woodman ME, Stevenson B, Bondada S. Interleukin-10 mediated autoregulation of murine B-1 B-cells and its role in Borrelia hermsii infection. PLoS ONE. 2010;5:e11445. doi: 10.1371/journal.pone.0011445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiorentino DF, Bond MW, Mosmann TR. Two types of mouse T helper cell. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. J Exp Med. 1989;170:2081–2095. doi: 10.1084/jem.170.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiorentino DF, Zlotnik A, Vieira P, Mosmann TR, Howard M, Moore KW, O'Garra A. IL-10 acts on the antigen-presenting cell to inhibit cytokine production by Th1 cells. J Immunol. 1991;146:3444–3451. [PubMed] [Google Scholar]
- 11.Couper KN, Blount DG, Riley EM. IL-10: the master regulator of immunity to infection. J Immunol. 2008;180:5771–5777. doi: 10.4049/jimmunol.180.9.5771. [DOI] [PubMed] [Google Scholar]
- 12.Schandene L, Alonso-Vega C, Willems F, Gerard C, Delvaux A, Velu T, Devos R, de Boer M, Goldman M. B7/CD28-dependent IL-5 production by human resting T cells is inhibited by IL-10. J Immunol. 1994;152:4368–4374. [PubMed] [Google Scholar]
- 13.Joss A, Akdis M, Faith A, Blaser K, Akdis CA. IL-10 directly acts on T cells by specifically altering the CD28 co-stimulation pathway. Eur J Immunol. 2000;30:1683–1690. doi: 10.1002/1521-4141(200006)30:6<1683::AID-IMMU1683>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 14.DiLillo DJ, Weinberg JB, Yoshizaki A, Horikawa M, Bryant JM, Iwata Y, Matsushita T, Matta KM, Chen Y, Venturi GM, Russo G, Gockerman JP, Moore JO, Diehl LF, Volkheimer AD, Friedman DR, Lanasa MC, Hall RP, Tedder TF. Chronic lymphocytic leukemia and regulatory B cells share IL-10 competence and immunosuppressive function. Leukemia. 2013;27:170–182. doi: 10.1038/leu.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bichi R, Shinton SA, Martin ES, Koval A, Calin GA, Cesari R, Russo G, Hardy RR, Croce CM. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6955–6960. doi: 10.1073/pnas.102181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schubert ML, Huckelhoven A, Hoffmann JM, Schmitt A, Wuchter P, Sellner L, Hofmann S, Ho AD, Dreger P, Schmitt M. Chimeric Antigen Receptor T Cell Therapy Targeting CD19-Positive Leukemia and Lymphoma in the Context of Stem Cell Transplantation. Hum Gene Ther. 2016;27:758–771. doi: 10.1089/hum.2016.097. [DOI] [PubMed] [Google Scholar]
- 17.Lamichhane P, Karyampudi L, Shreeder B, Krempski J, Bahr D, Daum J, Kalli KR, Goode EL, Block MS, Cannon MJ, Knutson KL. IL10 Release upon PD-1 Blockade Sustains Immunosuppression in Ovarian Cancer. Cancer research. 2017;77:6667–6678. doi: 10.1158/0008-5472.CAN-17-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saraiva M, O'Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol. 2010;10:170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 19.Ullman-Cullere MH, Foltz CJ. Body condition scoring: a rapid and accurate method for assessing health status in mice. Lab Anim Sci. 1999;49:319–323. [PubMed] [Google Scholar]
- 20.Ramsay AG, Johnson AJ, Lee AM, Gorgun G, Le Dieu R, Blum W, Byrd JC, Gribben JG. Chronic lymphocytic leukemia T cells show impaired immunological synapse formation that can be reversed with an immunomodulating drug. J Clin Invest. 2008;118:2427–2437. doi: 10.1172/JCI35017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golde WT, Gollobin P, Rodriguez LL. A rapid, simple, and humane method for submandibular bleeding of mice using a lancet. Lab Anim (NY) 2005;34:39–43. doi: 10.1038/laban1005-39. [DOI] [PubMed] [Google Scholar]
- 22.Plumb JA. Cell sensitivity assays: the MTT assay. Methods Mol Med. 2004;88:165–169. doi: 10.1385/1-59259-406-9:165. [DOI] [PubMed] [Google Scholar]
- 23.Keir ME, Latchman YE, Freeman GJ, Sharpe AH. Programmed death-1 (PD-1):PD-ligand 1 interactions inhibit TCR-mediated positive selection of thymocytes. J Immunol. 2005;175:7372–7379. doi: 10.4049/jimmunol.175.11.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Edwards JP, Mosser DM. Dynamic and transient remodeling of the macrophage IL-10 promoter during transcription. J Immunol. 2006;177:1282–1288. doi: 10.4049/jimmunol.177.2.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, Buchbinder A, Budman D, Dittmar K, Kolitz J, Lichtman SM, Schulman P, Vinciguerra VP, Rai KR, Ferrarini M, Chiorazzi N. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]
- 26.Fais F, Ghiotto F, Hashimoto S, Sellars B, Valetto A, Allen SL, Schulman P, Vinciguerra VP, Rai K, Rassenti LZ, Kipps TJ, Dighiero G, Schroeder HW, Jr, Ferrarini M, Chiorazzi N. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest. 1998;102:1515–1525. doi: 10.1172/JCI3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–1854. [PubMed] [Google Scholar]
- 28.Patten PE, Chu CC, Albesiano E, Damle RN, Yan XJ, Kim D, Zhang L, Magli AR, Barrientos J, Kolitz JE, Allen SL, Rai KR, Roa S, Mongini PK, MacCarthy T, Scharff MD, Chiorazzi N. IGHV-unmutated and IGHV-mutated chronic lymphocytic leukemia cells produce activation-induced deaminase protein with a full range of biologic functions. Blood. 2012;120:4802–4811. doi: 10.1182/blood-2012-08-449744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berg DJ, Davidson N, Kuhn R, Muller W, Menon S, Holland G, Thompson-Snipes L, Leach MW, Rennick D. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4(+) TH1-like responses. J Clin Invest. 1996;98:1010–1020. doi: 10.1172/JCI118861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alhakeem SS, Sindhava VJ, McKenna MK, Gachuki BW, Byrd JC, Muthusamy N, Bondada S. Role of B cell receptor signaling in IL-10 production by normal and malignant B-1 cells. Ann N Y Acad Sci. 2015;1362:239–249. doi: 10.1111/nyas.12802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sleiman SF, Langley BC, Basso M, Berlin J, Xia L, Payappilly JB, Kharel MK, Guo H, Marsh JL, Thompson LM, Mahishi L, Ahuja P, MacLellan WR, Geschwind DH, Coppola G, Rohr J, Ratan RR. Mithramycin is a gene-selective Sp1 inhibitor that identifies a biological intersection between cancer and neurodegeneration. J Neurosci. 2011;31:6858–6870. doi: 10.1523/JNEUROSCI.0710-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brightbill HD, Plevy SE, Modlin RL, Smale ST. A prominent role for Sp1 during lipopolysaccharide-mediated induction of the IL-10 promoter in macrophages. J Immunol. 2000;164:1940–1951. doi: 10.4049/jimmunol.164.4.1940. [DOI] [PubMed] [Google Scholar]
- 33.Milanini-Mongiat J, Pouyssegur J, Pages G. Identification of two Sp1 phosphorylation sites for p42/p44 mitogen-activated protein kinases: their implication in vascular endothelial growth factor gene transcription. The Journal of biological chemistry. 2002;277:20631–20639. doi: 10.1074/jbc.M201753200. [DOI] [PubMed] [Google Scholar]
- 34.Cantwell M, Hua T, Pappas J, Kipps TJ. Acquired CD40-ligand deficiency in chronic lymphocytic leukemia. Nat Med. 1997;3:984–989. doi: 10.1038/nm0997-984. [DOI] [PubMed] [Google Scholar]
- 35.Gorgun G, Holderried TA, Zahrieh D, Neuberg D, Gribben JG. Chronic lymphocytic leukemia cells induce changes in gene expression of CD4 and CD8 T cells. J Clin Invest. 2005;115:1797–1805. doi: 10.1172/JCI24176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dennis KL, Blatner NR, Gounari F, Khazaie K. Current status of interleukin-10 and regulatory T-cells in cancer. Curr Opin Oncol. 2013;25:637–645. doi: 10.1097/CCO.0000000000000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fluckiger AC, Durand I, Banchereau J. Interleukin 10 induces apoptotic cell death of B-chronic lymphocytic leukemia cells. J Exp Med. 1994;179:91–99. doi: 10.1084/jem.179.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kitabayashi A, Hirokawa M, Miura AB. The role of interleukin-10 (IL-10) in chronic B-lymphocytic leukemia: IL-10 prevents leukemic cells from apoptotic cell death. Int J Hematol. 1995;62:99–106. doi: 10.1016/0925-5710(95)00395-9. [DOI] [PubMed] [Google Scholar]
- 39.Kosmaczewska A, Ciszak L, Suwalska K, Wolowiec D, Frydecka I. CTLA-4 overexpression in CD19+/CD5+ cells correlates with the level of cell cycle regulators and disease progression in B-CLL patients. Leukemia. 2005;19:301–304. doi: 10.1038/sj.leu.2403588. [DOI] [PubMed] [Google Scholar]
- 40.Grzywnowicz M, Karczmarczyk A, Skorka K, Zajac M, Zaleska J, Chocholska S, Tomczak W, Giannopoulos K. Expression of Programmed Death 1 Ligand in Different Compartments of Chronic Lymphocytic Leukemia. Acta Haematol. 2015;134:255–262. doi: 10.1159/000430980. [DOI] [PubMed] [Google Scholar]
- 41.Sun Z, Fourcade J, Pagliano O, Chauvin JM, Sander C, Kirkwood JM, Zarour HM. IL10 and PD-1 Cooperate to Limit the Activity of Tumor-Specific CD8+ T Cells. Cancer research. 2015;75:1635–1644. doi: 10.1158/0008-5472.CAN-14-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kowalewski DJ, Schuster H, Backert L, Berlin C, Kahn S, Kanz L, Salih HR, Rammensee HG, Stevanovic S, Stickel JS. HLA ligandome analysis identifies the underlying specificities of spontaneous antileukemia immune responses in chronic lymphocytic leukemia (CLL) Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E166–175. doi: 10.1073/pnas.1416389112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saulep-Easton D, Vincent FB, Quah PS, Wei A, Ting SB, Croce CM, Tam C, Mackay F. The BAFF receptor TACI controls IL-10 production by regulatory B cells and CLL B cells. Leukemia. 2016;30:163–172. doi: 10.1038/leu.2015.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drennan S, D'Avola A, Gao Y, Weigel C, Chrysostomou E, Steele AJ, Zenz T, Plass C, Johnson PW, Williams AP, Packham G, Stevenson FK, Oakes CC, Forconi F. IL-10 production by CLL cells is enhanced in the anergic IGHV mutated subset and associates with reduced DNA methylation of the IL10 locus. Leukemia. 2017 doi: 10.1038/leu.2016.356. [DOI] [PubMed] [Google Scholar]
- 45.Curry JM, Eubank TD, Roberts RD, Wang Y, Pore N, Maity A, Marsh CB. M-CSF signals through the MAPK/ERK pathway via Sp1 to induce VEGF production and induces angiogenesis in vivo. PLoS One. 2008;3:e3405. doi: 10.1371/journal.pone.0003405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yun S, Dardik A, Haga M, Yamashita A, Yamaguchi S, Koh Y, Madri JA, Sumpio BE. Transcription factor Sp1 phosphorylation induced by shear stress inhibits membrane type 1-matrix metalloproteinase expression in endothelium. The Journal of biological chemistry. 2002;277:34808–34814. doi: 10.1074/jbc.M205417200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.