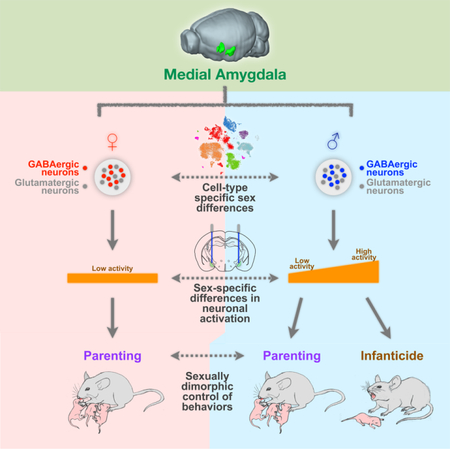
SUMMARY
Social behaviors, including behaviors directed toward young offspring, exhibit striking sex differences. Understanding how these sexually dimorphic behaviors are regulated at the level of circuits and transcriptomes will provide insights into neural mechanisms of sex-specific behaviors. Here, we uncover a sexually dimorphic role of the medial amygdala (MeA) in governing parental and infanticidal behaviors. Contrary to traditional views, activation of GABAergic neurons in the MeA promotes parental behavior in females, while activation of this population in males differentially promotes parental versus infanticidal behaviors in an activity level-dependent manner. Through single-cell transcriptomic analysis, we found that molecular sex differences in the MeA are specifically represented in GABAergic neurons. Collectively, these results establish crucial roles for the MeA as a key node in the neural circuitry underlying pup-directed behaviors and provide important insight into the connection between sex differences across transcriptomes, cells, and circuits in regulating sexually dimorphic behavior.
Graphical Abstract
IN BRIEF
The degree of activation of GABAergic neurons in the medial amygdala in male mice differentiates parenting vs infanticide.
INTRODUCTION
Social behaviors encompass a broad set of behaviors critical for the survival and well-being of an individual (Adolphs, 2010; Alexander, 1974; Chen and Hong, 2018). The diverse catalog and contextual display of social behaviors can greatly vary between the sexes (Dulac and Kimchi, 2007; Fernald, 2012; Stowers and Liberles, 2016; Yang and Shah, 2014), and disruption of social behaviors, which is a prominent feature in psychiatric disorders, often appears in a sexually dimorphic manner (Nestler and Hyman, 2010; Palanza, 2001). A major challenge in neuroscience lies in understanding how sex differences in the brain, from molecules to circuits, synergistically contribute to sex differences in behavioral displays.
Parental behaviors, like grooming and retrieval of young, are a sexually dimorphic set of social behaviors that are centered on the protection and survival of offspring (Numan and Sheehan, 1997; Wu et al., 2014). On the other hand, infanticide is a behavior focused on the killing of young offspring to increase the reproductive fitness of the infanticidal individual (Hrdy, 1979), and is also sexually dimorphic. In laboratory rodent species, virgin males are infanticidal while virgin females display parenting behaviors (Dulac et al., 2014). Although studies have implicated the medial preoptic area (MPOA) as a key region for regulating parental behaviors (Fang et al., 2018; Numan, 1974; Wei et al., 2018; Wu et al., 2014), little is known about the contribution of other brain regions beyond the MPOA in parenting (Marlin et al., 2015; Scott et al., 2015), and even less is known about brain regions regulating infanticidal behavior and its relationship to parenting (Tsuneoka et al., 2015).
The medial amygdala (MeA), located downstream of sensory organs involved in pheromonal detection, is a sexually dimorphic brain region involved in many innate social behaviors (Hong et al., 2014; Swanson, 2000; Unger et al., 2015). Previous studies have shown that large lesions of the MeA in female rats result in increased maternal behaviors (Fleming et al., 1980; Numan et al., 1993; Sheehan et al., 2001). These findings have led to a long-standing view that the MeA is not part of the main circuitry that promotes parenting behavior, but may be involved in suppressing parental behaviors (Kohl et al., 2017; Numan and Young, 2016). However, there has been no direct functional evidence linking activation of the MeA to the regulation of parenting in females. Also, in male mice, although the MeA is among brain regions that are activated during exposure to pup sensory cues (Kohl et al., 2017; Li et al., 2017; Tachikawa et al., 2013), there exists little direct evidence for a functional role of the MeA in parenting or infanticidal behavior in males.
Moreover, while previous studies have identified aspects of sex differences in the MeA (Cooke and Woolley, 2005; Morris et al., 2008; Xu et al., 2012), they largely have lacked cell-type specificity. A full understanding of these sex differences at a molecular and cellular level is still missing—it remains unclear whether the observed differences reflect distinctions in cell type composition (e.g. the presence or absence of a certain cell type) or quantitative variations in features of the same cell type(s) (e.g. gene expression).
Here, we establish a sexually dimorphic role for the MeA in regulating pup-directed behaviors. We found that activation of GABAergic neurons in the posterodorsal region of the MeA (MeApd) actually promotes parenting in females—contrary to traditional views. Interestingly, in males, GABAergic MeApd neurons regulate both parenting and infanticidal behavior in an activity level-dependent manner. In contrast to sex differences in the role of GABAergic neurons, glutamatergic neurons exert the same effect on both males and females in promoting self-grooming behavior. Finally, we provide a comprehensive characterization of molecular sex differences at the single-cell transcriptomic level, which supports the different roles of male and female GABAergic MeA neurons, but not glutamatergic neurons, in behavioral and circuit functions. These findings provide important insight into the connection between sex differences at the levels of molecules, cells, and circuits in regulating sexually dimorphic behaviors.
RESULTS
Activation of GABAergic neurons in the MeApd promotes parenting behavior in females
Although lesion studies have previously implicated the MeA in suppressing parenting behaviors, a causal role for the MeA in parenting behaviors through cell type-specific stimulation of MeA neurons has not been identified. To examine the function of the MeA in parenting, we utilized an optogenetic approach to stimulate neurons within the MeApd (a subregion of the MeA known to regulate social behavior). During pup exposure, while virgin male mice naturally exhibit infanticidal behavior, virgin female mice naturally exhibit several forms of pup-directed parenting behavior, including pup grooming, pup retrieval, and pup crouching, as previously described (Fang et al., 2018; Wu et al., 2014). We first focused on manipulating GABAergic neurons within the MeApd in virgin females by utilizing the genetic marker Vgat, since a previous study found that this subpopulation controls social behaviors in males (Hong et al., 2014). To specifically activate MeApd GABAergic neurons, we stereotaxically injected AAV encoding Cre-dependent channelrhodopsin (ChR2) into the MeApd of Vgat-cre female mice (Figures 1A and 1B). Following laser stimulation, Fos protein expression was highly co-localized with neurons expressing ChR2 (Figures 1C and 1D), indicating that laser stimulation was capable of inducing robust activation of MeApd GABAergic neurons.
Figure 1. Optogenetic Activation of MeApd GABAergic Neurons Promotes Parenting Behavior in Virgin Females.
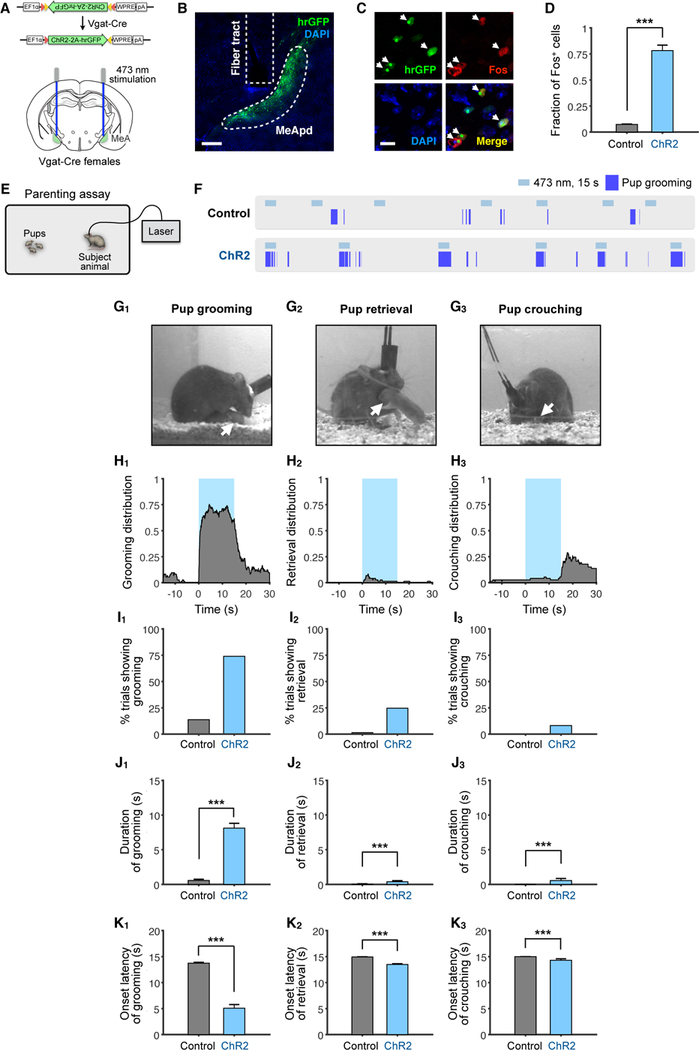
(A) Schematic of viral injection and fiber implantation strategy for ChR2 stimulation.
(B) Example image of injection site and viral expression. Scale bar = 200 µm.
(C) Co-localization of Fos expression and ChR2 cells (expressing hrGFP) following photostimulation. Arrowheads: co-localized cells. Scale bar = 50 µm.
(D) Quantification of Fos+/GFP+ cells in control and ChR2-injected animals. ChR2, 3 mice; Control, 2 mice.
(E) Schematic of parenting assay.
(F) Representative raster plots of control and ChR2 animals illustrating photostimulation-induced pup grooming.
(G) Example video frames of animals showing different behaviors. Arrows indicate location of pup.
(H) Distribution of pup grooming, retrieval, and crouching episodes (percentage of trials showing the indicated behavior at different time points) with respect to stimulation onset.
(I) Percentage of trials showing different photostimulation-induced behaviors.
(J) Average duration of different behaviors during stimulation.
(K) Average latency to different behaviors following onset of photostimulation. If behavior was not observed, latency was considered 15 seconds, which was the duration of stimulation. ChR2, n = 73 trials (6 mice); control, n = 138 trials (10 mice). Mean ± SEM. Wilcoxon rank-sum test. ***p < 0.001.
See also Figure S1.
To examine parenting behaviors, we utilized a pup interaction assay that involved placing pups with an adult animal, and scored different parenting behaviors displayed towards the pups (Figure 1E). Surprisingly, contrary to the traditional view that the MeA may inhibit parental behaviors, optogenetically stimulating MeApd Vgat+ neurons in virgin females remarkably promoted pup grooming in a time-locked manner (Figures 1F–1H, Movie S1), whereas stimulation of EYFP controls did not show pup grooming behavior (Figure 1F). Photostimulation triggered pup grooming in over 75% of stimulation trials with an average duration of 8.1 seconds, which was significantly different than EYFP controls (Figures 1I–1K). When only considering stimulation trials that showed an induction of behavior, the average latency was 1 second (Figure S1A). The pup grooming induced by photostimulation was similar to naturally occurring grooming behavior towards pups (Figure 1G, Movie S1), which included visible licking, holding pups with forelimbs, and head bobbing consistent with licking motions.
We next examined two additional maternal behaviors, pup retrieval and pup crouching (Figure 1G). Pup retrieval was triggered in 24.7% of photostimulation trials (Figure 1I), significantly higher than in controls, while the average duration of this behavior was lower than pup grooming. In addition, pup crouching was observed in 8.2% of photostimulation trials (Figure 1I). Interestingly, its occurrence was significantly increased immediately after the termination of photostimulation in ~40% of the trials (Figures 1H, S1B, S1C). To determine whether the increased crouching was due to either a primary, delayed effect of photostimulation, or due to a secondary effect from photostimulation-induced pup grooming, we examined the fraction of photostimulation trials in which crouching co-occurred with pup grooming during or following photostimulation, and trials with crouching alone but without pup grooming. We found that the majority of crouching events co-occurred with pup grooming (Figure S1D) and that the post-stimulation pup crouching followed stimulation-induced pup grooming, suggesting that the crouching was likely a secondary effect from the induced pup grooming. Taken together, our results suggest that activation of Vgat+ neurons in the MeApd of females promotes parenting behavior and predominantly pup grooming. The observation that pup retrieval and crouching frequently co-occurred with photostimulation-induced pup grooming suggests that GABAergic neuron activation leads to an increased parenting drive and that the photostimulation-induced pup grooming is likely a specific manifestation of parental behaviors.
GABAergic neurons in the MeApd are required for ongoing parenting behavior in females
To determine whether MeApd Vgat+ neurons are necessary for parenting behavior, we used optogenetics to inhibit activity of this population during natural parenting behavior in female animals. We generated animals virally expressing Cre-dependent halorhodopsin (eNpHR3) in the MeApd of Vgat-cre females (Figures 2A and 2B). In order to reliably test the effect of time-locked neuronal inhibition on pup grooming, we performed photostimulation in females that showed a baseline level of spontaneous pup grooming behavior.
Figure 2. GABAergic Neurons Are Required for Natural Pup Grooming in Females.
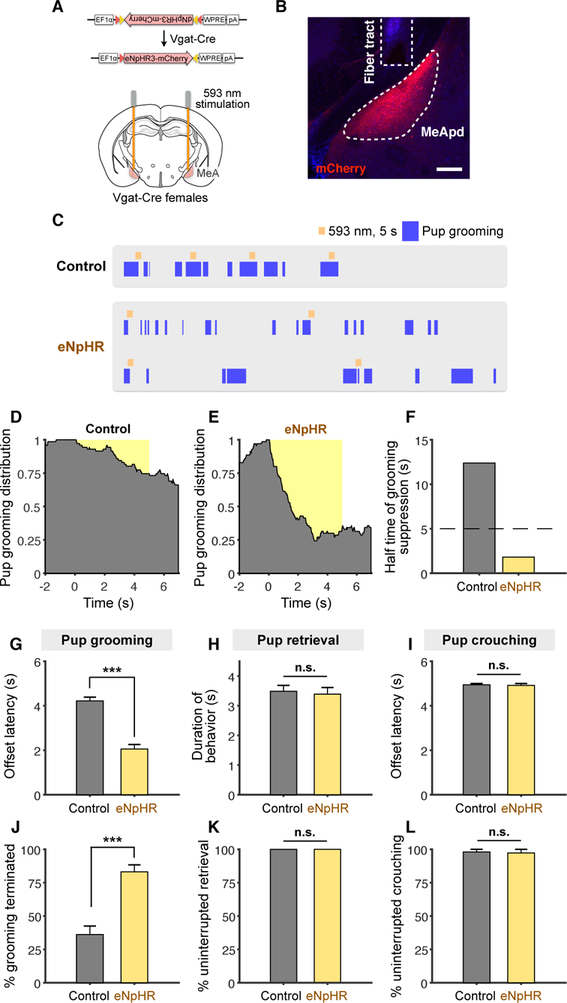
(A) Schematic of viral injection and fiber implantation strategy for eNpHR inhibition.
(B) Example image of injection site and viral expression. Scale bar = 200 µm.
(C) Representative raster plots of control and eNpHR animals illustrating suppression of pup grooming during photostimulation.
(D-E) Distribution of pup grooming episodes in control and eNpHR animals (percentage of trials showing pup grooming at different time points) with respect to stimulation onset.
(F) Half-time of pup grooming suppression in control and eNpHR animals (STAR Methods).
(G, I) Average time until termination of pup grooming (G) or crouching (I) following stimulation initiation. Trials that did not show any termination of behavior were considered as a 5 second offset, or the duration of stimulation.
(H) Average duration of pup retrieval during stimulation (STAR Methods).
(J-L) Average percentage of trials showing termination of pup grooming (J), uninterrupted retrieval (K), and uninterrupted crouching (L).
eNpHR, grooming, n = 70 trials; retrieval, n = 28 trials; crouching, n = 29 trials (6 mice); control, grooming, n = 71 trials; retrieval, n = 61 trials; crouching, n = 50 trials (7 mice). Mean ± SEM. Wilcoxon rank-sum test. n.s. not significant, ***p < 0.001.
See also Figure S1.
Optogenetic inhibition of MeApd Vgat+ neurons led to a reduction in pup grooming in virgin females (Figures 2C–2F, Movie S2). To characterize the temporal dynamics of the pup grooming behavior, we fit the temporal distribution of behavioral occurrence with an exponential decay function, and found that the half-time of pup grooming suppression in eNpHR animals was markedly shorter than that of EYFP controls (Figures 2C–2F). eNpHR animals also exhibited shorter average latency from initiation of stimulation to termination of pup grooming (Figure 2G) and greater percentage of trials showing termination of pup grooming than EYFP controls (Figure 2J). Moreover, we found that photoinhibition of MeApd Vgat+ neurons suppressed pup grooming in lactating mothers, similar to our results from virgin females (Figure S1F–S1G). These data suggest that optogenetic inhibition of MeApd Vgat+ neurons markedly suppresses ongoing pup grooming in both virgin and lactating females.
We found that optogenetic inhibition did not inhibit pup retrieval or pup crouching behavior (Figures 2H, 2I, 2K, 2L, S1E). Duration of pup retrieval and offset latency of pup crouching were similar between EYFP and eNpHR animals during photostimulation, and the percentage of trials with no interruptions in pup retrieval or pup crouching was similar between eNpHR and control animals, suggesting that Vgat+ neurons are not required for retrieval or crouching. Collectively, our results indicate that MeApd Vgat+ neurons in both virgin and parental females are required for pup grooming behavior.
Activation of MeApd GABAergic neurons in males triggers infanticidal behavior
Parenting behaviors in mice are highly dependent on sex and reproductive state; while females usually exhibit parental behaviors, virgin males engage in infanticidal behavior (Dulac et al., 2014). Previous studies have observed sex differences in gene expression, morphology, and electrophysiology of MeApd neurons, raising the possibility that MeApd neurons in males vs. females may control distinct behaviors towards pups. To test this, we activated MeApd Vgat+ neurons in virgin males (Figures 3A and 3B). Following photostimulation, virgin males initiated time-locked infanticidal behavior toward pups, with an average latency of 1.3 second (Figures 3C–3F). The majority of photostimulation trials had a behavior onset latency between 0–1 seconds (Figure 3G). These results suggest that MeApd Vgat+ neurons drive infanticidal behavior in males.
Figure 3. Activation of GABAergic Neurons Promotes Infanticidal Behavior in Virgin Males.
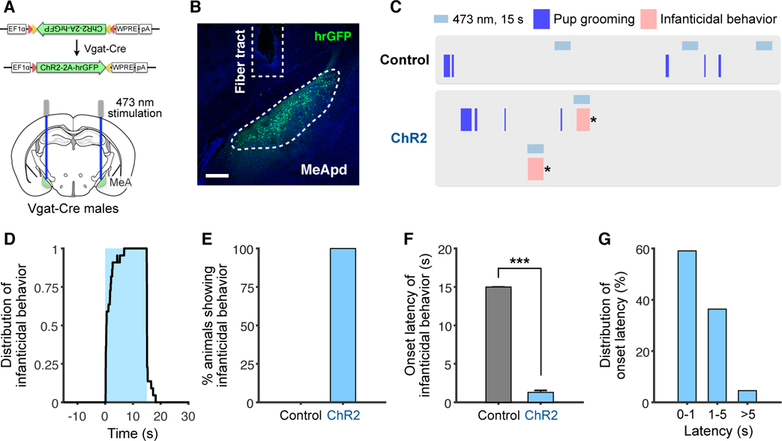
(A) Schematic of viral injection and fiber implantation strategy for ChR2 stimulation.
(B) Example image of injection site and viral expression. Scale bar = 200 µm.
(C) Representative raster plots of control and ChR2 animals illustrating promotion of infanticidal behavior during photostimulation. * indicates when the pup was removed.
(D) Distribution of infanticidal behavior episodes in ChR2 animals (percentage of trials showing infanticide at different time points) with respect to stimulation onset.
(E) Percentage of animals showing infanticidal behavior.
(F) Average latency until infanticidal behavior onset following stimulation initiation.
(G) Latency distribution of infanticidal behavior onset following stimulation initiation.
ChR2, n = 22 trials (5 mice); control, n = 26 trials (4 mice). Mean ± SEM. Wilcoxon rank-sum test. ***p < 0.001.
See also Figure S2.
While our findings suggested that MeApd Vgat+ neurons promote pup-directed behavior in both males and females, it was unclear whether this function was specific to the Vgat+ population or whether other neuronal populations within the MeApd were also capable of eliciting pup-directed behavior. To address this question, we investigated the role of the non-overlapping, glutamatergic neuronal population (expressing Vglut2) within the MeApd of males and females in pup-directed behaviors (Figure S2A). The Vglut2+ neurons have previously been shown to promote self-grooming in males (Hong et al., 2014), but its function in females is unknown. Moreover, its behavioral function in the presence of pups in both males and females has not been examined. We found that photostimulation of the Vglut2+ population in both males and females resulted in time-locked self-grooming behavior in the presence of pups, but did not trigger any pup-directed behavior (Figures S2B-S2D, S2G, S2H). The onset latency and latency distribution of self-grooming were comparable between males and females (Figures S2E, S2F, S2I, S2J), suggesting that activation of the Vglut2+ population exerts the same behavioral effects in males and females.
Collectively, these results indicate that the sex-specific regulation of pup-directed behaviors is specifically carried out by the MeApd GABAergic neurons and that the MeApd glutamatergic neurons do not exert sexually dimorphic behavioral functions.
GABAergic neurons are activated at different levels during infanticidal vs. parental behaviors
To understand how the MeApd regulates distinct pup-directed behaviors—parental vs. infanticidal behavior—in males vs. females, we next asked whether MeApd Vgat+ neurons are activated during naturally occurring parental and infanticidal behaviors. To examine the dynamics of neural activity, we utilized fiber photometry to record Ca2+ signals from MeApd Vgat+ neurons in freely-behaving animals when they engaged in different pup-directed behaviors. We injected AAV encoding Cre-dependent GCaMP6s, a fluorescent Ca2+ sensor (Chen et al., 2013), into the MeApd of Vgat-cre males and females (Figures 4A and 4B). We confirmed the injection sites and fiber implant sites through histology (Figure 4C).
Figure 4. GABAergic Neurons Are Active at Different Levels During Pup Grooming vs. Infanticidal Behavior.
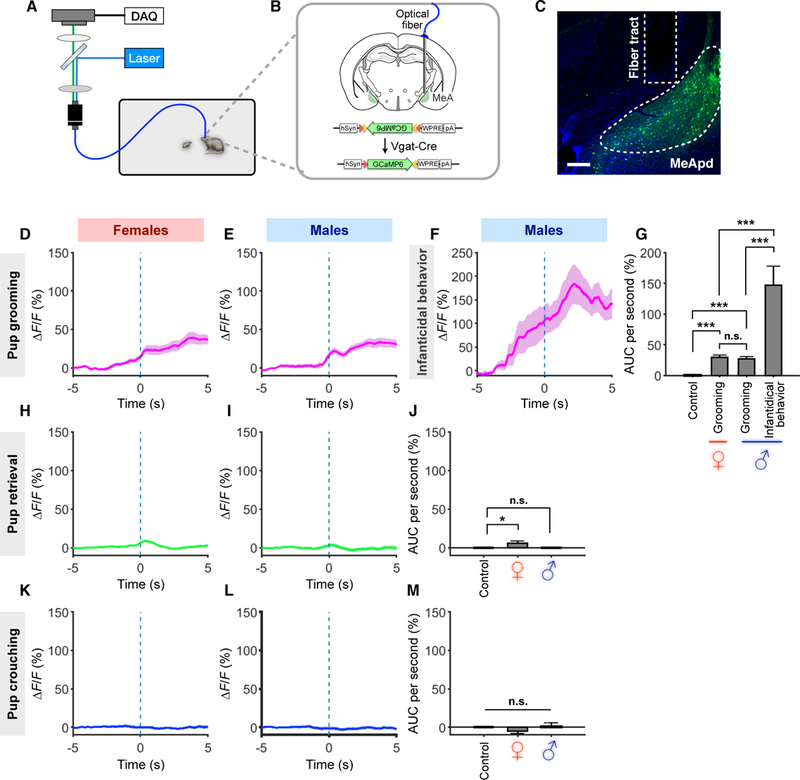
(A-B) Schematic of behavioral assay, injection strategy, and fiber implantation location for fiber photometry recording experiments.
(C) Representative injection site and fiber implantation for fiber photometry recording. Scale bar = 200 µm.
(D-F, H-I, K-L) Average Ca2+ signal changes during pup grooming in virgin females (D) and virgin males (E) and virgin males with infanticidal behavior (F), during pup crouching in virgin females (K) and virgin males (L), and during pup retrieval in virgin females (H) and virgin males (I). Mean ± SEM.
(G, J, M) Comparison of area under curve (AUC) per second during pup grooming and infanticidal behavior (G), pup retrieval (J), and pup crouching (M).
GCaMP6s virgin females, grooming, n = 16 trials; retrieval, n = 57; crouching, n = 40; GCaMP6s virgin males, grooming, n = 15; retrieval, n = 25; crouching, n = 35; infanticidal behavior, n = 7. EYFP control, grooming, n = 80; retrieval, n = 69; crouching, n = 204. (3~6 mice for each group). Mean ± SEM; one-way ANOVA with post-hoc Wilcoxon rank-sum with Bonferroni correction. n.s. not significant, *p < 0.05, ***p < 0.001.
See also Figure S3.
In GCaMP6-expressing virgin females but not EYFP controls, Ca2+ signal increased following onset of pup grooming (Figures 4D, 4G, S3A, S3C), indicating that Vgat+ neurons are activated during this behavior. In contrast, we did not observe significant increases in Ca2+ signal during pup crouching, but did observe a slight but significant increase in Ca2+ signal during retrieval (Figures 4H, 4K, 4J, 4M, S3J, S3K). As virgin females display similar parental behaviors as mothers, we next examined whether MeApd Vgat+ neurons are also activated during parenting behaviors in mothers. We found that MeApd Vgat+ neurons in maternal females are significantly activated during pup grooming, slightly but significantly activated during retrieval, and not activated during crouching; levels of activity between virgin females and mothers were not significantly different (Figures S3I–S3K). These results suggest that Vgat+ neurons in the MeApd are activated in both virgin and maternal females during parenting behaviors.
In GCaMP6-expressing virgin males but not EYFP controls, we observed a sharp increase in Ca2+ signal following the onset of infanticidal behavior (Figures 4F 4G, S3B), indicating that Vgat+ neurons are indeed activated during this behavior. In some virgin males, we occasionally observed spontaneous parental behaviors like pup grooming, crouching, and retrieval. We observed a significant increase in Ca2+ signal during grooming bouts but not during crouching or retrieval (Figures 4E, 4I, 4L, 4J, 4M, S3A, S3L–S3N). MeApd Vgat+ neurons in fathers also showed similar activation during pup grooming, but no activation in retrieval and crouching (Figures S3L–S3N; fathers do not display infanticidal behavior). Strikingly, the increase in Ca2+ signal during infanticidal behavior in virgin males was significantly higher than that during pup grooming in both males and females (both virgin and parental), while the increase in Ca2+ signal during pup grooming was not significantly different among virgin and parental males and females (Figure 4G, S3I, S3L). Post-hoc histology confirmed that injection site and fiber implantation of males showing infanticide and grooming were comparable (Figure S3H). Since grooming or infanticide usually follows brief bouts of pup sniffing, which showed a small, but not significant, difference in their average durations (Figure S3F), we further measured Ca2+ signal 2s after sniffing bouts had ended to minimize the remaining Ca2+ signals contributed by sniffing bouts. We found that Ca2+ signal during infanticidal behavior was still >5 times higher than that during pup grooming behavior (Figure S3G).
Together, these results support our findings that activity of Vgat+ neurons in the MeApd is involved in promoting pup-directed behaviors in both males and females and in both virgin and parental animals, and point to a possible link between different levels of Vgat+ neuronal activity and distinct displays of opposing pup-directed behaviors in virgin males vs. females.
GABAergic neurons promote infanticidal vs. parental behavior in an activation level-dependent manner
The above findings raised the possibility that two opposing pup-directed behaviors may be controlled by different levels of activity in MeApd Vgat+ neurons. Specifically, we hypothesized that lower levels of activation of MeApd Vgat+ neurons may result in pup grooming, while higher levels of activation may result in infanticidal behavior. To test this idea, we sought to optogenetically activate MeApd Vgat+ neurons at different stimulation intensities.
In virgin males, stimulation of MeApd Vgat+ neurons at high laser intensities (>2.5 mW/mm2) resulted in infanticidal behaviors for 100% of trials (Figures 5A–5C). Stimulation at low laser intensities (<1 mW/mm2), however, predominantly resulted in pup grooming (Figures 5A–5C, Movie S3). When photostimulating at medium (1–2.5 mW/mm2) laser intensities, the percentage of trials triggering infanticidal behavior remained high, with no pup grooming observed, but the average duration was lower and average latency was higher for infanticide than at higher laser intensities (Figures 5A–5C). EYFP controls showed no time-locked behaviors (Figures S4E1–2). Optogenetic activation of Vgat+ cells outside the MeApd did not result in any time-locked behaviors (Figure S4J), indicating that the identified behavioral functions are specific to the MeApd. These results support a functional role for different levels of activation of MeApd Vgat+ neurons in the opposing control of infanticidal behavior vs. pup grooming in virgin males.
Figure 5. GABAergic Neurons Promote Pup Grooming vs. Infanticidal Behavior in an Activity Level-Dependent Manner.
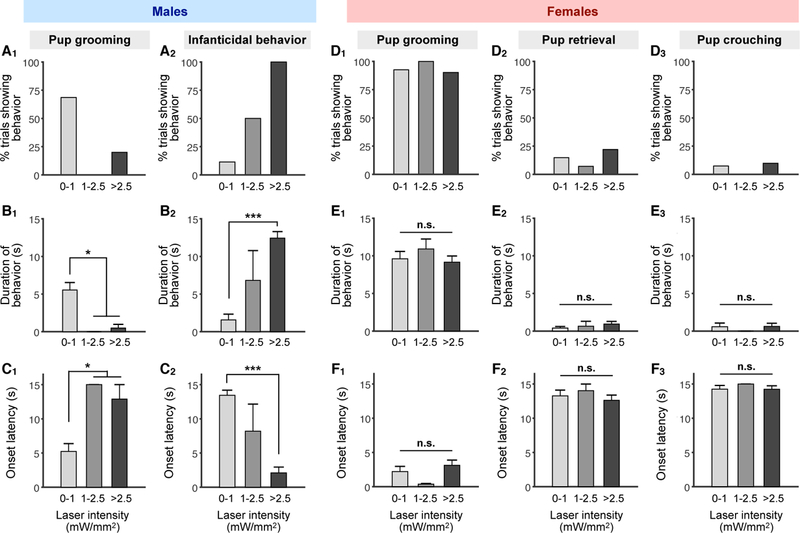
(A-C) Percentage of trials showing pup grooming and infanticidal behavior (A), average durations of different behaviors during stimulation epochs (B), and average onset latency of different behaviors following initiation of stimulation (C) in virgin males at different photostimulation intensities.
(D-F) Percentage of trials showing pup grooming, retrieval, and crouching (D), average duration of pup grooming, retrieval, and crouching during stimulation epochs (E), and average onset latency of pup grooming, retrieval, and crouching following initiation of stimulation (F) in virgin females at different stimulation intensities. Trials where no behavior was observed during the stimulation epoch were considered as a latency of 15 seconds, the duration of stimulation.
ChR2 males: 0–1 mW/mm2, n = 35 trials; 1–2.5 mW/mm2, n = 4; >2.5 mW/mm2, n = 5; ChR2 females: 0–1 mW/mm2, n = 27; 1–2.5 mW/mm2, n =14; >2.5 mW/mm2, n = 41 (n ≥ 5 mice for each group). Mean ± SEM; one-way ANOVA with post-hoc Wilcoxon rank-sum with Bonferroni correction. n.s. not significant, *p < 0.05, ***p < 0.001.
See also Figure S4.
By contrast, in virgin females, stimulation of Vgat+ neurons at high, medium, and low laser intensities only resulted in pup grooming, with no obvious difference in the percentage of trials, duration, or latency between stimulation intensities (Figures 5D–5F). There was also no obvious effect of stimulation intensity on pup retrieval or pup crouching (Figures 5D–5F). EYFP controls showed no time-locked behaviors (Figures S4F1–2). Like in virgin females, both low- and high-intensity stimulation of the Vgat+ neurons in lactating mothers resulted in pup grooming (Figure S4C, S4D, S4F3). Thus, unlike in virgin males, differing levels of activation in virgin and parental females does not result in different behaviors.
Lastly, we examined the effect of photostimulation of Vgat+ neurons in fathers, which normally display parental, but not infanticidal, behaviors. Since different levels of activation of Vgat+ neurons led to opposing behaviors in virgin males, we asked whether a stimulation level-dependent behavioral transition also occurs in fathers. Indeed, while a low-intensity stimulation resulted in grooming, increasing the stimulation intensity triggered infanticidal behavior in fathers (Figure S4A, S4B, S4G, S4E3). This experiment suggests that, although the MeApd GABAergic neurons in fathers are normally active at a low level during pup grooming (Figures 4G and S4B1), the circuit is capable of eliciting infanticidal behavior when activated at a higher level (Figures S4A and S4B2).
Collectively, these results demonstrate the functional relevance of scalable neuronal activation in MeApd Vgat+ neurons as a mechanism to control infanticidal behavior vs. pup grooming, and that this scalability is specific to males but not females.
MeA cell types are not different between males and females
The intriguing finding that Vgat+ neurons within the MeApd underlie highly sexually dimorphic displays of pup-directed behaviors raised the possibility that these neurons may differ in their molecular or cellular features between the sexes. To comprehensively understand the nature of sexual dimorphism in the MeA in specific cell populations, we performed single-cell RNA sequencing (scRNA-seq; Kolodziejczyk et al., 2015) of MeA cells from both virgin male and virgin female mice to identify any transcriptomic differences between the sexes at a single-cell resolution. Using scRNA-seq (Drop-seq; Macosko et al., 2015), we captured the single-cell transcriptomes of 44,437 cells (21,715 male cells and 22,722 female cells) containing a total of 16,961 genes from acutely dissected and dissociated MeA of adult males and females (Figures 6A and 6B). Our new dataset, which contains high-quality data for 10,164 neurons from both sexes, was greatly expanded beyond our previous dataset (~2,000 neurons from only males; Wu et al., 2017).
Figure 6. Cell Type Identities and Composition Do Not Differ Between Male and Female MeA.
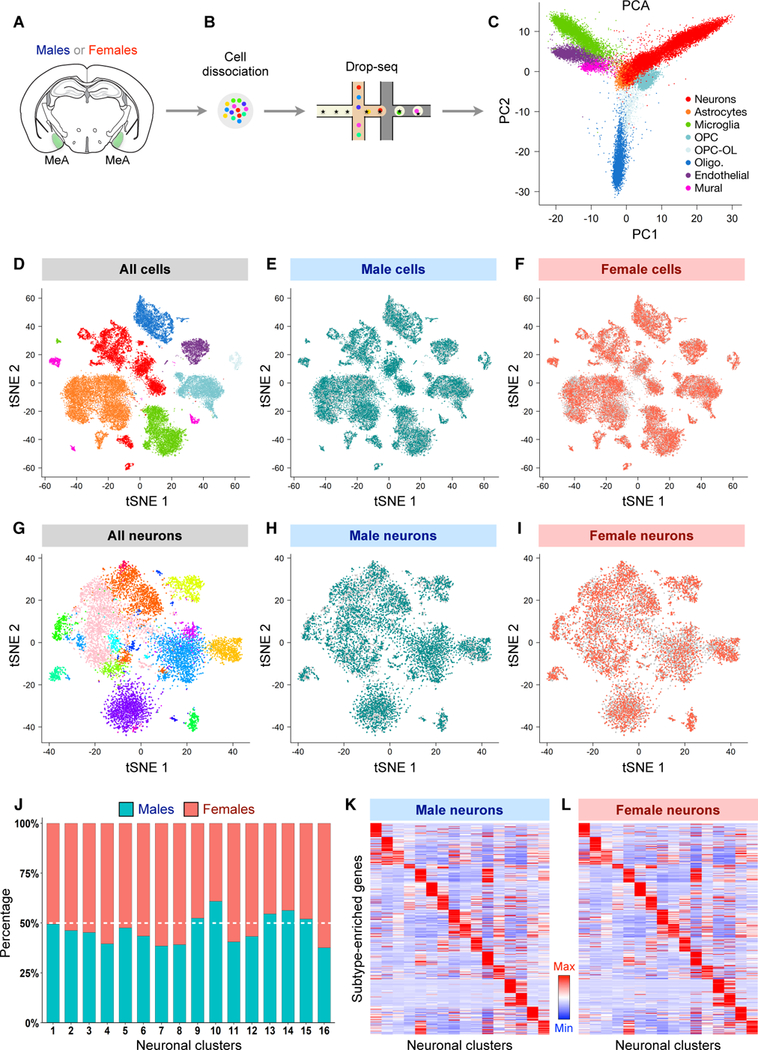
(A-B) Schematic showing MeA dissection, single-cell dissociation, and sequencing using Drop-seq in virgin males and virgin females.
(C) Separation of 44,437 MeA cells from both males (21,715 cells) and females (22,722 cells) by principal component (PC) 1 and PC 2.
(D-F) Two-dimensional tSNE visualization showing the distribution of all cells (D), cells from males (E), and cells from females (F) among major MeA cell types.
(G-I) Two-dimensional tSNE visualization showing the distribution of all neurons (G), male neurons (H), and female neurons (I) among MeA neuronal subtypes. In (C-I), each dot corresponds to a single cell. Cells are colored according to major cell types (C, D) or neuronal subtypes (G). OPC, oligodendrocyte precursor cells; Olig., oligodendrocytes; OPC-OL, a transitional cell state between OPCs and oligodendrocytes.
(J) Bar plots showing the percentage of male and female cells in each neuronal subtype. There was no significant difference between males and females for any subtype (Wilcoxon rank-sum test, FDR > 0.05, n = 6 male samples (12 mice) and 7 female samples (20 mice; STAR Methods).
(K-L) Heat maps showing similar expression pattern of select top subtype markers across MeA neuronal subtypes between males (K) and females (L). The same set of markers are shown in K and L, and expression level is averaged within each subtype and normalized for each row (gene) (STAR Methods).
See also Figures S5, S6, and S7, Tables S1 and S2.
One potential mechanism for sexual dimorphism at the behavioral level is that males and females may differ in the identity or proportion of MeA cells. To compare the composition of MeA cell types in males and females, we first classified the major cell types in both males and females using principal components analysis (PCA), dimensionality reduction by t-distributed stochastic neighbor embedding (tSNE), and graph clustering with the Louvain-Jaccard algorithm (Shekhar et al., 2016) (Figures 6C, 6D, S5A, S6A-S6P; STAR Methods). This first-level analysis identified the same categories of major cell types (neurons, astrocytes, microglia, etc.) between males and females (Figures 6D–6F, Table S1). In addition, comparison of the relative abundance of these cell types revealed no significant difference between males and females (Figure S5B), and the top major cell type markers also showed similar expression between the sexes (Figures S5C–S5L). Furthermore, male and female cells were largely homogenously intermingled in the principal component (PC) projections and tSNE map for all major cell types (Figures 6D–6F, S5A), indicating no clear separation in global gene expression pattern between the sexes.
We next focused on the 10,164 neurons from both males and females in our data set and further divided them into subtypes based on the expression pattern of all neuronally enriched genes (Table S2, STAR Methods). This analysis identified the same 16 neuronal subtypes in both males and females, with no overt separation in the overall gene expression pattern of male and female cells for all subtypes as shown on the tSNE map (Figures 6G–6I). Moreover, the top genes that were selectively enriched in each subtype were shared between males and females (Figures 6K and 6L, Table S2), and the proportion of each subtype was not significantly different between the sexes (Figure 6J). Together, our single-cell analysis suggests that sexual dimorphism in the MeA does not arise from differences in major cell type or neuronal subtype identity or relative abundance.
MeA GABAergic neurons exhibit greater molecular sex differences than glutamatergic neurons
Given our finding that sexually dimorphic displays of behavior are controlled by GABAergic, but not glutamatergic, neurons, we next focused on examining sex differences within neuronal subpopulations expressing GABAergic and glutamatergic markers. We found no significant difference in the relative abundance of GABAergic and glutamatergic neurons between males and females (Wilcoxon rank-sum test, p = 0.53; STAR Methods), ruling out this factor as a source of the observed behavioral dimorphism. To further evaluate the degree of molecular sex differences between GABAergic and glutamatergic neurons, we next compared the number of genes differentially expressed between males and females within these two populations. As expected, we detected sex-specific expression for a set of X- and Y-chromosome genes known to be exclusively expressed in male (i.e. Eif2s3y, Ddx3y, Kdm5d, Uty) or female cells (i.e. Xist, Tsix) (data not shown). Because of their ubiquitous expression, we excluded these genes in further analysis.
Using three different methods based on distinct statistical models (edgeR, limma, and DESeq2; STAR Methods), we consistently observed remarkably more differentially expressed genes in GABAergic neurons than in glutamatergic neurons, with 42–67 differentially expressed genes in GABAergic neurons and no or only a handful of differentially expressed genes in glutamatergic neurons (Figures 7A-7C, 7E, 7F, Tables S3, S4). This difference was not due to differences in transcriptome coverage in GABAergic neurons (Figures S6Q–S6T). The differentially expressed genes that we identified in GABAergic neurons not only included genes (Brs3 and Greb1) that were previously shown to be female- or male-enriched in the MeA at the tissue level (Xu et al., 2012) but also genes that have not been previously reported to be sexually different (Figure 7E, Table S4). To further verify sexually differential gene expression in GABAergic neurons, we performed dual-color fluorescent in situ hybridization (FISH) for Slc32a1 (encoding Vgat) and several candidate genes predicted to be differentially expressed in GABAergic neurons (Brs3, Greb1, Fam84a, and Ankrd55) as well as genes predicted to show no sex differences (Cck and Nos1), as negative controls (Figures 7G-7N, S7G–S7H). The FISH results were highly concordant with our scRNA-seq findings, revealing significant sex differences in the fluorescence signals of the four candidate genes in Vgat+ cells and no sex differences for the negative controls.
Figure 7. GABAergic Neurons Exhibit Greater Molecular Sex Differences than Glutamatergic Neurons.
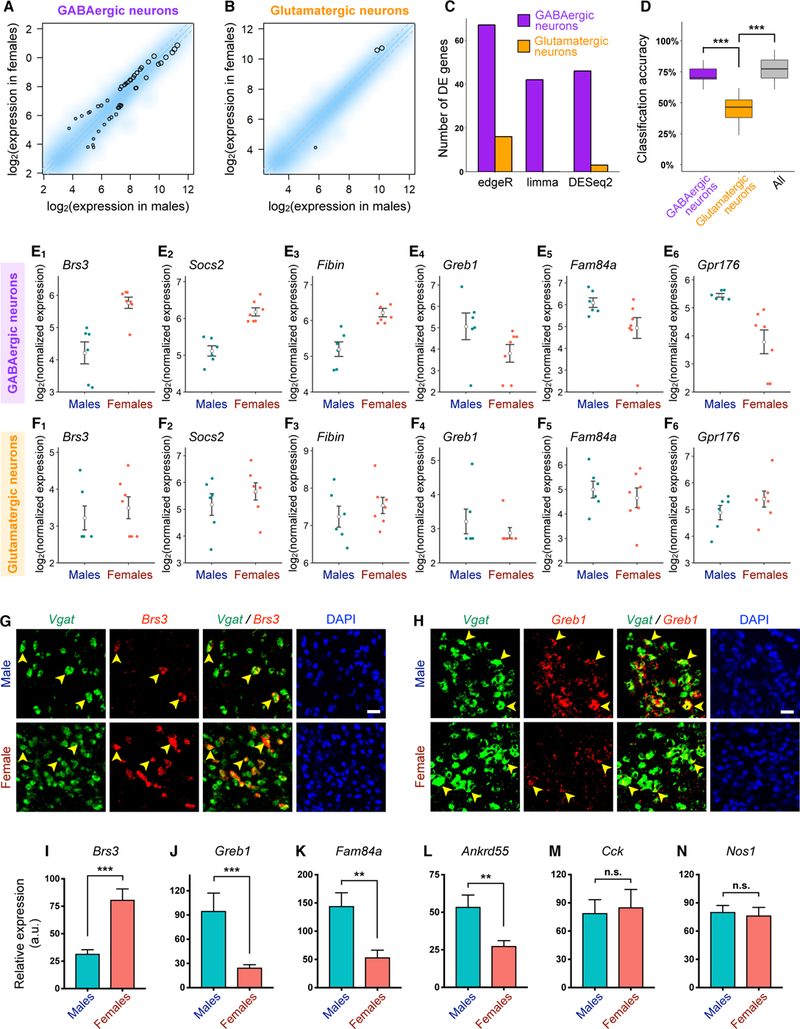
(A-B) Smoothed scatter plots showing expression of neuronally enriched genes in females versus males in GABAergic (A) and glutamatergic (B) neurons. Expression level was normalized for each sample (STAR Methods) and averaged across male and female samples. Color reflects density of genes. Black circles highlight genes significantly differentially expressed between males and females based on the DESeq2 method (STAR Methods). Circle size is proportional to mean expression level across all samples. Grey dotted lines = fold change cutoff of 1.3.
(C) Number of significantly differentially expressed genes in GABAergic and glutamatergic neurons using three different approaches (STAR Methods).
(D) Box plots showing accuracy of classification of male versus female samples using linear discriminative analysis of GABAergic, glutamatergic, or all neurons.
(E-F) Dot plots showing expression level of representative genes with significantly higher (E1-E3) or lower (E4-E6) expression in females versus males in GABAergic neurons (E) but not in glutamatergic neurons (F). Each dot represents the summed expression level of each gene over all GABAergic or glutamatergic neurons in an independent biological sample (n = 6 male samples (12 mice) and 7 female samples (20 mice; STAR Methods). Open circles and error bars: mean ± SEM.
(G-H) Representative double FISH images showing sex differences in the expression of Brs3 (G) and Greb1 (H) in Vgat+ cells (arrowheads) in the MeApd. Scale bar = 50 µm.
(I-N) Quantification of fluorescence intensity of probes specific for each transcript in Vgat+ cells in sections of male and female MeApd. n = 5–9 different sections from 3–5 mice for each sex (STAR Methods).
Mean ± SEM; Wilcoxon rank-sum test. n.s. not significant, **p < 0.01, ***p < 0.001.
The above analysis supports the notion that there are more genes significantly differentially expressed between the sexes in GABAergic neurons than in glutamatergic neurons. However, it remained a formal possibility that glutamatergic neurons may contain sub-threshold sex differences, which would not be detected as readily by our differential expression analysis. To corroborate the above findings at a transcriptomic level without relying on the identification of individual differentially expressed genes, we constructed statistical models using linear discriminative analysis (STAR Methods) to classify male vs. female samples based on the expression profile of all ~3,700 neuronally enriched genes. We found that the classification model trained on all neurons achieved a classification accuracy that was significantly higher than chance (Figure 7D). Interestingly, the classification model trained on GABAergic neurons achieved a high classification accuracy that was comparable to the model trained using all neurons (Figure 7D). In contrast, the classification model trained on glutamatergic neurons performed near chance levels and its accuracy was significantly lower than the model trained on GABAergic neurons (Figure 7D). Similar results were obtained with a different statistical method, support vector machine (data not shown). These results suggest that GABAergic neurons, but not glutamatergic neurons, contain sex differences in high-dimensional gene expression profiles that are sufficient to classify males vs. females.
To further investigate whether sex differences are represented in specific GABAergic subtypes, we performed further clustering analysis within GABAergic neurons to identify subtypes (Figure S7A). We identified 13 GABAergic subtypes, which exist in both males and females (Figures S7B and S7C), and confirmed that there were no sex differences in their relative abundance (Figure S7D). As the numbers of cells in individual GABAergic subtypes vary drastically between subtypes, preventing us from making a fair comparison of sex differences in gene expression across subtypes, we asked whether the sexually differentially expressed genes identified in all GABAergic neurons were enriched in specific GABAergic subtypes. We found that while these genes were distributed across different subtypes, many of them were enriched in a specific subset of GABAergic subtypes (Figures S7E and S7F). This suggests that different GABAergic subtypes might contribute differentially to molecular sex differences in GABAergic neurons.
Together, our scRNA-seq analysis identifies greater sex differences in MeA GABAergic neurons, which supports the distinct roles of male vs. female GABAergic neurons, but not glutamatergic neurons, in behavioral and circuit functions. This provides a potential molecular basis for the sexually dimorphic displays of pup-directed behaviors that this region controls.
DISCUSSION
In this study, we identify a major behavioral function for the MeA in controlling opposing pup-directed behaviors, parenting vs. infanticidal behavior, and uncover cell type-specific sex differences in the MeA at circuit and transcriptomic levels in the regulation of these behaviors. Using cell type-specific manipulations, we have shown that MeApd GABAergic neurons in females promote parenting behavior, whereas different activation levels of this population in males can distinctly control infanticidal vs. parenting behavior. Single-cell transcriptomic analysis reveals that GABAergic neurons exhibit greater molecular sex differences than glutamatergic neurons. Our study points toward a potential mechanism by which sexual dimorphism at the molecular, cellular, and circuit level are linked to regulate sexually dimorphic displays of behavior.
The functional role of the MeA in pup-directed behaviors
In female rodents, the MeA has long been thought to suppress maternal behaviors based on the finding that lesioning the MeA results in increased maternal behaviors and immediate early gene studies demonstrating increased neuronal activation in non-parental females during pup exposure (Fleming et al., 1980; Numan et al., 1993; Sheehan et al., 2001, 2000). In contrast to this view, our results demonstrate that GABAergic, but not glutamatergic, neurons within the female MeApd promote and are required for parental behavior in a time-resolved manner. This discrepancy may be due to differences in experimental sensitivity and specificity. Here, we bidirectional manipulated a specific neuronal MeApd subpopulation using optogenetics, which allows for finer temporal control of cell type-specific neuronal activity.
Moreover, while previous studies have found that the MeA is activated in males during exposure to pup sensory cues (Kohl et al., 2017; Li et al., 2017; Tachikawa et al., 2013), a defined function of the male MeA in pup-directed behaviors like infanticide had not been established. We found that the MeApd GABAergic subpopulation in males is capable of promoting infanticidal behavior. These findings add an extra layer of complexity to the sex-specific contribution of the MeA in parenting circuits and establishes the MeA as a key component for infanticidal behavior.
Neuronal subpopulations expressing Gal or Esr1 in the MPOA have been shown to regulate pup-directed behaviors (Fang et al., 2018; Kohl et al., 2018; Wei et al., 2018; Wu et al., 2014). While MPOA neurons were found to promote pup retrieval in a time-resolved manner, time-locked behavioral effects for other pup-directed behaviors, including pup grooming, were not observed. Moreover, no obvious difference was found in MPOA function between males and females. Thus, the function of MeApd GABAergic neurons and that of MPOA neurons differs in two key manners: MeApd GABAergic neurons 1) predominantly control pup grooming and infanticidal behavior in a time-resolved manner, and 2) this control is different between males and females. Clarifying the nature of the distinct roles between the MeA and the MPOA will be an important area of future study.
Scalable control of infanticidal vs. parental behavior in males
One intriguing finding from our study is that activation of male MeApd Vgat+ neurons can promote both infanticide and parenting, depending on the level of activation. Scalable control of distinct behaviors through manipulation of activity levels has been proposed as a mechanism to regulate distinct behavioral choices (Lee et al., 2014), but the underlying explanation for this phenomenon has remained unclear. In particular, without evidence that this occurs during natural behaviors, this scalability phenomenon could be interpreted as an artifactual behavioral phenotype. Here, using fiber photometry, we found that Vgat+ neurons display significantly higher activity during infanticidal behavior compared to pup grooming behavior, suggesting that scalable activation controlling different behaviors indeed occurs in vivo. Whether differences in activation level during natural behaviors arise from different levels of activity within the same number of neurons or from the recruitment of different numbers or subtypes of active neurons remains an open question (Lee et al., 2014).
In many animal species, males undergo drastic behavioral changes during the transition from virgin males to fathers, with one of the most prominent changes being the behavioral transition from infanticide to parenting (Dulac et al., 2014; Perrigo et al., 1992). The neural circuit mechanisms governing this transition have remained unclear. Our finding that MeApd GABAergic neurons exhibit stronger activation during infanticidal behavior compared to pup grooming mirrors previous observations that activation of the MeApd by pup sensory cues or following infanticide is higher in virgin males than fathers (Kohl et al., 2017; Tachikawa et al., 2013), suggesting that the differential activation of MeApd GABAergic neurons may underlie the behavioral transition from virgin males to fathers. Interestingly, despite the fact that in fathers the MeApd GABAergic neurons are normally active at a low level during pup grooming, the circuit is capable of eliciting infanticidal behavior when activated at a higher level. Our results open up new avenues for future studies on the precise mechanism and timing of this transition.
In females, we found that there was no transition of pup grooming to infanticide with differing levels of activation in both virgin and lactating females, indicating that this aspect of the female MeApd circuit differs from males. Because female lab mice do not normally show infanticidal behavior in the same context as males, the threshold for eliciting infanticidal behavior through activation of the female MeApd may be different than in males, and may require much higher levels of activation to override natural behaviors. Alternatively, circuit mechanisms necessary for infanticidal behavior might be absent in females.
Cell type-specific sexual dimorphism and its relation to behavior
The MeA has previously been shown to display sex differences (Cooke and Woolley, 2005; Xu et al., 2012), but a clear link between sex differences and the behavioral function of the MeA has been poorly understood. In our study, we uncovered a relationship across sex differences in the MeA at levels of gene expression, cell types, circuit function, and behavior. Our single-cell transcriptomic analysis combined with our circuit and behavioral approach offers a tantalizing look at the regulation of sexually dimorphic behaviors through sexually dimorphic functional organization of the brain.
A long-standing question concerns the underlying nature of sex differences in the brain at molecular and cellular levels. Sex differences could arise from 1) differences in cell-type identities, 2) differences in the relative abundance of cell types, or 3) differences in gene expression within the same cell type. Previous studies on molecular sex differences utilizing tissue-level RNA sequencing or microarrays were unable to distinguish between these possibilities. Moreover, although previous studies on sexually dimorphic brain function have found that a single gene that marks a behaviorally relevant neuronal population may differ between males and females (e.g. Scott et al., 2015; Yang et al., 2013), differential expression of a single gene does not necessarily mean differences in number of the corresponding cells.
Recent advances in single-cell transcriptomic profiling allow for the definition of cell types not through a single gene, but through the co-expression of a set of tens or hundreds of genes, providing a method to dissect the molecular and cellular organization of sexual dimorphism at the single-cell transcriptomic level. Through sequencing >44,000 single cells from the MeA, we found that the representation of sex differences at the single-cell transcriptomic level entailed no significant differences in the identity or relative abundance of major cell types and neuronal subtypes. Strikingly, sex differences in gene expression were predominantly observed in GABAergic neurons but not glutamatergic neurons. This is consistent with our behavioral and circuit data that GABAergic, but not glutamatergic, neurons control different behavioral functions between males vs. females. Our results offer a potential organizational structure between sexually dimorphic displays of behavior and molecular and cellular sex differences in the brain.
Genes with sexually different expression in the GABAergic neuronal population encompass a wide range of cellular functions, including cell adhesion (Vstm5, Robo1, Cbln2), ion channel function (Cacna1c, Kcnip4), and G-protein coupled receptor signaling (Gpr75, Gpr176, Galr1). One possibility is that the sex differences in gene expression contributes to underlying differences in the physiology and, consequently, behavioral function of these neurons; for example, these genes may modulate the excitability of male or female cells, thus setting different activation thresholds for infanticidal vs. parenting behaviors. Clarifying the functional link between gene expression and physiological or behavioral differences will be able to delineate the mechanism by which sex differences at the molecular level contribute to sexually dimorphic control of behavior.
Collectively, our findings establish major roles for the MeA in the sexually dimorphic, activity level-dependent control of parenting vs. infanticidal behavior and reveal the nature of cell type-specific sex differences through single-cell sequencing. These concepts open up new avenues of research in further understanding the neural mechanisms of opposing, sexually dimorphic behaviors. Our study demonstrates the power of combining circuit manipulations and single-cell transcriptomic analysis in bridging and synergizing findings across these different levels to achieve a unified comprehension of the neural control of behavior.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Weizhe Hong (whong@ucla.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals used for single-cell sequencing experiments were 8–10 week old wild-type virgin female and virgin male C57BL/6J purchased from Jackson Laboratories. Animals used for stereotaxic surgery and behavioral experiments were 8–10 week old VgatCre/+ and Vglut2Cre/+ (Vong et al., 2011) males and females from our breeding colony. Both genotypes were first purchased from Jackson Laboratories (stock No: 016962 and 016963) and backcrossed to C57BL/6J to generate a breeding colony. P1–P10 pups used for behavioral experiments were generated in our breeding colony. Animals were housed in 12 h light-dark cycle (10 p.m. – 10 a.m. light), with food and water available ad libitum. All experiments were performed during the dark cycle of the animals. Care and experimental manipulations of all animals were carried out in accordance with the NIH Guide for Care and Use of Laboratory Animals and approved by UCLA IACUC.
METHOD DETAILS
Viruses
AAV2-EF1α-DIO-eNpHR3-mCherry and AAV2-EF1α-DIO-EYFP were purchased from the University of North Carolina vector core. AAV2-EF1α-FLEX-ChR2-nuclear hrGFP and AAV5-syn-Flex-GCaMP6s were purchased from the University of Pennsylvania vector core.
Stereotaxic Surgeries
VgatCre/+ and Vglut2Cre/+ males and females were anesthetized with isoflurane and mounted on a stereotaxic device (Kopf instruments). Injections were carried out using a pulled, fine glass capillary (WPI). For ChR2 experiments, viruses were injected bilaterally into MeApd (ML ±2.00, AP −1.5–1.6, DV −5.15–5.25 from bregma) for males and females, with a total volume of 300 nL per side. For eNpHR3 experiments, viruses were injected bilaterally into MeApd at two coordinates for males and females (ML ±2.00, AP −1.5/1.7, DV −5.25 from bregma) with 350 nL at each site, for a total of 700 nL per side. A ferrule fiber-optic cannula (200 um core diameter, Newdoon) was then placed 0.4–0.5 mm above the virus injection site in the MeApd and fixed to the skull with dental cement (Parkell; Metabond) for optogenetic experiments. For fiber photometry experiments, a fiber was implanted 0.2 mm above the injection site. All control animals in this study were animals with the same genetic background injected with EYFP-expressing viruses.
For experiments involving ChR2 stimulation of cells adjacent to MeApd, viruses were injected bilaterally adjacent to MeApd (ML ±3.20, AP −1.5–1.6, DV −5.5 from bregma), and ferrule fiber-optic cannulas were then placed 0.5 mm above either the virus injection site or the normal coordinates for MeApd.
Optogenetics Experiments
VgatCre/+ animals were injected with ChR2, eNpHR3, or EYFP, at 8–10 weeks, and the viruses were allowed to incubate 3–5 weeks before behavioral testing or perfusion while the animal recovered in its home cage. After the recovery period, animals were tested for 2–3 weeks. A ferrule patch cord was coupled to the ferrule fiber implanted in the mouse using a zirconia split sleeve (Doric Lenses). Optic fibers were connected using an FC/PC adaptor (Doric Lenses) to a 473-nm blue laser or a 593-nm yellow laser (CNI Laser). An Arduino micro-controller board and a customized MATLAB program were used to control laser pulses. The MATLAB program sent commands through a serial connection to the Arduino micro-controller board, which in turn generated the TTL signals to the laser to produce illumination pulses. Stimulations were delivered depending on the animal’s behavior, with at least one minute between each stimulation trial. In particular, the probability to observe stimulation-induced grooming was lower when animals were on the opposite side of the cage or not facing the pups; therefore, we triggered stimulation during random bouts when the animal was in the proximity of pups. The same criteria were used for both EYFP and ChR2-expressing animals. For stimulation experiments, blue (473 nm) light was delivered in 20 ms pulses at 20 Hz for 15 seconds, at an intensity of 3 mW mm−2 at the fiber tip for all experiments other than those testing scalability. MeA GABAergic neurons have previously been shown to fire at ~20 Hz (Bian, 2013; Xie et al., 2016), and photostimulation at this frequency has been shown to reliably elicit action potentials in MeA GABAergic neurons (Hong et al., 2014). For experiments testing scalability, final output powers ranged from 0.1–5 mW mm−2. For inhibition experiments, yellow (593 nm) light was delivered continuously for 5 seconds at final output powers of 5–10 mW mm−2.
All behavior was conducted in a dark room illuminated by red light. Adult males and females were moved to a behavioral testing room in their home cage and allowed to adapt for >30 minutes for three days before testing. Cages were placed on a custom behavior rig with video acquisition capabilities prior to testing. Following patch cord attachment to the implanted fiber, animals were allowed to adapt for >5 minutes before placing P1–P10 pups into the home cage. Each behavioral session entailed a 10–30 minute session where the adult and pups were allowed to freely interact before pups were removed. If infanticidal behavior was observed both naturally or following stimulation, pups were immediately removed and sacrificed. Pup retrieval was encouraged by manually moving pups away from the nest.
Males and females were assessed for baseline levels of pup-directed behaviors. Individuals with a low baseline of pup-directed behavior were screened and selected for ChR2 experiments, since a high baseline of pup-directed behavior would prevent one from determining the specific effect of stimulation on behavior. Conversely, a relatively high baseline of pup-directed behavior was critical for inhibition experiments. The same criteria were used for selecting EYFP control animals for each functional manipulation approach.
For experiments with mothers and fathers, following stereotaxic surgery the animals were allowed to recover for 3 days before pairing with 1–2 individuals of the opposite sex. Following delivery of pups after ~3 weeks, mothers and fathers were tested as described above.
For behavioral analysis, we manually carried out frame-by-frame annotation of pup-directed behaviors and stimulation bouts. Pup grooming was defined as any combination of actions comprised of when 1) the adult’s mouth was in contact with the pup, 2) the adult showed characteristic head bobbing associated with licking, 3) the adult was grasping parts of the pup with its forelimbs, and 4) the adult had visible tongue protrusion contacting the pup. For infanticidal behavior, we quantified the first behavioral frame where the animal’s jaw made contact with the pup proceeded by biting of the pup. When the animal stopped contact with the pup, either naturally or after the stimulation epoch, we quantified this as the end frame of the infanticidal epoch. We immediately removed the pup and inspected it for visible wounds during the behavioral experiments following infanticidal behavior, and sacrificed the pup if it showed any wounds. To compare and combine data across all trials, trials from all animals were aligned to the onset of stimulation. Each frame during each trial was scored as showing the specified behavior or not. This approach allowed for comparison across animals and trials, and a “percentage of trials showing behavior” could be calculated.
In Figure 2H, the retrieval time during stimulation epochs was typically too short for many trials, making a comparison difficult. Thus, to measure disruption of pup retrieval, we examined the duration of the entire retrieval event, starting before the stimulation epoch. If there were any disruption caused by photostimulation, this duration should be shorter between control and eNpHR animals, which we did not observe. In Figure 2K-L, the percentage of uninterrupted retrieval and crouching is shown, as the percentage of trials showing interruption of either of these behaviors during photostimulation is zero or close to zero. For mother halo trial comparisons against GFP controls (Figure S1G), we subsampled trial data from virgin GFP females for statistical comparison.
To determine co-localization of ChR2-expressing cells and Fos activation, a train of blue light (473 nm, 30s on and 30 s off, 20 ms, 20 Hz, stimulated 30 times) was delivered to isolated individuals in their home cage. Animals did not undergo behavioral testing 24 hours prior to this, to avoid background Fos expression. The animals were sacrificed 1.5 hours following stimulation.
Fiber Photometry
Following AAV5-syn-Flex-GCaMP6s and AAV2-EF1α-DIO-EYFP (as control) virus injection, an optical fiber (200 µm O.D., 0.37 numerical aperture; Newdoon) was inserted in the MeApd. Virgin mice were individually housed for 2–3 weeks for recovery. Fluorescence signals was acquired with a fiber photometry system (Doric Lenses Inc, Quebec) with some modifications. We used a 473-nm blue laser (CNI Laser) to replace the LED driver for higher detection sensitivity. The analog voltage signals were digitalized at 100 Hz and recorded by a Micro 1401 digitizer and Spike2 software (CED, Cambridge, UK). The laser power was adjusted at the tip of the optical fiber to 15–30 uW, to minimize bleaching.
Adaptation procedures were the same as described for optogenetic experiments. Immediately before recording, we first connected a ferrule patch cord to the ferrule fiber implanted in the mouse using a zirconia split sleeve (Doric Lenses) for adaptation, and then preheated the laser for 30 minutes. Behaviors were recorded by a camera (Point Grey). Immediately before the start of a recording experiment the laser was turned on. 5–10 minutes later, a new pup was introduced at the corner of the cage furthest away from the resident and the resident was allowed to freely interact with the pup. The pup introduction was repeated for several trials with a trial interval of 3–5 minutes. This was done to normalize the pup presentation between parental and infanticidal males, since for infanticidal males we took the pup out after infanticide had occurred, and therefore allow for a suitable comparison between the two cohorts. For males with strong infanticidal behavior, pups were immediately removed and sacrificed when infanticidal behavior naturally occurred. For some parenting behaviors with a low spontaneous baseline, like pup retrieval and pup crouching, we co-housed the test animals with pups 24 hours before testing to increase the level of these behaviors.
For experiments with mothers and fathers, following stereotaxic surgery the animals were allowed to recover for 3 days before pairing with 1–2 individuals of the opposite sex. Following delivery of pups after ~3 weeks, mothers and fathers were tested largely as described above, except that animals were allowed to freely interact with pups during the entirety of the experiment, as previously described (Fang et al., 2018).
Photometry data were exported to MATLAB files for further analysis. Behavior was scored frame-by-frame and aligned to calcium signals through an Arduino-controlled light pulse. Similar to our optogenetics approaches, we then aligned the onset of each behavior from each animal and collapsed them to allow for grouped data analysis. We defined behavioral onset as the physical contact of the adult with the pup. The ∆F/F ratio was calculated by measuring fluorescence intensity before the behavioral onset and immediately after specific behaviors occurred. We derived the values of fluorescence change (∆F/F) by calculating (F−F0)/F0, where F0 is the baseline fluorescence signal averaged over a 2 s-long control time window, which was typically set between 5 s and 3 s before each behavior bout. ∆F/F values were presented with mean plots with a shaded area indicating SEM. To calculate the average response of ∆F/F values, we also calculated the area under curve (AUC) per second during 5s after each behavior bout. The aforementioned methods were applied for both GCaMP and EYFP groups. The traces presented in figures were based on trial mean, and the comparisons were done with animal mean.
For mother and father statistical comparisons against GFP controls, we compared against virgin GFP data; there was no obvious difference between virgin and mother or father GFP control GCaMP data (data not shown).
Immunohistology
Animals were sacrificed 4–6 weeks post-injection and perfused with 4% PFA. The brains were dissected out and fixed in 4% PFA for 2 hours at room temperature, rinsed with 1X PBS, then placed in 15%–30% sucrose overnight at 4°C. Brains were then rinsed in PBS, embedded in OCT, and frozen at −80°C until sectioning.
35 um sections were cut on a Leica CM1950 cryostat. Free-floating sections were washed in PBS 3× 5 minutes, followed by 1 hour blocking in 5% normal donkey serum (NDS). Primary antibody (goat anti-Fos; Santa Cruz Biotechnology sc-52-G, 1:400) was diluted in 0.1% PBS/Triton X-100 and 5% NDS and incubated overnight (>12 hours) at 4°C. Sections were then washed with PBS three times and incubated with secondary antibody (donkey anti-goat 568; Invitrogen 1:500) overnight at 4°C. Sections were then washed with PBS three times and mounted onto Superfrost Plus slides, dried >15 minutes room temperature, and coverslipped in 50% glycerol containing DAPI (1 ug/mL). Images were acquired using a confocal microscope (Zeiss LSM 880).
Generation of Single-Cell Suspensions
Single-cell dissociation was performed using the Act-seq method as previously described (Macosko et al., 2015; Wu et al., 2017). 8–10 week old male and female mice were anesthetized in an isoflurane chamber and decapitated. The brain was immediately dissected out and placed on ice-cold ACSF containing 124 mM NaCl, 2.5 mM KCl, 1.2 mM NaH2PO4, 24 mM NaHCO3, 5 mM HEPES, 13 mM glucose, 2 mM MgSO4, and 2 mM CaCl2, bubbled with a carbogen gas (95% O2 and 5% CO2) and with a pH of 7.3–7.4. The brain was sectioned in ice-cold ACSF on a vibratome (Leica VT1200) into 300 µm slices and immediately placed into an ice-cold, carbogen-bubbled recovery solution containing 93 mM N-methyl-D-glucamine, 2.5 mM KCl, 1.2 mM NaH2PO4, 30 mM NaHCO3, 20 mM HEPES, 25 mM glucose, 10 mM MgSO4, 0.5 mM CaCl2, 5 mM sodium ascorbate, 2 mM thiourea, 3 mM sodium pyruvate, and 45 µM actinomycin-D (Sigma-Aldrich, Cat# A1410), with a pH of 7.3–7.4 for 15 minutes. Following this, the MeA was microdissected in ice-cold ACSF using coordinates from Paxinos and Franklin’s the Mouse Brain in Stereotaxic Coordinates (~Bregma −1.06 mm–Bregma −1.94 mm) from three consecutive 300 µm sections, and then cut into < 1 mm chunks before dissociation. Tissue chunks were then transferred to a Petri dish containing 1 mg/mL pronase (Sigma-Aldrich, Cat# P6911) and 45 µM actinomycin-D in carbogen-bubbled ACSF, and digested at 34°C for 20 minutes in a chamber continuously aerated with carbogen. Following digestion, the pronase solution was exchanged for ice-cold, carbogen-bubbled ACSF containing 1% fetal bovine serum and 3 µM actinomycin-D, and dissociated through gentle trituration using Pasteur pipettes with polished tip openings of 600 µm, 300 µm, and 150 µm in diameter. After trituration, cells were filtered through a 20 µm filter, pelleted, washed with ice-cold ACSF, and resuspended in ACSF + 0.01% bovine serum albumin.
Brain tissues from two to three male or female mice were combined in each Drop-seq experiment (considered as an independent biological sample) to collect enough cells. In total six male samples and seven female samples were collected. Vaginal swabs were taken for each female to determine the stage of the estrous cycle; all females used were in diestrus.
Drop-Seq Procedure
Drop-seq was performed largely as previously described (Macosko et al., 2015; Wu et al., 2017). Cells were diluted to a final droplet occupancy of ~5% and barcoded beads were diluted to a final droplet occupancy of ~10%. Cells were processed for Drop-seq within ~15 minutes after collection. Flow rates of 3 mL/hr were used for the aqueous cell suspension and lysis buffer with beads, while a flow rate of 15 mL/hr was used for the droplet-generating oil. Following droplet generation from ~2 mL of aqueous volume (1 mL cells and 1 mL beads), droplets were broken, beads were harvested, and hybridized RNA was reverse transcribed. Populations of ~3,000 beads (~150 STAMPs [single-cell transcriptomes attached to microparticles]) were separately amplified with 13 PCR cycles. Single-cell sequencing libraries were generated by tagmentation using Nextera XT (Illumina, Cat# FC-131–1024) and sequenced on the Illumina NextSeq500 sequencer with 20 bp for read 1 and 63 bp for read 2.
Read Alignment and Generation of Digital Gene Expression Data
Paired-end sequenced reads were processed as previously described (Macosko et al., 2015; Wu et al., 2017). Bases 1–12 and 13–20 of read 1 were used to infer the cell barcode and unique molecular identifier (UMI), respectively. Read pairs were filtered for barcode and UMI quality, with any containing a base quality lower than 10 discarded. Any SMART adaptor sequences or polyA sequences longer than 6 bases were trimmed from the 5’ end and 3’ of read 2 respectively, and then aligned to the mm10 build of the mouse genome using STAR v2.5.0a (Dobin et al., 2012) with default settings. Uniquely mapped exonic reads were grouped by cell barcode and recorded. Barcode synthesis errors were corrected as previously described (Shekhar et al., 2016). A digital expression matrix of the number of UMIs per gene in each cell was created, with UMIs within an edit distance of 1 collapsed. Due to size restriction of supplemental materials, raw transcript number of 16,961 genes in 6,800 cells (which include 2,000 neurons and 200 cells from each of the other seven major cell types that are randomly sampled from males and females) are included in Table S1. The entire scRNA-seq dataset is available at GEO.
Dimensionality Reduction, Clustering, and Classification for Major Cell Types and Neuronal Subtypes
Cells that met the following criteria were used for the major cell type clustering analysis: gene number > 200, transcript number > 250, and the ratio between read number and transcript number > 3.5. Genes detected in > 20 cells with > 30 transcripts were used. Mitochondrial RNAs, tRNAs, and rRNAs were excluded. This resulted in a data set of 44,437 cells (21,715 male cells and 22,722 female cells) and 16,961 genes. Transcript counts were normalized to library size as described previously (Shekhar et al., 2016). Raw transcript count of each gene in each cell was divided by the total transcript number in that cell and then timed by the median of total transcript numbers in all cells in the data set. Dimensionality reduction and clustering analysis were then performed using log-transformed expression data (ln(normalized counts + 1)).
Principal component analysis (PCA) was performed using the fast.prcomp function in the gmodels R package. We used the top 5,000 most variable genes selected with the feature selection function in the BackSPIN algorithm (Zeisel et al., 2015). PCs 1–80 (Figure S4) were then used as input for t-distributed stochastic neighbor embedding (tSNE) to generate a two-dimensional non-linear embedding of the cells as previously described (Van Der Maaten and Hinton, 2008), with 2,000 iterations and a perplexity parameter of 30. To group cells into transcriptionally similar clusters, we applied a Louvain-Jaccard graph clustering algorithm (Shekhar et al., 2016), using the scores of the cells along PCs 1–80. This resulted in 39 clusters in our data.
To determine the major cell type for each cluster, we identified top genes enriched in each cluster. We performed comparisons between every single cluster and all the other clusters using a previously described non-parametric binomial test (Shekhar et al., 2016). p values were FDR adjusted using Benjamini-Hochberg correction. Differentially expressed genes that were detected in at least 10% of the cells in a cluster with a fold change of at least 2 and an FDR < 0.05 were then ranked by ln(fold change) × proportion of positive cells within that cluster to select top genes enriched in each cluster. We then assigned each cluster to a major cell type (neurons, astrocytes, microglia, oligodendrocyte precursor cells (OPCs), oligodendrocytes, a transitional state between OPCs and oligodendrocytes, endothelial cells, and mural cells) based on the presence/absence of known cell type markers within its top enriched genes. Clusters expressing markers of more than one major cell type, which likely represented cell doublets, were excluded from subsequent analysis (two clusters, 1,049 cells). To identify cell type-specific genes presented in Figures S5B and S5C, we compared each major cell type to the other cell types using the binomial test (Shekhar et al., 2016), and ranked the genes by fold change.
To identify neuronal subtypes, cells that were classified as neurons in the major cell type analysis were used for further clustering (excluding two female neurons in which one transcript of a Y chromosome gene was detected). Similar to previous studies (Wu et al., 2017; Zeisel et al., 2015), we selected neuronally enriched genes, using the binomial test to compare neurons to all the other cell types. 3,706 genes with fold change ≥ 1.5, FDR < 0.05, and expressed in > 10 neurons with > 15 transcripts were selected, resulting in a data set of 10,164 cells (4,743 cells from females and 5,421 cells from males) and 3,706 genes. We performed PCA using all the 3,706 genes and used PCs 1–40 for tSNE visualization (2,000 iterations, perplexity = 30). We then applied the Louvain-Jaccard graph clustering algorithm (Shekhar et al., 2016), using the scores of the cells along PCs 1–40. We next calculated the ratio between the numbers of cells expressing GABAergic (Gad1, Gad2, or Slc32a1) and glutamatergic (Slc17a6 or Slc17a7) markers in each cluster and considered clusters with a ratio > 4 or < 0.25 as predominantly GABAergic or glutamatergic, respectively. Clusters with a ratio between 0.25 and 4 were subjected to further subclustering analysis using PCA and the Louvain-Jaccard algorithm. This process was iterated until no predominantly GABAergic or glutamatergic clusters could be further isolated, and four clusters (41 cells) that had < 30 cells were removed. We then carried out an iterative merging process during which pairwise differential expression analysis was performed between all the clusters using the binomial test and two clusters were merged if the number of genes more highly expressed in one cluster than the other (fold change ≥ 1.5, FDR < 0.05) was < 5, eventually leading to 16 neuronal subclusters or subtypes. We further used the binomial test to identify subtype-enriched genes (fold change ≥ 2, FDR < 0.05, detected in ≥ 10% of neurons in a subtype) by comparing each neuronal subtype with all the other subtypes and ranking the genes by fold change.
For the subclustering of GABAergic neurons (neurons expressing Gad1, Gad2, or Slc32a1 but not Slc17a6 or Slc17a7), we performed PCA using all neuronally enriched genes and used PCs 1–20 for tSNE visualization (2,000 iterations, perplexity = 30). We then applied the Louvain-Jaccard graph clustering algorithm (Shekhar et al., 2016), using the scores of the cells along PCs 1–20. We further carried out an iterative merging process during which pairwise differential expression analysis was performed between all the clusters using the binomial test and two clusters were merged if the total number of differentially expressed genes (fold change ≥ 2, FDR < 0.05) was < 20, eventually leading to 13 neuronal subclusters.
Comparison of Cell Type Composition and Gene Expression between Male and Female MeA
For the comparison of cell type composition, the percentage of each major cell type or neuronal subtype among all cells or neurons was calculated for each male or female sample. Wilcoxon rank-sum test was used to compare the percentage between the six male and seven female samples and p values were FDR adjusted using Benjamini-Hochberg correction for the number of major cell types or neuronal subtypes tested.
For differential gene expression (DGE) analysis between male and female GABAergic or glutamatergic neurons, we defined GABAergic neurons as neurons expressing (≥ 1 transcript) GABAergic (Gad1, Gad2, or Slc32a1) markers but not glutamatergic (Slc17a6 or Slc17a7) markers (3,457 neurons) and glutamatergic neurons as neurons expressing glutamatergic markers but not GABAergic markers (2,256 neurons). Transcript number in all GABAergic or glutamatergic neurons within each sample was summed up for each neuronally enriched gene and differential gene expression analysis between six male samples and seven male samples was then performed (treating each biological sample as an independent replicate). Compared to treating single cells as independent replicates, this sample-level comparison is less likely to be biased by outlier cells, thus reducing noise and improving the reliability of the results. We used the edgeR (Robinson et al., 2009), limma (Ritchie et al., 2015), and DESeq2 (Love et al., 2014) R packages for DGE analysis. Briefly, for edgeR and limma, expression values were normalized for library size using the TMM method, and differential expression analysis was performed using the qCML method and the limma-trend method, respectively. For DESeq2, expression values were normalized for library size using the “ratio” method, and differential expression analysis was performed using the DESeq function. Genes with a fold change > 1.3, an expression level > the first quartile of the expression level of all neuronally enriched genes, and an FDR < 0.1 (for edgeR and limma) or 0.15 (for DESeq2) were considered to be significantly differentially expressed. Although our differential expression analysis was performed at the sample level, the slightly lower number of glutamatergic neurons compared to GABAergic neurons might lead to higher noise in the data of glutamatergic neurons, resulting in fewer differentially expressed genes detected in glutamatergic neurons. To exclude this possibility, we performed differential gene expression analysis (using edgeR and limma) between males and females after randomly downsampling GABAergic neurons to the same number as glutamatergic neurons. We found that the results remained largely the same, with markedly more differentially expressed genes in GABAergic neurons than in glutamatergic neurons (data not shown).
For the classification of male versus female samples based on gene expression data, the mixOmics R package (Rohart et al., 2017) was used. Briefly, the transcript number of each neuronally enriched gene was summed up across GABAergic, glutamatergic, or all neurons within each sample. X chromosome genes Xist, Tsix, and Xist_exon1, which were exclusively or predominantly expressed in females, and Y chromosome genes were excluded. Using the splsda function in the mixOmics package, a sparse partial least squares (PLS) analysis was first performed with sample sex as the response, followed by classification using a linear discriminative analysis (Rohart et al., 2017). Classification accuracy was calculated using five-fold cross-validation repeated 100 times. The optimal values for the sparsity parameters in splsda (including the optimal number of features for computing each PLS component and the optimal number of components to achieve the best classification performance) were determined through five-fold cross-validation repeated ten times using the tune.splsda function (Rohart et al., 2017). This ensured that the optimal performance of the models constructed using the three different data sets was compared and therefore the differences in performance were unlikely to be due to the choice of parameters. For the classification of male versus female samples using support vector machine (SVM), the same GABAergic, glutamatergic, and all-neuron data sets described above were used. SVM was performed using all genes in the data sets with a linear kernel using the e1071 R package. Classification accuracy was computed using bootstrap with 1,000 iterations.
Fluorescent In Situ Hybridization
Dual color FISH experiments were carried out as previously described (Wu et al., 2017). Antisense probes were generated using cDNA template isolated from brain slices containing MeA, with probe sequences taken from the Allen Brain Atlas (http://www.alleninstitute.org). Images were acquired using Zeiss LSM 880. For each group, we quantified 5–9 sections from 3–5 mice that specifically covered the MeApd area and quantified the fluorescence intensities from the gene of interest in all Vgat+ cells within the boundary of the MeApd using ImageJ. Briefly, fluorescence intensity was quantified by first calculating the average fluorescence intensity in each channel per image, and then subtracting this value to correct for background fluorescence. A mask was generated from the remaining fluorescent signal from Vgat, and the fluorescence intensities from the gene of interest present within this mask were then calculated. Significance was tested using Wilcoxon rank-sum test.
QUANITIFICATION AND STATISTICAL ANALYSIS
All statistical analysis for behavioral experiments was conducted using MATLAB (Mathworks), and is described in the respective Methods, Results, and Figure Legends above. The data were analyzed using Wilcoxon rank-sum test and one-way ANOVA. Significance was defined as α < 0.05 using two-tailed tests, except for ANOVA which is one-tailed. All statistical analysis for single-cell transcriptomic analysis was conducted in R, as described in detail above. Comparison of gene expression between major cell types or neuronal subtypes was performed using a previously described non-parametric binomial test. Comparison of the proportions of different cell types or subtypes between males and females was performed using Wilcoxon rank-sum test. Differential gene expression between the sexes was analyzed using the edgeR, limma, and DESeq2 R packages. Classification of male versus female samples based on gene expression data was performed using linear discriminative analysis with the mixOmics R package or SVM using the e1071 R package.
DATA AND SOFTWARE AVAILABILITY
Gene expression data are available in the Gene Expression Omnibus, GEO: GSE124061. The data, code, and behavior videos that support the findings of this study are available upon request to the corresponding author.
Supplementary Material
Movie S1. ChR2 Stimulation of MeApd Vgat+ Neurons Promotes Pup Grooming in Females, Related to Figure 1.
(A-C) Two-dimensional tSNE visualization showing the distribution of all GABAergic neurons (A), GABAergic neurons from males (B, green dots), and GABAergic neurons from females (C, red dots) among MeA GABAergic subtypes.
(D) Percentage of male and female cells in each GABAergic subtype. No significant difference in percentage was observed between males and females for any subtype (Wilcoxon rank-sum test, FDR > 0.05, n = 6 male samples and 7 female samples; STAR Methods).
(E-F) Heat maps showing the distribution of significantly differentially expressed genes upregulated in males (E) and females (F) across GABAergic subtypes. Many of these genes were enriched in a specific subset of subtypes, such as clusters 1, 7, 11, and 13. Expression level is averaged within each subtype and normalized (mean subtracted and divided by standard deviation) for each row (gene).
(G-H) Representative double FISH images showing similar levels of expression of Cck (G) and Nos1 (H) in Vgat+ cells between males and females in the MeApd. Scale bar = 50 µm.
Raw transcript number of 16,961 genes in 6,800 cells. 2,000 neurons and 200 cells from each of the other seven major cell types are randomly sampled from males and females. The cells are named as “A_B_C” where A is cell barcode, B is sample name, and C denotes cell type. The entire scRNA-seq dataset is available at GEO.
Gene IDs, fold change enrichment, FDR values, and cluster identities for genes enriched in each neuronal subtype. Fold change was calculated by comparing expression in one subtype against the average expression across all other cell types.
Raw transcript number of 3,706 neuronally enriched genes in GABAergic and glutamatergic neurons in the MeA. The neurons are named as “A_B_C” where A denotes GABAergic (“GABA”) or glutamatergic (“Glut”) neurons, B is sample name, and C is a randomly assigned cell number.
Sexually differentially expressed genes in GABAergic and glutamatergic neurons identified using edgeR, limma, and DESeq2 are listed.
Movie S2. eNpHR Inhibition of MeApd Vgat+ Neurons Suppresses Pup Grooming in Females, Related to Figure 2.
Movie S3. ChR2 Stimulation of MeApd Vgat+ Neurons in Males Promotes Pup Grooming at Lower Intensities, Related to Figure 5.
(A) Average onset latency of pup grooming only including stimulation trials that showed an induction of behavior.
(B) Percentage of trials showing crouching during or after stimulation epochs.
(C) Average duration of pup crouching post-stimulation.
(D) Percentage of trials showing pup crouching during or after stimulation with or without accompanying pup grooming. “Crouching only” are trials in which stimulation triggered crouching was not preceded by pup grooming, and “Crouching and grooming” are trials with pup grooming during stimulation epoch followed by crouching.
(E) Representative raster plots of EYFP (top) and eNpHR-expressing (bottom) animals illustrating no suppression of pup crouching during photostimulation bouts.
(F) Distribution of pup grooming episodes in eNpHR lactating female animals (percentage of trials showing pup grooming at different time points) with respect to photostimulation onset.
(G) Half-time of pup grooming suppression in control and eNpHR lactating female animals. Data shown is mean ± SEM; statistical significance (***p < 0.001) was evaluated by Wilcoxon rank-sum test.
Mother eNpHR, n = 30 trials (4 mice); female GFP, n = 26 trials (3 animals). Mean ± SEM. Statistical significance (***p < 0.001) was evaluated by Wilcoxon rank-sum test.
(A) Schematic of viral injection and fiber implantation strategy for ChR2 activation
(B) Representative raster plot of control and ChR2 females illustrating self-grooming during photostimulation bouts.
(C and G) Distribution of self-grooming episodes (percentage of trials showing self-grooming at different time points) with respect to photostimulation onset in females (C) and males (G).
(D and H) Percentage of females (D) and males (H) showing stimulation-triggered self-grooming.
(E and I) Onset latency of self-grooming following photostimulation initiation in females (E) and males (I).
(F and J) Latency distribution of self-grooming following photostimulation initiation in females (F) and males (J).
Female ChR2, n = 37 trials (4 mice); male ChR2, n = 39 trials (4 mice); female GFP, n = 40 trials (5 mice); male GFP, n = 36 trials (3 mice). Mean ± SEM; statistical significance (n.s. not significant, *p < 0.05, **p < 0.01, ***p < 0.001) was evaluated by Wilcoxon rank-sum test.
(A) Example fiber photometry traces during a single episode of pup grooming in virgin males (blue) and females (red).
(B) Example fiber photometry trace during a single episode of infanticide in virgin males.
(C-E) Fiber photometry recording of EYFP-expressing female animals during pup grooming (C), pup retrieval (D), and during pup crouching (E).
(F-G) Average time of sniffing prior to behaviors in males (F) and AUC 2 seconds following sniffing termination and during behaviors (G).
(H) Average distance of fiber tip from MeApd in animals showing pup grooming and infanticide. (I-K) AUC during pup grooming (I), retrieval (J), and crouching (K) in virgin females and mothers.
(L-N) AUC during pup grooming (L), retrieval (M), and crouching (N) in virgin males and fathers. Data shown is mean ± SEM. n ≥ 4 mice for each group. Statistical significance (n.s. not significant, *p < 0.05, **p < 0.01) was Wilcoxon rank-sum test (F and G) and one-way ANOVA followed by Wilcoxon rank-sum test with Bonferroni post hoc multiple comparisons correction (I-K and L-N) (STAR Methods)
(A) Percentage of trials showing pup grooming (A1) and infanticidal behavior (A2) at different photostimulation intensities in fathers.
(B) Average durations of pup grooming (B1) and infanticidal behavior (B2) during photostimulation epochs at different photostimulation intensities in fathers.
(C) Percentage of trials showing pup grooming (C1), pup retrieval (C2), and pup crouching (C3) at different photostimulation intensities in mothers.
(D) Average duration of pup grooming (D1), pup retrieval (D2), and pup crouching (D3) during photostimulation epochs at different photostimulation intensities in mothers.
(E) Average durations (E1) and onset latency (E2) of pup grooming and infanticidal behavior during photostimulation epochs in EYFP-expressing virgin males, and durations of pup grooming and infanticidal behavior in EYFP-expressing fathers (E3).
(F) Average durations (F1) and onset latency (F2) of different parental behaviors during photostimulation epochs in EYFP-expressing virgin females, and durations of different parental behaviors in EYFP-expressing mothers (F3).
(G) Representative raster plot from ChR2-expressing virgin males illustrating the transition between pup grooming and infanticide during photostimulation bouts at different stimulation intensities.
(H-I) Schematic showing injection and fiber implantation strategy for testing the specificity of MeApd activation in pup-directed behaviors using an off-target approach. (H) involves viral injection outside of MeA and fiber implantation inside MeA. (I) involves viral injection outside of MeA and fiber implantation outside of MeA.
(J) Percentage of trials showing infanticidal behavior in off-target injected animals. 1 = strategy in (H), 2 = strategy in (I).
For activity level-dependent experiments, ChR2 mothers: 0–1 mW/mm2, n = 46 trials; 1–2.5 mW/mm2, n = 10 trials; >2.5 mW/mm2, n = 30 trials (8 mice). ChR2 fathers: 0–1 mW/mm2, n = 45 trials; 1–2.5 mW/mm2, n = 9 trials; >2.5 mW/mm2, n = 11 trials (4 mice). GFP control mothers: n = 33 trials (4 mice). GFP control fathers, n = 17 trials (2 mice). GFP control virgin females, n = 88 trials (5 mice). GFP control virgin males, n = 40 trials (4 mice). For off-target experiments, n = 20 trials per condition (4 mice). Data shown is mean ± SEM. Statistical significance (n.s. not significant, ***p < 0.001) was evaluated by one-way ANOVA followed by Wilcoxon rank-sum test with Bonferroni post hoc multiple comparisons correction (B and D).
(A) Plots of principal components (PC) 1–50 of 44,437 MeA cells showing no obvious separation of male (green dots) and female (red dots) cells along any PC dimension.
(B) Percentage of male and female cells in each major cell type. There was no significant difference between males and females for any subtype (Wilcoxon rank-sum test, FDR > 0.05, n = 6 male samples and 7 female samples; STAR Methods).
(C-D) Heat map showing similar expression pattern of select top genes enriched in each major cell type in males (C) and females (D). Markers are selected based on data combining both male and female cells (STAR Methods). Expression level is averaged within each subtype and normalized (mean subtracted and divided by standard deviation) for each row (gene).
(D-K) Dot plots showing no sex difference in the expression level of representative major cell type markers for neurons (D), astrocytes (E), microglia (F), OPCs (G), OPC-OL (H), oligodendrocytes (I), endothelial cells (J), and mural cells (K). Each dot represents an independent biological sample (n = 6 male samples and 7 female samples). Expression level was normalized for library size for each sample (STAR Methods) and log2 transformed.
(A-P) Violin plots showing the distribution of total gene number (A-H) and total transcript number (I-P) in different major cell types in males and females.
(Q-T) Violin plots showing the distribution of total transcript number (Q, R) and total gene number (S, T) in GABAergic neurons and glutamatergic neurons in males (Q, S) and females (R, T). Dots represents single cells. Y-axes are on a log2 scale.
HIGHLIGHTS.
Medial amygdala controls parenting and infanticide in a sexually dimorphic manner
GABAergic but not glutamatergic neurons promote parenting behavior in females
Scalable activation of GABAergic neurons in males controls parenting vs infanticide
scRNA-seq reveals molecular sex differences specifically within GABAergic neurons
ACKNOWLEDGEMENTS
We would like to thank E. Marcus for critical comments on the manuscript, Y. Zuo and H. Li for technical assistance, X. Li for technical support on RNA-seq, and M. Mohr for helpful discussions. This work was supported in part by an NIH NINDS Neurobehavioral Genetics training grant NS048004 (to PBC), a Whitehall Foundation grant, a NARSAD Young Investigator grant, a Sloan Research Fellowship, a Searle Scholars Award, a Klingenstein-Simons Fellowship Award, a Brain Research Foundation grant, and a Packard Fellowship in Science and Engineering (to WH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Adolphs R (2010). Conceptual Challenges and Directions for Social Neuroscience. Neuron 65, 752–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander RD (1974). The Evolution of Social Behavior. Annu. Rev. Ecol. Syst 5, 325–383. [Google Scholar]
- Bian X. (2013). Physiological and morphological characterization of GABAergic neurons in the medial amygdala. Brain Res 1509, 8–19. [DOI] [PubMed] [Google Scholar]
- Chen P, and Hong W (2018). Neural Circuit Mechanisms of Social Behavior. Neuron 98, 16–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke BM, and Woolley CS (2005). Sexually Dimorphic Synaptic Organization of the Medial Amygdala. J. Neurosci 25, 10759–10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2012). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C, and Kimchi T (2007). Neural mechanisms underlying sex-specific behaviors in vertebrates. Curr. Opin. Neurobiol 17, 675–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C, O’Connell LA, and Wu Z (2014). Neural control of maternal and paternal behaviors. Science 345, 765–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y-Y, Yamaguchi T, Song SC, Tritsch NX, and Lin D (2018). A Hypothalamic Midbrain Pathway Essential for Driving Maternal Behaviors. Neuron 98, 192–207.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernald RD (2012). Social Control of the Brain. Annu. Rev. Neurosci 35, 133–151. [DOI] [PubMed] [Google Scholar]
- Fleming AS, Vaccarino F, and Luebke C (1980). Amygdaloid inhibition of maternal behavior in the nulliparous female rat. Physiol. Behav 25, 731–743. [DOI] [PubMed] [Google Scholar]
- Hong W, Kim D-W, and Anderson DJ (2014). Antagonistic Control of Social versus Repetitive Self-Grooming Behaviors by Separable Amygdala Neuronal Subsets. Cell 158, 1348–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdy SB (1979). Infanticide Among Animals: A Review, Classification, and Examination of the Implications for the Reproductive Strategies of Females. Ethol. Sociobiol 1, 13–40. [Google Scholar]
- Kohl J, Autry AE, and Dulac C (2017). The neurobiology of parenting: A neural circuit perspective. BioEssays 39, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl J, Babayan BM, Rubinstein ND, Autry AE, Marin-Rodriguez B, Kapoor V, Miyamishi K, Zweifel LS, Luo L, Uchida N, et al. (2018). Functional circuit architecture underlying parental behaviour. Nature 556, 326–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, and Teichmann SA (2015). The Technology and Biology of Single-Cell RNA Sequencing. Mol. Cell 58, 610–620. [DOI] [PubMed] [Google Scholar]
- Lee H, Kim DW, Remedios R, Anthony TE, Chang A, Madisen L, Zeng H, and Anderson DJ (2014). Scalable control of mounting and attack by Esr1+neurons in the ventromedial hypothalamus. Nature 509, 627–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Mathis A, Grewe BF, Osterhout JA, Ahanonu B, Schnitzer MJ, Murthy VN, and Dulac C (2017). Neuronal Representation of Social Information in the Medial Amygdala of Awake Behaving Mice. Cell 1–15. [DOI] [PMC free article] [PubMed]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Maaten LJP, and Hinton GE (2008). Visualizing high-dimensional data using t-sne. J. Mach. Learn. Res 9, 2579–2605. [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlin BJ, Mitre M, James AD, Chao MV, and Froemke RC (2015). Oxytocin enables maternal behaviour by balancing cortical inhibition. Nature 520, 499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JA, Jordan CL, and Breedlove SM (2008). Sexual Dimorphism in Neuronal Number of the Posterodorsal Medial Amygdala Is Independent of Circulating Androgens and Regional Volume in Adult Rats. J Comp Neurol 506, 851–859. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, and Hyman SE (2010). Animal models of neuropsychiatric disorders. Nat. Neurosci 13, 1161–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numan M (1974). Medial preoptic area and maternal behavior in the female rat. J. Comp. Physiol. Psychol 87, 746–759. [DOI] [PubMed] [Google Scholar]
- Numan M, and Sheehan TP (1997). Neuroanatomical circuitry for mammalian maternal behavior. Ann. N. Y. Acad. Sci 807, 101–125. [DOI] [PubMed] [Google Scholar]
- Numan M, and Young LJ (2016). Neural mechanisms of mother-infant bonding and pair bonding: Similarities, differences, and broader implications. Horm. Behav 77, 98–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numan M, Numan M, and English J (1993). Excitotoxic amino acid injections into the medial amygdala facilitate maternal behavior in virgin female rats. Horm. Behav 27, 56–81. [DOI] [PubMed] [Google Scholar]
- Palanza P (2001). Animal models of anxiety and depression: how are females different? Neurosci. Biobehav. Rev 25, 219–233. [DOI] [PubMed] [Google Scholar]
- Perrigo G, Belvin L, and Vom Saal FS (1992). Time and sex in the Male mouse: Temporal regulation of infanticide and parental behavior. Chronobiol. Int 9, 421–433. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2009). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohart F, Gautier B, Singh A, and Lê Cao K-A (2017). mixOmics: An R package for ‘omics feature selection and multiple data integration. PLOS Comput. Biol 13, e1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott N, Prigge M, Yizhar O, and Kimchi T (2015). A sexually dimorphic hypothalamic circuit controls maternal care and oxytocin secretion. Nature 525, 519–522. [DOI] [PubMed] [Google Scholar]
- Sheehan T, Paul M, Amaral E, Numan MJ, and Numan M (2001). Evidence that the medial amygdala projects to the anterior/ventromedial hypothalamic nuclei to inhibit maternal behavior in rats. Neuroscience 106, 341–356. [DOI] [PubMed] [Google Scholar]
- Sheehan TP, Cirrito J, Numan MJ, and Numan M (2000). Using c-Fos immunocytochemistry to identify forebrain regions that may inhibit maternal behavior in rats. Behav. Neurosci 114, 337–352. [DOI] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, et al. (2016). Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell 166, 1308–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers L, and Liberles SD (2016). State-dependent responses to sex pheromones in mouse. Curr. Opin. Neurobiol 38, 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson LW (2000). Cerebral hemisphere regulation of motivated behavior 886, 113–164. [DOI] [PubMed] [Google Scholar]
- Tachikawa KS, Yoshihara Y, and Kuroda KO (2013). Behavioral transition from attack to parenting in male mice: A crucial role of the vomeronasal system. J. Neurosci 158, 5120–5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuneoka Y, Tokita K, Yoshihara C, Amano T, Esposito G, Huang AJ, Yu LM, Odaka Y, Shinozuka K, McHugh TJ, et al. (2015). Distinct preoptic-BST nuclei dissociate paternal and infanticidal behavior in mice. EMBO J 34, 2652–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger EK, Burke KJ, Yang CF, Bender KJ, Fuller PM, and Shah NM (2015). Medial amygdalar aromatase neurons regulate aggression in both sexes. Cell Rep 10, 453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vong L, Ye C, Yang Z, Choi B, Chua S, and Lowell BB (2011). Leptin Action on GABAergic Neurons Prevents Obesity and Reduces Inhibitory Tone to POMC Neurons. Neuron 71, 142–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y-C, Wang S-R, Jiao Z-L, Zhang W, Lin J-K, Li X-Y, Li S-S, Zhang X, and Xu X-H (2018). Medial preoptic area in mice is capable of mediating sexually dimorphic behaviors regardless of gender. Nat. Commun 9, 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YE, Pan L, Zuo Y, Li X, and Hong W (2017). Detecting Activated Cell Populations Using Single-Cell RNA-Seq. Neuron 96, 313–329. [DOI] [PubMed] [Google Scholar]
- Wu Z, Autry AE, Bergan JF, Watabe-Uchida M, and Dulac CG (2014). Galanin neurons in the medial preoptic area govern parental behaviour. Nature 509, 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Fox GE, Liu J, Lyu C, Lee JC, Kuang H, Jacobs S, Li M, Liu T, Song S, et al. (2016). Brain Computation Is Organized via Power-of-Two-Based Permutation Logic. Front. Syst. Neurosci 10, 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Coats JK, Yang CF, Wang A, Ahmed OM, Alvarado M, Izumi T, and Shah NM (2012). Modular genetic control of sexually dimorphic behaviors. Cell 148, 596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CF, and Shah NM (2014). Representing sex in the brain, one module at a time. Neuron 82, 261–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CF, Chiang MC, Gray DC, Prabhakaran M, Alvarado M, Juntti SA, Unger EK, Wells JA, and Shah NM (2013). Sexually dimorphic neurons in the ventromedial hypothalamus govern mating in both sexes and aggression in males. Cell 153, 896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Manchado ABM, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, et al. (2015). Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. ChR2 Stimulation of MeApd Vgat+ Neurons Promotes Pup Grooming in Females, Related to Figure 1.
(A-C) Two-dimensional tSNE visualization showing the distribution of all GABAergic neurons (A), GABAergic neurons from males (B, green dots), and GABAergic neurons from females (C, red dots) among MeA GABAergic subtypes.
(D) Percentage of male and female cells in each GABAergic subtype. No significant difference in percentage was observed between males and females for any subtype (Wilcoxon rank-sum test, FDR > 0.05, n = 6 male samples and 7 female samples; STAR Methods).
(E-F) Heat maps showing the distribution of significantly differentially expressed genes upregulated in males (E) and females (F) across GABAergic subtypes. Many of these genes were enriched in a specific subset of subtypes, such as clusters 1, 7, 11, and 13. Expression level is averaged within each subtype and normalized (mean subtracted and divided by standard deviation) for each row (gene).
(G-H) Representative double FISH images showing similar levels of expression of Cck (G) and Nos1 (H) in Vgat+ cells between males and females in the MeApd. Scale bar = 50 µm.
Raw transcript number of 16,961 genes in 6,800 cells. 2,000 neurons and 200 cells from each of the other seven major cell types are randomly sampled from males and females. The cells are named as “A_B_C” where A is cell barcode, B is sample name, and C denotes cell type. The entire scRNA-seq dataset is available at GEO.
Gene IDs, fold change enrichment, FDR values, and cluster identities for genes enriched in each neuronal subtype. Fold change was calculated by comparing expression in one subtype against the average expression across all other cell types.
Raw transcript number of 3,706 neuronally enriched genes in GABAergic and glutamatergic neurons in the MeA. The neurons are named as “A_B_C” where A denotes GABAergic (“GABA”) or glutamatergic (“Glut”) neurons, B is sample name, and C is a randomly assigned cell number.
Sexually differentially expressed genes in GABAergic and glutamatergic neurons identified using edgeR, limma, and DESeq2 are listed.
Movie S2. eNpHR Inhibition of MeApd Vgat+ Neurons Suppresses Pup Grooming in Females, Related to Figure 2.
Movie S3. ChR2 Stimulation of MeApd Vgat+ Neurons in Males Promotes Pup Grooming at Lower Intensities, Related to Figure 5.
(A) Average onset latency of pup grooming only including stimulation trials that showed an induction of behavior.
(B) Percentage of trials showing crouching during or after stimulation epochs.
(C) Average duration of pup crouching post-stimulation.
(D) Percentage of trials showing pup crouching during or after stimulation with or without accompanying pup grooming. “Crouching only” are trials in which stimulation triggered crouching was not preceded by pup grooming, and “Crouching and grooming” are trials with pup grooming during stimulation epoch followed by crouching.
(E) Representative raster plots of EYFP (top) and eNpHR-expressing (bottom) animals illustrating no suppression of pup crouching during photostimulation bouts.
(F) Distribution of pup grooming episodes in eNpHR lactating female animals (percentage of trials showing pup grooming at different time points) with respect to photostimulation onset.
(G) Half-time of pup grooming suppression in control and eNpHR lactating female animals. Data shown is mean ± SEM; statistical significance (***p < 0.001) was evaluated by Wilcoxon rank-sum test.
Mother eNpHR, n = 30 trials (4 mice); female GFP, n = 26 trials (3 animals). Mean ± SEM. Statistical significance (***p < 0.001) was evaluated by Wilcoxon rank-sum test.
(A) Schematic of viral injection and fiber implantation strategy for ChR2 activation
(B) Representative raster plot of control and ChR2 females illustrating self-grooming during photostimulation bouts.
(C and G) Distribution of self-grooming episodes (percentage of trials showing self-grooming at different time points) with respect to photostimulation onset in females (C) and males (G).
(D and H) Percentage of females (D) and males (H) showing stimulation-triggered self-grooming.
(E and I) Onset latency of self-grooming following photostimulation initiation in females (E) and males (I).
(F and J) Latency distribution of self-grooming following photostimulation initiation in females (F) and males (J).
Female ChR2, n = 37 trials (4 mice); male ChR2, n = 39 trials (4 mice); female GFP, n = 40 trials (5 mice); male GFP, n = 36 trials (3 mice). Mean ± SEM; statistical significance (n.s. not significant, *p < 0.05, **p < 0.01, ***p < 0.001) was evaluated by Wilcoxon rank-sum test.
(A) Example fiber photometry traces during a single episode of pup grooming in virgin males (blue) and females (red).
(B) Example fiber photometry trace during a single episode of infanticide in virgin males.
(C-E) Fiber photometry recording of EYFP-expressing female animals during pup grooming (C), pup retrieval (D), and during pup crouching (E).
(F-G) Average time of sniffing prior to behaviors in males (F) and AUC 2 seconds following sniffing termination and during behaviors (G).
(H) Average distance of fiber tip from MeApd in animals showing pup grooming and infanticide. (I-K) AUC during pup grooming (I), retrieval (J), and crouching (K) in virgin females and mothers.
(L-N) AUC during pup grooming (L), retrieval (M), and crouching (N) in virgin males and fathers. Data shown is mean ± SEM. n ≥ 4 mice for each group. Statistical significance (n.s. not significant, *p < 0.05, **p < 0.01) was Wilcoxon rank-sum test (F and G) and one-way ANOVA followed by Wilcoxon rank-sum test with Bonferroni post hoc multiple comparisons correction (I-K and L-N) (STAR Methods)
(A) Percentage of trials showing pup grooming (A1) and infanticidal behavior (A2) at different photostimulation intensities in fathers.
(B) Average durations of pup grooming (B1) and infanticidal behavior (B2) during photostimulation epochs at different photostimulation intensities in fathers.
(C) Percentage of trials showing pup grooming (C1), pup retrieval (C2), and pup crouching (C3) at different photostimulation intensities in mothers.
(D) Average duration of pup grooming (D1), pup retrieval (D2), and pup crouching (D3) during photostimulation epochs at different photostimulation intensities in mothers.
(E) Average durations (E1) and onset latency (E2) of pup grooming and infanticidal behavior during photostimulation epochs in EYFP-expressing virgin males, and durations of pup grooming and infanticidal behavior in EYFP-expressing fathers (E3).
(F) Average durations (F1) and onset latency (F2) of different parental behaviors during photostimulation epochs in EYFP-expressing virgin females, and durations of different parental behaviors in EYFP-expressing mothers (F3).
(G) Representative raster plot from ChR2-expressing virgin males illustrating the transition between pup grooming and infanticide during photostimulation bouts at different stimulation intensities.
(H-I) Schematic showing injection and fiber implantation strategy for testing the specificity of MeApd activation in pup-directed behaviors using an off-target approach. (H) involves viral injection outside of MeA and fiber implantation inside MeA. (I) involves viral injection outside of MeA and fiber implantation outside of MeA.
(J) Percentage of trials showing infanticidal behavior in off-target injected animals. 1 = strategy in (H), 2 = strategy in (I).
For activity level-dependent experiments, ChR2 mothers: 0–1 mW/mm2, n = 46 trials; 1–2.5 mW/mm2, n = 10 trials; >2.5 mW/mm2, n = 30 trials (8 mice). ChR2 fathers: 0–1 mW/mm2, n = 45 trials; 1–2.5 mW/mm2, n = 9 trials; >2.5 mW/mm2, n = 11 trials (4 mice). GFP control mothers: n = 33 trials (4 mice). GFP control fathers, n = 17 trials (2 mice). GFP control virgin females, n = 88 trials (5 mice). GFP control virgin males, n = 40 trials (4 mice). For off-target experiments, n = 20 trials per condition (4 mice). Data shown is mean ± SEM. Statistical significance (n.s. not significant, ***p < 0.001) was evaluated by one-way ANOVA followed by Wilcoxon rank-sum test with Bonferroni post hoc multiple comparisons correction (B and D).
(A) Plots of principal components (PC) 1–50 of 44,437 MeA cells showing no obvious separation of male (green dots) and female (red dots) cells along any PC dimension.
(B) Percentage of male and female cells in each major cell type. There was no significant difference between males and females for any subtype (Wilcoxon rank-sum test, FDR > 0.05, n = 6 male samples and 7 female samples; STAR Methods).
(C-D) Heat map showing similar expression pattern of select top genes enriched in each major cell type in males (C) and females (D). Markers are selected based on data combining both male and female cells (STAR Methods). Expression level is averaged within each subtype and normalized (mean subtracted and divided by standard deviation) for each row (gene).
(D-K) Dot plots showing no sex difference in the expression level of representative major cell type markers for neurons (D), astrocytes (E), microglia (F), OPCs (G), OPC-OL (H), oligodendrocytes (I), endothelial cells (J), and mural cells (K). Each dot represents an independent biological sample (n = 6 male samples and 7 female samples). Expression level was normalized for library size for each sample (STAR Methods) and log2 transformed.
(A-P) Violin plots showing the distribution of total gene number (A-H) and total transcript number (I-P) in different major cell types in males and females.
(Q-T) Violin plots showing the distribution of total transcript number (Q, R) and total gene number (S, T) in GABAergic neurons and glutamatergic neurons in males (Q, S) and females (R, T). Dots represents single cells. Y-axes are on a log2 scale.
Data Availability Statement
Gene expression data are available in the Gene Expression Omnibus, GEO: GSE124061. The data, code, and behavior videos that support the findings of this study are available upon request to the corresponding author.


